Abstract
Background
As a rate‐limiting enzyme of glycolysis, pyruvate kinase muscle isozyme M2 (PKM2) participates in tumor metabolism and growth. The regulatory network of PKM2 in cancer is complex and has not been fully studied in bladder cancer. The 5‐methylcytidine (m5C) modification in PKM2 mRNA might participate in the pathogenesis of bladder cancer and need to be further clarified. This study aimed to investigate the biological function and regulatory mechanism of PKM2 in bladder cancer.
Methods
The expression of PKM2 and Aly/REF export factor (ALYREF) was measured by Western blotting, qRT‐PCR, and immunohistochemistry. The bioprocesses of bladder cancer cells were demonstrated by a series of experiments in vitro and in vivo. RNA immunoprecipitation, RNA‐sequencing, and dual‐luciferase reporter assays were conducted to explore the potential regulatory mechanisms of PKM2 in bladder cancer.
Results
In bladder cancer, we first demonstrated that ALYREF stabilized PKM2 mRNA and bound to its m5C sites in 3′‐untranslated regions. Overexpression of ALYREF promoted bladder cancer cell proliferation by PKM2‐mediated glycolysis. Furthermore, high expression of PKM2 and ALYREF predicted poor survival in bladder cancer patients. Finally, we found that hypoxia‐inducible factor‐1alpha (HIF‐1α) indirectly up‐regulated the expression of PKM2 by activating ALYREF in addition to activating its transcription directly.
Conclusions
The m5C modification in PKM2 mRNA in the HIF‐1α/ALYREF/PKM2 axis may promote the glucose metabolism of bladder cancer, providing a new promising therapeutic target for bladder cancer.
Keywords: PKM2, ALYREF, glycolysis, 5‐methylcytidine modification, bladder cancer, HIF‐1α
Our data suggested that m5C may play a critical role in hypoxia/glycolysis network and targeting HIF‐1α/ALYREF/PKM2 signaling may be a promising therapeutic strategy to treat bladder cancer.

1. BACKGROUND
Bladder cancer (BCa) is the 10th most common malignancy worldwide, with an estimated 549,000 new cases diagnosed and 200,000 deaths occurred in 2018 [1, 2]. Intrinsic biological invasiveness and specific radio‐ and chemo‐resistance of bladder tumors bring about high recurrence and progression rates in patients [3]. Thus, a deep understanding of its regulatory network is essential for improving treatment in BCa.
Aerobic glycolysis (also termed as the Warburg effect) is a phenomenon that most cancer cells enhance their glucose consumption and produce lactic acid even in the presence of oxygen [4]. Consumed glucose is transformed into various molecules that sustain nucleotide and triglyceride biosynthesis. Thus cancer cells, including BCa cells, utilize the nutrients ingested in the substances to sustain their uncontrolled growth [3, 5]. Pyruvate kinase muscle isozyme M2 (PKM2) is a rate‐limiting glycolytic enzyme that plays an important role in tumor metabolism and growth [6]. Recent studies showed that PKM2 was highly expressed in multiple cancers, and predicted poor prognosis [7, 8, 9]. PKM2 can promote the proliferation and migration of cancer cells in a variety of cancers [8, 10, 11]. The role of PKM2 in cancer remains controversial. Dayton et al. [12] found that PKM2 can prevent hepatocellular carcinoma by a non‐cell‐autonomous mechanism. Morita et al. [13] found that it was PKM1 rather than PKM2 to promote tumor growth. In BCa, PKM2 can promote the growth, migration, and cisplatin resistance of BCa cells and may become a poor prognostic factor of BCa patients [14, 15, 16, 17]. Our previous study confirmed that the expression levels of most glycolysis‐related genes, including PKM2, were higher in BCa tissues than in paired adjacent tissues, suggesting the oncogenic role of glycolysis and PKM2 in BCa [18].
Gene expression is regulated by complex genetic and epigenetic layers [19]. Hypoxia‐inducible factor‐1alpha (HIF‐1α), c‐myc, and epidermal growth factor receptor can up‐regulate the transcription of PKM2 and increase PKM2 protein synthesis [20, 21, 22]. Several microRNAs can be recruited in the 3′‐untranslated region (UTR) of PKM2 mRNA transcripts to eventually reduce PKM2 expression [23, 24, 25]. Post‐translational modifications, such as phosphorylation, acetylation, succinylation, and oxidation, can also regulate PKM2 expression [26]. Recently, it was found that RNA modification in mRNA levels played an important role in BCa development [27, 28, 29]. Besides the hot spot of N6‐methyladenosine (m6A), RNA modifications also include 5‐methylcytidine (m5C), pseudouridine, N4‐acetylcytidine, N1‐methyladenosine, 2′‐O‐methylations, N7‐methylguanosine, 5‐hydroxymethylcytidine cytidine and inosine [30]. However, the methylation modification of PKM2 is currently unclear. The epitranscriptome is a newly discovered layer of gene expression, characterized as post‐transcriptional modifications of RNAs [31, 32]. As the most common internal modification in eukaryotic mRNAs, m6A has been found to influence glycolysis in BCa [33], gastric cancer [34], and colorectal cancer [35]. However, the roles of other RNA modifications, such as m5C modification, in cancer glycolysis remain unclear.
m5C modification was first identified in transfer RNAs and ribosomal RNAs [36, 37, 38]. Subsequently, Squires et al. [39] identified 10,275 m5C sites in human RNAs. Huang et al. [40] identified several hundred exonic m5C sites located in human and mouse tissues. It was reported that m5C modification was enriched in CG‐rich regions and regions immediately following a translation initiation region of mRNAs. Dynamic, tissue‐specific, conserved m5C modification of mRNAs is mainly written by NOP2/Sun RNA methyltransferase family member 2 (NSUN2) and specifically read by Aly/REF export factor (ALYREF, also named THOC4) and Y‐box binding protein 1 (YBX1). ALYREF specifically reads m5C regions by the lysine residue K171. YBX1 can recruit ELAV‐like RNA‐binding protein 1 (ELAVL1) to regulate the stability of target mRNAs [27, 41]. However, the impact of ALYREF on gene expression is currently unclear. Chen et al. [27] found that m5C promoted the pathogenesis of BCa through stabilizing mRNAs. By analyzing the data in this study combined with the data reported by Yang et al. [41], we found that PKM2 mRNA may exist m5C modification and be bound by ALYREF. It seemed that m5C modification in PKM2 mRNA participated in the progression of BCa.
HIF‐1α is a transcription factor that responds to hypoxia. HIF‐1α is a member of the key proteins to regulate glycolysis. The stabilized HIF‐1α dimerizes with HIF‐1β and binds to hypoxia‐responsive elements (HREs), which are located in promoter regions of HIF‐1α target genes. Glycolytic enzymes HK2, phosphoglycerate kinase 1, and lactic dehydrogenase A can all be up‐regulated by HIF‐1α. PKM2 can also be activated by HIF‐1α [20, 42‐45]. However, whether m5C plays a significant role in the HIF‐1α/PKM2 axis is not clear.
In the present study, we further examined the biological function of PKM2 in BCa, investigated its potential regulatory mechanism, and attempted to provide potential therapeutic targets in the treatment of BCa.
2. MATERIALS AND METHODS
2.1. Clinical specimens
We obtained tumor tissues and adjacent normal tissues from BCa patients who underwent radical cystectomy at the First Affiliated Hospital of Nanjing Medical University between March 2010 and July 2013. The final follow‐up time was March 2021. All patients signed the informed consents. This study was approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University. Liquid nitrogen containers were used to store all samples. A total of 162 BCa tissue specimens were used to make tissue microarray (TMA). Fifty BCa tissues and their adjacent normal tissues were collected to extract RNA. Histopathological examination confirmed the bladder cancer diagnosis.
2.2. Cell lines
The BCa cell lines 5637 and T24 and renal cell line 293T were purchased from the type culture collection of the Chinese Academy of Sciences (Shanghai, China). 5637 and T24 cells were authenticated by short tandem repeat profiling (Supplementary data 1 and data 2). All cells were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gimini, Calabasas, CA, USA) and 1% penicillin‐streptomycin and incubated at 37°C with 5% CO2 in a humidified atmosphere. For hypoxia exposure, cells were incubated in an incubator chamber containing an aggregate of 94% N2, 5% CO2, and 1% O2. Total RNAs and proteins were extracted 24 and 48 hours after hypoxia exposure.
2.3. Transfection
Lentivirus vectors of ALYREF knockdown, NSUN2 knockdown, and ALYREF overexpression were purchased from Obio Technology Corp (Shanghai, China). ALYREF overexpression lentivirus was termed as ALYREF‐OE, and a negative control was termed as NC. ALYREF‐knockdown lentiviruses were termed as shALYREF#1 and shALYREF#2, NSUN2 knockdown lentivirus was termed as shNSUN2, and control lentivirus was termed as shNC. Lentivirus transfection was conducted as previously described [46]. Wild‐type ALYREF, K171A‐mutant ALYREF, PKM2, and control (green fluorescent protein, GFP) plasmids were synthesized by GeneCopoeia (Rockville, MD, USA). Plasmid transfection was conducted using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The transfections of lentivirus were stable, and the transfections of plasmids were transient.
2.4. Quantitative real‐time polymerase chain reaction (qRT‐PCR)
Trizol reagent (Invitrogen) was used to extract total RNAs from bladder cancer tissues,paired normal tissues, T24 cells, 5637 cells or 293T cells. HiScript II (Vazyme, Nanjing, Jiangsu, China) was used to synthesize cDNA. For mRNA expression analysis, quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed using SYBR Green Master Mix (Vazyme) on LightCycler 480 (Roche, USA). A typical cycling condition included 95°C for 5 min followed by 40 cycles at 95°C for 10 s, 60°C for 30 s. Melting curve analysis is acquired according to the instrument documentation. β‐actin was used as the internal standard control. Each sample was run in triplicate. Relative expression values for each gene were calculated using the 2−△△CT method. All primer sequences used here are listed in Supplymentary Table S1.
2.5. Western blotting
Cells or tissues were treated with Radio Immunoprecipitation Assay buffer (Beyotime, Shanghai, China) containing protease inhibitors (P8340, Sigma‐Aldrich, St. Louis., MO, USA). The concentration of proteins was measured by using a bicinchonininc acid kit (Beyotime). Extracted proteins were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% non‐fat milk and incubated with primary antibodies for anti‐PKM2 (1:1000, ab154816, Abcam, USA), anti‐ALYREF (1:800, GTX113917, GeneTex, Irvine, CA, USA), anti‐HIF‐1α (1:1000, #36169, Cell Signaling Technology, USA), anti‐NSUN2 (1:1000, 702036, ThermoFisher, Waltham, MA, USA), anti‐GAPDH (1:1000, HRP‐60004, Proteintech, Rosemont, IL, USA), or anti‐β‐actin (1:1000, 66009‐1‐lg, Proteintech). The membrane was then incubated with anti‐mouse secondary antibodies (1:5000, GB23301, Servicebio, Wuhan, Hubei, China,) or anti‐rabbit secondary antibodies (1:5000, GB23303, Servicebio). Protein levels were detected using an enhanced chemiluminescence system (Pierce, Rockford, IL, USA).
2.6. TMA and immunohistochemistry (IHC)
IHC staining of TMA was performed to evaluate the expression of ALYREF and PKM2 proteins. TMAs were stained with anti‐ALYREF antibody or anti‐PKM2 antibody, incubated with horseradish peroxidase‐conjugated goat anti‐rabbit antibody (GB23303, Servicebio), and then counterstained with hematoxylin. Images were obtained under a microscope (Nikon, Minato Ward, Tokyo, Japan). The percentage of positive cells was scored into 5 categories: 0 (0% positive cells), 1 (<10%), 2 (10% to 50%), 3 (50% to 80%), and 4 (>80%). The staining intensity was scored into 4 categories: 0, negative staining; 1, light staining; 2, moderate staining; and 3, strong staining. The two scores were multiplied to get a final score value ranging from 0 to 12. The final scores of 0‐7 were identified as low expression, and 8‐12 as high expression. Protein expression was evaluated by two pathologists blinded to the clinical data.
2.7. Immunofluorescence
5637 and T24 cells grown on the cover glasses were washed twice with phosphate buffer saline, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X‐100, blocked with milk, and then incubated with anti‐ALYREF antibody for 1 hour. After incubated with fluorescence‐labelled secondary antibodies (Servicebio), the slides were observed under a fluorescence microscope (ECLIPSE TI‐SR, Nikon). All steps were performed at room temperature.
2.8. Cell counting kit‐8 (CCK‐8) assay
Cells were cultivated on 96‐well plates, with 1000 or 1500 cells per well. Optical density (OD) value at 450 nm was detected by CCK‐8 assay (Dojindo, Kamimashiki‐gun, Kumamoto, Japan) after 1, 2, 3, 4, and 5 days as described in the manual. The cell proliferation rate was calculated using the formula: cell proliferation rate = OD of test group ‐ OD of control group / OD of control group × 100%.
2.9. Colony formation assay
Transfected cells were seeded into 6‐well plates (500 cells/well) and kept in DMEM containing 10% FBS for 7‐14 days. The colonies were fixed with 4% paraformaldehyde. After stained with crystal violet, the visible colonies were counted.
2.10. Xenograft model in nude mice
About 1 × 107 transfected T24 cells were injected subcutaneously into the axilla of BALB/c nude mice (4‐6 weeks old, 5 mice/group). The tumor volume was measured each week. Three or four weeks after injection, the mice were euthanized, and their tumors were isolated and fixed in 4% formalin for IHC analysis. Animal studies were approved by the Animal Research Ethics Committee of Nanjing Medical University.
2.11. RNA m5C dot blotting
After treated with RNase‐free DNase set (Qiagen, Hilden, Germany), the extracted mRNA was used for m5C dot blotting. mRNA concentration was measured using NanoDrop Nd‐1000 spectrophotometer (Agilent, Santa Clara, CA, USA). mRNA was denatured by heating at 65°C for 5 min. Then a Bio‐Dot apparatus (Bio‐Rad, Hercules, CA, USA) was used to transfer the mRNA onto a nitrocellulose membrane (Amersham, GE Healthcare, Pittsburgh, PA, USA). After cross‐linked by ultraviolet and blocked by milk, the membranes were incubated with anti‐m5C antibody (1:1000, ab10805, Abcam) overnight at 4°C. Later, the membranes were incubated with anti‐mouse antibody (1:3000, GB23301, Servicebio). Finally, we used the chemiluminescence system (Bio‐Rad) to visualize the membrane. The membrane stained with 0.02% methylene blue in 0.3 mol/L sodium acetate (pH 5.2) was used as a loading control to ensure consistency.
2.12. RNA‐sequencing (RNA‐seq)
Total RNAs were isolated from stable ALYREF‐knockdown 5637 cells and their corresponding controls. Then, RNA‐seq was performed by Allwegene company (Beijing, China) using the Illumina (San Diego, CA, USA) HiSeqTM2500 instrument. Alignment and transcript splicing analyses were performed using Tophat2 (http://ccb.jhu.edu/software/tophat/index.shtml) and Cufflinks software (http://cole‐trapnell‐lab.github.io/cufflinks), respectively. All genes were quantitatively analyzed by fragments per kilobase million values, and differentially expressed genes were identified with |log2(FoldChange)| > 1 and q value < 0.005. Finally, differentially expressed genes were functionalized by Gene Set Enrichment Analysis (GSEA) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) analysis.
2.13. RNA immunoprecipitation (RIP) assay
RIP assay was performed using the Magna RIP™ kit (Millipore). About 1 × 107 cells were lysed in RIP lysis buffer. Anti‐ALYREF (5 μg) or control IgG (5 μg) was incubated with magnetic beads for 2 h at 4°C. Mixed with 900 μL of RIP buffer (Sigma‐Aldrich), 100 μL of the supernatant of RIP lysate was added to the bead‐antibody complexes for incubation overnight at 4°C. Then the beads were incubated with proteinase K buffer for 30 min at 55°C, and RNA was finally extracted for qRT‐PCR analysis.
2.14. Methylated RNA immunoprecipitation (MeRIP) assay
Extracted from transfected T24 cells by Trizol reagent, total RNAs were then treated with RNase‐free DNase set (Qiagen). After immunoprecipitated with anti‐m5C antibody (1:1000), the chemically fragmented RNAs (∼100 nucleotides) were washed three times with RIP buffer. After incubating with elution buffer (Sigma‐Aldrich) for 1 h at 4°C, RNAs were eluted from the beads. After ethanol precipitation, input and eluted RNAs were obtained.
2.15. Luciferase reporter assay
293T cells were co‐transfected with plasmids containing 3′‐UTR of wild‐type or mutant fragments from PKM2 and plasmids with wild‐type ALYREF or mutant ALYREF using Lipofectamine 3000. After incubation for 48 hours, the luciferase activity was determined by using the dual‐luciferase reporter assay system (Promega, Madison, WI, USA). Finally, the relative luciferase activity was normalized to the renilla, and each experiment was repeated thrice.
2.16. RNA stability assay
To test RNA stability, transfected 5637 or T24 cells were exposed to 2 μg/mL actinomycin D (Act D, Sigma‐Aldrich), which can interfere with transcription and inhibit mRNA synthesis. Total RNAs were extracted at the times indicated, and PKM2 mRNA was analyzed by qRT‐PCR.
2.17. Glucose, lactic acid, and adenosine triphosphate (ATP) detection
We qualified glucose utilization, lactic acid production, and intracellular ATP production by using glucose assay kit, lactic acid assay kit, and ATP assay kit (Jiancheng Corporation, Nanjing, Jiangsu, China), respectively, as per the manufacturer's instructions.
2.18. Bioinformatic analysis
We downloaded the clinical data from the database of The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) and Gene Expression across Normal and Tumor tissue (http://medical‐genome.kribb.re.kr/GENT/). PROMO database [47] and JASPAR database [48] were used to identify potential transcription factors.
2.19. Statistical analysis
SPSS 19.0 software (IBM, Chicago, IL, USA) was used for statistical tests. Differences between two groups were analyzed by Student's t test or Chi‐square test, and P < 0.05 was considered significant. Survival was calculated from the date of diagnosis. Kaplan‐Meier curves and log‐rank test for significance were used for survival analysis. Univariate and multivariate analyses were conducted with Cox proportional hazards regression model. Statistical correlation was analyzed by Pearson's correlation coefficient analysis. All data are presented as the mean ± standard deviation (SD) from three different independent experiments.
3. RESULTS
3.1. PKM2 promoted glycolytic metabolism in BCa cells and acted as a poor prognostic factor in BCa patients
We firstly analyzed the TCGA database and observed that the expression of PKM2 was higher in BCa tissues than in normal tissues (Figure 1A) and that the high expression of PKM2 predicted poor overall survival (OS) of BCa patients (Figure 1B). Next, we found that PKM2 expression was up‐regulated in BCa tissues than in adjacent normal tissues from 50 patients by qRT‐PCR (Figure 1C). Meanwhile, we divided the 162 primary BCa surgical specimens into low PKM2 expression (n = 85) and high PKM2 expression groups (n = 77) by IHC analysis. Subsequent Kaplan‐Meier survival curves showed that high PKM2 expression predicted poor OS and recurrence‐free survival (RFS) in BCa patients (Figure 1D).
FIGURE 1.
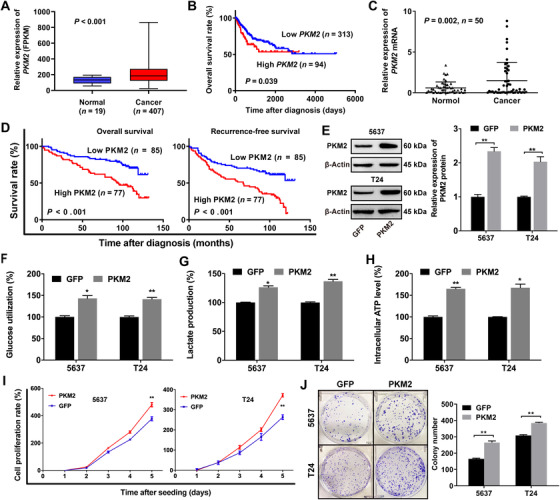
PKM2 promotes glucose metabolism in bladder cancer cells and predicts poor survival in BCa patients. (A) PKM2 is higher in BCa tissues than in normal tissues (TCGA database). (B) The OS curves show that PKM2 predicts poor prognosis for BCa (TCGA database). (C) PKM2 mRNA is up‐regulated in BCa tissues as compared with adjacent normal tissues by qRT‐PCR. (D) Kaplan‐Meier survival curves show that PKM2 up‐regulation is significantly associated with poor OS and RFS in BCa patients. (E) Validation of the overexpression efficacy of PKM2 in 5637 and T24 cell lines by Western blotting. (F,G,H) Effects of PKM2 overexpression on glucose utilization, lactic acid production, and intracellular ATP production measured by glucose assay, lactic acid assay, and intracellular ATP assay in 5637 and T24 cells. (I) PKM2 contributes to the proliferation of 5637 and T24 cells as measured by CCK‐8 assays. (J) PKM2 contributes to the colony formation of 5637 and T24 cells as measured by colony formation assays. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: PKM2, pyruvate kinase muscle isozyme M2; GFP, green fluorescent protein; BCa, bladder cancer; TCGA, The Cancer Genome Atlas; OS, overall survival; qRT‐PCR, quantitative real‐time polymerase chain reaction; RFS, recurrence‐free survival; ATP: adenosine triphosphate; FPKM, fragments per kilobase million values; CCK‐8, cell counting kit‐8.
To assess the tumor‐promoting function of PKM2 in BCa, we cloned PKM2‐coding regions into plasmids. Plasmid vectors (PKM2 or control) were transfected into 5637 and T24 cells. Western blotting was used to qualify the overexpression of PKM2 (Figure 1E). The glucose utilization, lactic acid detection, and intracellular ATP detection assays showed that PKM2 could promote glucose utilization (Figure 1F), lactic acid production (Figure 1G), and intracellular ATP production (Figure 1H) in BCa 5637 and T24 cells. We also found that PKM2 could promote BCa cell proliferation (Figure 1I) and colony formation (Figure 1J).
Consistent with previous studies [14, 49], these data indicated that PKM2 regulated glycolytic metabolism and played an oncogenic function in BCa.
3.2. ALYREF stabilized PKM2 mRNA by a m5C‐dependent manner
ALYREF RIP‐seq data from Yang et al. [41] indicated that PKM2 may be bound by ALYREF. They also identified m5C‐modified sites in PKM2 mRNA. Therefore, we designed primers in the CG‐enriched region of PKM2 3′‐UTR and conducted RIP assays. The data suggested that the anti‐ALYREF antibody could significantly enrich PKM2 3′‐UTR, compared with the anti‐IgG antibody (Figure 2A). NSUN2 catalyzes m5C methylation of mRNA. Then, ALYREF recognizes the methylated mRNA through the residue K171 and mediates its nuclear‐cytoplasmic shuttling [41]. Thus, we constructed a plasmid expressing a K171A ALYREF mutant (substitute alanine for lysine at position 171). Then, BCa cells were transfected with wild‐type or K171A mutant ALYREF. Further RIP assays revealed that the K171A mutation accounted for the dramatically reduced level of ALYREF binding to PKM2 3′‐UTR (Figure 2B). Dot blotting, m5C‐RNA MeRIP, and RIP assays demonstrated that NSUN2 knockdown caused a decreased m5C level of total RNA in T24 and 5637 cells (Figure 2C and Supplementary Figure S1), decreased enrichment of the 3′‐UTR of PKM2 in T24 cells (Figure 2D), and reduced binding efficiency of ALYREF to the 3′‐UTR of PKM2 mRNA (Figure 2E).
FIGURE 2.
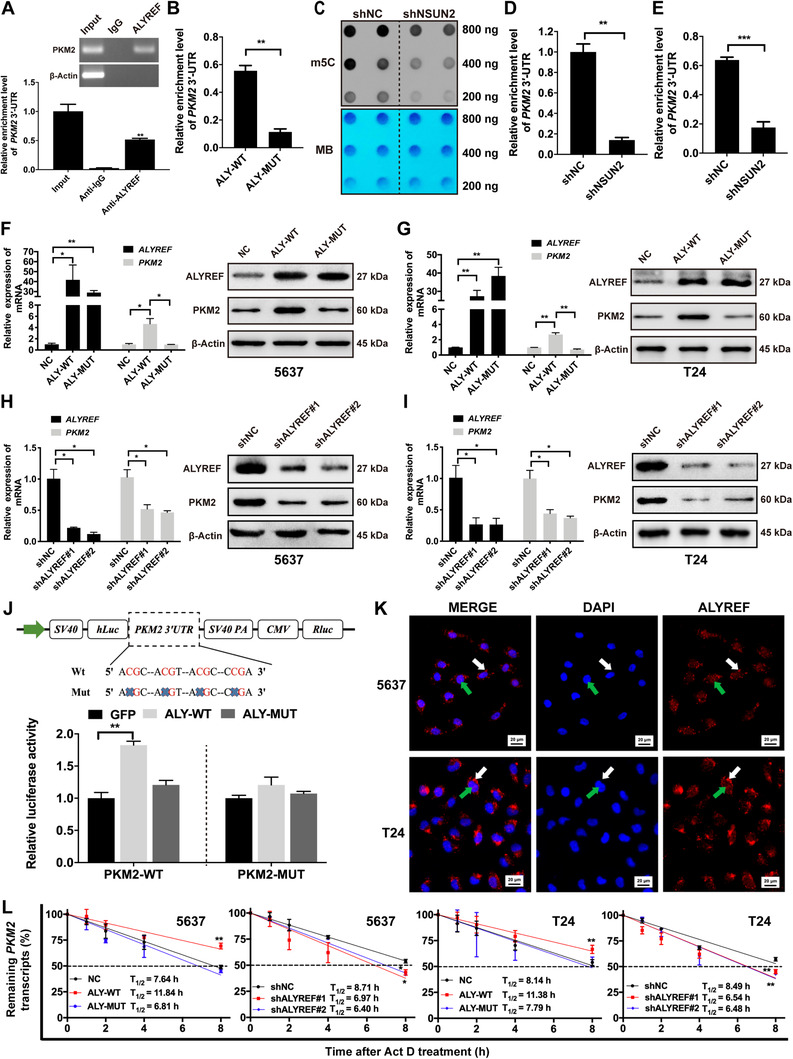
ALYREF directly binds to PKM2 mRNA and regulates its stability in an m5C‐dependent manner. (A) The RIP assays show that ALYREF interactwith 3′‐UTR of PKM2 in T24 cells. (B) The RIP assays were performed in T24 cells transfected with ALYREF wild type or K171A mutation. qRT‐PCR assays show that K171A mutation result in a significantly decreased level of ALYREF binding to 3′‐UTR of PKM2. (C) m5C‐modified RNAs were detected by dot blotting in T24 cells with or without NSUN2 knockdown. NSUN2 knockdown significantly decreases the m5C level of total RNAs. MB was used as a loading control. (D) NSUN2 knockdown dramatically decreases the m5C level of the 3′‐UTR of PKM2 in T24 cells by MeRIP assay. (E) RIP assays reveal the binding efficiency of ALYREF to PKM2 3’‐UTR is decreased following knockout of NSUN2 in T24 cells. (F, G) ALYREF overexpression promotes PKM2 expression, while ALYREF with K171A mutation fails to regulate PKM2 expression as qualified by qRT‐PCR and Western blotting. (H, I) ALYREF knockdown decreases PKM2 expression in 5637 and T24 cells as measured by qRT‐PCR and Western blotting. (J) Relative luciferase activity of the reporter carrying PKM2 3′‐UTR with either wild‐type or m5C site deletion co‐transfected with ALYREF (ALY‐WT), ALYREF K171A mutant (ALY‐MUT), or control vector (GFP) into 293T cells was detected and normalized to renilla luciferase activity. (K) ALYREF protein is located in both nucleus (green arrows) and cytoplasm (white arrows) of 5637 and T24 cells by immunofluorescence assays. (L) 5637 and T24 cells were treated with Act D. The half‐life of PKM2 mRNA can be shortened by ALYREF knockdown and lengthened by ALYREF overexpression. ALYREF with K171A mutation can not regulate the half‐life of PKM2 mRNA. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: ALYREF, Aly/REF export factor; PKM2, Pyruvate kinase muscle isozyme M2; m5C, 5‐methylcytidine; RIP, RNA immunoprecipitation; UTR, untranslated region; qRT‐PCR, quantitative real‐time polymerase chain reaction; NC, negative control; shNC, negative control of knockdown; NSUN2, NOP2/Sun RNA methyltransferase family member 2; shNSUN2, knockdown of NOP2/Sun RNA methyltransferase family member 2; shALYREF, knockdown of Aly/REF export factor; FPKM, fragments per kilobase million values; MB, methylene blue; SV40, simian virus 40; hLuc, fireflyer luciferase; PA, poly A; CMV, cytomegalovirus; Rluc, Renillar luciferase; PKM2‐WT, Pyruvate kinase muscle isozyme M2 wild type; PKM2‐WUT, Pyruvate kinase muscle isozyme M2 mutation; Act D, actinomycin D.
To further explore the influence of ALYREF on PKM2 expression, we conducted qRT‐PCR and Western blotting assays. Our data confirmed that ALYREF overexpression remarkably up‐regulated the expression of PKM2 (Figure 2F and 2G) and ALYREF knockdown dramatically reduced the expression of PKM2 (Figure 2H and 2I), while ALYREF with K171A mutation lost its ability to regulate mRNA and protein levels of PKM2 in 5637 and T24 cells (Figure 2F and 2G). We also conducted dual‐luciferase assays to elucidate whether the m5C sites of PKM2 3′‐UTR were required for ALYREF to increase PKM2 expression. We constructed both wild‐type and mutant PKM2 reporters. For the mutant forms of PKM2, we deleted the m5C consensus sequences. Our results revealed that forced expression of wild‐type ALYREF significantly increased the luciferase activity of PKM2 3′‐UTR reporter in 293T cells, compared with mutant ALYREF or empty vector, whereas mutations in the m5C sites partially abrogated the luciferase activity (Figure 2J). Meanwhile, we investigated the subcellular localization of ALYREF by immunofluorescence. The results revealed that plenty of ALYREF proteins were located in both the nucleus and cytoplasm of BCa cells (Figure 2K). Next, ALYREF knockdown/overexpression cells were treated with Act D. The results revealed that knockdown or overexpression of ALYREF shortened or prolonged the half‐life of PKM2 mRNA, while K171A‐mutant ALYREF failed to regulate it (Figure 2L). Furthermore, consistent with the effect of ALYREF knockdown, we found that knockdown of NSUN2, the m5C writer, also resulted in the down‐regulation of PKM2 levels (Supplementary Figure S2).
Together, all results showed that ALYREF can stabilize PKM2 mRNA by an m5C‐mediated pathway.
3.3. ALYREF promoted glycolysis and proliferation of BCa cells
We conducted RNA‐seq to study the role of ALYREF in BCa cells and identified 569 significantly changed genes in 5637 cells after ALYREF knockdown, including 439 down‐regulated genes and 130 up‐regulated genes (Figure 3A). GSEA showed that the glycolysis set was strongly reduced after ALYREF knockdown (Figure 3B). Next, we combined our RNA‐seq data (569 genes) with ALYREF RIP‐seq data (4489 genes with potential m5C sites) from Yang et al. [41] and found that 206 potential mRNAs might be bound and regulated by ALYREF (Figure 3C). PKM2, as the major participant in glycolysis metabolism, was down‐regulated in ALYREF‐knockdown 5637 cells. Glucose utilization, lactic acid detection, and intracellular ATP detection assays revealed that ALYREF knockdown significantly reduced glucose utilization, lactic acid production, and intracellular ATP levels, and ALYREF overexpression promoted glycolysis in 5637 and T24 cells (Figure 3D‐3F). ALYREF knockdown inhibited BCa cell proliferation significantly, whereas ALYREF overexpression showed the opposite effect as assessed by using CCK‐8 assays (Figure 3G). ALYREF knockdown inhibited and ALYREF overexpression promoted colony formation of in BCa cells (Figure 3H and 3I). K171A‐mutant ALYREF failed to regulate the proliferation and colony formation of BCa cells. In the subcutaneous implantation experiment, the xenograft model was constructed by using stably transfected T24 cells. The results showed that knockdown of ALYREF inhibited tumor volume and weight in mice, while ALYREF overexpression had the opposite effect (Figure 3J‐3L). Meanwhile, the expression of Ki67 (a cell proliferation marker) was decreased in ALYREF knockdown groups and increased in ALYREF overexpression groups significantly (Supplementary Figure S3). Furthermore, the TCGA database and qRT‐PCR assays showed a positive relationship between PKM2 expression and ALYREF expression in BCa tissues (Supplementary Figure S4A and S4B). This positive relationship was also observed in IHC analysis of the paraffin‐embedded tissues from BCa xenografts (Supplementary Figure S4C and S4D). The data strongly suggested that ALYREF can regulate glucose metabolism and be a tumor‐driver gene in BCa.
FIGURE 3.
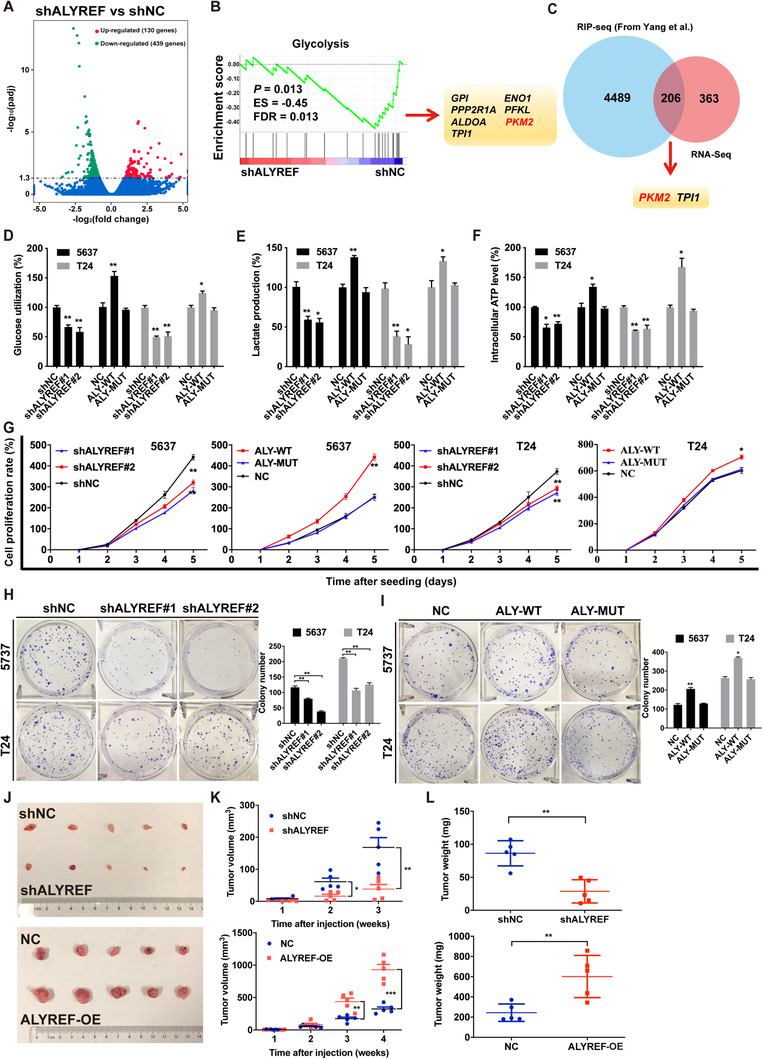
ALYREF promotes glycolysis and proliferation of BCa cells. (A) There are 569 genes significantly changed in ALYREF knockdown (shALYREF) compared to control (shNC) 5637 cells. The results were determined by RNA‐seq and shown by the volcano plot. (B) GSEA showes that the glycolysis set is strongly correlated with ALYREF. Significantly regulated genes are also presented. (C) A Venn diagram showes 206 overlapping genes between RNA‐seq data from the present study and ALYREF RIP‐seq data from Yang et al. [41]. (D, E, F) Effects of ALYREF knockdown, ALYREF wild‐type overexpression, or ALYREF mutation overexpression on glucose utilization, lactic acid production, and intracellular ATP level in 5637 and T24 cells. (G) Effects of ALYREF knockdown, ALYREF wild‐type overexpression, or ALYREF mutation overexpression on cell proliferation detected by CCK‐8 assays. (H) The efficiencies of cell colony formation in BCa cells with ALYREF knockdown. (I) Effects of ALYREF wild‐type or ALYREF mutation overexpression on cell proliferation measured by CCK‐8 assays. (J, K, L) Effects of ALYREF knockdown or overexpression on tumor volume and weight in nude mouse xenograft models. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: shALYREF, knockdown of Aly/REF export factor; NC, negative control; shNC, negative control of knockdown; ES, enrichment score; FDR, false discovery rate; GPI, glucose‐6‐phosphate isomerase; PPP2R1A, protein phosphatase 2 scaffold subunit Aalpha; ALDOA, aldolase, fructose‐bisphosphate A; TPI1, triosephosphate isomerase 1; ENO1, enolase 1; PFKL, phosphofructokinase, liver type; PKM2, Pyruvate kinase muscle isozyme M2; RIP‐seq, RNA Iimmunoprecipitation sequencing; ATP, adenosine triphosphate; ALY‐WT, Aly/REF export factor wild type; ALY‐MUT, Aly/REF export factor mutation; ALY‐OE, Aly/REF export factor overexpression; CCK‐8 assay, cell counting kit‐8 assay; BCa, bladder cancer.
3.4. ALYREF exerted a carcinogenic effect in BCa by up‐regulating PKM2 expression
To explore the synergenic carcinogenic effect of PKM2 with ALYREF in BCa, we co‐transfected plasmid vector (PKM2 or control) in ALYREF‐knockdown or control 5637 and T24 cells. The expression of PKM2 protein in transfacted 5637 and T24 cells was confirmed by Western blotting (Figure 4A). The overexpression of PKM2 largely rescued the inhibition of glucose utilization, lactic acid production, and intracellular ATP levels caused by ALYREF knockdown (Figure 4B). CCK‐8 assays (Figure 4C) and colony formation assays (Figure 4D) also showed that PKM2 can restore the cell proliferation and colony formation inhibited by ALYREF knockdown in BCa cells. These data suggested that PKM2 mediated the carcinogenic effect of ALYREF in BCa cells.
FIGURE 4.
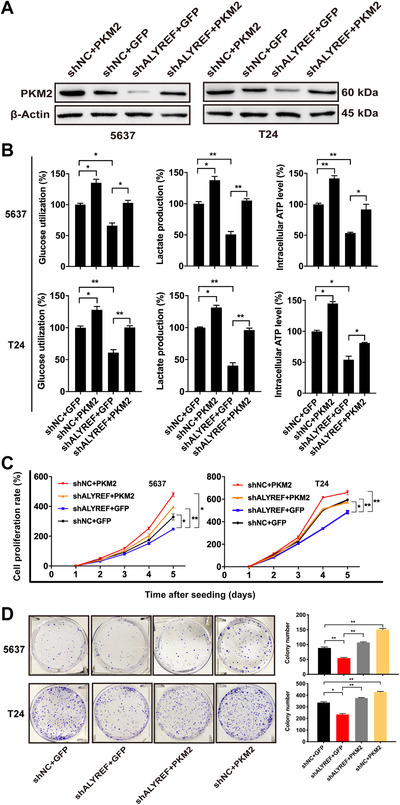
Overexpression of PKM2 reverses the inhibition of aerobic glycolysis and cell proliferation caused by ALYREF knockdown in BCa cells. (A) Western blotting of PKM2 expression in 5637 and T24 cells co‐transfected with lentivirus vector (ALYREF knockdown or control) and plasmid vector (PKM2 or control). (B) Glucose assay, lactic acid assay, and ATP assay showe that overexpression of PKM2 rescues inhibition of aerobic glycolysis caused by ALYREF knockdown in 5637 and T24 cells. (C) Overexpression of PKM2 rescues the cell proliferation affected by ALYREF knockdown in 5637 and T24 cells by CCK‐8 assays. (D) Overexpression of PKM2 rescues the colony formation affected by ALYREF knockdown in 5637 and T24 cells by colony formation assays. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: PKM2, Pyruvate kinase muscle isozyme M2; GFP, green fluorescent protein; shNC, negative control of knockdown; ALYREF, Aly/REF export factor; shALYREF, knockdown of Aly/REF export factor; BCa, bladder cancer.
3.5. ALYREF was up‐regulated in BCa tissues and predicted poor prognosis in BCa patients
Firstly, qRT‐PCR and Western blotting analysis demonstrated that ALYREF was significantly up‐regulated in BCa tissues, compared with matched adjacent normal tissues (Figure 5A and 5B). Next, we conducted IHC analysis of TMA (Figure 5C) and divided 162 primary BCa surgical specimens into low ALYREF expression (n = 123) and high ALYREF expression groups (n = 39). Subsequent statistical analysis showed that ALYREF expression in BCa patients was associated with tumor size and lymph node metastasis (Table 1). Kaplan‐Meier survival curves showed that high ALYREF expression predicted poor OS and RFS in BCa patients (Figure 5D). Meanwhile, TCGA database analysis also showed similar results that ALYREF was up‐regulated in BCa tissues (Supplementary Figure S5A) and predicted poor prognosis in BCa patients (Supplementary Figure S5B). Moreover, ALYREF was also found to be significantly up‐regulated in many other tumors available in the GENT database (Supplementary Figure S5C).
FIGURE 5.
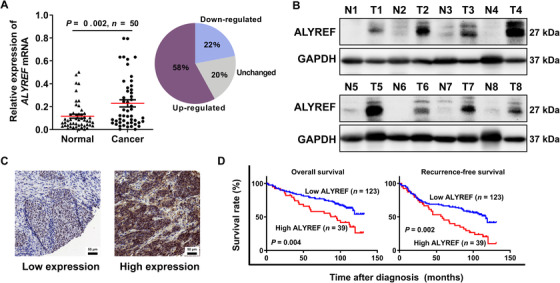
ALYREF is up‐regulated in BCa tissues and predicts poor prognosis for BCa patients. (A) Relative expression of ALYREF mRNA in 50 paired BCa tissues and adjacent normal tissues by qRT‐PCR. (B) The typical expression of ALYREF protein in 8 paired BCa tissues (T) and adjacent normal tissues (N) by Western blotting. (C) IHC staining of ALYREF. ALYREF protein is stained in brown and located in both nucleus and cytoplasm. (D) Kaplan‐Meier survival analysis shows that ALYREF up‐regulation predicts poor OS and RFS in BCa patients. The log‐rank test was used to compare differences between the two groups. Abbreviations: ALYREF, Aly/REF export factor; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; BCa, bladder cancer; CI, confidence interval; qRT‐PCR, quantitative real‐time polymerase chain reaction; IHC, immunohistochemistry; OS, overall survival; RFS, recurrence‐free survival.
TABLE 1.
Association between ALYREF and clinicopathologic characteristics of the 162 bladder cancer patients
| ALYREF expression [cases (%)] | ||||
|---|---|---|---|---|
| Parameter | Total [cases (%)] | Low | High | P value |
| Total | 162 | 123 (75.9) | 39 (24.1) | |
| Gender | 0.203 | |||
| Male | 125 (77.2) | 92 (74.8) | 33 (84.6) | |
| Female | 37 (22.8) | 31 (25.2) | 6 (15.4) | |
| Age at surgery (years) | 0.177 | |||
| ≥ 65 | 94 (58.0) | 75 (61.0) | 19 (48.7) | |
| < 65 | 68 (42.0) | 48 (39.0) | 20 (51.3) | |
| Pathological stage | 0.612 | |||
| pT2‐T4 | 41 (25.3) | 17 (13.8) | 24 (61.5) | |
| pTa‐T1 | 121 (74.7) | 106 (86.2) | 15 (38.5) | |
| Grade | 0.135 | |||
| High | 87 (53.7) | 62 (50.4) | 25 (64.1) | |
| Low | 75 (46.3) | 61 (49.6) | 14 (35.9) | |
| Tumor size (cm) | 0.017 | |||
| ≥ 4 | 65 (40.1) | 43 (35.0) | 22 (56.4) | |
| < 4 | 97 (59.9) | 80 (65.0) | 17 (43.6) | |
| Lymph node metastasis | 0.021 | |||
| Present | 47 (29.0) | 30 (24.4) | 17 (43.6) | |
| Absent | 115 (71.0) | 93 (75.6) | 22 (56.4) | |
| PKM2 expression | 0.044 | |||
| Low | 85 (52.5) | 70 (56.9) | 15 (38.5) | |
| High | 77 (47.5) | 53 (43.1) | 24 (61.5) | |
Abbreviations: ALYREF, Aly/REF export factor; PKM2, pyruvate kinase muscle isozyme M2.
3.6. HIF‐1α up‐regulated the ALYREF expression in BCa cells
To study whether ALYREF expression is regulated by certain transcription factors that bind to the ALYREF promoter, we utilized bioinformatic analysis (PROMO and JASPAR databases) to excavate potential regulatory factors and HIF‐1α got the highest score. The data also predicted the potential putative binding sites in the promoter region of ALYREF and identified a consensus HRE (5′‐ACGTGC‐3′, Figure 6A). Subsequently, to analyze the effect of HIF‐1α on the expression of ALYREF in human BCa cells, we exposed 5637 and T24 cells to 20% or 1% O2 for 24 h or 48 h. The mRNA and protein of ALYREF and PKM2 were significantly increased in 5637 and T24 cells, with the increased expression of HIF‐1α protein (Figure 6B and 6C). In addition, to determine the requirement of the HRE site for HIF‐1α to transactivate ALYREF promoter, we conducted luciferase reporter assays. The deletion mutation of HRE nearly abolished the transactivation of ALYREF promoter by hypoxia (Figure 6D). We also inserted the PKM2 promoter fragments encompassing the HRE in the reporter plasmids. The PKM2 HRE significantly increased relative luciferase activity in hypoxic 293T cells (Figure 6E).
FIGURE 6.
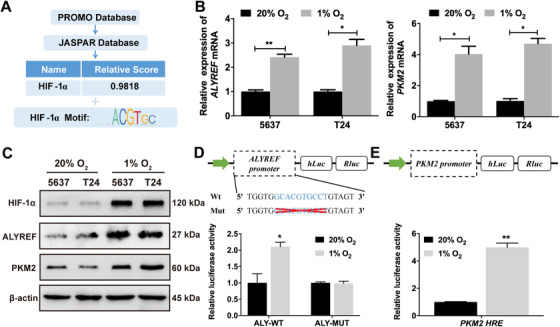
HIF‐1α up‐regulates ALYREF expressions in BCa cells. (A) Bioinformatic analysis identifies HIF‐1α as the potential transcription factor and ‘ACGTGC’ as the consensus hypoxia response element for ALYREF. (B) ALYREF and PKM2 mRNA levels are up‐regulated in 1% O2 group as compared with 20% O2 group by qRT‐PCR. 5637 and T24 cells were exposed to 20% or 1% O2 for 24 h. (C) HIF‐1α, ALYREF, and PKM2 proteins are up‐regulated in 5637 and T24 cells exposed to 1% O2 as compared with those exposed to 20% O2 for 48 h by Western blotting. (D) Relative luciferase activity was measured in 293T cells transfected with reporter plasmids containing either wild‐type or mutant ALYREF promoter fragments. Then cells were exposed to 20% or 1% O2 for 48 h. (E) Relative luciferase activity was measured in 293T cells transfected with reporter plasmids containing PKM2 promoter fragments. Then cells were exposed to 20% or 1% O2 for 48 h. The ratio of luciferase activity was determined. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: HRE, hypoxia‐responsive element. Abbreviations: BCa, bladder cancer; HIF‐1α, hypoxia‐ inducible factor‐ 1‐alpha; ALYREF, Aly/REF export factor; PKM2, Pyruvate kinase muscle isozyme M2; Wt, wild type; Mut, mutation; ALY‐WT, Aly/REF export factor wild type; ALY‐MUT, Aly/REF export factor mutation; hLuc, fireflyer luciferase; Rluc, Renillar luciferase.
4. DISCUSSION
In this study, we found that PKM2 was highly expressed in BCa tissues and related to the poor prognosis of BCa patients in the TCGA database and our clinical specimens. Further experiments showed that the highly expressed PKM2 promoted glycolysis and proliferation of BCa cells. These data further confirmed the carcinogenic role of PKM2 in BCa.
Using the ALYREF RIP‐seq data from Yang et al.[41], we discovered that PKM2 may have m5C sites and be bound by ALYREF. The role of m5C may be similar to that of m6A in mRNA metabolism at different stages, including splicing, maturation, export, stability, translation, and decay [50, 51]. ALYREF specifically recognizes the m5C sites catalyzed by NSUN2 and consequently promotes nucleus‐cytoplasm shuttling of mRNAs [41]. Therefore, we conducted RIP assays and confirmed that ALYREF could specifically recognize the 3′‐UTR of PKM2 mRNA through K171. Subsequently, we found that knockdown of NSUN2 reduced the m5C level of total RNA, including the m5C level of PKM2 mRNA 3'‐UTR, in BCa cells through dot blotting assays and MeRIP assays. Further RIP assays showed that NSUN2 knockdown dramatically reduced the binding efficiency of ALYREF to the 3′‐UTR of PKM2 mRNA. These data indicated that NSUN2 can methylate the 3′‐UTR of PKM2 mRNA and then ALYREF bound with it in BCa cells. Moreover, our qRT‐PCR and Western blotting experiments showed that ALYREF can enhance the expression of PKM2 mRNA through K171. Further dual‐luciferase reporter assays indicated that this function relied on the interaction between ALYREF and m5C sites of PKM2 3′‐UTR. Act D assays showed that ALYREF can stabilize PKM2 mRNA, and immunofluorescence showed that plenty of ALYREF proteins were located in both the nucleus and cytoplasm of BCa cells. We also found that knockdown of NSUN2 reduced the expression level of PKM2. These data showed that ALYREF could specifically recognize the m5C site of 3′‐UTR in PKM2 mRNA through K171, which increased the stability of methylated mRNA and ultimately led to increased protein levels.
At present, there is a lack of effective therapeutic targets and prognostic indicators for BCa [3]. Profound metabolic abnormalities are key mechanisms mediating BCa tumorigenesis and may provide potential therapeutic targets [52]. The expression level of PKM2 was up‐regulated by ALYREF, suggesting that ALYREF may also promote aerobic glycolysis of BCa, and subsequent data also validated our conjecture. Our RNA‐seq analysis showed that ALYREF potentially up‐regulated glucose metabolism in BCa. Then we studied the function of ALYREF in BCa. In vitro, we found that overexpression of ALYREF significantly promoted the glucose utilization, lactic acid production, intracellular ATP production, and proliferation of BCa cells; the opposite was true when ALYREF was knockdown. In vivo, we also found that ALYREF played an oncogenic function in BCa. Domínguez‐Sánchez et al. [53] observed that ALYREF was highly expressed in a broad range of tumors with 10 pairs of tissues separately, which was consistent with the data from the GENT database. This means that ALYREF may also play an oncogenic role in other cancers, besides BCa. Another study reported that the overexpression of ALYREF might be associated with oral cancer metastasis [54]. All these results strongly support that ALYREF promotes glycolytic metabolism and cell proliferation in BCa. However, K171A mutation almost abrogated the function of ALYREF, which suggested the biological function of ALYREF relied on its m5C reader activity.
Proliferating cells preferred aerobic glycolysis [4]. PKM2, as a rate‐limiting enzyme of glycolysis, is preferentially expressed in value‐added cells, including cancer cells [55, 56]. ALYREF was also highly expressed in proliferative cells [53]. Additionally, overexpression of PKM2 totally reversed the biological function affected by ALYREF knockdown in BCa cells. All these suggest that ALYREF plays its oncogenic roles in BCa and participates in a complex regulatory network of glycolysis mainly by the regulation of PKM2, providing new insights into molecular mechanisms of cancer cell metabolism.
Previous studies indicated that HIF‐1α bound to the HRE of PKM2 and directly activated its transcription [20]. We reached the consistent results that PKM2 expression was directly transactivated by HIF‐1α in BCa cells. Noticeably, database analysis and subsequent validation assays showed that ALYREF expression was also up‐regulated by HIF‐1α, which indicated HIF‐1α could indirectly up‐regulate the expression of PKM2 in an m5C‐ALYREF‐dependent manner. A proposed working model of the HIF‐1α/ALYREF/PKM2 axis in BCa is shown in Figure 7. Our research also has some limitations, such as lack of m5C‐RIP sequencing and further verification of hypoxia mechanism. A previous study reported that PKM2 can regulate HIF‐1α activity directly, which suggests that PKM2 may play a more complex role in BCa [57]. More detailed and systematic studies need to be explored in the future.
FIGURE 7.
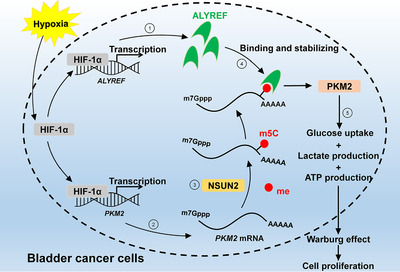
Working model: The role of HIF‐1α/ALYREF/PKM2 axis in BCa. Abbreviations: NSUN2, NOP2/Sun RNA methyltransferase family member 2; m5C, 5‐methylcytidine; me, methyl; BCa, bladder cancer.
5. CONCLUSIONS
In summary, our work clarified that PKM2 mRNA can be stabilized by ALYREF in an m5C‐dependent manner. ALYREF, an m5C RNA‐binding protein, can promote BCa cell proliferation by PKM2‐mediated glycolysis. As the high expression of PKM2 and ALYREF was associated with poor outcomes in BCa patients, ALYREF and its target genes PKM2 may be promising biomarkers to guide early diagnosis of BCa. Moreover, HIF‐1α could indirectly up‐regulate the expression of PKM2 by activating ALYREF in addition to directly activating its transcription. Above all, our study suggested that m5C may play a critical role in the hypoxia‐glycolysis network and HIF‐1α/ALYREF/PKM2 signaling may be a potential therapeutic target for BCa.
DECLARATIONS
STUDY APPROVAL
All animal experiments were approved by the Animal Management Committee of Nanjing Medical University. All procedures involving human participants in this study were approved by the Research Ethics Committee of The First Affiliated Hospital of Nanjing Medical University. All patients included in the present study signed informed consent.
AUTHOR CONTRIBUTIONS
QL and JFW designed the study and HWY put the design into practice. JZW, WZ, JH, XY, RZ, HY, WBY and HCL designed and completed the experiments. JT, PCL, and MG collected clinical tissues. JZW, WZ, JH, and XY analyzed the data and wrote the manuscript. QL, JFW, and HWY revised the paper. This manuscript was approved by all authors.
CONFLICT OF INTEREST
The authors declared no conflicts.
Supporting information
Supporing Infromation
Supporing Infromation
Supporing Infromation
ACKNOWLEDGMENTS
The present study was supported by grants from the National Natural Science Foundation of China (81602235, 81772711), the “333” project of Jiangsu Province (LGY2018055, LGY2016002), Key Provincial Talents Program of Jiangsu Province (ZDRCA2016006), the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801), the Provincial Initiative Program for Excellency Disciplines of Jiangsu Province (BE2016791).
Wang J‐Zi, Zhu W, Han J, Yang X, Zhou R, Lu H‐C, et al. The role of the HIF‐1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun. 2021;41:560–575. 10.1002/cac2.12158
Contributor Information
Qiang Lu, Email: doctorlvqiang@sina.com.
Ji‐Fu Wei, Email: weijifu@njmu.edu.cn.
Haiwei Yang, Email: haiweiyang@njmu.edu.cn.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. [DOI] [PubMed] [Google Scholar]
- 2. Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Communications (London, England). 2019;39(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Massari F, Ciccarese C, Santoni M, Iacovelli R, Mazzucchelli R, Piva F, et al. Metabolic phenotype of bladder cancer. Cancer Treat Rev. 2016;45:46‐57. [DOI] [PubMed] [Google Scholar]
- 4. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat. 2018;38:1‐11. [DOI] [PubMed] [Google Scholar]
- 6. Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, et al. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol Lett. 2016;11(3):1980‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu H, Tu W, Chen Y, Zhu M, Jin H, Huang T, et al. The combination of PKM2 overexpression and M2 macrophages infiltration confers a poor prognosis for PDAC patients. J Cancer. 2020;11(8):2022‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li TE, Wang S, Shen XT, Zhang Z, Chen M, Wang H, et al. PKM2 Drives Hepatocellular Carcinoma Progression by Inducing Immunosuppressive Microenvironment. Front Immunol. 2020;11:589997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Papadaki C, Manolakou S, Lagoudaki E, Pontikakis S, Ierodiakonou D. Correlation of PKM2 and CD44 Protein Expression with Poor Prognosis in Platinum‐Treated Epithelial Ovarian Cancer: A Retrospective Study. Cancers. 2020;12(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao H, Zhang L, Chen Y, Zhou C, Wang X, Wang D. PKM2 Promotes Breast Cancer Progression by Regulating Epithelial Mesenchymal Transition. Analytical Cellular Pathology (Amsterdam). 2020;2020:8396023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, et al. LncRNA‐FEZF1‐AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling. Clin Cancer Res. 2018;24(19):4808‐19. [DOI] [PubMed] [Google Scholar]
- 12. Dayton TL, Gocheva V, Miller KM, Israelsen WJ, Bhutkar A, Clish CB, et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 2016;30(9):1020‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morita M, Sato T, Nomura M, Sakamoto Y, Inoue Y, Tanaka R, et al. PKM1 Confers Metabolic Advantages and Promotes Cell‐Autonomous Tumor Cell Growth. Cancer Cell. 2018;33(3):355‐67.e7. [DOI] [PubMed] [Google Scholar]
- 14. Huang C, Huang Z, Bai P, Luo G, Zhao X, Wang X. Expression of pyruvate kinase M2 in human bladder cancer and its correlation with clinical parameters and prognosis. Onco Targets Therapy. 2018;11:2075‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Hao F, Nan Y, Qu L, Na W, Jia C, Chen X. PKM2 Inhibitor Shikonin Overcomes the Cisplatin Resistance in Bladder Cancer by Inducing Necroptosis. Int J Biol Sci.. 2018;14(13):1883‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Zhang F, Wu XR. Inhibition of Pyruvate Kinase M2 Markedly Reduces Chemoresistance of Advanced Bladder Cancer to Cisplatin. Sci Rep. 2017;7:45983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu Q, Hong B, Zhang L, Wang J. Pyruvate kinase M2 inhibits the progression of bladder cancer by targeting MAKP pathway. J Cancer Res Ther. 2018;14(Supplement):S616‐s21. [DOI] [PubMed] [Google Scholar]
- 18. Yang X, Cheng Y, Li P, Tao J, Deng X, Zhang X, et al. A lentiviral sponge for miRNA‐21 diminishes aerobic glycolysis in bladder cancer T24 cells via the PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 2015;36(1):383‐91. [DOI] [PubMed] [Google Scholar]
- 19. Boriack‐Sjodin PA, Ribich S, Copeland RA. RNA‐modifying proteins as anticancer drug targets. Nat Rev Drug Discov. 2018;17(6):435‐53. [DOI] [PubMed] [Google Scholar]
- 20. Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, et al. Pyruvate kinase M2 is a PHD3‐stimulated coactivator for hypoxia‐inducible factor 1. Cell. 2011;145(5):732‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kleszcz R, Paluszczak J, Krajka‐Kuźniak V, Baer‐Dubowska W. The inhibition of c‐MYC transcription factor modulates the expression of glycolytic and glutaminolytic enzymes in FaDu hypopharyngeal carcinoma cells. Adv Clin Exp Med. 2018;27(6):735‐42. [DOI] [PubMed] [Google Scholar]
- 22. Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates β‐catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu Q, Liu LZ, Yin Y, He J, Li Q, Qian X,et al. Regulatory circuit of PKM2/NF‐κB/miR‐148a/152‐modulated tumor angiogenesis and cancer progression. Oncogene. 2015;34(43):5482‐93. [DOI] [PubMed] [Google Scholar]
- 24. Guo M, Zhao X, Yuan X, Jiang J, Li P. MiR‐let‐7a inhibits cell proliferation, migration, and invasion by down‐regulating PKM2 in cervical cancer. Oncotarget. 2017;8(17):28226‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu CY, Chen J, Qin XH, You P, Ma J, Zhang J, et al.: Long non‐coding RNA NORAD promotes the prostate cancer cell extracellular vesicle release via microRNA‐541‐3p‐regulated PKM2 to induce bone metastasis of prostate cancer. J Exp Clin Cancer Res. 2021;40(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zahra K, Dey T, Ashish, Mishra SP , Pandey U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front Oncol. 2020;10:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, et al.: 5‐methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21(8):978‐90. [DOI] [PubMed] [Google Scholar]
- 28. Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, et al. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF‐κB/MYC signaling network. Oncogene. 2019;38(19):3667‐80. [DOI] [PubMed] [Google Scholar]
- 29. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri‐miR221/222 maturation in m6A‐dependent manner. Mol Cancer. 2019;18(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335(6076):1643‐6. [DOI] [PubMed] [Google Scholar]
- 31. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6‐methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15(5):313‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303‐d07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu H, Yang X, Tang J, Si S, Zhou Z, Lu J, et al. ALKBH5 Inhibited Cell Proliferation and Sensitized Bladder Cancer Cells to Cisplatin by m6A‐CK2α‐Mediated Glycolysis. Molecular Therapy Nucleic Acids. 2021;23:27‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3‐mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193‐205. [DOI] [PubMed] [Google Scholar]
- 35. Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, et al. m(6)A‐dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37(2):e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon‐domain modifications. EMBO Rep. 2008;9(7):629‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Helm M. Post‐transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34(2):721‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al.: Widespread occurrence of 5‐methylcytosine in human coding and non‐coding RNA. Nucleic Acids Res. 2012;40(11):5023‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang T, Chen W, Liu J, Gu N, Zhang R. Genome‐wide identification of mRNA 5‐methylcytosine in mammals. Nat Struct Mol Biol. 2019;26(5):380‐88. [DOI] [PubMed] [Google Scholar]
- 41. Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5‐methylcytosine promotes mRNA export ‐ NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8(9):705‐13. [DOI] [PubMed] [Google Scholar]
- 43. Semenza GL. HIF‐1 mediates the Warburg effect in clear cell renal carcinoma. J Bioenerg Biomembr. 2007;39(3):231‐4. [DOI] [PubMed] [Google Scholar]
- 44. Robey IF, Lien AD, Welsh SJ, Baggett BK, Gillies RJ. Hypoxia‐inducible factor‐1alpha and the glycolytic phenotype in tumors. Neoplasia (New York, NY). 2005;7(4):324‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu H, Forbes RA, Verma A. Hypoxia‐inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem. 2002;277(26):23111‐5. [DOI] [PubMed] [Google Scholar]
- 46. Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA circ‐ITCH inhibits bladder cancer progression by sponging miR‐17/miR‐224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species‐tailored searches. Bioinformatics. 2002;18(2):333‐4. [DOI] [PubMed] [Google Scholar]
- 48. Khan A, Fornes O, Stigliani A, Gheorghe M, Castro‐Mondragon JA, van der Lee R, et al. JASPAR 2018: update of the open‐access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46(D1):D260‐d66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tao T, Su Q, Xu S, Deng J, Zhou S, Zhuang Y, et al. Down‐regulation of PKM2 decreases FASN expression in bladder cancer cells through AKT/mTOR/SREBP‐1c axis. J Cell Physiol. 2019;234(3):3088‐104. [DOI] [PubMed] [Google Scholar]
- 50. Liu Z, Zhang J. Human C‐to‐U Coding RNA Editing Is Largely Nonadaptive. Mol Biol Evol. 2018;35(4):963‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS, et al.: RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C‐dependent manner. Cell Death Dis. 2020;11(4):270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cheng Y, Yang X, Deng X, Zhang X, Li P, Tao J, et al.: Metabolomics in bladder cancer: a systematic review. Int J Clin Exp Med. 2015;8(7):11052‐63. [PMC free article] [PubMed] [Google Scholar]
- 53. Dominguez‐Sanchez MS, Saez C, Japon MA, Aguilera A, Luna R. Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer. 2011;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saito Y, Kasamatsu A, Yamamoto A, Shimizu T, Yokoe H, Sakamoto Y, et al.: ALY as a potential contributor to metastasis in human oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2013;139(4):585‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dayton TL, Jacks T. Vander Heiden MG: PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17(12):1721‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mazurek S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43(7):969‐80. [DOI] [PubMed] [Google Scholar]
- 57. Palsson‐McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, et al. Pyruvate kinase M2 regulates Hif‐1α activity and IL‐1β induction and is a critical determinant of the warburg effect in LPS‐activated macrophages. Cell Metab. 2015;21(1):65‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporing Infromation
Supporing Infromation
Supporing Infromation


