Abstract
LRIG1, leucine-rich repeats and immunoglobulin-like domains protein 1, was discovered more than 20 years ago and has been shown to be downregulated or lost, and to function as a tumor suppressor in several cancers. Another well-reported biological function of LRIG1 is to regulate and help enforce the quiescence of adult stem cells (SCs). In both contexts, LRIG1 regulates SC quiescence and represses tumor growth via, primarily, antagonizing the expression and activities of ERBB and other receptor tyrosine kinases (RTKs). We have recently reported that in treatment-naïve human prostate cancer (PCa), LRIG1 is primarily regulated by androgen receptor (AR) and is prominently overexpressed. In castration-resistant PCa (CRPC), both LRIG1 and AR expression becomes heterogeneous and, frequently, discordant. Importantly, in both androgen-dependent PCa and CRPC models, LRIG1 exhibits tumor-suppressive functions. Moreover, LRIG1 induction inhibits the growth of pre-established AR+ and AR− PCa. Here, upon a brief introduction of the LRIG1 and the LRIG family, we provide an updated overview on LRIG1 functions in regulating SC quiescence and repressing tumor development. We further highlight the expression, regulation and functions of LRIG1 in treatment-naïve PCa and CRPC. We conclude by offering the perspectives of identifying novel cancer-specific LRIG1-interacting signaling partners and developing LRIG1-based anti-cancer therapeutics and diagnostic/prognostic biomarkers.
Keywords: LRIG1, Tumor suppressor, Prostate cancer, Stem cells, Cancer stem cells
1. Introduction: LRIG1 and the LRIG family
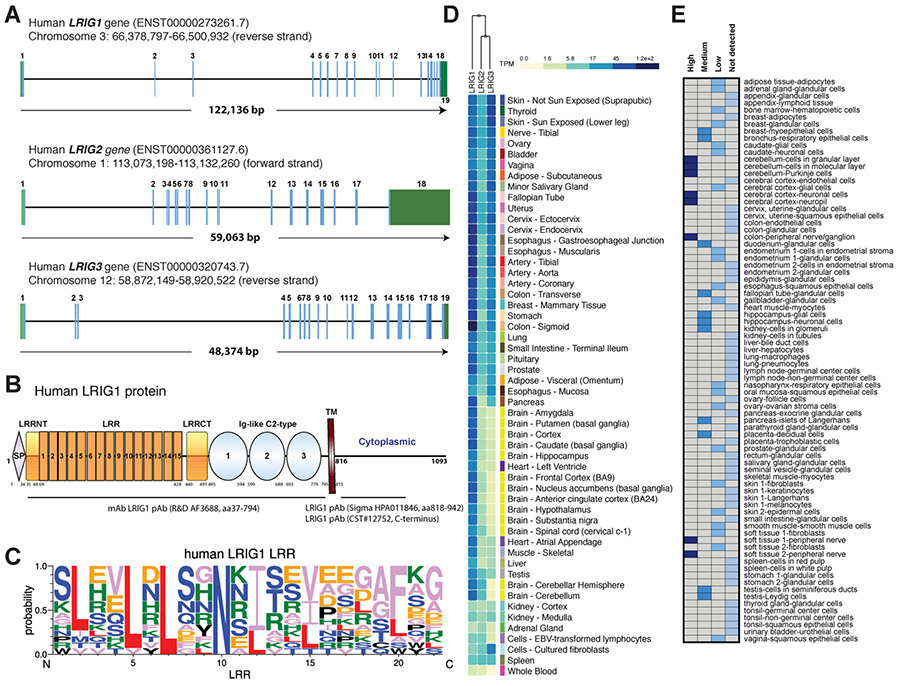
The LRIG (leucine-rich repeats and immunoglobulin-like domains) family of proteins are type I single-transmembrane proteins and consists of 3 members: LRIG1–3. Initially, a Drosophila transmembrane protein kekkon 1 was reported to participate in a negative feedback loop to regulate Gurken/EGFR (epidermal growth factor receptor) activity during oogenesis [1]. The human homolog of kekkon, located on chromosome 3q14, LRIG1 (formerly named LIG1), was later cloned (Fig. 1A) and found to share high similarity with the mouse Lrig1 gene sequence [2,3]. Additional members, the LR1G2 gene on chromosome 1p13 and LRIG3 on 12q13 (Fig. 1A) were subsequently reported [4,5]. The 3 vertebrate LRIG proteins share identical domain structures: an ectodomain (ECD) containing a signal peptide (SP), 15 tandem leucine-rich repeats (LRR), the C2-type immunoglobulin-like (Ig-like) domains, a single-pass transmembrane (TM) domain and a relatively short cytoplasmic tail (Fig. 1B). The 15 LRRs are 22-aa sequence motifs rich in consensus hydrophobic amino acid (aa) leucine (Fig. 1C). Structural domains composed of LRRs facilitate protein-protein interactions [6].
Fig. 1.
LRIG1 mRNA expression in normal human tissues. (A) Genomic structure of human LRIG1-3. The human LRIG1 gene, located on Chr3q14, spans 122,136 bp and consists of 19 exons. The LRIG2 and LRIG3 genes are smaller than the LRIG1 gene. The sizes of exons and introns are not drawn to scale. (B) Schematic of the human LRIG1 protein structure. SP, signal peptide; LRR, leucine-rich repeat; LRRNT, LRR N-terminal flanking; LRRCT, LRR C-terminal flanking; TM, transmembrane. Shown below are several commonly used anti-LRIG1 antibodies (mAb, monoclonal antibody; pAb, polyclonal antibody). (C) Sequence logos for human LRIG1 LRR amino acid alignments. (D) Heat map of LRIG1, LRIG2 and LRIG3 mRNA levels from GTEx. (E) Heat map presentation of the relative LRIG1 protein expression levels in human normal tissues based on immunohistochemistry staining (using the HPA011846; B) in samples from 44 normal tissue types of 144 individuals in the Human Protein Atlas. The box colors, dark blue, medium blue, blue and light blue, respectively, represent the “High, Medium, Low, Not Detected’ expression levels of the LRIG1 protein, as indicated on the top of the heat map.
Notwithstanding the significant structural similarity, the three LRIG proteins display distinct tissue expression patterns and biological functions. Based on the GTEx data, LRIG1 mRNA is widely expressed across most human tissues with high abundance in colon and brain (Fig. 1D). High LRIG1 protein expression is also detected in the brain and colon (Fig. 1E). Considerable evidence supports that LRIG1 is a negative regulator of ERBB receptor tyrosine kinases (RTKs), including EGFR/ERBB1/HER1, ERBB2/HER2/Neu, ERBB3/HER3, and ERBB4/HER4 [7, 8]. ERBB stimulation by the ligands such as EGF rapidly increases LRIG1 transcript and protein levels, and following the mutual recognition between the respective extracellular domains (of LRIG1 and ERBBs), LRIG1 promotes recruitment of the E3 ubiquitin ligase c-Cbl through its cytoplasmic tail. A segment spanning amino acids 900–936 within LRIG1 cytoplasmic domain was mapped as the Cbl docking site. Formation of these complexes triggers elevated ubiquitylation and accelerated lysosomal degradation of ERBB receptors resulting in the suppression of ERBB functions [7,8]. Alternatively, there is also evidence suggesting Cbl-independent LRIG1 regulation of EGFR [9]. In glioblastoma, EGFR variant III (EGFRvIII) with deletion of exons 2–7, is the most common oncogenic EGFR mutant, in which lack of dimerization arm and part of the ligand-binding domain results in impaired internalization and intercellular recycling. Restricted degradation makes EGFRvIII constitutively active, driving tumorigenesis and aggressiveness. However, EGFRvIII retains interactions with LRIG1 and LRIG1 destabilizes EGFRvIII in a Cbl-independent manner; hence, ectopic LRIG1 expression opposes the EGFRvIII oncogenic functions in glioblastoma cells [9]. In addition to ERBBs, LRIG1 also attenuates the activity of several other RTKs [10]. For instance, LRIG1 interacted with Met receptor independently of hepatocyte growth factor (HGF) stimulation and promoted Met receptor degradation independently of Cbl-mediated polyubiquitination [11]. By simultaneously regulating both Met and NeuT (an activated form of Her2), LRIG1 decreased expression of both receptors and opposed NeuT/Met collaboration in cellular invasion [11]. LRIG1 also functioned as a negative feedback regulator of Ret activation via physical interactions, restricting the Ret RTK activity and downstream signaling induced by GDNF in neuronal cells [12].
LRIG2 has been implicated in regulating neuron migration, axon regeneration and neural development. Binding of LRIG2 to Neogenin, a receptor of RGM (repulsive guidance molecules), negatively regulated ADAM-mediated proteolysis and ectodomain shedding of Neogenin in neurons, leading to an inhibition of neurite outgrowth [13]. Knockdown of Lrig2 promoted regeneration of distally crushed optic nerves, suggesting the therapeutic potential of LRIG2 inhibition in CNS regeneration [13]. LRIG2 mutations have been implicated in one human disease called urofacial syndrome (UFS; also called Ochoa syndrome), which is an extremely rare inherited autosomal recessive congenital disorder characterized by inverted smile and occult neuropathic bladder [14]. Biallelic mutations in LRIG2 were identified in three families affected by UFS [14]. Subsequent study in mice with homozygous Lrig2 mutations provided evidence that the gene had important roles for nerve patterning in lower-urinary-tract, and its mutations led to bladder outlet obstruction in UFS [15]. Pathogenic LRIG2 variants were also seen in a pediatric cohort with chronic kidney disease and UFS [15].
Lrig3 is required for lateral canal morphogenesis during inner ear development, as revealed by studies in mice with gene-trapping mutagenesis [16]. Homozygous Lrig3 mutant mice exhibited truncated lateral semicircular canal and craniofacial defects. Antagonistic interactions between Lrig3 and Netrin1, a key regulator of fusion plate formation, ensured the precise spatiotemporal regulation of three-dimensional architectural shaping in the inner ear organogenesis [16]. in vitro assays indicated that ectopic LRIG3 bound to and co-localized with EGFR, ERBB2 and ERBB4, but in contrast to LRIG1-induced ERBB downregulation, an upward trend of ERBB receptor levels was observed in LRIG3-ERBB interactions, implying diverse functions for LRIG members [17]. Indeed, further studies, based on LRIG and ERBB co-expression in HEK293 T cells, revealed distinct functions of the three LRIG proteins: LRIG1 decreased four ERBB receptors and LRIG2 showed no significant effects whereas LRIG3 increased expression of ERBB receptors [18]. Thus, LRIG1 and LRIG3 exhibited opposite functions, with LRIG3 enhancing the stability of ERBBs [18].
2. LRIG1 as a stem cell (SC) regulator and an enforcer of quiescence
2.1. LRIG1 regulates the epidermal and bulge SCs
Epidermis is the outmost protective layer of skin, providing a multilayered water-proof barrier against pathogens and dehydration. Earlier study showed that the Lrig1−/− mice developed abnormal skin on tail, face and ear, including thickening tail and facial alopecia at postnatal age of 3 weeks to 4 months [19]. Psoriasiform epidermal hyperplasia was observed in the affected tail skin. Lrig1 deletion in keratinocytes promoted hyperproliferation and terminal differentiation. Interestingly, LRIG1 was found to be down-regulated in human psoriatic lesions relative to the strong LRIG1-expressing epidermal basal cells and hair follicles in normal skin [19]. However, another study in human psoriatic epidermis and clinically matched normal skin showed no difference in the mRNA levels of LRIG1, −2, −3, but the distribution of LRIG proteins were altered in psoriatic lesions: in normal skin LRIG1 protein was detected in lower spinous layers and scattered cells in the basal layer whereas in psoriatic lesions, LRIG1 was less frequently localized on cell surface [20]. Regardless of the different LRIG1 expression patterns due to distinct antibody epitopes, both studies [19,20] consistently demonstrated an important role of LRIG1 in epidermal homeostasis.
Mammalian epidermis comprises the interfollicular epidermis (IFE), sebaceous glands (SG), and sweat glands. Distinct populations of epidermal SCs are responsible for constant renewal, regeneration and wound healing. Hair follicles (HFs) are pilosebaceous units associated to the IFE, beginning at the surface of the epidermis and extending into deep dermis. The HFs, which consist of the upper infundibulum, midregion isthmus and the lower segment including the bulge and hair germ, cycle through analgen (active growth), catagen (retraction) and telogen (resting) phases that are regulated by diverse SC populations [21-23].
IFE is the main component of epidermis maintained by resident SC populations. Transit amplifying (TA) cells arise from SCs and undergo finite times of division prior to undergoing terminal differentiation. Based on the expression of two human IFE SC markers, Dll1 (Delta like canonical Notch ligand 1) and MCSP (melanoma-associated chondroitin sulfate proteoglycan), Jensen et al. [24] separated the SCs (MCSP+, Dll1+) and TA cells (MCSP−, Dll1−) from human epidermal keratinocytes and performed comparative expression profiling, which identified 14 genes, including LRIG1, up-regulated in SCs compared with TA cells. Subsequently, LRIG1 was found to be preferentially expressed in the basal layer in human IFE, enriched in the same population considered as SCs, and play an important role in regulating the IFE SC quiescence [24].
Lrig1 is also one of the SC makers identified in the upper pilosebaceous unit in the mouse skin [25]. Lrig1 expression defined a distinct population of quiescent multipotent cells in the junctional zone in mouse HFs - the upper region of the isthmus adjacent to the lower infundibulum, and Lrig1-expressing cells contributed to all epidermal lineages in reconstitution experiments. However, during steady-state homeostasis, these cells in the junctional zone were only bipotent contributing to IFE and SG whereas they selectively expanded upwards into IFE in response to pro-proliferative stimulation [25].
To further elucidate the biological functions of Lrig1+ epidermal SCs, the same group conducted lineage-tracing studies by developing an Lrig1 knock-in mouse model, in which the eGFP-ires-CreERT2 cassette was inserted right following Lrig1 translation initiation codon to recapitulate the endogenous Lrig1 expression [26]. Interestingly, the Lrig1-marked SCs from the upper junctional zone were highly proliferative and contributed to infundibulum or SG replenishment, independently from IFE. Despite this compartmentalization, the boundaries were eliminated during wound repair when the progeny of Lrig1+ SCs from the junctional zone were detected within the IFE shortly after injury and contributed to long-term tissue maintenance [26].
Notably, a reciprocal feedback regulation between Lrig1 and Myc seemed to be important for the maintenance of SC quiescence in epidermis [25,26]. Myc activation promoted cell proliferation in the basal layer of mouse epidermis, which was antagonized by Lrig1. Immunofluorescence staining revealed inverse expression patterns of LRIG1 and MYC in the basal layer of human IFE, and in cultured human keratinocytes transduced with LRIG1, both MYC protein levels and cell proliferation were decreased. Correspondingly, increased proliferation emerged with up-regulation of Myc mRNA and protein in Lrig1-null epidermis [25,26]. Precisely how LRIG1 expression downregulated MYC remains unclear. Reciprocally, Myc transcriptionally activated Lrig1, and thus, Myc activation elevated Lrig1 mRNA whereas Myc knockdown downregulated Lrig1 expression [26]. It is worth noting that Blimp1 functions as another negative regulator of Myc in regulating the quiescence of SG SCs [27].
2.2. LRIG1 regulates the gastrointestinal (GI) tract SCs
The GI tract is composed of multiple cellular layers: mucosa, submucosa, muscularis externa and serosa. The mucosa of the small intestine forms numerous crypt-villus units, consisting of protruding finger-shaped villi and moat-like invaginated crypts. Villi are responsible for nutrient uptake and protection against environmental hazards and the crypts harbor self-renewing SCs. Six main epithelial cell lineages exist in the small intestine: enterocytes and Microfold cells in the absorptive linage, and Goblet, Paneth, enteroendocrine and Tuft cells in the secretory lineage. The intestinal SCs (ISCs) reside in the bottom of the crypt, nourished by the surrounding niche. There are two main ISC populations, the crypt base columnar (CBC) cells and +4 SCs (+4 position counting from the bottom of the crypt directly above Paneth cells) [28]. Lgr5 is a well-established CBC specific marker whereas Bmi1, Tert and HopX mark the +4 SCs [28]. Lineage mapping indicated that Lrig1 marked a distinct ISC population, with different global transcriptional profiling from Lgr5+ ISCs [29]. Lrig1 expression was traced from birth and Lrig1+ cells were competent for epithelium reconstitution after irradiation [29]. Lrig1 was found highly enriched in the SC niche of the small intestine and colon, and Lrig1-null mice exhibited enlarged crypt morphology, suggesting crypt hyperplasia due to Lrig1 loss [29]. EGF produced by surrounding Paneth cells activated EGFR signaling, and loss of Lrig1 enhanced ErbB signaling within the ISC compartment resulting uncontrolled intestinal epithelium turnover rate [30,31].
Interstitial cells of Cajal (ICC) are myofibroblasts located close to neurons in the muscular layers of the GI tract, controlling the GI smooth muscle activity. Lrig1 was also found to regulate ICC postnatal development, and Lrig1-marked smooth muscle progenitors could give rise to both ICC-DMP (deep muscular plexus) and ICC-SMP (submucosal plexus) [32].
The main anatomical sections of the stomach are the cardia, fundus, body and pyloric antrum. Resident SCs at multiple locations were the major sources for gastric tissue renewal [33-35]. Lrig1+ SCs gave rise to Dclk1-expressing chemosensory Tuft cells in normal gastric fundus [36]. Lineage-tracing studies suggested that Lrig1-expressing cells in isthmus were long-lived gastric epithelial progenitors, capable of differentiating into all gastric lineages. In response to chemically induced acute oxyntic atrophy, Lrig1+ progenitor cells committed to parietal cells and contributed to gastric mucosa regeneration [37]. Another study using a different Lrig1 reporter mouse model showed heterogeneous Lrig1 expression in distinct regions of stomach epithelium, and a population of Lrig1-expressing cells were capable of long-term tissue maintenance [38].
In oral epithelial SCs, Lrig1 marked the infrequently dividing cell population through mediating quiescence in homeostasis [39].
2.3. LRIG1 regulates neural SCs (NSCs)
Since Lrig1 was initially discovered in the attempt to isolate novel genes promoting neural differentiation in P19 embryonal carcinoma cells in vitro and Lrig1 showed a prominent expression in the brain [3], there have been great interests in studying the roles of LRIG1 in neural development. Neural stem cells (NSCs) are multipotent SCs resident in the central nervous system (CNS), generating neurons and glial cells. Mammalian adult NSCs are restricted to two major niches in the brain: the ventricular-subventricular zone (V-SVZ) of the lateral ventricles and the subgranular zone in the hippocampal dentate gyrus [40]. Lrig1 was reported as a negative regulator of Ret, thus inhibiting GDNF/Ret induced axonal growth of sympathetic neurons [12]. Studies in Lrig1-- deficient mice suggested that Lrig1 was an essential modulator of dendrite development and Lrig1 deficiency contributed to abnormal hippocampal dendritic development and cognitive deficits [12,41,42].
Balanced quiescence and activation are essential for long-term maintenance in many SC populations, including adult NSCs. The SVZ neurogenic niche harbors both quiescent and activated NSCs, and Lrig1 expression was enriched in quiescent NSCs compared to the activated counterparts [43-45]. Combining in silico analysis with public single-cell RNA-seq datasets and Lrig1 reporter mouse lines, a recent study identified Lrig1 as a NSC maker in V-SVZ neurogenic niche [46]. Lrig1 expression marked all spatial NSC subtypes, and the labeled, non-proliferative Lrig1+ SCs possessed neurogenic activity throughout adult life and contributed to the neurogenic progeny into olfactory bulb interneurons [46].
2.4. LRIG1 regulates other SCs
The cornea is the outmost transparent layer of the eye. Corneal integrity plays a critical role in vision: refracting the light and focusing on objects. The cornea is composed of epithelium, stroma and endothelium. Corneal epithelial SCs reside in the basal layer of the limbus in peripheral cornea (thus called limbal SCs; LSCs). LSC deficiency (LSCD) due to loss of LSCs results in chronic inflammation and impaired ability to repopulate the corneal epithelium. Limbal autografting of corneal epithelial SCs represents the best option for ocular surface rehabilitation in unilateral LSCD [47,48]. Cultured human ocular keratinocytes establish 3 typical clones, holoclones, meroclones and paraclones, with the holoclones exhibiting properties of SCs and paraclones and meroclones representing terminally differentiated and intermediate cells, respectively [49,50]. Gene expression profiling of human corneal keratinocytes revealed high LRIG1 expression in holoclone SCs and an important role in controlling the homeostasis of corneal keratinocyte stem/progenitor cells [51]. Genetic loss of Lrig1 developed pathological corneal phenotypes, and Lrig1 controlled the corneal cell-fate determination by negatively regulating Stat3-dependent inflammatory pathway during repair [51].
Meibomian glands (MG) are holocrine sebaceous glands lining the margin of the eyelids, secret lipid-rich meibum and prevent evaporation of the eye’s tear film. MG dysfunction is a common condition, characterized by reduced amount or quality of gland secretion. Proper function of MG relies on continual renewal of acinar epithelial cells. LRIG1 was identified as a biomarker for progenitor cells in human MG, and LRIG1 was expressed exclusively in the basal epithelial layer of acinar periphery, which gave rise to differentiated sebocytes in the central region [52].
Mammalian teeth contain exposed dental crown and buried root. Human teeth are "rooted", in which the root canal gradually becomes closed and unable to regrow in adulthood. In contrast, rodent incisors and molars are "rootless", with open root canal and ever-growing capacity. As gnawing wears away incisor substance, replenishment takes place every 40–50 days. This trait makes the mouse incisor a unique model to study adult SCs [53]. An unbiased gene co-expression analysis indicated that Lrig1 defined two distinct quiescent SC subpopulations in mouse incisor epithelium and the proximal mesenchyme [54].
3. LRIG1 as a pleiotropic tumor suppressor
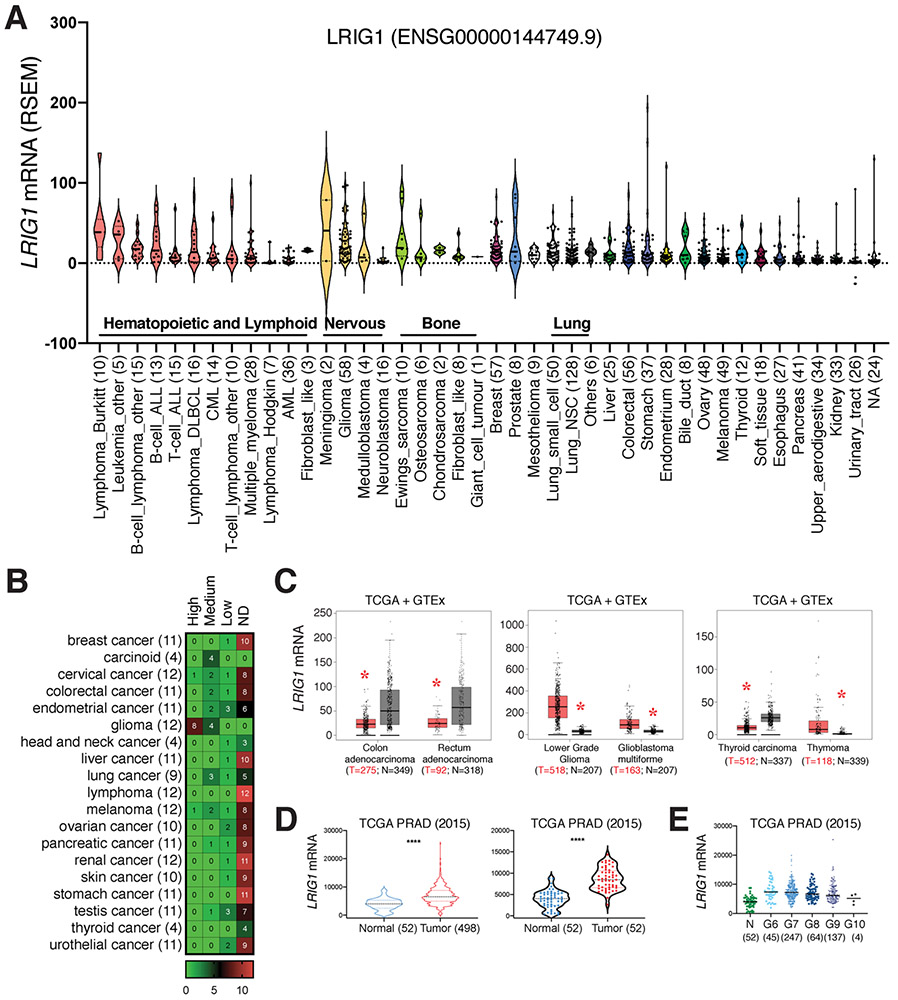
Sustaining proliferation is the most fundamental hallmark of cancer cells, which is largely enabled by growth factors and engagement of their cognate cell-surface receptors including RTKs. RTKs are key regulators of essential cellular processes including proliferation, differentiation, motility and metabolism. Though tightly controlled in normal tissue, RTK expression and functions become dysregulated during tumorigenesis resulting in oncogenic functions [55,56]. Remarkably, LRIG1 has been evinced as a negative regulator of multiple RTKs. LRIG1 is located on Chr3p14, a region commonly lost in many cancers [10,57, 58]. These observations suggest that LRIG1 likely functions as a tumor suppressor. In support, bioinformatics analysis revealed generally low expression of LRIG1 in cancer cells and patient tumors although LRIG1 overexpression was also observed in some cancers (Fig 2). Therefore, the majority of cultured human cancer cell lines in the CCLE (the Cancer Cell Line Encyclopedia) database express very low levels of LRIG1 mRNA (Fig. 2A). Most patient tumor specimens in the Protein Atlas also showed low levels of LRIG1 protein (Fig. 2B). Regardless of the LRIG1 expression levels, its tumor-suppressive functions would be predicted with better patient survival. Indeed, bioinformatics-based meta-analysis of genomic datasets across eight independent cohorts of five solid tumors including breast, lung, bladder, glioma and melanoma identified LRIG1 as one of the top four survival-related genes and as best correlated with good prognosis [59]. Below, we discuss LRIG1 as a pleiotropic tumor suppressor in different tumor systems.
Fig. 2. LRIG1 expression in human cancers and cancer cell lines.
(A) Low levels of LRIG1 mRNA expression in most cultured human cancer cell lines. Gene expression data was extracted from the CCLE Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle). Shown are the violin plots of the LRIG1 mRNA levels (RSEM) in the indicated cancer cell types (n indicated in parentheses). Median and quartiles as indicated. Each dot represents an individual cell line. (B) Heat map of LRIG1 protein expression in the indicated human cancers. The LRIG1 protein expression data was extracted from the Human Protein Atlas, based on immunohistochemistry staining of LRIG1 (HPA011846; see Fig. 1B) in samples from 189 cancer patients of 19 different cancer types (total n indicated in parentheses, scale bar represents case number). ND, not detected. The numbers in individual boxes represent the number of cases. Note that the majority of patient tumors expressed undetectable LRIG1 protein with the glioma as an exception. (C) LRIG1 mRNA expression is decreased in colorectal and thyroid cancers but increased in glioma and thymoma. Shown are the box plots of LRIG1 mRNA levels in tumor samples from TCGA and combined normal tissues from TCGA and GTEx (http://gepia.cancer-pku.cn). *p < 0.05 (ANOVA). (D) Increased LRIG1 mRNA levels in human PCa. Shown are LRIG1 mRNA levels in two different tumor-normal comparisons from the TCGA-PRAD dataset. ****p < 0.0001 (paired Student’s t-test). (E) In TCGA-PRAD dataset, LRIG1 mRNA levels are increased in PCa of all Gleason (G) grades compared to normal (N). p < 0.001 (paired Student’s t-test). However, LRIG1 mRNA levels gradually declined accompanying the increased tumor grade. p < 0.0001 (Jonckheere-Terpstra test).
3.1. LRIG1 as a tumor suppressor in GI tumors
The LRIG1 gene promoter region has been reported to be hypermethylated in human colorectal cancer (CRC) cell lines and primary tumor tissues [60,61], suggesting that LRIG1 might be downregulated in CRC cells. Indeed, the LRIG1 protein was undetectable in the majority of the 11 CRC cell lines in the Protein Atlas (Fig 2B). The LRIG1 mRNA levels were also downregulated in patient CRC (Fig. 2C).
Lrig1-null mice developed spontaneous duodenal adenomas with elevated ErbB1–3 and pErk1/2 levels [29]. Adenomatous polyposis coli (APC) is a tumor suppressor gene whose mutations result in the colon cancer predisposition syndrome called familial adenomatous polyps (FAP). Loss of even a single copy of the Apc gene in Lrig1-expressing colonic progenitor cells led to distal colonic tumors, as well as tumors along the GI tract [62]. In addition to the GI neoplasia, extracolonic FAP features including periampullary tumors, gastric abnormalities and retinal pigment epithelium defects were also detected [62]. Interestingly, loss of the second Apc allele in Lrig1+ cells resulted in dysplastic adenomas in the jejunum and distal colon [29,62]. Genomic profiling of the adenomas resulting from inducible loss of a single Apc allele in Lrig1+/− colonic stem cells revealed increased mutation and reduced DNA repair as well as increased inflammation [63]. Intra-tumoral genomic heterogeneity was observed in these hypermutated tumors although transcriptomes and splicing patterns (including loss of intron retention) were relatively uniform [63].
Lrig1 marks the gastric epithelial progenitor population capable of regenerating gastric mucosa upon injury [37,38]. Infection of carcinogenic Helicobacter pylori, the strongest known risk factor for gastric cancer, elevated Lrig1+ progenitor activity in a lineage-tracing mouse model, which was validated by the increased expression of LRIG1 in human gastric premalignant lesions [64]. LRIG1 expression was dynamically and negatively related to gastric carcinogenesis, with its expression up-regulated in spasmolytic polypeptide-expressing metaplasia and early stage of metaplasia but barely detectable in precancerous lesions [65]. An inverse correlation between LRIG1 and EGFR was also observed in different stages of tumorigenesis [65]. Of clinical significance, LRIG1 expression in gastric cancer patients was positively associated with relapse-free survival rate, suggesting LRIG1 as a potential prognostic biomarker [65].
3.2. LRIG1 as a tumor suppressor in skin (and other squamous) cancers
There are three major types of skin cancer: basal cell carcinoma (BCC), squamous cell carcinoma (SCC) and melanoma, with BCC being the most common. BCC preferentially arises from the basal SCs within hair follicle and mechanosensory niches [66]. These SCs were restricted in upper/lower bulge and the isthmus, and were marked by Lrig1 expression [66]. LRIG1 expression was also detected in most human BCC subtypes (superficial, nodular, adenoid and infiltrative) as well as in tumors of skin appendages (including porocarcinoma and SG carcinoma), and lower LRIG1 expression was observed in advanced and late-stage tumors [67]. Consistently, LRIG1 protein was not detectable in most skin cancer specimens in the Protein Atlas (Fig. 2B). In SG tumors, LRIG1 overexpression was observed in poorly differentiated carcinoma [68].
Cutaneous SCC is characterized by uncontrolled growth of squamous cells. SCC can become invasive, grow beyond the epidermis and spread to other organs. Immunohistochemical (IHC) analyses performed on patient cutaneous SCC revealed higher LRIG1 expression in well-differentiated low-grade regions [69]. Tumors with low LRIG1 expression showed more frequent metastasis while patients with high LRIG1 displayed survival benefit [69]. These clinical outcomes suggested LRIG1 as a promising prognostic indicator in cutaneous SCC [69]. These observations were confirmed in SCC cell lines and human oral SCCs [70]. An independent study in 128 invasive squamous cell cervical carcinomas also revealed more intensive LRIG1 expression in early-stage tumors and LRIG1 expression correlated with increased survival rate [71]. In human ocular surface squamous neoplasia, LRIG1 was expressed in benign but downregulated in malignant epithelium, and its expression correlated inversely with EGFR expression [72]. LRIG1 overexpression in human SCC H357 cells drastically reduced tumor incidence, and 76 % of SCC cell lines from multiple organs had loss of heterozygosity (LOH) of LRIG1, which was an early event in human SCC development [73].
Melanoma, developing in melanin-producing melanocytes, is the most aggressive type of skin cancer. LRIG1 inhibited growth of melanoma xenografts and mediated the inhibitory effect of isoliquiritigenin, a phenolic chemical found in licorice [74]. During hypoxia-evoked aggressive melanoma progression, human melanoma cell line A2058 emerged more aggressive phenotypes including enhanced invasion, migration, vasculogenic mimicry and epithelial-mesenchymal transition (EMT), which were antagonized by LRIG1 overexpression and aggravated by LRIG1 depression [75]. Upregulation of LRIG1 repressed the pEGFR and pERK activated by hypoxia stimulation, whereas silencing of LRIG1 enhanced metastasis that was counteracted by erlotinib, a selective EGFR inhibitor [75].
3.3. LRIG1 as a tumor suppressor in GBM and other brain tumors
There are two categories of brain tumors: primary brain tumors originating in the brain and secondary (metastatic) tumors beginning elsewhere and spreading to the brain. Two most common types of primary brain tumors are meningioma and glioma. A tissue microarray (TMA) analysis of 409 meningiomas revealed the correlation between cytoplasmic LRIG1 and LRIG2 expression with histological subtypes [76]. Cytoplasmic LRIG1 was detected in 67 % of meningioma samples, most frequently in benign subtypes. Since meningiomas are twice as common in women, a significant correlation was also observed between estrogen receptor (ER) status and LRIG1 expression [76].
Glioma develops in glial cells (astrocytes and oligodendrocytes), and based on histological features, gliomas are further classified into ependymoma, astrocytoma, oligodendroglioma, brainstem glioma, optic nerve glioma and mixed glioma. Gliomas are graded according to malignancy or aggressiveness on a scale of I to IV: biologically benign (grade I), low-grade gliomas (LGG, grade II) and high-grade gliomas (HGG, grade III- IV). Glioblastoma multiforme (GBM) is the most malignant Grade IV tumor, fast-growing and invasive [77]. Interestingly, LRIG1 mRNA levels were significantly elevated in both LGG and GBM (Fig. 2B, C; middle). An immunohistochemistry analysis of LRIG1–3 in 404 human astrocytic tumors revealed nuclear, perinuclear and cytoplasmic expression patterns [78]. Although cytoplasmic staining was the dominant pattern, the perinuclear LRIG1–3 staining showed better inverse correlation with tumor progression and was associated with lower histopathological grade and better prognosis [78]. Subsequent manipulation on LRIG1 expression demonstrated that LRIG1 inhibited glioma growth and aggressiveness by attenuating EGFR activation and downstream PI3K/AKT signaling pathway [79-81]. Similar effect was observed in pituitary adenoma [82]. Notably, LRIG1 destabilized EGFRvIII in GBM, dampened EGFRvIII oncogenic activity and inhibited EGFRvIII -driven GBM cell proliferation and invasion [9].
In platelet-derived growth factor B (PDGFB) induced murine gliomas, Lrig1 expression was comparable in both low-grade and high-grade gliomas vs. normal mouse brain tissues [83]. However, further ablation of a single or both Lrig1 alleles significantly increased the incidence of HGG, implicating Lrig1 as a haplo-insufficient tumor suppressor in gliomas [83]. LRIG1 overexpression in human glioblastoma cell line TB107 suppressed the invasion and malignancy, partially by inhibition of MET phosphorylation [83].
3.4. LRIG1 as a tumor suppressor in lung, breast and other cancers (excluding prostate cancer)
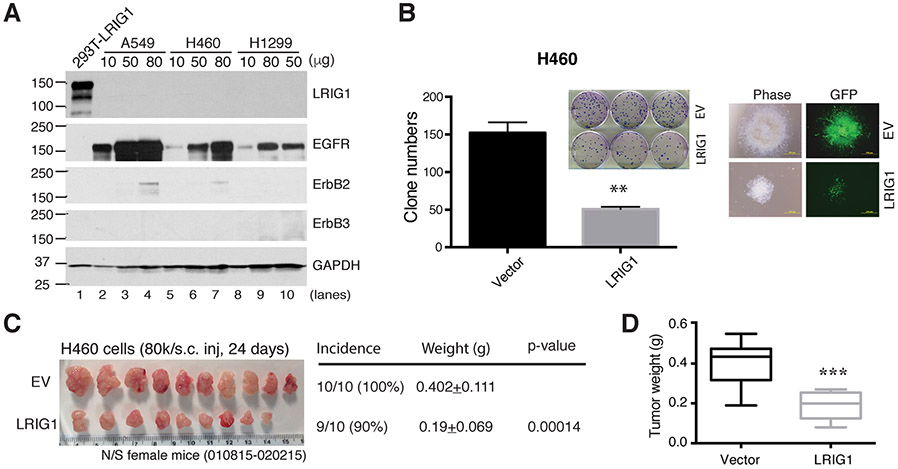
Non-small-cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for 80%–85% lung cancer diagnoses. Lrig1 is an important regulator of tissue homeostasis in murine airway as well as a tumor suppressor in lung cancer [73]. In Lrig1−/− murine tracheal epithelial cells (MTEC) and human lung adenocarcinoma and squamous cancer cell lines, LRIG1 expression was essential for cell-cell contact inhibition via interaction with EGFR and E-cadherin [73]. Ten human pre-invasive carcinoma samples showed reduced LRIG1 expression compared to matched local normal airway biopsies, and LOH at LRIG1 locus occurred in 75 % of 138 lung cancer cell lines and 40 % of pre-invasive lung cancer lesions, suggesting LRIG1 loss was associated with early tumorigenesis [73]. Independent studies in two NSCLC cohorts with 347 and 182 patients, respectively, indicated that high LRIG1 expression was correlated with better patient survival [84,85]. Hyperactivated EGFR signaling results in resistance to EGFR tyrosine kinase inhibitors (TKIs) in NSCLC patients harboring mutant EGFR. LRIG1 expression was downregulated in NSCLC cell lines with promoter hypermethylation, compared to normal human bronchial epithelial cell line BEAS-2B, and reduced LRIG1 was also observed in surgically resected primary NSCLC [86]. Transfection of LRIG1 in EGFR-mut NSCLC cell lines led to downregulation in expression and phosphorylation of mutant EGFR, as well as other RTKs including HER2, HER3, MET and IGF-1R [86]. Consistent with these reported tumor-suppressive functions of LRIG1, we also observed that lentiviral mediated LRIG1 overexpression in human lung cancer cells (Fig. 3A) inhibited clonal capacity in vitro (Fig. 3B) and tumor growth in vivo (Fig. 3C-D).
Fig. 3. LRIG1 overexpression inhibits lung cancer xenograft growth.
(A) WB of LRIG1 in lung cancer cells. A549, H460 and H1299 cells infected with pLVX-LRIG1 (LRIG1) or empty control (EV) lentivirus were used to prepare whole cell lysate in WB analysis of LRIG1 (a mAb against LRIG1 was used) and other proteins indicated. 293T-LRIG1 cells were used as positive control. Varying amounts of proteins were loaded in WB and GAPDH was used as a control. Note that that these 3 lung cancer cell lines do not express detectable endogenous LRIG1 and also lacked appreciable expression of ERBB2/ERBB3. (B) Clonal assays in H460 cells infected with pLVX-LRIG1 lentivirus (LRIG1) and the control empty lentivirus (EV) for 72 h and plated in 6-well plates (300 cells/well). Clones were counted 13 days after plating. Presented are the mean ± SD from triplicate cells. **p < 0.01 when compared with the corresponding EV controls (paired Student’s t-test). Shown on the right are representative images of the clones. (C-D) LRIG1 expression inhibits lung cancer xenograft growth. Shown in C are the tumor images and tumor incidence (# tumors/# injections), endpoint tumor weights (mean ± S.D) and the corresponding p-value (Student’s t-test). Shown in D is the boxplot of tumor weights (***p < 0.001).
In a study with 104 renal cell carcinoma (RCC) including clear cell and papillary RCC and chromophobe, decreased LRIG1 and ERBB2 expression was detected in clear cell RCC whereas up-regulation of EGFR and down-regulation of ERBB4 were present in all three RCC subtypes [87].
Breast cancer (BC) is a heterogenous malignancy classified by the expression of steroid hormone receptors estrogen receptor (ER), progesterone receptor (PR) and HER2. Strong evidence indicates that LRIG1 represents a critical negative growth regulator in BC [58]. Most BC specimens in the Protein Atlas expressed little LRIG1 [Fig. 2B], and highest LRIG1 expression was observed in luminal A and lowest in basal-like BC subtypes, respectively [58]. Importantly, high LRIG1 expression correlated with better clinical outcome and greater relapse-free survival [58]. In contrast, loss of LRIG1 protein was detected in 42 of 67 human breast tumors, with >2-fold decrease in high-grade tumors [88]. Significant downregulation of LRIG1 mRNA and an inverse correlation with tumor grade were confirmed from Oncomine database in silico analysis [88]. Lrig1 expression was suppressed in focal murine mammary adenocarcinomas driven by transgenic overexpression of activated ErbB2 (consistent with the observations in HER2+ human BC) and suppressed Lrig1 expression by constitutively activated ErbB2 signaling in BC cells in turn contributed to ErbB2 overexpression thus forming a feed-forward regulation loop [88].
ERα positive BC is the most common subtype, in which LRIG1 expression was enriched and higher LRIG1 expression was linked to better relapse-free survival [89]. Mechanistically, ERα transcriptionally activated LRIG1 expression via direct genomic binding, and ErbB2 activation attenuated ERα expression and antagonized ERα-driven LRIG1 transcription [89]. Basal-like BC are negative for ER, PR and HER2 and thus termed triple-negative BC or TNBC, which are highly invasive and metastatic. Endogenous LRIG1 expression was downregulated during EMT of human mammary epithelial cells, which provides a potential explanation for the lowest LRIG1 expression in basal-like BC [90]. LRIG1 depletion resulted in accelerated EMT whereas restoring LRIG1 expression opposed EMT, and reduced migration and invasion [90]. Earlier studies in 73 BCE employing fluorescence in situ hybridization (FISH) revealed increased LRIG1 copy numbers in 34 % tumors with coincidental increases in ERBB2 copy numbers [91,92]. However, a subsequent study in 971 stage I/II BC revealed 3.9 % gain and 8.9 % loss in LRIG1 copy numbers [93]. LRIG1 copy number loss was enriched in TNBC and HER2+ BC than in luminal A and luminal B BC whereas LRIG1 copy number gains showed no significant differences among the BC subtypes. Combined with an analysis in 1,576 BCE samples in public datasets, it was determined that LRIG1 loss was a critical risk factor for distant metastasis and death and LRIG1 loss in early-stage tumors predicted early and late relapse [93]. Nevertheless, a recent study using droplet digital polymerase chain reaction (ddPCR) in 423 primary invasive BC cytosol samples from Sweden, generated yet discordant conclusions [94]. In contrast to the above study [93], LRIG1 loss exhibited significant correlation with tumor grade and nodal status, and LRIG1 status was not significantly associated with 10-year patient survival [94]. The discordance in these studies [92-94] might be due to the differences between the different patient cohorts and analytical methods utilized.
4. LRIG1 as an oncogenic signaling-induced feedback tumor suppressor in prostate cancer (PCa)
4.1. LRIG1 expression and functions in untreated (primary) PCa
PCa is the most prevalent cancer and second-leading cause of cancer-related deaths for men in North America. Early-stage tumors are highly curable after prostatectomy and local therapy such as radiation. After biochemical recurrence and in advanced high-grade tumors, androgen-deprivation therapy (ADT; also called chemical castration) is the common treatment option. Eventually, the majority of PCa patients develop castration-resistant PCa (CRPC) with lethal metastasis. PCa development and progression are known to involve the activation of multiple the oncogenic (mitogenic) signaling pathways including androgen receptor (AR), ERBBs, MYC, STAT3 and others [95-98].
LRIG1 has been under-studied in PCa, compared to studies in other cancers such as breast and GI cancers and glioma. An earlier study demonstrated conflicting results of LRIG1 expression and clinical significance in two independent PCa patient cohorts, including a Swedish cohort with 355 patients and an American cohort with 293 patients [99]. In the Swedish cohort, mainly untreated, high LRIG1 expression was associated with worse patient survival whereas in the American cohort, all received prostatectomy, LRIG1 expression correlated with better survival [99].
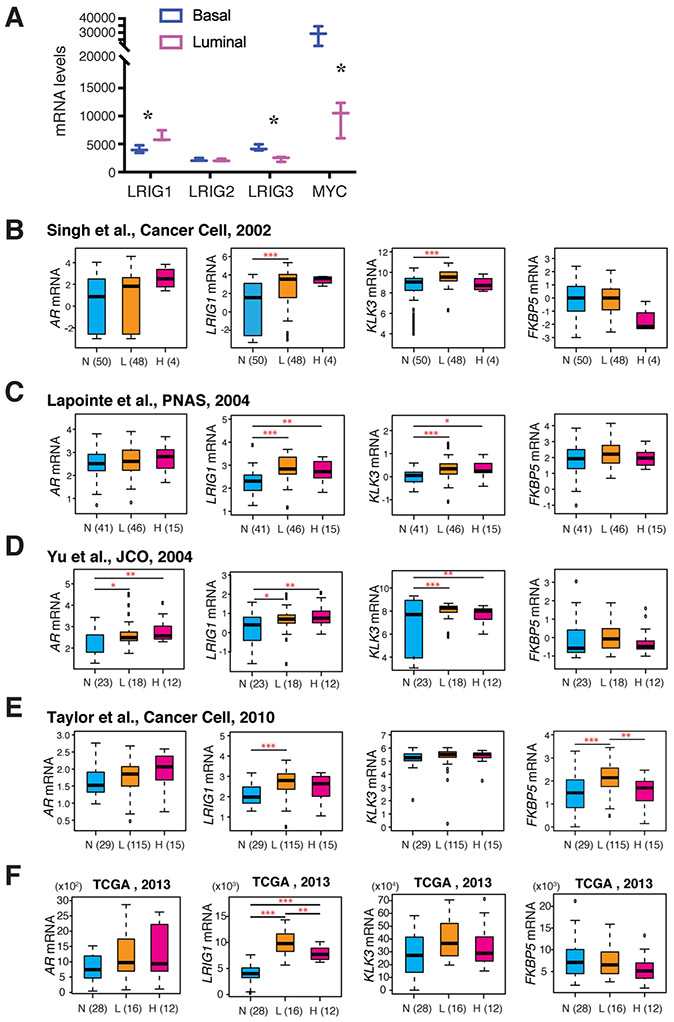
We have recently published a comprehensive study on LRIG1 expression, regulation and functions in PCa as well as in CRPC [100]. We found that LRIG1, which is preferentially expressed in the normal human prostate (NHP) luminal cells (Fig. 4A; [101]), showed much higher transcript levels in prostate tumors compared to normal or benign prostate tissues from a total of ~1200 PCa cases in Oncomine and TCGA PCa datasets [100]. Indeed, unlike CRC and thyroid cancer, which showed reduced LRIG1 mRNA (Fig. 2C), PCa in TCGA, most of which were untreated, showed LRIG1 upregulation (Fig. 2D-E, Fig. 4B-F; [100]). Importantly, higher LRIG1 mRNA levels in PCa were associated with prolonged overall survival [100]. LRIG1 protein was also upregulated in untreated PCa specimens compared to matched benign tissues, based on IHC analysis in three TMAs containing 306 prostate tumors and 307 normal prostate tissues [100].
Fig. 4. LRIG1 is expressed in normal human prostate luminal cells and upregulated in PCa.
(A) Contrasting mRNA expression patterns between LRIG1 and MYC and between LRIG1 and LRIG3 in normal human prostate (NHP) luminal and basal cells. Shown are the mRNA levels (normalized read_counts) for the indicated genes based on our RNA-seq profiling data (GSE67070). *p < 0.05 (paired Student’s t-test). (B-F). LRIG1 mRNA levels are up-regulated in low grade prostate tumor tissues (L) compared with the matched normal tissues (N), but slightly downregulated in high grade tumor tissues (H) in 4 representative Oncomine datasets (B-E) and in TCGA (F). Similar trend was observed in some datasets with PSA (KLK3) and FKBP5 mRNA levels. Patient numbers are indicated. Note that the AR mRNA levels were elevated in only one dataset (D). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Notably, the LRIG1 expression pattern exhibited a close positive correlation with AR expression in untreated PCa cell lines and xenografts, with low/no LRIG1 detected in AR− cells [100]. Functionally, LRIG1 re-expression in AR− PCa cells that express little endogenous LRIG1, such as PC3, Du145 and PPC-1, reduced both tumor incidence and tumor growth [100]. In contrast, knocking down endogenous LRIG1 promoted growth of AR+LRIG1+ PCa xenografts [100]. In both cases, tumor inhibition or promotion upon LRIG1 manipulation was associated mainly with changes in cell proliferation rather than apoptosis [100]. These xenograft studies indicate that LRIG1 exhibits tumor-suppressive properties in both AR+ and AR− PCa subtypes. Likewise, transgenic expression of LRIG1 in the mouse prostate (i.e., human LRIG1 transgene under the control of ARR2PB promoter) inhibited the development and growth of both Hi-Myc and TRAMP tumors, especially at early stages [100]. Importantly, inducible LRIG1 expression inhibited growth of pre-established AR− PCa models PC3 and Du145 [100] as well as AR+ LNCaP model (Fig. 5). These latter observations indicate therapeutic potential of LRIG1.
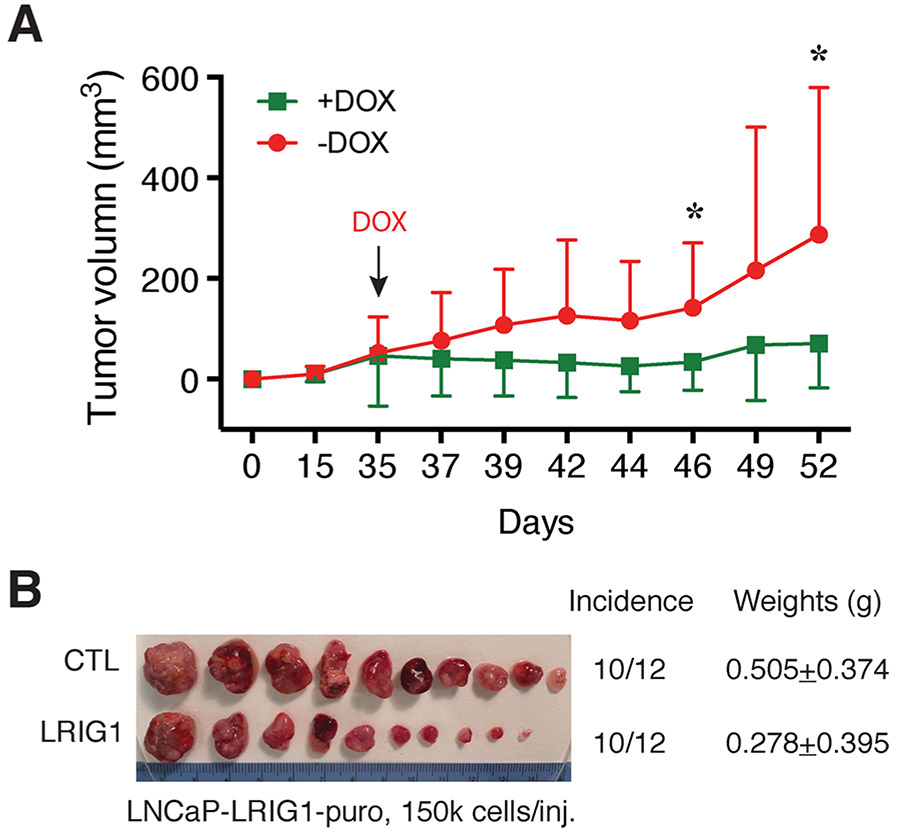
Fig. 5. LRIG1 induction inhibits the growth of AR+ LNCaP xenograft tumors.
LNCaP cells were infected with a doxycycline (DOX) inducible LRIG1-encoding lentiviral vector (MOI of 5) and selected with puromycin for ~2 weeks [100]. The LNCaP-LRIG1-puro cells were implanted subcutaneously (s.c) into two groups of male NOD/SCIDγ mice (12 mice/group), and, on day 35, one group of mice received DOX-supplemented chow (+DOX) and the other group the regular chow (−DOX). Tumor volumes were monitored and measured using a digital caliper and data presented in A (*p < 0.05; paired Student’s t-test). The experiment was terminated on day 52 and tumors harvested. Presented in B is the image of endpoint tumors with incidence and weights indicated. Note that the LRIG1-expressing tumors were nearly twice as smaller as the control (CTL) tumors (i.e., −DOX) although the p-value is not statistically significant due to big variations in tumor size.
4.2. AR represents the major transcriptional regulator of LRIG1 in untreated PCa
AR signaling is intimately involved in PCa carcinogenesis and progression. Our recent study [100], as well as studies from other groups [99,102,103] suggest that AR represents a critical transcriptional regulator of LRIG1. In addition to the aforementioned coordinated expression between AR and LRIG1 in PCa cell line and xenograft models [100], synthetic androgen R1881 or dihydrotestosterone (DHT) robustly stimulates LRIG1 expression in LNCaP and other androgen-responsive PCa cells [99,100,102]. LRIG1 mRNA levels highly correlated with the well-known AR target genes prostate specific antigen (PSA; KLK3) and, sometimes, FKBP5 (Fig. 4B-F; [100]). Multiple AR binding sites (ABS) were identified across the LRIG1 genomic region by chromatin immunoprecipitation sequencing (ChIP-seq) [100,103], and ChIP-qPCR targeting 4 major ABS revealed significant AR-binding activities in AR+ PCa cells, which were further enhanced by DHT [100].
4.3. LRIG1 signaling in treatment-naive PCa: interplays with AR, ERBB and MYC
As discussed above, in untreated PCa, AR represents a major transcriptional activator of LRIG1, which in turn represses AR-driven (i.e., AR+) prostate tumorigenesis. Another upstream activator of LRIG1 is the ERBB/ligand signaling [7,8]. In the normal human prostate, ERBB2 was most abundantly expressed at the mRNA levels followed by ERBB3 while EGFR was expressed at low levels and ERBB4 minimally expressed (Fig. 6A). There was a moderate correlation (i.e., R = 0.3 – 0.4) between LRIG1 and EGFR, and between LRIG1 and ERBB2, mRNA levels (Fig. 6A). In contrast, in PCa in the TCGA-PRAD dataset, most of which were untreated, ERBB3 was expressed at the highest levels among the four ERBBs and the upregulated ERBB3 significantly correlated with LRIG1 mRNA levels (Fig. 6B; [100]).
Fig. 6. Dynamic relationship between LRIG1 and ERBB family members.
(A) In normal prostate tissues, ERBB2 mRNA was expressed at highest levels followed by ERBB3 and EGFR while ERBB4 mRNA was barely detectable, and there was reasonable correlation between the ERBB2 and ERBB3 mRNA levels with LRIG1. Pearson correlation coefficient for linear regression was calculated based on expression data of the 5 genes in 232 normal prostate tissues from the GTEx project, with R and p-value indicated.
(B) In PCa, ERBB3 was expressed at the highest level and there was a significant correlation between ERBB3 mRNA levels with LRIG1. Pearson correlation coefficient for linear regression was calculated based on expression data in 498 PCa samples from TCGA project, with R and p-value indicated.
The above discussions suggest an intricate relationship among AR, LRIG1 and ERBBs in PCa [100,104]. AR not only directly regulates its own inhibitor LRIG1 [100], but it also transcriptionally regulates EGFR and ERBB3 in that the AR mRNA levels highly and moderately correlated with the mRNA levels of EGFR and ERBB3, respectively, and that there were strong and moderate AR-binding peaks at the EGFR and ERBB3, respectively, genomic regions [100]. LRIG1 then functionally, and posttranscriptionally, antagonizes AR as well as ERBBs. Indeed, LRIG1 re-expression or knockdown in PCa, repressed and induced, respectively, the protein levels of various (p)ERBBs in a model-dependent manner [100]. And, importantly, LRIG1 induction inhibited the growth of PC3 and Du145 tumors expressing an oncogenic rat ErbB2 mutant, Neu* [100].
Yet another upstream oncogenic activator of LRIG1 is MYC. The MYC gene is often amplified and MYC protein is overexpressed in the precursor lesions and early-stage PCa, and transgenic MYC expression is sufficient to induce cancer phenotypes in the mouse prostate [105]. Interestingly, in the NHP, MYC shows a contrasting expression pattern to LRIG1 in that it was mostly expressed in the basal cells (Fig. 4A; [101]). However, in PCa MYC protein is largely expressed in the luminal progenitor cells [106], which is why the ARR2PB-driven MYC expression could cause full-blown murine PCa [105]. Earlier studies in mouse epidermis revealed an intriguing reciprocal relationship between Lrig1 and Myc in that Myc appeared to transcriptionally activate Lrig1 and Lrig1 could also downregulate Myc protein [25,26]. We have demonstrated that transgenic LRIG1 expression inhibited MYC-driven prostate tumors and that in human PCa, knocking down endogenous LRIG1 increased c-MYC levels in LNCaP and VCaP cells while LRIG1 overexpression reduced c-MYC in Dul45 and PPC-1 cells [100].
4.4. LRIG1 expression, regulation and functions during PCa progression and in CRPC
Most primary PCa is diagnosed as localized multifocal disease, composed of genetically identical subclones harboring distinct mutations. Although genomic amplifications (alterations) of AR rarely occur in primary PCa [95], the inter-focal genetic heterogeneity contributes to metastasis and therapy resistance [95-97] and PCa progression is accompanied by further cellular and genomic diversification [107]. Intriguingly, the LRIG1 and ERBB3 mRNA expression followed a similar pattern, i.e., significantly increased in early-stage PCa (i.e., Gleason 6 and 7) and then gradually decreased with increasing Gleason grade (Fig. 2E; [100]). This may likely be related to the fact that AR signaling (not the AR expression) gradually becomes attenuated in advanced, high-grade prostate tumors [107].
ADT induces unprecedented cellular and molecular heterogeneity in patient tumors. For example, the expression levels and distribution patterns of AR in CRPC become highly heterogeneous in that a significantly increased fraction of CRPC cells or clones express little AR, i.e., AR−/lo or show cytoplasmic AR expression [98]. Accompanying the altered AR expression, LRIG1 expression also becomes heterogeneous and discordant (with AR) in CRPC (Fig. 7; [100]). In the 3 Oncomine datasets, the LRIG1 mRNA levels were increased, decreased or un-altered in CRPC compared to hormone-naïve tumors (Fig. 7A). In several whole-mount patient CRPC we analyzed, AR−/lo PCa cells became predominant whereas LRIG1 was persistently and highly expressed and discordant LRIG1 and AR expression patterns were observed showing AR+LRIG1+, AR−/loLRIG1−/lo and AR−/loLRIG1+ phenotypes (Fig. 7B; [100]). in vitro and in vivo modeling of castration in several PCa models also revealed dynamic (both concordant and divergent) changes in AR and LRIG1 (Fig. 7C-D). in vivo, the LNCaP-AI (androgen-independent; castration-resistant) tumors showed upregulation in AR and (slight) decreases in LRIG1 compared to androgen-dependent (AD) tumors (Fig. 7C; [100]). The LAPC4-AI tumors showed concordantly reduced AR and LRIG1 compared to LAPC4-AD tumors while both LAPC9 CE/AI tumors showed low levels of AR and LRIG1 (Fig. 7C). In contrast, the VCaP-AI tumors upregulated AR but downregulated LRIG1 (Fig. 7C). in vitro, a short-term castration of LNCaP cells, i.e., culturing in CDSS (charcoal dextran stripped serum) for 48 h, led to downregulation of LRIG1 without significant changes in AR (Fig. 7C). Long-term castration (i.e., from 1 week to 15 months) of LNCaP cells in vitro using 3 different regimens induced epigenetically-driven cyclic changes in AR protein, as reported earlier [108], which was accompanied by interesting alterations in LRIG1 (Fig. 7D). In LNCaP cells castrated for 1 week and 3 weeks, both AR and full-length 146-kD LRIG1 were lost but, strikingly, two smaller LRIG1 species migrating at ~115-kD and 100-kD [100] were detected (Fig. 7D). In LNCaP cells castrated for 10–15 months, divergent levels of AR re-appeared accompanied by the appearance of full-length LRIG1 (Fig. 7D).
Fig. 7. Persistent and increased LRIG1 expression in CRPC.
(A) Heterogeneous LRIG1 mRNA expression in CRPC. Shown are the relative LRIG1 mRNA levels in CRPC compared to corresponding hormone-naïve PCa in 3 Oncomine datasets. LRIG1 was upregulated in the Tomlins dataset (FC = 2.359; p = 0.009) and showed reduced trend in the Best dataset (FC= −1.635; p = 0.095) whereas LRIG1 did not change in Holzbeirlein dataset (FC = 1.029; p = 0.483). (B) Discordant LRIG1 and AR expression and persistently high LRIG1 expression in patient CRPC. Shown are matched IHC images of AR and LRIG1 in the whole-mount slides of 4 patient CRPC specimens (adapted with permission from [100]). Note that most CRPC cells lost AR expression but retained high levels of LRIG1. (C) Persistent LRIG1 expression in CRPC xenograft models. Whole cell lysates (60 μg/lane) prepared from 4 pairs of androgen-dependent (AD) and androgen-independent (AI; castration-resistant) xenograft tumors (lanes 1-8) and from 1 pair of in vitro castrated (i.e., CDSS for 48 h) LNCaP cells (lanes 9-10) were used in WB analysis of the molecules indicated. (D) Alterations of LRIG1 in an in vitro castration model. As detailed in [108], LNCaP cells were subjected to 3 regimens of long-term castration in culture, i.e., CDSS (charcoal dextran stripped serum), ENZA (enzalutamide; 10 μM), or CDSS plus bica (bicalutamide; 20 μM) for the time intervals indicated (w, week; m, month). Whole cell lysates (60 μg/lane) were used in WB analysis of the molecules indicated. *, cleaved ~110-kD and 100-kD LRIG1 ECD fragments. The arrow indicates the 60-kD ECD fragment.
These studies of LRIG1 and AR (Fig. 7; [100]) suggest that, although AR represents the MAJOR transcriptional regulator of LRIG1 in treatment-nai’ve PCa, AR continues to regulate LRIG1 expression in CRPC to certain levels. This would help explain the concordant AR+LRIG1+ and AR−/loLRIG1−/lo phenotypes observed in patient CRPC (Fig. 7A-B; [100]). Regardless of LRIG1 expression and its regulation in CRPC, LRIG1 still manifests tumor-inhibitory effects in CRPC, as overexpression of LRIG1 in LNCaP AI cells inhibited tumor development whereas LRIG1 knockdown in LAPC9 AI cells promoted tumor growth [100].
5. LRIG1 as a potential biomarker and anti-cancer therapeutic
That LRIG1 expression correlates with better patient survival in many cancers [59] including gastric cancer [65], cutaneous SCC [69], squamous cell cervical carcinoma [71], glioma [78], NSCLC [84,85], breast cancer [58,89,93] as well as PCa [100] suggests that the LRIG1 status and expression levels may be developed as a diagnostic and/or prognostic biomarker in the clinic. Our observations that LRIG1 induction inhibited the growth of pre-established AR+ and AR− PCa models (Fig. 5; [100]) indicate that LRIG1, as a tumor suppressor, may also be therapeutic. In fact, the ECD (ectodomain) of LRIG1 has been reported to display tumor-inhibitory and therapeutic effects in several cancers [109-111]. ECD shedding is the release of the soluble extracellular domain following proteolytic cleavage, an irreversible post-translational modification fundamental in cellular processes and pathologies [112]. Proteolytic shedding of the soluble LRIG1 ECD may have evolved as a critical regulatory mechanism for growth factor (particularly, ERBB) signaling. Besides the major predicted full-length LRIG1 protein of ~146-kD, minor bands with lower molecular weight (M.W), e.g., 110-kD (the predicted M.W of the LRIG1 ECD) and 60-kD, have been detected by antibodies against the LRIG1 ECD [100]. In tail skin lysates from wild-type mouse, an additional 60-kD band was detected, which was absent in Lrig1 knock-out mouse tissue. The 60-kD LRIG1 band was also detected in human tissue lysates from prostate, ileum, stomach and skin [112]. With ectopic expression of LRIG1 FLAG-tagged at the N-terminus, full-length LRIG1 was detected in cell lysates whereas the 110-kD and 60-kD bands were observed in cell culture supernatants [112]. Release of the putative LRIG1 ECD was enhanced by metalloprotease activators and abolished by metalloprotease inhibitors, strongly suggesting proteolytic processing of the LRIG1 protein at the ECD [112]. Interestingly, in the in vitro generated LNCaP-CRPC cells, we observed several lower M.W LRIG1 bands at 110-kD, 100-kD and 60-kD when AR was lost, which likely represented various LRIG1 ECD cleavage products (Fig. 7D). Regardless, expression of the LRIG1 ECD or truncated LRIG1 containing only LRRs (Fig. 1B), inhibited cancer cell growth in vitro and retarded the growth of patient-derived orthotopic xenografts [109-113]. As LRIG1 has been evinced to manifest strong inhibitory effects against (p)ERBBs [7,8,100], these studies [109-112] highlight the potential of developing the LRIG1 ECD (or part of the ECD) into novel peptide anti-cancer therapeutics. In support, soluble LRIG1 ECD has been demonstrated to exhibit pan-RTK inhibitory activity targeting ErbBs, Met, Ret, Ron, PDGFRα, IGF1R and AXL [110].
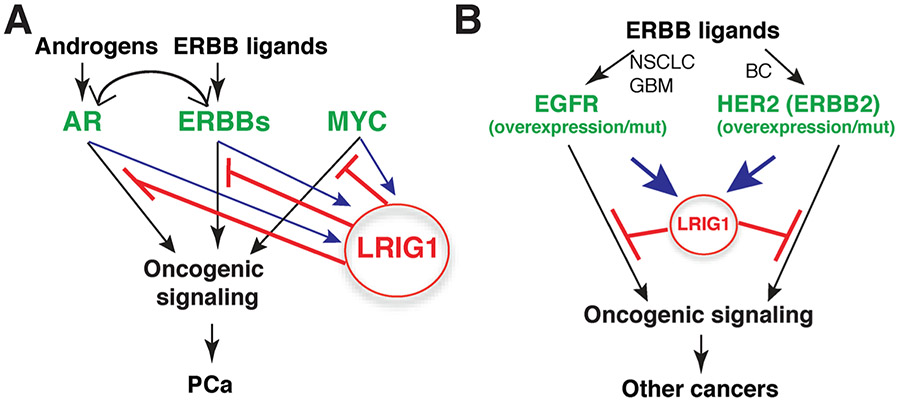
6. Conclusions and outstanding questions
Our discussions above render it clear that LRIG1 has evolved as a pleiotropic tumor suppressor feedback-induced by multiple oncogenic signals (Fig. 8; [100]). Therefore, in PCa, oncogenic signaling activated by androgen/AR, growth factors/ERBBs and MYC can all upregulate LRIG1, which in turn functions to antagonize tumorigenic process driven by these pathways (Fig. 8A; [100]). Likewise, in human cancers such as GBM and NSCLC, which are driven by EGFR overexpression and/or EGFR mutations, as well as in HER2+ breast cancer, ERBB activation by their cognate ligands leads to induction of LRIG1, which feedback retards the oncogenesis (Fig. 8B). There are still many outstanding questions. For example, are there biochemical mechanisms of LRIG1 in addition to neutralizing the ERBB and RTKs? In the context of reciprocal interactions between LRIG1 and MYC [100], how does LRIG1 negatively regulates MYC? In treatment-failed tumors such as CRPC, LRIG1 continues to be expressed at high levels but what transcription factors might be regulating LRIG1 in largely AR−/lo PCa cells? With respect to the LRIG1-ECD functioning as a pan-RTK therapeutic, is there a minimal domain (or motif) in the ECD that accounts for the major tumor-inhibitory effects? Answers to these, and other, questions will undoubtedly advance our understanding of the molecular mechanisms underpinning the tumor-suppressive functions of LRIG1 and facilitate the development of LRIG1-based biomarkers and therapeutics.
Fig. 8. LRIG1 as a pleiotropic feedback tumor suppressor.
(A) In PCa, oncogenic signaling from androgen/AR, ERBB/ligands and MYC induces LRIG1 expression and the upregulated LRIG1, in turn, antagonizes tumorigenesis driven by these pathways. Adapted with permission from [100]. (B) LRIG1 similarly functions as a feedback tumor suppressor in other ERBB-driven human cancers (see Text).
Acknowledgements
Work in the authors’ lab was supported, in part, by grants from the U. S National Institutes of Health (NIH) National Cancer Institute (NCI) R01CA237027, R01CA240290, R21CA237939, and R21CA218635, and Department of Defense (W81XWH-16-1-0575) (all to DGT), and by the Roswell Park Comprehensive Cancer Center (RPCCC) and NCI center grantP30CA016056. Q.L. was supported, in part, by the National Natural Science Foundation of China (No. 81802973). We acknowledge the support of RPCCC Shared Resources (SR) including BSGSR, ETM, FICSR, GSR, LASR, and PNSR. We also thank other Tang lab members for contributions to the LRIG1 project and for helpful discussions.
Footnotes
Declaration of Competing Interest
None.
References
- [1].Ghiglione C, Carraway KL 3rd, Amundadottir LT, et al. , The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis, Cell 96 (6) (1999) 847–856, 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- [2].Nilsson J, Vallbo C, Guo D, et al. , Cloning, characterization, and expression of human LIG1, Biochem. Biophys. Res. Commun 284 (5) (2001) 1155–1161, 10.1006/bbrc.2001.5092. [DOI] [PubMed] [Google Scholar]
- [3].Suzuki Y, Sato N, Tohyama M, et al. , cDNA doning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. LIG-1, a protein with leucine-rich repeats and immunoglobulin-like domains, J. Biol. Chem 271 (37) (1996) 22522–22527, 10.1074/jbc.271.37.22522. [DOI] [PubMed] [Google Scholar]
- [4].Holmlund C, Nilsson J, Guo D, et al. , Characterization and tissue-specific expression of human LRIG2, Gene 332 (2004) 35–43, 10.1016/j.gene.2004.02.002. [DOI] [PubMed] [Google Scholar]
- [5].Guo D, Holmlund C, Henriksson R, et al. , The LRIG gene family has three vertebrate paralogs widely expressed in human and mouse tissues and a homolog in Ascidiacea, Genomics 84 (1) (2004) 157–165, 10.1016/j.ygeno.2004.01.013. [DOI] [PubMed] [Google Scholar]
- [6].Kobe B, Kajava AV, The leucine-rich repeat as a protein recognition motif, Curr. Opin. Struct. Biol 11 (6) (2001) 725–732, 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- [7].Gur G, Rubin C, Katz M, et al. , LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation, EMBO J. 23 (16) (2004) 3270–3281, 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Laederich MB, Funes-Duran M, Yen L, et al. , The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases, J. Biol. Chem 279 (45) (2004) 47050–47056, 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- [9].Stutz MA, Shattuck DL, Laederich MB, et al. , LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII, Oncogene. 27 (43) (2008) 5741–5752, 10.1038/onc.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Neirinckx V, Hedman H, Niclou SP, Harnessing LRIG1-mediated inhibition of receptor tyrosine kinases for cancer therapy, Biochim. Biophys. Acta Rev. Cancer 1868 (1) (2017) 109–116, 10.1016/j.bbcan.2017.02.007. [DOI] [PubMed] [Google Scholar]
- [11].Shattuck DL, Miller JK, Laederich M, et al. , LRIG1 is a novel negative regulator of the Met receptor and opposes Met a Her2 synergy, Mol. Cell. Biol 27 (5) (2007) 1934–1946, 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ledda F, Bieraugel O, Fard SS, et al. , Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF, J. Neurosci 28 (1) (2008) 39–49, 10.1523/JNEUROSCI.2196-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].van Erp S, van den Heuvel DMA, Fujita Y, et al. , Lrig2 negatively regulates ectodomain shedding of axon guidance receptors by ADAM proteases, Dev. Cell 35 (5) (2015) 537–552, 10.1016/j.devcel.2015.11.008. [DOI] [PubMed] [Google Scholar]
- [14].Stuart HM, Roberts NA, Burgu B, et al. , LRIG2 mutations cause urofacial syndrome, Am. J. Hum. Genet 92 (2) (2013) 259–264, 10.1016/j.ajhg.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roberts NA, Hilton EN, Lopes FM, et al. , Lrig2 and Hpse2, mutated in urofacial syndrome, pattern nerves in the urinary bladder, Kidney Int. 95 (5) (2019) 1138–1152, 10.1016/j.kint.2018.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Abraira VE, Del Rio T, Tucker AF, et al. , Cross-repressive interactions between Lrig3 and netrin 1 shape the architecture of the inner ear, Development 135 (24) (2008) 4091–4099, 10.1242/dev.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Abraira VE, Satoh T, Fekete DM, et al. , Vertebrate Lrig3-ErbB interactions occur in vitro but are unlikely to play a role in Lrig3-dependent inner ear morphogenesis, PLoS One 5 (2) (2010) e8981, 10.1371/journal.pone.0008981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rafidi H, Mercado F 3rd, Astudillo M, et al. , Leucine-rich repeat and immunoglobulin domain-containing protein-1 (Lrig1) negative regulatory action toward ErbB receptor tyrosine kinases is opposed by leucine-rich repeat and immunoglobulin domain-containing protein 3 (Lrig3), J. Biol. Chem 288 (30) (2013) 21593–21605, 10.1074/jbc.M113.486050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Suzuki Y, Miura H, Tanemura A, et al. , Targeted disruption of LIG-1 gene results in psoriasiform epidermal hyperplasia, FEBS Lett. 521 (1-3) (2002) 67–71, 10.1016/s0014-5793(02)02824-7. [DOI] [PubMed] [Google Scholar]
- [20].Karlsson T, Mark EB, Henriksson R, et al. , Redistribution of LRIG proteins in psoriasis, J. Invest. Dermatol 128 (5) (2008) 1192–1195, 10.1038/sj.jid.5701175. [DOI] [PubMed] [Google Scholar]
- [21].Jaks V, Kasper M, Toftgård R, The hair follicle-a stem cell zoo, Exp. Cell Res 316 (8) (2010) 1422–1428, 10.1016/j.yexcr.2010.03.014. [DOI] [PubMed] [Google Scholar]
- [22].Schepeler T, Page ME, Jensen KB, Heterogeneity and plasticity of epidermal stem cells, Development. 141 (13) (2014) 2559–2567, 10.1242/dev.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Alonso L, Fuchs E, The hair cycle, J. Cell. Sci 119 (Pt 3) (2006) 391–393, 10.1242/jcs02793. [DOI] [PubMed] [Google Scholar]
- [24].Jensen KB, Watt FM, Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence, Proc. Natl. Acad. Sci. U.S.A 103 (32) (2006) 11958–11963, 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jensen KB, Collins CA, Nascimento E, et al. , Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis, Cell Stem Cell 4 (5) (2009) 427–439, 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Page ME, Lombard P, Ng F, et al. , The epidermis comprises autonomous compartments maintained by distinct stem cell populations, Cell Stem Cell 13 (4) (2013) 471–482, 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Horsley V, O’Carroll D, Tooze R, et al. , Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland, Cell. 126 (3) (2006) 597–609, 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gehart H, Clevers H, Tales from the crypt: new insights into intestinal stem cells, Nat. Rev. Gastroenterol. Hepatol 16 (1) (2019) 19–34, 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- [29].Powell AE, Wang Y, Li Y, et al. , The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor, Cell. 149 (1) (2012) 146–158, 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wong VW, Stange DE, Page ME, et al. , Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling, Nat. Cell Biol 14 (4) (2012) 401–408, 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sato T, van Es JH, Snippert HJ, et al. , Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts, Nature. 469 (7330) (2011) 415–418, 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kondo J, Powell AE, Wang Y, et al. , LRIG1 regulates ontogeny of smooth muscle-derived subsets of interstitial cells of Cajal in mice, Gastroenterology. 149 (2) (2015) 407–419, 10.1053/j.gastro.2015.04.018, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mills JC, Shivdasani RA, Gastric epithelial stem cells, Gastroenterology 140 (2) (2011) 412–424, 10.1053/j.gastro.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Barker N, Huch M, Kujala P, et al. , Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro, Cell Stem Cell 6 (1) (2010) 25–36, 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- [35].Stange DE, Koo BK, Huch M, et al. , Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium, Cell. 155 (2) (2013) 357–368, 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Choi E, Petersen CP, Lapierre LA, et al. , Dynamic expansion of gastric mucosal doublecortin-like kinase 1-expressing cells in response to parietal cell loss is regulated by gastrin, Am. J. Pathol. 185 (8) (2015) 2219–2231, 10.1016/j.ajpath.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Choi E, Lantz TL, Vlacich G, et al. , Lrig1+ gastric isthmal progenitor cells restore normal gastric lineage cells during damage recovery in adult mouse stomach, Gut. 67 (9) (2018) 1595–1605, 10.1136/gutjnl-2017-313874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schweiger PJ, Clement DL, Page ME, et al. , Lrig1 marks a population of gastric epithelial cells capable of long-term tissue maintenance and growth in vitro, Sci. Rep 8 (1) (2018) 15255, 10.1038/s41598-018-33578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Byrd KM, Piehl NC, Patel JH, et al. , Heterogeneity within stratified epithelial stem cell populations maintains the oral mucosa in response to physiological stress, Cell Stem Cell 25 (6) (2019) 814–829, 10.1016/j.stem.2019.11.005, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gage FH, Temple S, Neural stem cells: generating and regenerating the brain, Neuron. 80 (3) (2013) 588–601, 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- [41].Alsina FC, Hita FJ, Fontanet PA, et al. , Lrig1 is a cell-intrinsic modulator of hippocampal dendrite complexity and BDNF signaling, EMBO Rep. 17 (4) (2016) 601–616, 10.15252/embr.201541218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Trinchero MF, Buttner KA, Sulkes Cuevas JN, et al. , High plasticity of new granule cells in the aging Hippocampus, Cell Rep. 21 (5) (2017) 1129–1139, 10.1016/j.celrep.2017.09.064. [DOI] [PubMed] [Google Scholar]
- [43].Lugert S, Basak O, Knuckles P, et al. , Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging, Cell Stem Cell 6 (5) (2010) 445–456, 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- [44].Codega P, Silva-Vargas V, Paul A, et al. , Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche, Neuron. 82 (3) (2014) 545–559, 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Morizur L, Chicheportiche A, Gauthier LR, et al. , Distinct molecular signatures of quiescent and activated adult neural stem cells reveal specific interactions with their microenvironment, Stem Cell Reports 11 (2) (2018) 565–577, 10.1016/j.stemcr.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nam HS, Capecchi MR, Lrig1 expression prospectively identifies stem cells in the ventricular-subventricular zone that are neurogenic throughout adult life, Neural Dev. 15 (1) (2020) 3, 10.1186/s13064-020-00139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sacchetti M, Rama P, Bruscolini A, et al. , Limbal stem cell transplantation: clinical results, limits, and perspectives, Stem Cells Int. 2018 (2018), 8086269, 10.1155/2018/8086269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Daya SM, Conjunctival-limbal autograft, Curr. Opin. Ophthalmol 28 (4) (2017) 370–376, 10.1097/ICU.0000000000000385. [DOI] [PubMed] [Google Scholar]
- [49].Barrandon Y, Green H, Three clonal types of keratinocyte with different capacities for multiplication, Proc Natl Acad Sci U S A. 84 (8) (1987) 2302–2306, 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pellegrini G, Golisano O, Paterna P, et al. , Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface, J. Cell Biol 145 (4) (1999) 769–782, 10.1083/jcb.145.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nakamura T, Hamuro J, Takaishi M, et al. , LRIG1 inhibits STAT3-dependent inflammation to maintain corneal homeostasis, J. Clin. Invest 124 (1) (2014) 385–397, 10.1172/JCI71488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xie HT, Sullivan DA, Chen D, et al. , Biomarkers for progenitor and differentiated epithelial cells in the human meibomian gland, Stem Cells Transl. Med 7 (12) (2018) 887–892, 10.1002/sctm.18-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Diagnosis ∣ Severe prognathic malocclusion, Lab Anim. (NY) 36 (1) (2007) 22–23, 10.1038/laban0107-22.17885660 [DOI] [Google Scholar]
- [54].Seidel K, Marangoni P, Tang C, et al. , Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis, Elife. 6 (2017) e24712, 10.7554/eLife.24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell 144 (5) (2011) 646–674, 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [56].Du Z, Lovly CM, Mechanisms of receptor tyrosine kinase activation in cancer, Mol. Cancer 17 (1) (2018) 58, 10.1186/s12943-018-0782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hedman H, Nilsson J, Guo D, et al. , Is LRIG1 a tumour suppressor gene at chromosome 3p14.3? Acta Oncol. (Madr) 41 (4) (2002) 352–354, 10.1080/028418602760169398. [DOI] [PubMed] [Google Scholar]
- [58].Wang Y, Poulin EJ, Coffey RJ, LRIG1 is a triple threat: ERBB negative regulator, intestinal stem cell marker and tumour suppressor, Br. J. Cancer 108 (9) (2013) 1765–1770, 10.1038/bjc.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rouam S, Moreau T, Broët P, Identifying common prognostic factors in genomic cancer studies: a novel index for censored outcomes, BMC Bioinformatics 11 (2010) 150, 10.1186/1471-2105-11-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ljuslinder I, Golovleva I, Palmqvist R, et al. , LRIG1 expression in colorectal cancer, Acta Oncol. (Madr) 46 (8) (2007) 1118–1122, 10.1080/02841860701426823. [DOI] [PubMed] [Google Scholar]
- [61].Kou C, Zhou T, Han X, et al. , LRIG1, a 3p tumor suppressor, represses EGFR signaling and is a novel epigenetic silenced gene in colorectal cancer, Biochem. Biophys. Res. Commun 464 (2) (2015) 519–525, 10.1016/j.bbre.2015.06.173. [DOI] [PubMed] [Google Scholar]
- [62].Powell AE, Vlacich G, Zhao ZY, et al. , Inducible loss of one Apc allele in Lrig1-expressing progenitor cells results in multiple distal colonic tumors with features of familial adenomatous polyposis, Am. J. Physiol. Gastrointest. Liver Physiol 307 (1) (2014) G16–G23, 10.1152/ajpgi.00358.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Preston JL, Stiffler N, Epigenetic loss of heterozygosity of Apc and an inflammation-associated mutational signature detected in Lrig1+/−driven murine colonic adenomas, BMC Cancer 20 (1) (2020) 126, 10.1186/s12885-020-6616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wroblewski LE, Choi E, Petersen C, et al. , Targeted mobilization of Lrig1+ gastric epithelial stem cell populations by a carcinogenic Helicobacter pylori type IV secretion system, Proc. Natl. Acad. Sci. U.S.A 116 (39) (2019) 19652–19658, 10.1073/pnas.1903798116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yu S, Yang M, Lim KM, et al. , Expression of LRIG1, a negative regulator of EGFR, is dynamically altered during different stages of gastric carcinogenesis, Am. J. Pathol 188 (12) (2018) 2912–2923, 10.1016/j.ajpath.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Peterson SC, Eberl M, Vagnozzi AN, et al. , Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches, Cell Stem Cell 16 (4) (2015) 400–412, 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Quist SR, Eckardt M, Kriesche A, et al. , Expression of epidermal stem cell markers in skin and adnexal malignancies, Br. J. Dermatol 175 (3) (2016) 520–530, 10.1111/bjd.14494. [DOI] [PubMed] [Google Scholar]
- [68].Pünchera J, Barnes L, Kaya G, Lrig1 Expression in human sebaceous gland tumors, Dermatopathology Basel (Basel) 3 (2) (2016) 44–54, 10.1159/000446427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tanemura A, Nagasawa T, Inui S, et al. , LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: immunohistochemical analysis for 38 cases, Dermatol. Surg 31 (4) (2005) 423–430, 10.1111/j.1524-4725.2005.31108. [DOI] [PubMed] [Google Scholar]
- [70].Jensen KB, Jones J, Watt FM, A stem cell gene expression profile of human squamous cell carcinomas, Cancer Lett. 272 (1) (2008) 23–31, 10.1016/j.canlet.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lindström AK, Ekman K, Stendahl U, et al. , LRIG1 and squamous epithelial uterine cervical cancer: correlation to prognosis, other tumor markers, sex steroid hormones, and smoking, Int. J. Gynecol. Cancer 18 (2) (2008) 312–317, 10.1111/j.1525-1438.2007.01021.x. [DOI] [PubMed] [Google Scholar]
- [72].Nagata M, Nakamura T, Sotozono C, et al. , LRIG1 as a potential novel marker for neoplastic transformation in ocular surface squamous neoplasia, PLoS One 9 (4) (2014) e93164, 10.1371/journal.pone.0093164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lu L, Teixeira VH, Yuan Z, et al. , LRIG1 regulates cadherin-dependent contact inhibition directing epithelial homeostasis and pre-invasive squamous cell carcinoma development, J. Pathol 229 (4) (2013) 608–620, 10.1002/path.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xiang S, Chen H, Luo X, et al. , Isoliquiritigenin suppresses human melanoma growth by targeting miR-301b/LRIG1 signaling, J. Exp. Clin. Cancer Res 37 (1) (2018) 184, 10.1186/s13046-018-0844-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Li W, Zhou Y, LRIG1 acts as a critical regulator of melanoma cell invasion, migration, and vasculogenic mimicry upon hypoxia by regulating EGFR/ERK-triggered epithelial-mesenchymal transition, Biosci. Rep 39 (1) (2019), 10.1042/BSR20181165.BSR20181165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ghasimi S, Haapasalo H, Eray M, et al. , Immunohistochemical analysis of LRIG proteins in meningiomas: correlation between estrogen receptor status and LRIG expression, J. Neurooncol 108 (3) (2012) 435–441, 10.1007/s11060-012-0856-x. [DOI] [PubMed] [Google Scholar]
- [77].Louis DN, Perry A, Reifenberger G, et al. , The 2016 World Health Organization classification of tumors of the central nervous system: a summary, Acta Neuropathol. 131 (6) (2016) 803–820, 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- [78].Guo D, Nilsson J, Haapasalo H, et al. , Perinuclear leucine-rich repeats and immunoglobulin-like domain proteins (LRIG1-3) as prognostic indicators in astrocytic tumors, Acta Neuropathol. 111 (3) (2006) 238–246, 10.1007/s00401-006-0032-5. [DOI] [PubMed] [Google Scholar]
- [79].Ye F, Gao Q, Xu T, et al. , Upregulation of LRIG1 suppresses malignant glioma cell growth by attenuating EGFR activity, J. Neurooncol 94 (2) (2009) 183–194, 10.1007/s11060-009-9836-1. [DOI] [PubMed] [Google Scholar]
- [80].Xie R, Yang H, Xiao Q, et al. , Downregulation of LRIG1 expression by RNA interference promotes the aggressive properties of glioma cells via EGFR/Akt/c-Myc activation, Oncol. Rep 29 (1) (2013) 177–184, 10.3892/or.2012.2102. [DOI] [PubMed] [Google Scholar]
- [81].Mao F, Wang B, Xiao Q, et al. , A role for LRIG1 in the regulation of malignant glioma aggressiveness, Int. J. Oncol 42 (3) (2013) 1081–1087, 10.3892/ijo.2013.1776. [DOI] [PubMed] [Google Scholar]
- [82].Cheng SQ, Fan HY, Xu X, et al. , Over-expression of LRIG1 suppresses biological function of pituitary adenoma via attenuation of PI3K/AKT and Ras/Raf/ERK pathways in vivo and in vitro, J. Huazhong Univ. Sci. Technol. Med. Sci 36 (4) (2016) 558–563, 10.1007/s11596-016-1625-4. [DOI] [PubMed] [Google Scholar]
- [83].Mao F, Holmlund C, Faraz M, et al. , Lrig1 is a haploinsufficient tumor suppressor gene in malignant glioma, Oncogenesis. 7 (2) (2018) 13, 10.1038/s41389-017-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kvarnbrink S, Karlsson T, Edlund K, et al. , LRIG1 is a prognostic biomarker in non-small cell lung cancer, Acta Oncol. (Madr) 54 (8) (2015) 1113–1119, 10.3109/0284186X.2015.1021427. [DOI] [PubMed] [Google Scholar]
- [85].An Y, Zhao Z, Ou P, et al. , Expression of LRIG1 is associated with good prognosis for human non-small cell lung cancer, Bull. Sch. Med. Md 94 (47) (2015) e2081, 10.1097/MD.0000000000002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Torigoe H, Yamamoto H, Sakaguchi M, et al. , Tumor-suppressive effect of LRIG1, a negative regulator of ErbB, in non-small cell lung cancer harboring mutant EGFR, Carcinogenesis. 39 (5) (2018) 719–727, 10.1093/carcin/bgy044. [DOI] [PubMed] [Google Scholar]
- [87].Thomasson M, Hedman H, Ljungberg B, et al. , Gene expression pattern of the epidermal growth factor receptor family and LRIG1 in renal cell carcinoma, BMC Res. Notes 5 (2012) 216, 10.1186/1756-0500-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Miller JK, Shattuck DL, Ingalla EQ, et al. , Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer, Cancer Res. 68 (20) (2008) 8286–8294, 10.1158/0008-5472.CAN-07-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Krig SR, Frietze S, Simion C, et al. , Lrig1 is an estrogen-regulated growth suppressor and correlates with longer relapse-free survival in ERα-positive breast cancer, Mol. Cancer Res. 9 (10) (2011) 1406–1417, 10.1158/1541-7786.MCR-11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Yokdang N, Hatakeyama J, Wald JH, et al. , LRIG1 opposes epithelial-to-mesenchymal transition and inhibits invasion of basal-like breast cancer cells, Oncogene 35 (22) (2016) 2932–2947, 10.1038/onc.2015.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ljuslinder I, Maimer B, Golovleva I, et al. , Increased copy number at 3p14 in breast cancer, Breast Cancer Res. 7 (5) (2005) R719–R727, 10.1186/bcr1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ljuslinder I, Golovleva I, Henriksson R, et al. , Co-incidental increase in gene copy number of ERBB2 and LRIG1 in breast cancer, Breast Cancer Res. 11 (3) (2009) 403, 10.1186/bcr2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Thompson PA, Ljuslinder I, Tsavachidis S, et al. , Loss of LRIG1 locus increases risk of early and late relapse of stage I/II breast cancer, Cancer Res. 74 (11) (2014) 2928–2935, 10.1158/0008-5472.CAN-13-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Faraz M, Tellström A, Ardnor CE, et al. , LRIG1 gene copy number analysis by ddPCR and correlations to clinical factors in breast cancer, BMC Cancer 20 (1) (2020) 459, 10.1186/s12885-020-06919-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Boutros PC, Fraser M, Harding NJ, et al. , Spatial genomic heterogeneity within localized, multifocal prostate cancer, Nat. Genet 47 (7) (2015) 736–745, 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- [96].Gundem G, Van Loo P, Kremeyer B, et al. , The evolutionary history of lethal metastatic prostate cancer, Nature. 520 (7547) (2015) 353–357, 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hong MK, Macintyre G, Wedge DC, et al. , Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer, Nat. Commun 6 (2015) 6605, 10.1038/ncomms7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Li Q, Deng Q, Chao HP, et al. , Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses, Nat. Commun 9 (1) (2018) 3600, 10.1038/s41467-018-06067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Thomasson M, Wang B, Hammarsten P, et al. , LRIG1 and the liar paradox in prostate cancer: a study of the expression and clinical significance of LRIG1 in prostate cancer, Int. J. Cancer 128 (12) (2011) 2843–2852, 10.1002/ijc.25820. [DOI] [PubMed] [Google Scholar]
- [100].Li Q, Liu B, Chao HP, et al. , LRIG1 is a pleiotropic androgen receptor-regulated feedback tumor suppressor in prostate cancer, Nat. Commun 10 (1) (2019) 5494, 10.1038/s41467-019-13532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhang D, Park D, Zhong Y, et al. , Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer, Nat. Commun 7 (2016) 10798, 10.1038/ncomms10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Febbo PG, Lowenberg M, Thorner AR, et al. , Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer, J. Urol 173 (5) (2005) 1772–1777, 10.1097/01.ju.0000155845.44729.ba. [DOI] [PubMed] [Google Scholar]
- [103].Tan PY, Chang CW, Chng KR, et al. , Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival, Mol. Cell. Biol 32 (2) (2012) 399–414, 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Traish AM, Morgentaler A, Epidermal growth factor receptor expression escapes androgen regulation in prostate cancer: a potential molecular switch for tumour growth, Br. J. Cancer 101 (12) (2009) 1949–1956, 10.1038/sj.bjc.6605376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Ellwood-Yen K, Graeber TG, Wongvipat J, et al. , Myc-driven murine prostate cancer shares molecular features with human prostate tumors, Cancer Cell 4 (3) (2003) 223–238, 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- [106].Zhang D, Zhao S, Li X, et al. , Prostate luminal progenitor cells in development and cancer, Trends Cancer 4 (11) (2018) 769–783, 10.1016/j.trecan.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Tomlins SA, Mehra R, Rhodes DR, et al. , Integrative molecular concept modeling of prostate cancer progression, Nat. Genet 39 (1) (2007) 41–51, 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- [108].Rycaj K, Cho EJ, Liu X, et al. , Longitudinal tracking of subpopulation dynamics and molecular changes during LNCaP cell castration and identification of inhibitors that could target the PSA−/lo castration-resistant cells, Oncotarget 7 (12) (2016) 14220–14240, 10.18632/oncotarget.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Goldoni S, Iozzo RA, Kay P, et al. , A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity, Oncogene 26 (3) (2007) 368–381, 10.1038/sj.onc.1209803. [DOI] [PubMed] [Google Scholar]
- [110].Johansson M, Oudin A, Tiemann K, et al. , The soluble form of the tumor suppressor Lrig1 potently inhibits in vivo glioma growth irrespective of EGF receptor status, Neuro Oncol. 15 (9) (2013) 1200–1211, 10.1093/neuonc/not054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Neirinckx V, Hau AC, Schuster A, et al. , The soluble form of pan-RTK inhibitor and tumor suppressor LRIG1 mediates downregulation of AXL through direct protein-protein interaction in glioblastoma, Neurooncol Adv. 1 (1) (2019), 10.1093/noajnl/vdz024 vdz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Yi W, Holmlund C, Nilsson J, et al. , Paracrine regulation of growth factor signaling by shed leucine-rich repeats and immunoglobulin-like domains 1, Exp. Cell Res 317 (4) (2011) 504–512, 10.1016/j.yexcr.2010.11.005. [DOI] [PubMed] [Google Scholar]
- [113].Lichtenthaler SF, Lemberg MK, Fluhrer R, Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments, EMBO J. 37 (15) (2018) e99456, 10.15252/embj.201899456. [DOI] [PMC free article] [PubMed] [Google Scholar]