Abstract
Glycan analysis by mass spectrometry has rapidly progressed due to the interest in understanding the role of glycans in disease and tumor progression. Glycans are complex molecules that pose analytical challenges due to their isomeric compositions, labile character, and ionization preferences. This study sought to demonstrate infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) as a novel approach for the direct analysis of N-linked glycans. The glycoprotein bovine fetuin was chosen for this analysis as its glycome is well characterized and heavily composed of sialylated glycans. Native N-linked glycans produced by enzymatic cleavage (via PNGase F) of bovine fetuin were analyzed directly by IR-MALDESI in both positive and negative ionization mode. In this study, we detected 12 N-linked glycans in negative mode and 4 N-linked glycans in positive mode, a significant increase in the amount of underivatized glycans detected by other ionization sources. Importantly, all N-linked glycans detected contained at least one sialic acid residue which are known to be labile. This work represents a critical first step for N-linked glycan analysis by IR-MALDESI with future efforts directed at mass spectrometry imaging.
Keywords: IR-MALDESI, N-linked Glycans, Glycomics, Direct Analysis
Graphical Abstract
Liquid droplets of N-linked glycans digested from bovine fetuin glycoprotein ionized by IR-MALDESI.
INTRODUCTION
Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) is an ambient ionization source developed in 2006 combining laser desorption techniques with electrospray ionization (ESI).1,2 IR-MALDESI fires a mid-IR laser at sample material, desorbing neutral species into the gas-phase where they encounter an orthogonal electrospray plume. Neutral molecules partition into the plume and are post-ionized by charged electrospray droplets. Existing evidence shows characteristics of the ionization mechanism in IR-MALDESI resembles those of ESI, making IR-MALDESI a soft and versatile ionization source.3,4 IR-MALDESI has been demonstrated as an effective technique for the direct analysis of biological tissues for metabolites and lipids as well as liquid substrates for high-throughput screening of enzyme activity and biological buffers.5 A significant opportunity not yet pursed is the use of IR-MALDESI for the analysis of N-linked glycans.
Glycans are structurally diverse polysaccharides that pose many analytical challenges affecting the detection of diverse coverage in soft ionization techniques such as ESI and matrix-assisted laser desorption ionization (MALDI). The first challenge involves the labile nature of sialic acid residues, where sample preparation and ionization conditions cause sialic acid residues to dissociate from glycans. This results in misidentified glycans with reduced sialic acid content. Existing evidence shows that sialic acid dissociation is more prevalent in MALDI than ESI, likely as a result of large energy transfer from MALDI matrices during proton transfer.6 Many chemical derivatization reactions exist, such as esterification, in attempts to stabilize sialic acid residues.7 However these sample preparation methods are time-consuming and have variable reaction efficiencies. Another challenge for native glycan analysis is due to the negative charge on the carboxylic acid in sialic acid, which causes the preference for negative ionization. While ESI is capable of achieving sensitive measurements in negative mode, MALDI platforms primarily operate in positive mode due to low sensitivity in negative mode, thereby reducing the diversity of glycans detected by MALDI.8 Due to these limitations, an ESI-like ionization method would be more ideal for N-linked glycan profiling.
The ESI-like characteristics of IR-MALDESI suggest that it would be a good alternative to analyze N-linked glycans due to preservation of sialic acid residues during ionization and the increased sensitivity of negative mode ionization. IR-MALDESI is coupled to a high-resolution accurate mass spectrometer which will provide confident glycan identifications by mass and spectral accuracy. Additionally, IR-MALDESI has been coupled with ion mobility for isomer separation opening up future opportunities for glycan analyses.9 In this study, we report the first analysis of N-linked glycans by IR-MALDESI. The detection of N-linked glycans digested from bovine fetuin was compared between positive and negative ionization modes and putative N-linked glycan structures are reported. This work demonstrates the fundamental ionization of N-linked glycans by IR-MALDESI with the goal to pursue mass spectrometry imaging of biological tissues for changes in N-glycosylation.
METHODS
Digestion and Preparation of N-linked Glycans
N-linked glycans were cleaved from bovine fetuin (Sigma Aldrich, St. Louis, MO, USA) using methods previously described.10 100 mM ammonium bicarbonate (Acros Organics, Geel, Belgium) at pH = 8.3 was prepared for digest buffer. 250 μg of bovine fetuin was loaded onto a 10 kDa MWCO filter with 2 μL of 1 M dithiothreitol (Sigma Aldrich, St. Louis, MO, USA) and 200 μL of digest buffer for incubation at 56°C. The denatured proteins were alkylated with 50 μL of 1 M iodoacetamide (Sigma Aldrich, St. Louis, MO, USA) and incubated at 37°C to prevent reformation of disulfide bonds. The glycoprotein was concentrated onto the filter by centrifuging for 40 minutes. Three washes cycles with 100 μL digest buffer and 20 minutes of centrifugation were completed. 1000 units of PNGase F (Bulldog Bio, Portsmouth, NH, USA) were added to the filter to cleave N-linked glycans from the protein and incubated overnight at 37°C for 18 hours. Glycans were eluted by centrifugation for 20 minutes followed by three wash cycles as previously described. Samples were incubated in the −80°C freezer for 30 minutes and subsequently dried to completion in a vacuum concentrator at room temperature for 5 hours. The dried N-linked glycans were resuspended in 50 μL of optima LC/MS grade water (Fisher Scientific, Waltham, MA, USA) directly before IR-MALDESI analysis.
IR-MALDESI Analysis
5 μL of resuspended glycans were pipetted onto a Teflon microwell slide (Prosolia, Indianapolis, IN, USA). A mid-IR laser (JGM Associates, Burlington, MA, USA) operating at a wavelength of 2.97 μm was used for laser ablation with an energy of 0.4 mJ per burst.9,11 The electrospray solvent consisted of 50% acetonitrile (Fisher Scientific, Hampton, NH, USA) containing 1 mM acetic acid (Sigma Aldrich, St. Louis, MO, USA) for negative mode and 60% acetonitrile containing 0.1% formic acid (Sigma Aldrich, St. Louis, MO, USA) for positive mode. Stable electrospray was achieved with a flow rate of 2 μL/min at a voltage of 3.2 kV. IR-MALDESI was coupled to a Q Exactive HF-X (Thermo Fisher Scientific, Bremen, Germany) set to a mass resolving power of 240,000FWHM at m/z 200, analyzing between 150–1500 m/z in positive and negative ionization mode. Automatic gain control (AGC) was turned off and a fixed injection time of 50 ms was used. High mass measurement accuracy was achieved using lock masses for internal calibration.
Data Analysis
A list of N-linked glycans detected previously from bovine fetuin and reported in literature were compiled to search in the data.12–15 N-linked glycans were identified manually by searching for monoisotopic masses and confirming isotopic distributions in XCalibur. GlycoWorkbench was utilized to compare theoretical monoisotopic masses and search glycan databases by experimental m/z. Putative glycan structures were drawn using the SNGF nomenclature.16,17
RESULTS AND DISCUSSION
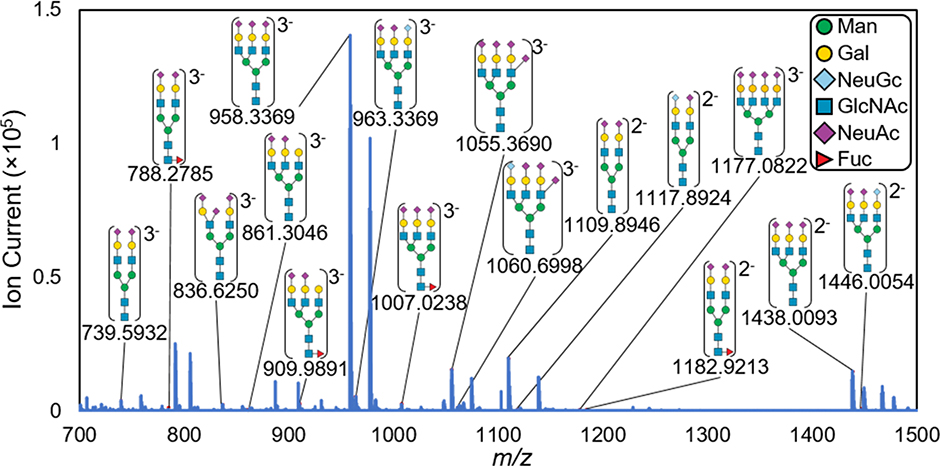
The aim of this work was to demonstrate: 1) the ionization of N-linked glycans by IR-MALDESI; 2) the detection of sialylated glycan species; and 3) the comparison of glycans detected in positive and negative ionization. To easily compare structure types, the detected glycan structures were putatively drawn according to literature-based characterization. As shown in Figure 1, each detected N-linked glycan contains at least one sialic acid residue and more than half the structures contain three and four sialic acid residues. Additionally, the most abundant glycan detected by IR-MALDESI is the trisialylated triantennary form. No dissociation of sialic acid was observed in this analysis, which is supported by the detected glycans being reported in literature as derivatized species. Other abundant peaks in the mass spectrum are unidentified multiply-charged ions that were not found in bovine fetuin literature and are listed in Supplemental Material (Table S1).
Figure 1.
IR-MALDESI mass spectrum of detected N-linked glycans in negative mode with their experimental m/z shown below each putative identification. These N-linked glycans were detected as glycosylamines which is discussed in more detail below. Putative structures were assigned based on accurate mass and literature-based characterizations and are heavily composed of sialic acid residues ( ).
).
Multiply-charged protonated and deprotonated N-linked glycans were detected by IR-MALDESI. The production of multiply-charged ions by IR-MALDESI will be directly beneficial for enhanced structural elucidation by MS/MS fragmentation in untargeted glycan analyses.18 From a list of 42 previously reported N-linked glycans, 12 were detected in negative mode and 4 were detected in positive mode (Table 1, Figure S1). The higher number of glycans detected in negative mode is attributed to the negative charge on sialic acid residues. Additionally, positive mode ionization collects a high number of ambient ions which fill the C-trap due to their constant flux. It is important to note that the 42 previously reported glycans were discovered using chemical derivatization methods. Analyses of native glycans have reported up to 5 and 6 native glycans by negative mode ESI and MALDI, respectively.13,14 Therefore, the 12 native N-linked glycans detected by IR-MALDESI represents a significant increase compared to previous reports in the literature. The glycans not detected by IR-MALDESI were primarily high-mannose glycans which are significantly less abundant than the sialylated glycans in bovine fetuin.
Table 1.
List of N-linked glycans detected as glycosylamines in both positive and negative mode ionization with high mass measurement accuracy (MMA) in parts per million (ppm). More glycans were detected in negative mode ionization. Putative identifications were made based on accurate mass and reported glycans in literature.
| Negative Mode | [M-2H+]2− | [M-3H+]3− | ||
| N-Linked Glycan | Theo m/z | MMA | Theo m/z | MMA |
| Gal2GlcNAc4Man3NeuAc2 | 1109.8922 | 2.1 | 739.5924 | 1.1 |
| Gal2GlcNAc4Man3NeuAc1NeuGc1 | 1117.8897 | 2.5 | ||
| Gal2GlcNAc4Man3Fuc1NeuAc2 | 1182.9212 | 0.1 | 788.2784 | 0.2 |
| Gal2GlcNAc4Man3NeuAc3 | 836.6242 | 1.0 | ||
| Gal3GlcNAc5Man3NeuAc2 | 861.3031 | 1.7 | ||
| Gal3GlcNAc5Man3Fuc1NeuAc2 | 909.9891 | 0.0 | ||
| Gal3GlcNAc5Man3NeuAc3 | 1438.0060 | 2.3 | 958.3349 | 2.1 |
| Gal3GlcNAc5Man3NeuAc2NeuGc1 | 1446.0035 | 1.3 | 963.6666 | 1.9 |
| Gal3GlcNAc5Man3Fuc1NeuAc3 | 1007.0209 | 2.9 | ||
| Gal3GlcNAc5Man3NeuAc4 | 1055.3667 | 2.2 | ||
| Gal3GlcNAc5NeuAc3NeuGc1 | 1060.6984 | 1.4 | ||
| Gal4GlcNAc6Man3NeuAc4 | 1177.0775 | 4.0 | ||
| Positive Mode | [M+2H+]2+ | [M+3H+]3+ | ||
| N-Linked Glycan | Theo m/z | MMA | Theo m/z | MMA |
| Gal2GlcNAc4Man3NeuAc1 | 966.3591 | 2.0 | ||
| Gal2GlcNAc4Man3NeuAc2 | 1111.9068 | 0.1 | ||
| Gal3GlcNAc5Man3NeuAc3 | 1440.0206 | 0.6 | 960.3495 | 0.2 |
| Gal3GlcNAc5Man3NeuAc4 | 1057.3813 | 0.4 | ||
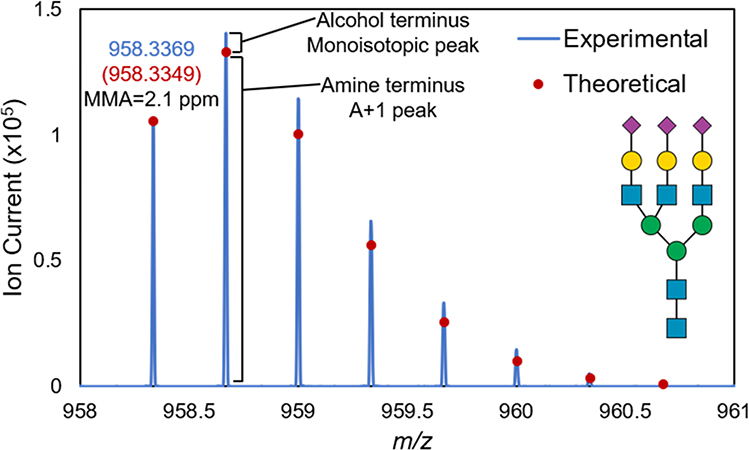
In this study, N-linked glycans were detected with a primary amine terminus. During enzymatic digestion, PNGase F cleaves N-linked glycans from asparagine residues. This produces glycosylamines which are subsequently hydrolyzed in slightly acidic conditions to form an alcohol terminus at the reducing end. The theoretical m/z of the alcohol terminus was approximately 10 ppm from the experimental A+1 peak for all observed glycans, leading us to investigate further. Due to the pH of the ammonium bicarbonate buffer (pH = 8.3) and a lack of an acidic quench in this experiment, N-linked glycans with a primary amine terminus were produced, as observed previously.19 The presence of the primary amine terminus was confirmed based on mass and spectral accuracy.20 Using Gal3GlcNAc5Man3NeuAc3 as an example in Figure 2, the mass accuracy of the monoisotopic peak was reduced to 2.1 ppm when considering the primary amine terminus. Additionally, the theoretical isotopic distribution of a primary amine terminus closely resembles the experimental distribution. Each isotopologue is slightly more abundant than each theoretical abundance due to the presence of both primary amine and alcohol termini, with the primary amine being the predominant form.
Figure 2.
Full experimental isotopic distribution of Gal3GlcNAc5Man3NeuAc3. Mass and spectral accuracy indicate this glycan as having a primary amine terminus. The monoisotopic peak in this distribution was observed at m/z 958.3369 where the expected alcohol terminus structure would have been at m/z 958.6629. Each isotopologue is slightly more abundant than the theoretical abundance due to the presence of both primary amine (major form) and alcohol termini (minor form).
CONCLUSIONS
The work presented here demonstrates the enhanced potential of IR-MALDESI for N-linked glycan analysis. Highly abundant and multiply-sialylated glycans were detected without stabilization by chemical derivatization. Additionally, we demonstrated the improved ionization of glycan species in negative mode compared to positive mode ionization. This work indicates IR-MALDESI as an alternative approach for profiling underivatized N-linked glycans by negative mode ionization. In future work, mass spectrometry imaging of N-linked glycans in biological tissues by IR-MALDESI will be pursued.
Supplementary Material
ACKNOWLEDGEMENTS
This work was performed in part by the Molecular Education, Technology, and Research Innovation Center (METRIC) at NC State University, which is supported by the State of North Carolina. The authors gratefully acknowledge the financial support received from the National Institutes of Health (R01GM087964) and North Carolina State University.
Footnotes
Supporting Information. Mass spectrum of N-linked glycan in positive ionization mode, unidentified peaks in negative ionization mode mass spectrum, complete list of N-linked glycans searched within the data
REFERENCES
- (1).Sampson JS; Hawkridge AM; Muddiman DC Generation and Detection of Multiply-Charged Peptides and Proteins by Matrix-Assisted Laser Desorption Electrospray Ionization (MALDESI) Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2006, 17 (12), 1712–1716. [DOI] [PubMed] [Google Scholar]
- (2).Bokhart MT; Muddiman DC Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry Imaging Analysis of Biospecimens. Analyst 2016, 141 (18), 5236–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dixon RB; Muddiman DC Study of the Ionization Mechanism in Hybrid Laser Based Desorption Techniques. Analyst 2010, 135 (5), 880–882. [DOI] [PubMed] [Google Scholar]
- (4).Tu A; Muddiman DC Internal Energy Deposition in Infrared Matrix-Assisted Laser Desorption Electrospray Ionization With and Without the Use of Ice as a Matrix. J. Am. Soc. Mass Spectrom. 2019, 30 (11), 2380–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nazari M; Ekelöf M; Khodjaniyazova S; Elsen NL; Williams JD; Muddiman DC Direct Screening of Enzyme Activity Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization. Rapid Commun. Mass Spectrom. 2017, 31 (22), 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Harvey DJ Structural Determination of N-Linked Glycans by Matrix-Assisted Laser Desorption/Ionization and Electrospray Ionization Mass Spectrometry. Proteomics 2005, 5 (7), 1774–1786. [DOI] [PubMed] [Google Scholar]
- (7).Reiding KR; Blank D; Kuijper DM; Deelder AM; Wuhrer M High-Throughput Profiling of Protein N-Glycosylation by MALDI-TOF-MS Employing Linkage-Specific Sialic Acid Esterification. Anal. Chem. 2014, 86 (12), 5784–5793. [DOI] [PubMed] [Google Scholar]
- (8).Wuhrer M; Deelder AM Negative-Mode MALDI-TOF/TOF-MS of Oligosaccharides Labeled with 2-Aminobenzamide. Anal. Chem. 2005, 77 (21), 6954–6959. [DOI] [PubMed] [Google Scholar]
- (9).Ekelöf M; Dodds J; Khodjaniyazova S; Garrard KP; Baker ES; Muddiman DC Coupling IR-MALDESI with Drift Tube Ion Mobility-Mass Spectrometry for High-Throughput Screening and Imaging Applications. J. Am. Soc. Mass Spectrom. 2020, 31 (3), 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hecht ES; McCord JP; Muddiman DC A Quantitative Glycomics and Proteomics Combined Purification Strategy. J. Vis. Exp. 2016, 2016 (109), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ekelöf M; Manni J; Nazari M; Bokhart M; Muddiman DC Characterization of a Novel Miniaturized Burst-Mode Infrared Laser System for IR-MALDESI Mass Spectrometry Imaging. Anal. Bioanal. Chem. 2018, 410 (9), 2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sun X; Tao L; Yi L; Ouyang Y; Xu N; Li D; Linhardt RJ; Zhang Z N-Glycans Released from Glycoproteins Using a Commercial Kit and Comprehensively Analyzed with a Hypothetical Database. J. Pharm. Anal. 2017, 7 (2), 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Palmisano G; Larsen MR; Packer NH; Thaysen-Andersen M Structural Analysis of Glycoprotein Sialylation-Part II: LC-MS Based Detection. RSC Adv. 2013, 3 (45), 22706–22726. [Google Scholar]
- (14).Selman MHJ; Hoffmann M; Zauner G; McDonnell LA; Balog CIA; Rapp E; Deelder AM; Wuhrer M MALDI-TOF-MS Analysis of Sialylated Glycans and Glycopeptides Using 4-Chloro-α-Cyanocinnamic Acid Matrix. Proteomics 2012, 12 (9), 1337–1348. [DOI] [PubMed] [Google Scholar]
- (15).Zhou H; Froehlich JW; Briscoe AC; Lee RS The Glycofilter: A Simple and Comprehensive Sample Preparation Platform for Proteomics, N-Glycomics and Glycosylation Site Assignment. Mol. Cell. Proteomics 2013, 12 (10), 2981–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ceroni A; Maass K; Geyer H; Geyer R; Dell A; Haslam SM GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J. Proteome Res. 2008, 7 (4), 1650–1659. [DOI] [PubMed] [Google Scholar]
- (17).Varki A; Cummings RD; Aebi M; Packer NH; Seeberger PH; Esko JD; Stanley P; Hart G; Darvill A; Kinoshita T; Prestegard JJ; Schnaar RL; Freeze HH; Marth JD; Bertozzi CR; Etzler ME; Frank M; Vliegenthart JFG; Lütteke T; Perez S; Bolton E; Rudd P; Paulson J; Kanehisa M; Toukach P; Aoki-Kinoshita KF; Dell A; Narimatsu H; York W; Taniguchi N; Kornfeld S Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25 (12), 1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cramer R; Corless S The Nature of Collision-Induced Dissociation Processes of Doubly Protonated Peptides: Comparative Study for the Future Use of Matrix-Assisted Laser Desorption/Ionization on a Hybrid Quadrupole Time-of-Flight Mass Spectrometer in Proteomics. Rapid Commun. Mass Spectrom. 2001, 15 (22), 2058–2066. [DOI] [PubMed] [Google Scholar]
- (19).Muzikar J; Mechref Y; Huang Y; Novotny MV Enhanced Post-Source Decay and Cross-Ring Fragmentation of Oligosaccharides Facilitated by Conversion to Amino Derivatives. Rapid Commun. Mass Spectrom. 2004, 18 (13), 1513–1518. [DOI] [PubMed] [Google Scholar]
- (20).Khodjaniyazova S; Nazari M; Garrard KP; Matos MPV; Jackson GP; Muddiman DC Characterization of the Spectral Accuracy of an Orbitrap Mass Analyzer Using Isotope Ratio Mass Spectrometry. Anal. Chem. 2018, 90 (3), 1897–1906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.