Abstract
Background/Aim: Oncotype DX recurrence score (RS) for breast cancer is a useful tool for determining chemotherapy indication but it is expensive and time-consuming. We determined whether four immuno-histochemical markers, namely human epidermal growth factor 2 (HER2), estrogen receptor (ER), progesterone receptor (PgR), and Ki-67, are predictive of an RS ≥26 in Japanese patients. Patients and Methods: The study included 95 Japanese patients evaluated for RS. A predictive model was created using logistic regression analysis. Results: The discriminant function was calculated as follows: p=1/{1+exp [−(4.611+1.2342×HER2−0.0813×ER− 0.0489 ×PgR+0.0857×Ki67)]}. Using a probability of 0.5 as the cutoff, the accuracy, sensitivity, specificity, positive predictive and negative predictive values were 90.5%, 72.2%, 94.8%, 76.4% and 93.5%, respectively. Conclusion: The model had a high negative predictive value in predicting RS ≥26 in Japanese patients, indicating that Oncotype DX testing may be omitted in patients with a negative result according to the predictive model.
Keywords: Breast cancer, recurrence score, predict, Japan
Oncotype DX recurrence score (RS) (Genomic Health, Redwood City, CA, USA), which is calculated based on the expression levels of 21 genes in paraffin-embedded specimens using reverse transcriptase-polymerase chain reaction assay, predicts the probability of distant recurrence in patients with estrogen receptor (ER)-positive/human epidermal growth factor receptor 2 (HER2)-negative breast cancer (1). Albeit a globally utilized tool, the RS results are not immediately available. Moreover, performing Oncotype DX testing for all patients with ER-positive/HER2-negative breast cancer is not medically economical.
RS is based on the measurement of the expression levels of 16 cancer-related genes and five reference genes. The cancer-related genes evaluated for RS can be categorized into five classes: Proliferation-related, invasion-related, HER2, ER, and ‘other’. The information obtained by Oncotype DX testing is more extensive than that obtained by routine immunohistochemical staining for the four markers, ER, progesterone receptor (PgR), HER2 and Ki-67, which are used in daily clinical practice to determine the intrinsic breast cancer subtype. Although it is not possible to predict RS using immunohistochemical staining for these four markers alone, the data obtained with the immunohistochemical approach might be sufficient to determine whether the RS is low or high. Several models for estimating RS have been reported to date (2); however, to the best of our knowledge no study has evaluated RS predictive models using data from Japanese patients with breast cancer.
The results of the Trial Assigning Individualized Options for Treatment demonstrated that endocrine treatment alone was non-inferior to chemotherapy plus endocrine treatment for invasive disease-free survival in patients with an RS<26 (3). Therefore, we aimed to establish a model based on the four immunohistochemical markers and to evaluate its ability to predict an RS of ≥26 in Japanese patients with breast cancer.
Patients and Methods
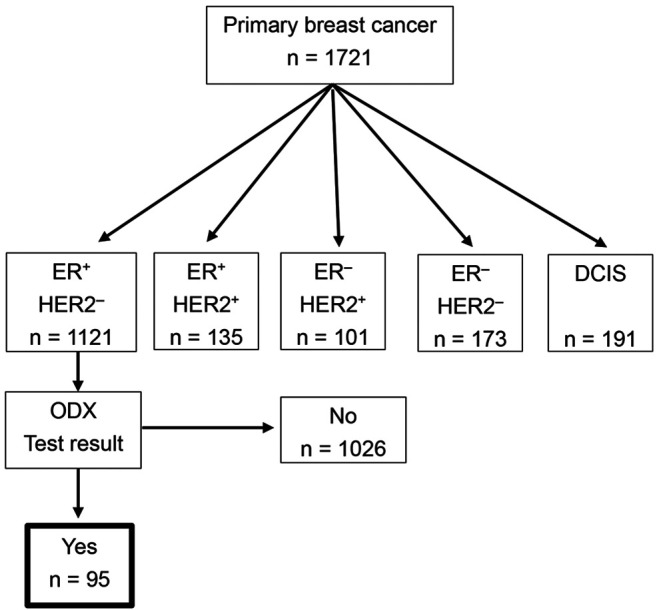
Patients. A total of 1721 patients underwent surgery due to primary breast cancer in Yokohama Rosai Hospital between April 2012 and May 2020. Of these, 1,121 patients were ER-positive/HER2-negative, and 95 patients among them who were submitted for Oncotype DX testing were enrolled in this retrospective study (Figure 1). The Oncotype DX testing was either recommended by the treating physician or requested by the patient. Clinicopathological information, including patient age, tumor size, lymph node status, nuclear grade, and immunohistochemical staining results for ER, PgR, HER2, and Ki-67 were collected from medical records. The study was approved by the Yokohama Rosai Hospital Ethics Committee (no. 2-31). Consent was obtained using an opt-out system.
Figure 1. Diagram of patients enrolled in this study. In total, 1,721 patients underwent surgery due to primary breast cancer at Yokohama Rosai Hospital between April 2012 and May 2020. Of these, 1,121 patients had ER-positive/HER2-negative disease, and 95 patients among them submitted to Oncotype DX testing were enrolled in this retrospective study. DCIS: Ductal carcinoma in situ; ODX: Oncotype DX.
Immunohistochemical staining. Core needle biopsy specimens were used for immunohistochemical staining, which was a part of the routine evaluation at the time of diagnosis. Surgical specimens were used in cases with inadequate information on immunohistochemical staining before surgery. Immunohistochemical staining for HER2 was performed using DAKO HercepTest II (DAKO, Denmark) and evaluated with DAKO Autostainer Link 48 platform (Agilent Technologies, Japan). Immunostaining for ER, PgR, and Ki-67 was evaluated by BenchMark ULTRA and Aperio Scanscope AT Turbo automated immunostainers using I-VIEW DAB universal kit (Roche Diagnostics KK, Japan). To determine the number of cells stained for ER, PgR, and Ki-67, five areas were randomly selected and approximately 1,000 cells were counted in each area to obtain average scores for each marker. Quantitative scores that indicated the proportion of positively stained cells in each marker were used for the analysis of immunohistochemical staining for ER, PgR, and Ki-67, whereas scores of 0+, 1+, and 2+ were used to analyze immunohistochemical staining for HER2. The criterion for HER2 negativity was the presence of an immunohistochemistry score of 0+, 1+, or 2+ with a fluorescence in situ hybridization score of <2.0 (4), and the criterion for ER positivity was an ER-positive cell rate of >1% (5).
Statistical analysis. Logistic regression analysis was performed for developing the predictive model. Partial regression coefficients and standard partial regression coefficients were calculated. A larger absolute value for a standard partial regression coefficient indicated a greater influence of the variable in the predictive model. Receiver-operating characteristic (ROC) curves were plotted, and areas under the ROC curves (AUCs) were calculated. Additionally, for further internal validation of our predictive model, the estimated AUC and 95% confidence interval (CI) were calculated with 1000 replications of bootstrap samples in our cohort. A p-value of <0.05 was considered to indicate statistical significance. All statistical analyses were performed using Bell Curve version 3.20 for Excel (Social Survey Research Information, Japan) and JMP version 15.0 (SAS Institute, Cary, NC, USA).
Results
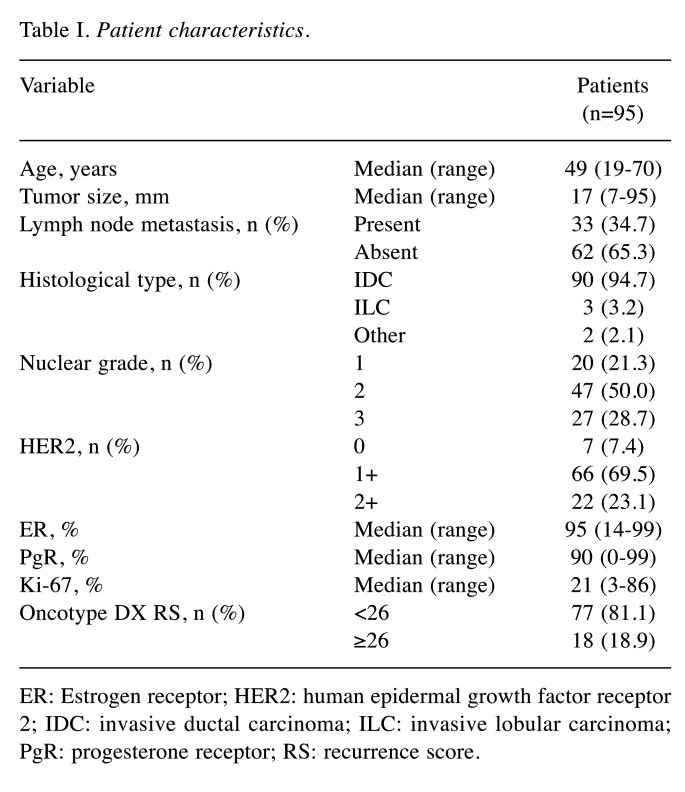
Patient characteristics. The characteristics of patients included in the study are shown in Table I. The study cohort included 33 (34.7%) patients with lymph node metastases; there were a maximum of three lymph node metastases, including micrometastases. There were 77 (81.1%) and 18 (18.9%) patients with an RS of <25 and ≥26, respectively. The median percentage of cells staining positively for ER, PgR, and Ki-67 were 95% (range=14-99%), 90% (range=0-99%), and 21% (range=3-86%), respectively.
Table I. Patient characteristics.
ER: Estrogen receptor; HER2: human epidermal growth factor receptor 2; IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma; PgR: progesterone receptor; RS: recurrence score.
Predictive model. A model based on HER2, ER, PgR, and Ki-67 for predicting an RS ≥26 was constructed in two steps as follows.
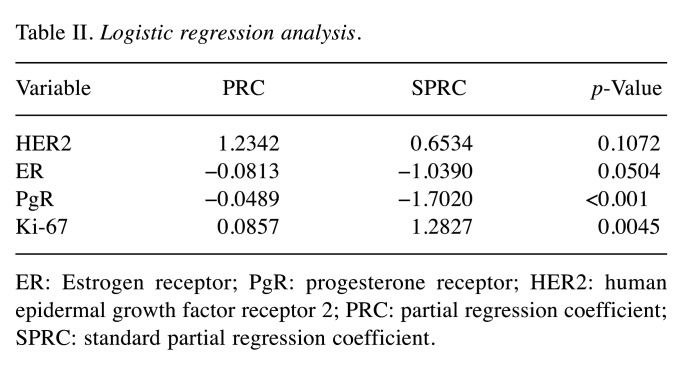
First step: Logistic regression analysis. The probability of RS ≥26 using four variables (HER2, ER, PgR, and Ki-67) was calculated using logistic regression analysis (Table II). Quantitative scores that indicated the proportion of positively stained cells for each marker were used for the analysis for ER, PgR and Ki-67, whereas scores of 0, 1, 2 were used for HER2.
Table II. Logistic regression analysis.
ER: Estrogen receptor; PgR: progesterone receptor; HER2: human epidermal growth factor receptor 2; PRC: partial regression coefficient; SPRC: standard partial regression coefficient.
The discriminant function was as follows: p=1/{1+exp[−(4.611+1.2342×HER2−0.0813×ER−0.0489×PgR+0.0857×Ki67)]}
The standard partial regression coefficient of PgR was −1.7020, making PgR the variable with the largest value among the four variables, followed by Ki-67 with 1.2827. Both PgR and Ki-67 were significant variables in the logistic regression analysis.
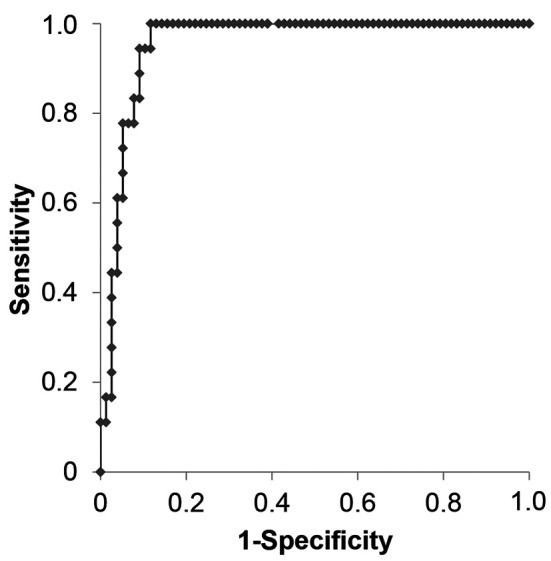
Second step: Analysis based on the ROC curve. The AUC of this predictive model was 0.956 (95% confidence interval=0.917-0.994, p<0.001; Figure 2). When using a probability of 0.5 as the cutoff value, the accuracy, sensitivity, specificity, and positive and (NPV) negative predictive values were 90.5%, 72.2%, 94.8%, 76.4%, and 93.5%, respectively.
Figure 2. Receiver-operating characteristic (ROC) curves of the predictive model. The area under the ROC curve (AUC) was 0.956.
Internal validation. For the internal validation of the predictive model, 1,000 bootstrapped replications were performed to resample the data. The result of the bootstrapping analysis was similar to that obtained with the original samples (AUC=0.949, 95% confidence interval=0.831-0.975).
Discussion
Oncotype DX, a useful tool for assessing the indication for chemotherapy in breast cancer, cannot be used in all patients in Japan because the test results take a long time and it is expensive for the patients, and thus there is a desperate need to establish a model predictive of the RS. Therefore, we developed a model based on four immunohistochemical markers in routine clinical use to predict Japanese patients with breast cancer which would have an RS ≥26. This predictive model will be useful for patients in countries where Oncotype DX is not immediately available, as in Japan.
Several predictive models for RS have been reported, and age, tumor size, nuclear grade, lymphovascular invasion, histological type, ER, PgR, HER2, and Ki-67 have been reported as clinicopathological factors associated with RS (2,6-11). In the present study, we specifically utilized four immunohistochemical markers that are routinely used to determine the intrinsic breast cancer subtype in clinical practice. Klein et al., who investigated the relationship between RS and a predictive score based on these four markers in a U.S. cohort using linear regression analysis, reported that the correlation coefficient was 0.594 (10). Importantly, in our model, the stepwise addition of other clinical parameters to these four markers in the logistic regression model did not further increase the accuracy rate (data not shown). Therefore, we used these four factors in our predictive model.
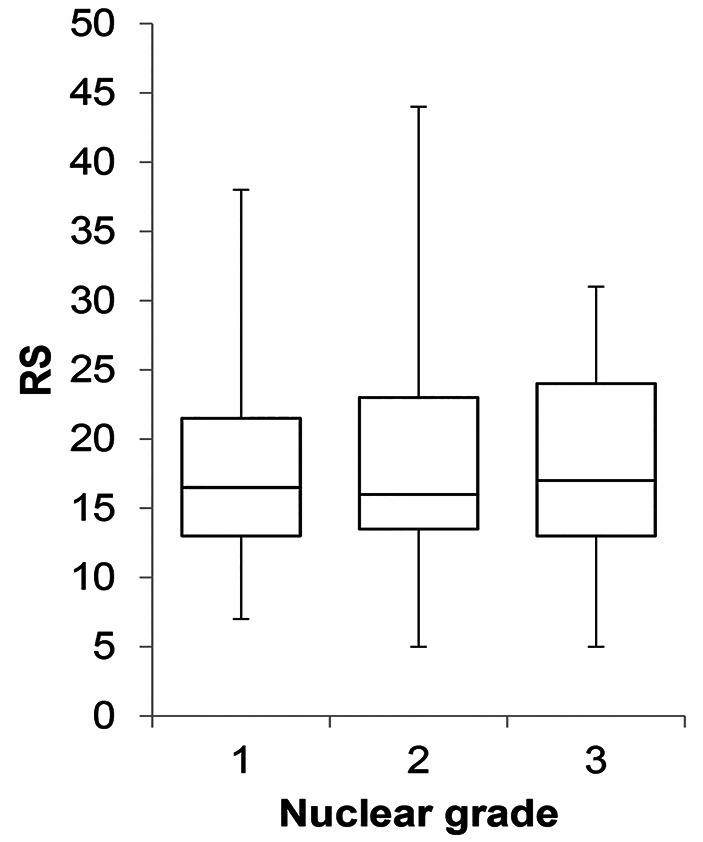
In the present study, PgR was a significant variable in the predictive model and the standard partial regression coefficient was highest for PgR among the four immunohistochemistry markers. Several studies reported that PgR was strongly associated with RS (6,7,12), and Kurozumi et al. reported that PgR negativity was a poor prognostic marker in patients with ER-positive/HER2-negative breast cancer (13), which the current study results are in agreement with. Conversely, some studies emphasized the correlation of RS with Nottingham histological grade and nuclear grade (7,9). In the present study, nuclear grade was not associated with RS (Figure 3) and was not included in the predictive model. One reason for this finding might be that grade classification is subjective and less reproducible (14). Sahebjam et al. also reported that there was no significant correlation between Nottingham grade and RS when the effect of Ki-67 was controlled (15), suggesting that the correlation between Nottingham grade and RS was most likely due to the effect of Ki-67.
Figure 3. Box plot of Oncotype DX recurrence score (RS) for the patients of this study. The RS was not found to depend on nuclear grade. The upper and lower bars represent maximum and minimum scores, respectively. The box represents the area from the third and first quartiles. The horizontal line represents the median score.
In a previous study, the AUC of a model predictive of RS ≥26 ranged from 0.81 to 0.90 (2,8,11,16). Although the accuracy depends on the cutoff value used, Orucevic et al. (2) and Lee et al. (11) reported overall accuracy rates of 92.7% and 56.4%, respectively, for their predictive models. In our predictive model, the overall accuracy rate was 90.5%, and this seemed to be good. Specifically, the NPV was high (93.5%), whereas the positive predictive value was low (76.4%). These results might be due to the low number of patients with an RS ≥26 in the present study cohort. Future studies including populations with high RSs may improve the positive predictive value of our predictive model. Our model had a high NPV, indicating that Oncotype DX testing may be omitted in patients with a negative result (i.e. a probability score below the cutoff of 0.5) based on our predictive model. Approximately 80% of all cases were negative by our predictive model; therefore, there would be considerable economic benefit by performing further testing by Oncotype DX only in patients who are predicted to have a positive result (i.e. a probability score above the cutoff of 0.5) using our predictive model. Kapadia et al. also reported that the indication for Oncotype DX testing should be narrowed to selected patients (17).
Regarding model validity, Steyerberg et al. reported that split-sample validation is inefficient and recommended bootstrapping for estimation of the internal validity of a predictive logistic regression model (18). We used this method for internal validation. Our result seemed good. However, this model was based on data from a single institution; thus, verification at other institutions is required.
Patients with lymph node-negative disease were analyzed in several studies evaluating predictive models for RS in breast cancer (2,8,11). However, in clinical practice, deciding on whether to administer chemotherapy to patients with ER-positive/HER2-negative breast cancer and metastasis in 1-3 lymph nodes remains difficult. Moreover, it remains unclear whether predictive models can be created by including both negative and positive cases of lymph node metastasis. Bello et al. reported that lymph node status did not reflect tumor biology based on the similar RS distribution between patients with and without lymph node metastasis (19). Therefore, it may be appropriate to develop a predictive model that includes patients with lymph node metastasis. The rate of lymph node metastasis was 34.7% in the present study; thus, our predictive model might be easier to apply to real-world clinical practice. In contrast, an RS cutoff of 26 may not be appropriate in patients with lymph node metastasis. Clinical trials on patients with lymph node metastasis indicated that prognosis was favorable with hormone therapy alone in patients with an RS≤17 (20,21). The results of the ongoing RxPONDER study (Rx for Positive Node, Endocrine Responsive Breast Cancer) should shed light on the unclear significance of an RS cutoff of 26 in patients with lymph node metastasis.
The present study also has several limitations. Firstly, the study was conducted at a single institution and included a small study population. Secondly, the predictive model was calculated based on the inclusion only of patients who underwent Oncotype DX testing and did not include all patients with ER-positive/HER2-negative breast cancer evaluated during the study period. Thirdly, the data on cohort prognosis based on the predictive model-based treatment were insufficient. Future prospective multi-center studies with larger cohorts are necessary to validate our predictive model.
We developed a model based on four immuno-histochemistry markers to predict RS ≥26 in Japanese patients; the developed predictive model had a high NPV, indicating that Oncotype DX testing may be omitted in patients with a negative result based on our predictive model. Such a model will be beneficial to the medical economy.
Conflicts of Interest
The Authors disclose no conflicts of interest in relation to this study.
Authors’ Contributions
SY designed the study, collected the samples and clinical data, analyzed statistics, and wrote the article. TC collected the samples and clinical data, designed the study, and revised the article. YS, FH, and HT collected the samples and clinical data. AY, KN, TI, and IE revised the article. TM analyzed the samples and statistics. All Authors read and approved the article.
References
- 1.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 2.Orucevic A, Bell JL, King M, McNabb AP, Heidel RE. Nomogram update based on TAILORx clinical trial results - Oncotype DX breast cancer recurrence score can be predicted using clinicopathologic data. Breast. 2019;46:116–125. doi: 10.1016/j.breast.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE Jr, Dees EC, Goetz MP, Olson JA Jr, Lively T, Badve SS, Saphner TJ, Wagner LI, Whelan TJ, Ellis MJ, Paik S, Wood WC, Ravdin PM, Keane MM, Gomez Moreno HL, Reddy PS, Goggins TF, Mayer IA, Brufsky AM, Toppmeyer DL, Kaklamani VG, Berenberg JL, Abrams J, Sledge GW Jr. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–121. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolff A, Hammond M, Hicks D, Dowsett M, Mcshane L, Allison K, Allred D, Bartlett J, Bilous M, Fitzgibbons P, Hanna W, Jenkins R, Mangu P, Paik S, Perez E, Press M, Spears P, Vance G, Viale G, Hayes D. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2019;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 5.Yip CH, Rhodes A. Estrogen and progesterone receptors in breast cancer. Future Oncol. 2014;10(14):2293–2301. doi: 10.2217/fon.14.110. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary LN, Jawa Z, Szabo A, Visotcky A, Chitambar CR. Relevance of progesterone receptor immunohistochemical staining to Oncotype DX recurrence score. Hematol Oncol Stem Cell Ther. 2016;9(2):48–54. doi: 10.1016/j.hemonc.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Thibodeau S, Voutsadakis IA. Prediction of Oncotype Dx recurrence score using clinical parameters: A comparison of available tools and a simple predictor based on grade and progesterone receptor. Hematol Oncol Stem Cell Ther. 2019;12(2):89–96. doi: 10.1016/j.hemonc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Yoo SH, Kim TY, Kim M, Lee KH, Lee E, Lee HB, Moon HG, Han W, Noh DY, Han SW, Kim TY, Im SA. Development of a nomogram to predict the recurrence score of 21-gene prediction assay in hormone receptor-positive early breast cancer. Clin Breast Cancer. 2020;20(2):98–107.e1. doi: 10.1016/j.clbc.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Gage M, Rosman M, Mylander W, Giblin E, Kim H, Cope L, Umbricht C, Wolff A, Tafra L. A Validated model for identifying patients unlikely to benefit from the 21-gene recurrence score assay. Clin Breast Cancer. 2019;15(6):467–472. doi: 10.1016/j.clbc.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein ME, Dabbs DJ, Shuai Y, Brufsky AM, Jankowitz R, Puhalla SL, Bhargava R. Prediction of the Oncotype DX recurrence score: use of pathology-generated equations derived by linear regression analysis. Mod Pathol. 2013;26(5):658–664. doi: 10.1038/modpathol.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SB, Kim J, Sohn G, Kim J, Chung IY, Kim HJ, Ko BS, Son BH, Ahn SH, Lee JW, Jung KH. A Nomogram for predicting the oncotype DX recurrence score in women with T1-3N0-1miM0 hormone receptor-positive, human epidermal growth factor 2 (HER2)-negative breast cancer. Cancer Res Treat. 2019;51(3):1073–1085. doi: 10.4143/crt.2018.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark BZ, Dabbs DJ, Cooper KL, Bhargava R. Impact of progesterone receptor semiquantitative immunohistochemical result on Oncotype DX recurrence score: a quality assurance study of 1074 cases. Appl Immunohistochem Mol Morphol. 2013;21(4):287–291. doi: 10.1097/PAI.0b013e31826f80c9. [DOI] [PubMed] [Google Scholar]
- 13.Kurozumi S, Matsumoto H, Hayashi Y, Tozuka K, Inoue K, Horiguchi J, Takeyoshi I, Oyama T, Kurosumi M. Power of PgR expression as a prognostic factor for ER-positive/HER2-negative breast cancer patients at intermediate risk classified by the Ki67 labeling index. BMC Cancer. 2017;17(1):354. doi: 10.1186/s12885-017-3331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalton LW, Page DL, Dupont WD. Histologic grading of breast carcinoma. A reproducibility study. Cancer. 1994;73(11):2765–2770. doi: 10.1002/1097-0142(19940601)73:11<2765::aid-cncr2820731119>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 15.Sahebjam S, Aloyz R, Pilavdzic D, Brisson ML, Ferrario C, Bouganim N, Cohen V, Miller WH Jr, Panasci LC. Ki 67 is a major, but not the sole determinant of Oncotype Dx recurrence score. Br J Cancer. 2011;105(9):1342–1345. doi: 10.1038/bjc.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orucevic A, Bell JL, McNabb AP, Heidel RE. Oncotype DX breast cancer recurrence score can be predicted with a novel nomogram using clinicopathologic data. Breast Cancer Res Treat. 2017;163(1):51–61. doi: 10.1007/s10549-017-4170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapadia S, Gudiwada SP, Kaji AH, Chlebowski RT, Venegas R, Ozao-Choy J, Dauphine C. Can Oncotype DX testing be omitted in invasive breast cancer patients with clinicopathologic factors predicting very high pretest probability of a concordant result. Breast J. 2020;26(11):2199–2202. doi: 10.1111/tbj.14068. [DOI] [PubMed] [Google Scholar]
- 18.Steyerberg EW, Harrell FE Jr, Borsboom GJ, Eijkemans MJ, Vergouwe Y, Habbema JD. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54(8):774–781. doi: 10.1016/s0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]
- 19.Bello DM, Russell C, McCullough D, Tierno M, Morrow M. Lymph node status in breast cancer does not predict tumor biology. Ann Surg Oncol. 2018;25(10):2884–2889. doi: 10.1245/s10434-018-6598-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL, Davidson NE, Sledge GW, Winer EP, Hudis C, Ingle JN, Perez EA, Pritchard KI, Shepherd L, Gralow JR, Yoshizawa C, Allred DC, Osborne CK, Hayes DF, Breast Cancer Intergroup of North America Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stemmer SM, Steiner M, Rizel S, Geffen DB, Nisenbaum B, Peretz T, Soussan-Gutman L, Bareket-Samish A, Isaacs K, Rosengarten O, Fried G, McCullough D, Svedman C, Shak S, Liebermann N, Ben-Baruch N. Clinical outcomes in ER+ HER2 -node-positive breast cancer patients who were treated according to the Recurrence Score results: evidence from a large prospectively designed registry. NPJ Breast Cancer. 2017;3:32. doi: 10.1038/s41523-017-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]