Significance
Surgical removal of pancreatic cancer remains the only option for a cure. To verify the extent of tumor removal, surgeons rely on pathologic evaluation of frozen sections of surgical margins. However, this process can be challenging, time consuming, and subjective. Here, we used the MasSpec Pen to rapidly distinguish pancreatic cancer from healthy pancreatic and bile duct tissues by generating classification models based on the molecular signatures acquired from tissue. We evaluated this technology in an operating room during pancreatic surgeries and used these classification models to predict on data obtained in vivo and ex vivo with high performance. Our results suggest that the MasSpec Pen platform has the potential to improve and expedite margin evaluation during pancreatic cancer surgery.
Keywords: pancreatic cancer, MasSpec Pen, surgical margin evaluation, mass spectrometry
Abstract
Intraoperative delineation of tumor margins is critical for effective pancreatic cancer surgery. Yet, intraoperative frozen section analysis of tumor margins is a time-consuming and often challenging procedure that can yield confounding results due to histologic heterogeneity and tissue-processing artifacts. We have previously described the development of the MasSpec Pen technology as a handheld mass spectrometry–based device for nondestructive tissue analysis. Here, we evaluated the usefulness of the MasSpec Pen for intraoperative diagnosis of pancreatic ductal adenocarcinoma based on alterations in the metabolite and lipid profiles in in vivo and ex vivo tissues. We used the MasSpec Pen to analyze 157 banked human tissues, including pancreatic ductal adenocarcinoma, pancreatic, and bile duct tissues. Classification models generated from the molecular data yielded an overall agreement with pathology of 91.5%, sensitivity of 95.5%, and specificity of 89.7% for discriminating normal pancreas from cancer. We built a second classifier to distinguish bile duct from pancreatic cancer, achieving an overall accuracy of 95%, sensitivity of 92%, and specificity of 100%. We then translated the MasSpec Pen to the operative room and predicted on in vivo and ex vivo data acquired during 18 pancreatic surgeries, achieving 93.8% overall agreement with final postoperative pathology reports. Notably, when integrating banked tissue data with intraoperative data, an improved agreement of 100% was achieved. The result obtained demonstrate that the MasSpec Pen provides high predictive performance for tissue diagnosis and compatibility for intraoperative use, suggesting that the technology may be useful to guide surgical decision-making during pancreatic cancer surgeries.
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal cancer, with a 5-y survival rate of 9% for all stages (1). Among patients with resectable tumors, surgical resection with microscopically negative margins is required for prolonged disease-free survival (2). Thus, differentiating normal tissue from tumor is the cornerstone of effective pancreatic oncologic surgery (3, 4). In practice, delineation of pancreatic resection margins is difficult and variable. Tumor margin status is commonly assessed intraoperatively by microscopic histopathological evaluation of frozen sections prepared from excised pancreatic neck and common bile duct margins prior to completion of the operation (5, 6). However, artifacts on tissue histology from frozen section preparation complicates histopathologic evaluation, further accentuated by the complex pathology of PDAC, with high desmoplastic stromal content and chronic pancreatitis (CP) at the pancreatic resection margins as well as inflammation induced by surgical manipulation of bile duct tissue–mimicking tumor (7, 8). Moreover, the reported accuracy of frozen section analysis is variable center-to-center and depends on the methodology, skillset, and subspecialty of the pathologist on call (9–12).
Several optical and molecular technologies are being developed with the expectation of improving pancreatic cancer detection during surgery (SI Appendix, Text) (13–15). In particular, mass spectrometry (MS) technologies are powerful for investigating molecular differences between normal and cancerous tissues with high sensitivity, chemical specificity, and speed and have also been explored for surgical use and margin evaluation (16). Desorption electrospray ionization (DESI) MS, for example, has been used to investigate molecular differences between normal and cancerous tissues ex vivo cancerous and normal tissue sections (17–19). In particular, Zare and coworkers applied DESI-MS imaging to analyze the metabolic and lipid profiles of adjacent histologic tissue sections of margin regions collected from pancreatic surgery (20). Other techniques (i.e., rapid evaporative ionization MS and laser-based MS devices) have been developed for in vivo cancer detection (21–24). However, these techniques have not yet been demonstrated for pancreatic cancer tissue detection.
We developed the MasSpec Pen system as a biocompatible handheld device coupled to a mass spectrometer for rapid (∼15 s) and nondestructive tissue analysis (25). Upon contact with a tissue and activation by a foot pedal, a discrete water droplet is formed and contained at the MasSpec Pen tip reservoir, allowing metabolites and lipids to be gently extracted into the water droplet, which is then transported to the mass spectrometer for analysis. To date, we have applied the MasSpec Pen in conjunction with least absolute shrinkage and selector operator (Lasso) penalized logistic regression to classify several tumor types ex vivo, achieving 96% overall accuracy in cross-validation (CV) (25–27). More recently, we translated the MasSpec Pen technology to an operating room (OR) to evaluate technical feasibility in open human surgeries and as a laparoscopic device in a robotic surgical procedure (28, 29).
Here, we describe the application and first use of the MasSpec Pen for the acquisition and statistical prediction of molecular data acquired from pancreatic and biliary tissues, including in vivo data of surgical margins collected in an OR from patients undergoing pancreatic surgery. Collectively, our results indicate that the MasSpec Pen technology may be useful to enhance surgical margin evaluation in pancreatic cancer procedures by providing near–real-time diagnostic feedback to surgeons and physicians.
Results
Molecular Analysis and Classification of Pancreatic Tissues.
We first used the MasSpec Pen to analyze 131 PDAC and normal banked human tissue samples to evaluate its potential for PDAC diagnosis and surgical margin evaluation within the primary organ. All patient demographic information obtained from the tissue banks is provided in Table 1. Rich molecular profiles including diverse metabolites, fatty acids (FA), and glycerophospholipid (GP) species such as phosphatidylethanolamines (PE), phosphatidylserines (PS), and phosphatidylinositols (PI) were detected in both tissue types (Fig. 1A and SI Appendix, Text). Following MasSpec Pen analyses, the tissue samples were stamped with surgical ink to precisely demarcate the region of analysis. The tissues were then flash frozen, sectioned, and hematoxylin and eosin (H&E) stained. Pathologic evaluation of the H&E-stained tissue sections was performed within the stamped tissue region, allowing identification of histologic features present within the tissue area analyzed. Normal pancreatic tissues were typically composed of lobular ducts, minimal stroma, and acinar cells. In contrast, PDAC tissues often presented regions of tumor cells in irregular ductal formation mingled with CP, desmoplastic stroma, and fibrosis.
Table 1.
Patient demographics for human pancreas and bile duct tissue samples and fresh tissue analyzed intraoperatively
| Variable | Banked data | OR data | ||
| Nontumor pancreas | PDAC | Bile duct | ||
| Total, no. | ||||
| Patients(s) | 53 | 58 | 23 | 18 |
| Samples | 54 | 77 | 26 | — |
| Analyses | 54 | 87 | 31 | 64 |
| No. of regions analyzed, no. of samples | ||||
| 1 | 54 | 68 | 21 | |
| 2 | 0 | 8 | 5 | |
| 3 | 0 | 1 | 0 | |
| Age of patients, median, y | 64 | 68.5 | 66 | 63.5 |
| Sex, no. of patients | ||||
| Male | 19 | 33 | 13 | 10 |
| Female | 34 | 25 | 10 | 8 |
| Race, no. of patients | ||||
| White | 43 | 47 | 18 | 15 |
| Black | 10 | 10 | 3 | 2 |
| Asian | 0 | 0 | 2 | 0 |
| Other | 0 | 1 | 0 | 1 |
| Tissue origin, no. of patients | ||||
| CHTN | 45 | 46 | 3 | 0 |
| BCM | 8 | 12 | 20 | 0 |
| OR at BCM (clinical study) | 0 | 0 | 0 | 18 |
| Reason for surgery, no. of patients | ||||
| PDAC | 0 | 58 | 12 | 7 |
| Benign tumors | 20 | 0 | 3 | 2 |
| Cysts and lesions (e.g., IPMN, N, PanIN, MCN, etc.) | 22 | 0 | 2 | 3 |
| Neuroendocrine tumor | 0 | 0 | 1 | 3 |
| Pancreatitis | 7 | 0 | 0 | 0 |
| Autopsy | 1 | 0 | 0 | 0 |
| Other | 2 | 0 | 4 | 3 |
| Unavailable | 1 | 0 | 1 | 0 |
In the columns labeled “Banked data,” normal, uninvolved pancreas tissues obtained from a patient undergoing surgery for PDAC resection are grouped under “PDAC,” while bile duct tissues, diagnosed postanalysis by pathology as normal or cancerous, are grouped under “Bile duct.” Note that samples of both uninvolved tissue and cancerous tissue were obtained from 24 patients. There is one patient from which both a sample of cancerous and bile duct tissue were obtained, thus demographic information is noted under “PDAC,” and sample-specific information is under “PDAC” or “Bile Duct.” Under “Reason for surgery,” the “Other” subcategory includes uninvolved tissues resected from patients with end-stage renal disease, trauma, pseudopapillary neoplasm, hepatocellular carcinoma, ampullary cancer, cholangiocarcinoma, cystic duct adenocarcinoma, and two patients whose data were unavailable.
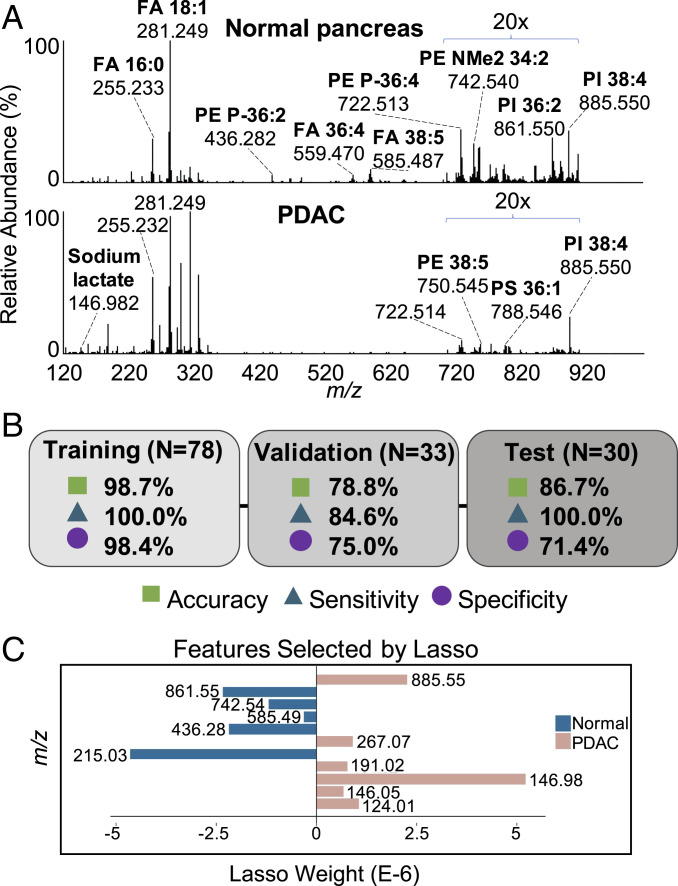
Fig. 1.
MasSpec Pen analyses of banked pancreas tissues and Lasso classification model. (A) Representative mass spectra generated from averaging all normal pancreas mass spectra (n = 63) and PDAC mass spectra (n = 15) included in the training set. (B) Overall accuracy, sensitivity, and specificity for the Lasso prediction results for training, validation, and independent test sets. (C) Features (m/z) selected by Lasso as characteristic of normal pancreas (negatively weighted values) and PDAC (positively weighted values). Tentative ion identifications be found in SI Appendix, Table S2.
We employed the Lasso method to build classification models for discriminating normal and PDAC based on a sparse set of m/z features characteristic of disease state (30). To maximize robustness of the Lasso model in detecting PDAC or normal pancreas, we used a training set of 78 tissue analyses with clear histologic regions of normal and tumor cells (>70%) within the area sampled. Using leave-one-out CV, Lasso enabled classification of normal pancreas from PDAC with a prediction accuracy of 98.7%, sensitivity of 100.0%, and specificity of 98.4% (Fig. 1B). Detailed prediction results are provided in SI Appendix, Table S1. Lasso selected 11 features as predictive of PDAC or normal pancreas (Fig. 1C), which agreed with mass spectral trends (Fig. 1A). Features with proposed ion identifications are listed in SI Appendix, Table S2.
We next evaluated the predictive performance of the model using a validation set of 33 mass spectra obtained from 30 tissues with complex tissue histology within the tissue region analyzed, including low epithelial tumor cell concentration, mixed cellular composition, or atypical molecular profiles. Within these complex samples, a total of 26 analyses were classified in agreement with postanalysis pathologic diagnosis (Fig. 1B and SI Appendix, Table S3). Of 13 mass spectra obtained from PDAC tissue analyses, 11 were classified correctly by our method including four tissues with tumor cell concentration ranging from 5 to 25%. Seven analyses were classified in disagreement with postpathologic evaluation in the validation set. Of these, three tissues presented extensive CP and fibrosis within the region of MasSpec Pen analysis, histologic features that are difficult to segregate from intermixed PDAC and normal cells. Although we did not include CP and fibrosis in our training set and thus their mass spectra were atypical, we chose to test our classifiers on these samples to explore the performance of our method. Another tissue, cancer sample P128T, presented a cluster of 10% tumor cells at the edge of the MasSpec Pen sampling area and was misclassified as normal. Although maximal experimental effort was put into precisely demarcating the tissue region analyzed by the MasSpec Pen, slight inaccuracies in the stamping system could have contributed to under sampling of tissue regions at the edge of the analyzed area and thus misclassification. Collectively, an accuracy of 78.8%, sensitivity of 84.6%, and specificity of 75.0% was achieved.
To further test the statistical classifier, we then used the model to predict on an independent test set of 14 normal pancreas and 16 PDAC tissues with clear histologic composition. Note that these data were collected on a different orbitrap mass spectrometer than the one used to collect training and validation set data. Overall, 26 tissues were classified in agreement with pathology, resulting in 86.7% accuracy, 100.0% sensitivity, and 71.4% specificity (Fig. 1B).
Detection and Diagnosis of PDAC in the Common Bile Duct Margins.
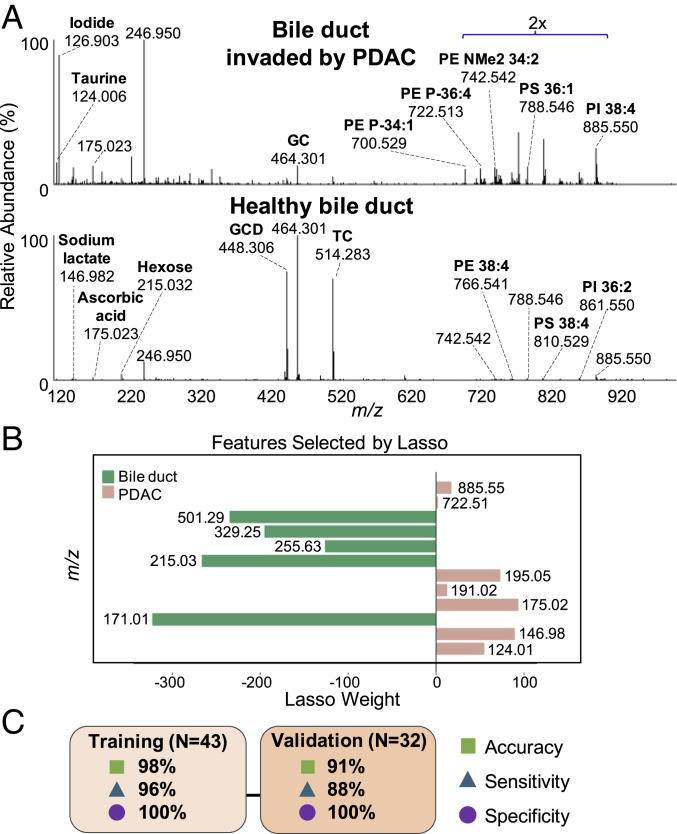
Intraoperative frozen section examination of the common bile duct margin is regularly performed in pancreatic cancer surgery to assess possible tumor involvement in addition to the transection margin of the pancreatic neck. As such, evaluation of the ability of the MasSpec Pen to identify PDAC within common bile duct tissue is essential for clinical use. To assess performance for detection of PDAC within the bile duct margin, we first analyzed 26 banked bile duct tissues with the MasSpec Pen. Five of the samples were analyzed in two distinct tissue regions, resulting in 31 total analyses. Normal bile duct tissue samples were composed of uniform epithelial cells embedded into surrounding pancreas tissue or alongside fibro-adipose tissue. Bile duct tissue samples with invasive PDAC presented with either normal histologic features with PDAC encroaching into the sampling area or as entirely infiltrated by PDAC. As shown in Fig. 2A, normal bile duct mass spectra presented high relative abundances of metabolites and bile acids (SI Appendix, Fig. S1), observed in the m/z range 400 to 550, such as deprotonated deoxyglycocholic (m/z 448.306) and glycocholic (m/z 464.301) acid. The molecular profile of bile duct tissue with invading PDAC presented higher relative abundances of metabolites, FAs, as well as many GP species similar to what was observed in primary PDAC tissues (Fig. 1A).
Fig. 2.
MasSpec Pen analyses of banked bile duct tissues and Lasso statistical classifier. (A) All bile duct (n = 16) in the training set and bile duct invaded by PDAC (n = 8) in the validation set were averaged to produce representative mass spectra. The relative abundance of the lipid mass range (m/z 700 to 900) was amplified by a factor of 2 in both mass spectra. (B) Plot of Lasso features (m/z) and corresponding weights. (C) Accuracy, sensitivity, and specificity results for training and validation sets. Abbreviations: GC (glycocholic acid), GCD (glycodeoxycholic acid), and TC (taurocholic acid).
The mass spectra obtained from 16 normal bile duct and 27 PDAC tissues were then used as a training set to build a classification model of normal bile duct versus PDAC. Lasso selected 12 m/z features as important predictors for disease classification (Fig. 2B), allowing discrimination of bile duct from PDAC with a leave-one-out CV prediction accuracy of 98% (Fig. 2C). Detailed prediction results are shown in SI Appendix, Table S4. Proposed ion identifications of Lasso features are listed in SI Appendix, Table S5. We then evaluated the predictive performance of the Lasso model on a validation set comprised of data acquired from normal bile duct (n = 7), bile duct with invading PDAC (n = 8), and primary PDAC tissues (n = 17). Comparisons of Lasso prediction results to postanalysis pathological diagnosis are provided in SI Appendix, Table S6. Overall, a prediction accuracy of 91%, sensitivity of 88%, and specificity of 100% was achieved in the validation set (Fig. 2C).
In Vivo and Ex Vivo Analysis of Human Tissue during Pancreatic Surgeries.
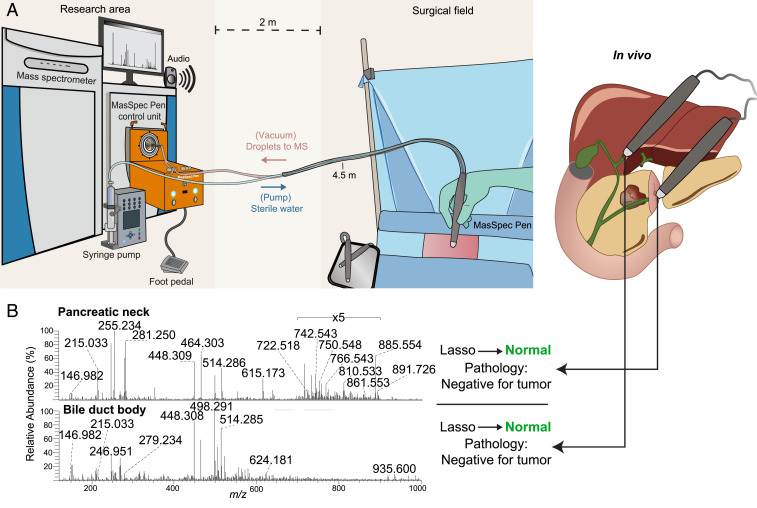
As a critical step toward testing and validating the MasSpec Pen for intraoperative tissue analysis and diagnosis, we translated the system to the OR for direct tissue analysis during surgical procedures. The MasSpec Pen was used by the surgeons and clinical staff for analysis of in vivo tissues and freshly excised specimens in 18 pancreatic surgeries, including distal pancreatectomy, pancreaticoduodenectomy (Whipple), and hepaticojejunostomy procedures (Fig. 3A and SI Appendix, Fig. S2). Patient demographic information for OR cases is included in Table 1. A total of 110 intraoperative analyses from healthy pancreas, bile duct, lymph node, liver, and pancreatic neck and bile duct margins were performed with the MasSpec Pen, including 61 in vivo and 49 on freshly excised tissues. The in vivo analyses were performed at the surgeon’s discretion within tissue regions of interest such as neck and bile duct margins and benign tissue regions. The ex vivo analyses of similar regions were performed by clinical research assistants also within surgical margins and normal tissue uninvolved by tumor. Note that no observable tissue damage occurred due to MasSpec Pen use as reported by the pathologic department, allowing for subsequent intra- and postoperative histopathologic evaluation of the tissues.
Fig. 3.
Clinical testing of the MasSpec Pen during pancreatic surgery. (A) Schematic of MasSpec Pen platform in an OR for surgical use and clinical testing. (B) Mass spectra obtained in vivo from the pancreatic neck margin and bile duct body from Patient 15 undergoing a Whipple procedure. Corresponding Lasso prediction results (performed after surgery) and final pathology notes are shown.
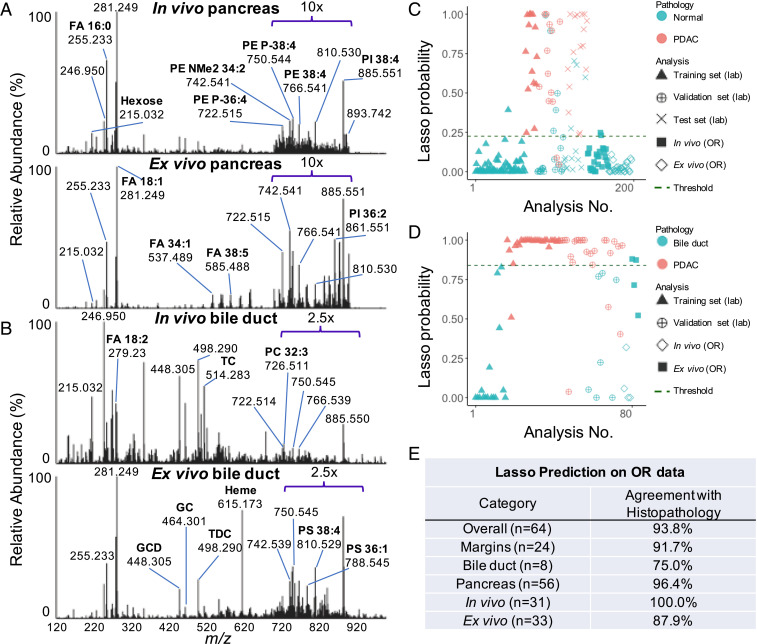
Molecular profiles acquired intraoperatively from pancreas and bile duct tissues (Fig. 4 A and B) appeared qualitatively similar to those acquired from banked tissues in the laboratory (Figs. 1A and 2A). Fig. 4A shows the average mass spectra for all the 29 in vivo and 27 ex vivo pancreatic tissue analyses performed in the OR. Lipid profiles of intraoperative pancreatic data were generally comparable to banked normal pancreas, with several GP species detected in high relative abundance. In addition, hexose (m/z 215.032), FA 16:0 (m/z 255.232), FA 18:1 (m/z 281.249), and FA dimers, such as oleic and arachidonic acid (m/z 585.488), were also detected intraoperatively at similar relative abundances to banked pancreatic samples. We noted higher relative abundances of chlorinated triacylglycerol (TG) species such as TG 52:2 (m/z 893.741) in intraoperative data collected from multiple tissue sites. Similarly, the average mass spectra (Fig. 4B) from four in vivo and four ex vivo bile duct tissue analyses obtained intraoperatively presented similar molecular profiles to laboratory data, including high relative abundances of bile acids and GP species (Fig. 2A). Generally, a relative increase in spectral noise and in relative abundances of ions from blood molecules and anesthetic agents were observed in all the intraoperative data when compared to data acquired in the laboratory using banked tissues. Within these, a prominent peak at m/z 615.171 was identified as deprotonated heme, which was likely detected due to residual blood on the tissue surface. In addition, we observed an increase in the ion abundance of the deprotonated metabolite (m/z 246.950) of a general anesthetic, sevoflurane, which we have previously characterized through MS/MS experiments (30). This peak was also observed in banked tissues, particularly in banked bile duct mass spectra (Fig. 2A).
Fig. 4.
Intraoperative molecular analysis and statistical prediction results of human pancreatic and bile duct tissues. (A) Representative mass spectra of pancreatic tissues collected intraoperatively using the MasSpec Pen. All data were averaged from all in vivo (n = 29) and ex vivo (n = 27) measurements to produce representative mass spectra. The relative abundance of the lipid mass range (m/z 700 to 900) in both spectra was amplified by a factor of 10 to facilitate data visualization. (B) Mass spectra averaged from all bile duct data acquired in vivo (n = 4) and ex vivo (n = 4). The relative abundance of the lipid mass range was amplified by a factor of 2.5 in both spectra. (C) Estimated Lasso probability and pathologic diagnosis associated with each individual pancreatic tissue analysis from banked samples analyzed in laboratory and OR data following statistical prediction with the normal pancreas versus PDAC classification model. The optimal probability threshold was determined to be 0.225, where a sample is classified as normal pancreas if its estimated probability is below this threshold, and a sample is classified as PDAC if its probability is above this threshold. (D) Estimated Lasso probability and pathologic diagnosis associated with each bile duct tissue analysis from banked samples and OR data following statistical prediction using the normal bile duct versus PDAC model. The Lasso threshold was found to be 0.839, and similarly, a sample is classified as normal bile duct if its estimated probability is below this threshold or PDAC if its probability is above this threshold. (E) Summary of statistical prediction results on intraoperative data compared with postoperative histopathology. Abbreviations: GC (glycocholic acid), GCD (glycodeoxycholic acid), TC (taurocholic acid), and TDC (taurodeoxycholic acid).
Statistical Prediction on Intraoperative Human Data.
Of the 110 analyses obtained intraoperatively, we evaluated the predictive performance of the classification models on 64 analyses from 16 cases as an independent test set. From the 46 analyses not classified, 32 were from data acquired on liver and lymph node tissues, which were not included in our models. In addition, there were 14 analyses of pancreatic or bile duct tissues that did not yield ion signal above suitable signal-to-noise ratios and were thus excluded, resulting in a final sample set of 64 in vivo and ex vivo analyses from pancreatic and bile duct tissues. Using the normal pancreas versus PDAC model, we predicted on 29 in vivo and 27 ex vivo mass spectra from normal tissue. To compare statistical performance of our classification models on intraoperative data collected in the OR to banked data acquired in the laboratory, we plotted the estimated Lasso probability values of each individual analysis from samples analyzed with the MasSpec Pen (Fig. 4 C and D). These estimated probabilities enable binary classification based on an optimized probability threshold, where samples with associated probabilities below the threshold are negative (i.e., normal), and samples with probabilities above the threshold are positive, (i.e., PDAC). The Lasso probabilities generated from statistical analysis of data from normal and cancerous pancreatic tissues analyzed in the laboratory as well as the OR are plotted in Fig. 4C. Similarly, for all banked and intraoperative bile duct analyses predicted on using the bile duct versus PDAC classifier, the resulting Lasso probabilities are plotted in Fig. 4D. For predicting on the intraoperative pancreatic data, 96.4% accuracy was achieved, with 54 of 56 predictions classified in agreement with final postoperative histopathology reports (Fig. 4E and Table 2). Note that when the MasSpec Pen was used to analyze suspected tumor regions as determined by surgical gross evaluation, the tumor tissue was presumably physically beneath benign tissue. Thus, final histopathology reports were used to assess agreement with Lasso prediction results for margins, and gross assessment of tissue was used to assess agreement with Lasso results for normal tissue analyses. Using the bile duct versus PDAC model to predict on three in vivo and five ex vivo normal bile duct analyses, 75.0% accuracy was achieved, as six of eight predictions agreed with postoperative histopathology reports (Fig. 4E and Table 2). Considering all 64 analyses, 93.8% overall agreement was achieved. Interestingly, incorporating intraoperative data acquired from in vivo and ex vivo normal pancreas (n = 9) and bile duct (n = 4) tissues into each respective training set with banked data improved prediction agreement on the remaining intraoperative pancreas (n = 47) and bile duct (n = 4) data to 100% for both models (SI Appendix, Table S7).
Table 2.
Comparison of preoperative diagnosis, surgical gross assessment, postoperative histopathologic report, and Lasso prediction for intraoperative MasSpec Pen analyses
| Case | Preoperative diagnosis | MSP analysis | Gross assessment by surgeon | Final histopathology report | Lasso results | |
| No. | Type | |||||
| 1 | Adenocarcinoma involving the uncinate process of the pancreas | 1 | IV | Suspected tumor | Pancreas needle biopsy: (−) for carcinoma | Normal ¥ |
| 2 | EV | Suspected tumor | Normal ¥ | |||
| 3 | EV | Suspected tumor | Normal ¥ | |||
| 2 | PDAC involving HOP | 1 | IV | Suspected tumor | Pancreas needle biopsy: (−) for carcinoma | Normal ¥ |
| 3 | Pancreatic cyst | 1 | EV | Normal pancreas | Pancreatic margin: (−) for cystic lesion | Normal ¥ |
| 4 | Bile duct cancer | 1 | IV | Bile duct margin | Only uncinate/SMA margin: (+) carcinoma | Bile duct § |
| 5 | PDAC involving HOP | 1 | IV | Suspected tumor | Only uncinate margin: (+) for carcinoma; mild/moderate CP | Normal ¥ |
| 2 | EV | Normal pancreas | Normal ¥ | |||
| 6 | Pancreatic mass | 1 | IV | Normal pancreas | All margins: (−) for carcinoma | Normal ¥ |
| 2 | IV | Normal pancreas | Normal ¥ | |||
| 3 | EV | Pancreatic surgical margin | Normal ¥ | |||
| 4 | EV | Pancreatic surgical margin | Normal ¥ | |||
| 7 | Malignant pancreatic neoplasm | 1 | IV | Suspected tumor | All margins: (−) for carcinoma | Normal ¥ |
| 2 | IV | Suspected tumor | Normal ¥ | |||
| 3 | IV | Transected pancreatic neck | Normal ¥ | |||
| 8 | Adenoma of ampulla of Vater | 1 | IV | Pancreatic neck margin | (−) for high-grade dysplasia or invasive carcinoma | Normal ¥ |
| 2 | EV | Pancreatic neck margin | Normal ¥ | |||
| 3 | EV | Pancreatic neck margin | Normal ¥ | |||
| 4 | EV | Pancreatic neck margin | Normal ¥ | |||
| 9 | Mucinous cystic lesion on HOP | 1 | IV | Pancreatic body | All margins: not involved by lesion | Normal ¥ |
| 2 | IV | Pancreatic neck | Normal ¥ | |||
| 3 | EV | Normal pancreas | Normal ¥ | |||
| 4 | EV | Normal pancreas | Normal ¥ | |||
| 5 | EV | Normal pancreas | Normal ¥ | |||
| 11 | Mass of pancreas | 1 | IV | Normal pancreas | All margins: (−) for cyst and carcinoma | Normal ¥ |
| 2 | IV | Normal pancreas | Normal ¥ | |||
| 3 | EV | Normal pancreas | Normal ¥ | |||
| 4 | EV | Normal pancreas | Normal ¥ | |||
| 5 | EV | Normal pancreas | Normal ¥ | |||
| 12 | Mass of pancreas | 1 | IV | Normal pancreas | All margins: (−) for carcinoma | Normal ¥ |
| 13 | Malignant neoplasm of pancreas, unspecified location of malignancy | 1 | IV | Normal pancreas | All margins: (−) for carcinoma | Normal ¥ |
| 2 | IV | Normal pancreas | Normal ¥ | |||
| 3 | EV | Pancreatic tumor margin | Normal ¥ | |||
| 4 | EV | Pancreatic tumor margin | Normal ¥ | |||
| 5 | EV | Posterior side of suspected mass | PDAC ¥ | |||
| 6 | EV | Posterior side of suspected mass | PDAC ¥ | |||
| 14 | PNET, recurrent, HOP | 1 | IV | Normal pancreas | Pancreatic head biopsy: (−) for carcinoma | Normal ¥ |
| 15 | Ampulla of Vater mass, mass of pancreas | 1 | IV | Pancreatic body | All margins: (−) for carcinoma | Normal ¥ |
| 2 | IV | Pancreatic neck margin | Normal ¥ | |||
| 3 | IV | Pancreatic neck margin | Normal ¥ | |||
| 4 | IV | Normal bile duct | Bile duct § | |||
| 5 | EV | Pancreatic neck margin | Normal ¥ | |||
| 16 | PNET, HOP | 1 | IV | Suspected tumor | All margins: (−) for carcinoma | Normal ¥ |
| 2 | IV | Suspected tumor | Normal ¥ | |||
| 3 | IV | Normal pancreas | Normal ¥ | |||
| 4 | IV | Normal pancreas | Normal ¥ | |||
| 5 | EV | Pancreatic neck margin | Normal ¥ | |||
| 6 | EV | Pancreatic neck margin | Normal ¥ | |||
| 7 | EV | Bile duct margin | Bile duct § | |||
| 8 | EV | Bile duct margin | PDAC § | |||
| 9 | EV | Normal pancreas | Normal ¥ | |||
| 17 | IPMN | 1 | IV | Normal pancreas | All margins: (−) for carcinoma | Normal ¥ |
| 2 | IV | Normal pancreas | Normal ¥ | |||
| 3 | IV | Normal bile duct | Bile duct § | |||
| 4 | IV | Normal bile duct | Bile duct § | |||
| 18 | Ampullary carcinoma | 1 | IV | Normal pancreas | All margins: (−) for carcinoma | Normal ¥ |
| 2 | IV | Normal pancreas | Normal ¥ | |||
| 3 | EV | Pancreatic neck margin | Normal ¥ | |||
| 4 | EV | Pancreatic neck margin | Normal ¥ | |||
| 5 | EV | Bile duct margin | PDAC § | |||
| 20 | PDAC; jejunal diverticulum; fatty liver versus cirrhosis | 1 | EV | Pancreatic neck margin | All margins: (−) for carcinoma | Normal ¥ |
| 2 | EV | Pancreatic neck margin | Normal ¥ | |||
| 3 | EV | Pancreatic neck margin | Normal ¥ | |||
| 4 | EV | Bile duct margin | Bile duct § | |||
Classification using the normal pancreas versus PDAC Lasso model is indicated with ¥, while statistical classification using the normal bile duct versus PDAC Lasso model is indicated with § in “Lasso predictions” column. AC, adenocarcinoma; HOP, head of pancreas; IPMN, intraductal papillary mucinous neoplasm; PAN-IN, pancreatic intraepithelial neoplasia; PNET, pancreatic neuroendocrine tumor.
Discussion
Precise pancreatic tumor resection achieving microscopically negative margins in the pancreatic neck and bile duct margins is significantly correlated with longer disease-free survival, but the complexities of PDAC tissue histology and pathologic assessment render this a challenging and time-consuming task via frozen sections (5, 6, 8, 31). We evaluated the usefulness of the MasSpec Pen technology in enabling accurate tissue identification and in assisting surgical decision-making by generating classification models for PDAC diagnosis and testing their performance on intraoperative data collected in vivo and on freshly excised tissues.
We used banked samples with clear histologic diagnoses to train a Lasso model to classify tissues as normal pancreas or PDAC, yielding a CV prediction accuracy of 98.7%, which is comparable to previously reported results for DESI-MS (98%) (20). We evaluated this model on a validation set comprised of tissues with low tumor cell concentration (5 to 25%) and complex pathologic diagnosis more closely representing pancreatic tissues routinely evaluated in clinical pathology (32). This method correctly classified four of six validation set samples presenting low tumor cell concentration along with stroma, CP, and fibrosis in the analyzed region, including two samples with 5% tumor cells and 10 to 20% tumor cells, respectively. A total of 10 samples contained CP, a histologic feature widely regarded as difficult to discriminate from PDAC via pathologic assessment (7). Although CP samples were not included in our training set, eight of the 10 samples that presented CP were correctly classified as normal pancreas by our method. Despite highly complex histologic composition (SI Appendix, Table S3), 78.8% agreement was attained based on strict normal or PDAC assignment by pathology, similar to the 81% validation accuracy achieved in the DESI-MS study (20), which excluded tissues containing CP and fibrosis altogether. In an independent test set of samples (n = 30), a classification agreement of 86.9% was achieved, with every PDAC sample (n = 16) classified correctly, highlighting the robustness of our method for tissue diagnosis even when data are collected with a different MS instrument. To enable bile duct margin assessment, we built a Lasso classifier using data obtained from normal bile duct tissues and primary PDAC tissues and validated the model on heterogeneous bile duct and PDAC samples as well as bile duct tissues presenting invading PDAC. For training and validation sets, 98 and 91% accuracies were achieved for discriminating bile duct from PDAC, respectively. In an effort to generate a robust Lasso model, we trained this classifier on primary PDAC tissue data given the higher concentration of cancer cells present in these tissues compared to the more heterogenous bile duct tissues comprised of bile duct epithelium mingled with PDAC cells, as well as the larger sample size available for PDAC tissues. Of the eight bile duct tissues with PDAC, seven were correctly predicted as PDAC. We expect to refine these statistical classifiers by incorporating bile duct tissues with invasive PDAC into the training set and testing performance with additional sample sets.
Notably, our classification models (Figs. 1C and 2B) consisted of metabolites and lipid species that play key biological roles related to cancer metabolism and biochemistry. For example, the chlorinated adduct of the hexose isomer (m/z 215.032) was selected as a feature indicative of normal tissue in both models. Although tandem MS does not allow identification of the structural isomer, we speculate that this biomolecule is glucose, as one of the main functions of the pancreas is maintaining glucose homeostasis (33). Other ions identified as deprotonated taurine (m/z 124.006) and chlorinated adduct of lactate (m/z 146.982) were selected as predictive of PDAC in both models. Lower levels of glucose but elevated levels of taurine and lactate in tissue have been associated with pancreatic cancer, implying a shift to aerobic glycolysis as per the Warburg effect, corroborating our results (34, 35). In the bile duct model, a molecule tentatively identified as glycerol-3-phosphate (m/z 171.006), an intermediate of glycolysis, was selected for bile duct characterization. Interestingly, a study by Kamphorst et al. reported depletion of glycerol-3-phosphate levels in PDAC compared to benign adjacent tissues (35). Several GPs such as PIs and PEs were weighted toward normal pancreatic and bile duct tissues, including PE P-16:0_20:4 (m/z 722.514), PE NMe2 18:2_16:0 (m/z 742.542), and PI 18:0_18:2 (m/z 861.550), while PI 18:0_20:4 (m/z 885.550) was weighted toward PDAC in both of our models. GPs are major constituents of cellular membranes and play diverse roles in signaling and metabolism (36, 37), many of which have been previously reported as potential biomarkers of PDAC using DESI-MS and metabolomics assays (SI Appendix, Text) (20, 38, 39).
To evaluate clinical utility, we then integrated the MasSpec Pen system into the pancreatic surgical workflow and showed compatibility for in vivo and ex vivo tissue analyses. No adverse effects, complications, or limitations to histologic tissue processing or diagnosis were identified due to MasSpec Pen analyses. Importantly, although an increase in the abundances of noise and peaks related to biological fluids and anesthetics was observed in the intraoperative data, the molecular species identified in banked tissues (Figs. 1A and 2A) were consistently detected in vivo and from freshly excised tissues (Fig. 4 A and B). Most notably, the statistical classifiers generated from banked tissue data were used to predict on data from 64 analyses performed with the MasSpec Pen intraoperatively as an independent test set, resulting in 93.8% agreement with final pathological reports (Fig. 4E). In particular, out of 24 analyses from surgical margins diagnosed as negative via final pathologic diagnosis, 22 were predicted as normal by our classifier, resulting in 91.7% agreement with pathology. Though only eight analyses were collected from bile duct intraoperatively, six analyses were predicted as normal, in congruence with final pathologic diagnosis. Yet, incorporating intraoperative data into the training sets further improved prediction performance to 100% agreement on withheld intraoperative data from both pancreatic and bile duct tissue, thus meriting further investigation as a statistical model development approach with additional data collection (SI Appendix, Table S7). These results are particularly encouraging considering that the MasSpec Pen analyses obtained in the OR were performed by the surgical staff without comparable technical experience as the research staff, who acquired data from banked tissues within the tightly controlled laboratory environment, thus underscoring the user friendliness of the device.
The intraoperative results achieved highlight the potential impact the MasSpec Pen could have in advancing surgical care for patients. For Patient 15, for example, MasSpec Pen analyses of the pancreatic neck margin and bile duct body were performed in vivo (three analyses of neck margin, one of bile duct) and ex vivo (one analysis of the neck margin) during the Whipple procedure (Fig. 3B). All five analyses from both regions were predicted by Lasso as normal tissues, which agreed with intraoperative frozen section and the final pathologic results (both margins were negative for carcinoma) (Table 2). For Patient 6, four mass spectra corresponding to two in vivo normal pancreas analyses and two ex vivo surgical margin analyses were predicted on using our classification model. Of these, all benign pancreas and margin analyses were classified as normal by Lasso, with margins confirmed as negative for carcinoma via histopathological diagnosis. We also tested consecutive analyses of the same tissue region to explore predictive power of our models on analytical replicates. For example, three repeat ex vivo analyses of the pancreatic neck margin were performed for Patient 20 undergoing a Whipple procedure for PDAC resection, with all three analyses predicted as normal, in agreement with final pathology reports (Table 2). For Patient 16, two repeat ex vivo analyses of the common bile duct margin, a region ultimately determined as negative for malignancy, were performed. The first analysis was classified as normal, in agreement with pathology, while the second analysis of the same region was classified as PDAC, which disagreed with final pathology. While further data collection and refinements to the technology are expected to improve model performance, our results suggest that the models built with banked data and the resulting molecular predictors can be used to predict on independent data acquired from in vivo and ex vivo tissues and ultimately may improve the efficacy of surgical intervention for patients with pancreatic cancer.
Note that there are limitations to the clinical findings in this study. First, our statistical classifiers should be expanded to include other surgical margins, such duodenal, retroperitoneal, and uncinate, and cancers that are surgically treated with a Whipple procedure, such as neuroendocrine tumors and cholangiocarcinoma. Clinically relevant histological features including CP and precursor lesions and the effect of neoadjuvant treatment such as chemotherapy or chemoradiation on molecular profiles should also be evaluated. Given the rich molecular profiles obtained from preliminary intraoperative analyses of benign regional lymph nodes (SI Appendix, Fig. S3), we will further explore the potential of the MasSpec Pen to diagnose lymph node metastases through increased data collection and subsequent model generation. Importantly, tumor tissue was not physically exposed in the cancer surgeries evaluated in this study (n = 13), and all transection margins analyzed were negative by final postoperative pathology. Thus, while 91.7% agreement was achieved for all the negative margins, the lack of intraoperative data from positive margins inhibited performance evaluation for positive margin assessment. Larger cohorts of patients and multicenter studies are warranted for rigorous validation of the method and statistical models.
Our study provides evidence that the MasSpec Pen technology can be implemented in the OR for use by surgical teams, allowing effective molecular analysis of pancreatic tissues in vivo and ex vivo. Nevertheless, several technical challenges should be addressed to further refine the technology for intraoperative use (SI Appendix, Text). We observed that poor-quality mass spectra with biological peaks below detection limits were acquired for 14 tissue analyses, which were consequently excluded from statistical analysis. We speculate that the decrease in spectral quality may be due to variabilities in the tissue sampling and extraction process and are currently testing various approaches to improve robustness and signal reproducibility for intraoperative tissue analysis in the OR. Note that in this pilot clinical study, only a few tissue regions were rapidly analyzed in vivo and on freshly excised tissues to evaluate feasibility and diagnostic performance. Yet, as the surface area of the pancreatic neck margin and the common bile duct margin is ∼225 to 600 mm2 and ∼25 to 400 mm2, respectively, consecutive analyses (sampling area 5.73 mm2, ∼15 s each) may be necessary (40, 41). Even so, MasSpec Pen analyses of larger tissue areas are still conceivably faster than the typical time needed for frozen section analysis of the excised specimen with expert pathology staffing, with the benefit of potentially providing real-time diagnostic feedback and preventing unnecessary excision of healthy tissues in vivo. We plan to conduct additional investigations on optimizing pen tip diameters greater than 4 mm to further accelerate margin evaluation, as well as! determining the depth of molecular extraction in tissue to more strictly assess the capabilities of the technology for surgical margin evaluation.
Our results provide compelling evidence that the MasSpec Pen and statistical classifiers may be valuable for assisting surgeons in making efficient, informed clinical decisions on margin status during pancreatic cancer surgery in vivo and in freshly excised tissues. We foresee that the MasSpec Pen may offer particular diagnostic utility at institutions with limited access to subspecialized pathologists with expertise in pancreatic cancer (9, 42). Following rigorous validation in further studies, this technology holds immense potential for more rapid and precise intraoperative margin assessment, which could ultimately lead to reduced re-excision rates, patient distress, and healthcare costs.
Materials and Methods
Banked Tissue Samples.
Our study adhered to the Standards for Reporting of Diagnostic Accuracy guidelines (43) for diagnostic studies (SI Appendix, Fig. S4). We obtained 157 deidentified, frozen tissue samples, including PDAC, normal pancreas, and normal bile duct from Cooperative Human Tissue Network (CHTN) and Baylor College of Medicine (BCM) under approved Institutional Review Board (IRB) protocol (No. 2018-06-0094). Tissue samples were stored at −80 °C prior to analysis.
MasSpec Pen Method Development for Pancreatic Cancer Detection.
The MasSpec Pen coupled to a Q Exactive Orbitrap mass spectrometer (Thermo Scientific) was used to perform analyses of the first four batches of samples, while a Q Exactive HF Orbitrap mass spectrometer (Thermo Scientific) was used to analyze the last batch. Experiments in the laboratory with banked tissues were performed using a MasSpec Pen with a pen tip reservoir diameter of 2.7 mm and tubing length of 3 m. Samples were thawed and analyzed in ambient conditions. Data were acquired in the negative ion mode from m/z 120 to 1,800 using water as the solvent. Given the heterogeneity of pancreatic tumors and low density of tumor cells, tumor samples with larger surface areas (6 to 10 mm, diameter) were analyzed in two to three distinct regions, resulting in 172 analyses from 157 tissues. Following MS analysis, an ink stamp was applied to the tissue sample to demarcate region of analysis for correlation to mass spectra. Samples were then immediately flash frozen in liquid nitrogen and sectioned at 5- to 10-µm thickness with a CryoStar NX50 (Thermo Scientific). Tentative ion identifications were based on high mass accuracy measurements and tandem MS analyses (SI Appendix, Fig. S5).
Intraoperative Clinical Testing during Pancreatic Surgeries.
The pilot clinical tests were conducted at a hospital affiliate of Baylor St. Luke’s Hospital under approved IRB protocol (No. 2018-06-0094) from BCM and The University of Texas (UT) at Austin. Development and optimization of the MasSpec Pen system and materials for clinical use has recently been described (28). A Q Exactive Orbitrap mass spectrometer (Thermo Scientific) equipped with a home-built MasSpec Pen interface was installed in an OR. Prior to each surgery, surgical staff placed up to three autoclaved MasSpec Pen devices on the surgical field. Tubing length was adjusted to 4.5 m to fit the OR environment. One MasSpec Pen was reserved for ex vivo analyses and placed outside of the surgical field for handling by research personnel. UT research personnel connected the ends of each tubing to a MasSpec Pen control box interfacing the mass spectrometer. In vivo MasSpec Pen analyses were conducted by the attending surgeon G.V.B. or W.E.F. Ex vivo analyses were conducted by C.P. or K.B. immediately following excision of tissue. Data were not communicated to surgical staff, and no statistical classification was performed during surgery.
Patients were deemed eligible for study participation on the basis of being scheduled to undergo pancreatic surgery, independently of our study, conducted by surgeons G.V.B. or W.E.F. Prior to surgery, eligible patients were identified and notified of the study by research personnel C.P. or K.B. Patients who agreed to participate in the study gave written consent prior to surgery (n = 20). While 20 patients provided consent, the MasSpec Pen was used in cases from 19 patients, with no MasSpec Pen analyses able to be performed for Case 19. Of the 19 cases where MasSpec Pen analyses were performed, note that data were not acquired for one surgery (Case 10) as the system was found to be below operational conditions. A total of 46 intraoperative analyses collected from in vivo and ex vivo tissues were excluded due to poor-quality mass spectra (i.e., low signal [signal-to-noise ratio for biological species below 3] and excess interfering peaks [n = 14]) or due to tissue type not being represented by statistical models (e.g., lymph node or liver analyses [n = 32]).
Statistical Analysis.
Statistical analysis was first performed by J.Q.L. and independently repeated and verified by statistician S.W. Mass spectra corresponding to each sample analysis were averaged and imported to RStudio for statistical analysis. Background peaks were subtracted. Bile acids were detected in high relative abundance in banked bile duct tissue, which could potentially compromise statistical classification as surgical incisions may result in bile leakage onto other tissue types. We thus removed prominent bile acid species in a preprocessing step prior to Lasso analysis to increase the robustness of the model. The resulting m/z values were then binned to the 0.01 m/z, then the data were normalized either by the nonzero median or total ion count. Logistic regression was performed via Lasso penalized regression (27) using the “glmnet” package in the CRAN R library.
Classification models were built using MasSpec Pen data obtained from histologically validated tissue sections, which were evaluated blindly by expert, board-certified pathologists S.D. and W.Y. Tissues with clear pathologic diagnoses of normal or concentrated tumor cells were used to train statistical classifiers, from which Lasso selected a subset of features, or m/z values, to discriminate normal from cancerous tissue. Leave-one-out CV was used to assess model performance, providing diagnostic metrics including accuracy, sensitivity, and specificity. The normal pancreas versus PDAC model was used to predict on data in a validation set using pancreatic tissues with mixed, complex histopathology and/or lower quality or atypical mass spectra and an independent test. The normal bile duct versus PDAC model was used to predict on data in a validation set comprised of more heterogeneous bile duct and primary PDAC tissues and bile duct tissues invaded by PDAC. Accuracy, sensitivity, specificity, and 95% Wilson CIs (44) were calculated based on agreement of Lasso prediction results with postanalysis pathological evaluation.
For predicting disease status on in vivo and fresh ex vivo data obtained in the OR, we compared Lasso prediction results of regions of normal tissue that were uninvolved by lesion with gross surgical assessment. Lasso predictions on resected tissue specimens including margin regions and suspected tumor were compared with final postoperative pathology reports.
Supplementary Material
Acknowledgments
We thank the CHTN and BCM for providing banked tissue samples. We are extremely grateful to Tim Hooper for his assistance. This work was supported by the National Cancer Institute of the NIH under Awards R00CA190783 and 1R33CA229068-01A1, the Gordon and Betty Moore Foundation through Grant GBMF8049 to L.S.E., and the Welch Foundation through Grant F-1895 to L.S.E. M.E.K. is grateful to the NSF for a graduate fellowship.
Footnotes
Competing interest statement: M.E.K., J.Z., R.J.D., J.Q.L., C.L.F., J.S., and L.S.E. are inventors in US Patent 10,643,832 and/or in other patent applications related to the MasSpec Pen technology licensed by the University of Texas to MS Pen Technologies, Inc. J.Z., J.S., and L.S.E. are shareholders in MS Pen Technologies, Inc. J.S., L.S.E., and C.L.F. serve as chief medical officer, chief scientific officer, and a consultant, respectively, for MS Pen Technologies, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104411118/-/DCSupplemental.
Data and Materials Availability
The anonymized mass spectra data for this study have been deposited in Dataverse (DOI: 10.7910/DVN/P4CY2Z).
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Chakraborty S., Singh S., Surgical resection improves survival in pancreatic cancer patients without vascular invasion—A population based study. Ann. Gastroenterol. 26, 346–352 (2013). [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner M., et al., Curative resection is the single most important factor determining outcome in patients with pancreatic adenocarcinoma. Br. J. Surg. 91, 586–594 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Metildi C. A., et al., Fluorescence-guided surgery allows for more complete resection of pancreatic cancer, resulting in longer disease-free survival compared with standard surgery in orthotopic mouse models. J. Am. Coll. Surg. 215, 126–135, discussion 135–136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han S. S., et al., Analysis of long-term survivors after surgical resection for pancreatic cancer. Pancreas 32, 271–275 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Tummala P., Howard T., Agarwal B., Dramatic survival benefit related to R0 resection of pancreatic adenocarcinoma in patients with tumor ≤25 mm in size and ≤1 involved lymph nodes. Clin. Transl. Gastroenterol. 4, e33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adsay N. V., et al., Chronic pancreatitis or pancreatic ductal adenocarcinoma? Semin. Diagn. Pathol. 21, 268–276 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Lechago J., Frozen section examination of liver, gallbladder, and pancreas. Arch. Pathol. Lab. Med. 129, 1610–1618 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Liu Y. J., et al., Frozen section interpretation of pancreatic margins: Subspecialized gastrointestinal pathologists versus general pathologists. Int. J. Surg. Pathol. 24, 108–115 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Kooby D. A., et al., Value of intraoperative neck margin analysis during Whipple for pancreatic adenocarcinoma: A multicenter analysis of 1399 patients. Ann. Surg. 260, 494–501, discussion 501–503 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Nelson D. W., Blanchard T. H., Causey M. W., Homann J. F., Brown T. A., Examining the accuracy and clinical usefulness of intraoperative frozen section analysis in the management of pancreatic lesions. Am. J. Surg. 205, 613–617, discussion 617 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Soer E., et al., Dilemmas for the pathologist in the oncologic assessment of pancreatoduodenectomy specimens: An overview of different grossing approaches and the relevance of the histopathological characteristics in the oncologic assessment of pancreatoduodenectomy specimens. Virchows Arch. 472, 533–543 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lwin T. M., et al., Near-infrared tumor-specific fluorescence imaging of pancreatic cancer in orthotopic mouse models using the da-Vinci firefly imaging system. J. Am. Coll. Surg. 225, S194–S195 (2017). [Google Scholar]
- 14.van Manen L., et al., Validation of full-field optical coherence tomography in distinguishing malignant and benign tissue in resected pancreatic cancer specimens. PLoS One 12, e0175862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandya A. K., et al., Evaluation of pancreatic cancer with Raman spectroscopy in a mouse model. Pancreas 36, e1–e8 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., Sans M., Garza K. Y., Eberlin L. S., Mass spectrometry technologies to advance care for cancer patients in clinical and intraoperative use. Mass Spectrom. Rev., 10.1002/mas.21664 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Eberlin L. S., et al., Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 72, 645–654 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberlin L. S., et al., Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc. Natl. Acad. Sci. U.S.A. 111, 2436–2441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee S., et al., Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. U.S.A. 114, 3334–3339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberlin L. S., et al., Pancreatic cancer surgical resection margins: Molecular assessment by mass spectrometry imaging. PLoS Med. 13, e1002108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balog J., et al., Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med. 5, 194ra93 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Woolman M., et al., Picosecond infrared laser desorption mass spectrometry identifies medulloblastoma subgroups on intrasurgical timescales. Cancer Res. 79, 2426–2434 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Fatou B., et al., In vivo real-time mass spectrometry for guided surgery application. Sci. Rep. 6, 25919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzafetas M., et al., The intelligent knife (iKnife) and its intraoperative diagnostic advantage for the treatment of cervical disease. Proc. Natl. Acad. Sci. U.S.A. 117, 7338–7346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., et al., Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci. Transl. Med. 9, eaan3968 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sans M., et al., Performance of the MasSpec Pen for rapid diagnosis of ovarian cancer. Clin. Chem. 65, 674–683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibshirani R., Regression shrinkage and selection via the lasso. J. R.. Stat. Soc. B Met 58, 267–288 (1996). [Google Scholar]
- 28.Zhang J., et al., Direct molecular analysis of in vivo and freshly excised tissues in human surgeries with the MasSpec Pen technology. medRxiv [Preprint] (2020). 10.1101/2020.12.14.20248101 (Accessed 28 June 2020). [DOI]
- 29.Keating M. F., et al., Integrating the MasSpec Pen to the da Vinci surgical system for in vivo tissue analysis during a robotic assisted porcine surgery. Anal. Chem. 92, 11535–11542 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Feider C. L., et al., Molecular imaging of endometriosis tissues using desorption electrospray ionization mass spectrometry. Sci. Rep. 9, 15690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatima J., et al., Pancreatoduodenectomy for ductal adenocarcinoma: Implications of positive margin on survival. Arch. Surg. 145, 167–172 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network , Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Röder P. V., Wu B., Liu Y., Han W., Pancreatic regulation of glucose homeostasis. Exp. Mol. Med. 48, e219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang A. S., et al., HR-MAS MRS of the pancreas reveals reduced lipid and elevated lactate and taurine associated with early pancreatic cancer. NMR Biomed. 27, 1361–1370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamphorst J. J., et al., Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson A. D., Thematic review series: Systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: A global approach to lipid analysis in biological systems. J. Lipid Res. 47, 2101–2111 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Santos C. R., Schulze A., Lipid metabolism in cancer. FEBS J. 279, 2610–2623 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Shu X., et al., Prospective metabolomics study identifies potential novel blood metabolites associated with pancreatic cancer risk. Int. J. Cancer 143, 2161–2167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beger R. D., Schnackenberg L. K., Holland R. D., Li D. H., Dragan Y., Metabonomic models of human pancreatic cancer using 1D proton NMR spectra of lipids in plasma. Metabolomics 2, 125–134 (2006). [Google Scholar]
- 40.Adsay N. V., et al., Whipple made simple for surgical pathologists: Orientation, dissection, and sampling of pancreaticoduodenectomy specimens for a more practical and accurate evaluation of pancreatic, distal common bile duct, and ampullary tumors. Am. J. Surg. Pathol. 38, 480–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lal N., Mehra S., Lal V., Ultrasonographic measurement of normal common bile duct diameter and its correlation with age, sex and anthropometry. J. Clin. Diagn. Res. 8, AC01–AC04 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts D. J., Pathology: Functionality in resource-poor settings. Arch. Pathol. Lab. Med. 137, 748–751 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Bossuyt P. M.et al. and STARD Group , STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. BMJ 351, h5527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agresti A. C., Coull B. A., Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52, 119–126 (1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The anonymized mass spectra data for this study have been deposited in Dataverse (DOI: 10.7910/DVN/P4CY2Z).