Viruses of bacteria, also known as bacteriophages, harbor the greatest diversity of DNA modifications identified to date. To fight against restriction endonucleases of their hosts, bacteriophages modify their genomic DNA through introduction of various moieties including amino acids, polyamines, and sugars (1, 2). A series of transformations are involved in DNA base modification. Followed by formation of hydroxymethyl pyrimidine nucleotides, which are utilized by DNA polymerase, replication and postreplicative modifications furnish installation of these moieties onto the DNA polymer (3–7). Burke et al. (8) show that bacteriophages resort to C5-cytosine methyltransferase (C5-MT) and 5-methylcytosine dioxygenase ten-eleven translocation enzyme (TET) as an alternative mechanism to postreplicatively form hydroxymethylcytosine on DNA. The bacteriophage TET enables site-specific hydroxylation of 5-methylcytosine (5mC), installed by C5-MT, to produce 5-hydroxymethylcytosine (5hmC). Through bioinformatic screening, the authors identify and characterize tailoring enzymes, such as glycosyltransferases, that collaborate with phage C5-MT and TET to further elaborate DNA at 5hmC site.
TET/base J-binding proteins (TET/JBPs) are present in all domains of life. They belong to iron(II) and 2-oxoglutarate (Fe/2OG)-dependent dioxygenases (9). Fe/2OG enzymes are known to catalyze diverse, but well-controlled oxidative modifications, such as hydroxylation, halogenation, epoxidation, among many others, at the expense of 2OG and molecular oxygen (10–13). In eukaryotes, TETs have been demonstrated to catalyze consecutive reactions to covert 5mC to 5-carboxylcytosine (5caC) though 5hmC and 5-formylcytosine (5fC), en route to demethylation of 5mC (Fig. 1) (14–16). In addition to 5mC, its oxidized forms (5hmC, 5fC, and 5caC) have also been demonstrated to be stable epigenetic marks that have regulatory functions in chromatin remodeling and gene expression (17, 18).
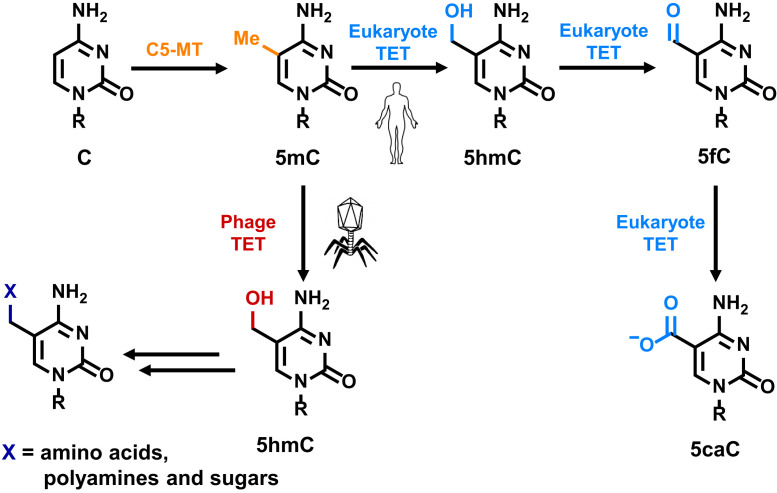
Fig. 1.
TETs (ten-eleven translocation enzymes) in bacteriophage and eukaryote have distinctive substrate specificity and reactivity. In eukaryote, methylated cytosine (5mC) is converted to 5-carboxylcytosine (5caC) though 5-hydroxymethylcytosine (5hmC) and 5-formylcytosine (5fC), while 5hmC is the only product (5mC → 5hmC) catalyzed by phage TETs.
Notably, the work carried out by Burke et al. (8) demonstrates that bacteriophage TETs along with C5-MT and tailoring enzymes serve a role in phage DNA hypermodification. Using an Escherichia coli expression system and liquid chromatography-mass spectrometry/mass spectrometry, Burke et al. provide evidence that phage TETs perform a single oxidation event to convert 5mC to 5hmC, but no further oxidation as observed in eukaryotic TETs (Fig. 1). Additionally, the authors divulge a GpC-centered specificity for these enzymes using next-generation sequencing approaches. The findings are supported in vitro by testing the hydroxylation activity of a purified TET. The choice of a palindromic dinucleotide sequence could be significant in maintaining information during DNA replication, similar to what is shown for CpG (hydroxy)methylation in eukaryotes. On the other hand, the distinct difference in oxidation cycling between the phage (5mC → 5hmC) and eukaryotic TETs (5mC → 5hmC → 5fC → 5caC) hints a need of C5-MT/TET in phage to increase the chemical diversity of their genomic DNA, which can be used to employ new functionalities in the arms race against bacteria. The active-site features of phage TETs that enforce the single-oxidation event and, at the same time, avoid further oxidation are yet to be determined. The insight will undoubtedly contribute to a deeper understanding of these important enzymes.
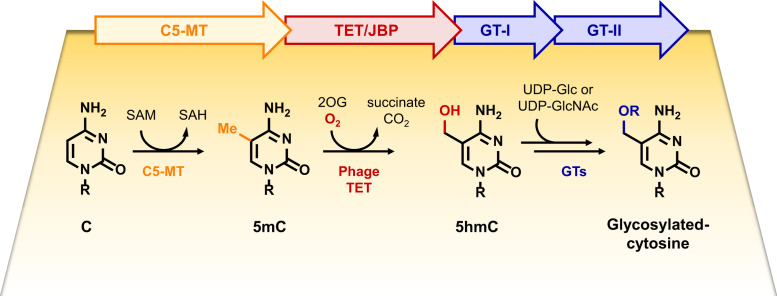
Computational analyses of the gene neighborhoods of phage C5-MT/TET suggest that 5hmC, generated via C5-MT and TET, can be utilized by other adjacent enzymes. The authors examined two biosynthetic gene clusters composed of two glycosyltransferases clustered with C5-MT and TET. Through both in vivo and in vitro studies, the authors discovered formation of glycosylated cytosines on DNA (Fig. 2). These observations confirm the interdependency of these clustered genes in context of glycosylated cytosine formation and suggest that phage cytosine hypermodification is furnished through a collaborative enzymatic transformation.
Fig. 2.
C5-glycosylated cytosines are assembled through a collaborative work of C5-MT, TET, and glycosyltransferases (GTs) in bacteriophage.
Chemical modification of canonical nucleobases is expected to expand the DNA functions, similar to what has been shown for posttranscriptional modifications in RNA and posttranslational modifications in proteins. The rational exploitation of “genome neighborhood” information in phage using computation-guided strategies for functional discovery, followed by biochemical analysis of the predicted biosynthetic pathways by Burke et al. reveals a strategy to expand the structural diversity of 5mC and uncovers a biological function of Fe(II)/2OG-dependent 5mC TET other than epigenetic regulation demonstrated in eukaryotic systems.
Footnotes
The authors declare no competing interest.
See companion article, “Phage-encoded ten-eleven translocation dioxygenase (TET) is active in C5-cytosine hypermodification in DNA,” 10.1073/pnas.2026742118.
References
- 1.Warren R. A., Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 34, 137–158 (1980). [DOI] [PubMed] [Google Scholar]
- 2.Weigele P., Raleigh E. A., Biosynthesis and function of modified bases in bacteria and their viruses. Chem. Rev. 116, 12655–12687 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kornberg S. R., Zimmerman S. B., Kornberg A., Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. J. Biol. Chem. 236, 1487–1493 (1961). [PubMed] [Google Scholar]
- 4.Zimmerman S. B., Kornberg S. R., Kornberg A., Glucosylation of deoxyribonucleic acid. II. α-Glucosyl transferases from T2- and T6-infected Escherichia coli. J. Biol. Chem. 237, 512–518 (1962). [PubMed] [Google Scholar]
- 5.Wilhelm K., Rüger W., Deoxyuridylate-hydroxymethylase of bacteriophage SPO1. Virology 189, 640–646 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Schellenberger U., Livi L. L., Santi D. V., Cloning, expression, purification, and characterization of 2′-deoxyuridylate hydroxymethylase from phage SPO1. Protein Expr. Purif. 6, 423–430 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Lee Y. J., et al., Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc. Natl. Acad. Sci. U.S.A. 115, E3116–E3125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke E. J., et al., Phage-encoded ten-eleven translocation dioxygenase (TET) is active in C5-cytosine hypermodification in DNA. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2026742118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer L. M., Tahiliani M., Rao A., Aravind L., Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 8, 1698–1710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam M. S., Leissing T. M., Chowdhury R., Hopkinson R. J., Schofield C. J., 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 87, 585–620 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Gao S. S., Naowarojna N., Cheng R., Liu X., Liu P., Recent examples of α-ketoglutarate-dependent mononuclear non-haem iron enzymes in natural product biosyntheses. Nat. Prod. Rep. 35, 792–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez S., Hausinger R. P., Catalytic mechanisms of Fe(II)- and 2-oxoglutarate-dependent oxygenases. J. Biol. Chem. 290, 20702–20711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J. M. Bollinger, Jr, et al., “Mechanisms of 2-oxoglutarate-dependent oxygenases: The hydroxylation paradigm and beyond” in 2-Oxoglutarate-Dependent Oxygenases, Hausinger R. P., Schofield C. J., Eds. (Royal Society of Chemistry, 2015), pp. 95–122. [Google Scholar]
- 14.He Y. F., et al., Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito S., et al., Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiti A., Drohat A. C., Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellinger M. W., et al., 5-Formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 19, 831–833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellén M., Ayata P., Dewell S., Kriaucionis S., Heintz N., MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]