Abstract
Purpose
Liver transplantation (LT) currently yields the best outcomes for hepatocellular carcinoma (HCC). However, tumor recurrence still occurs in some patients. Identifying markers that predict HCC recurrence after LT is an unmet medical need.
Methods
In this study, differential expression analysis was used to identify differentially expressed microRNAs (DEmiRs) between HCC and liver tissues in the The Cancer Genome Atlas database and in data from patients with recurrent or non-recurrent HCC in the GSE64989 dataset. The expression profiles of the overlap DEmiRs were used to construct an miRNA-based risk score to predict prognosis using Cox regression analysis. The target genes of the miRNAs of interest were predicted, and they were analyzed for functional enrichment. Furthermore, we used the miRNAs of interest to construct a competitive endogenous RNA (ceRNA) network of long non-coding RNAs (lncRNAs), miRs and mRNAs.
Results
Four up-regulated and three down-regulated miRNAs in HCC and recurrent HCC after LT were considered as candidate miRs. MiR-3200-3p and miR-3690 were selected to construct the miR-based risk score, which was found to be associated with poor overall survival and progression-free survival. Furthermore, it proved to be an independent prognostic factor after adjusting for other clinicopathological factors. The corresponding ceRNA networks of these two miRs that we constructed may help to understand their regulatory mechanisms in HCC.
Conclusion
We propose a risk score based on miR-3200-3p and miR-3690 that may be useful as a prognostic marker to predict HCC recurrence after LT. We generated a ceRNA network involving these miRNAs, which may help reveal their regulatory roles in HCC.
Keywords: hepatocellular carcinoma, liver transplantation, microRNA, recurrence, ceRNA network
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor in the world, causing approximately 700,000 deaths worldwide each year.1,2 The main pathogenic factors of HCC are obesity, type 2 diabetes mellitus (T2DM), non-alcoholic fatty liver, alcoholism, aflatoxin B1 and other genetic diseases.3,4 Although progress has been made in the treatment of liver cirrhosis, the incidence of liver cancer continues to rise.5 Liver transplantation (LT) can be an effective treatment for liver cirrhosis and cancer, and it is widely used to treat patients with early HCC.6 Although historically LT may not have been ideal for HCC, the treatment has advanced substantially.7 Studies show that the 5-year survival rate of patients who meet the Milan criteria and undergo LT can exceed 70%, with fewer than 15% suffering recurrence.8 HCC recurrence after LT typically leads to rapidly progressing disease, resulting in death. Thus, identifying markers that can predict HCC recurrence after LT is an unmet medical need.
It may be fruitful to search for such markers among microRNAs (miRs), which are short, non-coding RNAs consisting of 18–25 nucleotides that can bind to target mRNAs and suppress their translation or promote their degradation.9 Expression of miRs regulates a number of cellular processes that affect the hepatic international roughness index (IRI), including promotion of inflammation, cellular regeneration, and autophagy.10 By interacting with multiple target genes, miRs can influence many important biological processes, such as cell growth, tissue differentiation, cell proliferation, embryonic development and apoptosis.10 The dysregulation of miRs may be a common feature of HCC,11 and miRs are considered prognostic biomarks in HCC,15 as well as various other cancers.12–14 As potential biomarkers in various diseases, miRNAs have received special attention. Numerous studies have shown that miRs are extremely stable and easy to detect in various types of biological samples. Whether miR expression patterns are associated with HCC recurrence after LT is unclear.
In our present study, we aimed to define a risk score based on miR expression for assessing the likelihood of HCC recurrence after liver transplantation (RALT). Furthermore, we planned to explore the potential biological mechanisms of relevant miRs by constructing a potential competitive endogenous RNA (ceRNA) network in HCC recurrence.
Materials and Methods
Data Processing
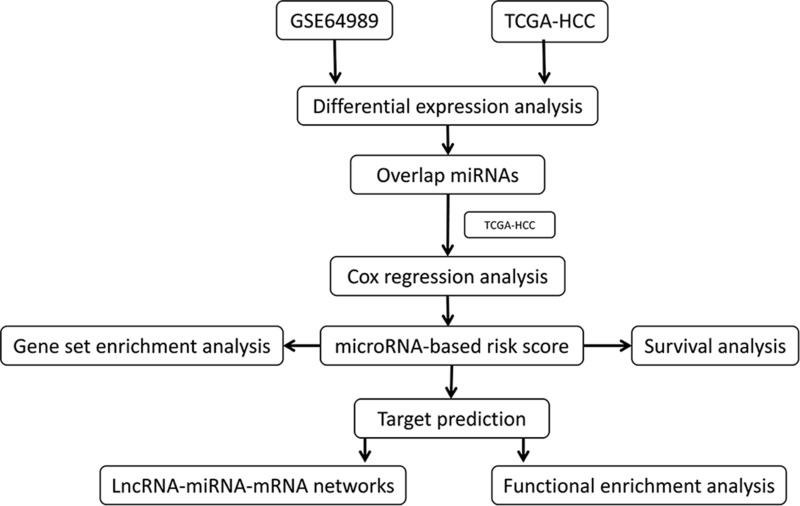
HCC mature miRs in the Cancer Genome Atlas (TCGA)16 were downloaded from the Xena server at the University of California at Santa Cruz (https://xenabrowser.net/datapages/, version 09–08-2017). The corresponding clinical information was also downloaded. The present study focused on HCC, so samples of mixed-type liver cancer were removed. In addition, the miRNA expression profile of GSE64989,17 based on the GPL16384 platform, was downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database, including HCC samples from 8 patients who experienced recurrence after LT and 10 patients who did not experience recurrence after LT. Sequences and symbol of mature miRs were obtained from miRbase version 22 (http://www.mirbase.org/). The miR symbols in GSE64989 were converted to be consistent with mature miR symbols in the mature miR data in the TCGA. The workflow of the present study is shown in Figure 1.
Figure 1.
Workflow of the present study.
Abbreviations: TCGA, The Cancer Genome Atlas; HCC, hepatocellular carcinoma; LncRNA, long non-coding RNA; miRNA, MicroRNA.
Screening of Differentially Expressed miRNAs
The limma package18 was used to screen for differentially expressed miRs (DEmiRs) in two comparison-pairs: HCC vs normal liver in the TCGA dataset, or HCC with recurrence after LT vs HCC without recurrence after LT in the GSE64989 dataset. The up- or down-regulated miRs overlapping between the two comparison-pairs were considered as RALT-related miRs, and the miRs with P < 0.05 were considered to be significant.
Univariate and Multivariate Cox Regression Analyses
Univariate and multivariate Cox regression analyses were performed based on the expression of the RALT-related miRs and overall survival (OS). The RALT-related miRs with P < 0.05 were considered significant in univariate Cox analysis and subjected to multivariate Cox analysis. The RALT-related miR-based risk score was created as:
Risk score = ExprmiR1 βmiR1 + ExprmiR2 βmiR2 + ExprmiR3 βmiR3 … …,
where β is the regression coefficient and Expr is the expression value of the miR. HCC patients were divided into low- or high-risk groups defined by the median risk score. The survival package19 was used to perform survival analysis of patients with high or low risk.
Target Prediction and Correlation Analysis
The targets of candidate miRs, including mRNAs and lncRNAs, were predicted using the miRNet web tool (https://www.mirnet.ca/).20 A ceRNA network of lncRNA-miRNA-mRNA was generated to satisfy the following two conditions:1 the lncRNA and mRNA were targeted by the same miR;2 the expression of lncRNA and mRNA positively correlated with each other. The lncRNA and mRNA targeted by the same miR were used to perform Pearson correlation analysis. P < 0.05 was considered significant. Cytoscape software21 was used to visualize the network.
Functional Enrichment Analysis
In order to further explore the biological processes and pathways involving candidate miRs, we used Metascape22 web tools to analyze gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment. P < 0.05 was considered statistically significant. Given that a miR may modulate the levels of hundreds of targets and affect various cellular processes, we explored the ultimate effects of the interactions of these cellular processes affected by a single miRNA using gene set enrichment analysis (GSEA).23,24 GSEA was performed using the JAVA program (http://www.broadinstitute.org/gsea) and the MSigDB c2.cp.kegg.v7.2.symbols.gmt gene set.
Statistical Analysis
All analyses were performed using R version 4.0.2 (https://www.r-project.org/). The unpaired t-test provided in the limma package was used to screen for DEmiRs. Kaplan-Meier survival analysis and the Log rank test were used to compare survival between low- and high-risk patients. All tests were two-tailed, and differences associated with P < 0.05 were considered to be statistically significant, unless otherwise stated.
Results
DEmiRs in HCC
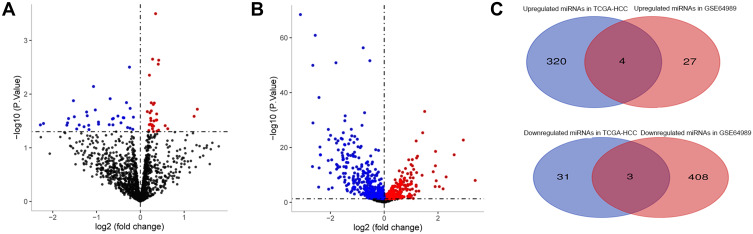
A total of 65 DEmiRs were identified in the comparison between HCC with recurrence and HCC without recurrence in the GSE64989 dataset, of which 31 were up-regulated and 34 were down-regulated (Figure 2A). A total of 735 DEmiRs were identified in the comparison between HCC and normal liver tissue in the TCGA dataset, of which 411 were down-regulated and 324 were up-regulated (Figure 2B). Seven miRs were identified as RALT-related: miR-3200-3p, miR-140-3p, miR-466, miR-3690, miR-335-5p, miR-382-5p, and miR-133b (Figure 2C).
Figure 2.
Differential expression analysis. (A) Volcano plot of differentially expressed microRNAs in the GSE64989 dataset. (B) Volcano plot of differentially expressed microRNAs in the TCGA hepatocellular carcinoma dataset. Red indicates up-regulated, blue indicates down-regulated, and gray indicates not significantly different. (C) Overlap of up-regulated miRNAs and down-regulated miRNAs in the two comparison-pairs (see Methods) were visualized on the Venn diagram. miRNA, microRNA.
The miR-Based Risk Score as a Prognostic Tool in HCC
Univariate and multivariate Cox regression analyses identified miR-3200-3p and miR-3690 as associated with poor OS (Table 1). Thus, an miR-based risk score was created as
Table 1.
Univariate and Multivariate Analyses of microRNAs Related to Hepatocellular Carcinoma Recurrence After Liver Transplantation
| microRNA | Univariate Cox Analysis | Multivariate Cox Analysis | ||||
|---|---|---|---|---|---|---|
| Beta | Hazard Ratio | P value | Beta | Hazard Ratio | P value | |
| hsa-miR-3200-3p | 0.198 | 1.218 | 0.002 | 0.172 | 1.187 | 0.011 |
| hsa-miR-140-3p | −0.029 | 0.971 | 0.862 | |||
| hsa-miR-466 | −0.049 | 0.953 | 0.604 | |||
| hsa-miR-3690 | 0.425 | 1.530 | 0.005 | 0.362 | 1.437 | 0.019 |
| hsa-miR-335-5p | 0.013 | 1.013 | 0.888 | |||
| hsa-miR-382-5p | 0.006 | 1.006 | 0.916 | |||
| hsa-miR-133b | 0.222 | 1.249 | 0.156 | |||
Risk score = ExprmiR-3200-3p* 0.172 + ExprmiR-3690* 0.362.
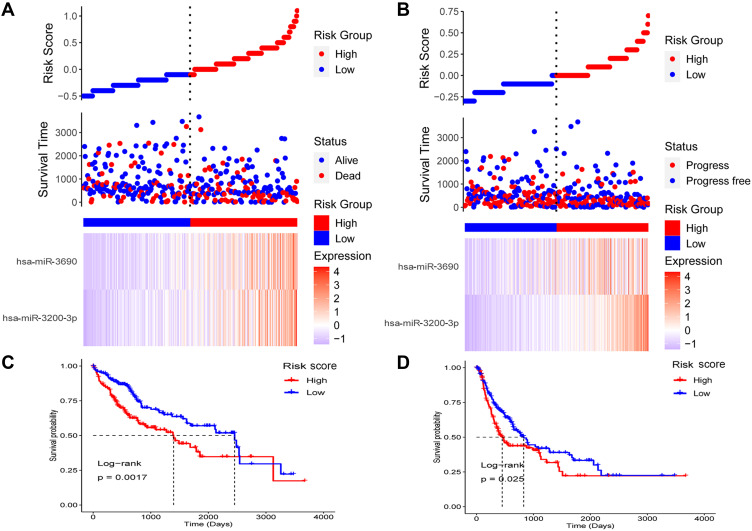
This risk score was associated with poor OS (HR = 2.718, P < 0.001) and poor progression-free survival (PFS) (HR = 1.720, P = 0.023). When patients with HCC were divided into high- or low-risk groups according to the median risk score (Figure 3A and B), the patients in the high-risk group had worse OS (Figure 3C) and PFS (Figure 3D). Furthermore, the miRNA-based risk score proved to be an independent prognostic factor, after adjusting for other clinicopathological characteristics (Table 2).
Figure 3.
The miRNA-based risk score as a prognostic biomarker in hepatocellular carcinoma (HCC). (A) Patients with HCC were divided into high- or low-risk groups. Their risk scores, survival, and expression of the two miRNAs are shown. (B) Patients with HCC were divided into high- or low-risk groups. Their risk scores, progression, and expression of the two miRNAs are shown. (C) Patients with HCC in the high-risk group showed worse overall survival. (D) Patients with HCC in the high-risk group showed worse progression-free survival.
Table 2.
Univariate and Multivariate Analyses of Clinicopathological Features and the microRNA-Based Risk Score
| Characteristic | Univariate Cox Analysis | Multivariate Cox Analysis | |||||
|---|---|---|---|---|---|---|---|
| Beta | HR | P value | Beta | HR | P value | ||
| Sex | Male | Ref. | |||||
| Female | 0.179 | 1.196 | 0.331 | ||||
| Age | < 65 yr | Ref. | |||||
| ≥ 65 yr | 0.189 | 1.209 | 0.289 | ||||
| T | T1-2 | Ref. | |||||
| T3-4 | 0.922 | 2.514 | <0.001 | 1.106 | 3.021 | 0.038 | |
| Tx/Not available | 0.432 | 1.541 | 0.668 | 0.258 | 1.295 | 0.811 | |
| N | N0 | Ref. | |||||
| N1 | 0.762 | 2.143 | 0.289 | 1.025 | 2.787 | 0.186 | |
| Nx/Not available | 0.434 | 1.543 | 0.023 | 0.360 | 1.433 | 0.211 | |
| M | M0 | Ref. | |||||
| M1 | 1.436 | 4.203 | 0.015 | 1.125 | 3.080 | 0.069 | |
| Mx/Not available | 0.435 | 1.545 | 0.026 | 0.267 | 1.306 | 0.363 | |
| AJCC stage | I–II | Ref. | |||||
| III–IV | 0.897 | 2.451 | <0.001 | −0.388 | 0.678 | 0.499 | |
| Not available | 0.925 | 2.523 | 0.002 | −0.170 | 0.844 | 0.691 | |
| Grade | G1-2 | Ref. | |||||
| G3-4 | 0.046 | 1.047 | 0.803 | ||||
| Not available | 0.462 | 1.586 | 0.434 | ||||
| AFP (ng/mL) | <20 | ||||||
| ≥20 | 0.551 | 1.734 | 0.014 | 0.278 | 1.321 | 0.241 | |
| Not available | 1.177 | 3.243 | <0.001 | 0.769 | 2.157 | 0.002 | |
| Vascular invasion | None | Ref. | |||||
| Macro | 0.714 | 2.042 | 0.060 | 0.138 | 1.148 | 0.734 | |
| Micro | 0.234 | 1.264 | 0.311 | 0.107 | 1.113 | 0.663 | |
| Not available | 0.993 | 2.699 | <0.001 | 0.525 | 1.690 | 0.032 | |
| Risk score | 1.000 | 2.718 | <0.001 | 1.039 | 2.827 | <0.001 | |
Abbreviations: HR, hazard rate; Ref., reference; AJCC, American Joint Committee on Cancer; AFP, alpha- fetoprotein.
Involvement of the Two RALT-Related miRs in Multiple Cancer-Related Biological Processes and Pathways
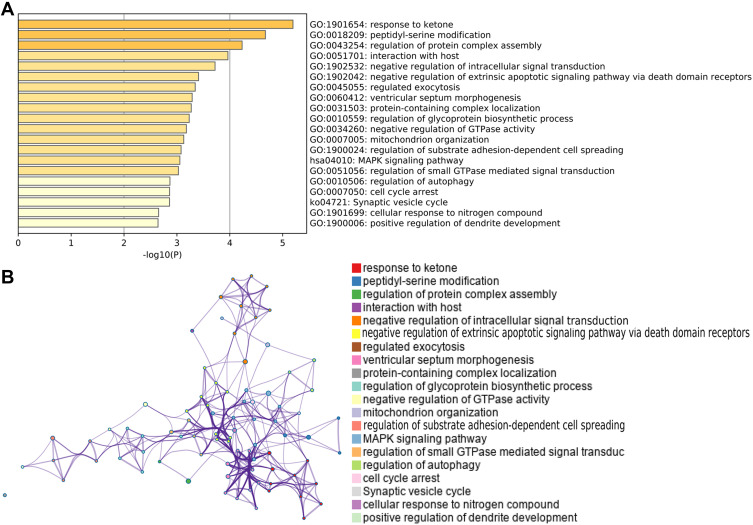
A total of 97 mRNAs and 51 lncRNAs were predicted to be targeted by the two miRs. Functional enrichment analysis of the mRNAs indicated that the two miRs may be involved in the response to ketone, peptidyl-serine modification, regulation of protein complex assembly, the MAPK signaling pathway, and the synaptic vesicle cycle (Figure 4A). The interactions of these GO terms are shown in Figure 4B.
Figure 4.
Functional enrichment analysis. (A) GO terms and KEGG pathways involving target genes of the miRNA-3200-3p and miRNA-3690. (B) Interactions among the GO terms and KEGG pathways. GO, Gene Ontology.
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
GSEA and ceRNA Network of lncRNA-miR-mRNA
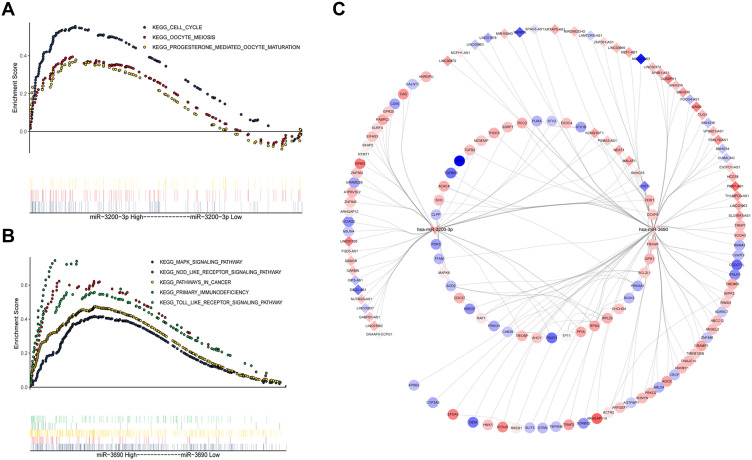
GSEA indicated that miR-3200-3p may be involved mainly in the cell cycle, oocyte meiosis and progesterone-mediated oocyte maturation (Figure 5A); while miR-3690 may be involved in NOD-like receptor signaling, MAPK signaling, Toll-like receptor signaling, primary immunodeficiency and pathways in cancer (Figure 5B). A ceRNA network of lncRNA-miR-mRNA was constructed according to established standards, and it comprised two miRs, 52 lncRNAs and 97 mRNAs (Figure 5C). This network may help us understand the pathways that regulate HCC.
Figure 5.
Results of gene set enrichment analysis and a competing endogenous RNA network of mRNA-miRNA-lncRNA. (A) Pathways affected by miRNA-3200-3p. (B) Pathways affected by hsa-miRNA-3690-3p. (C) The competing endogenous RNA network. Diamonds represent lncRNAs; V, miRNAs; and ellipses, mRNAs. Red and blue indicate up- or down-regulated, respectively, in hepatocellular carcinoma.
Discussion
Several previous studies reported miR-based signatures that may predict recurrence-free survival of HCC patients after LT. One such signature contained miR-19a, miR-223, miR-24, miR-886-5p, miR-247, and miR-126;25 another contained 67 miRs;26 and a third contained miR-214 and miR-3187, in addition to the Milan criteria.17 The heterogeneity of these signatures may reflect the clinical heterogeneity of HCC, as well as the small size of those studies. In the present work, we identified seven mature miRs that were up- or down-regulated not only in HCC in general, but also in HCC that recurred after LT. These seven miRs were defined as RALT-related miRs, and two of them (miR-3200-3p and miR-3690) emerged from Cox regression as suitable for defining a risk score. This score was associated with poor OS and PFS, suggesting that it may be a useful prognostic tool for risk stratification of HCC patients, and it may even predict tumor recurrence after LT.
The miR-3690 in our risk score may promote cell proliferation and cell cycle progression by altering DKK3 expression in human thyroid cancer,27 while miR-3200-3p has been identified as a target of NF-κB in TNFα- stimulated HeLa cells.28 Apart from these reports, we are unaware of other studies linking the two miRs in our risk score with cancer. Our analysis suggests that the targets of the two miRs may be involved in MAPK signaling and cell cycle arrest. Specifically, our GSEA indicated that miR-3200-3p may promote the cell cycle, while miR-3690 may promote the MAPK signaling pathway. Both the cell cycle and MAPK signaling play an important role in HCC and likely contribute to its recurrence. High expression of miR-3690 may also activate various HCC-related pathways, such as pathways in cancer and signaling pathways involving the NOD-like receptor29 and Toll-like receptor.30 Thus, these two miRs may be potential therapeutic targets in HCC.
Our ceRNA network suggests that miR-3200-3p and miR-3690 target six lncRNAs: KCNQ1OT1, NEAT1, SNHG15, MALAT1, PSMA3-AS1, and X1ST. KCNQ1OT1 is abnormally expressed in HCC, and it induces apoptosis. It can also inhibit metastasis and invasion by drug-resistant liver cancer cells.31 Up-regulation of NEAT1 may promote the development of liver cancer, making it a potential biomarker for the diagnosis and treatment of liver cancer.32 SNHG15 may be an independent predictor of poor prognosis in HCC.33 MALAT1, expressed in a wide range of cell types, may be useful on its own, or in combination with other markers, for predicting cancer outcome.34 PSMA3-AS1 has been linked to multiple myeloma,35 and it may promote esophageal cancer.36 We are unaware of reports linking PSMA3-AS1 to HCC progression. Similarly, we are unaware that X1ST has ever been linked to disease. In this way, the ceRNA network that we develop here may help elucidate how the two miRs in our risk score regulate HCC onset and progression.
Although the present study provides new insights into the relationships between miRs and the prognosis of HCC patients, it has some limitations. It is based on retrospective data, so the findings should be verified in prospective studies. The molecular mechanisms suggested in our bioinformatics analyses need to be tested in biological experiments.
Conclusion
We propose a risk score based on miR-3200-3p and miR-3690 as a prognostic marker in HCC, which may be useful for predicting HCC recurrence after LT. We also generated a ceRNA network that may help explain the regulatory pathways behind HCC.
Funding Statement
This work was supported by the Self-raised Scientific Research Funds of Medicine and Health of Guangxi Province (Grant NO.Z20211636).
Data Sharing Statement
The raw analyses from this study can be obtained from the corresponding author upon reasonable request.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Lurje I, Czigany Z, Bednarsch J, et al. Treatment strategies for hepatocellular carcinoma (-) a multidisciplinary approach. Int J Mol Sci. 2019;20(6):1465. doi: 10.3390/ijms20061465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadier B, Bulsei J, Nahon P, et al. Early detection and curative treatment of hepatocellular carcinoma: a cost-effectiveness analysis in France and in the United States. Hepatology. 2017;65(4):1237–1248. doi: 10.1002/hep.28961 [DOI] [PubMed] [Google Scholar]
- 3.Marengo A, Rosso C, Bugianesi E. Liver Cancer: connections with Obesity, Fatty Liver, and Cirrhosis. Annu Rev Med. 2016;67:103–117. doi: 10.1146/annurev-med-090514-013832 [DOI] [PubMed] [Google Scholar]
- 4.Gramantieri L, Fornari F, Callegari E, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12(6A):2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruix J, Sherman M; American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–1022. doi: 10.1002/hep.24199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutkowski P, Linecker M, DeOliveira ML, Mullhaupt B, Clavien PA. Challenges to liver transplantation and strategies to improve outcomes. Gastroenterology. 2015;148(2):307–323. doi: 10.1053/j.gastro.2014.08.045 [DOI] [PubMed] [Google Scholar]
- 7.Starzl TE, Marchioro TL, Vonkaulla KN, Hermann G, Brittain RS, Waddell WR. Homotransplantation of the Liver in Humans. Surg Gynecol Obstet. 1963;117:659–676. [PMC free article] [PubMed] [Google Scholar]
- 8.Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology. 2019;156(2):477–91 e1. doi: 10.1053/j.gastro.2018.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 10.Farid WR, Pan Q, van der Meer AJ, et al. Hepatocyte-derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl. 2012;18(3):290–297. doi: 10.1002/lt.22438 [DOI] [PubMed] [Google Scholar]
- 11.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47(6):1955–1963. doi: 10.1002/hep.22256 [DOI] [PubMed] [Google Scholar]
- 12.Gan Z, Zou Q, Lin Y, et al. Construction and validation of a seven-microRNA signature as a prognostic tool for lung squamous cell carcinoma. Cancer Manag Res. 2019;11:5701–5709. doi: 10.2147/CMAR.S191637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Lv Y, Liang R, et al. Four-miRNA signature as a prognostic tool for lung adenocarcinoma. Onco Targets Ther. 2018;11:29–36. doi: 10.2147/OTT.S155016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai X, Lu D, Lin Y, Lv Y, He L. A seven-miRNA expression-based prognostic signature and its corresponding potential competing endogenous RNA network in early pancreatic cancer. Exp Ther Med. 2019;18(3):1601–1608. doi: 10.3892/etm.2019.7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1A):A68–77. doi: 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liese J, Peveling-Oberhag J, Doering C, et al. A possible role of microRNAs as predictive markers for the recurrence of hepatocellular carcinoma after liver transplantation. Transpl Int. 2016;29(3):369–380. doi: 10.1111/tri.12733 [DOI] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barakat A, Mittal A, Ricketts D, Rogers BA. Understanding survival analysis: actuarial life tables and the Kaplan-Meier plot. Br J Hosp Med. 2019;80(11):642–646. doi: 10.12968/hmed.2019.80.11.642 [DOI] [PubMed] [Google Scholar]
- 20.Chang L, Zhou G, Soufan O, Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244–W51. doi: 10.1093/nar/gkaa467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 25.Han ZB, Zhong L, Teng MJ, et al. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol. 2012;6(4):445–457. doi: 10.1016/j.molonc.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barry CT, D’Souza M, McCall M, et al. Micro RNA expression profiles as adjunctive data to assess the risk of hepatocellular carcinoma recurrence after liver transplantation. Am J Transplant. 2012;12(2):428–437. [DOI] [PubMed] [Google Scholar]
- 27.Shen F, Gan XX, Deng XY, et al. MicroRNA-3690 promotes cell proliferation and cell cycle progression by altering DKK3 expression in human thyroid cancer. Oncol Lett. 2020;20(5):223. doi: 10.3892/ol.2020.12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou F, Wang W, Xing Y, Wang T, Xu X, Wang J. NF-kappaB target microRNAs and their target genes in TNFalpha-stimulated HeLa cells. Biochim Biophys Acta. 2014;1839(4):344–354. doi: 10.1016/j.bbagrm.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 29.Hu B, Ding GY, Fu PY, et al. NOD-like receptor X1 functions as a tumor suppressor by inhibiting epithelial-mesenchymal transition and inducing aging in hepatocellular carcinoma cells. J Hematol Oncol. 2018;11(1):28. doi: 10.1186/s13045-018-0573-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonnin M, Fares N, Testoni B, et al. Toll-like receptor 3 downregulation is an escape mechanism from apoptosis during hepatocarcinogenesis. J Hepatol. 2019;71(4):763–772. doi: 10.1016/j.jhep.2019.05.031 [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Zhao X, Ma X, Yuan Z, Hu M. KCNQ1OT1 contributes to sorafenib resistance and programmed deathligand1mediated immune escape via sponging miR506 in hepatocellular carcinoma cells. Int J Mol Med. 2020;46(5):1794–1804. doi: 10.3892/ijmm.2020.4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling ZA, Xiong DD, Meng RM, et al. LncRNA NEAT1 Promotes Deterioration of Hepatocellular Carcinoma Based on In Vitro Experiments, Data Mining, and RT-qPCR Analysis. Cell Physiol Biochem. 2018;48(2):540–555. doi: 10.1159/000491811 [DOI] [PubMed] [Google Scholar]
- 33.Zhang JH, Wei HW, Yang HG. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20(9):1720–1724. [PubMed] [Google Scholar]
- 34.Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–6768. doi: 10.2147/CMAR.S169406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu H, Han H, Song S, et al. Exosome-Transmitted PSMA3 and PSMA3-AS1 Promote Proteasome Inhibitor Resistance in Multiple Myeloma. Clin Cancer Res. 2019;25(6):1923–1935. doi: 10.1158/1078-0432.CCR-18-2363 [DOI] [PubMed] [Google Scholar]
- 36.Qiu BQ, Lin XH, Ye XD, et al. Long non-coding RNA PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by modulating the miR-101/EZH2 axis as a ceRNA. Aging. 2020;12(2):1843–1856. doi: 10.18632/aging.102716 [DOI] [PMC free article] [PubMed] [Google Scholar]