Abstract
Bearing a relatively large single-stranded RNA genome in nature, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) utilizes sophisticated replication/transcription complexes (RTCs), mainly composed of a network of nonstructural proteins and nucleocapsid protein, to establish efficient infection. In this study, we develop an innovative interaction screening strategy based on phase separation in cellulo, namely compartmentalization of protein-protein interactions in cells (CoPIC). Utilizing CoPIC screening, we map the interaction network among RTC-related viral proteins. We identify a total of 47 binary interactions among 14 proteins governing replication, discontinuous transcription, and translation of coronaviruses. Further exploration via CoPIC leads to the discovery of extensive ternary complexes composed of these components, which infer potential higher-order complexes. Taken together, our results present an efficient and robust interaction screening strategy, and they indicate the existence of a complex interaction network among RTC-related factors, thus opening up opportunities to understand SARS-CoV-2 biology and develop therapeutic interventions for COVID-19.
Keywords: SARS-CoV-2, RTC, nonstructural proteins, protein-protein interaction, CoPIC, high-order complexes, PRC2
Graphical abstract

The unprecedented COVID-19 pandemic is caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Xu et al. establish an innovative interaction screening strategy based on phase separation in cellulo and reveal comprehensive high-order complexes within the SARS-CoV-2 proteome, thus providing more effective therapeutic opportunities.
Introduction
The current pandemic of COVID-19 (coronavirus disease-2019), a respiratory disease leading to more than 164 million confirmed cases and 3.4 million deaths globally (World Health Organization, 2021), is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), an enveloped, positive-sense, single-stranded RNA betacoronavirus of the family Coronaviridae (Zhu et al., 2020). SARS-CoV-2 possesses a remarkably large RNA genome, with a genome size of approximately 30 kb, flanked by 5′ and 3′ untranslated regions. The genomic RNA at the 5′ end features the partly overlapping open reading frames (ORFs) 1a and 1ab, which occupy two-thirds of the capped and polyadenylated genome (Lu et al., 2020; Tan et al., 2005). The expression of ORF1ab requires a ribosomal frameshift near the end of ORF1a. In this manner, SARS-CoV-2 genome translation yields the large replicase polyproteins pp1a and pp1ab (Abduljalil and Abduljalil, 2020; Perlman and Netland, 2009). Pp1a and pp1ab go through extensive autoproteolytic processing, mediated by two ORF1a-encoded proteases and ultimately generating 16 nonstructural proteins (Nsps), which make up the primary constituents of the replication/transcription complexes (RTCs). These include a variety of key enzymes functioning for viral RNA synthesis, i.e., the primase Nsp8 (Kirchdoerfer and Ward, 2019; Konkolova et al., 2020), RdRp-Nsp12 (Cheng et al., 2005; Shannon et al., 2020), the helicase Nsp13 (Jang et al., 2020; Lee et al., 2010), the exoribonuclease Nsp14 (Minskaia et al., 2006), and the endoribonuclease NendoU-Nsp15 (Bhardwaj et al., 2004); several multi-spanning membrane proteins (Nsp3, Nsp4, and Nsp6) that presumably contribute to the formation of double-membrane vesicles (DMVs) (Cottam et al., 2011; Kanjanahaluethai et al., 2007; Oostra et al., 2007); proteases for polyprotein cleavage (Nsp5) (Stobart et al., 2013); and the accessory modulating factors hijacking host defense (Nsp1 and Nsp2) (Antonin et al., 2020; Cornillez-Ty et al., 2009). For the 3′ one-third of the genome, ORFs that encode four structural proteins and nine accessory proteins are transcribed to form a nested set of subgenomic mRNAs (Abduljalil and Abduljalil, 2020).
The synthesis of SARS-CoV-2 RNA within host cells depends on the efficient and correct assembly of RTCs under the coordinated cascade of transcription and replication. This process takes place in intracellular DMVs (Klein et al., 2020; Snijder et al., 2020), which are actually remodeled to serve as a platform for protein-protein interactions (PPIs) and provide a favorable environment for replication. Although SARS-CoV-2 Nsps (e.g., Nsp7, Nsp8, Nsp12, and Nsp13) are associated with the viral RTCs during infection (Yan et al., 2020), the molecular details of which Nsps and how they come together to form the viral RTCs are not clear. Notably, it is also reported that nucleocapsid (N) protein functions as the only structural protein shuttling inside/outside RTCs and facilitating the transcription and replication of coronavirus RNA (Cong et al., 2020; Surjit and Lal, 2008). Recently released structural information revealed the interaction of the C-terminal domain of N protein with the pore factor of DMVs (Khan et al., 2021). As a critical step toward understanding this process, it is necessary to investigate the interaction network among Nsps. Information to date in this regard has been scarce, with only a limited number of interactions identified (Imbert et al., 2008; Pan et al., 2008; von Brunn et al., 2007).
Although multiple methods have been developed to detect PPIs, few are suited for high-throughput protein interaction analysis in cells. The most commonly used methods for investigating PPIs of viruses, including coronaviruses (He et al., 2004; Pan et al., 2008; von Brunn et al., 2007), yeast two-hybrid (Y2H) screening, and coimmunoprecipitation (coIP), have their own limitations and both suffer severe false-positive and false-negative results (Berggård et al., 2007; Miernyk and Thelen, 2008; Phizicky and Fields, 1995). To alleviate these drawbacks, in this study we developed a method to study PPIs in living cells, namely compartmentalization of protein-protein interactions in cells (CoPIC). Essentially, CoPIC is the adapted version of a phase separation-based method for PPIs in vitro in our previous study, called phase-separated condensate-aided enrichment of biomolecular interactions in test tubes (CEBIT) (Zhou et al., 2020). One major requirement for CEBIT is the availability of large quantities of well-behaved purified materials, which is difficult to achieve for intrinsically ill-behaved subjects such as large dynamic protein complexes. Correspondingly in CoPIC, one interaction partner is fused with the scaffolds that drive the formation of phase-separated condensates, and the other partner (named the client) is fused with a fluorescent protein, and can be recruited to the condensate via specific interactions in cells. Overall, CoPIC emerges as a simple, sensitive, and efficient method to investigate PPIs in mammalian cells.
To elucidate molecular mechanisms of viral replication and transcription, there is a need to systematically examine possible intraviral interactions. Although relatively unexplored among the virological studies, intraviral interactions have significant and unexpected effects on host range, transmissibility, immunopathology, and therapeutic effectiveness (Dao et al., 2020; Häuser et al., 2012; Lee et al., 2011; Yin et al., 2019). We therefore established an interaction map for RTC-related viral proteins by CoPIC matrix screening and identified a total of 47 binary interactions among 14 proteins governing replication, discontinuous transcription, and translation of coronaviruses. Moreover, comprehensive high-order complexes within the SARS-CoV-2 proteome were also comprehensively deduced. Overall, increased awareness of intraviral interactions as described herein is a necessary step for achieving a better understanding of SARS-CoV-2 and thus providing more effective therapeutic opportunities.
Results
Establishment of robust scaffolds for phase separation in cells
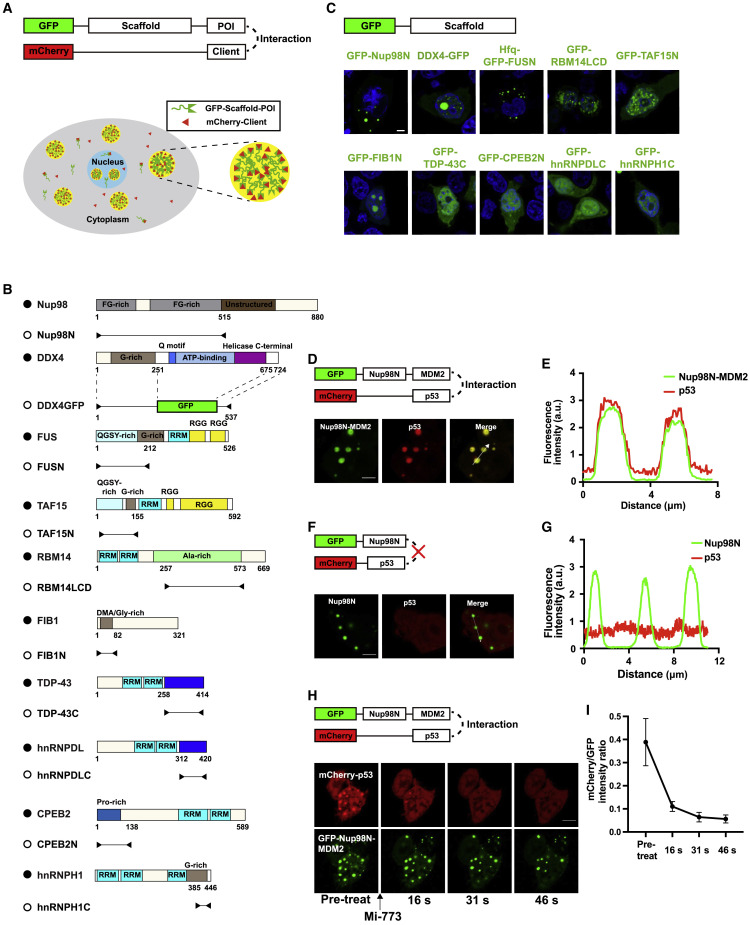
The design strategy of CoPIC is as follows: (1) a system (called the scaffold) capable of forming membraneless phase-separated compartments in cells is generated; (2) one component of the PPI of interest (POI) is fused with the scaffold and hence is enriched in the compartments; and (3) the recruitment of another component (called the client) into the compartments serves as the readout for the interaction (Figure 1 A). For simplicity, we chose phase separation-prone low-complexity domains (LCDs) as potential scaffolds to form the compartmental hubs for interactions (Malinovska et al., 2013; Wang et al., 2018; Weber and Brangwynne, 2012). To this end, we generated a series of expression vectors with a green fluorescent protein (GFP) fused with a number of LCD candidates (Figure 1B), derived from Nup98 (Schmidt and Görlich, 2015), FIB1 (Berry et al., 2015), hnRNPDL (Batlle et al., 2020), RBM14 (Hennig et al., 2015), TAF15 (Couthouis et al., 2011), TDP-43 (Saini and Chauhan, 2011), CPEB2N (Kato et al., 2012), and hnRNPH1 (Wang et al., 2018). In addition, several optimized LCD combinations were also designed, including Hfq-GFP-FUSN, in which the weak multivalent interactions of FUSN to phase separate are enhanced by the homo-hexamer of Hfq (Murakami et al., 2015; Someya et al., 2012), and DDX4GFP, of which the DEAD-box helicase domain within DDX4 was replaced by GFP as previously described with modifications (Nott et al., 2015).
Figure 1.
Establishment of a robust PPI-mediated recruitment system in cells
(A) Schematic diagram showing the strategy of CoPIC. The direct interaction between the protein of interest (POI) and the potential binding client is assessed by the enrichment of mCherry signals into the phase-separated compartments of the GFP-labeled scaffold.
(B) The domain structures of scaffold candidates tested in CoPIC.
(C) Characterization of scaffold candidates fused with GFP in HEK293 cells, with the nucleus stained by DAPI. Scale bar, 5 μm. All assays were performed in triplicate.
(D and F) Validation of the direct interaction between GFP-Nup98N-MDM2 and mCherry-p53 using CoPIC. The p53 fusion protein, as indicated by the mCherry signal, was recruited to the green compartment of GFP-Nup98N-MDM2 by the specific interaction (D). The co-expression of GFP-Nup98N and mCherry-p53 served as the control (F). Scale bars, 5 μm. All assays were performed in triplicate.
(E and G) Fluorescent intensity profiles of the lines with white arrows from (D) and (F).
(H) Treatment of cells co-expressing GFP-Nup98N-MDM2 and mCherry-p53 with Mi-773, an inhibitor of the MDM2/p53 interaction. Scale bar, 5 μm. All assays were performed in triplicate.
(I) Plot of the intensity ratio of mCherry versus GFP under Mi-773 treatment. Each data point was determined by three independent assays, and error bars represent standard deviations.
As shown by the fluorescent images in Figures 1C and S1A–S1G, GFP-Nup98N, DDX4GFP, or Hfq-GFP-FUSN formed green micrometer-scale membraneless spherical compartments while others exhibited poor performance of either irregularly shaped condensates or few condensates. Further analyses indicated higher sphericity of compartments of GFP-Nup98N and Hfq-GFP-FUSN (Figure S1H) while there was a higher proportion of spherical compartments in DDX4GFP (Figure S1I). All compartments formed by GFP-Nup98N, DDX4GFP, or Hfq-GFP-FUSN showed fluidity when examined by fluorescence recovery after photobleaching (FRAP) assays (Figures S1D–S1F). Collectively, we chose GFP-Nup98N, DDX4GFP, and Hfq-GFP-FUSN as potential protein scaffolds of CoPIC for further investigation.
Nup98N condensates compartmentalize client PPI-mediated recruitment
To investigate whether the aforementioned scaffold-derived compartments support client PPI-mediated recruitment, we chose a well-known PPI pair, MDM2/p53 (Momand et al., 1992), as an example for the test. First, we fused MDM2 to the C terminus of GFP-Nup98N, DDX4GFP, or Hfq-GFP-FUSN, yielding the scaffolding vectors GFP-Nup98N-MDM2, DDX4GFP-MDM2, or Hfq-GFP-FUSN-MDM2, respectively. Among the constructs, GFP-Nup98N-MDM2 and Hfq-GFP-FUSN-MDM2 formed robust compartments upon overexpression in HEK293 cells (Figures 1D and S1J), while DDX4GFP-MDM2 failed to form dynamic phase-separated compartments (Figure S1L). When GFP-Nup98N-MDM2 was co-expressed with the transactivation helix of p53 (15–29 aa, abbreviated to p53 hereafter) labeled with mCherry (mCherry-p53) in cells, the mCherry red fluorescence signal was enriched within the green compartments (Figures 1D and 1E). Moreover, the enrichment was MDM2-dependent, as mCherry-p53 was not recruited into GFP-Nup98N-derived compartments (Figures 1F and 1G). Similar results were obtained for Hfq-GFP-FUSN-MDM2 and mCherry-p53 (Figures S1J and S1K).
To further confirm that the enrichment of red fluorescence signal was due to the cognate interaction between MDM2 and p53, we treated the cells co-expressing GFP-Nup98N-MDM2 and mCherry-p53 with Mi-773, a potent inhibitor specifically targeting the MDM2/p53 interaction (Wang et al., 2014). Upon treatment with Mi-773, the mCherry signal within the puncta immediately started to decrease, and little remained by 46 s post-treatment (Figure 1H). Correspondingly, the ratio between the intensity of mCherry over that of GFP and Pearson’s correlation coefficient also decreased (Figures 1I and S1M). Notably, the rapid response to Mi-773 suggests the potential application of CoPIC in high-throughput screening of specific inhibitors for PPI, where the effectiveness of inhibitors is evaluated by the recruitment of clients into the compartments or not. Taken together, our results indicate that GFP-Nup98N can yield robust compartments for recruitment-based PPI detection in cells and thus works as the scaffold for engineered membraneless compartments used in CoPIC for subsequent studies.
Assessment of direct and indirect interactions within the PRC2 complex using CoPIC
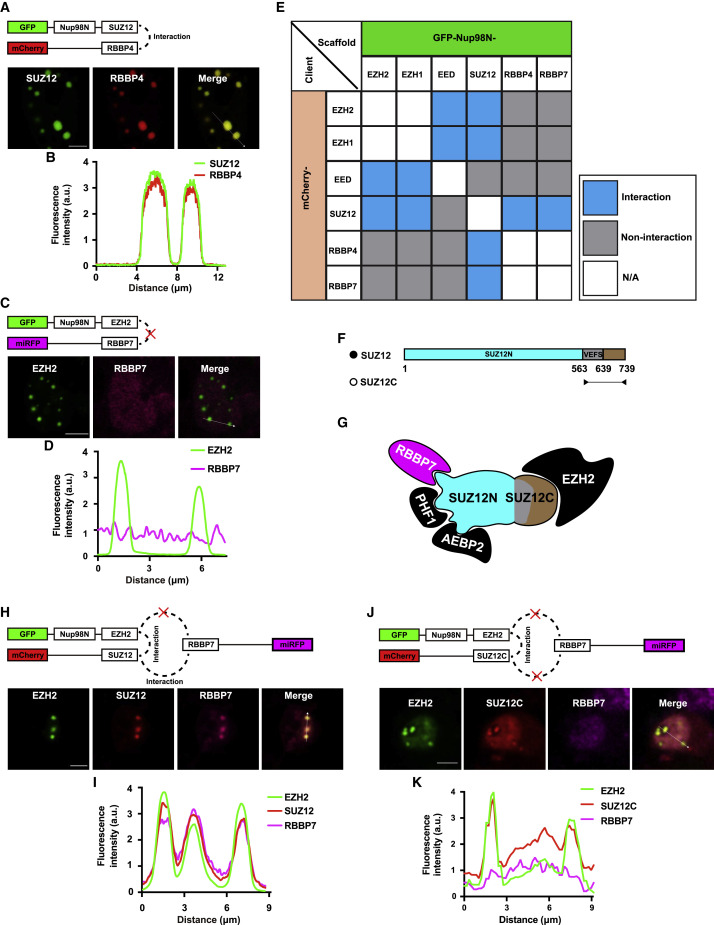
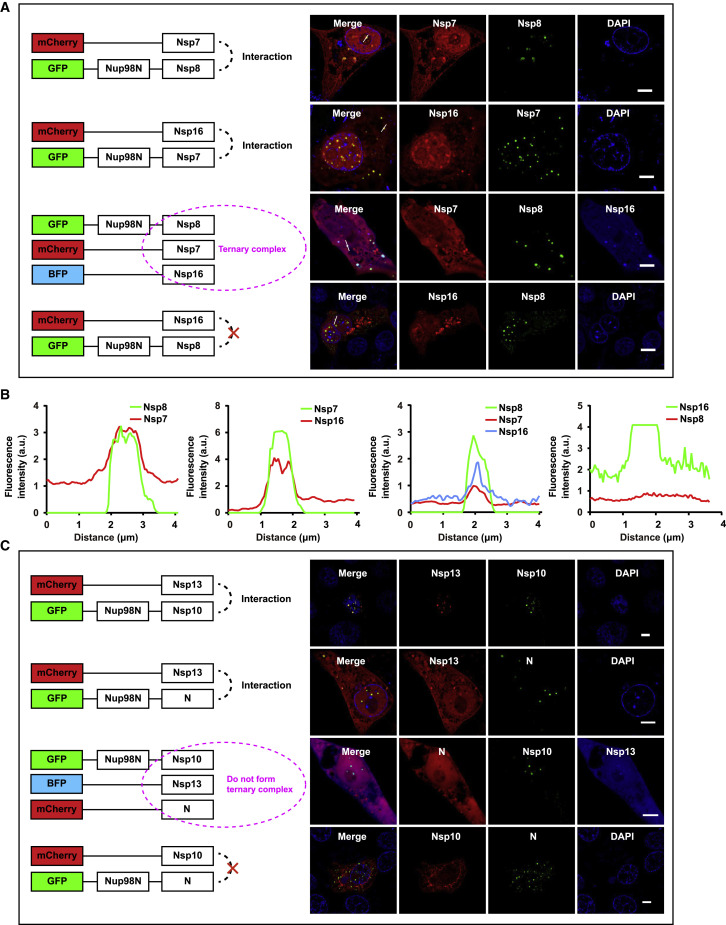
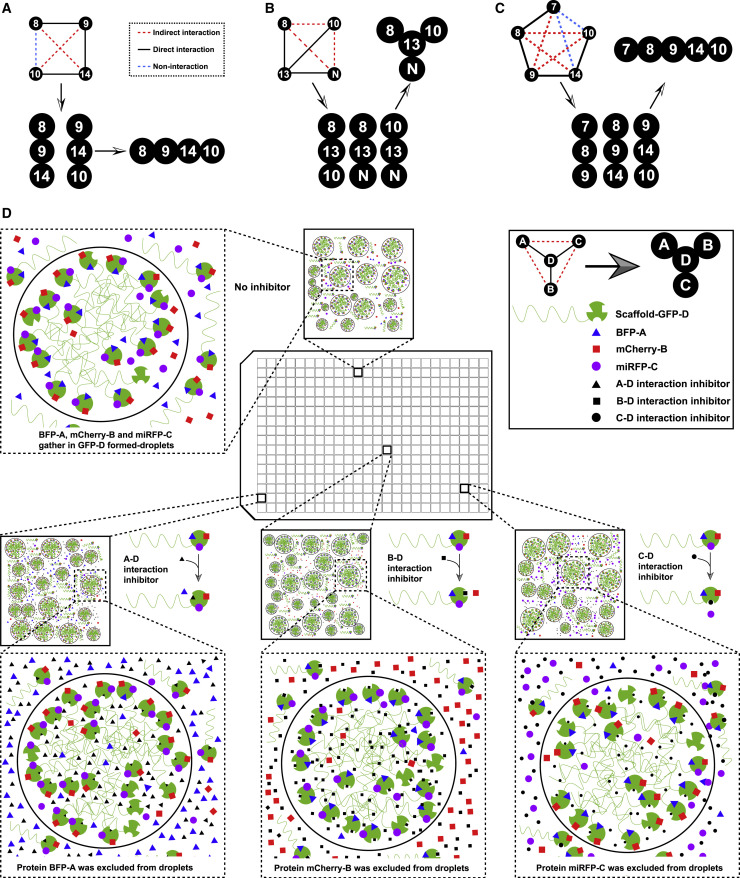
One of the most challenging obstacles in characterizing PPIs is to distinguish direct and indirect interactions. Next, we evaluated whether CoPIC can fulfill the task of discriminating direct or indirect interactions. To this end, we used the well-studied polycomb repressive complex 2 (PRC2) as the testing example. PRC2 is comprised of four core subunits including enhancer of zeste 1 or 2 (EZH1/2), embryonic ectoderm development (EED), suppressor of zeste 12 (SUZ12), and retinoblastoma-binding protein 4 or 7 (RBBP4/7) (Figure S2A) (Margueron and Reinberg, 2011). We generated vectors expressing all PRC2 core components fused with mCherry- or GFP-Nup98N. Based on extensive biochemical and structural studies in the literature, it is known that RBBP4/7 directly interacts with SUZ12 but not with EZH2 (Glancy et al., 2021; Tan et al., 2014). We performed binary co-expression experiments to assess the interaction relationship of these components using CoPIC. Consistently, the co-expression of mCherry-RBBP4 with GFP-Nup98N-SUZ12, but not that of miRFP-RBBP7 with GFP-Nup98N-EZH2, resulted in client enrichment in the compartments (Figures 2 A–2D).
Figure 2.
Characterization of direct and indirect interactions within the PRC2 complex using CoPIC
(A) CoPIC analysis of the positive interaction between SUZ12 and RBBP4. Scale bar, 5 μm. All assays were performed in triplicate.
(C) CoPIC analysis of the negative interaction between EZH2 and RBBP7. Scale bar, 5 μm. All assays were performed in triplicate.
(E) Summary of all pairwise interactions between PRC2 core subunits, including EZH1/2, SUZ12, EED, and RBBP4/7.
(F) Domain structures of SUZ12 and SUZ12C (C-terminal region of SUZ12, known to bind to EZH2 but not RBBP7).
(G) Schematic diagram showing the known factors binding to SUZ12.
(H) CoPIC analysis of indirect interaction of EZH2 with RBBP7 mediated by SUZ12. Scale bar, 5 μm. All assays were performed in triplicate.
(J) Verification of the bridging role of full-length SUZ12 but not SUZ12C in recruiting RBBP7 into EZH2-containing compartments. Scale bar, 5 μm. All assays were performed in triplicate.
(B, D, I, and K) Fluorescence intensity profiles of the lines with white arrows in (A), (C), (H), and (J), respectively, are shown.
Further pairwise interactions between all components of PRC2 were conducted. The results were consistent with the previous studies, where the interactions between EED and EZH1/2, between EZH1/2 and SUZ12, and between SUZ12 and RBBP4/7 were direct and all other interactions were indirect (Figure 2E). As a key component of PRC2, SUZ12 is reported to adopt an extended conformation with the C terminus binding EZH2 and the N terminus binding RBBP4/7 and two accessory proteins, PHF1 and AEBP2 (Figures 2F and 2G). Subsequent CoPIC assays utilizing the C-terminal part of SUZ12, SUZ12C, showed that only EZH2, but not RBBP7, PHF1, or AEBP2, was enriched in the GFP-Nup98N-SUZ12C compartments (Figure S2B). On the contrary, CoPIC assays utilizing the full-length SUZ12 showed substantial enrichment of the signals of RBBP7, PHF1, or AEBP2 within the compartments of GFP-Nup98N-SUZ12 (Figure S2B).
Because we demonstrated that CoPIC can robustly detect direct PPIs, we conducted further investigation into the indirect PPIs within the PRC2 complex. Among the mutual interactions of PRC2 complex components, there are two distinctive interaction patterns: (1) direct contact of RBBP4/7 with SUZ12 mediates the indirect interaction of RBBP4/7 with EZH2; and (2) the interaction of SUZ12 with EED requires the involvement of EZH2 (Ciferri et al., 2012; Kasinath et al., 2018). Next, we sought to characterize the typical indirect interactions via CoPIC and introduced a ternary co-expression system expressing GFP-Nup98N-EZH2, miRFP-RBBP7, and mCherry-SUZ12. Both miRFP and mCherry signals were enriched and co-localized within the compartments of GFP-Nup98N-EZH2, indicating the bridging role of SUZ12 in recruiting RBBP7 into EZH2-containing compartments (Figures 2H and 2I). On the contrary, the co-localization and enrichment were not observed with co-expression of mCherry-SUZ12C (Figures 2J and 2K). Similar results of CoPIC also verified the indirect interaction of SUZ12 with EED via EZH2 (Figure S2C). Taken together, these data indicate that indirect interactions can be unambiguously demonstrated by combining binary and ternary co-transfection experiments via CoPIC.
CoPIC screening of direct PPIs among SARS-CoV-2 RTC-related viral proteins
Taking advantage of the established CoPIC system, we screened PPIs among SARS-CoV-2 RTC-related viral proteins. The genes encoding selected viral proteins (Table S1; Figure S3A) were cloned into the expression vectors fused with fluorescent protein tags such as mCherry, BFP, or GFP-Nup98N. Considering the unique properties of Nsp4 and Nsp6 as transmembrane proteins and the difficulty to synthesize Nsp3, these three Nsps were not included in our analysis. First, the expression and subcellular localization of these proteins was examined in Vero E6 cells, which have been reported to be susceptible to SARS-CoV-2 replication in vitro (Chu et al., 2020). Confocal imaging was carried out to detect the individual proteins at 24 h post-transfection (p.t.). With adequate protein levels expressed in Vero E6 cells, representative images of cellular localization were taken with a confocal microscope. As shown in Figure S3B, most Nsps except Nsp1 and Nsp2 were localized both in the nucleus and cytoplasm. Additionally, the correct sizes of all constructs were further verified by western blot analyses (Figure S3C).
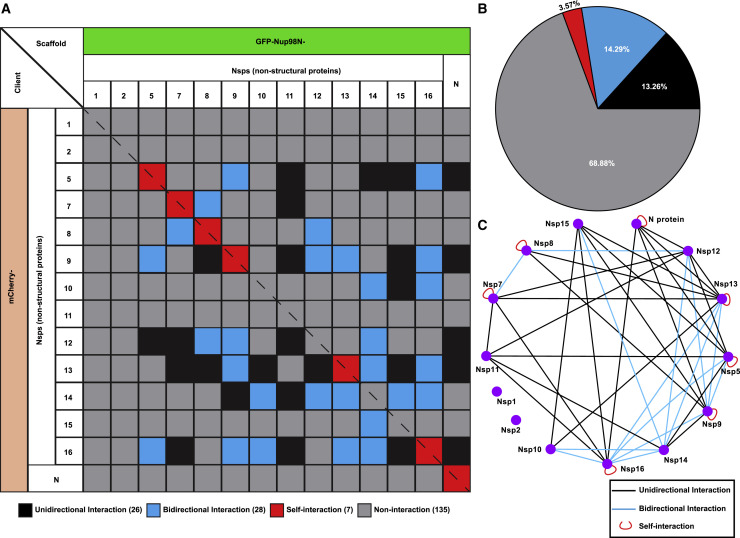
After verifying the proper expression of selected viral proteins, CoPIC screening was conducted for pairwise interactions between all combinations of the 14 proteins. Among the 196 protein pairs tested, a total of 47 interaction combinations were identified as positive and summarized in a pairwise matrix (Figure 3 A). In total, 26 interactions showed one directionality, implying a possible influence of fusion domains on the interactions. Nevertheless, 14 pairs of interactions were detected in both directions. In addition, self-interactions were observed for Nsp5, Nsp7, Nsp8, Nsp9, Nsp13, Nsp16, and N protein, suggesting that these proteins could form dimeric or multimeric complexes by interacting with themselves. However, CoPIC failed to detect the previously reported self-interactions for Nsp10 and Nsp15 (Ricagno et al., 2006; Su et al., 2006), a failure that could be due to an adverse effect of partner fusion on the self-interaction. Taken together, the SARS-CoV-2 RTC-related viral factors interactome-based CoPIC screening suggests concentrated interactions of ORF1b-encoded Nsps that were mainly connected by Nsp12, Nsp13, Nsp14, Nsp15, and Nsp16 (Figures 3B and 3C). The second-most connected proteins were Nsp5, Nsp7, Nsp8, and Nsp10, whereas none was identified for Nsp1 and Nsp2 (Figures 3B and 3C).
Figure 3.
CoPIC screening of intraviral PPIs within SARS-CoV-2 RTC-related factors
(A) Summary of intraviral interactions between selected factors associated with RTCs. The grids with black/blue/red fill represent unidirectional/bidirectional/self-interactions identified by CoPIC while the gray ones are negative interactions.
(B) Analysis of the proportion of each type of interaction for all pairwise interactions.
(C) Annotation for the interaction network with degree-sorted layout.
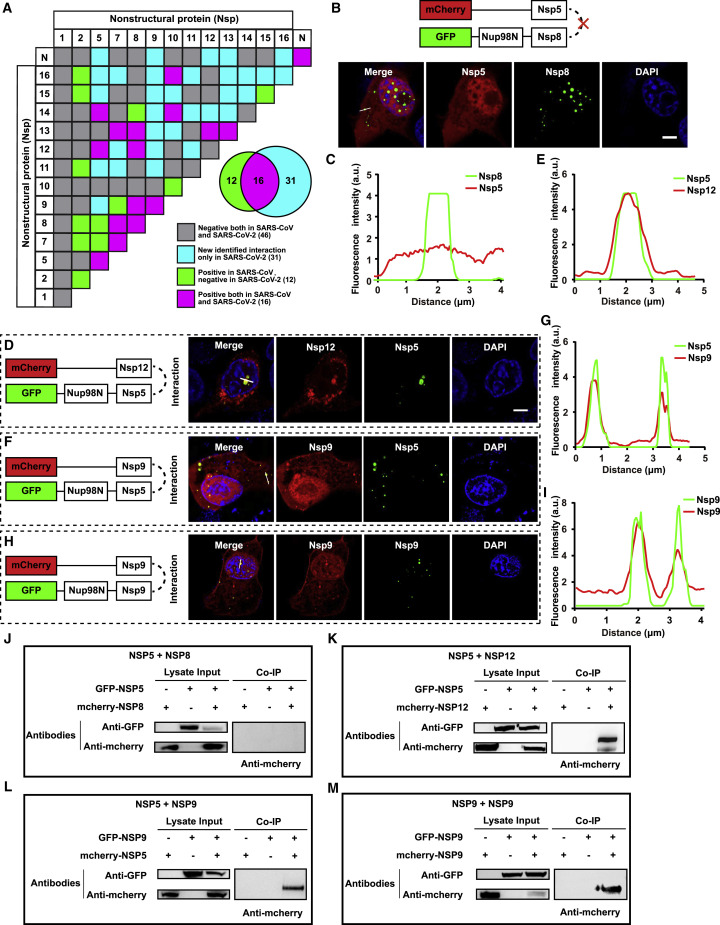
Although employing a similar expression strategy for replication, 31 interaction pairs in SARS-CoV-2 were identified in addition to 16 overlapped hits shared by SARS-CoV (Figure 4 A; Table S2). Among the discovered interaction combination in our study, co-expression of mCherry-Nsp9 and GFP-Nup98N-Nsp5 showed the enrichment of client proteins into the compartments and strong co-localization compared with the negative group (Nsp8-Nsp5) (Figures 4B, 4C, 4F, and 4G). The overlapped positive pairs in SARS-CoV and SARS-CoV-2, i.e., Nsp5-Nsp12 and Nsp9-Nsp9, were also well characterized via CoPIC analyses, implying the possible critical roles for maintaining fundamental functions in the coronavirus family (Figures 4D, 4E, 4H, and 4I). More positive pairwise interactions are exhibited in Figures S4 and S5.
Figure 4.
Analysis of interaction patterns between SARS-CoV and SARS-CoV-2
(A) Comparison of the intraviral interactions of SARS-CoV-2 (detected by CoPIC) with SARS-CoV (reported in the literature). The grids with magenta fill represent the positive interactions identified both in SARS-CoV and SARS-CoV-2; green indicates positive interactions in SARS-CoV while negative in CoPIC screening of SARS-CoV-2; cyan indicates the interactions only in SARS-CoV-2 identified by CoPIC; and gray indicates negative interactions both in SARS-CoV and SARS-CoV-2.
(B, D, F, and H) CoPIC analyses of the representative pairwise PPIs, in which Nsp5-Nsp8 is the negative interaction in SARS-CoV-2 identified by CoPIC, Nsp5-Nsp12 is a well-known interaction both in SARS-CoV and SARS-CoV-2, Nsp5-Nsp9 is an interaction in SARS-CoV-2 identified by CoPIC, and Nsp9-Nsp9 is a self-interaction. Scale bar, 5 μm. All assays were performed in triplicate.
(C, E, G, and I) Fluorescence intensity profiles of the lines with white arrows are shown.
(J–M) Coimmunoprecipitation analyses of the representative pairwise PPIs (B, D, F, and H).
A series of coimmunoprecipitation assays were performed to validate the selected candidates. Briefly, HEK293 cells were co-transfected with two expression plasmids fused with mCherry or GFP tag. At 48 h p.t., cells were lysed and proteins were immunoprecipitated with an anti-GFP monoclonal antibody (mAb). The expression of both tagged proteins was confirmed by western blotting using respective antibodies. As demonstrated in Figures 4J–4M, one of the positive candidate pairs could be immunoprecipitated with another in co-expression samples but not mCherry/GFP-tagged control alone expression samples, further verifying the specific interactions among the above combinations. More pairwise interactions validated by coIP are exhibited in Figure S5B.
Characterization of in-depth interactions within SARS-CoV-2
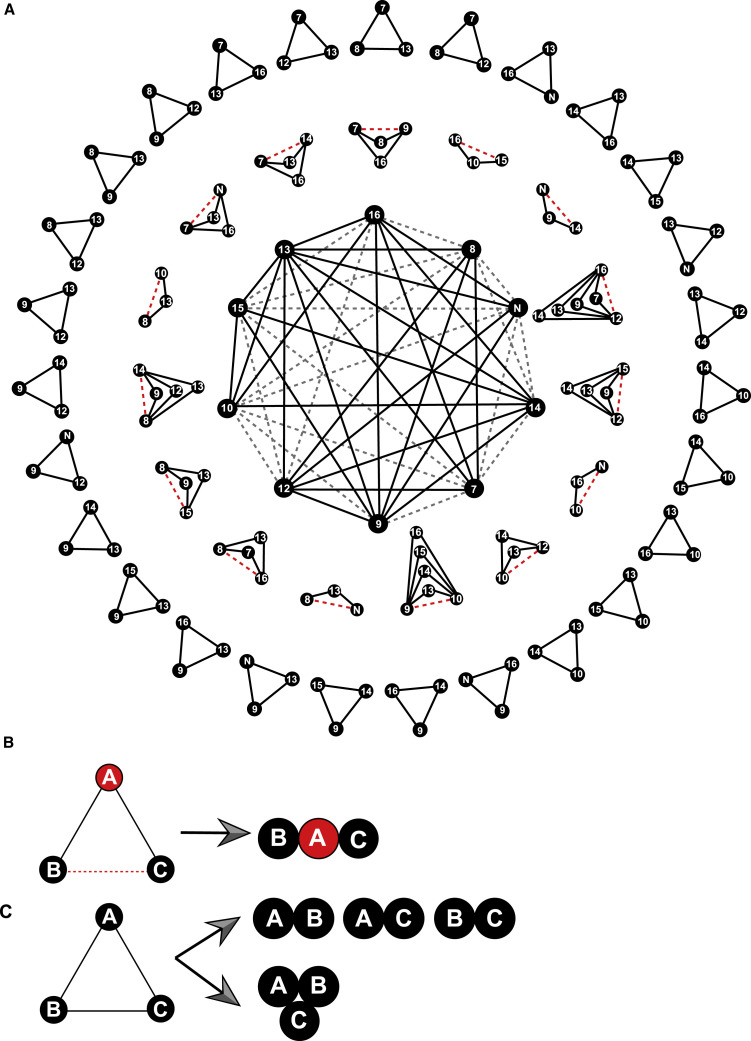
To fulfill a variety of critical functions during the viral life cycle, viral proteins tend to form binary, ternary, or larger complexes with different combinations. Considering that more than half of the interaction pairs turned out to be negative, we speculated that there might be extensive indirect interactions. The corresponding interaction patterns could be discerned, and the detailed strategy of selection is as follows: for virus-encoded proteins, A, B, and C, (1) A interacts with both B and C while B does not interact with C; and (2) it is not until the coexistence of A that B could interact with C, from which we can deduce that A, B, and C are capable of forming a ternary complex (Figures 5 A and 5B). Moreover, a large proportion of pairwise interactions with each other within multimeric complexes should also be mentioned (Figures 5A and 5C). Based on the above strategy, we designed the potential combinations and further investigated the detailed patterns of interactions. Among 137 potential protein combinations tested in our study, 31 positive pairs of indirect interactions were eventually identified (Figures 5 and S6). Taking the key pairs of Nsp7-Nsp8 as an example, there are recently published reports of structural analyses underlying the association of this combination with other Nsps, including Nsp12, Nsp9, and Nsp13 (Gao et al., 2020; Hillen et al., 2020; Wang et al., 2020; Yan et al., 2020). However, these interactions were mainly characterized by in vitro assays and have not been tested within mammalian cells. Herein, we performed CoPIC assays combined with coIP, which further supported the existence of high-order complexes. As shown in Figures S7A and S7B, a combination of CoPIC and coIP analyses showed a validated direct interaction between Nsp8 and Nsp12, Nsp7 and Nsp8, and Nsp7 and Nsp12, thus assembling into the core complex of RTCs to facilitate efficient replication. A ternary complex based on primase (Nsp8) and methyltransferase (Nsp16) was identified, whose formation was dependent on the involvement of Nsp7 (Figures 6 A and 6B). The contrary results were shown in the combination of Nsp10-N-Nsp13, in which Nsp13 failed to mediate the indirect interaction between Nsp10 and N (Figure 6C). Collectively, extensive high-order complexes within SARS-CoV-2 were identified via the unique advantage of the CoPIC system, providing clues for further functional studies.
Figure 5.
Mapping of extensive high-order interactions within SARS-CoV-2
(A) Network of selected viral proteins as the blueprint for mapping high-order complexes. The black solid lines are shown as the direct binary interactions while the red dashed lines are potential indirect interactions with the aid of extra intermediates. The triangles are the hypothetical or CoPIC-verified tertiary complexes and are listed in the outer ring and middle ring, respectively.
(B and C) Interpretation of the patterns of high-order complexes. Direct contacts of A with both B and C mediate the indirect interaction of B with C, thus forming tertiary complexes shown in the middle ring (B). On the contrary, the mode in the outer ring illustrates two possibilities (C): (1) a potential tertiary complex, in which all of the components can interact with each other, and (2) the combinations of multiple binary interactions.
Figure 6.
Characterization of representative indirect interactions of Nsp7-Nsp8-Nsp16 and Nsp10-N-Nsp13
(A) CoPIC analysis of the pairwise interactions within the positive combinations of Nsp7-Nsp8-Nsp16. Scale bars, 5 μm. All assays were performed in triplicate.
(B) Fluorescent intensity profiles of the lines with white arrows from (A).
(C) CoPIC analysis of the pairwise interactions within the negative combinations of Nsp10-N-Nsp13. Scale bars, 5 μm. All assays were performed in triplicate.
Validation of intraviral interactions in the context of infection
Although a large number of intraviral interactions were detected for SARS-CoV-2 in virtue of the large-scale screening analysis in the CoPIC system, ectopic expressions in the absence of virus infection make artificial defects inevitable. Moreover, note that almost all of the fusion proteins might lack an authentic N terminus as even C-terminally tagged expression plasmids in our assay would incorporate an N-terminal methionine while these proteins are generated through proteolytic cleavage and will lack methionine during the infection. Therefore, it would be helpful if the identified interactions were validated in the context of SARS-CoV2-infected cells.
The biosafety level-3 (BSL-3) classification of SARS-CoV-2, however, impedes research and antiviral development. Fortunately, the development of transcription and replication-competent SARS-CoV-2 virus-like-particles (trVLPs) (Ju et al., 2021) made it possible to demonstrate the potential interactions of Nsps derived from the proteolytic cleavage of the polyprotein. This trVLP expresses a reporter gene (GFP) replacing the viral N gene, which is required for viral genome packaging and virion assembly (SARS-CoV-2 GFP/ΔN trVLP). The complete viral life cycle can be achieved and exclusively confined in the cells ectopically expressing SARS-CoV-2 N protein.
Taking advantage of trVLP infection, we analyzed intraviral interaction patterns of N protein with a FLAG tag at the C terminus for detection based on the immunoprecipitation results of infected cell lysates. The precipitate was probed with different antibodies against Nsps. As demonstrated in Figure S7C, N protein precipitated not only a well-known partner Nsp3, but also other viral factors, including Nsp5, Nsp9, and Nsp16, consistent with the above results. Taken together, coIP validation under SARS-CoV-2 trVLP infection further supported the screening results, implying CoPIC as a simple and robust method for investigating PPIs in cells.
Analysis of the interaction hub concentrated on Nsp12-Nsp16
It is well known that ORF1ab of coronavirus is translated upon ribosomal frameshift inside ORF1a, implying significantly lower levels in producing viral proteins from Nsp12 to Nsp16 (Nsp12-Nsp16) than ORF1a-encoded products. Even so, Nsps, from Nsp12 to Nsp16, are thought to serve as a platform to recruit their cofactors (Nsp7 to Nsp10) to the central RTC, thus ensuring efficient replication and transcription. Consistent with this idea, the CoPIC screening demonstrated the concentrated interactions of Nsp12-Nsp16 with their partners (Figures 3, 4, and 5), suggesting the potential roles of intraviral interactions in the sophisticated regulation of enzyme activities.
Taking the canonical RNA-dependent RNA polymerase as an example, Nsp12 exhibits poorly processive RNA synthesis in vitro, contrasting with the efficient replication of coronavirus RNA genomes with a large size in vivo (Ahn et al., 2012; Cheng et al., 2005; te Velthuis et al., 2010). However, the involvement of the Nsp7/Nsp8 complex can increase the binding of Nsp12 to RNA, thus activating and conferring the processivity of Nsp12 to the RNA synthesis (Subissi et al., 2014). The Nsp7/Nsp8 complex increases the binding of Nsp12 to RNA, resulting in a larger stretch of nucleotide synthesized per binding event. Nsp8 works as RNA primase and Nsp7 might act to stabilize Nsp8 (Gao et al., 2020; Imbert et al., 2006; Yan et al., 2021; Zhai et al., 2005). Correspondingly, our CoPIC analysis combined with coIP confirmed direct interactions between Nsp12 and Nsp7/8 (Figure S7), consistent with the revealed atomic structure (Hillen et al., 2020).
For another presumed RTC catalytic core, Nsp13 helicase activity is also stimulated significantly in the presence of Nsp12 by a direct interaction identified both in SARS-CoV (Adedeji et al., 2012) and our study (Figures 3 and S5A). Cryo-electron microscopy observation revealed an architecture of SARS-CoV-2 mini RTCs assembled by the dynamic interactions of Nsp13 with the RdRp complex (Yan et al., 2020), whose reconstruction in vitro was also partially verified by the CoPIC analyses in the mammalian cells (Figures 6, S6, and S7). Likewise, in vitro studies showed that the extra addition of the Nsp10 protein enhances the weak Nsp14 ExoN activity (Baddock et al., 2020; Bouvet et al., 2012). In the case of Nsp16, a strong interaction of Nsp16 with Nsp10 was shown to trigger its 2′-O-methyltransferase (2′-O-MTase) activity (Bouvet et al., 2010; Imbert et al., 2008; Lin et al., 2020; Mahalapbutr et al., 2020). The direct interactions of both Nsp10-Nsp14 and Nsp10-Nsp16 were also validated by CoPIC (Figures 3 and S4), implying the importance of Nsp10 in two distinct regulatory mechanisms (Nsp14-ExoN and Nsp16-2′-O-MTase). Moreover, the CoPIC screening identified the interaction between Nsp14 and Nsp16 (Figures 3 and S4). There might be a tertiary complex of Nsp14-Nsp10-Nsp16, in which Nsp10-dependent activation of both enzymes may happen simultaneously. Nevertheless, more evidence would be required to confirm the non-limiting levels of Nsp10 to simultaneously activate the enzyme activities of Nsp14 and Nsp16.
Combined with the identified interaction of Nsp12 with Nsp14, an N 7-MTase that can methylate the first G of viral RNA (Ferron et al., 2018; Ma et al., 2015; Yan et al., 2021), we surmise that Nsp7/Nsp8/Nsp12/Nsp13/Nsp14 might tend to constitute a higher-order complex as the minimal viral replisome. If so, such assembly would be in charge of more capping activities, i.e., RTPase and N 7-MTase, as well as the unwinding of RNA secondary structures, RNA polymerization, and even mismatch excision by the 3′-to-5′ exoribonuclease activity of Nsp10-Nsp14. Under the last capping step, the Nsp10/Nsp16 complex would undergo methylation of cap-0 to cap-1 through its 2′-O-MTase activity (Krafcikova et al., 2020; Rosas-Lemus et al., 2020), and it might act periodically during Nsp7/Nsp8/Nsp12/Nsp13/Nsp14-mediated replication. Collectively, our interactome studies confirmed the significance and necessity of the key enzymes encoded by replicase ORF1b forming high-order complexes with the cofactors for efficient replication and transcription during the viral life cycle.
Exploration of potential higher-order complexes
Based on the screening by CoPIC, we constructed a comprehensive high-order interactome for SARS-CoV-2 RTC-related viral proteins (Figure 3). The resulting map reveals a focus on ORF1b-centered interactions that are mainly connected by Nsp12, Nsp13, Nsp14, Nsp15, and Nsp16, with associated enzymatic activities that are essential for viral RNA replication and protein translation. Further extensive interactions combined with the corresponding high-order complexes imply a crucial role for Nsps in recruiting core replication/transcription proteins for the assembly of the SARS-CoV-2 RTC and thus performing diverse functions during the viral life cycle. Integrated analyses of tertiary complexes identified by CoPIC revealed the various possibilities to trigger indirect interactions (Figure 6). Examples include the indirect interaction of Nsp7 with Nsp9 dependent on the involvement of intermediates including Nsp8, Nsp12, Nsp13, or Nsp16.
Since the coronavirus RTCs engage in a variety of RNA synthesis processes, one type of complex may be converted into another by the binding or release of specific protein partners. In this manner, such protein factors may either regulate the balance between different processes or directly form more sophisticated complexes with the core enzymes. Correspondingly, we derived several potential higher-order complexes comprising direct and indirect interactions. For example, the tandem mode of Nsp8-Nsp9-Nsp14-Nsp10 illustrates the cascaded linkage of indirect interactions Nsp8-Nsp9-Nsp14 and Nsp9-Nsp14-Nsp10, in which Nsp8 and Nsp10 bind to the different interfaces of the core Nsp9-Nsp14 and do not share any contacts (Figure 7 A). Multiple pairs of indirect interactions shared by the same intermediates result in a Y-shaped mode of Nsp8-Nsp10-Nsp13-N (Figure 7B), which emphasizes the mutual linkages of binding factors with intermediates in different interfaces with no interactive events between themselves (Figure 7B). On the contrary, a quinary tandem combination of Nsp7-Nsp8-Nsp9-Nsp14-Nsp10 comprising different intermediates was identified, exhibiting the successive chain (Nsp7-Nsp8-Nsp9, Nsp8-Nsp9-Nsp14, Nsp9-Nsp14-Nsp10) without forming a closed loop by end-to-end interaction (Figure 7C). More detailed high-order complexes are shown in Figure S8.
Figure 7.
The outlook for higher-order complexes and a potential HTS scheme for PPI inhibitors
(A–C) Simplified model systems deciphering the potential higher-order complexes.
(D) Schematic diagram of high-throughput screening for specific inhibitors against intraviral complexes.
The above high-order complexes based on CoPIC also exhibit good potential applications in the high-throughput screening for inhibitors against PPIs (Figure 7D). Simply put, the core intermediate (D) fused with the scaffold acts as the platform to recruit other client components (i.e., A to C) by different interfaces and thus form the high-order complex. The subsequent screening for effective inhibitors disrupting the pairwise interactions would be performed in combination with an imaging-based method, and the activity of each candidate would be determined by assessing the enrichment of a fluorophore-labeled client into condensates enriched with its binding partner. Taken together, these simplified model systems enable more precise clues for the formation of high-order complexes during the different stages of the viral life cycle and potential therapeutic opportunities against SARS-CoV-2 by repurposing specific inhibitors to disrupt the intraviral PPIs.
Discussion
With burgeoning knowledge about SARS-CoV-2 molecular architectures, it is essential to dissect the Nsp interaction network and thus understand the assembly of RTCs. In this regard, our study established an innovative interaction screening strategy based on phase separation in cellulo, that is, CoPIC. In the past years, various fluorescence-based methods for in cellulo visualization of protein-protein interactions have been introduced. Although it seems easy for visualization of PPIs to observe the colocalization of proteins tagged with different fluorescence, it is difficult to distinguish whether the proteins in the living cells have real interaction or just incidentally co-localized in the same place. Two relatively popular in vivo techniques for studying PPIs, fluorescence resonance energy transfer (Miyawaki, 2003; Sekar and Periasamy, 2003) and bimolecular fluorescence complementation (Kerppola, 2006), are based on the expression of fluorescently labeled proteins or fragments thereof. However, fluorescence resonance energy transfer requires costly instrumentation and advanced technical expertise, whereas bimolecular fluorescence complementation is based on the irreversible complementation and slow maturation of fluorophores, which does not allow real-time detection of PPIs. Even though alternative strategies based on the relocation of prey proteins have been developed, i.e., mitochondria-docking (Mito-docking) (Shao et al., 2019) and a differential cytolocalization assay (DCLA) (Blanchard et al., 2006), they are unlikely to distinguish between direct protein-protein interactions or those bridged by other peptides.
Since CoPIC detects the enrichment of client proteins in membraneless compartments versus the surrounding environment, discernible enrichment means the establishment of a concentration gradient. Therefore, only authentic interactions can overcome the associated entropy loss. As such, detection of nonspecific PPIs, i.e., false-positives, can be minimized. Nevertheless, if the binding partners to be recruited prefer the specific scaffold-derived condensates versus the surrounding environments, false-positive results can be generated. However, with proper controls, such false-positive hits can be detected, and a change of scaffold can be exploited to circumvent this issue. There is another caveat associated with CoPIC. If the partner to be recruited can also undergo phase separation, the potentially complicated interaction relationship between the scaffold phase and the client phase might hinder the straightforward recruitment of the clients. For example, for the PPI screening involving the N protein, there were hardly any positive interactions detected except for its self-association when N protein serves as the client; however, multiple PPIs were detected when N was fused with the scaffold, GFP-Nup98N (Figure 3A). We reasoned that the phase separation property of N protein (Iserman et al., 2020; Perdikari et al., 2020; Zhao et al., 2020) interferes with its recruitment into the phase-separated compartment of the scaffolds. To circumvent this caveat, binding partners with phase separation propensity should be fused with the scaffold, and those without phase separation propensity should serve as the clients.
In previous studies with the yeast two-hybrid system for identifying PPIs within SARS-CoV (Imbert et al., 2008; von Brunn et al., 2007), Nsp2 was shown to have a wide range of interactions with other viral proteins, including Nsp3N, Nsp6, Nsp8, Nsp11, and Nsp16, but these phenomena could not be observed in our CoPIC assays. Eliminating the misgivings on the inappropriate expression (Figure S3C), we surmise that the difference between the current results and previous data can be explained as follows. (1) The absence of protein modifications in the Y2H system. Since protein modifications that are likely to influence protein interactions might be different for certain proteins in the context of yeast and mammalian cells, our CoPIC system might exhibit better performance in reflecting genuine interactions for human viruses. Moreover, the results performed by mammalian two-hybrid assays against SARS-CoV (Pan et al., 2008) seemed not to have any overlap with those results from the Y2H system. (2) The poor reproducibility of the Y2H system. It was striking that none of the interactions revealed in the two independent studies (Imbert et al., 2008; von Brunn et al., 2007) was overlapping, although both used the Y2H system for the screening. (3) The potential interference of Nsp2 with the intracellular milieu. It has been shown that Nsp2 interacts with two host proteins, prohibitin 1 (PHB1) and PHB2 (Cornillez-Ty et al., 2009; Davies et al., 2020), which have been implicated in cellular functions, including cell-cycle progression, cell migration, cellular differentiation, apoptosis, and mitochondrial biogenesis. More notably, continuous investigations on host interactors for SARS-CoV-2 proteins further profile eukaryotic translation initiation factor 4E family member 2 (EIF4E2 or 4EHP) and GRB10 interacting GYF protein 2 (GIGYF2) as the partners of Nsp2 (Davies et al., 2020; Gordon et al., 2020), known to regulate translation initiation and endosome vesicle sorting. It is quite possible that the overexpression of Nsp2 may change the intracellular milieu and perturb host intracellular signaling, thus interfering with the recruitment into the Nup98 compartments.
Even with these caveats considered, CoPIC is a simple and efficient method for investigating PPIs in cells. In addition, CoPIC can be used to detect weak interactions since the signals are enriched within the compartments and the client proteins are overexpressed to high concentrations. Taking the interaction of Nsp14 with Nsp16 as an example, the negative pair demonstrated by the typical coIP assay turned out to exhibit a salient bidirectional colocalization within the compartment mediated by GFP-Nup98N (Figures 3, S4B, and S4C). Notably, CoPIC allows the compartmentalization of multiple PPIs simultaneously, which is hard to achieve by other approaches and is important for the dissection of direct and indirect PPIs, hence the potential high-order complexes (Watanabe et al., 2017; Zhang et al., 2018). Better yet, an extra benefit is that CoPIC is especially suitable for PPI detection in a high-throughput fashion using an imaging-based method, e.g., high-content imaging techniques.
When applied to investigate the complicated interaction within the SARS-CoV-2 proteome, the availability of CoPIC allowed the revelation of several salient findings: (1) SARS-CoV-2 shares conserved interactions with SARS-CoV and might have also evolved more interactions to facilitate its efficient replication/transcription; (2) most of the 16 Nsps are associated with viral RTC; and (3) the interactions are centered on ORF1b-encoded Nsps that were mainly connected by Nsp12, Nsp13, Nsp14, and Nsp16.
Although utilizing a similar expression strategy for efficient replication/transcription, SARS-CoV-2 exhibits a quite divergent pattern from other coronaviruses, sharing only a limited number of homologs (Figure 4A). By comparative analyses of the interactions between the two subspecies, SARS-CoV and SARS-CoV-2, in combination with independently published data (Li et al., 2021; Pan et al., 2008; von Brunn et al., 2007), the conserved interactions can be classified in the following categories: (1) complex formation among the scaffold proteins, where Nsp7 and Nsp8 support the RNA-dependent RNA polymerase function of nsp12, and nsp10 facilitates the methyltransferase activities of nsp14 and nsp16 by the respective associations; (2) interaction between RNA-dependent RNA polymerase (Nsp12) and other key enzymes, including primase (Nsp8), helicase (Nsp13), and guanine-N 7-MTase (Nsp14); (3) a connection of the main protease (Nsp5) to RdRp (Nsp12) and guanine-N 7-MTase (Nsp14); and (4) self-interaction of the accessory proteins such as Nsp5, Nsp7, Nsp8, Nsp9, Nsp13, and Nsp16, and structural protein N. These conserved interactions highlight the evolutionary convergence on the replication strategies at the protein-protein interaction level and point to its potentially critical role in regulating viral replication.
It is striking to identify 31 interactions among RTC-related factors in our study, several of which were confirmed using coIP (Figures 4 and 5). However, note that the negative results may be due to the limitations of the assay itself and do not mean that the interaction cannot occur during infection. Of these, Nsp1 and Nsp2 have the fewest binding partners, which might result from their effect on host viability and suppression of protein synthesis (Angeletti et al., 2020; Garmashova et al., 2006; Rao et al., 2020; Yuan et al., 2020). Among the identified interactions, perhaps the most interesting are those taking place between ORF1b-encoded core enzymes (Nsp12, Nsp13, Nsp14, Nsp15, and Nsp16), responsible for the efficient genome replication and transcription. Compared with other well-known core enzymes encoded by SARS-CoV-2, the role of Nsp15 in the viral cycle is still unknown. Nsp15, also named NendoU (nidovirus endoribonuclease, specific for U), preferentially cleaves the 3′ end of U and forms 2′-3′ cyclic phosphate ends (Kim et al., 2020). Previous reverse genetics analyses suggested the dispensable role of NendoU activity during viral replication (Ivanov et al., 2004). Mutations of the catalytic site with His and Lys residues in MHV were shown to promote a subtle defect in subgenomic mRNA accumulation, while mutation of a conserved Asp residue both in HCoV-229E and MHV almost abolished viral RNA replication (Ivanov et al., 2004; Kang et al., 2007). In addition, Nsp15 might be responsible for evading the host innate immune system, which is also independent of its endonuclease activity (Liu et al., 2019; Zheng and Perlman, 2018). Intriguingly, our CoPIC screening identified multiple binding partners, mainly concentrated on Nsp9, Nsp10, Nsp13, Nsp14, and Nsp16, suggesting the potential involvement of Nsp15 in multiple sophisticated processes. These findings lay the foundation for further functional studies.
In summary, the current work established a sensitive method for identifying PPIs in living cells and thus constructed an interaction network of SARS-CoV-2 RTC-related factors. Among the identified interactions in combination with the data obtained by other methods, the most intriguing and striking are those between the viral core enzymes and the corresponding high-order complexes. While the functional importance of most intraviral PPIs remains less well understood, our network provides a powerful resource to dissect their roles in mediating the genome replication/transcription processes of SARS-CoV-2. The extensive interaction landscape among intraviral proteomes seems to be the norm (Bartel et al., 1996; Choi et al., 2000; Dimitrova et al., 2003; Guo et al., 2001; McCraith et al., 2000; Zell et al., 2005), underscoring the crucial roles of PPIs in the viral life cycle. Hence, the disruption of these intraviral PPIs would likely provide therapeutic opportunities against SARS-CoV-2. Notably, we have been already actively engaging in high-throughput screening for drugs disrupting essential PPIs within SARS-CoV-2 using CoPIC.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mCherry (E5D8F) Rabbit mAb | Cell Signaling Technology | Cat #43590; RRID: AB_2799246 |
| GFP (D5.1) Rabbit mAb | Cell Signaling Technology | Cat #2956; RRID: AB_1196615 |
| DYKDDDDK Tag (D6W5B) Rabbit mAb | Cell Signaling Technology | Cat #14793, RRID: AB_2572291 |
| SARS-CoV-2 NSP3 Rabbit pAb | ABclonal Technology | Cat #A20236, RRID: AB_2891174 |
| SARS-CoV-2 3CLpro Rabbit pAb | ABclonal Technology | Cat #A20198, RRID: AB_2891175 |
| SARS-CoV-2 NSP9 Rabbit pAb | ABclonal Technology | Cat #A20308, RRID: AB_2891176 |
| SARS-CoV-2 NSP16 Rabbit pAb | ABclonal Technology | Cat #A20283, RRID: AB_2891177 |
| F(ab’)2-Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, HRP | Thermo Fisher Scientific | Cat #A24537; RRID: AB_2536005 |
| Bacterial and virus strains | ||
| Escherichia coli DH5α | Tiangen Biotech | CB101-02 |
| Chemicals, peptides, and recombinant proteins | ||
| Kanamycin | Inalco | Cat #1758-9316; CAS: 70560-51-9 |
| Ampicillin | Inalco | Cat #1758-9314; CAS: 69-52-3 |
| Phenylmethylsulfonyl fluoride (PMSF) | Amresco | Cat #0754; CAS: 329-98-6 |
| PBS | Solarbio | Cat P1020 |
| 4% Paraformaldehyde | Beyotime Biotechnology | Cat #P0099; CAS: 30525-89-4 |
| Hoechst 33258 | Beyotime Biotechnology | Cat #C1017; CAS: 23491-45-4 |
| DMEM | Hyclone | Cat #SH30022.01 |
| FBS | GIBCO | Cat #10099-141 |
| GFP-Trap Magnetic Agarose | Chromotek | Cat #gtma-100 |
| Protease inhibitor cocktail | Bimake | Cat #B14002 |
| MI-773 (SAR405838) | Selleck | Cat #S7649 |
| FuGENE® HD Transfection Reagent | Promega | Cat #E2311 |
| Rabbit IgG | Beyotime | Cat #A7016 |
| Critical commercial assays | ||
| Total Protein Extraction Kit for Animal Cultured Cells/Tissues | Invent | Cat #SD-001 |
| BCA Protein Assay Kit | Thermo Fisher Scientific | Cat #23227 |
| Chemiluminescence reagent | Thermo Fisher Scientific | Cat #34077 |
| Deposited data | ||
| Microscopy images | This paper | https://doi.org/10.17632/gtswbcb5nv.2 |
| Experimental models: Cell lines | ||
| Vero E6 | A gift from J.Z. Lab, Tsinghua University | N/A |
| Caco-2-NFlag | A gift from Q.D. Lab, Tsinghua University | N/A |
| HEK293 | Cell Bank/Stem Cell Bank, Chinese Academy of Sciences | Cat #GNHu 43 |
| Oligonucleotides | ||
| GFP-Nup98N (forward primer for PCR) | gtagcggcggctccctcgaaatgtttaa caaatcatttggaacacccttt |
N/A |
| GFP-Nup98-3.1 (reverse primer for PCR) | cctctagatgcatgctcgagttcctcttcc ttcttcttagggtctga |
N/A |
| GFP-Nup98-EZH1 (forward primer for PCR) | taagaagaaggaagaggaactcgagggcggt agcggtggcagcggtggtagcggcggctccat ggaaataccaaatcccccta |
N/A |
| GFP-Nup98-EZH1 (reverse primer for PCR) | acactatagaatagggccctctaga ctaaaggacgtcggtctccctctcg |
N/A |
| mCherry-EZH1 (forward primer for PCR) | cggtggtagcggcggctccctcga gatggaaataccaaatccccctacct |
N/A |
| mCherry-EZH1 (reverse primer for PCR) | acactatagaatagggccctctag actaaaggacgtcggtctccctctcg |
N/A |
| GFP-Nup98-RBBP4 (forward primer for PCR) | taagaagaaggaagaggaactcga gggcggtagcggtggcagcggtggtagcggcg gctccatggccgacaaggaagcagccttcg |
N/A |
| GFP-Nup98-RBBP4 (reverse primer for PCR) | acactatagaatagggccctctagact aggacccttgtccttctggatcc |
N/A |
| mCherry-RBBP4 (forward primer for PCR) | cggtggtagcggcggctccctcgag atggccgacaaggaagcagccttcg |
N/A |
| mCherry-RBBP4 (reverse primer for PCR) | acactatagaatagggccctctagac taggacccttgtccttctggatcc |
N/A |
| GFP-Nup98-EED (forward primer for PCR) | aagaaggaagaggaactcgagggc ggtagcggtggcagcggtggtagcggc ggctccatgtccgagagggaagtgtc |
N/A |
| GFP-Nup98-EED (reverse primer for PCR) | cactatagaatagggccctctagatgcat gttatcgaagtcgatcccagc |
N/A |
| mCherry-EED (forward primer for PCR) | aagaaggaagaggaactcgagggcggt agcggtggcagcggtggtagcggcggct ccatgtccgagagggaagtgtc |
N/A |
| mCherry-EED (reverse primer for PCR) | cactatagaatagggccctctagatgcat gttatcgaagtcgatcccagc |
N/A |
| BFP-EED (forward primer for PCR) | aagaaggaagaggaactcgagggcggta gcggtggcagcggtggtagcggcggctcc atgtccgagagggaagtgtc |
N/A |
| BFP-EED (forward primer for PCR) | cactatagaatagggccctctagat gcatgttatcgaagtcgatcccagc |
N/A |
| GFP-Nup98-EZH2-3.1 (forward primer for PCR) | gtagcggcggctccctcgagatgggc cagactgggaagaaatctgagaag |
N/A |
| GFP-Nup98-EZH2-3.1 (reverse primer for PCR) | cctctagatgcatgctcgagtcaagggatt tccatttctctttcgatgcc |
N/A |
| mCherry-EZH2-3.1 (forward primer for PCR) | gtagcggcggctccctcgagatgggcca gactgggaagaaatctgagaag |
N/A |
| mCherry-EZH2-3.1 (reverse primer for PCR) | cctctagatgcatgctcgagtcaagggat ttccatttctctttcgatgcc |
N/A |
| GFP-Nup98-RBBP7 (forward primer for PCR) | agaagaaggaagaggaactcgagggcggta gcggtggcagcggtggtagcggcggctccat ggcgagtaaagagatgtt |
N/A |
| GFP-Nup98-RBBP7 (reverse primer for PCR) | atagaatagggccctctagatgcatgttaa gatccttgtccctccagttc |
N/A |
| mCherry-RBBP7 (forward primer for PCR) | agaagaaggaagaggaactcgagggcgg tagcggtggcagcggtggtagcggcggctc catggcgagtaaagagatgtt |
N/A |
| mCherry-RBBP7 (reverse primer for PCR) | atagaatagggccctctagatgcatgttaag atccttgtccctccagttc |
N/A |
| MiRFP-RBBP7 (forward primer for PCR) | agaagaaggaagaggaactcgagggcggta gcggtggcagcggtggtagcggcggctccatg gcgagtaaagagatgtt |
N/A |
| MiRFP-RBBP7 (reverse primer for PCR) | atagaatagggccctctagatgcat gttaagatccttgtccctccagttc |
N/A |
| GFP-Nup98-SUZ12 (forward primer for PCR) | tggtagcggcggctccctcgagat ggcgcctcagaagcacggcggtgggg |
N/A |
| GFP-Nup98-SUZ12 (reverse primer for PCR) | ctatagaatagggccctctagatgcatg tcagagtttttgttttttgctctgt |
N/A |
| mCherry-SUZ12 (forward primer for PCR) | tggtagcggcggctccctcgagatgg cgcctcagaagcacggcggtgggg |
N/A |
| mCherry-SUZ12 (reverse primer for PCR) | ctatagaatagggccctctagatgcat gtcagagtttttgttttttgctctgt |
N/A |
| GFP-Nup98-AEBP2 (forward primer for PCR) | cggtggtagcggcggctccctcgaga tgtcgtcggatggggaacccctga |
N/A |
| GFP-Nup98-AEBP2 (reverse primer for PCR) | acactatagaatagggccctctagatt acctcttcaacctcttctgcggc |
N/A |
| mCherry-AEBP2 (forward primer for PCR) | cggtggtagcggcggctccctcgagat gtcgtcggatggggaacccctga |
N/A |
| mCherry-AEBP2 (reverse primer for PCR) | acactatagaatagggccctctagatt acctcttcaacctcttctgcggc |
N/A |
| GFP-Nup98-PHF1 (forward primer for PCR) | cggtggtagcggcggctccctcgaga tggcgcagcccccccggctgagcc |
N/A |
| GFP-NupN-PHF1 (reverse primer for PCR) | acactatagaatagggccctctagat cagaagatgccccctcctccccac |
N/A |
| mCherry-PHF1 (forward primer for PCR) | cggtggtagcggcggctccctcgaga tggcgcagcccccccggctgagcc |
N/A |
| mCherry-PHF1 (reverse primer for PCR) | acactatagaatagggccctctagatc agaagatgccccctcctccccac |
N/A |
| GFP-NupN-SUZ12C (forward primer for PCR) | gcggtggtagcggcggctccctcga gcacaatcgtctgtatttccatagt |
N/A |
| GFP-NupN-SUZ12C (reverse primer for PCR) | gcatttaggtgacactatagaatagggcc ctctagatcagagtttttgttttttgctctgttt |
N/A |
| mCherry-SUZ12C (forward primer for PCR) | gcggtggtagcggcggctccctcgagca caatcgtctgtatttccatagt |
N/A |
| mCherry-SUZ12C (reverse primer for PCR) | gcatttaggtgacactatagaatagggc cctctagatcagagtttttgttttttgctctgttt |
N/A |
| mCherry-p53 (forward primer for PCR) | gcggtggtagcggcggctccctcgaga gtcaggaaacattttcag |
N/A |
| mCherry-p53 (reverse primer for PCR) | cactatagaatagggccctctagagt tttcaggaagtagtttcca |
N/A |
| GFP-NupN-MDM2 (forward primer for PCR) | cggtggtagcggcggctccctcgagat gactgatggtgctgtaaccacca |
N/A |
| GFP-NupN-MDM2 (reverse primer for PCR) | gacactatagaatagggccctctagagtt caccaccaccaggttgcgata |
N/A |
| DDX4 (forward primer for PCR) | aatacgactcactatagggagacccaagct tgcggccgcatgggagatgaagattgggtggc cgaaatcaatccgc |
N/A |
| DDX4GFP 208-251-a (reverse primer for PCR) | ctccttctgcttctgacttccaagaattct ttccagagcctgttattacttcttcatttaaaccttt gtaaccacctcgttcacttccactgccact |
N/A |
| DDX4GFP 208-251-b (forward primer for PCR) | cagaagcagaaggaggagaaagtagtgat actcaaggaccaaaagtgacctacatacccc ctcctccaaaagtgagcaagggcga |
N/A |
| DDX4 675-724-a (reverse primer for PCR) | acccagctgtgttcaaagtgctcttgccctttctggta tcaactgatgcaaacacgtttcctcttgtactaccact gaagccaggaatgtatgtactaaaggcgtacagctcgtccat |
N/A |
| DDX4 675-724-b (reverse primer for PCR) | acactatagaatagggccctctagatgcatgtta ctcgagatcccatgactcatcatctactggattgg gagcttgtgaagaagaaaacccagctgtgttcaaa |
N/A |
| GFP-CPEB2 (forward primer for PCR) | gtggtagcggcggctccctcgaa atggggccgccgccgagc |
N/A |
| GFP-CPEB2 (reverse primer for PCR) | acactatagaatagggccctctag atgcatgttactcgagtcatgctgccggtacccca |
N/A |
| GFP-FIBN (forward primer for PCR) | gtggtagcggcggctccctcga aatggggatgaaaccgggc |
N/A |
| GFP-FIBN (reverse primer for PCR) | acactatagaatagggccctctag atgcatgttactcgagtactctgattgccacgttt |
N/A |
| GFP-hnRNPDLC (forward primer for PCR) | gtggtagcggcggctccctcgaa atggggcaacagcagcaac |
N/A |
| GFP-hnRNPDLC (reverse primer for PCR) | acactatagaatagggccctctagat gcatgttactcgagtgtacggctggtaattgt |
N/A |
| GFP-hnRNPH1C (forward primer for PCR) | gtggtagcggcggctccctcgaaat gggggccggtgccagc |
N/A |
| GFP-hnRNPH1C (reverse primer for PCR) | acactatagaatagggccctctagatg catgttactcgagtcatgccccagccgccgctcatac |
N/A |
| GFP-RBM14LCD (forward primer for PCR) | gtggtagcggcggctccctcgaaa tggggctgggcgttggc |
N/A |
| GFP-RBM14LCD (reverse primer for PCR) | acactatagaatagggccctctagatg catgttactcgagttggggtgctgttggcgtt |
N/A |
| GFP-TAF15_156 (forward primer for PCR) | gtggtagcggcggctccctcgaaa tggggatgagcgatag |
N/A |
| GFP- TAF15_156 (reverse primer for PCR) | acactatagaatagggccctctagatgcatgtt actcgagtattttcgcgctggctgtg |
N/A |
| GFP-TDP-43C (forward primer for PCR) | gtggtagcggcggctccctcgaaatg gggagcaatgccga |
N/A |
| GFP-TDP43-C (reverse primer for PCR) | acactatagaatagggccctctagatg catgttactcgagtattcataccccagccgct |
N/A |
| Hfq (forward primer for PCR) | cccaagcttgcggccgcatgaaacc gattaacattcaggat |
N/A |
| Hfq-GFP (reverse primer for PCR) | gctcactttcatggaaccgccggaacc gccttccagttccagctg |
N/A |
| Hfq-GFP-FUSN (forward primer for PCR) | cggtggtagcggcggctccctcgacatg gcctcaaacgattatacccaac |
N/A |
| Hfq-GFP-FUSN (reverse primer for PCR) | tttaggtgacactatagaatagggccctct agatgcatgctcgaggtcctgct gtccatagccaccgctg |
N/A |
| Hfq-GFP-FUSN-MDM2 (forward primer for PCR) | cggtggctatggacagcaggacctcgagat gactgatggtgctgtaacca |
N/A |
| Hfq-GFP-FUSN-MDM2 (reverse primer for PCR) | tgacactatagaatagggccctctagatta gttcaccaccaccaggttgc |
N/A |
| mCherry-PHF1(1-511) (forward primer for PCR) | cggtggtagcggcggctccctcgagatggc gcagcccccccggctgagcc |
N/A |
| mCherry-PHF1(1-511) (reverse primer for PCR) | atagaatagggccctctagatcatgagcgt ctaggaagacctgg |
N/A |
| Recombinant DNA | ||
| pCDNA3.1-mCherry-N | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-N | This paper | N/A |
| pCDNA3.1-BFP-N | This paper | N/A |
| pCDNA3.1-mCherry-Nsp1 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp1 | This paper | N/A |
| pCDNA3.1-BFP-Nsp1 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp2 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp2 | This paper | N/A |
| pCDNA3.1-BFP-Nsp2 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp5 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp5 | This paper | N/A |
| pCDNA3.1-BFP-Nsp5 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp7 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp7 | This paper | N/A |
| pCDNA3.1-BFP-Nsp7 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp8 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp8 | This paper | N/A |
| pCDNA3.1-BFP-Nsp8 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp9 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp9 | This paper | N/A |
| pCDNA3.1-BFP-Nsp9 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp10 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp10 | This paper | N/A |
| pCDNA3.1-BFP-Nsp10 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp11 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp11 | This paper | N/A |
| pCDNA3.1-BFP-Nsp11 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp12 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp12 | This paper | N/A |
| pCDNA3.1-BFP-Nsp12 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp13 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp13 | This paper | N/A |
| pCDNA3.1-BFP-Nsp13 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp14 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp14 | This paper | N/A |
| pCDNA3.1-BFP-Nsp14 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp15 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp15 | This paper | N/A |
| pCDNA3.1-BFP-Nsp15 | This paper | N/A |
| pCDNA3.1-mCherry-Nsp16 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-Nsp16 | This paper | N/A |
| pCDNA3.1-BFP-Nsp16 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N | This paper | N/A |
| pCDNA3.1-DDX4-GFP | This paper | N/A |
| pCDNA3.1-Hfq-GFP-FUSN | This paper | N/A |
| pCDNA3.1-GFP-RBM14LCD | This paper | N/A |
| pCDNA3.1-GFP-TAF15N | This paper | N/A |
| pCDNA3.1-GFP-FIB1N | This paper | N/A |
| pCDNA3.1-GFP-TDP-43C | This paper | N/A |
| pCDNA3.1-GFP-CPEB2N | This paper | N/A |
| pCDNA3.1-GFP-hnRNPDLC | This paper | N/A |
| pCDNA3.1-GFP-hnRNPH1C | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-EZH2 | This paper | N/A |
| pCDNA3.1-mCherry-EZH2 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-EZH1 | This paper | N/A |
| pCDNA3.1-mCherry-EZH1 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-EED | This paper | N/A |
| pCDNA3.1-mCherry-EED | This paper | N/A |
| pCDNA3.1-BFP-EED | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-SUZ12 | This paper | N/A |
| pCDNA3.1-mCherry-SUZ12 | This paper | N/A |
| pCDNA3.1-mCherry-SUZ12C | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-RBBP4 | This paper | N/A |
| pCDNA3.1-mCherry-RBBP4 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-RBBP7 | This paper | N/A |
| pCDNA3.1-mCherry-RBBP7 | This paper | N/A |
| pCDNA3.1-miRFP-RBBP7 | This paper | N/A |
| pCDNA3.1-GFP-Nup98N-MDM2 | This paper | N/A |
| pCDNA3.1-mCherry-p53 | This paper | N/A |
| Software and algorithms | ||
| NIS-Elements AR Analysis | Nikon Corporation Healthcare Business Unit | https://www.microscope.healthcare.nikon.com/en_AOM/products/software/nis-elements |
| Adobe Illustrator CC | Adobe Systems | https://www.adobe.com/products/illustrator.html?promoid=RL89NGY7&mv=other |
| Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/download.html |
| Cytoscape | Cytoscape Software | https://cytoscape.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Pilong Li (pilongli@mail.tsinghua.edu.cn).
Materials availability
This study did not generate new unique reagents. Plasmids generated in this study are available from the Lead Contact without restriction.
Experimental model and subject details
Cell lines
HEK293 cells were obtained from Cell Bank/Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China), and Vero E6 cells were kindly provided by Dr. Jianyang Zeng and Dr. Dan Zhao from the Institute for Interdisciplinary Information Sciences (Tsinghua University, Beijing, China). Both HEK293, Vero E6, and Caco-2-NFlag cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone, Inc.) supplemented with 10% fetal bovine serum (Invitrogen, Inc.) at 37°C with 5% CO2. FuGENE HD (Promega, Inc.) was used for transient transfection according to the manufacturer’s instructions.
SARS-CoV-2 virus-like particles
The transcription and replication-competent SARS-CoV-2 virus-like-particles (trVLP) were maintained by our laboratory.
Method details
Construction of recombinant plasmids
To construct CoPIC-related vectors, the LCD regions, including the N-terminal half of Nup98 (aa 1-515); the N-terminal half of FIB1 (aa 1-82); the N-terminal half of TAF15 (aa 1-155); the N-terminal half of CPEB2 (aa 2-138); the C-terminal half of TDP-43 (aa 258-414); the C-terminal half of hnRNPDL (aa 312-420); the C-terminal half of hnRNPH1 (aa 385-446); the low complexity region of RBM14 (aa 257-573); the N-terminal half of FUS (aa 1-212); Bacillus subtilis Hfq (BsHfq); full-length (FL) EZH1; FL EZH2; FL SUZ12 (aa 1-739); truncated SUZ12 (aa 561-739); FL EED; FL RBBP4; FL RBBP7; FL PHF1, and truncated AEBP2 (aa 209-503), were amplified using the cDNA derived from HEK293 cells as a template and then cloned into pCDNA3.1 expression vectors. The detailed primer sequences are listed in Key Resources Table. The ORFs of SARS-CoV-2 viral proteins were synthesized de novo and cloned into the pCDNA3.1 plasmid by Synbio Technologies (Monmouth Junction, NJ, USA) (General Biosystems).
Microscopy
Imaging was with a NIKON A1 microscope equipped with a 100 × oil immersion objective. NIS-Elements AR Analysis was used to analyze these images.
Fluorescence recovery after photobleaching (FRAP) measurements
In cellulo FRAP experiments were carried out with a NIKON A1 microscope equipped with a 100 × oil immersion objective. Droplets were bleached with the corresponding laser pulse (3 repeats, 70% intensity, dwell time 1 s). Recovery from photobleaching was recorded for the indicated time.
Validation of expression of fusion proteins
To validate the correct expression of SARS-CoV-2-encoded proteins fused with a mCherry tag, Vero E6 cells transfected with the indicated plasmids were harvested and extracted for lysates using Minute™ Total Protein Extraction Kit for Animal Cultured Cells/Tissues (Invent Biotech) according to the manufacturer’s instructions. The cell lysates were subjected to western blot analyses using the antibody against mCherry (1:1,000 dilution).
Co-immunoprecipitation assays
To confirm the interactions of viral protein pairs, Co-IP assays were performed. HEK293 cells (5 × 107) were co-transfected with the respective expression plasmids. Cells were harvested at 48 h p.t. and washed twice with chilled phosphate-buffered saline (PBS) supplemented with protease inhibitor cocktail (Bimake, Inc.). Cell lysates were obtained using Minute™ Total Protein Extraction Kit for Animal Cultured Cells/Tissues (Invent Biotech) according to the manufacturer’s instructions. Subsequently, the lysates were incubated with GFP-Trap Magnetic Agarose (ChromoTek, Inc.) at 4°C for 1h, with rotation. After incubation, beads were collected with a Magnetic Separator, and the unbounded samples were removed and the rest saved for analysis. After 5 times washing, the samples were boiled for 10 min and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and examined by western blot to determine if the mCherry-tagged protein was co-immunoprecipitated with the GFP-tagged protein.
To verify the interactions identified by CoPIC under infection, Caco-2-NFlag cells were infected with recombinant SARS-CoV-2 GFP/ΔN trVLP at an M.O.I of 0.05, harvested at 48 h post-infection (p.i.), and extracted for lysates using Minute™ Total Protein Extraction Kit for Animal Cultured Cells/Tissues (Invent Biotech) according to the manufacturer’s instructions. One-tenth volumes of the supernatants were preserved as the input controls, and the rest cells were divided into two equal parts and incubated overnight at 4°C with Protein A+G agarose beads (Beyotime Biotech) plus either anti-Flag rabbit mAb or rabbit IgG as a control. The beads were washed four times with lysis buffer and then precipitated for western blot analyses. The primary antibodies used were as follows: anti-Nsp3 (1:1,000 dilution), anti-3CLpro (1:1,000 dilution), anti-Nsp9 (1:1,000 dilution), anti-Nsp16 (1:1,000 dilution), and anti-Flag (1:1,000 dilution).
Western blotting
All antibodies used in the study are summarized in the Key Resources Table. Figures 4J–4M, S3C, S5B, S7B, and S7C were directly analyzed by western blot. The cell samples were resuspended using 2 × Laemmli loading buffer. Proteins were fractionated by SDS-PAGE and transferred to a PVDF membrane. The membranes were incubated overnight with primary antibodies, then with the corresponding peroxide-labeled IgG. Finally, enhanced chemiluminescence reagent was used to visualize the results.
Quantification and statistical analysis
Number of biological replicates (BR) is stated in each legend. Error bars represent SD. Graphpad Prism version 8 was used for statistical analysis and graph preparation. For confocal image analysis, NIS-Elements AR Analysis (Nikon, Inc.), ImageJ (National Institutes of Health), and Adobe Illustrator CC (Adobe Systems, Inc.) were used.
Acknowledgments
We acknowledge Prof. Zhiyong Lou (Tsinghua University, Beijing, China) for suggestions on the manuscript. We are also grateful to SLSTU-Nikon Biological Imaging Center (Center of Pharmaceutical Technology, Tsinghua University, Beijing, China) for imaging support and Zan Yin for illustrations. This work was supported by grants from the National Key Research and Development Program of China (2019YFA0508403 to P.L.), the National Natural Science Foundation of China (31800637 to J.W.; 31871443 to P.L.), and the Students Research Training Program of Tsinghua University (1911S0032 to H.L.). The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Author contributions
Conceptualization, P.L.; methodology, H.L. and J.W.; investigation, W.X., and G.P.; writing – original draft, W.X. and H.L.; writing – review & editing, G.P., J.W., X.J., Q.D., and P.L.; funding acquisition, P.L. and H.L.
Declaration of interests
The authors declare no competing interests.
Published: July 17, 2021; corrected online September 28, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109482.
Supplemental information
Data and code availability
All microscopy image data have been deposited on Mendeley Data via the following link: https://doi.org/10.17632/gtswbcb5nv.2. This study did not generate any unique code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abduljalil J.M., Abduljalil B.M. Epidemiology, genome, and clinical features of the pandemic SARS-CoV-2: a recent view. New Microbes New Infect. 2020;35:100672. doi: 10.1016/j.nmni.2020.100672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji A.O., Marchand B., Te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.-G., Choi J.-K., Taylor D.R., Oh J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92:584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin T., Janvier A., Schaeffer L., Sosnowski P., Lauriane K., Hammann P., Westhof E., Eriani G., Martin F. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2020;27:253–264. doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddock H.T., Brolih S., Yosaatmadja Y., Ratnaweera M., Bielinski M., Swift L., Cruz-Migoni A., Morris G.M., Schofield C.J., Gileadi O. Characterisation of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: Implications for its role in viral genome stability and inhibitor identification. bioRxiv. 2020 doi: 10.1101/2020.08.13.248211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P.L., Roecklein J.A., SenGupta D., Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- Batlle C., Yang P., Coughlin M., Messing J., Pesarrodona M., Szulc E., Salvatella X., Kim H.J., Taylor J.P., Ventura S. hnRNPDL phase separation is regulated by alternative splicing and disease-causing mutations accelerate its aggregation. Cell Rep. 2020;30:1117–1128.e5. doi: 10.1016/j.celrep.2019.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggård T., Linse S., James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. USA. 2015;112:E5237–E5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard D., Hutter H., Fleenor J., Fire A. A differential cytolocalization assay for analysis of macromolecular assemblies in the eukaryotic cytoplasm. Mol. Cell. Proteomics. 2006;5:2175–2184. doi: 10.1074/mcp.T600025-MCP200. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335:165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I.-R., Stenger D.C., French R. Multiple interactions among proteins encoded by the mite-transmitted wheat streak mosaic tritimovirus. Virology. 2000;267:185–198. doi: 10.1006/viro.1999.0117. [DOI] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C., Lander G.C., Maiolica A., Herzog F., Aebersold R., Nogales E. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., Thiel V., de Haan C.A.M., Reggiori F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94:e01925-19. doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Liao L., Yates J.R., 3rd, Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T.N.M., Kang S.-H., Bak A., Folimonova S.Y. A non-conserved p33 protein of citrus tristeza virus interacts with multiple viral partners. Mol. Plant Microbe Interact. 2020;33:859–870. doi: 10.1094/MPMI-11-19-0328-FI. [DOI] [PubMed] [Google Scholar]
- Davies J.P., Almasy K.M., McDonald E.F., Plate L. Comparative multiplexed interactomics of SARS-CoV-2 and homologous coronavirus nonstructural proteins identifies unique and shared host-cell dependencies. ACS Infect. Dis. 2020;6:3174–3189. doi: 10.1021/acsinfecdis.0c00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova M., Imbert I., Kieny M.P., Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. USA. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmashova N., Gorchakov R., Frolova E., Frolov I. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 2006;80:5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy E., Ciferri C., Bracken A.P. Structural basis for PRC2 engagement with chromatin. Curr. Opin. Struct. Biol. 2021;67:135–144. doi: 10.1016/j.sbi.2020.10.017. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Rajamäki M.-L., Saarma M., Valkonen J.P.T. Towards a protein interaction map of potyviruses: Protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 2001;82:935–939. doi: 10.1099/0022-1317-82-4-935. [DOI] [PubMed] [Google Scholar]
- Häuser R., Blasche S., Dokland T., Haggård-Ljungquist E., von Brunn A., Salas M., Casjens S., Molineux I., Uetz P. Bacteriophage protein-protein interactions. Adv. Virus Res. 2012;83:219–298. doi: 10.1016/B978-0-12-394438-2.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Iyer K.S., Ho D., Newcombe E.A. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. bioRxiv. 2020 doi: 10.1101/2020.04.27.063180. [DOI] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Snijder E.J., Dimitrova M., Guillemot J.-C., Lécine P., Canard B. The SARS-coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserman C., Roden C.A., Boerneke M.A., Sealfon R.S.G., McLaughlin G.A., Jungreis I., Fritch E.J., Hou Y.J., Ekena J., Weidmann C.A. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell. 2020;80:1078–1091.e6. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.-J., Jeong S., Kang D.Y., Sp N., Yang Y.M., Kim D.-E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10:4481. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X., Zhu Y., Wang Y., Li J., Zhang J., Gong M., Ren W., Li S., Zhong J., Zhang L. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog. 2021;17:e1009439. doi: 10.1371/journal.ppat.1009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Bhardwaj K., Li Y., Palaninathan S., Sacchettini J., Guarino L., Leibowitz J.L., Kao C.C. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinath V., Faini M., Poepsel S., Reif D., Feng X.A., Stjepanovic G., Aebersold R., Nogales E. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018;359:940–944. doi: 10.1126/science.aar5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.T., Zeb M.T., Ahsan H., Ahmed A., Ali A., Akhtar K., Malik S.I., Cui Z., Ali S., Khan A.S. SARS-CoV-2 nucleocapsid and Nsp3 binding: An in silico study. Arch. Microbiol. 2021;203:59–66. doi: 10.1007/s00203-020-01998-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkolova E., Klima M., Nencka R., Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J. Struct. Biol. 2020;211:107548. doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.-R., Kwon H.-M., Park K., Oh S., Jeong Y.-J., Kim D.-E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Salwinski L., Zhang C., Chu D., Sampankanpanich C., Reyes N.A., Vangeloff A., Xing F., Li X., Wu T.-T. An integrated approach to elucidate the intra-viral and viral-cellular protein interaction networks of a gamma-herpesvirus. PLoS Pathog. 2011;7:e1002297. doi: 10.1371/journal.ppat.1002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. (NY) 2021;2:99–112.e7. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Chen H., Ye F., Chen Z., Yang F., Zheng Y., Cao Y., Qiao J., Yang S., Lu G. Crystal structure of SARS-CoV-2 nsp10/nsp16 2′-O-methylase and its implication on antiviral drug design. Signal Transduct. Target. Ther. 2020;5:131. doi: 10.1038/s41392-020-00241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fang P., Fang L., Hong Y., Zhu X., Wang D., Peng G., Xiao S. Porcine deltacoronavirus nsp15 antagonizes interferon-β production independently of its endoribonuclease activity. Mol. Immunol. 2019;114:100–107. doi: 10.1016/j.molimm.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. USA. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalapbutr P., Kongtaworn N., Rungrotmongkol T. Structural insight into the recognition of S-adenosyl-l-homocysteine and sinefungin in SARS-CoV-2 Nsp16/Nsp10 RNA cap 2′-O-methyltransferase. Comput. Struct. Biotechnol. J. 2020;18:2757–2765. doi: 10.1016/j.csbj.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim. Biophys. Acta. 2013;1834:918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S., Holtzman T., Moss B., Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J.A., Thelen J.J. Biochemical approaches for discovering protein-protein interactions. Plant J. 2008;53:597–609. doi: 10.1111/j.1365-313X.2007.03316.x. [DOI] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev. Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Momand J., Zambetti G.P., Olson D.C., George D., Levine A.J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Murakami T., Qamar S., Lin J.Q., Schierle G.S.K., Rees E., Miyashita A., Costa A.R., Dodd R.B., Chan F.T.S., Michel C.H. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., Baldwin A.J. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: Involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39:e106478. doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E.M., Fields S. Protein-protein interactions: Methods for detection and analysis. Microbiol. Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Hoskins I., Garcia D., Tonn T., Ozadam H., Cenik E.S., Cenik C. Genes with 5′terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 NSP1 protein. bioRxiv. 2020 doi: 10.1101/2020.09.13.295493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.-P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxiv. 2020 doi: 10.1101/2020.04.17.047498. [DOI] [Google Scholar]
- Saini A., Chauhan V.S. Delineation of the core aggregation sequences of TDP-43 C-terminal fragment. ChemBioChem. 2011;12:2495–2501. doi: 10.1002/cbic.201100427. [DOI] [PubMed] [Google Scholar]
- Schmidt H.B., Görlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife. 2015;4:e04251. doi: 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar R.B., Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 2003;160:629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., He L., Deng F., Wang H., Vlak J.M., Hu Z., Wang M. Mito-docking: A novel in vivo method to detect protein-protein interactions. Small Methods. 2019;3:1900010. [Google Scholar]
- Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya T., Baba S., Fujimoto M., Kawai G., Kumasaka T., Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: Insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012;40:1856–1867. doi: 10.1093/nar/gkr892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart C.C., Sexton N.R., Munjal H., Lu X., Molland K.L., Tomar S., Mesecar A.D., Denison M.R. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 2013;87:12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 2006;80:7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Lal S.K. The SARS-CoV nucleocapsid protein: A protein with multifarious activities. Infect. Genet. Evol. 2008;8:397–405. doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.Z., Yan Y., Wang X.X., Jiang Y., Xu H.E. EZH2: Biology, disease, and structure-based drug discovery. Acta Pharmacol. Sin. 2014;35:161–174. doi: 10.1038/aps.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sun W., Zhao Y., McEachern D., Meaux I., Barrière C., Stuckey J.A., Meagher J.L., Bai L., Liu L. SAR405838: An optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Cancer Res. 2014;74:5855–5865. doi: 10.1158/0008-5472.CAN-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Choi J.-M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Seki T., Fukano T., Sakaue-Sawano A., Karasawa S., Kubota M., Kurokawa H., Inoue K., Akatsuka J., Miyawaki A. Genetic visualization of protein interactions harnessing liquid phase transitions. Sci. Rep. 2017;7:46380. doi: 10.1038/srep46380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.C., Brangwynne C.P. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2021. Coronavirus disease (COVID-19) dashboard. https://covid19.who.int/?adgroupsurvey={adgroupsurvey}&gclid=EAIaIQobChMInp_Ejqnm8QIV1To4Ch1L6AZ3EAAYASABEgIvx_D_BwE. [PubMed] [Google Scholar]
- Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Yin P., Zhao H., Zhang N., Jian X., Song S., Gao S., Zhang L. Intraviral interactome of Chikungunya virus reveals the homo-oligomerization and palmitoylation of structural protein TF. Biochem. Biophys. Res. Commun. 2019;513:919–924. doi: 10.1016/j.bbrc.2019.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., Shen Q., Wu S., Chen S., Lomakin I.B., Xiong Y. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell. 2020;80:1055–1066.e6. doi: 10.1016/j.molcel.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R., Seitz S., Henke A., Munder T., Wutzler P. Linkage map of protein-protein interactions of Porcine teschovirus. J. Gen. Virol. 2005;86:2763–2768. doi: 10.1099/vir.0.81144-0. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Huang H., Zhang L., Wu R., Chung C.I., Zhang S.Q., Torra J., Schepis A., Coughlin S.R., Kornberg T.B., Shu X. Visualizing dynamics of cell signaling in vivo with a phase separation-based kinase reporter. Mol. Cell. 2018;69:334–346.e4. doi: 10.1016/j.molcel.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Xu W., Zhang X., Wang X., Yuan E., Xiong Y., Wu S., Li S., Wu N., Tian T. Understanding the phase separation characteristics of nucleocapsid protein provides a new therapeutic opportunity against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.10.09.33273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Perlman S. Immune responses in influenza A virus and human coronavirus infections: An ongoing battle between the virus and host. Curr. Opin. Virol. 2018;28:43–52. doi: 10.1016/j.coviro.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Li W., Li J., Xie L., Wu R., Wang L., Fu S., Su W., Hu J., Wang J., Li P. Phase-separated condensate-aided enrichment of biomolecular interactions for high-throughput drug screening in test tubes. J. Biol. Chem. 2020;295:11420–11434. doi: 10.1074/jbc.RA120.012981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microscopy image data have been deposited on Mendeley Data via the following link: https://doi.org/10.17632/gtswbcb5nv.2. This study did not generate any unique code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.