Abstract
ATG8 family proteins are critical players in autophagy, a cytoprotective process that mediates degradation of cytosolic cargo. During autophagy, ATG8s conjugate to autophagosome membranes to facilitate cargo recruitment, autophagosome biogenesis, transport, and fusion with lysosomes for cargo degradation. In addition to these canonical functions, recent reports demonstrate that ATG8s are also delivered to single-membrane organelles, which leads to highly divergent degradative or secretory fates, vesicle maturation, and cargo specification. The association of ATG8s with different vesicles involves complex regulatory mechanisms still to be fully elucidated. Whether individual ATG8 family members play unique canonical or non-canonical roles also remains unclear. This review summarizes the many open molecular questions regarding ATG8s that are only beginning to be unraveled.
Keywords: LC3, GABARAP, non-canonical autophagy, LC3-associated phagocytosis, multivesicular bodies, unconventional secretion
ATG8s: bona fide components of macroautophagy
Autophagy is a conserved catabolic process with crucial roles in cellular and organismal homeostasis [1]. Autophagy facilitates the turnover of diverse cytoplasmic components by targeting them to acidic, hydrolase-containing lysosomes for degradation and recycling. During macroautophagy, one of three main forms of autophagy, cargo are engulfed into specialized double-membrane vesicles called autophagosomes that fuse with lysosomes to degrade cargo [2]. Seminal studies in yeast identified the core autophagy machinery (see Glossary) required for autophagy (ATGs) [3]. Arguably, the identification of the autophagy-related proteins ATG8s (Atg8 in yeast, and members of the LC3 and GABARAP families in mammals; Box 1) as integral components of autophagosomes [4–6], was one of the most important discoveries, as it allowed the monitoring of the autophagy process, thereby ushering the expansion of the autophagy field.
Box 1: The ATG8 family of proteins.
While yeast and other simple eukaryotes have a single ATG8 isoform, this family is diversified in mammals into two subfamilies: LC3 and GABARAP [91]. In humans, ATG8s are comprised of seven members, LC3A, LC3B, LC3B2, LC3C, GABARAP, GABARAPL1, and GABARAPL2. All ATG8s are small cytosolic proteins sharing a ubiquitin-like structural folding preceded by two N-terminal alpha-helices. The 3D-structure of these proteins defines hydrophobic pockets that comprise a docking surface known as the LC3-interating region (LIR)-docking site (LDS) (Figure I), which allows the binding of LIR-containing proteins, including members of the autophagy core machinery, cargo receptors, and adaptor proteins [19]. Recently, an additional protein-binding interface in ATG8s named ubiquitin-interacting motif (UIM), has also been described to participate in the recruitment of autophagy-related proteins [92]. Moreover, other interactors can bind to ATG8s through alternative regions. For example, Lamin B which interacts with LC3B through a sequence in the N-terminus of LC3B [93]. While ATG8s perform most of their autophagy-related functions once they are conjugated to autophagosome membranes (see below), only a very small number of ATG8 binding partners are known to preferentially bind to this form. ATG8s are synthesized as pro-proteins cleaved by the ATG4 family of proteases to expose a C-terminal glycine. Subsequently, ATG8s are conjugated to the lipid phosphatidylethanolamine (PE) in the growing phagophore and ultimately in autophagosome membranes via two ubiquitin-like conjugation systems [7]. In the first pathway, ATG7 (E1-like) and ATG10 (E2-like) catalyze ATG5-ATG12 conjugation, and this dimer recruits ATG16L1 to form the ATG5-ATG12:ATG16L1 complex. The second conjugation system involves ATG7 (E1-like) and ATG3 (E2-like) to mediate PE-lipidation of ATG8s. In this pathway, the ATG5-ATG12:ATG16L1 complex is proposed to confer an E3-like function that targets PE-lipidated ATG8s to the sites of autophagosome formation via interaction with members of the core autophagy machinery, such as FIP200 and WIPI [7]. ATG4 proteases can ultimately deconjugate ATG8s bound to the outer autophagosome membrane, which can be reused in another round of autophagy. In a so-called non-canonical pathway, the same conjugation systems via ATG16L1 can alternatively target lipidated ATG8s to single-membrane vesicles with degradative and/or secretory functions (see section on Regulatory mechanisms operating on ATG8 proteins that specify canonical versus non-canonical functions).
Box 1 Figure I: 3-D structure of LC3B.
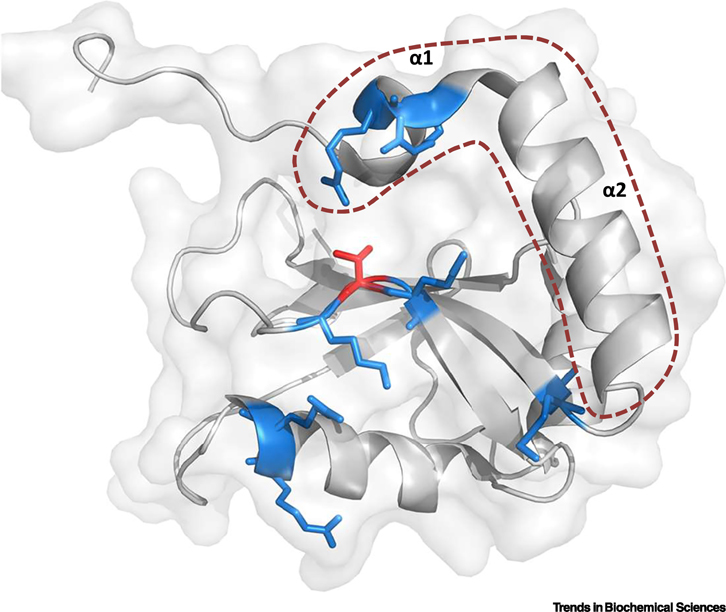
Crystal structure of LC3B showing the typical ubiquitin-like folding described for ATG8s (PDB ID 3VTU). The two consecutive N-terminal alpha helices are highlighted by an orange dotted line. Basic amino acids shown in blue form part of the LIR-docking site (LDS) and participate in the binding of LIR-containing proteins. Highlighted in red is LC3B Threonine 50, a key residue located within the LDS, whose phosphorylation regulates protein-protein interactions and is crucial for the autophagy process (see section on Regulatory mechanisms operating on ATG8 proteins that specify canonical versus non-canonical functions). Figure created with the software PyMOL.
ATG8s associate with the phagophore, the autophagosomal precursor, and remain attached to both the inner and the outer autophagosome membranes once the vesicle is fully sealed (Figure 1, Key Figure, Panel A). Upon lysosomal fusion, ATG8s on the outer membrane are deconjugated, while lysosomal hydrolases degrade the inner pool of ATG8s [1]. Notably, a third of all ATGs identified are dedicated to the processing and conjugation of ATG8 proteins to autophagosome membranes, reinforcing the importance of ATG8s in autophagy [7]. Inhibiting ATG8 conjugation leads to inefficient autophagosome closure, lysosomal fusion, and by extension, cargo degradation [8], while genetic deletion of ATG8s leads to smaller autophagosomes that are unable to fuse with lysosomes [9].
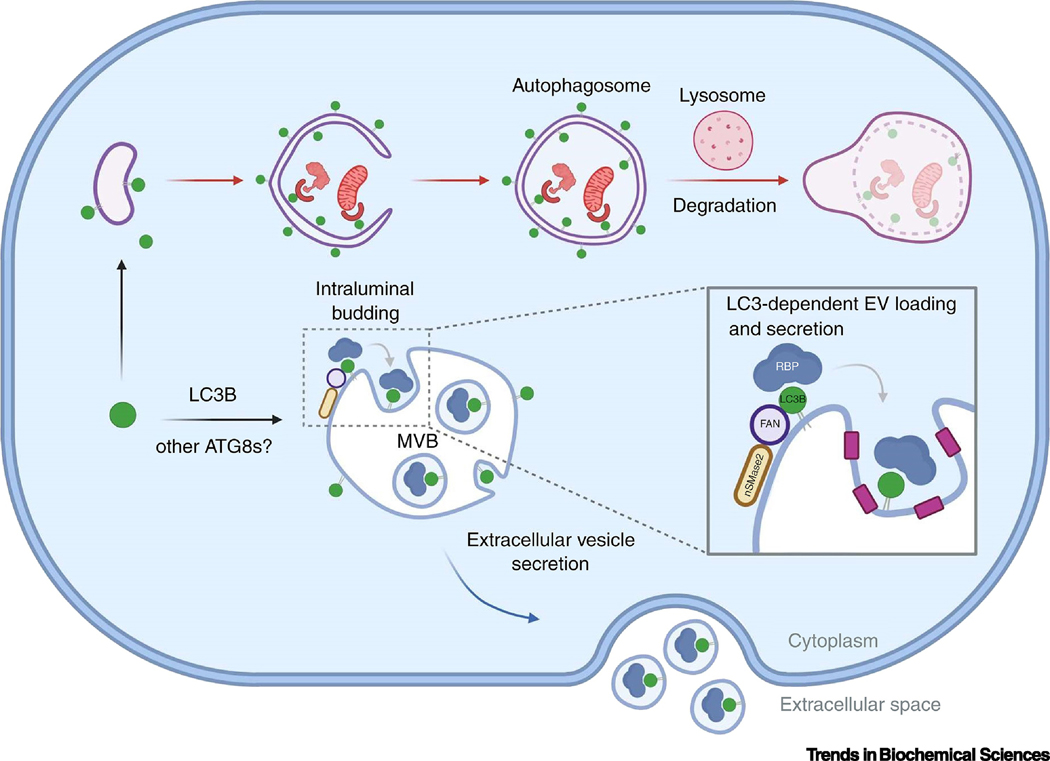
Figure 1. Key Figure: Overview of degradative and secretory roles of ATG8 proteins described in this review.
ATG8s (green circles) incorporate into intracellular vesicles with degrative fate, including autophagosomes (Panel A) and diverse vesicles of endosomal origin (Panels B and C). During macroautophagy (Panel A), ATG8s associate with the outer- and inner membrane of autophagosomes via canonical conjugation mechanisms (orange text boxes). In the inner membrane, ATG8s interact with receptors to recruit cytosolic cargo, such as defective mitochondria and protein aggregates. On the outer membrane, ATG8s bind adaptor proteins that bring the transport and fusion machinery, allowing directional retrograde transport of autophagosomes and fusion with lysosomes for cargo degradation. Via non-canonical conjugation mechanisms (purple boxes), ATG8 also incorporate into phagosomes containing components of extracellular origin such as bacteria or apoptotic bodies (Panel B). During this process, called LC3-associated phagocytosis (LAP) ATG8s associate to the cytosol-facing part of the membrane and may play roles in fusion and transport for lysosomal degradation. In addition, ATG8s associate to multivesicular bodies (MVB) to recruit receptors and facilitate cargo degradation in a process called endosomal microautophagy (eMI). Interestingly, ATG8s can also associate with secretory vesicles: small extracellular vesicles and secretory autophagosomes, via potentially non-canonical and canonical conjugation machineries, respectively. Within MVB, ATG8s participate in the inward budding of small extracellular vesicles and recruit RNA-binding proteins by direct interaction, to allow their subsequent secretion upon fusion of the MVB with the plasma membrane, during a process called LC3-dependent extracellular vesicle loading and secretion (LDELS) (Panel D). Within secretory autophagosomes, ATG8s may mostly participate in the recruitment of specific transport and fusion machinery, which can facilitate directional anterograde trafficking towards the plasma membrane for fusion and cargo release to the extracellular media. Some cargo, such as IL-1β can be incorporated in between the inner- and outer membranes of autophagosomes for unconventional secretion (Panel E). Finally, ATG8s incorporate into amyloid-β-containing endosomes (Panel C). During LC3-associated endocytosis (LANDO), endosomes containing amyloid-β are endocytosed (1), then amyloid-β is delivering to lysosomes for degradation (2) and finally fusion of endosomes with the plasma membrane allows the recycling of amyloid-β receptor back to the cell surface (3). See text for further details.
How do ATG8s facilitate autophagy? Interestingly, ATG8s have membrane tethering and fusogenic capacities in vitro conferred by two conserved N-terminal alpha helices [10], proposed to drive phagophore expansion within cells. In addition, seminal discoveries revealed that ATG8s act as nodes for protein-protein interactions [11], typically involving an LC3-interacting region (LIR)-docking site (LDS) located in the ATG8 protein [12] binding to a stretch of amino acids called the LIR of the interacting protein (Box 1). These protein-protein interactions allow ATG8s to recruit other autophagy machinery components at the phagophore to promote autophagosome formation [13]. ATG8 interactions are likely to differ between the inner and outer membranes of both developing and mature autophagosomes, thereby serving distinct functions. In the inner membrane, ATG8s interact with specialized proteins called cargo receptors, key mediators of selective autophagy (Figure 1). These proteins bind different cargos, from protein aggregates to damaged and surplus organelles and pathogens, which are sequestered and ultimately degraded [14]. In the outer membrane, ATG8s bind adaptor proteins with known roles in vesicle transport and fusion [15] (Figure 1), such as FYCO1 and JIP1, which promote directional transport of autophagosomes within cells via association to molecular motors [16,17], and PLEKHM1 which facilitates fusion of autophagosome-lysosome membranes via recruitment of components of the homotypic fusion and protein sorting (HOPS)-tethering complex [18].
Is there any functional specialization for the members of the ATG8 family, among all these different protein-binding events? In mammalian cells, LC3 and GABARAP members display overlapping interactors, as determined by affinity-purification assays. However, in general terms, GABARAP subfamily members appear mostly dedicated to the recruitment of core autophagy machinery components as well as proteins involved in fusion with lysosomes, evidenced by the preferential binding of GABARAPs to ULK1 and ATG13, and PLEKHM1 proteins, respectively [19]. In contrast, the LC3 subfamily preferential binds to cargo receptors, such as p62, and vesicle transport-related adaptor proteins, such as FYCO1 [19]. Recent studies of ATG8 deletion in mammalian cells reinforce that GABARAP members are essential for autophagosome-lysosome fusion, while LC3 members recruit specialized cargo [9]. Despite these general observations, certain cargo receptors, such as NIX, have enhanced binding to GABARAPs [19]. How the topologically different protein-protein interactions displayed by ATG8s in autophagosome membranes are organized and achieved remains unclear, although a recent publication started to shed light into interactions specific to the inner side of autophagosomes [20]. In the absence of results showing a preference for any ATG8 member for the inner versus the outer membrane of autophagosomes, we speculate that protein-protein interactions with individual ATG8 family members may be regulated via post-translational modifications, such as protein phosphorylation (see section on Regulatory mechanisms operating on ATG8 proteins that specify canonical versus non-canonical functions).
The key roles that ATG8s play in macroautophagy, described above, are known as their canonical functions, and by same means the process can be referred to as canonical autophagy. However, a growing number of studies are revealing alternative functions for ATG8s in other processes, which are also highly relevant in health and disease. These alternative roles are here referred to as non-canonical functions of ATG8s, and for simplicity, the pathways by extension as non-canonical autophagy, although they encompass a quite diverse array of functional processes. The physiological relevance and the molecular machinery implicated in non-canonical autophagy is currently an active topic of research. In the sections below, we describe the relevance of ATG8s in non-canonical pathways comprising: alternative degradative pathways, such as LC3-associated phagocytosis, LC3-associated endocytosis, endosomal microautophagy; the interplay between ATG8s and viruses; and the functions of ATG8s in unconventional secretory pathways via autophagosomes or small extracellular vesicles.
Recruitment of ATG8s to single-membrane vesicles with degradative fates
Besides the canonical roles that ATG8s play in macroautophagy, accumulating evidence demonstrates that ATG8s can also be incorporated into different single-membrane vesicles with degradative functions (Figure 1, Panels B and C). These include phagosomes, macropinosomes, endosomes, and entotic vacuoles [21,22]. These ATG8-positive vesicles ultimately fuse with lysosomes to degrade cargo, which in this case, is of extracellular origin. Interestingly, the formation of these single-membrane degradative vesicles is independent of the autophagy-initiation machinery involved with phagophore formation and relies instead on the invagination of the cell plasma membrane via different molecular pathways. However, the conjugation of ATG8s to membranes requires the same ubiquitin-like conjugation systems that operate during canonical macroautophagy, with ATG16L1 playing an essential role in recognition of these vesicles [23] (Box 1).
ATG8s play critical roles in phagosomes and endosomes. Phagosomes formed via the so-called LC3-associated phagocytosis (LAP) require the ATG8 conjugation machinery for their acidification and cargo degradation. In the absence of the conjugation machinery, inefficient cargo degradation leads to detrimental pro-inflammatory responses by phagocytic cells [24]. The ATG8 conjugation machinery also appears to be crucial in a mechanistically distinct process called LC3-associated endocytosis (LANDO) (Figure 1, Panel C). In microglia, LANDO allows the uptake of amyloid-β protein (Aβ) from the extracellular media to facilitate its degradation and the recycling of Aβ receptor in the cell surface, preventing plaque buildup in the neural tissue that may lead to Alzheimer’s disease [22].
To understand the potential functions that ATG8s play in these single-membrane vesicles, it is important to consider that ATG8s may associate exclusively with their cytosol-facing side (Figure 1), the only side accessible to the ATG8 conjugation machinery. We, therefore, speculate that ATG8s do not specify cargo within these vesicles because of their topological orientation, in contrast to autophagosomes. Because formation of phagosomes, macropinosomes, and endosomes can be ATG8-independent processes, and because ATG8 conjugation to phagosomes occurs only after the vesicle is fully formed [25], ATG8s are unlikely to play a primary role in the biogenesis of these vesicles. Rather, ATG8s may mediate the recruitment of transport- and fusion-related factors to facilitate lysosomal degradation of cargo. LC3-positive phagosomes are known to recruit the transport-related protein FYCO1, and subsequently acquire endolysosomal markers during the process of acidification [26]. An exception to the above-described functional roles and topological distribution of ATG8s within single-membrane vesicles with degradative fates may be endosomal microautophagy (eMI) (Figure 1, Panel B). This process allows the delivery of cytosolic cargo within endosomes via inward budding mediated by the endosomal complexes required for transport (ESCRT) machinery. LC3B and GABARAPL2 participate in this process, and likely facilitate the recruitment of cargo receptors including p62, NDP52, and NBR1 to deliver material into endosomal cisternae [27]. A similar mechanism seems to contribute to the selective turnover of lysosome membrane components, including the ion-channel Mucolipin 1 (also known as TRPML1) [28].
It is presently unclear whether functional specialization exists among ATG8 family members in autophagy-related degradative pathways. Most studies have focused on the incorporation of LC3 members into the membrane of single-membrane degradative vesicles, with less data implicating GABARAPs in these non-canonical processes. However, early evidence points to other members of the ATG8 family, such as GABARAPL2 associating with single-membrane vesicles of endosomal origin, and both LC3 and GABARAP proteins may play essential roles in these scenarios [29,30]. Whether and how the protein interactome of ATG8s at single-membrane vesicles may primarily include transport and fusion proteins remains to be elucidated.
ATG8s and viruses
Studies of the complex interplay between viruses and host cells have illuminated biochemical functions of the ATG8 family. ATG8s interact with viral components and are critical for host defense via mechanisms including virophagy and immune signaling; moreover, many viruses co-opt ATG8s to evade clearance and facilitate various steps in their respective life cycles. This evolutionary arms race reinforces ATG8s as important determinants of infection and pathogenesis and informs key cellular functions for this protein family in autophagy and autophagy-related processes.
ATG8s can facilitate host defense through the autophagic degradation of viral proteins, viral particles, and host-cell machinery that impacts virus replication. LC3B and likely other ATG8s interact with capsid proteins of select viruses via p62 to target invading virions for autophagic degradation [31,32]. In contrast, damage to endosomal membranes during viral entry recruits Galectin-8, NDP52, and LC3B to facilitate clearance of these compromised organelles and associated virions [33]. ATG8s may also restrict viral entry through the sequestration of viral proteins, such as Influenza (IAV) hemagglutinin, within amphisome-like structures or endosomal recruitment of TRIM5α [34,35]. However, it remains unclear whether these antiviral mechanisms involve classical autophagy or non-canonical functions of ATG8s [34,36].
The ATG8 family also modulates antiviral immune signaling. ATG8s are critical regulators of innate immune pattern recognition receptors (PRRs) and interferons (IFNs) production during viral infection. Although ATG8 proteins facilitate autophagic degradation of select PRRs, including RIG-I-like receptors (RLRs) and NOD-like receptor (NLRs), evidence also supports non-canonical regulation and functions during viral infection [37–39]. Early studies in phagocytes suggest a LAP-like process is involved in Toll-like receptor 9 (TLR9)-dependent IFN production during DNA virus infection [40][41]. More recently, cyclic GMP-AMP synthase (cGAS) – stimulator of interferon genes (STING) were shown to activate autophagy through mechanisms independent of ULK1 and BECN1 during infection [42]. This pathway also elicits negative feedback on IFN production, since STING is a target of autophagic degradation. Finally, ATG8s regulate inflammasome-mediated pathogen defense through autophagic degradation of inflammasome components, limiting IL-1β and IL-18 secretion [43,44], and under certain conditions also facilitate the release of IL-1β and other inflammatory molecules via non-canonical mechanisms [45] (see section on Functions of ATG8s in unconventional secretion).
IFNs upregulated by PRRs trigger diverse antiviral pathways within mammalian cells. These include expression of IFN-inducible GTPases such as immunity-related GTPases (IRGs) and guanylate-binding proteins (GBPs) [46]. In humans, all ATG8 isoforms bind immunity-related GTPase M (IRGM), which coordinates autophagy during microbial defense [42,47]. Furthermore, during murine norovirus infection, ATG5-ATG12:ATG16L1-dependent targeting of LC3B and possibly other ATG8s to membrane-associated replication complexes is critical for the recruitment of IRGs and GBPs that inhibit viral replication through mechanisms independent of classical autophagy [48].
At the same time, viruses have evolved mechanisms that subvert ATG8s to evade clearance and leverage their functions for various stages of the infection cycle. Generally, selective pressures have resulted in subversion mechanisms that undermine autophagosome formation or autophagosome-lysosome fusion, leaving ATG8-conjugation intact. ATG8-conjugation is frequently retained because viruses hijack the ATG8s and autophagosomal membranes for their replication. Seminal studies of poliovirus infection revealed that virus replication occurs on LC3B-positive autophagosomal membranes and is enhanced by autophagy activation, whereas genetic disruption of ATGs compromised virus production [49]. Furthermore, many RNA viruses specifically promote the accumulation of ATG8-positive membranes [50]. Human parainfluenza virus disrupts SNAP29-STX17 complexes critical for autophagosome-lysosome fusion to facilitate replication [51], whereas Zika virus requires ATG16L1 to form LC3B-positive ER-derived replication complexes [52]. Interestingly, select flaviviruses induce lipophagy to fulfill the energy requirements of viral replication and facilitate assembly [53,54]. There is also evidence that certain viruses utilize ATG8s independent of its lipidation status [55,56]. Coronaviruses (CoVs) require LC3B to form ER-derived replication membranes but virus production and LC3B membrane localization are largely unaffected in cells deficient for ATG8-conjugation [56]. The discovery that ATG8s facilitate the exocytosis of certain viruses provides an additional clue to the evolutionary convergence between viruses and the ATG8 family. Indeed, evidence supports that viruses hijack ATG8 proteins and associated membranes to facilitate their egress and transmission to neighboring cells (Box 2).
Box 2: ATG8 proteins in viral exocytosis.
Increasing evidence supports roles for ATG8 proteins in viral exocytosis. For example, poliovirus and coxsackievirus B (CVB3) are sequestered by autophagosome-like structures within infected cells and released in microvesicles bearing autophagosomal markers including lipidated LC3B, a process termed autophagosome-mediated exit without lysis (AWOL) (Figure I) [94,95]. Notably, the micro-vesicle envelope acquired during viral egress protects virions from immune recognition and facilitates transmission to neighboring cells. A number of enveloped viruses highjack ATG8 proteins and the autophagy pathway to facilitate membrane acquisition during egress. Epstein-Barr virus (EBV), human cytomegalovirus and varicella-zoster virus acquire ATG8-conjugated membranes during envelope acquisition in the cytosol (Figure I) [96–98]. Moreover, disruption of the ATG8-conjugation machinery severely impairs the production of EBV, resulting in the accumulation of viral dsDNA genomes in the cytosol of infected cells [96]. Flaviviruses including dengue virus (DENV) and hepatitis C virus (HCV) also highjack LC3B proteins and other ATGs to facilitate exocytic release through MVBs (Figure I). Although the underlying mechanisms that contribute to flavivirus exocytosis remain obscure, DENV infected cells release extracellular vesicles containing viral components and lipidated LC3B that mediate virus transmission [99]. Finally, IAV encodes an ion-channel protein, matrix protein 2 (M2), which inhibits autophagic clearance of the virus and facilitates viral egress by binding and targeting lipidated LC3B to the plasma membrane, functionally sequestering it from incorporation into autophagosomes (Figure I). Although the ATG8-conjugation machinery supports IAV filamentous budding and virion stability, infectious virus production is largely unaffected in ATG-deficient cells [100]. Collectively, these observations reinforce that viruses have evolved to hijack non-canonical functions of ATG8 proteins for their release outside host cells.
Box 2 Figure I: Viral subversion of ATG8 proteins for exocytosis.
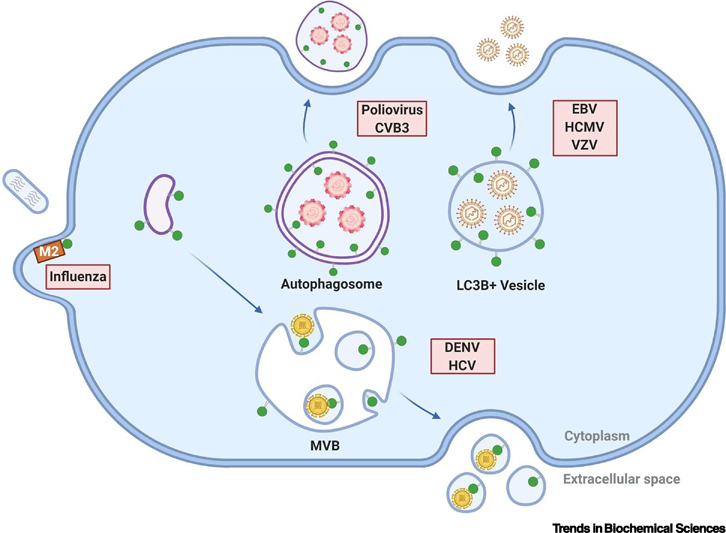
Viruses hijack LC3B and possibly other ATG8 family members (green circles) to facilitate egress and transmission between cells. Non-enveloped RNA viruses including poliovirus and coxsackievirus B (CVB3) are released in autophagosome-like vesicles bearing LC3B and phosphatidylserine. Enveloped double stranded DNA (dsDNA) viruses including Epstein-Barr virus (EBV), human cytomegalovirus (HCMV) and varicella zoster virus (VZV) are released via mechanisms regulated by ATGs and contain lipidated LC3B. Dengue virus (DENV) and hepatitis C virus (HCV) are released in small extracellular vesicles harboring lipid-modified LC3B that may originate from multivesicular bodies (MVBs). Finally, influenza virus filamentous budding involves matrix protein 2 (M2)-dependent targeting of LC3B to the plasma membrane; however, LC3B does not appear to be packaged into bud influenza virions. See text for more details.
Functions of ATG8s in unconventional secretion
Viral hijacking of the ATG8 proteins for exocytosis broaches roles for the autophagy machinery in cellular secretion and intercellular communication. Indeed, ATGs and specifically ATG8s play roles in unconventional secretion of proteins. While the precise molecular mechanisms of ATG8-dependent unconventional secretion are only beginning to be unraveled, genetic studies suggest these pathways generally fall into two categories: (1) mechanisms that employ autophagosome-like vesicles as a secretory intermediate, and (2) mechanisms that are functionally independent of autophagosome formation.
Extracellular secretion of ATG8-positive autophagosomes
ATG8-positive autophagosomal intermediates have been implicated in the unconventional secretion of proteins [57,58] (Figure 1, Panel E). Indeed, ATG8-positive vesicles facilitate the secretion of acyl-CoA-binding protein Acb1 in yeast (AcbA in Dictyostelium discoideum) and damage-associated inflammatory molecules in mammalian cells, including IL-1β, IL-18, and the high mobility group protein B1 (HMGB1) [45,59,60]. More recently, this list has grown to include ferritin, insulin-degrading enzyme (IDE), galectin-3, annexins A1 and A2, as well as the aggregation prone proteins amyloid-β and α-synuclein [58]. Nevertheless, the precise mechanisms by which autophagosomal intermediates capture specific cargo and deliver it outside the cell remains an area of active investigation.
In yeast, unconventional secretion of Acb1 requires ATG8, core ATGs in autophagosome formation (ATG1, ATG6, ATG9, ATG17) and Grh1 (the yeast orthologue of GRASP55 and GRASP65) [59,60]. In addition, secretion required multivesicular bodies (MVBs) biogenesis but independent of machinery that mediates autophagosome-vacuole fusion [59,60]. This led to a model where ATG8 and nascent autophagosomal membranes carrying Acb1 are captured at MVBs and released upon MVB fusion with the cell surface [57]. Although recent work suggests that GRASP function may be peripheral to this secretory pathway in mammals, autophagosome-like intermediates may be critical in the secretion of discrete cargo. Indeed, secretion of IL-1β requires core ATGs essential for autophagosome formation but proceeds via translocation of this secretory protein into the intermembrane space of LC3-positive double-membrane vesicles [45,61]. Recently, transmembrane protein TMED10 was identified as a protein channel that translocates mature IL-1β into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), an organelle implicated in autophagosome formation [62]. Moreover, genetic depletion of TMED10 impairs secretion of murine IL-1β in macrophage and neutrophil cell lines. Hence, IL-1β is loaded into vesicular precursors of autophagosomes prior to extracellular secretion. This secretion also requires Sec22b, a regulator of ER-Golgi protein trafficking, as well as soluble NSF-attachment protein receptors (SNAREs) implicated in vesicular fusion with the plasma membrane including SNAP23, SNAP29, and their cognate partners STX3 and STX4; STX17, a SNARE required for autophagosome-lysosome fusion, is dispensable for IL-1β release [63]. Collectively, these data suggest that secretory autophagosomes fuse with the plasma membrane as opposed to lysosomes in order to mediate IL-1β extracellular release. Clearly, it will be important to determine whether other targets of secretory autophagy are released through mechanisms similar to IL-1β or involve alternative ATG-dependent pathways that specialize in the secretion of distinct cargo.
The localization of LC3 to secretory autophagosomes intimates important roles for the ATG8 family in cellular secretion. Interestingly, many secretory autophagy cargos, including IL-1β and HMGB1, lack well-defined LIRs and are not known to bind ATG8-family members, suggesting that cargo is recruited to secretory autophagosomes via ancillary mechanisms independent of ATG8 proteins. The exception to this observation is ferritin, which interacts with LC3B via NCOA4 and, in addition to being secreted, is a target for autophagic degradation [63][64]. Remarkably, multiple ATG8-interacting proteins linked to autophagy, including p62, have been detected in conditioned media and plasma [65,66]. However, in the absence of functional studies, it remains unclear whether ATG8s and secretory autophagosomes are involved in release of these proteins. While ATG8 proteins may target a limited set of proteins for secretory autophagy, it is tempting to speculate that the main functions of the ATG8 family in secretion may relate to their critical role in autophagosome biogenesis, trafficking and fusion [19]. Indeed, recent studies demonstrating that LC3B phosphorylation by STK4 regulates the directional trafficking of autophagosomes, suggesting roles for this kinase in coordinating the itineraries of degradative and secretory autophagosomes [67] (see section on Regulatory mechanisms operating on ATG8 proteins that specify canonical versus non-canonical functions). Moreover, the essential functions of ATG8 proteins in membrane fusion events during autophagosome formation and maturation may also contribute to the fusion of secretory autophagosomes with the plasma membrane and/or endocytic compartments [9,68]. Notably, ATG8-associated membrane fusion appears independent of the luminal pH of donor and recipient compartments, and thus, theoretically competent to drive fusion with numerous cellular membranes beyond the lysosome [68]. Ultimately, this may be relevant to secretory autophagy in situations where lysosome function is compromised such as during infection, cancer and aging, enabling cells to expel material into the extracellular space.
ATG8s secreted via small extracellular vesicles
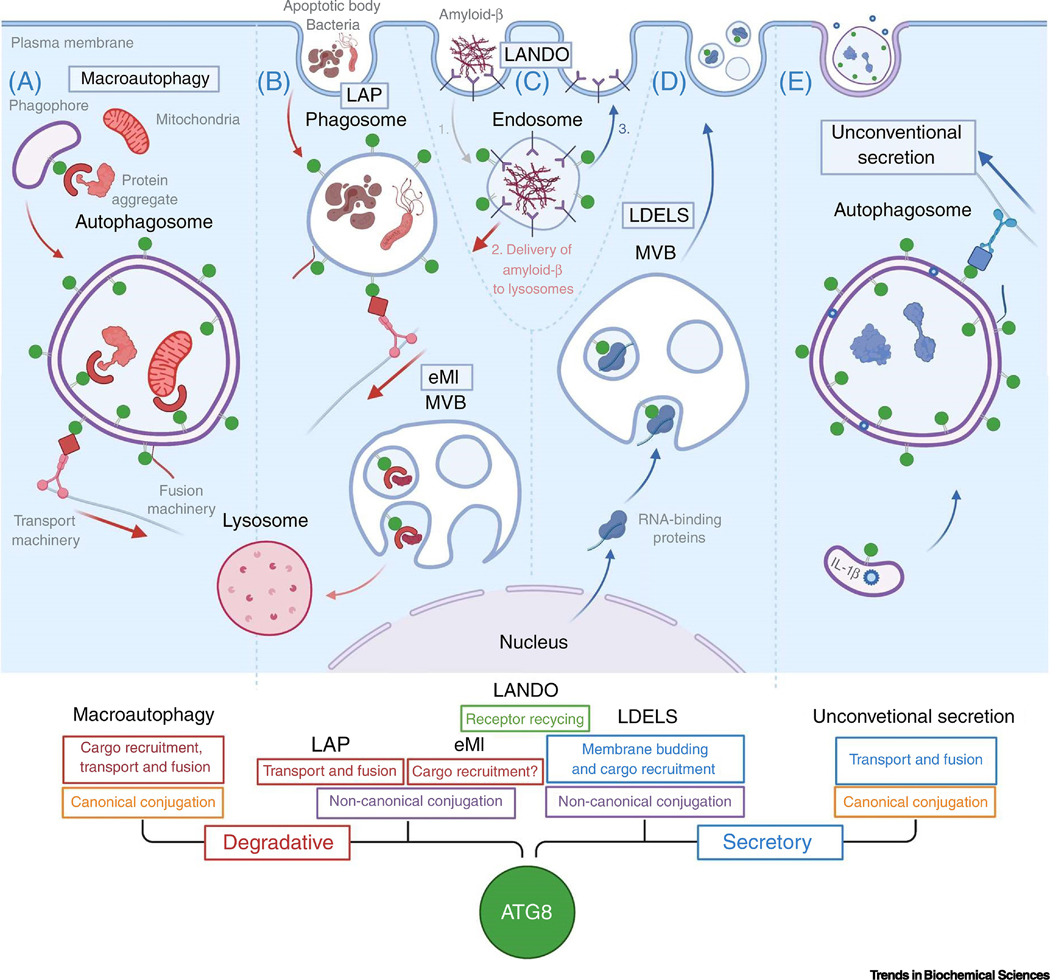
The autophagy machinery is also implicated in extracellular vesicle secretion (EVs) [69,70]. Notably, lipidated ATG8 proteins are highly enriched within a subset of EVs and specifies the loading of RNA-binding proteins and small non-coding RNAs through a process termed LC3-dependent EV loading and secretion (LDELS) [70] (Figure 1, Panel D and Figure 2). Furthermore, this novel EV-sorting pathway requires the LC3-conjugation machinery and neutral sphingomyelinase 2 (nSMase2)-dependent production of ceramide but is independent of ATG14 and FIP200/RBCC1, two ATGs required for canonical autophagy (Figure 2). Overall, genetic and cell biological data suggest budding of lipidated ATG8 at the limiting membrane of MVBs into small intraluminal vesicles (ILVs), which are released as EVs upon fusion of this organelle with the plasma membrane.
Figure 2. Proposed model of LC3-dependent extracellular vesicle loading and secretion (LDELS).
During LDELS, LC3B (green circles) and possibly other ATG8 proteins are delivered to the limiting membrane of multivesicular bodies (MVBs) where they specify the loading of cargoes including RNA binding proteins (RBPs; blue) into extracellular vesicles for secretion outside the cell. This pathway requires neutral sphingomylinase-2 (nSMase2; yellow) and LC3B-dependent recruitment of Factor-associated with nSMase2 activity (FAN; purple), which may facilitate intraluminal budding via localized ceramide production.
ATG8 proteins serve at least two important functions in LDELS. First, ATG8s recruit the Factor-associated with nSMase activation (FAN), through a conserved LIR, which facilitates EV biogenesis by stimulating nSMase2 production of ceramide that drives intraluminal budding and ILV formation (Figure 2) [70]. In contrast to many other EV biogenesis pathways, LDELS appears to be largely independent of ESCRT machinery, and only requires CHMP4B. The second critical function that ATG8 proteins serve in LDELS is sorting cargo such as RBPs into EVs for extracellular release [70] (Figure 1 and Figure 2); many of these RBPs have been identified as ATG8 interactors [11]. Consistent with these observations, LDELS also regulates the secretion of small non-coding RNAs including small nucleolar RNA (snoRNAs) and microRNAs (miRNAs). The molecular mechanisms and physiological roles of RBP-RNA secretion remain poorly understood. Interestingly, a number of RBPs and RNAs targeted by LDELS are implicated in viral regulation and innate immune signaling. For example, HNRNPK and LARP1 regulate HCV and DENV infection, respectively, whereas G3BP1 and snoRNAs have roles in activating double-stranded RNA (dsRNA)-dependent protein kinase (PKR) during infection and stress [71–74]. Collectively, these observations may suggest a role for LDELS in innate immune responses and stress signaling.
Analogous to LAP, LANDO, and eMI, the LDELS pathway involves delivery of LC3B proteins to single-membrane organelles of the endolysosomal system through mechanisms requiring the LC3-conjugation machinery but distinct from classical autophagy [27,75,76]. However, the function and fate of LC3B proteins within these pathways diverges dramatically. During LANDO and LAP, LC3B completely decorates the surface of endosomes and phagosomes [22,75]. In contrast, LC3B proteins are localized to subdomains at the limiting membrane of MVBs during LDELS, which subsequently undergo intraluminal budding to deliver LC3B into the lumen of these organelles in the form of ILVs [70]. Furthermore, during eMI, cytosolic targets are captured at MVBs via ESCRT-dependent intraluminal budding pathways [27]. Although LDELS and eMI both involve MVB intraluminal budding and ILV formation, LDELS results in the secretion of RBPs via EVs, whereas eMI primarily leads to cargo degradation. The molecular mechanisms specifying LC3B-dependent cargo loading in LDELS versus eMI remain to be elucidated.
Regulatory mechanisms operating on ATG8s to specify canonical versus non-canonical functions
Although the mechanisms that specify ATG8 proteins for classical versus non-canonical functions remain poorly understood, these operational decisions are likely the results of complex signaling pathways converging upon the conjugation machinery. Ultimately, ATG8 functions may be directed by two regulatory control points: (I) the conjugation of ATG8 to autophagosomes versus vesicles of alternative nature and (II) the specific ATG8 protein interactome at that precise location.
Many non-canonical autophagy pathways involve processes where ATG8 proteins are directly conjugated to lipids at single-membrane structures such as the plasma membrane or endolysosomal compartments. Recently, studies dissecting the functions of ATG16L1, which specifies the site of ATG8 lipidation [77], revealed that its FIP200-binding domain is required for classical autophagy but dispensable for LC3 lipidation at the plasma membrane and endolysosomes, whereas the WD40 domain is required for LC3 lipidation at non-canonical sites but dispensable for autophagy [23]. Moreover, recent biochemical studies indicate that ATG8s are conjugated to phosphatidylserine at these vesicles [78], providing a new potential molecular signature to differentiate these non-canonical processes. Although the molecular mechanisms underlying regulation of the different ATG16L1 domains during autophagy and non-canonical pathways remain unclear, this may be coordinated through interactions with accessory proteins [79].
The cargoes themselves may direct ATG8 conjugation to alternative vesicles to perform non-canonical functions. For example, binding of extracellular Aβ to receptors TREM2 and CD36 triggers the lipidation of LC3 at endosomes during LANDO [22]. Although the mechanisms that regulate LANDO are poorly understood, one can postulate that signals transmitted by TREM2 and CD36 in response to Aβ binding recruit the ATG8-conjugation machinery to endosomes to facilitate trafficking and lysosomal degradation of this aggregation-prone protein. Osmotic imbalances within endolysosomal compartments may also activate non-canonical autophagy and endolysosomal ATG8 lipidation through mechanisms dependent upon the V-ATPase complex [80]. Thus, signals from cargo at organelles or structures not traditionally thought to be targeted by autophagy can direct non-canonical functions of ATG8 proteins. Interestingly, RNA granules hitchhike on the surface of endolysosomes and the autophagy machinery is implicated in the turnover of these structures or components thereof [81,82]. Given these observations, it is tempting to speculate that RNA granules may be regulated through non-canonical functions of ATG8 proteins in processes such as eMI or LDELS.
During classical autophagy, ATGs are involved in protein interactions and post-translational modifications controlling discrete steps in this pathway. Emerging evidence suggests that similar mechanisms may control the non-canonical functions of ATG8 proteins. Phosphorylation regulates the binding of ATG8 interactors by modifying the affinity of the LIR-LDS-binding interface during autophagy. There are several examples of ATG8 interactors, which upon phosphorylation in the vicinity of their LIR, increase their binding affinity to ATG8s. ULK1-related phosphomimic mutations close to the LIRs of the proteins VPS34 or Beclin1, implicated in the initial steps of autophagosome formation, enhance the binding of VSP34 and Beclin1 to GABARAP [83]. Similarly, phosphorylation of the cargo receptor optineurin by the kinase TBK1 increases its binding affinity to LC3 to ultimately restrict Salmonella growth [84]. Direct phosphorylation of ATG8s also impacts their functions in autophagy, possibly via regulating multiple protein-protein interactions, in which could be a major functional regulatory switch. Interestingly, PKA phosphorylation of LC3 at Ser12 impairs recruitment autophagosomes, whereas phosphorylation of Thr6 and Thr29 by PKC does not appear to affect autophagy, yet the impact of these three modifications on non-canonical autophagy remains untested [85,86]. STK3/4 meditated phosphorylation of LC3B at Thr50, a residue located within LC3B LDS, is required for autophagosome-lysosome fusion and autophagy completion [87]. Furthermore, LC3B Thr50 can be phosphorylated by alternative kinases such as NEK9 to regulate selective autophagy [88], and the vicinity of this residue (K49 and K51) is susceptible to acetylation to regulate subcellular location of LC3B and starvation-induced autophagy [89].
These studies indicate that post-translational modification of ATG8 LDS may have important regulatory implications. Notably, recent studies demonstrate that LC3B phosphorylation at Thr50 by STK4 decreases the binding of the vesicle transport-related protein FYCO1 to promote the directional trafficking of autophagosomes toward lysosomal compartments in the perinuclear region [67]. Conversely, expression of a non-phosphorylatable LC3B-T50A mutant results in increased FYCO1 binding and an increased transport of autophagosomes towards the plasma membrane. Given these observations, it is tempting to speculate that LC3B phosphorylation at Thr50 may play a role not only in classical autophagy but also in ATG8-dependent secretory pathways via reprogramming key ATG8 protein-protein interactions related to vesicle transport. Finally, autophagosome fusion with lysosomes can also be regulated by phosphorylation. Specifically, TBK1-mediated phosphorylation prevents premature ATG4-mediated deconjugation of LC3C and GABARAP-L2 from autophagosome membranes to ultimately allow fusion with lysosomes [90]. Whether similar mechanisms operate to control fusion of ATG8-positive vesicles with plasma membranes during non-canonical pathways remains to be elucidated. Going forward, unraveling the mechanisms that direct classical autophagy versus non-canonical functions for ATG8 proteins remains a topic of immense interest in the field.
Concluding remarks
ATG8s are key core components of the macroautophagy machinery and commonly used as autophagosome markers and reporters of the macroautophagy process. Besides this traditional or canonical role, accumulating evidence supports that ATG8s also modulate alternative and diverse cell biological functions at other vesicles including phagosomes, endosomes, and macropinosomes, Furthermore, the ultimate fate of these vesicles can be either degradation or secretion. This review summarizes the alternative functions of ATG8s that are now beginning to be elucidated and hypothesizes how the functional repertoire of ATG8s may be regulated. We believe that to explain the functional divergence that ATG8s play in autophagosomes versus alternative vesicles with degradative or secretory fate, additional proteomic and imaging studies must address which ATG8 members are enriched at each of these vesicular membranes, and whether post-translational regulation plays a role on controlling protein-protein interactions and functional outcomes (see Outstanding Questions). All these novel findings and concepts have opened up a new and exciting line of research in the autophagy field.
Outstanding questions.
How do ATG8s carry out their diverse functions at different intracellular locations?
Do ATG8s perform similar canonical and non-canonical functions, or are there functional differences among individual family members?
What is the protein interactome of ATG8s in different vesicles and membrane topologies?
What is the relevance of ATG8s in non-canonical pathways in disease and aging?
Highlights.
Macroautophagy is a conserved cytoprotective process that facilitates degradation of damaged or unwanted cellular components and pathogens, collectively termed cargo
ATG8 proteins play key functions during macroautophagy upon conjugation to double-membrane vesicles, termed autophagosomes, which sequester cargo for lysosomal degradation
In addition to their canonical functions in macroautophagy, ATG8s also function in non-canonical pathways that do not involve autophagosome formation, and in such cases, ATG8s can be targeted to single-membrane vesicles with roles in divergent processes including cargo degradation and secretion
The canonical and non-canonical functions of ATG8 proteins are regulated, at least in part, by the autophagy conjugation machinery and possibly the unique interactome that ATG8s have at distinct cellular locations
Acknowledgments
We thank Dr. Sara Landeras Bueno for preparing Box 1 Figure I. Figure 1, Figure 2 and Box 2 Figure I were created with BioRender.com. J.N.T. was supported by a Fundación Ramon Areces Postdoctoral Fellowship, and an NIH K99/R00 pathway to independence grant (K99AG062774); A.M.L. was supported by a Banting Postdoctoral Fellowship from the Government of Canada (201409BPF-335868) and a Cancer Research Society Scholarship for Next Generation of Scientists; J.D. was supported by NIH grants AG057462, CA213775, and CA126792; and MH was supported by NIH grant GM117466.
Glossary
- Cargo receptors:
soluble or membrane-bound proteins that bind different kinds of cargo (including ubiquitinated proteins, protein aggregates, organelles, and pathogens) and bring them to autophagosomes; this latter step requires the interaction of cargo receptors with ATG8s associated with the inner membrane of autophagosomes. Examples of soluble cargo receptors include SQSTM1/p62, NBR1, NDP52, TAX1BP1, CALCOCO1, and NCOA4 (involved in the degradation of different kinds of cargo), and membrane bound receptors include FAM134B, TEX264, CCPG1, RTN3 (implicated in endoplasmic reticulum degradation) and NIX, BNIP3, FKBP8, FUNDC1 (implicated in mitochondria degradation)
- Core autophagy machinery:
group of cellular proteins facilitating the different steps of macroautophagy that lead to autophagosome formation and ultimately cargo degradation. These stages include the initiation step, controlled by the ULK complex (comprised of autophagy proteins ULK1/2, ATG13, ATG101, and FIP200); the nucleation of the autophagosome precursor called the phagophore, directed by the PI3K III nucleation complex (consisting of VPS15, VPS34, BECLIN1, ATG14, and NRBF2); the phagophore expansion, in which the proteins ATG2A, ATG2B, ATG9, the recently discovered VMP1 and TMEM41B and the PI3P-binding complex (WIPIs and DFCP1) participate; WIPIs (specifically WIPI2b via interaction with ATG16L1) facilitate the conjugation of ATG8s to the growing phagophore and ultimately the autophagosome membranes via the conjugation machinery (ATG3, ATG4, ATG7, ATG10, and a multimer comprised of ATG5, ATG12, ATG16L1 – the ATG5-ATG12:ATG16L1 complex). The final stages include fusion of the autophagosome with lysosomes and cargo degradation. Autophagosome-lysosome fusion is directed by two separate cognate SNARE complexes (STX17-SNAP29-VAMP8 and/or YKT6-SNAP29-STX7), the HOPS-tethering complex (see below), components of the PI3K III complex, ATG8 proteins and a number of recently defined tethering factors (EPG5, PLEKHM1, BRUCE, and GRASP55)
- Endosomes:
membrane vesicles formed upon invagination and budding of the cell plasma membrane that deliver extracellular materials into the cell
- Entotic vacuoles:
vesicles that engulf live neighboring cells and may play important roles in non-apoptotic cell death
- Endosomal complexes required for transport (ESCRT) machinery:
group of cytosolic protein complexes including ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III as well as accessory proteins that direct membrane remodeling during cytokinesis, membrane repair, and intraluminal vesicle biogenesis at late endosomes to form multivesicular bodies
- Endoplasmic reticulum-Golgi intermediate compartment (ERGIC):
an organelle in more complex eukaryotes that mediates the trafficking and sorting of cargo between the endoplasmic reticulum and the Golgi apparatus
- Extracellular vesicles (EVs):
heterogeneous mixture of small membrane-bound structures released from cells under both physiological and disease conditions that carry select biomolecules and potentially function as vehicles of intercellular communication
- Galectin-8:
member of the lectin protein family that have conserved carbohydrate recognition domains and bind β-galactosides
- Homotypic fusion and protein sorting (HOPS)-tethering complex:
heterohexameric complex comprising VPS11, VPS16, VPS18, VPS33, VPS39, and VPS41 that facilitates autophagosome-lysosome fusion via interactions with the SNARE Syntaxin-17 (STX17) and Rab7
- Interferons (IFNs):
group of secreted signaling proteins produced by host cells in response to viral infection and function in antiviral defense
- Lipophagy:
the autophagic process in which lipid droplets are delivered to the lysosome and degraded
- Macropinosomes:
large subtype of endosome formed upon rearrangement of the plasma membrane via rapid polymerization of actin fibers. Macropinosomes encapsulate large volume of liquid and solutes from the extracellular media
- Multivesicular bodies (MVBs):
specialized subtype of endosome containing intraluminal vesicles that are formed upon inward budding. Fusion of MVBs with the plasma membrane results in the secretion of extracellular vesicles
- NOD-like receptors (NLRs):
family of intracellular receptors that bind pathogen-associated and damage-associated molecular patterns and can oligomerize to form large signaling complexes termed inflammasomes which regulate the production of inflammatory cytokines and apoptosis
- Pattern recognition receptors (PRRs):
innate immune sensors that recognize conserved molecular patterns within pathogen or cell intrinsic biomolecules that are indicative of infection or damage
- Phagosomes:
specialized vesicles generated within phagocytic cells that can engulf pathogens or apoptotic bodies from the extracellular media and play critical roles in immunity
- RIG-I-like receptors (RLRs):
family of intracellular pattern recognition receptors including retinoic-acid inducible gene I (RIG-I) and melanoma differentiation-associated 5 (MDA5) involved in the recognition of viral RNA by the innate immune system
- Soluble NSF-attachment protein receptors (SNAREs):
group of proteins that facilitate the fusion of vesicles with target membranes
- Toll-like receptors (TLRs):
family of membrane receptors that recognize structurally conserved molecules derived from pathogens such as virus associated double-stranded RNA, unmethylated DNA and bacterial lipopolysaccharides. TLR activation triggers numerous signaling pathways including the production of interferons
- Unconventional secretion:
eukaryotic secretory proteins classically traffic to the cell surface or are released into the extracellular space along itineraries involving the endoplasmic reticulum (ER) and Golgi apparatus. However, there is an increasing list of proteins, lacking an N-terminal secretory signal sequence, that bypass these organelles and are released via so called unconventional mechanisms
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansen M. et al. (2018) Autophagy as a promoter of longevity: insights from model organisms. Nature Reviews Molecular Cell Biology 19, 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dikic I.and Elazar Z.(2018) Mechanism and medical implications of mammalian autophagy. Nature Reviews Molecular Cell Biology 19, 349–364 [DOI] [PubMed] [Google Scholar]
- 3.Tsukada M.and Ohsumi Y.(1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 [DOI] [PubMed] [Google Scholar]
- 4.Ichimura Y. et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 5.Kirisako T. et al. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabeya Y. et al. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lystad AH and Simonsen A.(2019) Mechanisms and Pathophysiological Roles of the ATG8 Conjugation Machinery. Cells 8(9), 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuboyama K. et al. (2016) The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041 [DOI] [PubMed] [Google Scholar]
- 9.Nguyen TN et al. (2016) Atg8 family LC3/GAB ARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol 215, 857–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatogawa H. et al. (2007) Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 11.Behrends C. et al. (2010) Network organization of the human autophagy system. Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen T.and Lamark T.(2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7(3): 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birgisdottir ÅB et al. (2013) The LIR motif - crucial for selective autophagy. J. Cell Sci 126, 3237–3247 [DOI] [PubMed] [Google Scholar]
- 14.Rogov V. et al. (2014) Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Molecular Cell 53(2):167–78 [DOI] [PubMed] [Google Scholar]
- 15.Søreng K. et al. (2018) Membrane Trafficking in Autophagy. In International Review of Cell and Molecular Biology 336pp. 1–92 [DOI] [PubMed] [Google Scholar]
- 16.Pankiv S. et al. (2010) FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 188, 253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu M. meng et al. (2014) LC3 Binding to the scaffolding protein jip1 regulates processive dynein-driven transport of autophagosomes. Dev. Cell 29, 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McEwan DG et al. (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 57, 39–54 [DOI] [PubMed] [Google Scholar]
- 19.Johansen T.and Lamark T.(2020) Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. Journal of Molecular Biology 432, 80–103 [DOI] [PubMed] [Google Scholar]
- 20.Le Guerroué F. et al. (2017) Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Mol. Cell 68, 786–796.e6 [DOI] [PubMed] [Google Scholar]
- 21.Florey O. et al. (2011) Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol 13, 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heckmann BL et al. (2019) LC3-Associated Endocytosis Facilitates β-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 178, 536–551.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fletcher K. et al. (2018) The WD 40 domain of ATG 16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 37:e97840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez J. et al. (2011) Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. U. S. A 108, 17396–17401 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Sanjuan MA et al. (2007) Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 [DOI] [PubMed] [Google Scholar]
- 26.Ma J. et al. (2014) Cutting Edge: FYCO1 Recruitment to Dectin-1 Phagosomes Is Accelerated by Light Chain 3 Protein and Regulates Phagosome Maturation and Reactive Oxygen Production. J. Immunol 192, 1356–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mejlvang J. et al. (2018) Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol 217, 3640–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C. et al. (2020) Selective Lysosome Membrane Turnover Is Induced by Nutrient Starvation. Dev. Cell 55, 289–297.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasai M. et al. (2017) Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat. Immunol 18, 899–910 [DOI] [PubMed] [Google Scholar]
- 30.Shui W. et al. (2008) Membrane proteomics of phagosomes suggests a connection to autophagy. Proc. Natl. Acad. Sci. U. S. A 105, 16952–16957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orvedahl A. et al. (2010) Autophagy Protects against Sindbis Virus Infection of the Central Nervous System. Cell Host Microbe 7, 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumpter R. et al. (2016) Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell 165, 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staring J. et al. (2017) PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 541, 412–416 [DOI] [PubMed] [Google Scholar]
- 34.Keown JR et al. (2018) A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5 and mammalian autophagy-related ATG8 proteins. J. Biol. Chem 293, 18378–18386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribeiro CMS et al. (2016) Receptor usage dictates HIV-1 restriction by human TRIM5α in dendritic cell subsets. Nature 540, 448–452 [DOI] [PubMed] [Google Scholar]
- 36.Imam S. et al. (2016) TRIM5α Degradation via Autophagy Is Not Required for Retroviral Restriction. J. Virol 90, 3400–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Y. et al. (2018) LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 37, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin S. et al. (2017) Tetherin Suppresses Type I Interferon Signaling by Targeting MAVS for NDP52-Mediated Selective Autophagic Degradation in Human Cells. Mol. Cell 68, 308–322.e4 [DOI] [PubMed] [Google Scholar]
- 39.Hou P. et al. (2020) A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 31(1):62–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HK et al. (2007) Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315, 1398–1401 [DOI] [PubMed] [Google Scholar]
- 41.Henault J. et al. (2012) Noncanonical Autophagy Is Required for Type I Interferon Secretion in Response to DNA-Immune Complexes. Immunity 37, 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S. et al. (2018) Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol 217, 997–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupfer C. et al. (2013) Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol 14, 480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi CS et al. (2012) Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol 13, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dupont N. et al. (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30, 4701–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilla-Moffett D. et al. (2016) Interferon-Inducible GTPases in Host Resistance, Inflammation and Disease. Journal of Molecular Biology 428, 3495–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chauhan S. et al. (2015) IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol. Cell 58, 507–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biering SB et al. (2017) Viral Replication Complexes Are Targeted by LC3-Guided Interferon-Inducible GTPases. Cell Host Microbe 22, 74–85.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson WT et al. (2005) Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3, 0861–0871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Münz C.(2017) Autophagy proteins in viral exocytosis and anti-viral immune responses. Viruses 9(10):288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding B. et al. (2014) Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe 15, 564–577 [DOI] [PubMed] [Google Scholar]
- 52.Cao B. et al. (2017) Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J. Exp. Med 214, 2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heaton NS and Randall G.(2010) Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JY et al. (2017) Hepatitis C Virus Induces the Localization of Lipid Rafts to Autophagosomes for Its RNA Replication. J. Virol 91(20):e00541–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abernathy E. et al. (2019) Differential and convergent utilization of autophagy components by positive-strand RNA viruses. PLoS Biol. 17(1):e2006926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reggiori F. et al. (2010) Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7, 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramani S.and Malhotra V.(2013) Non-autophagic roles of autophagy-related proteins. EMBO Reports 14, 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Claude-Taupin A. et al. (2017) Autophagy’s secret life: Secretion instead of degradation. , Essays in Biochemistry 61, 637–647 [DOI] [PubMed] [Google Scholar]
- 59.Manjithaya R. et al. (2010) Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol 188, 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duran JM et al. (2010) Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol 188, 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M. et al. (2015) Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion. Elife 4:e11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M. et al. (2020) A Translocation Pathway for Vesicle-Mediated Unconventional Protein Secretion. Cell 181, 637–652.e15 [DOI] [PubMed] [Google Scholar]
- 63.Kimura T. et al. (2017) Dedicated SNARE s and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 36, 42–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santana-Codina N.and Mancias JD (2018) The role of NCOA4-mediated ferritinophagy in health and disease. Pharmaceuticals 11(4):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeppesen DK et al. (2019) Reassessment of Exosome Composition. Cell 177, 428–445.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peinado H. et al. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nieto-Torres J. et al. (2020) LC3B phosphorylation regulates FYCO1 binding and directional transport of autophagosomes. bioRxiv DOI: 10.1101/2020.05.15.081638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weidberg H. et al. (2011) LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell 20, 444–454 [DOI] [PubMed] [Google Scholar]
- 69.Guo H. et al. (2017) Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev. Cell 43, 716–730.e7 [DOI] [PubMed] [Google Scholar]
- 70.Leidal AM et al. (2020) The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol 22, 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poenisch M. et al. (2015) Identification of HNRNPK as Regulator of Hepatitis C Virus Particle Production. PLoS Pathog. 11, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki Y. et al. (2016) Characterization of RyDEN (C19orf66) as an Interferon-Stimulated Cellular Inhibitor against Dengue Virus Replication. PLoS Pathog. 12(1):e1005357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reineke LC and Lloyd RE (2015) The Stress Granule Protein G3BP1 Recruits Protein Kinase R To Promote Multiple Innate Immune Antiviral Responses. J. Virol 89, 2575–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osama AY et al. (2015) Potential role for snoRNAs in PKR activation during metabolic stress. Proc. Natl. Acad. Sci. U. S. A 112, 5023–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez J. et al. (2015) Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol 17, 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Wei Y. et al. (2008) Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 4, 949–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujita N. et al. (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durgan J. et al. (2020) Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. bioRxiv doi: 10.1101/2020.05.14.096115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boada-Romero E. et al. (2013) TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 32, 566–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Florey O. et al. (2015) V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy 11, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao YC et al. (2019) RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell 179, 147–164.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buchan JR et al. (2013) XEukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Birgisdottir ÅB. et al. (2019) Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy 15, 1333–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wild P. et al. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cherra SJ et al. (2010) Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol 190, 533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang H. et al. (2010) Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem. Biophys. Res. Commun 395, 471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilkinson DS et al. (2015) Phosphorylation of LC3 by the Hippo Kinases STK3/STK4 Is Essential for Autophagy. Mol Cell 57, 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shrestha BK et al. (2020) NIMA-related kinase 9 –mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1. J. Biol. Chem 295, 1240–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang R. et al. (2015) Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466 [DOI] [PubMed] [Google Scholar]
- 90.Herhaus L. et al. (2020) TBK1-mediated phosphorylation of LC3C and GABARAP-L2 controls autophagosome shedding by ATG4 protease. EMBO Rep. 21(1):e48317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Slobodkin M.and Elazar Z.(2013) The Atg8 family: Multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 55, 51–64 [DOI] [PubMed] [Google Scholar]
- 92.Marshall RS et al. (2019) ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 177, 766–781.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dou Z. et al. (2015) Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bird SW et al. (2014) Nonlytic viral spread enhanced by autophagy components. Proc. Natl. Acad. Sci. U. S. A 111, 13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen YH et al. (2015) Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160, 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nowag H. et al. (2014) Macroautophagy proteins assist epstein barr virus production and get incorporated into the virus particles. EBioMedicine 1, 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buckingham EM et al. (2016) Exocytosis of Varicella-Zoster Virus Virions Involves a Convergence of Endosomal and Autophagy Pathways. J. Virol 90, 8673–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taisne C. et al. (2019) Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci. Rep 9(1):4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu YW et al. (2016) Autophagy-associated dengue vesicles promote viral transmission avoiding antibody neutralization. Sci. Rep 6:32243, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beale R. et al. (2014) A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 15, 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]