Abstract
Bacterial microcompartments are widespread organelles that play important roles in the environment and are associated with a number of human diseases. A key feature of bacterial MCPs is a selectively permeable protein shell that mediates the movement of substrates, products and cofactors in and out. Here we discuss current knowledge of selective transport across the protein shells of bacterial MCPs, including mechanisms, regulation and unanswered questions.
Keywords: Microcompartment, carboxysome, selective transport
Bacterial microcompartments (MCPs) are a widespread family of proteinaceous organelles. They are found in about 20% of bacteria and are used for autotrophic CO2 fixation as well as the catabolism of at least eight different carbon sources [1–4]. MCPs play important roles in the global carbon cycle and the ecology of diverse bacteria including a number of pathogens. MCPs are polyhedral in shape, about 100–400 nm in diameter and consist of metabolic enzymes encapsulated within a protein shell. Different types of MCPs encapsulate enzymes for the metabolism of different compounds. Some are used to concentrate enzymes together with their substrates, speeding up rate-limiting reactions and preventing counterproductive reactions with substrate analogs. Others sequester metabolic intermediates that are toxic to cytoplasmic constituents or would be lost to the environment by diffusion through the cell envelope (Figure 1).
Figure 1:
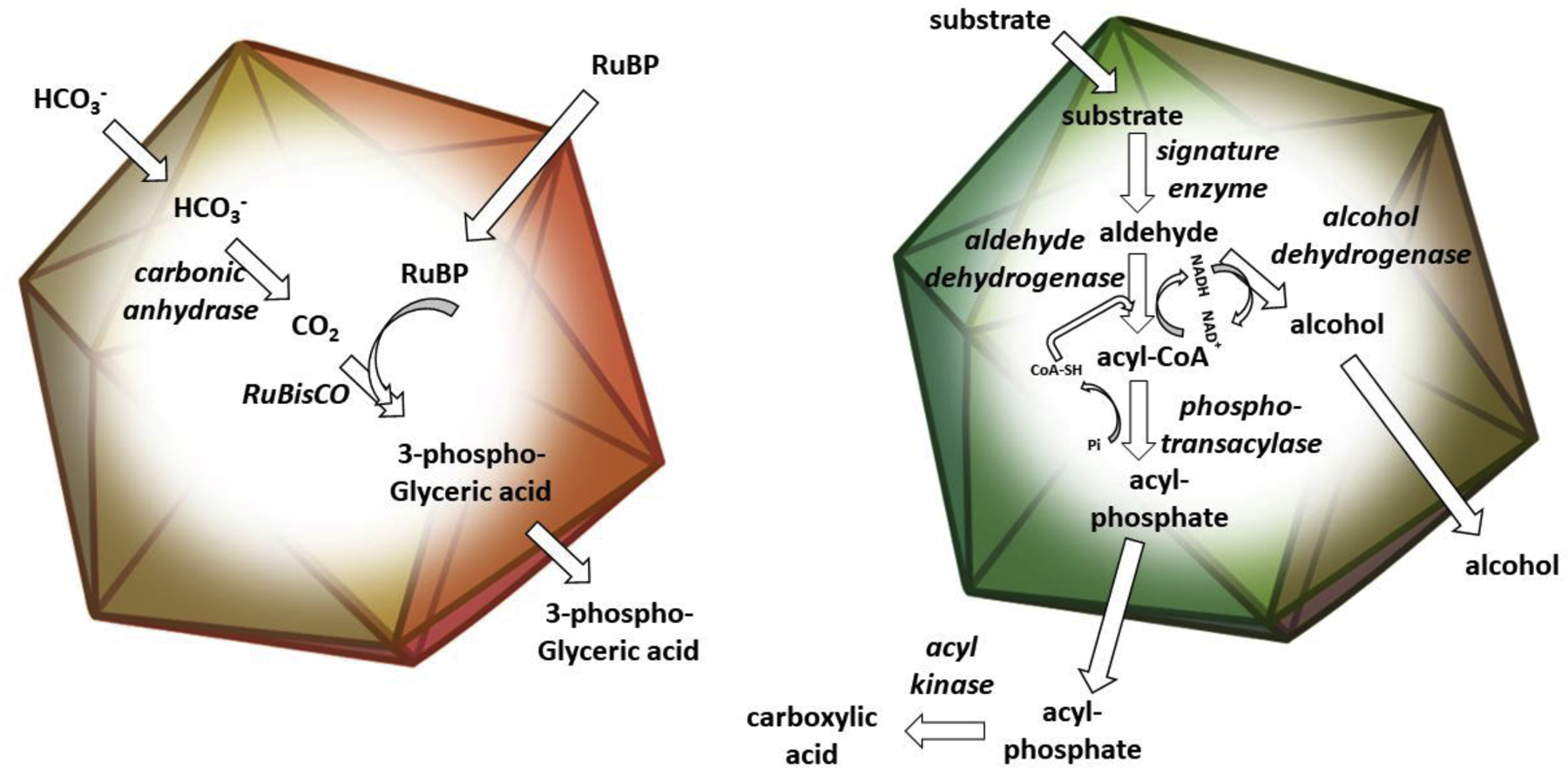
Schematic drawing of a carboxysome (left), and a catabolic MCP (right). Both carboxysomes and catabolic MCPs consist of metabolic enzymes encapsulated within a selectively permeable protein shell. The carboxysome (left panel) functions to enhance carbon fixation by concentrating CO2 around the catalytically inefficient RuBisCO enzyme (described further in the text). Catabolic MCPs are used to sequester intermediates that are toxic or poorly retained by the cell envelope (particularly aldehydes) during the metabolism of various carbon sources. A generic catabolic MCP is shown in the right panel. Enzymes are italicized.
A critical and defining component of MCPs are their protein shells [5,6]. The shells of MCPs control their internal environments by acting as selective barriers to the diffusion of small molecules (active transport has not been shown in MCPs) [2,7,8]. One particularly crucial function of the MCP shell is to impede the outward diffusion of toxic or poorly-retained metabolic intermediates (such as CO2 and short-chain aldehydes) that are generated internally within the MCP by the encapsulated enzymes. This effectively concentrates pathway intermediates within the MCP together with metabolic enzymes. In some cases, this is critical for increasing reaction rates, in other instances it is used to sequester toxic/poorly-retained intermediates that are further metabolized to non-toxic/well-retained compounds that subsequently exit the MCP and either enter central metabolism or leave the cell as waste products. In addition, there are indications that MCP shells impede the entry of compounds that would interfere with MCP metabolism via enzyme inactivation or by-reactions, in particular molecular oxygen. Perhaps most interesting however is that beyond confining and concentrating particular intermediates, MCP shells must also allow pathway substrates, products, enzymatic cofactors and possibly even electrons to pass inward and outward as necessary. How a protein shell controls the transport of multiple metabolites (and perhaps redox chemistry) is a key aspect of MCP physiology as well as an intriguing question in biochemistry.
Structural studies have provided many insights into how MCP shells function in selective molecular transport [2,5,7,8]. The shells of all known MCPs are polyhedral in shape and are primarily composed of proteins having Pfam domains 03319 or 00936. Proteins with domain 03319 are pentamers that comprise the vertexes of MCP shells (Figures 2 and 3). Members of this group, which are also known as bacterial microcompartment vertex (BMV) proteins have key structural roles but are not known to mediate molecular transport across the shell [9]. Proteins having Pfam domain 00936 are a family of diversified proteins, referred to as bacterial microcompartment (BMC) domain proteins. This protein family has key structural roles as well as varied roles in molecular transport [10]. BMC proteins form relatively flat, hexamers and pseudohexameric trimers that tile side-by-side into the mixed protein sheets that form the facets of MCP shells (Figures 2 and 3). Across genomes, BMC domain proteins show remarkable diversification (including gene fusions and rearrangements) that is apparently needed for efficient function of different MCP types [2,5,7,8]. Interestingly, given the nature of the variation seen, it is evident that transport requirements have substantially driven the divergence of BMC domain proteins. Canonical BMC domain proteins are homohexamers with each subunit having a single BMC domain [10]. Each hexamer has a central pore whose small size (4–7 Å) and electrostatic properties are thought to mediate selective molecular diffusion of smaller molecules such as MCP substrates and products (Figure 3). Many MCP operons include several paralogs of canonical hexamers with varied pore chemistries suggesting that different hexamers are used for transport of different compounds [6,7]. A second major class of shell protein has monomers comprised of two fused BMC domains [11–14]. This type of shell protein assembles into pseudohexameric trimers that have a similar overall shape to the canonical hexamers enabling the formation of mixed sheets from trimers and hexamers. In a number of instances, trimeric BMC domain proteins have iron sulfur clusters in place of the central pores suggesting a role in electron transfer across the MCP shell [13,14]. For a second type of BMC trimer, each monomer has two permuted BMC domains. In several instances, this class of BMC domain protein has been crystalized in two forms: one with a relatively large central pore and another where the central region of the protein is closed [11,15]. This suggests that some trimeric BMC domain proteins might act as dynamic gates for transport of larger substrates and enzymatic cofactors. Additionally, this type of BMC trimer been found in a stacked conformation that creates a central cavity suggesting molecular transport by an “airlock” mechanism [11,16]. Notably, the inferences made from structural studies of individual shell proteins are supported by structures of recombinant empty MCP shells which are icosahedra, with pentamers as the vertexes, hexamers and trimers forming the faces, and with the trimers stacked potentially forming “airlock” structures [17]. Thus, overall, structural biology has provided many insights into MCP functions and mechanisms.
Figure 2:
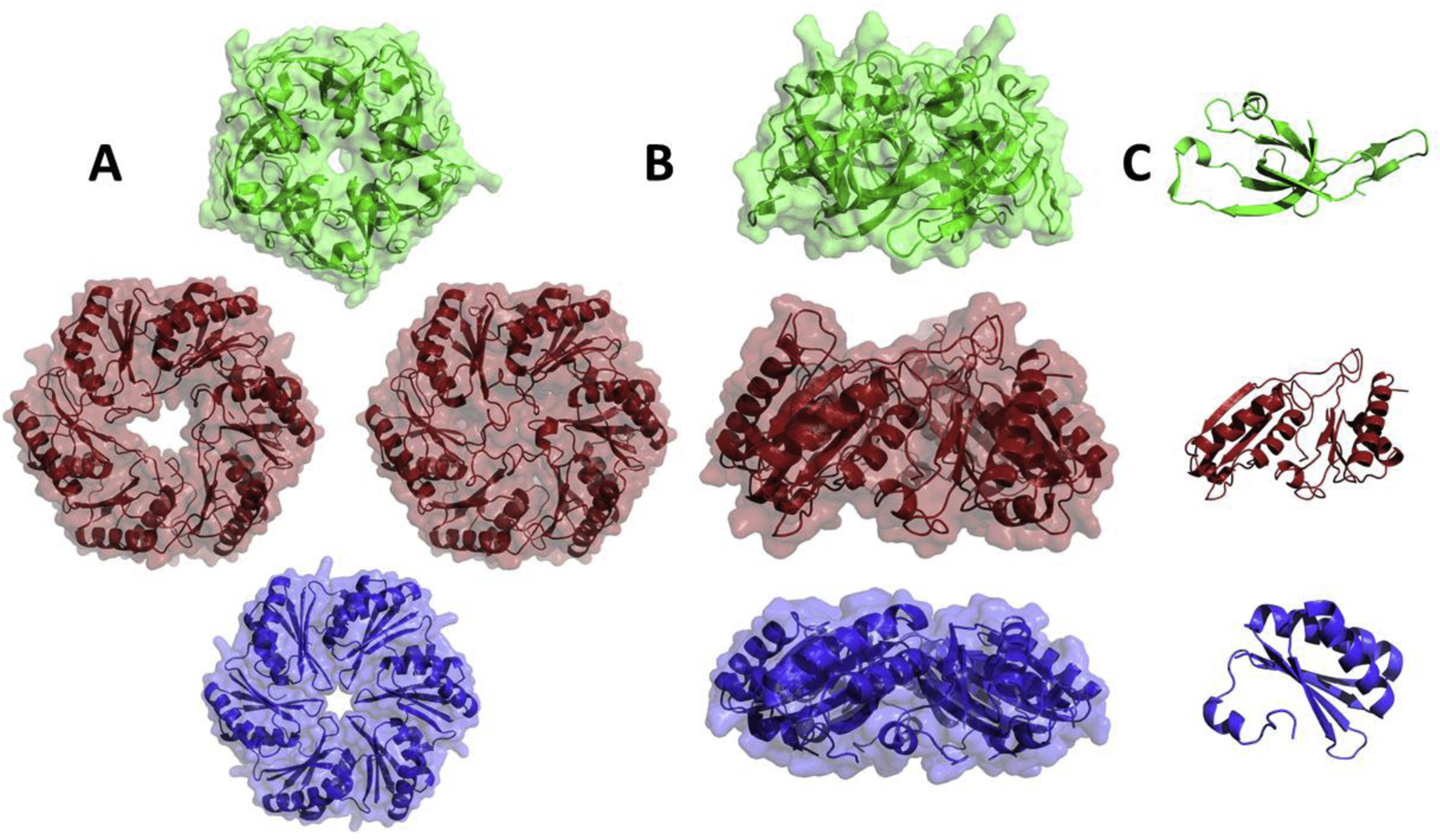
Representative BMC shell proteins. Top and side views of the homomeric ring (A and B) and the monomeric subunit (C). Top row: a pentameric shell protein from Synechocystis sp. PCC 6803 (PDB 2QW7, shown in green). This class of shell protein form the vertexes of the shell. Middle row: a permuted trimeric BMC domain shell protein from Escherichia coli K-12 shown in the closed and open conformations (PDB 3I82 and 3I87, shown in red). Member of this class of shell proteins are proposed to act as gated-pores for molecular transport. Bottom row: a canonical hexameric BMC domain shell protein from Salmonella enterica (PDB 3NGK, shown in blue). The central pores of canonical BMC hexamers are proposed to meditate the selective transport of MCP substrates and products.
Figure 3:
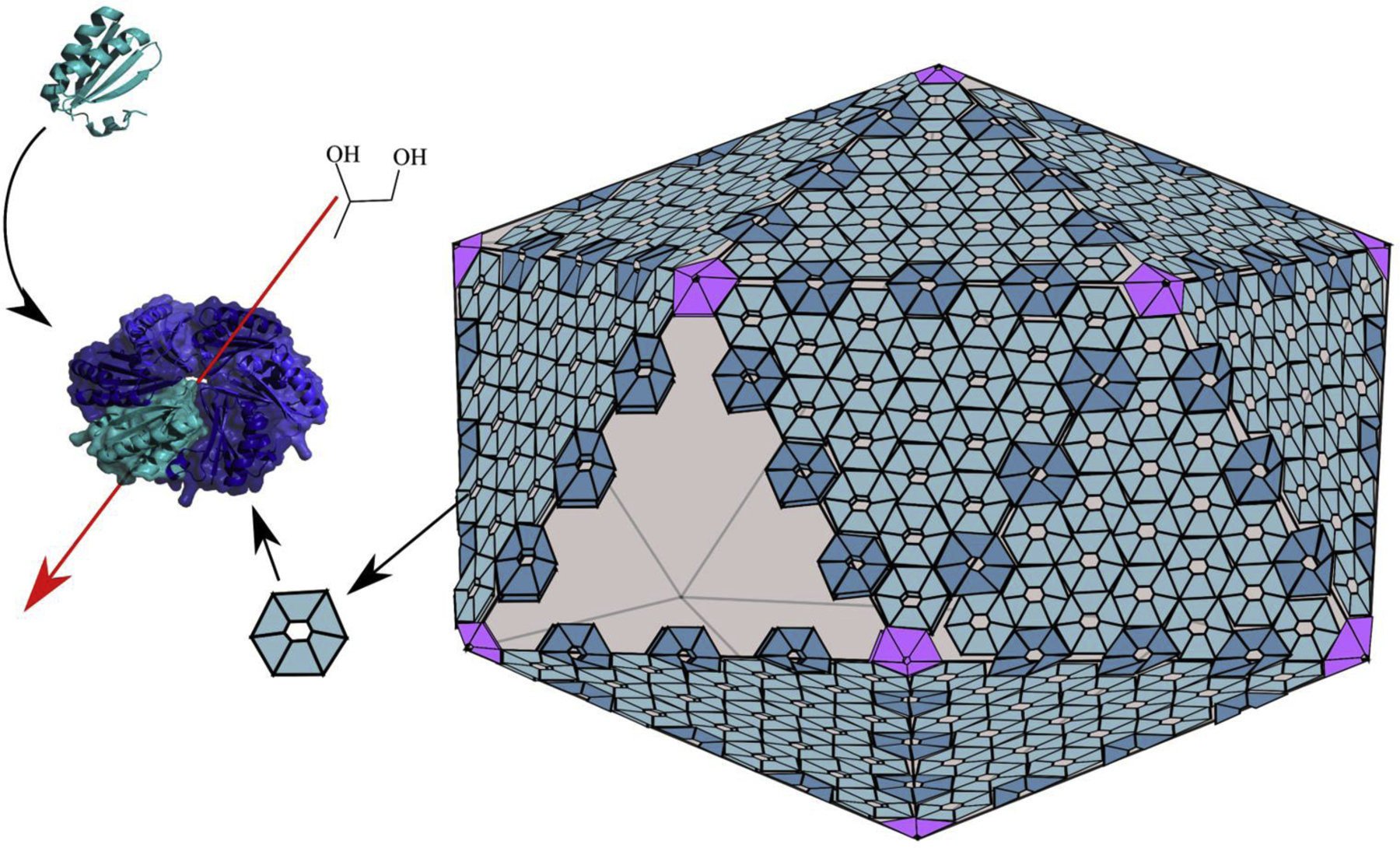
Generic diagram of the protein shells of bacterial MCPs. The shells of all known MCPs are built from a related set of proteins. The vertexes are occupied by pentamers having Pfam domain 03319. The facets of the shell are built from mosaic sheets of varied BMC domain proteins all of which include Pfam 00936. BMC domain proteins play key roles in molecular transport. For example, canonical hexamers (left) mediate the selective diffusion of MCP products and substrates (such as 1,2-propanediol) through there central pores. The overall shape and number of hexamers and other types of shell proteins are thought vary between MCPs.
With regard to a comprehensive understanding of MCP function, including molecular transport, our knowledge is most advanced for carboxysomes and the propanediol utilization (Pdu) MCP where insights from structural, physiological, genetic and computational studies combine to provide the most complete picture. There are two types of carboxysomes, α and β which differ in their phylogeny and composition [8]. Both types consist of Rubisco and carbonic anhydrase encased within a protein shell and are essential components of a CO2 concentrating mechanism (CCM) that improves the growth of autotrophic bacteria by the Calvin cycle [8]. The first step of the CCM is concentration of bicarbonate in the cytoplasm of the cell by active transport. Bicarbonate (and ribulose bisphosphate, RuBP) diffuse across the shell of the carboxysome and into the lumen. In the lumen, bicarbonate is converted to CO2 by carbonic anhydrase. The carboxysome shell acts as a barrier to the outward diffusion of CO2 resulting in elevated CO2 levels near Rubisco. Rubisco converts RuBP and CO2 to two molecules of 3-phosphoglycerate (3PGA) which diffuse outward into the cytoplasm and enter central metabolism. A high concentration of CO2 within the MCP near Rubisco speeds up its reaction rate (Rubisco is a slow enzyme that is rate limiting for the Calvin Cycle) and reduces photorespiration, a counterproductive by-reaction where O2 competes with CO2 as the substrate for Rubisco.
A great deal of work has gone into understanding how the carboxysome shell acts as a diffusion barrier to CO2 and also mediates the transport of Rubisco’s substrates and products. These studies have been carried out in several model systems and have investigated both α- and β-carboxysomes [6–8]. The role of the carboxysome shell as a barrier to outward CO2 diffusion was proposed by early mathematical models for the CCM [18]. Later, this idea was supported by genetic studies that showed carboxysome shell mutants require high CO2 for autotrophic growth and by studies that showed Rubisco activity is impaired in mutants unable to generate or retain CO2 internally within the carboxysome (carbonic anhydrase and shell protein mutants) [8,19–22]. It was also proposed that the carboxysome shell might serve as a barrier to the inward movement of O2 to further reduce photorespiration, although earlier work did not specifically test this idea. In addition, initial models did not address how larger molecules such as ribulose bisphosphate (RuBP) and 3-phosphoglyerate (the substrate and product of Rubisco) crossed the shell or how selective transport might be managed by a protein shell. In 2005, a seminal work by Yeates and co-workers reported the first structures of MCP shell proteins (carboxysome shell proteins CcmK2 and CcmK4). This study provided a number of insights into how MCP shells might assemble and act as selective permeability barriers [10]. The structures of CcmK2 and CcmK4 allowed identification of the BMC domain. Both the CcmK2 and CcmK4 proteins are relatively flat cyclic hexamers that tile into protein sheets suitable for shell formation. The primary openings in the sheets formed by CcmK2 and CcmK4 are pores through the centers of the hexamers and it was proposed that these pores might support molecular transport across the shell. It was also pointed out that the pores are lined with positive charges which might allow selective transport of bicarbonate (negatively charged) compared to CO2 and O2 (uncharged). The role of shell protein pores in transport is supported by the finding that a site-directed mutation in CcmK2 (S39A) impaired CO2 fixation and altered Rubisco kinetics [23]. Selective transport is supported by molecular dynamics studies that indicate HCO3− moves more easily through the central pores of α-carboxysome shell protein CsoS1A and β-carboxysome shell proteins CcmK2 and CcmK4 compared to CO2 or O2 [23,24]. The idea that the carboxysome shell acts as a barrier to O2 diffusion is supported by studies that suggest hydrogenase is protected from O2-inactivation when encapsulated within and engineered carboxysome shell [25]. With regard to the movement of substrates and products across the carboxysome shell, computational methods have suggested that 3PGA binding might induce a conformational change in CcmK2 pore that facilitates its transport [23]. On the other hand, however, it has been proposed that the permuted trimeric BMC domain proteins (discussed above) might act as “airlocks” for the transport of larger molecules such as 3PGA and RuBP [11,12,16]. Further, studies will be needed to work out the details.
Several studies have suggested that carboxysome permeability is regulated. Carboxysomes are typically comprised of 2–7 types of shell proteins depending on the species. Microarray studies have indicated that some shell proteins, particularly those of the β-carboxysome, are differentially regulated by environmental conditions such as oxidative stress or nitrogen limitation [26–28]. Given that different shell proteins often have different pore structures, changing their relative levels in carboxysomes could affect permeability. Furthermore, the interchangeability of shell subunits is supported by structural studies [29]. A recent study has also suggested post-transcriptional regulation of permeability where specialized BMC heterohexamers with alternative pore structures stack on the outer surface of the carboxysome, face-to-face with the hexamers that form the faces of the shell, forming a cap that alters metabolite flow through the shell [30]. This is supported by structural studies that showed β-carboxysome shell proteins CcmK3 and CcmK4 form mixed hexamers that stack to form dodecamers as well as by analyses that found structural properties that appear to favor heterohexamer staking over side-to-side tiling [30]. Overall, studies on carboxysomes indicate that multiple mechanisms are used to regulate shell permeability, but many details about these processes and their physiological relevance are unknown.
In addition to the carboxysome, substantial work has been done on molecular transport across the shell of the Pdu MCP. The Pdu MCP is used for the B12-dependent degradation of 1,2-propanediol as a carbon and energy source [31]. This MCP consists of 1,2-propanediol catabolic enzymes encapsulated within a protein shell [2]. The degradation of 1,2-propanediol starts with its diffusion across the shell and into the lumen of the MCP where it is first converted to propionaldehyde and subsequently to propionyl-phosphate and 1-propanol which exit the MCP and enter central metabolism, or leave the cell, respectively. The function of this MCP is to sequester propionaldehyde (a toxic intermediate) to prevent cellular toxicity and DNA damage which must be accomplished while allowing MCP substrates and products to traverse the shell [32]. In addition, the enzymes encapsulated within the shell of the Pdu MCP require NAD+, NADH, FAD, ATP, HS-CoA and coenzyme B12 [2] raising the question of how enzymatic cofactors are transported across the shell of the Pdu MCP which also restricts the outward diffusion of propionaldehyde.
The shell of the Pdu MCP is built from one BMV and seven BMC domain proteins [31]. Pdu BMC domain proteins include two canonical BMC hexamers (PduA and PduJ), a permuted hexamer (PduU), two permuted trimers (PduB and PduB’), one canonical trimer (PduT) and one canonical hexamer (PduK) with a ~70 amino acid C-terminal extension. Genetic, structural and computational studies have shown that the central pore of the PduA hexamer allows efficient transport of 1,2-propanediol (the substrate) into the Pdu MCP while restricting the efflux of propionaldehyde (a toxic intermediate) [33,34]. The central pore of the PduA protein is formed from six GSG motifs, one from each monomer, with serine S40 forming the constriction point [13] (Figure 4). Early structural studies pointed out that the pore of PduA is lined with numerous hydrogen-bond donors and acceptors which might result in the preferential diffusion of 1,2-propanediol over the less polar propionaldehyde [13]. Site-directed mutagenesis of residue S40 of PduA showed that its central pore is a major route of 1,2-propanediol uptake into the Pdu MCP and that this pore is designed to minimize propionaldehyde release [33]. Structural analyses of these same PduA S40 mutants supported a model in which pore permeability is controlled by the size of the pore and its electrostatic properties [33]. Further genetic studies indicated that residues near S40 (K37) also influenced transport and growth on 1,2-propanediol [35]. Molecular dynamics simulations indicated that the central pore of PduA preferentially allows the diffusion of 1,2-propanediol over propionaldehyde by about 3–10-fold [34]. Thus, the PduA shell protein, a major component of the shell of the Pdu MCP, is thought to allow the selective diffusion of 1,2-propanediol into the Pdu MCP.
Figure 4:
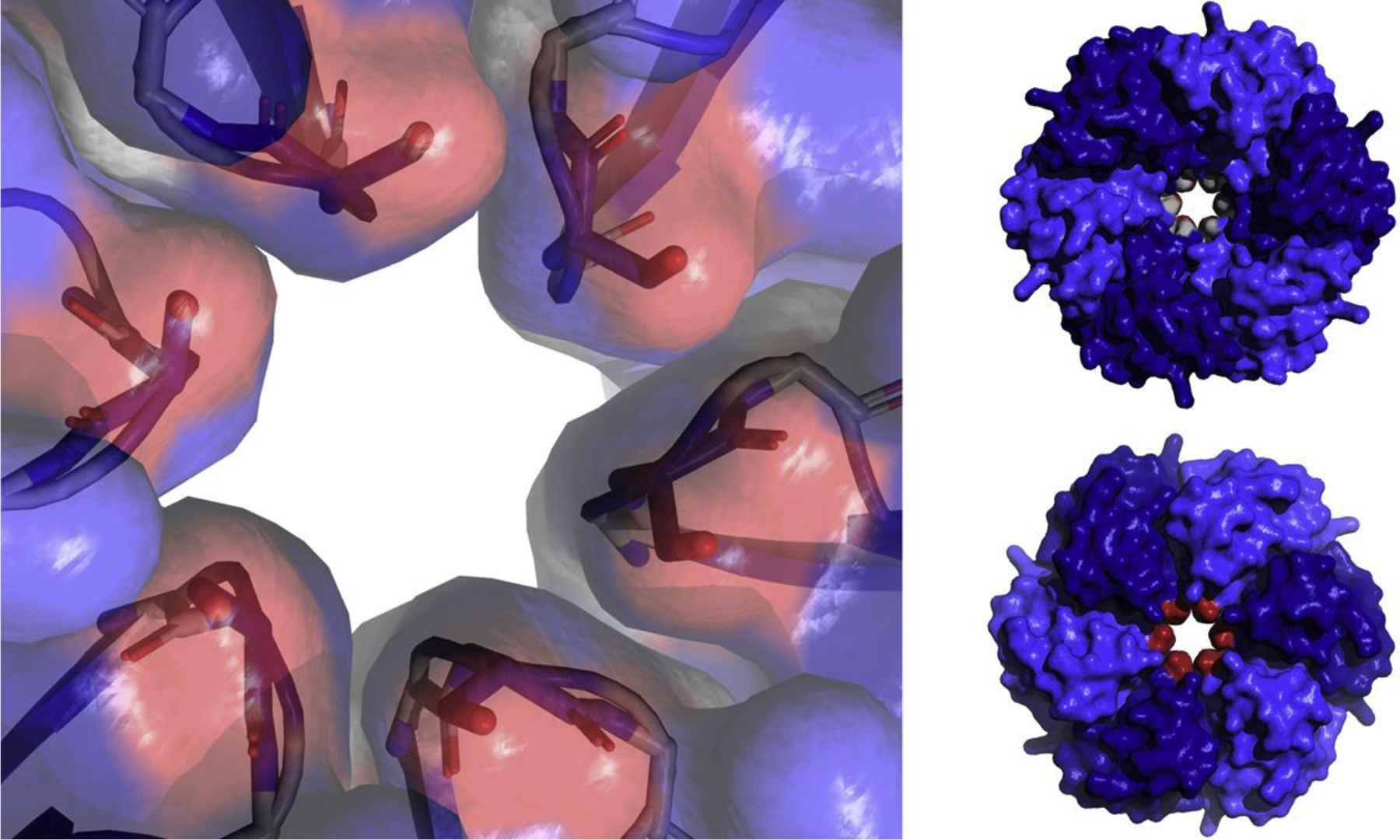
Close-up of the central pore of PduA (PDB 3NGK) showing the six serine 40 residues (red, one per subunit) which form the constriction point of the pore (left). Space filling models of the PduA hexamer from both sides showing the GSG motif which makes of the pore surface, the two glycines (shown in grey) can be seen in the pore (top right). The six serine 40 residues which form the constriction point (shown in red) can be seen in the pore (bottom right).
Given that PduA plays a key role in 1,2-propanediol transport into the Pdu MCP, it is surprising that PduJ has no apparent role in this process [36]. PduJ is a canonical BMC hexamer that is 82% identical to PduA in amino acid sequence. The pores of PduJ and PduA are essentially identical in structure and like PduA, PduJ is major shell constituent [2,37,38]. Yet, mutations in PduJ (equivalent to those that block the central pore of PduA), do not affect 1,2-propanediol uptake by the Pdu MCP [36]. This suggests that unknown factors influence the transport of 1,2-propanediol through the central pores of the PduA and/or PduJ shell proteins. In this regard, studies also showed that the chromosomal position of PduJ affects its function. When the pduA gene on the chromosome of S. enterica was replaced with the pduJ gene, the central pore of PduJ is then able to mediate 1,2-propaenediol transport based on site-directed mutagenesis of pore residues [36]. It was proposed that chromosomal position influences the arrangement of Pdu shell proteins within higher order complexes that make up the Pdu MCP in a way that affects molecular transport through their pores. Hence, studies indicate that transport through the central pores of BMC domain proteins is modulated by other factors that interact with BMC domain shell proteins.
A key question with regard to molecular transport across the shell of the Pdu MCP is how cofactors (NAD+, NADH, FAD, ATP, HS-CoA and coenzyme B12), which are required by the core enzymes, move across the shell. Two of the major components of the shell of the Pdu MCP are permuted trimers (PduB and PduB’). A number of proteins related in sequence to PduB and PduB’ have been crystallized in open and closed conformation suggestive of a gated central pore for the transport of larger molecules [11,12,39,40]}. In addition, as mentioned above, permuted trimers often occur in a stacked conformation that may allow them to function like an airlock [16,17]. Presumably, the airlock would open and close to allow cofactor transport in response to a signal. In the Eut MCP which is closely related to the Pdu MCP structural studies suggest the substrate (ethanolamine) allosterically regulates the pore of the EutL permuted trimer favoring a closed conformation [41]. Thus, MCP substrate might be used to promote pore closing. However, how these findings fit into MCP physiology has not been fully established and how a single type of shell protein mediates the transport of multiple cofactors is unclear. Thus, a number of questions about cofactor transport across the shell of the Pdu MCP are unanswered.
Studies have also shown that cofactors are recycled internally within MCPs and this has bearing on the presumed need for cofactor transport across the MCP shell. Genetic studies have shown that both NAD+ and HS-CoA are recycled internally within the Pdu (and Eut) MCP by the enzymatic reactions occurring therein [42–44]. This raises the possibility that cofactors could be encapsulated during MCP assembly then recycled internally eliminating the need for transport across the shell. Contrary to this idea, however, studies showed that Pdu MCPs genetically blocked for internal cofactor recycling still process 1,2-propanediol at about ½ the wild-type rate indicating cofactors could still be transported back and forth across the shell [42].
The shell of the Pdu MCP has three minor protein components (PduK, PduT and PduU) that are thought to fulfill specialized roles. PduT is a canonical trimer that has an iron-sulfur center occupying the central pore region [13,14]. Consequently, it was proposed that PduT is used for the transfer of electrons or iron-sulfur centers across the shell of the Pdu MCP. Studies have also indicated that PduT is associated with PduS (a B12 reductase) suggesting that PduT might transfer electrons across the shell for the reduction of B12 [14]. However, these ideas need further support. The two other minor shell components of the Pdu MCP are PduK and PduU. The central pore of PduU is capped by a β-barrel and PduK has a C-terminal extension of 70 amino acids [45]. However, the functions of these proteins are uncertain and there is no indication of a role in molecular transport across the shell.
Unanswered questions about molecular transport across MCP shells.
There are a number of puzzling questions surrounding molecular transport across MCP shells. As mentioned above, there is compelling evidence that the shell of the Pdu MCP restricts the outward diffusion of propionaldehyde. However, at the same time, the substrates and products of the Pdu MCP (1,2-propanediol, propionyl-phosphate and 1-propanol) must be efficiently transported across the shell and the lumen enzymes must be supplied with required cofactors: NAD+, NADH, FAD, ATP, HS-CoA and coenzyme B12. Most MCP loci (including the pdu locus) encode multiple canonical BMC hexamers with varied pore structures that might allow the selective transport of substrates and products, but the details have not yet been established. How the lumen enzymes are supplied with needed cofactors (which are much larger than propionaldehyde) is also incompletely understood. As mentioned, it has been proposed that permuted BMC trimers have larger gated pores for cofactor transport. However, for many MCP loci the number of BMC trimers present is insufficient for each cofactor to have its own transport protein [7,46]. Hence, trimeric gates would need to be relatively nonspecific to meet the cofactor requirements of the lumen enzymes. How this would occur at a molecular level is not clear. It has also been pointed out that some MCPs, such as type II choline utilization (Cut) MCPs lack trimeric BMC shell proteins. This highlights the question of how cofactor transport works in these cases and raises the possibility that different types of MCPs use different systems for cofactor homeostasis. Yet another interesting question is how the structure of the pore region of hexameric BMC domain proteins influence transport across the MCP shell. As mentioned, different shell proteins have varied pore structures consistent with selectivity for the varied substrates/products found in diverse MCPs, but the rules that govern transport have not been fully worked out. In general, it is thought that pore size and electrostatics determines substrate selectivity, but the details are still elusive. For example, shell proteins CsoS1 and CcmK2 from α- and β-carboxysome have very different pore lining residues but are thought to serve the same transport function. Lastly, we reiterate that a number of studies have raised the possibility that shell permeability is regulated. Regulation of transcription in response to changing environmental conditions could allow control of MCP shell composition and therefore permeability. Alternatively, protein factors might interact with shell proteins to modulate molecular transport. For example, despite the fact that the PduA and PduJ shell proteins are nearly identical only PduA mediates 1,2-propanediol transport. This suggests that other factors influence pore permeability of PduA or PduJ [36]. Studies have also suggested that some shell proteins might be capped by other specialized shell proteins, to alter shell permeability. In this case, the capping proteins could be regulated by changing environmental conditions [30]. Thus, overall, studies of molecular transport across MCPs shells have uncovered some key functional principles but there are still a number of outstanding questions about molecular mechanisms and control systems. The answers to these questions should provide us with a deeper understanding the design and operational principles of bacterial MCPs, protein-based containers that create specialized intracellular environments for metabolic optimization in part through the use of selectively permeable protein shells.
Highlights.
A key feature of bacterial microcompartments is a selectively permeable protein shell
The shells of microcompartments are able to retain toxic or volatile metabolites while allowing microcompartment substrates, products and cofactors to pass.
The central pores in microcompartment shell proteins control shell permeability based on their size, and their electrostatic and dynamic properties.
Molecular transport across microcompartment shells appears to be regulated at multiple levels, but the details are obscure.
Acknowledgements
This work was supported by grant AI081146 from the National Institutes of Health to T.A.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Abdul-Rahman F, Petit E, Blanchard JL: The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. Journal of Phylogenetics & Evolutionary Biology 2013, 01:1–7. [Google Scholar]
- [2].Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA: Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 2014, 78:438–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jorda J, Lopez D, Wheatley NM, Yeates TO: Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci 2013, 22:179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sutter M, Melnicki MR, Schulz F, Woyke T, Kerfeld CA: A catalog of the diversity and ubiquity of metabolic organelles in bacteria. bioRxiv 2021:2021.2001.2025.427685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yeates TO, Thompson MC, Bobik TA: The protein shells of bacterial microcompartment organelles. Current Opinion in Structural Biology 2011, 21:223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yeates TO, Jorda J, Bobik TA: The shells of BMC-type microcompartment organelles in bacteria. Journal of Molecular Microbiology and Biotechnology 2013, 23:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M: Bacterial microcompartments. Nature Reviews Microbiology 2018, 16:277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rae BD, Long BM, Badger MR, Price GD: Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev 2013, 77:357–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO: Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci 2013, 22:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO: Protein structures forming the shell of primitive bacterial organelles. Science 2005, 309:936–938. [DOI] [PubMed] [Google Scholar]
- [11].Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA: Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. Journal of Molecular Biology 2009, 392:319–333. [DOI] [PubMed] [Google Scholar]
- [12].Tanaka S, Sawaya MR, Yeates TO: Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 2010, 327:81–84. [DOI] [PubMed] [Google Scholar]
- [13].Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO: Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. Journal of Biological Chemistry 2010, 285:37838–37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pang A, Warren MJ, Pickersgill RW: Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster binding site. Acta Crystallographica Section D: Biological Crystallography 2011, 67:91–96. [DOI] [PubMed] [Google Scholar]
- [15].Tanaka S, Sawaya MR, Phillips M, Yeates TO: Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci 2009, 18:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA: The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment. Journal of Biological Chemistry 2013, 288:16055–16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sutter M, Greber B, Aussignargues C, Kerfeld CA: Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 2017, 356:1293–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kaplan A, Reinhold L: CO2 Concentrating Mechanisms in Photosynthetic Microorganisms. Annu Rev Plant Physiol Plant Mol Biol 1999, 50:539–570. [DOI] [PubMed] [Google Scholar]
- [19].Price GD, Badger MR: Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2 requiring phenotype: evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol 1989, 91:505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Price GD, Howitt SM, Harrison K, Badger MR: Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J. Bacteriol 1993, 175:2871–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC: CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem 2008, 283:10377–10384. [DOI] [PubMed] [Google Scholar]
- [22].Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S: The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 2009, 4:e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Faulkner M, Szabo I, Weetman SL, Sicard F, Huber RG, Bond PJ, Rosta E, Liu LN: Molecular simulations unravel the molecular principles that mediate selective permeability of carboxysome shell protein. Sci Rep 2020, 10:17501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mahinthichaichan P, Morris DM, Wang Y, Jensen GJ, Tajkhorshid E: Selective permeability of carboxysome shell pores to anionic molecules. J Phys Chem B 2018, 122:9110–9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li T, Jiang Q, Huang J, Aitchison CM, Huang F, Yang M, Dykes GF, He HL, Wang Q, Sprick RS, et al. : Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production. Nat Commun 2020, 11:5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Billis K, Billini M, Tripp HJ, Kyrpides NC, Mavromatis K: Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation. PLoS One 2014, 9:e109738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sommer M, Cai F, Melnicki M, Kerfeld CA: β-Carboxysome bioinformatics: identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J Exp Bot 2017, 68:3841–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sun Y, Wollman AJM, Huang F, Leake MC, Liu LN: Single-organelle quantification reveals stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell 2019, 31:1648–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Greber BJ, Sutter M, Kerfeld CA: The plasticity of molecular interactions governs bacterial microcompartment shell assembly. Structure 2019, 27:749–763.e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sommer M, Sutter M, Gupta S, Kirst H, Turmo A, Lechno-Yossef S, Burton RL, Saechao C, Sloan NB, Cheng X, et al. : Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of beta carboxysome shells. Plant Physiol 2019, 179:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC: The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. Journal of Bacteriology 1999, 181:5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sampson EM, Bobik TA: Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. Journal of Bacteriology 2008, 190:2966–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA: Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proceedings of the National Academy of Sciences of the United States of America 2015, 112:2990–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Park J, Chun S, Bobik TA, Houk KN, Yeates TO: Molecular dynamics simulations of selective metabolite transport across the propanediol bacterial microcompartment shell. Journal of Physical Chemistry B 2017, 121:8149–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Slininger Lee MF, Jakobson CM, Tullman-Ercek D: Evidence for improved encapsulated pathway behavior in a bacterial microcompartment through shell protein engineering. ACS Synthetic Biology 2017, 6:1880–1891. [DOI] [PubMed] [Google Scholar]
- [36].Chowdhury C, Chun S, Sawaya MR, Yeates TO, Bobik TA: The function of the PduJ microcompartment shell protein is determined by the genomic position of its encoding gene. Molecular Microbiology 2016, 101:770–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Havemann GD, Bobik TA: Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. Journal of Bacteriology 2003, 185:5086–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang M, Simpson DM, Wenner N, Brownridge P, Harman VM, Hinton JCD, Beynon RJ, Liu L-n: Decoding the stoichiometric composition and organisation of bacterial metabolosomes. Nature Communications 2020:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Larsson AM, Hasse D, Valegård K, Andersson I: Crystal structures of β-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore. Journal of experimental botany 2017, 68:3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pang A, Liang M, Prentice MB, Pickersgill RW: Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallographica Section D: Biological Crystallography 2012, 68:1642–1652. [DOI] [PubMed] [Google Scholar]
- [41].Thompson MC, Cascio D, Leibly DJ, Yeates TO: An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein. Protein Science 2015, 24:956–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cheng S, Fan C, Sinha S, Bobik TA: The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS ONE 2012, 7:e47144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu Y, Jorda J, Yeates TO, Bobik TA: The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. Journal of Bacteriology 2015, 197:2392–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Huseby DL, Roth JR: Evidence that a metabolic microcompartment contains and recycles private cofactor pools. Journal of Bacteriology 2013, 195:2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Crowley CS, Sawaya MR, Bobik TA, Yeates TO: Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure 2008, 16:1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stewart AM, Stewart KL, Yeates TO, Bobik TA: Advances in the world of bacterial microcompartments. Trends Biochem Sci 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]


