SUMMARY
Neurogenesis comprises many highly regulated processes including proliferation, differentiation and maturation. However, the transcriptional landscapes underlying brain development are poorly characterized. We describe a developmental single-cell catalog of ~220,000 zebrafish brain cells encompassing 12 stages from embryo to larva. We characterize known and novel gene markers for ~800 clusters and provide an overview of the diversification of neurons and progenitors across these timepoints. We also introduce an optimized GESTALT lineage recorder that enables higher expression and recovery of Cas9-edited barcodes to query lineage segregation. Cell type characterization indicates that most embryonic neural progenitor states are transitory and transcriptionally distinct from neural progenitors of post-embryonic stages. Reconstruction of cell specification trajectories reveals that late-stage retinal neural progenitors transcriptionally overlap cell states observed in the embryo. The zebrafish brain development atlas provides a resource to define and manipulate specific subsets of neurons and to uncover the molecular mechanisms underlying vertebrate neurogenesis.
eTOC Blurb
The complexity and dynamics of vertebrate brain development are poorly understood. Raj et al. have generated a single-cell atlas of zebrafish brain development. They document the expansion of neuronal diversity, analyze the transition from early to late progenitors, and reconstruct cellular trajectories and lineages.
Graphical Abstract

INTRODUCTION
The vertebrate brain develops from a limited pool of embryonic neural progenitor cells that cycle through rounds of proliferation, diversification, and terminal differentiation into an extensive catalogue of distinct neuronal and glial cell types. A central goal in developmental neurobiology is to investigate how neuronal complexity arises through molecular specification and commitment by studying the origins and fates of cells during development. Fundamental insights into these processes have been gained via classic approaches using genetic markers, perturbations and fate mapping (Cepko, 2014; Kretzschmar and Watt, 2012; Ma et al., 2017; Wamsley and Fishell, 2017; Wilson et al., 2002; Woo and Fraser, 1995; Woodworth et al., 2017). These approaches have recently been complemented by single-cell genomics technologies in the developing nervous system, including the spinal cord (Delile et al., 2019; Rosenberg et al., 2018); cortex (Nowakowski et al., 2017; Zhong et al., 2018); olfactory system (H. Li et al., 2017); cerebellum (Carter et al., 2018; Tambalo et al., 2020); retina (Clark et al., 2019; Hu et al., 2019; Xu et al., 2020); and whole animal (Farnsworth et al., 2020; Farrell et al., 2018; Wagner et al., 2018). These studies have provided transcriptome-level views of the rich heterogeneous states that cells progress through as they proliferate, migrate and differentiate. Nevertheless, existing datasets are limited in their scope as they focus on specific brain regions, survey limited timepoints or do not enrich for neural cell types, thereby missing transitions and cellular diversity. Thus, there is a need for a large-scale neurodevelopmental single-cell resource that profiles whole brain development across a range of closely-spaced embryonic and post-embryonic stages. In addition, such an atlas would help address fundamental questions about the dynamics of brain development. For example, it is poorly understood how embryonic neural progenitors are molecularly related to post-embryonic neural progenitors. Furthermore, the transcriptional programs that are activated or suppressed as neural progenitors become fate-restricted and differentiate are largely unknown.
Here we present resources to obtain global views of neurogenesis, cell type heterogeneity, specification trajectories and lineage relationships in the developing zebrafish brain. We generated a single-cell RNA-seq (scRNA-seq) atlas consisting of ~220,000 cells from 12 hours post fertilization (hpf) to 15 days post fertilization (dpf). We also created a new version of the scGESTALT CRISPR-Cas9 lineage recorder (McKenna et al., 2016; Raj et al., 2018b) with improved barcode capture and used it to query early lineage decisions. Using the cell type atlas, we analyzed the expansion of neuronal diversity, the loss of transitory embryonic progenitors, and the maintenance of distinct larval progenitor states. We reconstructed cell specification trajectories of the zebrafish retina and hypothalamus, revealing gene expression cascades and distinct specification programs. Collectively, the zebrafish brain development atlas reveals molecular and cellular changes at an unprecedented scale and resolution, and lays the foundation for the detailed analysis of neuronal diversification.
RESULTS
Building a developmental atlas of the zebrafish brain with single-cell transcriptomics
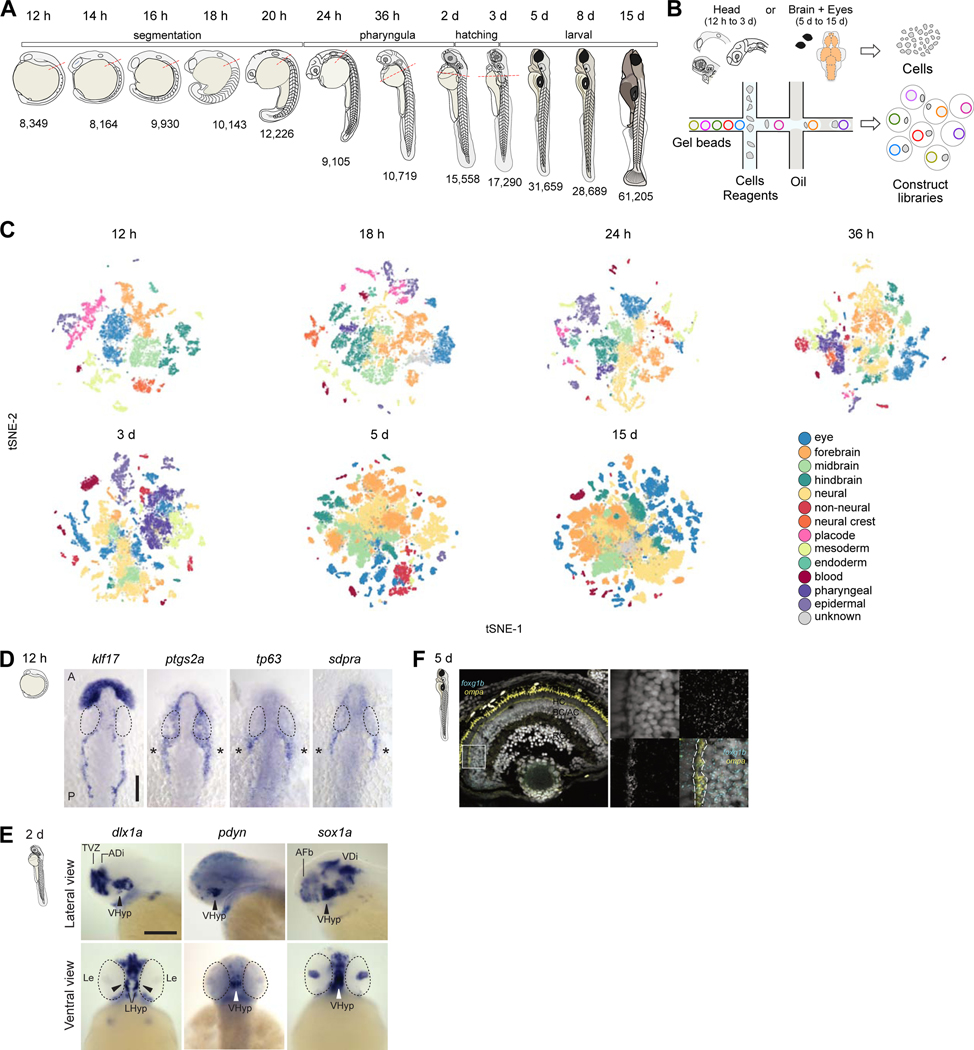
To reveal the landscape of cell states and cell types during brain development, we profiled 223,037 cells across 12 stages of zebrafish embryonic and larval development using the 10X Chromium scRNA-seq platform. Samples spanned from 12 hpf (shortly after gastrulation), when the embryo is undergoing early developmental patterning, to 15 dpf, when larvae are mature, exhibit complex behaviors, and are expected to exhibit substantial cell type diversity (Figure 1A). To enrich for brain cell types, we dissected the heads of animals from 12 hpf to 3 dpf, and the brains and eyes from 5 dpf to 15 dpf (Figure 1B). To determine cell type diversity in the head and brain of zebrafish, data from each stage was analyzed individually using Louvain clustering (Figure 1C and Figure S1). This approach identified a total of 815 cell clusters across all 12 timepoints (Table S1). To classify each cluster, we compared enriched gene markers with existing gene expression annotations in the ZFIN database and literature, as described previously (Raj et al., 2018b). Plotting expression of known cell type markers identified clusters corresponding to neural progenitors (sox19a), dozens of neuron subtypes (elavl3, gad2, slc17a6b), eye cells (foxg1b, lim2.4, pmela, ca14, gnat1, opn1mw1), radial glia (mfge8a, s100b), neural crest (sox10), oligodendrocytes (mbpa), blood cells (cahz, etv2, cd74a), cartilage (matn4, col9a2), pharyngeal arches (pmp22a, prrx1b, barx1), sensory placodes (dlx3b, six1b), and epidermal cells (epcam, cldni), among others. As expected, cell type complexity increased with developmental time. We validated new marker expression across several cell types identified in our dataset, such as sdpra in the trigeminal placode, sox1a in the hypothalamus, and ompa in the retina (Figure 1D-F). Our analysis also revealed groups of embryonic clusters that were absent or transcriptionally distinct from larval clusters, suggesting that many embryonic cell states are transitory. Several of these transitions are known developmental changes (e.g. loss of placodes and rhombomeres), but changes in neural progenitor cell states are poorly understood (see below).
Figure 1. Developmental compendium of zebrafish head and brain cell types.
A. Schematic of the developmental stages profiled. Red hatched line, head regions selected for enrichment of brain cells. Samples from 5 to 15 dpf were dissected to obtain brain and eye specifically. h, hours post fertilization; d, days post fertilization
B. Schematic of scRNA-seq using the 10X Genomics platform.
C. Cell type heterogeneity within each stage. Clusters at each stage were assigned to a region or tissue type based on known markers and color coded to reflect their classification. tSNE implementations: Barnes-Hut (12h to 3d), Fourier transform (5d and 15d).
D. In situ hybridization for novel markers in the trigeminal placode at 12 hpf. klf17 is expressed on the anterior polster and ventral mesoderm, delineating the border of the embryo. Trigeminal ganglia markers ptgs2a, tp63 and sdpra (cavin2a) are expressed bilaterally (asterisks) posterior to the eye. Eyes are delineated by dotted lines. A: Anterior; P: Posterior. Scale bar,100 μm.
E. In situ hybridization validation of novel marker sox1a in the hypothalamus at 2 dpf. Top panels, lateral view of brain; Bottom panels, ventral view of brain. dlx1a and pdyn are known hypothalamic markers. Eyes are delineated by dotted lines. VHyp: Ventral Hypothalamus; TVZ: Telencephalic Ventricular Zone; ADi: Anterior Diencephalon; AFb: Anterior Forebrain; VDi: Ventral Diencephalon; Le: Lens. Scale bar,200 μm.
F. smFISH validation of novel marker ompa in horizontal cells of the retina at 5 dpf. Left panel, retina section stained with DAPI (grey), pan-retinal foxg1b (cyan) and ompa (yellow). Strong yellow signal in photoreceptors represent autofluorescence. White box, area zoomed in for the right panels. Dotted lines, horizontal cell layer. PR, photoreceptor cells; HC, horizontal cells; BC, bipolar cells; AC, amacrine cells; RGC, retinal ganglion cells
See also Figure S1
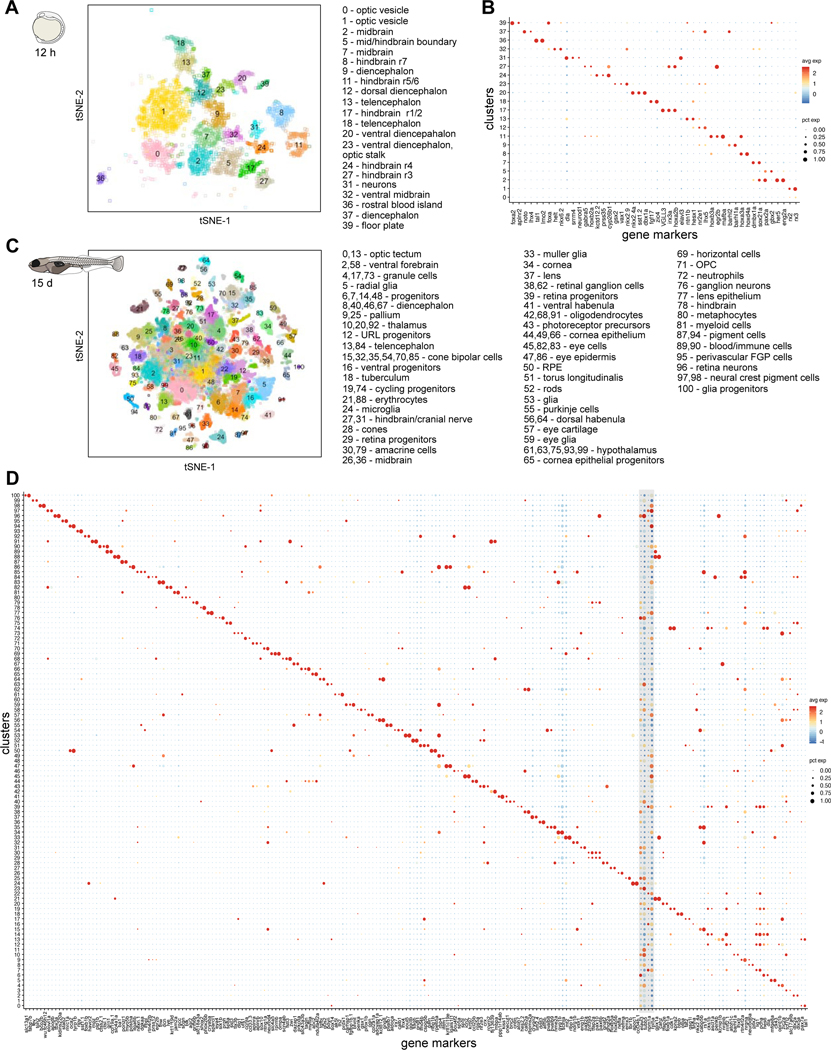
To enable direct comparison of cell types across our time course, we subsetted the 12 hpf dataset to only comprise neural populations and blood cells found in the brain, eliminating non-relevant head cells from earlier stages, such as mesoderm, placodes, and periderm. This approach resulted in an initial set of 21 clusters at 12 hpf (Figure 2A) that diversified into 98 clusters by 15 dpf (Figure 2C). Notably, most clusters could be uniquely identified using a minimal group of 2–3 enriched gene markers (Figure 2B, 2D). For example, at 12 hpf, the optic vesicle is identified by expression of rx2 and rx3; hindbrain rhombomeres 5/6 by hoxb3a and eng2b; and ventral diencephalon by nkx2.4a and dbx1a. Similarly, at 15 dpf, the cerebellar granule cells are marked by expression of oprd1b and zic2a; optic tectum by pax7a and tal1; and a new retinal cell type by kidins220a, foxg1b (exclusively detected in retinal cells) and tbx3a. We did not find minimal groups of marker genes to unambiguously define cycling progenitors, differentiating progenitors and newly born neurons, as many of these subtypes had similar expression signatures of pan neuronal or pan progenitor marker genes, such as elavl3 and tubb5 in neurons, and rpl5a and npm1a in progenitors (Figure 2D, grey box).
Figure 2. Brain cell type diversification from 12 hpf to 15 dpf.
A. tSNE plot of 12 hpf dataset. Only clusters corresponding to neural and blood cell types are shown. Inferred identities of each cluster are described.
B. Dot plot of gene expression pattern of select marker genes (columns) for each cluster (row). Dot size indicates the percentage of cells expressing the marker; color represents the average scaled expression level.
C. tSNE plot of 15 dpf dataset. Inferred identities of each cluster are described.
D. Dot plot of gene expression patterns of select marker genes for each cluster. Layout is same as (B). Grey box represents generic neuronal and progenitor genes. tSNE implementations: Barnes-Hut (A), Fourier transform (C)
At 12 hpf, the early demarcation of multiple brain regions is already apparent and by 15 dpf these regions expand and diversify further. For example, the optic vesicle at 12 hpf is defined by one cluster and is the origin of 18 retinal cell types at 15 dpf. Similarly, a single cluster of ventral diencephalon cells (expressing shha, nkx2.4a, nkx2.1, rx3) at 12 hpf develops into 7 major hypothalamus cell types at 15 dpf. An exception to this diversification is the loss of rhombomeres (r1-r7) in the hindbrain (Moens and Prince, 2002).
To further explore brain neuronal subtypes at 15 dpf, we analyzed the expression of transcription factors, neuropeptides and their receptors, and genes involved in neuronal physiology such as neurotransmitters, transporters, receptors, and channels (Chen et al., 2017; Pandey et al., 2018; Tiklová et al., 2019; Zeisel et al., 2018). Our results indicate that nearly all identified neuron subtypes can be distinguished from one another via the expression of individual or combinations of genes belonging to these categories (Figure 3A-C). For example, cluster 2 and 84 neurons are GABAergic forebrain neurons that express dlx2a and dlx5a, while cluster 84 neurons additionally express six3b, gria1a and gria2b.
Figure 3. Neuron subtype diversity at 15 dpf.
A-C. Violin plots of select marker gene expression in identified brain neuron subtypes at 15 dpf. Retinal neurons and nascent neurons are omitted from the analysis. Cluster numbers are indicated at the bottom along with their inferred spatial location in the brain. Cluster 76 has unknown spatial location. Detailed cluster descriptions are in Table S1 and can be explored interactively at https://github.com/brlauuu/zf_brain.
A. Expression of transcription factors.
B. Expression of neuropeptides and their receptors.
C. Expression of genes involved in neuron electrophysiology.
D. Matrix showing overlap of neuron subtypes identified at 15 dpf and earlier larval (5 and 8 dpf) or later juvenile [25 dpf (Raj et al., 2018b)] stages. The cluster number at 15 dpf is shown and an orange circle indicates that the subtype is detected in another stage.
We next asked if neuron clusters detected at 15 dpf are found at earlier larval stages, when most behavioral experiments are performed (Figure S2). 68% (23/34 clusters) and 74% (25/34 clusters) of 15 dpf neuron clusters have a closely matching counterpart at 5 dpf and 8 dpf (based on enriched marker gene expression), respectively (Figure 3D). Sampling issues might have prevented the identification of additional overlapping clusters, but our data indicate a large overlap between identified cell types from 5 to 15 dpf. These results suggest that the zebrafish brain already has considerable cell type diversity at early larval stages. Furthermore, 97% (33/34) of 15 dpf clusters overlapped with clusters identified in our previously described 23–25 dpf juvenile brain dataset (Raj et al., 2018b). Thus, by 15 dpf late larval stage, nearly all of the brain cell types that persist into the early juvenile stage have already been established. Notably, among cell types that are “missing” or under-represented at 15 dpf but readily detected at 23–25 dpf are cell types in the optic tectum, cerebellum and the torus longitudinalis, suggesting that these structures undergo further diversification after 15 dpf. In contrast, many cell types in the pallium, habenula (Pandey et al., 2018), hypothalamus and preoptic area are detected across these stages, suggesting that they develop earlier.
In summary, we generated a zebrafish brain development cell type atlas spanning 12 stages of brain organogenesis. The complete dataset can be explored using the accompanying app: https://github.com/brlauuu/zf_brain.
Neurogenic expansion during brain development
During development, cell composition shifts from predominantly progenitor populations to more differentiated cell types (Schmidt et al., 2013). To better characterize how differentiation varies during neuronal development, we first asked if our dataset captured the two neurogenic phases (primary and secondary) before and after 2 dpf that have been traditionally defined through histological analyses (Allende and Weinberg, 1994; Korzh et al., 1998; Mueller and Wullimann, 2003). We considered neural progenitors as non-differentiated neuronal precursor cells that may or may not be proliferating, and express a subset of classical progenitor markers e.g. sox19a, dla, s100b, and cell cycle genes. Since the brain is undergoing substantial molecular changes during these developmental windows, we defined the transcriptional programs and cells that exhibit these programs as progenitor cell states. We calculated the percentage of the dataset that corresponds to neural progenitor cells, neurons (expressing markers such as elavl3, elavl4) or other cell types across each timepoint in our dataset. Since the earlier stages (12 hpf to 3 dpf) also contained non-brain and non-eye cell types, we subsetted the early timepoints to only brain and eye cells. With increasing developmental time, we observed a progressive decrease in the fraction of the dataset comprising neural progenitor cells (from 53.8% to 18.3%) with a concomitant increase in neurons (from 4.5% to 58%) (Figure 4A). For example, we observed an initial increase in the number of distinct progenitor clusters from 12 hpf to 18 hpf (early embryo stages), while the number of neuron clusters remained low (Figure 4A, right panels). From 20 hpf to 3 dpf (intermediate stages), the total progenitor clusters decreased while neuron clusters started to increase. For example, neuronal clusters expanded from 11 at 20 hpf to 23 at 36 hpf. This burst coincides with the presumed timing of late-stage primary neurogenesis in zebrafish (Mueller and Wullimann, 2003). Notably, by 5 to 15 dpf (late larva stages), a second expansion of neuronal populations, corresponding to the secondary neurogenic phase (Mueller and Wullimann, 2003), had occurred (53 neuronal subtypes at 5 dpf). At 5 dpf, we detected cell types identified as early as 36 hpf (e.g. tal1+, gata3+ neurons in the optic tectum, and tfap2e+, barhl2+ neurons in the thalamus), as well as subtypes only observed during the second phase, such as nrgnb+ prkcda+ neurons in the forebrain and cone bipolar cell subtypes in the retina. Collectively, our dataset captures both phases of neurogenesis and reveals the diversification of neurons in multiple brain structures.
Figure 4. Developmental diversification of neurons and progenitors.
A. Area plot of the percentage of dataset at each timepoint corresponding to neural progenitors, neurons, and other cell types. Right panels, total number of clusters of progenitors and neurons at each stage.
B. tSNE plot of embryonic, intermediate and larval neural progenitors. All progenitor cells were analyzed together after subsetting from the whole dataset.
C-D. Heatmaps of select gene expression in early embryonic (C) and late larval (D) brain neural progenitors. Top panel, genes enriched in embryonic progenitors. Bottom panel, genes enriched in larval progenitors. TF, transcription factor. *pax6a is expressed in multiple regions
E. Heatmap of Pearson correlation values of 79 spatial markers in embryonic, intermediate and larval neural progenitors. Spatial markers were selected based on existing literature. Groups of co-varying genes in the midbrain and forebrain are highlighted with dashed boxes.
F. Plot of highly variable genes that co-vary with any of the selected 79 spatial markers in embryonic and larval progenitors. Co-variation was determined by Pearson correlation, with several thresholds (from stringent to relaxed) displayed along the x-axis.
See also Figure S3
Dampening of spatial and developmental signatures during the transition from embryonic to larval neural progenitors
We next analyzed our dataset to determine how cell states change during the transition from the embryonic to post-embryonic brain. The zebrafish brain undergoes lifetime constitutive neurogenesis due to the persistence of neural progenitor pools distributed along the brain’s axis (Schmidt et al., 2013). However, the embryonic origins and transcriptional programs that underlie their development are poorly understood. Furthermore, how the molecular identities of embryonic and post-embryonic neural progenitor cell states compare have not been well characterized. To address these questions, we asked how neural progenitor gene expression signatures globally change from embryo to larva. Based on the results described above, we defined early embryonic brain progenitors as neural cell transcriptional states from 12 hpf to 18 hpf, intermediate stage brain progenitors as neural cell transcriptional states from 20 hpf to 3 dpf, and larval brain progenitors as neural cell transcriptional states from 5 dpf to 15 dpf (Figure 4B, Figure S3). We determined the greatest sources of variation within these populations. For embryonic brain progenitors we found that the top 3 principal components comprise genes implicated in spatial and developmental patterning (Gibbs et al., 2017; Moens and Prince, 2002; Wilson et al., 2002; Wilson and Rubenstein, 2000). Cells exhibit characteristic anteroposterior and dorsoventral axial signatures (Figure 4C, top panel). For example, the telencephalon (anterior forebrain) is marked by foxg1a and emx3a expression, the midbrain by pax2a and eng2a, and the hindbrain is segmented into rhombomeres marked by distinct combinatorial patterns of egr2b and hox gene expression. Furthermore, all cells are in a highly proliferative state with strong expression of cell cycle genes such as pcna, mki67 and cdca7a. Collectively, the expression signatures are reflective of a developmental state during which the embryo is orchestrating a rapid expansion of neural progenitor populations concurrent with their acquisition of positional information and overt absence of differentiation (Schmidt et al., 2013; Stigloher et al., 2008).
In contrast, larval neural progenitors comprised two major groups: proliferating (expressing cell cycle genes pcna and top2a) and non-proliferating (depleted expression of cell cycle markers) (Figure 4D, bottom panel). Indeed, the top 3 principal components in the larval progenitors comprised genes that mark stem cells (PC1, PC3) and differentiation (PC2). The non-proliferating group is subdivided into radial glia (stem cells) and her2+ neural progenitors expressing proneural genes insm1b and scrt2. The proliferating group is subdivided into her2+ and scrt2- neural progenitor cells, her2- progenitors, her2+ and neurod1+ progenitor cells, and upper rhombic lip progenitors (localized to cerebellum) expressing atoh1c and oprd1b.
Strikingly, most larval progenitors were characterized by a reduced spatial signature (except for the cerebellar upper rhombic lip pool), such that cells were less enriched in region-specific transcription factors relative to embryonic progenitors (Figure 4D, top panel). For example, radial glia exist in multiple pools along the brain axis (Than-Trong and Bally-Cuif, 2015), but they formed a single cluster in our dataset (marked by expression of fabp7a, cx43, s100b and aqp1a.1). This result suggests that radial glia are largely transcriptionally similar. Although some expression of region-specific transcription factors was detected in larval progenitor clusters, these signatures were not sufficiently strong to resolve clusters as they were during embryonic stages.
To explore the apparent dearth of spatial signatures further, we calculated pairwise correlation scores for 79 transcription factors and signaling proteins with known spatial expression patterns in the forebrain and midbrain based on previously described histological analysis (ZFIN), and which were identified as gene markers for neuronal clusters in our dataset. These genes showed strongest correlations in embryonic progenitors, followed by intermediate stage progenitors, and were weakly correlated in larval progenitors (Figure 4E).
Since spatial signatures are encoded by a combinatorial code of genes with overlapping expression patterns, we asked whether the same subsets of genes co-varied with each of the 79 spatial markers across embryonic, intermediate, and larval neural progenitors. We found that intermediate stage progenitors showed overlap in co-varying genes with both embryonic and larval progenitors. For example, 44/79 genes had >40% overlap in their top 20 co-varying genes between embryonic and intermediate stage progenitors, and 23/79 genes had >40% overlap between intermediate and larval stage progenitors. In contrast, we found low overlap across embryonic and larval stages (3/79 genes had >40% overlap in their top 20 co-varying genes). Additionally, when we searched for genes that strongly co-varied with these 79 spatial markers (Pearson correlation >0.4), we found 38 genes during embryonic stages, 17 genes during intermediate stages, but only 4 genes during larval stages (Figure 4F).
Taken together, these results demonstrate that intermediate stage progenitors resemble a hybrid of early embryonic and late larval progenitor signatures. Furthermore, the overall spatial code between embryonic and larval progenitors are distinct, and the embryonic spatial code involves a larger collection of genes.
An optimized scGESTALT lineage recorder
A long-term goal in developmental neurobiology is to understand the lineage relationships of neurons. As a first step to derive lineage relationships of the cell types identified in the brain development atlas, we performed lineage recording experiments with scGESTALT. This lineage recorder enables simultaneous cell type and cell lineage identification by combining scRNA-seq with CRISPR-Cas9 barcode editing (McKenna et al., 2016; Raj et al., 2018b). To enable higher recovery of edited barcodes from single cells, we optimized the design and library preparation of the lineage recording cassette, including barcode editing of a transgene coding region and compatibility with the 10x platform (see Methods). To test the performance of this new recording cassette, we barcoded early embryonic lineage relationships by injecting Cas9 protein and target guide RNAs into 1-cell embryos (Figure 5A) and then isolated four 15 dpf larval brains. We recovered barcodes and transcriptional profiles of 5,794 cells total (barcode recovery rate 30–75% compared to 6–28% of our previous scGESTALT version (Raj et al., 2018b)). Edited barcodes showed no overlap between animals, displayed a diverse spectrum of repair products that spanned single and multiple sites, and were of varying clone sizes (Figure 5B-D, Figure S4A). These features closely resembled the editing patterns obtained with our previous recorders (McKenna et al., 2016; Raj et al., 2018b). Using the recovered barcodes and associated transcriptomes, we reconstructed lineage trees representing cell lineage segregations formed during early embryogenesis (for one example see Figure S4B). These lineage trees accompany our transcriptional cell type atlas and are available to explore at https://scgestalt.mckennalab.org/
Figure 5. Optimization of scGESTALT lineage recorder for better barcode recovery.
A. Schematic overview of CRISPR-Cas9 lineage recording. Optimized scGESTALT comprises a barcode cassette in the 3’end of DsRed transgene (single copy) and the medaka beta-actin promoter. Embryos were injected with Cas9 protein and DsRed sgRNAs and animals were profiled at 15 dpf by scRNA-seq.
B. Pairwise comparisons using cosine dissimilarity of barcode edit patterns from four (ZF1–4) edited 15 dpf larval brains.
C. Chord diagram of the nature and frequency of deletions within and between target sites. Each colored sector represents a target site. Links between target sites represent inter-site deletions; self-links represent intra-site deletions. Link widths are proportional to the edit frequencies.
D. Type of edit at each target site within the barcode from edited ZF1–4 larval brains.
E. Heat map of lineage relationships between non-retinal and retinal cell types in the eye. All clusters with >3 cells and all barcodes with >1 cell were used to determine if there is enrichment of cell type-specific barcodes across each cluster pair. Blue indicates significant enrichment and lineage segregation. Purple indicates no significant enrichment and no lineage segregation. Grey indicates insufficient sampling power and undefined lineage status. Cluster numbers are indicated (e.g. C45) and either cell type gene markers (e.g. cldna+) or the exact name of the cell type (e.g. cone bipolar cells) are indicated along the rows. Along the columns, the numbers within the brackets indicate the number of barcodes and number of cells, respectively, for that cluster.
F. Heat map of lineage relationships between brain regions and the retina. Neuron clusters that could be pseudospatially assigned to each region were used (See Table S1). Analysis, layout and color code are same as in E.
G. Heat map of lineage relationships between neuronal cell types in the forebrain and midbrain. Analysis, layout and color code are same as in E. Clusters were assigned to a brain region (e.g. pallium, hypothalamus), and for clusters where a more precise location could not be inferred a gene marker is indicated (e.g. pitx2+).
H. Heat map of lineage relationships between brain progenitor clusters. Analysis, layout and color code is same as in E. Cell type marker genes are indicated along with the cluster number. URL, upper rhombic lip
I. Bar plot of the proportion (based on Jaccard Index) of granule cell (cerebellum neurons) barcodes that are shared with each brain progenitor cluster. Cluster numbers are the same as in H.
Since the injection of editing reagents into 1-cell embryos saturates editing within 4–6 hours (McKenna et al., 2016), we expected early lineage divergences to be overrepresented in our dataset. We first asked if our recorder captured diverse multi-lineage tissue origins of the eye, which is derived from neuroectoderm, surface ectoderm and mesoderm (Figure 5E). Eye cell types were identified as clusters that contained cells from scRNA-seq samples comprising eye tissue exclusively. Retinal cell types were defined as clusters expressing the pan-retinal marker foxg1b (Figure 1F), whereas non-retinal cell types were depleted in foxg1b. We performed pairwise comparisons of all eye clusters with at least 4 independent barcodes (each with at least 2 cells). Since <1% of all cells were captured by scRNA-seq, we asked if there is cell type-specific barcode enrichment greater than expected by chance (“lineage segregation” in Figure 5E). For cluster pairs where we did not observe significant lineage segregation, we asked if this was due to a lack of sampling (“lineage status undefined”) or true lack of cell type-specific barcode enrichment (“no lineage segregation”). The latter case would indicate that two cell types shared a more recent common ancestor than cell types that segregated earlier. We found that multiple retinal and non-retinal cell types segregated from each other, as would be expected due to early separation of their tissue origins. Interestingly, however, a few non-retinal cell types (e.g. clusters 34, 44, 49) did not fully segregate from retinal cell types, suggesting that they shared a common progenitor. Furthermore, there was extensive lineage segregation between various non-retinal cell types (e.g. clusters 45, 47, 86). In contrast, we did not observe lineage segregation between the different retinal cell types, likely due to the termination of barcode editing prior to terminal divisions. The exception was cluster 28 (cones), which segregated from clusters 15 and 32 (cone bipolar cells) and 28 (retinal ganglion cells). Thus, lineage splits between retinal and non-retinal cell types, and within non-retinal subtypes preceded most splits within retinal subtypes.
Next, we asked if our recorder captured lineage divergences between neurons across brain regions and the retina. Although the hindbrain and retina formed distinct lineages early in development, forebrain and midbrain neurons continued to share progenitors across the same barcoding period (Figure 5F). Pairwise comparisons of all forebrain and midbrain clusters revealed examples of emerging segregation along multiple spatial axes (Figure 5G). For example, we saw evidence of dorsal-ventral split: cluster 9 pallium (dorsal) separated from cluster 25 sub-pallium (ventral). Furthermore, barcode enrichments confirmed rostral-caudal splits: cluster 64 habenula separated from clusters 9 and 25 pallium (telencephalon, rostral) and clusters 0 and 13 optic tectum (caudal). Overall, the lineage segregations agree with classic fate mapping experiments (Woo and Fraser, 1995) and correlate with the anteroposterior and dorsoventral gene expression signatures of early progenitors (Figure 4).
To query the lineage relationships of brain progenitor cell types, we performed pairwise comparisons of progenitor clusters at 15 dpf (Figure 5H). Notably, the upper rhombic lip (URL) progenitors (cluster 12) formed a separate lineage from all progenitor classes except cluster 74, a cycling progenitor subtype expressing pif1. Since URL progenitors give rise to granule cells in the cerebellum, we asked if the two cell types shared barcodes. We found that the proportion of barcode overlap was highest between granule cells and URL progenitors (Figure 5I). The URL progenitors formed a distinct cluster as early as 12 hpf (cluster 9) in our transcriptional dataset. Thus, URL progenitors become discrete in both lineage and transcriptional signature relatively early in development.
In summary, we present an optimized scGESTALT cassette with improved lineage barcode expression and recovery by scRNA-seq. The barcodes display high sequence diversity, which is important for generating large-scale distinct labels in a developing animal. The scGESTALT transgenic line is available as a resource for the community and can be paired with other transgenic lines for temporal, spatial or cell-type specific control of barcode editing (see Discussion).
Cell specification trajectories in the retina and hypothalamus
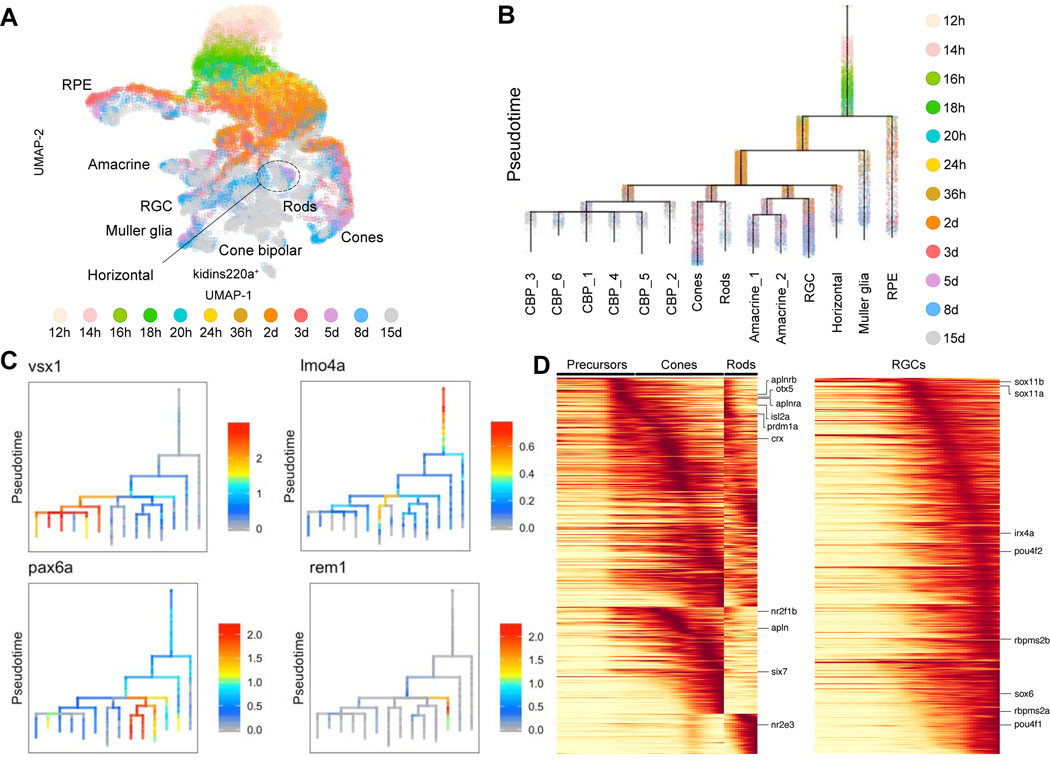
With the exception of a few model systems (Clark et al., 2019; Delile et al., 2019; Guo and Li, 2019; Holguera and Desplan, 2018; Kim et al., 2020; Tambalo et al., 2020), little is known about gene expression cascades that accompany the development of progenitors into terminally differentiated neurons. To address how different neuronal populations become molecularly specialized, we reconstructed gene expression trajectories from 12 hpf to 15 dpf. We first tested our approach on the subsetted retina dataset in which cell types expand from a single cluster at 12 hpf to 18 clusters at 15 dpf (Figure 2). UMAP embedding of the subsetted dataset revealed progressive paths from the embryonic state to defined cell types at 15 dpf (Figure 6A, Figure S5A). One outlier cluster that expressed kidins220a and whose progenitor state may not have been captured in our timepoints, was excluded from further analysis. Although UMAP represents continuity in the data, it does not order individual cells according to their relative developmental time (i.e. pseudotime). Therefore, we also used URD (Farrell et al., 2018) to construct a branching specification tree that represents the developmental trajectories in the retina at a higher resolution (Figure 6B, Figure S5B, Figure S6A-B). Many of the major branching features agreed with the UMAP representation. For example, the trajectories revealed the early segregation of RPE, shared branching of photoreceptor cells, a path towards multiple cone bipolar cell subtypes, and a common branchpoint between amacrine and retinal ganglion cells (RGC).
Figure 6. Cell specification trajectories in the hypothalamus.
A. UMAP visualization of retinal cell types. Retinal cells (based on clustering analysis) from 12 hpf to 15 dpf were subsetted from the full dataset and analyzed together. Cells are color coded by stage.
B. Cell specification tree of zebrafish retinal development. Trajectories were generated by URD and visualized as a branching tree. Cells are color coded by stage. 12 hpf cells were assigned as the root and 15 dpf differentiated cells were assigned as tips. CBP, cone bipolar cells (6 subtypes are numbered); RGC, retinal ganglion cells; RPE, retinal pigment epithelium
C. Expression of select genes are shown on the retina specification tree.
D. Heat maps of gene expression cascades of photoreceptor cell trajectories and retinal ganglion cell trajectories. Cells were selected based on high expression along trajectories leading to these cell types, compared to expression along opposing branchpoints. Red, high expression. Yellow, low expression
See also Figures S5 to S9
Plotting gene expression of known early regulators of eye development and terminal cell type markers on the URD tree supported the inferred specification branches (Figure 6C, Figure S7). For example, pax6a was most enriched in the amacrine and RGC branches, and vsx1 marked cone bipolar cells with fezf2 marking one specific subtype. Notably, our analysis also revealed previously unknown markers and characteristics of horizontal and amacrine cells. Zebrafish horizontal cells are GABAergic (gad2+, gad1b+), but unlike mammals where these cells do not express GABA membrane uptake transporters (Deniz et al., 2011), zebrafish cells expressed slc6a1l (likely a duplication of slc6a1 involved in GABA uptake from the synaptic cleft), suggesting that they may be capable of uptake. Additionally, whereas slc32a1 GABA transporter is expressed in mouse horizontal and amacrine cells (Cueva et al., 2002), we observed restriction of slc32a1 to amacrine cells and slc6a1l to horizontal cells. Finally, we detected several novel horizontal cell markers such as ompa and prkacaa (Figure 1F).
To discover the gene expression trajectories from precursors to different retinal cell types, we used differential gene expression approaches that characterize pseudotime-ordered molecular trajectories. This analysis revealed known and novel regulatory steps (Figure 6D, Figure S8). For example, RGC specification trajectories confirmed several known differentiation regulators including sox11a, sox11b, sox6, irx4a, and pou4f2 (Rheaume et al., 2018). Similarly, known regulators of photoreceptor differentiation such as isl2a (Fischer et al., 2011), prdm1a (Brzezinski et al., 2010), otx5 (Viczian et al., 2003), and crx (Shen and Raymond, 2004) were expressed early in our photoreceptor trajectories, while known regulators of cone versus rod fate, such as six7 (Ogawa et al., 2015), nr2f1b (Satoh et al., 2009), and nr2e3 (Chen et al., 2005) were expressed as those trajectories diverged. Furthermore, our analysis revealed novel transcription factors within the gene expression cascades. For example, we detected runx1t1, foxp1b, mef2aa in the RGC pathway; tfap2a in horizontal cell trajectory; and tbx3a and tbx2a in amacrine cell branches. Interestingly, among signaling pathways, we found that both apelin receptors (aplnra, aplnrb) were expressed in photoreceptor progenitors, while one of their ligands (apln) was expressed in differentiating cones; this suggests a potential cell autonomous role for apelin signaling in photoreceptor cells in addition to its role in preventing photoreceptor degeneration via vascular remodeling (McKenzie et al., 2012).
A surprising result from this analysis was that a Muller glia pathway was detected earlier in zebrafish than expected based on studies in mouse, where these cells are detected late (Centanin and Wittbrodt, 2014; Clark et al., 2019). We found a cluster of cells as early as 20 hpf (cluster 50) that expresses markers (e.g. cahz, rlbp1a) that are shared with the Muller glia cluster (cluster 33) at 15 dpf (Sup Table). smFISH analysis of Muller glia markers validated their expression at 36 hpf and 2 dpf (Figure S9). Similarly, in our transcriptional trajectories (Figure 6B), the Muller glia expression program is the earliest non-epithelial retinal program to diverge, commencing with the expression of several her-family transcription factors (her4, her12, and her15), then proceeding through a cascade of intermediate overlapping expression states such as onset of fabp7a, s100a10b, and later connexin genes that are characteristic of Muller glia fate (Figure S8). Cells from all timepoints can already be found in the early part of the Muller glia branch. These observations suggest that cells early in development transition from a naive progenitor state to a Muller glia-like transcriptional state, and do so continually during larval development.
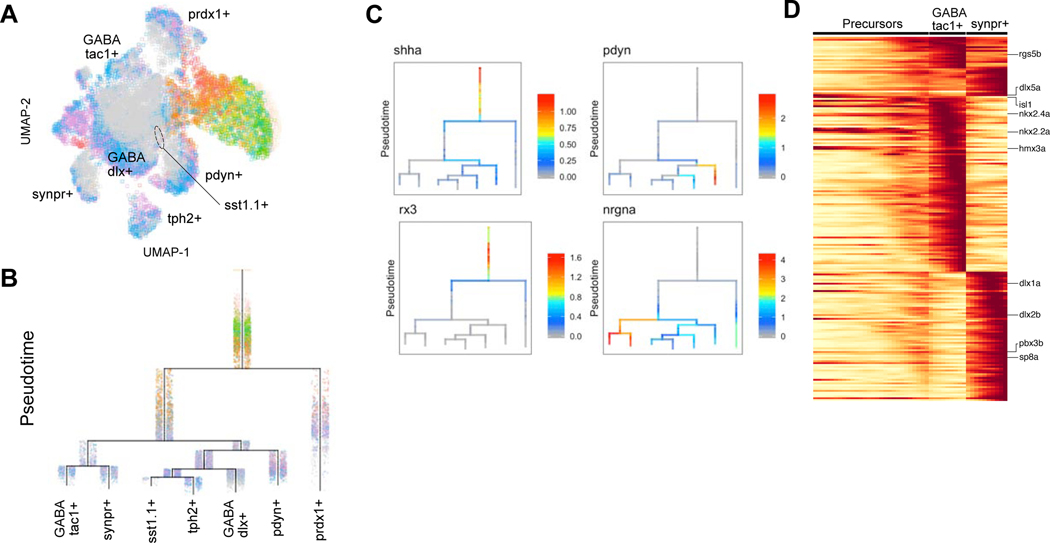
To extend our analysis to a central brain region, we reconstructed specification trajectories and expression cascades for hypothalamic neurons. These cells expanded from a single ventral diencephalon cluster at 12 hpf to 7 clusters at 15 dpf (Figure 7, Figure S6C-D, Figure S10). The earliest branchpoint denoted segregation of prdx1+ and prdx1- cells. Committed hypothalamic progenitors in the prdx1- trajectory gave rise to neuronal precursors expressing proneural transcription factors such as ascl1a, scrt2, insm1a and elavl3 (early neuron fate marker) (Figure S11). The specified cell types then matured over time and were characterized by expression of neuronal maturation markers such as tubb5, gap43, ywhag2, snap25a, scg2b and elavl4. The prdx1- group further diverged into two major groups: nrgna+ and nrgna- trajectories (Figure 7B, 7C). The nrgna+ branch segregated into GABAergic tac1+, synpr- subtype and GABAergic tac1+, synpr+ positive subtype (Figure 7D). The nrgna- branch subdivided into glutamatergic pdyn+ neurons and a GABAergic branch that further resolved to sst1.1+ and tph2+ neuron subtypes. We detected expression of known regulators of hypothalamus development in the early branches such as shha, rx3, nkx2.4b. We also identified new candidate regulators in later branches including nrgna in the synpr+ and synpr- trajectories, and sox1a, sox1b and sox14 in the pdyn+ trajectory (Figure S12, Figure 1E). The results in the retina and hypothalamus demonstrate that the brain development atlas can be used to reconstruct neuronal differentiation trajectories and define the underlying gene expression cascades
Figure 7. Cell specification trajectories in the hypothalamus.
A. UMAP visualization of hypothalamus cell types. Hypothalamus cells (based on clustering analysis) from 12 hpf to 15 dpf were subsetted from the full dataset and analyzed together. Cells are color coded by stage.
B. Cell specification tree of zebrafish hypothalamus development. Trajectories were generated by URD and visualized as a branching tree. Cells are color coded by stage. 12 hpf cells were assigned as the root and 15 dpf differentiated cells were assigned as tips.
C. Expression of select genes are shown on the hypothalamus specification tree.
D. Heat map of gene expression cascade of nrgna+ cell trajectories. Red, high expression. Yellow, low expression
See also Figures S6, S10 to S12
Differences in progenitor specification strategies between retina and hypothalamus
Pseudotime analysis represents cell trajectories in relative but not absolute time (Bendall et al., 2014; Trapnell et al., 2014). Therefore, comparing the developmental and pseudotime age of cells can define whether molecular states are unique to a given developmental stage or persist through development (Figure 6B, 7B). For example, mapping RGC and pdyn+ neurons from different developmental stages onto the pseudotime trajectory showed the expected maturation of these cell types with developmental age (Figure S13A, B). In addition, even at 15 dpf some RGC and pdyn+ neurons were still in an immature state, consistent with the continuous growth and differentiation in the zebrafish retina and brain (Centanin and Wittbrodt, 2014; Schmidt et al., 2013).
To systematically analyze the relationships of pseudotime state and developmental stage, we mapped differentiated cells, precursors and progenitors found in different pseudotime windows to their origin in developmental time. We found that the proportion of differentiated cells increased, whereas the number of early progenitors in both retina and hypothalamus decreased with developmental age. In contrast, precursor cells from an intermediate pseudotime window were present in embryo and larva. These precursor cells expressed genes that were an intermediate of progenitor [e.g. insm1a, her4.1 in hypothalamus (Xie and Dorsky, 2017); hes2.2, rx2 in retina] and early differentiation genes (e.g. tubb5, gap43 in hypothalamus; foxg1b in retina). In addition, a second class of retinal progenitors mapped to an earlier pseudotime trajectory but was also present from embryonic to late larval stages (Figure 8, Figure S13C). Comparison of these progenitors between 24–36 hpf and 15 dpf identified only 71 differentially expressed genes. The majority of these genes (56/71) increased in all cells of the retina between these stages, while a few (15/71) were only upregulated in the 15 dpf group. A similar population was not detected in the hypothalamus. These observations suggest that as the retina grows, some progenitor cell states observed in the embryo persist later in development without extensive maturation.
Figure 8. Progenitor differences between retina and hypothalamus.
Retinal and hypothalamus cells were divided into progenitor (purple), precursor (orange), and differentiated (blue) cells, as shown on the URD tree. The fraction of cells in each of these transcriptional states was then determined for three developmental periods (12–24 hpf, 36 hpf–3 dpf, and 5–15 dpf). In the retina, cells can be found in a progenitor state (light purple) that persists post-embryonically.
See also Figure S13
DISCUSSION
As the brain develops, embryonic neural progenitor pools transition through many cellular states as they become more committed, diversify into post-embryonic neural progenitors, and undergo terminal differentiation. Although regulators and transcriptional changes of this process have been identified (e.g. using specific driver lines and in situ detection of select genes), the global transcriptional networks mediating the sequential activation and maturation of neurogenic programs from embryo to later stages are largely unknown. To help address this question, we used scRNA-seq to generate a zebrafish brain development atlas. This resource supports the identification of marker genes, the comparison of cell types, and the dissection of cell specification and differentiation trajectories during vertebrate brain development.
Our data address how the transcriptional programs of neural progenitors vary and contribute to fate-restriction during development. Different models to explain these processes have been proposed. For example, neural progenitors of the medial and lateral mouse ganglionic eminence, which give rise to cortical interneurons, have been found to converge to a shared mitotic signature regardless of their region of origin, followed by expression of cardinal fate-specific transcription factors post-mitotically (Mayer et al., 2018). In contrast, the spinal cord has dedicated pools of domain-specific neural progenitors that retain domain-specific signatures (Delile et al., 2019; Jessell, 2000; Lee and Pfaff, 2001; Sagner and Briscoe, 2019). Our results indicate that early embryonic neural progenitors in the zebrafish brain are transcriptionally distinct from late larval neural progenitors. These cell state changes might reflect developmental shifts from an establishment program during gastrulation, where strong spatial patterning cues set up regional boundaries, to a maintenance program at late stages, where progenitors are geographically confined and express dampened regional restriction signatures. Although expression of some spatially-enriched transcription factors (e.g. pax6a, eng2a, nkx2.4a) and signaling proteins detected in embryonic progenitors are also detected in late progenitors, the overall signatures are different, as these factors co-vary with different sets of genes in larva relative to embryo.
The expression of pan-progenitor markers at larval stages raises the question of how neural progenitor pools remain or become fate restricted. There are several different scenarios that might address this question. First, it is conceivable that embryo and larva share a minimal core set of regionally-restricted transcription factors that are sufficient to ensure spatial restriction, despite differences in their relative expression levels and downstream targets. Spatial genes that are highly expressed in the embryo may be lowly expressed in the larva, and be sufficient to maintain regionally-restricted cell states. Second, cell-type specific transcription factors rather than spatially defined regulators might guide specification and differentiation at these stages, independent of positional information. Such signatures would be difficult to analyze via scRNA-seq, which is biased towards recovering highly expressed genes. Third, it is also possible that restrictions at the genomic level, such as chromatin accessibility, may ensure that cells maintain the signature of their spatial origin. Fate mapping experiments of early and late neural progenitors, profiling open chromatin states of neural progenitors, and transcriptome analyses that recover lowly expressed genes will provide further insight into these questions.
Our reconstruction of specification trajectories for cell types in the retina and hypothalamus revealed several findings. First, our data supports a multipotent progenitor model whereby multiple differentiated cell types can be traced to common post-embryonic progenitors. For example, all retinal neurons can be traced to an early pseudotime progenitor branch containing cells from larval stages, consistent with multipotency and fate stochasticity of zebrafish retinal progenitors (Boije et al., 2015; He et al., 2012). The early emergence of Muller glia observed in both the time course atlas and eye trajectory reconstruction is particularly interesting in light of clonal analyses. For example, single retinal progenitor cells in zebrafish give rise to clones comprised of neurons and one Muller glia cell (Rulands et al., 2018). This observation has been interpreted as evidence for a progenitor that first gives rise to neurons and then differentiates into a Muller glia cell. It is also conceivable based on our data that an early progenitor has Muller glia-like properties and divides, with one daughter expanding to give rise retinal neurons while the other daughter forms Muller glia. Second, our results reveal that whereas progenitor cell types in the rest of the brain appear molecularly distinct between the embryo and larva, there are progenitor cell states in the eye that are maintained from the embryo to larva (Figure 4 and Figure 7). A subset of 15 dpf retinal progenitors have similar transcriptional states as observed in the embryonic eye. This observation raises the possibility that a subset of long-term retinal progenitors may be “frozen” in an embryonic phase that could possibly underlie the multi-fate potential of these cells. An independent study of zebrafish retinal stem cells has proposed a similar conclusion (Xu et al., 2020). Collectively, these findings highlight differences in neurogenic programs in the central nervous system, and underscore the power of investigating multiple specification trajectories simultaneously.
Our results also highlight differences between zebrafish and mammalian neurogenesis. For example, we detected pan-neuronal transcriptional signatures (e.g. neurod1, ascl1a, insm1a, neurog1) in zebrafish radial glia and other progenitors at late stages of development, suggesting that neurons remain the principal output of these cells. This is consistent with fate mapping studies that have shown that zebrafish radial glia persist into adulthood and contribute to neurogenesis (Schmidt et al., 2013). In contrast, radial glia progenitor cells in the developing embryonic mouse brain shift from neurogenic to gliogenic programs (Mission et al., 1991; Schmechel and Rakic, 1979).
While developmental atlases and trajectories can help identify cellular differentiation paths, a full understanding of cell type specification requires lineage tracing experiments. To catalyze such approaches we introduced improvements to scGESTALT through a redesigned recorder cassette for optimized mRNA expression and library compatibility with the 10X Chromium scRNA-seq platform. The resulting higher recovery of barcodes allows more dense reconstruction of lineage trees. Our analysis revealed differences between the timing of segregation between different brain regions: neuronal lineages in the retina and hindbrain diverged earlier than the forebrain and midbrain. These results complement classic zebrafish fate maps of brain compartmentalization (Woo and Fraser, 1995) and recent analysis of clonal cells in forebrain and midbrain (Solek et al., 2017). Furthermore, our findings support early transcriptional and lineage segregation of cerebellar upper rhombic lip progenitors relative to other classes of progenitor cells. To query additional lineage divergences and combine with cellular trajectories, our optimized recorder can be readily adapted for barcoding lineages at developmental windows that correspond to different branches of the specification trees (Raj et al., 2018b) or combined with cell- or tissue-specific Cas9 driver lines to introduce lineage labels in populations of interest.
The resources presented here lay the groundwork for characterizing lineage histories and transcriptional changes underlying the development and diversification of the vertebrate brain. Future extensions include the generation of transgenic reporters to select populations of interest and perform deeper analyses of cell type heterogeneity and differentiation (Pandey et al., 2018). Cell specification trajectories can be extended to include additional subregions of the brain to generate increasingly complex trees and combined with other zebrafish scRNA-seq datasets (Cosacak et al., 2019; Farnsworth et al., 2020; Farrell et al., 2018; Lange et al., 2020; Pandey et al., 2018; Tambalo et al., 2020; Wagner et al., 2018; Xu et al., 2020) to trace complete trajectories from gastrulation to adulthood. Finally, it will be interesting to perform comparative studies by using our atlas in conjunction with datasets in other vertebrates (Hashikawa et al., 2020; La Manno et al., 2020; Tosches et al., 2018; Yan et al., 2020).
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bushra Raj (bushranraj@gmail.com).
Materials Availability
Zebrafish scGESTALT.2 transgenic line generated in this study is available via request from the Lead Contact or Alexander F. Schier. All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
The datasets generated during this study are available at GEO [accession code]. The processed data is available as a resource to explore at: https://github.com/brlauuu/zf_brain. The code used in this study is available at: https://github.com/brlauuu/zf_brain and Methods S1.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish husbandry
All zebrafish work was performed at the facilities of Harvard University, Faculty of Arts & Sciences (HU/FAS). This study was approved by the Harvard University/Faculty of Arts & Sciences Standing Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08. The HU/FAS animal care and use program maintains full AAALAC accreditation, is assured with OLAW (A3593–01), and is currently registered with the USDA. The stages profiled in this study correspond to 12 hpf, 14 hpf, 16 hpf, 18 hpf, 20 hpf, 24 hpf, 36 hpf, 2 dpf, 3 dpf, 5 dpf, 8 dpf and 15 dpf. At the developmental stages profiled in this study, the sex of the organism is not yet determined.
METHOD DETAILS
Chromogenic in situ hybridization
Embryos were dechorionated with forceps and then fixed in 4% PFA in 1X PBS (pH 7.4) overnight at 4°C. After fixation, embryos were dehydrated in methanol series (0%, 25%, 50%, 75% and 100% MetOH in PBSTween 0.3% (PBST)) and stored in 100% methanol at −20°C. Embryos were rehydrated by reversing the methanol series for 10 min in each step at room temperature (RT) and washed 2 × 5 min in PBST. To bleach pigment in 2 dpf fish, larvae were incubated for 10 min in bleaching solution (3% H2O2/0.5% KOH in ddH2O) at room temperature and washed 3 × 5 min in PBST (Thisse et al., 2004). For permeabilization, 2 dpf larvae were incubated with Proteinase K (10 μg/ml in PBST) for 2 min at RT and postfixed in 4% PFA in 1X PBS for 30 min at RT. Afterwards, embryos were washed 3 × 5 min in PBST at RT, prehybridized in HYB+ solution (50% Deionized Formamide (Amresco), 5X SSC (Ambion), 0.1% Tween-20, 5mg/ml Torula RNA (Sigma) in ddH2O) for 3 hours at 69°C, and hybridized overnight with the antisense probes diluted in HYB+ at 69°C. The rest of the steps were performed as described previously, by hand (Navajas Acedo et al., 2019). Before imaging, embryos were cleared using an increasing MetOH series. For imaging of 12 hpf embryos, the yolk was dissected away, and the embryos were flat mounted on a microscope slide and covered with a cover slip. Larvae were photographed on a Zeiss AxioZoom.V16.
The antisense probes were synthetized from DNA fragments amplified from TLAB zebrafish cDNA using the following primers: klf17 (Fw GAAGGAAAGACTGCATCCTGAC; Rv CTGCTGTCCCAAAATAGGAGTT), ptgs2a (Fw CGAGGACTATGTTCAGCACTTG; Rv TGCACATCGATCACAATACAAA), tp63 (Fw TGCTTTGCTAAATTGTGCTGTC; Rv ATTGCCGCTTATGAGAATCAAG), cavin2a (Fw GAGCCTTCTCGTGCTAACAAGT; Rv CAGGCATTTCAGTTCAATTTCA), sox1a (Fw AATCAAGACCGCGTAAAGAGAC; Rv TTTGGTGGAGTGTTTCTGAATG), pdyn (Fw AAGAGAACGCCATACTGAAAGG; Rv GCAGTTACGAATTGCCATGATA), dlx1a (Fw AAGGAGGAGAGGTTCGTTTCA; Rv AGTGTGTGTCAGCAGGTGTCTT).
smFISH staining and imaging
Zebrafish larvae were fixed with 4% PFA (Sigma-Aldrich, P6148) in PBS overnight at 4°C. They were then washed twice with PBS and subsequently incubated in 30% sucrose (Sigma-Aldrich, 84100) in PBS overnight at 4°C and then embedded in O.C.T. (Sakura, 4583) and stored at −80°C. 20 μm sections were cut with a cryostat (ThermoFisher, 957000) and captured onto polylysine (Sigma, P8920) -coated #1.5 coverslips.
Single-molecule FISH probe sets were generated as previously described and coupled to either Atto 647N NHS ester (Millipore Sigma #18373) (foxg1b, cahz) or Atto 550 NHS ester (Millipore Sigma # 92835) (ompa, rlbp1a) (Lord et al., 2019). Sectioned larvae were affixed to polylysine-coated #1.5 coverslips, and staining was carried out as previously described (Lord et al., 2019), with each coverslip contained in a well of a plastic 6-well plate. During the probe hybridization step, coverslips were placed upside-down onto a 100μl droplet of probe solution on Parafilm (Farack and Itzkovitz, 2020). Sample mounting was performed as previously described (Lord et al., 2019). Mounted samples were imaged on an Olympus spinSR spinning disk microscope fitted with a UPLAPO 60X/1.5 oil immersion objective using 0.3μm slices.
smFISH image processing
All image processing was performed in Fiji (Schindelin et al., 2012). Rolling-ball background subtraction (radius 25 pixels) was performed on smFISH channels before maximum intensity projections were produced from 30 slices (Figure 1F) or 50 slices (Sup Figure 9) of processed z-stacks. Channels were scaled individually, maximizing for visibility.
Optimization of scGESTALT lineage cassette
In our previous iteration of scGESTALT, the barcode capture rate by scRNA-seq was 6–28%. (Raj et al., 2018b), thereby limiting the density of lineage tree reconstruction. To improve recovery we adapted a different transgenic cassette (Yoshinari et al., 2012) for lineage recording. This cassette has the following modifications compared to our previous recorder: (1) The heat-shock inducible (hsp70l) promoter of the previous version is now replaced with a constitutive ubiquitous promoter (medaka beta-actin) to drive strong widespread expression of the barcode mRNA. Expression of the cassette was confirmed by fluorescence and the signal was more intense than that obtained with the heat shock promoter. Furthermore, this version eliminates the requirement to heat shock edited animals to express the barcode prior to scRNA-seq experiments. (2) We adapted the 3’ end of the DsRed open reading frame as a lineage recorder cassette with up to 8 sgRNA target sites positioned next to each other. This vastly improved expression of the construct compared to our previous version where the recording cassette was placed downstream of the DsRed open reading frame. (3) We made library preparation compatible with the 10X Genomics platform.
To generate scGESTALT.2 barcode founder fish, one-cell embryos were injected with zebrafish codon optimized Tol2 mRNA and pT2Olactb:loxP-dsR2-loxP-EGFP vector (gift from Atsushi Kawakami (Yoshinari et al., 2012) ). Potential founder fish were screened for widespread DsRed expression and grown to adulthood. Adult founder transgenic fish were identified by outcrossing to wild type fish and screening clutches of embryos for ubiquitous DsRed expression. Single copy scGESTALT.2 F1 transgenics were identified using qPCR, as described previously (McKenna et al., 2016; Pan et al., 2013; Raj et al., 2018b).
SgRNAs specific to sites 1–8 of the scGESTALT.2 array were generated by in vitro transcription using EnGen sgRNA Synthesis Kit as previously described (Raj et al., 2018a). The following oligonucleotides were used for sgRNA synthesis:
dsR-1 TTCTAATACGACTCACTATAGGCGTGAACTTCCCCTCCGAGTTTTAGAGCTAGA,
dsR-2 TTCTAATACGACTCACTATAGTGATGCAGAAGAAGACCATGTTTTAGAGCTAGA,
dsR-3 TTCTAATACGACTCACTATAGAGCGCCTGTACCCCCGCGAGTTTTAGAGCTAGA,
dsR-4 TTCTAATACGACTCACTATAGCCCTGAAGCTGAAGGACGGGTTTTAGAGCTAGA,
dsR-5 TTCTAATACGACTCACTATAGCTGCCCGGCTACTACTACGGTTTTAGAGCTAGA,
dsR-6 TTCTAATACGACTCACTATAGGACATCACCTCCCACAACGGTTTTAGAGCTAGA,
dsR-7 TTCTAATACGACTCACTATAGGAGCAGTACGAGCGCACCGGTTTTAGAGCTAGA,
dsR-lowX TTCTAATACGACTCACTATAGGAGTTCAAGTCCATCTACAGTTTTAGAGCTAGA
To initiate early barcode editing, single copy scGESTALT.2 F1 male transgenic adults were crossed to wildtype female adults and one-cell embryos were injected with 1.5 nl of Cas9 protein (NEB) and sgRNAs 1–8 in salt solution (8 μM Cas9, 100 ng/μl pooled sgRNAs, 50 mM KCl, 3 mM MgCl2, 5 mM Tris HCl pH 8.0, 0.05% phenol red). Since editing results in loss of DsRed signal, transgenic animals were distinguished from wild type animals by amplifying the scGESTALT.2 barcode by PCR using genomic DNA from the tail fin at 15 dpf. In the experiments presented in this study, early lineage decisions were barcoded by injecting reagents at the one-cell stage. It is worth noting that the scGESTALT.2 barcode can be readily paired with a two-step barcoding protocol. This would require the establishment of a second stable transgenic line for in vivo expression of Cas9 and a subset of sgRNAs matching the target sequences of the new barcode cassette to enable sequential barcoding at early and late stages. Such a line can be established using a similar step-by-step guidance that has been detailed previously (Raj et al., 2018a).
Processing of samples for scRNA-seq time course
Wild type embryos (12 hpf, 14 hpf, 16 hpf, 18 hpf, 20 hpf, 24 hpf, 36 hpf) and larvae (2 dpf, 3 dpf, 5 dpf, 8 dpf) were used for scRNA-seq analysis. Samples for 15 dpf had a mix of wild type and barcode edited larvae. Two of the 15 dpf samples consisted of only eye cells (no brain). Embryos from 12 hpf to 36 hpf were first de-chorionated by incubating in 1 mg/ml pronase (Sigma-Aldrich) at 28 C for 6–7 min until chorions began to blister, and then washed three times in ~200 ml of zebrafish embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 0.1% methylene blue) in a glass beaker. Embryos were de-yolked using two pairs of watchmaker forceps, and the heads were chopped just anterior of the spinal cord. All processing steps were done using 100 mm Petri dishes coated with Sylgard. (Raj et al., 2018a). Samples from 2 and 3 dpf were processed similarly to the embryos, except they were not de-chorionated as they had hatched out of the chorions. Larvae from 5 dpf to 15 dpf were dissected to remove whole brains and eyes as described previously (Raj et al., 2018a). The following numbers of embryos and larvae were used for each timepoint: 12 hpf – ~20 embryos; 14 hpf – ~20 embryos; 16 hpf – ~18 embryos; 18 hpf – ~18 embryos; 20 hpf – ~30 embryos; 24 hpf – ~30 embryos; 36 hpf – ~15 embryos; 2 dpf – ~30 larvae; 3 dpf – ~30 larvae; 5 dpf – ~25 larvae; 8 dpf – ~ 25 larvae; 15 dpf – ~15 larvae. Tissues were dissociated into single cells using the Papain Dissociation Kit (Worthington) as described previously (Raj et al., 2018a). Cells were resuspended in 50 μl to 150 μl of DPBS (Life Technologies) depending on anticipated amount of material, and counted using a hemocytometer. Samples were run on the 10X Genomics scRNA-seq platform according to the manufacturer’s instructions (Single Cell 3’ v2 kit). Libraries were processed according to the manufacturer’s instructions. Transcriptome libraries were sequenced using NextSeq 75 cycle kits.
scGESTALT.2 library prep
To generate scGESTALT.2 libraries, lineage edited 15 dpf samples post cDNA amplification and prior to fragmentation were split into two halves. One half was processed for transcriptome libraries as instructed by the manufacturer. The other half was processed for lineage libraries as follows. To enrich for scGESTALT.2 lineage barcodes, 5 μl of the whole transcriptome cDNA was PCR amplified using Phusion polymerase (NEB) and 10XPCR1_F (CTACACGACGCTCTT CCGATCT) and GP10X2_R (GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT GCTGCTTC ATCTACAAGGTGAAG). The reaction (98 C, 30 s; [98 C, 10 s; 67 C, 25 s; 72 C, 30 s] x 14–15 cycles; 72 C, 2 min) was cleaned up with 0.6X AMPure beads and eluted in 20 ul EB buffer (Omega). Finally, adapters and sample indexes were incorporated in another PCR reaction using Phusion polymerase and 10XP5Part1long (AATGATACGGCGACCACCGA GATCTACACTCTTTCC CTACACGACGCTCTTCCGATCT) and 10XP7Part2Ax (CAAGCAGAAGACGGCATACGAGAT-xxxxxxxx-GTGACTGGAGTTCAGACGTGT), where x represents index bases. These include A1: GGTTTACT; A2: TTTCATGA; A3: CAGTACTG; A4: TATGATTC. Thus, up to 4 scGESTALT.2 samples were multiplexed in a sequencing run. Libraries were sequenced using MiSeq 300 cycle kits and 20% PhiX spike-in. Sequencing parameters: Read1 250 cycles, Read2 14 cycles, Index1 8 cycles, Index2 8 cycles. Standard sequencing primers were used.
Bioinformatic processing of raw sequencing data and cell type clustering analysis
Transcriptome sequencing data were processed using Cell Ranger 2.1.0 according to the manufacturer’s guidelines. scGESTALT.2 sequencing data were processed with a custom pipeline (https://github.com/aaronmck/SC_GESTALT) as previously described (Raj et al., 2018b). The scGESTALT.2 barcode for each cell was matched to its corresponding cell type (tSNE cluster membership) assignment using the cell identifier introduced during transcriptome capture. Cells with fewer than 500 expressed genes, greater than 9% mitochondrial content or very high numbers of UMIs and gene counts that were outliers of a normal distribution (likely doublets/multiplets) were removed from further analysis. Clustering analysis was performed using the Seurat v2.3.4 package (Butler et al., 2018) as described previously (Raj et al., 2018b). For Figure 3 and Sup Figure 2, we selected the list of transcription factors, neuropeptides and their receptors, and genes involved in neuron electrophysiology from our enriched marker analysis and previous literature (Chen et al., 2017; Pandey et al., 2018; Tiklová et al., 2019; Zeisel et al., 2018).
Construction of lineage trees from GESTALT barcodes
All unique barcodes were then encoded into an event matrix and weights file, as described previously (McKenna et al., 2016; Raj et al., 2018b), and were processed using PHYLIP mix with Camin-Sokal maximum parsimony (Felsenstein, 1989). Individual cells were then grafted onto the leaves matching their barcode sequence. After the subtrees were attached, we repeatedly eliminated unsupported internal branching by recursively pruning parent-child nodes that had identical barcodes. Cell annotations are then added to the corresponding leaves. The resulting tree was converted to a JSON object, annotated with cluster membership, and visualized with custom tools using the D3 software framework.
QUANTIFICATION AND STATISTICAL ANALYSIS
Packages used
Seurat v2.3.4, URD 1.1.1, NMF, Destiny, R Stats
Lineage segregation analysis between cell types
We combined all barcodes obtained from 4 fish. For our analysis, we only considered barcodes with at least two cells, and we only analyzed cell types with at least 4 barcodes. To test segregation between any two cell types/clusters, we first retrieved all barcodes that were present in at least one of the two cell types. Then, we split these barcodes into two categories: “shared barcode” or “specific barcode”. A shared barcode was defined as one that contains cells from both cell types. In contrast, a specific barcode was defined as one that only contains cells from one of the two cell types. Our null hypothesis is that the two cell types come from the same ancestor at the time of Cas9 editing. Thus, we asked whether the number of observed specific barcodes can be explained by chance under the null hypothesis. If it cannot be explained by chance, it indicates that the two cell types have segregated.
To do so, we performed a randomization test as below:
We generated a pool of cells. The size of the pool is the total number of cells from the two cell types. The ratio of the two cell types in the pool is equal to the ratio observed in the real data. Under the null hypothesis, the pool of cells come from the same ancestor, so they would share the same barcode.
For each barcode, we randomly sampled the same number of cells of this barcode from the pool of cells.
We repeated this for all the barcodes, and then calculated the number of barcodes that only contain one cell type (i.e. “specific barcode”).
We repeated steps 2 and 3 5000 times.
We calculated how many times (for example n times) the number of specific barcodes from the random sampling process is greater than or equal to the number of specific barcodes from the real data.
The probability that the number of specific barcodes can be explained by chance under the null hypothesis is n/5000.
If the probability < 0.01 (pvalue < 0.01), we rejected the null hypothesis.
Next, for each cell type we split its corresponding pairwise comparison cell types into two categories: “with segregation” or “other”. For the “other” category, we considered two interpretations. First, it could signify that there is no segregation between the two cell types. Second, it could suggest that we did not recover enough cells with barcode information, such that there is not enough power to detect lineage segregation (low sampling). To distinguish between the two scenarios, for each cell type in the two categories, we calculated the ratio between the number of cells with barcodes and the number of all cells from scRNA-seq. If the ratio of one cell type from the “other” category is greater than or equal to the smallest ratio from the first category (“with segregation”), it indicates this cell type did not have low sampling issues. Thus, it supports the interpretation that there is no segregation between the queried cell types. Otherwise, we assign the cell type pair as “undefined” (i.e. insufficient sampling power to query lineage segregation).
Granule cell analysis
For each progenitor cell type, we used barcodes that did not include any cells from the other nine progenitor cell types. The Jaccard Index between each progenitor cell type and granule cell was calculated as below:
Analyzing dampened spatial correlations in progenitors
Progenitors were isolated by subsetting the data to include clusters expressing markers such as sox19a, her genes, pcna, mki67, fabp7a, gfap, id1, etc (Supplementary Table). Cells from 12 hpf – 18 hpf were considered embryonic progenitors, cells from 20 hpf – 3 dpf were considered intermediate progenitors, and cells from 5 dpf – 15 dpf were considered larval progenitors. Variable genes were calculated for embryonic, intermediate and larval progenitors separately using the FindVariableGenes function from Seurat v2.3.4 with parameters: x.low.cutoff = 0.015, x.high.cutoff = 3, y.cutoff = 0.7. Then, a list of 79 transcription factors with known spatial signatures was assembled by consulting previously described histological analysis (ZFIN) together with those that were identified as gene markers for neuronal clusters in our dataset.. Separately in the three progenitor groups, the pairwise Pearson correlation was calculated pairwise between all genes detected as variable in each progenitor group. For several thresholds between 0.2–0.8, the number of genes that correlated more strongly than the threshold with any of the 79 spatial transcription factors (excluding self-correlation) were determined. The strongest correlations were observed in the embryonic population, followed by the intermediate population, and for any threshold, more genes correlated with the spatial TFs in the embryonic progenitors than the larval progenitors.
Construction and analysis of branching transcriptional trajectories using URD
We built branching transcriptional trajectories from cells of the retina and hypothalamus to determine the molecular events that occur as cells diversify and differentiate in these tissues. First, cells from the retina and hypothalamus were isolated from each stage by determining clusters that belonged to these tissues by expression of marker genes.
Determination of variable genes
For URD trajectory analyses, a more restrictive set of variable genes was calculated on each subset of the data, as previously described (Farrell et al., 2018; Pandey et al., 2018) using the URD findVariableGenes function, with parameter diffCV.cutoff = 0.3. Briefly, a curve was fit that related each gene’s coefficient of variation to its mean expression level and represents the expected coefficient of variation resulting from technical noise, given a gene’s mean expression value; genes with much higher coefficients of variation likely encode biological variability and were used downstream.
Removal of outliers
Poorly connected outliers can disrupt diffusion map calculation and so were removed from the data. A k-nearest neighbor network was calculated between cells (Euclidean distance in variable genes) with 100 nearest neighbors. Cells were then removed based on either unusually high distance to their nearest neighbor or unusually high distance to their 20th nearest neighbor, given their distance to their nearest neighbor using the URD function knnOutliers (retina: x.max = 40, slope.r = 1.05, int.r = 4.3, slope.b = 0.75, int.b = 11.5; hypothalamus: x.max = 40, slope.r = 1.1, int.r = 3, slope.b = 0.66, int.b = 11.5).
Removal of doublets by NMF modules
To remove putative cell doublets (i.e. where two cells are encapsulated into a single droplet and processed as one cell), which can disrupt trajectory relationships, we removed cells that expressed multiple NMF (non-negative matrix factorization) modules characteristic of different expression programs, as previously described (Siebert et al., 2019). NMF modules were computed using a previously published NMF framework (https://github.com/YiqunW/NMF) (Farrell et al., 2018). The analysis was performed on log-normalized read count data for a set of variable genes using the run_nmf.py script with the following parameters: -rep 5 -scl “false” - miter 10000 -perm True -run_perm True -tol 1e-6 -a 2 -init “random” -analyze True. Several k parameters were evaluated for each tissue, and k was chosen to maximize the number of modules, while minimizing the proportion of modules defined primarily by a single gene (retina, k = 45; hypothalamus, k = ). Modules were used downstream that (a) had a ratio between their top-weighted and second-highest weighted gene of < 5, and (b) exhibited a strong cell-type signature, as determined by plotting on a UMAP representation and looking for spatial restriction. Pairs of modules that were appropriate for using to remove doublets (and that did not define transition states) were determined using the URD function NMFDoubletsDefineModules with parameters module.thresh.high = 0.4, and module.thresh.low = 0.15. Putative doublets were identified using the URD function NMFDoubletsDetermineCells with parameters frac.overlap.max = 0.03, frac.overlap.diff.max = 0.1, module.expressed.thresh = 0.33 and were then removed.
Choice of root and tips
Branching transcriptional trajectories in the retina and hypothalamus were constructed using URD 1.1.1 (Farrell 2018). Briefly, cells from the first stage of the time course (12 hpf) were selected as the ‘root’ or starting point for the tree. Terminal cell types comprised the clusters at 15 dpf from these tissues, with the exception of clusters that were clearly progenitor or precursors based on known gene expression (retina: 29, 39, 43). Additionally, in the retina, one cluster (96) was excluded because it did not seem that any related cell types had been recovered in previous stages.
Construction of branching transcriptional trajectories
A diffusion map was calculated using destiny (Haghverdi et al., 2015; 2016), using 140 (retina) or 100 (hypothalamus) nearest neighbors (approximately the square root of the number of cells in the data), and with a globally-defined sigma of 14 (retina) or 8 (hypothalamus) — slightly smaller than the suggested sigma from destiny. Pseudotime was then computed using the simulated ‘flood’ procedure previously described (Farrell et al., 2018), using the following parameters: n = 100, minimum.cells.flooded = 2. Biased random walks were performed to determine the cells visited from each terminal population in the data as previously described (Farrell et al., 2018), using the following parameters: optimal.cells.forward = 40, max.cells.back = 80, n.per.tip = 50000, end.visits = 1. The branching tree was then constructed using URD’s buildTree function with the following parameters: divergence.method = “ks” (hypothalamus) or divergence.method = “preference” (retina), save.all.breakpoint.info = TRUE, cells.per.pseudotime.bin = 40, bins.per.pseudotime.window = 5, p.thresh = 0.0001 (hypothalamus) or , p.thresh = 0.01 (retina), and min.cells.per.segment = 10. The resulting trees were then evaluated using known marker genes and branch regulators.
Finding genes that vary during differentiation
Genes were selected for inclusion in gene cascades based on their differential expression relative to other cell types in the tissue. See the Supplementary Analysis for the full set of commands used. Within each tissue, cells were first compared in large populations that defined major cell types (retina: cone bipolar cells, photoreceptors, amacrine cells, retinal ganglion cells, horizontal cells, Muller glia, retinal pigmented epithelium; hypothalamus: prdx1+ neurons, pdyn+ neurons, GABAergic dlx+ neurons, nrgna+ neurons). Comparisons were performed pairwise, and genes were considered differential in a population if they were upregulated compared to at least 2 (hypothalamus) or 3 (retina) other groups. Genes were considered differentially expressed based on their expression fold-change (retina: ≥1.32-fold change, hypothalamus: ≥ 1.41-fold change) and their performance as a precision-recall classifier for the two cell populations compared (≥ 1.1-fold better than a random classifier). Additionally, the aucprTestAlongTree function from URD was used to select additional genes by performing pairwise comparisons, starting from a terminal cell type and comparing at each branchpoint along the way, back to the root (Farrell et al., 2018). Genes were selected based on expression fold-change between branchpoints (hypothalamus: ≥1.74-fold upregulated; hypothalamus, populations with small cell numbers (GABAergic dlx+ cells): ≥1.51-fold upregulated; retina: ≥1.32-fold upregulated), their function as a precision-recall classifier between branchpoints (hypothalamus: ≥1.2-fold better than a random classifier; hypothalamus, populations with small cell numbers (GABAergic dlx+ cells): ≥1.15-fold better than a random classifier; retina: ≥1.1-fold better than a random classifier), their function as a precision recall classifier globally (i.e. between the entire trajectory leading to a cell type and the rest of the tissue): ≥1.03-fold better than a random classifier, and their upregulation globally (i.e. between the entire trajectory leading to a cell type and the rest of the tissue): ≥1.07-fold upregulated. Mitochondrial, ribosomal, and tandem duplicated genes were excluded. Cells were ordered according to pseudotime, split into groups of at least 25 cells that differ at least 0.005 in pseudotime, and the mean expression was determined with a 5-group moving window. A spline curve was fit to the mean expression vs. pseudotime relationship of selected genes, using the smooth.spline function from R’s stats package, with the parameter spar = 0.5. Genes were then sorted according to their peak expression in pseudotime, normalized to their max expression observed in the tissue, and plotted on a heatmap.
Analyzing progenitor populations
To determine whether retinal progenitors mature transcriptionally over time, we looked for genes that were differentially expressed between young and old progenitors. We chose cells that occupied the same region of the URD tree from either early (24 / 36 hpf) or late (15 dpf) stages. We looked for genes that were differentially expressed in 15 dpf progenitors that: (1) were 1.1-fold better as a precision-recall classifier than random, (2) changed ≥1.32-fold in expression, (3) were expressed in at least 20% of progenitors, (4) had a mean expression value ≥ 0.8, and (5) were more differentially expressed than equally sized cell populations chosen at random at least 99% of the time.
To determine whether cells were found in progenitor or precursor states long-term, we first defined progenitor and precursor states by cells’ assignment in the URD tree, cross-referenced with the expression of progenitor / precursor markers. We then determined how many cells from different stages fell into each of these different states.
Supplementary Material
Code used to perform retina and hypothalamus cell trajectory analysis using URD
Master table describing all identified clusters from 12 hpf to 15 dpf. Columns are as follows: Stage – developmental stage profiled. Cluster – corresponding cluster number. Match N-1 – cluster from prior stage that is the most similar to the current cluster. For example Row 45 (14 hpf cluster 0) is similar to 12 hpf cluster 0. Assigned Cell type/State – cell type annotation. Enriched markers – subset of enriched markers are shown for each cluster.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| PFA | Sigma-Aldrich | Cat#158127, Cat#P6148 |
| Hydrogen peroxide (H2O2) | Sigma-Aldrich | Cat#H1009 |
| KOH | Sigma-Aldrich | Cat#P5958 |
| Formamide, deionized | Amresco | Cat#4170848 |
| Proteinase K | Sigma-Aldrich | Cat#3115836001 |
| 30% sucrose | Sigma-Aldrich | Cat#84100 |
| O.C.T. | Sakura | Cat#4583 |
| poly-lysine | Sigma-Aldrich | Cat#P8920 |
| Atto 647N NHS ester | Millipore Sigma | Cat#18373 |
| Atto 550 NHS ester | Millipore Sigma | Cat#92835 |
| Papain Dissociation System | Worthington | Cat#LK003150 |
| Torula RNA | Sigma-Aldrich | Cat#83850 |
| 20X SSC | Invitrogen/Ambion | Cat#AM9770 |
| EnGen Cas9 NLS, S. pyogenes | NEB | Cat#M0646 |
| Critical Commercial Assays | ||
| 10X Reagents | 10X Genomics | Chromium Single Cell 3′v2 Reagent Kits |
| EnGen sgRNA Synthesis Kit, S. pyogenes |
NEB | Cat#E3322 |
| Deposited Data | ||
| Raw data files for scRNA-Seq | NCBI GEO | TBD |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Zebrafish scGESTALT.2 | This paper | N/A |
| Oligonucleotides | ||
| Recombinant DNA | ||
| pT2Olactb:loxP-dsR2-loxP-EGFP | gift from Atsushi Kawakami (Yoshinari et al., 2012) |
N/A |
| Software and Algorithms | ||
| Cell Ranger | 10X Genomics | https://support.10xgenomics.com/single-cell-geneexpression/software/pipelines/latest/installation |
| URD | Farrell et al., 2018 | https://github.com/farrellja/URD |
| NMF modules | Farrell et al., 2018 | https://github.com/YiqunW/NMF |
| Seurat v2.3.4 | Butler et al., 2018 | https://satijalab.org/seurat/ |
| R, RStudio | RStudio | https://rstudio.com |
| Other | ||
| scRNA-seq resource | This paper | https://github.com/brlauuu/zf_brain |
| scGESTALT lineage resource | This paper | https://scgestalt.mckennalab.org/ |
Highlights.
Developmental atlas defines zebrafish brain cell types from embryo to juvenile
Optimized scGESTALT CRISPR-Cas9 lineage recorder enables higher barcode capture
Comparison of early versus late neural progenitors identifies different landscapes
ACKNOWLEDGEMENTS
We thank members of the Schier lab for discussion and advice, the Bauer Core Facility (Harvard) and the Molecular Biology Core Facility (Dana Farber Cancer Institute) for sequencing services, the Harvard zebrafish facility staff for technical support, and the Imaging Core Facility of the Biozentrum for microscopy resources. We thank M. Shafer and J. Gagnon for comments on the manuscript, and H. Boije for comments on retinal lineages. This work was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research and 1K99HD098298 to B.R., 1K99HD091291 to J.A.F., R00HG010152 to A.M., R01HD85905, DP1HD094764, ERC 834788, an Allen Discovery Center grant, and a McKnight Foundation Technological Innovations in Neuroscience Award to A.F.S.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allende ML, Weinberg ES, 1994. The expression pattern of two zebrafish achaete-scute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev. Biol. 166, 509–530. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D, 2014. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boije H, Rulands S, Dudczig S, Simons BD, Harris WA, 2015. The independent probabilistic firing of transcription factors: a paradigm for clonal variability in the zebrafish retina. Developmental Cell 34, 532–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Lamba DA, Reh TA, 2010. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RA, Bihannic L, Rosencrance C, Hadley JL, Tong Y, Phoenix TN, Natarajan S, Easton J, Northcott PA, Gawad C, 2018. A single-cell transcriptional atlas of the developing murine cerebellum. Current Biology 28, 2910–2920.e2. [DOI] [PubMed] [Google Scholar]
- Centanin L, Wittbrodt J, 2014. Retinal neurogenesis. Development 141, 241–244. [DOI] [PubMed] [Google Scholar]
- Cepko C, 2014. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 15, 615–627. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J, 2005. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 25, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Wu X, Jiang L, Zhang Y, 2017. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep 18, 3227–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, Gronostajski RM, Fertig EJ, Goff LA, Blackshaw S, 2019. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosacak MI, Bhattarai P, Reinhardt S, Petzold A, Dahl A, Zhang Y, Kizil C, 2019. Single-cell transcriptomics analyses of neural stem cell heterogeneity and contextual plasticity in a zebrafish brain model of amyloid toxicity. Cell Rep 27, 1307–1318.e3. [DOI] [PubMed] [Google Scholar]
- Cueva JG, Haverkamp S, Reimer RJ, Edwards R, Wässle H, Brecha NC, 2002. Vesicular gamma-aminobutyric acid transporter expression in amacrine and horizontal cells. J. Comp. Neurol. 445, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delile J, Rayon T, Melchionda M, Edwards A, Briscoe J, Sagner A, 2019. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 146, dev173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz S, Wersinger E, Schwab Y, Mura C, Erdelyi F, Szabó G, Rendon A, Sahel J-A, Picaud S, Roux MJ, 2011. Mammalian retinal horizontal cells are unconventional GABAergic neurons. J. Neurochem. 116, 350–362. [DOI] [PubMed] [Google Scholar]
- Farack L, Itzkovitz S, 2020. Protocol for single-molecule fluorescence in situ hybridization for intact pancreatic tissue. STAR Protocols 1, 100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth DR, Saunders LM, Miller AC, 2020. A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 459, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF, 2018. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, 1989. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics, Vol. 5 (1989), pp. 164–166 5, 164–166. [Google Scholar]
- Fischer AJ, Bongini R, Bastaki N, Sherwood P, 2011. The maturation of photoreceptors in the avian retina is stimulated by thyroid hormone. Neuroscience 178, 250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs HC, Chang-Gonzalez A, Hwang W, Yeh AT, Lekven AC, 2017. Midbrain-Hindbrain Boundary Morphogenesis: At the Intersection of Wnt and Fgf Signaling. Front. Neuroanat. 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Li JYH, 2019. Defining developmental diversification of diencephalon neurons through single cell gene expression profiling. Development 146, dev174284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Buettner F, Theis FJ, 2015. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31, 2989–2998. [DOI] [PubMed] [Google Scholar]
- Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ, 2016. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848. [DOI] [PubMed] [Google Scholar]
- Hashikawa Y, Hashikawa K, Rossi MA, Basiri ML, Liu Y, Johnston NL, Ahmad OR, Stuber GD, 2020. Transcriptional and spatial resolution of cell types in the mammalian habenula. Neuron 106, 743–758.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA, 2012. How variable clones build an invariant retina. Neuron 75, 786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguera I, Desplan C, 2018. Neuronal specification in space and time. Science 362, 176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wang X, Hu B, Mao Y, Chen Y, Yan L, Yong J, Dong J, Wei Y, Wang W, Wen L, Qiao J, Tang F, 2019. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 17, e3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM, 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29. [DOI] [PubMed] [Google Scholar]
- Kim DW, Washington PW, Wang ZQ, Lin SH, Sun C, Ismail BT, Wang H, Jiang L, Blackshaw S, 2020. The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development. Nat Comms 11, 4360–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzh V, Sleptsova I, Liao J, He J, Gong Z, 1998. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev. Dyn. 213, 92–104. [DOI] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM, 2012. Lineage tracing. Cell 148, 33–45. [DOI] [PubMed] [Google Scholar]
- La Manno G, Siletti K, Furlan A, Gyllborg D, Vinsland E, Langseth CM, Khven I, Johnsson A, Nilsson M, Lönnerberg P, Linnarsson S, 2020. Molecular architecture of the developing mouse brain. bioRxiv 135C, 2020.07.02.184051. doi: 10.1101/2020.07.02.184051 [DOI] [PubMed] [Google Scholar]
- Lange C, Rost F, Machate A, Reinhardt S, Lesche M, Weber A, Kuscha V, Dahl A, Rulands S, Brand M, 2020. Single cell sequencing of radial glia progeny reveals the diversity of newborn neurons in the adult zebrafish brain. Development 147, dev185595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-K, Pfaff SL, 2001. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat. Neurosci. 4, 1183–1191. [DOI] [PubMed] [Google Scholar]
- Li H, Horns F, Wu B, Xie Q, Li J, Li T, Luginbuhl DJ, Quake SR, Luo L, 2017. Classifying drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell 171, 1206–1220.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord ND, Carte AN, Abitua PB, Schier AF, 2019. The pattern of Nodal morphogen signaling is shaped by co-receptor expression. bioRxiv 125, 2019.12.30.891101. doi: 10.1101/2019.12.30.891101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Shen Z, Yu Y-C, Shi S-H, 2017. Neural lineage tracing in the mammalian brain. Curr. Opin. Neurobiol. 50, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Hafemeister C, Bandler RC, Machold R, Batista Brito R, Jaglin X, Allaway K, Butler A, Fishell G, Satija R, 2018. Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J, 2016. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JAG, Fruttiger M, Abraham S, Lange CAK, Stone J, Gandhi P, Wang X, Bainbridge J, Moss SE, Greenwood J, 2012. Apelin is required for non-neovascular remodeling in the retina. Am. J. Pathol. 180, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mission JP, Takahashi T, Caviness VS, 1991. Ontogeny of radial and other astroglial cells in murine cerebral cortex. Glia 4, 138–148. [DOI] [PubMed] [Google Scholar]
- Moens CB, Prince VE, 2002. Constructing the hindbrain: insights from the zebrafish. Dev. Dyn. 224, 1–17. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF, 2003. Anatomy of neurogenesis in the early zebrafish brain. Brain Res. Dev. Brain Res. 140, 137–155. [DOI] [PubMed] [Google Scholar]
- Navajas Acedo J, Voas MG, Alexander R, Woolley T, Unruh JR, Li H, Moens C, Piotrowski T, 2019. PCP and Wnt pathway components act in parallel during zebrafish mechanosensory hair cell orientation. Nat Comms 10, 3993–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, Ounadjela JR, Shuga J, Wang X, Lim DA, West JA, Leyrat AA, Kent WJ, Kriegstein AR, 2017. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Shiraki T, Kojima D, Fukada Y, 2015. Homeobox transcription factor Six7 governs expression of green opsin genes in zebrafish. Proc. Biol. Sci. 282, 20150659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, Schier AF, 2013. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Shekhar K, Regev A, Schier AF, 2018. Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-seq. Current Biology 28, 1052–1065.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Gagnon JA, Schier AF, 2018a. Large-scale reconstruction of cell lineages using single-cell readout of transcriptomes and CRISPR-Cas9 barcodes by scGESTALT. Nat Protoc 13, 2685–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF, 2018b. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 36, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, Trakhtenberg EF, 2018. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat Comms 9, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, Pun SH, Sellers DL, Tasic B, Seelig G, 2018. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulands S, Iglesias-Gonzalez AB, Boije H, 2018. Deterministic fate assignment of Müller glia cells in the zebrafish retina suggests a clonal backbone during development. Eur. J. Neurosci. 48, 3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, Briscoe J, 2019. Establishing neuronal diversity in the spinal cord: a time and a place. Development 146, dev182154. [DOI] [PubMed] [Google Scholar]
- Satoh S, Tang K, Iida A, Inoue M, Kodama T, Tsai SY, Tsai M-J, Furuta Y, Watanabe S, 2009. The spatial patterning of mouse cone opsin expression is regulated by bone morphogenetic protein signaling through downstream effector COUP-TF nuclear receptors. J. Neurosci. 29, 12401–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmechel DE, Rakic P, 1979. A Golgi study of radial glial cells in developing monkey telencephalon: morphogenesis and transformation into astrocytes. Anat. Embryol. 156, 115–152. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Strähle U, Scholpp S, 2013. Neurogenesis in zebrafish - from embryo to adult. Neural Development 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y-C, Raymond PA, 2004. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Dev. Biol. 269, 237–251. [DOI] [PubMed] [Google Scholar]
- Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, Juliano CE, 2019. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365, eaav9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solek CM, Feng S, Perin S, Weinschutz Mendes H, Ekker M, 2017. Lineage tracing of dlx1a/2a and dlx5a/6a expressing cells in the developing zebrafish brain. Dev. Biol. 427, 131–147. [DOI] [PubMed] [Google Scholar]
- Stigloher C, Chapouton P, Adolf B, Bally-Cuif L, 2008. Identification of neural progenitor pools by E(Spl) factors in the embryonic and adult brain. Brain Res. Bull. 75, 266–273. [DOI] [PubMed] [Google Scholar]
- Tambalo M, Mitter R, Wilkinson DG, 2020. A single cell transcriptome atlas of the developing zebrafish hindbrain. Development 147, dev184143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than-Trong E, Bally-Cuif L, 2015. Radial glia and neural progenitors in the adult zebrafish central nervous system. Glia 63, 1406–1428. [DOI] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill J-P, Thisse C, 2004. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519. [DOI] [PubMed] [Google Scholar]
- Tiklová K, Björklund ÅK, Lahti L, Fiorenzano A, Nolbrant S, Gillberg L, Volakakis N, Yokota C, Hilscher MM, Hauling T, Holmström F, Joodmardi E, Nilsson M, Parmar M, Perlmann T, 2019. Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat Comms 10, 581–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G, 2018. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA, 2003. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development 130, 1281–1294. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM, 2018. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamsley B, Fishell G, 2017. Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat. Rev. Neurosci. 18, 299–309. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Brand M, Eisen JS, 2002. Patterning the zebrafish central nervous system. Results Probl Cell Differ 40, 181–215. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Rubenstein JL, 2000. Induction and dorsoventral patterning of the telencephalon. Neuron 28, 641–651. [DOI] [PubMed] [Google Scholar]
- Woo K, Fraser SE, 1995. Order and coherence in the fate map of the zebrafish nervous system. Development 121, 2595–2609. [DOI] [PubMed] [Google Scholar]
- Woodworth MB, Girskis KM, Walsh CA, 2017. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet. 18, 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Dorsky RI, 2017. Development of the hypothalamus: conservation, modification and innovation. Development 144, 1588–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Tang X, Jin M, Zhang H, Du L, Yu S, He J, 2020. Unifying developmental programs for embryonic and postembryonic neurogenesis in the zebrafish retina. Development 147, dev185660. [DOI] [PubMed] [Google Scholar]
- Yan W, Peng Y-R, van Zyl T, Regev A, Shekhar K, Juric D, Sanes JR, 2020. Cell Atlas of the human fovea and peripheral retina. Sci Rep 10, 9802–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari N, Ando K, Kudo A, Kinoshita M, Kawakami A, 2012. Colored medaka and zebrafish: Transgenics with ubiquitous and strong transgene expression driven by the medaka β-actinpromoter. Develop. Growth Differ. 54, 818–828. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S, 2018. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Le Sun, Pan N, Xu X, Tang F, Zhang J, Qiao J, Wang X, 2018. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Code used to perform retina and hypothalamus cell trajectory analysis using URD
Master table describing all identified clusters from 12 hpf to 15 dpf. Columns are as follows: Stage – developmental stage profiled. Cluster – corresponding cluster number. Match N-1 – cluster from prior stage that is the most similar to the current cluster. For example Row 45 (14 hpf cluster 0) is similar to 12 hpf cluster 0. Assigned Cell type/State – cell type annotation. Enriched markers – subset of enriched markers are shown for each cluster.
Data Availability Statement
The datasets generated during this study are available at GEO [accession code]. The processed data is available as a resource to explore at: https://github.com/brlauuu/zf_brain. The code used in this study is available at: https://github.com/brlauuu/zf_brain and Methods S1.