Significance
To generate modality-specific percepts (e.g., pain vs. itch), the brain must interpret the activity that arises from the output cells (projection neurons) of the dorsal horn of the spinal cord, which receive and then transmit pain- or itch-provoking stimuli. However, neither the extent to which projection neurons relay modality-specific messages nor the molecular makeup of these neurons is clear. Here, we report that the projection neurons are, in fact, diverse, not only molecularly but also functionally. By focusing on genes expressed by projection neurons within the neurokinin-1 receptor (NK1R) and non–NK1R-expressing populations, we also demonstrate considerable functional heterogeneity among these subsets, including nociceptive (pain)-specific, pruriceptive (itch)-specific, and polymodal (both pain and itch) subsets of projection neurons.
Keywords: pain, itch, projection neurons, dorsal horn, RNA-seq
Abstract
A remarkable molecular and functional heterogeneity of the primary sensory neurons and dorsal horn interneurons transmits pain- and or itch-relevant information, but the molecular signature of the projection neurons that convey the messages to the brain is unclear. Here, using retro-TRAP (translating ribosome affinity purification) and RNA sequencing, we reveal extensive molecular diversity of spino- and trigeminoparabrachial projection neurons. Among the many genes identified, we highlight distinct subsets of Cck+-, Nptx2+-, Nmb+-, and Crh+-expressing projection neurons. By combining in situ hybridization of retrogradely labeled neurons with Fos-based assays, we also demonstrate significant functional heterogeneity, including both convergence and segregation of pain- and itch-provoking inputs into molecularly diverse subsets of NK1R- and non–NK1R-expressing projection neurons.
Recent studies described a plethora of neurochemically distinct primary afferent and spinal cord interneuron populations that are tuned to discrete pain and itch stimulus modalities (1–5). Importantly, however, the generation of modality-specific percepts does not arise from the brain’s analysis of the activity of interneurons, but rather from activity of different projection neuron populations, which must be interpreted at supraspinal loci. It is, therefore, critical to address the extent to which the primary afferent and interneuron heterogeneity and specificity extends to the projection neurons in the dorsal horn of the spinal cord and in its medullary homolog, the trigeminal nucleus caudalis (TNC) (6).
Anatomical studies identified three morphological classes of lamina I projection neurons, and there is some, albeit controversial, evidence for the correlation of morphology with the electrophysiological profile, ascending projection, and receptor expression of these cells (for review see ref. 1). There is certainly considerable evidence for functional heterogeneity. For example, many lamina I projection neurons respond only to noxious mechanical and/or thermal stimulation (7–12). Another population, in primates (10, 13) and cats (8, 14), responds only to innocuous cooling. Lastly, there is a significant population of wide dynamic range (WDR) neurons that respond to both innocuous and noxious stimuli in the deep dorsal horn (9, 15–18). Subsets of the WDR and nociceptive-specific neurons are pruriceptive (19–24), with separate populations, in primates, responding to the pruritogens, histamine, and cowage (21, 22).
From a molecular perspective, however, the projection neurons are often regarded as relatively homogeneous, despite evidence to the contrary. Until recently, the neurokinin 1 receptor (NK1R) was the predominant marker of projection neurons in lamina I and the lateral spinal nucleus (LSN) (25, 26). However, two recent studies introduced genes, namely Phox2a (27) and Gpr83 (28), that define large populations of projection neurons. Importantly, behavioral studies in mice in which Phox2a was deleted (27), or the neurons that express Gpr83 were stimulated (28), demonstrated that those projection neurons, many of which target the brainstem parabrachial nucleus, contribute to the transmission of different pain modalities. There is also evidence for subsets of NK1R neurons, based on projection target (29) or molecular makeup (29–32). Most recently, using unbiased single-cell transcriptomics, Häring et al. (33) identified an excitatory neuron cluster (Glut15) that includes NK1R-expressing spinoparabrachial neurons. This cluster included Lypd1, a forebrain protein implicated in anxiety disorders (34). However, as Lypd1 labels ∼95% of spinoparabrachial neurons (33), it likely does not define a functionally distinct subset.
Here we first identified molecularly distinct subpopulations of projection neurons and next asked whether these subpopulations are also functionally heterogeneous. The approach that we took involved a sequential series of filtering steps. First, we performed projection neuron-centric RNA sequencing (RNA-seq). Using retro-TRAP (translating ribosome affinity purification), we purified lateral parabrachial (LPb)-projecting neurons from the spinal cord and TNC and generated RNA-seq datasets of candidate projection neuron genes. As many of these genes are also expressed by spinal cord and TNC interneurons, in the next filtering step we verified the projection neuron hits by combining retrograde tracing and multiplexed in situ hybridization. The latter studies were qualitative in nature, but they identified genes that establish molecular heterogeneity of projection neurons. Lastly, we performed functional studies using TRAP2 mice (targeted recombination in active populations) (35), in which a reporter is expressed in activated neurons. In the same mice, we documented activated neurons in the more traditional manner by immunostaining for the Fos protein. Our studies evaluated responsiveness to pain- and itch-provoking stimuli and demonstrated functional heterogeneity in the molecular diverse projection neuron population. Taken together, we report both algogen and pruritogen convergence as well as preliminary evidence for labeled-line properties of molecularly diverse subsets of NK1R- and non–NK1R-expressing projection neurons.
Results
Selective Purification and Profiling of Projection Neuron RNA.
To purify and sequence RNA specifically from projection neurons, we injected a replication-deficient, retrograde herpes simplex virus (HSV)-based viral vector encoding a green fluorescent protein (GFP)-tagged large ribosomal subunit protein L10 (HSV-GFPL10) into the LPb of wild-type mice, which induced expression of GFP-L10 in spino- and trigeminoparabrachial projection neurons (Fig. 1A). In a parallel study, to selectively target LPb-projecting neurons that express the NK1R, we injected an HSV-based viral vector encoding a Cre-recombinase–dependent HA-tagged L10 (HSV-flex-HAL10) into the LPb of NK1R-Cre mice. Data obtained from animals injected with HSV-GFPL10 or HSV-flex-HAL10 are hereafter referred to as the “PN” or “NK” dataset, respectively. To increase the total number of labeled projection neurons, in the following studies we combined spinal cord and trigeminal tissue. Two weeks after the viral injections, we recorded GFP- and HA-tagged ribosomes in projection neurons throughout the spinal cord and TNC in wild-type (Fig. 1B) and in NK1R-Cre (Fig. 1C) mice, respectively.
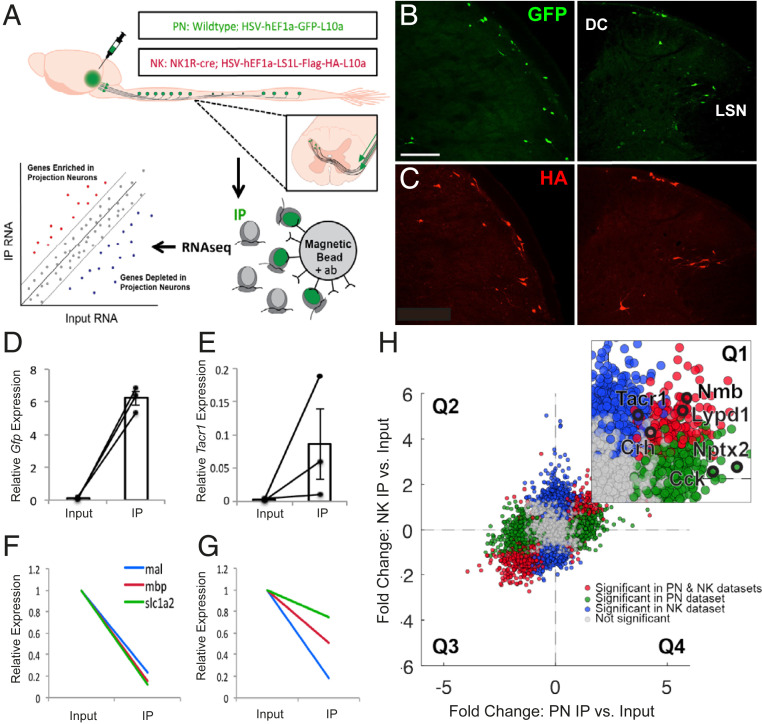
Fig. 1.
Selective purification and profiling of projection neurons reveal candidate genes expressed by projection neurons. (A) Experimental design. (B and C) Representative images of GFP (B) and HA (C) immunofluorescence in nucleus caudalis (Left) and spinal cord (Right) from wild-type and NK1R-Cre mice, respectively, illustrate projection neurons GFP or HA-tagged ribosomal protein. (Scale bar 100 μm.) (D and E) qPCR results showing enrichment of Gfp (D) and Tacr1 (E) in IP relative to input samples, in PN and NK experiments, respectively. Data are normalized to Rpl27 and represented as mean ± SEM. (F and G) qPCR shows depletion of glial genes in IP relative to input samples in PN (F) and NK (G) experiments. Data are normalized to Rpl27 and input relative expression and represented as mean ± SEM. (H) RNA sequencing shows differential expression data of IP relative to input fold change for PN experiments vs. NK experiments. Quadrant 1 (Q1) contains genes enriched in both datasets; Q2 contains genes depleted in both; Q3 and Q4 contain genes differentially altered in PN vs. NK datasets. Inset shows enlarged Q1 with genes of interest highlighted in black. Genes significantly enriched or depleted in both PN and NK datasets are highlighted in red. Genes significantly changed in PN, but not NK dataset, are highlighted in green, while genes significantly changed in NK, but not PN, datasets are highlighted in blue. All significant differences P < 0.05.
Confirmation of specific immunolabeling of projection neurons was followed by the immunoprecipitation of GFP- or HA-tagged ribosomes and associated mRNA from spinal cord and trigeminal tissue. Specificity of the immunoprecipitation (IP) was confirmed by qPCR. In all IP vs. input libraries we recorded enrichment of Gfp (∼64.6-fold; Fig. 1D and SI Appendix, Fig. S1A) and Tacr1 (∼26.5-fold; Fig. 1E and SI Appendix, Fig. S1B). Depletion of glial markers (36) in the PN libraries (mbp: ∼6.4-fold; mal: ∼3.9-fold; slc1a2: ∼7.0-fold) and in the NK libraries (mbp: ∼4.2-fold; mal: ∼5.4-fold; slc1a2: ∼1.2-fold) (Fig. 1 F and G and SI Appendix, Fig. S1C) further confirmed specificity of the IPs.
Candidate Projection Neuron Genes.
After qPCR confirmation that the IPs were specific for projection neurons, as a first filtering step, we performed bulk ribosomal RNA-seq on all IP (GFP or HA-tagged ribosomes) and input samples (dorsal spinal cord and TNC). To compare the PN and NK datasets obtained by RNA sequencing and differential expression analysis (SI Appendix, Fig. S1 F and G and Datasets S1 and S2) we plotted the gene fold changes within each dataset against the changes in the other (Fig. 1H). Data points in quadrants one (Q1) and three (Q3) represent transcripts that are enriched or depleted, respectively, in projection neurons in both datasets; data points in quadrants two (Q2) and four (Q4) represent transcripts that are differentially enriched or depleted. As ∼90% of all lamina I spinoparabrachial neurons express the NK1R (37, 38), we expected both datasets to be largely overlapping and thus the majority of data points should lie in Q1 and Q3. This was indeed what we observed. On the other hand, and somewhat unexpectedly, as Phox2A had only been recorded during development (27), we observed significant enrichment of Phox2a (∼4.8-fold) in the NK1R subset of projection neurons, but not in the non-NK1R subset. We presume that our detection of the actively translated Phox2A message in the adult using RNA-seq is a more sensitive detection method than immunohistochemistry. This association suggests that the Phox2a population of projection neurons is functionally comparable to the NK1R-expressing subset. Importantly, the transcripts in Q4 provide a valuable (albeit limited) list of genes likely expressed by the non–NK1R-expressing projection neurons, for which there are currently no or only very limited marker genes identified (Dataset S3). Lastly, using qPCR we confirmed enrichment of the candidate gene hits from the RNA-seq datasets (SI Appendix, Fig. S1 D and E).
Because the first filter (RNA sequencing and confirmation by qPCR) identified pan-neuronal as well as projection neuron genes, in a second filter step we performed fluorescent in situ hybridization (ISH) to narrow down the list of hits to those more discretely expressed by projection neurons. As a result, our focus is on several genes that are enriched in the PN or NK RNA-seq and qPCR datasets (Q1) (Fig. 1H and SI Appendix, Fig. S1 D–G). Importantly, we prioritized genes that have previously been implicated in pain and/or itch processing, but not described with respect to projection neuron neurochemistry. Because the TNC contains significantly greater numbers of projection neurons than in the spinal cord, our analyses began in the TNC.
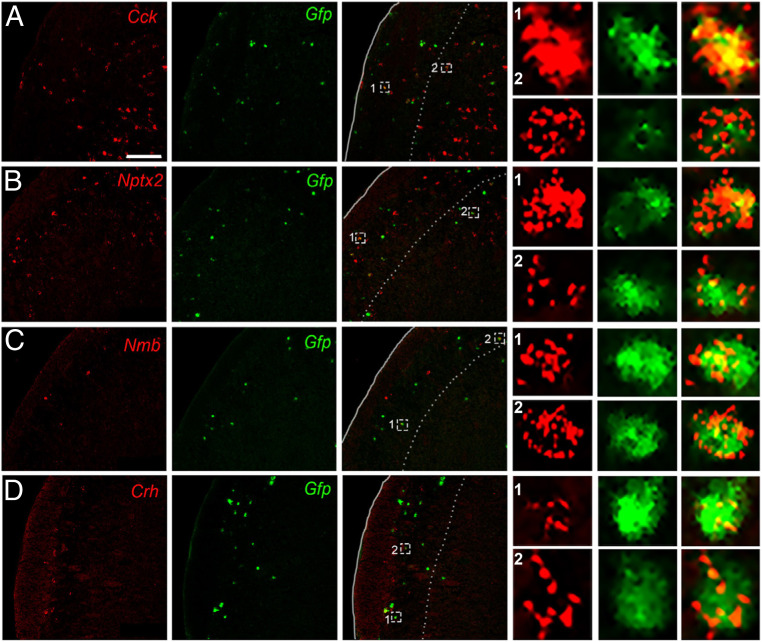
Cck encodes cholecystokinin, a peptide expressed in dorsal horn neurons (33, 39–43) and has been implicated as an antiopioid (44, 45). To confirm our RNA-seq and qPCR findings of Cck enrichment in projection neurons, we retrogradely labeled LPb-projecting neurons with HSV-GFPL10 and performed double fluorescent ISH for Gfp and Cck in the TNC. Fig. 2A illustrates Cck-expressing/Gfp-positive neurons (i.e., projection neurons) in laminae III and IV, and interestingly, many fewer in lamina I. However, based on the high number of Cck-positive/Gfp-negative neurons, we conclude that the majority of the Cck-expressing cells are interneurons. We observed similar patterns of double labeling in the spinal cord dorsal horn.
Fig. 2.
In situ hybridization confirms candidate projection neuron genes identified by RNA-seq. Representative TNC sections illustrate colabeling of Cck (A), Nptx2 (B), Nmb (C), and Crh (D) mRNA (red) with Gfp-tagged trigeminoparabrachial projection neurons (green). Insets show enlarged examples of individual cells positive for both candidate gene and Gfp. (Scale bars, 100 μm.)
The neuronal pentraxin 2 (Nptx2) gene encodes a secreted protein involved in excitatory synaptogenesis. Nptx2 has been implicated in various neuropsychiatric disorders (46) and pain processing (47). Here we recovered Gfp-expressing LPb-projecting neurons that coexpress Nptx2, predominantly in laminae I and III/IV (Fig. 2B). As for Cck, however, the high number of Nptx2-positive/Gfp-negative neurons indicates that Nptx2-expresssing interneurons predominate.
Neuromedin B (Nmb), a member of the bombesin-like family of peptides, is robustly expressed in sensory neurons and in scattered dorsal horn neurons (48) and has been implicated in both pain and itch processing (48–51). We found that Nmb is expressed sparsely in the TNC, predominantly in superficial laminae and a subset of these were Gfp-labeled LPb-projecting neurons (Fig. 2C).
Lastly, we characterized corticotropin-releasing hormone (Crh), which is a major contributor to the hypothalamic-pituitary-adrenal axis-induced stress response and has been implicated in both peripheral and central pain processing (52, 53). We recorded Crh/Gfp-labeled LPb-projecting neurons (Fig. 2D) in superficial TNC. Taken together, these results identify genes not previously associated with LPb-projecting neurons. Interestingly, although Häring et al. (33) reported that Crh is expressed in the Glut15 cluster, which includes spinoparabrachial neurons, this was not the case for Cck, Nptx2, and Nmb.
Molecular Heterogeneity of the NK1R-Expressing Dorsal Horn Neurons.
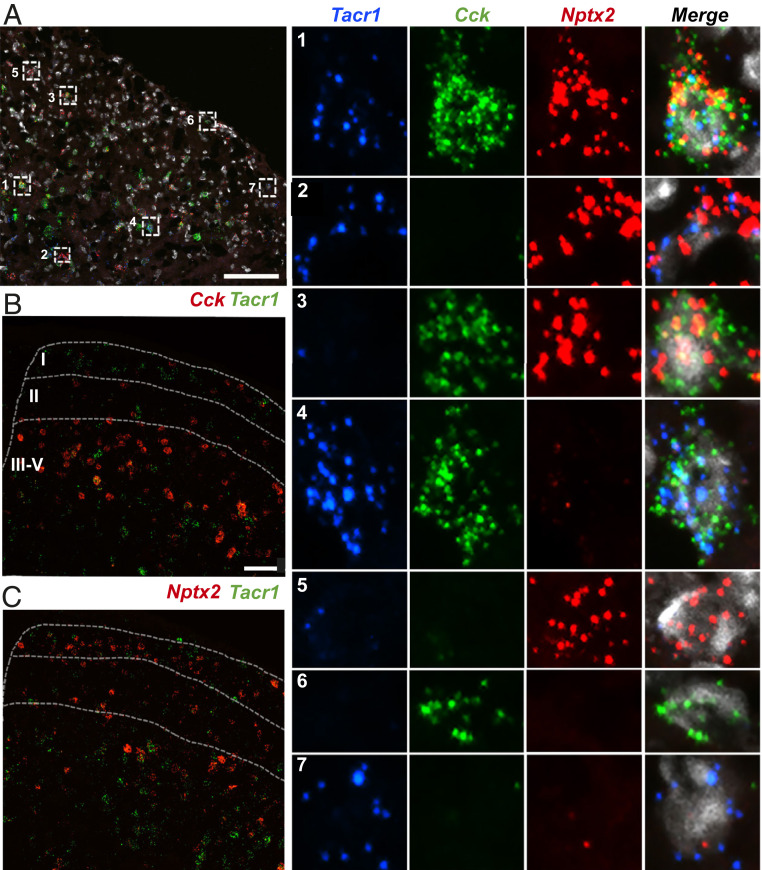
To determine the extent to which the projection neuron-associated genes identified by RNA-seq are expressed in subsets of the NK1R-expressing neurons, or whether these genes define unique populations, we next performed double- and triple-fluorescent ISH for Tacr1 and each of the enriched genes (Fig. 3 and SI Appendix, Fig. S2). We recognize that although the NK1R is expressed by a large number of interneurons (54), it is still considered a reliable marker of projection neurons. Recognizing this limitation, in a subsequent experiment, we introduced Retrobeads to directly mark the projection neurons (see below). Fig. 3A illustrates that for each gene tested, we recorded cells that coexpressed the enriched gene and Tacr1 and others that only expressed the gene or Tacr1. Interestingly, although we observed Tacr1-expressing neurons that coexpressed Cck in the deep dorsal horn, only rarely did we find Cck and Tacr1 coexpressed in lamina I neurons (Fig. 3B and SI Appendix, Fig. S2A). In both superficial and deep dorsal horn, we observed subsets of Tacr1-expressing neurons that coexpress Nptx2 (Fig. 3C and SI Appendix, Fig. S2B), Nmb (SI Appendix, Fig. S2C), and Crh (SI Appendix, Fig. S2D). In all cases, we observed many neurons that solely expressed Tacr1, or that were positive for the candidate gene, but not Tacr1 (SI Appendix, Fig. S2 A–D).
Fig. 3.
Molecular heterogeneity of dorsal horn Tacr1-expressing neurons. (A) Representative section of lumbar spinal cord after triple ISH for Tacr1 (blue), Cck (green), Nptx2 (red), and DAPI (white). Insets show examples of enlarged single neurons with different gene expression combinations, including triple-labeled (A, 1), double-labeled (A, 2 to 4) and single-labeled (A, 5 to 7) cells. (B and C) Representative sections of lumbar spinal cord after double ISH for Tacr1 (green) and (B) Cck (red) or (C) Nptx2 (red). (Scale bars, 100 μm.)
We also investigated whether Cck and Nptx2 defined nonoverlapping subpopulations of NK1R-expressing neurons. Using triple-fluorescent ISH for Tacr1, Cck, and Nptx2, we, in fact, observed neuron subtypes that express every combination of the genes (Fig. 3A). Specifically, some neurons triple-labeled for Tacr1, Cck, and Nptx2 (Fig. 3 A, 1); others coexpress two of the genes (Fig. 3 A, 2–4), and others only express one of the three (Fig. 3 A, 5–7). Based on these results, we conclude that there are at least four subsets of Tacr1-expressing neurons: Tacr1+Cck+Nptx2+ (Fig. 3 A, 1), Tacr1+Cck+Nptx2− (Fig. 3 A, 4), Tacr1+Cck−Nptx2+ (Fig. 3 A, 2), and Tacr1+Cck−Nptx2− cells (Fig. 3 A, 7). Clearly, the NK1R-expressing projection neuron population is not at all homogeneous. Rather, Tacr1 is but one marker of a molecularly heterogeneous population of projection neurons.
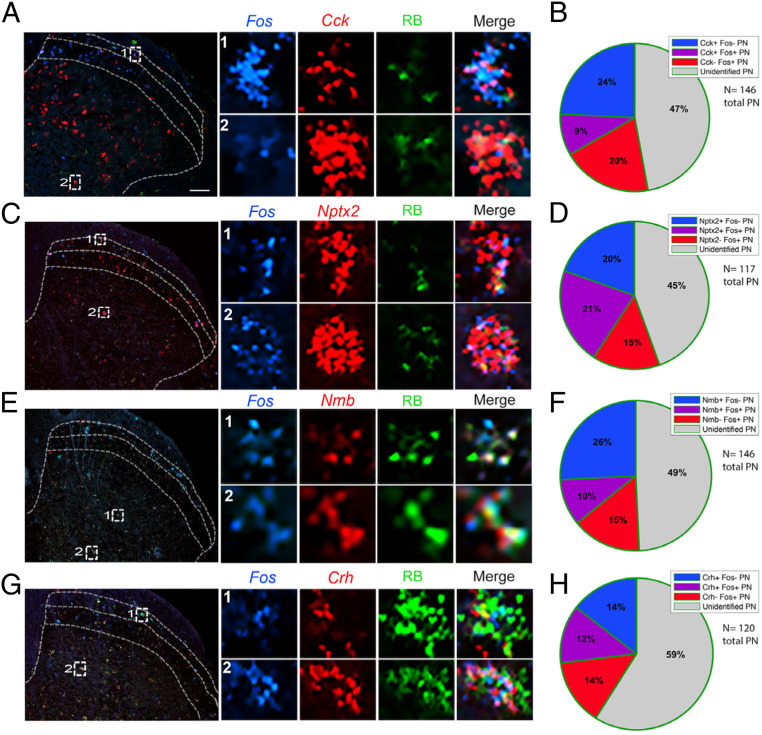
Fig. 4.
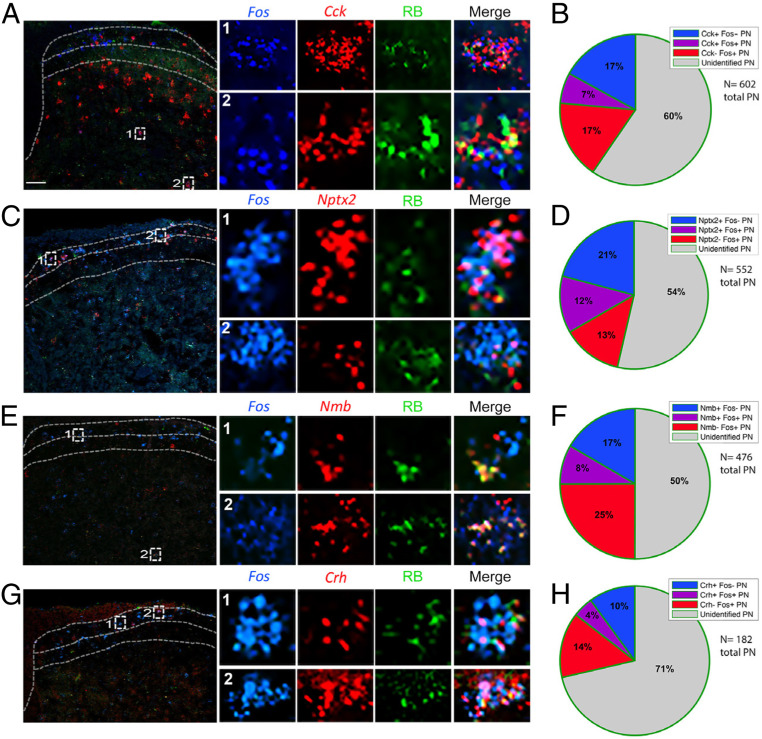
A subset of molecularly defined projection neurons responds to noxious heat. (A, C, E, and G) Representative images of lumbar spinal cord illustrate Retrobead-labeled spinoparabrachial neurons (green) coexpressing Cck (A), Nptx2 (C), Nmb (E), or Crh (G) (red), and the Fos immediate-early gene (blue), after hindpaw stimulation with noxious heat (50 °C). Insets show enlarged examples of triple-labeled cells. (B, D, F, and H) Pie charts illustrate percentages of projection neurons that express Fos after noxious heat stimulation, percentage of projection neurons that express Cck (B), Nptx2 (D), Nmb (F), or Crh (H), as well as the percentage of projection neurons that express both Fos and respective genes. Quantification includes data from two to four mice per gene and three to six lumbar spinal cord sections per mouse. (Scale bars, 100 μm.)
Fig. 5.
A subset of molecularly defined projection neurons responds to pruritic (chloroquine) stimulation. (A, C, E, and G) Representative images of trigeminal nucleus caudalis illustrate Retrobead-labeled trigeminoparabrachial neurons (green) coexpressing Cck (A), Nptx2 (C), Nmb (E), or Crh (G) (red) and the Fos immediate-early gene (blue), after chloroquine injection into the cheek. Insets show enlarged examples of triple-labeled cells. (B, D, F, and H) Pie charts illustrate percentages of projection neurons that express Fos after chloroquine injection, percentage of projection neurons that express Cck (B), Nptx2 (D), Nmb (F), or Crh (H), as well as the percentage of projection neurons that express both Fos and respective genes. Quantification includes data from two to four mice per gene and three to six lumbar spinal cord sections per mouse. (Scale bars, 100 μm.)
Fig. 7.
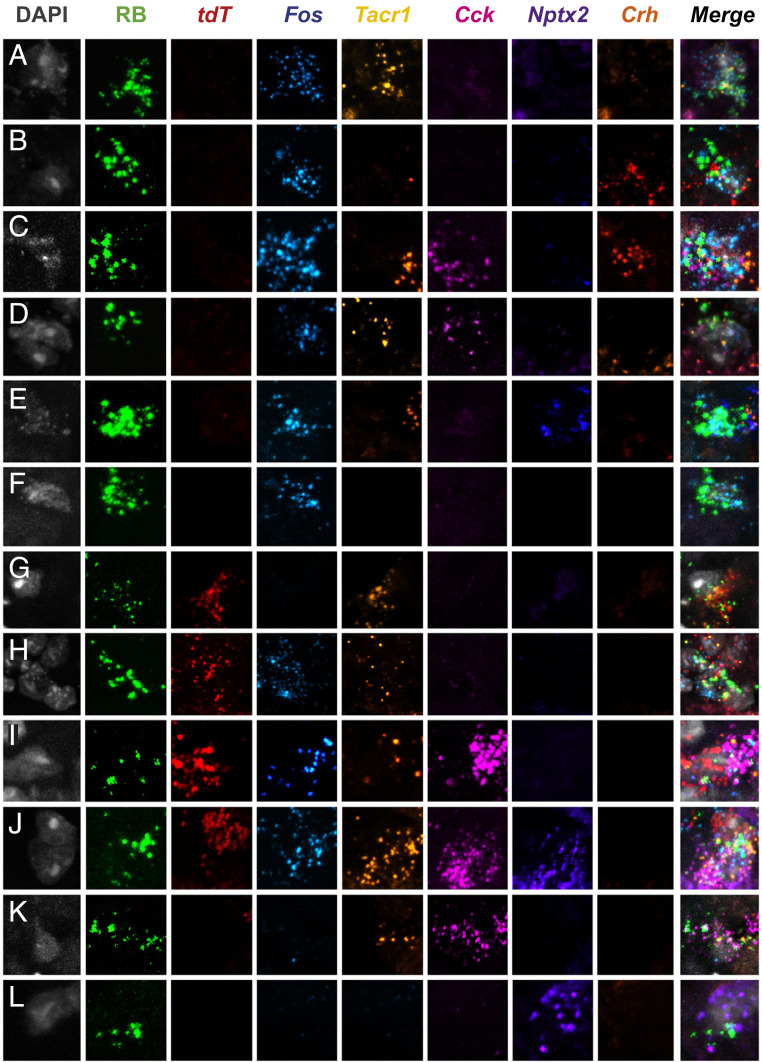
Highly multiplexed in situ hybridization reveals subsets of molecularly and functionally diverse projection neurons in the superficial dorsal horn. (A–L) Representative superficial dorsal horn section illustrates Retrobead (RB)-labeled projection neurons from heat (Fos) and chloroquine (tdT)-stimulated TRAP2 mice. A subset of projection neurons is activated by heat (50 °C), but not chloroquine (A–F); other subsets are activated by chloroquine only (G), both heat and chloroquine (H–J), or neither stimulus (K and L). Each functionally defined subset includes projection neurons that express varying combinations of the genes Tacr1, Cck, Nptx2, and Crh.
Recently, several studies reported that substance P (Tac1), which targets the NK1R, is also expressed by a subset of the NK1R-expressing projection neurons (29, 32). We confirmed these findings. Specifically, our RNA-seq and qPCR analysis not only identified Tac1 as a gene expressed by NK1R projection neurons (approximately fourfold enrichment by RNA-seq), but using ISH, we also demonstrated significant coexpression of Tac1 and Tacr1 mRNA in a subset of projection neurons labeled with Retrobeads (SI Appendix, Fig. S3A). As to other reported genes expressed in projection neurons, Häring et al. (33) delineated several that cluster with Tacr1 in the dorsal spinal cord, including Lypd1 and Elavl4. We confirmed these findings by RNA-seq (Fig. 1H) and by ISH in spinoparabrachial projection neurons (SI Appendix, Fig. S3B).
Pain- and Itch-Provoking Stimuli Engage Subsets of Molecularly Defined Projection Neurons.
We next used Fos mRNA expression to monitor the responsiveness of the retrogradely labeled (Retrobead) projection neuron subsets to an algogenic/painful stimulus (submerging one hindpaw in 50 °C water) or a pruritic/itch-provoking stimulus (cheek injection of chloroquine [CQ], 5.0 µg/µL). To prevent movement- and scratching-provoked Fos, the mice were anesthetized throughout the experiment. Twenty minutes after stimulation the mice were killed and we performed triple-labeled ISH in lumbar spinal cord and TNC. The total cell counts for each experiment were obtained by combining data from two to four mice with three to six sections each, and included Retrobead-labeled cells from superficial and deep laminae of the dorsal horn and TNC, as well as the LSN.
Pain-relevant projection neurons.
As expected, Fos induction was most pronounced ipsilateral to the stimulus (SI Appendix, Fig. S4A). We recorded the greatest number of heat-induced Fos+ neurons in superficial laminae (I/II) and Cck+ projection neurons in laminae III/IV, and in both regions we observed double-labeled Fos- and Cck-positive projection neurons (Fig. 4A). On average, about 33% of all projection neurons in lumbar cord were Cck positive, and 9% expressed Cck and were noxious-heat responsive, i.e., Fos positive (Fig. 4B), indicating that ∼27% of the Cck-projection neurons responded to noxious heat. For the Nptx2 population, we observed 41% of projection neurons that were Nptx2+ and 21% that coexpressed Nptx2 and Fos after noxious heat stimulation (Fig. 4 C and D), indicating that ∼50% of the Nptx2-expressing projection neurons respond to the noxious heat stimulus. As both Nmb (Fig. 4 E and F) and Crh (Fig. 4 G and H) have highly restricted expression patterns, as expected, we only recorded a few cells per section that were positive for either gene. In this limited number, we found that 36% of the projection neurons were Nmb positive and 10% of all projection neurons were both Nmb and Fos expressing, indicating that ∼27% of the limited number of Nmb+ projection neurons are “pain” responsive. Finally, for the Crh population, we recorded 26% of projection neurons expressing the gene, and 12% of projection neurons that expressed Crh responded to noxious heat, suggesting that roughly 50% of Crh-positive projection neurons respond to noxious heat.
Itch-relevant projection neurons.
Similar to what we observed with the heat stimulus, we found that chloroquine-mediated Fos induction was most pronounced in the superficial laminae of the TNC, ipsilateral to the stimulus (SI Appendix, Fig. S4B). We found that 24% of the projection neuron population in the TNC expressed Cck and 29% of these Cck+ projection neurons were activated by pruritic stimulation (Fig. 5A); many were in deep dorsal horn. In other words, only 7% of all projection neurons coexpressed Fos and Cck (Fig. 5B). We found that 33% of TNC projection neurons express Nptx2 (Fig. 5C); 36% of these projection neurons responded to chloroquine. The latter neurons comprised 12% of all projection neurons (Fig. 5D). In contrast to the chloroquine responsive Cck+ population, the Nptx2+ responsive neurons predominated in lamina I. As for the Nmb population of TNC projection neurons, we found that 25% of projection neurons express Nmb (Fig. 5E) and 8% coexpressed Nmb and Fos, i.e., 32% of Nmb-expressing projection neurons responded to chloroquine (Fig. 5F). Lastly, we observed that only 14% of the TNC projection neurons express Crh (Fig. 5G) and 4% coexpresed Crh and Fos, indicating that 28% of Crh-positive projection neurons in the TNC respond to the pruritogen (Fig. 5H). To summarize, we report here that approximately one-third of each dorsal horn and TNC population of Cck-, Nptx2-, Crh-, and Nmb-expressing projection neurons responds to heat and/or chloroquine. The extent of convergence is addressed in the next set of experiments.
Pain- and Itch-Provoking Stimuli Converge upon Projection Neurons.
To visualize neurons in the mouse that were activated by two stimuli separated in time, we next used the TRAP2 (Targeted Recombination in Active Populations) (35) (Fig. 6 and SI Appendix, Figs. S5–S7). After exposure to a stimulus delivered within a specific time frame, neurons that are activated by that stimulus permanently express tdTomato (tdT), a reporter that is expressed in a Cre- and Fos-dependent manner. In preliminary time course studies, we determined that a larger number of TRAPed neurons could be detected 3 h after injection of 4-OH-tamoxifen (100 mg/kg). The studies were performed in mice that were injected with Fluoro-Gold (FG) into the LPb 1 wk prior to the first (TRAPed) stimulation, which allowed for analysis of convergent activation in projection neurons.
Fig. 6.
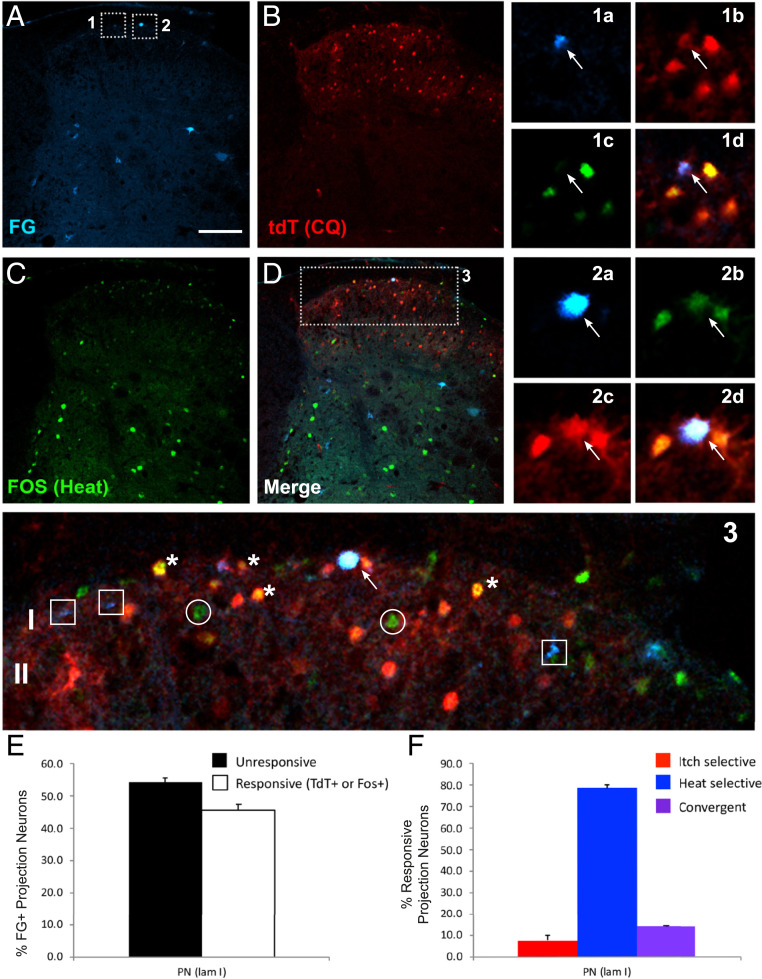
Activation of retrogradely labeled projection neurons first trapped by chloroquine and later immunostained for Fos in response to noxious heat. (A–D) Representative images of lumbar spinal cord from TRAP2-tdTomato mice illustrate Fluorogold-labeled spinoparabrachial projection neurons (A: FG; blue), chloroquine (CQ)-activated tdTomato (B: tdT; red), and heat-activated Fos (C: green)-immunoreactive neurons. Insets 1 and 2 are magnified to the Right (a–d) and 3 Below. The arrows in 1a–d point to a projection neuron (FG+) that was activated by CQ (tdT+), but not heat (Fos−) and in 2a–d, to a projection neuron that was activated by both stimuli (tdT+/Fos+). The image Below (3) highlights examples of single-labeled (squares), double-labeled (asterisks), and triple-labeled (arrow) neurons. (Scale bar, 100 μm). (E) Histograms illustrate the percentage of lamina I projection neurons (PN) that responded (white bars) or did not respond (black bars) to one or both stimuli. (F) Histograms illustrate the percentage of lamina I projection neurons (PN) that responded only to CQ (red bars), only to heat (blue bars), or to both stimuli (purple bars).
An important feature of our protocol is that we anesthetized the mice in order to reduce movement (scratching, paw withdrawal, or ambulation)-evoked tdT expression. Specifically, we anesthetized the 4-OH-tamoxifen–injected mice and then applied a noxious heat stimulus (submerging the mouse hindpaw five times in 50 °C water for 30 s, at 30-s intervals) or injected chloroquine (200 μg) into the hindpaw. One week later, at which point the 4-OH-tamoxifen window was closed, and again under anesthesia, we injected the mice that previously received the heat stimulus with chloroquine, whereas mice that previously received chloroquine were administered the heat stimulus. All mice were killed 90 min later and lumbar spinal cord was processed for immunocytochemical demonstration of the Fos protein and concurrent tdT- expression.
As expected, tdT fluorescence (endogenous) and Fos immunoreactivity were most pronounced in the dorsal horn ipsilateral to the stimulus (SI Appendix, Fig. S5). Demonstrating convergence, we found that many of the FG-positive projection neurons were also tdT- and Fos-positive (Fig. 6 A–D and SI Appendix, Figs. S6 and S7). These triple-labeled cells (i.e., FG+, Fos+, tdT+) predominated in lamina I. By contrast, double-labeled, presumptive interneurons, i.e., FG−, Fos+, tdT+, were prevalent in lamina II. Other projection neurons in laminae I and V responded only to the noxious (heat) stimulus, i.e., were tdT positive and Fos negative (SI Appendix, Fig. S6B), while others responded only to the pruritogen, i.e., were tdT negative and Fos positive (SI Appendix, Fig. S6A). Somewhat surprisingly, counts of single- and double-labeled projection neurons demonstrated that the majority of projection neurons did not respond to either stimulus, even though they were located in sections that contained large numbers of activated neurons (SI Appendix, Fig. S7E). There are several possible explanations for this result. First, many of the double-labeled neurons are likely interneurons, which may be less susceptible than projection neurons to the anesthetic used in this experiment. Furthermore, the retrograde tracer likely only marked a subset of the total projection neuron population and therefore some of the tdTomato- and/or Fos-positive neurons may include Fluorogold-negative projection neurons. As to specificity and convergence, although the heat-only responsive neurons predominated, notably in laminae I and II, projection neurons responsive only to chloroquine were readily recorded in all laminae, regardless of the stimulus order (Fig. 6 B and C and SI Appendix, Fig. S7 B, C, and F). Taken together, we conclude that subsets of projection neurons transmit both pain- and itch-relevant messages to the brain. These are intermingled with others that are more selective to one or the other stimulus.
Molecularly Distinct Projection Neuron Populations Are Functionally Heterogeneous.
To integrate the molecular heterogeneity findings demonstrated above with the functional heterogeneity revealed in the TRAP2 studies, we next performed a highly multiplexed, albeit nonquantitative ISH analysis, on lumbar dorsal horn tissue from TRAP2 mice stimulated alternatively with pain- and itch-provoking stimuli. In these experiments, after the 4-OH-tamoxifen, we first “TRAP”ed itch-responsive neurons by injecting chloroquine into the hindpaw, and 1 wk later we stimulated pain-responsive neurons by submerging the same hindpaw in 50 °C water. To identify projection neurons, these studies were performed in mice that were injected with Retrobeads into the LPb 1 wk prior to the chloroquine. We killed the mice 15 to 30 min after the second stimulus and performed multiplexed ISH for tdT (itch-responsive neurons), Fos (pain-responsive neurons), Tacr1, Cck, Nptx2, and Crh (Figs. 7 and 8 and SI Appendix, Figs. S8 and S9). In both the superficial (Fig. 7) and deep dorsal horn (SI Appendix, Fig. S9 A–G), as well as the LSN (SI Appendix, Fig. S9 H and I) we observed projection neurons that responded only to the pain-provoking stimulus, i.e., are Fos positive and tdT negative (Fig. 7 A–F and SI Appendix, Fig. S9 D, E, and I). Some projection neurons responded to both the pain- and itch-provoking stimuli (Fig. 7 H–J and SI Appendix, Fig. S9 A–C and H) and others did not respond to either (Fig. 7 K and L and SI Appendix, Fig. S9 F and G). Less frequently, we observed neurons that responded only to the itch-provoking stimulus (Fig. 7G). Importantly, each of these activated projection neurons expressed varying combinations of the genes. As expected, most of the projection neurons express the NK1R and one or more of the genes; however, by integrating the TRAP2 analysis, we now demonstrate that these neurons are also functionally heterogeneous, including itch- and pain-responsive subsets. Lastly, we found that the non–NK1R-expressing projection neurons are also molecularly diverse, but in this limited population we did not observe examples of convergence of pain and itch inputs onto these cells (Fig. 8). Overall, we conclude that despite limited evidence for functional labeled lines, the molecular diversity of the projection neurons that respond to both pain- and itch-provoking inputs provides a basis for the transmission of distinct functional messages to the brain.
Fig. 8.
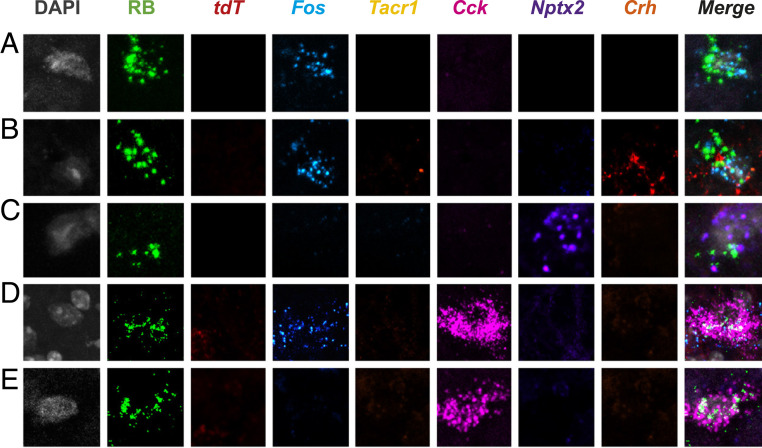
NK1R-negative projection neurons are also molecularly heterogeneous. Representative Retrobead (RB)-labeled projection neurons in superficial (A–C) and deep dorsal horn (D and E) of TRAP2 mice that are negative for Tacr1 express different combinations of Cck, Nptx2, and Crh. The illustrated projection neurons respond either only to heat (50 °C; i.e., Fos mRNA+; A, B, and D) or neither to heat nor chloroquine (Fos mRNA− and tdT mRNA−; C and E).
Discussion
There is general agreement that primary sensory neurons and dorsal horn interneurons are heterogeneous, responding to different degrees selectively to pain- and or itch-provoking stimuli. Surprisingly, despite considerable evidence that the projection neurons that transmit pain and itch are diverse in regards to location, projection targets, morphology, and electrophysiological properties, reference is often made to a rather homogeneous molecular and functional population of projection neurons. Based on the present ribosomal profiling of the LPb-projecting neurons we conclude that there are, in fact, molecularly and functionally distinct subpopulations of projection neurons that can be distinguished based on gene expression, spatial location in the dorsal horn and TNC, and responsiveness to pain- and/or itch-provoking stimulation.
Technical Considerations.
As noted above, we designed the initial filtering steps to identify neuronal genes that are expressed by projection neurons. Given the nature of the input samples, which included all spinal cord and TNC cells, glial and neuronal, we recognize that many of the RNA-seq hits, although clearly enriched in neurons relative to glia, also included genes expressed by interneurons. However, the subsequent filter steps, taken together with a comparison of results with the Häring et al. (33) database, directed our attention to genes that, although not exclusively expressed in projection neurons, unequivocally defined subtypes. Furthermore, because there is considerable variability in retrograde labeling, it was never our intention to quantify the percentage of projection neurons that express each gene. Rather these filtering steps identified genes that informed the subsequent in situ and functional studies that demonstrated molecular and functional heterogeneity of the projection neurons.
Although our results clearly indicate that there is convergence of noxious and pruritic inputs onto subsets of projection neurons, for several technical reasons, it was more difficult to provide evidence for labeled lines, i.e., segregation of pain and itch inputs onto distinct subsets of projection neurons. First, as noted above, retrograde labeling of spinal cord projection neurons using viruses and tracers can vary considerably from mouse to mouse and, therefore, it is very likely that we missed subsets of projection neurons. As a result, numbers of projection neurons in the samples were undoubtedly underestimated. Second, although the tamoxifen inducibility of the TRAP2 mouse allows for temporal and spatial recombination, it was still difficult to find the right balance between labeling large numbers of active neurons (achieved with higher doses of tamoxifen) vs. nonselective labeling (due to movement and walking rather than to “pain” or “itch”). Although it was possible to mitigate the latter by stimulating the mice under anesthesia, recovery from anesthesia occurs before the end of the tamoxifen window. Therefore, some walking-induced labeling will be detected. Finally, because induction of the Fos activity marker was easier and more consistent to achieve than induction of the tdTomato activity reporter (due to its tamoxifen dependence), their patterns of expression were not identical and made it impossible to conclude for example that a Fos-positive (heat-mediated) but tdT-negative (CQ-mediated) neuron corresponded to a neuron that was activated by heat but not CQ. Indeed, the lack of tdT labeling could have been the result of insufficient tamoxifen-mediated recombination. On the other hand because Fos is much more sensitive than tdTomato, we could confidently conclude that a tdTomato-positive, but Fos-negative neuron corresponded to a neuron that responded selectively to chloroquine. In other words, the findings argue strongly for some specificity in the responsiveness of subsets of projection neurons.
NK1R Is Not the Ideal Projection Neuron Marker.
A common trend in the literature is to equate pain and itch projection neurons with NK1R expression. Although Todd and colleagues determined that 80% and ∼90% of lamina I projection neurons in the rat (38, 55) and mouse (32), respectively, express the NK1R, fewer than half of deep dorsal horn projection neurons express the receptor (32, 55). Studies that used the NK1R to define the functional contribution of those projection neurons also have limitations. For example, Mantyh et al. (25) and Carstens et al. (26) found that ablating NK1R-expressing neurons reduces behaviors indicative of both pain and itch. At first glance this suggests that pain and itch inputs converge on NK1R-expressing projection neurons. Based on our present findings, we found that a population of NK1R-expressing projection neurons can transmit both inputs; however, we also found that subpopulations of NK1R-expressing neurons likely transmit modality-specific information to the brain. We conclude, therefore, that the NK1R is not the ideal marker to interrogate specificity at the level of the projection neurons.
Projection Neurons that Do Not Express the NK1R.
Although most superficial dorsal horn projection neurons express the NK1R, about 10 to 20% of the lamina I projection neurons do not (1). The same is true for the majority of projection neurons in laminae III to V, which despite considerable literature implicating them in pain processing, have largely been ignored (6). In the present study, we compared RNA-seq data of projection neurons from wild-type mice (i.e., all LPb-projecting neurons of the spinal cord dorsal horn and TNC) with those from NK1R-Cre mice (i.e., NK1R-expressing projection neurons). Using differential analysis, we generated a list of genes that are significantly enriched in the PN dataset and depleted in the NK dataset. We hypothesize that these genes are selectively enriched in the non–NK1R-expressing projection neurons. Although pursuit of these genes is not the subject of the present study, we present them as a resource for future investigation. Additionally, our RNA-seq data provide a preliminary confirmation of a recent identification of Gpr83 expression in the non–NK1R-expressing projection neurons (28). Specifically, although modest, we found a ∼1.7-fold enrichment of Gpr83 in the PN dataset, but no enrichment in the NK dataset (SI Appendix, Files S1 and S2). On the other hand, we did not find enrichment of two other reported markers of the non–NK1R-expressing projection neurons, namely the glycine α1 (GlyRα1) and GluR4 AMPA receptor subunits (56, 57). Finally, our studies using Hiplex ISH revealed examples of molecularly diverse non-NK1R–expressing projection neurons. In contrast to our findings of algogen and pruritogen convergence on NK1R projection neurons, however, we did not find comparable convergence in what admittedly was a limited sample of non-NK1R projection neurons. For the same reason we are hesitant to conclude that there is greater specificity of modality transmission along the non-NK1R population.
Molecular Properties of Projection Neurons vs. Existing Dorsal Horn Neuronal Transcriptomics.
Recent single-cell and single-nucleus RNA-sequencing studies reported on the molecular heterogeneity of spinal cord neurons. Sathyamurthy et al. (58) identified 16 dorsal horn excitatory neuron types, but interestingly, they concluded that none are defined uniquely by NK1R expression. In fact, they found expression of Tacr1 across several clusters and made no mention of projection neurons. In contrast, Häring et al. (33) delineated 15 dorsal excitatory neuron categories and, by integrating a retrograde tracing approach, concluded that spinoparabrachial neurons are concentrated within one of the 15 clusters of excitatory neurons (Glut 15). Our study differs considerably from these studies in approach, experimental procedures, and most importantly, in conclusions. Thus, to specifically characterize the molecular heterogeneity of projection neurons, we used bulk profiling of isolated projection neuron ribosomes from dorsal spinal cord and TNC tissue. Not surprisingly, we concur with Häring et al.’s identification of Glut 15, one of the clusters defined by NK1R enrichment, as a projection neuron population. Specifically, as predicted by Häring et al. (33), we identified Crh, Lypd1, and Elavl4 in projection neurons, all of which are included in the Glut 15 cluster. Importantly, although Häring et al. proposed Lypd1 as a novel marker for projection neurons, because Lypd1 is expressed in almost 95% of NK1R projection neurons, it cannot be used to define subtypes. In contrast, we identified several genes expressed in more discrete subsets of projection neurons. Importantly, several of these genes belong to excitatory populations other than the Glut 15 cluster. For example, our finding of Cck+, Nptx2+, and Nmb+ projection neurons demonstrates that there are spinoparabrachial neurons within the Glut 2, 3, 7, 9, and 11 through 14 excitatory clusters. On the other hand, as many of these genes are also expressed in sensory neurons and dorsal horn interneurons, we could not exclusively associate a particular candidate gene with a functional subset of projection neurons. It follows that these genes cannot be used as sole markers of projection neurons. Future studies of their relative functional contributions will require complex intersectional approaches.
Specificity vs. Convergence and the Generation of Pain and Itch Percepts.
Using techniques that selectively stimulate or ablate subsets of neurons (e.g., DREADDS, intersectional knockouts, optogenetics) recent studies have provided considerable evidence for a labeled-line view of pain and itch transmission, at least at the level of primary sensory neurons and dorsal horn interneurons (3–5, 59). On the other hand, with some exceptions (10, 13), electrophysiological data overwhelmingly suggest polymodality at all levels of the neuraxis (60). The latter implies that pain- and itch-provoking inputs either converge on modality-indiscriminate circuits, a conclusion that ignores results from many studies, or that a population code is in effect, one that results from cross-talk among different labeled lines. In fact, with regard to pain, itch, and thermal processing, several groups described combinatorial coding at the level of the DRG (61) and spinal cord (60, 62). Indeed, Sun et al. (60) reported that GRP interneurons, generally considered to exclusively contribute to the processing of itch messages, also contribute to pain processing, depending on noxious stimulus intensity. Our findings are consistent with a population code model of pain and itch processing. We speculate that a similar code underlies the processing of pain and itch messages by the molecularly diverse population of projection neurons that receive convergent modality inputs. To what extent a labeled line component integrates with the output of projection neurons that use a population code in the generation of pain and itch and most importantly how the brain interprets these messages remains to be determined.
Methods
Animals.
Animal experiments were approved by the University of California, San Francisco (UCSF) Institutional Animal Care and Use Committee and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals (63). We puchased the following mice from The Jackson Laboratory: Male and female C57BL/6J mice (stock no. 000664) and TRAP2 mice (stock no. 030323; see also ref. 35). NK1R-Cre mice were provided by Xinzhong Dong, Johns Hopkins University, Baltimore, MD (64). All mice were adults (6 to 12 wk old) and housed on a 12 h light/dark schedule.
Stereotaxic Injections.
The lateral parabrachial nucleus (LPb, coordinates (in millimeters): ±1.3 from midline, −5.34 from Bregma, −3.6 from skull) of anesthetized (ketamine (60 mg/kg)/xylazine (8.0 mg/kg) mice was injected bilaterally with 0.5 μL of a herpes simplex-based viral vector expressing the large ribosomal subunit protein L10a (a gift from Zachary Knight, UCSF, San Francisco, CA). Wild-type mice received a GFP-tagged L10a (HSV-hEF1a-GFP-L10a) viral vector and NK1R-Cre mice, a Cre-recombinase–dependent HA-tagged L10a (HSV-hEF1a-LS1L-Flag-HA-L10a) viral vector. Mice were killed 2 to 3 wk after surgery and their tissues dissected for immunoprecipitation. To retrogradely label LPb-projecting neurons of the spinal cord dorsal horn and TNC, wild-type mice received 0.5 μL of green Retrobeads (Lumafluor), 2% FG (Fluorochrome), or the HSV-hEF1a-GFP-L10a.
Immunoprecipitation, RNA, and cDNA Preparation.
To immunoprecipitate tagged ribosomes and their associated mRNA, we followed the protocol described by Ekstrand et al. (36). See SI Appendix for minor modifications applied to the protocol. cDNA was prepared with the Ovation RNA-seq V2 kit (Nugen) and a portion was set aside for analysis by qPCR (see below for methods used). Libraries for RNA-seq were prepared with the remaining cDNA using the Ovation Ultralow Library System (NuGen).
qPCR.
We quantified mRNA levels with the Bio-Rad CFX Connect System using TaqMan Gene Expression Assay (Applied Biosystems). All TaqMan values were normalized to ribosomal protein Rpl27. Fold-enrichment plots from TaqMan data were obtained by dividing the IP RNA value for each gene by the input RNA value (IP/input).
Sequencing and Bioinformatic Analysis.
RNA sequencing was performed on an Illumina HiSEq. 4000 sequencer using 50-bp single-end reads. We sequenced the following samples: GFP-IP experiment (four IP replicates paired with four input replicates, which were obtained from pooling four mice per replicate); and HA-IP experiment (three IP replicates paired with three input replicates, obtained from pooling four mice per replicate). RNA-seq data were processed in Galaxy and further analyzed with Microsoft Excel and MATLAB (R2015b). RNA STAR (v 2.6.0b) was used to align the reads. Htseq-count (v 0.9.1) and DESeq2 (v 1.18.1) were used for transcript abundance estimation and differential expression testing, respectively. The University of California Santa Cruz GRCm38 (mm10 build) was used for gene annotation.
ISH.
We followed the Advanced Cell Diagnostics protocol for RNAscope ISH on fresh TG tissue (Advanced Cell Diagnostics, multiplex fluorescent assay, cat. no. 320850 and Hiplex 12 ancillary kit, cat. no. 324140). See SI Apendix for full protocols. For the complete list of probes used, see SI Appendix, Table S1.
For Fos induction studies, mice were injected 2 wk prior to stimulation with HSV-hEF1a-GFP-L10a or green Retrobeads into the LPb. For pruritic stimulation, under anesthesia, we injected chloroquine (CQ; 500 μg in 100 μL) into the left cheek, and performed ISH in the TNC for Fos and each candidate gene. For noxious heat stimulation, under anesthesia, we submerged the left hindpaw in 50 °C water for 30 s and performed ISH on the lumbar spinal cord. All mice were injected intraperitoneally (i.p.) with an anesthetizing dose of Avertin (1.25%) 20 to 30 min before stimulation and killed 15 to 30 min after stimulation with a lethal Avertin dose.
TRAP2 Assay.
TRAP2 mice (35) were crossed with mice that ubiquitously express tdTomato (tdT) after tamoxifen-induced Cre recombination (Ai14 mice; The Jackson Laboratory, stock no. 007914). Adult male TRAP2-tdT mice first received 2% FG into the LPb (n = 3 per group). One week later, the TRAP2-tdT mice received an i.p. injection of 4-hydroxy-tamoxifen (100 mg/kg, dissolved in oil; Sigma). Three hours later, the TRAP2-tdT mice were stimulated, under anesthesia, contralateral to the FG injections: one group of mice received 200 μg of chloroquine (in 25 μL) into the hindpaw (CQ group) and in another group the entire hindpaw was dipped five times for 30 s each, into a 50 °C waterbath, with 30 s between each stimulus (heat group). One week later, under anesthesia, the CQ group received the heat stimulus and the heat group received the CQ stimulus. Ninety minutes later, all mice received an overdose of Avertin (2.5% and lumbar spinal cord tissue was processed for immunohistochemistry or ISH.
Imaging and Image Analysis.
All images were taken with an LSM 700 confocal microscope (Zeiss) and Zeiss Zen software (2010). The same parameters were used for all images within an experiment.
We analyzed 12-μm sections (L3–L5 lumbar spinal cord or TNC) from two to four animals per condition (three to six sections per animal). We used a custom MATLAB script to count cells positive for the retrograde labels, the candidate genes, and/or Fos. We also used MATLAB to manually draw a border between superficial and deep dorsal horn or TNC and used a custom MATLAB script to count the percentage of projection neurons in each region that express each gene.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant NSR35097306 (A.I.B.), Open Philanthropy (A.I.B.), and the NSF Graduate Research Program (R.W.). We thank Dr. Zachary Knight for providing significant advice in the design of experiments.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105732118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Todd A. J., Neuronal circuitry for pain processing in the dorsal horn. Nat. Rev. Neurosci. 11, 823–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braz J., Solorzano C., Wang X., Basbaum A. I., Transmitting pain and itch messages: A contemporary view of the spinal cord circuits that generate gate control. Neuron 82, 522–536 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaMotte R. H., Dong X., Ringkamp M., Sensory neurons and circuits mediating itch. Nat. Rev. Neurosci. 15, 19–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch S. C., Acton D., Goulding M., Spinal circuits for touch, pain, and itch. Annu. Rev. Physiol. 80, 189–217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peirs C., Seal R. P., Neural circuits for pain: Recent advances and current views. Science 354, 578–584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wercberger R., Basbaum A. I., Spinal cord projection neurons: A superficial, and also deep, analysis. Curr. Opin. Physiol. 11, 109–115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen B. N., Perl E. R., Spinal neurons specifically excited by noxious or thermal stimuli: Marginal zone of the dorsal horn. J. Neurophysiol. 33, 293–307 (1970). [DOI] [PubMed] [Google Scholar]
- 8.Han Z.-S., Zhang E.-T., Craig A. D., Nociceptive and thermoreceptive lamina I neurons are anatomically distinct. Nat. Neurosci. 1, 218–225 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Bester H., Chapman V., Besson J. M., Bernard J. F., Physiological properties of the lamina I spinoparabrachial neurons in the rat. J. Neurophysiol. 83, 2239–2259 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Craig A. D., Krout K., Andrew D., Quantitative response characteristics of thermoreceptive and nociceptive lamina I spinothalamic neurons in the cat. J. Neurophysiol. 86, 1459–1480 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Hachisuka J., et al., Semi-intact ex vivo approach to investigate spinal somatosensory circuits. eLife 5, 1–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hachisuka J., Koerber H. R., Ross S. E., Selective-cold output through a distinct subset of lamina I spinoparabrachial neurons. Pain 161, 185–194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dostrovsky J. O., Craig A. D., Cooling-specific spinothalamic neurons in the monkey. J. Neurophysiol. 76, 3656–3665 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Light A. R., Sedivec M. J., Casale E. J., Jones S. L., Physiological and morphological characteristics of spinal neurons projecting to the parabrachial region of the cat. Somatosens. Mot. Res. 10, 309–325 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Wall P. D., The laminar organization of dorsal horn and effects of descending impulses. J. Physiol. 188, 403–423 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig A. D., Kniffki K. D., Spinothalamic lumbosacral lamina I cells responsive to skin and muscle stimulation in the cat. J. Physiol. 365, 197–221 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willis W. D. Jr., Pain pathways in the primate. Prog. Clin. Biol. Res. 176, 117–133 (1985). [PubMed] [Google Scholar]
- 18.Hylden J. L. K., Hayashi H., Dubner R., Bennett G. J., Physiology and morphology of the lamina I spinomesencephalic projection. J. Comp. Neurol. 247, 505–515 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Carstens E., Responses of rat spinal dorsal horn neurons to intracutaneous microinjection of histamine, capsaicin, and other irritants. J. Neurophysiol. 77, 2499–2514 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Simone D. A., et al., Comparison of responses of primate spinothalamic tract neurons to pruritic and algogenic stimuli. J. Neurophysiol. 91, 213–222 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Davidson S., et al., The itch-producing agents histamine and cowhage activate separate populations of primate spinothalamic tract neurons. J. Neurosci. 27, 10007–10014 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson S., et al., Pruriceptive spinothalamic tract neurons: Physiological properties and projection targets in the primate. J. Neurophysiol. 108, 1711–1723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moser H. R., Giesler G. J. Jr., Characterization of pruriceptive trigeminothalamic tract neurons in rats. J. Neurophysiol. 111, 1574–1589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen N. A., Giesler G. J. Jr., Response characteristics of pruriceptive and nociceptive trigeminoparabrachial tract neurons in the rat. J. Neurophysiol. 113, 58–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantyh P. W., et al., Inhibition of hyperalgesia by ablation of lamina I spinal neurons expressing the substance P receptor. Science 278, 275–279 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Carstens E. E., Carstens M. I., Simons C. T., Jinks S. L., Dorsal horn neurons expressing NK-1 receptors mediate scratching in rats. Neuroreport 21, 303–308 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roome R. B., et al., Phox2a defines a developmental origin of the anterolateral system in mice and humans. Cell Rep. 33, 108425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi S., et al., Parallel ascending spinal pathways for affective touch and pain. Nature 587, 258–263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang T., et al., Identifying the pathways required for coping behaviours associated with sustained pain. Nature 565, 86–90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blomqvist A., Mackerlova L., Spinal projections to the parabrachial nucleus are substance P-immunoreactive. Neuroreport 6, 605–608 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Gamboa-Esteves F. O., McWilliam P. N., Batten T. F., Substance P (NK1) and somatostatin (sst2A) receptor immunoreactivity in NTS-projecting rat dorsal horn neurones activated by nociceptive afferent input. J. Chem. Neuroanat. 27, 251–266 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Cameron D., et al., The organisation of spinoparabrachial neurons in the mouse. Pain 156, 2061–2071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Häring M., et al., Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat. Neurosci. 21, 869–880 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Tekinay A. B., et al., A role for LYNX2 in anxiety-related behavior. Proc. Natl. Acad. Sci. U.S.A. 106, 4477–4482 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen W. E., et al., Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekstrand M. I., et al., Molecular profiling of neurons based on connectivity. Cell 157, 1230–1242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Khater K. M., Todd A. J., Collateral projections of neurons in laminae I, III, and IV of rat spinal cord to thalamus, periaqueductal gray matter, and lateral parabrachial area. J. Comp. Neurol. 515, 629–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spike R. C., Puskár Z., Andrew D., Todd A. J., A quantitative and morphological study of projection neurons in lamina I of the rat lumbar spinal cord. Eur. J. Neurosci. 18, 2433–2448 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Gutierrez-Mecinas M., et al., Expression of cholecystokinin by neurons in mouse spinal dorsal horn. J. Comp. Neurol. 527, 1857–1871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leah J., Menétrey D., de Pommery J., Neuropeptides in long ascending spinal tract cells in the rat: Evidence for parallel processing of ascending information. Neuroscience 24, 195–207 (1988). [DOI] [PubMed] [Google Scholar]
- 41.Antunes Bras J. M., et al., Effects of peripheral axotomy on cholecystokinin neurotransmission in the rat spinal cord. J. Neurochem. 72, 858–867 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Elde R., Schalling M., Ceccatelli S., Nakanishi S., Hökfelt T., Localization of neuropeptide receptor mRNA in rat brain: Initial observations using probes for neurotensin and substance P receptors. Neurosci. Lett. 120, 134–138 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Hökfelt T., Holmberg K., Shi T. J. S., Broberger C., CCK-ergic mechanisms in sensory systems. Scand. J. Clin. Lab. Invest. Suppl. 234, 69–74 (2001). [PubMed] [Google Scholar]
- 44.Faris P. L., Komisaruk B. R., Watkins L. R., Mayer D. J., Evidence for the neuropeptide cholecystokinin as an antagonist of opiate analgesia. Science 219, 310–312 (1983). [DOI] [PubMed] [Google Scholar]
- 45.Watkins L. R., Kinscheck I. B., Mayer D. J., Potentiation of morphine analgesia by the cholecystokinin antagonist proglumide. Brain Res. 327, 169–180 (1985). [DOI] [PubMed] [Google Scholar]
- 46.Chang S., et al., NPTX2 is a key component in the regulation of anxiety. Neuropsychopharmacology 43, 1943–1953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miskimon M., et al., Selective expression of Narp in primary nociceptive neurons: Role in microglia/macrophage activation following nerve injury. J. Neuroimmunol. 274, 86–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleming M. S., et al., The majority of dorsal spinal cord gastrin releasing peptide is synthesized locally whereas neuromedin B is highly expressed in pain- and itch-sensing somatosensory neurons. Mol. Pain 8, 52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su P.-Y., Ko M.-C., The role of central gastrin-releasing peptide and neuromedin B receptors in the modulation of scratching behavior in rats. J. Pharmacol. Exp. Ther. 337, 822–829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan L., et al., Distinct roles of NMB and GRP in itch transmission. Sci. Rep. 7, 15466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishra S. K., Holzman S., Hoon M. A., A nociceptive signaling role for neuromedin B. J. Neurosci. 32, 8686–8695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lariviere W. R., Melzack R., The role of corticotropin-releasing factor in pain and analgesia. Pain 84, 1–12 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Hargreaves K. M., Dubner R., Costello A. H., Corticotropin releasing factor (CRF) has a peripheral site of action for antinociception. Eur. J. Pharmacol. 170, 275–279 (1989). [DOI] [PubMed] [Google Scholar]
- 54.Al Ghamdi K. S., Polgár E., Todd A. J., Soma size distinguishes projection neurons from neurokinin 1 receptor-expressing interneurons in lamina I of the rat lumbar spinal dorsal horn. Neuroscience 164, 1794–1804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marshall G. E., Shehab S. A. S., Spike R. C., Todd A. J., Neurokinin-1 receptors on lumbar spinothalamic neurons in the rat. Neuroscience 72, 255–263 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Polgár E., Al-Khater K. M., Shehab S., Watanabe M., Todd A. J., Large projection neurons in lamina I of the rat spinal cord that lack the neurokinin 1 receptor are densely innervated by VGLUT2-containing axons and possess GluR4-containing AMPA receptors. J. Neurosci. 28, 13150–13160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puskár Z., Polgár E., Todd A. J., A population of large lamina I projection neurons with selective inhibitory input in rat spinal cord. Neuroscience 102, 167–176 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Sathyamurthy A., et al., Massively parallel single nucleus transcriptional profiling defines spinal cord neurons and their activity during behavior. Cell Rep. 22, 2216–2225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kupari J., Häring M., Agirre E., Castelo-Branco G., Ernfors P., An atlas of vagal sensory neurons and their molecular specialization. Cell Rep. 27, 2508–2523.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun S., et al., Leaky gate model: Intensity-dependent coding of pain and itch in the spinal cord. Neuron 93, 840–853.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F., et al., Sensory afferents use different coding strategies for heat and cold. Cell Rep. 23, 2001–2013 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Ran C., Hoon M. A., Chen X., The coding of cutaneous temperature in the spinal cord. Nat. Neurosci. 19, 1201–1209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 64.Green D. P., Limjunyawong N., Gour N., Pundir P., Dong X., A mast cell- specific receptor mediates neurogenic inflammation and pain. Neuron 101, 412–420.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.