Abstract
The coronavirus disease 2019 (COVID-19) global pandemic has turned into the largest public health and economic crisis in recent history impacting virtually all sectors of society. There is a need for effective therapeutics to battle the ongoing pandemic. Repurposing existing drugs with known pharmacological safety profiles is a fast and cost-effective approach to identify novel treatments. The COVID-19 etiologic agent is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a single-stranded positive-sense RNA virus. Coronaviruses rely on the enzymatic activity of the replication–transcription complex (RTC) to multiply inside host cells. The RTC core catalytic component is the RNA-dependent RNA polymerase (RdRp) holoenzyme. The RdRp is one of the key druggable targets for CoVs due to its essential role in viral replication, high degree of sequence and structural conservation and the lack of homologues in human cells. Here, we have expressed, purified and biochemically characterised active SARS-CoV-2 RdRp complexes. We developed a novel fluorescence resonance energy transfer-based strand displacement assay for monitoring SARS-CoV-2 RdRp activity suitable for a high-throughput format. As part of a larger research project to identify inhibitors for all the enzymatic activities encoded by SARS-CoV-2, we used this assay to screen a custom chemical library of over 5000 approved and investigational compounds for novel SARS-CoV-2 RdRp inhibitors. We identified three novel compounds (GSK-650394, C646 and BH3I-1) and confirmed suramin and suramin-like compounds as in vitro SARS-CoV-2 RdRp activity inhibitors. We also characterised the antiviral efficacy of these drugs in cell-based assays that we developed to monitor SARS-CoV-2 growth.
Keywords: coronavirus, COVID-19, nsp12, RNA-dependent RNA polymerase
Introduction
The coronavirus disease 2019 (COVID-19) outbreak caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is considered to be the most severe global public health crisis since the influenza pandemic in 1918 [1,2]. As of February 2021, there were more than 100 million confirmed cases and 2.5 million deaths globally [3]. Remdesivir is currently the only antiviral drug approved for COVID-19 treatment. Remdesivir is a nucleoside analogue that acts as a small molecule inhibitor of an essential coronavirus enzyme complex, the RNA-dependent RNA polymerase (RdRp) [4]. Nucleoside analogues are one of the largest and most effective classes of small molecule drugs against viruses like human immunodeficiency virus, hepatitis B virus and herpesviruses [5,6]. The efficacy of remdesivir as a COVID-19 treatment remains controversial. While one clinical trial found remdesivir to shorten recovery time in COVID-19 patients, other clinical trials found no effect of remdesivir on recovery time or mortality rate [7–10]. Besides remdesivir, several other clinically approved nucleoside analogue compounds are currently under study for treating COVID-19 [11]. Molnupiravir [12,13] and AT-527 [14] are the most promising candidates so far, while ritonavir [15], ribavirin [16], favipiravir [17] and sofosbuvir [6,18] have not demonstrated significant antiviral effect against SARS-CoV-2 in the laboratory or clinical settings.
Coronaviruses (CoVs) are enveloped single-stranded positive-sense RNA viruses that belong to the order Nidovirales[19]. CoV replication and transcription occur in the cytoplasm of infected cells and are mediated by the replication–transcription complex (RTC) [19,20]. The core RTC is composed of (i) the RdRp — a trimeric complex of the catalytic subunit nonstructural protein (nsp) 12 and two accessory factors nsp7 and nsp8 [21,22]; (ii) the nsp13 helicase [23-25] and (iii) several RNA-processing enzymes such as nsp14, a bifunctional enzyme with 3′-to-5′ exoribonuclease (ExoN) and N7-methyltransferase activities [26–28]. The replication of CoVs is more complex than that of other RNA viruses due to their unusually large genomes [29]. Replication of these large genomes requires enhanced RdRp processivity, which is promoted by the two nsp12 accessory subunits, nsp7 and nsp8 [21,22,29]. Replication of large genomes also involves enzymatic functions that could decrease the high error rate typical of viral RNA polymerases to avoid detrimental fitness effects [30]. CoVs encode a 3′-to-5′ ExoN (nsp14) that can remove mis-incorporated nucleotides [31–36]. This potential replication proofreading function may diminish the effectiveness of immediate chain terminator nucleoside analogues, since they can be removed by the nsp14 exonuclease when added to the nascent RNA chain [32,37]. On the other hand, remdesivir works as a delayed chain terminator: its addition to the growing RNA molecule results in termination only after incorporation of up to three additional nucleotides and hence shows some resistance to excision by the nsp14 exonuclease [38,39].
RdRp has three characteristics that when combined position it as a key drug target for CoVs: (i) it is essential for viral replication; (ii) it has a high level of structural conservation among CoVs; and (iii) it lacks a counterpart in human cells [19]. However, due to the proofreading activity of nsp14 exonuclease, the potential of nucleoside analogues as antiviral drugs for COVID-19 treatment may be limited. Instead, identifying RdRp inhibitors which are not nucleoside analogues from currently approved or experimental drugs may lead to earlier and faster clinical trials, as these drugs have known pharmacokinetic and pharmacodynamic profiles and drug regimens [40].
Here, we describe the expression and purification of active SARS-CoV-2 RdRp holoenzyme (nsp12/nsp7/nsp8). We also developed a fluorescence resonance energy transfer (FRET)-based strand displacement assay suitable for high-throughput screening (HTS) to identify potential RdRp inhibitors using a custom chemical library of over 5000 compounds. We demonstrate that several non-nucleoside analogues potently block RdRp activity in vitro and one of them, GSK-650394, potently inhibits SARS-CoV-2 infectivity in a cell-based model of viral infection.
Results
Protein expression and purification
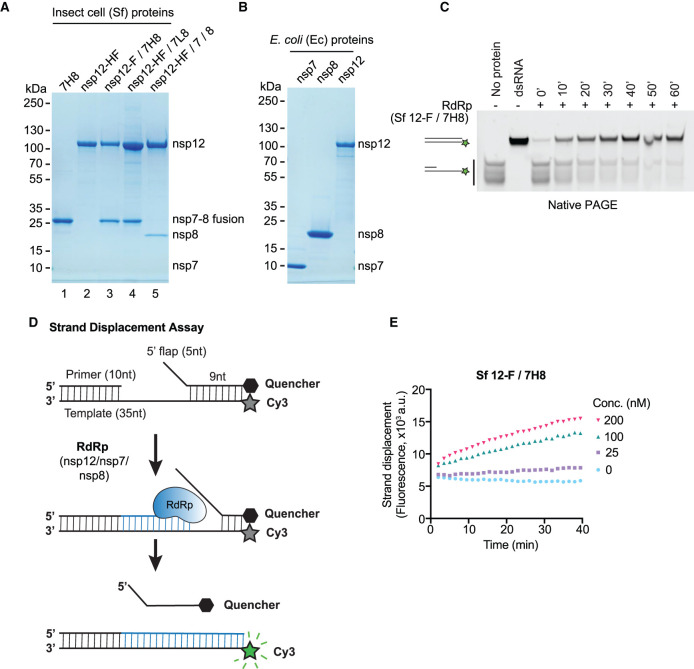
Coronavirus RdRp constitutes the catalytic core of the RTC and is composed of nsp12 in complex with two copies of nsp8 and one copy of nsp7 (nsp12/nsp82/nsp7) [41]. To maximise the chances of generating active RdRp in sufficient amounts for HTS, we followed two protein expression strategies. First, we chose a eukaryotic expression system and expressed proteins in baculovirus-infected insect cells (Spodoptera frugiperda, Sf; Figure 1A). Using a similar approach to previous work with SARS-CoV-1 RdRp, we expressed and purified nsp12/nsp7–nsp8 complex in which the C-terminus of nsp12 was tagged with 3xFlag and nsp7 and nsp8 were fused with a His6 tag linker (7H8, similar to [22]). Purification by 3xFlag affinity, heparin affinity, and size exclusion chromatography yielded a near-homogeneous protein preparation (Figure 1A, lane 3: nsp12-F/7H8). To be able to obtain better yields, we transferred the His6-tag to the catalytic subunit nsp12 and produced a complex containing a His6-3xFlag tag on nsp12 (Sf nsp12-HF) and a neutral 6 amino acid (Gly-Gly-Ser)2-linker between nsp7 and nsp8 (7L8). Purification of this complex by affinity to Ni-NTA agarose, ion exchange and size exclusion chromatography resulted in ∼100× higher protein yields (Supplementary Table S1), albeit with less stoichiometric nsp7–nsp8 fusion protein (Figure 1A, lane 4: nsp12-HF/7L8). We also tried co-expression of individual nsp7 and nsp8 subunits with tagged nsp12. Purification of this complex resulted in sub-stoichiometric amounts of the nsp7 and nsp8 subunits (Figure 1A, lane 5: nsp12-HF/7/8). We additionally purified the nsp7-His6-nsp8 fusion protein and nsp12-His6-3xFlag in isolation (Figure 1A, lane 1: 7H8, lane 2: nsp12-HF) obtaining very good yield for 7H8 (Supplementary Table S1). In a second approach, we expressed the three proteins individually in Escherichia coli as N-terminal His-SUMO fusion proteins (Figure 1B). In this system, the affinity tag and SUMO fusion can be removed after affinity purification by a SUMO-specific protease [42], leaving behind the same N-terminus as would be generated by viral protease-mediated polyprotein cleavage in infected cells. We expressed nsp7, nsp8 and nsp12 using this system and purified the proteins by affinity to Ni-NTA agarose, fusion protein removal, ion exchange and size exclusion chromatography (Figure 1B).
Figure 1. Development of a FRET-based SARS-CoV-2 RdRp strand displacement assay.
(A) Purified SARS-CoV-2 RdRp proteins expressed in baculovirus-infected insect cells (Spodoptera frugiperda, Sf) analysed by SDS–PAGE and Coomassie staining. 7H8: nsp7-His6-nsp8, nsp12-HF: nsp12-His6-3xFlag, nsp12-F/7H8: nsp12-3xFlag/nsp7-His6-nsp8, nsp12-HF/7L8: nsp12-His6-3xFlag/nsp7-GGSGGS-nsp8, nsp12-HF/7/8: nsp12-His6-3xFlag/nsp7/nsp8. (B) Bacterially expressed and purified SARS-CoV-2 nsp7, nsp8 and nsp12 proteins analysed by SDS–PAGE and Coomassie staining. The proteins were expressed as 14His-SUMO fusion proteins in E. coli. 14His-SUMO was removed by a SUMO-specific protease during purification generating native N-termini. (C) Gel-based primer-extension assay to test RNA-dependent RNA synthesis using the RdRp preparation Sf nsp12-F/7H8. The substrate consists of a 10 nt RNA primer annealed to the 3′ end of a 35 nt RNA template. The 5′ end of the template strand is labelled with a Cy3 fluorophore. Reaction products were analysed by native PAGE and visualisation of Cy3 fluorescence. Formation of duplex RNA by RdRp was observed over time. Controls: a preformed Cy3-labelled dsRNA with the same size as the reaction product (dsRNA), the primed substrate (no protein). (D) Schematic diagram illustrating the FRET-based RdRp strand displacement assay. The RNA substrate is composed of a Cy3 fluorophore-containing template strand, an annealed primer and an annealed quencher strand with a 5′ flap. RdRp activity synthesises RNA by extending the primer strand and displaces the quencher strand. The displaced quencher strand can no longer anneal to fully synthesised duplex RNA leading to an increase in Cy3 fluorescent signal. (E) FRET-based strand displacement assay using the indicated concentrations of Sf nsp12-F/7H8.
Development of a FRET-based strand displacement assay for RdRp
To assess RNA-dependent RNA polymerisation activity we generated a primed RNA substrate by annealing a 35 nucleotide (nt) RNA template that contained a Cy3 fluorophore on its 5′ end with a 10 nt primer strand complementary to the 3′-end of the template. After incubation of RdRp with primed substrate and NTPs, reaction products were separated by native PAGE (Figure 1C). We tested in this assay the activity of the RdRp preparation from insect cells containing C-terminally tagged nsp12 and a co-expressed nsp7–nsp8 fusion protein with a His6-linker (Sf nsp12-F/7H8). This RdRp preparation was able to extend the primed RNA substrate to generate duplex RNA in a time-dependent manner, confirming RNA-dependent RNA polymerisation activity (Figure 1C).
We developed a variation of this primer-extension assay that can measure activity directly in solution, without requiring running products in a gel (Supplementary Figure S1A,B). As in Figure 1C, the Cy3-labelled primed RNA substrate is extended by the RdRp complex and generates duplex RNA. Then, after 60 min, we added a 14 nt RNA strand partially complementary to the template strand with a quencher molecule at its 3′ end. If RdRp has extended the primer, this quencher strand will not anneal to the template strand and will not be able to quench Cy3 fluorescence (Supplementary Figure S1A). We tested nsp12-F/7H8 in this assay and found that Cy3 fluorescence was greatly increased when RdRp was included in the reaction and the presence of Mn2+ enhanced RdRp activity compared with Mg2+ alone (Supplementary Figure S1B), which is in line with a published SARS-CoV-1 nsp12 enzymatic characterisation [43].
None of the primer-extension assays described above (Figure 1C and Supplementary Figure S1A and B) are amenable to accurate HTS as they involve multiple steps and rely only on end point values. Therefore, we designed a FRET-based assay suitable for HTS based on RNA synthesis-coupled strand displacement activity (Figure 1D). Strand displacement refers to the ability of certain DNA/RNA polymerases to displace downstream non-template strands from the template strand while polymerising nucleotides [44,45] (Figure 1D). The RNA substrate was constructed by annealing the primed 35 nt RNA template with the 14 nt quencher strand (Figure 1D). This structure places the Cy3 fluorophore in close proximity to the quencher localised on the opposite strand. As RdRp elongates the primer, it displaces the downstream quencher strand producing a fluorescent signal. As the final product is an RNA duplex, the quencher strand is prevented from reannealing (Figure 1D). When Sf nsp12-F/7H8 was incubated with the strand displacement substrate, fluorescence increased near-linearly with time and was dependent on enzyme concentration (Figure 1E). The presence of Mn2+ was not required but again greatly enhanced RdRp activity compared with Mg2+ alone (Supplementary Figure S1C–E). Unless stated otherwise, 2 mM Mn2+ was included in subsequent experiments. The fluorescence increase was dependent on NTPs (Supplementary Figure S1F) suggesting that (i) there were no contaminating nucleases in the reactions that could also have resulted in freeing the Cy3 fluorophore from the quencher and ii) RdRp polymerisation was driving strand displacement of the quencher strand.
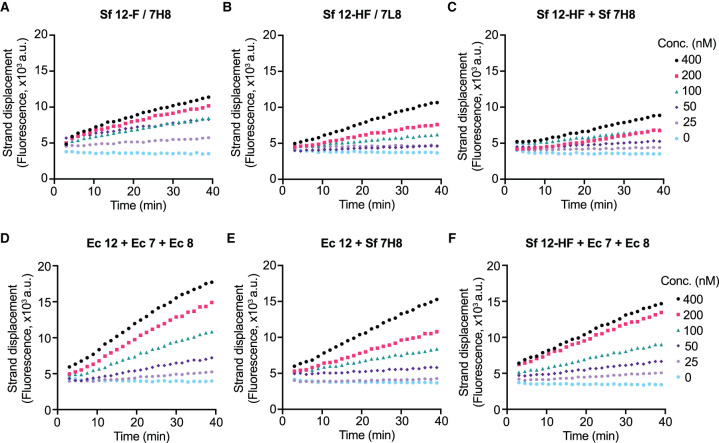
We tested our different RdRp preparations from Figure 1A,B using the FRET-based strand displacement assay in a concentration range of 25–400 nM (Figure 2). The C-terminally tagged nsp12 co-expressed with nsp7–nsp8 fusion protein with His linker (Sf nsp12-F/7H8, Figure 2A) was slightly more active than Sf nsp12-F/7L8 (Figure 2B), which we suspect is due to a lower stoichiometry of the 7L8 fusion protein (Figure 1A, lanes 3 and 4). The RdRp complex in which nsp7 and nsp8 were co-expressed individually with nsp12 (Sf nsp12-HF/7/8) did not show measurable activity (Supplementary Figure S2A), probably because nsp7 and nsp8 were sub-stoichiometric (Figure 1A, lane 5). Pre-incubating individually expressed Sf nsp12-HF and Sf 7H8 fusion protein (1 : 3 ratio, Figure 2C) resulted in slightly less activity than the co-purified Sf nsp12-F/7H8 (Figure 2A). Pre-incubating the three individually expressed proteins from E. coli (Ec) nsp12, nsp7, and nsp8 at a ratio of 1 : 3 : 6 produced the highest activity (Figure 2D). Mixing Ec nsp12 with insect cell expressed 7H8 at a ratio of 1 : 3 (Sf 7H8, Figure 2E) and mixing Sf nsp12-HF with Ec nsp7 and Ec nsp8 at a ratio of 1 : 3 : 6 (Figure 2F) also produced moderate activity. All together, these results suggest that either co-purification of nsp12 with a nsp7–nsp8 fusion protein or mixing and preincubation of individually purified proteins, regardless of insect cell or bacterial expression, can generate active RdRp complexes.
Figure 2. Activity of different RdRp preparations in the strand displacement assay.
RdRp was mixed at the indicated concentrations with 100 nM RNA substrate and 300 µM of each NTP and Cy3 fluorescence was recorded. Enzyme preincubation was performed where indicated at 22°C for 30 min at the indicated ratios (5 µM nsp12). (A) Sf nsp12-F/7H8. (B) Sf nsp12-HF/7L8. (C) Sf nsp12-HF after preincubation with Sf 7H8 (1 : 3 ratio). (D) Ec nsp12 after preincubation with Ec nsp7 and Ec nsp8 (1 : 3 : 6 ratio). (E) Ec nsp12 after preincubation with Sf 7H8 (1 : 3 ratio). (F) Sf nsp12-HF after preincubation with Ec nsp7 and Ec nsp8 (1 : 3 : 6 ratio).
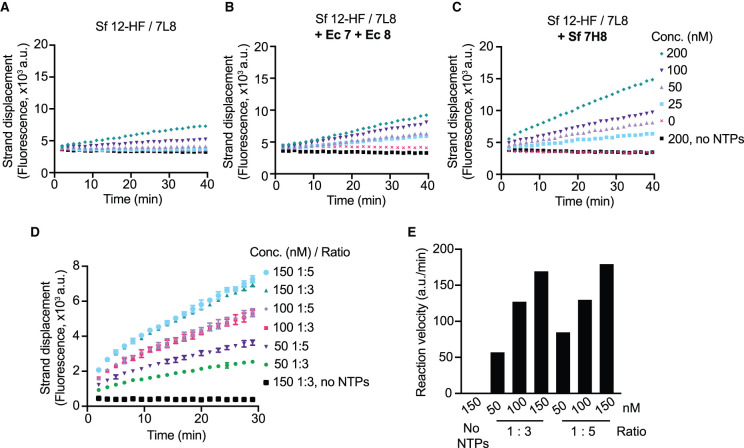
Best protein yields were obtained in insect cells when nsp12 was co-expressed with 7L8 fusion protein (Supplementary Table S1), and therefore, we optimised the HTS assay conditions using this RdRp preparation (Sf nsp12-HF/7L8). The 7L8 fusion protein was sub-stoichiometric (Figure 1A, lane 4), so we tested if the activity of Sf nsp12-HF/7L8 could be increased by further addition of nsp12 cofactors: Ec nsp7 and Ec nsp8 or Sf 7H8. We preincubated the proteins at high concentrations (10 µM for Sf nsp12-HF/7L8 either with Ec nsp7 and Ec nsp8 in 1 : 3 : 6 ratio, or with Sf 7H8 in 1 : 6 ratio) for 30 min and found they increased the activity of Sf nsp12-HF/7L8 (Figure 3A–C), with Sf 7H8 fusion protein addition showing the highest activity (Figure 3C). We also mixed Sf nsp12-HF/7L8 and Sf 7H8 at a ratio of 1 : 3 or 1 : 5 at different concentrations and found both ratios conferred similar activity at 100 nM and 150 nM Sf nsp12-HF/7L8 (Figure 3D,E). A final concentration of 150 nM RdRp complex in a 1 : 3 ratio was chosen and used for the high-throughput screen and all subsequent experiments. Finally, the presence of 5% DMSO, which is contributed by the chemical library in the HTS assay, did not affect RdRp activity (Supplementary Figure S3A).
Figure 3. Optimisation of assay conditions for HTS.
(A–C) Strand displacement assay using insect cell expressed (Sf) nsp12-HF/7L8 at the indicated concentrations alone (A) or after pre-incubating at 22°C for 30 min with Ec nsp7 and Ec nsp8 (ratio 1 : 3 : 6) (B) or with Sf 7H8 (ratio 1 : 6) (C). As control, a condition without NTPs using 200 nM RdRp was included. (D) Optimisation experiment to decide on RdRp concentration using HTS conditions (100 nM strand displacement substrate, 300 µM NTPs). Sf nsp12-HF/7L8 was preincubated at 22°C for 30 min with Sf 7H8 at either 1 : 3 or 1 : 5 ratio as specified and tested in the strand displacement assay at the indicated concentrations. Under these conditions, the unquenched Cy3 template alone had an average fluorescence value of 50 × 103 a.u. (E) Reaction velocities extracted from the curves shown in (D).
Chemical library screen design and results
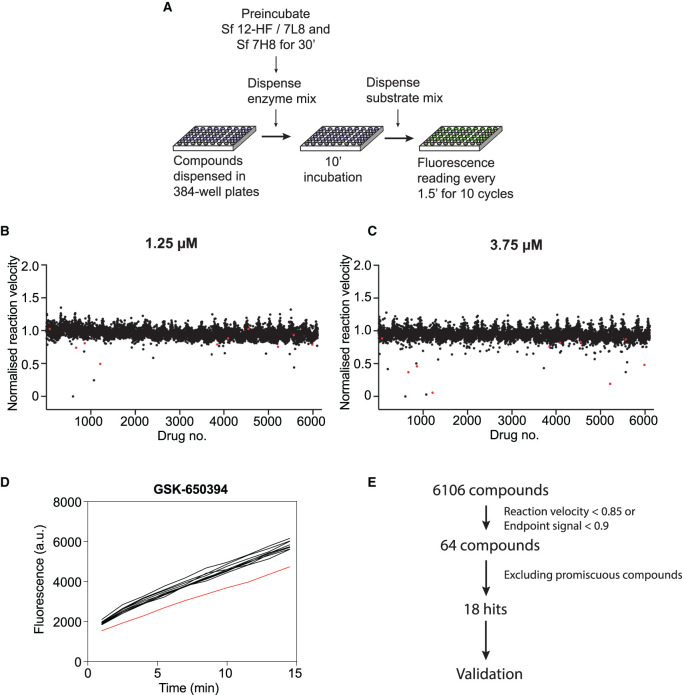
The optimised strand displacement assay was used to screen a custom library of over 5000 compounds for inhibitors of SARS-CoV-2 RdRp activity in a 384-well format (see Zeng et al. [46] for contents and description of the library). The compounds were incubated with RdRp at room temperature for 10 min, and then reactions were started by substrate addition (Figure 4A). Fluorescent signals were recorded in 90 s intervals for 10 cycles, which covered the linear phase of the reactions (Supplementary Figure S4A), and fluorescence increase over time was used to calculate the reaction velocity for each well. The library was screened at two compound concentrations, 1.25 µM (Figure 4B) and 3.75 µM (Figure 4C). Reactions that showed >15% reduction in the velocity (Figure 4B,C) or >10% reduction in the endpoint signal (Supplementary Figure S4C,D) were manually inspected (example in Figure 4D). 64 compounds were selected as primary hits. Compounds were then discounted if: (i) the compound also scored as a strong hit in parallel HTSs of other SARS-CoV-2 enzymes (with the exception of sulphated naphthylamine derivatives, see below) [47–52]; (ii) the compound was reported or strongly predicted to be a promiscuous inhibitor due to colloidal aggregation (LogP value >3.6 and >85% similarity to a known aggregator) [53,54]; (iii) the compound was reported or predicted to be a nucleic acid intercalator. This analysis resulted in the selection of 18 compounds for further validation experiments (Supplementary Table S2). Notably, 5 compounds of the 18 selected primary hits could be categorised as sulphated naphthylamine derivatives (suramin, NF 023, PPNDS, Evans Blue and Diphenyl Blue). These compounds were also identified as inhibitors of SARS-CoV-2 nsp13 helicase in a parallel screen (Zeng et al. [46]). Suramin and suramin-like molecules are polyanionic compounds that likely bind to positively charged patches found in a diverse array of proteins [55–61], including nucleic acid binding proteins like helicases [62] and viral polymerases [63–67]. In vitro validation experiments described in Zeng et al. showed that all five compounds inhibit SARS-CoV-2 nsp13 helicase with IC50 values between 0.5 and 6 µM and SARS-CoV-2 RdRp with IC50 values between 0.5 and 5 µM. They are not predicted to have aggregation tendencies (Supplementary Table S2) and their nsp13 inhibition was shown to be insensitive to detergent (Zeng et al. [46]), making nonspecific inhibition due to colloidal aggregation unlikely [68]. Suramin, as the main representative drug of this group, was selected to be included in further in vitro validation experiments as part of this study. Ultimately, 14 compounds were chosen for further validation experiments.
Figure 4. High-throughput chemical library screen against SARS-CoV-2 RNA-dependent RNA polymerase.
(A) Logistics of the screen. A custom chemical library consisting of over 5000 compounds was screened against RdRp activity using the FRET-based strand displacement assay in a 384-well format. RdRp was prepared by preincubation of Sf nsp12-HF/7L8 and Sf 7H8 in a 1 : 3 ratio for 30 min at room temperature. RdRp was dispensed into compound-containing 384-well plates and incubated for 10 min. Reactions were started by the addition of a substrate mix and florescence monitored in 90 s intervals. (B,C) Results of the HTS screen performed at 1.25 µM (B) and 3.75 µM (C) compound concentration. The normalised reaction velocity plotted against the compound number is shown. Validated hits are shown in red. (D) Kinetic curves with >15% reduction in reaction velocity or >10% reduction in fluorescent signal at the endpoint were inspected manually. As example, kinetic data for compound GSK-650394 is shown (red curve, data from surrounding wells in black). (E) Summary of the HTS hit selection strategy. From over 5000 compounds tested in the screen, 64 were considered primary hits after manual inspection of HTS reactions, which showed a reduction in reaction velocity below 85%, or a reduction in endpoint signal below 90%. Out of these, 46 primary hits were eliminated as they likely represent nonspecific modes of enzymatic inhibition such as colloidal aggregation or interference with the substrate structure. As part of this analysis promiscuous compounds that were identified as hits in other SARS-CoV-2 HTS [46,48–52] were removed with the exception of five suramin and suramin-like compounds, which were also identified in the SARS-CoV-2 nsp13 helicase HTS (Zeng et al. [46]). In vitro validation of the effect of suramin and suramin-like compounds on the activity of SARS-CoV-2 helicase and SARS-CoV-2 RdRp can be found in Zeng et al. [46]. A total of 18 compounds (including five suramin and suramin-like compounds) were selected as hits, of which 14 were included in further in vitro validation in this work.
Validation of hits
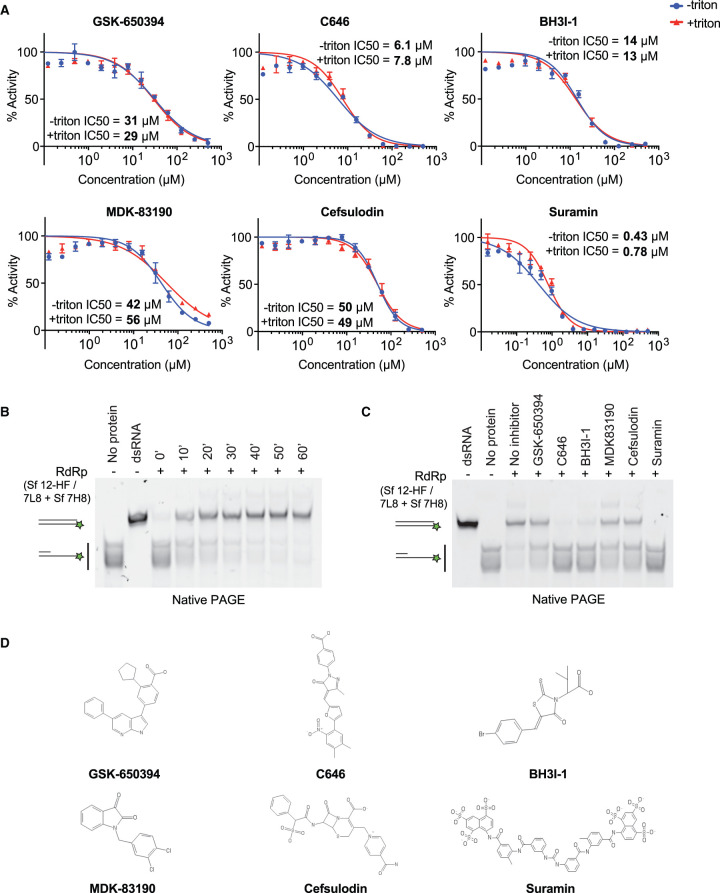
To confirm and quantify RdRp inhibition by the selected compounds in vitro, we performed compound titration experiments to determine the drug concentration at which half maximal inhibition is achieved (IC50). We carried these experiments out in the presence and in the absence of the non-ionic detergent Triton X-100 to rule out unspecific inhibition due to compound aggregation [68]. We found that six compounds showed only weak or no clear RdRp inhibition (Supplementary Figure S5A, Supplementary Table S3). Another two compounds showed interference with the fluorescent substrate by quenching (Supplementary Figure S5A, Supplementary Table S3). Therefore, these compounds were excluded from further analysis. The remaining 6 compounds inhibited RdRp with IC50 values of 0.43–56 µM and showed no or only minor sensitivity to detergent, indicating they might be bona fide RdRp inhibitors (Figure 5A, Table 1).
Figure 5. In vitro validation of selected compounds identified as RdRp inhibitors.
(A) Concentration–response curves of selected compounds using the strand displacement assay. The experiment was performed using 150 nM RdRp, 100 nM RNA substrate and 300 µM of each NTP in the presence (+Triton) or absence (−Triton) of 0.01% Triton X-100. Quenching controls are shown in Supplementary Figure S5B. IC50 values were calculated using Prism software. (B,C) Native gel-based assays using a primed RNA substrate as in Figure 1C. Reactions were started by mixing 300 nM RdRp complex (formed by preincubation of Sf nsp12-HF/7L8 and Sf 7H8 at 1 : 3 ratio) with 50 nM RNA substrate and 1 mM NTPs. Reaction products were analysed by native PAGE and visualisation of Cy3 fluorescence. (B) RdRp reactions were incubated for the indicated amounts of time. (C) Validation of selected compounds using 30-min RdRp reactions. RdRp complex was incubated with 50 µM of the indicated compounds for 10 min before reactions were started by substrate addition. Controls: a preformed dsRNA with the same size as the reaction product (dsRNA), the primed substrate (no protein). (D) Chemical structures of selected RdRp inhibitors.
Table 1. Inhibitory activity of compounds against SARS-CoV-2 RdRp in vitro.
| Compound name | IC50 (µM) | ||
|---|---|---|---|
| No detergent | + 0.01% Triton X-100 | Data figure | |
| Suramin1 | 0.43 | 0.78 | 4 |
| C646 | 6.1 | 7.8 | 4 |
| BH3I-1 | 14 | 13 | 4 |
| GSK-650394 | 31 | 29 | 4 |
| MDK-83190 | 42 | 56 | 4 |
| Cefsulodin | 50 | 49 | 4 |
For a further characterisation of suramin and the suramin-related compounds NF 023, PPNDS, Evans Blue, Diphenyl Blue see Zeng et al. [46].
We tested the six compounds in the gel-based polymerisation assay which monitors RNA synthesis more directly using the same primed RNA substrate as in Figure 1C. The RdRp preparation used here (Sf 12-HF/7L8 + Sf 7H8, ratio 1 : 3) also showed time-dependent RNA duplex formation (Figure 5B). We performed a 30-min reaction with the compounds at 50 µM and found they inhibited RdRp consistent with their IC50 values from the kinetic assay. Suramin, C646 and BH3I-1 inhibited RNA duplex formation almost completely, and the compounds with higher IC50 values, GSK-650394, MDK83190 and cefsulodin showed partial inhibition (Figure 5C).
Viral inhibition assays
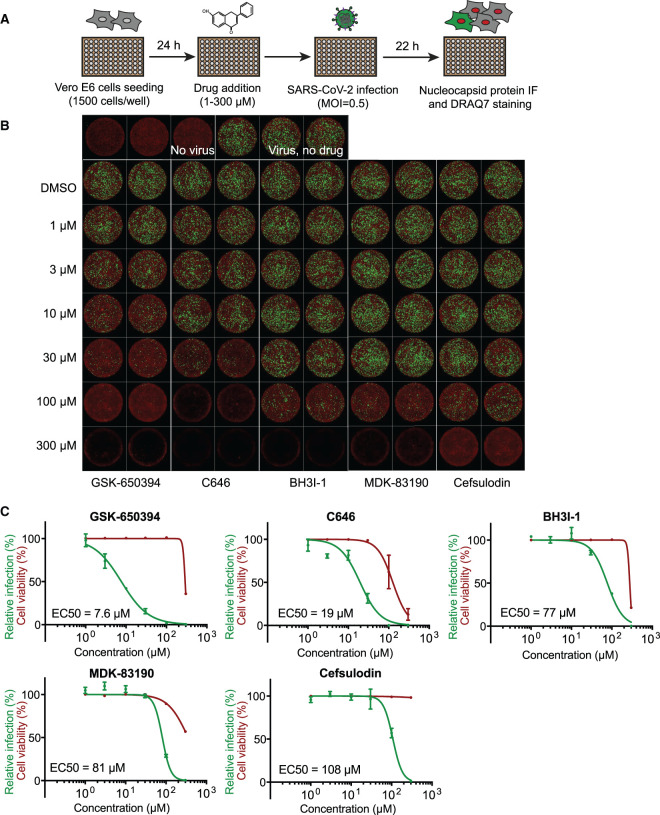
The validation experiments indicated that six compounds are genuine SARS-CoV-2 RdRp inhibitors in biochemical assays (Figure 5A and Table 1). We, therefore, evaluated their potential antiviral activity against SARS-CoV-2 in Vero E6 cells. Cells were treated with the compounds and then infected with SARS-CoV-2. After 22 h, viral plaques were analysed by viral nucleocapsid (N) protein immunofluorescence (Figure 6A). We tested the five novel RdRp inhibitors for their potential antiviral activity using this assay (Figure 6B). Dose–response curves and the half-maximal effective concentration (EC50) for each compound was calculated (Figure 6C). Two of them, GSK-650394 and C646, presented a dose-dependent reduction in the levels of SARS-CoV-2 N protein (green) in Vero E6 cells with low micromolar EC50 values (EC50 = 7.6 µM and EC50 = 19 µM, respectively, Figure 6B,C). While GSK-650394 showed cytotoxicity only at the highest concentration of 300 µM, C646 showed cytotoxicity at 100 µM. Suramin antiviral activity in Vero E6 cells following the same aforementioned protocol has been evaluated by us in the accompanying manuscript Zeng et al. and an EC50 of 9.9 µM against SARS-CoV-2 was observed (Zeng et al. [46]).
Figure 6. Antiviral activity of selected compounds against SARS-CoV-2 in Vero E6 cells.
(A) Viral inhibition experiments workflow graphical representation. In brief, 24 h after Vero E6 cells were seeded in a 96-well format, they were treated with selected drugs at specified concentrations. Then, cells were infected with a SARS-CoV-2 isolate at a MOI of 0.5 PFU per cell. Twenty-two hours later, cells were fixed and analysed by immunofluorescence staining and imaging. (B) SARS-CoV-2 antiviral activity of GSK-650394, C646, BH3I-1, MDK-83190 and cefsulodin. Representative overlaid images of viral N protein immunofluorescence (green) and DNA dye DRAQ7 staining (red). (C) Dose–response curve analysis. Viral infection values represent the area of viral plaques visualised by viral nucleocapsid protein staining (green curves) and cell viability was measured in the same experiment as the area of cells stained with the DNA dye DRAQ7 (red curves). Data are plotted as percentage relative to DMSO only control wells (100%). Data represent the mean and standard deviation (SD) of 3 replicates. Areas were calculated using FIJI and EC50 values were calculated using Prism software.
Combination therapy, the use of two or more antivirals with different modes of action, is a known strategy for limiting drug resistance as well as achieving better physiological outcomes [69]. The latter is usually caused by reduced toxicity due to the use of lower doses in combination protocols compared with monotherapy doses [69]. Consequently, a potential synergy between RdRp inhibitors with different mechanisms of action (non-nucleoside analogues identified here and a nucleoside analogue, remdesivir) was assessed [39,70,71]. Following the viral inhibition protocol described in Figure 6A, we performed dose–response experiments for each compound in the presence of a fixed concentration of 0.5 µM remdesivir, which as a single agent inhibited viral infection by less than 10%. Under these assay conditions, none of the validated inhibitors showed significant synergy with remdesivir (Supplementary Figure S6).
Discussion
The RTC of CoVs catalyses the synthesis of all genomic RNAs (viral replication) and sub-genomic RNAs (viral transcription). The core of SARS-CoV-2 RTC is the RdRp holoenzyme, composed of nsp12, the catalytic subunit, together with its cofactors nsp7 and nsp8 [20-22,72]. SARS-CoV-2 RdRp is the target of remdesivir, the only antiviral approved for human use to treat COVID-19 by regulatory agencies [70]. Remdesivir is a broad-spectrum nucleoside analogue first used for the treatment of Ebola infections [4]. Remdesivir's moderate success in COVID-19 treatment is still a matter of controversy as new clinical trials suggest that it is not as effective as first thought [7-10]. CoVs harbour a unique RNA-replication proofreading trait conferred by the 3′-to-5′ exonuclease activity of nsp14 [30], offering a possible explanation as to why the development or repurposing of effective nucleoside analogues, with the exception of remdesivir, has been unsuccessful [32,37]. Therefore, the characterisation of non-nucleoside analogue-type drugs that target SARS-CoV-2 RdRp might be needed more than ever. Moreover, COVID-19 is the third known transfer, after SARS-CoV in 2003 and MERS-CoV in 2012, of an animal coronavirus to a human population in only 20 years and therefore it is highly probable that new zoonotic outbreaks will occur in the future [73]. Thus, it is vital to develop more broad-spectrum antiviral strategies, together with vaccination programs, to curb future cross-species transmissions.
Here, we have expressed, purified and characterised a diverse array of SARS-CoV-2 RdRp complexes. We used two expression hosts, a bacterial system (E. coli) and a baculovirus-insect cell system and showed that recombinant proteins from both systems individually expressed or expressed as a complex lead to active proteins. We characterised different constructs of nsp12, nsp7 and nsp8 and tested activities of RdRp complexes formed with different stoichiometries of the subunits. We showed that the SARS-CoV-2 RdRp can strand displace downstream RNA encountered during synthesis. Based on this characteristic of SARS-CoV-2 RdRp, we designed a novel and robust FRET-based assay that allows real-time monitoring of RdRp activity in vitro. We utilised this assay to screen a custom chemical library to identify compounds that inhibit SARS-CoV-2 RdRp enzymatic activity.
Our study identified GSK-650394, C646 and BH3I-1 as well as confirmed suramin and suramin-like compounds as in vitro inhibitors of SARS-CoV-2 RdRp activity. These compounds do not have typical chemical structures of nucleoside analogues and thus likely inhibit RdRp through a different mechanism of action from remdesivir. Further work is required to understand their mode of inhibition. Cryo-electron microscopy structures of SARS-CoV-2 RdRp have been presented by several groups [21,39,41,72,74,75]. Future compound-protein structures may help reveal the inhibitory mechanisms of the RdRp inhibitors identified in this study.
GSK-650394, suramin and C646 showed also significant inhibition on viral growth in cell-based assays with an EC50 of 7.6 µM, 9.9 µM and 19 µM, respectively (Figure 6B,C) [46]. GSK-650394 is an inhibitor of SGK-1 kinase [76] that plays an important role in a diverse range of cellular processes such as stress response, ion transport, inflammation and cell proliferation [77]. GSK-650394 has been reported to impede the growth of cellular models of cancers [76,78] and has also been implicated in the inhibition of replication of influenza A virus, an enveloped negative-sense ssRNA virus, in human A549 cells [79]. In our study, GSK-650394 showed a low micromolar EC50 of 7.6 µM in inhibiting SARS-CoV-2 in cell-based assays, with effects on cell death only at the highest concentration tested (300 µM). Suramin is an antiparasitic drug first introduced by Bayer in 1916 that is still used to treat sleeping sickness and river blindness [80]. Suramin has been repurposed for many diverse applications including the inhibition of purinergic signaling, treatment of viral infections and treatment for advanced malignancies reflecting its multitude of targets [55]. Suramin is a polyanionic compound and probably binds tightly to positively charged patches found in DNA or RNA binding proteins [55,56]. This is consistent with our findings, as suramin and suramin analogues inhibit in vitro both SARS-CoV-2 RdRp and SARS-CoV-2 nsp13 helicase (Zeng et al. [46]). C646 is a competitive inhibitor of the histone acetyltransferase p300 that amongst other effects results in a reduction of pro-inflammatory gene expression in cells and animal models [81–84]. C646 was also shown to suppress the replication of different strains of influenza A viruses in A549 cells with an EC50 of 16 μM by affecting several steps of the viral life cycle [85]. Murine models of influenza infection treated with C646 also showed a significant reduction in viral replication with no associated toxicity [85].
Remdesivir under our experimental conditions had an EC50 of 1.8 μM, which is consistent with reported values in Vero E6 cells [71,86]. Interestingly, this is not far from the EC50 value we obtained for GSK-650394 (Figure 6). Further in vitro, cell-based and pre-clinical studies are needed to confirm and characterise these novel RdRp inhibitors. Moreover, they can be used as promising lead compounds to design improved drug candidates.
In summary, we have identified small molecule non-nucleoside analogue inhibitors of SARS-CoV-2 RdRp that also inhibited viral replication in cell-based assays. We provide evidence that GSK-650394, suramin and C646 could be considered as drug candidates that deserve further evaluation, as these compounds were found to exhibit antiviral activity against SARS-CoV-2 in a relevant cell culture model at non-cytotoxic concentrations. This screening strategy can be expanded to other small molecule compound libraries, and together with the biochemical insights regarding SARS-CoV-2 RdRp expression and purification described here, it should accelerate the search for improved antivirals that target SARS-CoV-2 RdRp holoenzyme.
Materials and methods
Protein expression
The following proteins were expressed in baculovirus-infected insect cells: nsp7-His6-nsp8 (Sf 7H8), nsp12-His6-3xFlag (Sf nsp12-HF), nsp12-3xFlag/nsp7-His6-nsp8 (Sf nsp12-F/7H8), nsp12-His6-3xFlag/nsp7-(GGS)2-nsp8 (Sf nsp12-HF/7L8), and nsp12-His6-3xFlag/nsp7/nsp8 (Sf nsp12-HF/7/8). The coding sequences of SARS-CoV-2 nsp7, nsp8 and nsp12 (NCBI reference sequence NC_045512.2) were codon-optimised for S. frugiperda and synthesised (GeneArt, Thermo Fisher Scientific). Baculoviral expression constructs were generated using the biGBac vector system [87]. An empty polyhedrin (polh) expression cassette from pLIB was inserted into the vectors pBIG1a, pBIG1b, and pBIG1c to generate pBIG1a(polh), pBIG1b(polh), and pBIG1c(polh).
Nsp12 was subcloned into pBIG1a(polh) either to contain a C-terminal His6-3xFlag tag (protein sequence: M-nsp12-GGSHHHHHHGSDYKDHDGDYKDHDIDYKDDDDK, pBIG1a_nsp12-His6-3xFlag, Addgene ID: 169182) or to contain a C-terminal 3xFlag tag (protein sequence: M-nsp12-GGSDYKDHDGDYKDHDIDYKDDDDK, pBIG1a_nsp12-3xFlag). Nsp7 and nsp8 were either subcloned individually into pBIG1b(polh) and pBIG1c(polh) to not contain a tag (pBIG1b_nsp7, Addgene ID: 169178 and pBIG1c_nsp8, Addgene ID: 169179), or into pBIG1b(polh) to be expressed as fusion protein with a His6-linker (protein sequence: M-nsp7-HHHHHH-nsp8, pBIG1b_nsp7-His6-nsp8, Addgene ID: 169180) or with a (GGS)2-linker (protein sequence: M-nsp7-GGSGGS-nsp8, pBIG1b_nsp7-Linker-nsp8, Addgene ID: 169181). Co-expression constructs were generated by subcloning expression cassettes from pBIG1 vectors into pBIG2abc vectors: pBIG1a_nsp12-3xFlag, pBIG1b_nsp7-His6-nsp8 and empty pBIG1c were used to generate pBIG2abc_nsp12-3xFlag/nsp7-His6-nsp8 (nsp12-F/7H8, Addgene ID: 169183). The vectors pBIG1a_nsp12-His6-3xFlag, pBIG1b_nsp7-Linker-nsp8, and empty pBIG1c were used to generate pBIG2abc_nsp12-His6-3xFlag/nsp7-Linker-nsp8 (nsp12-HF/7L8, Addgene ID: 169184). The vectors pBIG1a_nsp12-His6-3xFlag, pBIG1b_nsp7 and pBIG1c_nsp8 were used to generate pBIG2abc_nsp12-His6-3xFlag/nsp7/nsp8 (nsp12-HF/7/8, Addgene ID: 169185). Baculoviruses were generated and amplified in Sf9 insect cells (Thermo Fisher Scientific) using the EMBacY baculoviral genome [88]. For protein expression, Sf9 cells were infected with baculovirus and collected 48 h after infection, flash-frozen and stored at −70°C.
The following proteins were expressed in E. coli (Ec): 14His-SUMO-nsp7 (Ec 7, vector: pK27Sumo_His-SUMO-nsp7, Addgene ID: 169186), 14His-SUMO-nsp8 (Ec 8, vector: pK27Sumo_His-SUMO-nsp8, Addgene ID: 169187) and 14His-SUMO-nsp12 (Ec 12, vector: pK27Sumo_His-SUMO-nsp12, Addgene ID: 169188). The coding sequences of SARS-CoV-2 nsp7, nsp8 and nsp12 were codon-optimised for E. coli and synthesised (Genewiz). Nsp7, nsp8 and nsp12 were subcloned into the pK27Sumo expression vector [42] to code for Ulp1-cleavable 14His-SUMO fusion proteins. For expression, the vectors were transformed into T7 Express lysY competent E. coli cells (New England Biolabs). Expression cultures were grown in LB containing 50 µg/ml kanamycin to an OD600 of 0.5 and expression was induced with 0.4 mM IPTG at 16°C. After overnight incubation, cells were collected, flash-frozen and stored at −70°C.
Protein purification
All proteins were purified at 4°C. The proteins Sf nsp12-F/7H8 and Sf nsp12-HF/7/8 were purified by FLAG affinity, Heparin affinity and size exclusion chromatography. Insect cell pellets containing these proteins were resuspended in Flag buffer (50 mM HEPES pH 7.4, 150 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT) supplemented with protease inhibitors (Roche Complete Ultra EDTA-free tablets, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, 1 mM AEBSF) and lysed by Dounce homogenisation. After centrifugation at 39 000 g, 60 min, 4°C, the protein was purified from the cleared lysate by affinity to Anti-FLAG M2 Affinity gel (Sigma–Aldrich) and eluted in Flag buffer containing 0.5 mg/ml 3xFLAG peptide. The Flag eluate was diluted 1 : 6 with dilution buffer (50 mM HEPES pH 7.4, 5 mM MgCl2, 10% glycerol, 1 mM DTT) and loaded onto a HiTrap Heparin HP column (GE Healthcare) equilibrated in Heparin buffer A (50 mM HEPES pH 7.4, 25 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT). The protein was eluted with a gradient to Heparin buffer B (Heparin buffer A containing 500 mM KCl). Protein-containing fractions were concentrated and further purified by gel filtration on a Superdex 200 Increase 10/300 GL column (GE Healthcare) in Heparin buffer A containing 150 mM KCl. The protein was concentrated, flash-frozen and stored at −70°C.
The proteins Sf 7H8, Sf nsp12-HF, and Sf nsp12-HF/7L8 were purified by Ni-NTA affinity, ion exchange, and size exclusion chromatography. Insect cell pellets containing these proteins were resuspended in His-tag buffer (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.02% Nonidet P40 substitute, 20 mM imidazole) containing protease inhibitors (Roche Complete Ultra EDTA-free tablets, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, 1 mM AEBSF) and lysed using a Dounce homogeniser. The lysate was cleared by centrifugation at 39 000 g, 60 min, 4°C and the supernatant added to Ni-NTA Agarose (Invitrogen) equilibrated in His-tag buffer. The beads were washed with His-tag buffer and protein was eluted with His-elution buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.02% Nonidet P40 substitute, 200 mM imidazole). The eluate was diluted 1 : 5 with MonoQ buffer A (20 mM Tris–HCl pH 8.0, 2 mM MgCl2, 10% glycerol, 0.02% Nonidet P40 substitute, 1 mM DTT) and loaded onto a MonoQ 5/50 GL column (GE Healthcare) equilibrated with MonoQ buffer A containing 50 mM NaCl. After gradient elution to 1 M NaCl, protein-containing fractions were concentrated and further purified by gel filtration using a Superdex 200 Increase 10/300 GL column (GE Healthcare). 7H8 was purified in SEC-150 buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.02% Nonidet P40 substitute, 1 mM DTT). The proteins nsp12-HF and nsp12-HF/7L8 were purified in SEC-300 buffer (25 mM HEPES pH 7.5, 300 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.02% Nonidet P40 substitute, 1 mM DTT). The proteins were concentrated, flash-frozen and stored at −70°C.
The proteins 14His-SUMO-nsp7, 14His-SUMO-nsp8, and 14His-SUMO-nsp12 were purified by Ni-NTA affinity, proteolysis by His-Ulp1, removal of cleaved 14His-SUMO and His-Ulp1 by Ni-NTA, ion exchange and size exclusion chromatography. E. coli pellets containing these proteins were resuspended in His-SUMO buffer (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.05% Nonidet P40 substitute, 30 mM imidazole) containing protease inhibitors (Roche Complete EDTA-free tablets, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, 1 mM AEBSF) and were lysed by sonication. The lysate was cleared by centrifugation at 39 000 g, 60 min, 4°C and the protein was purified from the supernatant using Ni-NTA Agarose (Invitrogen). The beads were washed with His-SUMO buffer and protein was eluted with His-SUMO elution buffer (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.05% Nonidet P40 substitute, 400 mM imidazole). The SUMO-specific protease His-Ulp1 was added to the eluates to a concentration of 0.02 mg/ml and the proteins were dialysed overnight using His-SUMO dialysis buffer (25 mM Tris–HCl pH 7.5, 250 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.02% Nonidet P40 substitute, 1 mM DTT). Cleaved 14His-SUMO and His-Ulp1 were removed by affinity to Ni-NTA Agarose (Invitrogen). The flowthrough was diluted 1 : 5 with MonoQ buffer A and loaded onto a MonoQ 5/50 GL column (GE Healthcare), which was equilibrated with MonoQ buffer A containing 50 mM NaCl. After gradient elution to 1 M NaCl, all fractions were analysed, and protein-containing fractions concentrated. The proteins were further purified by gel filtration using a Superdex 200 Increase 10/300 GL column (GE Healthcare). nsp7 and nsp8 were purified in SEC-150 buffer, and nsp12 was purified in SEC-300 buffer. The proteins were concentrated, flash-frozen and stored at −70°C.
FRET-based strand displacement assay
A FRET-based fluorescence-quenching assay was designed to monitor strand displacement activity of the RdRp complex. The assay uses a tripartite RNA substrate. HPLC-purified RNA oligonucleotides were purchased from Eurofins Genomics or Integrated DNA Technologies with the following sequences:
Cy3 strand: 5′-Cy3-GCACUUAGAUAUGACUCGUUCUGCAGGCCAGUUAA-3′
Quencher strand: 5′-CUGCGUCUAAGUGC-(AbRQ)-3′
Primer strand: 5′-UUAACUGGCC-3′
The Cy3 strand, the quencher strand and the primer strand were annealed at 1 : 2 : 2 ratio at a concentration of 10 µM : 20 µM : 20 µM respectively by heating the oligonucleotide mix to 75°C for 5 min and gradually cooling it down to 4°C over 50 min in annealing buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 20 mM NaCl and 5 mM MgCl2). A reaction was typically started at room temperature by adding 10 µl of a 2× substrate solution containing the RNA substrate and NTPs in reaction buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM DTT, 5 mM MgCl2, 2 mM MnCl2, 0.1 mg/ml BSA, 0.01% Triton X-100) to 10 µl of a 2× enzyme solution containing the RdRp complex in reaction buffer. A typical reaction contained a final concentration of 100 nM RNA substrate, 300 µM of each NTP, and 150 nM RdRp. Upon displacement of the quencher strand by the RdRp complex during synthesis, Cy3 is no longer quenched and thus able to fluoresce. The fluorescent signal was read at 545 nm (excitation) and 575 nm (emission) with 10 nm bandwidth using a Tecan Spark microplate reader.
FRET-based polymerisation assay
The Cy3 template strand and the primer were annealed at 1 : 2 ratio using the protocol described above. A reaction was typically started at room temperature by adding 10 µl of a 2× substrate solution containing the RNA substrate and NTPs in reaction buffer to 10 µl of a 2× enzyme solution containing the RdRp complex in reaction buffer. The quencher strand was added at 2-fold concentration (relative to the Cy3 strand) after a 60-min reaction. The quencher strand was allowed to anneal with any unpolymerized template RNA for 35 min at room temperature before the fluorescent signal was read.
Gel-based polymerisation assay
The Cy3 template strand and the primer were annealed at 1 : 2 ratio using the protocol described above. A reaction was typically started at room temperature by adding 5 µl of a 2× substrate solution containing the RNA substrate and NTPs in reaction buffer to 5 µl of a 2× enzyme solution containing the RdRp complex in reaction buffer. The reaction was stopped by adding 5 µl stopping buffer (20 mM EDTA, 0.5% SDS, 2 mg/ml proteinase K, 10% glycerol) and incubating at 37°C for 5 min. Reaction products were then analysed by running on a 20% non-denaturing PAGE gel in 0.5× TBE buffer (Life Technologies) for 20 min at room temperature and 200 V. An Amersham Imager 600 was used to image Cy3 fluorescence. To test compound inhibition, a compound volume of 0.5 µl at a concentration of 1 mM was incubated with 5 µl 2× enzyme solution for 10 min before starting the reaction.
RdRp high-throughput screening assay
The screening plates containing the custom library of compounds were prepared and stored as described in (Zeng et al. [46]). For the HTS assay, first, 10 µM of Sf nsp12-HF/7L8 complex was incubated with 30 µM Sf 7H8 for 30 min on ice to form the RdRp complex. Then the RdRp complex was diluted in reaction buffer, before being dispensed at 10 µl volume to the plates to incubate with compounds for 10 min at room temperature. To start the reaction, 10 µl 2× substrate mix (600 µM NTPs, 200 nM RNA substrate) in reaction buffer was dispensed and the plates were spun briefly before being transferred to a Tecan Spark microplate reader. The fluorescent signal was first read at 1 min of reaction and then read every 1.5 min for 10 cycles. The screening was performed twice, one with a final compound concentration of 1.25 µM and one with 3.75 µM.
Data analysis
MATLAB was used to process data. The slope of fluorescent signal increase over time was calculated using a linear regression model for each compound as the reaction velocity (V). Then V for each compound is normalised against the DMSO controls in each plate as following:
where mean(V_DMSO) is the average of velocity values of DMSO controls in a particular plate.
The endpoint signal of each compound was also normalised against DMSO controls. Both the normalised reaction velocity and endpoint signal were used for hit selection. Compounds that gave >15% reduction in the reaction velocity or >10% reduction in the endpoint signal were further inspected. A kinetic curve (example in Figure 4D), in which the y-axis shows fluorescent signals, and the x-axis shows timepoints, was plotted for each of the compounds that passed the threshold. Ten wells that were before and after the compound well were plotted together in the same graph. We manually inspected the curves and confirmed that 64 compounds gave obvious inhibition.
The Z-score was also calculated for each compound (Supplementary Figure S4B) as following:
where mean(Normalised_V_all) is the average of normalised velocity values of all compounds and SD(Normalised_V_all) is the standard deviation of normalised velocity values of all compounds.
To assess the quality of the screen, the Z-factor was calculated for each plate as following:
where SD(V_DMSO) is the standard deviation of reaction velocity values of all DMSO controls in a particular plate, SD(background) is the standard deviation of reaction velocity values of background wells containing no enzyme, and mean(background) is the average of reaction velocity values of background wells. The average Z-factor for the screen is 0.71, indicating the screen is good, as described by Zhang et al. [89].
The method used for predicting drug aggregation propensity is described in [54] and can be accessed in http://advisor.bkslab.org/.
Tested drugs
Compounds selected for in vitro experimental validation and cell-based studies were purchased and resuspended following the manufacturer's instructions. See Supplementary Table S4 for more details.
Viral inhibition assay
The production of the SARS-CoV-2 virus isolate used in these assays and the in-house generation of the SARS-CoV-2 nucleocapsid (N) recombinant antibody is described elsewhere (Biochem J, nsp13 paper). The conditions for the viral inhibition assay were the following: 15.000 Vero E6 cells (NIBC, U.K.) resuspended in DMEM containing 10% FBS were seeded into each well of 96-well imaging plates (Greiner 655090) and cultured overnight at 37°C and 5% CO2. The next day, 5× solutions of compounds were generated by dispensing 10 mM stocks of compounds into a V-bottom 96-well plate (Thermo 249946) and back filling with DMSO to equalise the DMSO concentration in all wells using an Echo 550 (Labcyte) before resuspending in DMEM containing 10% FBS. The assay plates with seeded Vero E6 cells had the media replaced with 60 µl of fresh growth media, then 20 µl of the 5× compounds were stamped into the wells of the assay plates using a Biomek Fx automated liquid handler. Finally, the cells were infected by adding 20 µl of SARS-CoV-2 with a final MOI of 0.5 PFU/cell. Twenty-two hours post-infection, cells were fixed, permeabilised, and stained for SARS-CoV-2 N protein using Alexa488-labelled-CR3009 antibody and cellular DNA using DRAQ7 (Abcam). Whole-well imaging at 5× was carried out using an Opera Phenix (PerkinElmer) and fluorescent areas and intensity calculated using the Phenix-associated software Harmony (PerkinElmer).
Acknowledgement
We thank Anne Early for assistance.
Abbreviations
- COVID-19
coronavirus disease 2019
- CoVs
coronaviruses
- FRET
fluorescence resonance energy transfer
- HTS
high-throughput screening
- Nsp
nonstructural protein
- RdRp
RNA-dependent RNA polymerase
- RTC
replication–transcription complex
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
Data Availability
All data associated with this paper has been deposited in FigShare (https://figshare.com/).
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research U.K. (FC001066), the U.K. Medical Research Council (FC001066), and the Wellcome Trust (FC001066). This work was also funded by a Wellcome Trust Senior Investigator Award (106252/Z/14/Z) to J.F.X.D. FW and BC have received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement Nos. 844211 and 895786. J.Z. has received funding from a Ph.D. fellowship awarded by Boehringer Ingelheim Fonds.
Open Access Statement
Open access for this article was enabled by the participation of The Francis Crick Institute in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
CRediT Author Contribution
John F. Diffley: Conceptualization, resources, formal analysis, supervision, funding acquisition, methodology, project administration, writing — review and editing. Agustina P. Bertolin: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing — original draft, project administration, writing — original draft, writing — review and editing. Florian Weissmann: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing — original draft, writing — review and editing. Jingkun Zeng: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing — original draft, writing — review and editing. Viktor Posse: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing — review and editing. Jennifer C. Milligan: Conceptualization, resources, validation, investigation, methodology. Berta Canal: Conceptualization, resources, investigation, writing — review and editing. Rachel Ulferts: Conceptualization, investigation, writing — review and editing. Mary Wu: Investigation. Lucy S. Drury: Resources, investigation. Michael Howell: Resources, supervision. Rupert Beale: Resources, supervision.
Supplementary Material
References
- 1.Zhou, P., Yang, X.L., Wang, X.G., Hu, B., Zhang, L., Zhang, W.et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu, F., Zhao, S., Yu, B., Chen, Y.M., Wang, W., Song, Z.G.et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World-Health-Organization. COVID19 - Numbers at a glance, https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 28th February 2021
- 4.Agostini, M.L., Andres, E.L., Sims, A.C., Graham, R.L., Sheahan, T.P., Lu, X.et al. (2018) Coronavirus susceptibility to the antiviral remdesivir (GS-5734) Is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9, e00221-18 10.1128/mBio.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan, P.C., Stevens, S.K. and Deval, J. (2018) Nucleosides for the treatment of respiratory RNA virus infections. Antivir. Chem. Chemother. 26, 2040206618764483 10.1177/2040206618764483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zandi, K., Amblard, F., Musall, K., Downs-Bowen, J., Kleinbard, R., Oo, A.et al. (2020) Repurposing nucleoside analogs for human coronaviruses. Antimicrob. Agents Chemother. 65, e01652-20 10.1128/AAC.01652-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, Y., Zhang, D., Du, G., Du, R., Zhao, J., Jin, Y.et al. (2020) Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395, 1569–1578 10.1016/S0140-6736(20)31022-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spinner, C.D., Gottlieb, R.L., Criner, G.J., Arribas Lopez, J.R., Cattelan, A.M., Soriano Viladomiu, A.et al. (2020) Effect of remdesivir vs standard care on clinical status at 11 days in patients With moderate COVID-19: a randomized clinical trial. J. Am. Med. Assoc. 324, 1048–1057 10.1001/jama.2020.16349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman, J.D., Lye, D.C.B., Hui, D.S., Marks, K.M., Bruno, R., Montejano, R.et al. (2020) Remdesivir for 5 or 10 days in patients with severe Covid-19. N. Engl. J. Med. 383, 1827–1837 10.1056/NEJMoa2015301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO-Solidarity-Trial-Consortium, Pan, H., Peto, R., Henao-Restrepo, A.M., Preziosi, M.P., Sathiyamoorthy, V.et al. (2021) Repurposed antiviral drugs for Covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 384, 497–511. 10.1056/NEJMoa2023184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonis, A., Theobald, S.J., Fatkenheuer, G., Rybniker, J. and Malin, J.J. (2021) A comparative analysis of remdesivir and other repurposed antivirals against SARS-CoV-2. EMBO Mol. Med. 13, e13105 10.15252/emmm.202013105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agostini, M.L., Pruijssers, A.J., Chappell, J.D., Gribble, J., Lu, X., Andres, E.L.et al. (2019) Small-Molecule antiviral beta-d-N (4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 93, e01348-19 10.1128/JVI.01348-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheahan, T.P., Sims, A.C., Zhou, S., Graham, R.L., Pruijssers, A.J., Agostini, M.L.et al. (2020) An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12, eabb5883 10.1126/scitranslmed.abb5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steven, S., Good, J.W., Jung, K.H., La Colla, P., Collu, G., Moussa, A.et al. (2020) AT-527 is a potent in vitro replication inhibitor of SARS-CoV-2, the virus responsible for the COVID-19 pandemic. bioRxiv 2020.08.11.242834 10.1101/2020.08.11.242834 [DOI] [Google Scholar]
- 15.RECOVERY Trial (https://www.recoverytrial.net/results) [Internet] 2020
- 16.Choy, K.T., Wong, A.Y., Kaewpreedee, P., Sia, S.F., Chen, D., Hui, K.P.Y.et al. (2020) Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 178, 104786 10.1016/j.antiviral.2020.104786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udwadia, Z.F., Singh, P., Barkate, H., Patil, S., Rangwala, S., Pendse, A.et al. (2020) Efficacy and safety of favipiravir, an oral RNA-dependent RNA polymerase inhibitor, in mild-to-moderate COVID-19: a randomized, comparative, open-label, multicenter, phase 3 clinical trial. Int. J. Infect. Dis. 103, 62–71 10.1016/j.ijid.2020.11.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacramento NF-R, C.Q., Temerozo, J.R., de Paula Dias Da Silva, A., da Silva Gomes Dias, S., dos Santos da Silva, C., Ferreira, A.C.et al. (2020) The in vitro antiviral activity of the anti-hepatitis C virus (HCV) drugs daclatasvir and sofosbuvir against SARS-CoV-2. J. Antimicrob. Chemother. online ahead of print 10.1093/jac/dkab072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.V'Kovski, P., Kratzel, A., Steiner, S., Stalder, H. and Thiel, V. (2020) Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170. 10.1038/s41579-020-00468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snijder, E.J., Decroly, E. and Ziebuhr, J. (2016) The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 96, 59–126 10.1016/bs.aivir.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchdoerfer, R.N. and Ward, A.B. (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10, 2342 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subissi, L., Posthuma, C.C., Collet, A., Zevenhoven-Dobbe, J.C., Gorbalenya, A.E., Decroly, E.et al. (2014) One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl Acad. Sci. U.S.A. 111, E3900–E3909 10.1073/pnas.1323705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov, K.A. and Ziebuhr, J. (2004) Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5'-triphosphatase activities. J. Virol. 78, 7833–7838 10.1128/JVI.78.14.7833-7838.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov, K.A., Thiel, V., Dobbe, J.C., van der Meer, Y., Snijder, E.J. and Ziebuhr, J. (2004) Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78, 5619–5632 10.1128/JVI.78.11.5619-5632.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seybert, A., Hegyi, A., Siddell, S.G. and Ziebuhr, J. (2000) The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6, 1056–1068 10.1017/S1355838200000728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen, Y., Cai, H., Pan, J., Xiang, N., Tien, P., Ahola, T.et al. (2009) Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl Acad. Sci. U.S.A. 106, 3484–3489 10.1073/pnas.0808790106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouvet, M., Imbert, I., Subissi, L., Gluais, L., Canard, B. and Decroly, E. (2012) RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl Acad. Sci. U.S.A. 109, 9372–9377 10.1073/pnas.1201130109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minskaia, E., Hertzig, T., Gorbalenya, A.E., Campanacci, V., Cambillau, C., Canard, B.et al. (2006) Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl Acad. Sci. U.S.A. 103, 5108–5113 10.1073/pnas.0508200103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorbalenya, A.E., Enjuanes, L., Ziebuhr, J. and Snijder, E.J. (2006) Nidovirales: evolving the largest RNA virus genome. Virus Res. 117, 17–37 10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogando, N.S., Ferron, F., Decroly, E., Canard, B., Posthuma, C.C. and Snijder, E.J. (2019) The curious case of the nidovirus exoribonuclease: its role in RNA synthesis and replication fidelity. Front. Microbiol. 10, 1813 10.3389/fmicb.2019.01813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snijder, E.J., Bredenbeek, P.J., Dobbe, J.C., Thiel, V., Ziebuhr, J., Poon, L.L.et al. (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004 10.1016/S0022-2836(03)00865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, E.C., Blanc, H., Surdel, M.C., Vignuzzi, M. and Denison, M.R. (2013) Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 9, e1003565 10.1371/journal.ppat.1003565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo, Y. and Deutscher, M.P. (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29, 1017–1026 10.1093/nar/29.5.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckerle, L.D., Lu, X., Sperry, S.M., Choi, L. and Denison, M.R. (2007) High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81, 12135–12144 10.1128/JVI.01296-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckerle, L.D., Becker, M.M., Halpin, R.A., Li, K., Venter, E., Lu, X.et al. (2010) Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6, e1000896 10.1371/journal.ppat.1000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham, R.L., Becker, M.M., Eckerle, L.D., Bolles, M., Denison, M.R. and Baric, R.S. (2012) A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 18, 1820–1826 10.1038/nm.2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferron, F., Subissi, L., De Morais AT, S., Le, N.T.T., Sevajol, M., Gluais, L.et al. (2018) Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl Acad. Sci. U.S.A. 115, E162–EE71 10.1073/pnas.1718806115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordon, C.J., Tchesnokov, E.P., Woolner, E., Perry, J.K., Feng, J.Y., Porter, D.P.et al. (2020) Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 295, 6785–6797 10.1074/jbc.RA120.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kokic, G., Hillen, H.S., Tegunov, D., Dienemann, C., Seitz, F., Schmitzova, J.et al. (2021) Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 12, 279 10.1038/s41467-020-20542-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashburn, T.T. and Thor, K.B. (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 10.1038/nrd1468 [DOI] [PubMed] [Google Scholar]
- 41.Hillen, H.S., Kokic, G., Farnung, L., Dienemann, C., Tegunov, D. and Cramer, P. (2020) Structure of replicating SARS-CoV-2 polymerase. Nature 584, 154–156 10.1038/s41586-020-2368-8 [DOI] [PubMed] [Google Scholar]
- 42.Stein, A., Ruggiano, A., Carvalho, P. and Rapoport, T.A. (2014) Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 158, 1375–1388 10.1016/j.cell.2014.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn, D.G., Choi, J.K., Taylor, D.R. and Oh, J.W. (2012) Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 157, 2095–2104 10.1007/s00705-012-1404-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho, M.W., Richards, O.C., Dmitrieva, T.M., Agol, V. and Ehrenfeld, E. (1993) RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J. Virol. 67, 3010–3018 10.1128/JVI.67.6.3010-3018.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maga, G., Villani, G., Tillement, V., Stucki, M., Locatelli, G.A., Frouin, I.et al. (2001) Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl Acad. Sci. U.S.A. 98, 14298–14303 10.1073/pnas.251193198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng, J., Weissmann, F., Bertolin, A.P., Posse, V., Posse, V.et al. (2021) Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase. Biochem. J. 478, 2405–2423 10.1042/BCJ20210201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng, J., Weissmann, F., Bertolin, A.P., Posse, V., Canal, B., Ulferts, R.et al. (2021) Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp13 helicase. Biochem. J. 478, 2405–2423 10.1042/BCJ20210201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canal, B., McClure, A.W., Curran, J.F., Wu, M., Ulferts, R., Weissmann, F.et al. (2021) Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp14/nsp10 exoribonuclease. Biochem. J. 478, 2445–2463 10.1042/BCJ20210198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canal, B., Fujisawa, R., McClure, A.W., Deegan, T., Wu, M., Ulferts, R.et al. (2021) Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp15 endoribonuclease. Biochem. J. 478, 2465–2479 10.1042/BCJ20210199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu, S., Mak, T., Ulferts, R., Wu, M., Deegan, T., Fujisawa, R.et al. (2021) Identification of SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of the nsp14 RNA cap methyltransferase. Biochem. J. 478, 2481–2497 10.1042/BCJ20210219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milligan, J.C., Zeisner, T.U., Papageorgiou, G., Joshi, D., Soudy, C., Ulferts, R.et al. (2021) Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp5 main protease. Biochem. J. 478, 2499–2515 10.1042/BCJ20210197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim, C.T., Tan, K.W., Wu, M., Ulferts, R., Armstrong, L.A., Ozono, E.et al. (2021) Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp3 papain-like protease. Biochem. J. 478, 2517–2531 10.1042/BCJ20210244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorne, N., Auld, D.S. and Inglese, J. (2010) Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol. 14, 315–324 10.1016/j.cbpa.2010.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irwin, J.J., Duan, D., Torosyan, H., Doak, A.K., Ziebart, K.T., Sterling, T.et al. (2015) An aggregation advisor for ligand discovery. J. Med. Chem. 58, 7076–7087 10.1021/acs.jmedchem.5b01105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiedemar, N., Hauser, D.A. and Maser, P. (2020) 100 years of suramin. Antimicrob. Agents Chemother. 64, e01168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, K., Chong, R.A., Yu, Q., Bai, J., Spratt, D.E., Ching, K.et al. (2016) Suramin inhibits cullin-RING E3 ubiquitin ligases. Proc. Natl Acad. Sci. U.S.A. 113, E2011–E2018 10.1073/pnas.1601089113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su, L., Bryan, N., Battista, S., Freitas, J., Garabedian, A., D'Alessio, F.et al. (2020) Identification of HMGA2 inhibitors by alphaScreen-based ultra-high-throughput screening assays. Sci. Rep. 10, 18850 10.1038/s41598-020-75890-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, M., Sooreshjani, M.A., Mikek, C., Opoku-Temeng, C. and Sintim, H.O. (2018) Suramin potently inhibits cGAMP synthase, cGAS, in THP1 cells to modulate IFN-beta levels. Future Med. Chem. 10, 1301–1317 10.4155/fmc-2017-0322 [DOI] [PubMed] [Google Scholar]
- 59.Zhu, W., Xu, M., Chen, C.Z., Guo, H., Shen, M., Hu, X.et al. (2020) Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol. Transl. Sci. 3, 1008–1016 10.1021/acsptsci.0c00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren, P., Zheng, Y., Wang, W., Hong, L., Delpeyroux, F., Arenzana-Seisdedos, F.et al. (2017) Suramin interacts with the positively charged region surrounding the 5-fold axis of the EV-A71 capsid and inhibits multiple enterovirus A. Sci. Rep. 7, 42902 10.1038/srep42902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hosoi, Y., Matsumoto, Y., Tomita, M., Enomoto, A., Morita, A., Sakai, K.et al. (2002) Phosphorothioate oligonucleotides, suramin and heparin inhibit DNA-dependent protein kinase activity. Br. J. Cancer 86, 1143–1149 10.1038/sj.bjc.6600191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basavannacharya, C. and Vasudevan, S.G. (2014) Suramin inhibits helicase activity of NS3 protein of dengue virus in a fluorescence-based high throughput assay format. Biochem. Biophys. Res. Commun. 453, 539–544 10.1016/j.bbrc.2014.09.113 [DOI] [PubMed] [Google Scholar]
- 63.Tarantino, D., Pezzullo, M., Mastrangelo, E., Croci, R., Rohayem, J., Robel, I.et al. (2014) Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase. Antiviral Res. 102, 23–28 10.1016/j.antiviral.2013.11.016 [DOI] [PubMed] [Google Scholar]
- 64.Croci, R., Pezzullo, M., Tarantino, D., Milani, M., Tsay, S.C., Sureshbabu, R.et al. (2014) Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds. PLoS One 9, e91765 10.1371/journal.pone.0091765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mastrangelo, E., Pezzullo, M., Tarantino, D., Petazzi, R., Germani, F., Kramer, D.et al. (2012) Structure-based inhibition of norovirus RNA-dependent RNA polymerases. J. Mol. Biol. 419, 198–210 10.1016/j.jmb.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 66.Ferla, S., Netzler, N.E., Ferla, S., Veronese, S., Tuipulotu, D.E., Guccione, S.et al. (2018) In silico screening for human norovirus antivirals reveals a novel non-nucleoside inhibitor of the viral polymerase. Sci. Rep. 8, 4129 10.1038/s41598-018-22303-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Netzler, N.E., Enosi Tuipulotu, D., Eltahla, A.A., Lun, J.H., Ferla, S., Brancale, A.et al. (2017) Broad-spectrum non-nucleoside inhibitors for caliciviruses. Antiviral Res. 146, 65–75 10.1016/j.antiviral.2017.07.014 [DOI] [PubMed] [Google Scholar]
- 68.Feng, B.Y. and Shoichet, B.K. (2006) A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 1, 550–553 10.1038/nprot.2006.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Lazikani, B., Banerji, U. and Workman, P. (2012) Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 30, 679–692 10.1038/nbt.2284 [DOI] [PubMed] [Google Scholar]
- 70.Beigel, J.H., Tomashek, K.M., Dodd, L.E., Mehta, A.K., Zingman, B.S., Kalil, A.C.et al. (2020) Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 383, 1813–1826 10.1056/NEJMoa2007764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M.et al. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, Q., Wu, J., Wang, H., Gao, Y., Liu, Q., Mu, A.et al. (2020) Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell 182, 417–428.e13 10.1016/j.cell.2020.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, L.F. and Anderson, D.E. (2019) Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 34, 79–89 10.1016/j.coviro.2018.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin, W., Mao, C., Luan, X., Shen, D.D., Shen, Q., Su, H.et al. (2020) Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368, 1499–1504 10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen, J., Malone, B., Llewellyn, E., Grasso, M., Shelton, P.M.M., Olinares, P.D.B.et al. (2020) Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 182, 1560–1573.e13 10.1016/j.cell.2020.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sherk, A.B., Frigo, D.E., Schnackenberg, C.G., Bray, J.D., Laping, N.J., Trizna, W.et al. (2008) Development of a small-molecule serum- and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Res. 68, 7475–7483 10.1158/0008-5472.CAN-08-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Di Cristofano, A. (2017) SGK1: the dark side of PI3K signaling. Curr. Top. Dev. Biol. 123, 49–71 10.1016/bs.ctdb.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang, X., Lan, C., Zhou, J., Fu, W., Long, X., An, Y.et al. (2017) Development of a new analog of SGK1 inhibitor and its evaluation as a therapeutic molecule of colorectal cancer. J. Cancer 8, 2256–2262 10.7150/jca.19566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alamares-Sapuay, J.G., Martinez-Gil, L., Stertz, S., Miller, M.S., Shaw, M.L. and Palese, P. (2013) Serum- and glucocorticoid-regulated kinase 1 is required for nuclear export of the ribonucleoprotein of influenza A virus. J. Virol. 87, 6020–6026 10.1128/JVI.01258-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.PDK, H. (2020) The drugs of sleeping sickness: their mechanisms of action and resistance, and a brief history. Trop. Med. Infect. Dis. 5, 14 10.3390/tropicalmed5010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bowers, E.M., Yan, G., Mukherjee, C., Orry, A., Wang, L., Holbert, M.A.et al. (2010) Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. 17, 471–482 10.1016/j.chembiol.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu, X.Y., Huang, C.S., Li, Q., Chang, R.M., Song, Z.B., Zou, W.Y.et al. (2012) P300 exerts an epigenetic role in chronic neuropathic pain through its acetyltransferase activity in rats following chronic constriction injury (CCI). Mol. Pain 8, 84 10.1186/1744-8069-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santer, F.R., Hoschele, P.P., Oh, S.J., Erb, H.H., Bouchal, J., Cavarretta, I.T.et al. (2011) Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol. Cancer Ther. 10, 1644–1655 10.1158/1535-7163.MCT-11-0182 [DOI] [PubMed] [Google Scholar]
- 84.van den Bosch, T., Boichenko, A., Leus, N.G.J., Ourailidou, M.E., Wapenaar, H., Rotili, D.et al. (2016) The histone acetyltransferase p300 inhibitor C646 reduces pro-inflammatory gene expression and inhibits histone deacetylases. Biochem. Pharmacol. 102, 130–140 10.1016/j.bcp.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao, D., Fukuyama, S., Sakai-Tagawa, Y., Takashita, E., Shoemaker, J.E. and Kawaoka, Y. (2015) C646, a novel p300/CREB-binding protein-specific inhibitor of histone acetyltransferase, attenuates influenza A virus infection. Antimicrob. Agents Chemother. 60, 1902–1906 10.1128/AAC.02055-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pruijssers, A.J., George, A.S., Schafer, A., Leist, S.R., Gralinksi, L.E., Dinnon, III, K.H.et al. (2020) Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 32, 107940 10.1016/j.celrep.2020.107940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weissmann, F., Petzold, G., VanderLinden, R., Huis In ‘t Veld, P.J., Brown, N.G., Lampert, F.et al. (2016) biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl Acad. Sci. U.S.A. 113, E2564–E2569 10.1073/pnas.1604935113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trowitzsch, S., Bieniossek, C., Nie, Y., Garzoni, F. and Berger, I. (2010) New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 172, 45–54 10.1016/j.jsb.2010.02.010 [DOI] [PubMed] [Google Scholar]
- 89.Zhang, J.H., Chung, T.D. and Oldenburg, K.R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this paper has been deposited in FigShare (https://figshare.com/).