Abstract
Aberrant activation of the RAS superfamily is one of the critical factors in carcinogenesis. Among them, KRAS is the most frequently mutated one which has inspired extensive studies for developing approaches to intervention. Although the cognition toward KRAS remains far from complete, mounting evidence suggests that a variety of post-translational modifications regulate its activation and localization. In this review, we summarize the regulatory mode of post-translational modifications on KRAS including prenylation, post-prenylation, palmitoylation, ubiquitination, phosphorylation, SUMOylation, acetylation, nitrosylation, etc. We also highlight the recent studies targeting these modifications having exhibited potent anti-tumor activities.
Keywords: oncogene, KRAS, post-translational modification, prenylation, postprenylation, palmitoylation, ubiquitination, phosphorylation, SUMOylation, acetylation, nitrosylation, cancer therapy
Introduction
RAS was the first mutated gene identified in human cancer over three decades ago, and numerous studies have confirmed mutant RAS as an essential driving factor for tumor initiation and maintenance [1]. The three RAS genes encode four protein isoforms, namely, HRAS, NRAS, KRAS4A, and KRAS4B, which share a high degree of similarity in their primary sequence, protein structure and biochemical properties [2]. The full-length RAS proteins consist of a G domain (the N-terminal 1–165 aa) and a C-terminal HVR. There are two regions within the G domain, switch I (SI) and switch II (SII), which are responsible for conformational changes during GDP–GTP cycling as well as binding with downstream effectors, including RAF proteins, phosphatidylinositol 3-kinase (PI3K) and ral guanine nucleotide dissociation stimulator (RALGDS) [3]. Contrary to the G domain, the HVR is different in each isoform [4].
RAS proteins function as binary molecular switches that cycle between ‘on’ and ‘off’ states depending on their binding to guanosine triphosphate (GTP) or guanosine diphosphate (GDP). The conversion between the two states is controlled by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) [4]. GEFs and GAPs are large, multidomain proteins that interact with various molecules to regulate the activation of RAS [5]. Generally, GEFs such as Son-of-Sevenless homologue 1(SOS1) catalyze the exchange from the GDP- to GTP-bound form, while neurofibromin of GAPs terminates the active state by stimulating the hydrolysis of GTP [6]. Furthermore, the dependence of RAS proteins on GEFs and GAPs as well as the activation of downstream effectors are governed by another principle: translocation [4]. RAS proteins act as cytosolic precursors, undergoing a series of posttranslational modifications, membrane association, and subcellular trafficking to the plasma membrane, where the recruited GEFs and GAPs switch them on and off [6]. In the plasma membrane, RAS proteins encounter downstream effectors, which supports their oncogenic activity [7]. Indeed, the interplay of RalGDS with RAS in the plasma membrane determines the subsequent activation of RAS-like proto-oncogene A(RalA) and RAS-like proto-oncogene B(RalB), which demonstrates that membrane recruitment is a pivotal step in these activation processes [8]. Once activated, RAS proteins play a causal role in human diseases, including cancer, RASopathies, capillary malformations, and psychiatric and neurodevelopmental disorders [9].
Analysis of the current data confirmed that 19% of cancer patients harbored mutant RAS, while KRAS was the most dominant mutation (~75%) [10]. Among these human cancers with mutant RAS, the most common cases include pancreatic adenocarcinoma, colon adenocarcinoma, rectum adenocarcinoma, and lung adenocarcinoma (Table 1). Conventionally, mutant RAS proteins are thought to be deficient in GAP-mediated GTP hydrolysis and/or increased intrinsic nucleotide exchange rates, resulting in abnormal accumulation of RAS in the GTP-bound form. Accumulation of activated RAS is associated with proliferation, survival, transformation, autophagy and apoptosis [11, 12]. These events allow aberrant cells to flourish at the expense of normally functioning cells, which drives tumorigenesis (Figs. 1 and 2). However, among the somatic gain-of-function mutations of RAS proteins, KRAS itself is the most frequently mutated, yet it is the least available to target [13].
Table 1.
Frequency of mutation in RAS isoforms in selected human cancers.
| Cancer type | KRAS (%) | NRAS (%) | HRAS (%) | Total (%) |
|---|---|---|---|---|
| PAAD | 75 | <1 | <1 | 76 |
| COAD | 43 | 5 | 2 | 53 |
| READ | 42 | 11 | 0 | 53 |
| UCEC | 25 | 8 | 2 | 35 |
| LUAD | 28 | <1 | <1 | 29 |
| SKCM | 2 | 28 | 2 | 33 |
| UCS | 12 | 2 | 0 | 14 |
| THYM | 1 | 3 | 9 | 13 |
| THCA | 1 | 8 | 3 | 12 |
| STAD | 9 | 1 | 1 | 11 |
| PCPG | 0 | 0 | 10 | 10 |
The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
PAAD pancreatic adenocarcinoma, COAD colon adenocarcinoma, READ rectum adenocarcinoma, UCEC uterine Corpus endometrial carcinoma, SKCM skin cutaneous melanoma, LUAD lung adenocarcinoma, UCS uterine carcinosarcoma, THYM thymoma, THCA thyroid carcinoma, STAD stomach adenocarcinoma, PCPG pheochromocytoma and paraganglioma.
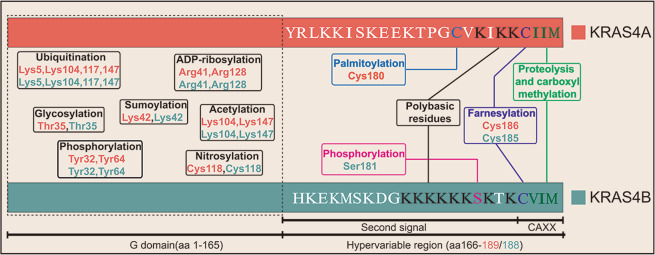
Fig. 1. PTMsof KRAS proteins.
A diagram of the full length of KRAS4A and KRAS4B is drawn, which contains the G domain and hyper variable region (HVR). To distinguish KRAS4A and KRAS4B, different colors are utilized, and the amino acid sequence of the HVR is indicated with abbreviations. For the G domain, the PTMs between KRAS4A and KRAS4B are similar, including ubiquitination, glycosylation, sumoylation, phosphorylation, acetylation, nitrosylation, and ADP-ribosylation. Within the HVR, specific modifications and related residues share the same color.
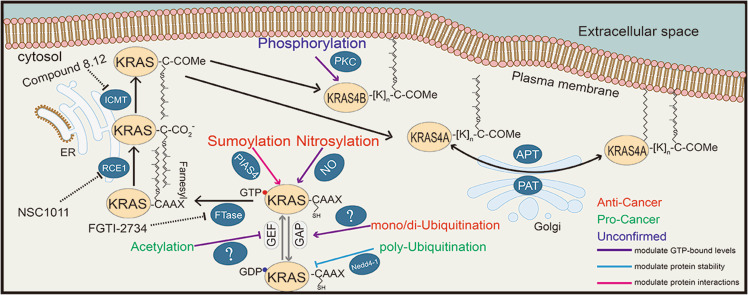
Fig. 2. PTMs regulate the activation and carcinogenesis of KRAS.
A series of modifications occur during the activation or inactivation of KRAS. Each known enzyme that is responsible for a particular process is exhibited. KRAS proteins undergo consecutive modifications to assure their attachment to the plasma membrane and subsequent activation. Meanwhile, there are other modifications that regulate their activation and carcinogenic potency. These processes are indicated by three font colors, where red represents anticancer, green denotes pro-cancer, and brown indicates unconfirmed processes. In addition, the consequence of each modification is also depicted by lines with different colors, where the purple line represents the impact on the level of the GTP-bound state, the yellow line indicates the impact on protein stability, and the pink line denotes the impact on interactions with effectors. GEF guanine nucleotide exchange factors, GAP GTPase-activating proteins, FTase farnesyltransferase, NO nitric oxide, PIAS4 protein inhibitor of activated STAT 4, RCE1 RAS-converting enzyme 1, ICMT isoprenylcysteine carboxyl methyltransferase, PKC protein kinase C, APT acyl protein thioesterase, and PAT protein acetyltransferases.
Despite decades of tenacious efforts, effective anti-KRAS therapies in the clinic remain elusive. Attempts to inhibit RAS by interfering with GTP/GDP binding have proven fruitless because of the lack of well-defined hydrophobic pockets on the surface of RAS proteins and the picomolar affinity of GTP/GDP to KRAS, as well as their high intracellular concentration [14]. RAS proteins were once considered to be undruggable, which ignited greater enthusiasm to pursue alternative strategies [15]. One is to target upstream and downstream effectors, such as pharmacological inhibition of extracellular regulated protein kinases (ERK) and mitogen-activated protein kinase (MEK) [16]. Moreover, there have been breakthroughs in regulating RAS directly, especially agents toward KRASG12C, for which the mutation on codon 12 with cysteine might be a target for inhibitors to overcome the lack of a binding pocket. AMG 510 is the first agent targeting KRASG12C that entered clinical trial, in which covalent binding with mutant cysteine disrupted both SI and SII, impaired the nucleotide exchange from GDP to GTP, and impeded the interaction with RAF. However, the dependence on mutant cysteine also limits its clinical usage [17]. In addition, the concept of synthetic lethality prompted insight into the indirect regulation of RAS [18]. Another pivotal and feasible approach is to focus on posttranslational modifications (PTMs), which allow RAS to associate with membranes and to be subsequently activated. RAS proteins undergo a series of PTMs to ensure their biological functions, which provides a broader intervention approach [19]. Due to the unique structure, direct targeting of modified RAS seems extremely difficult. However, the modifications of RAS proteins are catalyzed by relevant enzymes, which offers an important opportunity to explore efficient and selective inhibitors to interfere with RAS-driven cancer.
Targeting these modifications not only interferes with the association with the plasma membrane but also modulates the interaction with effectors and regulates protein stability, which has been explored for years as vulnerabilities in RAS oncogenesis and oncogene addiction in cancer [20, 21]. This review will summarize the PTMs of KRAS and associated targeting strategies.
Posttranslational modifications of KRAS
Regulation of KRAS by plasma membrane targeting
With the recognition that the association of KRAS with the inner face of the PM is pivotal for its oncogenic activity, early studies determined that a series of modifications modulate this association.
Prenylation, proteolysis and methylation
RAS proteins contain a CAAX tetrapeptide in the C-terminus that possesses a series of modifications named CAAX processing, affording their attachment to specific proteins and membranes [22]. First, the CAAX motiffunctions as the substrate for cytosolicprenyltransferases, farnesyltransferase (FTase) and geranylgeranyltransferase 1 (GGTase1), which allow RAS proteins to have weak affinity for plasma membranes [23]. If the amino acid in the X position is serine or methionine, exactly as in RAS proteins, FTase modifies the cysteine-containing 15-carbon farnesyl isoprenoid. When the CAAX motif ends with leucine, GGTase1 adds a20‑carbon polyisoprene lipid to the cysteine residue [24, 25]. Although CAAX prenylation is considered to be immediate and unregulated, small GTP-binding protein GDP-dissociation stimulator (SmgGDS), classified as a guanine nucleotide exchange factor, can regulate the farnesylation of multiple small GTPases [26]. Moreover, SmgGDS-607 was discovered to promote the farnesylation of HRAS by accelerating protein release from FTase [27]. In addition, regulation of KRAS prenylation via interactions with the noncoding, small nucleolar RNAs SNORD50A and SNORD50B was reported [28]. Zoledronic acid (ZA), a nitrogen-containing bisphosphonate, can disrupt the activity of RAS through prenylation [29]. Recently, the prenyl-binding protein phosphodiesterase δ(PDEδ) was demonstrated to bind with KRAS4B and influence its trafficking [30]. Moreover, inhibition of PDEδ with molecules such as deltarasin or deltazinone hindered oncogenic signaling and tumorigenic growth [31]. It is worth noting that in the presence of FTase inhibitor, KRAS is capable of modification by GGTase1, which allows its full biological function [32].
Next, prenylated RAS proteins shift to the surface of the endoplasmic reticulum (ER) and interact with RAS-converting enzyme 1 (RCE1). Prenylation seems to be a prerequisite for the second step; one reason is that membrane transport mediates the colocalization of RAS with RCE1, and another is substrate specificity [33]. Then, RCE1 removes the last three amino acids AAX by proteolysis, which converts prenylcysteine into the new C-terminus [34]. Finally, the C-terminal prenylcysteine serves as a substrate for isoprenylcysteine carboxyl methyltransferase (ICMT), which catalyzes the methylation of RAS proteins, neutralizes the negative charge, prevents plasma membrane repulsion and thereby increases membrane affinity [35]. Of the steps in CAAX processing, only the carboxyl methylation catalyzed by ICMT is reversible (although no specific esterase has been identified).The result of this processing is to remodel the globular hydrophilic region in the C-terminus and render it hydrophobic, aiding RAS protein insertion into the plasma membrane and ful filling their biological activity.
Palmitoylation and depalmitoylation
Nevertheless, in addition to the CAAX processing of RAS proteins, another element called “second signals” within the HVR sequence is needed, which work together to promote the association with the plasma membrane and subcellular localization. In the case of NRAS or HRAS, the “second signal” is palmitoylation of one or two cysteine residues [36]. ForKRAS4B, this occurs at a polybasic region (PBR) that is upstream of the CAAX motif, which consists of eight lysine residues. These positively charged residues interact with the negatively charged phospholipid headgroups of the plasma membrane by electrostatic binding [37]. In brief, the association of KRAS4B with the plasma membrane depends on its farnesylation and positively charged PBR. For KRAS4A, a combination of cysteine palmitoylation at cysteine and lysine residues as in KRAS4B is required [38, 39]. Evidence demonstrated that palmitoylation afforded RAS proteins 100-fold higher affinity for membranes than that of the prenylated protein alone [40]. For KRAS4A, palmitoylation occurs on cysteine 180 at the C-terminus via thioester linkages and is referred to as S-palmitoylation [41]. Unlike other isoforms, although palmitoylation of KRAS4A dramatically increases the efficiency of plasma membrane targeting, it is not absolutely required because the polybasic regions provide weak affinity for the plasma membrane [38]. The protein palmitoylation machinery consists of a family of 25 protein acyltransferases (PATs) each of which possesses a DHHC sequence in its catalytic site but differs in subcellular localization and substrate specificity [42]. Recently, a protein complex of DHHC domain-containing 9(DHHC9) and Golgi complex-associated protein of 16 kDa(GCP16) was identified as a PAT capable of modifying HRAS, which colocalized with palmitoylated RAS on the Golgi apparatus [43]. Although the DHHC9/DHHC9 complex modifies HRAS, its knockdown fails to block HRAS palmitoylation, suggesting that multiple PATs have activity toward RAS [44]. Considering the substrate promiscuity of PATs, it seems complicated to characterize another special PAT specific to KRAS4A.
In addition to palmitoylation, the distribution of RAS proteins is also mediated by depalmitoylation, in which the palmitoylation/depalmitoylation cycle guarantees that RAS proteins shuttle easily between the plasma membrane and Golgi apparatus. Palmitoylation seems labile owing to the weak stability of the thioester bond at physiological pH, but existing evidence demonstrates that palmitoylation is reversed by cellular thioesterases [45]. The first non-lysosomal proteins found to have thioesterase activity toward RAS were acylproteinthioesterase 1 and 2 (APT1 and 2) [46]. Interestingly, other regulators of RAS depalmitoylation exist in addition to APTs. For example, FKBP12, a 12 kDa FK506-binding protein, mediates depalmitoylation of HRAS on a proline near the palmitoylated cysteines [47]. However, the full repertoire, specificity and activity of cellular thioesterases that depalmitoylate RAS remain largely uncharacterized [48].
Phosphorylation
Compared with KRAS4A, KRAS4B does not require further posttranslational modification as a second signal beyond CAAX progressing to associate with the plasma membranes; however, the phosphorylation on tyrosine32, tyrosine64, andserine181 is also associated with its biological functions [49]. It has been reported that phosphorylation of KRAS4B on serine 181 within the polybasic region is mediated by protein kinase Cs (PKCs) and cGMP-dependent protein kinase 2 (PKG2), diminishing the affinity for plasma membrane due to partial neutralization of positively charged residue [50]. Phosphorylation promotes rapid dissociation of KRAS from the plasma membrane and association with intracellular membranes, including the outer membrane of mitochondria. Moreover, phosphorylated KRAS was found to translocate from the plasma membrane to the ER, where it interacted with the inositol trisphosphate receptor (IP3R) and thereby attenuated cell growth by blocking the interaction of inositol 1,4,5-trisphosphatereceptor (IP3R) with the pro-survival effector Bcl-XL [51]. In addition, phosphorylation on serine 181 might act as a regulator to modulate oncogenic properties of KRAS in vivo. One report showed that this phosphorylation inhibited tumor initiation by blocking the interaction with calmodulin, thereby abrogating the suppression of noncanonical Wnt signaling [52]. However, a conflicting report demonstrated that the tumorigenicity capacity of KRAS required phosphorylation on serine181, a phospho-mimetic mutation of KRAS with enhanced tumorigenesis [53]. Therefore, whether phosphorylation of KRAS on serine 181 is required for oncogenesis needs further confirmation.
Regulation of KRAS by nucleotide exchange
KRAS acts as a molecular switch whose active or inactive state is controlled by GEF or GAP. Interestingly, in addition to somatic mutation, several other posttranslational modifications also modulate the nucleotide exchange from GTP binding to GDP binding.
Acetylation
It was recently shown that KRAS canbe acetylated on lysine104, which decreased GEF-mediated nucleotide exchange and increased the level of the inactive GDP-bound state, attenuating the transforming activity of KRAS [54]. Results from a molecular dynamics model demonstrated that acetylation at lysine104 disturbed the switch IIregion via electrostatic interaction, which disrupted the interaction of KRAS and GEFs [55]. By extension, histone deacetylase 6 (HDAC6) and sirtuin 2 (SIRT2) were identified as deacetylases that regulated the level of acetylated KRAS, and interference with either enzyme strikingly impaired the survival of cancer cells expressing mutant KRAS [56]. Additionally, a later study discovered that lysine147 acts as a novel substrate for SIRT2-mediated deacetylation, whose acetylation status strongly affects the oncogenic properties of KRAS [57]. It is important to note that lysine104 and lysine147 also function as sites for mono/diubiquitination, as discussed below, and whether there is a competitive relationship between acetylation and ubiquitination should be explored in further research. In addition to lysine104 and lysine147, which are located in the C-terminus, a recent study reported that acetylation also occurred in the N-terminus. The results from mass spectrometric characterization and structural analysis showed that the acetyl group of the N-terminus might maintain the stability of the switch regions and the N-terminus [58].
Nitrosylation
The vital function of reactive oxygen species in signal transduction prompted further investigations into direct modification of RAS [59]. Evidence showed that nitrosylation of cysteine 118 was mediated by nitric oxide (NO) and appeared in all RAS isoforms, which led to a profound potentiation of GDP/GTP exchange through conformational change. Therefore, this nitrosylation increases the level of the active GTP-bound state, promoting the activation of RAS proteins [60, 61]. More interestingly, nitrosylation of KRAS was demonstrated to participate in the positive regulation of inducible nitric oxide synthase (iNOS), which enhanced the production of NO and thereby increased the level of nitrosylation [62]. Moreover, endothelial nitric oxide synthase (eNOS) also facilitates the nitrosylation and activation of KRAS, whose inhibition strongly inhibits tumor initiation and maintenance [63]. Hence, this evidence supports the association of redox regulation with activation of KRAS, which might offer new insight into the regulation of RAS proteins.
Ubiquitination
Modification of proteins by ubiquitin, the so-called ubiquitination, is regulated by ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin ligase (E3) and deubiquitylation enzymes, and is of high importance for the stability, activity and localization of proteins [64]. Studies ascertained that lysine104, lysine117, and lysine147are sites for mono/diubiquitination, acting as reversible triggers for signal initiation. Mono-ubiquitination of KRAS at lysine147 impairs GAP-mediated GTP hydrolysis, promoting GTP loading and enhancing affinity to downstream effectors such as PI3K. Mono/diubiquitination on lysine117 accelerates nucleotide exchange and thereby enhances activation [55, 65]. However, neither GAP-mediated GTP hydrolysis nor nucleotide exchange is changed under monoubiquitination at lysine104 of KRAS [55]. Considering the location of lysine104 near the GEF binding region as well as the point mutation experiment, ubiquitination at lysine104 might alter GEF-mediated catalysis [66]. Nevertheless, the E3 ligases responsible for the ubiquitination of KRAS at lysine147, lysine117 and lysine104 remain to be determined. Most recently, the inactivation of leucine zipper-like transcription regulator 1(LZTR1) was found to inhibit KRAS ubiquitination, which provided an unexpected layer of KRAS regulation [67].
SUMOylation
SUMOylation (small ubiquitin-like modification), analogous to ubiquitination, also regulates many biological functions of proteins by conjugating small ubiquitin-like modifiers to lysine residues in a series of enzymatic reactions. It is not surprising that SUMOylation is required for KRAS-driven tumorigenesis, and inhibition of the SUMO pathway by depletion of ubiquitin-conjugating enzyme 9 (UBC9) attenuates the growth of KRAS mutant colorectal cancer cells [68]. SUMOylation of KRAS at lysine42 seemed to be related to its oncogenesis by regulating the initiation and activation of downstream effects [69]. Further study identified that the SUMO-protein ligase PIAS4 is responsible for the SUMOylation of KRAS, which can be removed by SUMO-specific peptidase 1/2 (SENP1/2), demonstrating that this process is reversible [70]. However, the stoichiometry and physiologic significance of these modifications still need to be ascertained.
Others
In addition to subcellular localization and nucleotide exchange, several posttranslational modifications also affect the oncogenic activity of KRAS in other ways, such as protein interaction and protein stability. One indisputable discovery is that the phosphorylation on tyrosine32 and tyrosine64 by Src leads to a conformational change of switch I and II regions, which inhibits the association of KRAS with downstream effectors [71]. Moreover, this phosphorylation can be reversed by protein tyrosine phosphatase non-receptor type 11(PTPN11), which was found to be required for the growth of mutant KRAS-driven cancers [72]. In addition, phosphorylated KRAS proteins have been found to form distinct nanoclusters at the plasma membrane, which favors activation of RAF and PI3K [73]. It is well known that polyubiquitination involves the ubiquitin-proteasome system, which regulates intracellular protein levels and functions [74]. Later, evidence demonstrated that it was the ubiquitin ligase Nedd4-1 that targeted KRAS for polyubiquitination at lysine5 and degradation [75]. Interestingly, the deubiquitylation enzyme OTUB1 inhibits KRAS monoubiquitination by interfering with several E2s, which is independent of its catalytic activity [76]. Another ubiquitin ligase that might be responsible for KRAS is β-transducing repeat-containing protein (β-TrCP1). A recent study showed that a complex of E2 and E3 maintains the protein stability of KRAS by degrading β-TrCP1 [77]. E3 ligase ring finger protein 7 (RNF7) functions paradoxically in KRAS-driven tumors, but whether it modulates KRAS ubiquitination and activity remains to be validated [78–80].
Many bacterial factors, such as Exoenzyme S (ExoS) and Lethal Toxin (LT),also modify RAS proteins [81]. The first identified bacterial toxin inactivating RAS is LT produced by Clostridiumsordeli, which acts as a glucosyltransferase that utilizes uridine diphosphate-glucose (UDPG) to modify threonine35 of KRAS and thereby inhibits MAPK signaling [82, 83]. The adenosine diphosphate (ADP)-ribosyl transferase ExoSproduced by Pseudomonas aeruginosacatalyzes the ADP-ribosylation of RAS onarginine41 and arginine128. The modification onarginine41 attenuates GTP loading and inhibits the interaction of RAS with effectors [84].
Targeting the PTM of KRAS
RAS proteins undergo a series of continuous posttranslational modifications, as described above, and these processes regulate their attachment to the plasma membrane and their subcellular localization, thereby regulating the activation and oncogenic capacity of KRAS. Therefore, each of the steps in KRAS protein “maturation” might represent a potential target for therapy.
Inhibition of prenylation
Considering that farnesylation functions as a prerequisite for subsequent modifications, including proteolysis and methylation, and RAS proteins lacking farnesylation are unable to attach to the plasma membrane, interfering with farnesylation inspired intense research for anti-KRAS therapy [85]. The intervention of farnesylation can be mediated in two ways: one is using mimetic polypeptides of the CAAX motif to compete with KRAS for FTase, another is utilizing an analog of farnesyl pyrophosphate (FPP) that is involved in farnesylation to compete for binding to FTase [86]. Together, these peptides or small molecule inhibitors used to impair the catalytic activity of FTase during farnesylation of KRAS are termed farnesyltransferase inhibitors (FTIs). Among these mimetic polypeptides, L-744832 might be available for combination therapy, which augmented radiation sensitivity in pancreatic cancer cell lines at 5 or 10 μM, and L-744832 together with everolimus, an mTOR inhibitor, induced apoptotic cell death in non-Hodgkin lymphomas [87, 88]. At the same time, several nonpeptide molecular inhibitors targeting FTase selectively were also widely designed, some of which, including BMS-214662, lonafarnib, and tipifarnib, have been evaluated in clinical trials [89]. Moreover, two of them, lonafarnib and tipifarnib, entered the Phase III clinical trial stage and were approved for the treatment of advanced solid cancer with mutant KRAS, such as pancreatic cancer, non-small cell lung cancer, and colon cancer. Unexpectedly, data analysis of clinical trials showed that neither monotherapy nor combination therapy with lonafarnib and tipifarnib was effective in improving outcomes [90]. Looking back, the discrepancy between laboratory findings and clinical data revealed that KRAS4B, the major isoform expressed in human tumors, underwent alternative geranylgeranylation by the related GGTase1 in the presence of FTIs, which maintained its membrane association and biological function [32]. Overall, the effectiveness of monotherapy with FTIs for KRAS-driven solid cancer has been less than satisfactory.
To overcome GGTase1-mediated resistance to FTIs, a method of dual inhibition of FTase and GGTase1 was designed to target the prenylation process of KRAS [24]. It was observed that dual deletion of FTase and GGTase1 impaired tumor development in KRAS-driven lung cancer [91]. Moreover, L-778123, a mimetic polypeptide of CAAX, was discovered to have a potential inhibitory effect toward FTase (in vitro IC50 = 2 nM) and GGTase1 (in vitro IC50 = 98 nM). However, recent studies showed that although the prenylation of HDJ2 and Rap1A (both FTase and GGTase1 substrates) was inhibited by L-778123, KRAS, the intended target of L-778123, was not inhibited. The antitumor effect of L-778123 might largely rely on other substrates of FTI or GGTase1, such as Ras Homolog Family Member B (RhoB). The lower efficiency in suppressing the prenylation level of KRAS might hamper the clinical trial of L-778123 [92, 93]. In addition, FTase and GGTase1 catalyze a broad range of substrates such as RHO family proteins, which are essential for normal physiological function, thus limiting the usability of this method due to high toxicity and narrow therapeutic window [94]. Excitingly, FGTI-2734, a new mimetic polypeptide of CAAX designed to dually inhibit FTase and GGTase1, inhibited the membrane localization of KRAS in pancreatic, lung, and colon human cancer cell lines, exhibiting strong antitumor activity by inducing apoptosis in a xenograft model derived from pancreatic cancer patients. Most importantly, FGTI-2734 has little effect on the growth of tumors independent of mutant KRAS, which warrants further preclinical and clinical studies [95]. This evidence supports the notion that FGTI-2734 might be a promising KRAS-targeting agent that deserves further development, and the strategy to target both FTase and GGTase1 deserves further study.
Finally, given that FPP is produced from the mevalonate pathway, which, along with its subsequent metabolic product geranylgeranyl pyrophosphate (GGPP), functions as an element for farnesylation or geranylgeranylation, impeding the mevalonate pathway might represent an indirect approach to disturb the prenylation process [96]. Therefore, the traditional reagents statins that inhibit the mevalonate pathway are generally considered to exert suppressive effects on tumorigenesis [97, 98]. However, a recent study suggested that the tumor-suppressive function of statins might be mechanistically unrelated to the prenylation process of KRAS [99]. Another emerging inhibitor of the mevalonate pathway is zoledronic acid (ZA), which acts as a bisphosphonate-based inhibitor. ZA targets the farnesyl pyrophosphate synthetase (FPPS) of the mevalonate pathway and impairs RAS membrane localization, thereby hindering the growth of breast cancer cells [100]. These inhibitors had shown anticancer activity but were limited due to poor pharmacokinetic properties and high affinity to bone mineral [101]. Moreover, the discovery of allosteric non-bisphosphonate FPPS inhibitors boosted the strategy to interfere with the mevalonate pathway [102].
Inhibition of postprenylation
After prenylation, KRASA proteins are modified by RCE1 and ICMT, which mediate the process of proteolysis and methylation, together called postprenylation processes. Given that either farnesylated or geranylgeranylated KRAS undergoes postprenylation processes, there might be less worry about the problem of selectivity. Therefore, a feasible approach to target prenylated KRAS is inhibition of RCE1 or ICMT [103]. Indeed, deletion of RCE1 in a mouse model resulted in incorrect cellular localization of the KRAS protein. Nevertheless, compared with FTI treatment, the reduction of KRAS-driven transformation of fibroblasts with conditional elimination of RCE1 was small [104]. Interestingly, evidence showed that a skin cancer cell line with deletion of RCE1 was strikingly sensitive to FTIs [105]. Whether there is a viable synergistic effect between RCE1 inhibitors and FTIs merits further confirmation. Although RCE1 disruption induced a modest impact on tumorigenesis, the results from genetic manipulation of ICMT showed that, except for the effect on mislocalization, deletion of ICMT strongly impeded the transformation capacity of fibroblasts expressing oncogenic KRAS [106]. As discussed above, previous studies defined the effect of targeting RCE1 and ICMT by eliminating genes, which encouraged researchers to discover small molecule inhibitors of the two enzymes.
Among the agents that had inhibitory effects toward RCE1, NSC1011 was considered the best compound, which had been characterized to induce incorrect cellular localization of RAS proteins in a human colon cancer cell line [107]. Furthermore, many updated RCE1 inhibitors were generated after the analysis of the structure-activity relationship (SAR) of NSC1011, and these new agents exhibited low cell toxicity and were more effective at mislocalizing KRAS proteins [108]. However, deletion of RCE1 in a mouse model caused the development of lethal cardiomyopathy and exacerbated the development of KRAS-induced myeloproliferative disease [109, 110]. Accordingly, it is necessary to assess the safety and confirm the availability of agents developed to target RCE1 in preclinical models.
In terms of the inhibitors of ICMT, according to the difference among structural characteristics, they could be divided into analogs of substrate or synthetic agents [111]. In detail, the methyl group donor S-adenosylmethionine (AdoMet) is involved in the methylation process of KRAS, and together with its metabolite S-adenosylhomocysteine (AdoHcy) constitutes a feedback loop that can negatively regulate the activity of ICMT [112, 113]. Therefore, one approach to interfere with ICMT was using the analogs of AdoHcy or inhibiting the activity of AdoHcy hydrolase; all of which increased the level of AdoHcy, resulting in inhibition of ICMT. Indeed, these compounds have antitumor activity, but the lack of selectivity and specificity limits their potential [114, 115]. Moreover, another approach to couple with the conception of analogs is developing agents to compete with prenylated KRAS for methylation, for example, S-farnesyl-thiopropionic acid-triazole compounds, which exhibited antitumor effects at micromolar concentrations in a pancreatic cancer cell line [116]. Among these synthetic compounds, cysmethynil is the most potent compound characterized so far and is a derivative of indole. Indeed, cysmethynil induces them is localization of RAS, suppresses its activity, induces cell death through autophagy and reduces the growth of xenograft tumors in human colon and prostate cancer cells [117]. Unfortunately, the demerits in pharmacokinetic properties such as poorwater solubility make it inappropriate for clinical use [118]. Recently, further SAR analysis of cysmethynil identified a novel derivative named compound 8.12, which is capable of impairing tumorigenesis of prostate and liver cancer cells with improved pharmacokinetic properties [119].
Although inhibition of RCE1 and ICMT represents a potential avenue for KRAS functional blockade and many agents are undergoing preclinical testing, there might also be some challenges. A report demonstrated that the RAS pathway was still activated with the inhibition of RCE1 and ICMT, in which mislocalized RAS could transduce activation signals from other locations [120]. Of note, targeting the processes of prenylation and postprenylation resulted in the generation of a few inhibitors that are currently under development at various stages (Table 2).
Table 2.
Selected inhibitors targeting PTMs of KRAS.
 |
Inhibition of palmitoylation/depalmitoylation
Compared with the other isoforms such as HRAS or NRAS, KRAS, especially one of its splice variants KRAS4B, does not need palmitoylation or depalmitoylation to ensure its affinity to the plasma membrane and subsequent activation [121]. It is not absolutely necessary for KRAS4A to be palmitoylated since the polybasic regions can support weak efficiency for plasma membrane targeting [38]. Accordingly, inhibition of palmitoylation or depalmitoylation to affect the activity of KRAS is generally considered an inferior target. However, the palmitoylation–depalmitoylation cycle of KRAS4A enables it to translocate to the mitochondrial membrane so that it encounters hexokinase 1 (HK1) [122]. Early evidence showed that the survival of cancer cells expressing oncogenic KRAS was dependent on the separation of glucose and glutamine metabolism [123]. In return, high glucose can induce KRAS mutation through O-GlcNAcylation of some enzymes, which might provide persistent activation of the RAS pathway. In addition, the effect of oncogenic induced senescence (OIS) by KRAS is suppressed by O-GlcNAcylation, which facilitates the development of lung cancer [124, 125]. These results suggest that there might be positive feedback between oncogenic KRAS and glycometabolism. Thus, it will be interesting to confirm the interplay between KRAS and glycometabolism, and human cancer with high expression of KRAS4A might be sensitive to a particular metabolic process, which can be utilized for treatment.
Intervention of ubiquitination
In addition to oncogenic mutations, stability regulation of KRAS also affects its activity and carcinogenicity, meaning that the degradation of oncogenic KRAS in a particular way is feasible to achieve treatment goals. There is one well-characterized ubiquitin-proteasome system in which E3 together with E1 and E2 catalyzes the binding of specific residues with ubiquitin, mediating the degradation of proteins through the 26S proteasome as well as their subcellular localization and functional state [126]. In this system, the most crucial player is ubiquitin ligase, which has the property of specific recognition of the substrate, and its abnormal regulation is associated with the development of cancer [127]. Thus, finding and targeting the E3 ligase is another alternative method to treat oncogenic KRAS-driven cancer [128]. Most inspiringly, a recent study reported that the ubiquitin ligase WD repeat domain 76 (WDR76) acts as a tumor suppressor, impeding the proliferation, transformation, metastasis and invasion of hepatoma cells through the degradation of HRAS by polyubiquitination [129]. Aberrantly activated KRAS underwent a conformational change, which inhibited the ubiquitin ligase Nedd4-1-mediated polyubiquitination and degradation, suppressing the development of cancer [75]. However, there is a problem: while some ubiquitin ligases function as tumor suppressors through the degradation of oncogenic KRAS, developing strategies to activate these ubiquitin ligases excessively might be inconvenient. Therefore, targeting other E3 ligases that are responsible for mono/diubiquitination or SUMOylation seems more easily implemented. Moreover, some artificially engineered ubiquitin ligases, such as RC-U and recombinant chimeric proteins, have been produced to induce specific degradation of KRAS via the ubiquitin-proteasome system, which was efficient in suppressing pancreatic cancer cell growth in vitro and in vivo [130, 131]. Hence, there is hope for a specific and druggable E3 ligase of KRAS, which might need further efforts.
Others
In addition to the modifications discussed above, phosphorylation, acetylation and nitrosylation can also regulate the oncogenic function of KRAS proteins. Targeting KRAS phosphorylation on serine181 is an intriguing possibility. Phosphorylation by PKC promotes dissociation of KRAS from the plasma membrane and association with intracellular membranes, where phosphorylated KRAS interacts with Bcl-XL. The PKC agonist bryostatin-1 exhibits antitumor activity in vitro and in vivo in a serine181 dependent manner, suggesting an approach to therapy of KRAS-dependent tumors with agents that stimulate phosphorylation of serine181 [132]. Acetylation at lysine104 and lysine147 by SIRT2 and HDAC6 increases the level of active KRAS with a GTP-bound state, and deletion of SIRT2 enhances tumor growth in a KRAS-driven pancreatic cancer cell line [57]. However, the specific enzyme that mediates acetylation of KRAS needs to be confirmed, which might be another feasible target for treatment. Although nitrosylation promotes activation of all RAS isoforms, the key details in this process are not completely clear, and determining the interplay of redox regulation with RAS activation will provide another intervention strategy.
Direct targeting of KRAS
Similarly, direct targeting of KRAS has remained challenging due to the specific features of its molecular structure. However, the strategy to target KRAS directly is still under investigation and has branched off in different directions. One method is to prevent the formation of the KRAS–GTP complex, in which competing GTP analogs are designed to compete with nucleotide binding to RAS. However, the actual inhibition of KRAS activation by these analogs was lower than expected owing to the high affinity of GTP to KRAS and high cellular GTP concentrations [133]. Another strategy to prevent KRAS–GTP complex formation is to inhibit the interaction of KRAS with GEFs. The best known GEF is SOS1; therefore, SOS1 inhibitors were synthesized to block the KRAS-SOS1 interaction. Studies showed that these inhibitors suppressed SOS1-mediated nucleotide exchange and inhibited RAS activation, resulting in inhibition of cell proliferation and downregulation of RAS signaling [134]. However, the binding activity of these inhibitors to KRAS is weak, and it is unknown whether the inhibitors have similar effects on the KRAS mutational setting [135].
At the same time, with deeper insight into KRAS biology, several small molecule inhibitors selectively targeting the KRASG12C mutation with similar covalent binding mechanisms have emerged as the most promising approach and are now reaching advanced stages of clinical investigation. The inherently reactive characteristic of cysteine, which is found at codon 12 of KRASG12C, can be used to develop covalent small molecule inhibitors. Covalently targeting active site cysteines is a widely used strategy in drug discovery [136]. Moreover, wild-type KRAS lacks cysteines in the active site, so KRASG12C can be specifically targeted by this covalent approach. Among the developed inhibitors, AMG510, MRTX849 and ARS-3248 covalently bind to KRASG12C at the cysteine, locking KRASG12C in an inactive state and inhibiting KRAS-dependent signaling [137–139]. These inhibitors have been tested in clinical trials to treat patients with non-small cell lung cancer (NSCLC) or solid tumors and exhibit impressive antitumor activity with no obvious dose-limiting toxicity. Although covalent KRASG12C inhibitors are regarded as the most promising approach, there also exist several concerns [140]. One is that although KRASG12C is the most common mutant variant in NSCLC accounting for ~40% of all KRAS mutant tumors and ~13% of all lung adenocarcinomas, it is present in only ~3% of colorectal cancer cases and a small subset of patients with pancreatic, endometrial and urothelial cancers, which might be applied to lower populations than expected [141, 142]. In addition, the therapeutic potential of these inhibitors might be impaired by intrinsic resistance mechanisms. Therefore, the combination of these inhibitors with other agents, such as programmed cell death protein 1 (PD-1), MEK, SHP2 or ErbB inhibitors, is evaluated in preclinical and clinical trials [143]. Moreover, combination therapy of these covalent inhibitors with FTIs or other inhibitors that target PTMs of KRAS might be worth further investigation.
Conclusion and prospects
After almost four decades of extensive research concentrated on the RAS superfamily of GTPases, this complex process has provided an increasingly comprehensive characterization of the goal: anti-RAS therapy. Although our understanding is still incomplete, we can at least recognize the fact that RAS proteins are substrates for a wide variety of PTMs, which play essential roles in their biological function. Given that these modifications are catalyzed by specific enzymes, each step of these processes might represent a potential target for cancer treatment. Given that among the RAS superfamily members, KRAS is the most frequently mutated in human cancers, especially in those that urgently need improved therapies, such as pancreatic cancer, the focus in this review is firmly on KRAS. However, why KRAS is the most frequently mutated in general and why the frequency of mutant RAS proteins varies depending on the tissue of origin are questions that remain to be answered.
The KRAS proteins are modified by prenylation, palmitoylation, ubiquitination, phosphorylation, SUMOylation, acetylation and nitrosylation. It is clear from the successful study above that PTMs of KRAS are common in various tumors and expand the scalability of KRAS regulation. There are some unique advantages. In fact, the relevant enzymes are the key regulators of PTMs. Compared to KRAS itself, these enzymes have well-defined binding pockets that are more accessible to be targeted by small molecules. Although substrate specificity might be the major barrier to the clinical use of enzyme inhibitors, the role of one particular enzyme in tumorigenesis varies, dictated by the substrates it interacts with. Moreover, discovering and identifying a specific enzyme that is responsible for a particular modification during the maturation processes of KRAS may provide a more promising therapeutic strategy with fewer adverse reactions. Therefore, targeting the relevant enzyme-mediated KRAS PTMs represents a powerful and efficient strategy for the treatment of KRAS-driven cancer types. Evidence suggests that therapeutics designed to target enzymes involved in these modifications decrease the carcinogenicity of KRAS. Among these indispensable modifications, the targeting processes of prenylation and postprenylation might be more reasonable, as these physiological behaviors mediate KRAS protein subcellular trafficking and membrane localization. Although inhibition of one particular procedure was insufficient to inhibit tumorigenesis, combination therapy such as dual inhibition of FTase and GGTase1 could be more effective. Other options, such as dual inhibition of FTase and RCE1, if possible, might be worth further investigation. A significant challenge in combination therapy is to reduce toxic and side effects due to general inhibition of protein processing. Hopefully, the discovery of FGTI-2734, a dual FTase and GGTase1 inhibitor, encourages researchers to continue exploring the dual inhibition strategy, which effectively inhibits KRAS membrane localization and growth of mutant KRAS-dependent tumors at nontoxic doses.
In addition, combination therapy with inhibition of PTMs and other treatment strategies has attracted much attention and research. For instance, treatment with FTI augments the sensitivity of cancer cells to radiotherapy. Moreover, targeting modifications, including prenylation and postprenylation, blocks KRAS proteins in the inactive state of GDP binding, while activated KRAS proteins also undergo other modifications, such as ubiquitination and sumoylation, whose inhibition with specific enzyme inhibitors is worth the effort. Moreover, the development of a proteolysis targeting chimera (PROTAC) and hydrophobic tagging approach using specific recognition of enzymes with substrates and the ubiquitination process might offer another feasible method to degrade related enzymes and proteins directly. In essence, targeting the PTMs of KRAS described above means targeting plasma localization. Beyond that, there are still other ways to drug KRAS. Some examples include targeting downstream effector signaling, such as the mitogen-activated protein kinase (MAPK) cascade, utilizing synthetic lethal interaction partners, such as serine/threonine kinase 33 (STK33) and TANK-binding kinase 1 (TBK1), targeting KRAS-mediated metabolic processes, and incorporating immunization therapy. Another emerging hotspot is the development of inhibitors of mutant KRAS. Among these inhibitors, AMG510 and MRTX849 are now attracting considerable attention. Although the latest analyses of clinical data show that these inhibitors are less optimal than expected, the combination strategy with inhibitors of PTM is worth investigating. These promising developments broaden the outlook of anti-KRAS therapies. In summary, although many obstacles stand in the path to success, the continuous research efforts and constantly developed novel strategies have made researchers confident in the ability to defeat the oncogene KRAS.
Competing interests
The authors declare no competing interests.
Contributor Information
Jin-jian Lu, Email: jinjianlu@um.edu.mo.
Hong Zhu, Email: hongzhu@zju.edu.cn.
References
- 1.Cox AD, Der CJ. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papke B, Der CJ. Drugging RAS: know the enemy. Science. 2017;355:1158–63.. doi: 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 3.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–304.. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 4.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77.. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–57.. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NagDas SK, Winfrey VP, Olson GE. Identification of ras and its downstream signaling elements and their potential role in hamster sperm motility1. Biol Reprod. 2002;67:1058–66.. doi: 10.1095/biolreprod67.4.1058. [DOI] [PubMed] [Google Scholar]
- 8.Ferro E, Trabalzini L. RalGDS family members couple Ras to Ral signalling and that’s not all. Cell Signal. 2010;22:1804–10.. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Tajan M, Paccoud R, Branka S, Edouard T, Yart A. The RASopathy family: consequences of germline activation of the RAS/MAPK pathway. Endocr Rev. 2018;39:676–700. doi: 10.1210/er.2017-00232. [DOI] [PubMed] [Google Scholar]
- 10.Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–74.. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74.. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25:620–7. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–92.. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Disco. 2014;13:828–51.. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazo JS, Sharlow ER. Drugging undruggable molecular cancer targets. Annu Rev Pharmacol Toxicol. 2016;56:23–40. doi: 10.1146/annurev-pharmtox-010715-103440. [DOI] [PubMed] [Google Scholar]
- 16.Merchant M, Moffat J, Schaefer G, Chan J, Wang X, Orr C, et al. Combined MEK and ERK inhibition overcomes therapy-mediated pathway reactivation in RAS mutant tumors. PLoS One. 2017;12:e0185862. doi: 10.1371/journal.pone.0185862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51.. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell. 2017;168:890–903. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Disco. 2007;6:541–55.. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 20.Michael JV, Goldfinger LE. Concepts and advances in cancer therapeutic vulnerabilities in RAS membrane targeting. Semin Cancer Biol. 2019;54:121–30.. doi: 10.1016/j.semcancer.2017.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu P, Wang Y, Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B. 2019;9:871–9. doi: 10.1016/j.apsb.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–91.. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Seabra MC, Reiss Y, Casey PJ, Brown MS, Goldstein JL. Protein farnesyltransferase and geranylgeranyltransferase share a common alpha subunit. Cell. 1991;65:429–34.. doi: 10.1016/0092-8674(91)90460-G. [DOI] [PubMed] [Google Scholar]
- 24.Fu HW, Casey PJ. Enzymology and biology of CaaX protein prenylation. Recent Prog Horm Res. 1999;54:315–42. [PubMed] [Google Scholar]
- 25.Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J Mol Biol. 2004;343:417–33.. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 26.Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP, et al. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J Biol Chem. 2010;285:35255–66.. doi: 10.1074/jbc.M110.129916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Torres D, Fierke CA. The chaperone SmgGDS-607 has a dual role, both activating and inhibiting farnesylation of small GTPases. J Biol Chem. 2019;294:11793–804.. doi: 10.1074/jbc.RA119.007438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A, et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48:53–8. doi: 10.1038/ng.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai X, Xia H, Zhou S, Tang Q, Bi F. Zoledronic acid enhances the efficacy of the MEK inhibitor trametinib in KRAS mutant cancers. Cancer Lett. 2019;442:202–12.. doi: 10.1016/j.canlet.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Dharmaiah S, Bindu L, Tran TH, Gillette WK, Frank PH, Ghirlando R, et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEdelta. Proc Natl Acad Sci U S A. 2016;113:6766–75. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papke B, Murarka S, Vogel HA, Martin-Gago P, Kovacevic M, Truxius DC, et al. Identification of pyrazolopyridazinones as PDEdelta inhibitors. Nat Commun. 2016;7:11360. doi: 10.1038/ncomms11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–64.. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 33.Otto JC, Kim E, Young SG, Casey PJ. Cloning and characterization of a mammalian prenyl protein-specific protease. J Biol Chem. 1999;274:8379–82.. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 34.Manolaridis I, Kulkarni K, Dodd RB, Ogasawara S, Zhang Z, Bineva G, et al. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature. 2013;504:301–5. doi: 10.1038/nature12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hancock JF, Cadwallader K, Marshall CJ. Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B) EMBO J. 1991;10:641–6. doi: 10.1002/j.1460-2075.1991.tb07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–9. doi: 10.1016/0092-8674(90)90294-O. [DOI] [PubMed] [Google Scholar]
- 37.Silvius JR, Bhagatji P, Leventis R, Terrone D. K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol Biol Cell. 2006;17:192–202. doi: 10.1091/mbc.e05-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A. 2015;112:779–84.. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;121:421–7. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- 40.Shahinian S, Silvius JR. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry. 1995;34:3813–22.. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder H, Leventis R, Rex S, Schelhaas M, Nagele E, Waldmann H, et al. S-Acylation and plasma membrane targeting of the farnesylated carboxyl-terminal peptide of N-ras in mammalian fibroblasts. Biochemistry. 1997;36:13102–9. doi: 10.1021/bi9709497. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–27.. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, et al. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–8. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 44.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–71.. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS) J Biol Chem. 1998;273:15830–7. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 46.Rusch M, Zimmermann TJ, Burger M, Dekker FJ, Gormer K, Triola G, et al. Identification of acyl protein thioesterases 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M. Angew Chem Int Ed Engl. 2011;50:9838–42. doi: 10.1002/anie.201102967. [DOI] [PubMed] [Google Scholar]
- 47.Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, et al. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol Cell. 2011;41:173–85.. doi: 10.1016/j.molcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabaczar S, Czogalla A, Podkalicka J, Biernatowska A, Sikorski AF. Protein palmitoylation: palmitoyltransferases and their specificity. Exp Biol Med. 2017;242:1150–7. doi: 10.1177/1535370217707732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–6. doi: 10.1016/S0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 50.Cho KJ, Casteel DE, Prakash P, Tan L, van der Hoeven D, Salim AA, et al. AMPK and endothelial nitric oxide synthase signaling regulates K-Ras plasma membrane interactions via cyclic GMP-dependent protein kinase 2. Mol Cell Biol. 2016;36:3086–99.. doi: 10.1128/MCB.00365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung PJ, Tsai FD, Vais H, Court H, Yang J, Fehrenbacher N, et al. Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc Natl Acad Sci U S A. 2013;110:20593–8. doi: 10.1073/pnas.1306431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, et al. K-Ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell. 2015;163:1237–51. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 53.Barcelo C, Paco N, Morell M, Alvarez-Moya B, Bota-Rabassedas N, Jaumot M, et al. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Res. 2014;74:1190–9. doi: 10.1158/0008-5472.CAN-13-1750. [DOI] [PubMed] [Google Scholar]
- 54.Yang MH, Nickerson S, Kim ET, Liot C, Laurent G, Spang R, et al. Regulation of RAS oncogenicity by acetylation. Proc Natl Acad Sci U S A. 2012;109:10843–8. doi: 10.1073/pnas.1201487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker R, Wilkerson EM, Sumita K, Isom DG, Sasaki AT, Dohlman HG, et al. Differences in the regulation of K-Ras and H-Ras isoforms by monoubiquitination. J Biol Chem. 2013;288:36856–62.. doi: 10.1074/jbc.C113.525691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang MH, Laurent G, Bause AS, Spang R, German N, Haigis MC, et al. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol Cancer Res. 2013;11:1072–7. doi: 10.1158/1541-7786.MCR-13-0040-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song HY, Biancucci M, Kang HJ, O’Callaghan C, Park SH, Principe DR, et al. SIRT2 deletion enhances KRAS-induced tumorigenesis in vivo by regulating K147 acetylation status. Oncotarget. 2016;7:80336–49.. doi: 10.18632/oncotarget.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dharmaiah S, Tran TH, Messing S, Agamasu C, Gillette WK, Yan W, et al. Structures of N-terminally processed KRAS provide insight into the role of N-acetylation. Sci Rep. 2019;9:10512. doi: 10.1038/s41598-019-46846-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lander HM, Ogiste JS, Teng KK, Novogrodsky A. p21ras as a common signaling target of reactive free radicals and cellular redox stress. J Biol Chem. 1995;270:21195–8. doi: 10.1074/jbc.270.36.21195. [DOI] [PubMed] [Google Scholar]
- 60.Lander HM, Ogiste JS, Pearce SF, Levi R, Novogrodsky A. Nitric oxide-stimulated guanine nucleotide exchange on p21ras. J Biol Chem. 1995;270:7017–20.. doi: 10.1074/jbc.270.13.7017. [DOI] [PubMed] [Google Scholar]
- 61.Lander HM, Hajjar DP, Hempstead BL, Mirza UA, Chait BT, Campbell S, et al. A molecular redox switch on p21(ras). Structural basis for the nitric oxide-p21(ras) interaction. J Biol Chem. 1997;272:4323–6. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- 62.Lee M, Choy JC. Positive feedback regulation of human inducible nitric-oxide synthase expression by Ras protein S-nitrosylation. J Biol Chem. 2013;288:15677–86.. doi: 10.1074/jbc.M113.475319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim K-H, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–9. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mansour M. Ubiquitination: friend and foe in cancer. Int J Biochem Cell Biol. 2018;101:80–93. doi: 10.1016/j.biocel.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 65.Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, et al. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci Signal. 2011;4:ra13. doi: 10.1126/scisignal.2001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin G, Kistler S, George SD, Kuhlmann N, Garvey L, Huynh M, et al. A KRAS GTPase K104Q mutant retains downstream signaling by offsetting defects in regulation. J Biol Chem. 2017;292:4446–56.. doi: 10.1074/jbc.M116.762435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bigenzahn JW, Collu GM, Kartnig F, Pieraks M, Vladimer GI, Heinz LX, et al. LZTR1 is a regulator of RAS ubiquitination and signaling. Science. 2018;362:1171–7. doi: 10.1126/science.aap8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu B, Swatkoski S, Holly A, Lee LC, Giroux V, Lee CS, et al. Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc Natl Acad Sci U S A. 2015;112:1724–33. doi: 10.1073/pnas.1415569112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi BH, Philips MR, Chen Y, Lu L, Dai W. K-Ras Lys-42 is crucial for its signaling, cell migration, and invasion. J Biol Chem. 2018;293:17574–81.. doi: 10.1074/jbc.RA118.003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi BH, Chen C, Philips M, Dai W. RAS GTPases are modified by SUMOylation. Oncotarget. 2018;9:4440–50.. doi: 10.18632/oncotarget.23269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kano Y, Gebregiworgis T, Marshall CB, Radulovich N, Poon BPK, St-Germain J, et al. Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation. Nat Commun. 2019;10:224. doi: 10.1038/s41467-018-08115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med. 2018;24:954–60.. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 73.Barcelo C, Paco N, Beckett AJ, Alvarez-Moya B, Garrido E, Gelabert M, et al. Oncogenic K-ras segregates at spatially distinct plasma membrane signaling platforms according to its phosphorylation status. J Cell Sci. 2013;126:4553–9. doi: 10.1242/jcs.123737. [DOI] [PubMed] [Google Scholar]
- 74.Sumita K, Yoshino H, Sasaki M, Majd N, Kahoud ER, Takahashi H, et al. Degradation of activated K-Ras orthologue via K-Ras-specific lysine residues is required for cytokinesis. J Biol Chem. 2014;289:3950–9. doi: 10.1074/jbc.M113.531178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng T, Wang Q, Fu J, Lin Q, Bi J, Ding W, et al. Impeded Nedd4-1-mediated Ras degradation underlies Ras-driven tumorigenesis. Cell Rep. 2014;7:871–82.. doi: 10.1016/j.celrep.2014.03.045. [DOI] [PubMed] [Google Scholar]
- 76.Baietti MF, Simicek M, Abbasi Asbagh L, Radaelli E, Lievens S, Crowther J, et al. OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol Med. 2016;8:288–303. doi: 10.15252/emmm.201505972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shukla S, Allam US, Ahsan A, Chen G, Krishnamurthy PM, Marsh K, et al. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated beta-TrCP1 degradation. Neoplasia. 2014;16:115–28. doi: 10.1593/neo.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, Tan M, Jia L, Wei D, Zhao Y, Chen G, et al. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J Clin Invest. 2014;124:835–46.. doi: 10.1172/JCI70297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie C-M, Wei D, Zhao L, Marchetto S, Mei L, Borg J-P, et al. Erbin is a novel substrate of the Sag-βTrCP E3 ligase that regulates KrasG12D-induced skin tumorigenesis. J Cell Biol. 2015;209:721–37.. doi: 10.1083/jcb.201411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan M, Xu J, Siddiqui J, Feng F, Sun Y. Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis. Mol Cancer. 2016;15:81. doi: 10.1186/s12943-016-0567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aktories K. Bacterial protein toxins that modify host regulatory GTPases. Nat Rev Microbiol. 2011;9:487–98.. doi: 10.1038/nrmicro2592. [DOI] [PubMed] [Google Scholar]
- 82.Just I, Selzer J, Hofmann F, Green GA, Aktories K. Inactivation of Ras by Clostridium sordellii lethal toxin-catalyzed glucosylation. J Biol Chem. 1996;271:10149–53.. doi: 10.1074/jbc.271.17.10149. [DOI] [PubMed] [Google Scholar]
- 83.Genth H, Just I. Functional implications of lethal toxin-catalysed glucosylation of (H/K/N)Ras and Rac1 in Clostridium sordellii-associated disease. Eur J Cell Biol. 2011;90:959–65.. doi: 10.1016/j.ejcb.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 84.Ganesan AK, Vincent TS, Olson JC, Barbieri JT. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J Biol Chem. 1999;274:21823–9. doi: 10.1074/jbc.274.31.21823. [DOI] [PubMed] [Google Scholar]
- 85.Kato K, Cox AD, Hisaka MM, Graham SM, Buss JE, Der CJ. Isoprenoid addition to Ras protein is the critical modification for its membrane association and transforming activity. Proc Natl Acad Sci U S A. 1992;89:6403–7. doi: 10.1073/pnas.89.14.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Basso AD, Kirschmeier P, Bishop WR. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J Lipid Res. 2006;47:15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- 87.Alcock RA, Dey S, Chendil D, Inayat MS, Mohiuddin M, Hartman G, et al. Farnesyltransferase inhibitor (L-744,832) restores TGF-β type II receptor expression and enhances radiation sensitivity in K-ras mutant pancreatic cancer cell line MIA PaCa-2. Oncogene. 2002;21:7883–90.. doi: 10.1038/sj.onc.1205948. [DOI] [PubMed] [Google Scholar]
- 88.Mendes J, Gonçalves AC, Alves R, Jorge J, Pires A, Ribeiro A, et al. L744,832 and everolimus induce cytotoxic and cytostatic effects in non-Hodgkin lymphoma cells. Pathol Oncol Res. 2016;22:301–9. doi: 10.1007/s12253-015-9998-4. [DOI] [PubMed] [Google Scholar]
- 89.Marin-Ramos NI, Ortega-Gutierrez S, Lopez-Rodriguez ML. Blocking Ras inhibition as an antitumor strategy. Semin Cancer Biol. 2019;54:91–100. doi: 10.1016/j.semcancer.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 90.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31.. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu M, Sjogren AK, Karlsson C, Ibrahim MX, Andersson KM, Olofsson FJ, et al. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc Natl Acad Sci U S A. 2010;107:6471–6. doi: 10.1073/pnas.0908396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lobell RB, Liu D, Buser CA, Davide JP, DePuy E, Hamilton K, et al. Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol Cancer Ther. 2002;1:747–58. [PubMed] [Google Scholar]
- 93.Du W, Prendergast GC. Geranylgeranylated RhoB mediates suppression of human tumor cell growth by farnesyltransferase inhibitors. Cancer Res. 1999;59:5492–6. [PubMed] [Google Scholar]
- 94.Subramani PA, Narala VR, Michael RD, Lomada D, Reddy MC. Molecular docking and simulation of curcumin with geranylgeranyl transferase1 (GGTase1) and farnesyl transferase (FTase) Bioinformation. 2015;11:248–53.. doi: 10.6026/97320630011248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kazi A, Xiang S, Yang H, Chen L, Kennedy P, Ayaz M, et al. Dual farnesyl and geranylgeranyl transferase inhibitor thwarts mutant KRAS-driven patient-derived pancreatic tumors. Clin Cancer Res. 2019;25:5984–96.. doi: 10.1158/1078-0432.CCR-18-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–69.. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 97.Demierre M-F, Higgins PDR, Gruber SB, Hawk E, Lippman SM. Statins and cancer prevention. Nat Rev Cancer. 2005;5:930–42.. doi: 10.1038/nrc1751. [DOI] [PubMed] [Google Scholar]
- 98.Wong WW, Dimitroulakos J, Minden MD, Penn LZ. HMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosis. Leukemia. 2002;16:508–19.. doi: 10.1038/sj.leu.2402476. [DOI] [PubMed] [Google Scholar]
- 99.Yu R, Longo J, van Leeuwen JE, Mullen PJ, Ba-Alawi W, Haibe-Kains B, et al. Statin-induced cancer cell death can be mechanistically uncoupled from prenylation of RAS family proteins. Cancer Res. 2018;78:1347–57.. doi: 10.1158/0008-5472.CAN-17-1231. [DOI] [PubMed] [Google Scholar]
- 100.Senaratne SG, Mansi JL, Colston KW. The bisphosphonate zoledronic acid impairs Ras membrane [correction of impairs membrane] localisation and induces cytochrome c release in breast cancer cells. Br J Cancer. 2002;86:1479–86.. doi: 10.1038/sj.bjc.6600297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gnant M, Mlineritsch B, Schippinger W, Luschin-Ebengreuth G, Pöstlberger S, Menzel C, et al. Endocrine therapy plus zoledronic acid in premenopausal breast cancer. N Engl J Med. 2009;360:679–91.. doi: 10.1056/NEJMoa0806285. [DOI] [PubMed] [Google Scholar]
- 102.Jahnke W, Rondeau J-M, Cotesta S, Marzinzik A, Pellé X, Geiser M, et al. Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery. Nat Chem Biol. 2010;6:660–6. doi: 10.1038/nchembio.421. [DOI] [PubMed] [Google Scholar]
- 103.Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–12.. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 104.Bergo MO, Wahlstrom AM, Fong LG, Young SG. Genetic analyses of the role of RCE1 in RAS membrane association and transformation. Methods Enzymol. 2008;438:367–89. doi: 10.1016/S0076-6879(07)38026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, et al. Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol Cell Biol. 2002;22:171–81.. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bergo MO, Gavino BJ, Hong C, Beigneux AP, McMahon M, Casey PJ, et al. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest. 2004;113:539–50.. doi: 10.1172/JCI200418829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Manandhar SP, Hildebrandt ER, Jacobsen WH, Santangelo GM, Schmidt WK. Chemical inhibition of CaaX protease activity disrupts yeast Ras localization. Yeast. 2010;27:327–43. doi: 10.1002/yea.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mohammed I, Hampton SE, Ashall L, Hildebrandt ER, Kutlik RA, Manandhar SP, et al. 8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells. Bioorg Med Chem. 2016;24:160–78.. doi: 10.1016/j.bmc.2015.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bergo MO, Lieu HD, Gavino BJ, Ambroziak P, Otto JC, Casey PJ, et al. On the physiological importance of endoproteolysis of CAAX proteins: heart-specific RCE1 knockout mice develop a lethal cardiomyopathy. J Biol Chem. 2004;279:4729–36.. doi: 10.1074/jbc.M310081200. [DOI] [PubMed] [Google Scholar]
- 110.Aiyagari AL, Taylor BR, Aurora V, Young SG, Shannon KM. Hematologic effects of inactivating the Ras processing enzyme Rce1. Blood. 2003;101:2250–2. doi: 10.1182/blood-2002-07-2250. [DOI] [PubMed] [Google Scholar]
- 111.Marin-Ramos NI, Balabasquer M, Ortega-Nogales FJ, Torrecillas IR, Gil-Ordonez A, Marcos-Ramiro B, et al. A potent isoprenylcysteine carboxylmethyltransferase (ICMT) inhibitor improves survival in Ras-driven acute myeloid leukemia. J Med Chem. 2019;62:6035–46.. doi: 10.1021/acs.jmedchem.9b00145. [DOI] [PubMed] [Google Scholar]
- 112.Shi YQ, Rando RR. Kinetic mechanism of isoprenylated protein methyltransferase. J Biol Chem. 1992;267:9547–51. doi: 10.1016/S0021-9258(19)50125-9. [DOI] [PubMed] [Google Scholar]
- 113.Diver MM, Pedi L, Koide A, Koide S, Long SB. Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT. Nature. 2018;553:526–9. doi: 10.1038/nature25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wnuk SF, Yuan CS, Borchardt RT, Balzarini J, De Clercq E, Robins MJ. Anticancer and antiviral effects and inactivation of S-adenosyl-L-homocysteine hydrolase with 5ʼ-carboxaldehydes and oximes synthesized from adenosine and sugar-modified analogues. J Med Chem. 1997;40:1608–18.. doi: 10.1021/jm960828p. [DOI] [PubMed] [Google Scholar]
- 115.Winter-Vann AM, Kamen BA, Bergo MO, Young SG, Melnyk S, James SJ, et al. Targeting Ras signaling through inhibition of carboxyl methylation: an unexpected property of methotrexate. Proc Natl Acad Sci U S A. 2003;100:6529–34.. doi: 10.1073/pnas.1135239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bergman JA, Hahne K, Song J, Hrycyna CA, Gibbs RA. S-Farnesyl-thiopropionic acid triazoles as potent inhibitors of isoprenylcysteine carboxyl methyltransferase. ACS Med Chem Lett. 2012;3:15–9. doi: 10.1021/ml200106d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang M, Tan W, Zhou J, Leow J, Go M, Lee HS, et al. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem. 2008;283:18678–84.. doi: 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- 118.Winter-Vann AM, Baron RA, Wong W, dela Cruz J, York JD, Gooden DM, et al. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc Natl Acad Sci U S A. 2005;102:4336–41.. doi: 10.1073/pnas.0408107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lau HY, Ramanujulu PM, Guo D, Yang T, Wirawan M, Casey PJ, et al. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol Ther. 2014;15:1280–91.. doi: 10.4161/cbt.29692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–50.. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 121.Lin DTS, Davis NG, Conibear E. Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochem Soc Trans. 2017;45:913–21.. doi: 10.1042/BST20160303. [DOI] [PubMed] [Google Scholar]
- 122.Amendola CR, Mahaffey JP, Parker SJ, Ahearn IM, Chen W-C, Zhou M, et al. KRAS4A directly regulates hexokinase 1. Nature. 2019;576:482–6. doi: 10.1038/s41586-019-1832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu CM, Tien SC, Hsieh PK, Jeng YM, Chang MC, Chang YT, et al. High glucose triggers nucleotide imbalance through O-GlcNAcylation of key enzymes and induces KRAS mutation in pancreatic cells. Cell Metab. 2019;29:1334–49.e10. doi: 10.1016/j.cmet.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 125.Taparra K, Wang H, Malek R, Lafargue A, Barbhuiya MA, Wang X, et al. O-GlcNAcylation is required for mutant KRAS-induced lung tumorigenesis. J Clin Invest. 2018;128:4924–37.. doi: 10.1172/JCI94844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nandi D, Tahiliani P, Kumar A, Chandu D. The ubiquitin-proteasome system. J Biosci. 2006;31:137–55.. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 127.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–54.. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Senft D, Qi J. Ronai ZeA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jeong WJ, Park JC, Kim WS, Ro EJ, Jeon SH, Lee SK, et al. WDR76 is a RAS binding protein that functions as a tumor suppressor via RAS degradation. Nat Commun. 2019;10:295. doi: 10.1038/s41467-018-08230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ma Y, Gu Y, Zhang Q, Han Y, Yu S, Lu Z, et al. Targeted degradation of KRAS by an engineered ubiquitin ligase suppresses pancreatic cancer cell growth in vitro and in vivo. Mol Cancer Ther. 2013;12:286–94.. doi: 10.1158/1535-7163.MCT-12-0650. [DOI] [PubMed] [Google Scholar]
- 131.Pan T, Zhang Y, Zhou N, He X, Chen C, Liang L, et al. A recombinant chimeric protein specifically induces mutant KRAS degradation and potently inhibits pancreatic tumor growth. Oncotarget. 2016;7:44299–309.. doi: 10.18632/oncotarget.9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-Xl on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–93.. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 133.Becher I, Savitski MM, Savitski MF, Hopf C, Bantscheff M, Drewes G. Affinity profiling of the cellular kinome for the nucleotide cofactors ATP, ADP, and GTP. ACS Chem Biol. 2013;8:599–607. doi: 10.1021/cb3005879. [DOI] [PubMed] [Google Scholar]
- 134.Hillig RC, Sautier B, Schroeder J, Moosmayer D, Hilpmann A, Stegmann CM, et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS–SOS1 interaction. Proc Natl Acad Sci U S A. 2019;116:2551–60.. doi: 10.1073/pnas.1812963116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A. 2012;109:5299–304.. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gehringer M, Laufer SA. Emerging and re-emerging warheads for targeted covalent inhibitors: applications in medicinal chemistry and chemical biology. J Med Chem. 2019;62:5673–724.. doi: 10.1021/acs.jmedchem.8b01153. [DOI] [PubMed] [Google Scholar]
- 137.Fakih M, O’Neil B, Price TJ, Falchook GS, Desai J, Kuo J, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors (Abstract) J Clin Oncol. 2019;37:3003. doi: 10.1200/JCO.2019.37.15_suppl.3003. [DOI] [Google Scholar]
- 138.Papadopoulos KP, Ou S-HI, Johnson ML, Christensen J, Velastegui K, Potvin D, et al. A phase I/II multiple expansion cohort trial of MRTX849 in patients with advanced solid tumors with KRAS G12C mutation. J Clin Oncol. 2019;37:TPS3161–TPS61. doi: 10.1200/JCO.2019.37.15_suppl.TPS3161. [DOI] [Google Scholar]
- 139.Janes MR, Zhang J, Li L-S, Hansen R, Peters U, Guo X, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172:578–89.e17. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 140.Bar-Sagi D, Knelson EH, Sequist LV. A bright future for KRAS inhibitors. Nat Cancer. 2020;1:25–7. doi: 10.1038/s43018-019-0016-8. [DOI] [PubMed] [Google Scholar]
- 141.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pr. 2009;205:858–62.. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 142.Scheffler M, Ihle MA, Hein R, Merkelbach-Bruse S, Scheel AH, Siemanowski J, et al. K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J Thorac Oncol. 2019;14:606–16.. doi: 10.1016/j.jtho.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 143.Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRAS(G12C) inhibition. Clin Cancer Res. 2020;26:1633–43. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]