Abstract
Ribonucleotides are frequently incorporated into DNA and can be used as a marker of DNA replication enzymology. To investigate on a genome-wide scale, how E. coli pol V accesses undamaged chromosomal DNA during the SOS response, we mapped the location of ribonucleotides incorporated by steric gate variants of pol V across the entire E. coli genome. To do so, we used strains that are deficient in ribonucleotide excision repair (ΔrnhB), deficient in pol IV DNA polymerase, constitutively express all SOS-regulated genes [lexA(Def)] and constitutively “activated” RecA* (recA730). The strains also harbor two steric gate variants of E. coli pol V (Y11A, or F10L), or a homolog of pol V, (pol VR391-Y13A). Ribonucleotides are frequently incorporated by the pol V-Y11A and pol VR391-Y13A variants, with a preference to the lagging strand. In contrast, the pol V-F10L variant incorporates less ribonucleotides and no strand preference is observed. Sharp transitions in strand specificity are observed at the replication origin (oriC), while a gradient is observed at the termination region. To activate RecA* in a recA+ strain, we treated the strains with ciprofloxacin and genome-wide mapped the location of the incorporated ribonucleotides. Again, the pol V-Y11A steric gate variant exhibited a lagging strand preference. Our data are consistent with a specific role for pol V in lagging strand DNA synthesis across the entire E. coli genome during the SOS response.
Keywords: SOS response, DNA polymerase V, Steric gate mutant, R391, Ribonucleotide incorporation, Ribonucleotide Excision Repair
1. Introduction
Most damage-induced mutagenesis in E. coli is dependent upon DNA polymerase V (pol V) [1,2]. Pol V is encoded by the umuDC genes and is induced as part of the stress-inducible SOS response [3]. As part of the SOS response, the RecA recombinase is induced and forms nucleoprotein filaments [4]. This is often referred to as “activated” RecA, or RecA*. In this so-called activated form, RecA* mediates the self-cleavage of the transcriptional repressor of the SOS response, LexA, leading to the expression of all LexA-regulated genes, including UmuD and UmuC. Dimeric UmuD is not active for mutagenesis until it undergoes a similar RecA* cleavage reaction to generate UmuD’2 [5–7]. UmuD’2 interacts with UmuC to form heterotrimeric UmuD’2C = pol V [8]. However, pol V exhibits weak activity in vitro [9]. Its activity is, however, significantly increased in vitro in the presence of RecA* [10]. It is believed that activation occurs when a single RecA monomer is transferred from the 3′ end of a RecA nucleoprotein filament along with ATP to UmuD’2C to generate a UmuD’2C-RecA-ATP complex, that is the biologically active form of pol V (pol V Mut) [11].
In addition to promoting DNA damage-induced translesion synthesis (TLS), in certain genetic backgrounds, pol V Mut confers a considerable spontaneous mutator activity to E. coli [12,13]. Studies using lacZ reporter assays suggest that much of this mutator activity is targeted to the lagging DNA strand, at least in the context of the lacZ allele [14]. We were interested in determining whether this strand bias would extend to the entire E. coli genome. To do so, we generated E. coli strains in which pol V would be expected to be highly active [recA730 lexA51(Def)] and also deficient in the major ribonucleotide excision repair pathway, due to a deletion of RNase HII (ΔrnhB) [15]. We then utilized so-called “steric gate” mutant variants of pol V that have altered abilities to incorporate ribonucleotides during replication of undamaged DNA [16] and have used the HydEn-seq method [17] to track pol V-dependent misincorporated ribonucleotides across the E. coli genome. In the presence of RecA* (genetically activated recA730, or recA+ treated with ciprofloxacin), pol V variants with an increased ability to incorporate ribonucleotides were tracked to the lagging strand across the entire genome. In particular, there was a sharp strand transition at the single E. coli origin of replication (oriC). The lagging strand bias is clearly pol V-dependent, since the bias disappeared in the pol V-F10L variant that exhibits decreased ribonucleotide incorporation [16]. Our study, therefore, provides the first evidence that under appropriate activating conditions, pol V has access to the entire undamaged E. coli genome and particularly, the lagging strand.
2. Materials and methods
2.1. Bacterial strains and plasmids
All bacterial strains and plasmids used in this study are listed in Table 1.
Table 1.
Strains and plasmids used in this study.
| Strains/plasmids | Relevant genotype/ characteristic | Reference |
|---|---|---|
| RW838a | E. coli: lexA51(Def) recA730 Δ(umuDC)596::ermGT ΔdinB61::ble ΔrnhB782::kan | [15] |
| RW916a | E. coli: lexA51(Def) Δ(umuDC)596::ermGT ΔdinB61::ble ΔrnhB782::kan | LGIc stocks |
| RW988a | E. coli: Δ(umuDC)596::ermGT ΔdinB61::ble ΔrnhB782::kan ΔpolB::ΩSpc | LGI stocks |
| JW0178b | E. coli: ΔrnhB782::kan | E. coli Genetic Stock Center |
| pRW134 | pGB2, umuD’C (pol V wild-type) | [25] |
| pJM963 | pGB2, umuD’C-Y11A (pol V steric gate) | [16] |
| pJM964 | pGB2, umuD’C-F10L (pol V variant) | [16] |
| pJM1282 | pGB2, rumA′ B-Y13A (pol VR391 steric gate) | [20] |
Full genotype: thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44 galK2 hisG4 rpsL31 xyl-5 mtl-1 argE3 thi-1 sulA211.
Full genotype: Δ(araD-araB)567 ΔlacZ4787 rph-1 Δ(rhaD-rhaB)568 hsdR514.
LGI: Laboratory of Genomic Integrity.
2.2. Bacterial growth conditions
Bacterial cells were streaked out from frozen stocks on LB agar plates containing kanamycin (50 μg/mL), spectinomycin (25 μg/mL), and zeocin (25 μg/mL). 4–6 colonies were diluted in 2–3 mL LB media containing antibiotics and incubated at 37 °C overnight. The overnight culture was then diluted into 25 mL LB media containing antibiotics to have OD600 = 0.05. Where noted, ciprofloxacin was added to the media to a final concentration of 30 ng/mL. Cells were grown 2–3 hours (OD600 = 0.4–0.6) and then centrifuged to harvest the cell pellet. The saved pellet was used for DNA extraction.
2.3. HydEn-seq method
The HydEn-seq method and sequence data analyses have been previously described [17]. We used E. coli K12, DH10B reference genome build 2008-03-17 from the NCBI for sequence alignment. The strand bias plots were analyzed as previously described [18].
2.4. Data availability
The sequencing data of this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo with the accession no. GSE158570. Custom scripts are available upon request to the corresponding author.
3. Results
3.1. Genome-wide tracking of ribonucleotides in the E. coli genome by HydEn-seq method
To track the replication enzymology of pol V, we used the HydEn-seq method that has been previously used to track S. cerevisiae DNA polymerases α, ε, δ, and η [17,19]. In our current study, we have tracked incorporated ribonucleotides by pol V steric gate mutants Y11A, and F10L, and pol VR391-Y13A. These alleles were chosen based upon previous observations regarding their ability to misincorporate ribonucleotides into the E. coli genome. F10 is adjacent to the steric gate and the F10L substitution results in an increase in ribonucleotide discrimination, compared to wild-type pol V [16]; Y11A effectively removes any ribonucleotide selectivity by pol V [16]; and pol VR391-Y13A is a steric gate mutant of pol VR391, which is a potent homolog of E. coli pol V [20]. We expected that ribonucleotide misincorporation would be highest in pol VR391-Y13A and lowest in pol V-F10L. DNA libraries were prepared from the various strains and subject to HydEn-seq analysis. We used Illumina NextSeq 500 system to run paired-end sequencing. After alignment with the E. coli reference genome, we mapped the location of ribonucleotides incorporated into the genome (5′ DNA end after hydrolysis). We used the wild-type strain for background subtraction. To visualize the data, we have provided plots obtained from averaged values of three different biological replicates, or libraries (Fig. 1). The plots of pol V-Y11A (Fig. 1A, top) and pol VR391-Y13A (Fig. 1A, middle) support the hypothesis that both pols misincorporate ribonucleotides into the E. coli genome. On the other hand, the plot of the F10L variant was almost flat (Fig. 1A, bottom). This observation is, therefore, consistent with the previous in vitro study [16] indicating this variant does not incorporate ribonucleotides into the E. coli genome.
Fig. 1.
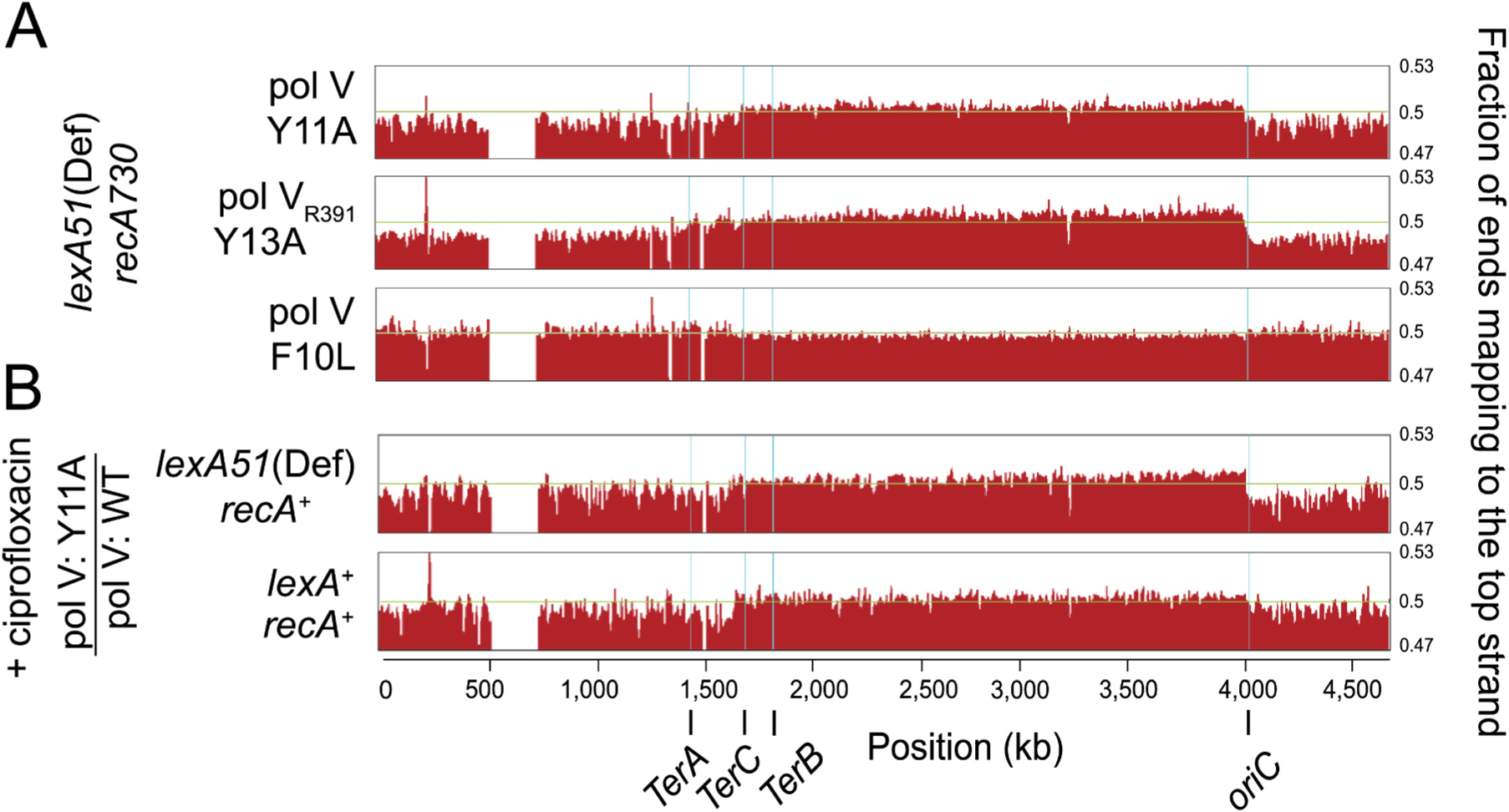
Genome-wide tracking of pol V strand-specific ribonucleotide incorporation. Pol V-Y11A (A, top), pol VR391-Y13A (A, middle), pol V-F10L (A, bottom), and pol V-Y11A treated with ciprofloxacin (B) are shown with the fraction of end reads mapped to the top strand in bins of 10,000 bp after background subtraction using the wild-type strain. The 0.47 and 0.53 numbers shown on the right side of the graphs are vertical viewing range in UCSC genome browser. For better illustration, three individual data points at positions 1,270,000, 1,360,000, and 1,370,000 were removed when they were out of the specified range. The numbers marked with the ticks below the graphs show the chromosomal position in the E. coli genome. Replication origin, oriC and replication terminators, TerA, TerB, and TerC are marked with a light blue vertical line in the graph. The light green horizontal line represents a top strand specificity of 0.5. The plots are the average values of three independent biological replicates or libraries. In ciprofloxacin experiments (B), E. coli cells with wild-type recA genetic background were grown in the presence of ciprofloxacin with a final concentration of 30 ng/mL for approximately 3 h. The blank area in the graph represents the locations in the genome with repetitive sequences that are not possible to be aligned at a single location to the reference genome.
3.2. Identification of DNA replication origin and termination region in the E. coli genome
Previously, it was shown that the HydEn-seq method could determine the location of DNA replication origins and termination regions in the yeast [17]. In E. coli, there is only one replication origin, oriC at chromosomal position 3,925,744–3,925,975 [21]. However, there are several termination sites in the termination region. Three main termination sites are named TerA, TerB, and TerC at chromosomal positions ~1430000, ~1770000, ~1690000 [22]. DNA replication starts at oriC in two opposite directions and ends in the termination region [23]. We used UCSC genome browser to display the data and track the chromosomal positions of oriC, TerA, TerB, and TerC sites (Fig. 1). This analysis revealed clear signal transitions that occur at oriC in pol V-Y11A (Fig. 1A, top and B), and especially in pol VR391-Y13A plot (Fig. 1A, middle). Transitions at the termination region were much less obvious with either mutant compared to the oriC.
3.3. pol V-Y11A and pol VR391-Y13A participate in genome replication with a lagging strand preference during the SOS response
For the obtained data shown in Fig. 1, we used wild-type strain for background subtraction and calculated the fraction of reads that are mapped to the leading and lagging strands. Interestingly, Y11A and Y13A steric gate variants show higher strand specificity for the lagging strand (Fig. 1A, top and middle). Since the F10L mutant does not incorporate ribonucleotides at a very high level, we were unable to observe a strand preference for F10L (Fig. 1A, bottom). The lagging strand preference allows us to investigate where in the genome pol V might have access. As the signals (y-axis) are distributed all over the genome from the replication origin to the termination region, we conclude pol V has access to the entire E. coli genome when it is highly activated in a recA730 lexA51(Def) background. This is a novel and prominent finding of this study.
3.4. Ciprofloxacin induces pol V-Y11A to misincorporate ribonucleotides with a lagging strand preference
In the previous experiments, pol V was highly activated due to the presence of RecA* encoded by recA730. We wanted to determine if a similar pol V strand specificity might be observed in recA+ cells that were “stressed” by exposure to the antibiotic ciprofloxacin. Therefore, we treated recA+ lexA+ and recA+ lexA51(Def) strains harboring pol V-Y11A and pol V wild-type plasmids with 30 ng/mL ciprofloxacin (Fig. 1B). Ciprofloxacin will eventually kill the cells and it is not possible for cells to form colonies overnight in 30 ng/mL Ciprofoxacin. However, as shown by Henrikus et al. [24], exposure of cells to 30 ng/mL ciprofloxacin for up to 3 h is a strong inducer of the SOS response (as measured by many end-points), which is why we chose the 30 ng/mL ciprofloxacin for 3 h regime for our studies.
We used treated pol V wild-type for background subtraction. Similar to the previous data shown in Fig. 1A, pol V-Y11A plots, the strains treated with ciprofloxacin showed a lagging strand preference (Fig. 1B). The strand bias was much less evident in the recA+ lexA+ strain treated with ciprofloxacin (Fig. 1B, bottom) and this is likely due to the overall lower steady-state levels of pol V induced after treatment compared to fully derepressed levels in the lexA51(Def) allele. These observations, therefore, confirm a pol V lagging strand bias in both recA+ lexA+ and recA+ lexA51(Def) backgrounds.
3.5. Template switch is sharper at oriC compared to the termination region
Previously, the HydEn-seq method revealed the template switch sharpness during S. cerevisiae DNA replication [17]. To investigate this aspect in our current study, we focused on the single oriC and termination region in E. coli. We retrieved the data for pol V-Y11A, pol VR391-Y13A, and pol V-F10L. We prepared new plots by using an origin-specific window of ~300 kbp versus a terminus-specific window of ~500 kbp and calculated the slope of transitions (black lines in Fig. 2A–F). Both pol V Y11A and pol VR391-Y13A showed much stronger strand transitions at oriC compared to the termination region. This observation is consistent with the fact that DNA replication initiates only from one single site (oriC) [21] but terminates at a number of termination sites [22]. Next, we calculated the transition slope of oriC and termination region for pol V-Y11A in the ciprofloxacin experiments (Fig. 2, G–J). In these graphs, pol V-WT was used for background subtraction. Strains with the lexA51(Def) allele showed sharper lagging strand transitions at oriC (Fig. 2G) compared to lexA+ (Fig. 2H). The calculations from Fig. 2A–J are shown in Fig. 2K.
Fig. 2.
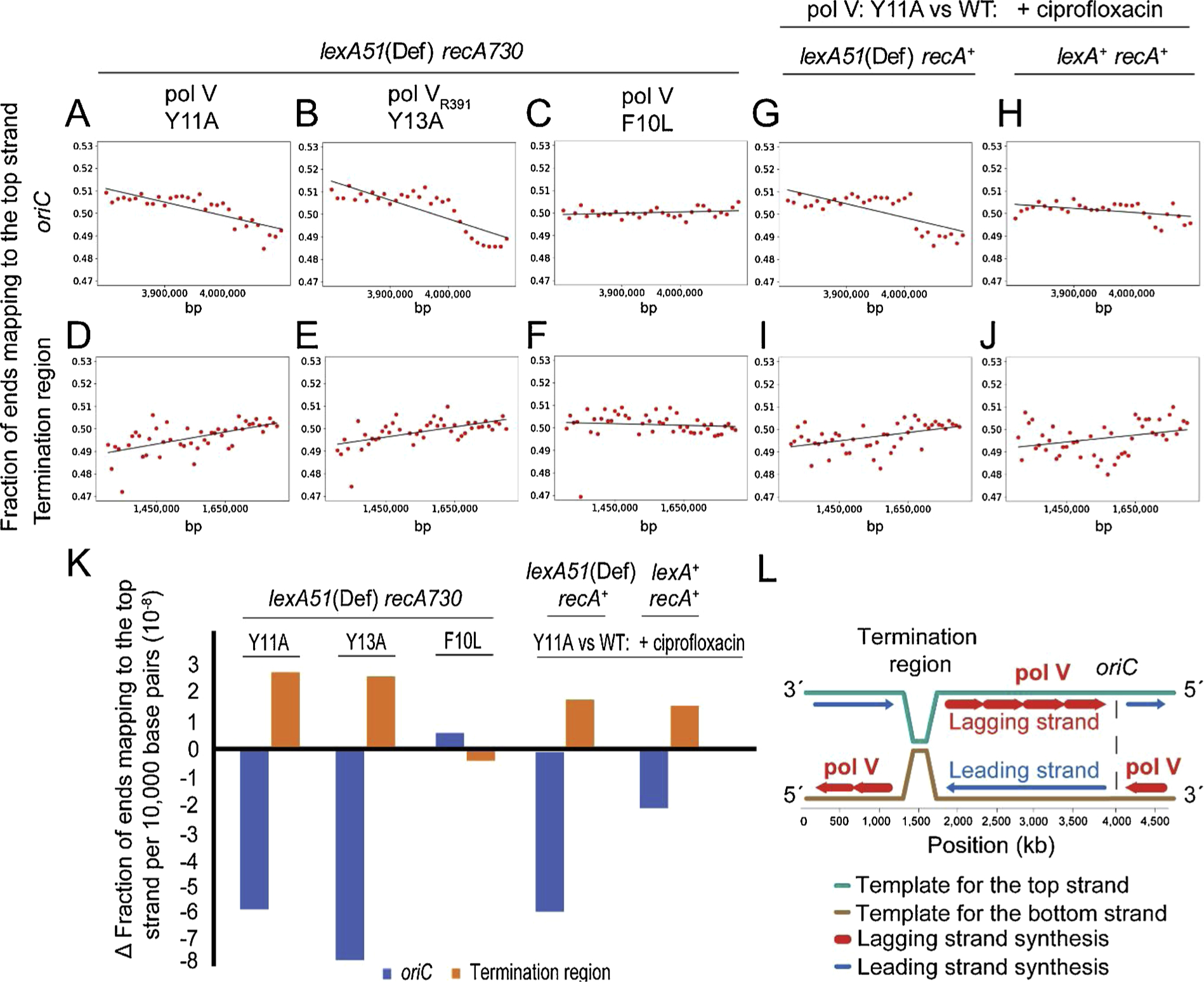
DNA strand specificity switch at oriC and termination region. HydEn-seq data of pol V-Y11A (A and D), pol VR391-Y13A (B and E), pol V-F10L (C and F), and ciprofloxacin experiments (G–J) were plotted at the DNA replication origin, oriC (A–C, G and H) and termination region (D–F, I and J) with the window size of ~300 kbp for origin and ~500 kbp for termination region in x-axis (A–J). Red dots in the scatter plots are the averaged data of three independent libraries similar to Fig. 1. The black line represents the data slope within the window. (K) The calculated slopes were used to generate a bar chart with the slopes value. The blue and brown colors represent oriC and termination region slopes, respectively. The x-axis shows different steric gates and the y-axis represents the slopes. (L) A schematic diagram of pol V replication enzymology in E. coli. The figure shows E. coli genome with one replication origin (oriC) (marked with dashed black vertical lines) and a termination region (buckets). Pol V participates in the whole genome replication with lagging strand preference (red arrows vs. blue arrows). The x-axis shows the chromosomal position in E. coli genome.
4. Discussion
The present study provides a framework to investigate the chromosomal distribution of DNA synthesis reaction in E. coli by a translesion DNA polymerase, pol V. By using a series of lacZ alleles that have been inserted into the E. coli chromosome in opposite directions, the Fijalkowska and Schaaper laboratories have elegantly shown that the majority of pol V-dependent spontaneous mutagenesis occurs on the lagging strand during genome duplication [14]. However, a limitation of the lacZ reversion assay is that it monitors the reversion of a single nucleotide in a fixed sequence context. To examine whether pol V replication occurs on the lagging strand genome-wide, we have taken advantage of pol V variants with mutations at the steric gate residue, and which are known to readily incorporate ribonucleotides into the E. coli genome [16,20], and have used HydEn-seq methodology [17] to map the location of pol V-dependent ribonucleotide misincorporation across the entire 4.6 Mbp genome.
While HydEn-seq detects a lagging strand preference for pol V, it is unable to display a similar leading-over-lagging preference as seen for pol η or other replicative DNA polymerases [17,19]. We speculate that even though the steric gate variant incorporates ribonucleotides with a preference for the lagging strand, the relative contribution of pol V to replication of the entire genome is minor.
Previously, Walsh et al., showed pol VR391-Y13A generates higher levels of SOS-induced spontaneous mutagenesis than pol V-Y11A, as it binds more repeatedly to the undamaged genome and for a longer time [20]. Accordingly, we saw a clear genome-wide lagging strand bias for pol VR391-Y13A.
The strand preference in our study might be a structural result of the pol V gating defects. However, our conclusion is consistent with previous observations with wild-type pol V using the lacZ reporter assay [14].
Based on the data from HydEn- seq method (Figs. 1 and 2), we propose a model of how pol V can gain access to the E. coli genome in vivo (Fig. 2L). In our model, SOS-induced pol V participates in the genome replication with a lagging strand preference (red arrows). However, as we subtracted the lagging and leading strands signals of the wild-type enzyme from the steric gate mutant, we cannot exclude the possibility that pol V also participates to some extent in leading strand synthesis. The lagging strand preference seen in our model is consistent with previous studies in which the lacZ reporter system was used to investigate the role of pol V during SOS mutagenesis [14], but now extends to the entire E. coli genome and multiple sequence contexts. As previously discussed [14], pol V may have better access to the lagging strand because the strand is replicated discontinuously and therefore provides more 3′ termini. The lagging strand also generates more single-stranded DNA and is a better target for RecA*, which is required for maximal activation of pol V in vivo.
Supplementary Material
Acknowledgments
We would like to thank the Genomics Core Facility at the University of Gothenburg for sequencing.
Funding source
This study was supported by the Swedish Research Council (www.vr.se) [2018-05121 to A.R.C.] and NICHD/NIH Intramural Research Program. The study sponsors had no involvement in the study design; collection, analysis and interpretation of data; the writing of the manuscript; or the decision to submit the manuscript for publication
Abbreviations:
- pol V
DNA polymerase V
- TLS
Translesion DNA synthesis
- RER
Ribonucleotide Excision Repair
Footnotes
Declaration of Competing Interest
The authors declare no competing interest.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.dnarep.2021.103075.
References
- [1].Kato T, Shinoura Y, Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light, Mol. Gen. Genet 156 (1977) 121–131. [DOI] [PubMed] [Google Scholar]
- [2].Steinborn G, Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis, Mol. Gen. Genet 165 (1978) 87–93. [DOI] [PubMed] [Google Scholar]
- [3].Bagg A, Kenyon CJ, Walker GC, Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli, Proc. Natl. Acad. Sci. U. S. A 78 (1981) 5749–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF, A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V, Crit. Rev. Biochem. Mol. Biol 45 (2010) 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shinagawa H, Iwasaki H, Kato T, Nakata A, RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis, Proc. Natl. Acad. Sci. U. S. A 85 (1988) 1806–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Burckhardt SE, Woodgate R, Scheuermann RH, Echols H, UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA, Proc. Natl. Acad. Sci. U. S. A 85 (1988) 1811–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nohmi T, Battista JR, Dodson LA, Walker GC, RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation, Proc. Natl. Acad. Sci. U. S. A 85 (1988) 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Woodgate R, Rajagopalan M, Lu C, Echols H, UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD’, Proc. Natl. Acad. Sci. U. S. A 86 (1989) 7301–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF, UmuD’(2)C is an error-prone DNA polymerase, Escherichia coli pol V, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schlacher K, Cox MM, Woodgate R, Goodman MF, RecA acts in trans to allow replication of damaged DNA by DNA polymerase V, Nature 442 (2006) 883–887. [DOI] [PubMed] [Google Scholar]
- [11].Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF, The active form of DNA polymerase V is UmuD’(2)C-RecA-ATP, Nature 460 (2009) 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V, RecA protein of Escherichia coli has a third essential role in SOS mutator activity, J. Bacteriol 172 (1990) 3030–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fijalkowska IJ, Dunn RL, Schaaper RM, Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity, J. Bacteriol 179 (1997) 7435–7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maliszewska-Tkaczyk M, Jonczyk P, Bialoskorska M, Schaaper RM, Fijalkowska IJ, SOS mutator activity: unequal mutagenesis on leading and lagging strands, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 12678–12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R, Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V, PLoS Genet. 8 (2012), e1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R, Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity, Nucleic Acids Res. 40 (2012) 6144–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, Malc EP, Mieczkowski PA, Fargo DC, Smith DJ, Kunkel TA, Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation, Nat. Struct. Mol. Biol 22 (2015) 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Orebaugh CD, Lujan SA, Burkholder AB, Clausen AR, Kunkel TA, Mapping ribonucleotides incorporated into DNA by hydrolytic end-sequencing, Methods Mol. Biol 1672 (2018) 329–345. [DOI] [PubMed] [Google Scholar]
- [19].Kreisel K, Engqvist MKM, Kalm J, Thompson LJ, Bostrom M, Navarrete C, McDonald JP, Larsson E, Woodgate R, Clausen AR, DNA polymerase η contributes to genome-wide lagging strand synthesis, Nucleic Acids Res. 47 (2019) 2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Walsh E, Henrikus SS, Vaisman A, Makiela-Dzbenska K, Armstrong TJ, Lazowski K, McDonald JP, Goodman MF, van Oijen AM, Jonczyk P, Fijalkowska IJ, Robinson A, Woodgate R, Role of RNase H enzymes in maintaining genome stability in Escherichia coli expressing a steric-gate mutant of pol VR391, DNA Repair (Amst.) 84 (2019), 102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Oka A, Sugimoto K, Takanami M, Hirota Y, Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication, Mol. Gen. Genet 178 (1980) 9–20. [DOI] [PubMed] [Google Scholar]
- [22].Hill TM, Arrest of bacterial DNA replication, Ann. Rev. Microbiol 46 (1992) 603–633. [DOI] [PubMed] [Google Scholar]
- [23].Bramhill D, Kornberg A, A model for initiation at origins of DNA replication, Cell 54 (1988) 915–918. [DOI] [PubMed] [Google Scholar]
- [24].Henrikus SS, Henry C, McGrath AE, Jergic S, McDonald JP, Hellmich Y, Bruckbauer ST, Ritger ML, Cherry ME, Wood EA, Pham PT, Goodman MF, Woodgate R, Cox MM, van Oijen AM, Ghodke H, Robinson A, Single-molecule live-cell imaging reveals RecB-dependent function of DNA polymerase IV in double strand break repair, Nucleic Acids Res. 48 (2020) 8490–8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Szekeres ES Jr., Woodgate R, Lawrence CW, Substitution of mucAB or rumAB for umuDC alters the relative frequencies of the two classes of mutations induced by a site-specific T-T cyclobutane dimer and the efficiency of translesion DNA synthesis, J. Bacteriol 178 (1996) 2559–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data of this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo with the accession no. GSE158570. Custom scripts are available upon request to the corresponding author.


