Significance
Oligodendrocyte precursor cells (OPCs) retain the capacity to remyelinate axons upon demyelinating injury. However, mode of cell division and differentiation dynamics of individual OPCs in deep brain structures, such as the corpus callosum, remains unknown. Using in vivo two-photon imaging in a focal model of demyelination, we show that OPCs undergo several rounds of symmetric and asymmetric cell divisions before producing a subset of daughter cells that differentiates into myelinating oligodendrocytes. The data presented here characterize the behavior of OPC clones and delineate the cellular principles that lead to remyelination.
Keywords: remyelination, OPC, corpus callosum, intravital imaging, oligondendrocyte
Abstract
Oligodendrocyte precursor cells (OPCs) retain the capacity to remyelinate axons in the corpus callosum (CC) upon demyelination. However, the dynamics of OPC activation, mode of cell division, migration, and differentiation on a single-cell level remain poorly understood due to the lack of longitudinal observations of individual cells within the injured brain. After inducing focal demyelination with lysophosphatidylcholin in the CC of adult mice, we used two-photon microscopy to follow for up to 2 mo OPCs and their differentiating progeny, genetically labeled through conditional recombination driven by the regulatory elements of the gene Achaete-scute homolog 1. OPCs underwent several rounds of symmetric and asymmetric cell divisions, producing a subset of daughter cells that differentiates into myelinating oligodendrocytes. While OPCs continue to proliferate, differentiation into myelinating oligodendrocytes declines with time, and death of OPC-derived daughter cells increases. Thus, chronic in vivo imaging delineates the cellular principles leading to remyelination in the adult brain, providing a framework for the development of strategies to enhance endogenous brain repair in acute and chronic demyelinating disease.
In the central nervous system (CNS), oligodendrocytes form myelin sheaths around axons, allowing for the rapid transduction of electrical impulses and providing metabolic support (1, 2). Myelin remodeling occurs throughout life, is regulated by neuronal activity, and has been associated with learning and circuit plasticity in mice and humans (3–10). However, a number of neurological diseases, multiple sclerosis (MS) among others, leads to impaired oligodendrocyte function and an eventual loss of myelin sheaths, causing demyelination (1, 11). Demyelination is associated with progressive axonal degeneration and neurological decline (2). Whereas substantial progress has been made over the last decade to reduce myelin loss in acute phases of demyelinating diseases such as MS, regenerative approaches to enhance remyelination remain scant (1, 12, 13). In rodents, remyelination is achieved via activation of oligodendrocyte precursor cells (OPCs) that are capable of generating new functional oligodendrocytes (1, 14–17). Snapshot-based genetic lineage tracing experiments showed that OPCs expand within areas of damage through a combination of migration and proliferation in response to demyelinating injury (1, 18). Recruited OPCs differentiate into oligodendrocytes and myelin sheaths are eventually restored within lesions. Remyelination also occurs in humans in the context of demyelinating diseases, in which differentiated oligodendrocytes possibly also contribute to myelin repair (4, 19, 20). However, myelin recovery in humans often remains incomplete. Thus, new therapeutic approaches to enhance myelin repair are needed (1).
Understanding OPC dynamics on a single-cell level could facilitate the development of remyelination-promoting strategies with the aim to prevent disease progression or to ameliorate symptoms. Recently, OPC kinetics, oligodendrocyte differentiation, and myelin formation in the healthy and injured adult mouse brain have begun to be studied in superficial layers of the cortex using in vivo two-photon imaging (7, 9, 21–23). However, myelination and remyelination in gray and white matter follows different dynamics (24). The clonal behavior of OPCs during remyelination in gray and white matter structures remains largely unknown (1). For example, the mode of cell division, self-renewal potential, and successive rounds of division of individual OPCs is unclear. We here used in vivo two-photon imaging to analyze OPC behavior and subsequent remyelination in the corpus callosum (CC), a white matter area connecting the two brain hemispheres that is commonly affected in demyelinating disease (11). Combining lysophosphatidylcholin (LPC, also called lysolecithin)–induced demyelination and repeated in vivo imaging (25, 26), we characterize the cellular principles of OPC behavior upon demyelination, allowing for insights that may guide future strategies to enhance regenerative brain repair.
Results
Chronic Imaging of OPCs and Daughter Cells in the Adult Brain.
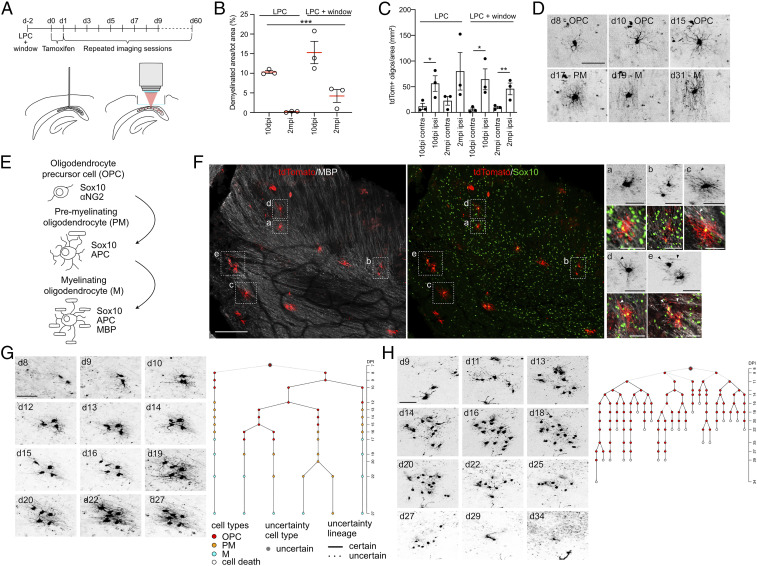
To study the behavior of OPC clones during remyelination in the CC, we genetically targeted OPCs using mice expressing a tamoxifen-inducible Cre recombinase under the control of the endogenous Achaete-scute homolog 1 (Ascl1) promoter crossed with a tdTomato reporter mouse line (Ascl1-tdTomato mice) (15, 27). To induce focal demyelination in the CC, mice were injected with LPC in the posterior CC, an approach commonly used to study remyelination (26). Mice were implanted with a transcortical window allowing for optical access using two-photon microscopy (Fig. 1A). To avoid dense labeling of astroglial cells that up-regulate Ascl1 immediately after lesion (28, 29), tamoxifen was injected 2 and 3 d after LPC injection to induce sparse labeling of Ascl1-targeted OPCs (Fig. 1A and SI Appendix, Fig. S1 A–E). Given that the posterior parts of the CC were imaged, virtually no tdTomato+ neuronal cells were detected under the window area (SI Appendix, Fig. S1 F and G). Labeled OPC clones were imaged every day up to 2 mo (Fig. 1A). Implantation of the window and the imaging protocol did not alter the extent of demyelination and remyelination, as measured using the initial extent and subsequent recovery of myelin basic protein (MBP)–labeled areas in experimental mice and controls (Fig. 1B and SI Appendix, Fig. S1 H–K). Inflammation was induced at 10 d after surgery in both LPC and LPC plus window conditions, as revealed by the microglia marker IBA1, but strongly decreased with time, as shown previously (SI Appendix, Fig. S2 A and B) (30). As expected, tdTomato+ cells were recruited to areas of demyelination early (10 d) after LPC with mature, tdTomato+ oligodendrocytes being present in remyelinated regions 2 mo after LPC (Fig. 1C and SI Appendix, Fig. S1 H–K).
Fig. 1.
Chronic imaging of OPCs and their progeny in the CC. (A) Experimental setup for demyelinating lesion induction and chronic in vivo imaging of Ascl1-tdTomato OPCs over up to 60 d. (B) Percentage of demyelinated area over the total area in LPC-treated control mice and LPC+window–imaged mice at 10 dpi and 2 mpi. Each circle represents a mouse. (C) Quantification of tdTomato+ cells belonging to oligodendrocytes lineage in LPC-treated control mice and LPC+window–imaged mice at different time points. Each dot represents a mouse. Note that implantation of a transcortical window does not affect response to LPC. (D) Representative example (collapsed z-stacks) of an imaged OPC going throughout the entire process of maturation until remyelinating oligodendrocyte. The morphological classification according to the schematic in E is reported. (E) Schematic showing the different stages of OPC differentiation to mature remyelinating oligodendrocyte and stage specific markers. (F) Representative confocal image of a horizontal section of an LPC-injected CC after 10 d of in vivo imaging (tdTomato/red; MBP/white; Sox10/green). (Right) Magnification of representative examples (collapsed z-stacks) of in vivo imaged cells showing different stages of differentiation that were confirmed by post hoc immunohistochemical analysis (tdTomato+, Sox10+, MBP+). “a” and “b”: OPC; “c,” “d,” and right cell in “e”: PM; left cell in “e”: M. The morphological classification is made according to the schematic in E. Arrowheads indicate MBP+ myelin sheaths. (G) Example pictures (collapsed z-stacks) of a representative OPC and its progeny imaged over the course of 27 d exhibiting survival and differentiation to remyelinating oligodendrocytes. (Right) Lineage tree deduced from tracking the OPC in the example pictures. (H) Example pictures (collapsed z-stacks) of a representative OPC and its progeny imaged over the course of 34 d showing the failure of that clone to generate surviving progeny. (Right) Lineage tree deduced from tracking the OPC in the example pictures. Values are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 were considered significant. (Scale bars, 50 μm [D, 2-photon F, staining F magnification b, g, and h], 100 μm [staining F], and 25 μm [staining F magnification a, c, d, and e].) d, days after Tamoxifen injection.
Using intravital imaging, Ascl1-targeted cells in individual clones were classified based on morphological criteria, such as cell body size and number/shape of processes, as OPCs, premyelinating oligodendrocytes (PM), and myelinating oligodendrocytes (M) (Fig. 1 D and E and SI Appendix, Table S1). Classification criteria applied to imaging data were validated using post hoc immunohistochemistry after the last imaging session using stage-specific markers of the oligodendrocytic lineage (Sox10+ MBP− for OPCs; Sox10+ MBP+ only in one or few processes for PM; Sox10+ MBP+ for M) (Fig. 1F and SI Appendix, Fig. S2C). The proportions of both tdTomato+ OPCs and more differentiated cells quantified using post hoc immunohistochemistry for SOX10/MBP, classification based on morphology using in vivo imaging, and expression of NG2/APC resulted comparable among the three experimental conditions (SI Appendix, Fig. S2D). However, morphological criteria did not allow for separating molecularly distinct OPCs, committed oligodendrocyte cells (COPs), and, in some cases, newly formed oligodendrocytes (NFOLs) that are referred to as OPCs hereafter (SI Appendix, Table S1) (6, 31, 32). A code was assigned to each cell and lineage trees were generated (Fig. 1 G and H and SI Appendix, Fig. S3 and Movies S1 and S2). We analyzed a total of 177 clones, with observation periods between 27 and 64 d (SI Appendix, Fig. S3).
Cell Division Characteristics of OPCs upon Focal Demyelination.
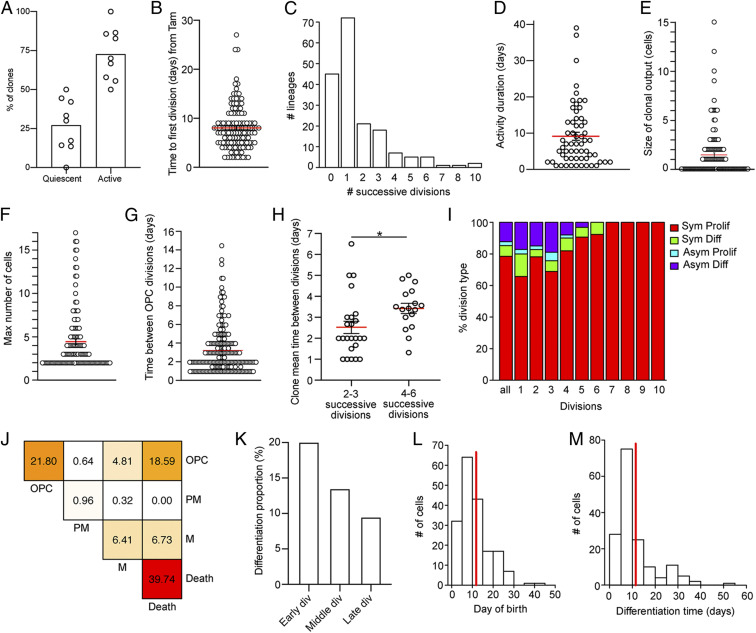
The majority of targeted OPCs entered the cell cycle and generated daughter cells (Fig. 2A and SI Appendix, Fig. S4A), with most cells being activated within 2 d after their first observation, on average 8 d after tamoxifen administration (Fig. 2B and SI Appendix, Fig. S4B). On average, OPCs went through 1.62 rounds of cell division before differentiating or dying. However, subsets of clones were able to show 10 consecutive cell divisions (Fig. 2C). Activity duration of clones (i.e., time from first to last division in the clone) was on average 9.13 d (Fig. 2D). The observed clonal output of OPCs was highly variable with single cells generating as many as 15 cells or no surviving daughter cell (Fig. 2E). On average, 4.36 cells were produced by active OPCs, with only a subset (average of 1.55 cells per clone) showing survival (Fig. 2 E and F).
Fig. 2.
In vivo imaging reveals dynamics of OPC activation and differentiation. (A) Percentage of clones not dividing during the entire imaging time (quiescent) and clones that divided at least once (active). Each circle represents a mouse (n = 177 clones from nine mice). (B) Time until the first division (in days) of the first OPC of the clone calculated from Tamoxifen injection. Each circle represents a clone (mean ± SEM: 8.01 ± 0.39; n = 131 cells). (C) Bar plot showing the maximum number of successive divisions in recorded lineages (mean ± SEM: 1.62 ± 0.14; n = 177 clones). (D) Duration (days) of the proliferating activity measured as the time between the first and last division in each clone. Each circle represents an active clone. Clones with activity duration = 0 (1 division) are excluded (mean ± SEM: 9.13 ± 1.11; n = 60 clones). (E) Final number of cells per active clone. Each circle represents a clone. Quiescent clones excluded (mean ± SEM: 1.55 ± 0.22; n = 132 clones). (F) Maximum number of cells per active clone. Each circle represents a clone. Quiescent clones are excluded (mean ± SEM: 4.36 ± 0.3; n = 132 clones). (G) Average time (in days) between OPC divisions. Time until first division is not considered. Each circle represents a division with certain lineage transitions (mean ± SEM: 3.22 ± 0.17; n = 221 divisions). (H) Mean division time (days) per clone. Each circle represents a clone. Clones are grouped in low dividing and high dividing. (I) Percentage of different division types considering all divisions together and single consecutive divisions. Clones with OPCs at last imaging time point that were born less than 10 d before are excluded (Sym Prolif generate OPC + OPC; Sym Diff generate M + M; Asym Prolif generate OPC + PM; and Asym Diff generate OPC + M. All n = 311 divisions [div]; 1 n = 35 div; 2 n = 87 div; 3 n = 74 div; 4 n = 50 div; 5 n = 32 div; 6 n = 13 div; 7 n = 10 div; 8 n = 5 div; 9 n = 4 div; and 10 n = 1 div). (J) Heat map representing the frequencies of modes of division of OPCs considering all divisions together. If a cell dies, its fate is reported with “Death” independently of its phenotypical stage at last time point it was observed. Clones with OPCs at last imaging time point that were born less than 10 d before are excluded (n = 312 divisions). (K) Differentiation proportion binning cells according to their birth division in the clone. It is calculated as the percentage of differentiating cell born at the considered bin over the total amount of cell born at that bin. Clones with OPCs at last imaging time point that were born less than 10 d before are excluded (Early div: 0 to 2. Cells depicted in “Division 0” correspond to the first single cells of the tree found during imaging sessions; Middle div: 3 to 4; Late div: 5 to 7. n = 993 cells). (L) Day of birth of differentiating cells (mean = 11.8; n = 182 cells). (M) Differentiation time (days) of differentiating cells calculated from birth date (mean = 11.5; n = 161 cells). Values are shown as mean ± SEM. *P < 0.05.
A fraction of OPCs showed rapid cell divisions whereas other cells returned to relative quiescence after initial divisions, with interdivision intervals lasting up to 14.5 d (Fig. 2G). The time interval between cell divisions was not different between early and late divisions (SI Appendix, Fig. S4C). However, OPC clones with low number of cell divisions divided significantly faster than clones with large clonal expansion, indicating a behavioral difference between OPCs generating a large or small number of daughter cells (Fig. 2H and SI Appendix, Fig. S4D). Active OPCs were capable to divide symmetrically (generating two progenitors or differentiating cells) or asymmetrically (generating one progenitor and one differentiating daughter) (Fig. 2I). Overall, a small percentage of divisions generated differentiating cells (Fig. 2 I and J), and only 16.46% of all cells that were generated differentiated to mature oligodendrocytes. Interestingly, differentiation proportion decreased for cells born at late divisions (Fig. 2 I and K and SI Appendix, Fig. S4 E and F). On average, differentiating OPCs were born at 11.8 d after induction (Fig. 2L) and took 11.5 d to reach the mature remyelinating stage (Fig. 2M).
Cell Death of OPC-Derived Cells in the Injured CC.
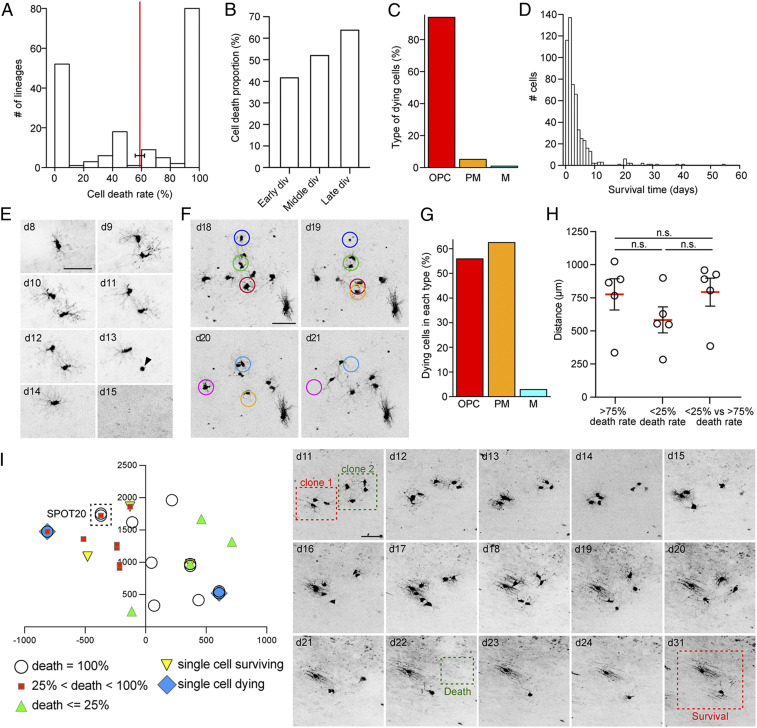
Given the ability to follow single clones over time, intravital imaging allows for analyses of OPC-derived cells that become eliminated. The average cell death rate within clonal lineages was 58.9%. Cell death rate was highly variable among clones: the majority of the clones showed either a cell death rate below 25% or a complete loss of all OPC-generated cells (Fig. 3A). Overall, 47.24% of all cells produced died. Cell death occurred throughout the imaging period (SI Appendix, Fig. S5A), but there was an increase of relative death rate for cells born at late divisions within clones (Fig. 3B and SI Appendix, Fig. S5B). Phenotypically, the majority of dying cells were OPCs (including COPs and NFOLs; SI Appendix, Table S1), and cell death occurred early after last mitosis with an average of 3.88 d (Fig. 3 C and F). The probability of dying was comparable between OPCs and PMs; however, when cells differentiated to remyelinating oligodendrocytes, chances of cell death were low (Fig. 3G and SI Appendix, Fig. S5C). No significant differences were found between distances among clones with similar or opposite cell death rates (Fig. 3 H and I). Furthermore, clones with diverse outcomes were found in the same imaging field of view (SPOT) or in closely neighboring SPOTs, suggesting that distinct cell death rates of clones cannot be explained solely by the position of clones within specific areas of the LPC-induced lesion (Fig. 3I and SI Appendix, Fig. S5 D and E). Furthermore, loss of cells due to migration out of the field of view seems highly unlikely given the size of imaged areas and migratory speed of OPCs (Fig. 3F).
Fig. 3.
Dynamics of cell death of single OPC clones. (A) Frequency of cell death in all chronically imaged lineages. The cell death rate per lineage is calculated as the percentage of cell death among cells after last division (mean ± SEM = 58.85 ± 3.25; n = 177 clones). (B) Cell death proportion according to their birth division within the clone. It is calculated as the percentage of dying cell born at the considered bin over the total amount of cell born at that bin. Clones with OPCs at last imaging time point that were born less than 10 d before are excluded (Early div: 0 to 2. Cells depicted in “Division 0” correspond to the first single cells of the tree found during imaging sessions; Middle div: 3 to 5; Late div: 6 to 10. n = 1,033 cells). (C) Percentage of OPCs, PMs, and mature oligodendrocytes in the dying cells group. Only certain cell types are considered (n = 530 cells). (D) Time point (days) of cell death after last cell division (n = 533 cells). (E) Example pictures (collapsed z-stacks) of a representative OPC and its progeny imaged over the course of 15 d. Both daughter cells die. The arrowhead depicts the apoptotic body of one of the OPC before its removal. (F) Example pictures (collapsed z-stacks) of representative dying cells. The measure of the radius of the circles surrounding the cells corresponds to the observed mean maximum speed of migration of dying OPCs per day. (G) Percentage of dying cells in each cell type group. Clones with OPCs at last imaging time point that were born less than 10 d before are excluded (n = 1,033 cells). (H) Distance (micrometers) among clones whose cell death rate is higher than 75%, among clones whose cell death rate is lower than 25%, or between clones whose cell death rate is lower than 25% and those whose cell death rate is higher than 75%. Each circle represents a mouse. Note that the lack of significant differences between groups means that clones are homogeneously distributed below the window. (I) Representative example of the position of in vivo imaged clones below the transcortical window. (Right) Example pictures (collapsed z-stacks) of clones in the same SPOT (SPOT20) that show different outcomes (i.e., survival versus cell death). Values are shown as mean ± SEM. n.s., not significant. (Scale bars, 50 μm [E, F, and I].)
Migratory Behavior of OPC-Derived Cells in the CC.
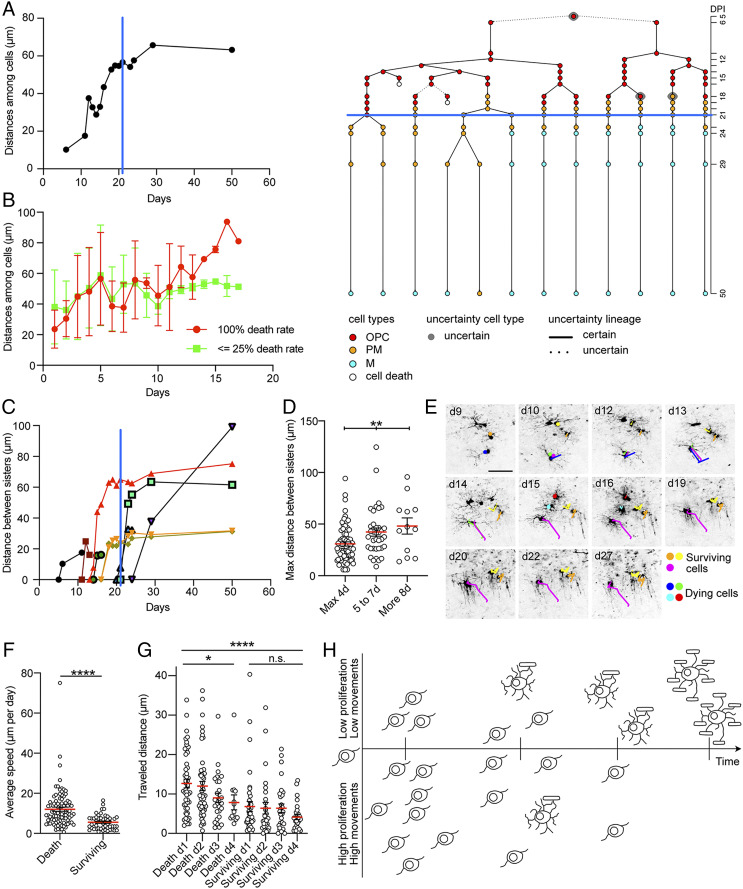
To study the spatial rearrangement of cells over time, changes in distances among cells within clones were measured (Materials and Methods). Cells increased the relative distance among them until their morphologies further differentiated: when OPC-derived cells started to differentiate, clones became more stable, and spatial rearrangements were strongly reduced (Fig. 4A and SI Appendix, Fig. S6A). Interestingly, the reorganization of cells over time was comparable in clones showing high or low cell death rate (Fig. 4B and SI Appendix, Fig. S6B). In vivo imaging allowed for studying spatial relationships among sister cells. Again, we found that sister cells pulled away from each other as soon as they were generated after division. The distance between sister cells showed a gradual increase over time and stabilized once cells differentiated into mature oligodendrocytes (Fig. 4C). Sister OPC pairs were grouped according to the time they could be followed before they differentiated, died, or divided again. The maximum distance reached between sister OPCs was significantly higher in pairs tracked for longer time periods (i.e., differentiating slower) compared to OPC pairs that quickly differentiated (i.e., tracked for shorter periods; Fig. 4D). Continuous imaging also enabled the quantification of individual cell migration over time. To probe for a possible relationship between cell fate and migratory behavior, we tracked movements of cells at the OPC stage (Fig. 4E and Movie S3). Cells that died showed a higher average and maximum speed (measured as micrometers moved per day) when compared to surviving cells (Fig. 4F and SI Appendix, Fig. S6 C–E). Distances traveled were different at each single day after birth and at each single day before death or differentiation between dying and surviving cells (Fig. 4G and SI Appendix, Fig. S6F). Migratory speed was not different within surviving cells across days after birth and before differentiation. Instead, dying cells moved faster in the first 2 d after birth compared to following days (Fig. 4G and SI Appendix, Fig. S6F). These findings were confirmed when progenitors with different fate belonging to the same clone were compared (SI Appendix, Fig. S6G).
Fig. 4.
Migratory behavior of OPC-derived clones in the CC. (A) Median distance (micrometers) among cells over time (days) in the clone represented in the lineage tree on the right. The blue lines in the graph and on the lineage tree depict the day cells start to differentiate. The rearrangement of the cells in the clone slows down when they differentiate. (B) Represented are mean and SD at each observed time point of the median distance (micrometers) among cells in clones with 100% death rate (red) and clones whose death rate is lower that 25% (green) over time (days). Clones are tracked until at least one OPC is present in the clone. The overlap of the two groups indicates that the cells of the clones of the two groups share the same tendency of increasing the distances among them over time (100% death rate n = 15 clones; <25% death rate n = 8 clones). (C) Distance (micrometers) between sister cells in the lineage represented in A over time (days). The blue line depicts the day cells start to differentiate. (D) Maximum distance (micrometers) reached between sister cells. Sister pairs are binned according to the number of days they have been tracked. Sister pairs are tracked until at least one of them is at OPC stage. Each circle represents a pair of sisters. (E) Example pictures (collapsed z-stacks) of a representative OPC clone imaged over the course of 27 d. The colored lines highlight the cumulative distance traveled by single cells over time. Yellow, orange, and magenta depict surviving cells. Blue, green, cyan, and red depict dying cells. (F) Average speed (micrometers per day) of dying and surviving cells calculated tracking the actual migration of single OPCs over time (minimum of 3 d tracking per cell). Each circle represents a single cell. (G) Traveled distance (micrometers) at each day after birth in dying and surviving cells. Each circle represents a single cell. (H) Schematic showing the differential behavior of surviving versus dying OPCs: surviving cells are born at earlier divisions of the clones and migrate only short distances. In contrast, dying cells are produced at later stages of clonal expansion and exhibit a higher propensity to move in the surrounding environment. The values are shown as mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001 were considered significant. n.s., not significant. (Scale bars, 50 μm [E].)
Discussion
We here used LPC-induced focal demyelination and in vivo imaging of genetically targeted OPCs and their progeny to describe the cellular dynamics of OPC-mediated remyelination in the CC. Longitudinal observation of OPCs and their progeny for up to 2 mo reveals functional inter- and intraclonal heterogeneity and provides insights into the cell division capacity and the migration/differentiation dynamics of OPCs and their daughter cells in vivo (Fig. 4H).
LPC-induced demyelination has been extensively used to study remyelination in the CC and revealed the potential for callosal OPCs to successfully remyelinate (26). LPC-mediated demyelination avoids confounding factors (e.g., a sustained immune response that is present in inflammatory models of MS) such as experimental autoimmune encephalitis. In LPC-induced lesions, remyelination is completed within weeks in young animals providing an excellent model to dissect the progressive steps that lead to myelin repair (26). Previous studies characterized remyelination analyzing activated OPCs and remyelinating oligodendrocytes on a population level, providing evidence of OPC heterogeneity in the rodent and human brain (33–35). Thus, analyzing clonal behavior may be helpful to understand the biology of remyelination on a single cell level. Genetic approaches such as MADM and barcoding would allow for the reconstruction of individual lineages (36); however, static clonal lineage tracing inherently suffers from relatively poor temporal resolution. Intravital imaging can provide insights, as recently pioneered in the context of remyelination in the zebrafish spinal cord (37). By using sparse genetic labeling combined with in vivo imaging, we here follow single clones over time in the mammalian CC during the entire process of remyelination from initial OPC recruitment to remyelinating oligodendrocytes.
Our data show that the majority of the clones remained quiescent or divided only few times. However, some OPCs were highly proliferative, with the ability to divide up to 10 consecutive times and produce large clones of more than 15 daughter cells. Notably, our data show that OPCs show distinct modes of cell division: from symmetric proliferative to symmetric differentiating and also asymmetric cell division, in which the OPC is self renewed while the other daughter cell differentiates. Interestingly, large clones showed longer times between consecutive divisions compared to low proliferating clones. Only 16.46% of OPC-derived cells differentiated into mature, remyelinating oligodendrocytes, with OPCs born at early divisions showing a higher probability to survive and to terminally differentiate. Furthermore, cell death was associated with distinct cell division histories of different clones, with higher probability of death when generated at later divisions. Thus, we could characterize behavioral heterogeneity of OPC clones in terms of proliferation and differentiation. Heterogeneity of OPC behavior has been previously associated with their developmental origin: dorsally derived OPCs dominate remyelination of the CC compared to ventrally derived cells despite a strong proliferation response of the latter ones (38). Furthermore, besides labeling of resident OPCs in the CC, the Ascl1 promoter is also active in oligodendrocyte-producing stem cells in the adult subventricular zone (SVZ) (15). Indeed, SVZ-derived cells have been shown to contribute to remyelination in the CC, while showing reduced remyelination potential compared to parenchymal OPCs (16, 17, 39). Based on the genetic labeling strategy used here, it is not possible to determine whether the OPCs that were followed through intravital imaging represent locally activated OPCs, whether they were recruited from the SVZ, or both. Future experiments using, for example, more restricted genetic labeling approaches or strategies allowing for spatially precise labeling (e.g., using virus-based approaches) will aim to provide a more detailed understanding of the origin and potential clonal differences between local and SVZ-derived OPCs in the CC. Furthermore, region-specific differences in the degree of LPC-induced demyelination and LPC-triggered inflammation may influence OPC behavior such as rounds of proliferation, differentiation, and also cell death. To not only follow individual OPCs but also the process of de- and remyelination in the adult CC it will be essential to combine monitoring of demyelination (using for example fluorescent measures of MBP expression).
Interestingly, we observed a high percentage of cell death of newly generated cells of the oligodendroglial linages, similar to previous observations during brain development and in the uninjured adult gray matter (7, 40–42). However, in contrast to healthy adult gray matter or early development we observed a higher percentage of dying OPCs in the remyelinating white matter, with the majority of cells dying very rapidly after the last cell division. Given the fast elimination of newly formed cells, it seems unlikely that they had reached a differentiated stage before death, as proposed during development or in intact gray matter (7, 40–42). The observed difference at which developmental stage cells dies may be due to the diverse and nonpermissive demyelinated environment. In addition, differentiation kinetics may be distinct between developmental tissues and lesioned adult white matter, leading to a more protracted differentiation (37). The classification based on morphology of cells using two-photon imaging as done here may be also partially imprecise, as cells we referred to here as OPCs will also include COPs and NFOLs (6, 31, 32). Thus, additional studies using intravital imaging of demyelination focusing on early cellular events in the gray matter and white matter will be needed to conclusively compare remyelination response between white and gray matter (23).
Interestingly, previous studies used in vivo imaging to show that adult OPCs in the cortex exhibit self-repulsion (i.e., mutually restricting their movement and growth) under physiological conditions (21). We observed this phenomenon also in the injured CC as cells belonging to the same clone pulled away from each other to expand the distance among them over time. In line with previous observations (7), migratory behavior was restricted to progenitors: differentiated progeny stopped migration and stabilized their position when reaching a PM state. Surprisingly, successfully differentiating progenitors moved shorter distances per day compared to dying cells. Given that surviving progenitors showed lower migration compared to dying cells very early on, it seems unlikely that reduced migration is due to early axonal contacts and differentiation. Different migratory behavior was also evident when comparing cells within clones that showed different fates (differentiation versus cell death), indicating that cell movements may not only be influenced by the environment (as sister cells may share similar environmental conditions) but may also depend on cell intrinsic properties.
In the future, in vivo imaging experiments will need to delineate the critical steps of OPC activation and subsequent differentiation when remyelination is impaired, in aged animals in which myelin repair becomes less efficient for example (43–45). Furthermore, novel approaches minimizing required removal of overlying cortex, with the use of three-photon microscopy, for example, may allow to reach the CC from the surface of the brain, excluding that window implantation may affect cellular behavior.
The data presented here characterize the in vivo behavior of OPC clones in response to focal demyelination and provide the entry point to address the challenging questions concerning the failure of remyelination in aged rodents and, ultimately, in demyelinating disease.
Materials and Methods
Transgenic Animals, Demyelination Procedure, and Chronic Transcortical Window Implantation.
The surgical intervention for demyelinating an area of the CC and placing the transcortical window was carried out in Ascl1CreERT2/tdTom mice aged 9 to 12 wk (25). Demyelination was locally induced as described in previous studies (46). A stereotaxic injection of 2 μL of a solution of 1% lysolecithin (LPC; Sigma) in 0.9% NaCl was performed in deeply anesthetized mice. The skin was opened, and after drilling a hole in the skull, LPC was injected unilaterally into the CC at −1.3 mm dorso/ventral from brain surface and −2.5 mm anterior/posterior, and −0.9 mm lateral from Bregma. Mice were kept under anesthesia, and 1 h and 30 min after LPC injection, the procedure for the implantation of the transcortical window was started. The surgery was carried out as explained in previous studies (25). Briefly, above the demyelinated CC (−2.5 anterior/posterior/–1.5 mediolateral mm from Bregma), the cranial bone is locally removed (circle 3 mm Ø). A cylinder (3 mm Ø) of cortical tissue that has been punched out using biopsy punches (Miltex) until a defined depth (1.5 mm deep from skull, level of the CC) was aspirated using a blunt needle (22 g) connected to an air pump until white matter tracks were visible. The area was cleaned, and the window (3-mm Ø stainless steel cannula, 1.5-mm height, and covered by a 3-mm Ø glass coverslip, Warner Instruments) was inserted, secured in place using the stereotactic arm, and stably fixed to the cranial bone with ultraviolet (UV)-curable dental cement (Ivoclar Vivadent). Animal experiments were approved by the Cantonal Commission for Animal Experimentation of the Canton of Zurich, Switzerland, in accordance with national and cantonal regulations.
In Vivo Imaging of OPCs.
Sparse labeling of OPCs was achieved by intraperitoneal (i.p.) injection of Tamoxifen (Tam; 180 mg/kg bodyweight; Sigma) at 2 to 3 d after surgery. Imaging fields of view containing identified OPCs were selected to obtain 10 to 20 SPOTs per mouse. Chronic two-photon imaging was performed starting 3 d after surgery (for details please refer to the SI Appendix, Supplementary Materials and Methods) (25). All SPOTs were checked on a daily basis, and no z-stack was acquired if no changes occurred. The time spent under the microscope for the mouse was kept minimal (<1 h/d).
Image Processing and Lineage Analysis.
Every lineage was annotated with a code (SI Appendix, Supplementary Materials and Methods) by a blinded researcher. Assembling of lineage trees was performed using R software using a custom-made script, and an igraph package was used for trees visualization. The majority of the analysis of the behavior of the clones was performed using a custom-made R script utilizing the lineage trees code. Multiple parameters values were computed considering all the clones/cells or part of them according to the analysis (SI Appendix, Supplementary Materials and Methods). For the analysis of the rearrangements of the cells in the clone, the value of the distance among cells was obtained using a Fiji custom-made macro. To measure the migration of single cells, pictures were manually aligned, and the manual-tracking plugin in Fiji was used. Cells were tracked during the OPC stage (SI Appendix, Supplementary Materials and Methods).
Immunohistochemistry and Distribution of Imaged Clones within the CC.
Immunohistochemical stainings were performed as described before (25). For the analysis of the distribution of imaged clones within the damaged CC under the window, coordinates were assigned to each clone taking into account its center. The distance between each clone whose death rate was >75%, and all the other clones fulfilling the same death rate criterium was measured. In the same way, the distance between each clone whose death rate was <25%, and all the other clones fulfilling the same death rate criterium was measured. Finally, the distance between clones whose death rate was <25%, and clones whose death rate was >75% was measured. Mice with at least three clones per condition were analyzed.
Statistical Analysis and Reproducibility.
The quantifications of immunohistochemical stainings were performed on a minimum of three animals, using six CC horizontal sections per brain. The demyelinated area was measured considering the MBP− area surrounded by MBP+ tissue within the CC and normalized using the total CC area. The analysis of the distribution of the imaged clones were performed using five mice. The analysis of the distance among cells in the clones was performed using clones from three animals. The data for the migration analysis were taken from seven mice. The statistical analysis of the data was performed using Graphpad Prism (version 8). A detailed report of statistical tests used, test results, sample sizes, and P values can be found in SI Appendix, Tables S2–S5 and in the figure legends for supplementary figures. First, normality was tested, and when data did not follow a normal distribution according to the Shapiro–Wilk test, the nonparametric Mann–Whitney U test or Wilcoxon matched-pairs test were performed. Alternatively, a paired or an unpaired two-tailed Student’s t test was used for mean comparison depending on the nature of the analysis (within or between subjects). In case of comparison of three or more groups, one-way ANOVA or Kruskal–Wallis tests were performed. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered significant. All the applied statistics were conducted on data from three or more biologically independent experimental replicates.
Supplementary Material
Acknowledgments
This work was supported by the European Research Council (STEMBAR to S.J.), the Swiss National Science Foundation (BSCGI0_157859 and 310030_196869 to S.J.), and the Zurich Neuroscience Center. We thank F. Helmchen and D. Chichung Lie for comments on the manuscript and U. Suter for providing the APC antibody.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025795118/-/DCSupplemental.
Data Availability
Raw data was deposited to the Image Data Resource (accession no. idr0113). All study data are included in the article and/or supporting information.
References
- 1.Franklin R. J. M., Ffrench-Constant C., Regenerating CNS myelin—From mechanisms to experimental medicines. Nat. Rev. Neurosci. 18, 753–769 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Nave K. A., Werner H. B., Myelination of the nervous system: Mechanisms and functions. Annu. Rev. Cell Dev. Biol. 30, 503–533 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Young K. M., et al., Oligodendrocyte dynamics in the healthy adult CNS: Evidence for myelin remodeling. Neuron 77, 873–885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung M. S., et al., Dynamics of oligodendrocyte generation and myelination in the human brain. Cell 159, 766–774 (2014). [DOI] [PubMed] [Google Scholar]
- 5.McKenzie I. A., et al., Motor skill learning requires active central myelination. Science 346, 318–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao L., et al., Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nat. Neurosci. 19, 1210–1217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes E. G., Orthmann-Murphy J. L., Langseth A. J., Bergles D. E., Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nat. Neurosci. 21, 696–706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson E. M., et al., Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science 344, 1252304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz J., Klein M. C., Behrens T. E., Johansen-Berg H., Training induces changes in white-matter architecture. Nat. Neurosci. 12, 1370–1371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bengtsson S. L., et al., Extensive piano practicing has regionally specific effects on white matter development. Nat. Neurosci. 8, 1148–1150 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Reich D. S., Lucchinetti C. F., Calabresi P. A., Multiple sclerosis. N. Engl. J. Med. 378, 169–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croxford A. L., Spath S., Becher B., GM-CSF in neuroinflammation: Licensing myeloid cells for tissue damage. Trends Immunol. 36, 651–662 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Martin R., Sospedra M., Rosito M., Engelhardt B., Current multiple sclerosis treatments have improved our understanding of MS autoimmune pathogenesis. Eur. J. Immunol. 46, 2078–2090 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Zawadzka M., et al., CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell 6, 578–590 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakatani H., et al., Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination. J. Neurosci. 33, 9752–9768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing Y. L., et al., Adult neural precursor cells from the subventricular zone contribute significantly to oligodendrocyte regeneration and remyelination. J. Neurosci. 34, 14128–14146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazanis I., et al., Subependymal zone-derived oligodendroblasts respond to focal demyelination but fail to generate myelin in young and aged mice. Stem Cell Rep. 8, 685–700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei F., et al., Accelerated remyelination during inflammatory demyelination prevents axonal loss and improves functional recovery. eLife 5, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter S. F., Leavitt J. A., Rodriguez M.. Direct observation of myelination in vivo in the mature human central nervous system. A model for the behaviour of oligodendrocyte progenitors and their progeny. Brain 120(Pt 11), 2071–2082 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Duncan I. D., Brower A., Kondo Y., Curlee J. F. Jr, Schultz R. D., Extensive remyelination of the CNS leads to functional recovery. Proc. Natl. Acad. Sci. U.S.A. 106, 6832–6836 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes E. G., Kang S. H., Fukaya M., Bergles D. E., Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci. 16, 668–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacmeister C. M., et al., Motor learning promotes remyelination via new and surviving oligodendrocytes. Nat. Neurosci. 23, 819–831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orthmann-Murphy J., et al., Remyelination alters the pattern of myelin in the cerebral cortex. eLife 9, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baxi E. G., et al., Lineage tracing reveals dynamic changes in oligodendrocyte precursor cells following cuprizone-induced demyelination. Glia 65, 2087–2098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilz G. A., et al., Live imaging of neurogenesis in the adult mouse hippocampus. Science 359, 658–662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blakemore W. F., Franklin R. J., Remyelination in experimental models of toxin-induced demyelination. Curr. Top. Microbiol. Immunol. 318, 193–212 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Kim E. J., Ables J. L., Dickel L. K., Eisch A. J., Johnson J. E., Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS One 6, e18472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Back S. A., et al., Hyaluronan accumulates in demyelinated lesions and inhibits oligodendrocyte progenitor maturation. Nat. Med. 11, 966–972 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Gadea A., Schinelli S., Gallo V., Endothelin-1 regulates astrocyte proliferation and reactive gliosis via a JNK/c-Jun signaling pathway. J. Neurosci. 28, 2394–2408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baydyuk M., et al., Tracking the evolution of CNS remyelinating lesion in mice with neutral red dye. Proc. Natl. Acad. Sci. U.S.A. 116, 14290–14299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marques S., et al., Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasuga Y., Fudge A. D., Zhang Y., Li H., Characterization of a long noncoding RNA Pcdh17it as a novel marker for immature premyelinating oligodendrocytes. Glia 67, 2166–2177 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Falcão A. M., et al., Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat. Med. 24, 1837–1844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spitzer S. O., et al., Oligodendrocyte progenitor cells become regionally diverse and heterogeneous with age. Neuron 101, 459–471.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jäkel S., et al., Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J., Shen Z., Yu Y. C., Shi S. H., Neural lineage tracing in the mammalian brain. Curr. Opin. Neurobiol. 50, 7–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marisca R., et al., Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci. 23, 363–374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crawford A. H., Tripathi R. B., Richardson W. D., Franklin R. J. M., Developmental origin of oligodendrocyte lineage cells determines response to demyelination and susceptibility to age-associated functional decline. Cell Rep. 15, 761–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menn B., et al., Origin of oligodendrocytes in the subventricular zone of the adult brain. J. Neurosci. 26, 7907–7918 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L. O., et al., Spatiotemporal control of CNS myelination by oligodendrocyte programmed cell death through the TFEB-PUMA axis. Cell 175, 1811–1826 e1821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapp B. D., Nishiyama A., Cheng D., Macklin W., Differentiation and death of premyelinating oligodendrocytes in developing rodent brain. J. Cell Biol. 137, 459–468 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barres B. A., et al., Cell death and control of cell survival in the oligodendrocyte lineage. Cell 70, 31–46 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Neumann B., Segel M., Chalut K. J., Franklin R. J., Remyelination and ageing: Reversing the ravages of time. Mult. Scler. 25, 1835–1841 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann B., et al., Metformin restores CNS remyelination capacity by rejuvenating aged stem cells. Cell Stem Cell 25, 473–485.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruckh J. M., et al., Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell 10, 96–103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nait-Oumesmar B., et al., Progenitor cells of the adult mouse subventricular zone proliferate, migrate and differentiate into oligodendrocytes after demyelination. Eur. J. Neurosci. 11, 4357–4366 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data was deposited to the Image Data Resource (accession no. idr0113). All study data are included in the article and/or supporting information.