Abstract
BACKGROUD & AIMS:
Gastric cancer (GC) is the third leading cause of global cancer mortality. Adenosine-to-inosine RNA editing is a recently described novel epigenetic mechanism involving sequence alterations at the RNA but not DNA level, primarily mediated by ADAR (adenosine deaminase that act on RNA) enzymes. Emerging evidence suggests a role for RNA editing and ADARs in cancer, however, the relationship between RNA editing and GC development and progression remains unknown.
METHODS:
In this study, we leveraged on the next-generation sequencing transcriptomics to demarcate the GC RNA editing landscape and the role of ADARs in this deadly malignancy.
RESULTS:
Relative to normal gastric tissues, almost all GCs displayed a clear RNA misediting phenotype with ADAR1/2 dysregulation arising from the genomic gain and loss of the ADAR1 and ADAR2 gene in primary GCs, respectively. Clinically, patients with GCs exhibiting ADAR1/2 imbalance demonstrated extremely poor prognoses in multiple independent cohorts. Functionally, we demonstrate in vitro and in vivo that ADAR-mediated RNA misediting is closely associated with GC pathogenesis, with ADAR1 and ADAR2 playing reciprocal oncogenic and tumor suppressive roles through their catalytic deaminase domains, respectively. Using an exemplary target gene PODXL (podocalyxin-like), we demonstrate that the ADAR2-regulated recoding editing at codon 241 (His to Arg) confers a loss-of-function phenotype that neutralizes the tumorigenic ability of the unedited PODXL.
CONCLUSIONS:
Our study highlights a major role for RNA editing in GC disease and progression, an observation potentially missed by previous next-generation sequencing analyses of GC focused on DNA alterations alone. Our findings also suggest new GC therapeutic opportunities through ADAR1 enzymatic inhibition or the potential restoration of ADAR2 activity.
Keywords: Transcriptome, Editome, RNA Editing, ADARs
Gastric adenocarcinoma (gastric cancer [GC]) is a deadly malignancy highly prevalent in Asia and the third leading cause of global cancer mortality.1,2 A heterogeneous disease arising from the complex interplay of host factors and environmental risk agents, such as Helicobactepylori, GCs have been shown to exhibit a wide spectrum of molecular aberrations.3 At the DNA level, we and others have shown that GCs can exhibit distinct patterns of chromosomal amplifications and deletions involving oncogenes and tumor-suppressor genes (ERRB2, FGFR2, and RB1), gene fusions (eg CD44-SLC1A2, CLDN18-ARHGAP26), microsatellite instability, and somatic mutations in genes, such as TP53, ARID1A, and RhoA.3–10 Clinically, however, few of these molecular alterations have significantly impacted the treatment of GC patients to date, with the exceptions of traztuzumab treatment in ERBB2-positive GC and ramucirumab (an anti-angiogenic therapy) in advanced GC.11,12 As such, there remains a compelling need to identify novel molecular targets and oncogenic processes operative in GC, which might be exploited for therapy.
Besides DNA sequence alterations, epigenetic alterations are also emerging as major players in GC pathogenesis, involving DNA methylation, histone modifications (acetylation, methylation), and expression of GC-associated microRNAs or long non-coding RNAs.13–15 Pertinent to this study, RNA editing is another recently described epigenetic mechanism in which sequence alterations are introduced into the transcripts of expressed RNAs, while leaving the underlying DNA sequence intact and unmodified. The most frequent type of RNA editing in humans involves adenosine to inosine (A-to-I) editing, and is primarily mediated by ADAR (adenosine deaminase that act on RNA) enzymes. Molecularly, A-to-I editing has been shown to increase both transcript and proteome diversities, as inosine residues are recognized as guanosine by the general cellular machinery. Previous studies have shown that RNA editing can contribute to transcriptomic and phenotypic diversity, through protein recoding, alternative splicing, altered microRNA regulation, and changes in transcript localization, expression, and degradation.16,17 Genome-wide studies have also suggested that RNA editing is pervasive, with >85% of RNAs likely to be edited in noncoding and/or coding sequences.18,19
Recently, our group has demonstrated important roles for ADAR-mediated RNA editing in human cancer, including liver and esophageal malignancies.20–22 To date, however, the role of ADARs and the landscape of RNA edited targets (the “editome”) in GC remains unknown. We hypothesized that since prior sequence alteration studies of GC have largely focused on genetic variation at the DNA level,3,7 important oncogenic contributions from RNA editing (“RNA mutations”) might have been missed. In this study, we addressed this knowledge gap by leveraging high-throughput transcriptome sequencing (RNA-Seq) of primary GCs and cell lines to dissect the relationship between RNA editing and GC progression and prognosis. We found evidence of widespread RNA misediting and ADAR deregulation occurring in GCs relative to normal gastric tissues, and a significant correlation between ADAR deregulation and GC patient survival. Using an exemplary target gene PODXL (podocalyxin-like), we discovered an ADAR2-regulated recoding RNA editing event causing an amino acid substitution from histidine (His) to arginine (Arg) at codon 241 of PODXL, conferred a loss-of-function phenotype that neutralizes the tumorigenic ability of the unedited PODXL. These results highlight RNA editing as an important pathogenic mechanism in gastric carcinogenesis, and ADAR enzymes as potential GC therapeutic targets.
Materials and Methods
The detailed Materials and Methods can be found in the Supplementary Materials.
Targeted RNA Editing Analysis
Direct sequencing was performed on polymerase chain reaction (PCR) products, and the editing frequency was calculated using software ImageJ (http://rsb.info.nih.gov/ij/). The reliability of this method was further verified by cloning of individual sequences as described previously.20 PCR products were subcloned into the T-easy vector (Promega, Madison, WI), and approximately 50 individual plasmids were sequenced for each sample. For each sample, 2–3 independent reverse transcription PCR reactions were performed.
Statistical Analyses
Unless otherwise indicated, the data are presented as mean ± SD of 3 independent experiments. The SPSS statistical package for Windows (version 16; SPSS Inc, Chicago, IL) was used to perform the data analyses. The ADAR1 or ADAR2 expression levels in any 2 groups of samples were compared using the Mann-Whitney U test. The editing levels of editing sites between 2 preselected groups were compared using the Mann-Whitney U test. An unpaired 2-tailed Student t test was used to compare the number of invaded cells, foci, and tumor volume between any 2 preselected groups. A P value <.05 was considered to be statistically significant.
Results
Global Identification of Adenosine-to-Inosine/Guanosine (A-to-I or A-to-G) RNA Editing Sites in Gastric Cancer by RNA-Seq
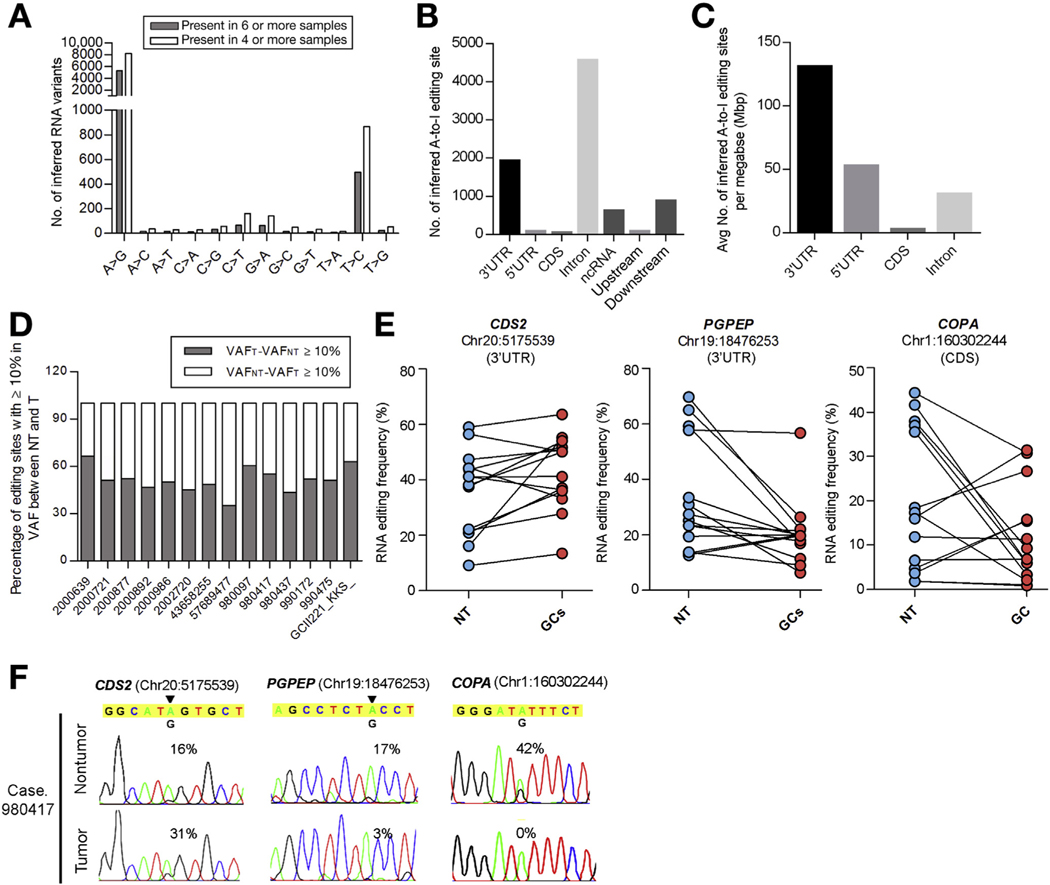
We performed high-throughput RNA-Seq of 14 matched pairs of gastric tumors and non-tumor (NT) gastric samples, generating a mean of 170.3-million reads that could be uniquely aligned to the human genome (hg19). The aligned reads provided substantial coverage (a mean of 68.1%) for the vast majority of human messenger RNA (mRNA) transcripts annotated in the UCSC genome browser (Supplementary Table 1). In order to perform a comprehensive high-quality analysis, we applied a 2-phase method for analyzing RNA editing sites in GC genomes. Relative to the number of RNA variants distributed in different genomic regions (coding sequences [CDS], untranslational regions [UTRs], introns, upstream/downstream, intergenic and pseudo/noncoding RNAs) and detected in one single sample, there was a drastic drop in those present in multiple samples (ranging from 2 to 10) (Supplementary Figure 1A). In particular, concomitant with a decrease in the number of RNA variants in CDS, a 5-fold increase in the proportion of A-to-G variants was observed, among all types of RNA variants present in ≥6 samples compared with those only detected in one sample (Supplementary Figure 1B). This may help to improve the lower technical validation rate of A-to-I (G) sites in CDS (<40%) than that in 3’ UTRs (90%–95%), as reported previously.20 In addition, approximately 96.2% (5076 of 5276) of A-to-I(G) RNA editing sites detected in ≥6 samples by our RNA-Seq were annotated in the Rigorously Annotated Database of A-to-I RNA Editing.23 To avoid false-positive results due to the DNA contamination, 4 matched pairs of tumors and NT samples (2000639, 2000721, 2000986, and 980417) were selected for whole genome sequencing. Approximately 96% of DNA variants have been sufficiently removed from the list of RNA variants that are present in ≥6 samples, suggesting the overall contamination from DNA variants was at an extremely low level and it was unlikely to affect the RNA editing analysis significantly (Supplementary Figure 1C). Using this reliable computational pipeline, we went on to check the distribution of 12 known types of inferred RNA variants, there was a significant enrichment of the A-to-G variation, consistent with previous studies reporting A-to-I editing as the most common type of RNA editing in humans18,24,25 (Figure 1A). We observed approximately 2-fold more editing sites in introns than those in 3’ UTRs, taking into account the genomic length of different regions, A-to-I editing was found to be most enriched in 3’ UTRs (Figure 1B and C), as reported previously.26–29
Figure 1.
RNA editome analysis of GC samples by RNA-Seq. (A) Distribution of 12 types of RNA editing sites in GC samples. Data represents a mean number of inferred RNA editing sites detected in the indicated number of samples. (B) Distribution of inferred A-to-I editing sites in each genomic region. (C) Distribution of A-to-I editing sites based on incidence per unit length (mega-base pair [Mbp]). (D) Percentage of A-to-I editing sites (present in ≥4 samples) demonstrating ≥10% increase or decrease in variation frequency (VAF) between gastric tumors (VAFT) and their matched non-tumor (VAFNT) gastric tissues. (E) Editing frequencies of 3 representative editing sites in 14 matched pairs of GCs and NT gastric tissues. (F) Sequence chromatograms of the indicated transcripts in a representative GC case (980417). An arrowhead indicates the editing position and value indicates the editing frequency of the corresponding editing site. Sequence chromatograms of the corresponding genomic DNA (gDNA) and complementary DNA (cDNA) sequences in GC cells are shown in Supplementary Figure 2.
Certain tumor types (brain, prostate, lung) have been reported to exhibit RNA editing deficiencies (hypoediting), while other tumor types (liver, esophageal squamous) are associated with RNA editing dysregulation (misediting).27,30,31 In GCs, we observed a clear misediting phenotype relative to normal tissues, as reflected by significant of both hypo- and hyperedited sites in tumors (Figure 1D). For example, in GC 990172, using a cut-off threshold of ≥10% variation frequency (VAF) between GCs and NT gastric tissues, approximately 52% and 48% of A-to-I edited sites demonstrated a decrease or increase (1690 and 1567 sites), respectively (Figure 1D). Figure 1 provides representative examples of editing sites in 3’ UTRs of 2 genes, CDP-diacylglycerol synthase 2; hyperedited, and pyroglutamyl-peptidase; hypoedited, and the CDS of a gene called COPA (coatomer protein complex, subunit α; hypoedited) occurring in the majority of GCs (Figure 1E and F, Supplementary Figure 2). Taken together, our results suggest that GC is neither a hypo- nor a hyperedited cancer, but instead a misedited cancer.
ADAR1 Enzyme Mediates A-to-I RNA Editing in Gastric Cancer
A-to-I RNA editing in humans is primarily mediated by ADAR enzymes. In vertebrates, 3 ADARs (ADAR1, ADAR2, and ADAR3) have been identified. ADAR1 and ADAR2 catalyze all currently known A-to-I editing sites, while ADAR3 is expressed specifically in the brain and has no documented deaminase activity. Notably, ADAR1 exists in 2 isoforms (p110 and p150), where the shorter p110 isoform is believed to act on host nuclear pre-RNAs, and the longer p150 may act primarily on exogenous viral RNAs16,25,32 and play a role in antiviral defense against viruses that replicate in the cytoplasm.33,34 For this study, we focused on the ADAR1 p110 isoform and ADAR2, which also acts on nuclear pre-mRNAs.
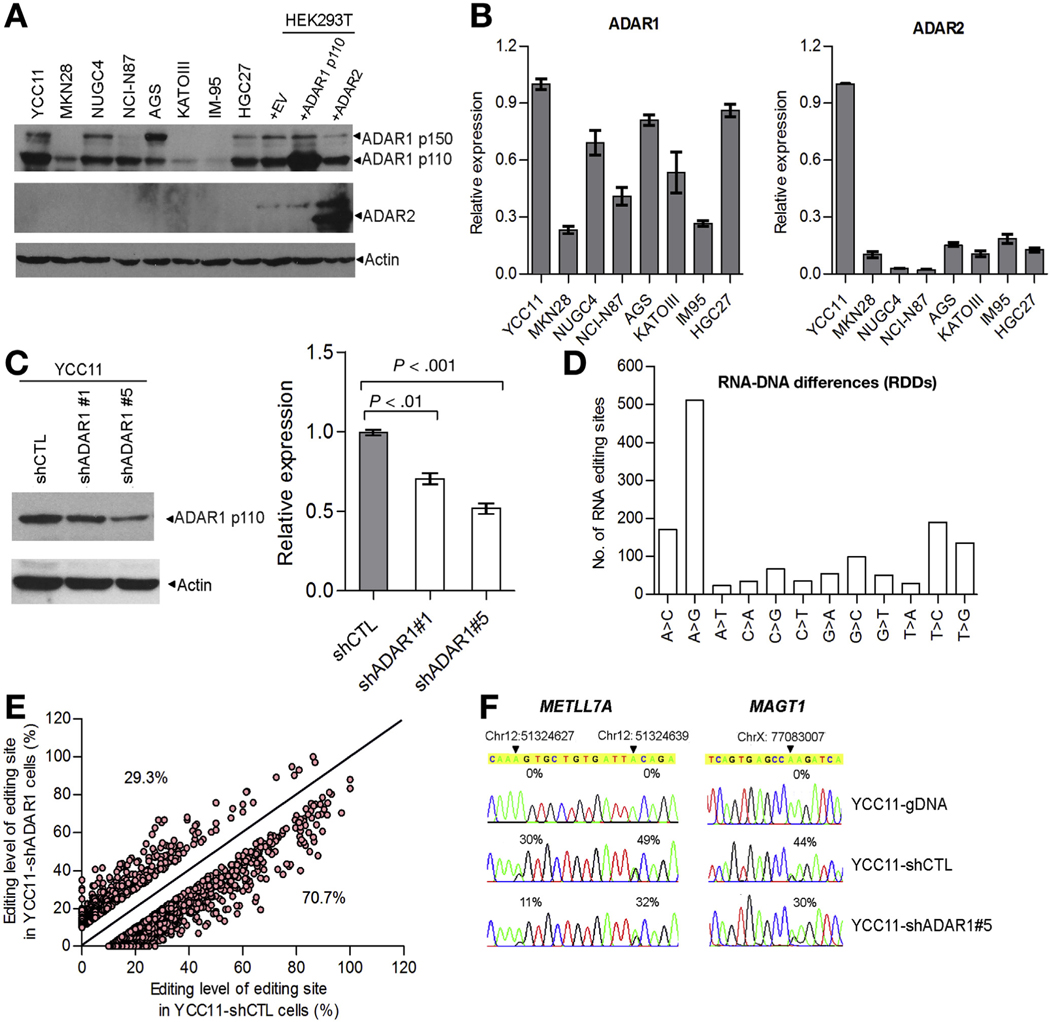
To determine whether ADAR deaminase activity might account for RNA editing alterations in GC, we functionally tested the effect of ADAR knockdown on A-to-I editing rates in GC cells. We determined expression levels of ADAR1 and ADAR2 among 8 GC cell lines (Figure 2A and B). ADAR1 was expressed at variable levels across all cell lines, with YCC11 cells displaying the highest endogenous ADAR1 expression. In contrast to ADAR1, there was a 4-cycle increase in the DCt value of ADAR2, as detected by quantitative real-time PCR (Supplementary Figure 3), indicating the endogenous level of ADAR2 in GC cell lines was approximately 16-fold lower than that of ADAR1. As such, while ADAR2 transcripts could be detected in the GC cells by quantitative real-time PCR, ADAR2 protein was undetectable in all GC lines (Figure 2A). This latter finding precluded assessment of ADAR2 knockdown on RNA editing rates.
Figure 2.
ADAR1-mediated A-to-I RNA editing in GC. (A) Western blot analysis of ADAR1 and ADAR2 among 8 GC cell lines. Antibody specificity was verified by overexpressing either ADAR1 p110 or ADAR2 expression construct in HEK293T cells. b-actin (Actin) was the loading control. (B) Quantitative real-time PCR (qPCR) measurement of ADAR1 (left panel) and ADAR2 (right panel) in the indicated cell lines. The relative expression of ADAR1/2 in the indicated cell line is shown in a bar chart (mean ± SD of 3 independent experiments). (C) ADAR1 p110 expression in YCC11 cells transiently transfected with 2 specific short hairpin RNAs (shRNAs) against ADAR1 gene (shADAR1 #1 and #5) or scrambled shRNA (shCTL) (left panel) was determined at protein and mRNA levels by Western blotting (left panel) and qPCR (right panel), respectively. (D) Mean number of different types of RNA–DNA differences (RDDs) in the indicated cells. (E) Scatter plots represent the editing frequencies of editing sites in YCC11-shCTL (x-axis) and YCC11-shADAR1#5 (y-axis). The editing sites below the diagonal line are those with the lower editing frequency in YCC11-shADAR1#5 cells compared to YCC11-shCTL. (F) Sequence chromatograms of METLL7A and magnesium transporter 1 transcripts and their matching genomic DNA (gDNA) sequences in the indicated cells. An arrowhead indicates the editing position and value indicates the editing frequency of the corresponding editing site.
We selected YCC11 cells for transient transfections with short-hairpin RNAs against ADAR1 (shADAR1#1 and #5) (Figure 2C), followed by exome-sequencing (Exome-Seq) and RNA-Seq analyses (Supplementary Table 2). For both control and knockdown cells, we compared exome-wide DNA and mRNA sequences distributed across 12 categories of RNA–DNA differences corresponding to candidate RNA editing sites. As expected, the majority of RNA–DNA differences were A-to-G sites (Figure 2D). Of these, ADAR1 knockdown caused approximately 71% of the A-to-I sites to exhibit decreased editing rates (Figure 2E). For example, we observed marked decreases in the editing frequencies of 3 A-to-I editing sites in 3’ UTRs of methyltransferase-like 7A and magnesium transporter 1 gene after ADAR1 knockdown (Figure 2F). These data suggest that there is a direct relationship between ADAR1 expression level and A-to-I editing rates in GC.
Genomic Gain/Loss Leads to the Differential Expression of ADAR1/2 in Primary Gastric Cancer and Their Clinical Correlates
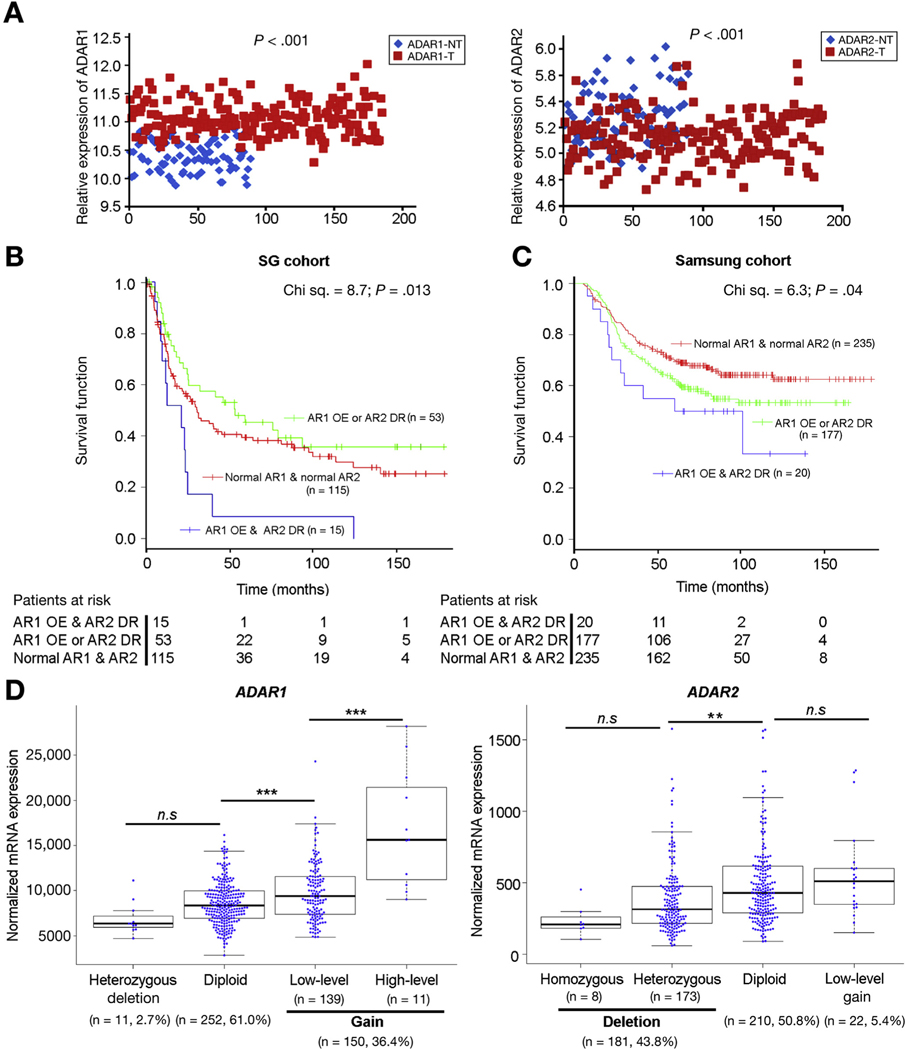
We sought to determine whether ADAR1 and ADAR2 might be deregulated in independent and larger cohorts of primary GCs. First, we analyzed ADAR expression levels in a GC microarray cohort of 185 gastric tumor and 89 NT gastric tissues (Singaporean [SG] cohort, 72 matched pairs of gastric tumor and NT gastric tissues).35 We found that ADAR1 was significantly overexpressed in primary GCs; whereas ADAR2 was significantly down-regulated (ADAR1: P < .001; ADAR2: P < .001, Mann-Whitney U test, Figure 3A). Intriguingly, patients with ADAR1 overexpression (OE) and ADAR2 down-regulation (DR), demonstrated the shortest overall survival (OS) times compared with patients who had either ADAR1 OE or ADAR2 DR (but not both), or to patients who had neither ADAR1 OE nor ADAR2 DR (P = .013, log-rank test; Figure 3B).
Figure 3.
Clinical implication of the differentially expressed ADARs in GCs. (A) Dot plots represent the relative ADAR1 (left panel) and ADAR2 (right panel) expression levels in a GC cohort (SG cohort) of 185 tumor and 89 NT gastric tissues (including 72 matched pairs), using the previously published Affymetrix Human Genome U133 Plus 2.0 Array data35 (Mann-Whitney U test). (B) Kaplan-Meier overall survival plot comparing patients demonstrating ADAR1 overexpression and ADAR2 DR (AR1 OE and AR2 DR) (blue line; n=15), AR1 OE or AR2 DR (green line; n = 53) and normal ADAR1 and ADAR2 expression in tumors (normal AR1 and AR2) (red line; n = 115; P = .013, log-rank test), in the SG cohort. (C) Kaplan-Meier overall survival plot comparing patients with AR1 OE and AR2 DR (blue line; n = 20), AR1 OE or AR2 DR (green line; n = 177) and normal ADAR1 and ADAR2 expression in tumors (normal AR1 and AR2) (red line; n = 235; P = .04, log-rank test), in the Samsung cohort.36 (D) Copy number variation and mRNA expression analyses of the ADAR1 and ADAR2 gene of TCGA GC datasets show the mRNA expression levels of ADAR1 and ADAR2 are correlated with the copy number changes in 413 GC patients. ns, no significance; **P < .01, *** P < .001, Mann-Whitney U test.
Second, to validate this clinical observation, we then analyzed an independent microarray cohort of 432 GCs profiled on a completely separate microarray platform. Once again, patients with ADAR1 OE and ADAR2 DR had the shortest OS times compared with patients who had ADAR1 OE or ADAR2 DR and patients who had neither ADAR1 OE nor ADAR2 DR (P = .04, log-rank test; Figure 3C; Samsung cohort).36 In univariate Cox analyses, statistically significant predictors for patient OS were sex, American Joint Committee on Cancer tumor staging, site of tumor, and the differentially expressed ADARs in tumors for the SG cohort (Supplementary Table 3). As for the Samsung cohort, Lauren’s classification, American Joint Committee on Cancer tumor staging and differentially expressed ADARs were statistically significant predictors of OS (Supplementary Table 4). In multivariate Cox analyses, only American Joint Committee on Cancer tumor staging could independently predict survival of GC patients in 2 cohorts (Supplementary Tables 3 and 4).
To identify the underlying mechanism for the differential expression of ADAR1 and ADAR2 in GC patients, our analyses of The Cancer Genome Atlas GC datasets (both single nucleotide polymorphism array and RNA-Seq data) demonstrated that the genomic gain of the ADAR1 gene and deletion of the ADAR2 gene observed in approximately 36.4% (n = 150) and 43.8% (n = 181) of 413 GC patients led to a significant increase in ADAR1 mRNA level and a significant decrease in ADAR2 mRNA expression in GCs, respectively (Figure 3D).
All these data suggested that, due to the genomic gain or loss, RNA editing enzymes ADAR1 and ADAR2 were differentially expressed in gastric tumors, and that ADAR1 overexpression concomitant with ADAR2 DR is associated with poor prognosis for GC patients.
RNA Editing Dysregulation Is Associated With Gastric Cancer Pathogenesis
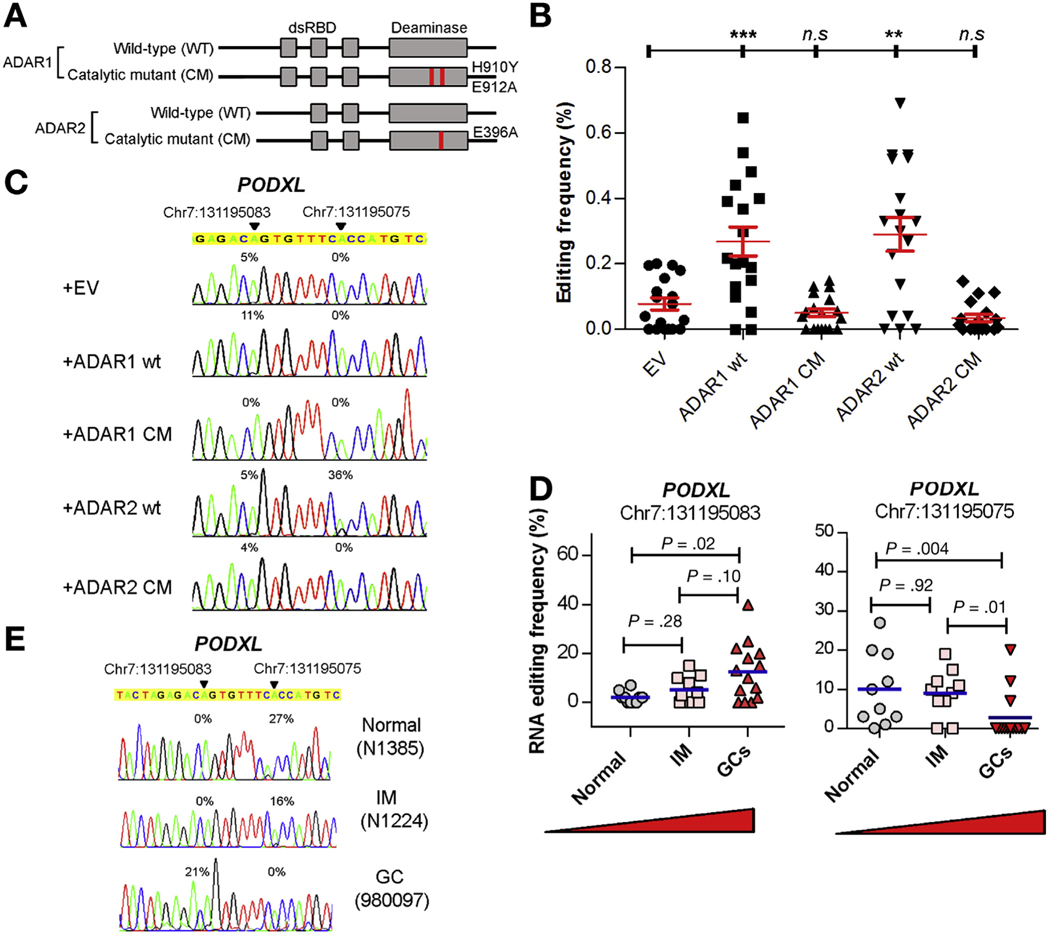
To identify RNA editing sites specifically regulated by ADAR1 or ADAR2, we leveraged on the wild-type (wt) ADAR expression constructs and constructed ADAR catalytic mutants (ADAR1 CM and ADAR2 CM) depleted of deaminase function by introducing specific point mutations (ADAR1: H901Y and E912A; ADAR2: E396A)37 (Figure 4A). The ADAR wt and mutated variants were subjected to RNA editing analysis in a GC cell line MKN28 and HEK293T (human embryonic kidney cells). Upon introduction of ADAR1/2 wt or CM mutant into cells, there was no significant difference in editing frequencies of 20 verified A-to-I editing sites between empty vector (EV) and ADAR1 or ADAR2 CM-transfected cells, indicating catalytic mutants are truly devoid of deaminase function (Figure 4B and Supplementary Figure 4A and B). Intriguingly, 2 editing sites (Chr7:131195083 and 131195075) in exon 3 of the PODXL gene were found to be specifically regulated by ADAR1 and ADAR2 protein, respectively (Figure 4C and Supplementary Figure 4B).
Figure 4.
RNA editing is dysregulated during the pathogenesis of GC. (A) Schematic diagram of catalytic mutants exhibiting point mutations in the deaminase domain. (B) Dot plots represent editing frequencies of 20 verified A-to-I RNA editing sites in MKN28 cells transiently transfected with the indicated expression constructs. ns, no significance; **P < .01, ***P < .001, Mann-Whitney U test. (C) Sequence chromatograms of the PODXL transcript in the indicated cell groups. (D) Dot plots represent editing levels of 2 editing sites within exon 3 of PODXL gene specifically regulated by either ADAR1 (left) or ADAR2 (right) in normal stomach (n = 10), 10 intestinal metaplasia (n = 10), and GC tissues (n = 14) (Mann-Whitney U test). (E) Sequence chromatograms of the PODXL transcript in the indicated samples. An arrowhead indicates the editing position and value indicates the editing frequency of the corresponding editing site (C, E).
In order to investigate whether the PODXL transcript could be edited by both ADAR1 and ADAR2, we overexpressed either individual ADAR1/2, or both ADARs in HEK293T cells, due to its high transfection efficiency than MKN28 cells. As shown in the Supplementary Figure 4C, there was no obvious difference in the expression levels of ADARs among different groups after the overexpression, and the PODXL transcripts can either be edited by ADAR1 or ADAR2 alone, but not regulated by both enzymes simultaneously (Supplementary Figure 4D).
We proceeded to measure the editing frequencies of these 2 ADAR1 or ADAR2-specific sites as readouts of overall RNA editing levels in 10 normal stomach samples, 10 gastric tissues with premalignant intestinal metaplasia, and 14 gastric tumors (the same batch as described previously). Intriguingly, the editing level of the ADAR1-specific editing site (Chr7:131195083) gradually increased during the pathogenesis of GC from normal stomach, premalignant stomach to clinically verified GC. Reciprocally, the editing frequency of the ADAR2-specific editing site gradually decreased during the progression to GC (Figure 4D and E). These data suggest that dysregulations in RNA editing caused by the differentially expressed ADARs are closely associated with the pathogenesis of GC.
ADAR1 and ADAR2 Have Oncogenic and Tumor Suppressive Roles, Respectively, Through Their Catalytic Deaminase Domains
Previous findings have suggested that ADAR enzymes may possess biologic activities beyond their deaminase domain, such as enhancement of microRNA processing and gene silencing via forming ADAR1/Dicer complex.38 To functionally test the role of the editing function of ADARs during gastric carcinogenesis, we introduced the wt and catalytic mutants of ADAR1 (p110) or ADAR2 into MKN28 cells expressing the relative low levels endogenous ADARs among 8 cell lines, using a lentiviral system (Figure 2A and B; Supplementary Figure 5A).
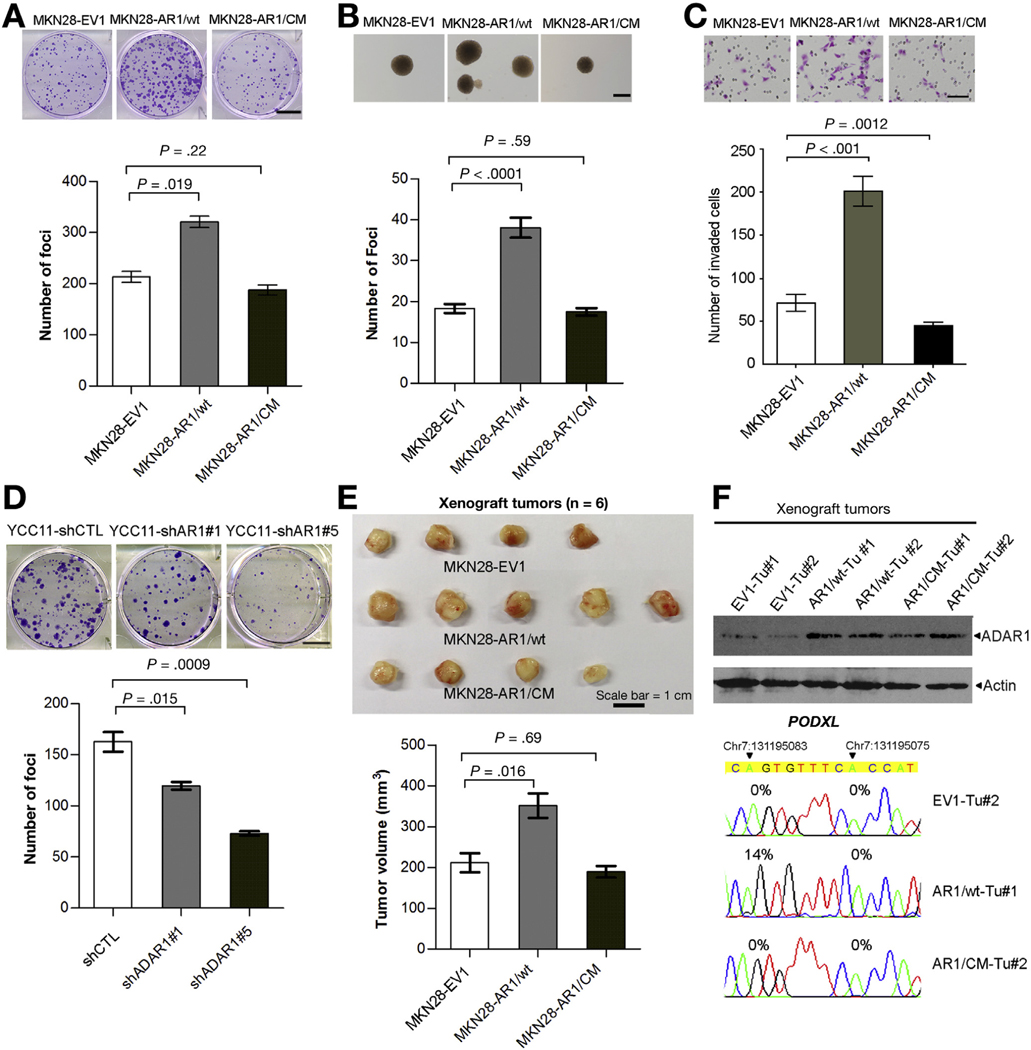
For ADAR1, we found that in in vitro tumorigenicity assays, cells transduced with wt ADAR1 lentivirus (MKN28AR1/wt) had higher frequencies of focus formation and colony formation in soft agar (anchorage-independent assay) than cells transduced with the control (MKN28-EV1) or ADAR1 catalytic mutant lentivirus (MKN28-AR1/CM) (Figure 5A and B). In addition, MKN28-AR1/wt cells had the increased invasive capabilities (Figure 5C). To ensure that these findings were not confined to a single GC line, we also silenced ADAR1 in YCC11 cells (Supplementary Figure 5B), and similarly observed a decrease in tumorigenicity, as detected by the foci formation assay (Figure 5D). Xenograft studies in mice further demonstrated that the volume of tumors induced by MKN28-AR1/wt cells was significantly larger than those induced by MKN28-EV1 or MKN28-AR1/CM cells at 12 weeks post injection (end point) (Figure 5E). Western blot analysis of ADAR1 confirmed ADAR1 overexpression in representative xenograft tumors (n = 2 of each group) at the end point, and we confirmed that the editing frequency of the ADAR1-specific site in exon 3 of the PODXL gene was much higher than that in tumors derived from MKN28-EV1 or MKN28-AR1/CM cells (Figure 5F).
Figure 5.
ADAR1 functions as an oncogene in GC through an RNA editing–dependent mechanism. (A) Quantification of foci formation induced by the indicated stable cell lines. Scale bar = 1 cm. (B) Quantification of colony formation in soft agar (anchorage-independent assay) induced by the indicated stable cell lines. Scale bar = 200 μm. (C) Quantification of invaded cells induced by the indicated stable cell lines. Scale bar = 100 μm. (D) Quantification of foci formation induced by the indicated stable cell lines. Scale bar = 1 cm. (A–D) All data are shown as the mean ± SD of triplicate wells with the same experiment and representative of 3 independent experiments, and statistical significance was determined by unpaired, 2-tailed Student’s t test. (E) Upper panel: tumors derived from the indicated stable cell lines 12 weeks after subcutaneous injection (n = 6 mice per group, scale bar = 1 cm); lower panel: volumes of tumors derived from the indicated cell lines at the end point. Data are presented as mean ± SD (unpaired 2-tailed Student’s t test). (F) Upper panel: Western blot analysis of ADAR1 protein in 6 representative xenograft tumors derived from the indicated stable cell lines. Lower panel: Sequence chromatograms of the PODXL transcript in the indicated tumor samples. An arrowhead indicates the editing position and value indicates the editing frequency of the corresponding editing site.
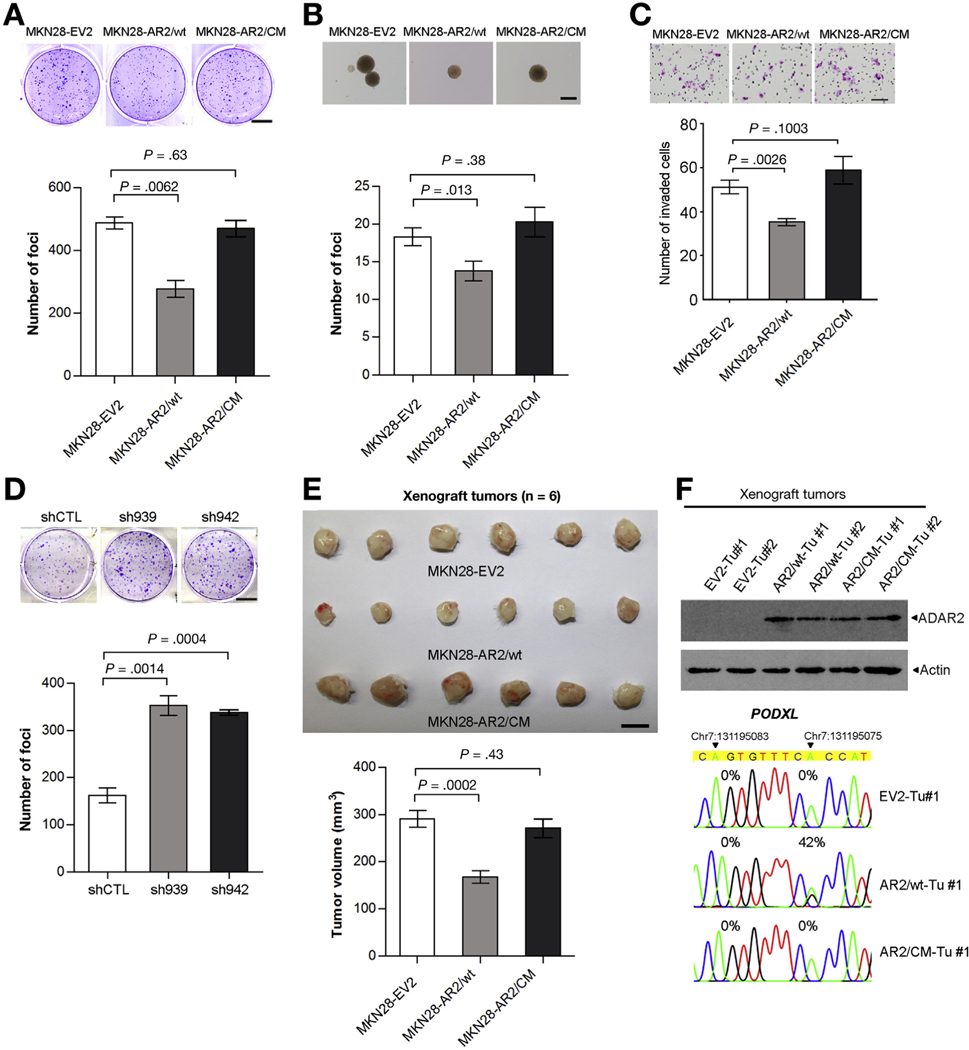
For ADAR2, the same strategy was utilized to investigate the role of ADAR2 in gastric carcinogenesis. MKN28 cells transduced with wt ADAR2 lentivirus (MKN28-AR2/wt) demonstrated markedly less tumorigenic abilities than cells transduced with the control (MKN28-EV2) or ADAR2 catalytic mutant lentivirus (MKN28-AR2/CM), as evidenced by foci formation and anchorage-independent assays (Figure 6A and B, Supplementary Figure 6A). In addition, MKN28-AR2/wt cells had the decreased invasive capabilities (Figure 6C). Silencing ADAR2 by introducing 2 specific short-hairpin RNAs (sh939 and sh942) in a GC cell line AGS, which has been stably transduced with wt ADAR2 lentivirus (AGS-AR2) dramatically increased the tumorigenicity as detected by the foci formation assay (Figure 6D and Supplementary Figure 6B). Similar results were also obtained in vivo using the xenograft tumor-formation assay (Figure 6E). At the molecular level, tumors induced by MKN28-AR2/wt cells displayed much higher editing levels of the ADAR2-specifc editing site within exon 3 of the PODXL gene than tumors derived from MKN28-EV2 or MKN28-AR2/CM cells (Figure 6F).
Figure 6.
ADAR2 functions as a tumor suppressor gene in GC, through an RNA editing–dependent mechanism. (A) Quantification of foci formation induced by the indicated stable cell lines. Scale bar = 1 cm. (B) Quantification of colony formation in soft agar (anchorage-independent assay) induced by the indicated stable cell lines. Scale bar = 200 μm. (C) Quantification of invaded cells induced by the indicated stable cell lines. Scale bar = 100 μm. (D) Quantification of foci formation induced by the indicated stable cell lines. Scale bar = 1 cm. (A–D) All data are shown as the mean ± SD of triplicate wells with the same experiment and representative of 3 independent experiments, and statistical significance was determined by unpaired, 2-tailed Student’s t test. (E) Upper panel: tumors derived from the indicated stable cell lines 16 weeks after subcutaneous injection (n = 6 mice per group, scale bar = 1 cm); lower panel: volumes of tumors derived from the indicated cell lines at the end point. Data are presented as mean ± SD (unpaired 2-tailed Student’s t test). (F) Upper panel: Western blot analysis of ADAR2 protein in 6 representative xenograft tumors derived from the indicated stable cell lines. Lower panel: Sequence chromatograms of the PODXL transcript in the indicated tumor samples. An arrowhead indicates the editing position and value indicates the editing frequency of the corresponding editing site.
In summary, these functional data demonstrate that ADAR1 and ADAR2 may have opposite effects on GC carcinogenesis—ADAR1 has anoncogenic ability, while ADAR2 functions asa tumor suppressor gene, and that these effects are mediated through their catalytic deaminase domains.
Functional Alternation of PODXL Transcript as a Result of the A-to-I RNA Editing During Gastric Cancer Progression
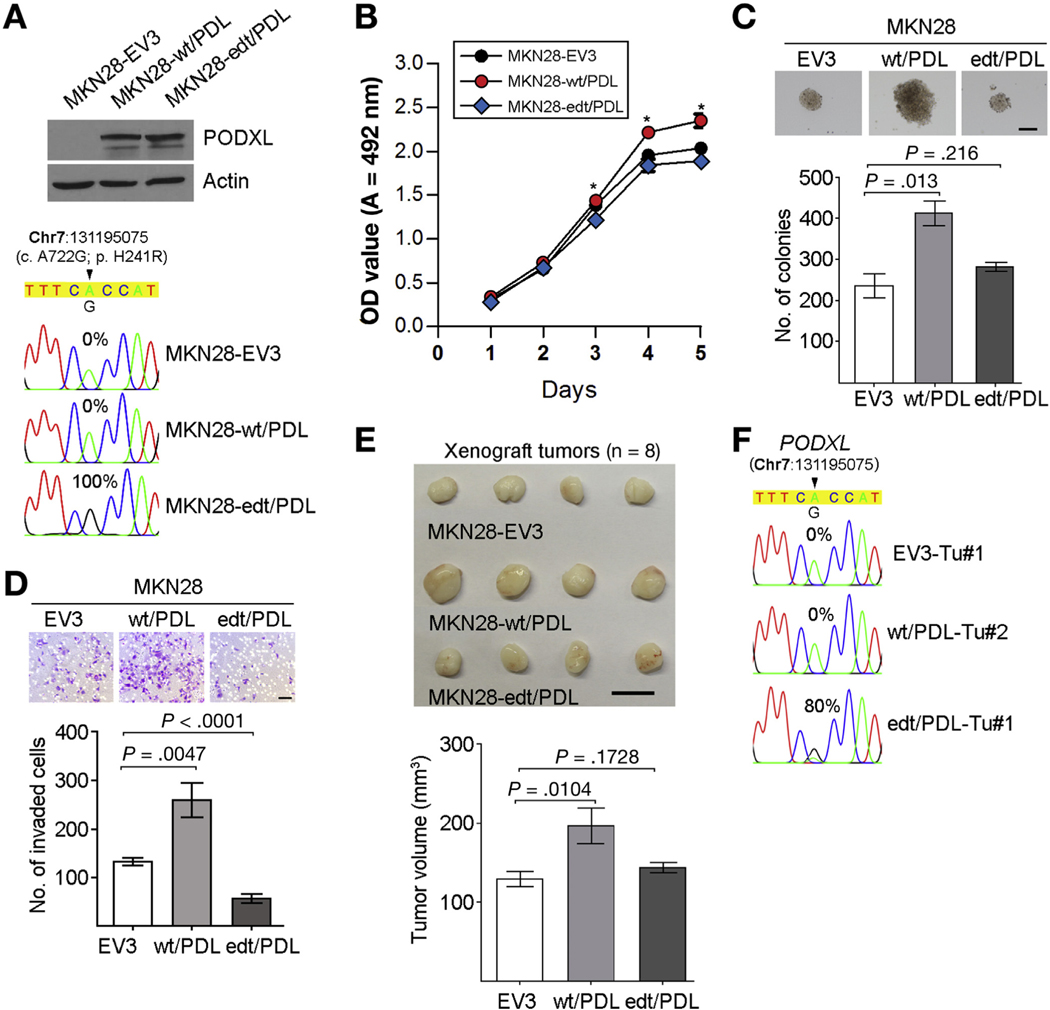
As mentioned, the PODXL gene encodes a protein that undergoes an amino acid substitution from His to Arg at codon 241, which is specifically regulated by ADAR2. We went on to study whether this recoding editing could cause its functional alteration. We introduced the wt (wt/PDL) or edited PODXL (edt/PDL) expression construct into MKN28 cells, which are devoid of endogenous edited transcripts and showing the lowest expression of PODXL among 8 GC cell lines (Supplementary Figure 7). MKN28 cells transduced with the wt/PDL lentivirus (MKN28-wt/PDL) with 100% of PODXL transcripts unedited had accelerated growth rates, higher frequencies of colony formation in soft agar, and increased invasive capability than cells transduced with the edt/PDL (MKN28-edt/PDL) or control lentivirus (MKN28EV3) (Figure 7A–D). Xenograft studies in mice demonstrated that the volume of tumors induced by MKN28-edt/PDL cells was significantly small than those induced by MKN28-wt/PDL cells at 12 weeks post injection (end point) (Figure 7E), as a consequence of the persistent expression of the edited PODXL (Figure 7F).
Figure 7.
Recoding editing of PODXL confers “loss-of-function” phenotype. (A) Upper panel: Western blot analysis of PODXL protein in the indicated cell lines. Lower panel: Sequence chromatograms of the PODXL transcript in the indicated cell lines. (B) Cell growth rates of the indicated cell lines were compared by XTT assays. Data are expressed as the mean ± SD of triplicate wells within the same experiment (*P < .05). Three independent assays were conducted. (C) Quantification of colony formation in soft agar induced by the indicated stable cell lines. Scale bar = 200 μm. (D) Quantification of invaded cells induced by the indicated stable cell lines. Scale bar = 100 μm. (C, D) All data are shown as the mean ± SD of triplicate wells with the same experiment and representative of 3 independent experiments, and statistical significance was determined by unpaired, 2-tailed Student’s t test. (E) Upper panel: tumors derived from the indicated stable cell lines 12 weeks after subcutaneous injection (n = 8 mice per group, scale bar = 1 μm); lower panel: volumes of tumors derived from the indicated cell lines at the end point. Data are presented as mean ± SD (unpaired two-tailed Student’s t test). (F) Sequence chromatograms of the PODXL transcript in the indicated tumor samples. (A, F) An arrowhead indicates the editing position and value indicates the editing frequency of the corresponding editing site. edt, edited.
All of these data indicate that the recoding editing of PODXL neutralizes the tumorigenic ability of the wt PODXL and confers “loss-of-function” phenotypes during GC progression, suggesting that PODXL is likely to be one of downstream editing targets that are responsible for the tumor-suppressive function of ADAR2.
Discussion
This study reports a comprehensive transcriptome-wide analysis of the GC RNA editing landscape, and a clinically significant relationship between ADAR-mediated RNA editing and GC pathogenesis and progression. Previous molecular studies of GC have focused primarily on characterizing aberrations at the DNA level, such as mutations, copy number amplifications, the corresponding editing site. edt, edited. and fusion genes.3–10 Alternatively, while GC gene expression studies using microarrays and RNA sequencing have been reported, no study to date has reported a systematic analysis of RNA edited sequence variation in primary GCs. Our data suggest that dysregulation of RNA editing is pervasive in GC, approaching levels reported for other types of epigenetic dysregulation including DNA methylation and histone modification. Our data support growing evidence that A-to-I RNA editing is an important contributor to cancer pathogenesis in several tumor types.21,22,39–46
Until recently, most of the scientific literature involving Ato-I RNA editing has been confined to studies in the central nervous system. For example, several ion channels and receptors, such as glutamate-gated channels, the voltage-gated Kv1.1 potassium channel, and the serotonin 5-HT2C receptor, have been reported to be RNA edited. In these cases, A-to-I editing was found to occur at precise locations within exons, changing key amino acids crucial for protein function.47 In recent years, however, the availability of next-generation sequencing-based RNA-Seq to demarcate entire transcriptomes has revealed a more global role for RNA editing in both normal development and diseases. Accumulating evidence now increasingly supports a role for ADAR-mediated A-to-I RNA editing in cancer development and progression. Interestingly, whether RNA editing is oncogenic or tumor suppressive appears to be influenced by tumor type. In brain tumors, all 3 ADAR RNA editing enzymes (ADAR1, 2, 3) are down-regulated, leading to a global reduction in A-to-I editing (hypo-editing or under-editing), which is functionally required for brain tumor development.27 In contrast, few recent studies utilizing the RNA-Seq data from The Cancer Genome Atlas (TGCA) project48 reported that thyroid carcinoma, head and neck squamous cell carcinoma, lung adenocarcinoma, and breast invasive carcinoma, demonstrated over-editing patterns (elevated editing levels) in tumors.44,45 As for GC, Han et al44 reported a total of 26,389 informative A-to-I editing sites in 285 stomach adenocarcinoma and 33 normal samples. However, <100 editing sites demonstrating differential editing levels (under-edited or over-edited) between matched tumors and normal samples were identified.44 In this study, we found 1203 A-toI editing sites distributed across 923 genes were recurrently and significantly under- or over-edited in GCs (data not shown), and observed a “misedited” phenotype arising through an imbalance of ADAR1 and ADAR2 activity, which was similar to hepatocellular carcinoma and esophageal squamous cell carcinoma as reported by us previously.20–22
Two findings in our study are of potential near-term clinical and translational significance. First, by comparing editing levels at different stages of gastric lesions along the normal to cancer continuum, we observed increasing levels of RNA editing dysregulation from normal gastric tissues, to premalignant intestinal metaplasia, and finally clinically verified GC samples. These results raise the possibility that measuring editing levels in patient gastric samples, particularly at the premalignant stage, may identify individuals at risk for subsequent GC development. Similar molecular “risk scores” have been proposed at the DNA methylation level.13 Second, due to the genomic gain of the ADAR1 gene located at chromosome 1q21 and the loss of the ADAR2 gene at chromosome 21q, we found that patients with developed GCs demonstrated the highest levels of ADAR imbalance, predicting the poorest clinical prognosis. Other than the ADAR1 copy number gain, the type I interferon response related to chronic inflammatory state has been reported to cause the increased editing levels in breast cancers.49 Altogether, measuring ADAR1 and ADAR2 levels in gastric tumors might facilitate the ability of clinicians to ascribe clinical prognosis to individual patients. Such patients with poor prognoses might also be targeted for enrollment into molecularly targeted clinical trials.
Our study also provides direct in vitro and in vivo evidence that ADAR1 and ADAR2 catalytic activity are functionally required for their oncogenic and tumor-suppressive roles, respectively. These results highlight ADAR1 as a potential therapeutic target in GC. For example, inhibition of ADAR1 might occur through genetic silencing via short-hairpin RNAs or CRISPR (clustered regularly interspaced short palindromic repeats) technologies.50 One potential concern for targeting ADARs is that this can cause widespread off-target effects, as ADARs have thousands of editing substrates. As an alternative, it is possible that one could specifically target key RNA editing events by reinstating a specific hyper-edited or hypo-edited transcript by introducing a specific RNA binding peptide51 or locked nucleotide acids52 into cells.
At present, it is unclear from our data whether the oncogenic or tumor-suppressive effects of ADAR1 and ADAR2, respectively, are due to a widespread deregulation of global editing, or due to the disruption of specific downstream genes. In hepatocellular carcinoma, a recent study described that increased editing of antizyme inhibitor 1 promoted tumor initiation by increasing both invasiveness and tumorigenic potential,22 while RNA edited rat sarcoma homologue family member Q was functionally relevant for invasiveness and recurrence in colorectal cancer.43 In this study, an ADAR2-induced recoding editing event (p.His241Arg) of PODXL, which was found to be associated with GC pathogenesis, neutralizes the tumorigenic capability of the wt PODXL, suggesting that PODXL is likely to be one of downstream editing targets that are responsible for the tumor-suppressive function of ADAR2, and the introduction of ADAR2-regulated sites, such as PODXL, could reverse the tumorigenic behavior of GC cells. Besides, we found that the PODXL transcripts can either be bound/edited by ADAR1 or ADAR2, but not regulated by both enzymes simultaneously. Intriguingly, after the overexpression of both enzymes, we observed an obvious decrease in the proportion of the PODXL transcripts having either ADAR1- or ADAR2-regulated site edited between cells transfected with ADAR1 or ADAR2 alone and both enzymes (ADAR1: 14% vs 8%; ADAR2: 28% vs 22%), suggesting a mutual interference of their specific editing activities at the PODXL transcript. One possible mechanism could be the ADAR1/ADAR2 heterodimerization might interfere with ADAR1 or ADAR2 specific editing activity, which has been reported by several studies.30,53 As the majority of GC cases showed the ADAR1 overexpression in tumors, it might further affect the specific editing activity of ADAR2 at the PODXL transcript via the formation of ADAR1/ADAR2 heterodimer, leading to more aggressive phenotypes. This is also consistent with our finding that GC patients demonstrating the most differentially expressed ADARs, as reflected by both ADAR1 overexpression and ADAR2 DR in tumors, had a poorer prognosis than those have either ADAR1 overexpression or ADAR2 DR. All these findings suggest the expression of RNA editing enzymes has to be precisely regulated, as a correct balance between ADAR1 and ADAR2 is critical for specific editing activity. Hence, the possibility of aberrant editing in cancer progression remains that the ADAR1/ADAR2 heterodimerization may contribute to the ADAR1 hyperactivity or editing defects caused by ADAR2.
In conclusion, GC is one of the most common and deadliest cancers worldwide, with few effective treatment options and is particularly prevalent in Asian countries.54 Identifying molecular aberrations in GC can improve our understanding of gastric carcinogenesis, identify strategies for subdividing patients into biologically and clinically relevant subgroups, and highlight novel therapeutic opportunities. Most mutational studies of GC have focused at the DNA level. Our study demonstrates that differentially expressed ADARs in gastric tumors with consequent A-to-I RNA editing dysregulation, may serve as a second layer of “somatic mutations” and a novel contributor to GC.
Supplementary Material
Acknowledgments
The authors thank Won Ki Kang, Sung Kim, and Hyun Cheol Cheong for contributing to clinical data and reagents associated with this project. We acknowledge the contributions of the Duke-NUS Genome Biology Facility for genomic profiling services.
Funding
This work was supported by the National Research Foundation Singapore, and the Singapore Ministry of Education under its Research Centres of Excellence initiative, NMRC Clinician Scientist-Individual Research Grant New Investigator Grant (CS-IRG NIG, grant number: NMRC/CNIG/1117/2014); NMRC Clinician Scientist-Individual Research Grant (CS-IRG, grant number: NMRC/CIRG/1412/2014); NUS Young Investigator Award (NUS YIA, grant number: NUSYIA_FY14_P22), NUS Start-up Fund (Ref number: NUHSRO/2015/095/SU/01); and core grants from Duke-NUS, GIS, and NMRC/TCR/009-NUHS/2013. The latter was supported through the National Medical Research Council/National Research Foundation Translational and Clinical Research (TCR) Flagship Program. This research is also supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s Tier 3 grants (MOE2014-T3–1-006).
Abbreviations used in this paper:
- ADAR
adenosine deaminase that act on RNA
- A-to-G
adenosine to guanosine
- A-to-I
adenosine to inosine
- CDS
coding sequences
- CM
catalytic mutant
- DR
down-regulation
- edt
edited
- EV
empty vector
- GC
gastric cancer
- mRNA
messenger RNA
- NT
non-tumor
- OE
overexpression
- OS
overall survival
- PCR
polymerase chain reaction
- PODXL
podocalyxin-like
- Ras
Rat sarcoma
- SG
Singaporean
- TCGA
The Cancer Genome Atlas
- UTR
untranslational region
- VAF
variation frequency
- wt
wild-type
Footnotes
Accession code: The RNA-Seq data were deposited in the following repository: Repository/DataBank Accession: European Genome-phenome Archive, accession no: EGAS00001001128. Databank URL: https://ega.crg.eu/.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2016.06.043.
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Hartgrink HH, Jansen EP, van Grieken NC, et al. Gastric cancer. Lancet 2009;374:477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 3.Tan P, Yeoh KG. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology 2015; 149:1153–1162.e3. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573–582. [DOI] [PubMed] [Google Scholar]
- 5.Kimura Y, Noguchi T, Kawahara K, et al. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Mod Pathol 2004; 17:1328–1337. [DOI] [PubMed] [Google Scholar]
- 6.Strickler JG, Zheng J, Shu Q, et al. p53 mutations and microsatellite instability in sporadic gastric cancer: when guardians fail. Cancer Res 1994;54:4750–4755. [PubMed] [Google Scholar]
- 7.McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 2014;11:664–674. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Lee SE, Kang SY, et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer 2013; 119:1627–1635. [DOI] [PubMed] [Google Scholar]
- 9.Tao J, Deng NT, Ramnarayanan K, et al. CD44-SLC1A2 gene fusions in gastric cancer. Sci Transl Med 2011; 3:77ra30. [DOI] [PubMed] [Google Scholar]
- 10.Yao F, Kausalya JP, Sia YY, et al. Recurrent fusion genes in gastric cancer: CLDN18-ARHGAP26 induces loss of epithelial integrity. Cell Rep 2015;12:272–285. [DOI] [PubMed] [Google Scholar]
- 11.Boku N. HER2-positive gastric cancer. Gastric Cancer 2014;17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs CS, Tomasek J, Yong CJ, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014; 383:31–39. [DOI] [PubMed] [Google Scholar]
- 13.Zouridis H, Deng N, Ivanova T, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med 2012;4:156ra140. [DOI] [PubMed] [Google Scholar]
- 14.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010;17:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SY, Kook MC, Kim YW, et al. CpG island hyper-methylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch 2010;457:415–422. [DOI] [PubMed] [Google Scholar]
- 16.Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet 2010;26:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi L, Chan TH, Tenen DG, et al. RNA editome imbalance in hepatocellular carcinoma. Cancer Res 2014; 74:1301–1306. [DOI] [PubMed] [Google Scholar]
- 18.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2004;2:e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol 2006;7:919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan TH, Lin CH, Qi L, et al. A disrupted RNA editing balance mediated by ADARs (adenosine deaminases that act on RNA) in human hepatocellular carcinoma. Gut 2014;63:832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin YR, Qiao JJ, Chan TH, et al. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res 2014;74:840–851. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Li Y, Lin CH, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 2013;19:209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res 2014; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li JB, Levanon EY, Yoon JK, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 2009;324:1210–1213. [DOI] [PubMed] [Google Scholar]
- 25.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 2002;71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Z, Cheng Y, Tan BC, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 2012;30:253–260. [DOI] [PubMed] [Google Scholar]
- 27.Paz N, Levanon EY, Amariglio N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 2007;17:1586–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramaswami G, Lin W, Piskol R, et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods 2012;9:579–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picardi E, Manzari C, Mastropasqua F, et al. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci Rep 2015;5:14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cenci C, Barzotti R, Galeano F, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem 2008;283:7251–7260. [DOI] [PubMed] [Google Scholar]
- 31.Maas S, Patt S, Schrey M, et al. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A 2001;98:14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev 2001;14:778–809; table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato S, Wong SK, Lazinski DW. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J Virol 2001;75:8547–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong SK, Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci U S A 2002;99:15118–15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y, Grabsch H, Ivanova T, et al. Comprehensive genomic meta-analysis identifies intra-tumoural stroma as a predictor of survival in patients with gastric cancer. Gut 2013;62:1100–1111. [DOI] [PubMed] [Google Scholar]
- 36.Lee J, Sohn I, Do IG, et al. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One 2014;9:e90133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai F, Drakas R, Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem 1995;270:17098–17105. [DOI] [PubMed] [Google Scholar]
- 38.Ota H, Sakurai M, Gupta R, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013;153:575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Q, Crews LA, Barrett CL, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A 2013;110:1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choudhury Y, Tay FC, Lam DH, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest 2012;122:4059–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemlich Y, Greenberg E, Ortenberg R, et al. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Invest 2013;123:2703–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoshan E, Mobley AK, Braeuer RR, et al. Reduced adenosine-to-inosine miR-455–5p editing promotes melanoma growth and metastasis. Nat Cell Biol 2015; 17:311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han SW, Kim HP, Shin JY, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med 2014;211:613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han L, Diao L, Yu S, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell 2015;28:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paz-Yaacov N, Bazak L, Buchumenski I, et al. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep 2015;13:267–276. [DOI] [PubMed] [Google Scholar]
- 46.Patil V, Pal J, Somasundaram K. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget 2015;6:43452–43471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nat Rev Genet 2001;2:869–878. [DOI] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fumagalli D, Gacquer D, Rothe F, et al. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep 2015;13:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carroll D. A CRISPR approach to gene targeting. Mol Ther 2012;20:1658–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schirle NT, Goodman RA, Krishnamurthy M, et al. Selective inhibition of ADAR2-catalyzed editing of the serotonin 2c receptor pre-mRNA by a helix-threading peptide. Org Biomol Chem 2010;8:4898–4904. [DOI] [PubMed] [Google Scholar]
- 52.Mizrahi RA, Schirle NT, Beal PA. Potent and selective inhibition of A-to-I RNA editing with 2’-O-methyl/locked nucleic acid-containing antisense oligoribonucleotides. ACS Chem Biol 2013;8:832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallo A, Keegan LP, Ring GM, et al. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J 2003;22:3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wohrer SS, Raderer M, Hejna M. Palliative chemotherapy for advanced gastric cancer. Ann Oncol 2004;15:1585–1595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.