Abstract
F-BAR domains, like all BAR domains, are dimeric units that oligomerize and bind membranes. F-BAR domains are generally coupled to additional domains that function in protein binding or have enzymatic activity. Because of their crescent-shape and ability to oligomerize, F-BAR domains have been traditionally viewed as membrane deformation modules. However, multiple independent studies have provided no evidence that certain F-BAR domains are able to tubulate membrane. Instead, a growing body of literature featuring structural, biochemical, biophysical, and microscopy-based studies supports the idea that the F-BAR domain family can be unified only by their ability to form oligomeric assemblies on membranes to provide platforms for molecular assembly.
Keywords: BAR domain, F-BAR domain, actin cytoskeleton, membrane linkers, oligomeric assemblies, membrane-bound platforms, EM, super-resolution microscopy
All F-BAR domains bind, but not all bend membrane
Bin/Amphiphysin/Rvs (BAR) superfamily proteins are critical components of membrane-linked processes in eukaryotic cells, including endocytosis, cytokinesis, and motility. These proteins are defined by the namesake BAR domain, which dimerizes to form a crescent-shaped bundle of six alpha-helices that directly binds membrane (Box 1). BAR dimers have the propensity to self-associate into oligomeric assemblies [1]. BAR proteins are also usually modular and contain additional domains that generally link components of the actin cytoskeleton to membrane via the BAR domain [1–3]. Their membrane-binding properties, tendency to oligomerize, banana-shape of the dimers, and localization of some BAR proteins at sites of membrane curvature in cells led to the view that BAR domains are curvature generating, stabilizing, and/or sensing modules (Figure 1A).
Box 1: Classifications of BAR domains.
The BAR domain forms a helix-bundle dimer of crescent shape. Positively charged residues are enriched on the concave or convex surface of the crescent and participate in binding negatively charged membrane lipids, particularly at the plasma membrane [9]. Most BAR domains form homodimers, although the highly related srGAPs can heterodimerize [44]. One BAR domain dimer is small compared to the membrane, and thus the protein surface of an oligomerized BAR domain is considered to be necessary for any membrane shaping activity such as tubulation [9].
BAR domains are classified into the classical BAR subfamily, including N-BARs, and also the F-BAR and I-BAR domain subfamilies (reviewed in [1]). The F-BAR subfamily, the subject of this review, is distinguished from classical BAR domains by their conserved primary sequence that is generally longer (~300 amino acids) than that of the classical BAR domain (~200 amino acids). F-BAR dimers also generally form a more extended structure, having a length of ~20 nm, and less curved shape compared to canonical BARs. The earliest F-BAR domain structures of FBP17, CIP4, and FCHo2 revealed that the arc depths of these dimers were approximately three times more shallow than those of BAR dimers [8], but there is variation in shape within the subfamilies and more recent studies have revealed that some F-BAR domain dimers, including those of S. pombe Cdc15 and H. sapiens GAS7 and FCHSD2, are nearly flat [14, 18, 40].
Homo sapiens has approximately 50 classical BAR domain proteins and 20 F-BAR proteins. In human tissues, the widely expressed F-BAR domain proteins are FBP17, CIP4, TOCA-1, PACSIN2/3, GAS7, FCHSD2, FES, srGAP1, and srGAP2, by mRNA frequency according to the National Center for Biotechnology Information (NCBI) [54]. Yeast have considerably fewer with approximately 16 and 10 classical BAR domain proteins in S. pombe and S. cerevisiae, respectively, and 7 F-BAR domain-containing proteins in each yeast species [55, 56].
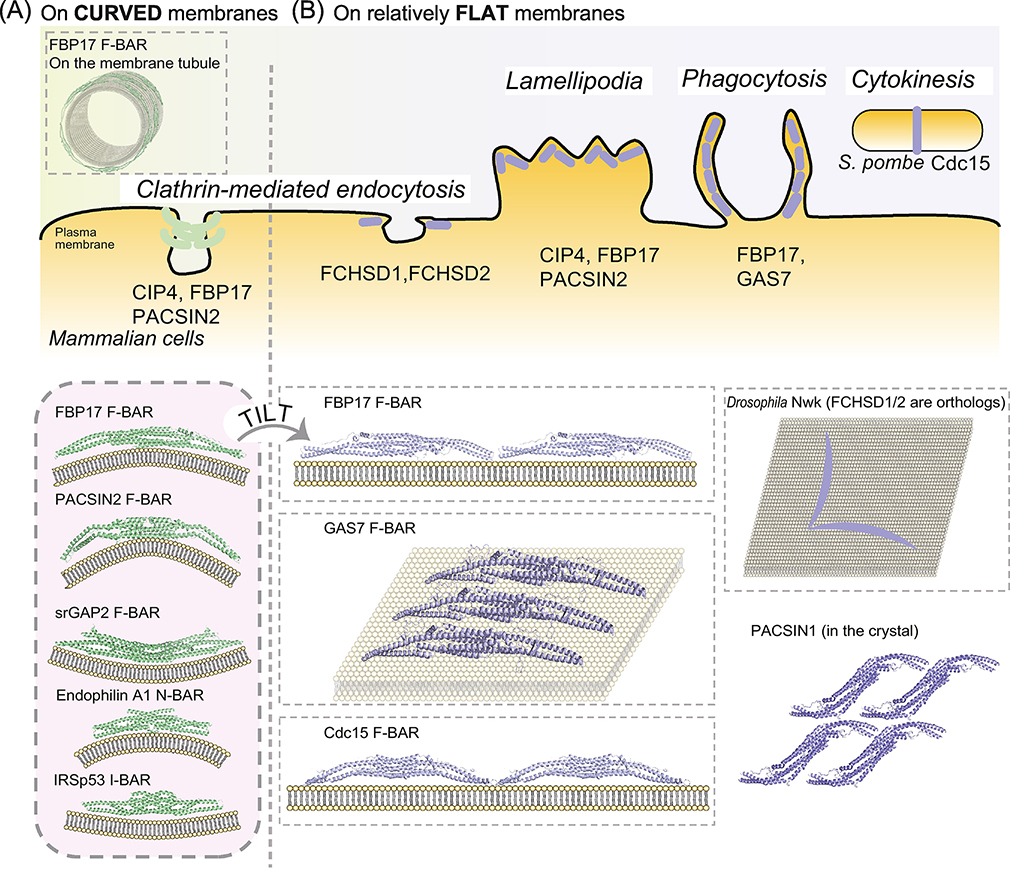
Figure 1: Assembly of F-BAR domains on flat membranes and their subcellular structures.
(A) Representative mode of F-BAR domain oligomerization on curved membrane. The structures are of the F-BAR domain of FBP17 (PDB ID: 2EFL) and PACSIN2 (PDB ID:3ABH) for membrane invaginations, the inverse F-BAR domain of srGAP2 (PDB ID: 5I6J) for protrusions, the N-BAR domain of endophilin A1 (PDB ID: 2D4C) for membrane invagination, and the I-BAR domain of IRSp53 (PDB ID: 1WDZ) for membrane protrusion. The spiral-like assembly of the FBP17 F-BAR domain by tip-to-tip contacts [6, 9] is also illustrated.
(B) Membrane binding of F-BAR domains on relatively flat membranes. CIP4 (PDB ID: 2EFK) and FBP17 (PDB ID: 2EFL) F-BAR domains arrange on the flat membrane in a tilted configuration in comparison with that on tubules, where the lateral surface of the F-BAR domain is the membrane binding surface. [9] Tip-to-tip contact was inferred from the arrangement of the F-BAR in the crystal [6]. GAS7b (PDB ID: 6IKN) assembles on the membrane in a similar manner, where the assembly is flat with one positively charged surface tilted toward the membrane [14].
S. pombe Cdc15 F-BAR (PDB ID: 6XJ1) assembles linear oligomers on flat membranes [40]. D. melanogaster Nwk F-BAR, an orthologue of mammalian FCHSD1/2, adopts a zig-zag assembly [32]. PACSIN1 F-BAR (PDB ID: 3HAI) forms a sheet-like assembly in the crystal lattice, suggesting it binds membrane with the positively charged surface [24]. The dashed boxes indicate models with experimental evidence on membranes in vitro.
Indeed, the earliest in vitro studies of BAR domains demonstrated their ability to tubulate when concentrated on membrane [4]. The purified human amphiphysin BAR domain was the first reported to deform liposomes made of total brain lipids (Folch fraction) in vitro, and electron microscopy (EM) was used to visualize its ability to drive vesicle tubulation through extensive self-association [4]. Further evidence for amphiphysin’s ability to deform membrane was obtained by overexpressing its BAR domain in cultured HeLa and COS-7 cells, which resulted in plasma membrane tubulation. These two assays – adding purified BAR domains to liposomes and over-expressing BAR domains in cultured cells – became standard for assaying BAR domain tubulation activity (Box 2).
Box 2. Methods for studying F-BAR activity on membranes: liposome and cell-based assays.
For liposome tubulation assays, the experimenter generates liposomes of the composition of their choosing. Choice of lipids and buffer will affect the properties of the liposomes, such as fluidity and tension [57]. Proteins, preferably with tags for purification removed, are then added to liposomes. Next, an appropriate visualization method is selected. For liposomes, transmission electron microscopy (EM) provides the necessary resolution to visualize F-BAR-generated tubules. Cryo-EM can provide finer resolution and allow for reconstruction of individual F-BAR domains on membrane, as was demonstrated for CIP4 F-BAR [9]. This approach provides not only information about if an F-BAR domain generates tubules, but also the F-BAR oligomer structure on the tubule. Alternatively, generation of giant unilamellar vesicles (GUVs), which are > 1 micron in diameter and used to model cell size, allows for conventional light microscopy visualization [58]. Lipids coupled to a fluorescent dye should be incorporated for membrane visualization and F-BAR domains should be linked to a fluorophore, e.g. GFP, which could potentially interfere with membrane binding and/or deformation.
One advantage of liposome tubulation assays is having experimental control of membrane properties and protein concentrations. However, experimenters may find that F-BAR domains only tubulate liposomes of certain compositions in specific buffers. Tubulation activity has not been detected for some F-BAR domains, which may raise concern that cellular regulation not recapitulated in vitro is required for tubulation or that optimal liposome and buffer composition has not been identified. Because it is not practical to test every lipid and buffer condition, cell-based assays may be a logical alternative.
In the cell-based assay, over-expression of F-BAR domains coupled to a fluorophore are driven from mammalian expression vectors in cultured cells; COS and HeLa cells are typically used and tubulation can be visualized by fluorescence microscopy. A negative control is required, and a membrane marker or dye should be utilized to assess if any resultant tubules are membrane-coated. This assay is advantageous because cells contain membranes of varying compositions and curvatures, which can be difficult to recreate in vitro. However, coupling a fluorophore to an F-BAR domain may interfere with membrane binding and/or oligomerization, which would inhibit tubulation. These properties however can be assessed in vitro as a complement to the cell-based assay. If a membrane-binding and oligomerization-competent F-BAR domain does not tubulate any cellular membrane in this assay, it is difficult to rationalize what a physiological function of membrane tubulation concocted under specific conditions in vitro would be.
The Fes/Cip4 homology BAR (F-BAR) subfamily is distinguished from classical BAR domains based on sequence and structure; F-BAR dimers form a more extended and less curved shape compared to BAR dimers, as first observed for the FBP17 and FCHo1/2 F-BAR domains [5, 6] in comparison with amphiphysin and endophilin BAR domains [7] (Box 1). These differences in dimer shape led to the idea that F-BAR domains may generate different degrees of membrane curvature than classical BARs [8]. There is supporting evidence for this hypothesis, as F-BARs cluster in spatially separate areas when overexpressed simultaneously with classical BAR domains in cultured cells [9].
Like classical BARs, a majority of F-BAR domains tubulate membrane in standard assays [5, 6, 8]. However, comprehensive examination of human F-BAR family members, including GAS7 and FCHSD1/2, under a variety of conditions of membrane composition, tension, and with or without tags for domain visualization did not detect membrane tubulation in the two standard assays, suggesting that some F-BAR domains do not tubulate membranes at all, but simply bind it (Table 1) [10]. This suggests that membrane curvature generation may not be a unifying function of the family (Figure 1B).
Table 1:
F-BAR domains that do not tubulate membrane in classical assays.
| Protein | Organism | Construct | Results | Ref | |
|---|---|---|---|---|---|
| Liposome assay | Cell-based assay | ||||
| Cdc15 | fission yeast | GFP-Cdc15 F-BAR (aa 19–312) Cdc15 F-BAR (aa 19–312) | No tubulation detected on liposomes (69% DOPC, 15% DOPE, 10% DOPS, 5% PI4P, 1% rhodamine-PE) in a variety of buffer conditions and concentrations | No tubulation detected when over-expressed in COS-7 cells | [10] |
| FCHSD1/2/Nwk | human | GFP-FCHSD1 F-BAR (aa 1–376) | No tubulation detected on liposomes (69% DOPC, 15% DOPE, 10% DOPS, 5% PI4P, 1% rhodamine-PE) in a variety of buffer conditions and concentraos | No tubulation detected when over-expressed in COS-7 cells | [10] |
| GFP-FCHSD2 F-BAR (aa 1–396) | No tubulatic n detected on liposomes (69% DOPC, 15% DOPE, 10% DOPS, 5% PI4P, 1% rhodamine-PE) in a variety of buffer conditions and concentrations | No tubulation detected when over-expressed in COS-7 cells | [10] | ||
| FCHSD2 F-BAR (aa 1–468) EGFP | Not reported | Induced protrusions when over-expressed in HeLa cells | [18] | ||
| fly | Nwk(aa 1–428)-EGFP Nwk(aa 1–428) | Did not tubulate, but induced scallops on liposomes (55% DOPC, 20% POPE, 14.5% DOPS, 10% brain PI(4,5)P2, 0.5% NBD-POPE). | Induced protrusions when over-expressed in HEK293T and D. melanogaster S2 cells | [17] | |
| Fer | human | GFP-Fer F-BAR (aa 1–287) | No tubulation detected on liposomes (69% DOPC, 15% DOPE, 10% DOPS, 5% PI4P, 1% rhodamine-PE) in a variety of buffer conditions and concentrations | No tubulation detected when over-expressed in COS-7 cells | [10] |
| GFP-Fer F-BAR (aa 1–300) | Not reported | No tubulation detected when over-expressed in COS-7 cells | [51] | ||
| GFP-Fer | Not reported | Induced lamellipodia formation when over-expressed in COS-7 cells | [52] | ||
| Fes | human | GFP-Fes F-BAR (aa 1–288) | No tubulation detected on liposomes (69% DOPC, 15% DOPE, 10% DOPS, 5% PI4P, 1% rhodamine-PE) in a variety of buffer conditions and concentrations | No tubulation detected when over-expressed in COS-7 cells | [10] |
| GAS7 | human | GFP-GAS7 F-BAR (aa 138–476) | No tubulation detected on liposomes (69% DOPC, 15% DOPE, 10% DOPS, 5% PI4P, 1% rhodamine-PE) in a variety of buffer conditions and concentrations | No tubulation detected when over-expressed in COS-7 cells | [10] |
| GFP-GAS7 F-BAR (aa 166–476) GAS7 F-BAR (aa 166–476) GFP-GAS7b GAS7b GAS7cb | No tubulation detected on liposomes (20% PC, 20% PE, 60% PS, 0.2% rhodamine-PE; 40% PC, 40% PE, 20% PS, 5% PIP3) | No tubulation detected when over-expressed in HeLa cells or macrophages | [14] | ||
| Hof1 | budding yeast | Hof1 F-BAR (aa 1–300)-GFP | Not reported | No tubulation detected when over-expressed in HeLa cells | [53] |
| RHOGAP4 | human | GFP-RhoGAP4 F-BAR(aa 1–381) | No tubulation detected on liposomes (69% DOPC, 15% DOPE, 10% DOPS, 5% PI4P, 1% rhodamine-PE) in a variety of buffer conditions and concentrations | No tubulation detected when over-expressed in COS-7 cells | [10] |
Here, we review the structures and oligomerization strategies of F-BAR domains, and how these properties are likely to dictate and constrain their biological functions on membranes. As in classical BARs like amphiphysin and endophilin, most F-BAR proteins contain additional domains, typically an SH3 domain(s), that can link the membrane-bound F-BAR to actin networks and/or promote actin polymerization locally [11]. However, there are now several examples of F-BAR domains directly serving as the bridging component themselves, and we discuss the emerging evidence that oligomerization therefore allows F-BAR domains to polymerize protein interaction networks that promote the structure and function of actin-based networks.
F-BAR oligomerization strategies inform function
F-BAR domains use varying modalities to oligomerize. For example, crystal packing contacts implicate residues at the tips of the F-BARs FBP17 and CIP4 in mediating oligomerization [6]. Cryo-EM of CIP4 F-BAR assembled on membrane allowed for single-particle reconstruction, which provided further evidence of tip-to-tip assembly of dimers in vitro [9]. This analysis also revealed that lateral contacts can occur between neighboring F-BAR dimers, facilitating membrane tubulation in vitro and resulting in spiral filaments with the concave faces of the assembled F-BAR domains contacting membrane (Figure 1A). Disrupting CIP4 F-BAR domain tip-to-tip or lateral oligomerization reduces tubule formation in standard assays [6, 9].
The Schizosaccharomyces pombe Imp2 F-BAR domain crystalized as a dimer of dimers, with residues at the tip of one dimer contacting the core of a neighboring dimer, resulting in a helical assembly when extended mathematically with the concave membrane-binding surfaces on the interior [12]. This oligomerization strategy is consistent with Imp2 F-BAR domain’s ability to tubulate vesicles in vitro, and mutation of oligomerization residues renders Imp2 tubulation-deficient [12]. It is logical that F-BAR domains like CIP4 and Imp2 that can assemble into spiral structures are able to tubulate membrane in vitro.
However, F-BAR domains also assemble on flat membranes using different oligomerization strategies (Figure 1). FBP17 F-BAR dimers make tip-to-tip contacts when assembled on a flat membrane in vitro and imaged by EM [9]. Intriguingly, in this instance, F-BAR domains contact the membrane with the lateral side of the crescent, rather than the canonical concave surface, occluding lateral oligomeric contact sites observed on membrane tubules. Although this flat assembly has not been detected in vivo, it may be relevant to FBP17 action at lamellipodia membranes [13]. In another example of sheet-like assembly, the mammalian GAS7 F-BAR domain forms flat filamentous oligomers (FFO) through lateral interactions of the F-BAR dimers on monolayers in vitro [14]. Similar to the orientation of FBP17 F-BAR domains observed on a flat membrane, the configuration of GAS7 dimers in the FFO suggests that residues on the lateral side of the GAS7 F-BAR contact the membrane. This oligomerization strategy of the GAS7 F-BAR domain into a flat sheet may explain why tubulation was not observed in the two standard assays [10, 14].
The S. pombe Cdc15 F-BAR domain forms tip-to-tip linear oligomers as visualized by EM, mediated by electrostatic interactions between reciprocally charged residues at dimer tips [10, 15]. A straight linear assembly of Cdc15 F-BAR domains would be consistent with their inability to deform membrane in vitro and also with the function of Cdc15 at a flat membrane along the cell wall of rod-shaped S. pombe [16]. Drosophila melanogaster Nervous wreck (Nwk) F-BAR domain is also non-tubulating and assembles tip-to-tip on monolayer membranes in vitro. Nwk dimers come together in a V-shape, resulting in zig-zag assemblies as visualized by EM [17]. This arrangement may explain the scalloped shapes of Nwk-bound membranes in vitro and when Nwk is overexpressed in S2 cells [17]. In the Cdc15 and Nwk F-BAR examples, the evidence suggests that the concave faces are utilized for membrane-binding when assembled in an oligomer.
Mammalian GAS7 and S. pombe Cdc15 exemplify how F-BAR domain structure and oligomerization strategy are connected to biological function. Consistent with the shallow curvature and sheet-like assembly of the GAS7 F-BAR domain, GAS7 localizes to the flat membrane at the base of the phagocytic cup in macrophages as revealed by super-resolution microscopy (Box 3) [14]. Furthermore, mutations that disrupt GAS7 F-BAR domain oligomerization inhibit phagocytosis, indicating the functional requirement for this GAS7 oligomerization mechanism. The mammalian FCHSD2 F-BAR dimer also has shallow curvature and localizes to the flat membrane at the base of clathrin-coated pits during endocytosis [18]. It is unknown what oligomeric FCHSD2 looks like in this context, but it will be interesting to see if FCHSD2 also assembles into flat sheets like the GAS7 F-BAR domain or in zig-zags like its orthologue Nwk [17]. S. pombe Cdc15 is a scaffolding protein of the cytokinetic ring, linking protein partners to the plasma membrane. In vitro studies demonstrated that oligomerization allows high avidity Cdc15 membrane-binding, suggesting that oligomerization allows efficient scaffolding of the cytokinetic ring in cells [10]. In support of this, Cdc15 oligomerization mutants are more dynamic and less stably associated with the membrane compared to wildtype and display cytokinetic ring instability. The shallow curvature of the Cdc15 F-BAR dimer, the tip-to-tip oligomerization strategy, and the inability to tubulate membrane in vitro are all consistent with function at a flat membrane in vivo.
Box 3: Super-resolution microscopy approaches.
Super-resolution microscopy provides better accuracy in localizing molecules than conventional microscopy methods through methods that bypass the diffraction limit of light. Super-resolution techniques include stimulated emission depletion microscopy (STED) [59, 60], structured illumination microscopy (SIM) [61], and the single molecule localization microscopy (SMLM) methods, stochastic optical reconstruction microscopy (STORM) [62] and photoactivated localization microscopy (PALM) [63]. SMLM techniques provide accurate positional information by selectively activating a subset of photoswitchable fluorophores or dyes coupled to the target at a time. When this subset of probes are extinguished, the sequence is repeated, allowing for acquisition of a large dataset of accurate position coordinates. SMLM methods can provide resolution of ~20 nm, which is equivalent to the length of a single F-BAR domain dimer. Thus, these methods provide the necessary resolution to model F-BAR domain assemblies [14].
At flat membranes, instead of driving membrane curvature, F-BAR domain oligomerization may function to generate high avidity membrane-binding and concentrate binding partners at the membrane. For instance, Cdc15 scaffolds a network of proteins via its F-BAR and SH3 domains that promote completion of cytokinesis [19, 20]. In mammalian cells, oligomerized F-BAR domains likely function similarly to create a high-density of binding modules for partners that promote actin polymerization for endocytosis, as well as podosome and phagocytic cup formation [18, 21–23].
While it is now appreciated that F-BAR domains oligomerize in a variety of modes, the next frontier is linking the structure of F-BAR dimers and their higher-order oligomeric assemblies to functions in vivo. For example, it is unclear how helical structures formed by CIP4 and FBP17 F-BAR domains in vitro relate to their functions at flat membranes, such as those of lamellipodia [13]. While the F-BAR domain of PACSIN/Syndapin can generate membrane tubules in vitro, crystal lattice contacts suggest that PACSIN1 assembles into sheets with contacts between the tip of one dimer and the core of another, with the concave face of each dimer facing the same direction toward the membrane [24]. Whether such an assembly could generate membrane tubulation in vivo, and if this flat assembly is utilized in cells is unknown [24]. In the case of S. pombe Imp2, although the F-BAR domain forms helical assemblies and tubulates membrane in vitro, structure-function analysis showed that mutations that disrupt oligomerization have little consequence for Imp2’s function in cytokinesis in vivo [12]. Thus, caution is warranted when drawing conclusions about the physiological relevance of membrane deformation from in vitro assays performed with supra-physiological concentrations of F-BAR domains.
F-BAR domains as protein interaction modules
F-BAR domains are typically connected to other domains that have protein interaction and/or enzymatic activities that in turn link F-BAR proteins to the actin cytoskeleton (reviewed in [11, 25]). Hence, F-BAR domains might be viewed simply as membrane-targeting modules, which merely localize other domains to their site(s) of action. Indeed, substitutions of an essential F-BAR domain in yeast with various yeast and human F-BAR domains support cell viability, suggesting that F-BAR domains are remarkably interchangeable membrane-binding modules [26]. However, there is mounting evidence that a key F-BAR domain function is mediating protein-protein interactions in addition to protein-membrane interactions, and these dual properties can allow F-BAR domains to form interaction platforms that tether complex actin-based structures to membrane.
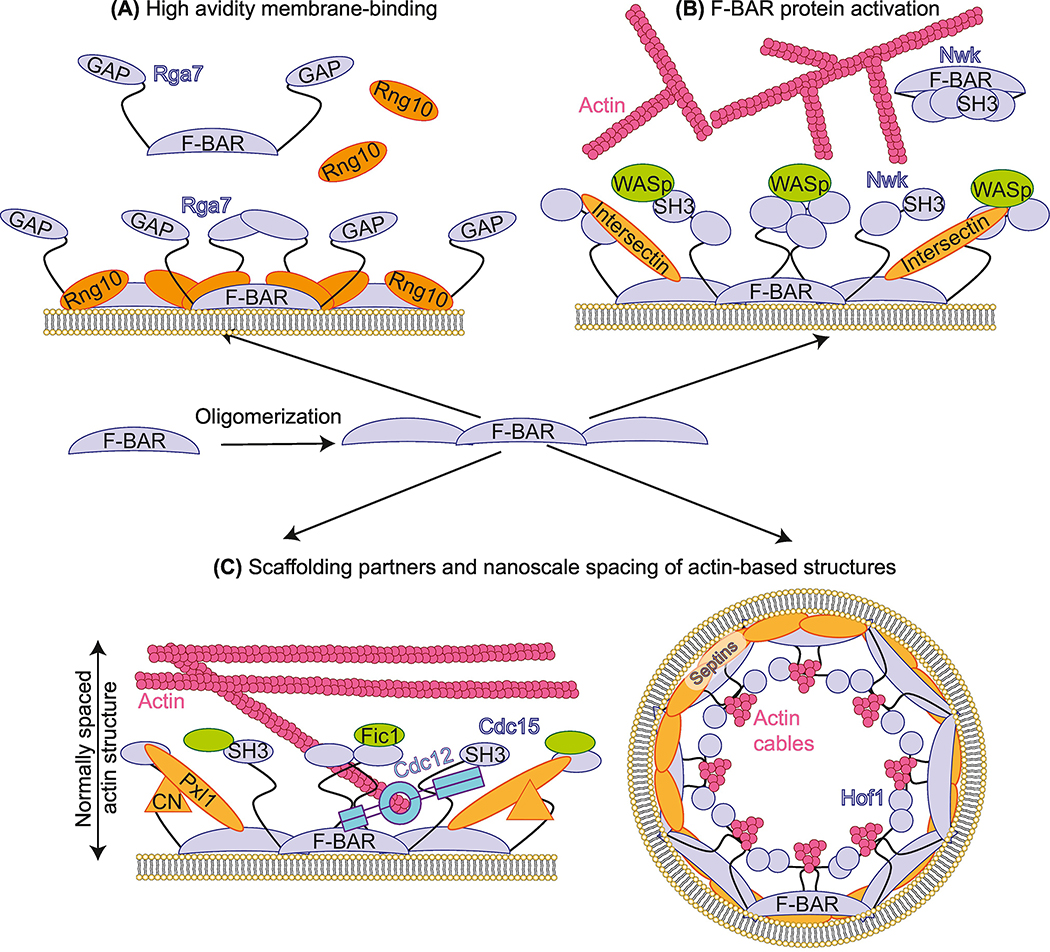
The list of reported F-BAR domain binding partners is expanding, as is insight into the functional consequences of such interactions in different biological contexts. For some F-BAR domains, a partner is required for robust localization to a site of action within the cell. An example of this type of relationship is the cooperation between S. pombe cytokinetic F-BAR protein Rga7 and coiled-coil protein Rng10 [27] (Figure 2A). Although the Rga7 F-BAR domain binds membrane in vitro, it does so with low affinity unless in complex with Rng10, and in cells Rga7 does not localize to the cell division site without Rng10, or vice-versa.
Figure 2: Through oligomerization, F-BAR domains polymerize protein interaction networks on membranes.
(A) Interaction between Rga7 F-BAR domain and coiled-coil protein Rng10 is required for high avidity membrane binding in vitro and in vivo in S. pombe [27]. (B) Coordinated binding by Intersectin/Dap160 and WASp is part of a multi-step model for Nwk/FCHSD2 activation at synaptic membranes [33].
(C) F-BAR domains scaffold protein partners directly at membrane to support the structure of actin-based networks. Left: in S. pombe, Cdc15 F-BAR domain utilizes opposite faces to bind membrane and actin nucleator Cdc12 simultaneously. The Cdc15 F-BAR domain scaffolds additional binding partners that dictate the functional nanoscale spacing of cytokinetic ring components [40]. Right: Possible arrangement of Hof1 F-BAR domain at the membrane looking down the bud neck in S. cerevisiae. Through its F-BAR domain and intrinsically disordered region, Hof1 and septins dictate the nanoscale spacing of actin cables [43]. Schematics not drawn to scale.
In platelets and megakaryocytes, the F-BAR domain of PACSIN2 binds the actin-binding protein Filamin A, and deletion or mutation of Filamin A disrupts PACSIN2 localization [28]. In epithelial cells, PACSIN2 F-BAR domain binds to polycystin-1 to facilitate its localization at lamellipodia membranes [29]. Similarly, the cytoskeleton-associated protein pyrin binds the PSTPIP1 F-BAR domain [30]. When expressed together in cultured cells, pyrin directs PSTPIP1 to inflammasomes, while PSTPIP1 does not localize there on its own [31]. Testing if Filamin A or pyrin modulates the affinity of PACSIN or PSTPIP1, respectively, for membrane would provide important mechanistic insights.
Binding partners also regulate F-BAR protein properties other than membrane binding. The D. melanogaster Nwk promotes membrane remodeling at synapses by activating WASp and directing actin assembly (Figure 2B). Nwk’s membrane and actin remodeling activities are autoinhibited by interactions between its N-terminal F-BAR and C-terminal SH3 domains [32]. Full activation of Nwk requires association with membrane, WASp, and Dap160/intersectin, a partner that binds both the F-BAR domain and a C-terminal SH3 domain of Nwk [33]. Multi-step activation of Nwk may ensure it promotes actin assembly only at the appropriate place and time.
In a reciprocal mechanism to that described for Nwk, some F-BAR domains influence the activity or localization of their partners. For example, during cytokinesis in Saccharomyces cerevisiae, the Hof1 F-BAR domain binds and inhibits Chs4, a chitin synthase III activator, which synthesizes the secondary septum at the division site [34]. Because Chs4 arrives at the bud neck during the early stages of cytokinesis, inhibition by the Hof1 F-BAR domain prevents premature septum synthesis [34]. The Hof1 F-BAR domain also inhibits actin cable formation by binding the formin Bnr1 to block actin nucleation [35]. This regulation ensures normal actin cable morphology and secretion [36].
F-BAR domains also serve to anchor proteins directly to membrane. In S. pombe, the Cdc15 F-BAR domain binds the cytokinetic formin Cdc12, promoting its recruitment to the division site where it nucleates the F-actin of the cytokinetic ring [37] (Figure 2C). Acting as a bridge between the cytoskeleton and the plasma membrane through F-BAR domain binding to Cdc12 is likely a key mechanism by which Cdc15 anchors the cytokinetic ring. The human F-BAR protein PSTPIP1 similarly localizes to the plasma membrane of the cytokinetic cleavage furrow [38].
In some cases, the F-BAR domain residues that mediate protein-protein interactions have been identified, but there does not appear to be a unifying mechanism of F-BAR domain–protein engagement. The PACSIN2 F-BAR domain associates with F-actin in vitro, but the residues implicated in this interaction are on the concave side of the F-BAR domain, which is also the membrane-binding face [39]. This suggests that the PACSIN2 F-BAR domain would be unable to bind membrane and F-actin simultaneously, and the physiological function of this interaction is unclear. For the Filamin A–PACSIN2 association, the residues important for binding are at the tips of the F-BAR dimer [28]. Because membrane binding is generally coincident with F-BAR dimer tip-to-tip oligomerization, determining if PACSIN2 can oligomerize on membranes in vivo and bind Filamin A simultaneously will be critical to understanding the physiological function of this association.
A completely different interaction mechanism has been identified between the formin Cdc12 and the Cdc15 F-BAR domain in S. pombe. Cdc12 binds the cytosolic face of the F-BAR dimer [40]. This positioning allows the F-BAR domain to simultaneously bind the membrane and formin, positioning Cdc12 for F-actin nucleation at the division site. A homology model of the PSTPIP1 F-BAR domain based on the FBP17 F-BAR domain structure reveals that negatively charged residues implicated in pyrin association are likely also on the convex side of the F-BAR domain [31]. Given these parallels, it will be interesting to determine if PSTPIP1 F-BAR domain also sandwiches itself between the membrane and a protein partner. Considering that most F-BAR domains use their concave faces for membrane-binding and tips for oligomerization, it is plausible that utilization of the convex face for protein binding will emerge as a general F-BAR feature.
F-BAR domains as platforms for complex actin-based structures
While there are now multiple examples of F-BAR domains acting as membrane-bound scaffolds for other proteins, there is emerging evidence that the impact of these interactions extends away from the membrane. For S. pombe Cdc15, mutating the cytosolic scaffolding face of the F-BAR domain not only reduces recruitment of formin Cdc12 to the cytokinetic ring, but also results in loss of paxillin-like protein Pxl1 and protein phosphatase calcineurin [40]. Ordinarily, the components of the cytokinetic ring are stratified into layers arranged from the plasma membrane toward the cell interior [41]. Surprisingly, disruption of the protein interaction network organized by the Cdc15 F-BAR domain collapses this architecture such that even components normally hundreds of nanometers away localize closer to the membrane [40].
Although the mechanism explaining how the Cdc15 F-BAR domain influences the nanoscale architecture of the cytokinetic ring is not yet clear, it is likely through binding partners. Pxl1 binds the Cdc15 F-BAR domain, but it also binds the Cdc15 C-terminal SH3 domain [40]. Because the Cdc15 F-BAR and SH3 domains occupy distinct spatial zones within the cytokinetic ring [41], it may be that Pxl1 displaces the Cdc15 SH3 domain away from the plasma membrane. Although this has not yet been tested, as described above there is precedent for the existence of proteins that bind multiple sites within an F-BAR protein. Dap160 interacts with both the F-BAR and SH3 domains of Nwk [33] and pyrin requires both the F-BAR domain and SH3 domain to bind PSTPIP1 [30]. Although the PSTPIP1 SH3 domain is dispensable for pyrin-mediated PSTPIP1 localization to the inflammasome [31], it may need to be engaged by pyrin for PSTPIP1 function. Determining if proteins generally modulate the overall conformation and function of F-BAR proteins via multivalent interactions will inform how they are regulated at actin-based structures.
Another possible explanation for the cytokinetic ring defects in cdc15 F-BAR scaffolding mutants is mis-regulated signaling in the absence of calcineurin. PSTPIP1 also facilitates signaling, by bringing a phosphatase and a substrate into proximity. PSTPIP1 binds the protein tyrosine phosphatase PEST through its F-BAR domain and the c-abl kinase through its C-terminal SH3 domain [42]. These binding events promote efficient c-abl dephosphorylation by PEST and thus regulate c-abl kinase activity. To test if Cdc15 works analogously to bring calcineurin and its substrates together, the full cohort of calcineurin substrates at the cytokinetic ring will first need to be identified.
F-BAR domains act as platforms for building actin-based structures in other biological contexts. The FCHSD2 F-BAR domain localizes to the base of clathrin coated pits, and is important for coordinating actin polymerization at these structures [18]. Similarly, the coordinated interactions of Dap160 and WASp with Nwk promote F-actin assembly during synaptic endocytosis [33]. In S. cerevisiae, Hof1 plays a role in linking F-actin cables to the septin network at the bud neck, which is crucial for proper nanoscale spacing of actin cables [43].
Concluding remarks
Multiple investigations have defined a subset of F-BAR domains that appear incapable of tubulating membrane under a wide variety of conditions (Table 1). We have proposed here that the unique sequence and structure of each domain dictates its oligomerization strategy and this, in turn, determines its ability to tubulate or deform membrane. The structure of the oligomer would also determine whether the domain could sense a particular membrane curvature and by binding it, perhaps stabilize it. In each of the studies we reference, not every possible lipid composition or membrane tension condition was tested in vitro and it can be argued that the ideal condition to allow membrane tubulation has yet to be defined. However, it is worth noting that the non-tubulating F-BAR domains were overproduced in intact cells that contain a rich assortment of membranes of varying composition and curvature and yet, tubulation was not observed. Therefore, the most parsimonious explanation for these observations is that the oligomeric structures of these F-BAR domains do not permit curvature sufficient for them to wrap around membrane tubules. Nonetheless, it remains possible that sequences adjacent to the F-BAR domain fragments tested, binding partners, or post-translational modifications may alter the core domain structure, even the oligomeric assembly strategy, and allow different membrane interaction properties to be revealed. Heterodimerization may also impact oligomeric structures in the one defined case of srGAP family F-BAR domains [44]. Returning to in vitro liposome tubulation assays with binding partners and knowledge of post-translational modifications will inform on these possibilities.
Though membrane tubulation ability varies among the F-BAR domains, the ability to oligomerize appears to be a unifying domain characteristic. However, in few cases do we have a complete picture of how the structure of the F-BAR dimer and its oligomerization mode support physiological function. To tease this apart, the strategy of oligomerization must be determined for each F-BAR domain. Then, investigators can perform appropriate structure-function analyses to test the role of oligomerization. The field also lacks information about the structure of F-BAR oligomers in their native contexts. It will be important to understand the size of oligomeric units in vivo and how many are required for F-BAR function. Super-resolution imaging approaches will need to be used to observe and measure the assemblies and to count the number of F-BAR dimers in each (Box 3).
F-BAR oligomerization likely serves to polymerize protein interaction networks on membranes. We presented emerging evidence that F-BAR domains themselves, rather than adjacent domains, can act as protein interaction hubs. Although instances of protein binding to F-BAR domains have been reported, further research is required to determine how extensive this mechanism of F-BAR domain function is. Certainly, the existence of distinct binding partners could explain why some F-BAR domains cannot fulfill the functions of others as would be expected if F-BAR domains were merely membrane-targeting modules [26]. Therefore, it will be informative to define the full cohort of F-BAR domain binding partners using genetic and biochemical approaches.
Given that there are now examples of a single F-BAR domain binding multiple proteins, a comprehensive understanding of interactors will open further questions regarding how multiple partners are coordinated. For instance, is multi-factor regulation by two or more proteins a common mode of F-BAR protein activation and/or regulation? Are F-BAR proteins differentially regulated spatially or temporally depending on which F-BAR domain partner is engaged? Alternatively, can a particular F-BAR domain bind two partners simultaneously? In addition, considering the examples in which F-BAR domains modulate the architectures of F-actin structures through multiple binding partners (Figure 2), future studies will need to consider the potential long-range consequences of disrupting F-BAR domain interactions.
Although we have focused on the F-BAR subfamily, the themes we have explored here extend more broadly to the BAR superfamily. For example, of the subfamily of sorting nexins that contain BAR domains (SNX-BARs) and function in endosomal sorting, only a subset can tubulate membrane [45]. Moreover, the Pinkbar (planar intestinal and kidney-specific BAR domain protein) I-BAR domain dimer has shallow curvature and cannot tubulate membrane [46] and the yeast Ivy1p I-BAR (inverse BAR) forms straight linear filaments in vitro, indicating that similar to the F-BAR family, there is diversity in oligomerization modes utilized by I-BAR proteins [47]. In analogy to the F-BAR domains discussed here, some BAR domains have binding partners [48–50], suggesting that a function in concentrating protein interaction networks at membranes may be a unifying characteristic of the larger superfamily.
Outstanding questions.
How does each F-BAR domain oligomerize?
What is the size and shape of each F-BAR domain oligomer?
Does the mode of F-BAR domain oligomerization confer function?
What is the full cohort of direct binding protein partners of each F-BAR domain and are they unique?
How do F-BAR domains support the structure and function of actin-based structures in different biological contexts?
Highlights.
F-BAR domains are membrane-binding modules that oligomerize and couple actin-based structures to membranes.
Although most F-BAR domains are observed to tubulate membranes in vitro, a subset of F-BAR domains lacking membrane tubulation activity have been identified, suggesting that membrane-tubulation activity does not unify F-BAR family function.
F-BAR domains employ diverse schemes of oligomerization and the oligomerization mode of a particular F-BAR domain dictates whether or not it can generate membrane curvature.
F-BAR domain oligomers polymerize protein interaction networks on membranes in part through recruitment of direct F-BAR domain binding proteins. Disruption of these membrane-proximal interactions can impact the organization of an entire actin-based structure.
Modes of F-BAR domain oligomerization and binding partners are specialized depending on cellular context.
Acknowledgements
We thank Gould lab members for helpful comments on this manuscript. C.E.S. was supported by NIH T32GM008554 and American Heart Association 17PRE33410245. This study was supported by grants from JSPS (KAKENHI 20H03252), from JST JST CREST (JPMJCR1863), and from the Uehara Memorial Foundation to S.S. and NIH R35GM131799 to K.L.G.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carman PJ and Dominguez R (2018) BAR domain proteins—a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophysical Rev 10, 1587–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suetsugu S et al. (2010) Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol 21, 340–9. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura T et al. (2018) Membrane re-modelling by BAR domain superfamily proteins via molecular and non-molecular factors. Biochem Soc Trans 46, 379–389. [DOI] [PubMed] [Google Scholar]
- 4.Itoh T and De Camilli P (2006) BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta 1761, 897–912. [DOI] [PubMed] [Google Scholar]
- 5.Henne WM et al. (2007) Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure 15, 839–52. [DOI] [PubMed] [Google Scholar]
- 6.Shimada A et al. (2007) Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129, 761–72. [DOI] [PubMed] [Google Scholar]
- 7.Peter BJ et al. (2004) BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 303, 495–499. [DOI] [PubMed] [Google Scholar]
- 8.Frost A et al. (2007) F-BAR Proteins Join the BAR Family Fold. Structure 15, 751–753. [DOI] [PubMed] [Google Scholar]
- 9.Frost A et al. (2008) Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald NA et al. (2015) Oligomerization but Not Membrane Bending Underlies the Function of Certain F-BAR Proteins in Cell Motility and Cytokinesis. Dev Cell 35, 725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald NA and Gould KL (2016) Linking up at the BAR: Oligomerization and F-BAR protein function. Cell Cycle 15, 1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald NA et al. (2016) The Tubulation Activity of a Fission Yeast F-BAR Protein Is Dispensable for Its Function in Cytokinesis. Cell Rep 14, 534–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsujita K et al. (2015) Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat Cell Biol 17, 749–58. [DOI] [PubMed] [Google Scholar]
- 14.Hanawa-Suetsugu K et al. (2019) Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7. Nat Commun 10, 4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts-Galbraith RH et al. (2010) Dephosphorylation of F-BAR protein Cdc15 modulates its confromation and stimulates its scaffolding activity at the cell division site. Mol Cell 39, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fankhauser C et al. (1995) The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell 82, 435–44. [DOI] [PubMed] [Google Scholar]
- 17.Becalska AN et al. (2013) Formation of membrane ridges and scallops by the F-BAR protein Nervous Wreck. Mol Biol Cell 24, 2406–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almeida-Souza L et al. (2018) A Flat BAR Protein Promotes Actin Polymerization at the Base of Clathrin-Coated Pits. Cell 174, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren L et al. (2015) The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell 26, 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts-Galbraith RH et al. (2009) The SH3 domains of two PCH family members cooperate in assembly of the schizosaccharomyces pombe contractile ring. J Cell Biol 184, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho HYH et al. (2004) Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell 118, 203–216. [DOI] [PubMed] [Google Scholar]
- 22.Takano K et al. (2008) EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J 27, 2817–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuboi S et al. (2009) FBP17 Mediates a Common Molecular Step in the Formation of Podosomes and Phagocytic Cups in Macrophages. J Biol Chem 284, 8548–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q et al. (2009) Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci U S A 106, 12700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suetsugu S and Gautreau A (2012) Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol 22, 141–150. [DOI] [PubMed] [Google Scholar]
- 26.Mangione MC et al. (2019) The intrinsically disordered region of the cytokinetic F-BAR protein Cdc15 performs a unique essential function in maintenance of cytokinetic ring integrity. Mol Biol Cell 30, 2790–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y et al. (2019) The F-BAR Domain of Rga7 Relies on a Cooperative Mechanism of Membrane Binding with a Partner Protein during Fission Yeast Cytokinesis. Cell Rep 26, 2540–2548.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Begonja AJ et al. (2015) FlnA binding to PACSIN2 F-BAR domain regulates membrane tubulation in megakaryocytes and platelets. Blood 126, 80–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao G et al. (2014) Polycystin-1 regulates actin cytoskeleton organization and directional cell migration through a novel PC1-Pacsin 2-N-Wasp complex. Hum Mol Genet 23, 2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoham NG et al. (2003) Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci U S A 100, 13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waite AL et al. (2009) Pyrin Modulates the Intracellular Distribution of PSTPIP1. PLoS One 4, e6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanishneva-Konovalova TB et al. (2016) Coordinated autoinhibition of F-BAR domain membrane binding and WASp activation by Nervous Wreck. Proc Natl Acad Sci U S A 113, E5552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Signore SJ et al. (2020) Autoregulation clamps the synaptic membrane-remodeling machinery and promotes productive actin-dependent endocytosis. bioRxiv, 2020.03.06.981076. [Google Scholar]
- 34.Oh Y et al. (2017) Hof1 and Chs4 Interact via F-BAR Domain and Sel1-like Repeats to Control Extracellular Matrix Deposition during Cytokinesis. Curr Biol 27, 2878–2886.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garabedian MV et al. (2018) Integrated control of formin-mediated actin assembly by a stationary inhibitor and a mobile activator. J Cell Biol 217, 3512–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graziano BR et al. (2014) The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol Biol Cell 25, 1730–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willet AH et al. (2015) The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol 208, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spencer S et al. (1997) PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol 138, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostan J et al. (2014) Direct interaction of actin filaments with F-BAR protein pacsin2. EMBO Rep 15, 1154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snider CE et al. (2020) Opposite Surfaces of the Cdc15 F-BAR Domain Create a Membrane Platform That Coordinates Cytoskeletal and Signaling Components for Cytokinesis. Cell Rep 33, 108526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald NA et al. (2017) Nanoscale architecture of the Schizosaccharomyces pombe contractile ring. Elife 6, e28865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong F et al. (2000) Cytoskeletal Protein PSTPIP1 Directs the PEST-Type Protein Tyrosine Phosphatase to the c-Abl Kinase to Mediate Abl Dephosphorylation. Mol Cell 6, 1413–1423. [DOI] [PubMed] [Google Scholar]
- 43.Garabedian MV et al. (2020) A septin-Hof1 scaffold at the yeast bud neck binds and organizes actin cables. Mol Biol Cell 31, 1988–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coutinho-Budd J et al. (2012) The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J Cell Sci 125, 3390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Weering JR et al. (2012) Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J 31, 4466–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pykalainen A et al. (2011) Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol 18, 902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh Y et al. (2016) Yeast Ivy1p Is a Putative I-BAR-domain Protein with pH-sensitive Filament Forming Ability in vitro. Cell Struct Funct 41, 1–11. [DOI] [PubMed] [Google Scholar]
- 48.Chen P-W et al. (2020) The BAR domain of the Arf GTPase-activating protein ASAP1 directly binds actin filaments. J Biol Chem 295, 11303–11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen P-W et al. (2016) The Arf GTPase-activating Protein, ASAP1, Binds Nonmuscle Myosin 2A to Control Remodeling of the Actomyosin Network. J Biol Chem 291, 7517–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasilina A et al. (2019) The ArfGAP ASAP1 Controls Actin Stress Fiber Organization via Its N-BAR Domain. iScience 22, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsujita K et al. (2006) Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol 172, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itoh T et al. (2009) The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci Signal 2, ra52. [DOI] [PubMed] [Google Scholar]
- 53.Moravcevic K et al. (2015) Comparison of Saccharomyces cerevisiae F-BAR Domain Structures Reveals a Conserved Inositol Phosphate Binding Site. Structure 23, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan Wah Hak L et al. (2018) FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat Cell Biol 20, 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cherry JM et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40, D700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood V et al. (2012) PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40, D695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitamata M et al. (2020) The roles of the diversity of amphipathic lipids in shaping membranes by membrane-shaping proteins. Biochem Soc Trans 48, 837–851. [DOI] [PubMed] [Google Scholar]
- 58.Ganzinger KA and Schwille P (2019) More from less – bottom-up reconstitution of cell biology. J Cell Sci 132, jcs227488. [DOI] [PubMed] [Google Scholar]
- 59.Hell SW and Wichmann J (1994) Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett 19, 780–2. [DOI] [PubMed] [Google Scholar]
- 60.Klar TA et al. (2000) Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci U S A 97, 8206–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gustafsson MG (2000) Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 198, 82–7. [DOI] [PubMed] [Google Scholar]
- 62.Rust MJ et al. (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3, 793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Betzig E et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–5. [DOI] [PubMed] [Google Scholar]