Abstract
Late gadolinium enhanced (LGE) cardiac magnetic resonance (CMR) imaging is the current gold standard for assessing myocardium viability for patients diagnosed with myocardial infarction, myocarditis or cardiomyopathy. This imaging method enables the identification and quantification of myocardial tissue regions that appear hyper-enhanced. However, the delineation of the myocardium is hampered by the reduced contrast between the myocardium and the left ventricle (LV) blood-pool due to the gadolinium-based contrast agent. The balanced-Steady State Free Precession (bSSFP) cine CMR imaging provides high resolution images with superior contrast between the myocardium and the LV blood-pool. Hence, the registration of the LGE CMR images and the bSSFP cine CMR images is a vital step for accurate localization and quantification of the compromised myocardial tissue. Here, we propose a Spatial Transformer Network (STN) inspired convolutional neural network (CNN) architecture to perform supervised registration of bSSFP cine CMR and LGE CMR images. We evaluate our proposed method on the 2019 Multi-Sequence Cardiac Magnetic Resonance Segmentation Challenge (MS-CMRSeg) dataset and use several evaluation metrics, including the center-to-center LV and right ventricle (RV) blood-pool distance, and the contour-to-contour blood-pool and myocardium distance between the LGE and bSSFP CMR images. Specifically, we showed that our registration method reduced the bSSFP to LGE LV blood-pool center distance from 3.28mm before registration to 2.27mm post registration and RV blood-pool center distance from 4.35mm before registration to 2.52mm post registration. We also show that the average surface distance (ASD) between bSSFP and LGE is reduced from 2.53mm to 2.09mm, 1.78mm to 1.40mm and 2.42mm to 1.73mm for LV blood-pool, LV myocardium and RV blood-pool, respectively.
Keywords: Image registration, Late gadolinium enhanced MRI, Cine cardiac MRI, Deep learning
1. Introduction
Myocardial infarction, cardiomyopathy and myocarditis represent common cardiac conditions associated with significant morbidity and mortality worldwide [1]. The assessment of myocardium viability for patients who experienced any of these diseases is critical for diagnosis and planning of optimal therapies. Accurate quantification of the compromised myocardium is a crucial step in determining the part of the heart that may benefit from therapy [20]. LGE CMR imaging is the most widely used technique to detect, localize and quantify the diseased myocardial tissue, also referred to as scar tissue. During the typical LGE CMR image acquisition protocol, a gadolinium-based contrast agent is injected into a patient, and the MR images are acquired 15–20min post-injection. In the LGE CMR images, the compromised LV myocardial regions appear much brighter than healthy tissue, due to the trapping and delayed wash-out of the contrast agent from the diseased tissue regions. As a concrete example, in case of myocardial infarction, LGE-MR imaging helps assess the transmural extent of the infarct, which helps predict the success of recovery following revascularization therapy and also provides additional insights about other potential complications associated with the disease [11].
In a clinical set-up, radiologists and cardiologists visually assess the viability of the myocardium based on the LGE CMR images. However, the gadolinium-based contrast agent reduces the contrast between the myocardium and the LV blood-pool. Although useful to identify scarred myocardium regions, LGE CMR images do not allow accurate delineation between the LV blood-pool and the LV myocardium. On the other hand, the bSSFP cine CMR images provide excellent contrast between myocardium and blood-pool (Fig. 1), and can be successfully employed to identify the myocardium and blood-pool, but they cannot show the scarred regions. Therefore, the LGE and bSFFP CMR images show complementary information pertaining to the heart, but neither image type, on its own, enables the extraction, quantification and global visualization of all desired features: LV blood pool, LV myocardium, and scarred regions.
Fig. 1.
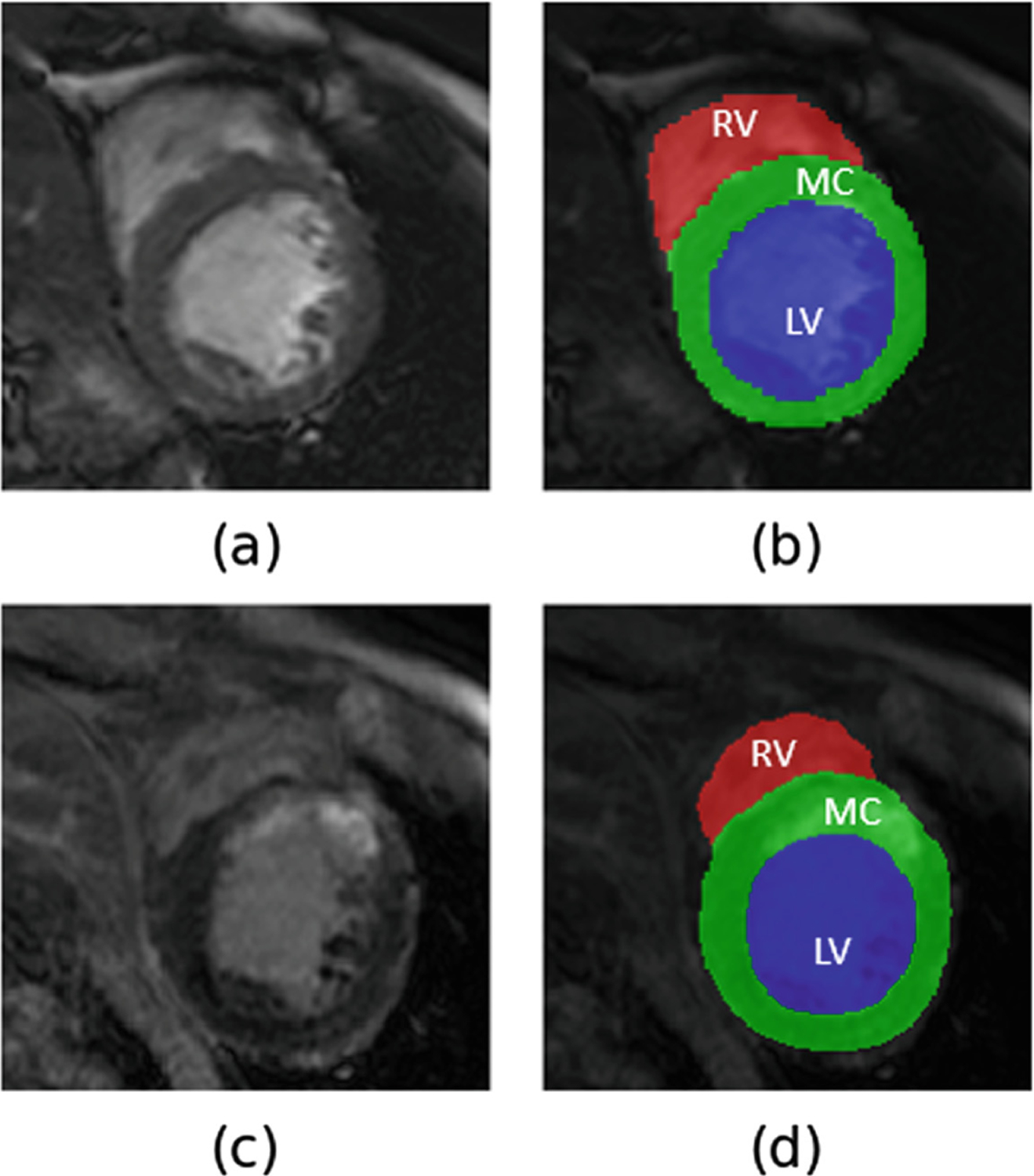
Example of (a) bSSFP cine CMR image with its (b) manual annotations - LV blood-pool (LV) in blue, LV myocardium (MC) in green and RV blood-pool (RV) in red overlayed on it and (c) LGE CMR image with its (d) manual annotations overlayed on it.
In the recent 2019 MS-CMRSeg challenge [21,22], participants were provided with the segmentation labels for LV blood-pool, LV myocardium, and RV blood-pool available for the bSSFP cine images to segment the same cardiac chambers from the LGE images of the same patients. Several studies proposed the generation of synthetic LGE images from the bSSFP images using cycleGAN [15], histogram matching [14], shape transfer GAN [17] or style transfer networks like MUNIT [2]. They use these synthetic LGE images and the annotations provided for the bSSFP cine images to train various U-Net architectures to segment LV blood-pool, LV myocardium and RV blood-pool from the actual LGE images. These methods result in good segmentation performance, however, they are time consuming, since they rely on a two-step process: training the adversarial networks to generate synthetic LGE images from the bSSFP images, followed by the training of the U-Net architectures (or its variants) on these synthetic LGE images to segment the cardiac chambers from the original LGE images.
An alternative approach to “learning” features from one image type and using them to segment the other image type is to segment the complementary features from the cine MRI and LGE-MRI images, then co-register the images and use the appropriate registration transformation to propagate the segmentation labels from the cine MRI into the LGE-MRI space or vice versa. Chenoune et al. [3] rigidly register 3D delay-enhanced images with the cine images using mutual information as the similarity measure. Wei et al. [20] use pattern intensity as similarity measure leading to accurate affine registration of cine and LGE images. More recently, Guo et al. [8] proposed employing rigid registration to initially align the cine images with the multi-contrast late enhanced CMR images, followed by deformable registration to further refine the alignment. In summary, all these works employ traditional approaches to iteratively optimize the registration cost function for a given image pair.
With the advent of deep learning, several groups proposed the utilization of neural networks to train image registration algorithms using similarity measures like normalized mutual information (NMI), normalized cross correlation (NCC), local Pearson correlation coefficient (LPC), sum of squared intensity difference (SSD) and sum of absolute difference (SAD) [12]. While the cost functions can be optimized by training large datasets using neural networks, these unsupervised registration methods do not perform significantly better than the traditional approaches, as the similarity measures used are the same [13]. Hence, while these unsupervised machine learning-based registration help in speeding up the registration process compared to the traditional unsupervised registration algorithms, they do not necessarily improve registration accuracy beyond that achieved using traditional unsupervised approaches.
In this paper, we propose a supervised deep learning based registration approach to register bSSFP cine CMR images to its corresponding LGE CMR images using a STN-inspired CNN. Some literature suggests that supervised registration techniques entail the use of the displacement field for training [7,16]. Our proposed method, on the other hand, only uses several segmentation labels to guide the registration. Hence, here we refer to it as a supervised registration, although, according to the literature nomenclature mentioned above, it could also be classified as a segmentation-guided registration.
We train the network on the 2019 MS-CMRSeg challenge dataset using the provided manual annotations (required only during training) of the LV blood-pool, LV myocardium, and RV blood-pool and compute a dual loss cost function that combines the benefits of both the Dice loss and cross-entropy loss. We compare the accuracy of our proposed rigid and affine supervised deep learning-based registration to the accuracy of previously disseminated unsupervised deep-learning-based rigid and affine registrations.
Our proposed method aims to address the limitations associated with the aforementioned methods as follows: (i) we fully exploit the information pertaining to the various regions of interest (RoIs) of the cardiac anatomy (i.e., LV blood-pool, LV myocardium and RV blood-pool) and devise a robust ROI-guided registration technique that improved registration accuracy beyond the previous unsupervised techniques; (ii) our method requires minimal preprocessing, specifically it only relies on several segmentation labels that could be obtained using manual annotation or using available and previously validated accurate, automatic segmentation techniques [5,6,9,18,19]; and (iii) does not require the need to train additional adversarial networks to generate synthetic LGE-MRI images [2,14,15,17], therefore reducing network training time without compromising registration accuracy.
2. Methodology
2.1. Data
The dataset used in this paper was made available through the 2019 MS-CMRSeg challenge [21,22]. The available data consisted of LGE, T2-weighted, and bSSFP cine MRI images acquired from 45 patients who had been diagnosed with cardiomyopathy. In this work, we utilize the LGE and bSSFP cine MRI images for registration. The manual annotations of the LV blood-pool, LV myocardium and RV blood-pool were performed by trained personnel using ITK-SNAP and corroborated by expert cardiologists. Both the bSSFP and LGE MRI images were acquired at end-diastole. The bSSFP images were acquired using a TR and TE of 2.7ms and 1.4ms, respectively, and consisted of 8–12 slices with an in-plane resolution of 1.25mm × 1.25mm and a slice thickness of 8–13mm. The LGE-MRI images were acquired using a T1-weighted inversion-recovery gradient-echo pulse sequences with a TR and TE of 3.6ms and 1.8ms, respectively, and consisted of 10–18 slices featuring an in-plane resolution of 0.75mm × 0.75mm and a 5mm slice thickness.
To account for the differences in slice thickness, image sizes and in-plane image resolution between the bSSFP cine CMR and LGE CMR images, all the images are resampled to a slice thickness of 5mm (using spline interpolation), in-plane image resolution of 0.75mm × 0.75mm and then resized to 224 × 224 pixels.
2.2. Spatial Transformer Network (STN) Architecture
The STN consists of three parts - a localisation network, a parameterised sampling grid (grid generator) and a differentiable image sampler. The localisation network function floc() can be any fully connected network or convolutional neural network that takes in an input feature map I ∈ RW×H×C with width W height H and channels C through a number of hidden layers, and outputs θ, where θ = floc(I), and contains the parameters of the transformation Tθ.
In an effort to explain the proposed algorithm, let us consider a regular grid G consisting of a set of points with source coordinates (, ). This grid G acts as input to the grid generator and the transformation Tθ is applied on it i.e. Tθ(G). This operation results in a set of points with target coordinates (, ) which is altered to translate, scale, rotate, skew etc. the input image depending on the values of the θ. Depending on the target coordinates (, ), the differentiable image sampler generates a transformed output feature map O ∈ RW×H×C׳ [10].
2.3. STN-Based Registration of Cine bSSFP and LGE MRI Images
In our experiments, we concatenate the bSSFP cine CMR image (224 × 224) with its corresponding LGE CMR image (224 × 224) and input the resulting 224×224×2 tensor into a CNN which is analogous to the localisation network. For a 2D affine registration transformation, the output θ of the CNN is a six-dimensional vector that results in the transformation matrix Tθ,
| (1) |
For a rigid registration transformation, the output θ of the CNN is three-dimensional i.e. θ = [x,y,z] where z is the rotation parameter and (x,y) are the translation parameters. This results in the transformation matrix Tθ,
| (2) |
The grid generator outputs a sampling grid Tθ(G) and the differentiable image sampler transforms the ground truth (GT) map of the LGE CMR image. The Dice loss and cross-entropy loss are computed using the GT map of the bSSFP cine CMR image and the transformed GT map of the LGE CMR image. The computed loss is then used to back-propagate the CNN (Fig. 2).
Fig. 2.
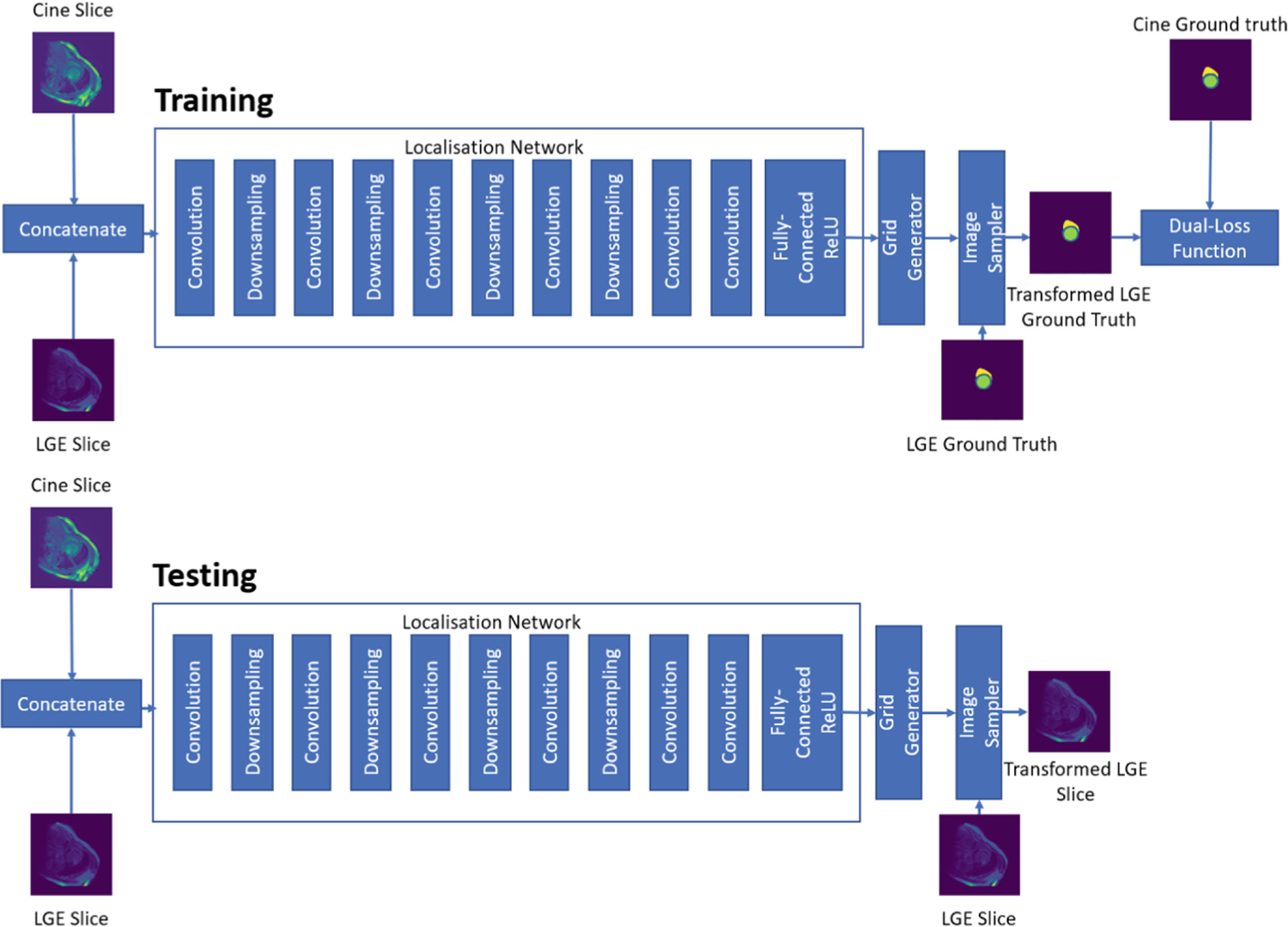
Overview of supervised registration of bSSFP cine CMR images and LGE CMR image using STN. In the training network, the GT LGE image is fed into the image sampler and the dual-loss function is computed using the transformed GT features from the LGE and cine bSSFP images. In the testing network, the LGE image slice is fed into the image sampler and the output consists of a spatially transformed LGE image slice.
2.4. Network Training
In this paper, we compare the registration accuracy of four different registration methods - unsupervised rigid, unsupervised affine, supervised rigid and supervised affine.
The unsupervised registration methods are trained using the CNN shows in Fig. 2, using NMI as a cost function. Our proposed supervised registration methods, which could also be classified as segmentation-guided image registration techniques, are trained using the following dual-loss function:
| (3) |
where is the cross-entropy loss, is the Dice loss and α allows us to modulate the effect of the Dice loss and cross-entropy loss on the overall dual-loss function.
In our experiments, of the total 45 bSSFP and LGE MRI datasets, we use 35 datasets for training, leaving 5 datasets for validation and the remaining 5 datasets for testing. During training, we augment both the bSSFP cine CMR and the LGE CMR images randomly on the fly using a series of translation, rotation and gamma correction operations. We train our networks using the Adam optimizer with a learning rate of 1e-5 and a gamma decay of 0.99 every alternate epoch for fine-tuning for 100 epochs on a machine equipped with NVIDIA RTX 2080 Ti GPU with 11GB of memory.
3. Results
To evaluate our registration, we identify the LV and RV blood-pool centres as the centroid of the segmentation masks of the LV and RV blood-pool corresponding to both the bSSFP cine CMR and LGE CMR images. The Euclidean distance between the blood-pool centers i.e. center distance (CD) from these two images is compared to the blood-pool CD of the bSSFP cine CMR image and its corresponding transformed LGE CMR image. We also quantify our registration accuracy using average surface distance (ASD), a popular evaluation metric for registration, between the LV blood-pool, LV myocardium and RV blood-pool masks of bSSFP cine CMR image and its corresponding LGE CMR image, before and after registration.
In Table 1, we show the mean CD and mean ASD before registration, after unsupervised rigid registration, unsupervised affine registration, supervised rigid registration and supervised affine registration of the test dataset. Figure 3 shows the comparison of CD and ASD of all the four above-mentioned registration approaches. We can observe that the CD is significantly reduced in both the supervised registration algorithms (rigid and affine) for LV blood-pool (p-value < 0.005) and RV blood-pool (p-value < 0.05). We can also observe that the ASD is significantly reduced for the LV blood-pool (p-value < 0.05), LV myocardium (p-value < 0.05) and RV blood-pool (p-value < 0.005) for both the rigid and affine supervised registration methods. However, the changes in the CD and ASD after unsupervised registration (both rigid and affine) is not very significant, compared to before registration.
Table 1.
Summary of registration evaluation. Mean (std-dev) center-to-center distance (CD) and average surface distance (ASD) for LV blood-pool (LV), LV myocardium (MC) and RV blood-pool (RV).
| LV CD (mm) | LV ASD (mm) | MC ASD (mm) | RV CD (mm) | RV ASD (mm) | |
|---|---|---|---|---|---|
| Before registration | 3.28 (1.83) | 2.53 (1.23) | 1.78 (0.78) | 4.36 (3.79) | 2.42 (1.20) |
| Unsupervised rigid | 3.12 (1.79) | 2.53 (1.13) | 2.17 (1.58) | 2.48 (3.78) | 2.45 (1.26) |
| Unsupervised affine | 2.78 (1.65) | 2.44 (1.58) | 1.85 (1.56) | 3.87 (3.75) | 2.30 (1.23) |
| Supervised rigid | 2.22 (1.08) ** | 2.14 (1.20)* | 1.42 (1.48)* | 2.69 (2.51)* | 1.72 (1.06) ** |
| Supervised affine | 2.27 (1.38)** | 2.09 (1.14) * | 1.40 (1.12) * | 2.52 (2.66) * | 1.73 (1.02)** |
Statistically significant differences between the registration metrics before and after registration were evaluated using the Student t-test and are reported using * for p < 0.05 and ** for p < 0.005.
The best evaluation metrics achieved are labeled in bold.
Fig. 3.
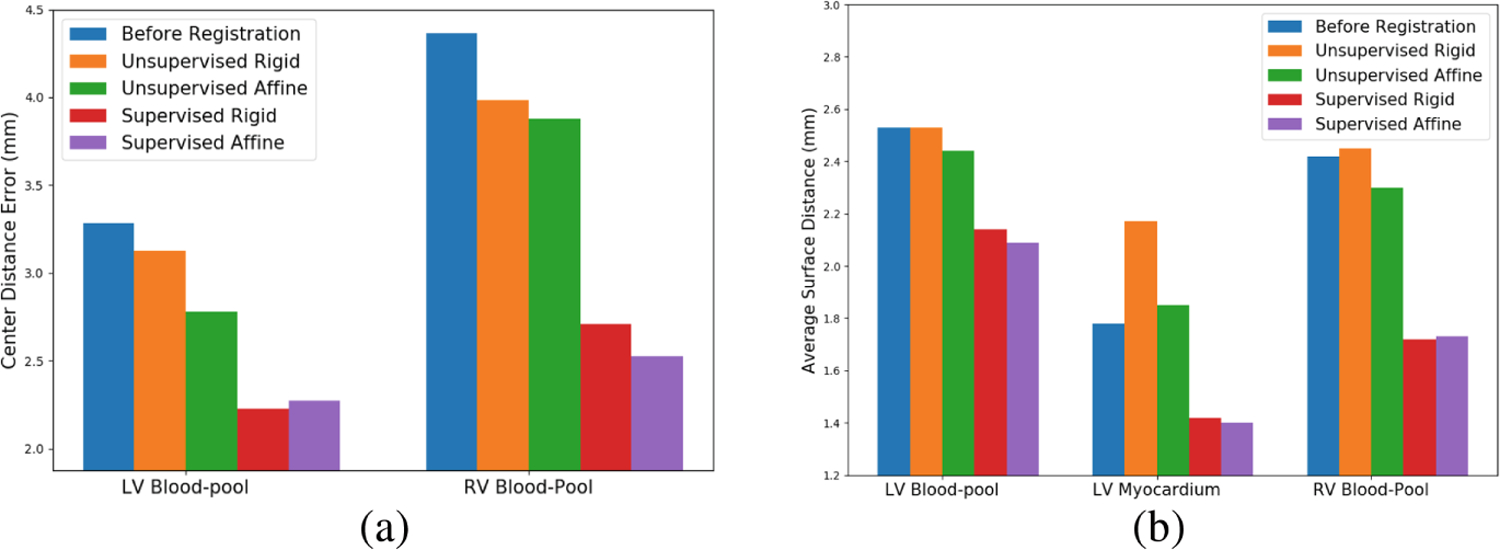
Comparison of (a) mean CD and (b) mean ASD values before registration, after unsupervised rigid registration, unsupervised affine registration, supervised rigid registration and supervised affine registration
Figure 4 shows an example of the manual annotations of LV blood-pool (green), LV myocardium (blue) and RV blood-pool (yellow) of a bSSFP cine CMR image overlayed on its corresponding LGE CMR image before registration and on the corresponding transformed LGE CMR image after unsupervised rigid, unsupervised affine, supervised rigid and supervised affine registration. Figure 4 also shows that when the LGE CMR image and its associated hyper-enhanced regions (marked by the enclosed pink contour) is overlaid onto the bSSFP cine CMR image and its associated labels, the hyper-enhanced regions erroneously appear as part of the LV blood-pool, instead of the LV myocardium. Nevertheless, following supervised registration, the hyper-enhanced regions correctly align with the LV myocardium, where they truly belong. Lastly, Fig. 4 also helps the reader visually appreciate the performance of each registration algorithm by showing the LV and RV blood-pool center-to-center distance before and after each registration algorithm is applied.
Fig. 4.
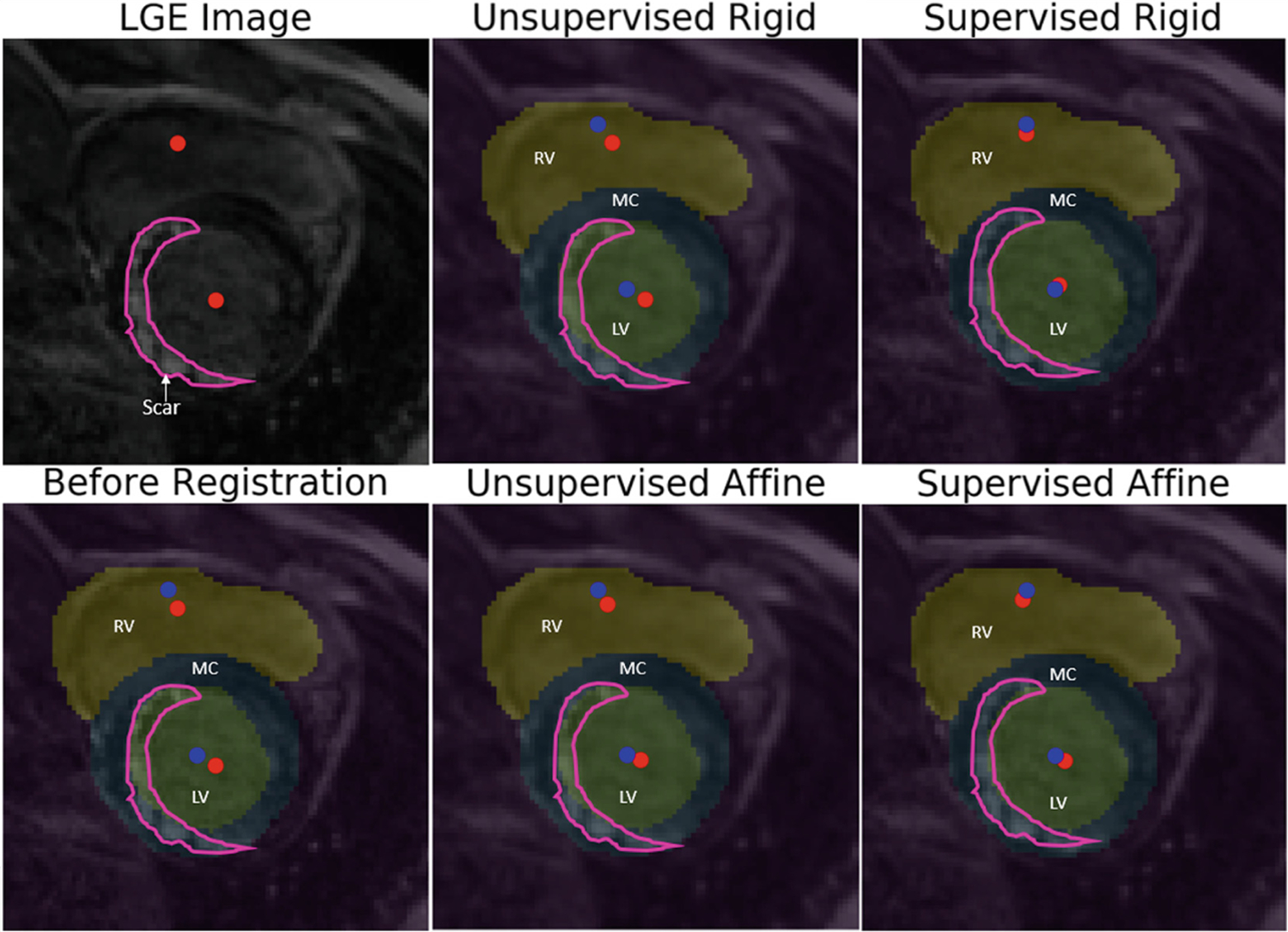
Panel 1–1: Unregistered LGE CMR image and associated hyper-enhanced regions marked by pink contour and LV and RV blood-pool centers marked by red dots; Panel 2–1: before registration (CD: 2.72mm, ASD: 2.24mm); overlaid unregistered LGE CMR image and features (from Panel 1–1) onto the bSFFP image showing the RV and LV blood-pools and their centers (marked by blue dots) and the LV myocardium (MC) marked on the bSSFP image (Note: The hyper-enhanced regions enclosed by pink contour erroneously appear over the LV blood-pool, not the LV myocardium, where they truly belong); Panel 1–2: overlaid LGE CMR image onto the bSSFP image following unsupervised rigid registration (CD: 2.56mm, ASD: 2.20mm); Panel 2–2: unsupervised affine registration (CD: 2.52mm, ASD: 2.18mm); Panel 1–3: supervised rigid registration (CD: 1.56mm, ASD: 1.64mm); and Panel 2–3: supervised affine registration(CD: 1.68mm, ASD: 1.76mm). (Note: The accurate overlay of the hyper-enhanced regions marked by the pink contour over the LV myocardium, as well as significantly improved LV and RV blood-pool center-to-center distance following supervised registration in Panel 1–3 and Panel 2–3).
4. Discussion
In this paper, we present a STN inspired CNN architecture to register the bSSFP cine CMR images to its corresponding LGE CMR images in a supervised manner using a dual-loss (weighted Dice loss and weighted cross-entropy loss) cost function. Our experiments show a statistically significant reduction of the CD between the bSSFP cine CMR images and the LGE CMR images in LV blood-pool from 3.28mm before registration to 2.22mm after supervised rigid registration and 2.27mm after supervised affine registration, and in RV blood-pool from 4.36mm before registration to 2.69mm after supervised rigid registration and 2.52mm after supervised affine registration. We also observed a statistically significant improvement in the ASD between the bSSFP and LGE MRI images in LV blood-pool from 2.53mm before registration to 2.14mm after supervised rigid registration and 2.09mm after supervised affine registration, in LV myocardium from 1.78mm before registration to 1.42mm after supervised rigid registration and 1.40mm after supervised affine registration, and in RV blood-pool from 2.42mm before registration to 1.72mm after supervised rigid registration and 1.73mm after supervised affine registration. These results are achieved with minimal pre-processing i.e. resampling all the images to a slice thickness of 5mm (using spline interpolation), pixel spacing of 0.75mm × 0.75mm and then resizing them to a common resolution of 224×224 pixels. Another major advantage of our proposed method is the time required for training (80s to train each epoch).
Our proposed supervised method outperforms both the unsupervised rigid and unsupervised affine registration methods. The registration results of unsupervised methods are obtained by training our network using NMI as cost-function. We also experimented with structural similarity image measure (SSIM) loss as a cost function, which yielded similar results. The manual annotations of LV blood-pool, LV myocardium and RV blood-pool used during supervised training enables the network to focus on registering the images accurately around the regions of interest, improving the overall registration accuracy.
We also experimented using only the manual annotations of LV blood-pool and LV myocardium, however the rotational transformation of the registration fails due to the circular nature of the LV blood-pool and LV myocardium. Hence, one potential drawback is the need for annotations of cardiac structures for training. While the LV blood-pool, LV myocardium and RV blood-pool labels used here were available through the challenge, there exist numerous sufficiently accurate and robust cardiac image segmentation methods, such as [5,6,9,18,19], that can be leveraged to annotate the desired structures from the bSSFP CMR images to provide sufficient rotational asymmetry for optimal registration. Lastly, another minor drawback is the possibility of losing certain critical information around the cardiac structures during image resampling prior to registration, but such challenges have always been faced in image registration, an example being atlas construction from images featuring different in-plane resolution and slice thickness.
We would like to address the insignificant difference between the supervised rigid and supervised affine registration approach. Note that both the bSSFP cine CMR and the LGE CMR images are acquired during the same imaging exam, using the same scanner, without changing the patient position, and while also employing ECG gating for end-diastole image capture, resulting in similar shapes and sizes of the cardiac structures.
This study was conducted using LGE and bSSFP cine MRI images from patients diagnosed with cardiomyopathy. Nevertheless, the proposed methods are useful for registering any gadolinium-enhanced and cine MRI images of any patient provided their cardiac conditions is visible and appropriate for gadolinium-enhanced imaging during diagnosis. Such conditions include, but are not limited to, myocarditis or myocardial infarction, or other diseases that show hyper-enhancement of the compromised myocardial regions.
To reduce the reliance on manual annotations to conduct the RoI-guided registration, in our future work we plan to investigate the use of previously validated machine learning-based segmentation techniques [5,6,9,18,19] to automatically extract the required ROI labels (as manual annotated labels are not typically available for large datasets), then proceed with the proposed segmentation-guided registration. As a further mitigation strategy, we also plan to devise and test the effectiveness of using a CNN-based weakly-supervised registration method.
Another area of improvement for our future work is to correct for slice misalignment during MR image acquisition prior to stacking up the segmented and registered image slices in the effort to build 3D models that help quantify and visualize the compromised myocardial regions using 3D maps, leveraging the methods by Dangi et al. [4].
5. Conclusion
In this work, we show that the proposed STN based RoI-guided CNN can be used to register bSSFP cine CMR sequence and LGE CMR sequence accurately and in a time-efficient manner. Our proposed method outperforms unsupervised deep learning algorithms trained using popular similarity metrics such as NMI.
Acknowledgement.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R35GM128877 and by the Office of Advanced Cyber-infrastructure of the National Science Foundation under Award No. 1808530.
References
- 1.Benjamin EJ, et al. : Heart disease and stroke statistics-2017 update: a report from the american heart association. Circulation 135(10), e146–e603 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, et al. : Unsupervised multi-modal style transfer for cardiac MR segmentation. arXiv preprint arXiv:1908.07344 (2019)
- 3.Chenoune Y, et al. : Rigid registration of delayed-enhancement and cine cardiac MR images using 3D normalized mutual information. In: 2010 Computing in Cardiology, pp. 161–164. IEEE; (2010) [Google Scholar]
- 4.Dangi S, Linte CA, Yaniv Z: Cine cardiac MRI slice misalignment correction towards full 3D left ventricle segmentation. In: Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling, vol. 10576, p. 1057607. International Society for Optics and Photonics; (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dangi S, Linte CA, Yaniv Z: A distance map regularized CNN for cardiac cine MR image segmentation. Med. Phys 46(12), 5637–5651 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dangi S, Yaniv Z, Linte CA: Left ventricle segmentation and quantification from cardiac cine MR images via multi-task learning. In: Pop M, et al. (eds.) STACOM 2018. LNCS, vol. 11395, pp. 21–31. Springer, Cham: (2019). 10.1007/978-3-030-12029-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosovitskiy A, et al. : FlowNet: learning optical flow with convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2758–2766 (2015) [Google Scholar]
- 8.Guo F, Li M, Ng M, Wright G, Pop M: Cine and multicontrast late enhanced MRI registration for 3D heart model construction. In: Pop M, et al. (eds.) STACOM 2018. LNCS, vol. 11395, pp. 49–57. Springer, Cham: (2019). 10.1007/978-3-030-12029-0_6 [DOI] [Google Scholar]
- 9.Hasan SK, Linte CA: CondenseUNet: a memory-efficient condensely-connected architecture for bi-ventricular blood pool and myocardium segmentation. In: Medical Imaging 2020: Image-Guided Procedures, Robotic Interventions, and Modeling, vol. 11315, p. 113151J. International Society for Optics and Photonics; (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaderberg M, Simonyan K, Zisserman A, et al. : Spatial transformer networks. In: Advances in Neural Information Processing Systems, pp. 2017–2025 (2015)
- 11.Juan LJ, Crean AM, Wintersperger BJ: Late gadolinium enhancement imaging in assessment of myocardial viability: techniques and clinical applications. Radiol. Clin 53(2), 397–411 (2015) [DOI] [PubMed] [Google Scholar]
- 12.Khalil A, Ng SC, Liew YM, Lai KW: An overview on image registration techniques for cardiac diagnosis and treatment. Cardiol. Res. Pract 2018, 1437125 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MCH, Oktay O, Schuh A, Schaap M, Glocker B: Image-and-spatial transformer networks for structure-guided image registration. In: Shen D, et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 337–345. Springer, Cham: (2019). 10.1007/978-3-030-32245-8_38 [DOI] [Google Scholar]
- 14.Liu Y, Wang W, Wang K, Ye C, Luo G: An automatic cardiac segmentation framework based on multi-sequence MR image. arXiv preprint arXiv:1909.05488 (2019)
- 15.Campello VM, Martín-Isla C, Izquierdo C, Petersen SE, Ballester MAG, Lekadir K: Combining multi-sequence and synthetic images for improved segmentation of late gadolinium enhancement cardiac MRI. In: Pop M, et al. (eds.) STACOM 2019. LNCS, vol. 12009, pp. 290–299. Springer, Cham: (2020). 10.1007/978-3-030-39074-7_31 [DOI] [Google Scholar]
- 16.Sokooti H, de Vos B, Berendsen F, Lelieveldt BPF, Išgum I, Staring M: Nonrigid image registration using multi-scale 3D convolutional neural networks. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 232–239. Springer, Cham: (2017). 10.1007/978-3-319-66182-7_27 [DOI] [Google Scholar]
- 17.Tao X, Wei H, Xue W, Ni D: Segmentation of multimodal myocardial images using shape-transfer GAN. arXiv preprint arXiv:1908.05094 (2019)
- 18.Upendra RR, Dangi S, Linte CA: An adversarial network architecture using 2D U-Net models for segmentation of left ventricle from cine cardiac MRI. In: Coudière Y, Ozenne V, Vigmond E, Zemzemi N (eds.) FIMH 2019. LNCS, vol. 11504, pp. 415–424. Springer, Cham: (2019). 10.1007/978-3-030-21949-9_45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upendra RR, Dangi S, Linte CA: Automated segmentation of cardiac chambers from cine cardiac MRI using an adversarial network architecture. In: Medical Imaging 2020: Image-Guided Procedures, Robotic Interventions, and Modeling, vol. 11315, p. 113152Y. International Society for Optics and Photonics; (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei D, Sun Y, Chai P, Low A, Ong SH: Myocardial segmentation of late gadolinium enhanced MR images by propagation of contours from cine MR images. In: Fichtinger G, Martel A, Peters T (eds.) MICCAI 2011. LNCS, vol. 6893, pp. 428–435. Springer, Heidelberg: (2011). 10.1007/978-3-642-23626-6_53 [DOI] [PubMed] [Google Scholar]
- 21.Zhuang X: Multivariate mixture model for cardiac segmentation from multi-sequence MRI. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 581–588. Springer, Cham: (2016). 10.1007/978-3-319-46723-8_67 [DOI] [Google Scholar]
- 22.Zhuang X: Multivariate mixture model for myocardial segmentation combining multi-source images. IEEE Trans. Pattern Anal. Mach. Intell 41(12), 2933–2946 (2018) [DOI] [PubMed] [Google Scholar]


