Abstract
As one of the most studied ribonucleic acid (RNA) modifications in eukaryotes, N6‐methyladenosine (m6A) has been shown to play a predominant role in controlling gene expression and influence physiological and pathological processes such as oncogenesis and tumor progression. Writer and eraser proteins, acting opposite to deposit and remove m6A epigenetic marks, respectively, shape the cellular m6A landscape, while reader proteins preferentially recognize m6A modifications and mediate fate decision of the methylated RNAs, including RNA synthesis, splicing, exportation, translation, and stability. Therefore, RNA metabolism in cells is greatly influenced by these three classes of m6A regulators. Aberrant expression of m6A regulators has been widely reported in various types of cancer, leading to cancer initiation, progression, and drug resistance. The close links between m6A and cancer shed light on the potential use of m6A methylation and its regulators as prognostic biomarkers and drug targets for cancer therapy. Given the notable effects of m6A in reversing chemoresistance and enhancing immune therapy, it is a promising target for combined therapy. Herein, we summarize the recent discoveries on m6A and its regulators, emphasizing their influences on RNA metabolism, their dysregulation and impacts in diverse malignancies, and discuss the clinical implications of m6A modification in cancer.
Keywords: cancer therapy, chemoresistance, immunotherapy, m6A methylation, oncogenesis, prognostic biomarkers, RNA epigenetics, RNA metabolism
As one of the best studied ribonucleic acid (RNA) modifications in eukaryotes, N6‐methyladenosine (m6A) plays a predominant role in controlling gene expression and further influences physiological and pathological processes, including oncogenesis and tumor progression. Here we review the functions of m6A and its regulators in RNA metabolism control, their oncogenic or tumor suppressive roles in diverse malignancies, as well as the application of m6A methylation in cancer diagnosis and therapeutics.

1. BACKGROUND
Analogous to deoxyribonucleic acid (DNA) and protein, RNA has more than 100 chemical modifications, which tremendously propels our understanding on gene expression control [1]. The most remarkable RNA modification is N6‐methyladenosine (m6A), methylated adenosine at the N6 position, which was first discovered in the 1970s [2, 3]. Although m6A is one of the most abundant messenger RNA (mRNA) modifications in mammals, its significance was not fully acknowledged until the identification of fat mass and obesity‐associated protein (FTO) as a demethylase and the advent of transcriptome‐wide m6A mapping techniques that depicts the full scope of m6A profile (Figure 1) [4, 5]. Next‐generation sequencing (NGS) revealed that the distribution of m6A on mRNA is widespread and not random. The consensus sequence RRACH (R indicates guanosine (G) or adenosine (A), while H indicates A, cytidine (C) or uridine (U)) and the enrichment in certain regions (3’ untranslated region and coding sequence) are common characteristics of the m6A epitranscriptome [4, 5]. Owing to the high abundance and reversible feature of m6A, more attention has been gained to the wide‐ranging regulation of m6A in physiological and pathological processes, especially in oncogenesis and tumor progression. Given the important roles of m6A in cancer, we discuss the functions of m6A and its regulators in RNA metabolism control, their oncogenic or tumor‐suppressive roles in diverse malignancies, as well as the potential application of m6A methylation in cancer diagnosis and therapeutics.
FIGURE 1.
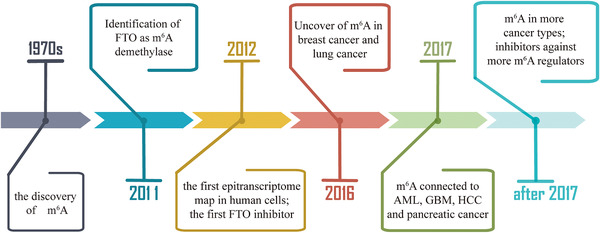
The timeline of RNA epigenetics. m6A was first discovered in the 1970s. In 2011, FTO was identified as an m6A demethylase. In 2012, the antibody‐based transcriptome‐wide sequencing method was developed to obtain m6A profiling in the human transcriptome. The first FTO inhibitor was found in the same year. Association of m6A with cancer began to be reported in breast cancer and lung cancer in 2016, and the cancer types expanded to AML, GBM, HCC, and pancreatic cancer in 2017. Up to now, m6A has been found to play critical roles in most cancer types, and inhibitors against more m6A regulators are in development. Abbreviations: RNA, ribonucleic acid; m6A, N6‐methyladenosine; FTO, fat mass and obesity‐associated protein; AML, acute myeloid leukemia; GBM, glioblastoma; HCC, hepatocellular carcinoma
2. m6A AND ITS REGULATORS IN RNA METABOLISM
The m6A modification is critical for RNA fate decision as it can influence almost all aspects of RNA metabolism, including synthesis (i.e. transcription), splicing, nuclear exportation, translation, and degradation. In this section, we summarize m6A regulators and their functions in RNA metabolism (Figure 2 and Table 1).
FIGURE 2.
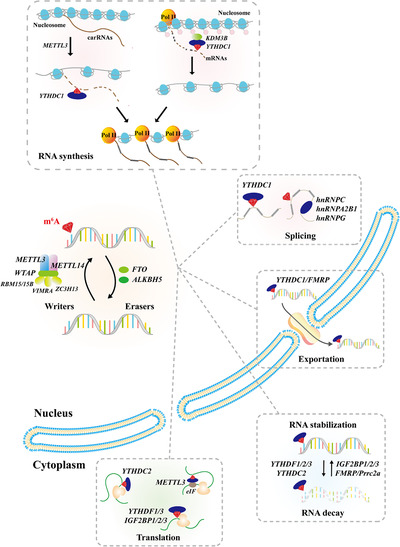
The functions of m6A and its machinery in RNA metabolism. The m6A modification is installed by m6A methyltransferases (Writers), consisting of METTL3/14, WTAP, VIRMA, RBM15/15B, and ZC3H13, and is removed by RNA demethylases (Erasers), including FTO and ALKBH5. The m6A reader proteins, including YTHDF1/2, YTHDF1/2/3, IGF2BP1/2/3, hnRNPA2B1/C/G, FMRP, Prrc2a and METTL3, work with m6A to participate in RNA synthesis, splicing, exportation, translation, and degradation. Abbreviations: m6A, N6‐methyladenosine; RNA, ribonucleic acid; METTL3/14, methyltransferase‐like 3/14; WTAP, Willms tumor 1 associated protein; VIRMA, Vir like m6A methyltransferase associated; RBM15/15B, RNA Binding Motif Protein 15/15B; ZC3H13, Zinc Finger CCCH‐Type Containing 13; FTO, fat mass and obesity‐associated protein; ALKBH5, alkB homolog 5; YTHDF1/2/3, YTH domain‐containing protein 1/2/3; IGF2BP1/2/3, insulin‐like growth factor 2 mRNA‐binding protein 1/2/3; hnRNPA2B1/C/G, heterogeneous nuclear ribonucleoproteins B1/C/G; FMRP, Fragile X mental retardation protein; Prrc2a, Proline‐Rich Coiled‐Coil 2A; carRNAs, chromosome‐associated regulatory RNAs; Pol II, polymerase II; mRNAs, messenger RNAs; eIF, eukaryotic translation initiation factor
TABLE 1.
The function of m6A in RNA metabolism
| RNA metabolism | m6A readers | Function | Mechanism | References |
|---|---|---|---|---|
| RNA synthesis | YTHDC1 | Enhance transcription | Regulate carRNAs and H3K9me2 to affect chromatin structure | [45, 46] |
| Splicing | YTHDC1 | Mediate alternative splicing | Recruit splicing factor SRSF3 and prevent SRSF10 | [23] |
| hnRNPA2B1 | Mediate alternative splicing | Function as splicing factor by itself | [33] | |
| hnRNPG | Mediate alternative splicing | m6A‐switch mechanism | [31] | |
| hnRNPC | Mediate alternative splicing | m6A‐switch mechanism | [30] | |
| Nuclear exportation | YTHDC1 | Promote exportation | Facilitate NXF1‐mediated export | [24] |
| FMRP | Promote exportation | Facilitate XPO‐mediated export | [34, 35] | |
| RNA stability | YTHDF2 | RNA decay | Recruit CCR4‐NOT deadenylase complex | [25, 36] |
| YTHDF3 | RNA decay | [27] | ||
| YTHDF1 | RNA decay | |||
| YTHDC2 | RNA decay | RNA decay | [37] | |
| IGF2BPs | Stabilize RNA | Recruit huR, PABPC1, MATR3 | [29] | |
| FMRP | Stabilize RNA | [38, 40] | ||
| PRRC2A | Stabilize RNA | [39] | ||
| Translation | YTHDF1 | Enhance translation | Facilitate cap‐dependent ribosome recruitment | [26] |
| YTHDF3 | Enhance translation | Interact with YTHDF1 | [27, 28] | |
| YTHDC2 | Enhance translation | couple active translation with prevention of mRNA decay | [37] | |
| METTL3 | Enhance translation | Interact with eIF3h and form a loop machinery | [41, 42] | |
| IGF2BPs | Enhance translation | Couple active translation with prevention of mRNA decay | [29] |
2.1. m6A regulators
The m6A modification on mRNA is installed by the m6A methyltransferase complex (MTC, also known as m6A “writers”). A heterodimer consisting of methyltransferase‐like 3 (METTL3) and methyltransferase‐like 14 (METTL14) constitutes the core of MTC, in which METTL3 is the catalytic subunit while METTL14 mediates substrate RNA recognition and binding [6, 7, 8, 9]. Other essential components of the MTC complex, including Willms tumor 1 associated protein (WTAP), RNA Binding Motif Protein 15 (RBM15), RNA Binding Motif Protein 15B (RBM15B), Zinc Finger CCCH‐Type Containing 13 (ZC3H13), and Vir like m6A methyltransferase associated (VIRMA), anchor MTC to target RNAs [10, 11, 12, 13, 14, 15].
The m6A modification is reversible and can be removed by m6A demethylases (also known as m6A “erasers”). As the first characterized RNA m6A demethylase, FTO also has oxidative demethylation activity towards multiple other types of DNA and RNA methylations, including m3T, m3U, m6Am, and m1A [16, 17]. Nonetheless, m6A is the major physiological substrate of FTO [16]. The alkB homolog 5 (ALKBH5) is the second m6A eraser which specifically demethylates RNA m6A [18].
The effect of m6A on gene expression is mediated by the m6A binding proteins, also known as m6A “readers”, which selectively interact with methylated RNAs and affect RNA metabolism. There are three well‐known families of m6A readers, YT521‐B homology (YTH) domain family, insulin‐like growth factor 2 mRNA‐binding proteins (IGF2BPs), and heterogeneous nuclear ribonucleoproteins (HNRNPs) [19, 20, 21, 22]. Members of the YTH domain family, including YTH domain‐containing protein 1 (YTHDC1), YTH domain‐containing protein 2 (YTHDC2), YTH domain‐containing family protein 1 (YTHDF1), YTH domain‐containing family protein 2 (YTHDF2), and YTH domain‐containing family protein 3 (YTHDF3), have been identified as direct m6A readers harboring m6A binding pockets [23, 24, 25, 26, 27, 28]. YTHDC1 is localized in the nucleus and regulates RNA splicing and nuclear exportation [23, 24] while cytoplasmic YTHDF1, YTHDF2, YTHDF3, and YTHDC2 modulate RNA decay and translation cooperatively [25, 26, 27, 28]. IGF2BPs, on the other hand, preferentially recognize and bind to m6A methylated mRNAs to promote their stability and translation [29]. Unlike these two families of m6A readers, the HNRNP family members, including heterogeneous nuclear ribonucleoprotein C (hnRNPC) and heterogeneous nuclear ribonucleoprotein G (hnRNPG), recognize their targets through an “m6A switch” mechanism in which methylated A on the opposite side of a U‐tract alters the structure and accessibility of hairpin RNAs [30, 31].
In short, writers and erasers work together to modulate m6A dynamics and maintain its homeostasis in cells, while the activity of readers allows m6A to exert its influence in each step of the RNA life cycle.
2.2. m6A‐mediated precursor mRNA (pre‐mRNA) splicing
Splicing is a fundamental step of gene expression regulation by removing introns and joining exons co‐transcriptionally. The alternative selection of exons results in the production of multiple mRNA variants and ultimately diverse protein products from a single gene, contributing to proteome diversity. The influence of m6A on alternative splicing was described by Dominissini et al. [5] and was further supported by studies showing that METTL3, WTAP, FTO and ALKBH5 all modulated alternative splicing [10, 18, 32]. The m6A methylated pre‐mRNAs indeed undergo alternative splicing through the activity of YTHDC1, heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNPA2B1), or an “m6A switch” mechanism. As a founding member of the YTH domain family, YTHDC1 binds methylated pre‐mRNAs and promotes exon inclusion by recruiting splicing factor serine/arginine‐rich splicing factor 3 (SRSF3) and repelling serine/arginine‐rich splicing factor (SRSF10) [23]. Similarly, hnRNPA2B1 binds to m6A‐bearing RNA and modulates a subset of METTL3‐ and m6A‐mediated alternative splicing events [33]. In the “m6A switch” mechanism, m6A affects RNA structure and enhances the accessibility of hnRNPC and hnRNPG to the flanking U‐tract, while loss of m6A or hnRNPC/hnRNPG can alter the splicing pattern of neighbor exons [30, 31].
2.3. m6A‐mediated RNA nuclear exportation
Fully spliced mRNAs are allowed to translocate from the nucleus to the cytoplasm, which is under tight control. The interaction of YTHDC1 and nuclear export adaptor protein SRSF3 facilitates RNA binding to nuclear RNA export factor 1 (NXF1) which assists in nuclear translocation [24]. Fragile X mental retardation protein (FMRP), also known as FMR1, is also required for the Exportin 1 (XPO1)‐mediated nuclear export of methylated mRNAs [34, 35]. The functional studies of these reader proteins support the earlier observation that accumulation of polyadenylated (polyA) RNA in the cytoplasm is associated with an increase in m6A methylation by ALKBH5 silencing [18], and the notion that m6A is a determinant for the subcellular location of mRNAs.
2.4. m6A‐modulated RNA stability
The steady level of mRNA is established by a balance between its production and degradation, thus, the stability of mRNA is of great importance on modulating mRNA metabolism and gene expression. Members of the YTH domain family play a crucial role in controlling mRNA turnover. As the first well‐defined m6A reader, YTHDF2 mediates the instability of the transcriptome in an m6A dependent manner [25]. The C‐terminal domain of YTHDF2 selectively binds m6A‐marked RNA while the N‐terminal domain mediates the anchoring of YTHDF2‐bound mRNA to RNA degradation site and the recruitment of carbon catabolite repression‐negative on TATA‐less (CCR4‐NOT) deadenylase complex, leading to the shortening of mRNA half‐life [25, 36]. Interestingly, a coordinated functional interaction among YTHDF proteins was reported, in which YTHDF3 could affect decay and translation of m6A‐modified RNA with combined efforts of YTHDF2 and YTHDF1, respectively [27]. YTHDC2 also plays an essential role in the translation and decay of methylated mRNA [37]. Considering the above‐mentioned m6A readers that mediate RNA degradation, mRNAs with declined m6A modification are supposed to be more stable. However, the opposite phenomenon was observed in a portion of mRNAs, especially for the transcripts of some oncogenes, suggesting an alternative mechanism of m6A‐dependent regulation on RNA half‐life. Before long, IGF2BP family proteins were identified as a new class of m6A readers that enhance mRNA stability by interacting with mRNA stabilizers, such as ELAV like RNA binding protein 1 (ELAVL1, also known as huR), poly(a) binding protein cytoplasmic 1 (PABPC1) and Matrin 3 (MATR3), and thereby, influencing gene expression [29]. In addition to IGF2BPs, FMRP and Proline‐Rich Coiled‐Coil 2A (PRRC2A) have been reported to bind to m6A marked mRNAs and play a role in maintaining mRNA stability, further demonstrating that m6A could function as a double‐edged sword in controlling mRNA half‐life [29, 38‐40].
2.5. m6A‐mediated RNA translation
Protein translation, a process in which the genetic codes are translated into amino acid sequences, is also tightly controlled. m6A has been widely reported to be involved in translation regulation. In the canonical cap‐dependent translation, YTHDF1 facilitates cap‐dependent ribosome recruitment to mRNA by forming a loop structure mediated by eukaryotic translation initiation factor 4G (eIF4G) and the interaction of YTHDF1 with eukaryotic translation initiation factor 3 (eIF3) [26]. YTHDF3 was later proven to have a coordinated translation‐promoting function with YTHDF1 [27, 28]. In addition, both IGF2BP proteins and YTHDC2 couple active translation with the prevention of mRNA decay [29, 37]. Notably, METTL3 plays a methyltransferase‐independent function to promote translation by interacting with eukaryotic translation initiation factor 3h (eIF3h) and forming mRNA loop machinery [41, 42]. It was also reported that m6A in 5′ untranslated region (5’UTR) of mRNAs or the body of circular RNAs (circRNAs) could promote translation in a cap‐independent manner [43, 44].
2.6. m6A‐associated RNA synthesis
Although it was thought that m6A mainly affects gene expression post‐transcriptionally, emerging evidence has shown that m6A carries a lot of weight in transcriptional control. Liu et al. [45] reported that METTL3 methylated chromosome‐associated regulatory RNAs (carRNAs), while YTHDC1 mediated the nuclear degradation of the methylated carRNAs. Loss of m6A methylation via Mettl3 knockout in mouse embryonic stem cells increased carRNAs levels and therefore facilitated chromatin accessibility and transcription activity [45]. Moreover, m6A on mRNAs could facilitate the open state of corresponding chromatin regions through YTHDC1‐mediated recruitment of histone H3 lysine 9 dimethylation (H3K9me2) demethylase lysine demethylase 3B (KDM3B), leading to the removal of the repressive H3K9me2 histone mark and the promotion of transcription [46].
In summary, m6A methylation has been widely associated with every aspect of RNA metabolism and gene expression regulation, attributing to the extensive research on the identification of m6A regulators and the exploration of their functions. We have been updated by rapidly expanding research in this field. For instance, the three homologs of YTHDF, YTHDF1, YTHDF2 and YTHDF3, were deemed to play distinct roles in controlling RNA decay and translation [25, 26, 27, 28]. However, a similar function of these three proteins on RNA decay and the compensation effect among them have been revealed very recently in certain cell contexts and bioprocesses, such as the ovarian development of zebrafish and the early development of mice [47, 48, 49]. Fully studying the interaction network of m6A regulators, including YTHDF proteins and others, under different contexts will give us a comprehensive insight into the effects of m6A modification on RNA metabolism.
3. ABERRANT m6A METHYLATION IN HUMAN CANCERS
Given the importance of m6A in controlling RNA metabolism, aberrant methylation usually causes dysregulation of gene expression, including activation of oncogenes and repression of tumor suppressors, which plays fundamental roles in the initiation, development, and progression of various cancer types (Figure 3 and Table 2).
FIGURE 3.
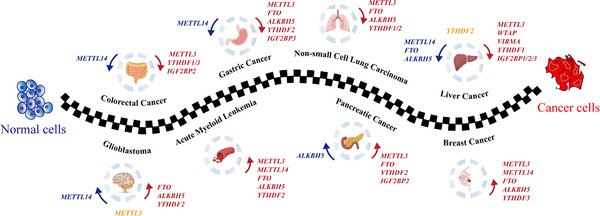
The roles of m6A regulators on tumorigenesis. The proteins promoting tumorigenesis are in red, the ones with tumor‐suppressive roles are in blue, while the ones with controversial function are in orange. Abbreviations: m6A, N6‐methyladenosine. METTL3/14, methyltransferase‐like 3/14; YTHDF1/2/3 YTH domain‐containing protein 1/2/3; IGF2BP1/2/3, insulin‐like growth factor 2 mRNA‐binding protein 1/2/3; FTO, fat mass and obesity‐associated protein; ALKBH5, alkB homolog 5; WTAP, Willms tumor 1 associated protein; VIRMA, Vir like m6A methyltransferase associated
TABLE 2.
The roles of m6A regulators in human cancer
| Cancer types | Regulators | Target genes | Roles | References |
|---|---|---|---|---|
| Liver cancer | METTL3 | SOCS2, SNAIL, LINC00958 | Oncogene | [64, 214, 215] |
| METTL14 | pri‐miR‐126 | Tumor suppressor | [62] | |
| WTAP | ETS1 | Oncogene | [65] | |
| VIRMA | ID2, GATA3 | Oncogene | [66, 67] | |
| ALKBH5 | LYPD1 | Tumor suppressor | [68] | |
| FTO | GNAO1 | Tumor suppressor | [69] | |
| YTHDF1 | Oncogene | [63, 73] | ||
| YTHDF2 | EGFR, IL11, SRPINE2 | Controversial | [70, 71, 72] | |
| IGF2BP1/2/3 | MYC, FSCN1, TK1, MARCRSL1 | Oncogene | [29] | |
| IGF2BP1 | SRF | Oncogene | [74] | |
| Non‐small Cell Lung Carcinoma | METTL3 | YAP, MALAT1, EGFR, TAZ, BRD4 | Oncogene | [41, 42, 120] |
| FTO | MZF1, USP7 | Oncogene | [121, 122] | |
| ALKBH5 | UBE2C, FOXM1, YAP | Oncogene | [123, 124, 125] | |
| YTHDF1 | KEAP1 | Oncogene | [126] | |
| YTHDF2 | 6PGD | Oncogene | [127] | |
| Gastric Cancer | METTL3 | ZMYM1, HDGF, SEC62, ARHGAP5‐AS1 | Oncogene | [76, 77, 78, 79, 80, 81] |
| METTL14 | Tumor suppressor | [82] | ||
| FTO | Oncogene | [82] | ||
| ALKBH5 | NEAT1 | Oncogene | [83] | |
| YTHDF2 | Oncogene | [216] | ||
| IGF2BP3 | HDGF | Oncogene | [77] | |
| Colorectal Cancer | METTL3 | SOX2, pri‐miR‐1246 | Oncogene | [97, 98] |
| METTL14 | pri‐miR‐375, lncRNA XIST, SOX4 | Tumor suppressor | [94, 95, 96] | |
| YTHDF1 | Oncogene | [99, 100] | ||
| YTHDF3 | GAS5 lncRNA | Oncogene | [101] | |
| IGF2BP2 | SOX2, MYC | Oncogene | [98, 102] | |
| Glioblastoma | METTL3 | ADAM19, SRSFs, SOX2 | Controversial | [105, 106, 107] |
| METTL14 | ADAM19 | Tumor suppressor | [107] | |
| FTO | Oncogene | [107] | ||
| ALKBH5 | FOXM1 | Oncogene | [108] | |
| YTHDF2 | MYC, VEGFA | Oncogene | [109] | |
| Acute Myeloid Leukemia | METTL3 | MYC, BCL2, PTEN, SP1, SP2 | Oncogene | [57, 58] |
| METTL14 | MYB, MYC | Oncogene | [59] | |
| FTO | ASB2, RARA, MYC, CEBPA | Oncogene | [51, 52] | |
| ALKBH5 | AXL, TACC3 | Oncogene | [55, 56] | |
| YTHDF2 | TNFRSF2 | Oncogene | [60] | |
| Pancreatic Cancer | METTL3 | pri‐miR‐25 | Oncogene | [90, 91, 152] |
| FTO | MYC | Oncogene | [89] | |
| ALKBH5 | KCNK15‐AS1, WIF1, PER1 | Tumor suppressor | [86, 87, 88] | |
| YTHDF2 | YAP | Oncogene | [92] | |
| IGF2BP2 | DANCR | Oncogene | [93] | |
| Breast Cancer | METTL3 | HBXIP, BCL2, AK4 | Oncogene | [111, 217, 218] |
| METTL14 | transforming growth factor β signaling pathway genes | Oncogene | [113] | |
| FTO | BNIP3, miR‐181b‐3p | Oncogene | [117, 118] | |
| ALKBH5 | NANOG | Oncogene | [114‐116, 219] | |
| YTHDF3 | ST6GALNAC5, GJA1, EGFR | Oncogene | [119] |
3.1. Acute myeloid leukemia
Acute myeloid leukemia (AML) is known for its devastating outcome and low 5‐year overall survival rate (<40%) in patients aged under 60 years old. It is originated from a disordered clone of hematopoietic stem and progenitor cells (HSPCs), leading to the blockage of myeloid differentiation and the production of leukemic stem cells (LSCs) with self‐renewal capacity that dominates the initiation of AML and the development of drug resistance [50]. The impact of FTO on AML when first discovered linked m6A RNA modification to AML [51]. Specifically, the overexpression of FTO promoted oncofusion proteins‐induced leukemogenesis through the demethylation of ankyrin repeat and socs box containing 2 (ASB2) and retinoic acid receptor alpha (RARA) mRNA transcripts [51]. The oncogenic function of FTO could be selectively inhibited by R‐2‐hydroxyglutarate (R‐2HG) in isocitrate dehydrogenase (IDH) wild‐type AML cells or by small molecular inhibitors FB23‐2 and CS1/2 in a broad panel of AML cells [52, 53], resulting in the suppression of cell growth, LSC maintenance and immune evasion [52, 54]. Another m6A demethylase ALKBH5 was also recently found to play oncogenic roles in AML through the KDM4C (lysine demethylase 4C)‐ ALKBH5‐AXL (tyrosine‐protein kinase receptor UFO) and ALKBH5‐m6A‐TACC3 (transforming acidic coiled‐coil containing protein 3) axes [55, 56].
On the other hand, the m6A methyltransferase machinery has also been linked to AML. Barbieri et al. [57] reported that METTL3 was recruited to transcriptional start sites by CCAAT enhancer‐binding protein zeta (CEBPZ), thus, promoted m6A deposition in the coding region and enhanced the translation of associated mRNA transcripts which helped to maintain leukemic state. Depletion of METTL3 in AML cells restrained translation of c‐MYC, B‐cell lymphoma 2 (BCL2), and phosphatase and tensin homolog (PTEN) through m6A‐mediated effects, leading to accelerated cell differentiation and apoptosis coupled with lower proliferative ability [58]. As another core component of the MTC, METTL14 also plays a critical role in leukemogenesis. The m6A modification and stability of transcriptional activator Myb (MYB) and MYC transcripts are under tight control by the SPI1 (transcription factor PU.1)‐METTL14 axis during normal hematopoiesis, while elevated expression of METTL14 leads to myeloid malignancy and enhanced self‐renewal capacity of leukemia stem cells by m6A‐mediated stabilization of MYB and MYC oncogenic transcripts [59].
Apart from m6A erasers and writers, the relationship between an m6A reader, YTHDF2, and leukemogenesis has also been uncovered. Paris et al. [60] reported that a decrease in Ythdf2 resulted in higher stability of the tumor necrosis factor receptor superfamily member 2 (Tnfrsf2) and more apoptosis of LSCs. They further found that depletion of YTHDF2 could promote hematopoietic stem cell (HSC) expansion, making YTHDF2 an additional promising anti‐leukemia target. Although m6A writers, erasers and readers are clearly associated with AML, how these regulators cooperate in the network is to be elucidated.
3.2. Liver cancer
As the sixth commonly diagnosed cancer, liver carcinoma is the fourth cause of tumor‐associated death globally [61]. The current existing challenge of liver cancer lies in the detection of late‐stage disease, recurrence, and distant metastasis. Therefore, growing efforts are being made to further understand the underlying mechanisms of liver cancer development and progression from various aspects, including RNA epigenetics.
By examination of m6A level in paired tumor and adjacent tissues, Ma et al. [62] found that m6A levels of polyA RNAs were decreased in hepatocellular carcinoma (HCC), the most common type of primary liver cancer. Further, they found that downregulation of METTL14 was associated with metastasis and could serve as a prognostic factor in HCC. Mechanistically, METTL14 could interact with the primary microRNA (miRNA) processing protein microprocessor complex subunit DGCR8 (DGCR8) and regulate primary miRNA‐126 processing in an m6A‐dependent manner [62]. In contrast, METTL3 is highly expressed in HCC and promotes HCC tumorigenicity and progression by regulating the suppressor of cytokine signaling 2 (SOCS2) and Snail family transcriptional repressor 1 (Snail1) mRNAs homeostasis [63, 64]. In addition to mRNA, the dysregulation of RNA methylation on the long non‐coding RNAs (lncRNAs) also contributes to the oncogenic function of METTL3. An HCC specific lncRNA, LINC00958, was stabilized by METTL3‐mediated m6A modification and was found to facilitate HCC lipogenesis and progression through the sponging of miRNA‐3619‐5p, and thus upregulated hepatoma‐derived growth factor (HDGF) expression [64]. A nanoplatform delivering LINC00958 small interfering RNA (siRNA) was then developed for anti‐HCC purposes [64]. As regulatory components of MTC, WTAP guides HuR‐mediated ETS Proto‐Oncogene 1 (ETS1) instability in an m6A‐dependent pattern [65] while VIRMA (also named KIAA1429) mediates the installation of m6A on the mRNA of DNA‐binding protein inhibitor ID‐2 (ID2) and the antisense lncRNA of GATA binding protein 3 (GATA3) [66, 67], thereby contributing to liver cancer development.
Besides MTC, the functions of other m6A modulators have been revealed in liver cancer as well. Declined ALKBH5 caused more m6A on LY6/PLAUR domain containing 1 (LYPD1), and the latter was recognized and stabilized by IGF2BP1, resulting in a more malignant HCC phenotype [68]. Sirtuin 1 (SIRT1)‐induced FTO SUMOylation (small ubiquitin‐related modifier, SUMO) leads to the degradation of FTO protein, alleviating FTO‐mediated G protein subunit alpha O1 (GNAO1) demethylation and increasing its expression, which has been shown to promote hepatocarcinogenesis [69].
Controversial roles of YTHDF2 have been reported in HCC. Yang et al. [70] reported that YTHDF2 was essential for HCC cell survival. By contrast, Hou et al. [71] reported that low expression of YTHDF2 could provoke inflammation, vascular reconstruction, and metastatic progression in HCC. This function could be blocked by hypoxia‐inducible factor (HIF)‐2α, revealing a molecular ‘rheostat’ role of YTHDF2 in the epitranscriptome and HCC progression [71]. Coincidentally, Zhong et al. [72] found YTHDF2 could suppress HCC cell proliferation by destabilizing the epidermal growth factor receptor (EGFR) mRNA and was inhibited by the hypoxia environment of HCC. For YTHDF1, its function in promoting the translation of Snail mRNA and driving epithelial‐to‐mesenchymal transition (EMT) seems to be consistent with the poor prognosis associated with its high expression level in HCC patients [63, 73]. The role of another reader protein, IGF2BP1, was also described in HCC, where IGF2BP1 protected serum response factor (SRF) mRNA from miRNA‐mediated decay in an m6A dependent manner, supporting IGF2BPs as oncogenic drivers in cancer [29, 74]. Overall, the above‐mentioned research lay the foundation for treating liver cancer from the RNA epigenetics view.
3.3. Gastrointestinal carcinoma
Gastric cancer is the fifth most‐diagnosed neoplasm globally, with approximately 1 million patients being newly diagnosed each year [75]. Considering its rapid progression and tendency to metastasis, scientists have been trying to find out intrinsic mechanisms of gastric cancer, and progress has been made in revealing the relationship between m6A regulators and the metastatic property of gastric cancer. For instance, METTL3 installs m6A on zinc finger MYM‐type containing 1 (ZMYM1) to increase its stability, and ZMYM1 recruits the C‐terminal‐binding protein (CtBP)/ lysine‐specific histone demethylase 1 (LSD1)/ REST corepressor 1 (CoREST) complex to repress E‐cadherin (also named cadherin‐1, CDH1) transcription, thus strengthening the EMT program and metastasis [76]. The activation of METTL3 transcription increases m6A modification on HDGF mRNA, facilitating the binding of IGF2BP3. Both of the secreted and nuclear HDGF contribute to gastric tumorigenesis and development [77]. The oncogenic role of METTL3 in gastric cancer was also demonstrated by other researchers [78, 79, 80, 81]. METTL14, in contrast, has a tumor‐suppressive function, and the knockdown of it activates the Wnt/ PI3K (phosphoinositide 3 kinase)‐ Akt (protein kinase B) signaling to promote tumor progression [82]. Other m6A‐related proteins, including IGF2BP3 and ALKBH5, are both shown to play oncogenic roles in the development of gastric carcinoma [77, 83].
Pancreatic cancer has the lowest survival rate (9%) among all cancer types [84, 85]. Providing insights into the development of pancreatic cancer from the aspect of RNA epigenetics is also of great significance. Three studies suggested a tumor‐suppressive role of ALKBH5 in pancreatic cancer. He et al. [86] reported that ALKBH5 inhibited pancreatic cancer motility by regulating the m6A level of antisense RNA 1 of KCNK15 (KCNK15‐AS1) lncRNA. Tang et al. [87] found that ALKBH5 was downregulated in pancreatic ductal adenocarcinoma (PDAC) cells and its overexpression sensitized cells to chemotherapy, with Wnt inhibitory factor 1 (WIF‐1) being identified as the target of ALKBH5. Recently, another research demonstrated that ALKBH5 led to demethylation of period circadian regulator 1 (PER1) mRNA and lifted PER1 level in a YTHDF2‐dependent manner, thereby reactivating the ATM (A‐T mutated)‐CHK2 (serine/threonine‐protein kinase)‐P53 (tumor protein 53)/CDC25C (cell division cycle 25C) pathway [88]. In contrast to ALKBH5, other m6A regulators, including FTO, METTL3, YTHDF2 and IGF2BP2, were all shown to exhibit oncogenic roles in pancreatic cancer by promoting cell proliferation, EMT, invasion, or chemo‐ and radio‐resistance [89, 90, 91, 92, 93].
As one of the most common types of carcinoma, colorectal cancer (CRC) is known for its increasing incidence globally [61]. m6A modification has been found to be involved in the pathogenesis of CRC in recent years. Reduction of METTL14 was found to be correlated with unfavorable prognosis of CRC patients. Mechanistically, less m6A modification on the oncogenic lncRNA (X inactive specific transcript) XIST or SRY‐box transcription factor 4 (SOX4) mRNA due to low level of METTL14 inhibited YTHDF2 binding, preventing the decay of XIST or SOX4, and resulting in the malignant phenotype [94, 95]. In addition, the processing of primary miRNA‐375 was inhibited in CRC with decreased expression of METTL14, which contributed to CRC progression [96]. Interestingly, the demethylation of histone H3 lysine 4 trimethylation (H3K4me3) on the METTL14 promoter is responsible for the repression of METTL14 transcription in CRC [95]. In contrast, METTL3 generally contributes to tumor development in CRC [97, 98]. For example, METTL3 was reported to have not only stemness‐inducing function but also tumorigenesis and metastasis‐promoting activity in CRC [98]. SRY‐box transcription factor 2 (SOX2), the downstream target of METTL3, was recognized and stabilized by IGF2BP2 in an m6A dependent manner [98]. In terms of m6A readers, YTHDF1 was regulated by MYC and promoted CRC development [99, 100], while YTHDF3, a well‐known target of Yes‐associated protein (YAP), formed a feedback loop by mediating the degradation of lncRNA growth arrest specific 5 (GAS5). The latter could facilitate YAP nuclear translocation, phosphorylation, and ubiquitin‐dependent decay in CRC [101]. In addition, long intergenic noncoding RNA for IGF2BP2 stability (LINRIS) was found able to inhibit ubiquitination of IGF2BP2 at lysine 139 and prevent its degradation via the autophagy‐lysosome pathway, thus promoting tumorigenesis through the LINRIS‐IGF2BP2‐MYC axis in CRC [102].
3.4. Glioblastoma
Glioblastoma (GBM) is a type of commonly occurred and aggressive brain tumor, in which m6A modification has been demonstrated to play a role as well [103, 104]. METTL3 is a predictive prognostic marker in GBM and plays a role in glioma stem‐like cells (GSCs) maintenance by depositing m6A modification in the SOX2 3’ untranslated region (3’UTR) region and leading to the overexpression of SOX2 [105]. Consistently, Li et al. [106] found that elevated expression of METTL3 correlated with the clinical aggressiveness of malignant gliomas. m6A modification of the splicing factor serine/arginine‐rich splicing factor (SRSF) decreased upon METTL3 knockdown, leading to YTHDC1‐dependent nonsense‐mediated mRNA decay of the SRSF transcripts and alternative splicing isoform switches in glioblastoma [106]. However, an opposite role of METTL3 in GBM has also been reported, in which METTL3 and METTL14 were considered as tumor suppressors by targeting metallopeptidase domain 19 (ADAM19), suggesting more in‐depth studies are remained to be done [107]. High forkhead box M1 (FOXM1) level caused by ALKBH5‐induced m6A reduction on FOXM1 mRNA contributes to GSC and tumorigenesis, while antisense RNA of FOXM1 (FOXM1‐AS) enhances the binding of ALKBH5 and FOXM1 [108]. More recently, Dixit et al. [109] reported a dependency of GSCs on YTHDF2, which surprisingly stabilized MYC and vascular endothelial growth factor A (VEGFA) transcripts in an m6A‐dependent manner, distinct from the well‐recognized role of YTHDF2 in mediating mRNA decay.
3.5. Breast cancer
As a highly heterogeneous neoplasm, breast cancer is the second cause of tumor‐associated death for women globally [110]. A positive feedback loop of HBXIP (Hepatitis B X‐interacting protein)/let‐7g (lethal‐7g)/METTL3/HBXIP, in which m6A modification was involved in gene expression regulation, was demonstrated to drive the aggressiveness of breast cancer [111]. In another study [112], the depletion of METTL3 induced adenylate kinase 4 (AK4) overexpression, reactive oxygen species (ROS) reduction, and less resistance of MCF‐7 cells to tamoxifen. METTL14 and ALKBH5 were shown to promote breast cancer growth and invasion by regulating m6A levels of key EMT and angiogenesis‐associated transcripts. Interestingly, the authors reported that METTL14 and ALKBH5 controlled each other's expression and inhibited YTHDF3, and the latter could in turn block RNA demethylase activity, forming a writer‐eraser‐reader collaborative loop [113]. The role of ALKBH5 in breast cancer has also been reported by other groups [114, 115, 116]. Notably, Zhang et al. [114, 116] found that the expression of ALKBH5 could be stimulated by HIF‐1α and HIF‐2α upon exposure to hypoxia, which increased breast cancer stem cells by reducing m6A modification on NANOG mRNA and increased NANOG protein level. Similar to ALKBH5, FTO also promotes breast cancer progression, with BCL2 interacting protein 3 (BNIP3) and miR‐181b‐3p being identified as targets of FTO [117, 118]. In addition, Chang et al. [119] revealed the involvement of YTHFD3‐mediated epitranscriptomic regulation in breast cancer brain metastasis. YTHDF3 overexpression was able to promote the translation of m6A‐modified ST6 N‐acetylgalactosaminide alpha‐2,6‐sialyltransferase 5 (ST6GALNAC5), gap junction protein alpha 1 (GJA1), and EGFR, which are related to brain metastasisFurther, they found that the overexpression of YTHDF3 was the combined consequence of increased gene copy number and the autoregulation of YTHDF3 cap‐independent translation by binding to m6A residues within its own 5’UTR.
3.6. Other cancers
Although advanced detection techniques and combined treatment have been used, lung carcinoma, especially non‐small cell lung cancer (NSCLC), is still the main cause of cancer‐associated death globally [61]. Most of the m6A modulators, including METTL3 [41, 42, 120], FTO [121, 122], ALKBH5 [123, 124, 125], YTHDF1 [126] and YTHDF2 [127], were demonstrated to be oncogenic in NSCLC, a main subtype accounted for 80%‐85% of lung cancer. It's worth mentioning that the oncogenic function of METTL3 is attributed to its methyltransferase‐dependent and ‐independent activities, suggested by recent studies [41, 42, 120]. As a writer, METTL3 installs m6A on mRNAs, such as YAP, and lncRNAs, such as metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1), and promotes the invasion and metastasis of NSCLC via the activation of the YAP pathway [120]. On the other hand, METTL3 functions like a reader to recognize m6A‐modified mRNAs and promote the production of oncoproteins, such as EGFR, tafazzin (TAZ), mitogen‐activated protein kinase 2 (MAPK2), DNA methyltransferase 3A (DNMT3A), and bromodomain‐containing protein 4 (BRD4) [41, 42]. Whether the oncogenic roles of METTL3 rely on its writer or reader activity in other cancer types is unclear and needs to be elucidated. In contrast to METTL3, ALKBH5 inhibits tumor growth and metastasis by lessening YAP level in a YTHDFs dependent manner and impairing YAP function with the help of the miRNA‐107/LATS2 (large tumor suppressor kinase 2) axis in lung cancer [125].
Diffuse large B‐cell lymphoma (DLBCL) is a subtype of lymphoid malignancy with heterogenous characteristics in clinical manifestation, pathology, and biology [128]. METTL3 was found to be upregulated in DLBCL tissues and promoted DLBCL progression by depositing m6A methylation on pigment epithelium‐derived factor (PEDF) transcript, though the detailed mechanism needs to be further studied [129]. A PIWI‐interacting RNA (piRNA), piRNA‐30473, was illustrated to have oncogenic activity in DLBCL through an m6A‐dependent manner [130]. Further, it was found that piRNA‐30473 could increase WTAP level, which facilitated m6A modifications on downstream targets, such as hexokinase 2 (HK2).
Ovarian cancer and endometrial cancer are highly aggressive gynecologic cancers [131, 132]. m6A regulators, including ALKBH5 [133], METTL3 [134], IGF2BP1 [74] and YTHDF1 [135], were suggested to be critical factors in promoting ovarian cancer. For instance, multi‐omics analysis has been used to explore the crucial component of m6A‐related modulators in ovarian cancer and identified a subunit of eIF3, eIF3C, as a direct YTHDF1 target [135]. Interestingly, the protein but not the RNA level of eIF3C was increased and positively correlated with the protein level of YTHDF1 in ovarian cancer patients, suggesting that modification of eIF3C mRNA could be more relevant to its role in cancer. Up to 70% of endometrial cancers exhibited m6A hypomethylation, possibly attributed to either a hotspot R298P (R is arginine, P is proline) mutation in METTL14 or a decline of METTL3 level [136]. The changes of these two key m6A modulators advanced endometrial tumor development via the AKT pathway.
Overall, dysregulation of m6A modifiers in cancer is frequently observed and plays crucial roles in cancer initiation, development, and drug resistance, through modulating/recognizing m6A on critical target transcripts.
4. FACTORS AFFECTING m6A IN CANCER
There is no doubt that m6A regulators dominate the layer of epitranscriptomic regulation; nonetheless, internal or external factors are able to regulate m6A incorporation in different contexts, especially in cancer. Here, we sum up factors that have an impact on m6A in cancer.
4.1. Genetic and epigenetic factors
Noncoding RNAs (ncRNAs) are a kind of RNA traditionally regarded as molecules that are not translated but have regulatory activities in gene expression. Accumulating data have shown that m6A methylation affects the production and/or functions of ncRNAs, including lncRNAs, circular RNAs, and miRNAs [12, 30, 44, 64, 94, 137‐140]. On the other hand, ncRNAs also play a role in m6A‐mediated gene expression regulation. For instance, ALKBH5 acted as an oncogene in GSCs by demethylating FOXM1 mRNA. Interestingly, this process was strengthened by FOXM1‐AS, a lncRNA antisense to FOXM1 [108]. The discovery of FOXM1‐AS as a pivotal modulator in ALKBH5‐dependent GSC proliferation emphasizes the role of ncRNA in GSC. RNA‐binding regulatory peptide (RBRP) is a peptide encoded by lncRNA LINC00266‐1, and its interaction with the m6A reader IGF2BP1 intensified the function of IGF2BP1, thus, reinforced the expression of MYC and the process of tumorigenesis. More importantly, higher RBRP level in patients was associated with shorter overall survival, confirming its oncogenic effect and the potential applications as a therapeutic target in treating cancers [141]. In colorectal cancer, the inhibition of miRNA‐455‐3p rescued β‐catenin depletion‐induced reduction of heat shock transcription factor 1 (HSF1) m6A modification and METTL3 interaction [142]. Taken together, ncRNAs exist as critical modulators of m6A‐dependent gene expression control, and more of their regulatory roles and mechanisms remain to be explored.
The RNA methylation also has crosstalk with histone modifications. To be specific, m6A peaks are enriched in the region of histone H3 lysine 36 trimethylation (H3K36me3) and are declined with the reduction of H3K36me3. Mechanistically, METTL14 recognizes and binds to H3K36me3, after which MTC interacts with RNA Polymerase II and further installs m6A to actively transcribed pre‐mRNA [143]. Histone acetyltransferase P300 (EP300)‐mediated histone H3 lysine 27 acetylation (H3K27ac) activates METTL3 transcription which stimulates m6A modification on HDGF mRNA and enhances its stability, and finally leads to tumor growth and liver metastasis in human gastric cancer [77]. Similarly, lysine demethylase 5c (KDM5C)‐induced H3K4me3 demethylation in the promoter of METTL14 attenuates METTL14 transcription, resulting in reduced m6A deposition on SOX4 and the upregulation of the tumor suppressor SOX4 in colorectal cancer [95].
The posttranslational modifications on m6A regulators have also been identified and were found to play crucial roles in controlling the activity of the m6A machinery and therefore, the epitranscriptome. In HCC, SIRT1 activated the SUMO E3 ligase RAN binding protein 2 (RANBP2) which mediated SUMOylation and degradation of FTO, and resulted in more m6A on GNAO1, an anti‐tumor molecule in HCC [69]. METTL3 could be activated by ATM‐mediated phosphorylation at serine 43 and localized to double‐strand break sites, and YTHDC1 was subsequently recruited due to METTL3‐induced m6A deposition. Interference with this METTL3‐m6A‐YTHDC1 axis enhanced the sensitivity of cancer cells to DNA damage‐based therapy [144].
To sum up, the complicated network connecting m6A modification and other genetic and epigenetic factors integrates comprehensive information from various sources and strengthens gene expression control more accurately.
4.2. Environmental exposure affects m6A methylation
In addition to internal factors, external exposure also has an influence on m6A methylation. Human carcinogens in different content elicit detrimental effects to human bodies in genotoxic or non‐genotoxic ways [145]. Evidence has shown that cigarette smoke causes oncogenic mutations and epigenetic changes [146, 147]. Tobacco smoking can alter miRNA encoding genes [148, 149, 150]. Genes with aberrant levels further participate in a myriad of pathological processes, including tumorigenesis and tumor progression [151]. Cigarette smoke condensate induced hypo‐methylation in METTL3 promoter caused METLL3 overexpression and subsequently more m6A modification which promoted maturation of miRNA‐25. The latter activated the AKT‐p70S6 kinase pathway and played an oncogenic role in pancreatic cancer [152].
Reduced global m6A level was observed in A549 lung epithelial cells in response to sodium arsenite and particulate matter, and change in m6A level was associated with the concentration of environmental toxicants [153]. In contrast, chronic exposure of human bronchial epithelial cells to sodium arsenite‐induced malignant phenotype with increased m6A modification which was synergistically regulated by m6A modulators [154]. Dynamic m6A incorporation was found in chemical carcinogen‐induced cellular transformation, in which the METTL3‐m6A‐CDCP1 (CUB domain‐containing protein 1) axis contributed a lot to cell proliferation and progression, consistent with the effect of chemical carcinogenesis [155].
As a well‐known oncogenic virus, Epstein–Barr virus (EBV) is the culprit of about 2% of all malignancies via regulating numerous host cell activities. Lang et al. [156] have observed the interplay between EBV and m6A decoration. EBV nuclear antigen 3C (EBNA3C), the viral‐encoded latent oncoprotein, was upregulated by METTL14‐mediated m6A modification, and could in turn activate METTL14 transcription and directly interact with METTL14 to promote its protein stability. Therefore, METTL14 appears to be an important factor in EBV‐induced oncogenesis. In addition, m6A modification plays a role in the lifecycle and infection of the hepatitis virus which predominantly contributes to chronic liver diseases and the tumorigenesis of HCC [157, 158]. It was found that Hepatitis B virus (HBV) pregenomic RNA (pgRNA) was m6A modified in the RRACH motif within the epsilon stem‐loop and bound by YTHDF2/3 proteins [157]. Blocking m6A methylation by either silencing METTL3 and METTL14 or mutating this adenosine base to cytosine affected the stability of pgRNA and suppressed reverse transcription. The infection of Hepatitis C virus (HCV), a single‐stranded RNA virus, was proven to be regulated by m6A modification as well [158]. The m6A machinery in host cells is present not only in the nucleus but also in the cytoplasm where they can modify the HCV RNA. Silencing of METTL3 and METTL14 in Huh7 hepatoma cells increased the production of infectious HCV particles and the percentage of HCV‐positive cells, while depletion of FTO inhibited HCV particle production and infection. Taken together, these studies indicate the important roles of m6A modification during the pathogenesis, development, and progression of virus‐related cancers, implicating that modulation of m6A modification could serve as prevention or therapeutic strategies in virus‐related cancers.
5. CLINICAL IMPLICATIONS OF m6A IN CANCERS
A growing body of research on m6A methylation reveals a new layer of epigenetic regulation in oncogenesis and provides implications for the use of m6A in innovative and effective diagnostic and therapeutic approaches.
5.1. Implications of m6A in cancer diagnosis and prognosis
Effective biomarkers, along with sensitive and specific detection methods, will greatly contribute to the early diagnosis of cancers, thus, improve the survival of patients. Recently, m6A methylation and its regulators have become emerging biomarkers for cancer diagnosis and prognosis [21, 159]. Owing to metabolic reversibility, high abundance and stability, methylated nucleosides could be accessible in biological fluids (e.g., serum and urine) or circulating cells [160, 161]. Huang et al. [161] developed a liquid chromatography‐electrospray ionization tandem mass spectrometric (LC‐ESI‐MS/MS) method to determine m6A level in single cells and found increased RNA m6A methylation in circulating tumor cells from the blood of lung cancer patients. Pei et al. [162] also detected elevated m6A level in the peripheral blood leukocyte from non‐small cell lung cancer patients by flow cytometry, indicating the potential use of m6A as a non‐invasive biomarker. Furthermore, m6A regulators, including METTL3 [77, 163‐169], WTAP [170, 171, 172, 173], FTO [174, 175, 176, 177], IGF2BPs [178, 179, 180, 181, 182, 183, 184], and YTHDFs [73, 185‐192], have been proven to associate with favorable or unfavorable prognosis in different types of cancers (detailed in Section 3). It should be noted that prognosis is not simply associated with the expression of a certain gene but a comprehensive signature of multiple m6A regulators in cancers, including lung cancer [163, 164], pancreatic cancer [166, 193], and HCC [194]. Despite that m6A and its regulators exhibit powerful potential as biomarkers, it is still challenging for clinical application due to the heterogeneity of m6A in patients and the lack of assays to detect site‐specific m6A from low‐input clinical samples. Future single‐cell sequencing techniques might provide powerful assistance in solving such problems.
5.2. m6A modification and chemosensitivity
Drug resistance is the major cause of therapeutic failure and recurrence in chemotherapy. Recent studies have indicated that m6A modification was associated with drug response and chemoresistance [21, 159]. Mutations of receptor tyrosine kinases, such as BCR‐ABL (breakpoint cluster region, BCR; tyrosine‐protein kinase, ABL), c‐kit proto‐oncogene (KIT) and fms like tyrosine kinase 3 (FLT3) frequently occur in leukemia and are effective therapeutic targets in the clinic [195, 196, 197]. The tolerance of tyrosine kinase inhibitors (TKIs), a big challenge in leukemia treatment, is mediated by m6A demethylation resulting from elevated FTO in leukemia cells [198]. Decreased ALKBH5 was found in gemcitabine‐treated patient‐derived xenograft (PDX) model and predicted poor clinical outcome in PDAC, while overexpression of ALKBH5 could sensitize PDAC to chemotherapy [87]. Moreover, METTL3‐induced m6A installation was found to contribute to oxaliplatin resistance in colon cancer [199], cisplatin resistance in NSCLC [120] and tamoxifen resistance in breast cancer [112]. Therefore, silencing of METTL3 could reverse drug resistance in the above scenarios [112, 120, 199], and could enhance the sensitivity of DNA damage‐based therapy in vivo and in vitro [144]. Collectively, robust evidence unveils the participation of m6A modulators in drug resistance, shedding light on the application of these regulators as predictive markers in chemotherapy or drug targets in combination with chemotherapy.
5.3. The m6A modification and cancer immunotherapy
Although immunotherapy has been considered as a promising treatment in defeating cancer, lacking durable effects in some groups of patients limits its efficacy [200, 201, 202]. Intriguingly, the absence of YTHDF1 in mice enhances antigen‐specific cluster of differentiation 8 (CD8)‐positive T‐cell anti‐tumor reaction due to promoted tumor antigen cross‐presentation in classical dendritic cells (cDCs) [203]. As a result, the therapeutic efficacy of programmed death‐ligand 1 (PD‐L1) checkpoint blockade is enhanced in YTHDF1‐deficient mice [203]. What's more, the decline of FTO also improves the low response of melanoma cells to interferon‐gamma and enhances the reaction to anti‐PD‐1 (programmed cell death protein 1, PD1) blockade in mice [204]. These studies suggest that YTHDF1 and FTO might be potential drug targets in combination with immunotherapy.
5.4. Targeting m6A and its regulators in cancer therapy
Given the benefits of targeting m6A methylation in cancer therapy, as discussed above, researchers never cease exploring effective inhibitors of m6A enzymes. The most well representative one is the development of small‐molecule agents targeting FTO. Initially, a natural compound named Rhein was found to bind to FTO catalytic domain and competitively inhibited the recognition of m6A substrate [205]. An ascorbic acid analog was then designed in 2014 to inhibit the 2‐oxoglutarate‐dependent hydroxylase activity of FTO and elevate m6A level [206]. Later, meclofenamic acid (MA) and an acylhydrazine compound, FTO inhibitor 12, have been identified to have inhibitory activity on FTO over ALKBH5 [207, 208]. By targeting FTO, R‐2HG and FB23‐2 exhibited promising inhibitory effects in the treatment of AML [52, 53]. More recently, two potent agents, CS1 and CS2, have been developed by high‐throughput screening from over 260,000 compounds and showed anti‐tumor effects in multiple cancers by suppressing the self‐renewal of cancer stem cells and immune evasion [54]. The inhibitors for m6A writers and readers are also of great interest to the researchers. By screening a library of 4000 analogs and derivatives of S‐adenosyl‐methionine (SAM), UZH1a has been found to be an effective METTL3 inhibitor and could modulate transcriptomic m6A signal in the MOLM13 leukemia cells; however, its in vivo effect still needs to be elucidated [209]. As a small molecule inhibitor, BTYNB was reported to disrupt the association between IGF2BP1 and target RNA, which resulted in the decrease of E2 factor (E2F)‐driven cell cycle transition and inhibition of tumor progression [210]. With more and more druggable m6A targets being proven with proof‐of‐concept evidence to combat cancers, the identification of specific inhibitors and the application of these inhibitors in the clinic, especially in combination with other therapies, are of great importance and in urgent need. Instead of altering the transcriptome‐wide m6A level, “m6A editing” is a CRISPR (clusters of regularly interspaced short palindromic repeats)‐CAS9 (CRISPR‐associated protein‐9)‐based method to mediate programmable RNA methylation or demethylation at a specific locus. To achieve site‐specific removal of m6A, Liu et al. [211] engineered m6A ‘erasers’ by fusing catalytic dead Cas9 (dCas9) with ALKBH5 or FTO, while Li et al. [212] chose the RNA‐targeting CRISPR‐Cas system, dCas13, to engineer ALKBH5. Recently, dcas13 fusions with truncated METTLE3 or modified METTL3‐METTL14 complex have been established to direct site‐specific m6A incorporation [213]. To date, these new techniques allow precise manipulation of a single methylation site, in the hope of evaluating the exact function of a single methylation site and targeting of single m6A for cancer treatment.
6. PERSPECTIVES AND CONCLUSION
Fast‐growing research in the RNA epigenetics field delineates a comprehensive picture about how m6A methylation is tightly controlled by its enzymes (writers and erasers) and works with reader proteins to participate in almost every step of RNA metabolism. Hypo‐ or hyper‐methylation might lead to aberrant gene expression, abnormal cellular function, and diseases, such as cancer. The direct links between m6A and various cancers not only provide an insight into the mechanism of tumorigenesis but also are valuable for guiding clinical applications in fighting against cancers. Considering the broad effects of m6A in strengthening the anti‐cancer effect of chemotherapy and immunotherapy, the combination of m6A‐targeting agents with traditional chemotherapeutic drugs or PD‐1/PD‐L1 inhibitors holds great therapeutic promise. However, there is still a debate whether targeting the total abundance/level of m6A methylation (i.e., targeting enzymes) or targeting gene‐/site‐specific m6A methylation is a better choice, which warrants more proof‐of‐concept studies. Overall, m6A modification is a rising star in the epigenetic field and holds therapeutic promise for a broad range of cancer.
7.
Abbreviations
RNA, ribonucleic acid; m6A, N6‐methyladenosine; DNA, deoxyribonucleic acid; mRNA, messenger RNA; FTO, fat mass and obesity‐associated protein; NGS, Next‐generation sequencing; G, guanosine; A, adenosine; C, cytidine; U, uridine; MTC, methyltransferase complex; METTL3, methyltransferase‐like 3; METTL14, methyltransferase‐like 14; WTAP, Willms tumor 1 associated protein; RBM15, RNA Binding Motif Protein 15; RBM15B, RNA Binding Motif Protein 15B; ZC3H13, Zinc Finger CCCH‐Type Containing 13; VIRMA, Vir like m6A methyltransferase associated; ALKBH5, alkB homolog 5; YTH, YT521‐B homology; IGF2BPs, insulin‐like growth factor 2 mRNA‐binding proteins; HNRNPs, heterogeneous nuclear ribonucleoproteins; YTHDC1, YTH domain‐containing protein 1; YTHDC2, YTH domain‐containing protein 2; YTHDF1, YTH domain‐containing family protein 1; YTHDF2, YTH domain‐containing family protein 2; YTHDF3, YTH domain‐containing family protein 3; hnRNPC, heterogeneous nuclear ribonucleoprotein C; hnRNPG, heterogeneous nuclear ribonucleoprotein G; hnRNPA2B1, heterogeneous nuclear ribonucleoproteins A2/B1; SRSF3, serine/arginine‐rich splicing factor 3; SRSF10, serine/arginine‐rich splicing factor; NXF1, nuclear RNA export factor 1; FMRP, Fragile X mental retardation protein; XPO1, Exportin 1; polyA, polyadenylated; ELAVL1, ELAV like RNA binding protein 1; PABPC1, poly(a) binding protein cytoplasmic 1; MATR3, Matrin 3; PRRC2A, Proline Rich Coiled‐Coil 2A; eIF4G, eukaryotic translation initiation factor 4G; eIF3, eukaryotic translation initiation factor 3; eIF3h, eukaryotic translation initiation factor 3h; 5’UTR, 5′ untranslated region; circRNAs, circular RNAs; carRNAs, chromosome‐associated regulatory RNAs; H3K9me2, histone H3 lysine 9 dimethylation; KDM3B, lysine demethylase 3B; AML, acute myeloid leukemia; HSPCs, hematopoietic stem and progenitor cells; LSCs, leukemic stem cells; ASB2, ankyrin repeat and socs box containing 2; RARA, retinoic acid receptor alpha; R‐2HG, R‐2‐hydroxyglutarate; IDH, isocitrate dehydrogenase; KDM4C, lysine demethylase 4C; AXL, tyrosine‐protein kinase receptor UFO; TACC3, transforming acidic coiled‐coil containing protein 3; CEBPZ, CCAAT enhancer binding protein zeta; BCL2, B‐cell lymphoma 2; PTEN, phosphatase and tensin homolog; SPI1, transcription factor PU.1; Tnfrsf2, tumor necrosis factor receptor superfamily member 2; HSC, hematopoietic stem cell; HCC, hepatocellular carcinoma; miRNA, microRNA; DGCR8, microprocessor complex subunit DGCR8; SOCS2, cytokine signaling 2; Snail1, snail family transcriptional repressor 1; lncRNAs, long non‐coding RNAs; HDGF, hepatoma‐derived growth factor; siRNA, small interfering RNA; ETS1, HuR‐mediated ETS Proto‐Oncogene 1; ID2, DNA‐binding protein inhibitor ID‐2; GATA3, GATA binding protein 3; LYPD1, LY6/PLAUR domain containing 1; SIRT1, Sirtuin 1; SUMO, small ubiquitin‐related modifier; GNAO1, G protein subunit alpha O1; HIF, hypoxia‐inducible factor; EGFR, epidermal growth factor receptor; EMT, epithelial‐to‐mesenchymal transition; SRF, serum response factor; ZMYM1, zinc finger MYM‐type containing 1; CtBP, C‐terminal‐binding protein; LSD1, lysine‐specific histone demethylase 1; CoREST, REST corepressor 1; PI3K, phosphoinositide 3 kinase; Akt, protein kinase B; KCNK15‐AS1, antisense RNA 1 of KCNK15; PDAC, pancreatic ductal adenocarcinoma; WIF‐1, Wnt inhibitory factor 1; PER1, period circadian regulator 1; ATM, A‐T mutated; CHK2, serine/threonine‐protein kinase; P53, tumor protein 53; CDC25C, cell division cycle 25C; CRC, colorectal cancer; SOX4, SRY‐box transcription factor 4; H3K4me3, histone H3 lysine 4 trimethylation; SOX2, SRY‐box transcription factor 2; YAP, Yes‐associated protein; GAS5, growth arrest specific 5; LINRIS, long intergenic noncoding RNA for IGF2BP2 stability; GBM, Glioblastoma; GSCs, glioma stem‐like cells; 3’UTR, 3’ untranslated region; SRSF, serine/arginine‐rich splicing factor; ADAM19, ADAM metallopeptidase domain 19; FOXM1, forkhead box M1; FOXM1‐AS, antisense RNA of FOXM1; VEGFA, vascular endothelial growth factor A; HBXIP, Hepatitis B X‐interacting protein; let‐7g, lethal‐7g; AK4, adenylate kinase 4; ROS, reactive oxygen species; BNIP3, BCL2 interacting protein 3; ST6GALNAC5, ST6 N‐acetylgalactosaminide alpha‐2,6‐sialyltransferase 5; GJA1, gap junction protein alpha 1; NSCLC, non‐small cell lung cancer; MALAT1, metastasis associated lung adenocarcinoma transcript 1; TAZ, tafazzin; MAPK2, mitogen‐activated protein kinase 2; DNMT3A, DNA methyltransferase 3A; BRD4, bromodomain‐containing protein 4; ATS2, large tumor suppressor kinase 2; DLBCL, Diffuse large B‐cell lymphoma; PEDF, pigment epithelium‐derived factor; piRNA, PIWI‐interacting RNA; HK2, hexokinase 2; ncRNAs, noncoding RNAs; RBRP, RNA‐binding regulatory peptide; HSF1, heat shock transcription factor 1; H3K36me3, histone H3 lysine 36 trimethylation; EP300; histone acethyltransferase P300; H3K27ac, histone H3 lysine 27 acetylation; KDM5C, lysine demethylase 5c; RANBP2, RAN binding protein 2; CDCP1, CUB domain containing protein 1; EBV, Epstein–Barr virus; EBNA3C, EBV nuclear antigen 3C; HBV, Hepatitis B virus; pgRNA, pregenomic RNA; HCV, Hepatitis C virus; LC‐ESI‐MS/MS, liquid chromatography electrospray ionization tandem mass spectrometric; BCR, breakpoint cluster region; ABL, tyrosine‐protein kinse; KIT, c‐kit proto‐oncogene; FLT3, fms like tyrosine kinase 3; TKIs, tyrosine kinase inhibitors; PDX, patient‐derived xenograft; CD8, cluster of differentiation 8; cDCs, classical dendritic cells; PD‐L1, programmed death‐ligand 1; PD1, programmed cell death protein 1; MA, meclofenamic acid; SAM, S‐adenosyl‐methionine; E2F, E2 factor; CRISPR, clusters of regularly interspaced short palindromic repeats; CAS9, CRISPR‐associated protein‐9; dCas9, catalytic dead Cas9;
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
SHD, HYW and HLH wrote the manuscript. SHD made the figures. SHD, YTW, YDL, HYW and HLH revised and approved the final manuscript.
ACKNOWLEDGMENTS
This work was supported in part by the grant 2021A1515010425 (H.H.) from Guangdong Basic and Applied Basic Research Foundation, and the National Key R&D Program of China 2020YFA0112403 (H.W.) from the Ministry of Science and Technology of the People's Republic of China.
Dong S, Wu Y, Liu Y, Weng H, Huang H. N6‐methyladenosine Steers RNA Metabolism and Regulation in Cancer. Cancer Commun. 2021;41:538–559. 10.1002/cac2.12161
Contributor Information
Hengyou Weng, Email: huanghl1@sysucc.org.cn.
Huilin Huang, Email: weng_hengyou@grmh-gdl.cn.
REFERENCES
- 1. Roundtree IA, Evans ME, Pan T and He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169(7):1187‐1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71(10):3971‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perry RP, Kelley DE. Existence of methylated messenger RNA in mouse L cells. Cell. 1974;1(1):37‐42. [Google Scholar]
- 4. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, Salmon‐Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature. 2012;485(7397):201‐6. [DOI] [PubMed] [Google Scholar]
- 6. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol. 2014;10(2):93‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63(2):306‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)‐adenosine methylation by the METTL3‐METTL14 complex. Nature. 2016;534(7608):575‐8. [DOI] [PubMed] [Google Scholar]
- 9. Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6‐methyladenosine methyltransferase. Cell Res. 2014;24(2):177‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8(1):284‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST‐mediated transcriptional repression. Nature. 2016;537(7620):369‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self‐Renewal. Mol Cell. 2018;69(6):1028‐1038.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA‐binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32(5‐6):415‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol. 2011;7(12):885‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, et al. Oxidative demethylation of 3‐methylthymine and 3‐methyluracil in single‐stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582(23‐24):3313‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616‐624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608‐624. [DOI] [PubMed] [Google Scholar]
- 21. Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non‐coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37(3):270‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou KI, Pan T. An additional class of m(6)A readers. Nat Cell Biol. 2018;20(3):230‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61(4):507‐519. [DOI] [PubMed] [Google Scholar]
- 24. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N(6)‐methyladenosine methylated mRNAs. Elife. 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)‐methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161(6):1388‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)‐methyladenosine‐modified RNA. Cell Res. 2017;27(3):315‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al.. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27(3):444‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al.. Recognition of RNA N(6)‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)‐methyladenosine‐dependent RNA structural switches regulate RNA‐protein interactions. Nature. 2015;518(7540):560‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN, et al. Regulation of Co‐transcriptional Pre‐mRNA Splicing by m(6)A through the Low‐Complexity Protein hnRNPG. Mol Cell. 2019;76(1):70‐81.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO‐dependent demethylation of N6‐methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A‐Dependent Nuclear RNA Processing Events. Cell. 2015;162(6):1299‐308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsu PJ, Shi H, Zhu AC, Lu Z, Miller N, Edens BM, et al. The RNA‐binding protein FMRP facilitates the nuclear export of N (6)‐methyladenosine‐containing mRNAs. J Biol Chem. 2019;294(52):19889‐19895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP Modulates Neural Differentiation through m(6)A‐Dependent mRNA Nuclear Export. Cell Rep. 2019;28(4):845‐854.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A‐containing RNA through direct recruitment of the CCR4‐NOT deadenylase complex. Nat Commun. 2016;7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)‐methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115‐1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, et al. Fragile X mental retardation protein modulates the stability of its m6A‐marked messenger RNA targets. Hum Mol Genet. 2018;27(22):3936‐3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29(1):23‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N(6)‐methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24(10):870‐878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62(3):335‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez‐Moya J, et al. mRNA circularization by METTL3‐eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561(7724):556‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coots RA, Liu XM, Mao Y, Dong L, Zhou J, Wan J, et al. m(6)A Facilitates eIF4F‐Independent mRNA Translation. Mol Cell. 2017;68(3):504‐514.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)‐methyladenosine. Cell Res. 2017;27(5):626‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, et al. N (6)‐methyladenosine of chromosome‐associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367(6477):580‐586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y, Xia L, Tan K, Ye X, Zuo Z, Li M, et al. N(6)‐Methyladenosine co‐transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. 2020;52(9):870‐877. [DOI] [PubMed] [Google Scholar]
- 47. Kontur C, Jeong M, Cifuentes D, Giraldez AJ. Ythdf m(6)A Readers Function Redundantly during Zebrafish Development. Cell Rep. 2020;33(13):108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zaccara S, Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A‐Modified mRNA. Cell. 2020;181(7):1582‐1595 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lasman L, Krupalnik V, Viukov S, Mor N, Aguilera‐Castrejon A, Schneir D, et al. Context‐dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev. 2020;34(19‐20):1373‐1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373(12):1136‐52. [DOI] [PubMed] [Google Scholar]
- 51. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)‐Methyladenosine RNA Demethylase. Cancer Cell. 2017;31(1):127‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R‐2HG Exhibits Anti‐tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172(1‐2):90‐105.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small‐Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35(4):677‐691.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38(1):79‐96.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang J, Li Y, Wang P, Han G, Zhang T, Chang J, et al. Leukemogenic Chromatin Alterations Promote AML Leukemia Stem Cells via a KDM4C‐ALKBH5‐AXL Signaling Axis. Cell Stem Cell. 2020;27(1):81‐97.e8. [DOI] [PubMed] [Google Scholar]
- 56. Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self‐Renewal in Acute Myeloid Leukemia. Cell Stem Cell. 2020;27(1):64‐80.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan‐Zambrano G, Robson SC, et al. Promoter‐bound METTL3 maintains myeloid leukaemia by m(6)A‐dependent translation control. Nature. 2017;552(7683):126‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)‐methyladenosine (m(6)A)‐forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369‐1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell. 2018;22(2):191‐205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25(1):137‐148.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. 68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 62. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) ‐methyladenosine‐dependent primary MicroRNA processing. Hepatology. 2017;65(2):529‐543. [DOI] [PubMed] [Google Scholar]
- 63. Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10(1):2065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64. Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6A‐mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A‐HuR‐dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cheng X, Li M, Rao X, Zhang W, Li X, Wang L, et al. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco Targets Ther. 2019;12:3421‐3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, et al. KIAA1429 contributes to liver cancer progression through N6‐methyladenosine‐dependent post‐transcriptional modification of GATA3. Mol Cancer. 2019;18(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R, et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A‐guided epigenetic inhibition of LYPD1. Mol Cancer. 2020;19(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu X, Liu J, Xiao W, Zeng Q, Bo H, Zhu Y, et al. SIRT1 Regulates N(6) ‐Methyladenosine RNA Modification in Hepatocarcinogenesis by Inducing RANBP2‐Dependent FTO SUMOylation. Hepatology. 2020. [DOI] [PubMed] [Google Scholar]
- 70. Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. MicroRNA‐145 Modulates N(6)‐Methyladenosine Levels by Targeting the 3'‐Untranslated mRNA Region of the N(6)‐Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017;292(9):3614‐3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252‐261. [DOI] [PubMed] [Google Scholar]
- 73. Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21(4):859‐868. [DOI] [PubMed] [Google Scholar]
- 74. Müller S, Glaß M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 promotes SRF‐dependent transcription in cancer in a m6A‐ and miRNA‐dependent manner. Nucleic Acids Res. 2019;47(1):375‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F, Gastric cancer. Lancet. 2020;396(10251):635‐648. [DOI] [PubMed] [Google Scholar]
- 76. Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3‐mediated N6‐methyladenosine modification is critical for epithelial‐mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3‐mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193‐1205. [DOI] [PubMed] [Google Scholar]
- 78. Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, et al. Impaired autophagic degradation of lncRNA ARHGAP5‐AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10(6):383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin S, Liu J, Jiang W, Wang P, Sun C, Wang X, et al. METTL3 Promotes the Proliferation and Mobility of Gastric Cancer Cells. Open Med (Wars). 2019;14:25‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. He H, Wu W, Sun Z, Chai L. MiR‐4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A‐caused stabilization of SEC62. Biochem Biophys Res Commun. 2019;517(4):581‐587. [DOI] [PubMed] [Google Scholar]
- 81. Liu T, Yang S, Sui J, Xu SY, Cheng YP, Shen B, et al. Dysregulated N6‐methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020;235(1):548‐562. [DOI] [PubMed] [Google Scholar]
- 82. Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K‐Akt signaling in gastric cancer. Cancer Med. 2019;8(10):4766‐4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75(3):379‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 85. Wang ZQ, Zhang F, Deng T, Zhang L, Feng F, Wang FH, et al. The efficacy and safety of modified FOLFIRINOX as first‐line chemotherapy for Chinese patients with metastatic pancreatic cancer. Cancer Commun. (Lond), 2019;39(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non‐Coding RNA KCNK15‐AS1 Methylation. Cell Physiol Biochem. 2018;48(2):838‐846. [DOI] [PubMed] [Google Scholar]
- 87. Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, et al. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF‐1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A‐YTHDF2‐dependent manner. Mol Cancer. 2020;19(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tang X, Liu S, Chen D, Zhao Z, Zhou J. The role of the fat mass and obesity‐associated protein in the proliferation of pancreatic cancer cells. Oncol Lett. 2019;17(2):2473‐2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, et al. The epitranscriptome m6A writer METTL3 promotes chemo‐ and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52(2):621‐629. [DOI] [PubMed] [Google Scholar]
- 91. Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, et al. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol Res Pract. 2019;215(11):152666. [DOI] [PubMed] [Google Scholar]
- 92. Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. YTH domain family 2 orchestrates epithelial‐mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16(23):2259‐2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6‐methyladenosine reader. Cell Death Differ. 2020;27(6):1782‐1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down‐regulating oncogenic long non‐coding RNA XIST. Mol Cancer. 2020;19(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14‐mediated N6‐methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, et al. METTL14 Suppresses CRC Progression via Regulating N6‐Methyladenosine‐Dependent Primary miR‐375 Processing. Mol Ther. 2020;28(2):599‐612. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR‐1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38(1):393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m(6)A‐IGF2BP2‐dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene c‐Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9(7):7476‐7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bai Y, Yang C, Wu R, Huang L, Song S, Li W, et al. YTHDF1 Regulates Tumorigenicity and Cancer Stem Cell‐Like Activity in Human Colorectal Carcinoma. Front Oncol. 2019;9:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164(3):550‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149(1):36‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3‐mediated m(6)A modification in glioma stem‐like cells maintenance and radioresistance. Oncogene. 2018;37(4):522‐533. [DOI] [PubMed] [Google Scholar]
- 106. Li F, Yi Y, Miao Y, Long W, Long T, Chen S, et al. N(6)‐Methyladenosine Modulates Nonsense‐Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019;79(22):5785‐5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA Methylation Regulates the Self‐Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18(11):2622‐2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem‐like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31(4):591‐606.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021;11(2):480‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019. 69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 111. Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al.. HBXIP‐elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let‐7g. Cancer Lett. 2018;415:11‐19. [DOI] [PubMed] [Google Scholar]
- 112. Liu X, Gonzalez G, Dai X, Miao W, Yuan J, Huang M, et al. Adenylate Kinase 4 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an m(6)A‐Based Epitranscriptomic Mechanism. Mol Ther. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, et al. Cross‐talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci Adv. 2018;4(10):eaar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF‐dependent and ALKBH5‐mediated m⁶A‐demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113(14):E2047‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fry NJ, Law BA, Ilkayeva OR, Carraway KR, Holley CL, Mansfield KD. N(6)‐methyladenosine contributes to cellular phenotype in a genetically‐defined model of breast cancer progression. Oncotarget. 2018;9(58):31231‐31243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, et al. Hypoxia‐inducible factors regulate pluripotency factor expression by ZNF217‐ and ALKBH5‐mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7(40):64527‐64542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6‐methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xu Y, Ye S, Zhang N, Zheng S, Liu H, Zhou K, et al. The FTO/miR‐181b‐3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond). 2020;40(10):484‐500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 Induces the Translation of m(6)A‐Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1‐miR‐1914‐3p‐YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121. Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, et al.. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512(3):479‐485. [DOI] [PubMed] [Google Scholar]
- 122. Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502(4):456‐464. [DOI] [PubMed] [Google Scholar]
- 123. Chao Y, Shang J, Ji W. ALKBH5‐m(6)A‐FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem Biophys Res Commun. 2020;521(2):499‐506. [DOI] [PubMed] [Google Scholar]
- 124. Guo J, Wu Y, Du J, Yang L, Chen W, Gong K, et al.. Deregulation of UBE2C‐mediated autophagy repression aggravates NSCLC progression. Oncogenesis. 2018;7(6):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al.. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs‐mediated YAP expression and inhibiting miR‐107/LATS2‐mediated YAP activity in NSCLC. Mol Cancer. 2020;19(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, et al. YTHDF1 links hypoxia adaptation and non‐small cell lung cancer progression. Nat Commun. 2019;10(1):4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6‐phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2020;41(5):541‐550. [DOI] [PubMed] [Google Scholar]
- 128. Miao Y, Medeiros LJ, Li Y, Li J, Young KH. Genetic alterations and their clinical implications in DLBCL. Nature Reviews Clinical Oncology. 2019;16(10):634‐652. [DOI] [PubMed] [Google Scholar]
- 129. Cheng Y, Fu Y, Wang Y, Wang J. The m6A Methyltransferase METTL3 Is Functionally Implicated in DLBCL Development by Regulating m6A Modification in PEDF. Front Genet. 2020;11:955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Han H, Fan G, Song S, Jiang Y, Qian C, Zhang W, et al. piRNA‐30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood. 2020. [DOI] [PubMed] [Google Scholar]
- 131. Bowtell DD, Böhm S, Ahmed AA, Aspuria PJ, Bast RC, Jr. , Beral V, et al. Rethinking ovarian cancer II: reducing mortality from high‐grade serous ovarian cancer. Nat Rev Cancer. 2015;15(11):668‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Brooks RA, Fleming GF, Lastra RR, Lee NK, Moroney JW, et al. Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin. 2019;69(4):258‐279. [DOI] [PubMed] [Google Scholar]
- 133. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR‐7 and BCL‐2. J Exp Clin Cancer Res. 2019;38(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151(2):356‐365. [DOI] [PubMed] [Google Scholar]
- 135. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(7):3816‐3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20(9):1074‐1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. m(6)A‐induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, et al. N6‐Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46(8):3906‐3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m(6)A(m) in the 5' cap controls mRNA stability. Nature. 2017;541(7637):371‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)‐methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zhu S, Wang JZ, Chen D, He YT, Meng N, Chen M, et al. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11(1):1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Song P, Feng L, Li J, Dai D, Zhu L, Wang C, et al. β‐catenin represses miR455‐3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer. 2020;19(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co‐transcriptionally. Nature. 2019;567(7748):414‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Zhang C, Chen L, Peng D, Jiang A, He Y, Zeng Y, et al. METTL3 and N6‐Methyladenosine Promote Homologous Recombination‐Mediated Repair of DSBs by Modulating DNA‐RNA Hybrid Accumulation. Mol Cell. 2020;79(3):425‐442.e7. [DOI] [PubMed] [Google Scholar]
- 145. Luch A. Nature and nurture ‐ lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5(2):113‐25. [DOI] [PubMed] [Google Scholar]
- 146. Clark SJ, Molloy PL. Smoke‐Induced Changes to the Epigenome Provide Fertile Ground for Oncogenic Mutation. Cancer Cell. 2017;32(3):278‐280. [DOI] [PubMed] [Google Scholar]
- 147. Vaz M, Hwang SY, Kagiampakis I, Phallen J, Patil A, O'Hagan HM, et al. Chronic Cigarette Smoke‐Induced Epigenomic Changes Precede Sensitization of Bronchial Epithelial Cells to Single‐Step Transformation by KRAS Mutations. Cancer Cell. 2017;32(3):360‐376.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Xi S, Inchauste S, Guo H, Shan J, Xiao Z, Xu H, et al. Cigarette smoke mediates epigenetic repression of miR‐217 during esophageal adenocarcinogenesis. Oncogene. 2015;34(44):5548‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Teschendorff AE, Yang Z, Wong A, Pipinikas CP, Jiao Y, Jones A, et al. Correlation of Smoking‐Associated DNA Methylation Changes in Buccal Cells With DNA Methylation Changes in Epithelial Cancer. JAMA Oncol. 2015;1(4):476‐85. [DOI] [PubMed] [Google Scholar]
- 150. Xi S, Xu H, Shan J, Tao Y, Hong JA, Inchauste S, et al. Cigarette smoke mediates epigenetic repression of miR‐487b during pulmonary carcinogenesis. J Clin Invest. 2013;123(3):1241‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lu J, Getz G, Miska EA, Alvarez‐Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834‐8. [DOI] [PubMed] [Google Scholar]
- 152. Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR‐25‐3p maturation via N(6)‐methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10(1):1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Cayir A, Barrow TM, Guo L, Byun HM. Exposure to environmental toxicants reduces global N6‐methyladenosine RNA methylation and alters expression of RNA methylation modulator genes. Environ Res. 2019;175:228‐234. [DOI] [PubMed] [Google Scholar]
- 154. Gu S, Sun D, Dai H, Zhang Z. N(6)‐methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite‐transformed cells. Toxicol Lett. 2018;292:1‐11. [DOI] [PubMed] [Google Scholar]
- 155. Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X, et al. Dynamic m(6)A mRNA methylation reveals the role of METTL3‐m(6)A‐CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38(24):4755‐4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Lang F, Singh RK, Pei Y, Zhang S, Sun K, Robertson ES. EBV epitranscriptome reprogramming by METTL14 is critical for viral‐associated tumorigenesis. PLoS Pathog. 2019;15(6):e1007796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, et al. N6‐methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci U S A. 2018;115(35):8829‐8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. N6‐Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe. 2016;20(5):654‐665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Huang H, Weng H, Deng X, Chen J. RNA Modifications in Cancer: Functions, Mechanisms, and Therapeutic Implications. Annual Review of Cancer Biology. 2020;4(1):221‐240. [Google Scholar]
- 160. Gehrke CW, Kuo KC, Waalkes TP, Borek E. Patterns of urinary excretion of modified nucleosides. Cancer Res. 1979;39(4):1150‐3. [PubMed] [Google Scholar]
- 161. Huang W, Qi CB, Lv SW, Xie M, Feng YQ, Huang WH, et al. Determination of DNA and RNA Methylation in Circulating Tumor Cells by Mass Spectrometry. Anal Chem. 2016;88(2):1378‐84. [DOI] [PubMed] [Google Scholar]
- 162. Pei Y, Lou X, Li K, Xu X, Guo Y, Xu D, et al. Peripheral Blood Leukocyte N6‐methyladenosine is a Noninvasive Biomarker for Non‐small‐cell Lung Carcinoma. Onco Targets Ther. 2020;13:11913‐11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Zhu J, Wang M, Hu D. Deciphering N(6)‐Methyladenosine‐Related Genes Signature to Predict Survival in Lung Adenocarcinoma. Biomed Res Int. 2020. 2020:2514230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of METTL3 as an Adverse Prognostic Biomarker in Hepatocellular Carcinoma. Dig Dis Sci. 2020. [DOI] [PubMed] [Google Scholar]
- 165. Qu N, Qin S, Zhang X, Bo X, Liu Z, Tan C, et al. Multiple m(6)A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer. 2020;20(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Hou J, Wang Z, Li H, Zhang H, Luo L. Gene Signature and Identification of Clinical Trait‐Related m(6) A Regulators in Pancreatic Cancer. Front Genet. 2020;11:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Jiang L, Zhang M, Wu J, Wang S, Yang X, Yi M, et al. Exploring diagnostic m6A regulators in endometriosis. Aging (Albany NY). 2020;12(24):25916‐25938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Li N, Zhan X. Identification of pathology‐specific regulators of m(6)A RNA modification to optimize lung cancer management in the context of predictive, preventive, and personalized medicine. Epma j. 2020;11(3):485‐504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, et al. N(6)‐methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer. 2019;18(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Li BQ, Huang S, Shao QQ, Sun J, Zhou L, You L, et al. WT1‐associated protein is a novel prognostic factor in pancreatic ductal adenocarcinoma. Oncol Lett. 2017;13(4):2531‐2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Xi Z, Xue Y, Zheng J, Liu X, Ma J, Liu Y. WTAP Expression Predicts Poor Prognosis in Malignant Glioma Patients. J Mol Neurosci. 2016;60(2):131‐6. [DOI] [PubMed] [Google Scholar]
- 172. Galardi S, Michienzi A, Ciafrè SA. Insights into the Regulatory Role of m(6)A Epitranscriptome in Glioblastoma. Int J Mol Sci. 2020;21(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zhuang J, Lin C, Ye J. m(6) A RNA methylation regulators contribute to malignant progression in rectal cancer. J Cell Physiol. 2020;235(9):6300‐6306. [DOI] [PubMed] [Google Scholar]
- 174. Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of Demethylase Genes, FTO and ALKBH1, Is Associated with Prognosis of Gastric Cancer. Dig Dis Sci. 2019;64(6):1503‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Guan K, Liu X, Li J, Ding Y, Li J, Cui G, et al. Expression Status And Prognostic Value Of M6A‐associated Genes in Gastric Cancer. J Cancer. 2020;11(10):3027‐3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wang JY, Chen LJ, Qiang P. The Potential Role of N6‐Methyladenosine (m6A) Demethylase Fat Mass and Obesity‐Associated Gene (FTO) in Human Cancers. Onco Targets Ther. 2020;13:12845‐12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38(4):2285‐2292. [DOI] [PubMed] [Google Scholar]
- 178. Yan X, Wan H, Hao X, Lan T, Li W, Xu L, et al. Importance of gene expression signatures in pancreatic cancer prognosis and the establishment of a prediction model. Cancer Manag Res. 2019;11:273‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Guo W, Huai Q, Wan H, Guo L, Song P, Gao S, et al. Prognostic Impact of IGF2BP3 Expression in Patients with Surgically Resected Lung Adenocarcinoma. DNA Cell Biol. 2021;40(2):316‐331. [DOI] [PubMed] [Google Scholar]
- 180. He X, Li W, Liang X, Zhu X, Zhang L, Huang Y, et al. IGF2BP2 Overexpression Indicates Poor Survival in Patients with Acute Myelocytic Leukemia. Cell Physiol Biochem. 2018;51(4):1945‐1956. [DOI] [PubMed] [Google Scholar]
- 181. Cao J, Mu Q, Huang H. The Roles of Insulin‐Like Growth Factor 2 mRNA‐Binding Protein 2 in Cancer and Cancer Stem Cells. Stem Cells Int. 2018. 2018:4217259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Dahlem C, Barghash A, Puchas P, Haybaeck J, Kessler SM. The Insulin‐Like Growth Factor 2 mRNA Binding Protein IMP2/IGF2BP2 is Overexpressed and Correlates with Poor Survival in Pancreatic Cancer. Int J Mol Sci. 2019. 20(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Wang X, Guan D, Wang D, Liu H, Wu Y, Gong W, et al. Genetic variants in m(6)A regulators are associated with gastric cancer risk. Arch Toxicol. 2021;95(3):1081‐1088. [DOI] [PubMed] [Google Scholar]
- 184. Lederer M, Bley N, Schleifer C, Hüttelmaier S. The role of the oncofetal IGF2 mRNA‐binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3‐12. [DOI] [PubMed] [Google Scholar]
- 185. Xu D, Shao J, Song H, Wang J. The YTH Domain Family of N6‐Methyladenosine “Readers” in the Diagnosis and Prognosis of Colonic Adenocarcinoma. Biomed Res Int. 2020. 2020:9502560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Liu M, Zhou S, Wang J, Zhang Q, Yang S, Feng J, et al. Identification of genes associated with survival of breast cancer patients. Breast Cancer. 2019;26(3):317‐325. [DOI] [PubMed] [Google Scholar]
- 187. Liu T, Yang S, Cheng YP, Kong XL, Du DD, Wang X, et al. The N6‐Methyladenosine (m6A) Methylation Gene YTHDF1 Reveals a Potential Diagnostic Role for Gastric Cancer. Cancer Manag Res. 2020;12:11953‐11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zhang B, Gu Y, Jiang G. Expression and Prognostic Characteristics of m(6) A RNA Methylation Regulators in Breast Cancer. Front Genet. 2020;11:604597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, et al. N6‐methyladenosine‐related Genomic Targets are Altered in Breast Cancer Tissue and Associated with Poor Survival. J Cancer. 2019;10(22):5447‐5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Shao XY, Dong J, Zhang H, Wu YS, Zheng L. Systematic Analyses of the Role of the Reader Protein of N (6)‐Methyladenosine RNA Methylation, YTH Domain Family 2, in Liver Hepatocellular Carcinoma. Front Mol Biosci. 2020;7:577460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Wang JY, Lu AQ. The biological function of m6A reader YTHDF2 and its role in human disease. Cancer Cell Int. 2021;21(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Liu S, Li G, Li Q, Zhang Q, Zhuo L, Chen X, et al. The roles and mechanisms of YTH domain‐containing proteins in cancer development and progression. Am J Cancer Res. 2020;10(4):1068‐1084. [PMC free article] [PubMed] [Google Scholar]
- 193. Meng Z, Yuan Q, Zhao J, Wang B, Li S, Offringa R, et al. The m(6)A‐Related mRNA Signature Predicts the Prognosis of Pancreatic Cancer Patients. Mol Ther Oncolytics. 2020;17:460‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Hu C, Yu M, Li C, Wang Y, Li X, Ulrich B, et al. miR‐550‐1 functions as a tumor suppressor in acute myeloid leukemia via the hippo signaling pathway. Int J Biol Sci. 2020;16(15):2853‐2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer. 2018;17(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Rossari F, Minutolo F, Orciuolo E. Past, present, and future of Bcr‐Abl inhibitors: from chemical development to clinical efficacy. J Hematol Oncol. 2018;11(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Larrosa‐Garcia M, Baer MR. FLT3 Inhibitors in Acute Myeloid Leukemia: Current Status and Future Directions. Mol Cancer Ther. 2017;16(6):991‐1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Yan F, Al‐Kali A, Zhang Z, Liu J, Pang J, Zhao N, et al. A dynamic N(6)‐methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018;28(11):1062‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zhang Y, Kang M, Zhang B, Meng F, Song J, Kaneko H, et al. m(6)A modification‐mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer. 2019;18(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 200. Yarchoan M, Johnson BA, 3rd , Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, et al. Personalized RNA mutanome vaccines mobilize poly‐specific therapeutic immunity against cancer. Nature. 2017;547(7662):222‐226. [DOI] [PubMed] [Google Scholar]
- 202. Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti‐tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(7743):270‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al.. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti‐PD‐1 blockade. Nat Commun. 2019;10(1):2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell‐active N6‐methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134(43):17963‐71. [DOI] [PubMed] [Google Scholar]
- 206. Zheng G, Cox T, Tribbey L, Wang GZ, Iacoban P, Booher ME, et al. Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem Neurosci. 2014;5(8):658‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43(1):373‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Toh JDW, Sun L, Lau LZM, Tan J, Low JJA, Tang CWQ, et al. A strategy based on nucleotide specificity leads to a subfamily‐selective and cell‐active inhibitor of N(6)‐methyladenosine demethylase FTO. Chem Sci. 2015;6(1):112‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Bedi RK, Huang D, Eberle SA, Wiedmer L, Sledz P, Caflisch A. Small‐Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem. 2020;15(9):744‐748. [DOI] [PubMed] [Google Scholar]
- 210. Müller S, Bley N, Busch B, Glaß M, Lederer M, Misiak C, et al. The oncofetal RNA‐binding protein IGF2BP1 is a druggable, post‐transcriptional super‐enhancer of E2F‐driven gene expression in cancer. Nucleic Acids Res. 2020;48(15):8576‐8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Liu XM, Zhou J, Mao Y, Ji Q, Qian SB. Programmable RNA N(6)‐methyladenosine editing by CRISPR‐Cas9 conjugates. Nat Chem Biol. 2019;15(9):865‐871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Li J, Chen Z, Chen F, Xie G, Ling Y, Peng Y, et al. Targeted mRNA demethylation using an engineered dCas13b‐ALKBH5 fusion protein. Nucleic Acids Res. 2020;48(10):5684‐5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Wilson C, Chen PJ, Miao Z, Liu DR. Programmable m(6)A modification of cellular RNAs with a Cas13‐directed methyltransferase. Nat Biotechnol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6‐methyladenosine methyltransferase‐like 3 promotes liver cancer progression through YTHDF2‐dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254‐2270. [DOI] [PubMed] [Google Scholar]
- 215. Li J, Chen F, Peng Y, Lv Z, Lin X, Chen Z, et al. N6‐Methyladenosine Regulates the Expression and Secretion of TGFβ1 to Affect the Epithelial‐Mesenchymal Transition of Cancer Cells. Cells. 2020;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Zhang J, Pi J, Liu Y, Yu J, Feng T. [Knockdown of YTH N(6)‐methyladenosine RNA binding protein 2 (YTHDF2) inhibits proliferation and promotes apoptosis in MGC‐803 gastric cancer cells]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2017;33(12):1628‐1634. [PubMed] [Google Scholar]
- 217. Wang H, Xu B, Shi J. N6‐methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl‐2. Gene. 2020;722:144076. [DOI] [PubMed] [Google Scholar]
- 218. Liu X, Gonzalez G, Dai X, Miao W, Yuan J, Huang M, et al. Adenylate Kinase 4 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an m(6)A‐Based Epitranscriptomic Mechanism. Mol Ther. 2020;28(12):2593‐2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6‐methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19(1):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


