Abstract
Background/ Aims:
Allopurinol can cause HLA class I associated life-threatening severe skin reactions. However, HLA risk and association with clinical features in allopurinol hepatotoxicity are unknown.
Methods:
11 of 17 patients with suspected allopurinol hepatotoxicity enrolled into the Drug Induced Liver Injury Network were adjudicated as definite, highly likely or probable. High resolution HLA sequencing was undertaken in cases and compared to population and other DILI controls.
Results-
Median age was 60 years, 54% were male, and 63% African- American, 27% Caucasian, and 9% Hispanic. Patients presented at a median of 52 days after starting allopurinol, all were hospitalized and 6 were jaundiced. The median peak ALT, alkaline phosphatase, and total bilirubin were 525 U/L, 521 U/L, and 7.8 mg/dL, respectively, with a median R ratio of 2.7 at onset. During follow-up, 9 patients were treated with corticosteroids including 5 of the 6 with suspected DRESS. Three patients died including two from liver failure at 38 and 45 days after onset, and the remaining 8 recovered. Three HLA alleles were found to be overrepresented in allopurinol cases, particularly in African Americans: HLA- B * 58:01, which has been previously linked to severe skin reactions, and HLA-B*53:01 and HLA-A*34:02 all of which are more frequently found in African-Americans than European-Americans or Latinos.
Conclusions:
Allopurinol hepatotoxicity is associated with systemic hypersensitivity, a short latency to onset, African American race and 3 HLA risk alleles, HLA-B*58:01, HLA-B*53:01 and HLA-A*34:02. HLA- 58:01 testing may help confirm a diagnosis of hepatotoxicity in allopurinol treated patients.
Keywords: Hyperuricemia, genetic polymorphisms, liver injury, gout, human leukocyte antigen
Lay summary
Allopurinol is commonly used to treat gout but can be associated with potentially severe skin and liver reactions in a minority of patients. The majority of the 11 patients with allopurinol related liver injury in the current study had evidence of a hypersensitivity reaction and jaundice when they presented. During follow-up, nine patients were treated with corticosteroids and 3 died of their illness. Going forward, testing for the 3 human leukocyte antigen Class I genes identified in this study, HLA-B*58:01, HLA-B*53:01, HLA-A*34:02, may help doctors more rapidly diagnose and treat future patients.
Introduction
Allopurinol is a xanthine oxidase inhibitor that is widely used to treat gout (1). Chronic therapy with allopurinol is associated with minor serum aminotransferase elevations in 2% to 6% of treated patients, which usually resolve spontaneously or with drug discontinuation (2,3). Allopurinol is also a rare but well known cause of potentially severe acute liver injury that frequently presents with prominent hypersensitivity features such as those seen with drug reaction with eosinophilia and systemic symptoms (DRESS). This includes fever, widespread rash, eosinophilia, atypical lymphocytosis, lymphadenopathy and other organ involvement with a typical latency of 2 to 6 weeks (2–4).
Allopurinol hypersensitivity syndrome has a high case fatality rate, either from acute liver failure or complications of other allergic manifestations such as Stevens-Johnson syndrome and toxic epidermal necrolysis (SJS/TEN), vasculitis, pancreatitis and renal dysfunction. African-American and Asian race as well as preexisting renal disease appear to be particular risk factors for severe cutaneous adverse drugs reactions (SCAR) such as SJS/TEN and DRESS associated with allopurinol (2–4). The mechanism of allopurinol hepatotoxicity is believed to be a hypersensitivity reaction and many cases resemble those of anticonvulsant hypersensitivity. Recently, several cases have been associated with reactivation of human herpesvirus-6, Epstein Barr virus (EBV) or cytomegalovirus (CMV) infections (4–7). Allopurinol SCAR have been closely linked to HLA-B*58:01 particularly in Asian populations, but also to a lesser extent in Caucasians where HLA- B*58:01 appears to account for approximately 60% of allopurinol associated SCAR (4–7).
The Drug Induced Liver Injury Network (DILIN) has been conducting studies of the presenting features, risk factors, and outcomes of patients with drug induced liver injury (DILI) since 2003 (8). In addition, mechanistic studies exploring the potential genetic susceptibility in DILI patients have been undertaken using genome wide association (GWA) studies and targeted human leukocyte antigen (HLA) genotyping. In the current study, we describe the presenting features, clinical outcomes and liver histopathology in a well-characterized cohort of consecutive patients with suspected allopurinol hepatotoxicity. We also compared HLA alleles associated with allopurinol DILI cases to those associated with drug-induced DILI cases in DILIN and population controls.
METHODS
DILIN Prospective study:
Patients with liver injury were enrolled into the ongoing DILIN prospective, observational registry study (8). Liver injury onset was defined as the first date after a subject met one of the predefined laboratory criteria of either a serum aspartate aminotransferase (AST) or alanine aminotransferase (ALT) level that exceeded 5 times the upper limit of normal (ULN) (or 5 times pretreatment baseline if baseline abnormal), a serum alkaline phosphatase (ALP) that exceeded 2 times the ULN (or 2 times pretreatment baseline if baseline abnormal), a total bilirubin of 2.5 mg/dl or above, or an international normalized ratio (INR) greater than 1.5 on two consecutive blood draws. All study participants provided written informed consent and were enrolled within 6 months of DILI onset.
A detailed medical history was obtained at the baseline study visit and additional laboratory and radiological testing were performed to more fully characterize the DILI event and exclude competing etiologies via testing for hepatitis A, B, C, E, HIV, anti-nuclear antibody (ANA) and smooth muscle antibody (SMA) titers, CMV, and EBV infection at the local laboratory. Cases without available hepatitis E virus testing or without HCV RNA results were tested centrally using stored serum samples. Enrolled patients were seen for a follow-up study visit 6 months after initial enrollment and those with evidence of chronic injury were asked to return for further study visits up to 48 months (8). Chronic DILI was defined as any patient having a persistently elevated serum AST, ALT, or ALP level, histological evidence of liver injury, or clinical evidence of portal hypertension at 6 months after DILI onset (8).
The severity of the DILI episode was categorized on a 5-point scale from mild (1), moderate (2), moderate-hospitalized (3), severe (4), and fatal (5), where a fatal score was assigned only if the patient died or had liver transplant due to DILI within six months of onset (8).
While self-reported races and ethnicity were available, we used genome wide single nucleotide polymorphism (SNP) genotypes to infer individual’s ethnicity. Genome-wide SNP data were generated for samples in DILIN over time by different Illumina platform including Infinium HumanCoreExome BeadChip 1Mduo array, Multi-Ethnic Genotyping Array (MEGA), and Expanded MEGA (MEGAEX). For each genotyping platform, we computed principal components (PCs) for the combined SNP data of DILIN and all racial groups of the 1000 genomes project, and then designated individual genetic ethnicity based on the PC clusters and their boundaries as described previously (9). If a subject’s race could not be inferred by PC criteria due to the uncertainty of cluster assignment or lack of genome-wide SNP data, self-reported race was used.
Liver histopathology:
Available liver biopsies were reviewed by a single expert liver histopathologist (DEK). All samples were scored for multiple histological features as well as an overall pattern of liver injury (10).
Causality assessment:
The causal relationship between the liver injury episode and allopurinol use was evaluated in a standardized fashion by the DILIN causality committee. A DILIN expert opinion causality score ranging from 1 (Definite > 95% likelihood), 2 (Highly Likely 75%−95% likelihood), 3 (Probable 50%−74% likelihood), 4 (Possible 25%−49% likelihood) to 5 (Unlikely < 25% likelihood) was assigned by consensus agreement of committee members for all of the retrospective and prospective DILIN cases. In subjects with 2 or more implicated drugs, an overall causality score was assigned to the case and then an individual causality score for each drug was assigned. In this study, cases with a causality score of 1, 2 or 3 were referred to as high confidence DILI cases and included in the analysis. The previously published RegiSCAR Score with a range of 0 to 6 points was used to identify patients with DRESS (11).
HLA sequencing:
HLA deep sequencing for the HLA Class I and II genes was performed for DILIN cases at the Vanderbilt Immunogenomics, Microbial Genetics and Single Cell Technologies core (IMGSCT), a laboratory accredited by the American Society for Histocompatibility and Immunogenetics (ASHI). Specific HLA loci were PCR amplified using sample specific MID-tagged primers that amplify polymorphic exons from Class I (A, B, C Exons 2 and 3) and Class II (DQ, Exons 2 and 3; DRB and DPB1, Exon 1) MHC genes. MID tagged primers were optimized to minimize allele dropouts and primer bias. Amplified DNA products from unique MID tagged products (up to 48 MIDs) were pooled in equimolar ratios for library preparation. Libraries were quantified using the KAPA library quantitation kit (Kapa Biosystems) and High sensitivity D1000 screentape on an Agilent 2200 Tapestation (Agilent) for concentration and size distribution. Normalized libraries were sequenced on the Illumina MiSeq platform using the MiSeq V3 600-cycle kit (2X300bp reads). Sequences were separated by MID tags and alleles called using an in house accredited HLA allele caller software pipeline that minimizes the influence of sequencing errors. Alleles were called using the IMGT HLA allele sequence database release v3270 (March 24, 2017), IIID HLA analysis suite release v3.11, and IIID allele caller release v2.7 HLA analyse reporting software that performs comprehensive allele balance and contamination checks on the final dataset.
Controls:
Two control groups were used in the HLA analysis. One is the population controls using the GWAS dataset of PAGE: The Charles Bronfman Institute for Personalized Medicine BioMe, from NCBI dbGAP.(phs000925.v1.p1), hereafter, referred to as BioMe. The BioMe GWAS dataset consists of 8,780 subjects with a diverse racial background, and SNP genotyping was based on Illumina MEGA platform. Similarly, genome wide SNP data were used to infer individual race as described above. Of 8,780 BioMe subjects, 5,816 were African Americans. The 4-digit HLA alleles in Class I and Class II genes were imputed by GWAS SNPs for each racial group, respectively, using the HLA genotype imputation with attributable bagging (HiBAG) program [12]. The second control group is referred to as disease control group, which consisted of all other DILI cases not caused by allopurinol (other drugs or herbal and dietary supplements) from the DILIN cohort. This included 1,512 DILI cases, of which 177 were African Americans. The comparison of allopurinol cases to non-allopurinol DILI cases in DILIN allows us to assess if the HLA risk alleles identified are more specific for allopurinol DILI.
Statistical analysis:
Descriptive statistics were computed for demographic and patient characteristics by frequency (percentage) for categorical variables and mean, standard deviation (SD), median, and the range for continuous variables. Although 10 of 11 allopurinol cases have genome wide genotype data available, there was inadequate statistical power to perform GWAS in this small dataset. Alternatively, we checked if any of the allopurinol DILI cases carried the minor allele of rs2476601 in PTPN22, the GWAS signal published previously for DILI (13). Since the distribution of HLA alleles often differs by racial and ethnicity groups, we focused on the HLA analysis in African-American subset as 7 of 11 allopurinol DILI cases are African-Americans. However, considering the limited number of allopurinol cases, we also followed up the top interesting HLA alleles using all cases in the dataset. Fisher exact tests were performed to test the allele frequency (AF) difference between allopurinol cases and the two control groups for each HLA allele. In addition, the population AF of each HLA allele in African Americans were obtained from BeTheMatch (https://bioinformatics.bethematchclinical.org/) and Allele Frequency Net Database (AFND, http://www.allelefrequencies.net/) to serve as the reference. Due to the small number of cases, candidate HLA alleles were not selected using the conventional criteria for false discovery rate that corrects for multiple testing. Instead, we focused more on the differences in allele frequency between groups. Therefore, we determined the candidate HLA alleles for allopurinol induced DILI if the HLA allele met the following two criteria: (1) the absolute difference of AF in cases and population AF is >15%; (2) p-value < 0.05 from the Fisher exact test for the comparison of allopurinol cases to BioMe controls.
For the HLA genes that harbor the candidate HLA alleles we identified above, we followed up with allelic cluster and haplotype analysis. First, the NetMHCpan-2.8 algorithm in MHCcluster [14] was used to identify clusters of HLA alleles in the gene of interest based on their predicted peptide-binding specificity. We then tested the association between each allelic cluster and allopurinol DILI. That is, HLA alleles in the same cluster were coded as the same marker. We then tested each cluster frequency difference between cases and controls by the Fisher exact test. Second, haplotype association was tested for two HLA gene combinations for the genes of interest using the score statistics in the haplo.stats program [15]. Clusters or haplotypes meeting p<0.05 were considered significant.
Results
Patient Population-
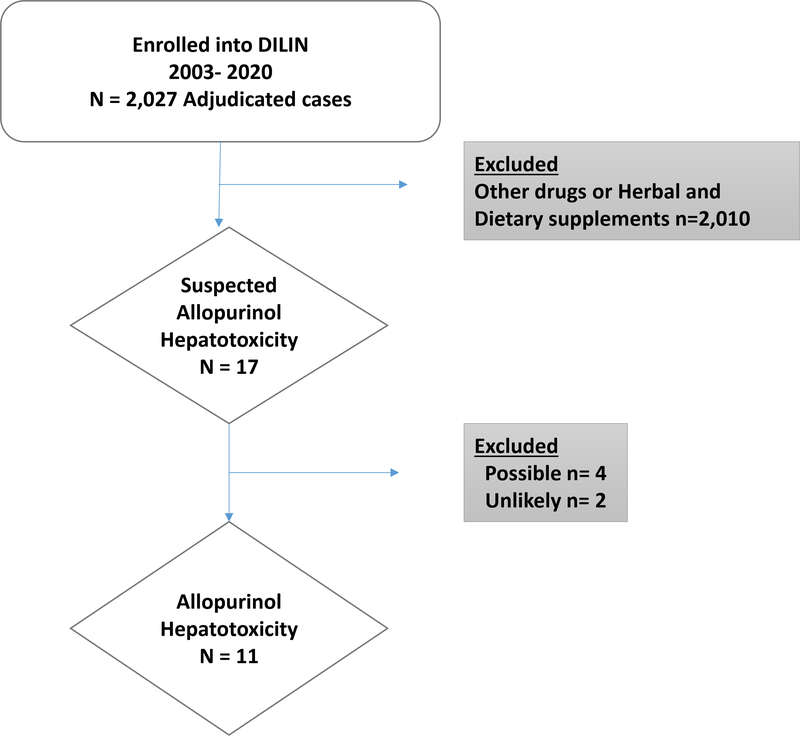
Between January 2003 and May 2020, 2,220 cases were enrolled in the DILIN Prospective Study of which 2027 had undergone 6 months of follow up and formal adjudication for causality. In 17 cases, allopurinol was potentially implicated, but only 11 cases were judged to be definite (n=4), highly likely (n=4) or probably (n=3) related to allopurinol. In the other 6 cases, allopurinol was considered only possible (n=4) or unlikely (n=2) the cause; these cases were not included in the analysis (Figure 1)
Figure 1.
Overview of study population. Amongst the 2027 adjudicated DILIN cases, there were 17 cases of suspected allopurinol hepatotoxicity. Eleven were categorized as definite/ highly likely/ probable allopurinol hepatotoxicity while 6 cases were attributed to other medications or disease processes.
The 11 confirmed cases of allopurinol hepatotoxicity included 6 men and 5 women, with a median age of 60 years (range: 27 to 74 years), and a median BMI of 33.7 kg/m2 (range: 24.1 to 42.1 kg/m2). Self-reported race was consistent with the genetically inferred race which was 7 African Americans, 3 European Americans, and 1 White Hispanic. (Table 1).
TABLE 1.
Clinical features in 11 allopurinol hepatotoxicity patients
| Clinical feature | Allopurinol cases N=11 | African American N=7 | White or Caucasian n=4 |
|---|---|---|---|
| Age (yrs) | 60 (27 to 74) | 67 (42 to 74) | 38 (27 to 52) |
| Male sex | 6 (54%) | 2 (29%) | 4 (100%) |
| Hispanic | 1 (9%) | 0 (0%) | 1 (25%) |
| BMI (Kg/m2) | 33.7 (24.1– 42.1) | 34.6 (24.1–40.7) | 30.4 (26.4–42.1) |
| Diabetes | 5 (46%) | 3 (43%) | 2 (50%) |
| Time to onset (days) | 52 (12 to 89) | 52 (12 to 89) | 49 (37 to 69) |
| Daily allopurinol dose (mg) | 200 (100 to 600) | 175 (100, 600) | 300 (100, 300) |
| GFR (mg/dl) | 70 (10.2 to 168.6) | 36 (10.2, 116.4) | 124 (70, 169) |
| Blood test results at DILI onset | |||
| AST (U/L) | 226 (105, 591) | 180 (105, 485) | 368 (168, 591) |
| ALT (U/L) | 362 (84, 716) | 331 (84, 651) | 532 (135, 716) |
| Alk P (U/L) | 252 (150, 1176) | 220 (150, 829) | 466 (252, 1176) |
| Total bilirubin (mg/dL) | 1.4 (0.5, 15.4) | 1.4 (0.5, 15.4) | 1.5 (0.5, 3.9) |
| INR | 1.1 (1.0, 1.4) | 1.1 (1.0, 1.4) | 1.1 (1.1, 1.1) |
| R ratio | 2.7 (0.4, 13.8) | 4.0 (0.4, 13.8) | 2.2 (1.5, 3.3) |
| Peak Blood test results | |||
| AST (U/L) | 412 (122, 1174) | 212 (122, 731) | 622 (310, 1174) |
| ALT (U/L) | 525 (173, 3388) | 381 (173, 2075) | 767 (522, 3388) |
| Alk P (U/L) | 521 (157, 1415) | 288 (157, 1105) | 1242 (252, 1415) |
| Total bilirubin (mg/dL) | 7.8 (1.0, 50.3) | 7.8 (1.0, 45.7) | 9.4 (2.4, 50.3) |
| INR | 1.4 (1.1, 7.3) | 1.4 (1.1, 2.2) | 1.2 (1.1, 7.3) |
| Corticosteroids given | 9 (82%) | 5 (71%) | 4 (100%) |
| DRESS | 6 (54%) | 4 (57%) | 2 (50%) |
| Death within 6 months | 3 (27%) | 3 (43%) | 0 (0%) |
| DILIN Severity score | |||
| Mild | 3 (28%) | 2 (29%) | 1 (25%) |
| Moderate | 0 (0%) | 0 (0%) | 0 (0%) |
| Moderate –hospitalization | 4 (36%) | 3 (43%) | 1 (25%) |
| Severe | 2 (18%) | 0 (0%) | 2 (50%) |
| Fatal | 2 (18%) | 2 (29%) | 0 (0%) |
GFR= glomerular filtration rate obtained prior to treatment
Data reported as Median (range) or N (%)
The median time from drug start to onset of liver injury was 52 days with a range of 12 to 89 days. The daily dose of allopurinol prescribed ranged from 100 to 600 mg/day, with a median dose of 200 mg/day. Other prescription drugs were implicated in 4 cases but none were considered probable. Hypersensitivity features were frequent with rash observed in 91%, fever in 45% and eosinophilia in 45% of patients. Medical co-morbidities included type 2 diabetes in 46% and occasional alcohol use 18%. The median estimated glomerular filtration rate (GFR) at the time allopurinol was started was 70 mL/min and was below 50 mL/min in 3 patients (range 10.2 to 168.6).
At onset, all 11 patients were symptomatic and required hospitalization and 6 were jaundiced. The median initial ALT was 362 U/L (range 84 to 716), Alk P 252 U/L (range 150 to 1176), total bilirubin 1.4 mg/dL (0.5 to 15.4) and R ratio 2.7 (0.4 to 13.8). The median peak bilirubin was 7.8 mg/dL but in only 3 patients did the INR rise above 1.5. Only 3 (27%) patients had a low titer anti-nuclear antibody and none had detectable smooth muscle antibody. The course of liver injury varied, with 5 patients presenting first with rash and fever and later developing liver biochemical abnormalities after allopurinol had been discontinued.
Clinical outcomes-
During follow-up, 9 patients were treated with corticosteroids, in 7 of whom there was subsequent clinical and biochemical improvement. Two corticosteroid treated patients had progressive liver injury and died from liver failure at 38 and 45 days after presentation. Of the remaining 2 patients who did not receive corticosteroids, 1 died 6 months after presentation from multi-organ failure and 1 improved spontaneously. No patient had evidence of chronic liver injury at 6 month follow-up. Severity scores ranked 3 patients as mild (1+: anicteric), 4 as moderate (3+: jaundiced and hospitalized), 2 as severe (4+: evidence of hepatic failure) and 2 as fatal (5+: death within 6 months of onset).
DRESS-
Although 10 patients had rash and 7 fever, 6 were judged to have DRESS syndrome (cases 1,3,5,7,9,10) as judged by a RegiSCAR score ≥4. RegiSCAR scores were 6 or more indicative of definite DRESS in 3 patients and 4 or 5 indicative of probable DRESS in 3 patients (Supplemental Table 1). Two patients underwent skin biopsy which contributed to the diagnosis (Cases 3 and 7). No patient had the diagnosis of Stevens-Johnson syndrome or toxic epidermal necrolysis. Testing for human herpes virus-6 was done in one patient and was negative. One patient (case 9) had CMV-DNA detectable by PCR at 9 days after DILI onset, but a liver biopsy on day 8 show no evidence of viral inclusions or changes typical of CMV hepatitis.
Liver Histopathology-
7 patients underwent liver biopsy at a median of 8 days after DILI onset (Table 2). A review of 6 available biopsies demonstrated three patterns of injury that had been previously established with allopurinol DILI: granulomatous hepatitis in one patient, acute hepatitis with necrosis in one patient and cholestasis with mild inflammation in three patients. The sixth patient showed cholestatic injury with mild duct paucity superimposed on underlying chronic hepatitis C. One biopsy was no longer available for review but was reported to show granulomatous hepatitis. Three of the six available biopsies for review showed increased numbers of eosinophils in portal areas. Two cases showed evidence of ductopenia.
TABLE 2.
Liver histopathology findings*
| Days from onset | R ratio/ Bilirubin at biopsy | Total inflammation score | Granulomas | Eosinophils | Cholestasis | Ductopenia | Pattern of Injury | |
|---|---|---|---|---|---|---|---|---|
| CASE 2 | 5 | 3.3/2.0 | 14 | + | + | − | None | Granulomatous hepatitis |
| CASE 4 | 6 | 1.3/12.8 | 9 | − | + | + | Mild | Chronic cholestatic hepatitis on Chronic hepatitis C |
| CASE 6 | 4 | 4.5/5.3 | 11 | − | + | − | None | Acute hepatitis |
| CASE 7 | 29 | 3.9/1.0 | 4 | − | − | + | None | Cholestatic hepatitis |
| CASE 9 | 8 | 1.6/15.4 | 3 | − | + | + | None | Cholestatic hepatitis |
| CASE 11 | 9 | 1.5/0.5 | 4 | − | − | + | Marked | Cholestatic hepatitis |
Case 1 was not available for central review. By description it showed granulomatous hepatitis and extramedullary hematopoiesis.
Genetic and HLA associations:
All 10 allopurinol DILI cases carried GG genotype at SNP rs2476601 in the PTPN22 gene, where G is the major allele, which is not surprising due to the low frequency of the minor allele, the A nucleotide (AF=0.1 for European Americans and 0.015 for African Americans from gnomAD (https://gnomad.broadinstitute.org/)).
HLA typing demonstrated an excess distribution of three class I HLA types: HLA-A*34:02, HLA-B*53:01 and HLA-B*58:01 with allele frequencies that were 2 to 18 fold higher than both population and disease controls (Table 3, upper row, Supplemental Table 2). HLA-B*53:01 was present in 5 while HLA-A*34:02 and HLA-B*58:01 were present in 4 of the 11 patients. Thus, all except two subjects had at least one of the 3 candidate risk alleles (Table 4). These 3 alleles are rare in European American and US Hispanic populations (Allele frequency (AF) <0.01 for all three alleles in both populations) and are several-fold more frequent among African Americans (AF 0.034, 0.118 and 0.038). Nevertheless, limitation of the analysis to the 7 African American patients with allopurinol hepatotoxicity still revealed a significantly higher frequency of the 3 alleles than among racially matched population and DILIN disease controls (Table 3: lower row). All six cases of DRESS syndrome had one of the 3 alleles and 4 had HLA-B*58:01. The two subjects without even one of these alleles were both European Americans without DRESS in whom the causality score was only probable (Table 4).
Table 3:
Summary of HLA Alleles with Potential Association with Allopurinol Hepatotoxicity
| Cases & controls: | Allopurinol cases | BioMe controls | DILIN controls | ||||
|---|---|---|---|---|---|---|---|
| All subjects | Sample sizes | n=11 | n=8,780 | n=1,512 | |||
| HLA allele | AF | count | AF | p value: Cases vs BioMe AF | AF | p value: Cases vs DILIN AF | |
| A*34:02 | 0.182 | 4 | 0.023 | 0.001 | 0.010 | 8.22E-05 | |
| B*53:01 | 0.227 | 5 | 0.085 | 0.035 | 0.026 | 0.0002 | |
| B*58:01 | 0.182 | 4 | 0.036 | 0.008 | 0.016 | 0.0005 | |
| African Americans | Sample size | n=7 | n=5,816 | n=177 | |||
| HLA allele | AF | count | AF | p value: Cases vs BioMe AF | AF | p value: Cases vs DILIN AF | |
| A*34:02 | 0.214 | 3 | 0.033 | 0.010 | 0.057 | 0.050 | |
| B*53:01 | 0.357 | 5 | 0.120 | 0.020 | 0.184 | 0.155 | |
| B*58:01 | 0.214 | 3 | 0.046 | 0.024 | 0.020 | 0.005 | |
Note: p-values were from Fisher exact tests to compare allopurinol Cases to BioMe controls and to DILIN controls
TABLE 4.
HLA Genotyping results for HLA-A and HLA-B
| HLA-A | HLA-B | Race or ethnicity | DRESS | Causality score | |||
|---|---|---|---|---|---|---|---|
| Case # | Allele 1 | Allele 2 | Allele 1 | Allele 2 | |||
| Case 1 | A*02:01 | A*02:01 | B*39:06 | B*44:03 | European American | No | 3 |
| Case 2 | A*02:01 | A*34:02 | B*35:01 | B*51:01 | Hispanic | Yes | 1 |
| Case 3 | A*02:02 | A*33:03 | B*53:01 | B*58:01 | African American | Yes | 1 |
| Case 4 | A*33:03 | A*74:01 | B*35:01 | B*53:01 | African American | No | 2 |
| Case 5 | A*03:01 | A*34:02 | B*53:01 | B*58:02 | African American | Yes | 2 |
| Case 6 | A*34:02 | A*68:02 | B* 15:10 | B*41:02 | African American | No | 2 |
| Case 7 | A*30:02 | A*68:02 | B*07:02 | B*58:01 | African American | Yes | 1 |
| Case 8 | A*01:01 | A*02:05 | B*13:02 | B*58:01 | European American | Yes | 1 |
| Case 9 | A*02:01 | A*26:01 | B*53:01 | B*58:01 | African American | Yes | 3 |
| Case 10 | A*01:01 | A*34:02 | B*37:01 | B*53:01 | African American | Yes | 2 |
| Case 11 | A*02:01 | A*02:01 | B*08:01 | B*56:01 | European American | No | 3 |
Note: The candidate alleles identified are highlighted in bold.
Analysis of functional clustering of HLA class I alleles based on their predicted binding specificity (Supplemental Figure 1) showed significant clustering of HLA-B alleles based on shared peptide binding specificities. HLA-B*56:01, B*51:01, B*35:01, and B*53:01 formed the first large cluster, and HLA-B*53:01 and B*58:01 were in the 2nd cluster. However, since there were no instances of HLA-B*56:01 and HLA-B*51:01 in allopurinol cases, the first cluster analysis was limited to HLA-B*35:01 and HLA-B*53:01. The HLA-B*35:01 and HLA-B*53:01 cluster did not show significant association with allopurinol DILI in African Americans when compared to either BioMe controls (cluster frequency: 0.43 vs. 0.21, p=0.051) or DILIN controls (cluster frequency: 0.43 vs. 0.25, p=0.128). On the other hand, the HLA-B*53:01 and HLA-B*58:01 cluster was significantly associated with allopurinol DILI in comparison to either BioMe controls (cluster frequency: 0.57 vs. 0.17, p= 6.67 ×10−4) or DILIN controls (cluster frequency: 0.57 vs. 0.20, p=0.004) in African-American subset and remained significant when all subjects were included (Table 5).
Table 5:
Allelic cluster association between allopurinol cases and controls from BioMe cohort, and between allopurinol cases and disease controls from DILIN.
| Analysis subset | Clusters | Cases AF | BioME controls AF | p value: Cases vs. BioMe AF | DILIN Controls AF | P values: Case vs. DILIN AF |
|---|---|---|---|---|---|---|
| African Americans | B*35:01,B*53:01 | 0.43 | 0.21 | 0.051 | 0.25 | 0.128 |
| B*53:01,B*58:01 | 0.57 | 0.17 | 6.67×10−4 | 0.20 | 0.004 | |
| All subjects | B*35:01,B*53:01 | 0.32 | 0.17 | 0.085 | 0.10 | 0.004 |
| B*53:01,B*58:01 | 0.41 | 0.12 | 0.0007 | 0.04 | 1.54×10−7 |
Note: p-values were from Fisher exact test by comparing the allele counts in allopurinol cases to controls from BioMe, and to DILIN cases due to other drugs or herbal (DILIN controls) for all alleles listed in the cluster.
Haplotype analysis was performed for HLA-A*34:02 with either HLA-B*53:01 or HLA-B*58:01 in the African American subset. Haplotype HLA-A*34:02 – HLA-B*53:01 showed higher frequency in allopurinol cases (0.14) than in both BioMe population controls (0.005, p=8.79×10−15) and DILIN controls (0.012, p=3.01×10−16) suggesting linkage disequilibrium between the two class I alleles. However, in contrast, the HLA-A*34:02-HLA-B*58:01 haplotype was not found in the 11 allopurinol cases and was in lower frequency (0.0004) in BioMe population controls, which implies that HLA-A*34:02 is not in strong linkage disequilibrium with HLA-B*58:01 and that the association of allopurinol liver injury with HLA-A*34:02 and HLA-B*58:01 are likely independent of each other.
Of note, none of the 5 liver injury cases with exposure to allopurinol that were excluded from this case series because of a low causality score for allopurinol carried HLA-B* 58:01, HLA-B*53:01 or HLA-A* 34:02 (Supplementary Table 3).
Discussion
Allopurinol is a xanthine oxidase inhibitor that is commonly used to treat and prevent gout and hyperuricemia. In calendar year 2017, over 14 million prescriptions of allopurinol were dispensed in the United States (16). In the current study, we report 11 cases of well documented acute liver injury associated with allopurinol use that were enrolled into the DILIN Prospective Study over a 16-year period. All of the cases developed evidence of liver injury within the first 3 months of treatment and were severe with all 11 patients requiring hospitalization and the majority developing jaundice. Similar to previous reports showing over-representation of allopurinol associated SJS/TEN in African Americans and Asians, there appeared to be an overrepresentation of African Americans compared to the overall DILIN cohort (60% vs 13%) (20). However, the frequency of alcohol use, diabetes, and concomitant medications as well as patient age and gender were similar to other cases enrolled into DILIN (Data not shown).
In comparison to prior studies, most patients with allopurinol induced liver injury had a mixed or cholestatic profile at presentation (4–7). This difference may have been due to the inclusion of an older group of patients. However, most of the patients had prominent hypersensitivity features at presentation with rash noted in 10 cases, fever in 5 and eosinophilia in 5 Interestingly, 4 of the 6 patients who presented with prominent features of a DRESS reaction had initially normal liver biochemistries and the liver enzymes increased only after their hospital admission (Cases #1,3,5,7). Subsequently 2 of these delayed onset cases were confirmed to be DRESS via skin biopsy. (Case 7, Case 3). The peak bilirubin in 3 of the 4 cases was less than 2.0 mg/dL. One patient (Case #1) had a history of multiple congenital anomalies and hypogammaglobulinemia as well suggestive of an immunodeficiency syndrome. In addition to a delayed onset of his granulomatous hepatitis, he also subsequently developed evidence of glomeruloneprhritis. In addition to these 4, two others (Case #9 and #10) had DRESS with high RegiSCAR scores and received corticosteroid therapy. Ultimately, 9 patients in all were treated with corticosteroids. Three patients died 38, 45 and 182 days after DILI onset, and in two cases death was considered to be liver-related.
Liver histopathology demonstrated a spectrum of abnormalities (Table 2). One patient had a picture of granulomatous hepatitis (Case #1) in the setting of extramedullary hematopoiesis while 3 patients showed cholestasis with mild inflammation. Two patients had evidence of ductopenia despite the fact that the biopsies were only obtained 6 and 9 days after DILI onset. These histopathological features are compatible with prior reports which report three patterns of injury that are typical for allopurinol: granulomatous hepatitis (particularly with fibrin ring granulomas), cholestatic hepatitis, and acute hepatitis with necrosis (17)
HLA analysis revealed an established association between HLA-B*58:01 and allopurinol hypersensitivity skin reactions (18). In addition, potential new associations are demonstrated for allopurinol DILI with HLA-B*53:01 and HLA-A*34:02 based on their higher frequencies in the cases than BioMe controls in African Americans. Although we did not have a large enough sample size to assess their effect in other racial groups, the only Hispanic patient carried both HLA-A*34:02 and HLA-B*53:01, and one of three European American patients carried HLA-B*58:01. The results of our haplotype analysis implied that the effect of HLA-B*53:01 on DILI is likely due to being on the same haplotype with HLA-A*34:02, but the effect of HLA-B*58:01 is likely independent from HLA-A*34:02. While we cannot rule out other ethnicity, our data supports the potential role of HLA-B*58:01 as a risk factor for allopurinol DILI in African Americans.
In the comparison with other non-allopurinol DILIN cases (disease controls), the HLA-A*34:02 and HLA-B*58:01 showed significantly higher AF in allopurinol DILI cases amongst African Americans. In particular, the AF for HLA-B *58:01 was 10-fold higher in allopurinol DILI cases than in non-allopurinol DILI cases. This is an encouraging observation as these two HLA alleles may specifically increase the risk of allopurinol induced liver injury. However, it should be noted that in our comparison to the non-allopurinol DILI cases in DILIN (N=1512 in Table 3), the AF may be biased due to the different proportion of European Americans between allopurinol cases and DILIN disease controls (27% vs. 70%). This could explain the reason why more significant results were found in the full dataset than in the smaller African-American subset (e.g, HLA-B*34:02: p=8.22×10−5 vs 0.05).
Prior studies have demonstrated a strong association of HLA-B *58:01 and allopurinol associated SJS/TEN in both Asians and Caucasians with an odds ratio of 80 to 100 (18, 19). Furthermore, epidemiological evidence suggests that African Americans and Asians are more likely to be hospitalized for allopurinol associated SCAR (20). The pathogenesis of allopurinol-induced SCAR such as SJS/TEN and DRESS are consistent with a delayed immune mediated reaction. Due to its high positive predictive value, pretreatment genotyping for HLA-B *58:01 has been introduced in some Southeast Asian countries in an effort to avoid use of the drug in these high risk patients and has proven to be effective and reduce the incidence of severe skin reactions in some Asian countries (21). However, up to 45% of patients of European ancestry with allopurinol associated SJS/TENS do not carry HLA-B*58:01 (3). Our current data now demonstrate a promising association between the HLA-B*58:01 allele and allopurinol hepatotoxicity in African Americans. In terms of the suspected mechanism, allopurinol is metabolized to its active metabolite, oxypurinol which is then cleared by the kidney. As a result, it is recommended that the dose of allopurinol be adjusted in patients with renal impairment (22). We note that several of the patients in our study had impaired renal function and received high daily doses of allopurinol (Table 1, supplemental Table 1). A recent study demonstrated that oxypurinol can activate HLA-B*58:01 restricted CD8+ T cells (23).
Limitations of our study include the relatively small number of allopurinol DILI cases available for genetic testing. For this reason, our selection of top HLA alleles was based on the objective criteria of allele frequency difference and nominal significance threshold without considering the correction of multiple testing. However, the association signals were supported in the subset of African-American cases. Furthermore, we did not have access to an independent validation cohort to confirm our genetic associations. However, identification of a replication cohort may prove difficult since allopurinol accounted for only one of 133 cases of acute liver failure attributed to drugs in the Acute Liver Failure Study Group Registry (23) and for only one of 661 cases of acute liver failure due to drugs reported to the U.S. liver transplant registry (24). Furthermore, no cases of allopurinol hepatotoxicity were reported in two large European DILI registries (26, 27). However, our findings are in line with prior studies demonstrating a strong association between HLA-B*58:01 and allopurinol associated SCAR (4–7).
In summary, allopurinol hepatotoxicity is characterized by a short latency with prominent features of systemic hypersensitivity and a mixed or cholestatic serum enzyme profile at presentation. The majority of patients were treated with corticosteroids which appeared to speed recovery from the symptoms and signs of hypersensitivity but which had unclear effects on the course of the liver injury as 2 of the 9 treated subjects died. These results also demonstrate that African Americans, who are at a higher risk of severe cutaneous adverse reactions from allopurinol, are similarly at higher risk of liver injury from this agent. This overrepresentation may be due to the higher frequency of HLA risk alleles among African Americans compared to European Americans as well as to an increased frequency of comorbidities such as chronic kidney disease. Interestingly, African Americans do not appear to be at higher risk of gout, despite the over-representation of hypertension and chronic kidney disease (20). Since allopurinol was recently recommended as the first line agent for patients with hyperuricemia and gout including those with renal impairment additional cases of allopurinol DILI will likely be seen (28).
Overall, 50% of subjects who had a RegiSCAR score of 4 or more for DRESS in this cohort carried HLA-B*58:01 and two-thirds were African American. The strong genetic association with HLA-B*58:01 in both African Americans and the overall population strongly implicated HLA-B*58:01 in the pathogenesis of allopurinol DILI. Currently, HLA-B*58:01 has a positive predictive value of 1–3% for allopurinol SCAR which given the overall prevalence of disease would be expected to be lower for allopurinol DILI. This suggests that using HLA-B*58:01 testing in isolation, more than 1000 allopurinol treated patients would need to be tested to prevent one case of either SCAR or DILI. Furthermore the negative predictive value of HLA-B*58:01 is not 100% meaning that cases of DILI will arise that do not carry HLA-B*58:01. Febuxostat is an alternative to allopurinol however it is more expensive and carries a black box warning for increased risk of all cause and cardiac associated death (28). Therefore, in the current environment, allopurinol will likely to continue to be used as a first line agent for gout and hyperuricemia. From our data, testing for HLA-B*58:01 and potentially other class I risk alleles may be of value in assessing allopurinol treated patients with unexplained liver injury.
Supplementary Material
Supplemental Table 1- Detailed clinical data of 11 high causality Allopurinol hepatotoxicity cases
Supplemental Table 2: HLA- allele frequencies in several large US populations
Supplemental Table 3: HLA Genotyping results for HLA-A and HLA-B in excluded low causality cases
Supplemental Figure 1- HLA-B allele cluster analysis in African Americans. The color key was based on the distance between allele binding specificity. Red color (near 0) indicates high similarity on binding specificity between alleles, and white (near 1) indicates completely different binding specificity. Allele clusters tested for association with allopurinol DILI are marked by rectangles.
Funding acknowledgement:
This work performed by investigators of the Drug Induced Liver Injury Network (DILIN) is structured as an U01 cooperative agreement supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health (NIH) with funds provided by the following grants: U01-DK065176 (Duke), U01-DK065201 (UNC), U01-DK065184 (Michigan), U01- DK065211 (Indiana), U01DK065193 (UConn), U01-DK065238 (UCSF/CPMC), U01-DK083023 (UTSW), U01-DK083027 (TJH/ UPenn), U01-DK082992 (Mayo), U01-DK083020 (USC). Additional support is provided by CTSA grants: UL1 RR025761 (Indiana), UL1TR000083 (UNC), UL1 RR024134 (UPenn), UL1 RR024986 (Michigan), UL1 RR024982 (UTSW), UL1 RR024150 (Mayo) and by the Intramural Research Program of the NIH, National Cancer Institute.
We are grateful to all patients and study coordinators who participated in the DILIN study. We also thank NCBI/dbGaP at http://www.ncbi.nlm.nih.gov/gap for the approval of our application to obtain the control dataset. The BioMe dataset was from the Charles Bronfman Institute for Personalized Medicine (IPM) BioMe BioBank at the Icahn School of Medicine at Mount Sinai (New York). Phenotype data collection was supported by The Andrea and Charles Bronfman Philanthropies. Funding support for genotyping, which was performed at The Center for Inherited Disease Research (CIDR), was supported by the NIH (U01HG007417). The GWAS data were from dbGaP accession number phs000925.v1.p1.
Conflicts of Interest: Dr. Fontana has received research support from Gilead, BMS, and Abbvie and consulted for Sanofi. Dr Phillips has no disclosures relevant to this manuscript. Within the past three years she has served as a consultant to Biocryst Pharma and Janssen Pharmaceuticals; She serves as Section Editor for Drug Allergy for UpToDate. She is co-director of IIID Pty Ltd that holds a patent for a method for identification and determination of hypersensitivity to abacavir and she folds a patent for detection of human leukocyte antigen A*32:01 in connection with diagnosing vancomycin associated drug reaction with eosinophilia and systemic symptoms without any financial renumeration. Dr’s Barnhart, Hoofnagle, Kleiner, Li, and Saeed have no reported conflicts.
Abbreviations
- AF
Allele frequency
- Alk P
Alkaline phosphatase
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- CF
Carriage frequency
- CMV
Cytomegalovirus
- EBV
Epstein Barr Virus
- DILI
Drug induced liver injury
- DILIN
Drug induced liver injury network
- DRESS
Drug reaction and eosinophilia and systemic symptoms
- GFR
Glomerular filtration rate
- GWA
Genome wide association
- HLA
Human leukocyte antigen
- INR
International normalized ratio
- SCAR
Severe cutaneous adverse reaction
- SNP
Single nucleotide polymorphism
- SJS
Stevens Johnson Syndrome
- TEN
Toxic epidermal necrolysis
- ULN
Upper limit of normal
Footnotes
Ethics approval statement
The study protocol and consent forms were reviewed and approved by local institutional review boards and a central data safety and monitoring board. All study participants provided written informed consent.
ClinicalTrials.gov number: NCT00345930.
References
- 1.Becker MA, Schumacher HR Jr, Wortmann RL, et al. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N Engl J Med 2005; 353: 2450–61 [DOI] [PubMed] [Google Scholar]
- 2.Vanderstigel M, Zafrani ES, Lejonc JL, Schaeffer A, Portos JL. Allopurinol hypersensitivity syndrome as a cause of hepatic fibrin-ring granulomas. Gastroenterol 1986; 90: 188–90. [DOI] [PubMed] [Google Scholar]
- 3.Khoo BP, Leow YH. A review of inpatients with adverse drug reactions to allopurinol. Singapore Med J 2000; 41: 156–60 [PubMed] [Google Scholar]
- 4.Hung SI, Chung WH, Liou LB, et al. HLA-B*58:01 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci 2005; 102: 4134–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaniwa N, Saito Y, Aihara M, et al. ; JSAR research group. HLA-B locus in Japanese patients with anti-epileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics 2008; 9: 1617–22. [DOI] [PubMed] [Google Scholar]
- 6.Lonjou C, Borot N, Sekula P, et al. ; RegiSCAR study group. A European study of HLA-B in Stevens-Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet Genomics 2008; 18: 99–107. [DOI] [PubMed] [Google Scholar]
- 7.Somkrua R, Eickman EE, Saokaew S, Lohitnavy M, Chaiyakunapruk N. Association of HLA-B*58:01 allele and allopurinol-induced Stevens Johnson syndrome and toxic epidermal necrolysis: a systematic review and meta-analysis. BMC Med Genet 2011; 12: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontana RJ, Watkins PB, Bonkovsky HL, Chalasani N, Davern T, Serrano J, Rochon J. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009; 32:55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YJ, Phillips E, Dellinger A, et al. HLA-B *14:01 and HLA-B *35:01 are associated with trimethoprim-sulfamethoxazole induced liver injury. Hepatology 2020. accepted: doi: 10.1002HEP.31258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleiner DE, Chalasani NP, Lee WM, et al. Hepatic histological findings in suspected drug-induced liver injury: systematic evaluation and clinical associations. Hepatology. 2014;59(2):661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kardaum SH, Sidoroff A, Valeyrie- Allanore L, et al. Variability in the clinical pattern of cutaneous side-effects of drugs with systemic symptoms: does a DRESS syndrome really exist? Br J Dermatol 2007; 156: 609–611. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, Weir BS. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J 2014;14:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirulli ET, Nicoletti P, Abramson K, Andrade RJ, Bjornsson ES, Chalasani N, et al. A Missense variant in PTPN22 is a risk factor for Drug-induced liver injury. Gastroenterology 2019; 156: 1707–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoof I, Peters B, Sidney J, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009;61:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002;70:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.https://clincalc.com. Drug usage Statistics, United States, 2007–2017. Accessed on October 4, 2020.
- 17.Chawla SK, Patel HD, Parrino GR, Soterakis J, Lopresti PA, D’Angelo WA. Allopurinol hepatotoxicity: Case report and literature review. Art & Rheumatism 1977; 20: 1546–1550. [DOI] [PubMed] [Google Scholar]
- 18.Somjrua R, Eickman EE, Saokaew S, Lohntnavy M, Chaiyakunapruk N. Association of HLA-B * 58:01 allelle and allopurinol induced stevens Johnson Syndrome and TEN: A systematic review and meta-analysis. BMC Med Genetics 2011: 12: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu R, Cheng Y, Zhu L, et al. Impact of HLA- B *58:01 allele and allopurinol-induced cutaneous adverse drug reactions: Evidence from 21 pharmacogenetic studies. Oncotarget 2016: 7: 81870–81879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu N, Rai SK, Terkeltaub R, Kim SC, Menendez ME, Choi HK. Racial disparities in the risk of Stevens-Johnson Syndrome and toxic epidermal necroloysis as urate-lowering drug adverse Events in the United States. Sem Arth Rheum 2016; 46: 253258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko TM, Tsai CY, Chen SY, et al. Use of HLA-B * 58:01 genotyping to prevent allopurinol induced severe cutaneous adverse reactions in Taiwan: National prospective cohort study. BMJ 2015; 351: h4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turnheim K, Krivanek P, Oberbauer R. Pharmacokinetics and pharmacodynamics of allopurinol in the elderly and young subjects. Br J Clin Pharmacol 1999; 48: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun J, Marcaida MJ, Eriksson KK, Jamin H, Fontana S, Pichler WJ, Yerly D. Oxypurinol directly and immediately activates the drug-specific T-cells via the preferential use of HLA-B *58:01. J Immunology 2014: 192: 2984–2993. [DOI] [PubMed] [Google Scholar]
- 24.Reuben A, Tillman H, Fontana RJ, et al. Outcomes in Adults with Acute Liver Failure between 1998 and 2013. Ann Intern Med 2016: 164: 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo MW, Galanko JA, Shrestha R, Fried MW, Watkins PW. Liver Transplantation for acute liver failure from drug induced liver injury in the United States. Liver Transplantation 2004; 10: 1018–10123 [DOI] [PubMed] [Google Scholar]
- 26.Bjornsson ES, Bergmann OM, Bjornsson HK, et al. Incidence, presentation, and outcomes in patients with drug induced liver injury in the General population of Iceland. Gastroenterology 2013; 144: 1419. [DOI] [PubMed] [Google Scholar]
- 27.Andrade R, Lucena MI, Fernandez MC, et al. Drug induced liver injury: an analysis of 461 incidences submitted to the Spanish Registry over a 10-year period. Gastroenterology 2005: 129: 512. [DOI] [PubMed] [Google Scholar]
- 28.Fitzgerald JD, Dalbeth N, Mikulus T, et al. 2020 American College of Rheumatology Guideline for the Management of Gout. Arthritis Care & Research 2020; 72: 744–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1- Detailed clinical data of 11 high causality Allopurinol hepatotoxicity cases
Supplemental Table 2: HLA- allele frequencies in several large US populations
Supplemental Table 3: HLA Genotyping results for HLA-A and HLA-B in excluded low causality cases
Supplemental Figure 1- HLA-B allele cluster analysis in African Americans. The color key was based on the distance between allele binding specificity. Red color (near 0) indicates high similarity on binding specificity between alleles, and white (near 1) indicates completely different binding specificity. Allele clusters tested for association with allopurinol DILI are marked by rectangles.