Abstract
Background
Lipid phosphate phosphatases (LPP) are critical for regulating the production and degradation of phosphatidic acid (PA), an essential signaling molecule under stress conditions. Thus far, the LPP family genes have not been reported in rapeseed (Brassica napus L.).
Results
In this study, a genome-wide analysis was carried out to identify LPP family genes in rapeseed that respond to different stress conditions. Eleven BnLPPs genes were identified in the rapeseed genome. Based on phylogenetic and synteny analysis, BnLPPs were classified into four groups (Group I-Group IV). Gene structure and conserved motif analysis showed that similar intron/exon and motifs patterns occur in the same group. By evaluating cis-elements in the promoters, we recognized six hormone- and seven stress-responsive elements. Further, six putative miRNAs were identified targeting three BnLPP genes. Gene ontology analysis disclosed that BnLPP genes were closely associated with phosphatase/hydrolase activity, membrane parts, phosphorus metabolic process, and dephosphorylation. The qRT-PCR based expression profiles of BnLPP genes varied in different tissues/organs. Likewise, several gene expression were significantly up-regulated under NaCl, PEG, cold, ABA, GA, IAA, and KT treatments.
Conclusions
This is the first report to describe the comprehensive genome-wide analysis of the rapeseed LPP gene family. We identified different phytohormones and abiotic stress-associated genes that could help in enlightening the plant tolerance against phytohormones and abiotic stresses. The findings unlocked new gaps for the functional verification of the BnLPP gene family during stresses, leading to rapeseed improvement.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07862-1.
Keywords: Abiotic stress, Gene structure, Gene ontology, miRNA, Phytohormone, Lipid phosphate phosphatases, Rapeseed
Background
Phospholipids exist in the cellular membranes of an organism. Most of them are structural, while a few serve as lipid-signaling molecules. Phosphatidic acid (PA) acts as a signaling compound and precursor for all phospholipids [1–3]. In plants, PA can be formed via three different pathways [1–3]. The PA abundance in plants is defined as the balance between enzymes responsible for PA synthesis and degradation. Phosphatidic acid kinase catalyzes PA phosphorylation to yield diacylglycerol pyrophosphate (DGPP). Phosphatidic acid phosphatase (PAP) is another key enzyme to keep a PA’s appropriate balance [4]. PAP can be divided into two types depending on the requirement of magnesium ion (Mg2+): (1) conventional PAPs, i.e., PAP1, the Mg2+-dependent PA phosphatase activities, and catalyzes PA dephosphorylation to generate diacylglycerol (DAG); and (2) PAP2, the Mg2+-independent PA phosphatase activities, named as lipid phosphate phosphatases (LPPs). These LPPs dephosphorylates PA to DAG but also dephosphorylates DGPP to PA [4]. In short, LPPs are members of the PAP superfamily and catalyze the dephosphorylation of phosphorous lipids, which play a vital role in numerous physiological functions, including cell migration, proliferation, and differentiation [3, 4].
Recently, significant progress has been made in the PAP superfamily. For instance, four PAP members (APP1, DPP1, LPP1, and PAH1) have been investigated in yeast. Where APP1 and PAH1 are Mg2+-dependent, and DPP1 and LPP1 are Mg2+-independent PAPs. Notably, PAH1 is the major regulator of triacylglycerol(s) (TAG) content [5, 6], and DPP1 and LPP1 play a crucial role in controlling the signal transduction of PA, DAG, and DGPP [7–9]. Plants also contain multiple PAP isoforms such as PAP1 (PAH1 and PAH2), similar to yeast PAH1, PAPs responsible for galactolipid synthesis [10], and transiently increased the PA and DGPP synthesis under multiple stresses in plants. In agreement, LPPs were found to be responsible for switching these signals on/off under stress conditions [11]. The LPP-mediated DAG production significantly affects the invasion and growth of Magnaporthe oryzae [5, 12]. In another study, four PAP2/LPP genes were cloned in Arabidopsis thaliana, similar to yeast LPPs [4, 13]. Northern blot analysis revealed that AtLPP1 was more likely to be expressed in leaves and roots, while the expression of AtLPP2 was recognized in all the tested tissues of A. thaliana [13]. Genotoxic (gamma-ray or UV-B) and elicitor treatments transiently induced the AtLPP1 and AtLPP2 expression levels involved in abscisic acid (ABA) signal transduction and stomatal movement [14, 15]. Physiological analysis showed that PA accumulation triggers early signal transduction actions that lead to ABA responses during seed germination and regulate the stomatal movement [14, 15]. Interestingly, PA is involved in ABA signaling, and thus AtLPP2 also serves as negative regulators in ABA-induced seed germination inhibition [15]. The HvLPP1/2 genes are involved in ABA sensitivity and breaking dormancy in barley (Hordeum vulgare L.) [16]. According to the literature, LPPs enzymes are involved in lipid synthesis and thus regulate plants’ growth. For example, VuPAPa and VuPAPb may be involved in membrane lipid modification, observed in cowpea (Vigna unguiculata L.) plants under drought stress [17]. The knock-down of NtLPP4 inhibited PA degradation and promoted pollen tube growth in tobacco (Nicotiana tabacum) plants [18].
Rapeseed (Brassica napus L.) is considered the second most important oilseed crop and serves as a primary oil source for human consumption and animal feed meals [19]. Numerous environmental stresses adversely affect rapeseed growth, productivity, and seed quality, ultimately reducing the final yield [19]. To date, LPP family genes are yet to be reported in rapeseed. The complete rapeseed genome sequence allows the identification and analysis of LPP genes in the rapeseed genome. Hence, a genome-wide comprehensive study has been performed to identify putative rapeseed LPP family genes. Additionally, their phylogenetic relationships, synteny analysis, gene structures, conserved motifs, cis-elements, miRNA regulator prediction, functional annotation have been characterized to get insights into the BnLPP genes. Moreover, the expression profiles in different tissues/organs and under numerous hormone and abiotic stresses have been extensively assessed.
Results
Identification and characterization of LPP gene family in Brassica napus L
In the current study, 11 BnLPPs genes were obtained containing the complete PAP2 functional domain (Table 1). Six genes were positioned in the A subgenome, and five genes were positioned in the C subgenome (Table 1). Detailed characteristics of 11 BnLPP genes are presented in Table 1. Briefly, coding DNA sequences (CDS) length ranged from 918 to 1089 bp with 2–8 exons, and the protein length ranged from 305 to 362 amino acids for BnLPP2A/BnLPP4A/BnLPP4B, and BnLPP3A/BnLPP3B, respectively. The protein molecular weight (MW) ranged from 34.7 kDa (BnLPP4A and BnLPP4B) to 40.5 kDa (BnLPP3A), and isoelectric points (pI) varied from 6.13 (BnLPP2A) to 8.64 (BnLPP1B). The subcellular location prediction revealed that 10 BnLPP proteins were positioned in the plasma membrane, while BnLPP1C was located in the endoplasmic reticulum. Meanwhile, 4 Brassica oleracea (BoLPP1A-BoLPP4), 6 Brassica rapa (BraLPP3B-BraLPP2B), and 4 Arabidopsis thaliana (AtLPP1-AtLPP4) LPP genes were also identified (Additional file 2).
Table 1.
The characteristics of 11 BnLPPs in Brassica napus L
| Gene name | Gene ID | Genomic position (bp) | CDS length (bp) | Exon | Protein length (aa) | MW | pI | Predicted Pfam domain | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| BnLPP1A | BnaA09g18500D | A09:11,485,093–11,486,357 - | 984 | 2 | 327 | 36.9 | 8.35 | PAP2 | PM |
| BnLPP1B | BnaC09g20440D | C09:17,421,172–17,423,263 - | 972 | 2 | 323 | 36.6 | 8.64 | PAP2 | PM |
| BnLPP1C | BnaA06g35100D | A06:23,178,693–23,179,882 + | 951 | 2 | 316 | 35.5 | 7.20 | PAP2 | ER |
| BnLPP2A | BnaC08g39060D | C08:34,977,648–34,979,784 - | 918 | 7 | 305 | 35.6 | 6.13 | PAP2 | PM |
| BnLPP2B | BnaA09g45250D | A09:30,966,399–30,968,509 - | 939 | 7 | 312 | 35.2 | 6.18 | PAP2 | PM |
| BnLPP3A | BnaC05g48240D | C05:42,805,948–42,809,057 + | 1089 | 8 | 362 | 40.5 | 6.13 | PAP2 | PM |
| BnLPP3B | BnaA05g33490D | A05:22,673,065–22,676,351 + | 1089 | 8 | 362 | 40.4 | 6.23 | PAP2 | PM |
| BnLPP3C | BnaC03g33070D | C03:20,223,052–20,225,854 + | 966 | 6 | 321 | 36.1 | 6.53 | PAP2 | PM |
| BnLPP3D | BnaA03g28040D | A03:13,723,633–13,726,024 + | 966 | 6 | 321 | 36.1 | 6.56 | PAP2 | PM |
| BnLPP4A | BnaA05g21920D | A05:16,840,547–16,842,185 - | 918 | 6 | 305 | 34.7 | 8.46 | PAP2 | PM |
| BnLPP4B | BnaC05g35130D | C05:34,426,978–34,428,721 - | 918 | 6 | 305 | 34.7 | 8.46 | PAP2 | PM |
In the genomic position, the positive (+) and negative (-) sign indicates the existence of gene on the positive and negative strand of that specific markers, respectively
CDS coding DNA sequences, bp base pair, MW molecular weight, pI isoelectric points, PM plasma membrane, ER endoplasmic reticulum
Multiple sequence alignment and phylogenetic analysis of BnLPP gene family
To understand the sequence characteristics, we performed a multiple sequence alignment analysis of the 11 BnLPP proteins using DNAMAN software with the default parameters. The four different A. thaliana LPP proteins (AtLPP1-AtLPP4) from each group were randomly selected as representatives for further comparison. The transmembrane structure and conversed domain structures of BnLPPs are displayed in Fig. 1. It was predicted that all the LPP proteins contained six membrane-spanning hydrophobic regions, named TM1-6 by TMHMM [19]. Our results showed that the PAP2 domains were highly conserved and commonly contained three consensus domains (denoted by a red bar), i.e., KX6RP (domain 1), PSGH (domain 2), and SRX5HX3D (domain 3). Notably, the conserved amino acids in the PAP2 domain were found to be essential for enzymatic activity. Thus, alteration in these amino acids may cause severe gene function losses [20].
Fig. 1.
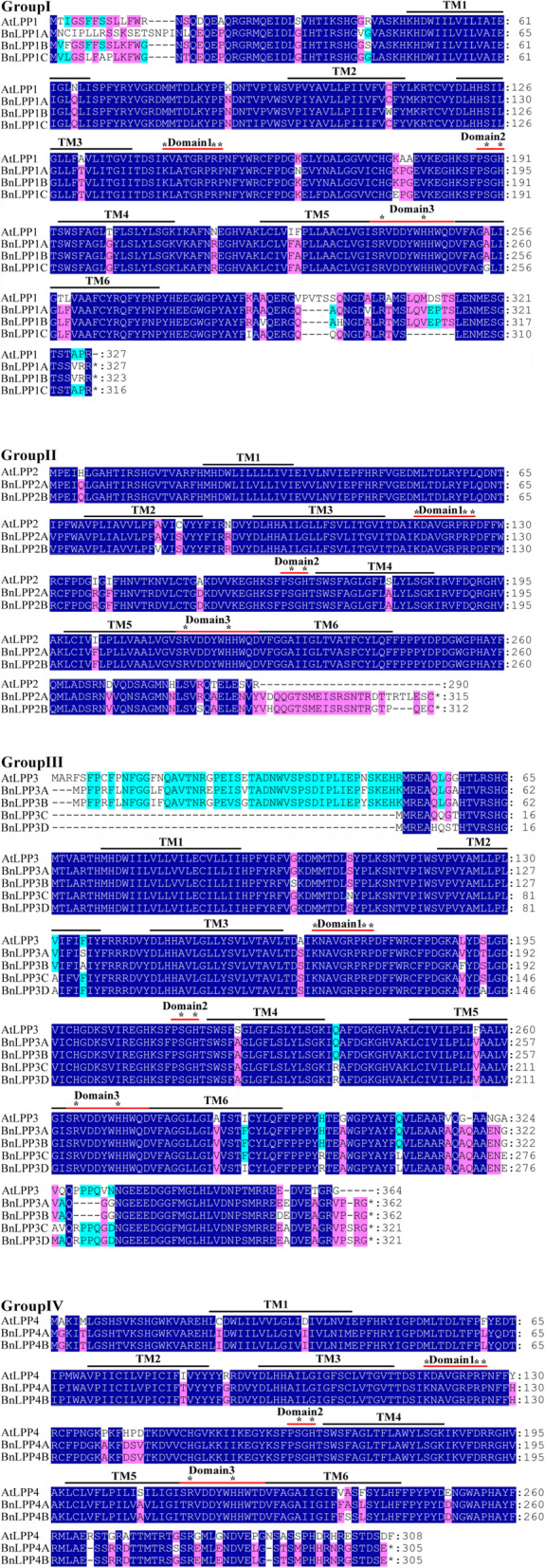
Alignment of multiple BnLPPs and selected AtLPPs protein sequences
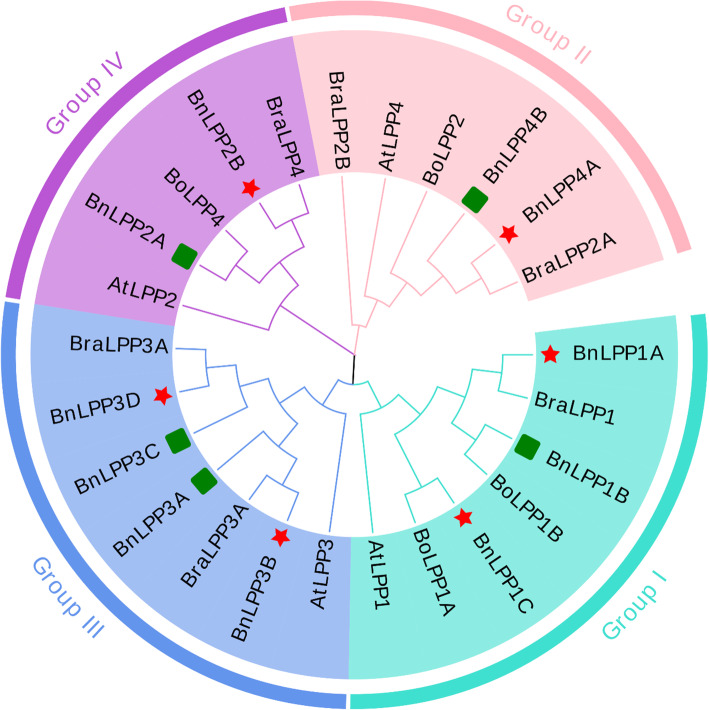
To determine the BnLPP genes family’s evolutionary relationships with (A) thaliana and the (B) napus ancestor species, based on the neighbor-joining (NJ) method, an unrooted phylogenetic tree was constructed between 25 LPP genes (11 from B. napus, 6 from B. rapa, 4 from B. oleracea and 4 from (A) thaliana). The phylogenetic analysis indicated that the 25 LPPs were grouped into four groups (Group I, II, III, and IV) (Fig. 2). Our results showed that Group I contained 7 LPPs members (3 BnLPPs, 1 BraLPP, 2 BoLPPs, and 1 AtLPP), Group II contained 6 LPPs members (2 BnLPPs, 2 BraLPPs, 1 BoLPP, and 1 AtLPP), Group III contained 7 LPPs members (4 BnLPPs, 2 BraLPPs, and 1 AtLPP), and Group IV contained 5 LPPs members (2 BnLPPs, 1 BraLPP, 1 BoLPP, and 1 AtLPP) (Fig. 2). Overall, LPPs grouping into the same sub-group may have similar functions. Notably, all LPPs members were evenly distributed in four groups; however, no BoLPPs belonged to Group III (Fig. 2). Moreover, it was found that the BnLPPs have close phylogenetic relationships with their ancestors’ species in each group. Arabidopsis and Brassicas have a common ancestor, but AtLPP3 had no (B) oleracea homologous gene in Group III, indicating that a few genes were lost during the Brassica species’ evolution.
Fig. 2.
A phylogenetic tree of 25 LPPs from B. napus, B. oleracea, B. rapa, and A. thaliana. All LPPs genes were divided into four groups based on the high bootstrap values and the phylogenetic tree’s topology. Overall, 11 BnLPPs from B. napus, 6 BraLPPs from B. rapa, 4 BoLPPs from B. oleracea, and 4 AtLPPs from A. thaliana were clustered into four groups (Group I-IV) based on high bootstrap values signified with different background colors. The red star and green rectangle indicate that these genes belong to the A and C subgenome, respectively
In the aligned amino acids, invariant ones were marked with black, and the conserved ones were marked with blue (3/3, 4/4, and 5/5), purple (2/3, 3/4, and 4/5), and cyan (2/4 and 3/5). Black bars represented the six transmembrane regions, and red bars represented the three domains of the phosphatase motif. Asterisks represented the conserved amino acid residues.
Gene structure and conserved motif composition of BnLPPs gene family
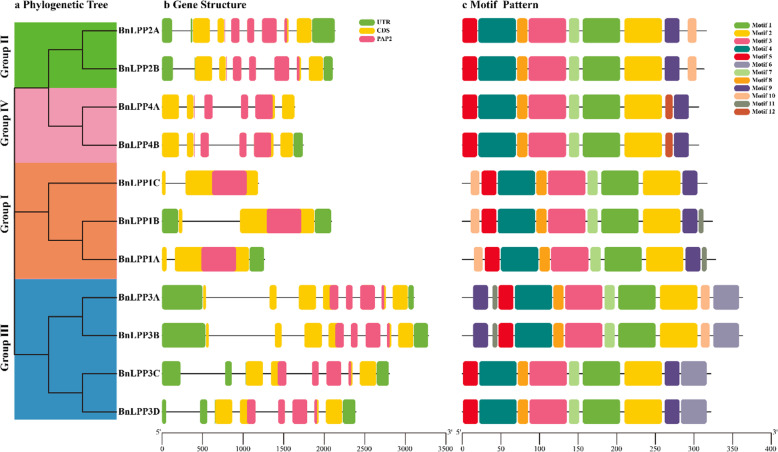
The exon-intron configurations of BnLPPs genes were examined to acquire further insights into the probable structural evolution of BnLPP family genes. Our results display that the number of exons of BnLPPs ranged from 2 to 8 (Table 1; Fig. 3). We also found that similar structures usually exist in the same group, e.g., the group I members have one intron and two exons. Likewise, groups II, III, and IV contained three or four introns in their respective PAP2 domains except for group I (Fig. 3a and b). Mainly, groups II and IV had a diverse intron/exon association pattern. These results showed that members within a group had a similar intron/exon pattern, consistent with the clusters of BnLPPs.
Fig. 3.
Phylogenetic relationships, gene structure, and architecture of conserved protein motifs in BnLPPs. a A phylogenetic tree based on the BnLPPs sequences. According to phylogenetic relationships, 11 BnLPPs were clustered into four groups (I-IV) and represented with different colors. b The exon-intron structure of BnLPPs. Green boxes indicate UTR regions, yellow boxes indicate exons, blackish-grey lines indicate introns, and pink boxes indicate PAP2 domain. c The motif composition of BnLPPs. Different colored boxes display different motifs. The details of each motif were presented in Additional file 3. The bottom scale shows the protein length
Furthermore, we investigated the full-length protein sequences of 11 BnLPPs to recognize their conserved motifs. Generally, 12 conserved motifs were identified, and motifs 1, 2, 3, 4, 5, 7, and 8 were found to be widely distributed. Interestingly, BnLPPs in the same group tends to have similar motif composition (Fig. 3c). For example, motif 12 was specific to group IV, while motif 6 was specific to group III (Fig. 3a). The similar motif arrangements in subgroups indicated the protein structure was conserved within a specific subfamily. Overall, the results reveal that members inside a group had identical gene structures, constant with their phylogenetic relationships. The group classifications’ stability was convincingly maintained by studying conserved motif compositions, gene structures, and phylogenetic relationships, showing that BnLPP proteins have very conserved amino acid residues, and members within the group may have analogous functions.
Chromosomal distribution and synteny analysis of BnLPP genes
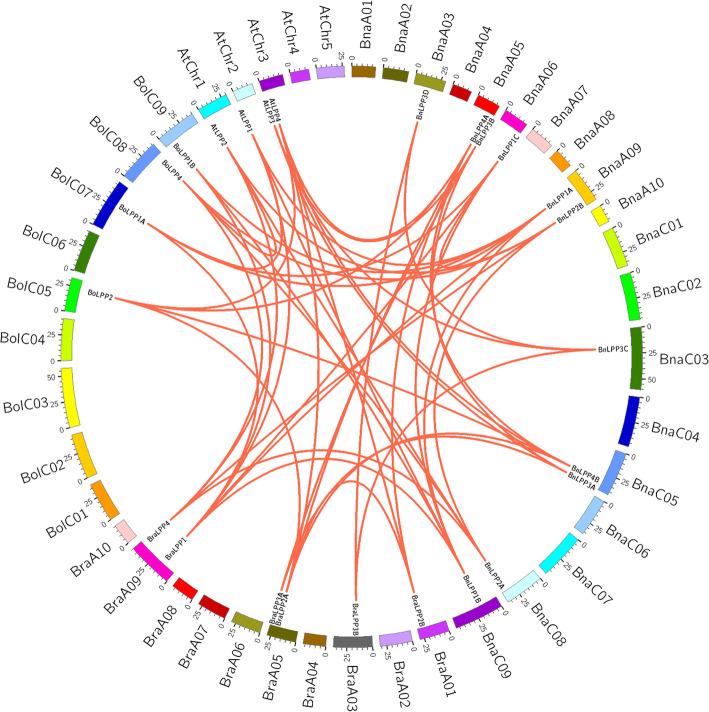
The expansion of new gene family members in plant genome evolution is partly attributed to tandem and segmental duplication [21], and the corresponding events were studied in BnLPPs. The chromosomal location of 11 BnLPPs was evaluated, and the result shows that 8 out of the 19 chromosomes had BnLPP genes (Table 1). Briefly, chromosomes A05, A09, and C05 harbored 2 BnLPPs, whereas other chromosomes (A03, A06, C03, C08, and C09) possess only one BnLPP gene (Table 1). However, despite A05 and C05 possess gene clusters (BnLPP4A and BnLPP3B, and BnLPP4B and BnLPP3A), no tandem duplication events were found in these regions (Fig. 4; Additional file 4). Additionally, we also identified 6 and 4 LPPs genes in the B. rapa and B. oleracea genomes, respectively (Additional file 2). Our findings show that these genes were similar to those in the A and C sub-genomes of B. napus.
Fig. 4.
Synteny analysis of LPPs in A. thaliana, B. rapa, B. olerecea, and B. napus. The red lines represented the syntenic LPP pairs between the two genomes. The chromosome number was shown at the bottom of each chromosome
Collinearity analysis revealed orthologs (speciation events) among the B. napus, B. rapa, B. oleracea, and A. thaliana LPP genes (Fig. 4). There was a tripling in Brassica species after diversion from their common ancestor with (A) thaliana [21]. Therefore, one AtLPP should theoretically correspond to three orthologs in (B) rapa and B. oleracea. However, more than one homologous gene of AtLPP1, AtLPP2, and AtLPP4 in B. rapa and B. oleracea and two-four homologous genes in the B. napus genomes (in both A and C subgenome) have been predicted in different groups (Fig. 2; Additional file 4). Interestingly, AtLPP3 has no homologous genes in B. oleracea, but four homologous genes in B. napus, located on A- (2) and C-subgenome (2) (Additional file 4). The synteny between BraLPPs, BoLPPs, and AtLPPs homologs genes was less than expected (4:6:4), indicating that duplicated genes might have been lost during evolution. Additionally, all BnLPPs genes were found to be associated with twelve and eight syntenic gene pairs, particularly between B. rapa and B. oleracea LPP genes. These results indicate that allotetraploidy was the main force for the rapid expansion of the LPP gene family in B. napus. Moreover, all LPP genes were obtained by whole-genome duplication (WGD; polyploidy) and segmental duplication events, and there was no putative tandem duplication. Overall, our results indicate that the LPP gene family’s expansion in the B. napus genome was mainly due to WGD and segmental duplication.
The ratio of Ka and Ks is an important index to evaluate repeated events’ positive selection pressure [21, 22]. The Ka/Ks of duplication BnLPPs varied from 0.0707 to 0.1712, and the mean value was 0.1012. All the duplicated BnLPPs gene pairs had the Ka/Ks values were less than 1 (Additional file 5), suggesting a strong purifying selective pressure occurred during the evolution of BnLPPs.
Cis-Elements in the promoters of BnLPPs
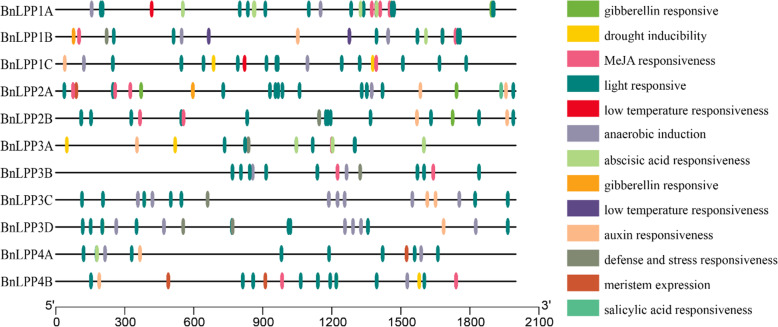
In order to explore gene function and regulation patterns, we studied the cis-elements in the region of 2000 bp upstream of the initiation codon of each BnLPPs. Our results revealed three major classes of cis-elements, i.e., stress-, hormone-, and light-responsive elements. Overall, 13 putative cis-elements were predicted in the BnLPPs promoter (Fig. 5). Among them, six hormone-responsive [(abscisic acid (ABA), auxin, methyl jasmonate (MeJA), gibberellin (GA), and salicylic acid (SA)], and remaining were associated with drought stress, low-temperature stress, defense, anaerobic induction, and meristem expression (Fig. 5). Relatively more light-responsive cis-elements were observed in the BnLPPs promoters (Additional file 6). As shown in Fig. 5, most of the hormone- and stress-responsive elements were specific to some genes highlighting their crucial role in hormone and stress response mechanisms.
Fig. 5.
Cis-elements that are related to different stress and hormone responses in the putative promoters of BnLPPs. Cis-elements with similar functions were displayed in the same color. Different color boxes show different identified cis-elements
Functional annotation analysis of BnLPP genes
To further discriminate the BnLPP genes’ functions, we implemented gene ontology (GO) annotation and enrichment analysis based on three classes, i.e., biological process (BP), molecular function (MF), and cellular component (CC). These GO terms boost our understanding of the precise gene functions. The GO annotation outcomes revealed numerous significantly enriched terms (Additional file 7). For instance, the GO-BP enrichment results revealed seven enriched terms, including cellular process (GO:0009987), phosphorus metabolic process (GO:0006793), phosphate-containing compound metabolic process (GO:0006796), dephosphorylation (GO:0016311), etc. (Additional file 7). The GO-CC enrichment outcomes discovered 13 enriched terms such as obsolete membrane part (GO:0044425), cell periphery (GO:0071944), an integral component of membrane (GO:0016021), obsolete plasma membrane part (GO:0044459), etc. (Additional file 7). Nearly all GO-CC terms are consistent with the subcellular localization of the BnLPP proteins. Likewise, GO-MF enrichment findings exposed eight enriched terms, including phosphatidate phosphatase activity (GO:0008195), phosphoric ester hydrolase activity (GO:0042578), phosphatase activity (GO:0016791), catalytic activity (GO:0003824), etc. (Additional file 7). In short, GO enrichment outcomes validate the functional role of BnLPP genes in numerous biological, cellular, and molecular processes that were associated with phosphatase activity, hydrolase activity, membrane parts, phosphorus metabolic process, and dephosphorylation.
Genome-wide analysis of miRNA targeting BnLPP genes
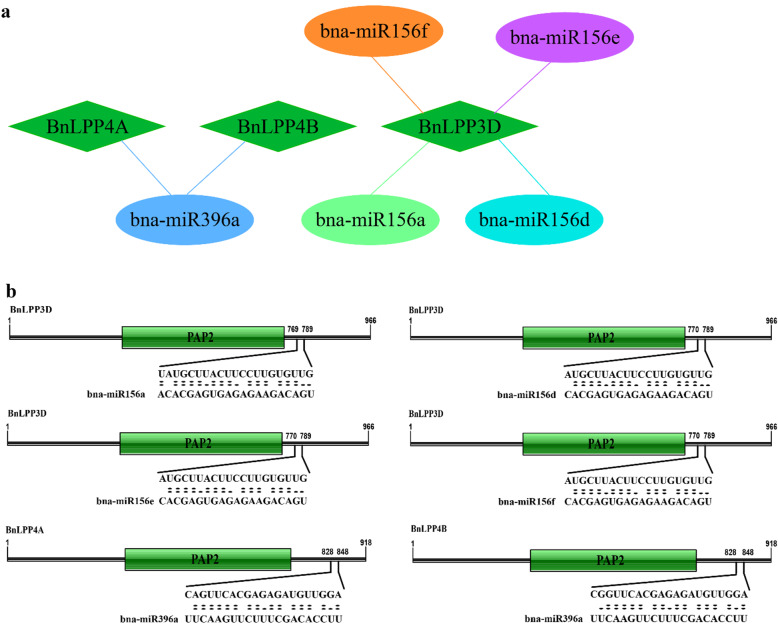
In recent years, numerous researchers have discovered that microRNA (miRNA)-mediated regulation is accompanying plants’ stress responses. Thus, to increase our knowledge of miRNAs connected with BnLPP gene regulation, we identified six putative miRNAs targeting three BnLPP genes (Fig. 6a). Prediction of miRNAs target sites is illustrated in Fig. 6b and Additional file 8. Our results showed that four members of the bna-miR156 family targeted one gene (BnLPP3D), and two members of the bna-miR396 family targeted two genes (BnLPP4A and BnLPP4B) (Fig. 6; Additional file 8). Predominantly, BnLPP3D was targeted by four miRNAs (bna-miR156a, bna-miR156d, bna-miR156e, and bna-miR156f).
Fig. 6.
a A network diagram of the regulatory relationships between the presumed miRNAs and particular BnLPP genes. b Prediction of miRNA target sites. Thick black lines represent the CDS of BnLPPs. PAP2 domain is shown in green boxes. miRNA complementary sites (black filling) with the nucleotide positions of BnLPPs are specified
Expression patterns of BnLPPs in different tissues
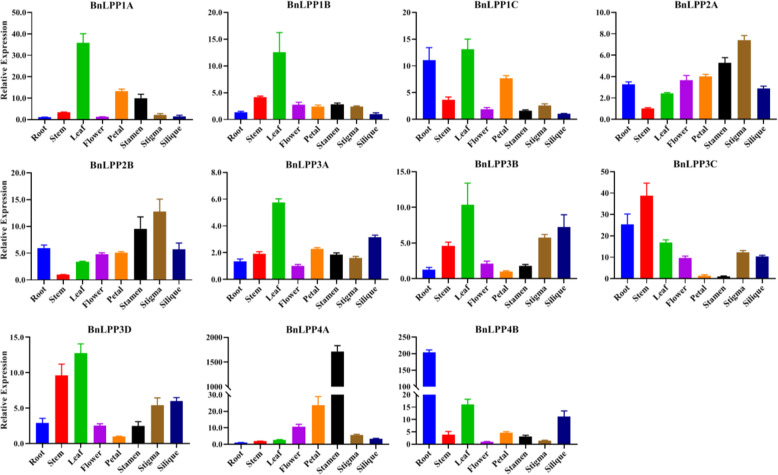
To demonstrate the expression patterns of the BnLPP genes, we examined eight tissues/organs (roots, stem, leaves, flower, petal, stamen, stigma, and silique) of rapeseed at various growth phases by qRT-PCR (Fig. 7). The expression profiles of BnLPP genes varied in the various tissues/organs. For instance, the expression of 6 genes (BnLPP1A, BnLPP1B, BnLPP1C, BnLPP3A, BnLPP3B, and BnLPP3D) were higher in leaves compared to other tissues (Fig. 7). The expression levels of BnLPP2A and BnLPP2B were higher in stigma. The expression of BnLPP4A was higher in stamens. The expression level of BnLPP4B was higher in roots, and BnLPP3C showed higher expression in stems. Our results advise that these candidate genes may play dynamic roles in advancing B. napus developmental processes.
Fig. 7.
BnLPPs expression profilesin differenttissues. The data were normalized toβ-actin,and the standard deviation was represented by vertical bars
Expression patterns of BnLPP genes under different stress and phytohormone treatments
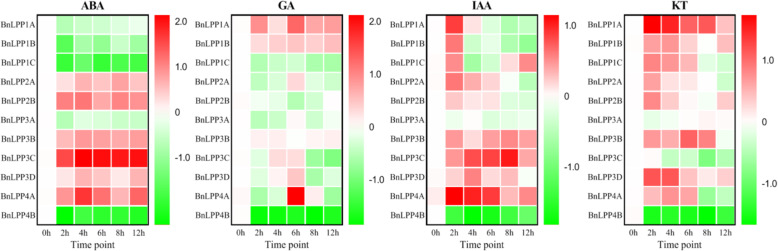
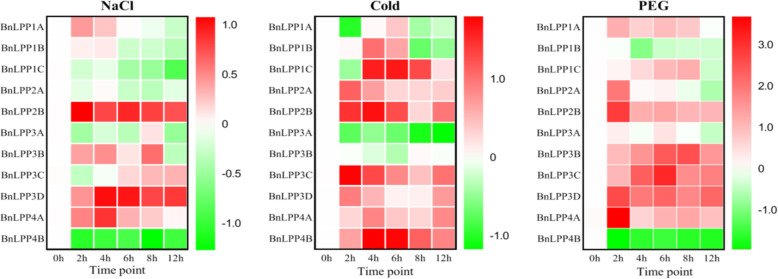
Under stress conditions, B. napus growth and developmental process greatly influenced physiological, biochemical, and molecular mechanisms. Hence, qRT-PCR was used to examine 11 BnLPP genes’ expression patterns at different time points after PEG, NaCl, and cold (Fig. 8), ABA, GA, IAA, and KT treatments (Fig. 9). In general, the expression of some BnLPPs was significantly induced by various treatments. For example, BnLPP1A was significantly responsive to NaCl, GA, IAA, and KT treatments. All treatments except GA induced the expression of BnLPP2B. Meantime, most BnLPPs (BnLPP1A/2B/4A/3B/3C/3D) were induced by one specific treatment. For instance, six BnLPPs (BnLPP1A/2B/4A/3B/3C/3D) were significantly up-regulated by NaCl treatment, and nine genes (BnLPP1B/1C/2A/2B/4A/4B/3B/3C/3D) were up-regulated by cold stress. Salt stress decreased the expression levels of BnLPP1C and BnLPP4B. Several genes indicated opposite expression patternsunder different hormone conditions. For instance, BnLPP1C was significantly increased by IAA or KT and inhibited by ABA or GA treatments. In short, results suggest that these genes may be vital for enlightening tolerance to numerous stresses, i.e., hormone and abiotic stress conditions.
Fig. 8.
Expression profiles of 11 BnLPPs under abiotic stress.The datawerenormalized to β-actin. The heatmap showed the change ratio of the BnLPPsexpression levels under NaCl, cold,and PEG stress. The red color displays high, and the green color displays low expression levels
Fig. 9.
Expression profiles of 11 BnLPP genes in response to different hormones. The data were normalized to the expression of β-actin. The heatmap showed the change ratio of the BnLPPs expression levels under ABA, GA, IAA, and KT. The red color displays high, and the green color displays low expression levels
Discussion
Rapeseed is an allotetraploid that practiced widespread genome repetition and integration events [23]. Nevertheless, rapeseed yield is affected by numerous environmental cues [19]. LPP plays a vital role in catalyzing phosphorous lipids’ dephosphorylation, which plays an active part in several physiological processes, including cell migration, proliferation, and differentiation [3, 4]. So far, only a few numbers of LPP genes have been recognized in plants. A genome-wide analysis is a crucial method for clarifying the biological functions of the LPP family associates in a particular plant species. Over the past few years, LPP family genes have been examined in different plant species, such as A. thaliana [13–15], barley [16], cowpea [17], and tobacco [18]. So far, there is no comprehensive study of the LPP gene family in rapeseed. The accessibility of the whole rapeseed genome permits the genome-wide characterization of the LPP family genes, which may further be used for rapeseed improvement.
In the present study, a total of 11 BnLPP genes (six on A- and five on C-subgenome) were identified in the rapeseed genome (Table 1), which surpasses the number of genes identified from closely related species (4 BoLPPs, 6 BraLPPs, and 4 AtLPPs genes) (Additional file 2). This may be due to the rapeseed genome duplication (A and C subgenomes). Notably, AtLPP2 and AtLPP4 had only one corresponding B. rapa and B. oleracea orthologous gene. However, AtLPP3 had no orthologous in B. oleracea, and two AtLPP3 orthologous were determined in B. rapa. Further, the B. oleracea was compared with its sister specie, i.e., B. rapa to reveal numerous chromosome rearrangements and asymmetrical gene loss in duplicated genomic blocks. The results show that the relative rates of gene loss/retention were consistent with asymmetrical evolution between B. oleracea, B. rapa, and (A) thaliana genomes. This evidence supports the role of asymmetrical evolution in the evolutionary success of polyploidy as well as their phenotypic novelty and adaptation [24, 25]. Further, the results indicated that most duplicated LPP genes were lost after Brassica genome triplication, which was more than 35 % of lost genes in the Brassica lineage via a deletion mechanism [24]. There might be a possibility that functionally redundant LPP copies were lost after genome triplication, while some important copies remained. Theoretically, there should be ten LPP genes in (B) napus, which was originated through hybridizing between B. oleracea and B. rapa.
According to phylogenetic analysis, 11 BnLPPs were classified into four groups. Thus, LPP genes from (A) thaliana were classified into all four groups together with BnLPPs, BraLPPs, and BoLPPs genes (Fig. 2), suggesting the comparable evolutionary events between four closely related species. According to Fig. 2, several BnLPPs genes were closely associated with BraLPPs, and BoLPPs, indicating that these genes might have originated by segmental duplication [25]. These arrangements in the phylogenetic tree were further established by the gene structure analysis (Fig. 3). Furthermore, the motifs designs were also similar within the group organization (Fig. 3). Particularly, group IV possesses diverse gene structures and motifs designs, signifying that BnLPPs have very conserved protein structures. The comparative analysis of LPPs between (B) napus and its parents revealed that the B. napus genome is not a simple sum of B. oleracea and B. rapa. However, it undergoes many losses and duplications after hybridization, which benefited natural selection and crop domestication [23].
Almost all BnLPPs were predicted to be localized in the plasma membrane except BnLPP1C, specifically localized in the endoplasmic reticulum. These outcomes are consistent with previous studies that all PAP must bind to the membrane as a phospholipid substrate location [26]. Further, these results were also supported by the GO enrichment analysis (Additional file 7). Based on similarities among protein sequences, BnLPPs were classified into four groups. Although the functional studies on plant’s LPPs are still lacking; thus, their function could be deduced through expression patterns. In B. napus, the BnLPPs family members can be expressed in all kinds of plant tissues, indicating that LPP is an important enzyme for plant development. However, the expression profiles of BnLPP genes varied in the various tissues. For example, the expression of six BnLPPs was higher in leaves than in other tissues (Fig. 7), which is consistent with the expression level of AtLPP1 [13–15]. Notably, some genes showed higher expression levels in leaves, stigma, stem, roots, and stamens, suggesting that BnLPPs might play tissue-specific roles in the rapeseed development.
Analysis of the cis-acting elements indicated that BnLPPs might respond to different stress and hormone signaling (Fig. 5). Therefore, the expression patterns of BnLPPs were evaluated under drought, salinity, and cold stress. Under drought stress, nine genes were up-regulated, eight genes were up-regulated under cold stress, and six genes showed higher expression against salinity. Interestingly, a few same genes also showed higher expression under different stress conditions, e.g., BnLPP3D and BnLPP3D. On the contrary, BnLPP4B was down-regulated under drought and salinity but was significantly increased under cold stress. In a recent study, AtLPP1 was induced under various stresses, including DNA damage, G-protein activation, and oxidative stress, while the AtLPP2 gene was constitutively expressed under the same stress conditions [13]. In our study, BnLPP2A and BnLPP2B, homologous to AtLPP2, were induced under drought, suggesting that BnLPPs gain new functions during evolution.
Likewise, the cis-elements analysis showed that BnLPP genes are involved in responding to various phytohormones [Fig. 5]. Phytohormones play significant roles in regulating stress responses in plants [27]. Our results confirmed that there might be cross-talk between the hormone signaling transduction and BnLPPs. Therefore, we analyzed the expression profiles of 11 BnLPPs under ABA, IAA, GA, and KT treatment. All the BnLPPs responded to ABA treatment, revealing that BnLPPs were widely involved in ABA’s signaling pathways. Hormone treatments induced the expression of BnLPP1A and BnLPP3D. On the other hand, the expression of BnLPP1A and BnLPP3D were not influenced by hormones. Our results showed that the functional gene diversification occurred during gene duplication, and the homologous genes might have different functions. Particularly, the expression level of BnLPP4B was decreased under hormones, and the other genes (homologous to BnLPP4B) showed opposite expression trends. The expression of AtLPP2 was decreased by ABA treatment, which participates in regulating seed germination and stomatal movements [14, 15]. In the current study, the expression of both BnLPP2A and BnLPP2B was increased by ABA, suggesting that AtLPP2 and BnLPP2A/2B might have different functions ABA signaling pathways.
miRNAs, a group of single-stranded non-coding micro RNAs, have been involved in post-transcriptional gene regulation [28, 29]. Recently, numerous miRNAs have been recognized by genome-wide examination in rapeseed that corresponds to diverse environmental cues [30–33]. Hence, we identified six putative miRNAs (belonging to two families, i.e., bna-miR156 and bna-miR396) targeting three BnLPP genes (Fig. 6). Interestingly, both miRNAs families respond to different stress conditions and developmental tissues in numerous plant species [34–37]. To our best knowledge, for the first time, we predicted that numerous miRNAs target LPP genes. These predictions open new windows for future investigations at a post-transcriptional level.
Conclusions
In the present study, we identified 11 BnLPPs, four BoLPPs, six BraLPPs, and four AtLPPs genes via genome-wide analysis. Based on similarities, these genes were clustered into four groups (Group I-Group IV). For better understandings, chromosome distribution, gene duplication, gene structure, conserved motifs, cis-elements, GO annotation, and miRNA prediction analysis have also been performed. Furthermore, the expression pattern in different tissues and under diverse abiotic stress and phytohormone stimuli. The results revealed that the loss and duplication events occurred during the expansion of BnLPPs family genes. Besides, BnLPPs showed differential tissue-specific, abiotic stress, and hormone-responsive expression patterns. The abiotic stress and hormone-related cis-elements were also identified in BnLPPs, appropriately explaining the induction of the BnLPPs by stress and hormone. For the first time, we have predicted that two miRNAs families were targeting the three BnLPP genes. In conclusion, the extensive data collected in the present study can be used for upcoming functional analysis of BnLPP genes in rapeseed growth, development, response to hormone and abiotic stresses.
Methods
Identification of LPP genes in rapeseed
To identify potential LPPs in rapeseed, the genome sequences of B. napus were obtained from the Genoscope database (https://www.genoscope.cns.fr/brassicanapus/), and a local database was constructed using NCBI-blast-2.7.1 + software. BlastP search was performed (e-value 1e-5) using four LPP amino acid sequences of Arabidopsis thaliana [AtLPP1 (At2g01180), AtLPP2 (At1g15080), AtLPP3 (At3g02600), and AtLPP4 (At3g18220)] as a query as described previously [13]. Meanwhile, HMMER 3.0 [38] was used to search LPPs with the PAP2 domain (PF01569) obtained from the Pfam database [39], and the value of cutoff was 0.01. Pfam protein database (http://pfam.xfam.org/) further confirmed the PAP2 domain in candidate genes, and the redundant putative LPPs were excluded manually. Eleven BnLPP genes were finally identified in the rapeseed genome. The information of sequence lengths, molecular weights, isoelectric points, and predicted subcellular location was obtained from the ExPasy website (http://web.expasy.org/protparam/) [40].
In addition, to reveal the evolutionary relationships of LPP genes in different plant species, potential LPP genes from two plant species (B. oleracea and B. rapa) were also identified using the same method as described above. The genome sequences of the B. oleracea and B. rapa were downloaded from the JGI Phytozome v12.1 [41] database.
Sequence analysis and structural characterization
Subcellular localization of BnLPP proteins was predicted using the WoLF PSORT server [42]. Multiple Expectation Maximization for Motif Elicitation (MEME) was used to analyze the conserved B. napus and A. thaliana protein sequences [43]. For conserved motifs discovery, the maximum number of motifs was set to 12, the classic mode and others default parameters were selected. Tbtools (V 1.068) software was used to display the BnLPPs gene structures [44].
Phylogenetic analysis
MapChart V2.1 [45] was used to obtain the gene chromosomal locations. Multiple Collinearity Scan toolkit (MCScanX) [46] was used to detect gene duplication events. The syntenic relationship of BnLPPs with other analyzed genomes was displayed by the Circos [47] and JCVI tools [48]. KaKs_Calculator 2.0 [49] was used to calculate non-synonymous (ka) and synonymous (ks) substitution of all the duplicated BnLPPs.
To investigate the phylogenetic relationships of BnLPP genes, a total of 25 LPP proteins with PAP2 domains were identified from four plant species, and their multiple sequence alignment was carried out by ClustalW [50]. MEGA X was used to construct a phylogenetic tree via the neighbor-joining method with 1000 bootstrap replicates [51]. For a better image, the tree was displayed via EvoVIEW 2.0 [52].
Promoter sequence analysis
The sequences of 2 Kb upstream from the start codon were downloaded from Genoscope and defined as the promoter of each BnLPPs. Then, PlantCARE [53] was used to predict cis-elements, and displayed using Tbtools [44].
Prediction of putative miRNA targeting BnLPP genes and functional annotation analysis
In the current study, the BnLPPs CDS sequences were used to predict targeted miRNAs using the psRNATarget server (http://plantgrn.noble.org/psRNATarget/home) with default parameters, except maximum expectation (E) = 5.0. We selected the targeted sites with high degrees of complementarity shown as shown in Fig. 6. Cytoscape (V3.8.2, https://cytoscape.org/download.html) software was used to create the interaction network between the prophesied miRNAs and the equivalent target BnLPP genes.
For functional annotation analysis, the BnLPP protein sequences were submitted to the eggNOG website (http://eggnog-mapper.embl.de/) for GO annotation with the set “Orthology restrictions” as “Transfer annotations from one-to-one orthology only”, and other default parameters.
Plant materials and treatments
In the present study, Westar, a rapeseed variety, was provided by the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences (CAAS), China, and was used for stress treatments. Healthy seeds were selected and were sterilized with a 10 % hypochlorous acid solution for 5 min. The seeds were germinated in a chamber (25 ℃, day/night, 16 h/8 h light/dark cycle) until the radicle’s length was reached 5 mm. These germinated seeds were used for the stress treatments. For salinity and drought treatments, the seeds were subjected to 150 mM NaCl and 15 % PEG-6000 solution on water-saturated filter paper, respectively. For cold stress, the seeds were subjected to 4 ℃ on water-saturated filter paper. For phytohormone treatment, the seeds were cultured in Murashige and Skoog (MS) nutrient solution with 100 µM abscisic acid (ABA), gibberellin (GA), kinetin (KT), and indole-3-acetic acid (IAA). The samples were harvested after 0 (CK), 2, 4, 6, 8, and 12 h of the treatments and were stored at − 80 ℃ for subsequent analysis.
The seeds were germinated as described above. They were then transplanted into nutritious soil in plant growth chambers at a temperature of 22 °C, a light intensity of 3,000 lx, and a photoperiod of 16 h light/8 h dark. Different tissues such as root, stem, leaf, flower, petal, stamen, stigma, and silique were harvested from the plants. All samples were frozen in liquid nitrogen and stored at − 80 °C until total RNA extraction and qRT-PCR analysis.
RNA isolation and quantitative real-time PCR
RNA extraction and the cDNA synthesis were carried out using TransZol Up Plus RNA and cDNA Synthesis SuperMix (TransGen Biotech, China) according to manufacturer instructions, respectively. The qRT-PCR was carried out in ABI StepOne (Applied Biosystems, America) using the SYBR Green Supermix. β-actin was used as an internal control, and all the primers used in the present study are listed in Additional file 1. The qRT-PCR procedure was performed as follows: 94 °C for 10 min, followed by 40 cycles of 94 °C for 15 s, 60 °C for 30 s, 72 °C for 10 s. Each qRT-PCR reaction was carried out with three biological triplicates, and the data were examined using the 2−△△CT method as described previously [54, 55]. For tissue-specific expression profiling, we followed the same method as described earlier.
Supplementary Information
Additional file 1: Table S1. Sequences of the primers used in this study.
Additional file 2: Table S2. The protein sequences of the LPP gene family in (A) thaliana, (B) oleracea, B. rapa, and B. napus.
Additional file 3: Table S3. Analysis and distribution of conserved motifs in BnLPP proteins.
Additional file 4: Table S4. Segmentally and tandemly duplication and one-to-one orthologues relationships between B. napus, B. rapa, B. oleracea, and A. thaliana.
Additional file 5: Table S5. Ka/Ks analysis of BnLPP gene pairs.
Additional file 6: Table S6.Cis-elements in the promoters of putative BnLPP genes.
Additional file 7: Table S7. The GO enrichment analysis of BnLPP genes.
Additional file 8: Table S8. The prediction of miRNAs target sites.
Acknowledgements
The authors are grateful to all lab members for their useful suggestions, support, and encouragement. Further, we would like to thank the anonymous reviewers for their constructive comments, valuable recommendations, and feedback, which facilitated advance the work. Further, WS and AR would like say thanks to CAAS for providing PhD Scholarhip and the research environment.
Abbreviations
- LPPs
Lipid phosphate phosphatases
- PAP
Phosphatidic acid phosphatase
- ABA
Abscisic acid
- GA
Gibberellin
- IAA
Indole-3-acetic acid
- KT
Kinetin
- GO
Gene ontology
Authors’ contributions
WS, LZ, and XZ initiated and designed the experiment. WS, AR, LZ, AG, XD, YC, and XZ performed the experiments and analyzed the data. WS, AR, and XZ analyzed the data and wrote the manuscript. YC, YL, and XZ proofread and revised the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program (2017YFD0101700), the Agriculture Science and Technology Innovation Program of CAAS, and the Hubei Agriculture Science and Technology Innovation Center.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. However, most of the data is available in additional files. The sequences of Brassica napus, Brassica oleracea, and Brassica rapa are available in the GENOSCOPE database (https://wwwdev.genoscope.cns.fr/brassicanapus/data/) and Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html##).
Declarations
Ethics approval and consent to participate
Not applicable. This study has not directly involved humans and animals. We comply with the Convention on the Trade in Endangered Species of Wild Fauna and Flora. Further, we also have appropriate permissions from the Oil Crops Research Institute, Chinese Academy of Agricultural Sciences (CAAS), Wuhan, China, to use the Westar (a rapeseed cultivar) seed specimens in the present study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wei Su and Ali Raza equally contributed to this work.
References
- 1.Kennedy EP. The Biosynthesis of Phospholipids. Am J Clin Nutr. 1958;6(3):216–220. doi: 10.1093/ajcn/6.3.216. [DOI] [PubMed] [Google Scholar]
- 2.Arisz SA, Testerink C, Munnik T. Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta. 2009;1791(9):869–75. [DOI] [PubMed]
- 3.Munnik T, Testerink C. Plant phospholipid signaling: “in a nutshell.” J Lipid Res. 2009;50(Supplement):S260–S265. [DOI] [PMC free article] [PubMed]
- 4.Carman GM, Han G-S. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31(12):694–9. [DOI] [PMC free article] [PubMed]
- 5.Carman GM. Discoveries of the phosphatidate phosphatase genes in yeast published in the Journal of Biological Chemistry. J Biol Chem. 2019;294(5):1681–1689. doi: 10.1074/jbc.TM118.004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chae M, Han G-S, Carman GM. The Saccharomyces cerevisiae Actin Patch Protein App1p Is a Phosphatidate Phosphatase Enzyme. J Biol Chem. 2012;287(48):40186–40196. doi: 10.1074/jbc.M112.421776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu W-I, Liu Y, Riedel B, Wissing JB, Fischl AS, Carman GM. Purification and Characterization of Diacylglycerol Pyrophosphate Phosphatase from Saccharomyces cerevisiae. J Biol Chem. 1996;271(4):1868–1876. doi: 10.1074/jbc.271.4.1868. [DOI] [PubMed] [Google Scholar]
- 8.Toke DA, Bennett WL, Oshiro J, Wu W-I, Voelker DR, Carman GM. Isolation and Characterization of the Saccharomyces cerevisiae LPP1 Gene Encoding a Mg2+-independent Phosphatidate Phosphatase. J Biol Chem. 1998;273(23):14331–14338. doi: 10.1074/jbc.273.23.14331. [DOI] [PubMed] [Google Scholar]
- 9.Han G-S, Johnston CN, Chen X, Athenstaedt K, Daum G, Carman GM. Regulation of the Saccharomyces cerevisiae DPP1-encoded Diacylglycerol Pyrophosphate Phosphatase by Zinc. J Biol Chem. 2001;276(13):10126–10133. doi: 10.1074/jbc.M011421200. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura Y, Tsuchiya M, Ohta H. Plastidic Phosphatidic Acid Phosphatases Identified in a Distinct Subfamily of Lipid Phosphate Phosphatases with Prokaryotic Origin. J Biol Chem. 2007;282(39):29013–29021. doi: 10.1074/jbc.M704385200. [DOI] [PubMed] [Google Scholar]
- 11.Kooijman EE, Testerink C. Phosphatidic Acid: An Electrostatic/Hydrogen-Bond Switch? In: Lipid Signaling in Plants. Edited by Munnik T. Berlin: Springer; 2010:203–22. 10.1007/978-3-642-03873-0_14.
- 12.Sadat MA, Jeon J, Mir AA, Choi J, Choi J, Lee Y-H. Regulation of Cellular Diacylglycerol through Lipid Phosphate Phosphatases Is Required for Pathogenesis of the Rice Blast Fungus, Magnaporthe oryzae. PLoS One. 2014;9(6):e100726. doi: 10.1371/journal.pone.0100726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierrugues O, Brutesco C, Oshiro J, Gouy M, Deveaux Y, Carman GM, Thuriaux P, Kazmaier M. Lipid Phosphate Phosphatases in Arabidopsis: regulation of the atlpp1 gene in response to stress. J Biol Chem. 2001;276(23):20300–20308. doi: 10.1074/jbc.M009726200. [DOI] [PubMed] [Google Scholar]
- 14.Paradis S, Villasuso AL, Aguayo SS, Maldiney R, Habricot Y, Zalejski C, Machado E, Sotta B, Miginiac E, Jeannette E. Arabidopsis thaliana lipid phosphate phosphatase 2 is involved in abscisic acid signalling in leaves. Plant Physiol Biochem. 2011;49(3):357–362. doi: 10.1016/j.plaphy.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Katagiri T, Ishiyama K, Kato T, Tabata S, Kobayashi M, Shinozaki K. An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. Plant J. 2005;43(1):107–117. doi: 10.1111/j.1365-313X.2005.02431.x. [DOI] [PubMed] [Google Scholar]
- 16.Barrero JM, Talbot MJ, White RG, Jacobsen JV, Gubler F. Anatomical and Transcriptomic Studies of the Coleorhiza Reveal the Importance of This Tissue in Regulating Dormancy in Barley. Plant Physiol. 2009;150(2):1006–1021. doi: 10.1104/pp.109.137901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.França MG, Matos AR, D’Arcy-Lameta A, Passaquet C, Lichtlé C, Zuily-Fodil Y, Pham-Thi AT. Cloning and characterization of drought-stimulated phosphatidic acid phosphatase genes from Vigna unguiculata. Plant Physiol Biochem. 2008;46(12):1093–1100. doi: 10.1016/j.plaphy.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Pleskot R, Pejchar P, Bezvoda R, Lichtscheidl IK, Wolters-Arts M, Marc J, Zárský V, Potocký M. Turnover of Phosphatidic Acid through Distinct Signaling Pathways Affects Multiple Aspects of Pollen Tube Growth in Tobacco. Front Plant Sci. 2012;3:54–54. doi: 10.3389/fpls.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raza A. Eco-physiological and Biochemical Responses of Rapeseed (Brassica napus L.) to Abiotic Stresses: Consequences and Mitigation Strategies. J Plant Growth Regul. 2020. 10.1007/s00344-020-10231-z.
- 20.Toke DA, McClintick ML, Carman GM. Mutagenesis of the phosphatase sequence motif in diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. Biochemistry. 1999;38(44):14606–13. [DOI] [PubMed]
- 21.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology. 2004;4(1):10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genetics. 2002;18(9):486. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 23.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 24.Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, et al. Comparative Genomics of Brassica oleracea and Arabidopsis thaliana Reveal Gene Loss, Fragmentation, and Dispersal after Polyploidy. Plant Cell. 2006;18(6):1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Liu Y, Yang X, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kok BPC, Venkatraman G, Capatos D, Brindley DN. Unlike Two Peas in a Pod: Lipid Phosphate Phosphatases and Phosphatidate Phosphatases. Chem Rev. 2012;112(10):5121–5146. doi: 10.1021/cr200433m. [DOI] [PubMed] [Google Scholar]
- 27.Wani SH, Kumar V, Shriram V, Sah SK. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016;4(3):162–176. doi: 10.1016/j.cj.2016.01.010. [DOI] [Google Scholar]
- 28.Cui C, Wang JJ, Zhao JH, Fang YY, He XF, Guo HS, Duan CG. A Brassica miRNA Regulates Plant Growth and Immunity through Distinct Modes of Action. Mol Plant. 2020;13:231–245. doi: 10.1016/j.molp.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Liu D, Zhang X, Chen D, Cheng Y, Shen F. Plant microRNAs in cross-kingdom regulation of gene expression. Int J Mol Sci. 2018;19:2007. doi: 10.3390/ijms19072007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Chen L, Zhang X, Liu T, Niu S, Wen J, Yi B, Ma C, Tu J, Fu T. Identification of miRNAs that regulate silique development in Brassica napus. Plant Sci. 2018;269:106–117. doi: 10.1016/j.plantsci.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Fu Y, Mason AS, Zhang Y, Lin B, Xiao M, Fu D, Yu H. MicroRNA-mRNA expression profiles and their potential role in cadmium stress response in Brassica napus. BMC Plant Biol. 2019;19:1–20. doi: 10.1186/s12870-018-1600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Körbes AP, Machado RD, Guzman F, Almerao MP, de Oliveira LFV, Loss-Morais G, Turchetto-Zolet AC, Cagliari A, dos Santos Maraschin F, Margis-Pinheiro M. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PloS One. 2012;7:e50663. doi: 10.1371/journal.pone.0050663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen E, Zou J, Hubertus Behrens F, Chen L, Ye C, Dai S, Li R, Ni M, Jiang X, Qiu J. Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J Exp Bot. 2015;66:7241–7253. doi: 10.1093/jxb/erv420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao-kang LI, Hong-fang LIU, Jing-ling LIU, Ming ZHENG, Wei HUA. Sequence identification and bioinformatics of bna-miR156 gene family and target genes in rapeseed (Brassica napus L.) Chinese J Oil Crop Sci. 2018;40(2):162. [Google Scholar]
- 35.Ding Y, Ye Y, Jiang Z, Wang Y, Zhu C. MicroRNA390 is involved in cadmium tolerance and accumulation in rice. Front Plant Sci. 2016;7:235. doi: 10.3389/fpls.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Jian H, Wang T, Wei L, Li J, Li C, Liu L. Identification of microRNAs actively involved in fatty acid biosynthesis in developing Brassica napus seeds using high-throughput sequencing. Front Plant Sci. 2016;7:1570. doi: 10.3389/fpls.2016.01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jian H, Wang J, Wang T, Wei L, Li J, Liu L. Identification of rapeseed microRNAs involved in early stage seed germination under salt and drought stresses. Front Plant Sci. 2016;7:658. doi: 10.3389/fpls.2016.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(suppl_2):W29–W37. [DOI] [PMC free article] [PubMed]
- 39.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018;47(D1):D427–D432. [DOI] [PMC free article] [PubMed]
- 40.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(W1):W597–W603. [DOI] [PMC free article] [PubMed]
- 41.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(D1):D1178–D1186. [DOI] [PMC free article] [PubMed]
- 42.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(suppl_2):W585–W587. [DOI] [PMC free article] [PubMed]
- 43.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(suppl_2):W202–W208. [DOI] [PMC free article] [PubMed]
- 44.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Voorrips RE. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed]
- 47.Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320(5875):486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 49.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genomics Proteomics Bioinformatics. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li K-B. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics. 2003;19(12):1585–1586. doi: 10.1093/bioinformatics/btg192. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Z, Zhang H, Gao S, Lercher MJ, Chen WH, Hu S. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44(W1):W236–W241. [DOI] [PMC free article] [PubMed]
- 53.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Chen Y, Fang H, Shi H, Chen K, Zhang Z, Tan X. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol Genet Genomics. 2014;289:1023–1035. doi: 10.1007/s00438-014-0853-1. [DOI] [PubMed] [Google Scholar]
- 55.Raza A, Wei S, Ang G, Mehmood SS, Hussain MA, Nie W, Yan L, Zou X, Zhang X. Catalase (CAT) Gene Family in Rapeseed (Brassica napus L.): Genome-Wide Analysis, Identification, and Expression Pattern in Response to Multiple Hormones and Abiotic Stress Conditions. Int J Mol Sci. 2021;22(8):4281. doi: 10.3390/ijms22084281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequences of the primers used in this study.
Additional file 2: Table S2. The protein sequences of the LPP gene family in (A) thaliana, (B) oleracea, B. rapa, and B. napus.
Additional file 3: Table S3. Analysis and distribution of conserved motifs in BnLPP proteins.
Additional file 4: Table S4. Segmentally and tandemly duplication and one-to-one orthologues relationships between B. napus, B. rapa, B. oleracea, and A. thaliana.
Additional file 5: Table S5. Ka/Ks analysis of BnLPP gene pairs.
Additional file 6: Table S6.Cis-elements in the promoters of putative BnLPP genes.
Additional file 7: Table S7. The GO enrichment analysis of BnLPP genes.
Additional file 8: Table S8. The prediction of miRNAs target sites.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. However, most of the data is available in additional files. The sequences of Brassica napus, Brassica oleracea, and Brassica rapa are available in the GENOSCOPE database (https://wwwdev.genoscope.cns.fr/brassicanapus/data/) and Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html##).