Abstract
Background:
LATS1 (Large Tumor Suppressor, isoform 1) is a gene that forms a complex with the cyclin-dependent kinase, CDK1, and regulates cell cycle progression. Genetic modifications lead to a loss in the activity of LATS1 gene. OSCC is the most commonly emerging cancer caused by genetic as well as epigenetic changes. Epigenetics changes vary from one population to another because these are influenced by dietary factors and environmental factors. Tobacco chewing and smoking has been reported as major risk factors in OSCC. No report was found in the previous literature showing promoter hypermethylation of LATS1 gene.
Methods:
A total of 50 OSCC patients and 20 normal individuals were recruited in this study. Blood samples (50) from OSCC patients and blood samples (20) from healthy individuals as controls were used in the present study. Isolation of genomic DNA was carried out from blood using the standard phenol-chloroform extraction. Further Isolated DNA was modified with sodium bisulfite using the agarose bead method and finally, the methylation studies of LATS1 gene were carried out using Methylation-Specific PCR (MSP-PCR).
Results:
19 out of 50 patients (38.0%) were found to be methylated for LATS1 gene.; a statistically significant result was obtained (p -value= < 0.05) with an odds ratio of 0.37 in cases compared to controls. The status of methylation of LATS1 genes was also found to be statistically significantly associated with smokers and tobacco chewers (p-value = < 0.05). The methylation of LATS1 gene showed a significant risk of developing OSCC in patients.
Conclusion:
These results suggest that the LATS1 gene may provide a better alternative as a diagnostic biomarker. This is the first report on the promoter hypermethylation of LATS1 gene in OSCC patients among the North Indian population.
Key Words: Oral squamous cell carcinoma (OSCC), epigenetic changes, LATS2 gene, promoter hypermethylation
Introduction
Cancer is the most common, vicious, and dangerously increasing diseases of the world today, associated with high morbidity and mortality. Oral Squamous Cell Carcinoma (OSCC) is the sixth most frequent type of cancer observed in the Indian population. Survival rates of patients with advanced OSCC have not increased significantly in recent years (Al-Swiahb et al., 2010). Oral cancer is a blended interaction between Genetic, epigenetic, and environmental factors. Development of OSCC is a multistep process resulting due to chronic inflammation and Genetic factors such as changes in oncogenes and Tumor Suppressor Genes (Hanahan and Weinberg, 2011). Tobacco consumption and smoking is the leading cause of OSCC. The most important etiologic factors did not seem to be the sole cause of cancer. Epigenetic mechanisms could also contribute to the silencing of Tumor-Suppressor Genes. Epigenetics relates to the heritable alterations in Gene expression that do not result from changes in Gene nucleotide sequence. Epigenetic modifications play a crucial role in cell cycle regulation, and differentiation, along with these modifications, inhibit Genes from Genetic mutations (Jha et al., 2016). DNA methylation was the earliest epigenetic changes to be seen in cancer cells. It plays a significant role within the regulation of the Gene expression and chromatin design by providing a stable Gene silencing mechanism (Mahmood and Rabbani, 2019). Promoter Methylation induced Gene silencing leads to cancer initiation and progression. In mammals, DNA methylation is directly linked with the covalent modification of the cysteine residue within the CpG dinucleotides. Methylation regions rich in CpG promoters inhibit DNA transcription by altering the binding of histone complexes (Lu et al., 2006). The DNA methylation markers can be used in cancer diagnostics for both disease classification and detection because epigenetic changes occur very early in the oncogenic process (Ehrlich, 2019). A Tumor Suppressor Genes, when functioning normally, inhibit the growth of Tumors; losses of their regular functions (inactivation) are needed. For Tumorigenesis. Allelic failures, Gene variations, and cryptic deletions are usually recognized as mechanisms required for the inactivation of Tumor Suppressor Genes. Tumor Suppressor Genes can be classified into two groups: Genes that directly control Tumor growth by inhibiting cell proliferation or increasing cell death (“gatekeepers”) and genes whose inactivation causes genetic variability, which in turn leads to mutations that increase neoplasm growth (Hisaoka et al., 2020).
LATS1 (Large Tumor Suppressor Gene) is a Ser/Thr kinase associated with the Tumor Suppressor pathway called the Hippo-LATS/Warts pathway that represses tumor outgrowth by controlling cell proliferation, cell growth, and cell death (Cao et al., 2020). LATS1 Gene is linked with mitotic events in mammalian cells, and that loss of its function interrupts normal cell cycle regulation, leading to the development of Tumors (Chan et al., 2005). In mammalian cells, LATS1 Gene interacting with Cdc2 in the mitotic phase and negatively regulates Cdc2 activity (Hao et al., 2008). LATS1 plays a vital role in controlling cell fate by promoting the degradation of YAP/TAZ proteins by phosphorylation-mediated ubiquitination and regulating a tumor-suppressive transcriptional factor p53, cell cycle checkpoint regulators, and the centrosomal protein phosphatase CDC25B (Nozaki et al., 2019). Overexpression and demethylation of the LATS1 gene inhibited cell proliferation, downregulated YAP expression, induced cell apoptosis, and cell cycle arrest in renal cell carcinoma cells (Williams and Stoeber 2012). LATS1 phosphorylates angiomotin to suppress cytokinesis, cell migration, and angiogenesis by inhibiting F-actin binding and polymerization via negative modulation of LIMK1 (Dai et al., 2013; Yang et al., 2006). LATS1 acts as a Tumor Suppressor Gene Promoter methylation targeting LATS1 inactivation in mice leads to the development of many tumors, such as ovarian cancer and sarcomas (St John et al., 1999). Cytoplasmic LATS1 gene expression was positively correlated with lymph node metastasis (Luo et al., 2020). LATS1 is downregulated in several human cancers such as astrocytoma, colorectal cancer, lung cancer, breast cancer, and glioma due to hypermethylation. LATS1 overexpression not only significantly inhibits migration, invasion cell growth and also delays cell cycle progression from G2/M to G1 in vitro glioma U251 cells (Ji et al., 2012). LATS1 knockout mice showed the development of mitotic defects, chromosomal misalignment, mouse embryonic fibroblasts (MEFs), multipolar spindle formation, centrosomal overduplication, cytokinesis failure, and chromosomal bridging (Wu et al., 2018). It was shown that LATS1 overexpression with upregulated p27 expression stimulated YAP phosphorylation inhibited cell proliferation in cervical cancer cells (Deng et al., 2017). The malignant transformation was strongly correlated with low membrane expression of LATS1 in the cervical epithelium (Zhou et al., 2014). LATS1 overexpression was observed to repress growth, induce apoptosis, and mesenchymal to epithelial transition (MET). Changes in the LATS1 function might be of pathological significance in human Tumorigenesis (Kamikubo et al., 2003). Based on the above background, the objectives of this study were to estimate the promoter hypermethylation status of LATS1 genes in circulating blood of OSCC patients and to determine their association with clinicopathological features of OSCC.
Materials and Methods
Sample Collection
Blood samples (50) were collected with the informed consent of patients diagnosed with OSCC after taking the necessary ethical clearance from Dharamshila Cancer Hospital & Research Centre, New Delhi. The blood samples (20) from healthy individuals (as controls) were also obtained. Only patients diagnosed with OSCC were considered in the patients group, while individuals not suffering from any cancer were considered as controls. The samples were further used for DNA extraction.
DNA Exraction
The DNA isolation was carried out from blood as well as serum samples. One part of this study was carried out on blood samples, and for the other part of this study, serum was separated from the blood by centrifuging at 2,200 rpm. Leucocyte or White Blood Cells (WBCs) collected from blood samples were lysed in digestion buffer (10 mM Tris-HCl, pH 8.0, 10 mM EDTA, 150 mM NaCl and 2% SDS) containing proteinase K (0.2 mg/ml). DNA was then purified using the standard phenol-chloroform extraction and ethanol precipitation (Hoque et al., 2004).
Sodium Bisulfite Modification
This method allows the particular study of methylation in a specific region by converting all non-methylated cytosine into uracil, while methylated cytosines remain unchanged. Isolated DNA was modified with sodium bisulfite using the agarose bead method (Tiwari et al., 2009).
Purity of DNA
To quantify the amount of DNA, Nanodrop ND-1000 spectral photometer was used. The quality of DNA was checked using agarose gel electrophoresis.
Methylation-Specific PCR (MS-PCR)
The methylation status of the LATS1 gene was detected using MS-PCR assay for bisulfite-converted DNA (Herman et al., 1996). The amplification of the bisulfite-treated DNA was done through methylation-specific – PCR; MS-PCR was carried out using specific primers for methylation and unmethylation for the LATS1 genes (Table 1). MSP was performed to check the promoter hypermethylation of the LATS1 gene in OSCC patients (Figure 2). Photomicrograph representing 2.5% agarose gel electrophoresis of PCR amplified LATS1 gene that was carried out in 1X TAE buffer at a voltage supply of 80 V.
Table 1.
Sequence of the Primers for LATS1 Gene
| Name of Gene | Sequences (5′–3′) | Annealing tem (°C) | Product size |
|---|---|---|---|
| LATS1 M | F: GGAGTTT CGTTTTGTC | 55 °C | 138 bp |
| R: CGACGTAATAACG AACGCCTA | |||
| LATS1 U | F: TAGGTTGGAGTGTGGTGGT | 56 °C | 121 bp |
| R: CCCAACATAATAACAAACACCT |
M, Methylated; U, Unmethylated
Figure 2.
MSB (Methylation Specific Band) and UMSB (Unmethylation Specific Band) of LATS1 in Blood Samples of OSCC Patients. Lane No- 1, Molecular Marker 100 base pair; Lane No- 2,4,6, MSB (Methylation Specific Band) in the blood samples of OSCC patients; Lane No-3,5,7, UMSB (Unmethylation Specific Band) in the blood samples of OSCC patients
The following steps and temperature conditions were used in the process: the initial denaturation was carried out at 95°C for 10 mins. The PCR cycles consisting of denaturation at 95°C for 40 sec, annealing temperature for LATS1: M = 55°C, U =56°C for 40 sec and elongation step was done at 72°C for 40 sec for 35 cycles. The final extension was carried out at 72°C for 10 min.
Statistical Analysis
The association between the hypermethylation of the LATS1 gene and risk of OSCC was estimated by computing odds ratios (ORs) and 95% confidence intervals (CI) using the Chi-square test which included several potential confounding variables. Statistical analysis was performed using Stat Calc (Statistical Calculator) of Epiinfo tool version 7.2. P<0.05 was considered to indicate a statistically significant difference.
Results
Isolation of DNA
The DNA isolated from the blood and serum samples were run on the 1% agarose gel and visualized under the gel documentation unit. This was carried out to check the quality of DNA (Figure 1). The isolated DNA was also quantified using Nanodrop ND-1000 spectral photometer.
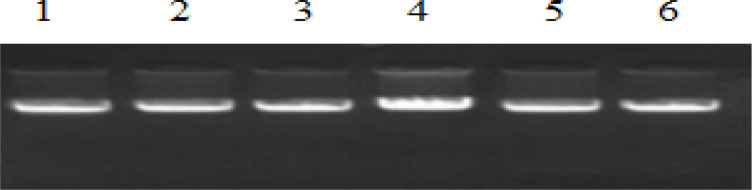
Figure 1.
Qualitative Analysis of DNA (Isolated DNA from Blood Samples of OSCC Patients and Controls). Lane No- 1-4, DNA of OSCC patients; Lane No- 5-6, DNA of healthy individuals (controls).
MS PCR Result of LATS1 GENE
Methylation Specific PCR was performed to check the promoter hypermethylation of LATS1 Gene in OSCC patients among the North Indian population (Figure 2) and (Figure 3).
Figure 3.
Methylation-Specific PCR Including Negative Control and Positive Control. Lane No- 1, Molecular marker 100 base pair; Lane No- 2, Negative control (H2O); Lane No-3, positive control (in vitro methylation)
Status of promoter hypermethylation
Hypermethylation of LATS1 was observed in 38% of OSCC patients. 19 out of 50 samples were found to be promoter hypermethylated through MSP (Methylation Specific PCR) in OSCC patients (Figure 2). The statistical analysis of promoter hypermethylation of LATS1 Gene was found to be significant (p-value= 0.017) with an odds ratio of 0.37 (Table 2). Hypermethylation of the LATS1 gene was observed in Serum samples, although the intensity of MSB was found to be less in serum samples (Figure 4).
Table 2.
Frequency of Methylation and Unmethylation of LATS1 Gene with the Relative Risk of OSCC in Patients and Healthy Controls
| Gene | Methylation status | Cases (n=50) | Control (n=20) | OR | 95%CI | P-value |
|---|---|---|---|---|---|---|
| LATS1 | M | 19 (38.00 %) | 0 (00.00 %) | 0.37 | 0.16–0.84 | 0.017 |
| U (ref) | 31 (62.00 %) | 20 (100.00 %) |
OR, odds ratio; 95% CI, 95 % confidence interval; ref, reference
Figure 4.
Comparision of the Methylation Status LATS1 Gene in Blood and Serum Samples of OSCC Patients. MSB (Methylation Specific Band) of LATS1 in serum and paired blood samples of OSCC patients along with ladder (Size 138 bp). Lane No- 1: Molecular marker 100 base pair; Lane No- 2,4,6, MSB (Methylation Specific Band) in serum samples of OSCC patients. Lane No- 3,5,7, MSB (Methylation Specific Band) in paired blood samples of OSCC patients
Correlation between Clinicopathological Information and Methylation Status in OSCC
In this study total 50 samples were taken, out of which 30 were tobacco chewers and similarly in case of smokers 30 samples were taken and out of which 20 were smokers. The methylation of LATS1 gene showed a significant risk of developing OSCC in patients (p-value<0.05) (Table 2). LATS1 was observed to be methylated 50% in tobacco chewers and 66.6% in smokers among patients. It was not observed among controls. The hypermethylation of the promoter region of the LATS1 gene was also found to be statistically significant in tobacco chewers (Table 3) and smokers (Table 4) as the p-value was found to be < 0.05.
Discussion
Oral squamous cell carcinoma (OSCC) is one of the most popular types of oral neoplasm, accounting for over 90 % of all mouth fatalities and 38 % of head and neck neoplasms. In fact, despite advances in the area of oral cancer detection, prevention, and multimodality approaches, the overall 5-year survival for OSCC remains to be modest at best. The common significant factor influencing OSCC survival after treatment is the grade of the Tumor at diagnosis. Hence, to promote long-term results, immediate detection in combined with primary and secondary control strategies is critical. A methyl group is covalently joined to cytosine C5. When DNA is treated with bisulfite, unmethylated cytosines are converted to uracil, but methylated cytosines are protected. In this phenomenon, the promoter region of the gene is hypermethylated which causes global hypomethylation to the gene. When these methyl groups are segregated from the cytosine nucleotide then the expression of the sequence is altered. This is called a reversal of methylation. This causes the expression to express which is delayed due to methylation. Epigenetic changes vary from population to population as they are mainly dependent upon the environmental and dietary factors which differ greatly (Jha et al., 2017).
Large Tumor Suppressor gene 1 (LATS1) are key tumor suppressor genes in the cell cycle regulation and DDR signaling (Najafi et al., 2016). Previous studies have indicated the promoter methylation profiles of P14ARF, MGMT, CDH1, APC, ATM, P15INK4b, P16INK4a, FADD, FAS, ERK and RAF1 in OSCC (Kordi-Tamandani et al., 2010b; Rigi-Ladiz et al., 2011; Kordi-Tamandani et al., 2012; Saberi et al., 2014; Kordi-Tamandani et al., 2014). To the best of our knowledge, no published work is there from India on the promoter hypermethylation of the selected gene. So, the present research also focused on correlating the methylation status of LATS2 gene with these risk factors like tobacco chewing and smoking in OSCC patients among the North Indian population. In the present study, the DNA was isolated from the collected blood samples of a cancer patient (OSCC) and control samples (patients not suffering from cancer). Moreover, isolation of DNA was followed by sodium bisulfite modification by agarose bead method and E Z DNA Gold Methylation Kit (Zymo Research, US), after which MSP was performed. A statistically significant difference in methylation of LATS2 (p-value0.05) between patient and control samples was observed. The methylation of LATS2 showed the risk of developing OSCC in patients. It has been proved that the outcome novel methylation markers may be used for designing drugs that modify methylation statuses in OSCC (Mikeska and Craig, 2014.) Hence, based upon the present study LATS2 could be used as a diagnostic biomarker in OSCC patients among north Indian population. Investigations on broader sample size are required to declare the clinical applicability of LATS2 hypermethylation in larger groups of patients. Early estimation of LATS2 hypermethylation might allow the identification of subgroups of patients with poor diagnosis, who might require a diverse therapeutic strategy. Therefore, a future study needs to be carried out to explore the status of this potential biomarker in the clinical control of OSCC and to estimate whether it can contribute to personalized treatment strategies.
Conclusion and Future Perspectives
Promoter methylation of Tumor Suppressor genes is a critical factor in the carcinogenesis of OSCC.The Study of DNA methylation is a beneficial procedure for evaluating the biological characteristics of oral cancers and may be a useful diagnostic biomarker. Hence, the present study was intended to investigate the methylation status of LATS2 gene and to correlate the methylation status of these genes with the risk of OSCC statistically. The risk of OSCC was also calculated and was found to be significant. This confirms that LATS2 hypermethylation is an important step in the OSCC patients among north India population and it can probably be used as a diagnostic biomarker in OSCC patients. Further the study on reversal of hypermethyation of LATS 2 gene and its reactivation needs to be carried out and can provide a good breakthrough in the study involving cancer therapy. Thus research needs to be carried out on a large scale as the sample size in the present study was small to draw any complete statistical conclusion. However this study is very important as this is the first study on the methylation status of this gene in OSCC among the North Indian population.
Acknowledgements
We are very much thankful to the Institute of Applied Medicines and Research,Ghaziabad, India for providing a good platform to carry out the research work.
Author Contribution Statement
None.
Ethical Approval and Consent to Participate
The present study was authorized and approved by the Dharamshila Cancer Hospital & Research Centre, New Delhi.. Informed consent of patients were also obtained.
Conflict of interest
We declare there is no conflict of interest.
References
- Al-Swiahb JN, Chen CH, Chuang HC, et al. Clinical, pathological and molecular determinants in squamous cell carcinoma of the oral cavity. Future Oncol. 2010;6:837–50. doi: 10.2217/fon.10.35. [DOI] [PubMed] [Google Scholar]
- Cao X, Wang C, Liu J, Zhao B. Regulation and functions of the Hippo pathway in stemness and differentiation. Acta Biochim Biophys Sin. 2020;52:736–48. doi: 10.1093/abbs/gmaa048. [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, et al. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–86. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, et al. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem. 2013;288:34041–51. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Zhang W, Liu S, et al. LATS1 suppresses proliferation and invasion of cervical cancer. Mol Med Rep. 2017;15:1654–60. doi: 10.3892/mmr.2017.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics. 2019;14:1141–63. doi: 10.1080/15592294.2019.1638701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N, Aylon Y. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ. 2017;24:1488–1501. doi: 10.1038/cdd.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel H, Singhal S, Mathur R et al. Promoter Hypermethylation of LATS2 Gene in Oral Squamous Cell Carcinoma (OSCC) Among North Indian Population. Asian Pac J Cancer Prev. 2020;21:1283–7. doi: 10.31557/APJCP.2020.21.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujar H, Weisenberger DJ, Liang G. The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes (Basel) 2019;10:172. doi: 10.3390/genes10020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- Hisaoka M, Tanaka A, Hashimoto H. Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma. Lab Invest. 2002;82:1427–35. doi: 10.1097/01.lab.0000032381.68634.ca. [DOI] [PubMed] [Google Scholar]
- Hoque MO, Begum S, Topaloglu O, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–7. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- Jha M, Patel S, Gupta S, et al. Promoter hypermethylation of tumor suppressor genes in lung cancer. Res J Pharm Biol Chem Sci. 2016;7:3166–74. [Google Scholar]
- Ji T, Liu D, Shao W, et al. Decreased expression of LATS1 is correlated with the progression and prognosis of glioma. J Exp Clin Cancer Res. 2012;31 doi: 10.1186/1756-9966-31-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikubo Y, Takaori-Kondo A, Uchiyama T, Hori T. Inhibition of cell growth by conditional expression of kpm, a human homologue of Drosophila warts/lats tumor suppressor. J Biol Chem. 2003;278:17609–14. doi: 10.1074/jbc.M211974200. [DOI] [PubMed] [Google Scholar]
- Kordi-Tamandani DM, Ladies MA, Hashemi M, et al. Analysis of p15INK4b and p16INK4a gene methylation in patients with oral squamous cell carcinoma. Biochem Genet. 2012;5-6:448–53. doi: 10.1007/s10528-011-9489-6. [DOI] [PubMed] [Google Scholar]
- Kordi-Tamandani DM, Moazeni-Roodi AK, Rigi-Ladiz MA, et al. Promoter hypermethylation and expression profile of MGMT and CDH1 genes in oral cavity cancer. Arch Oral Biol. 2010;10:809–14. doi: 10.1016/j.archoralbio.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Kordi-Tamandani DM, Saberi E, Jamali S, Ladiz MA. ERK and RAF1 genes: analysis of methylation and expression profiles in patients with oral squamous cell carcinoma. Br J Biomed Sci. 2014;3:100–3. doi: 10.1080/09674845.2014.11669972. [DOI] [PubMed] [Google Scholar]
- Lin XY, Zhang XP, Wu JH, Qiu XS, Wang EH. Expression of LATS1 contributes to good prognosis and can negatively regulate YAP oncoprotein in non-small-cell lung cancer. Tumour Biol. 2014;35:6435–43. doi: 10.1007/s13277-014-1826-z. [DOI] [PubMed] [Google Scholar]
- Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Mol Cancer Res. 2006;4:221–3. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- Luo SY, Kwok HH, Yang PC, et al. Expression of large tumour suppressor (LATS) kinases modulates chemotherapy response in advanced non-small cell lung cancer. Transl Lung Cancer Res. 2020;9:294–305. doi: 10.21037/tlcr.2020.03.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood N, Rabbani SA. DNA methylation readers and cancer: Mechanistic and Therapeutic Applications. Front Oncol. 2019;9:489. doi: 10.3389/fonc.2019.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi M, Kordi-Tamandani DM, Arish M. Evaluation of LATS1 and LATS2 promoter methylation with the risk of pterygium formation. J Ophthalmol. 2016;2016:5431021. doi: 10.1155/2016/5431021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki M, Yabuta N, Fukuzawa M, et al. LATS1/2 kinases trigger self-renewal of cancer stem cells in aggressive oral cancer. Oncotarget. 2019;10:1014–30. doi: 10.18632/oncotarget.26583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigi-Ladiz MA, Kordi Tamandani DM, Torkamanzehi A. Analysis of hypermethylation and expression profiles of APC and ATM genes in patients with oral squamous cell carcinoma. Clin Epigenetics. 2011;1:6. doi: 10.1186/1868-7083-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi E, Kordi-Tamandani DM, Jamali S, Rigi-Ladiz MA. Analysis of methylation and mRNA expression status of FADD and FAS genes in patients with oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2014;6:e562–8. doi: 10.4317/medoral.19805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John MA, Tao W, Fei X, et al. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–6. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Miyoshi Y, Takahata C, et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–5. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Tiwari SK, Manoj G, Prasanth K, et al. Simplified and versatile method for bisulfite-based DNA methylation analysis of small amounts of DNA. J Clin Lab Anal. 2009;23:172–4. doi: 10.1002/jcla.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226:352–64. doi: 10.1002/path.3022. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhao Z, Zhang H, et al. LATS1 inhibits metastasis and epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Int J Clin Exp Pathol. 2018;11:2053–63. [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu K, Hao Y, et al. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol. 2006;6:609–17. doi: 10.1038/ncb1140. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Tao F, Cheng Y, et al. Up-regulation of ITCH is associated with down-regulation of LATS1 during tumorigenesis and progression of cervical squamous cell carcinoma. Clin Invest Med. 2014;37:384–94. doi: 10.25011/cim.v37i6.22243. [DOI] [PubMed] [Google Scholar]
- Zuberi M, Dholariya S, Khan I, et al. Epigenetic Silencing of DAPK1and p16INK4a Genes by CpG Island Hypermethylation in Epithelial Ovarian Cancer Patients. Ind J Clin Biochem. 2020 doi: 10.1007/s12291-020-00888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]