Abstract
Many allergens feature hydrophobic cavities that allow the binding of primarily hydrophobic small‐molecule ligands. Ligand‐binding specificities can be strict or promiscuous. Serum albumins from mammals and birds can assume multiple conformations that facilitate the binding of a broad spectrum of compounds. Pollen and plant food allergens of the family 10 of pathogenesis‐related proteins bind a variety of small molecules such as glycosylated flavonoid derivatives, flavonoids, cytokinins, and steroids in vitro. However, their natural ligand binding was reported to be highly specific. Insect and mammalian lipocalins transport odorants, pheromones, catecholamines, and fatty acids with a similar level of specificity, while the food allergen β‐lactoglobulin from cow's milk is notably more promiscuous. Non‐specific lipid transfer proteins from pollen and plant foods bind a wide variety of lipids, from phospholipids to fatty acids, as well as sterols and prostaglandin B2, aided by the high plasticity and flexibility displayed by their lipid‐binding cavities. Ligands increase the stability of allergens to thermal and/or proteolytic degradation. They can also act as immunomodulatory agents that favor a Th2 polarization. In summary, ligand‐binding allergens expose the immune system to a variety of biologically active compounds whose impact on the sensitization process has not been well studied thus far.
Keywords: group 2 house dust mite allergens, lipocalin, nsLTP, PR‐10, serum albumin
1. INTRODUCTION
The EAACI and WAO nomenclature task force has defined an allergen as an antigen that causes an allergic disease.1, 2 The task force did not further characterize allergens as being either harmless or noxious environmental substances. Such a bias was intentionally avoided based on the fact that allergens can fall into either of these categories, or they may belong to two additional ones defined below. The vast majority of allergens induce the synthesis of specific IgE (sIgE) which depends on the Th2 polarization of naïve T helper cells during a type 2 immune response. While type 1 immune responses that target replicating microbial pathogens have been studied in great detail, elucidation of the mechanisms leading to type 2 immune responses to multicellular parasites, venoms, and allergens is lagging behind. However, this area of research is starting to gain momentum.3
The ability of an allergen to initiate the very first steps that will eventually result in a type 2 immune response can be based on its “rather harmless” interaction with innate immune receptors present on epithelial cells, as has been shown for invertebrate tropomyosins.4 The interaction of an allergen with a host organism can also result in damage to innate receptors or other constituent parts of its cells. Such has been shown for airborne allergenic proteases from the mold Alternaria alternata or from house dust mite, which cause damage to the respiratory epithelium resulting in a type 2 immune response.5 On the far end of the spectrum, allergenic toxins such as phospholipases A2 present in venoms of stinging and hematophagous insects or predatory animals can cause necrotic cell death that leads to the induction of a strong type 2 response.6
There is another category of allergens that requires either components from the matrix7 or bound ligands8 to act as adjuvants and whose presence is essential for the induction of a type 2 immune response.9 Certain lipids from the Brazil nut matrix are necessary for inducing signaling pathways that ultimately result in the synthesis of sIgE against the major Brazil nut allergen Ber e 1, a seed storage 2S albumin.10 The major birch pollen allergen Bet v 1 by itself neither stimulated dendritic cells in vitro nor induced Th2 polarization in vivo; it required components from the pollen matrix present in a birch pollen extract to manifest a Th2 polarization.11 Pru p 3, a non‐specific lipid transfer protein (nsLTP) and major allergen of peach, harbors a derivative of camptothecin bound to phytosphingosine as a ligand.12 The phytosphingosine part of the ligand was responsible for the activation of antigen‐presenting cells.13 Moreover, mice exposed to Pru p 3 plus ligand developed more Pru p 3‐sIgE than when exposed to Pru p 3 alone.
In addition to their immunological adjuvanticity, ligands can also increase the stability of allergens against proteolytic degradation. The birch pollen‐derived E1‐phytoprostane, recently identified as another Bet v 1 ligand, was shown to enhance the resistance of Bet v 1 to the proteolytic processing by endolysosomal extracts, which was proposed to directly influence its allergenicity.14 Likewise, the binding of fatty acids by the cockroach allergen Bla g 1 was found to significantly enhance the allergen's thermostability while inhibiting cleavage by cathepsin S, an endosomal protease essential for antigen processing and presentation.15 These topics are discussed in more details below.
This review summarizes our current knowledge on allergenic proteins that bind small molecule ligands. It also gives an overview on which types of ligands may bind to allergens and what their role in inducing a type 2 response is or might be. Thus, the intention is to indicate new avenues of scientific inquiry into the sensitizing capacities of allergens that are neither damaging nor toxic to host cells by themselves (Box 1).
BOX 1. Major milestone discoveries.
Availability of experimental molecular structures for a range of allergens with hydrophobic cavities16, 17
Establishment of biophysical and biochemical methods to study ligand‐protein interaction and its impact on local structural changes18, 19, 20, 21, 22, 23
Identification of group 2 house dust mite (HDM) allergens as ligand‐binding proteins16, 17
Discovery of the natural ligand of Pru p 312
Elucidation of the role of the phytosphingosin part of the Pru p 3 ligand in the sensitization process27
Discovery of the natural ligand of Cor a 129
Allergens from the PR‐10 family bind a broader spectrum of ligands in vitro than in vivo25, 26, 30
Ligand enhances proteolytic resistance of Bet v 1 and inhibits endolysosomal cathepsin S protease activity14
2. SERUM ALBUMINS
Serum albumins (SAs) are members of a highly conserved protein family that includes numerous allergens.31, 32, 33 SAs are clinically relevant allergens that originate from animals, where they are major blood components. SAs are also present in milk, muscle, and epithelia. It was demonstrated that ~30% of individuals allergic to animal dander show IgE reactivity toward SAs.34 A mature SA molecule is composed of approximately 585 amino acid residues that fold into three distinctive domains of similar size and whose structure is stabilized by several disulfide bridges (Figure 1, Equ c 3).35
FIGURE 1.
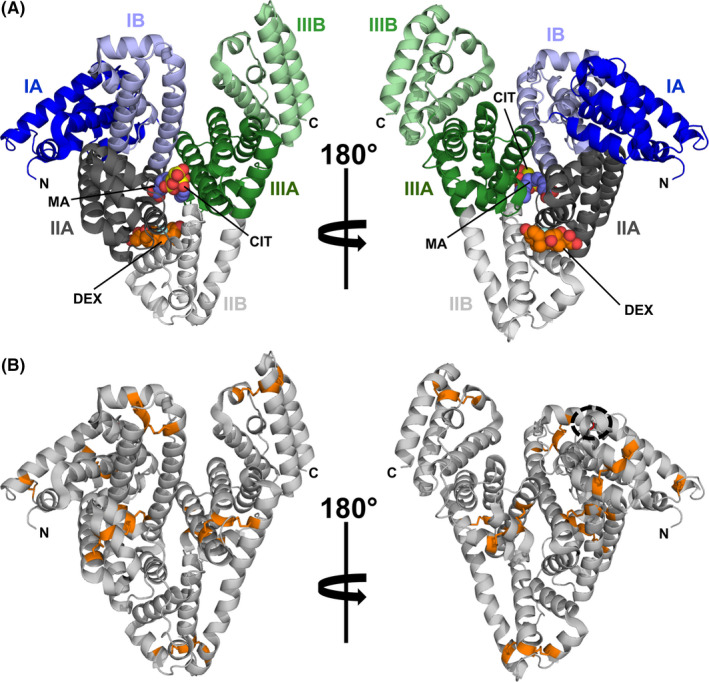
Model of equine serum albumin (ESA) in cartoon representation (PDB code: 6XK0). (A) Domain assignment as described by Sugio et al35 ESA binds simultaneously dexamethasone (DEX, carbon atoms shown as orange spheres), citric acid (CIT, carbon atoms shown as yellow spheres), and myristic acid (MA, carbon atoms shown as purple spheres).159 (B) Model of ESA with Cys residues and disulfide bridges marked in orange. Position of a single Cys residue, which is not participating in the formation of a disulfide bridge, is marked with a black dashed circle
The molecular architecture of SAs allows these molecules to adopt multiple conformations that facilitate simultaneous binding of various ligands (Figure 1),36 making SAs highly capable small‐molecule carrier proteins.37, 38 SAs bind many endogenous and exogenous compounds.39 In addition, SAs play a very important role in the transport of metal cations.40, 41, 42 Structural studies revealed the presence of many ligand‐binding sites. Currently, nine fatty acid‐binding sites have been identified, as well as several sites that are responsible for binding drugs and various metabolites or hormones.43
The most relevant allergenic SAs originate from mammals and birds.32 The allergenic mammalian SAs have very high sequence identities and similarities, cat and dog SAs being the most similar to the human homologue (approximately 83% identities). Even avian SAs display quite high sequence identities (>45%) and similarities (>60%) to human SA (HSA). Therefore, it is not surprising that SAs, due to their high sequence identities, show significant levels of cross‐reactivity.33, 41, 44 To date, there are eight SAs whose structures have been determined: bovine (BSA, Bos d 6), canine (CSA, Can f 3), equine (ESA, Equ c 3), feline (FSA, Fel d 2), HSA, goat (GSA), ovine (OSA),and rabbit (RSA).35, 41, 45, 46, 47, 48
While the ligand‐binding properties of human serum albumin are best studied, there is also a significant body of literature on interactions of small‐molecule compounds with BSA and ESA.39, 43, 49 Structural studies clearly show that SAs bind a broad spectrum of compounds that are biologically active, some of which may affect the human immune system. For example, SAs bind steroid hormones, saturated and unsaturated fatty acids, thyroxine, vitamin D, and flavonoid metabolites, as well as many exogenous compounds like drugs (Table 1).37, 38, 39, 43, 51 It was also shown that small‐molecule compounds like carbohydrates may bind SAs through the amino groups of lysine residues that are located on the surface of these proteins.49, 52 It was shown that derivatives of carbohydrates, such as the food additive D‐mannitol, can form stable covalent derivatives with SAs, leading to the haptenization of these proteins.52 In fact, D‐mannitol is a member of a large group of small molecule compounds that form covalent bonds with proteins like SAs. The interaction of the small molecule compounds with SA does not always require an aqueous solution, but the proteins may be modified by compounds present in air. One such example is the modification of HSA by vapors of hexamethylene diisocyanate leading to immunogenic protein derivatives.53
TABLE 1.
Natural compounds binding to allergens. The table lists only compounds for which experimental confirmation is available
| Types of compounds | Ligands | Allergen family | Allergens | References |
|---|---|---|---|---|
| Fatty acids | Arachidic acid | PR‐10 | Ara h 8, Bet v 1 | 25, 65 |
| Lauric acid | Lipocalin | Bos d 5 | 157 | |
| Lauric acid | nsLTP | Pru p 3 | 27, 158 | |
| Myristic acid | SA | Equ c 3 | 159 | |
| Myristic acid | PR‐10 | Bet v 1 | 25 | |
| Oleic acid | nsLTP | Cor a 8, Jug r 3, Mal d 3, Ole e 7, Pru p 3, Zea m 14 | 27, 102, 104, 107, 108 | |
| Oleic acid | PR‐10 | Bet v 1 | 25 | |
| Palmitic acid | nsLTP | Pru p 3, Zea m 14 | 27, 160 | |
| Palmitic acid | PR‐10 | Ara h 8, Bet v 1 | 25, 65 | |
| Stearic acid | PR‐10 | Bet v 1 | 25 | |
| Fatty acids (C12 to C22) | Bla g 1 | Bla g 1 | 15, 28 | |
| Lauric acid | Uteroglobin | Fel d 1 | 152 | |
| Lipo‐oligosaccharides | Lipoteichoic acid | Bla g 1 | Bla g 1 | 15 |
| LPS | NPC2 | Der f 2 | 16 | |
| LPS | NPC2 | Der p 2 | 121 | |
| LPS | nsLTP | Par j 1 | 161 | |
| Lipopeptides | Polymyxin B | LBP | Der p 7 | 134 |
| Oxylipins | Phytoprostane B1 | PR‐10 | Bet v 1 | 14 |
| Phytoprostane E1 | PR‐10 | Bet v 1 | 14 | |
| Phytoprostane F1 | PR‐10 | Bet v 1 | 14 | |
| Ricinoleic acid | nsLTP | Zea m 14 | 107 | |
| Phospholipids | Dipalmitoylphosphatidylcholine | nsLTP | Ole e 7 | 108 |
| Dipalmitoylphosphatidylglycerol | nsLTP | Ole e 7 | 108 | |
| Dipalmitoylphosphatidylserine | nsLTP | Ole e 7 | 108 | |
| Phosphatidylinositol | Bla g 1 | Bla g 1 | 28 | |
| Phosphatidylserine | Bla g 1 | Bla g 1 | 28 | |
| Phosphatidylcholine | Bla g 1 | Bla g 1 | 28 | |
| Phosphotidylethanolamine | Bla g 1 | Bla g 1 | 15, 28 | |
| Phosphotidylglycerol | Bla g 1 | Bla g 1 | 15, 28 | |
| Sphingosine derivatives | 10‐hydroxy‐camptothecin linked to phytosphingosine | nsLTP | Pru p 3 | 13 |
| Flavonoids and flavonoid derivatives | Apigenin | PR‐10 | Ara h 8, Bet v 1, Que a 1 | 65 |
| Daidzein | PR‐10 | Ara h 8, Bet v 1, Que a 1 | 65 | |
| Epicatechin | PR‐10 | Ara h 8, Bet v 1, Que a 1 | 65 | |
| Genistein | PR‐10 | Ara h 8, Bet v 1, Que a 1 | 65 | |
| Myricetin | PR‐10 | Fra a 1 | 26 | |
| Naringenin | PR‐10 | Bet v 1, Cor a 1 | 25, 63 | |
| Quercetin‐3‐O‐glucuronide | PR‐10 | Fra a 1 | 162 | |
| Q3O‐(Glc)‐Gala | PR‐10 | Cor a 1 | 29 | |
| Quercetin‐3‐O‐sophoroside | PR‐10 | Bet v 1 | 24 | |
| Catecholamines and trace amines | Norepinephrine | OBPb | Aed al 2 | 163 |
| Octopamine | Lipocalin | Bla g 4 | 92 | |
| Tyramine | Lipocalin | Blag 4 | 92 | |
| Odorants and pheromones | Limonene‐1,2‐epoxidec | Lipocalin | Rat n 1 | 88 |
| 2‐(sec‐butyl)thiazole | Lipocalin | Mus m 1 | 89 | |
| Steroids | Cholesterol | NPC2 | Der p 2 | 122 |
| Dehydroergosterol | PR‐10 | Bet v 1 | 25 | |
| Deoxycholate | PR‐10 | Bet v 1 | 64 | |
| Testosterone | SA | Equ c 3 | 39 | |
| Stigmasterol | PR‐10 | Ara h 8, Bet v 1, Cor a 1, Que a 1 | 65 | |
| Progesterone | PR‐10 | Ara h 8, Cor a 1, Que a 1 | 65 | |
| Androstenone | Uteroglobin | Fel d 1 | 152 | |
| Cytokinins | IPAd | PR‐10 | Bet v 1 | 65 |
| Kinetin | PR‐10 | Bet v 1 | 25 | |
| Zeatin | PR‐10 | Bet v 1, Pru p 1 | 65, 164 | |
| Gibberellins | Gibberellin A3 | PR‐10 | Vig r 6 | 165 |
| Hydroxycinnamic acids | Caffeic acid | PR‐10 | Ara h 8, Cor a 1 | 65 |
| Ferulic acid | PR‐10 | Ara h 8, Cor a 1 | 65 | |
| Stilbenoids | Resveratrol | PR‐10 | Ara h 8, Bet v 1, Cor a 1 | 62, 65 |
Q3O‐(Glc)‐Gal, quercetin‐3‐O‐(2“‐O‐β‐D‐glucopyranosyl)‐β‐D‐galactopyranoside.
Odorant binding protein.
It is not clear whether the ligand in the Rat n 1 structure (PDB code: 2AG2) was properly identified.
IPA, N‐isopentenyladenosine.
In summary, SAs are able to simultaneously bind many diverse compounds and it can be assumed that some small molecule compounds are always bound to these proteins. Therefore, the human immune system is exposed to SAs that are tightly associated with small molecules that may have immunomodulatory properties. One may speculate that the small molecule binding properties of SAs are responsible for these proteins to potentially become allergens despite the fact that mammalian SAs are very similar to HSA.33
3. PR‐10 ALLERGENS
Class 10 pathogenesis‐related proteins (PR‐10s) are part of a plant's immune defense against pathogens or abiotic stress.54, 55 PR‐10s include a wide variety of clinically relevant pollen and food allergens that possess a conserved three‐dimensional structure.29, 56, 57 Typically they contain a hydrophobic cavity which can accommodate different ligands (Table 1).56 PR‐10s are formed by a seven‐stranded antiparallel β‐sheet and a long C‐terminal α‐helix enclosed by two shorter helices arranged in a V‐shape (Figure 2A).58 Due to the high structural similarity of PR‐10 allergens, IgE antibodies can cause cross‐reactivity via a range of different exposure scenarios.59 Natural PR‐10 allergens are usually comprised of a mixture of different isoallergens (>67% amino acid sequence identity) and variants (>90% identity).60, 61 They were shown in vitro to bind a variety of small molecules, that is, flavonoids, cytokinins, and steroids, indicating functions in UV protection, transport of small molecules, and regulation of germination.25, 62, 63, 64, 65
FIGURE 2.
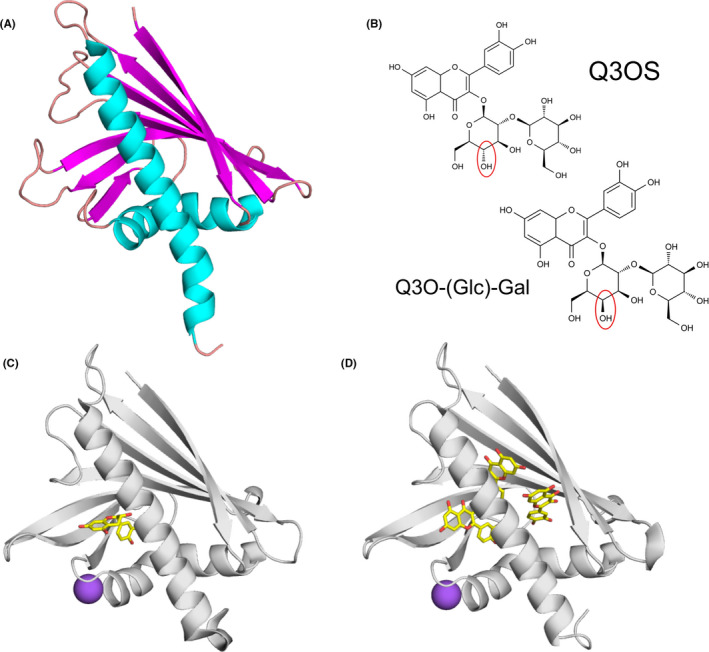
Allergens from the PR‐10 family and their ligands. (A) Cartoon representation of Bet v 1 (PDB code: 6R3C) with secondary structure elements marked with distinct colors (α‐helices in cyan, β‐strands in magenta, and loops in salmon). (B) Natural ligands of Bet v 1.0101 (Q3OS) and Cor a1.0401 (Q3O‐(Glc)‐Gal).29 (C) Crystal structure of Ara h 8.0101 in complex with epicatechin (PDB code: 4MA6).62 Epicatechin is shown in stick representation. Purple sphere represents Na+. (D) Crystal structure of Ara h 8.0101 in complex with quercetin (PDB code: 6AWS)
The glycosylated flavonoid derivative quercetin‐3‐O‐sophoroside (Q3OS) was identified as a natural ligand of Bet v 1.0101, the most abundant birch pollen isoallergen.26 Another quercetin derivative, quercetin‐3‐O‐(2´´‐O‐β‐D‐glucopyranosyl)‐β‐D‐galactopyranoside (Q3O‐(Glc)‐Gal), was recently found to be a natural ligand of Cor a 1, the major hazel allergen.29 Although the two ligands differ only in the orientation of one OH group in the sugar moiety (glucose vs. galactose) (Figure 2B), Cor a 1 selectively binds its own ligand but not Q3OS and vice versa. Of all isoallergens known, only Cor a 1.0401 and Bet v 1.0101 showed binding of the identified compounds.24, 26, 29 Since non‐glycosylated flavonoids are bound rather promiscuously, the varying sugar moieties appear to be responsible for the binding specificity. Interestingly, RNAi‐mediated silencing of the Fra a 1 genes in strawberry resulted in white strawberries due to a decrease of different glycosylated metabolites involved in flavonoid biosynthesis.66 These results suggest that glycosylated flavonoids might also be natural ligands of other PR‐10 allergens.
Other PR‐10 allergens that were characterized in terms of their interactions with ligands include proteins originating from peanut and kiwi fruit. There are two Ara h 8 isoforms from peanut in the official WHO/IUIS allergen database (www.allergen.org), Ara h 8.01 and Ara h 8.02. Although they share only 55% amino acid sequence identity, both were shown to bind the same small molecule ligands (apigenin, genistein, quercetin, daidzein, progesterone, arachidic acid, palmitic acid, and resveratrol; Table 1).65 Ara h 8.01 can also bind epicatechin and three molecules of quercetin (Figure 2) without undergoing an extensive structural change. In the case of Ara h 8, it was demonstrated that lipid binding increased its thermal and proteolytic stability.67 Act d 11 from kiwi fruit belongs to the major latex protein/ripening‐related protein family (MLP/RRP) and has the same overall fold as Bet v 1.68, 69 The crystal structure of Act d 11 purified from its natural source revealed the presence of an unknown ligand that is likely to contain an indole or a similar ring structure. While the identity of the ligand is still unknown, Act d 11 should be considered a “dressed allergen” that carries small molecule compounds capable of interacting with the human immune system.63
IgE‐binding assays and mediator release assays with Bet v 1 variants showed no increased binding affinities in the presence of ligand.14, 26 Furthermore, no increased activation of dendritic cells could be observed.14 However, pollen‐derived ligands like phytoprostane E1 enhanced the thermostability of Bet v 1 and increased its proteolytic resistance against the endolysosomal proteases cathepsin S and legumain.14 Preliminary data with Api g 1.0101 from celery indicated that the protease stability of Api g 1.0101 against trypsin was higher in the presence of the flavonoid aglycon apigenin.70 It has been shown that increased thermostability affects immunogenicity and allergenicity.71 Furthermore, reduced proteolytic processing of allergens could result in low loading and density of class II MHC peptide complexes, which in turn favors Th2 polarization.72
4. LIPOCALINS
Lipocalin allergens are the most important group of respiratory animal allergens and include a number of allergens with high sensitization rates among allergic individuals.73 In addition, the lipocalin Bos d 5 is a major allergen in bovine milk, which is the common trigger of food allergy in childhood.74 Lipocalins form a large protein family and are found in animals, plants, and bacteria. They are small single‐domain proteins of 150–180 amino acid residues. The amino acid sequence identities between different lipocalins are low but the fold is conserved, consisting typically of 8 antiparallel β‐strands forming a β‐barrel, and one α‐helix (Figure 3). Inside the β‐barrel, there is a central cavity which can usually accommodate one ligand (Table 1). The major function of lipocalins is to transport poorly water‐soluble ligands.75, 76 Over 20 lipocalin allergens have been characterized, the majority of which are respiratory allergens except for the food allergen Bos d 5.73
FIGURE 3.
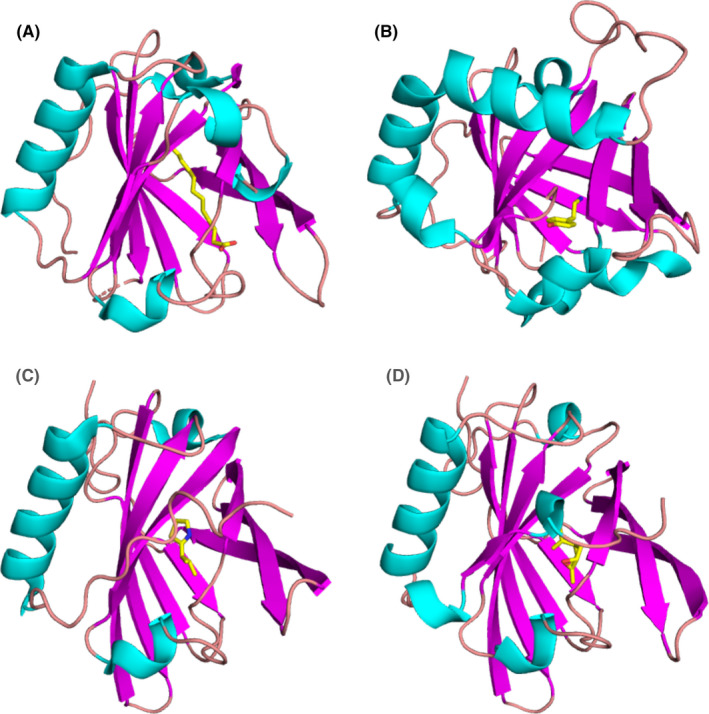
Allergens from the lipocalin family. (A) Binding of lauric acid to Bos d 5 monomer according to the crystal structure (PDB code: 4IB6).157 Lauric acid is shown in stick representation. (B) Bla g 4 in complex with tyramine (PDB code: 4N7C).92 (C) Mus m 1 in complex with a pheromone (PDB code: 1MUP).89 (D) Complex of Rat n 1 with limonene‐1,2‐epoxide (PDB code: 2A2G)88
Bos d 5 or β‐lactoglobulin occurs at the high concentration of 0.1 mM in bovine milk.77, 78 The ligand‐binding specificity of Bos d 5 is not very high as many different ligands, primarily fatty acids, have been reported for this allergen. The binding affinity for short chain fatty acids is weak (KDs are in mM range) but affinity is higher for long‐chain fatty acids (KDs are usually in µM range).79 A low or moderate binding affinity to Bos d 5 allows dissociation of ligands from the complex after transport. The impact of ligand binding on allergenicity or immunogenicity of Bos d 5 has been investigated in few studies. It has been proposed that retinoic acid or iron‐quercetin would bind to T‐cell and B‐cell receptors and suppress immunogenicity and allergenicity of Bos d 5.80, 81 Based on a study in a mouse model, it was also suggested that ligand‐bound Bos d 5 was able to protect against allergic sensitization to unrelated birch pollen allergens.82
Crystal structures of at least nine inhalant lipocalin allergens have been determined. The structures for Bos d 2, Can f 2, Can f 4, Can f 6, and Equ c 1 do not contain any ligands but the binding sites have been analyzed.83, 84, 85, 86, 87 The crystal structure of Rat n 1 contains limonene‐1,2‐epoxide88 and Mus m 1 harbors a thiazole derivative(Figure 3).89 All of these structures display a closed binding site, suggesting that a conformational change of the protein is required for ligand association and dissociation.90 The binding of tyramine and octopamine to Bla g 4 was especially well‐characterized by X‐ray crystallography (Figure 3) and isothermal titration calorimetry.91, 92 The possible effect of ligand binding to respiratory allergens has not been studied in detail. The Toll‐like receptor 4 (TLR4) agonist lipopolysaccharide was suggested to bind to Can f 6 and enhance innate immune signaling.93
The role of ligands for the allergenicity of lipocalins requires further investigations. However, some general remarks can be made. The food allergen Bos d 5 seems to have a lower ligand‐binding specificity and affinity compared to respiratory allergens. The binding affinity of the ligand is an important consideration. Many experiments such as basophil activation tests are performed at nanomolar allergen concentrations. If the allergen concentration is much lower than the KD for ligand binding, the number of complexed proteins and ligands is very low even in the presence of a clear excess of the ligand. The wide range of ligand‐binding capacities of Bos d 5 may indicate a general transport role for clearance of harmful compounds.75, 94
5. NON‐SPECIFIC LIPID TRANSFER PROTEINS
Non‐specific lipid transfer proteins (nsLTPs) belong to the prolamin superfamily and are ubiquitous plant proteins. They have been classified as pathogenesis‐related proteins as their expression is upregulated by biotic and abiotic stress.95, 96, 97 nsLTPs are categorized either as LTP1 (approximately 9 kDa) or LTP2 (approximately 7 kDa). nsLTPs are small soluble basic proteins that share a common fold based on four α‐helices stabilized by four conserved disulfide bridges. This compact structure enables nsLTPs to be highly resistant to heat and enzymatic degradation. A common feature of nsLTPs is a tunnel‐like hydrophobic cavity with a ligand‐binding site. This ligand‐binding pocket is able to accommodate and transport a wide variety of lipids, from phospholipids to fatty acids, as well as sterols and prostaglandin B2 (PGB2), among others (Table 1; Figure 4).98, 99 Accordingly, the lack of ligand specificity of nsLTPs resides in the high plasticity and flexibility of their lipid‐binding cavity, which is able to accommodate anything from one or two fatty acids to single‐ or double‐chain lipids.100
FIGURE 4.
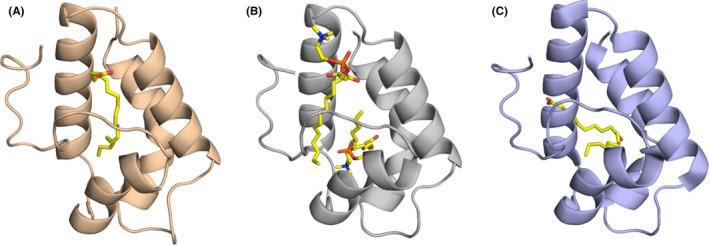
Representative structures of nsLTPs showing binding to different ligands. (A) Maize nsLTP in complex with α‐linolenic acid (PDB code: 1FK6).107 (B) Wheat nsLTP in complex with 1‐myristoylglycero‐3‐phosphorylcholine (PDB code: 1BWO).166 (C) Barley nsLTP forming a covalent complex with (12E)‐10‐oxo‐12‐octadecenoic acid (PDB code: 3GSH)167
A considerable number of allergenic nsLTPs from pollen and plant foods have been identified. Pru p 3 from peach, the first identified allergenic LTP1, is regarded as a major allergen that affects more than 50% of peach allergic patients, inducing symptoms that range from mild to severe.101 Pru p 3 displayed preferential binding to unsaturated short chain fatty acids such as oleic acid27, 102, 103 and phytosphingosine.13 A structural model was developed for Jug r 3, the LTP1 from walnut, by using an NMR interaction study and data‐driven docking calculations to assess the effect of ligand binding of oleate.104 Applying the WaterLOGSY NMR method, binding of oleic acid versus stearic acid was compared and the preferred binding of oleic acid was confirmed. Furthermore, the OLE‐binding sites were identified including the C‐terminal region of helix alpha 2, most of helix alpha 3,2 the loop between helices alpha 3 and alpha 4 and the C‐terminal loop. Regarding the ligand its hydrophobic tail is inside the cavity, while the carboxylate part is surface exposed. Overall, the internal cavity from Jug r 3 seems to be flexible depending on the ligand binding. In that context, the C‐terminal loop undergoes a conformational change upon lipid binding resulting to a more surface exposed position as compared to unliganded Jug r 3. This local conformational change leads to an increased IgE binding as shown by antibody binding assays and basophil activation tests. These data are in line with reports from Pru p 3, where the same area was previously identified being part of an immunodominant IgE epitope, which was less accessible for IgE antibodies in the apoform, versus the holoform providing increased IgE binding.104, 105
The prevalence of sIgE to Ole e 7, an LTP1 and minor allergen from Olea europaea pollen increases up to 50% in populations exposed to high levels of olive pollen.106 Interestingly, oleic acid, an unsaturated fatty acid with a C18 chain, showed the greatest binding affinity to Ole e 7 in accordance with other nsLTPs from maize, peach, apple, hazelnut, and walnut.27, 102, 104, 107 Ole e 7 preferentially binds negatively charged phospholipids such as phosphatidylserine and phosphatidylglycerol, with cholesterol able to compete with this binding.108 While the direct effect of this interaction on allergenicity has yet to be observed, its implications on Ole e 7 interfacial behavior could facilitate sensitization.
Ole e 7, like other aeroallergens, enters the body through the upper airways, reaching the mucosal surface and making contact with two lipid‐based barriers: the pulmonary surfactant located on the outer side of the mucus and the luminal plasma membrane of airway epithelial cells. Ole e 7 was shown to adsorb to air‐liquid interfaces and effectively interact with pre‐formed lipid monolayers composed of negatively charged phospholipids and cholesterol.108 Ole e 7 seemed to reverse the inhibitory effect on the surface adsorption of the pulmonary surfactant manifested by plasma proteins such as HSA and other clinically relevant allergens like Ole e 1.109 This suggests that Ole e 7 transfers surfactant lipid components to the interface whose impact on the structure and function of the pulmonary epithelium has yet to be determined.
In conclusion, the interaction of nsLTPs with ligands affects the local protein conformation and in turn the recognition by IgE antibodies, relevant for the allergic effector phase. Whether that ligand interaction is also important for the allergic sensitization phase still remains to be defined, as it was also shown that ligand binding may result in an increased susceptibility of nsLTPs to gastroduodenal proteolysis.110
6. GROUP 2 HOUSE DUST MITE ALLERGENS OF THE NIEMANN‐PICK TYPE C2
Proteins belonging to the ML superfamily have ML (MD‐2‐related lipid‐recognition) domains, which have been identified in multiple proteins of unknown biological function, associated with interaction with lipids, in plants, animals, and fungi. House dust mite (HDM) allergens from group 2 are some of the most important mite allergens, eliciting IgE antibody responses in 70%–90% of mite‐allergic patients.111, 112, 113 Sensitization to these allergens is associated with the development of allergic diseases, such as asthma, rhinitis, and atopic dermatitis. A longitudinal study of the evolution of the IgE response to a panel of twelve HDM allergens during the first two decades of life showed that Der p 2 was the earliest recognized allergen.113 This study also confirmed the importance of Der p 2 as a major allergen, together with Der p 1 and Der p 23. In addition to its immunogenic properties, Der p 2 can aggravate respiratory airway disease by adjuvant‐like activation of lung epithelial cells.114
Der p 2 and Der f 2 from the species Dermatophagoides pteronyssinus and D. farinae, respectively, are small approximately 14 kDa proteins that form a single immunoglobulin‐fold domain consisting of two three‐stranded antiparallel β‐sheets (Figure 5A).17, 115, 116, 117 The X‐ray crystal (but not the NMR) structure of Der p 2 revealed a large internal hydrophobic cavity, defined by the two β‐sheets, that was able to bind lipidic ligands.17, 116 Similarly for Der f 2, the separation and angle between the two sheets in the first NMR and crystal structures (“closed”)118, 119 were narrower than those described in a more recent X‐ray crystal structure (“open”).120 Der f 2 was shown to bind LPS (lipopolysaccharide) with nanomolar affinity.16 LPS bound to a cluster of basic residues at one edge of the pocket entrance that attracts negatively charged LPS, with its acyl chains inserted between the β‐sheets. To bind LPS, the internal cavity of Der f 2 must open wider than the “open” state described above.16 Der p 2 also bound LPS, although with low affinity.121 Recently, both recombinant allergens were reported to bind many lipids promiscuously.122 They preferentially bound cholesterol among eleven different lipids tested with a liposome pulldown assay.122
FIGURE 5.
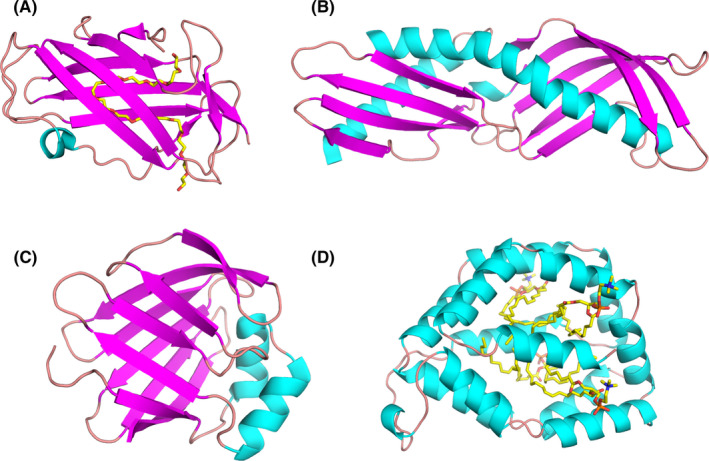
Models of ligand‐binding proteins from less common allergen families. (A) Crystal structure of Der f 2 with polyethylene glycol (PEG; PDB code: 1XWV). While PEG originates from the solution used during crystallization, its presence reveals a large cavity that is used by Der p 2 to bind hydrophobic ligands.120 (B) Structure of Der p 7 (PDB code: 3H4Z).134 This allergen was shown to bind the bacterial lipopeptide polymyxin B. (C) Solution structure of Der f 13 (PDB code: 2A0A).168 Der f 13 is a member of the fatty acid‐binding protein family. (D) Structure of Bla g 1 in complex with lipids including phosphatidic acid28
Group 2 HDM allergens belong to the ML superfamily of proteins, which bind specific lipids and play important roles in lipid recognition and metabolism.123 The ML superfamily also includes MD‐2, a member of the myeloid differentiation factor‐2‐related lipid‐recognition protein family, and the human cholesterol‐binding Niemann‐Pick type C2 (NPC2) protein. Hence, the binding of cholesterol by Der p 2122 is intriguing because the structure of Der p 2 is more closely related to the human NPC2 protein than to MD‐2.121, 124 As group 2 HDM allergens also share structural homology with MD‐2, a study investigated whether they could functionally act as MD‐2.125 MD‐2 is the LPS‐binding component of TLR4 signaling complex. Association of the MD‐2‐LPS complex to the ectodomain of TLR4 triggers the signaling cascade.126 Der p 2 was proven to have functional homology with MD‐2, by facilitating signaling through direct interactions with the TLR4 complex, and reconstituting LPS‐driven TLR4 signaling in the absence of MD‐2. Experimental allergic asthma was induced in wild type and MD‐2‐deficient, but not TLR4‐deficient, mice, by airway sensitization and challenge with Der p 2.125 This study supported the idea of the importance of lipid ligands for the innate immune responses of group 2 mite allergens. While this paper presents a convincing case for Der p 2‐MD‐2 molecular mimicry, some biochemical studies are suggested for follow‐up.127 For example all of the Der p 2–TLR4‐binding experiments were done with immunoprecipitations,125 which does not directly measure affinity. MD‐2 has a 0.8 nM affinity for TLR4128 and Der p 2 lacks the DDD motif important for MD‐2 recognition of TLR4.127 Therefore, it is not clear how Der p 2 could compete for TLR4 in a physiological setting. Second, the mimicry study utilized a Der p 2 Y91A mutant that was claimed to abolish LPS binding.125 This is unlikely to be highly effective as this residue is not highly conserved in mites.16 Biochemical studies establishing the affinity of Der p 2 binding to TLR4 and the affinity of Der p 2 Y91A binding to LPS would be useful to understand the physiological relevance of the proposed mimicry.
In summary, these studies indicate that various hydrophobic ligands of mite group 2 allergens can be accommodated in the binding pocket via conformational changes,16, 121 highlighting the relevance of molecular flexibility of these allergens. The flexibility of group 2 HDM allergens has also been reported to affect their antigenic surface.129 Several studies have shown interesting aspects of group 2 HDM allergens related to their capacity to induce adaptive immunity.130, 131, 132 Recently, anti‐Der p 2 human IgE monoclonal antibodies were isolated by hybridoma technology that have the correct pairing of the heavy and light chains as it occurs in vivo. Their associated epitopes were identified by immunoassays and NMR analyses.131
7. OTHER LIGAND‐BINDING ALLERGENS
HDM group 7 and 13 allergens have low to medium sensitization rates among HDM sensitized individuals.133 They are known to have lipophilic properties and have been shown to mimic molecules involved in innate immunity.134, 135 The structure of Der p 7 (Figure 5B) was observed to be similar to lipopolysaccharide‐binding protein (LBP) that interacts with Toll‐like receptors after binding lipopolysaccharide and other bacterially derived lipid ligands.134 Such lipid‐binding features are common among many allergens,136 and allergenicity resulting from functional mimicry of a TLR complex protein has been demonstrated in an animal model.125
Rather than binding LPS, Der p 7 binds with weak affinity to the bacterial lipopeptide polymyxin B, a nine‐residue cyclic peptide with a lipid tail from the Gram‐positive bacterium Bacillus polymyxa.134 The natural ligand for Der p 7 has not yet been identified, but the structural relation to a protein in the TLR pathway and the ability to bind bacterially derived lipid ligands are suggestive of a mechanism that would lead toward allergic sensitization to these proteins. This remains to be formally demonstrated for Der p 7.
Der p 13 (Figure 5C), which belongs to the fatty acid‐binding protein (FABP) family, was shown to selectively bind fatty acids and induce airway epithelial cell activation in vitro through TLR2‐MyD88‐NF‐kappaB and MAPK‐dependent mechanisms.135 Similar observation were made for Der p 5.137 More recently, it was demonstrated that HDM group 13 allergens (specifically Der p 13 and Blo t 13) are sensed by serum amyloid A1 (SAA1) that promotes pulmonary type 2 immunity.138 SAA1 interacts directly with allergenic mite FABPs, which then activates the N‐formyl peptide receptor 2 (FPR2). In addition to the direct interaction with SAA1, the authors speculate that, because of its lipid‐binding properties, Der p 13 can strip away the retinol molecules that typically stabilize SAA1 hexamers. This then drives epithelial cells to release interleukin (IL)‐33 in a SAA1‐dependent manner. It was observed that the SAA1–FPR2–IL‐33 axis was upregulated in nasal epithelial cells from patients with chronic rhinosinusitis.138 From an allergy standpoint, this is curious because Der p 13 is typically considered a minor allergen due to low prevalence of sIgE in mite‐allergic patients. However, it is highly expressed at levels on par with the major allergen Der p 2, so it is likely an abundant mite protein.139 The interpretation of these results appears to be that the binding and adjuvant properties of Der p 13 seem to be more consequential than the ability to stimulate IgE to itself.
Bla g 1 (Figure 5D), also a lipid‐binding protein, is not a major allergen for cockroach allergic patients,140, 141, 142 as is the case for house dust mite (eg, Der p 1, Der p 2, and Der p 23) and birch pollen (eg, Bet v 1).143 Bla g 1 is highly expressed in female cockroaches exclusively by midgut cells with a production that is modulated in relation to food intake.144, 145, 146 A role of Bla g 1 in digestion and nutrient absorption has been suggested.147 Bla g 1‐encoding DNA has an interesting genetic structure.148 Multiple tandem repeats of the gene are found in five different open reading frames, each with some homology to the currently described allergen.149 It is currently not known whether all of these homologous proteins are allergens. The protein structure of Bla g 1 corresponding to a tandem repeat is spherical with twelve α‐helices that enclose a large cavity capable of enclosing up to four diacyl‐phospholipids.15, 28 Recombinant Bla g 1 contained a variety of phospholipids whose nature depended on the expression system (Figure 5D), whereas natural Bla g 1 bound a mixture of the fatty acids palmitate, oleate, and stearate. When loaded with these natural fatty acids, Bla g 1 was both more proteolytically resistant and thermally stable than when loaded with lipids with shorter or longer fatty acid chains.150 The stabilizing effect of the ligands anticorrelated with the generation of known T‐cell epitopes using an in vitro assay.15 The implication is that the stabilizing property of the lipids could modulate the generation of T‐cell epitopes and subsequent allergenicity. As precedent, isoforms of Bet v 1 that display enhanced stability are able to avoid premature processing in the endosome, yielding a stronger Th2 response than less allergenic variants.71 This is a distinctly different role for the lipids in stabilizing Bla g 1, as opposed to the immunomodulatory properties of LPS, for example. It is possible that Bla g 1 ligands could also have immunomodulatory properties which have not been fully explored. Lastly, Bla g 1 has a very high affinity for phosphatidylcholine (PC) that exceeds the affinity for the natural fatty acids.15 PC is a major component of lung surfactant so it is possible that Bla g 1 would exchange fatty acids for PC disrupting the natural balance of surfactant in the lung, leading to barrier defects. The delivery of lipid adjuvants and the disruption of natural barriers are both suggested to lead to allergic disease.
Fel d 1, which is the most important cat allergen, has a fold similar to the one observed for uteroglobin.151 Both proteins possess several helices and an internal cavity that is able to accommodate hydrophobic molecules. It was shown that the cat allergen could bind lipids or steroids.152
8. THE IMPACT OF ALLERGEN‐BOUND LIGANDS ON THE IMMUNE RESPONSE
The fact that several types of allergens bind hydrophobic ligands has prompted the hypothesis that these ligands play a crucial role in inducing anallergic response.8, 153 The binding of lipids to food allergens effects their degradation in the GI tract and their passage through epithelial barriers impacting on the sensitization process.136 Len c 3, an nsLTP from lentils, otherwise sensitive to heating and digestion, increased its stability when binding lyso‐palmitoyl phosphatidylglycerol, a ligand—the authors argue—the allergen picks up during the cooking process.154 A derivative of the alkaloid camptothecin bound to phytosphingosine was identified as the ligand of the major peach allergen Pru p 3, another nsLTP.12 The lipid ligand induced the maturation of monocyte‐derived dendritic cells and the proliferation of peripheral blood mononuclear cells.13 The immunological activity of the ligand which resides in the phytosphingosine tail was mediated by Cd1d activation of iNKT cells. In a mouse model of peach anaphylaxis, the complex of Pru p 3 and the ligand induced higher levels of sIgE than the allergen alone.
Detailed studies of the respiratory allergen Bet v 1, the prototypic PR‐10 protein, revealed the impact of its ligands on the sensitizing process. A pharmaceutical‐grade recombinant Bet v 1 was produced in Escherichia coli for the formulation of a sublingual tablet to treat birch pollen allergy.155 This highly purified rBet v 1, free from any ligand, was unable to induce secretion of pro‐inflammatory or effector cytokines by human blood dendritic cells or mononuclear cells. Remarkably, Bet v 1‐depleted birch pollen extract was still able to induce Th2 polarization while purified rBet v 1 did not induce dendritic cell differentiation.11 Bet v 1 binds various classes of ligands in its large hydrophobic cavity including fatty acids, cytokinins, or flavonoids.156 Immunomodulatory pollen‐associated lipid mediators such as leukotriene‐like molecules and phytoprostanes when codelivered with the allergen were proposed to play a key role in Th2 polarization.7 Phytoprostane E1 has also been identified as a new ligand of Bet v 1.14 This pollen‐derived ligand was shown to enhance the proteolytic resistance of Bet v 1 and to inhibit endolysosomal cathepsin protease activity. Diminished proteolytic processing of antigens results in low loading and density of MHCII‐peptide complexes which favors Th2 polarization.72
9. CONCLUSIONS
Allergens belonging to protein families that include SAs, PR‐10s, nsLTPs, lipocalins, NCP2, LBPs, and FABPs represent a significant fraction of officially registered allergens. These proteins bind diverse compounds, including fatty acids, lipo‐oligosaccharides, phospholipids, oxylipins, catecholamines, trace amines, steroids, flavonoids, and different plant hormones, as well as odorants and pheromones. The studies of the complex mixtures of molecules to which the human body and its immune system are exposed to are just starting. Therefore, we are convinced that not only allergenic proteins but also the small molecular compounds accompanying them play important and still not well‐understood roles in allergic sensitization and diseases (Box 2).
BOX 2. Future research perspectives.
More studies on the effect of allergen natural ligands on stability and antigen processing that affects allergenicity
Identification of small molecules acting as ligands with an impact on the allergic sensitization phase
Identification of small molecules acting as ligands with an impact on the allergic effector phase
Identification of small ligand molecules ligands with an intrinsic immunogenic capacity
Better understanding why isoallergens show different ligand‐binding preferences
CONFLICT OF INTEREST
Dr. Chew reports grants from the National University of Singapore, Singapore Ministry of Education Academic Research Fund, Singapore Immunology Network, National Medical Research Council (NMRC) (Singapore), Biomedical Research Council (BMRC) (Singapore), and the Agency for Science Technology and Research (A*STAR) (Singapore), during the conduct of the study; and consulting fees from Sime Darby Technology Centre; First Resources Ltd; Genting Plantation, and Olam International, outside the submitted work. Dr. Pomés is employed by Indoor Biotechnologies, Inc. and is the contact PI of the 2R01AI077653‐10A1 Award from the NIH/NIAID that funds research presented in this article. She declares that her research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Dr. Rouvinen reports contract research between University of Eastern Finland and Desentum Ltd, outside the submitted work. Dr Rouvinen is also a shareholder in Desentum Ltd. All other authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
HB initiated this project. All authors contributed to the writing of the manuscript.
ACKNOWLEDGMENTS
The authors thank Andrea O'Malley and Alexander Foo for comments on the manuscript. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number 2R01AI077653‐10A1 (AP and MC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was further supported in part by the National Institute of Environmental Health Sciences Z01‐ES102906 (GAM), by the Danube Allergy Research Cluster funded by the Country of Lower Austria (KHS and HB), and by the U.S. Department of Agriculture, Agricultural Research Service (BKH).
REFERENCES
- 1.Johansson S, Bieber T, Dahl R, et al. Revised nomenclature for allergy for global use: report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol. 2004;113(5):832‐836. [DOI] [PubMed] [Google Scholar]
- 2.Johansson SG, Hourihane JO, Bousquet J, et al. A revised nomenclature for allergy. An EAACI position statement from the EAACI nomenclature task force. Allergy. 2001;56(9):813‐824. [DOI] [PubMed] [Google Scholar]
- 3.von Moltke J, Pepper M. Sentinels of the type 2 immune response. Trends Immunol. 2018;39(2):99‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gour N, Lajoie S, Smole U, et al. Dysregulated invertebrate tropomyosin‐dectin‐1 interaction confers susceptibility to allergic diseases. Sci Immunol. 2018;3(20):eaam9841. 10.1126/sciimmunol.aam9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiffers C, Hristova M, Habibovic A, et al. The transient receptor potential channel vanilloid 1 is critical in innate airway epithelial responses to protease allergens. American J Resp Cell Mol Biol. 2020;63(2):198‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palm NW, Rosenstein RK, Yu S, Schenten DD, Florsheim E, Medzhitov R. Bee venom phospholipase A2 induces a primary type 2 response that is dependent on the receptor ST2 and confers protective immunity. Immunity. 2013;39(5):976‐985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pointner L, Bethanis A, Thaler M, et al. Initiating pollen sensitization ‐ complex source, complex mechanisms. Clin Transl Allergy. 2020;10:36. 10.1186/s13601-020-00341-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas WR. Allergen ligands in the initiation of allergic sensitization. Current Allergy Asthma Rep. 2014;14(5):432. 10.1007/s11882-014-0432-x [DOI] [PubMed] [Google Scholar]
- 9.Dahl A. Pollen lipids can play a role in allergic airway inflammation. Front Immunol. 2018;9:2816. 10.3389/fimmu.2018.02816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirotti L, Florsheim E, Rundqvist L, et al. Lipids are required for the development of Brazil nut allergy: the role of mouse and human iNKT cells. Allergy. 2013;68(1):74‐83. [DOI] [PubMed] [Google Scholar]
- 11.Aglas L, Gilles S, Bauer R, et al. Context matters: TH2 polarization resulting from pollen composition and not from protein‐intrinsic allergenicity. J Allergy Clin Immunol. 2018;142(3):984‐987.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubells‐Baeza N, Gómez‐Casado C, Tordesillas L, et al. Identification of the ligand of Pru p 3, a peach LTP. Plant Mol Biol. 2017;94(1–2):33‐44. [DOI] [PubMed] [Google Scholar]
- 13.Tordesillas L, Cubells‐Baeza N, Gómez‐Casado C, et al. Mechanisms underlying induction of allergic sensitization by Pru p 3. Clin Exp Allergy. 2017;47(11):1398‐1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soh WT, Aglas L, Mueller GA, et al. Multiple roles of Bet v 1 ligands in allergen stabilization and modulation of endosomal protease activity. Allergy. 2019;74(12):2382‐2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foo ACY, Thompson PM, Perera L, et al. Hydrophobic ligands influence the structure, stability, and processing of the major cockroach allergen Bla g 1. Sci Rep. 2019;9(1):18294. 10.1038/s41598-019-54689-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichikawa S, Takai T, Yashiki T, et al. Lipopolysaccharide binding of the mite allergen Der f 2. Genes Cells. 2009;14(9):1055‐1065. [DOI] [PubMed] [Google Scholar]
- 17.Derewenda U, Li J, Derewenda Z, et al. The crystal structure of a major dust mite allergen Der p 2, and its biological implications. J Mol Biol. 2002;318(1):189‐197. [DOI] [PubMed] [Google Scholar]
- 18.Baranauskiene L, Kuo TC, Chen WY, Matulis D. Isothermal titration calorimetry for characterization of recombinant proteins. Curr Opin Biotechnol. 2019;55:9‐15. [DOI] [PubMed] [Google Scholar]
- 19.Di Cera E. Mechanisms of ligand binding. Biophys Rev. 2020;1(1):011303. 10.1063/5.0020997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furukawa A, Konuma T, Yanaka S, Sugase K. Quantitative analysis of protein‐ligand interactions by NMR. Prog Nucl Magn Reson Spectrosc. 2016;96:47‐57. [DOI] [PubMed] [Google Scholar]
- 21.Gossert AD, Jahnke W. NMR in drug discovery: a practical guide to identification and validation of ligands interacting with biological macromolecules. Prog Nucl Magn Reson Spectrosc. 2016;97:82‐125. [DOI] [PubMed] [Google Scholar]
- 22.Monzingo AF. Crystallographic studies of steroid‐protein interactions. Adv Exp Med Biol. 2019;1135:27‐45. [DOI] [PubMed] [Google Scholar]
- 23.Nusrat S, Khan RH. Exploration of ligand‐induced protein conformational alteration, aggregate formation, and its inhibition: a biophysical insight. Prep Biochem Biotechnol. 2018;48(1):43‐56. [DOI] [PubMed] [Google Scholar]
- 24.Seutter von Loetzen C, Hoffmann T, Hartl M, et al. Secret of the major birch pollen allergen Bet v 1: identification of the physiological ligand. Biochem J. 2014;457(3):379‐390. [DOI] [PubMed] [Google Scholar]
- 25.Mogensen JE, Wimmer R, Larsen JN, Spangfort MD, Otzen DE. The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J Biol Chem. 2002;277(26):23684‐23692. [DOI] [PubMed] [Google Scholar]
- 26.Seutter von Loetzen C, Jacob T, Hartl‐Spiegelhauer O, et al. Ligand recognition of the major birch pollen allergen Bet v 1 is isoform dependent. PLoS One. 2015;10(6):e0128677. 10.1371/journal.pone.0128677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubiela P, Aina R, Polak D, et al. Enhanced Pru p 3 IgE‐binding activity by selective free fatty acid‐interaction. J Allergy Clin Immunol. 2017;140(6):1728‐1731.e10. [DOI] [PubMed] [Google Scholar]
- 28.Mueller GA, Pedersen LC, Lih FB, et al. The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment. J Allergy Clin Immunol. 2013;132(6):1420‐1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob T, von Loetzen CS, Reuter A, et al. Identification of a natural ligand of the hazel allergen Cor a 1. Sci Rep. 2019;9(1):8714. 10.1038/s41598-019-44999-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurlburt BK, McBride J, Pote S, Chruszcz M, Maleki SJ. Ligand binding preferences of pathogenesis‐related class 10 (PR‐10) allergens. J Allergy Clin Immun. 2016;137(2):Ab268. [Google Scholar]
- 31.Chruszcz M, Kapingidza AB, Dolamore C, Kowal K. A robust method for the estimation and visualization of IgE cross‐reactivity likelihood between allergens belonging to the same protein family. PLoS One. 2018;13(11):e0208276. 10.1371/journal.pone.0208276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chruszcz M, Mikolajczak K, Mank N, Majorek KA, Porebski PJ, Minor W. Serum albumins‐unusual allergens. Biochim Biophys Acta. 2013;1830(12):5375‐5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spitzauer S. Allergy to mammalian proteins: at the borderline between foreign and self? Int Archives Allergy Immunol. 1999;120(4):259‐269. [DOI] [PubMed] [Google Scholar]
- 34.Spitzauer S, Pandjaitan B, Söregi G, et al. IgE cross‐reactivities against albumins in patients allergic to animals. J Allergy Clin Immunol. 1995;96(6 Pt 1):951‐959. [DOI] [PubMed] [Google Scholar]
- 35.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999;12(6):439‐446. [DOI] [PubMed] [Google Scholar]
- 36.Litus EA, Permyakov SE, Uversky VN, Permyakov EA. Intrinsically disordered regions in serum slbumin: what are they for? Cell Biochem Biophys. 2018;76(1–2):39‐57. [DOI] [PubMed] [Google Scholar]
- 37.Quinlan GJ, Martin GS, Evans TW. Albumin: biochemical properties and therapeutic potential. Hepatology. 2005;41(6):1211‐1219. [DOI] [PubMed] [Google Scholar]
- 38.Varshney A, Sen P, Ahmad E, Rehan M, Subbarao N, Khan RH. Ligand binding strategies of human serum albumin: how can the cargo be utilized? Chirality. 2010;22(1):77‐87. [DOI] [PubMed] [Google Scholar]
- 39.Czub MP, Venkataramany BS, Majorek KA, et al. Testosterone meets albumin ‐ the molecular mechanism of sex hormone transport by serum albumins. Chem Sci. 2019;10(6):1607‐1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bal W, Christodoulou J, Sadler PJ, Tucker A. Multi‐metal binding site of serum albumin. J Inorg Biochem. 1998;70(1):33‐39. [DOI] [PubMed] [Google Scholar]
- 41.Majorek KA, Porebski PJ, Dayal A, et al. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol Immunol. 2012;52(3–4):174‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Handing KB, Shabalin IG, Kassaar O, et al. Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins. Chem Sci. 2016;7(11):6635‐6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handing KB, Shabalin IG, Szlachta K, Majorek KA, Minor W. Crystal structure of equine serum albumin in complex with cetirizine reveals a novel drug binding site. Mol Immunol. 2016;71:143‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liccardi G, Asero R, D'Amato M, D'Amato G. Role of sensitization to mammalian serum albumin in allergic disease. Current Allergy Asthma Rep. 2011;11(5):421‐426. [DOI] [PubMed] [Google Scholar]
- 45.Bujacz A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. 2012;68(Pt 10):1278‐1289. [DOI] [PubMed] [Google Scholar]
- 46.Yamada K, Yokomaku K, Kureishi M, Akiyama M, Kihira K, Komatsu T. Artificial blood for dogs. Sci Rep. 2016;6:36782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokomaku K, Akiyama M, Morita Y, Kihira K, Komatsu T. Core‐shell protein clusters comprising haemoglobin and recombinant feline serum albumin as an artificial O2 carrier for cats. J Mater Chem B. 2018;6(16):2417‐2425. [DOI] [PubMed] [Google Scholar]
- 48.Bujacz A, Talaj JA, Zielinski K, Pietrzyk‐Brzezinska AJ, Neumann P. Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5‐diiodosalicylic acid. Acta Crystallogr D Struct Biol. 2017;73(Pt 11):896‐909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czub MP, Handing KB, Venkataramany BS, Cooper DR, Shabalin IG, Minor W. Albumin‐based transport of nonsteroidal anti‐inflammatory drugs in mammalian blood plasma. J Med Chem. 2020;63(13):6847‐6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gecibesler IH, Aydin M. Plasma protein binding of herbal‐flavonoids to human serum albumin and their anti‐proliferative activities. An Acad Bras Cienc. 2020;92(1):e20190819. 10.1590/0001-3765202020190819 [DOI] [PubMed] [Google Scholar]
- 51.Pajares MA, Zimmerman T, Sanchez‐Gomez FJ, et al. Amoxicillin inactivation by thiol‐catalyzed cyclization reduces protein haptenation and antibacterial potency. Front Pharmacol. 2020;11:189. 10.3389/fphar.2020.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hegde VL, Venkatesh YP. Generation of antibodies specific to D‐mannitol, a unique haptenic allergen, using reductively aminated D‐mannose‐bovine serum albumin conjugate as the immunogen. Immunobiology. 2007;212(2):119‐128. [DOI] [PubMed] [Google Scholar]
- 53.Wisnewski AV, Stowe MH, Cartier A, et al. Isocyanate vapor‐induced antigenicity of human albumin. J Allergy Clin Immunol. 2004;113(6):1178‐1184. [DOI] [PubMed] [Google Scholar]
- 54.Jain S, Kumar A. The pathogenesis related class 10 proteins in plant defense against biotic and abiotic stresses. Adv Plants Agric Res. 2015;3(1):00077. 10.15406/apar.2015.02.00077 [DOI] [Google Scholar]
- 55.Walter MH, Liu JW, Grand C, Lamb CJ, Hess D. Bean pathogenesis‐related (PR) proteins deduced from elicitor‐induced transcripts are members of a ubiquitous new class of conserved PR proteins including pollen allergens. Mol Gen Genet. 1990;222(2–3):353‐360. [DOI] [PubMed] [Google Scholar]
- 56.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. 10.1186/1471-2148-8-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandes H, Michalska K, Sikorski M, Jaskolski M. Structural and functional aspects of PR‐10 proteins. FEBS J. 2013;280(5):1169‐1199. [DOI] [PubMed] [Google Scholar]
- 58.Gajhede M, Osmark P, Poulsen FM, et al. X‐ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nature Struct Biol. 1996;3(12):1040‐1045. [DOI] [PubMed] [Google Scholar]
- 59.Eriksson NE, Formgren H, Svenonius E. Food hypersensitivity in patients with pollen allergy. Allergy. 1982;37(6):437‐443. [DOI] [PubMed] [Google Scholar]
- 60.Radauer C, Nandy A, Ferreira F, et al. Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences. Allergy. 2014;69(4):413‐419. [DOI] [PubMed] [Google Scholar]
- 61.Pomés A, Davies JM, Gadermaier G, et al. WHO/IUIS Allergen Nomenclature: providing a common language. Mol Immunol. 2018;100:3‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hurlburt BK, Offermann LR, McBride JK, Majorek KA, Maleki SJ, Chruszcz M. Structure and function of the peanut panallergen Ara h 8. J Biol Chem. 2013;288(52):36890‐36901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kofler S, Asam C, Eckhard U, Wallner M, Ferreira F, Brandstetter H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen bet v 1. J Mol Biol. 2012;422(1):109‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marković‐Housley Z, Degano M, Lamba D, et al. Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J Mol Biol. 2003;325(1):123‐133. [DOI] [PubMed] [Google Scholar]
- 65.McBride JK, Cheng H, Maleki SJ, Hurlburt BK. Purification and characterization of pathogenesis related class 10 panallergens. Foods. 2019;8(12):609. 10.3390/foods8120609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muñoz C, Hoffmann T, Escobar NM, et al. The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol Plant. 2010;3(1):113‐124. [DOI] [PubMed] [Google Scholar]
- 67.Petersen A, Rennert S, Kull S, et al. Roasting and lipid binding provide allergenic and proteolytic stability to the peanut allergen Ara h 8. Biol Chem. 2014;395(2):239‐250. [DOI] [PubMed] [Google Scholar]
- 68.Chruszcz M, Ciardiello MA, Osinski T, et al. Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening‐related proteins. Mol Immunol. 2013;56(4):794‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D'Avino R, Bernardi ML, Wallner M, et al. Kiwifruit Act d 11 is the first member of the ripening‐related protein family identified as an allergen. Allergy. 2011;66(7):870‐877. [DOI] [PubMed] [Google Scholar]
- 70.Rib‐Schmidt C. Stabilitätsanalyse und physiologische Charakterisierung des Hauptsellerieallergens Api g 1. Master Thesis, University of Bayreuth; 2016. [Google Scholar]
- 71.Machado Y, Freier R, Scheiblhofer S, et al. Fold stability during endolysosomal acidification is a key factor for allergenicity and immunogenicity of the major birch pollen allergen. J Allergy Clin Immunol. 2016;137(5):1525‐1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freier R, Dall E, Brandstetter H. Protease recognition sites in Bet v 1a are cryptic, explaining its slow processing relevant to its allergenicity. Sci Rep. 2015;5:12707. 10.1038/srep12707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hilger C, Kuehn A, Hentges F. Animal lipocalin allergens. Current Allergy Asthma Rep. 2012;12(5):438‐447. [DOI] [PubMed] [Google Scholar]
- 74.Restani P, Ballabio C, Di Lorenzo C, Tripodi S, Fiocchi A. Molecular aspects of milk allergens and their role in clinical events. Anal Bioanal Chem. 2009;395(1):47‐56. [DOI] [PubMed] [Google Scholar]
- 75.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318(Pt 1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grzyb J, Latowski D, Strzalka K. Lipocalins ‐ a family portrait. J Plant Physiol. 2006;163(9):895‐915. [DOI] [PubMed] [Google Scholar]
- 77.Kontopidis G, Holt C, Sawyer L. Invited review: beta‐lactoglobulin: binding properties, structure, and function. J Dairy Sci. 2004;87(4):785‐796. [DOI] [PubMed] [Google Scholar]
- 78.Sawyer L, Kontopidis G. The core lipocalin, bovine beta‐lactoglobulin. Biochim Biophys Acta. 2000;1482(1–2):136‐148. [DOI] [PubMed] [Google Scholar]
- 79.Le Maux S, Bouhallab S, Giblin L, Brodkorb A, Croguennec T. Bovine beta‐lactoglobulin/fatty acid complexes: binding, structural, and biological properties. Dairy Sci Technol. 2014;94:409‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hufnagl K, Ghosh D, Wagner S, et al. Retinoic acid prevents immunogenicity of milk lipocalin Bos d 5 through binding to its immunodominant T‐cell epitope. Sci Rep. 2018;8(1):1598. 10.1038/s41598-018-19883-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roth‐Walter F, Afify SM, Pacios LF, et al. Cow's milk protein beta‐lactoglobulin confers resilience against allergy by targeting complexed iron into immune cells. J Allergy Clin Immunol. 2021;147:321‐334.e4. [DOI] [PubMed] [Google Scholar]
- 82.Afify SM, Pali‐Schöll I, Hufnagl K, et al. Bovine holo‐beta‐lactoglobulin cross‐protects against pollen allergies in an innate manner in BALB/c mice: potential model for the farm effect. Front Immunol. 2021;12:611474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clayton GM, White J, Lee S, Kappler JW, Chan SK. Structural characteristics of lipocalin allergens: Crystal structure of the immunogenic dog allergen Can f 6. PLoS One. 2019;14(9):e0213052. 10.1371/journal.pone.0213052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lascombe MB, Gregoire C, Poncet P, et al. Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE‐reactive epitopes. J Biol Chem. 2000;275(28):21572‐21577. [DOI] [PubMed] [Google Scholar]
- 85.Madhurantakam C, Nilsson OB, Uchtenhagen H, et al. Crystal structure of the dog lipocalin allergen Can f 2: implications for cross‐reactivity to the cat allergen Fel d 4. J Mol Biol. 2010;401(1):68‐83. [DOI] [PubMed] [Google Scholar]
- 86.Niemi MH, Rytkonen‐Nissinen M, Janis J, Virtanen T, Rouvinen J. Structural aspects of dog allergies: the crystal structure of a dog dander allergen Can f 4. Mol Immunol. 2014;61(1):7‐15. [DOI] [PubMed] [Google Scholar]
- 87.Rouvinen J, Rautiainen J, Virtanen T, et al. Probing the molecular basis of allergy. Three‐dimensional structure of the bovine lipocalin allergen Bos d 2. J Biol Chem. 1999;274(4):2337‐2343. [DOI] [PubMed] [Google Scholar]
- 88.Chaudhuri BN, Kleywegt GJ, Bjorkman J, Lehman‐McKeeman LD, Oliver JD, Jones TA. The structures of alpha 2u‐globulin and its complex with a hyaline droplet inducer. Acta Crystallogr. 1999;55(Pt 4):753‐762. [DOI] [PubMed] [Google Scholar]
- 89.Böcskei Z, Groom CR, Flower DR, et al. Pheromone binding to two rodent urinary proteins revealed by X‐ray crystallography. Nature. 1992;360(6400):186‐188. [DOI] [PubMed] [Google Scholar]
- 90.Ferrari E, Corsini R, Burastero SE, Tanfani F, Spisni A. Thermal stability, ligand binding and allergenicity data of Mus m 1.0102 allergen and its cysteine mutants. Data Brief. 2020;29:105355. 10.1016/j.dib.2020.105355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan YW, Chan SL, Ong TC, et al. Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes. J Biol Chem. 2009;284(5):3148‐3157. [DOI] [PubMed] [Google Scholar]
- 92.Offermann LR, Chan SL, Osinski T, et al. The major cockroach allergen Bla g 4 binds tyramine and octopamine. Mol Immunol. 2014;60(1):86‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herre J, Grönlund H, Brooks H, et al. Allergens as immunomodulatory proteins: the cat dander protein Fel d 1 enhances TLR activation by lipid ligands. J Immunol. 2013;191(4):1529‐1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Flower DR. Beyond the superfamily: the lipocalin receptors. Biochim Biophysica Acta. 2000;1482(1–2):327‐336. [DOI] [PubMed] [Google Scholar]
- 95.Hoffmann‐Sommergruber K. Plant allergens and pathogenesis‐related proteins. What do they have in common? Int Arch Allergy Immunol. 2000;122(3):155‐166. [DOI] [PubMed] [Google Scholar]
- 96.Kader JC. Lipid‐Transfer Proteins in Plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:627‐654. [DOI] [PubMed] [Google Scholar]
- 97.van Loon LC, van Strien EA. The families of pathogenesis‐related proteins, their activities, andcomparative analysis of PR‐1 type proteins. Physiol Mol Plant Pathol. 1999;55:85‐97. [Google Scholar]
- 98.Salminen TA, Blomqvist K, Edqvist J. Lipid transfer proteins: classification, nomenclature, structure, and function. Planta. 2016;244(5):971‐997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scheurer S, Schulke S. Interaction of non‐specific lipid‐transfer proteins with plant‐derived lipids and its impact on allergic sensitization. Front Immunol. 2018;9:1389. 10.3389/fimmu.2018.01389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gonzalez‐Klein Z, Cuevas‐Zuviria B, Wangorsch A, et al. The key to the allergenicity of lipid transfer protein (LTP) ligands: a structural characterization. Biochim Biophys Acta Mol Cell Biol Lipids. 2021;1866:158928. 10.1016/j.bbalip.2021.158928 [DOI] [PubMed] [Google Scholar]
- 101.Hoffmann‐Sommergruber K, Pfeifer S, Bublin M. Applications of molecular diagnostic testing in food allergy. Curr Allergy Asthma Rep. 2015;15(9):56. 10.1007/s11882-015-0557-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aina R, Dubiela P, Geiselhart S, et al. Distinct lipid transfer proteins display different IgE‐binding activities that are affected by fatty acid binding. Allergy. 2019;74(4):827‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cuevas‐Zuviria B, Garrido‐Arandia M, Diaz‐Perales A, Pacios LF. Energy landscapes of ligand motion inside the tunnel‐like cavity of lipid transfer proteins: the case of the Pru p 3 allergen. Int J Mol Sci. 2019;20(6):1432. 10.3390/ijms20061432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dubiela P, Del Conte R, Cantini F, et al. Impact of lipid binding on the tertiary structure and allergenic potential of Jug r 3, the non‐specific lipid transfer protein from walnut. Sci Rep. 2019;9(1):2007. 10.1038/s41598-019-38563-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garcia‐Casado G, Pacios LF, Diaz‐Perales A, et al. Identification of IgE‐binding epitopes of the major peach allergen Pru p 3. J Allergy Clin Immunol. 2003;112(3):599‐605. [DOI] [PubMed] [Google Scholar]
- 106.Villalba M, Rodriguez R, Batanero E. The spectrum of olive pollen allergens. From structures to diagnosis and treatment. Methods. 2014;66(1):44‐54. [DOI] [PubMed] [Google Scholar]
- 107.Han GW, Lee JY, Song HK, et al. Structural basis of non‐specific lipid binding in maize lipid‐transfer protein complexes revealed by high‐resolution X‐ray crystallography. J MolBiol. 2001;308(2):263‐278. [DOI] [PubMed] [Google Scholar]
- 108.Oeo‐Santos C, López‐Rodríguez JC, García‐Mouton C, et al. Biophysical and biological impact on the structure and IgE‐binding of the interaction of the olive pollen allergen Ole e 7 with lipids. Biochim Biophys Acta Biomembr. 2020;1862(6):183258. 10.1016/j.bbamem.2020.183258 [DOI] [PubMed] [Google Scholar]
- 109.Lopez‐Rodriguez JC, Barderas R, Echaide M, et al. Surface activity as a crucial factor of the biological actions of Ole e 1, the main aeroallergen of olive tree (Olea europaea) Pollen. Langmuir. 2016;32(42):11055‐11062. [DOI] [PubMed] [Google Scholar]
- 110.Abdullah SU, Alexeev Y, Johnson PE, et al. Ligand binding to an allergenic aipid transfer protein enhances conformational flexibility resulting in an increase in susceptibility to gastroduodenal proteolysis. Sci Rep. 2016;6:30279. 10.1038/srep30279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heymann PW, Chapman MD, Aalberse RC, Fox JW, Platts‐Mills TA. Antigenic and structural analysis of group II allergens (Der f II and Der p II) from house dust mites (Dermatophagoides spp). J Allergy Clin Immunol. 1989;83(6):1055‐1067. [DOI] [PubMed] [Google Scholar]
- 112.Mueller GA, Randall TA, Glesner J, et al. Serological, genomic and structural analyses of the major mite allergen Der p 23. Clin Exp Allergy. 2016;46(2):365‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Posa D, Perna S, Resch Y, et al. Evolution and predictive value of IgE responses toward a comprehensive panel of house dust mite allergens during the first 2 decades of life. J Allergy Clin Immunol. 2017;139(2):541‐549.e8. [DOI] [PubMed] [Google Scholar]
- 114.Osterlund C, Gronlund H, Polovic N, Sundstrom S, Gafvelin G, Bucht A. The non‐proteolytic house dust mite allergen Der p 2 induce NF‐kappaB and MAPK dependent activation of bronchial epithelial cells. Clin Exp Allergy. 2009;39(8):1199‐1208. [DOI] [PubMed] [Google Scholar]
- 115.Ichikawa S, Hatanaka H, Yuuki T, et al. Solution structure of Der f 2, the major mite allergen for atopic diseases. J Biol Chem. 1998;273(1):356‐360. [DOI] [PubMed] [Google Scholar]
- 116.Mueller GA, Benjamin DC, Rule GS. Tertiary structure of the major house dust mite allergen Der p 2: sequential and structural homologies. Biochemistry. 1998;37(37):12707‐12714. [DOI] [PubMed] [Google Scholar]
- 117.Inohara N, Nunez G. ML – a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem Sci. 2002;27(5):219‐221. [DOI] [PubMed] [Google Scholar]
- 118.Ichikawa S, Takai T, Inoue T, et al. NMR study on the major mite allergen Der f 2: its refined tertiary structure, epitopes for monoclonal antibodies and characteristics shared by ML protein group members. J Biochem. 2005;137(3):255‐263. [DOI] [PubMed] [Google Scholar]
- 119.Suzuki M, Tanaka Y, Korematsu S, Mikami B, Minato N. Crystal structure and some properties of a major house dust mite allergen, Derf 2. Biochem Biophys Res Comm. 2006;339(2):679‐686. [DOI] [PubMed] [Google Scholar]
- 120.Johannessen BR, Skov LK, Kastrup JS, et al. Structure of the house dust mite allergen Der f 2: implications for function and molecular basis of IgE cross‐reactivity. FEBS Lett. 2005;579(5):1208‐1212. [DOI] [PubMed] [Google Scholar]
- 121.Keber MM, Gradisar H, Jerala R. MD‐2 and Der p 2 ‐ a tale of two cousins or distant relatives? J Endotoxin Res. 2005;11(3):186‐192. [DOI] [PubMed] [Google Scholar]
- 122.Reginald K, Chew FT. The major allergen Der p 2 is a cholesterol binding protein. Sci Rep. 2019;9(1):1556. 10.1038/s41598-018-38313-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Winkler MBL, Kidmose RT, Szomek M, et al. Structural insight into eukaryotic sterol transport through Niemann‐Pick type C proteins. Cell. 2019;179(2):485‐497.e18. [DOI] [PubMed] [Google Scholar]
- 124.Liao JX, Yin ZX, Huang XD, Weng SP, Yu XQ, He JG. Cloning and characterization of a shrimp ML superfamily protein. Fish Shellfish Immunol. 2011;30(2):713‐719. [DOI] [PubMed] [Google Scholar]
- 125.Trompette A, Divanovic S, Visintin A, et al. Allergenicity resulting from functional mimicry of a Toll‐like receptor complex protein. Nature. 2009;457(7229):585‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jerala R. Structural biology of the LPS recognition. Int J Med Microbiol. 2007;297(5):353‐363. [DOI] [PubMed] [Google Scholar]
- 127.Mueller GA, Min J, Foo ACY, Pomes A, Pedersen LC. Structural analysis of recent allergen‐antibody complexes and future directions. Curr Allergy Asthma Rep. 2019;19(3):17. 10.1007/s11882-019-0848-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Han J, Kim HJ, Lee SC, et al. Structure‐based rational design of a Toll‐like receptor 4 (TLR4) decoy receptor with high binding affinity for a target protein. PLoS One. 2012;7(2):e30929. 10.1371/journal.pone.0030929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Glesner J, Kapingidza AB, Godzwon M, et al. A human IgE antibody binding site on Der p 2 for the design of a recombinant allergen for immunotherapy. J Immunol. 2019;203(9):2545‐2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kulwanich B, Thanyaratsrisakul S, Jirapongsananuruk O, Hales BJ, Thomas WR, Piboonpocanun S. Effects of Ser47‐point mutation on conformation structure and allergenicity of the allergen of Der p 2, a major house dust mite allergen. Allergy Asthma Immunol Res. 2019;11(1):129‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mueller GA, Glesner J, Daniel JL, et al. Mapping human monoclonal IgE epitopes on the major dust mite allergen Der p 2. J Immunol. 2020;205(8):1999‐2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reginald K, Chew FT. Conformational IgE epitope mapping of Der p 2 and the evaluations of two candidate hypoallergens for immunotherapy. Sci Rep. 2018;8(1):3391. 10.1038/s41598-018-21792-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kidon MI, Chin CW, Kang LW, et al. Mite component‐specific IgE repertoire and phenotypes of allergic disease in childhood: the tropical perspective. Pediatr Allergy Immunol. 2011;22(2):202‐210. [DOI] [PubMed] [Google Scholar]
- 134.Mueller GA, Edwards LL, Aloor JJ, et al. The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins. J Allergy Clin Immunol. 2010;125(4):909‐917.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Satitsuksanoa P, Kennedy M, Gilis D, et al. The minor house dust mite allergen Der p 13 is a fatty acid‐binding protein and an activator of a TLR2‐mediated innate immune response. Allergy. 2016;71(10):1425‐1434. [DOI] [PubMed] [Google Scholar]
- 136.Jappe U, Schwager C, Schromm AB, et al. Lipophilic allergens, different modes of allergen‐lipid interaction and their impact on asthma and allergy. Front Immunol. 2019;10:122. 10.3389/fimmu.2019.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pulsawat P, Soongrung T, Satitsuksanoa P, et al. The house dust mite allergen Der p 5 binds lipid ligands and stimulates airway epithelial cells through a TLR2‐dependent pathway. Clin Exp Allergy. 2019;49(3):378‐390. [DOI] [PubMed] [Google Scholar]
- 138.Smole U, Gour N, Phelan J, et al. Serum amyloid A is a soluble pattern recognition receptor that drives type 2 immunity. Nat Immunol. 2020;21(7):756‐765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ogburn RN, Randall TA, Xu Y, et al. Are dust mite allergens more abundant and/or more stable than other Dermatophagoides pteronyssinus proteins? J Allergy Clin Immunol. 2017;139(3):1030‐1032 e1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Satinover SM, Reefer AJ, Pomes A, Chapman MD, Platts‐Mills TA, Woodfolk JA. Specific IgE and IgG antibody‐binding patterns to recombinant cockroach allergens. J Allergy Clin Immunol. 2005;115(4):803‐809. [DOI] [PubMed] [Google Scholar]
- 141.Glesner J, Filep S, Vailes LD, et al. Allergen content in German cockroach extracts and sensitization profiles to a new expanded set of cockroach allergens determine in vitro extract potency for IgE reactivity. J Allergy Clin Immunol. 2019;143(4):1474‐1481.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pomés A, Glesner J, Calatroni A, et al. Cockroach allergen component analysis of children with or without asthma and rhinitis in an inner‐city birth cohort. J Allergy Clin Immunol. 2019;144(4):935‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pomes A, Mueller GA, Randall TA, Chapman MD, Arruda LK. New insights into cockroach allergens. Curr Allergy Asthma Rep. 2017;17(4):25. 10.1007/s11882-017-0694-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gore JC, Schal C. Gene expression and tissue distribution of the major human allergen Bla g 1 in the German cockroach, Blattella germanica L. (Dictyoptera: Blattellidae). J Med Entomol. 2004;41(5):953‐960. [DOI] [PubMed] [Google Scholar]
- 145.Cabrera A, Randall TA, Ogburn RN, et al. Are allergens more abundant and/or more stable than other proteins in pollens and dust? Allergy. 2020;75(5):1267‐1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gore JC, Schal C. Expression, production and excretion of Bla g 1, a major human allergen, in relation to food intake in the German cockroach, Blattella germanica . Med Vet Entomol. 2005;19(2):127‐134. [DOI] [PubMed] [Google Scholar]
- 147.Suazo A, Gore C, Schal C. RNA interference‐mediated knock‐down of Bla g 1 in the German cockroach, Blattella germanica L., implicates this allergen‐encoding gene in digestion and nutrient absorption. Insect Mol Biol. 2009;18(6):727‐736. [DOI] [PubMed] [Google Scholar]
- 148.Pomes A, Melen E, Vailes LD, Retief JD, Arruda LK, Chapman MD. Novel allergen structures with tandem amino acid repeats derived from German and American cockroach. J Biol Chem. 1998;273(46):30801‐30807. [DOI] [PubMed] [Google Scholar]
- 149.Randall TA, Perera L, London RE, Mueller GA. Genomic, RNAseq, and molecular modeling evidence suggests that the major allergen domain in insects evolved from a homodimeric origin. Genome Biol Evol. 2013;5(12):2344‐2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Foo ACY, Thompson PM, Mueller GA. Removal and replacement of endogenous ligands from lipid‐bound proteins and allergens. J Vis Exp. 2021;(168). 10.3791/61780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kaiser L, Grönlund H, Sandalova T, et al. The crystal structure of the major cat allergen Fel d 1, a member of the secretoglobin family. J Biol Chem. 2003;278(39):37730‐37735. [DOI] [PubMed] [Google Scholar]
- 152.Bienboire‐Frosini C, Durairaj R, Pelosi P, Pageat P. The major cat allergen Fel d 1 binds steroid and fatty acid semiochemicals: acombined in silico and in vitro study. Int J Mol Sci. 2020;21(4):1365. 10.3390/ijms21041365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bublin M, Eiwegger T, Breiteneder H. Do lipids influence the allergic sensitization process? J Allergy Clin Immunol. 2014;134(3):521‐529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Finkina EI, Melnikova DN, Bogdanov IV, et al. Impact of different lipid ligands on the stability and IgE‐binding capacity of the lentil allergen Len c 3. Biomolecules. 2020;10(12):1668. 10.3390/biom10121668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nony E, Bouley J, Le Mignon M, et al. Development and evaluation of a sublingual tablet based on recombinant Bet v 1 in birch pollen‐allergic patients. Allergy. 2015;70(7):795‐804. [DOI] [PubMed] [Google Scholar]
- 156.Aglas L, Soh WT, Kraiem A, Wenger M, Brandstetter H, Ferreira F. Ligand binding of PR‐10 proteins with a particular focus on the Bet v 1 allergen family. Curr Allergy Asthma Rep. 2020;20(7):25. 10.1007/s11882-020-00918-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Loch JI, Bonarek P, Polit A, Swiatek S, Dziedzicka‐Wasylewska M, Lewinski K. The differences in binding 12‐carbon aliphatic ligands by bovine beta‐lactoglobulin isoform A and B studied by isothermal titration calorimetry and X‐ray crystallography. J Mol Recognit. 2013;26(8):357‐367. [DOI] [PubMed] [Google Scholar]
- 158.Pasquato N, Berni R, Folli C, et al. Crystal structure of peach Pru p 3, the prototypic member of the family of plant non‐specific lipid transfer protein pan‐allergens. J Mol Biol. 2006;356(3):684‐694. [DOI] [PubMed] [Google Scholar]
- 159.Shabalin IG, Czub MP, Majorek KA, et al. Molecular determinants of vascular transport of dexamethasone in COVID‐19 therapy. IUCrJ. 2020;7(6):1048‐1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shin DH, Lee JY, Hwang KY, Kim KK, Suh SW. High‐resolution crystal‐structure of the nonspecific lipid‐transfer protein from maize seedlings. Structure. 1995;3(2):189‐199. [DOI] [PubMed] [Google Scholar]
- 161.Bonura A, Corinti S, Schiavi E, et al. The major allergen of the Parietaria pollen contains an LPS‐binding region with immuno‐modulatory activity. Allergy. 2013;68(3):297‐303. [DOI] [PubMed] [Google Scholar]
- 162.Casanal A, Zander U, Munoz C, et al. The strawberry pathogenesis‐related 10 (PR‐10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates. J Biol Chem. 2013;288(49):35322‐35332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Calvo E, Mans BJ, Ribeiro JM, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc Nat Acad Sci. 2009;106(10):3728‐3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zubini P, Zambelli B, Musiani F, Ciurli S, Bertolini P, Baraldi E. The RNA hydrolysis and the cytokinin binding activities of PR‐10 proteins are differently performed by two isoforms of the Pru p 1 peach major allergen and are possibly functionally related. Plant Physiol. 2009;150(3):1235‐1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ruszkowski M, Sliwiak J, Ciesielska A, Barciszewski J, Sikorski M, Jaskolski M. Specific binding of gibberellic acid by cytokinin‐specific binding proteins: a new aspect of plant hormone‐binding proteins with the PR‐10 fold. Acta Crystallogr. 2014;70(Pt 7):2032‐2041. [DOI] [PubMed] [Google Scholar]
- 166.Charvolin D, Douliez JP, Marion D, Cohen‐Addad C, Pebay‐Peyroula E. The crystal structure of a wheat nonspecific lipid transfer protein (ns‐LTP1) complexed with two molecules of phospholipid at 2.1 A resolution. Eur JBiochem/FEBS. 1999;264(2):562‐568. [DOI] [PubMed] [Google Scholar]
- 167.Bakan B, Hamberg M, Larue V, Prange T, Marion D, Lascombe MB. The crystal structure of oxylipin‐conjugated barley LTP1 highlights the unique plasticity of the hydrophobic cavity of these plant lipid‐binding proteins. Biochem Biophys Res Comm. 2009;390(3):780‐785. [DOI] [PubMed] [Google Scholar]
- 168.Chan SL, Ong ST, Ong SY, Chew FT, Mok YK. Nuclear magnetic resonance structure‐based epitope mapping and modulation of dust mite group 13 allergen as a hypoallergen. J Immunol. 2006;176(8):4852‐4860. [DOI] [PubMed] [Google Scholar]


