Abstract
Combining bioorthogonal chemistry with the use of proteins engineered with adhesive and morphogenetic solid-binding peptides is a promising route for synthesizing hybrid materials with the economy and efficiency of living systems. Using optical sensing of chloramphenicol as a proof of concept, we show here that a GFP variant engineered with zinc sulfide and silica-binding peptides on opposite sides of its β-barrel supports the fabrication of protein-capped ZnS:Mn nanocrystals that exhibit the combined emission signatures of organic and inorganic fluorophores. Conjugation of a chloramphenicol-specific DNA aptamer to the protein shell through strain-promoted azide–alkyne cycloaddition and spontaneous concentration of the resulting nanostructures onto SiO2 particles mediated by the silica-binding sequence enables visual detection of environmentally and clinically relevant concentrations of chloramphenicol through analyte-mediated inner filtering of sub-330 nm excitation light.
Graphical Abstract
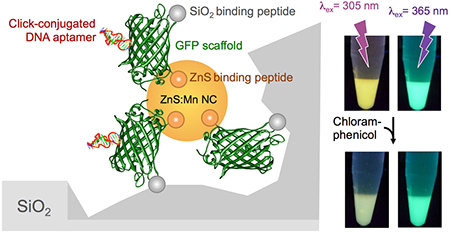
Hybrid materials comprising protein, nucleic acid, and inorganic species hold promise for a broad range of technological applications,(1–4) but the synthesis of individual components and their assembly into user-specified functional architectures can be difficult, time-consuming, and inefficient. Proteins endowed with the ability to template inorganic synthesis and assemble dissimilar materials through genetic installation of solid binding peptides (SBPs)(5, 6) at defined structural locations may provide a streamlined and green alternative to this challenge, particularly when used in combination with bioorthogonal click chemistry schemes.(7)
We selected superfolder green fluorescent protein(8) (sfGFP, Figure 1A) as a scaffold to explore protein-enabled synthesis and assembly of a prototypical multicomponent architecture for the following reasons. First, sfGFP exhibits improved thermostability and enhanced function (fluorescence) relative to A. victoria GFP.(8) Second, the ninth loop of the protein (residues 172–173) is permissive for the insertion of extraneous sequences up to 20 residues long (Figure S1)(9) and lies opposite to the N- and C-termini, which are available for the construction of genetic fusions (Figure 1A). Third, the 20 lysine residues of sfGFP are primarily located on the sides of its β-barrel with outward projecting side chains available for NHS chemistry (Figure S2).
Figure 1.

(A) Ribbon structure and electrostatic surface of sfGFP. Loop 9, N- and C-termini are labeled. (B) Schematic structure of sfGFP-CT43 and sfGFP::Car9-CT43.The CT43 and Car9 SBPs are depicted as orange and green spheres, respectively. (C) SDS-PAGE analysis of sfGFP-CT43 (lane 1) and sfGFP::Car9-CT43 (lane 2) purified by ion exchange and silica affinity chromatography, respectively. Lane M contains molecular mass markers.
In a bid to combine organic and inorganic fluorophores without relying on colloidal chemistry to synthesize quantum dots, or on ligand exchange and conjugation steps to decorate their surface with a fluorescent protein,(10–12) we set out to endow sfGFP with the ability to template the fabrication of luminescent Mn-doped ZnS nanocrystals. To this end, we fused the CT43 ZnS-binding peptide (AGDSSGVDSRSV)(13) to its C-terminus with the expectation that the resulting fusion (sfGFP-CT43; Figure 1B) would cap the growth of ZnS:Mn particles nucleated in a precursor electrolyte to ~4 nm, as observed with other protein frameworks.(13–16) We further engineered Car9 (DSARGFKKPGKR), an SBP that recognizes the edge of carbon nanostructures and the surface of silica in both linear and disulfide-constrained conformations,(17–19) within loop 9 of sfGFP-CT43 to produce a chimera that would combine biological fluorescence with nanocrystal biosynthesis and silica-binding activities (sfGFP::Car9-CT43; Figures 1B and S1).
Previously, we reported that genetically appended Car9 extensions enable rapid (<15 min) affinity protein purification on inexpensive silica beds.(18)Figure 1C (lane 2) shows that sfGFP::Car9-CT43 could be purified in this fashion, confirming that Car9 remains capable of binding silica as a loop 9 insert. While the purity of sfGFP::Car9-CT43 was slightly less than that of sfGFP-CT43 isolated by traditional ion exchange chromatography (Figure 1C, lane 1), such a rapid, high-yield, and low-cost purification process is desirable for protein-enabled materials science applications. As expected of a fully permissive site,(9) insertion of Car9 within loop 9 of sfGFP-CT43 did not affect the fluorescent properties of the sfGFP chromophore (Figure S3).
Using 5 μM of sfGFP-CT43 or sfGFP::Car9-CT43 and optimized conditions for protein-aided nanocrystal synthesis(16) yielded particles 14.5 ± 1.5 nm and 15 ± 1 nm in hydrodynamic diameter, respectively (Figure 2A–B, insets). As expected, UV–visible spectra exhibited the 335 nm absorption edge of quantum-confined ZnS:Mn particles (Figure 2A–B).(15)
Figure 2.

UV–visible spectra of ZnS:Mn nanocrystals synthesized in the presence of sfGFP-CT43 (A) or sfGFP::Car9-CT43 (B). Insets show particle size distributions determined by DLS. Emission spectra of sfGFP-CT43@ZnS:Mn (C) and sfGFP::Car9-CT43@ZnS:Mn nanocrystals (D) were recorded at 305 or 365 nm excitation. Insets in panel D show the appearance of solutions under 305 nm (right) or 365 nm (left) illumination.
We used excitation scans to identify an excitation wavelength (λex = 305 nm) that maximized nanocrystal to sfGFP emission (Figure S4). Photoluminescence emission spectra collected under these conditions (Figure 2C–D) exhibited two peaks corresponding to the contributions of the sfGFP chromophore (λex ≈ 510 nm) and of the 4T1 → 6A1 transition of Mn2+ (λex = 590 nm).(20) We further found that λex = 365 nm was appropriate to isolate the contribution of sfGFP at commensurate emission intensities (Figure 2C–D). Both excitation wavelengths are conveniently available on standard UV transilluminators, enabling direct visualization of the goldenrod color of sfGFP-CT43@ZnS:Mn and sfGFP::Car9-CT43@ZnS:Mn solutions illuminated at 305 nm relative to their bright green appearance at 365 nm (Figure 2D, insets).
We exploited these optical properties to determine if sfGFP::Car9-CT43@ZnS:Mn particles could be enriched on silica substrates as a result of the presence of the Car9 insert on the outer region of the protein corolla (Figure 3A). To this end, an excess of colloids was incubated with 35–70 μm silica particles for 5 min and beads were washed with buffer. Illumination at 305 and 365 nm confirmed colocalization of the goldenrod and green colors with the silica phase. For comparison, ZnS:Mn nanocrystals fabricated with sfGFP-CT43 were quantitatively removed by washing (Figure 3B). Thus, locating the Car9 silica-binding sequence opposite the CT43 zinc sulfide capping sequence allows for a functional sfGFP scaffold to effectively couple particles of different length scales and compositions with no requirement for additional chemistry.
Figure 3.

(A) Schematic depiction of the immobilization of sfGFP::Car9-CT43@ZnS:Mn particles on porous silica microbeads. (B) ZnS:Mn nanocrystals synthesized with sfGFP-CT43 or sfGFP::Car9-CT43 were mixed with silica beads (left), washed (right), and photographed under 305 nm (top) or 365 nm (bottom) light.
Traditional fluorescent sensors connect a sensory domain to a fluorescent molecule so that analyte binding to the sensory region alters the photophysical properties of the fluorescent fragment.(21–24) There is however growing interest in sensing strategies based on the inner filter effect (IFE) that exploit an overlap between the absorption band of a target analyte and the excitation band of a fluorophore to correlate the extent of fluorescence quenching with analyte concentration. IFE sensors have the advantages of simplicity, robustness, and amenability to use in solid formats.(25–29)
Like many marine toxins and aquatic pollutants,(30, 31) the antibiotic chloramphenicol strongly absorbs in the 300 nm region of the spectrum but is nearly transparent above 350 nm (Figure S5). It is therefore well suited to evaluate the potential of sfGFP-decorated ZnS:Mn nanocrystals in IFE sensing since it should quench both inorganic and protein emission by absorbing excitation light at 305 nm, but leave sfGFP emission unaffected when excitation is conducted at 365 nm. Indeed, we observed a progressive decrease in emission intensities of both sfGFP and nanocrystals peaks when a colloidal solution of sfGFP::Car9-CT43@ZnS:Mn was supplied with increasing concentrations of chloramphenicol and excited at 305 nm (Figure 4A–B). Stern–Volmer plots relating the ratio of unquenched to quenched fluorescence intensities (Fo/F) to quencher concentration(32) were linear for both fluorophores, with slopes (Stern–Volmer constants) of Ksv = 5.2 × 103 M−1 for sfGFP and Ksv = 8.5 × 103 M−1 for the nanocrystals (Figure 4C). Using an sfGFP fluorescence lifetime of To = 2.53 ns,(33) we calculated a bimolecular quenching rate constant kq = Ksv /To of ~2 × 1012 M−1 s−1 for the protein component of the assembled structure. The corresponding kq for the nanocrystal component was ~5 × 106 M−1 s−1 due to the extended fluorescence lifetime of the 4T1 → 6A1Mn2+ transition (To = 1.9 ms).(34) Finally, addition of 100 μg/mL (330 μM) chloramphenicol caused less than a 3% change in sfGFP emission intensity when the solution was excited at 365 nm (Figure S6), providing an opportunity for “built-in” diagnostics, calibration, or ratiometric measurements.
Figure 4.

Influence of chloramphenicol on sfGFP::Car9-CT43@ZnS:Mn emission. (A) Emission spectra were collected at λex = 305 nm in the presence of increasing chloramhenicol concentrations (0 to 100 μg/mL). (B) Appearance of the solutions under 305 nm (top) or 365 nm (bottom) illumination. (C) Stern–Volmer plots for protein (green) and nanocrystals (orange).
While these results indicate that ZnS:Mn nanocrystals stabilized by sfGFP::Car9-CT43 are suitable for IFE-based chloramphenicol detection, there is no analyte specificity and a spectrophotometer is required. To address the first limitation, we conjugated the chloramphenicol-specific CA7 DNA aptamer(35) to the protein shell of ZnS:Mn nanocrystals using high-efficiency strain-promoted azide–alkyne cycloaddition “click” chemistry (Figure S7).(36) Briefly, a synthetic CA7 oligonucleotide modified with a 5′ amine was reacted with an N-hydroxysuccinimide (NHS)-activated bicyclononyne (BCN) in DMF to yield BCN-CA7, while sfGFP::Car9-CT43-capped ZnS:Mn nanocrystals were reacted with an NHS-activated azide in PBS to derivatize primary amines in solvent-exposed lysine side chains with azido functionality. (Lysines in the Car9 sequence and the protein N-terminus should project toward the nanocrystal surface, and these amines should be unavailable for NHS-based coupling; see Figure S2.) Finally, we determined the ratio at which the two solutions should be mixed to form aptamer-conjugated assemblies without inducing nanocrystal precipitation, affecting optical properties (Figure S8), or interfering with silica binding (Figure 5).
Figure 5.

ZnS:Mn nanocrystals synthesized with sfGFP::Car9-CT43 and derivatized or not with the CA7 aptamer were concentrated on silica. Washed particles were exposed to increased chloramphenicol concentrations and photographed under UV illumination.
The unique combination of properties enabled the construction of an inexpensive sensor for visual detection of chloramphenicol in which sfGFP::Car9-CT43@ZnS:Mn nanoparticles are easily handled by virtue of their concentration on silica particles, and the CA7 aptamer enhances IFE efficiency by increasing quencher concentration in the vicinity of nanocrystals. Indeed, as little as 2 μg/mL (6.2 μM) chloramphenicol caused a change in emission intensity that could be detected with the naked eye when composite particles were visualized on a transilluminator operated at 305 nm (Figure 5). This is far below the 10 μg/mL (30.1 μM) threshold reported to be environmentally relevant in biogeochemical processes,(37)and at the lower end of the 2–15 μg/mL range at which Enterobacteriaceae are susceptible to its bacteriostatic action.(38) By contrast, unaided detection of as high as 32 μg/mL (100 μM) chloramphenicol was not possible with particles lacking the aptamer. Yet, the emission of the two colloidal solutions was similar and unaffected by the presence of chloramphenicol under 365 nm light, providing a built-in control for sensor integrity and enabling ratiometric quantification (Figure 5 and S9).
In summary, we have shown that genetic installation of morphogenetic (CT43) and adhesive (Car9) SBPs in a biologically active protein scaffold (sfGFP) can be combined with click chemistry (CA7 aptamer conjugation) to enable the fabrication of a functional hybrid architecture under ambient, environmentally friendly, and “single pot” reaction conditions. Using IFE-based chloramphenicol detection as proof of concept, we demonstrated the usefulness of such structures for the visual detection of environmentally and clinically relevant concentrations of the antibiotic chloramphenicol. Bearing in mind that ZnS:Mn nanocrystals are unstable at pH ≤ 5,(15, 16) this system has potential for field deployment and could be expanded to other antibiotics, UV-absorbing aquatic pollutants, and marine toxins by employing different aptamers. It should also be possible to sense compounds absorbing in different spectral regions by tuning the absorption/emission characteristics of the inorganic and organic fluorophores through compositional/size change and genetic engineering, respectively. More broadly, other SBPs, protein frameworks, and bioorthogonal reaction schemes (e.g., Staudinger ligation and tetrazine/trans-cyclo octene)(39) could be used for the production of static and dynamic architectures of different compositions and stoichiometries for applications in sensing, imaging, plasmonics, and photonics.
Supplementary Material
Acknowledgment
This work was supported by NIH Award 1U19ES019545-01 and ONR Award BRC-11123566 (to F.B.) and startup funds from the University of Washington (C.A.D.). We thank Brandon Coyle for constructing sfGFP::Car9-Car15.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Brian J. F. Swift, Department of Chemical Engineering, Box 351750, University of Washington, Seattle, Washington 98195, United States
Jared A. Shadish, Department of Chemical Engineering, Box 351750, University of Washington, Seattle, Washington 98195, United States
Cole A. DeForest, Department of Chemical Engineering, Box 351750, University of Washington, Seattle, Washington 98195, United States
François Baneyx, Department of Chemical Engineering, Box 351750, University of Washington, Seattle, Washington 98195, United States.
References
- 1.Uchida M; Klem MT; Allen M; Suci P; Flenniken M; Gillitzer E; Varpness Z; Liepold LO; Young M; Douglas T Adv. Mater 2007, 19, 1025–1042 DOI: 10.1002/adma.200601168 [DOI] [Google Scholar]
- 2.Maity B; Ueno T Chem. Rec 2016, DOI: 10.1002/tcr.201600122 [DOI] [PubMed] [Google Scholar]
- 3.Samanta A; Medintz IL Nanoscale 2016, 8, 9037–95 DOI: 10.1039/C5NR08465B [DOI] [PubMed] [Google Scholar]
- 4.Niemeyer CM Angew. Chem., Int. Ed 2010, 49, 1200–16 DOI: 10.1002/anie.200904930 [DOI] [PubMed] [Google Scholar]
- 5.Baneyx F; Schwartz DT Curr. Opin. Biotechnol 2007, 18, 312–317 DOI: 10.1016/j.copbio.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 6.Care A; Bergquist PL; Sunna A Trends Biotechnol. 2015, 33, 259–68 DOI: 10.1016/j.tibtech.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 7.Sapsford KE; Algar WR; Berti L; Gemmill KB; Casey BJ; Oh E; Stewart MH; Medintz IL Chem. Rev 2013, 113, 1904–2074 DOI: 10.1021/cr300143v [DOI] [PubMed] [Google Scholar]
- 8.Pedelacq JD; Cabantous S; Tran T; Terwilliger TC; Waldo GS Nat. Biotechnol 2006, 24, 79–88 DOI: 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- 9.Abedi MR; Caponigro G; Kamb A Nucleic Acids Res. 1998, 26, 623–30 DOI: 10.1093/nar/26.2.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palui G; Aldeek F; Wang W; Mattoussi H Chem. Soc. Rev 2015, 44, 193–227 DOI: 10.1039/C4CS00124A [DOI] [PubMed] [Google Scholar]
- 11.Biju V Chem. Soc. Rev 2014, 43, 744–64 DOI: 10.1039/C3CS60273G [DOI] [PubMed] [Google Scholar]
- 12.Medintz IL; Mattoussi H Phys. Chem. Chem. Phys 2009, 11, 17–45 DOI: 10.1039/B813919A [DOI] [PubMed] [Google Scholar]
- 13.Zhou W; Schwartz DT; Baneyx F J. Am. Chem. Soc 2010, 132, 4731–4738 DOI: 10.1021/ja909406n [DOI] [PubMed] [Google Scholar]
- 14.Swift BJ; Baneyx F PLoS One 2015, 10, e0124916 DOI: 10.1371/journal.pone.0124916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou W; Baneyx F ACS Nano 2011, 5, 8013–8018 DOI: 10.1021/nn2024896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W; Swift BJF; Baneyx F Chem. Commun 2015, 51, 3515–3517 DOI: 10.1039/C4CC09531F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coyle BL; Rolandi M; Baneyx F Langmuir 2013, 29, 4839–4846 DOI: 10.1021/la4000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyle BL; Baneyx F Biotechnol. Bioeng 2014, 111, 2019–26 DOI: 10.1002/bit.25257 [DOI] [PubMed] [Google Scholar]
- 19.Coyle BL; Baneyx F Chem. Commun 2016, 52, 7001–4 DOI: 10.1039/C6CC02660E [DOI] [PubMed] [Google Scholar]
- 20.Sooklal K; Cullum BS; Angel M; Murphy CJ J. Phys. Chem 1996, 100, 4551–4555 DOI: 10.1021/jp952377a [DOI] [Google Scholar]
- 21.Ng SM; Koneswaran M; Narayanaswamy R RSC Adv. 2016, 6, 21624–21661 DOI: 10.1039/C5RA24987B [DOI] [Google Scholar]
- 22.Carter KP; Young AM; Palmer AE Chem. Rev 2014, 114, 4564–601 DOI: 10.1021/cr400546e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou D Biochem. Soc. Trans 2012, 40, 635–9 DOI: 10.1042/BST20120059 [DOI] [PubMed] [Google Scholar]
- 24.Wu J; Liu W; Ge J; Zhang H; Wang P Chem. Soc. Rev 2011, 40, 3483–95 DOI: 10.1039/c0cs00224k [DOI] [PubMed] [Google Scholar]
- 25.Yang S; Wang C; Liu C; Wang Y; Xiao Y; Li J; Li Y; Yang R Anal. Chem 2014, 86, 7931–8 DOI: 10.1021/ac5019292 [DOI] [PubMed] [Google Scholar]
- 26.Lin M; Zou HY; Yang T; Liu ZX; Liu H; Huang CZ Nanoscale 2016, 8, 2999–3007 DOI: 10.1039/C5NR08177G [DOI] [PubMed] [Google Scholar]
- 27.Shao N; Zhang Y; Cheung S; Yang R; Chan W; Mo T; Li K; Liu F Anal. Chem 2005, 77, 7294–303 DOI: 10.1021/ac051010r [DOI] [PubMed] [Google Scholar]
- 28.Kim H; Lee BI; Byeon SH Chem. Commun 2015, 51, 725–728 DOI: 10.1039/C4CC08543D [DOI] [PubMed] [Google Scholar]
- 29.Huang S; Zhu FW; Xiao Q; Su W; Sheng JR; Huang CS; Hu BQ RSC Adv. 2014, 4, 46751–46761 DOI: 10.1039/C4RA08169B [DOI] [Google Scholar]
- 30.Gawley RE; Mao H; Haque MM; Thorne JB; Pharr JS J. Org. Chem 2007, 72, 2187–91 DOI: 10.1021/jo062506r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trerayapiwat K; Ricke N; Cohen P; Poblete A; Rudel H; Eustis SN Environ. Sci. Process Impacts 2016, 18, 1068–77 DOI: 10.1039/C6EM00233A [DOI] [PubMed] [Google Scholar]
- 32.Lakowicz JR Principles of fluorescence spectroscopy, 3rd ed.; Springer: New York, 2006; p xxvi, 954 pp. [Google Scholar]
- 33.Cotlet M; Goodwin PM; Waldo GS; Werner JH ChemPhysChem 2006, 7, 250–60 DOI: 10.1002/cphc.200500247 [DOI] [PubMed] [Google Scholar]
- 34.Bol AA; Meijerink A Phys. Rev. B: Condens. Matter Mater. Phys 1998, 58, R15997–R16000 DOI: 10.1103/PhysRevB.58.R15997 [DOI] [Google Scholar]
- 35.Mehta J; Van Dorst B; Rouah-Martin E; Herrebout W; Scippo ML; Blust R; Robbens J J. Biotechnol 2011, 155, 361–9 DOI: 10.1016/j.jbiotec.2011.06.043 [DOI] [PubMed] [Google Scholar]
- 36.DeForest CA; Tirrell DA Nat. Mater 2015, 14, 523–31 DOI: 10.1038/nmat4219 [DOI] [PubMed] [Google Scholar]
- 37.Roose-Amsaleg C; Laverman AM Environ. Sci. Pollut. Res 2016, 23, 4000–12 DOI: 10.1007/s11356-015-4943-3 [DOI] [PubMed] [Google Scholar]
- 38.Rahal JJ Jr.; Simberkoff MS Antimicrob. Agents Chemother 1979, 16, 13–18 DOI: 10.1128/AAC.16.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sletten EM; Bertozzi CR Angew. Chem., Int. Ed 2009, 48, 6974–98 DOI: 10.1002/anie.200900942 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


