Significance
How genes direct the development of the nervous system is a fundamental question in neurobiology. Here, we describe the gene networks that delineate cell fate and review the cell adhesion molecules used to guide axons and dendrites to the correct targets, and we discuss the mechanisms that cells use to migrate to their final destinations in the Drosophila visual system. We also draw comparisons between the fly and mammalian nervous systems that share common mechanisms used to guide their development. This allows us to suggest that principles emerging from the study of neuronal diversity, specification, and neural wiring in Drosophila are applicable to many systems and to most organisms.
Keywords: neural development, visual system, retina, Drosophila, patterning
Abstract
Like other sensory systems, the visual system is topographically organized: Its sensory neurons, the photoreceptors, and their targets maintain point-to-point correspondence in physical space, forming a retinotopic map. The iterative wiring of circuits in the visual system conveniently facilitates the study of its development. Over the past few decades, experiments in Drosophila have shed light on the principles that guide the specification and connectivity of visual system neurons. In this review, we describe the main findings unearthed by the study of the Drosophila visual system and compare them with similar events in mammals. We focus on how temporal and spatial patterning generates diverse cell types, how guidance molecules distribute the axons and dendrites of neurons within the correct target regions, how vertebrates and invertebrates generate their retinotopic map, and the molecules and mechanisms required for neuronal migration. We suggest that basic principles used to wire the fly visual system are broadly applicable to other systems and highlight its importance as a model to study nervous system development.
The visual system is integral to the detection and processing of environmental stimuli such as food, mates, or predators. Like auditory and somatosensory neurons, the visual system maintains point-to-point correspondence in space between sensory receptors and downstream processing centers, a phenomenon known as retinotopy for the visual system (1). Retinotopy facilitates the study of neural circuit development and function as it allows one to extrapolate general principles about visual system assembly by focusing on a single subunit. This, combined with the visual system’s accessibility, has made it one of the best-studied sensory modalities.
Drosophila has a long history as a genetic model organism used to study visual system development. Immunohistochemistry and molecular genetic experiments performed in Drosophila have unearthed some of the developmental mechanisms that build the visual system. Methodologies such as Golgi staining (2), highly specific enhancer trap lines (3, 4), single-cell RNA sequencing (5–8), and electron microscopy reconstructions (9–13) have allowed for careful morphological and molecular characterization of visual system components, providing both the roster of neuronal types as well as the identity of their synaptic partners (i.e., the connectome). Importantly, the neural stem cells of the Drosophila optic lobe (called neuroblasts) are generated as a wave of differentiation sweeps over a neuroepithelium, allowing one to simultaneously observe and compare neuroblasts of different ages at a single time point (14–16). Many concepts underlying Drosophila visual system development are readily applicable to other systems, such as the vertebrate visual system (17) and cortex (18).
Nervous system development follows a number of reproducible steps. Cell types must be specified and generated in the correct proportions. Neurons must target the correct (optic) ganglia and segregate their axons/dendrites into the correct target regions. Neurons of the same type must distribute their arbors across the topographic map. They must assemble themselves with stereotypic/columnar organization and project to the correct layers, and finally, upon reaching proximity to their synaptic partners, neurons must make the correct connections. All of these steps must be coordinated under precise spatiotemporal control.
Below, we describe the major concepts that have emerged from the study of the Drosophila visual system, with a specific emphasis on how the medulla optic ganglion is generated. The development of the retina (19–22), the formation of the optic neuroepithelia (23, 24), and the formation of neuropil layers (25, 26) have been discussed at length in numerous reviews and therefore, will not be mentioned further.
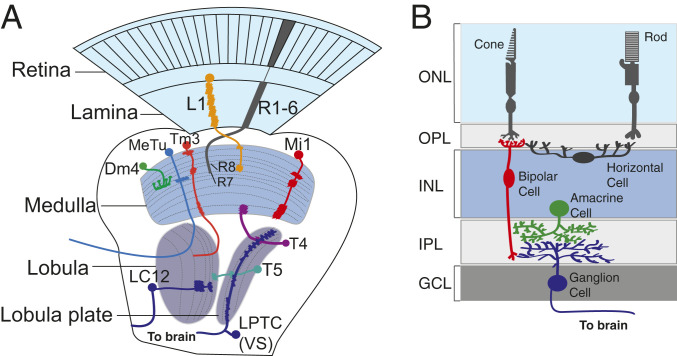
Optic Lobe Organization: A Primer
The visual system is the largest component of the fly nervous system; of the 100,000 neurons in the Drosophila brain, about two-thirds are devoted to sight (27). The visual system consists of the retina and four optic ganglia or neuropils: the lamina, medulla, lobula, and lobula plate (2). The retina consists of 800 unit-eyes or ommatidia, each of which is composed of eight photoreceptors (R1 to R8) (28); R1 to R6 specialize in image formation and dim light vision, while R7 and R8 are required for color vision and the detection of the vector of polarized light (29–31). Photoreceptors R1 to R6 project their axons to 800 lamina cartridges, while R7 and R8 project their axons to the 800 columns of the medulla (2, 32) (Fig. 1A). The medulla is the most complex neuropil in the optic lobe and consists of ∼100 distinct cell types organized into 10 layers (2, 32, 33). Medulla-intrinsic (Mi) neurons span the entire medulla, while Dm and Pm medulla neurons localize their arbors solely in the distal and proximal medulla, respectively (Fig. 1A). The 800 columns in the lobula are organized into six layers; the lobula plate has four. The medulla is connected to the lobula complex by transmedullary Tm/TmY neurons (Fig. 1A). The lobula plate is connected to the motion detectors in the medulla by T4 and TmY neurons and to the lobula by T5 neurons (2) (Fig. 1A). The lobula plate processes broad field motion (34, 35), while distinct visual features captured by the visual system are coded into lobula columnar (LC) neurons that send their projections to different optic glomeruli in the central brain (35, 36) (Fig. 1A). Lobula plate tangential system (LPTC) neurons such as vertical system (VS) neurons also connect the lobula plate to the central brain (Fig. 1A), and some medulla neurons also project their arbors to optic glomeruli in the central brain (37, 38). While the structure of the fly visual system does not appear to be evolutionarily conserved with mammals, there is some level of convergence (Fig. 1B). Like the fly, the mammalian retina contains light- and color-detecting photoreceptors that connect to regularly arranged interneurons. These photoreceptors synapse onto bipolar cells that, like the Drosophila lamina and Mi/Tm neurons, detect increments (on) or decrements (off) of light. Horizontal cells (like Drosophila lamina amacrine cells and several distal medulla neurons) integrate the input from multiple photoreceptor cells to bipolar cells. Like Dm, Pm, and medulla tangential neurons in the fly, amacrine cells are broadly arborizing neurons that modulate bipolar cell to retinal ganglion cell (RGC) signaling. Finally, bipolar cells synapse onto feature-detecting RGCs that, like fly LC neurons, project their axons to higher-order processing centers (17).
Fig. 1.
Drosophila and mammalian visual system organization. (A) Visual input in Drosophila is captured by photoreceptors divided into ∼800 ommatidia. Outer photoreceptor axons (R1 to R6) project to the cartridges of the lamina, while inner photoreceptor axons (R7 to R8) project to the medulla. Lamina neurons (e.g., L1) also project their axons to the medulla (orange–yellow). Medulla neurons can be divided into numerous classes. Mi neurons (Mi1; red) project their arbors throughout the entire medulla. Transmedullary neurons (e.g., Tm3; orange) connect the medulla to the lobula. Distal medulla (e.g., Dm4; green) neurons are multicolumnar and project arbors across multiple medulla columns. The lobula and lobula plate neuropils are responsible for processing different aspects of vision. T4 neurons (purple) connect the lobula plate to the proximal medulla, while T5 neurons (teal) connect the lobula/lobula plate, which processes broad field motion. LC (e.g., LC12) neurons (dark blue) project within the lobula and send an arbor to the central brain to process various visual features. LPTCs (e.g., VS neurons) are sensitive to wide-field motion and project their arbors to the central brain, as do medulla tubercule (bright blue) neurons. (B) Input to the mammalian visual system is captured by photoreceptors, which are categorized as dim light–sensing rods or bright light/color–sensing cones. Rod and cone photoreceptors synapse onto rod or cone bipolar cells (red), respectively. Horizontal cells (dark gray) integrate the input from multiple photoreceptor cells to bipolar cells. Bipolar cells (red) make synapses with feature-detecting RGCs (blue). RGCs project neurites to higher-order processing centers. Amacrine cells (green) modulate bipolar to RGC signaling. Like Dm neurons in the fly, they are broadly arborizing. Müller glia are integral to visual system processing but are not shown in the figure. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer.
Cell Fate Specification and Visual System Development
Proper visual system function requires the specification of distinct neural types in the correct numbers. The Drosophila adult visual system consists of roughly 60,000 neurons of about 200 different cell types (2, 6, 7, 33). During early larval development, the optic lobe primordium splits in half to form two crescent-shaped neuroepithelia: the outer proliferation center (OPC) and the inner proliferation center (IPC) (27, 39, 40). The OPC produces mainly neurons of the lamina and the medulla (27, 41), while the IPC generates some neurons of the lobula complex but also, the C neurons (which connect the lamina to the medulla) and the T neurons (which connect the medulla/lobula plate to the lobula) (27, 42, 43).
During larval development, a neurogenic wave moves from the medial edge of the OPC toward the lateral edge, causing the conversion of neuroepithelial cells into neuroblasts (14–16, 44). Medulla neuroblasts divide multiple times asymmetrically to regenerate themselves and produce a ganglion mother cell (GMC), a transit-amplifying cell that divides asymmetrically just once to generate two different neurons or glia in a Notch-dependent manner (45, 46). The diversity of neurons produced in the Drosophila medulla is regulated by the coordination of temporal and spatial patterning, as well as by Notch-mediated binary fate choice. The role of these mechanisms in regulating medulla neuron fate is described below and is compared with similar mechanisms used in vertebrates.
Temporal Patterning Generates Neural Diversity.
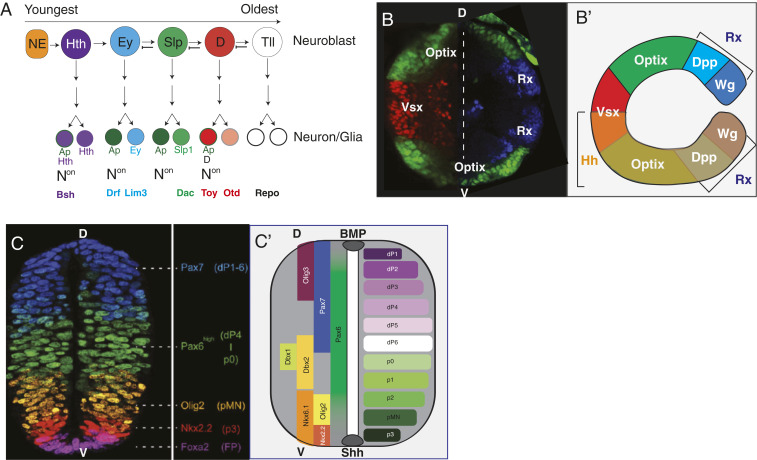
Production of a nervous system with the correct numbers and types of cells requires large-scale coordination of cell fate specification. The sequential expression of transcription factors in a temporal cascade in neural precursors (known as temporal patterning) is a widely used strategy for generating neural diversity. In the Drosophila visual system, OPC neuroblast clones generate long columnar chains of neurons spanning the medulla cortex, hinting that an individual OPC neuroblast divides multiple times to generate neurons of distinct fates (47, 48). Antibody screens identified the temporal transcription factors (tTFs) Homothorax (Hth), Eyeless (Ey), Sloppy Paired (Slp), Dichaete (D), and Tailless (Tll), which are sequentially expressed in rows of three to five neuroblasts in a temporal sequence (49, 50) (Fig. 2A). The tTFs Ey, Slp, D, and to an extent, Tll are required for proper sequence progression, and OPC tTFs cross-regulate each other. With the exception of Hth, each of them turns off the expression of the preceding factor (49, 50) and is required to activate the expression of the following one (49) (Fig. 2A). Consequently, tTFs exhibit partially overlapping expression patterns. As neuroblasts express each tTF over multiple divisions, simultaneous expression of two tTFs may be sufficient to delineate additional temporal windows (49). In the final divisions, Tll+ neuroblasts express the transcription factor Prospero (which is normally expressed only in GMCs) to instruct them to undergo a terminal cell division that produces two glial cells. Similar to other lineages in flies and vertebrates, OPC-derived glia and neurons do not arise from distinct classes of neuroblasts but rather, from the same progenitors, with glia arising in the terminal neuroblast divisions (49, 51, 52). Thus, temporal patterning can generate more than a dozen temporal windows with a combination of tTFs and provides the first layer of regulation to specify at least as many distinct neurons.
Fig. 2.
Spatiotemporal patterning/Notch signaling provide neuronal diversity in the visual system. (A) Drosophila OPC medulla neuroblasts express transcription factors in a temporal cascade whose output defines the identities of neurons born from those cells. Notchon/off signaling in the daughters of dividing Ganglion Mother Cells further diversifies neuronal fate. (B, B′) The OPC neuroepithelium can be subdivided into multiple regions based on the expression of transcription factors: Rx (blue) is expressed at the tips, Optix (red) is expressed in the middle, and Vsx (green) is expressed in the center. Image was generated by splicing two images (one of Vsx and Optix and the other with Optix and Rx) together. Growth factor Hh (B′, orange) is expressed in the ventral OPC, whereas the Rx domain can be subdivided into Wg and Dpp expressing regions. Dotted line: splice site. (C) Immunofluorescence image of spatially regulated transcription factor (TF) expression, which regionalizes progenitors to allow for the generation of different classes of spinal cord neurons. (Adapted with permission from ref. 99.) (C′) Bone morphogenetic protein (BMP) morphogen signaling activates the expression of basic helix-loop-helix (bHLH) TFs such as Olig3 (violet) to pattern the six classes of neurons in the dorsal spinal cord. Shh signaling suppresses the expression of Class I homeodomain TFs (e.g., Pax7, Dbx1, Dbx2, and Pax6) while activating the expression of Class II factors (e.g., Nkx2.2). The intersection of these gene expression patterns demarcates five progenitor classes in the ventral spinal cord.
Temporal patterning is used for neuronal specification in other contexts. The best-studied model for temporal patterning is the Drosophila embryonic ventral nerve cord (VNC), which is generated from 30 different neuroblasts per hemisegment (53). As neuroblasts age, they generate a temporally patterned subset of neurons and glia. The transcription factors Hunchback (Hb), Krüppel (Kr), Pdm1/2, Castor, and Grainy head are expressed sequentially by each neuroblast and specify this patterning (54). As with OPC neuroblasts, temporal series progression is cell intrinsic (55) and is mediated by inhibition of the previous factor by the following one (56). However, unlike the Drosophila visual system, the absence of these tTFs is not sufficient to stop the progression of the temporal cascade. For example, transition out of the first temporal state (Hb) is not dependent on Kr expression but rather, appears to be due to Hb depletion in a cell cycle–dependent manner (55). At the end of the VNC neuron temporal series, neuroblasts are decommissioned either by apoptosis or by entering quiescence (57).
In the vertebrate visual system, retinal progenitor cells (RPCs) sequentially produce all retinal cells via biased temporal patterning. RPCs undergo three types of divisions: symmetric, asymmetric, and differentiative (58–60). Live imaging and cultured cell experiments involving individual RPCs suggest that multipotent RPCs toggle between these division modes to generate all retinal cell types using stochastic, biased, cell-intrinsically regulated divisions (61–63). Despite the stochasticity inherent in RPC divisions, a crude temporal map exists. The widely overlapping temporal windows produce in a relatively stable order RGCs, horizontal cells, cones, amacrine cells, rods, bipolar cells, and finally, Müller glia. Furthermore, each cell type is composed of dozens of subtypes that appear to be produced in a temporal order; for example, GABAergic amacrine cells are consistently generated 2 to 3 d before glycinergic amacrine cells, and cone bipolar cells are born before rod bipolar cells (64, 65). Molecularly, the Atonal homolog Math5 is necessary in RPCs during their terminal division to promote early-born RGC fate and inhibit later-born (cone and amacrine) cell fate (66, 67). Temporal patterning is also regulated by the heterochronic pathway microRNA let-7, as well as the microRNAs (miRNAs) mir-126 and mir-9; these miRNAs increase their expression in later progenitors to promote the expression of the conserved heterochronic gene Lin-28b and the immunoglobulin superfamily (IgSF) protein protogenin (68). Overexpression of these genes generates an excess of later-born neuron types, indicating that these miRNAs are sufficient to advance the developmental clock in RPCs. Classical temporal patterning genes, such as Hb and Castor orthologs (which specify Drosophila VNC early vs. late neural fate), are also used to regulate mammalian retinal neuron fate. Expression of the early factor Ikaros/Hb in RPCs (as well as in mature neurons) is required to produce retinal neurons from early windows, while Casz1/Castor is required for late windows (69, 70). Recent evidence also suggests that the temporal factor Pou2f1/2(Pdm) is required in RPCs for the specification of cones produced in the intermediate period. Ikaros overexpression increases Pou2f1 expression, and Pou2f1 expression represses Casz1 (71); in addition to Pou2f1/2, the Forkhead box transcription factor FOXN4 (which resembles the tTF Slp) also activates Casz expression and represses Ikzf1 expression to promote mid- to late cell types within the early temporal windows (amacrine, horizontal, cone, and rod cells) at the expense of RGC production (72). This suggests that these two genes collaborate to specify an intermediate temporal window. Thus, an evolutionarily conserved temporal series of genes appears to regulate temporal patterning of retinal fate.
The patterning of the vertebrate cortex also resembles the temporal patterning seen in the Drosophila nervous system. The cerebral cortex is organized into six horizontal layers with columnar organization. Like the OPC neuroepithelium, radial glial progenitors (RGPs) first undergo symmetric divisions to expand the progenitor pool and then, divide asymmetrically as stem cells to sequentially produce cortical neurons (73). 3H-Thymidine labeling experiments showed that excitatory projection neurons residing in deeper cortical layers are born first, while upper layers are generated later (74). Recent clonal labeling experiments corroborate this and suggest that the members of a single cortical column are born from a single RGP that divides on average eight times to produce one to two neurons per layer. Clones are smaller and only produce superficial-layer neurons when induced later in development, indicating that, like neuroblasts, RGPs have a fixed output (75). Cultured RGPs and heterochronic transplants of older RGPs in younger cortices suggest that most RGP temporal patterning is cell intrinsic (76, 77). However, recent scRNAseq experiments using tagged neurons and their progenitors showed that a class of RGPs (later-born apical RGPs) born at E15.5 can be reprogrammed to an earlier RGP fate in a Wnt-dependent manner when transplanted to E12 animals (78, 79). However, later-born intermediate progenitors do not exhibit this plasticity. Intriguingly, ectopic expression of the tTF Ikaros/Hb is sufficient to specify early temporal fate in RGPs (and possibly, in neurons) as ectopic Ikaros expression in RGPs results in greater numbers of deep-layer cortical neurons; however, ikaros knockouts have no cortical defects, indicating that the gene is sufficient but not necessary (80).
So far, none of the temporal patterning genes expressed in the Drosophila optic lobe appear to be utilized to promote temporal fate in vertebrates. Despite this, the homologs of Hth, Ey, Slp, D, and Tll (i.e., Meig, Pax6, FoxG1, Pax2, and Tlx, respectively) are expressed in the developing vertebrate eye and show similar regulatory relationships. For example, the Hth homolog Meis activates Ey/Pax6 expression to promote lens development (81), and in slp/foxg1 mutants, Ey/Pax6 expression rises, increasing the size of the ciliary margin in the nasal retina. Pax6 is also required to up-regulate FoxG1 expression required for nasotemporal patterning of the optic vesicles (82). Finally, the Tll homolog Tlx represses Pax2/D expression in the developing eye (83). Thus, although Drosophila optic lobe tTFs have not been shown to function in the same manner in vertebrates, it is intriguing that their genetic relationships are conserved.
Notch Signaling Increases Medulla Neuronal Diversity.
After spatiotemporal patterning is used to determine the fate of neuronal progeny, Notch signaling is used in the following asymmetric single-GMC division to diversify the fates of the two GMC daughter cells. Notch signaling canonically acts through lateral inhibition; increased expression of the transmembrane ligand Delta on one cell causes an increase in Notch transmembrane receptor concentration in the neighboring cell, thus increasing its ability to respond to and signal through Delta and thereby, allowing two adjacent cells to adopt distinct fates, one Notchon and one Notchoff (84). The asymmetry in the Notch signal may also be regulated by asymmetric distribution of Notch inhibitors (such as Numb) into one side of a progenitor, thereby causing an increase in Notch expression in only one daughter cell (85, 86). This Notchon/Notchoff decision is utilized in nearly all GMCs across the nervous system and generates two neurons that acquire different fates (Fig. 2A); this effectively doubles the number of neural types produced at each temporal window. In a number of lineages, one of the Notchon or Notchoff sister cells is fated to undergo programmed cell death, resulting in a “hemilineage” where just one cell type is produced per GMC division (46, 87, 88).
In the vertebrate retina, asymmetric Notch signaling is also used to diversify cell fate. Two mechanisms regulate the asymmetry of Notch inheritance in RPCs. As in invertebrates, Numb is symmetrically inherited in symmetrically dividing cells but is asymmetrically inherited in asymmetrically dividing cells. The role of Numb here is somewhat subtle; numb mutants do not exhibit gross cell fate defects but cause a change in the ratios of cell types produced (89). Similarly, overexpression of Notch itself produces excess Müller glial cells (90, 91). Aside from asymmetric partitioning of Numb in the daughter cells, the ubiquitin ligase Mindbomb acts in the neighboring retinal pigmented neuroepithelia (RPE) to promote asymmetric Notch localization in RPCs. Mindbomb expression in the RPE degrades Notch to enhance apical RPE Delta localization, which itself induces apical RPC Notch localization (92). Thus, asymmetric inheritance of Notch in RPCs alters the ratios of retinal cell types produced.
Spatial Patterning of the Drosophila Medulla.
The diversity generated by binary combinations of temporal factors and Notch-dependent asymmetric GMC divisions can only account for the specification of less than half of the 100 medulla cell types (49). The remaining diversity relies on the contribution of spatial patterning or information imparted by neuroblast birth region. The OPC neuroepithelium is spatially divided into several regions defined by the expression of different transcription factors or signaling molecules that will instruct the fate of neurons born from each region. However, the expression of most of these genes remains restricted to the neuroepithelium and is turned off in neuroblasts; thus, it remains to be determined how neurons remember their spatial origins. The Chx10 homolog Vsx1 is expressed at the center of the OPC crescent (89), the SIX-family transcription factor Optix is expressed in the regions flanking Vsx (90), and the retinal homeobox Rx is expressed at the tips (41) (Fig. 2B). The Rx region is further subdivided into two subregions that express the signaling molecules Wingless (Wg) or Decapentaplegic (Dpp/BMP); Hedgehog (Hh) is expressed solely in the ventral half of the OPC (Fig. 2B′). Thus, one can define at least eight distinct OPC spatial domains (Fig. 2B). These genes are expressed in their respective domains as early as the embryonic optic anlage and appear to correspond to compartments of restricted lineages inherited from early patterning of the embryo (41, 89, 90). For instance, a lineage trace for Hh marks the OPC ventral domain, although Hh is no longer expressed in the OPC during neuroblast formation (41, 91). Vsx, Optix, and Rx are expressed in a mutually exclusive manner, and ectopic expression of Vsx and Rx causes loss of Optix expression (41). Although these are the only spatial factors that have been described, neuron labeling experiments during larval development suggest that more spatial subdivisions exist. For example, one Dm8 neuron subtype arises from roughly one-third of the Optix region, indicating that this neuroepithelial domain may have spatial subdivisions (92).
In all OPC subregions except the Wg region (where it differs slightly), the tTF cascade remains the same, although different spatial regions produce different neurons; this suggests that spatial factors modify the outcome of a common temporal pattern (41, 46). How do spatial and temporal patterning intersect to produce the 100 medulla neuron types? Medulla neurons can be divided into two separate subtypes; unicolumnar neurons exist at a 1:1 ratio of neurons to columns and restrict their arbors to a single column, while there are anywhere between 5 and 800 each of multicolumnar neurons, which project their arbors across multiple medulla columns (41). Fate mapping experiments identifying neurons from the Hth window show that neurons with a 1:1 ratio of neurons to columns arise from each of the roughly 800 medulla neuroblasts, while less numerous neurons are only produced in smaller numbers by neuroblasts from restricted spatial domains. This suggests that some neurons ignore spatial information, while others do not. Differences in response to spatial patterning also appear to be partly mediated by differential Notch activity in the two daughters of the GMC; unicolumnar Mi1 neurons generated by all neuroblasts are Non, while multicolumnar Pm3 neurons (generated only by Vsx1 neuroblasts) are Noff (41). Thus, Notch signaling allows neurons to utilize or ignore spatial inputs, providing a downstream mechanism by which spatial information specifies neurons at lower stoichiometry. Although this model describes broadly how different numbers of neurons are generated, it fails to account for more granular regulation of cell number. Thus, future experiments will detail the mechanisms underlying the regulation of cell number.
Like OPC neuroblasts, Drosophila embryonic VNC neuroblasts integrate temporal patterning with spatial information. VNC neuroblasts are arranged into rows and columns, each of which expresses a distinct spatial factor (93, 94). Spatial and temporal factors do not act independently; rather, it has been shown that spatial factors modify chromatin accessibility in the neuroepithelium so that the same temporal factor (Hb in this case) binds distinct transcriptional targets in each neuroblast. This allows each neuroblast to have its own transcriptional output and therefore, to produce diverse cell fates (95). Interestingly, temporal factors also appear to epigenetically prime neural stem cells to promote rod fate in the vertebrate retina. Proteomic experiments suggest that the tTF Casz1 interacts with the NuRD histone deacetylase complex and Polycomb repressors to suppress glial fate and promote rod fate. Treatment of Casz1 overexpression lines with histone deacetylase inhibitors decreases rod production, indicating that Casz1 exerts its function on chromatin remodeled by nucleosome remodeling and deacetylase (NuRD) (96). In the vertebrate cortex, tTFs also use Polycomb-mediated chromatin remodeling to encourage radial glial cells to transition from a more cell-intrinsically directed developmental state to one that is more receptive to external cues (79). It is unknown the extent to which OPC neuroepithelial cells are epigenetically primed by spatiotemporal factors.
In the developing vertebrate spinal cord, morphogenetic gradients establish regions of spatially subdivided transcription factor expression that promote cell fate. BMP secreted from the roof plate promotes specification of dorsal sensory interneurons, while secretion of Sonic Hedgehog (Shh) from the floor plate specifies motoneuron/ventral interneuron fate (Fig. 2 C and C′). Downstream of morphogen gradients, transcription factors are expressed in slightly overlapping spatial regions to generate distinct neural subdomains (97). A set of transcription factors (including the bHLH factors Olig3, Ascl1, and Ngn1/2) spatially patterns the six classes of neurons in the dorsal spinal cord, while Shh activates (Class II; e.g., Nkx2.2) and represses (Class I; e.g., Pax7, Dbx1, Dbx2, and Pax6) two classes of Homeodomain transcription factors in discrete patterns to demarcate five progenitor classes in the ventral spinal cord (Fig. 2 C and C′) (98, 99). As in Drosophila, spatial patterning appears to regulate tTF expression to further define the fate of newborn neurons. In all spatial domains, Onecut-family TFs define the earliest temporal windows, the Pdm-homolog Pou2f2 and Zfhx2-4 promote middle-born fate, and the factors Nfia/b/x and Neurod2/6 are expressed last. Like Drosophila mushroom body neuroblasts, which use extrinsic Activin signaling to help time temporal transitions, the vertebrate spinal cord utilizes TGF-β (transforming growth factor β) to regulate the length of each window (100–102). Thus, spatial patterning promotes the regulation of neural fates in the vertebrate spinal cord.
Neuron Targeting and Visual System Assembly
Do Pioneer Neurons Dictate Neuropil Targeting?
After visual system neurons are generated, they need to innervate the correct neuropils to find their target neurons in the proper layers and columns. For the most part, the neuronal types composing each neuropil are generated in proximity to where they will act, thereby simplifying neural ganglia assembly. The construction of each neuropil requires the arrival of pioneer axons and the fasciculation of follower axons (103, 104). In the case of laminated neuropils such as the medulla, pioneer neurons appear to serve a dual function; they might first delimit where the neuropil itself will be placed and may later serve as placeholders for “protolayer” formation during development. These protolayers will later split into more permanent layers that will compose the final laminated structure of optic lobe neuropils as more neurons intercalate within each neuropil during pupation (105).
Which neurons act as the pioneers for each optic ganglion? Photoreceptors appear to be required to pioneer the lamina neuropil, as the lamina is entirely missing in the absence of photoreceptor innervation; in fact, photoreceptors not only define the lamina but also, induce the differentiation and diversity of its five neuron types (106–109). Photoreceptors and lamina glia likely serve together as the placeholders for lamina neuropil formation; glia require photoreceptor input to migrate, while R1 to R6 require the same glial cells to tell them where to stop (110, 111). Unlike the lamina, the medulla neuropil and lobula complex remain relatively intact in the absence of photoreceptor innervation (106, 112), and clones inducing lamina neuron death have limited impact on medulla formation (112). However, mutants lacking photoreceptors (i.e., sine oculis mutants) lack a separation between the proximal and distal medulla, indicating that photoreceptors or lamina neurons may be required for the assembly of certain medulla components (106). Similarly, the Mi neuron Mi1 appears to be responsible for building the medulla layers M9 and M10 (i.e., proximal medulla), as clones removing Mi1 neurons lead to a complete disorganization of these layers (112). As Mi1 is one of the firstborn neurons in the medulla, it is a good candidate for demarcating the physical placement of the neuropil (41, 47, 49).
Which neurons pioneer the lobula complex? Like Mi1 neurons, Tm/TmY transmedullary neurons are generated at early time points and are the only lobula neurons that exist at a 1:1 ratio of neurons to columns (2). Thus, they might be good candidates for lobula pioneers, although this has never been shown (113). Finally, the lobula plate is likely pioneered by T4/T5 neurons, as mutants that prevent specification of subsets of T4 or T5 neurons result in the removal of the entire corresponding lobula plate layers where these neurons normally project. Furthermore, lobula plate tangential cells, which are the synaptic partners of T4/T5, fail to target in the absence of T4/T5 (43, 114, 115). Therefore, T4/T5 neurons are required for both the formation and layering of the lobula plate neuropil.
Although it is not known which neurons pioneer the vertebrate visual system, studies in the mouse visual system suggest that later-born neurons may use pioneer neurons to reach their proper targets. Earlier-born RGCs initially overshoot their targets in the visual system and transiently sample multiple sites. Upon making contact with their synaptic partners, they remove inappropriately targeting cells through axon retraction and programmed cell death. In contrast, later-born RGCs project their axons directly to their target sites and do not overshoot them (116). This suggests that later-born RGCs have axon guidance input from pioneer axons that allows them to target more accurately than earlier-born cells.
Glial Cells Enforce Neuropil Placement and Separation.
Neurons produced by the same lineages most often target their axons to the same optic ganglia. During Drosophila visual system development, OPC neuroblasts produce neurons belonging to the lamina and medulla neuropils (although some LC neurons are also generated from the tip of the OPC crescent) (41, 46, 49). Lobula complex–targeting neurons (e.g., C2, C3, T4, T5, and others) are born from the IPC (42, 43, 114, 115), and the remaining lobula complex neurons are born from central brain neuroblasts (117). The close proximity of the OPC and IPC puts the developing lamina and lobula complex neurons in direct apposition during larval development (118). As a result, the presence of physical and molecular boundaries—set up by rows of glial cells—is required to prevent their mixing.
One way in which glial cells prevent neuropil mixing is by secreting Slit, which is also secreted by medulla neurons. Glia-expressed Slit prevents Robo-expressing IPC neurons from mixing with the lamina precursor cells in the OPC; mutants in these genes cause lobula complex neurons born from the IPC (C or T neurons) to inappropriately invade the developing larval lamina and bisect the lamina and medulla neuropils (118, 119).
Netrin signaling from the IPC is also required to prevent the fusion of neighboring neuropils; disruption of Netrin signaling causes IPC neuroepithelial cells to invade the OPC, causing a fusion of the lobula and medulla neuropils during adulthood (120). Netrin is secreted by the IPC, while the Netrin receptors Frazzled and Unc5 are expressed in lamina glial cells that border the OPC. Thus, glia express Netrin receptors as a buffer to prevent neuroepithelial mixing. Mathematical modeling integrating Slit and Netrin signaling suggests that Netrin acts as both an attractant (through Frazzled) and repellent (through Unc5) to ensure the correct amount of spacing between the OPC and IPC, thereby preventing neuropil mixing (120). Thus, glia act as a barrier around the developing neuropils to maintain their placement and separation during development (121).
Cell Migration in the Visual System.
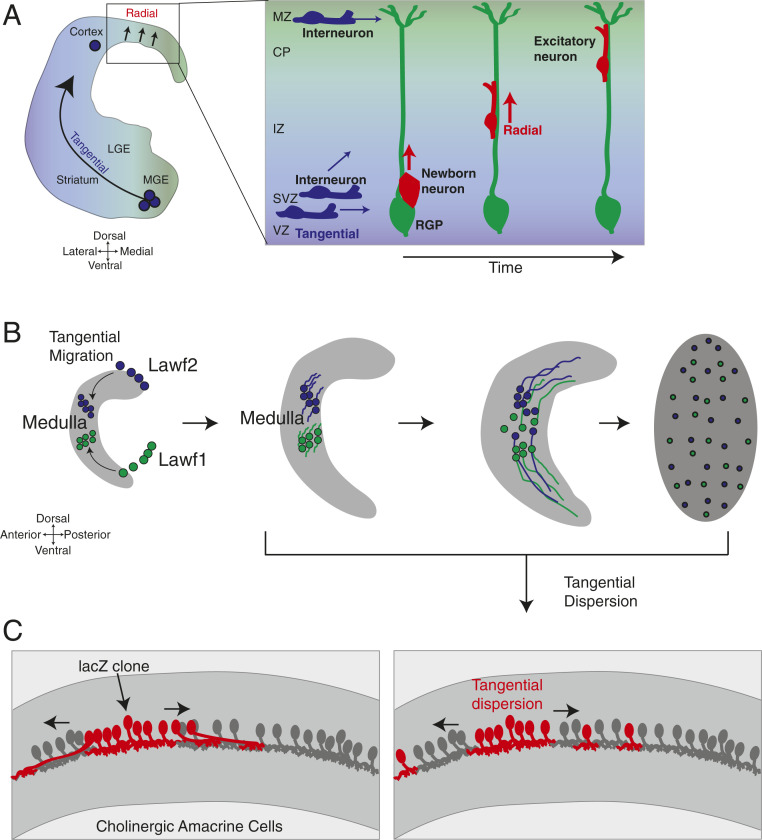
Following cell fate specification, some neurons migrate long distances to reach their final locations. Most cell migration studies in vertebrates entail two types of migration: radial migration, which involves parallel movement of neurons along a glial fiber (e.g., the movement of excitatory cortical projection neurons along RGPs in the cerebral cortex), and tangential migration, which involves the long-distance migration of neurons tangential to the cortical layers (e.g., migration of inhibitory interneurons in the cerebral cortex) (Fig. 3A) (122, 123). Morphologically, migrating neurons exhibit two major hallmarks: leading process extension (in which a neurite is polarized in the direction of movement) and nucleokinesis (in which the nucleus translocates into the leading process and drags the cell body in the direction of movement) (122).
Fig. 3.
Cell migration is a conserved phenomenon utilized across numerous neural types and many species. (A) Inhibitory interneurons born in the Medial Ganglionic Eminence (MGE) of the ventral telencephalon migrate tangentially into the cortical wall before dispersing in the cortical plate. (Inset) Excitatory cortical neurons are born from RGPs and radially migrate along glial fibers to reach their appropriate cortical layer. (B) Like inhibitory interneurons, developing Lawf neurons in Drosophila migrate tangentially to reach the medulla cortex. Later, Lawf1 and Lawf2 tangentially disperse across the medulla to reach their final positions. (C) Mosaic clones of ChAT-expressing mouse retinal amacrine cells (red) tangentially disperse away from their siblings (Right) to find their final positions; as a result, they are found intermingled among transgene-negative (gray) amacrine cells (186). Adapted from ref. 186, with permission from Elsevier. ChAT, choline acetyltransferase; CP, cortical plate; IZ: intermediate zone; LGE, lateral ganglionic eminence; MZ, marginal zone; SVZ, subventricular zone; VZ, ventricular zone.
Neurons of the Drosophila visual system also undergo cell migration, although very few of them do it. A subclass of lamina neurons (lamina wide field 1/2 [Lawf1/2]) and glia (epithelial/marginal [eg/mg]) generated from the same common progenitor migrate tangentially from their place of birth (from the lamina region at the tip of the OPC crescent) along the entire innermost concentric zone of the medulla cortex to reach the center of the medulla (110, 124–127) (Fig. 3B). This tangential migration appears to utilize the polarized extension of a leading process, indicating that they may use some of the same molecular mechanisms as vertebrate neurons to tangentially migrate. Early experiments suggested that guidance cues for eg/mg glia migration emanate from photoreceptors (128). However, Lawf neurons appear to use different mechanisms to migrate, as mutants lacking the ventral portion of the eye show disrupted eg/mg glial migration but show normal specification and migration of Lawf1/2 (124). Slit and Robo appear to be important chemotactic cues for Lawf migration. Slit shows expression in Lawf cells, while Robo3 shows expression in a concentric ring of medulla neurons (which includes Mi1 neurons) along the migratory path (127). It is believed that Slit acts as a ligand to generate repulsive cell–cell interactions to promote proper migration. In line with this hypothesis, slit and robo3 mutants exhibit disrupted Lawf migration. Similarly, Slit/Robo signaling is also required to regulate filopodia formation required for radial migration of vertebrate cortical neurons (129).
Later in pupal development, Lawf1/2 neurons undergo another type of cell movement using an unknown mechanism; their arbors spread along the entire retinotopic map before the cell bodies distribute evenly across the entire medulla to their final positions (124). This form of noncanonical cell movement resembles another form of cell movement—tangential dispersion—observed in vertebrate amacrine and horizontal cells. These cells first project their arbors to their final locations and then later in development, tangentially disperse their somas to match the spacing of their arbors (130, 131) (Fig. 3C). Amacrine neurons can migrate significant distances—as much as 145 μm away from their birthplace. Like canonically migrating neurons, tangentially dispersing somas extend small, laterally projecting neurites that provide directionality to the moving soma, suggesting that this is indeed an active migration process, although little is known about how this is regulated (131). Medulla multicolumnar neurons also appear to undergo tangential dispersion. These neurons project their neurites from their restricted places of birth to their final destinations during early pupation but later, begin to disperse their cell bodies throughout the dorsoventral (DV) axis of the medulla at around 24 h into pupation (41, 47, 48, 105). Although it is not known whether these neurites extend a leading process, this dispersion also appears to be an active process as morphogenetic movements in the optic lobes (i.e., lamina rotation) do not appear to play a role in this migration (48). Molecules responsible for tangential migration have not yet been identified, so the tractable genetics of Drosophila may identify conserved regulators used in this process.
Establishment of Retinotopy.
Topographic maps are found in all sensory systems and ensure that the quality and/or spatial origins of inputs from the environment are preserved when sensory neurons transmit their information to higher processing centers (1). The organization of these systems into columns or discrete units allows organisms to interpret sensory information about the location or type of stimuli. In the visual system, photoreceptors preserve the representation of the visual field and project their axons retinotopically to downstream processing centers such as the medulla (fly) or to the neurons in the outer plexiform layer of the retina (mouse). Vertebrate RGCs must also maintain their organization in the superior colliculus/tectum. For retinotopic assembly to occur in the visual system, photoreceptors and downstream visual system neurons must maintain the correct anteroposterior (AP) and DV axes of origin of the retinal image.
In the fly, photoreceptors project their axons to the lamina and medulla in an ordered arrangement to promote retinotopic circuit assembly. In the AP axis of the medulla, birth order regulates the targeting order of photoreceptor axons. As photoreceptors are recruited sequentially with the posterior-to-anterior movement of the morphogenetic furrow, they project their axons to the corresponding regions of the lamina (outer photoreceptors) and medulla (R7 and R8 color photoreceptors) (27, 132). Medulla photoreceptor entry appears to be partly mediated by cell adhesion molecules. At the anterior edge of the medulla, the youngest R8 neurites express a pulse of the transcription factor Sequoia, which leads to elevated levels of the cell adhesion molecule Capricious (133). The same is true for the cell adhesion molecules NCadherin (NCad) and Flamingo, which are also transiently expressed in R8 axons as they enter the medulla, with strongest expression in the youngest axons (134, 135). R8s retinotopically sort themselves as they enter the medulla by comparing cell adhesion molecule levels among earlier-born neurons. In addition to cell adhesion, paracrine Insulin receptor signaling feeds onto Dock/Pak to promote axon guidance and thus, retinotopy in the medulla; dock/pak mutants display gross axon guidance defects, leading to photoreceptor axon disorganization and loss of retinotopy (136–138).
Recruitment of neuroblasts by the passing of the neurogenic wave in the OPC must be synchronized with the morphogenetic furrow sweeping across the eye disc, thereby allowing for retinotopic assembly of photoreceptors and medulla neurons into corresponding columns. Similarly, in the lobula plate, memory trace experiments suggest that T4/T5 neurons are generated sequentially and are added to the lobula plate in correspondingly posterior-to-anterior retinal positions, indicating that birth order also acts to establish retinotopy in one axis of the lobula plate (43).
Less is known about how retinotopy along the DV axis is achieved. Genetic screens in Drosophila have identified Wnt4 as one of the signaling pathways required for the DV targeting of photoreceptors in the lamina. DWnt4 is expressed in the ventral lamina, while ventral photoreceptors are selectively responsive to Wnt4 signaling (139, 140). However, the disruption of retinotopy in DWnt4 mutants is not dramatic, and the mechanism of action by which these genes regulate retinotopic mapping is not known.
In vertebrates, the retinotopic organization of the visual system is set up by two orthogonal Ephrin gradients in the superior colliculus/tectum; EphrinA is expressed in a gradient along the AP axis, while EphrinB is expressed in a lateromedial gradient. Corresponding gradients of their cognate Eph receptors exist in the retina: EphA is expressed in a nasotemporal gradient, and EphB is expressed in a DV gradient. These gradients act in a concentration-dependent manner to promote growth cone repulsion, thereby providing a coordinate system to direct retinotectal mapping (141). However, unlike in vertebrates, Drosophila Eph does not appear to play an important role in the establishment of retinotopy, indicating that other molecules/mechanisms may be used (142).
Column Formation and Spacing.
Each neuropil in the visual system is organized into discrete layers and columns. The retinotopic organization of the Drosophila visual system into roughly 800 columns allows for faithful parallel transmission of spatial information across distinct layers. To generate columnar units, a number of challenges must be addressed. First, the number of columns must be the same from neuropil to neuropil; this is likely dictated by photoreceptor number. Supporting this model, Dm8 medulla neurons are generated in excess and are pared back to match the number of photoreceptor columns via programmed cell death (92). Second, neurons in each column must assemble in the correct position to ensure proper circuit function. Finally, neurites must properly restrict their size to prevent redundant sampling yet still allow for intracolumnar communication.
Differential cell adhesion regulates the internal organization of axons and dendrites within each column. The differential adhesion hypothesis postulates that less adhesive cells are relegated to the outside of a fascicle (or cartridge/column), while more adhesive cells move toward the center (143). This mechanism is used in Caenorhabditis elegans sensory neurons to properly organize dendrites in the amphid sense organ (144) and is also used to regulate photoreceptor/lamina cartridge organization. Differential adhesion is mediated by the cell adhesion molecules NCad and Flamingo, which promote the correct photoreceptor/lamina cell placement within a lamina cartridge (134, 135, 145, 146). Mutants in these genes generate cartridges with incorrect numbers of axons (134, 135, 147). Each cell adhesion molecule acts in a slightly different capacity, as Flamingo and NCad are differentially localized across the lamina cartridge and expressed at different levels in different cell types (148). Thus, changing levels of various adhesion molecules is sufficient to explain photoreceptor and lamina cell placement within a lamina cartridge (Fig. 4A). Similarly, NCad levels are also necessary and sufficient to regulate photoreceptor axon placement within medulla columns (112).
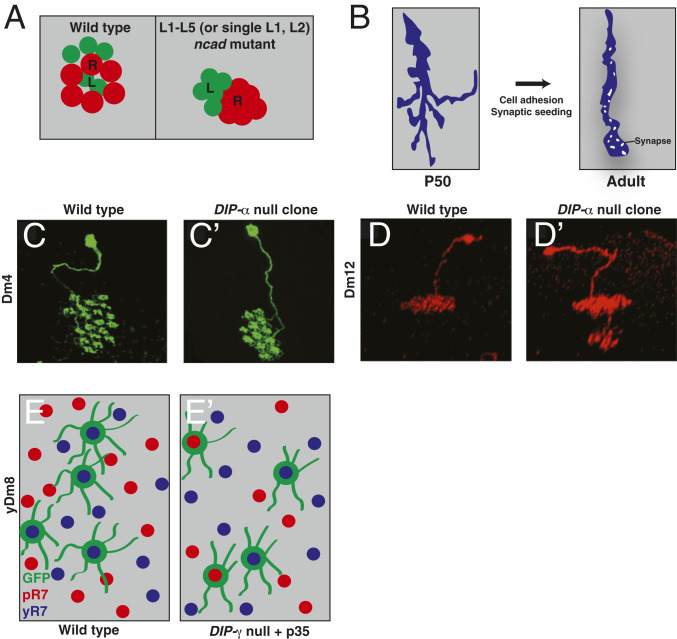
Fig. 4.
Cell adhesion molecules promote different aspects of nervous system assembly. (A) In the Drosophila lamina, differential adhesion promoted by high levels of NCad (lamina neurons [L]; green) or lower levels (photoreceptors [R]; red) directs lamina and neurons to their appropriate positions within a cartridge, internal for lamina neurons, periphery for photoreceptors. NCad mutants (Right) push lamina neurons from the cartridge center to the periphery. (B) Filopodial extensions from Drosophila photoreceptor axons are highly dynamic and explore their surroundings during pupation (Left). Later, cell adhesion molecules and synaptic components exponentially increase the stability of the axon, leading to column refinement and synaptogenesis (Right). (C–E) IgSF-containing DIPs/Dprs promote synaptogenesis by promoting adhesion between synaptic partners. DIP-mutant neurites target fewer columns (C and C′), target inappropriate sublayers (D and D′), or target the incorrect partners (E and E′). C, C', D, and D' are reprinted from ref. 161, with permission from Elsevier. E and E' are adapted from ref. 92, with permission from AAAS. GFP, green fluorescent protein; P50, 50% pupariation; pR7, pale photoreceptor R7; yR7, yellow photoreceptor R7.
In addition to molecules that regulate differential adhesion, Wnt signaling, likely acting in a planar cell polarity (PCP) capacity, also regulates neuron placement within a medulla column. In the medulla, recent scRNAseq data suggest that two Wnt ligands are produced in two juxtaposed ventral or dorsal domains in several distinct pairs of Tm neurons that can only be distinguished by the expression of Wnt4 in ventral regions vs. Wnt10 in dorsal neurons (7). Dwnt4 and Dwnt10 mutants exhibit relatively mild defects; there are instances of medulla column fusion, as well as defects in medulla column orientation, indicating that they likely act to promote proper placement of neurons within a medulla column. Similarly, mutants in known PCP proteins such as Fz2 and Vang cause similar defects (149). These defects are nonautonomous, indicating that the gradient set up by Wnts acting outside their cells of origin regulates the angle of orientation of a medulla column.
Proper column formation also requires that the neurites of unicolumnar neurons restrict their projections within a single column or tile. Neurite tiling in the optic lobe is controlled via the regulation of filopodial extension, which occurs in two phases. First, axons make actin-mediated exploratory branches across multiple columns, as observed in photoreceptor and lamina neurites (136, 137, 150–152) (Fig. 4B). Neurites then undergo a restriction phase in which they limit their axons to a single column. This restriction, in part, requires proper synapse formation, as neurons that fail to make synapses maintain their exploratory phase in search of a partner. One group of proteins required for restrictive phase initiation is the heparan sulfate proteoglycans Dally-like (Dlp) and Syndecan (Sdc), which act with their partner leukocyte common antigen–related (LAR) to promote synaptogenesis (153). dlp, sdc, and lar mutant photoreceptors invade neighboring medulla columns, but sdc and dlp mutants present distinct phenotypes, indicating that they work in parallel rather than in the same pathway (153). LAR also interacts with another protein, Liprin-α, to promote proper tiling in the lamina. liprin-α mutant R1 to R6 cells form irregular and fused cartridges (154), indicating that cell adhesion molecules required for synapse formation also promote proper column restriction. In addition to synaptogenesis proteins, cell adhesion molecules themselves promote neurite binding to synaptic partners and thus, column restriction. The down syndrome cell adhesion molecules Dscam2 and Dscam4 bind homophilically to promote proper column restriction in the lamina and in the medulla (155, 156). Additional cell adhesion molecules, such as the fly homolog of the mammalian IgSF9 gene Turtle (Tutl) and its binding partner Borderless, are also required both cell autonomously and nonautonomously to promote R7 tiling within a medulla column (157). Thus, different combinations of cell adhesion molecules promote photoreceptor tiling in the visual system.
Finally, growth factor signaling also regulates the tiling and arborization of neighboring neurons within a medulla column. In parallel to the cell adhesion molecule Tutl, the R7-expressed Activin receptor Baboon regulates R7 arbor size in the medulla (158) but also restricts the dendritic arbor size of the R7 target Dm8 and the R8 target Tm20 (159, 160). Also in parallel to activin, Insulin signaling from neighboring L5 cells activates the Tor pathway to expand the dendritic field size of Dm8 neurons (160). Thus, various combinations of cell adhesion molecules and signaling pathways regulate the receptive field size of unicolumnar and multicolumnar neurites in the visual system.
Although unicolumnar and lamina monopolar cells have been shown to use cell adhesion molecules and paracrine signaling to scale arbor size, less is known about how broadly arborizing multicolumnar neurons determine their arbor size and the extent of their branch overlap. Some work has implicated DIPs (Dpr-interacting proteins) and Dprs (defective proboscis response) in the regulation of arbor size (see below) (161). However, more work is needed to better understand the relationship between receptive field size, cell number, tiling, and neuronal function.
Although the vertebrate retina is not classically organized into columns, retinal neurons are also laid out with regular spacing as a retinal mosaic. Likewise, the neurons of the vertebrate cortex are organized into functional columns, whose composition also reflects their lineage. Recent work shows that RGP clones made at E18 produce about five neurons belonging to the same columnar region in the five different layers. Electrophysiology experiments suggest that many of these sister cells are synaptically connected (162). Molecularly, NCad has been shown to be important for layer formation (163), and in zebrafish, the IgSF protocadherin is required for proper columnar organization of the retina (164); however, the molecular basis of column formation in vertebrates has yet to be explored in detail.
Circuit Formation and Visual System Development
Guidance of neurons to the correct neuropil, column, and layer sufficiently restricts neurite location to allow for circuit assembly between pairs of neurons. One intellectual framework that is often referred to is Sperry’s chemoaffinity hypothesis, which postulates that “identification tags” localized on interacting neurons allow neurites to bind each other to promote proper nervous system targeting (165). It is possible to interpret Sperry’s work as the idea that neurons use a molecular code consisting of thousands of molecules to shape synaptic targeting. However, Sperry himself noted that this was unlikely to be the mechanism that shapes neurite targeting; instead, he felt that multiple gradients of small numbers of guidance molecules could be used in a “continuous series of decisions based on differential affinities” to specify “latitude and longitude” within the visual system (165). Sperry’s model has validity as, for instance, gradients of Ephrins explain retinotopy in the vertebrate retina.
However, the model has decreased utility when used to describe synaptic targeting. Hassan and Hiesinger (166) recently posited that a smaller number of molecules could help stochastically wandering neurites to match up to their partners in a sufficiently biased manner to ensure that neurons consistently make the correct connections. Indeed, biophysical principles such as wiring economy and volume exclusion are sufficient to model the assembly of lamina cartridges in the Drosophila optic lobe (167, 168). However, the reality is likely somewhere in between: that a few dozen molecules (rather than a few or a thousand) enact pattern formation rules to increase the probability that neurons make the correct connections. The following paragraphs describe different cell adhesion molecules, each of which biases neurons to form synapses with the correct partners.
As alluded to above, for synaptogenesis to occur, neuronal growth cones must stabilize at the correct layer in proximity to their synaptic partners. This is partly regulated by filopodial stabilization/growth cone restriction. As previously mentioned, R7 growth cones are highly dynamic during early pupation but restrict their targeting later in pupation as synaptogenesis initiates (150, 151) (Fig. 4B). Thus, genes previously shown to regulate layer formation in fact do so by filopodial stabilization. For example, NCad, Netrin, and LAR were initially considered to be guidance molecules (150–152, 154, 169, 170). However, defects in the mutants are secondary to filopodial defects, as mutants initially exhibit normal targeting but aberrant axon stabilization, leading to eventual retraction. Not only does filopodial stabilization favor synapse formation, but synapse formation itself stabilizes filopodia, resulting in a positive feedback loop that promotes synaptogenesis past a certain threshold (152). Thus, proper filopodial stabilization is essential to synaptogenesis initiation.
Other cell adhesion molecules likely act in a similar way to stabilize targeting and allow for synaptic seeding. The immunoglobulin (Ig) family protein Sidekick exhibits homophilic interactions and is required for layer targeting in the vertebrate retina (171). Mouse Sidekicks (Sdk) 1 and 2 are expressed in nonoverlapping sublayers and are localized in areas adjacent to pre- and postsynaptic machinery, indicating that they promote synapse formation. Supporting this, ectopic expression of Sdk is sufficient to misdirect neurons to the incorrect layer. Although Drosophila sdk is also expressed in specific layers in the optic lobe, it has not been shown to promote synaptogenesis. However, Sdk does appear to act homophilically within photoreceptors to regulate the placement of immature lamina neurons between photoreceptor columns to properly space lamina cartridges (172). Not all proteins required for synaptogenesis do so through neurite stabilization. For example, the cell adhesion molecule Kirre is necessary in the pupal lamina for synaptogenesis of L4 cells to L2 and other L4 cells, yet it is not required for neurite shape (173). It is unknown whether Kirre acts homophilically or whether it acts heterophilically: for instance, with its heterophilic binding partner, Rst, which is expressed in lamina precursor cells during larval development (174). Perhaps the two might bind each other within the lamina to regulate synaptogenesis.
Perhaps the best-studied cell surface proteins required for circuit assembly in Drosophila are the superfamily of immunoglobulin-containing Dprs, which bind to their cognate family DIPs. Members of this family were identified in a molecular assay designed to detect pairs of interacting proteins (175). Each DIP, which has three Ig domains, interacts with a number of Dprs, each of which possesses two Ig domains (176, 177); some of these proteins are also capable of homophilic binding (177, 178). As DIPs and Dprs show high sequence similarity among each other, specificity of DIP–Dpr interactions appears to be mediated via exclusion/negative constraints (177, 179). RNA sequencing (RNAseq) data suggest that DIPs and Dprs are differentially expressed in some corresponding synaptic partners just prior to synaptogenesis (177, 178). However, many synaptic partners in the medulla do not express matching DIPs and Dprs (177, 178).
Although multiple DIP/dpr mutants show little to no defects in circuit assembly, some DIP/Dpr pairs appear to be required for dendrite size, layer targeting, and synapse formation in the visual system. For example, DIPα-mutant Dm4 neuron clones target their neurites to fewer photoreceptor columns than in wild-type animals, as they are unable to compete with wild-type neurites (Fig. 4 C and C′). Conversely, whole-animal DIPα-mutant Dm4s target more columns. Similarly, ectopic dpr10 expression in the incorrect medulla layer is sufficient to mistarget neurons that express its binding partner DIPα (Dm4 and Dm12), and removal of DIPα also causes Dm12 neurons to target the incorrect layer(Fig. 4 D and D′) (161). In regard to synaptogenesis, DIPγ is necessary for a subset of Dm8 neurons (yDm8s) to properly target their Dpr11-expressing synaptic partners (i.e., yR7s) (Fig. 4 E and E′) (92, 176, 180). In both DIPα and DIPγ mutants, neurons that fail to partner with the correct synaptic targets are culled by programmed cell death (92, 161, 176, 180). DIPs and Dprs can also prevent ectopic synapse formation. DIPγ/β double-mutant clones possess similar numbers of synapses compared with wild-type lamina neurons, but the synapses are more diffusely localized along the neurite, suggesting that they are necessary to prevent promiscuous synapse formation (181). Thus, DIPs and Dprs promote distinct aspects of synapse formation in different optic lobe cell types.
The Beaten-path (Beat) and Sidestep (Side) proteins are another family of Ig cell surface protein pairs that appear to be involved in matching (175). Both sets of partner proteins were implicated in axon pathfinding in the Drosophila neuromuscular junction. Beat is required in motor axons, while its partner Side is an attractant expressed in embryonic muscles (182). beat and/or side mutants cause failure of motor neurons to defasciculate and enter the appropriate muscle domains; misexpression of Side is sufficient to mistarget motor axons to inappropriate muscle segments (183). Further analysis suggests that, similar to DIPs/Dprs, Beats/Sides are part of a large family with multiple partners that show specific interactions (184, 185). However, at least in the adult, these genes are very widely expressed across different neural types, indicating that Beats and Sides likely do not act in a “Sperry”-like one-to-one code but rather, act in combination to bias neurons to synapse with the correct partners.
Conclusions/Areas of Future Study
The study of Drosophila optic lobe development has unearthed a variety of mechanistic principles that seem to apply to the development of other systems. First, tTFs specify the production of discrete cell types. Spatial segregation of transcription factors in progenitors can also prime temporal factors to generate different cell fates. Spatial transcription factors and tTFs are used to a similar effect to pattern the Drosophila VNC but also, the vertebrate spinal cord and cerebral cortex. Birth order ensures the proper retinotopic assembly of neuropils along the AP axis of the fly eye, although much is left to be discovered regarding the regulation of DV retinotopy. Chemotactic cues secreted by glial cells help to separate groups of neurons into discrete neuropils. Differential adhesion allows for the assembly of distinct cell types into discrete columns, and paracrine signaling restricts the dendrite size of neurons that project their arbors across multiple columns. Finally, cell adhesion molecules stabilize neurites to allow for assembly of synaptic seeding factors but also, prevent promiscuous seeding of synaptic components.
Although conceptual advances in understanding nervous system assembly have been made in the last few decades, there are many exciting discoveries to be made on the horizon. In the next decade, we will understand the cell lineages of visual system neurons, and we will better understand how spatial factors epigenetically prime neurons to adopt their cell fates. We will also understand more about how neurons are generated in the correct numbers. Finally, we will discover how neurons target their axons and dendrites to specific regions in the brain and how neurons form retinal mosaics within the medulla. The answers to these questions will identify processes that will be of relevance in helping us understand how nervous systems are assembled in other biological contexts.
Acknowledgments
We thank Isabel Holguera-Lopez, Yu-Chieh David Chen, Neset Özel, Rana El-Danaf, and Sergio Cordoba for their comments on the manuscript and L. Zipursky, Gwenvael Le Dréau, and Elisa Martí for sharing images. We are supported by funding from National Eye Institute Grants F32 EY028012 (to J.M.) and R01 EY017916 (to C.D.).
Footnotes
The authors declare no competing interest.
Data Availability
There are no data underlying this work.
References
- 1.Luo L., Flanagan J. G., Development of continuous and discrete neural maps. Neuron 56, 284–300 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Fischbach K.-F., Dittrich A. P. M., The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475 (1989). [Google Scholar]
- 3.Jenett A., et al., A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeiffer B. D., et al., Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 9715–9720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macosko E. Z., et al., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konstantinides N., et al., Phenotypic convergence: Distinct transcription factors regulate common terminal features. Cell 174, 622–635.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Özel M. N., et al., Neuronal diversity and convergence in a visual system developmental atlas. Nature 589, 88–95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurmangaliyev Y. Z., Yoo J., Valdes-Aleman J., Sanfilippo P., Zipursky S. L.; Transcriptional Programs of Circuit Assembly in the Drosophila Visual System , Transcriptional programs of circuit assembly in the Drosophila visual system. Neuron 108, 1045–1057.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Meinertzhagen I. A., O’Neil S. D., Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J. Comp. Neurol. 305, 232–263 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Takemura S. Y., Lu Z., Meinertzhagen I. A., Synaptic circuits of the Drosophila optic lobe: The input terminals to the medulla. J. Comp. Neurol. 509, 493–513 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takemura S. Y., et al., A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Z., et al., A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takemura S. Y., et al., Synaptic circuits and their variations within different columns in the visual system of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 112, 13711–13716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasugi T., Umetsu D., Murakami S., Sato M., Tabata T., Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development 135, 1471–1480 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Egger B., Boone J. Q., Stevens N. R., Brand A. H., Doe C. Q., Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egger B., Gold K. S., Brand A. H., Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development 137, 2981–2987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanes J. R., Zipursky S. L., Design principles of insect and vertebrate visual systems. Neuron 66, 15–36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holguera I., Desplan C., Neuronal specification in space and time. Science 362, 176–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ready D. F., A multifaceted approach to neural development. Trends Neurosci. 12, 102–110 (1989). [DOI] [PubMed] [Google Scholar]
- 20.Treisman J. E., Heberlein U., Retinal differentiation in Drosophila: Formation of the eye field and control of differentiation. Curr. Top. Dev. Biol. 39, 119–158 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Cagan R., Principles of Drosophila eye differentiation. Curr. Top. Dev. Biol. 89, 115–135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y.-C., Desplan C., Gene regulatory networks during the development of the Drosophila visual system. Curr. Top. Dev. Biol. 139, 89–125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apitz H., Salecker I., A challenge of numbers and diversity: Neurogenesis in the Drosophila optic lobe. J. Neurogenet. 28, 233–249 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Brand A. H., Livesey F. J., Neural stem cell biology in vertebrates and invertebrates: More alike than different? Neuron 70, 719–729 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Kerwin S. K., et al., Regulated alternative splicing of Drosophila Dscam2 photoreceptor synapses. Genetics 208, 717–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo J., McQueen P. G., Shi B., Lee C. H., Ting C. Y., Wiring dendrites in layers and columns. J. Neurogenet. 30, 69–79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofbauer A., Campos-Ortega J. A., Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Rouxs Arch. Dev. Biol. 198, 264–274 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Ready D. F., Hanson T. E., Benzer S., Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53, 217–240 (1976). [DOI] [PubMed] [Google Scholar]
- 29.Heisenberg M., Buchner E., The role of retinula cell types in visual behavior of Drosophila melanogaster. J. Comp. Physiol. 117, 127–162 (1977). [Google Scholar]
- 30.Hu K. G., Stark W. S., Specific receptor input into spectral preference in Drosophila. J. Comp. Physiol. 121, 241–252 (1977). [Google Scholar]
- 31.Wernet M. F., et al., Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115, 267–279 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Bausenwein B., Dittrich A. P. M., Fischbach K. F., The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 267, 17–28 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Morante J., Desplan C., The color-vision circuit in the medulla of Drosophila. Curr. Biol. 18, 553–565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borst A., Fly visual course control: Behaviour, algorithms and circuits. Nat. Rev. Neurosci. 15, 590–599 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Barnhart E. L., Wang I. E., Wei H., Desplan C., Clandinin T. R., Sequential nonlinear filtering of local motion cues by global motion circuits. Neuron 100, 229–243.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu M., et al., Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. eLife 5, e21022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tai C.-Y., Chin A.-L., Chiang A.-S., Comprehensive map of visual projection neurons for processing ultraviolet information in the Drosophila brain. J. Comp. Neurol. 529, 1988–2013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timaeus L., Geid L., Sancer G., Wernet M. F., Hummel T., Parallel visual pathways with topographic versus nontopographic organization connect the Drosophila eyes to the central brain. iScience 23, 101590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nassif C., Noveen A., Hartenstein V., Early development of the Drosophila brain. III. The pattern of neuropile founder tracts during the larval period. J. Comp. Neurol. 455, 417–434 (2003). [DOI] [PubMed] [Google Scholar]
- 40.White K., Kankel D. R., Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. Dev. Biol. 65, 296–321 (1978). [DOI] [PubMed] [Google Scholar]
- 41.Erclik T., et al., Integration of temporal and spatial patterning generates neural diversity. Nature 541, 365–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apitz H., Salecker I., A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system. Nat. Neurosci. 18, 46–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinto-Teixeira F., et al., Development of concurrent retinotopic maps in the fly motion detection circuit. Cell 173, 485–498.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasugi T., Sugie A., Umetsu D., Tabata T., Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development 137, 3193–3203 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Doe C. Q., Chu-LaGraff Q., Wright D. M., Scott M. P., The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451–464 (1991). [DOI] [PubMed] [Google Scholar]
- 46.Bertet C., et al., Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell 158, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasegawa E., et al., Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development 138, 983–993 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Morante J., Erclik T., Desplan C., Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development 138, 687–693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X., et al., Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456–462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki T., Kaido M., Takayama R., Sato M., A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev. Biol. 380, 12–24 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Hartenstein V., Posakony J. W., Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development 107, 389–405 (1989). [DOI] [PubMed] [Google Scholar]
- 52.Turner D. L., Cepko C. L., A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131–136 (1987). [DOI] [PubMed] [Google Scholar]
- 53.Doe C. Q., Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219–240 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Isshiki T., Pearson B., Holbrook S., Doe C. Q., Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Grosskortenhaus R., Pearson B. J., Marusich A., Doe C. Q., Regulation of temporal identity transitions in Drosophila neuroblasts. Dev. Cell 8, 193–202 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Averbukh I., Lai S.-L., Doe C. Q., Barkai N., A repressor-decay timer for robust temporal patterning in embryonic Drosophila neuroblast lineages. eLife 7, e38631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maurange C., Cheng L., Gould A. P., Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891–902 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Rapaport D. H., Wong L. L., Wood E. D., Yasumura D., LaVail M. M., Timing and topography of cell genesis in the rat retina. J. Comp. Neurol. 474, 304–324 (2004). [DOI] [PubMed] [Google Scholar]
- 59.He J., et al., How variable clones build an invariant retina. Neuron 75, 786–798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen-Ba-Charvet K. T., Rebsam A., Neurogenesis and specification of retinal ganglion cells. Int. J. Mol. Sci. 21, 451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voinescu P. E., Kay J. N., Sanes J. R., Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J. Comp. Neurol. 517, 737–750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomes F. L. A. F., et al., Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development 138, 227–235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cayouette M., Barres B. A., Raff M., Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron 40, 897–904 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Godinho L., et al., Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron 56, 597–603 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Cepko C., Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 15, 615–627 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Brown N. L., Patel S., Brzezinski J., Glaser T., Math5 is required for retinal ganglion cell and optic nerve formation. Development 128, 2497–2508 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang S. W., et al., Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15, 24–29 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.La Torre A., Georgi S., Reh T. A., Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E2362–E2370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elliott J., Jolicoeur C., Ramamurthy V., Cayouette M., Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60, 26–39 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Mattar P., Ericson J., Blackshaw S., Cayouette M., A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron 85, 497–504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Javed A., et al., Pou2f1 and Pou2f2 cooperate to control the timing of cone photoreceptor production in the developing mouse retina. Development 147, dev188730 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Liu S., et al., Foxn4 is a temporal identity factor conferring mid/late-early retinal competence and involved in retinal synaptogenesis. Proc. Natl. Acad. Sci. U.S.A. 117, 5016–5027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cai L., Hayes N. L., Takahashi T., Caviness V. S. J. Jr, Nowakowski R. S., Size distribution of retrovirally marked lineages matches prediction from population measurements of cell cycle behavior. J. Neurosci. Res. 69, 731–744 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Angevine J. B. Jr, Sidman R. L., Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192, 766–768 (1961). [DOI] [PubMed] [Google Scholar]
- 75.Gao P., et al., Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frantz G. D., McConnell S. K., Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron 17, 55–61 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Shen Q., et al., The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 9, 743–751 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Oberst P., et al., Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature 573, 370–374 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Telley L., et al., Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 364, eaav2522 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Alsiö J. M., Tarchini B., Cayouette M., Livesey F. J., Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 110, E716–E725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang X., Friedman A., Heaney S., Purcell P., Maas R. L., Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 16, 2097–2107 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bäumer N., et al., Pax6 is required for establishing naso-temporal and dorsal characteristics of the optic vesicle. Development 129, 4535–4545 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Yu R. T., et al., The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc. Natl. Acad. Sci. U.S.A. 97, 2621–2625 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lai E. C., Notch signaling: Control of cell communication and cell fate. Development 131, 965–973 (2004). [DOI] [PubMed] [Google Scholar]
- 85.Rhyu M. S., Jan L. Y., Jan Y. N., Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 (1994). [DOI] [PubMed] [Google Scholar]
- 86.Bultje R. S., et al., Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron 63, 189–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Truman J. W., Moats W., Altman J., Marin E. C., Williams D. W., Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development 137, 53–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spana E. P., Doe C. Q., Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 (1996). [DOI] [PubMed] [Google Scholar]
- 89.Erclik T., Hartenstein V., Lipshitz H. D., McInnes R. R., Conserved role of the Vsx genes supports a monophyletic origin for bilaterian visual systems. Curr. Biol. 18, 1278–1287 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Gold K. S., Brand A. H., Optix defines a neuroepithelial compartment in the optic lobe of the Drosophila brain. Neural Dev. 9, 18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang T., Mazotta J., Dumstrei K., Dumitrescu A., Hartenstein V., Dpp and Hh signaling in the Drosophila embryonic eye field. Development 128, 4691–4704 (2001). [DOI] [PubMed] [Google Scholar]
- 92.Courgeon M., Desplan C., Coordination between stochastic and deterministic specification in the Drosophila visual system. Science 366, eaay6727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chu-LaGraff Q., Doe C. Q., Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science 261, 1594–1597 (1993). [DOI] [PubMed] [Google Scholar]
- 94.Isshiki T., Takeichi M., Nose A., The role of the msh homeobox gene during Drosophila neurogenesis: Implication for the dorsoventral specification of the neuroectoderm. Development 124, 3099–3109 (1997). [DOI] [PubMed] [Google Scholar]
- 95.Sen S. Q., Chanchani S., Southall T. D., Doe C. Q., Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci. eLife 8, e44036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mattar P., et al., A Casz1-NuRD complex regulates temporal identity transitions in neural progenitors. Sci. Rep. 11, 3858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hernandez-Miranda L. R., Müller T., Birchmeier C., The dorsal spinal cord and hindbrain: From developmental mechanisms to functional circuits. Dev. Biol. 432, 34–42 (2017). [DOI] [PubMed] [Google Scholar]
- 98.Briscoe J., Pierani A., Jessell T. M., Ericson J., A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445 (2000). [DOI] [PubMed] [Google Scholar]
- 99.Le Dréau G., Martí E., Dorsal-ventral patterning of the neural tube: A tale of three signals. Dev. Neurobiol. 72, 1471–1481 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Sagner A., et al., Temporal patterning of the central nervous system by a shared transcription factor code. https://www.biorxiv.org/content/10.1101/2020.11.10.376491v1.full.pdf (10 November 2020).
- 101.Rossi A. M., Desplan C., Extrinsic activin signaling cooperates with an intrinsic temporal program to increase mushroom body neuronal diversity. eLife 9, e58880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dias J. M., Alekseenko Z., Applequist J. M., Ericson J., Tgfβ signaling regulates temporal neurogenesis and potency of neural stem cells in the CNS. Neuron 84, 927–939 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Lin D. M., Auld V. J., Goodman C. S., Targeted neuronal cell ablation in the Drosophila embryo: Pathfinding by follower growth cones in the absence of pioneers. Neuron 14, 707–715 (1995). [DOI] [PubMed] [Google Scholar]
- 104.Hidalgo A., Brand A. H., Targeted neuronal ablation: The role of pioneer neurons in guidance and fasciculation in the CNS of Drosophila. Development 124, 3253–3262 (1997). [DOI] [PubMed] [Google Scholar]
- 105.Ngo K. T., Andrade I., Hartenstein V., Spatio-temporal pattern of neuronal differentiation in the Drosophila visual system: A user’s guide to the dynamic morphology of the developing optic lobe. Dev. Biol. 428, 1–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fischbach K. F., Neural cell types surviving congenital sensory deprivation in the optic lobes of Drosophila melanogaster. Dev. Biol. 95, 1–18 (1983). [DOI] [PubMed] [Google Scholar]
- 107.Huang Z., Kunes S., Hedgehog, transmitted along retinal axons, triggers neurogenesis in the developing visual centers of the Drosophila brain. Cell 86, 411–422 (1996). [DOI] [PubMed] [Google Scholar]
- 108.Huang Z., Shilo B. Z., Kunes S., A retinal axon fascicle uses spitz, an EGF receptor ligand, to construct a synaptic cartridge in the brain of Drosophila. Cell 95, 693–703 (1998). [DOI] [PubMed] [Google Scholar]
- 109.Fernandes V. M., Chen Z., Rossi A. M., Zipfel J., Desplan C., Glia relay differentiation cues to coordinate neuronal development in Drosophila. Science 357, 886–891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perez S. E., Steller H., Migration of glial cells into retinal axon target field in Drosophila melanogaster. J. Neurobiol. 30, 359–373 (1996). [DOI] [PubMed] [Google Scholar]
- 111.Poeck B., Fischer S., Gunning D., Zipursky S. L., Salecker I., Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron 29, 99–113 (2001). [DOI] [PubMed] [Google Scholar]
- 112.Trush O., et al., N-cadherin orchestrates self-organization of neurons within a columnar unit in the Drosophila medulla. J. Neurosci. 39, 5861–5880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shinomiya K., et al., The organization of the second optic chiasm of the Drosophila optic lobe. Front. Neural Circuits 13, 65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Apitz H., Salecker I., Spatio-temporal relays control layer identity of direction-selective neuron subtypes in Drosophila. Nat. Commun. 9, 2295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mora N., et al., A temporal transcriptional switch governs stem cell division, neuronal numbers, and maintenance of differentiation. Dev. Cell 45, 53–66.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Osterhout J. A., El-Danaf R. N., Nguyen P. L., Huberman A. D., Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell Rep. 8, 1006–1017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu H. H., et al., Clonal development and organization of the adult Drosophila central brain. Curr. Biol. 23, 633–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tayler T. D., Robichaux M. B., Garrity P. A., Compartmentalization of visual centers in the Drosophila brain requires Slit and Robo proteins. Development 131, 5935–5945 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Caipo L., et al., Slit neuronal secretion coordinates optic lobe morphogenesis in Drosophila. Dev. Biol. 458, 32–42 (2020). [DOI] [PubMed] [Google Scholar]
- 120.Suzuki T., et al., Netrin signaling defines the regional border in the Drosophila visual center. iScience 8, 148–160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pereanu W., Shy D., Hartenstein V., Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 283, 191–203 (2005). [DOI] [PubMed] [Google Scholar]
- 122.Cooper J. A., Cell biology in neuroscience: Mechanisms of cell migration in the nervous system. J. Cell Biol. 202, 725–734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Corbin J. G., Nery S., Fishell G., Telencephalic cells take a tangent: Non-radial migration in the mammalian forebrain. Nat. Neurosci. 4 (suppl.), 1177–1182 (2001). [DOI] [PubMed] [Google Scholar]
- 124.Chen Z., et al., A unique class of neural progenitors in the Drosophila optic lobe generates both migrating neurons and glia. Cell Rep. 15, 774–786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kaphingst K., Kunes S., pattern formation in the visual centers of the Drosophila brain: Wingless acts via decapentaplegic to specify the dorsoventral axis. Cell 78, 437–448 (1994). [DOI] [PubMed] [Google Scholar]
- 126.Yoshida S., et al., DPP signaling controls development of the lamina glia required for retinal axon targeting in the visual system of Drosophila. Development 132, 4587–4598 (2005). [DOI] [PubMed] [Google Scholar]
- 127.Suzuki T., et al., formation of neuronal circuits by interactions between neuronal populations derived from different origins in the Drosophila visual center. Cell Rep. 15, 499–509 (2016). [DOI] [PubMed] [Google Scholar]
- 128.Suh G. S. B., et al., Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron 33, 35–46 (2002). [DOI] [PubMed] [Google Scholar]
- 129.Guerrier S., et al., The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Galli-Resta L., Resta G., Tan S. S., Reese B. E., Mosaics of islet-1-expressing amacrine cells assembled by short-range cellular interactions. J. Neurosci. 17, 7831–7838 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huckfeldt R. M., et al., Transient neurites of retinal horizontal cells exhibit columnar tiling via homotypic interactions. Nat. Neurosci. 12, 35–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clandinin T. R., Feldheim D. A., Making a visual map: Mechanisms and molecules. Curr. Opin. Neurobiol. 19, 174–180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kulkarni A., Ertekin D., Lee C.-H., Hummel T., Birth order dependent growth cone segregation determines synaptic layer identity in the Drosophila visual system. eLife 5, e13715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee C.-H., Herman T., Clandinin T. R., Lee R., Zipursky S. L., N-cadherin regulates target specificity in the Drosophila visual system. Neuron 30, 437–450 (2001). [DOI] [PubMed] [Google Scholar]
- 135.Lee R. C., et al., The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat. Neurosci. 6, 557–563 (2003). [DOI] [PubMed] [Google Scholar]
- 136.Garrity P. A., et al., Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell 85, 639–650 (1996). [DOI] [PubMed] [Google Scholar]
- 137.Hing H., Xiao J., Harden N., Lim L., Zipursky S. L., Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell 97, 853–863 (1999). [DOI] [PubMed] [Google Scholar]
- 138.Song J., Wu L., Chen Z., Kohanski R. A., Pick L., Axons guided by insulin receptor in Drosophila visual system. Science 300, 502–505 (2003). [DOI] [PubMed] [Google Scholar]
- 139.Sato M., Umetsu D., Murakami S., Yasugi T., Tabata T., DWnt4 regulates the dorsoventral specificity of retinal projections in the Drosophila melanogaster visual system. Nat. Neurosci. 9, 67–75 (2006). [DOI] [PubMed] [Google Scholar]
- 140.Berger J., et al., Systematic identification of genes that regulate neuronal wiring in the Drosophila visual system. PLoS Genet. 4, e1000085 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seabrook T. A., Burbridge T. J., Crair M. C., Huberman A. D., Architecture, function, and assembly of the mouse visual system. Annu. Rev. Neurosci. 40, 499–538 (2017). [DOI] [PubMed] [Google Scholar]
- 142.Boyle M., Nighorn A., Thomas J. B., Drosophila Eph receptor guides specific axon branches of mushroom body neurons. Development 133, 1845–1854 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Steinberg M. S., Takeichi M., Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc. Natl. Acad. Sci. U.S.A. 91, 206–209 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yip Z. C., Heiman M. G., Ordered arrangement of dendrites within a C. elegans sensory nerve bundle. elife 7, e35825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Prakash S., Caldwell J. C., Eberl D. F., Clandinin T. R., Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat. Neurosci. 8, 443–450 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schwabe T., Borycz J. A., Meinertzhagen I. A., Clandinin T. R., Differential adhesion determines the organization of synaptic fascicles in the Drosophila visual system. Curr. Biol. 24, 1304–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen P. L., Clandinin T. R., The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron 58, 26–33 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schwabe T., Neuert H., Clandinin T. R., A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell 154, 351–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Han X., et al., DWnt4 and DWnt10 regulate morphogenesis and arrangement of columnar units via Fz2/PCP signaling in the Drosophila brain. Cell Rep. 33, 108305 (2020). [DOI] [PubMed] [Google Scholar]
- 150.Akin O., Zipursky S. L., Frazzled promotes growth cone attachment at the source of a Netrin gradient in the Drosophila visual system. eLife 5, 1–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Özel M. N., Langen M., Hassan B. A., Hiesinger P. R., Filopodial dynamics and growth cone stabilization in Drosophila visual circuit development. eLife 4, e10721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Özel M. N., et al., Serial synapse formation through filopodial competition for synaptic seeding factors. Dev. Cell 50, 447–461.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rawson J. M., et al., The heparan sulfate proteoglycans Dally-like and Syndecan have distinct functions in axon guidance and visual-system assembly in Drosophila. Curr. Biol. 15, 833–838 (2005). [DOI] [PubMed] [Google Scholar]
- 154.Choe K. M., Prakash S., Bright A., Clandinin T. R., Liprin-α is required for photoreceptor target selection in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 11601–11606 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Millard S. S., Flanagan J. J., Pappu K. S., Wu W., Zipursky S. L., Dscam2 mediates axonal tiling in the Drosophila visual system. Nature 447, 720–724 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tadros W., et al., Dscam proteins direct dendritic targeting through adhesion. Neuron 89, 480–493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ferguson K., Long H., Cameron S., Chang W.-T., Rao Y., The conserved Ig superfamily member Turtle mediates axonal tiling in Drosophila. J. Neurosci. 29, 14151–14159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ting C.-Y., et al., Tiling of r7 axons in the Drosophila visual system is mediated both by transduction of an activin signal to the nucleus and by mutual repulsion. Neuron 56, 793–806 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ting C.-Y., et al., Photoreceptor-derived activin promotes dendritic termination and restricts the receptive fields of first-order interneurons in Drosophila. Neuron 81, 830–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Luo J., et al., Antagonistic regulation by insulin-like peptide and activin ensures the elaboration of appropriate dendritic field sizes of amacrine neurons. eLife 9, e50568 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu S., et al., Interactions between the Ig-superfamily proteins DIP-α and dpr6/10 regulate assembly of neural circuits. Neuron 100, 1369–1384.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yu Y. C., Bultje R. S., Wang X., Shi S. H., Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature 458, 501–504 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Poskanzer K., Needleman L. A., Bozdagi O., Huntley G. W., N-cadherin regulates ingrowth and laminar targeting of thalamocortical axons. J. Neurosci. 23, 2294–2305 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cooper S. R., et al., Protocadherins control the modular assembly of neuronal columns in the zebrafish optic tectum. J. Cell Biol. 211, 807–814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sperry R. W., Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc. Natl. Acad. Sci. U.S.A. 50, 703–710 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hassan B. A., Hiesinger P. R., Beyond molecular codes: Simple rules to wire complex brains. Cell 163, 285–291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rivera-Alba M., et al., Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain. Curr. Biol. 21, 2000–2005 (2011). Correction in: Curr. Biol. 22, 172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Langen M., et al., The developmental rules of neural superposition in Drosophila. Cell 162, 120–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Clandinin T. R., et al., Drosophila LAR regulates R1-R6 and R7 target specificity in the visual system. Neuron 32, 237–248 (2001). [DOI] [PubMed] [Google Scholar]
- 170.Ting C.-Y., et al., Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development 132, 953–963 (2005). [DOI] [PubMed] [Google Scholar]
- 171.Yamagata M., Weiner J. A., Sanes J. R., Sidekicks: Synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell 110, 649–660 (2002). [DOI] [PubMed] [Google Scholar]
- 172.Astigarraga S., et al., Drosophila Sidekick is required in developing photoreceptors to enable visual motion detection. Development 145, dev158246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lüthy K., et al., The irre cell recognition module (IRM) protein Kirre is required to form the reciprocal synaptic network of L4 neurons in the Drosophila lamina. J. Neurogenet. 28, 291–301 (2014). [DOI] [PubMed] [Google Scholar]
- 174.Sugie A., Umetsu D., Yasugi T., Fischbach K. F., Tabata T., Recognition of pre- and postsynaptic neurons via nephrin/NEPH1 homologs is a basis for the formation of the Drosophila retinotopic map. Development 137, 3303–3313 (2010). [DOI] [PubMed] [Google Scholar]
- 175.Özkan E., et al., An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Carrillo R. A., et al., Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins. Cell 163, 1770–1782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cosmanescu F., et al., Neuron-subtype-specific expression, interaction affinities, and specificity determinants of DIP/dpr cell recognition proteins. Neuron 100, 1385–1400.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tan L., et al., Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163, 1756–1769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sergeeva A. P., et al., DIP/Dpr interactions and the evolutionary design of specificity in protein families. Nat. Commun. 11, 2125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Menon K. P., Kulkarni V., Takemura S. Y., Anaya M., Zinn K., Interactions between Dpr11 and DIP-γ control selection of amacrine neurons in Drosophila color vision circuits. eLife 8, e48935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu C., et al., Control of synaptic specificity by establishing a relative preference for synaptic partners. Neuron 103, 865–877.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fambrough D., Goodman C. S., The Drosophila beaten path gene encodes a novel secreted protein that regulates defasciculation at motor axon choice points. Cell 87, 1049–1058 (1996). [DOI] [PubMed] [Google Scholar]
- 183.Sink H., Rehm E. J., Richstone L., Bulls Y. M., Goodman C. S., Sidestep encodes a target-derived attractant essential for motor axon guidance in Drosophila. Cell 105, 57–67 (2001). [DOI] [PubMed] [Google Scholar]
- 184.Li H., et al., Deconstruction of the beaten Path-Sidestep interaction network provides insights into neuromuscular system development. eLife 6, 1–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pipes G. C., Lin Q., Riley S. E., Goodman C. S., The beat generation: A multigene family encoding IgSF proteins related to the beat axon guidance molecule in Drosophila. Development 128, 4545–4552 (2001). [DOI] [PubMed] [Google Scholar]
- 186.Reese B. E., Galli-Resta L., The role of tangential dispersion in retinal mosaic formation. Prog. Retin. Eye Res. 21, 153–168 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.