Abstract
Innate immune responses are tightly regulated by various pathways to control infections and maintain homeostasis. One of these pathways, the inflammasome pathway, activates a family of cysteine proteases called inflammatory caspases. They orchestrate an immune response by cleaving specific cellular substrates. Canonical inflammasomes activate caspase-1, whereas non-canonical inflammasomes activate caspase-4 and -5 in humans and caspase-11 in mice. Caspases are highly specific enzymes that select their substrates through diverse mechanisms. During inflammation, caspase activity is responsible for the secretion of inflammatory cytokines and the execution of a form of lytic and inflammatory cell death called pyroptosis. This review aims to bring together our current knowledge of the biochemical processes behind inflammatory caspase activation, substrate specificity, and substrate signalling.
Keywords: caspase, inflammasome, innate immunity, protease signalling
Introduction
Proteases are central enzymes that mediate numerous signalling roles to ensure cellular functions and organismal homeostasis [1]. Discovered more than 20 years ago, caspases are key signalling proteases that control various cell death processes and have been linked to inflammation and non-cell death-related functions [2–5].
Inflammatory caspases are a caspase subset activated by cellular platforms called inflammasomes [6–8]. Albeit mediating inflammasome signalling, our understanding of the biochemistry and the cellular processes governed by inflammatory caspases is limited. This mini-review aims to bring together our understanding of the mechanisms regulating inflammatory caspases activation, signalling and regulation.
Caspases…what's in the name?
The term caspase [2,9] is derived from the cysteine catalytic site used by the protease, and its rare specificity for cleavage at the carboxy-terminal side of Aspartic acid residues (D); cysteine-dependent aspartate-specific proteases. Caspases use a catalytic dyad composed of an histidine (H237 in caspase-1) and a cysteine (Cys285 in caspase-1) [2]. Initially discovered in Caenorhabditis elegans (C.elegans) [3,5], the role of caspases in development and innate immunity have since been characterised in a wide range of multicellular organisms. Recent work also clarifies important functions of caspases outside these processes, including proliferation, migration, and differentiation [10–13].
Caspases have a conserved modular organisation: a N-terminal domain (of variable length and function), a large catalytic subunit, and a small catalytic subunit [2] (Figure 1). These domains are separated by flexible linkers sensitive to proteolysis, the interdomain linker (IDL) and the recruitment domain linker (RDL). To date, twelve caspases have been identified in humans and ten in mice [2]. A classification system for caspases was developed, dividing each caspase into two main groups in accordance with their function, structure, and activation mechanism (Table 1).
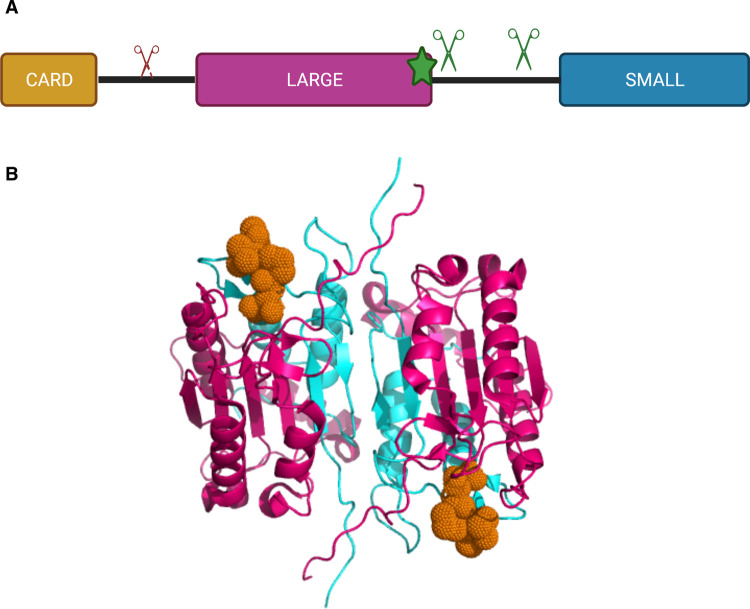
Figure 1. Structural organisation of an inflammatory caspase.
(A) Inflammatory caspases are composed of a CARD domain and a catalytic subunit, divided into a large and a small subunit. Caspases use a catalytic cysteine (shown as a star) to support their catalytic activity. Scissors represent inhibitory (red) and activating (green) self-processing sites. (B) Crystal structure of caspase-1 bound to the active-site inhibitor VX765 (PDB: 6PZP). The large subunit (pink) and the small subunit (cyan) of a caspase-1 dimer interacting with the active site inhibitor VX-765 (orange).
Table 1. Human caspases functions and activation mechanism overview.
| Caspase | Function/activation mechanism |
|---|---|
| 1 | Inflammation/dimerisation |
| 2 | Apoptosis/dimerisation |
| 3 | Apoptosis (Executioner)/dimerisation |
| 4 | Inflammation/dimerisation |
| 5 | Inflammation/dimerisation |
| 6 | Apoptosis (Executioner)/cleavage |
| 7 | Apoptosis (Executioner)/cleavage |
| 8 | Apoptosis (Initiator)/dimerisation |
| 9 | Apoptosis (Initiator)/dimerisation |
| 10 | Apoptosis (Initiator)/dimerisation |
| 12 | Unclear, catalytically inactive |
| 14 | Keratinocyte differentiation/dimerisation |
The first caspase group, the apoptotic caspases, can be further subdivided into initiator and executioner caspases based on their role within the apoptotic pathway. The initiator caspases (caspase-8, -9, and -10), are monomeric and contain a long homotypic N-terminal domain required for recruitment to their respective activation platform. The initiator caspases can then be further divided into whether they participate in the extrinsic or intrinsic apoptotic pathway. The extrinsic pathway involves caspase-8 and -10, which are activated by complexes like the death-inducing signalling complex (DISC) following binding of death ligands to their cognate receptors [14,15]. The intrinsic apoptotic pathway involves caspase-9, activated by the apoptosome following sensing of various intracellular signal like DNA-damage [16,17]. Once activated, the initiator caspases activate the executioner apoptotic caspases (caspase-3, -6 and -7) by cleaving their IDL [2,18].
Executioner caspases contain a short pro-domain (<30 residues) and are synthesised as inactive dimeric zymogens. Cleavage of executioner caspases by the initiator caspases allows full activation and therefore the cleavage of specific substrates to execute apoptotic cell death [19].
The second main caspase group is the inflammatory caspases [20]. The inflammatory caspases are encoded by three genes in humans (CASP1, CASP4, CASP5), and two in mice (casp1, casp11), clustered on a single locus, chromosome 11 in humans, and on chromosome 9 in mice. In mice, caspase-12 is also considered an inflammatory caspase [21] but roles of caspase-12 in inflammasome signalling have been debated [22]. Humans express a truncated and inactive version of caspase-12 [23,24], therefore this caspase will not be discussed further. The best characterised inflammatory caspase is caspase-1. This caspase, originally named IL-1β converting enzyme (ICE), was identified whilst studying the protease involved in the processing of the proIL-1β cytokine [4,25–27]. Shortly after, caspase-4, -5 and -11 [28] were linked to cell death and endotoxin responses. Caspase-1 is activated by a signalling complex called the canonical inflammasome, whereas caspase-4 and -11 (and potentially -5) are activated by the non-canonical inflammasome. Caspase-1 and caspase-4 are constitutively expressed in most cell types whereas caspase-5 expression is interferon inducible [29]. A recent study identified a gain-of-function mutation in CCAAT enhancer–binding protein epsilon (CEBPE) causes an autoinflammatory inflammasomopathy that leads to constitutive caspase-5 expression [30].
A few caspases fall out of the traditional classification due to their unique functions. An example of this is caspase-14, a caspase involved in keratinocyte differentiation [31]. Caspase-2 is another caspase member that has been linked with apoptotic processes and with innate immune functions, however does not comfortably fit into the current classification system [32].
The remainder of this review will focus on the mechanisms that govern inflammatory caspase activity.
Inflammatory caspase activation
Caspase-1
Inflammatory caspases exist as monomers under the cellular resting state and require dimerisation to become active. This dimerisation step is tightly regulated and is mediated by large, multi-protein signalling platforms called inflammasomes [6]. Inflammasomes are composed of a pattern recognition receptor (PRR) which senses danger-associated molecular patterns (DAMPs) or pathogen-associated molecular patterns (PAMPs), an adaptor protein (Apoptosis-associated speck-like protein (ASC)) and an inflammatory caspase [7,8].
PRR-activating inflammasomes are diverse in nature and recognise or respond to a multitude of chemically different ligands (summarised in Table 2).
Table 2. Inflammasome-forming pattern recognition receptors.
| Pattern recognition receptor | Examples of activating stimuli |
|---|---|
| NLRP1 | Proteases from pathogens [39,40] |
| viral dsRNA [41] | |
| Toxoplasma gondii [42] | |
| NLRP3 | Pore-forming toxins (Hemolysin, Candidalysin) [43,44] |
| B-glucan [45] | |
| ATP [46] | |
| Urate crystal [47] | |
| NLRP6 | Lipoteichoic acid (LTA) [48] |
| NLRP7 | Lipopeptide [49] |
| NLRC4/NAIP | T3SS proteins, Flagellin [50] |
| AIM2 | Cytosolic DNA [51,52] |
| Pyrin | Rho GTPase inactivation [53] |
| GBPs | LPS [54–56] |
Inflammasome-activating PRRs contain either a Pyrin domain (PYD) or Caspase activation and recruitment domain (CARD), both of which belong to the death-fold domain family [33]. The presence of these homotypic domains is a unifying feature of inflammasome-activating PRRs, which can therefore be divided into PYD-containing (NLRP3, NLRP6, NLRP7, AIM2, Pyrin) or CARD-containing (NLRP1, NLRC4, CARD8) PRR. Following PRR activation and oligomerisation, these domains undergo homotypic domain–domain interactions (PYD–PYD or CARD–CARD) that allow the recruitment of the adaptor protein ASC. ASC is a 22 kD adaptor protein, containing both a PYD and CARD domain. ASC–PYD oligomerisation leads to the formation of ASC filaments, and interactions between these filaments through ASC–CARD leads to the formation of the ASC speck, with a single cellular focus of ∼1 µm [34,35]. The ASC speck recruits caspase-1 monomers through CARD–CARD homotypic interactions, increasing local caspase-1 concentration therefore promoting caspase-1 dimerisation and allowing its activation [36]. Caspase dimerisation occurs through an interface located in the small catalytic subunit [2]. Dimerisation of caspase-1 induces basal activity and allows for the processing of the interdomain linker (IDL), which leads to structural reorganisation and stabilises the active site to generate a fully active caspase-1 species called p33/p10 (Figure 1) [36]. This activation mechanism, shared with initiator caspases, is known as proximity-induced dimerisation [19]. The caspase-1 p33/p10 species has the ability to process its established substrates (IL-1β, IL-18 and GSDMD) to mediate cell death and cytokine secretion (Figure 3). Caspase-1 subsequently cleaves its RDL to generate the p20/p10 species and dissociate from the inflammasome and become inactive [36]. Dimeric full-length caspase-1 is also partially active and can mediate cell death, but not cytokines processing [37].
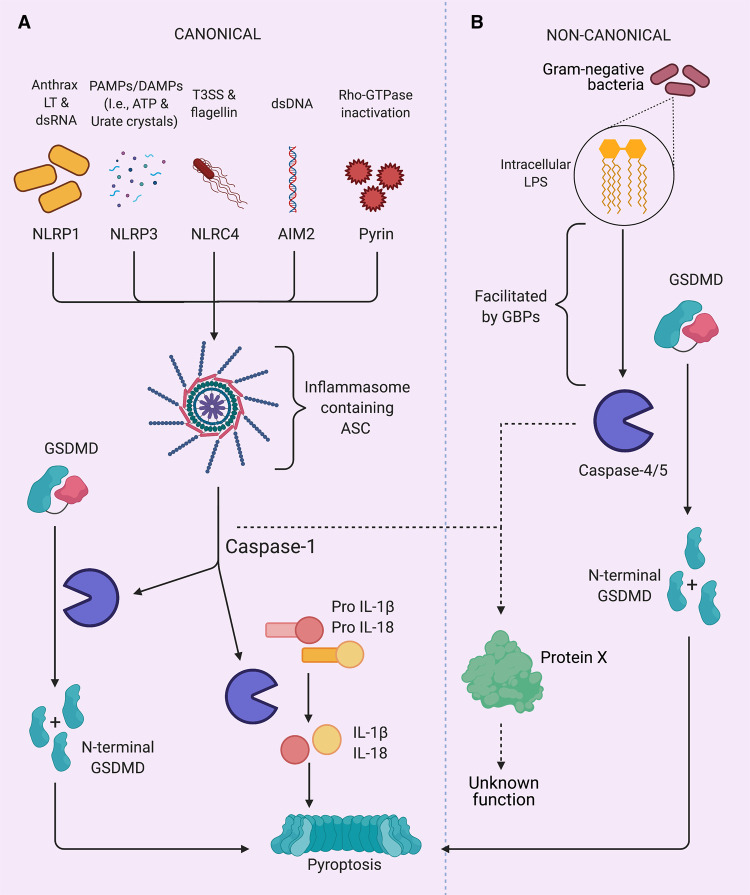
Figure 3. Canonical vs non-canonical inflammatory caspases signalling.
(A) PRRs recognise recognise DAMPs and PAMPs to activate the canonical inflammasome. This results in the activation of caspase-1 which cleaves GSDMD leading to pore formation and pyroptosis. Caspase-1 also activates pro-inflammatory cytokines IL-18 and IL-1b, which are release during pyroptosis and other substrates with unknown functional consequences (protein X). (B) Intracellular LPS from Gram negative bacteria activates the non-canonical inflammasome. GBPs aid the activation of caspase-4 and -5 by LPS. Caspase-4 and -5 cleave GSDMD, inducing pyroptosis, and other substrates with unknown functional consequences (protein X).
The ASC speck can also recruit and activate caspase-8 to trigger apoptosis [38].
Caspase-4 and -5
Inflammatory caspase-4 and -5 in humans, and caspase-11 in mice, are activated by the non-canonical inflammasome. Caspase-5 is conserved only in a few species (humans and great apes) and is believed to be the consequence of a genetic duplication. Until recently, these caspases were believed to directly binding lipopolysaccharide to trigger direct caspase dimerisation and activation [57]. However, recent studies identified cellular factors that facilitate the presentation of hydrophobic bacterial LPS to these caspases. Interferon-inducible guanylate binding proteins (GBPs) [58] recognise the outer section of LPS on cytosolic bacteria and allow for the assembly of an inflammasome directly on the bacteria. In human epithelial cells, this assembly platform is composed of GBP1, 2, 3 and 4 [54–56,59]. Outer membrane vesicles [60] can also activate the non-canonical inflammasome in a GBP-dependant manner [61]. In mice, GBPs facilitate the recruitment and localisation of IRGB10 to the membrane of invading pathogens, resulting in the destruction of the pathogen membrane and subsequent release of LPS and DNA, activating the non-canonical and AIM2 inflammasomes, respectively [62,63]. Humans do not express a functional orthologue of IRGB10. Caspase-4 and -5 active species are yet to be fully characterised; however, studies suggest that the p32/p10 form of caspase-4 could be the active species [64]. Studies into caspase-11 have supported this, showing that caspase-11 needs to be cleaved at the IDL to generate a fully active species [65,66]. Caspase-4 and -11 have also been suggested to be activated by the NLRP6 inflammasome downstream of Lipoteichoic acid (LTA) recognition [48]. However, the molecular basis of this process is not fully understood. Fatty acids and oxidised lipids have also been suggested to be endogenous ligands for the non-canonical inflammasome with cell-specific outcomes [67–69].
The cellular context controlling caspase-5 activation remain elusive. Specific LPS structures (e.g. Outer membrane vesicles from Pseudomonas aeruginosa) [70] and NLRP1 [6] were suggested to activate caspase-5. However, features enabling caspase-5 activation (instead of caspase-4) are subject of current investigations.
Caspase specificity
Inflammatory caspases are highly specific proteases that cleave defined protein substrates to orchestrate the innate immune responses. In the following section, we will discuss how caspases achieve this specificity through diverse mechanisms (Figure 2).
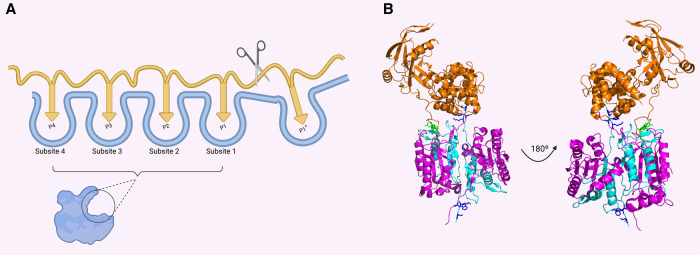
Figure 2. Substrate recognition mechanism by caspases.
(A) Representation of the caspase substrate-binding pocket. Four subsites on the caspase (Subsite 4 to Subsite 1) recognise the classical tetrapeptide on the substrate (P4 to P1). (B) Structure of the complex between caspase-11 (pink and cyan) and Gasdermin D (PDB: 6VIE) demonstrates that caspases can use exosite to recognise specific substrates. The structure shows the tetrapeptide recognise by the substrate-binding pocket (green) and the exosite (blue) that binds an additional region on Gasdermin D.
Primary specificity
Caspases natural substrate must be present in the same cellular compartment as the caspase and their cleavage site present specific features. First, the P1′ position is usually occupied by a small aliphatic amino acid. Secondly, the cleavage site is usually located in a solvent-exposed flexible structural element and is accessible to the protease active site. Finally, caspase substrate often displays an optimal primary cleavage sequence [71].
The substrates primary cleavage sequence bind through the caspase substrate-binding pocket, which can generally accommodate four amino acids from the substrate. According to the Schechter-Berger nomenclature [72], caspases recognise an aspartate in position P1 and several different amino acids in position P4 to P2 (Figure 2). Using small peptide library, the preferred substrate sequence for each caspase has been defined. Inflammatory caspases prefer aromatic or hydrophobic amino acids in position P4, glutamic acids in position P3, and small aliphatic amino acids in position P2 [73].
Recently, caspases were shown to cleave artificial substrates containing glutamic acid and phosphorylated serine in position P1, expanding the potential sequence of substrates [74].
Although the focus has been towards the primary tetrapeptide, multiple studies have highlighted the influence of extended subsites on caspase specificity. The Salvesen group identified extended subsites in caspase-11 and caspase-5 substrates that increase cleavage of selected substrates [75,76]. Extended subsites have also been suggested in other caspases [32].
The ability of certain caspases to cleave these primary sequences can also be influenced by post-translational modifications of the substrates. For example, phosphorylation of the substrates primary tetrapeptide has been shown to influence the ability of apoptotic caspases to cleave certain substrates [74,77]. However, its influence on inflammatory caspase specificity is currently unclear.
Caspase specificity to small peptides led to the development of caspase inhibitors. However, these inhibitors display relative specificity as they can target other caspases [78]. For example, an inhibitor derived from caspase-8 favourite recognition sequence will also inhibit efficiently other caspases [78].
Determinants outside the substrate-binding pocket have also been shown to influence caspase specificity [79].
Exosites
Exosites are structural motifs that allow binding of substrates independently of the primary substrate binding pocket. Although insufficient to allow substrate cleavage on their own, exosites enhance the cleavage efficiency of specific substrates and are used by various proteases to achieve protease specificity.
Exosites were first observed in apoptotic caspases. Caspase-7 harbours an exosite in its N-terminal domain that consist of four lysines [79]. Recently, this exosite has been suggested to bind RNA, facilitating RNA-binding protein cleavage by caspase-7 [80]. Similar sequences have been found in other apoptotic caspases, such as caspase-6 [81].
Until recently, no structure of caspases with their protein substrates were available. However, two recent papers outline unprecedent details on how exosites allow substrate specificity by inflammatory caspases.
Shao's and Tsiao's lab reveal the structure of caspase-1, -4 and -11 bound to the C-terminal fragment of GSDMD [82] or the full-length GSDMD [83]. Their structures reveal that the interaction between the loop L2 and L2’ of the caspase IDL creates a new binding site for the C-terminal domain of GSDMD, increasing caspase affinity for GSDMD and reducing the impact of a defined primary sequence (positions P4–P1 according to Schechter–Berger nomenclature [72]) for GSDMD to be cleaved by inflammatory caspases (Figure 2).
So far, GSDMD is the only inflammatory caspases substrate that has been clearly shown to bind on an inflammatory caspase exosite. However, recent evidence suggests that similar sites are also present for IL-1β [84].
Developing inhibitors that target exosites instead of the primary substrate-binding pocket bear the promise of more specific inhibitors.
Inflammatory caspases substrates and signalling
Caspase substrate cleavage has three functional consequences: a loss-of-functions, a gain of functions or a no-consequence effect. Caspase cleavage may also affect protein stability and target specific substrates to proteasomal degradation [85,86].
Proteomic studies have identified substrates for caspase-1 in mice [87,88] and humans [89] but so far failed to successfully identify caspase-4 and-5 substrates. Table 3 summarise substrates that have been identified by forward and reverse proteomics and shows that caspases are involved in multiple pathways (Figure 3).
Table 3. Inflammatory caspases substrates.
| Substrate | Uniprot ID | Cleavage site (P4–P1′) | Function | Caspase | Gain/Loss | Conserved | Reference |
|---|---|---|---|---|---|---|---|
| IL-1a | P01583 | Ile/Ala/Asn/Asp(104)/Ser | Pro-inflammatory cytokine | 5 | G | M/H | [95] |
| IL-1B | P01584 | Phe/Glu/Ala/Asp(27)/Gly | Pro-inflammatory cytokine | 1/5 | G | D/M/H | [4,89] |
| IL1B | P01584 | Tyr/Val/His/Asp(116)/Ala | Pro-inflammatory cytokine | 1 | G | D/M/H | [4,89] |
| IL-18 | O95256 | Leu/Glu/Ser/Asp(36)/Tyr | Pro-inflammatory cytokine | 1/4/5 | G | M/H | [75,111] |
| IL-37 | Q9NZH6 | Trp/Glu/Lys/Asp(20)/Glu | Anti-inflammatory cytokine | 1 | G | H | [94] |
| casp3 | P42574 | Ile/Gly/Thr/Asp(175)/Ser | Apoptotic pathway | 1/4 | G | M/H | [112,113] |
| casp7 | P55210 | Ile/Gln/Ala/Asp(198)/Ser | Apoptotic pathway | 1 | G | D/M/H | [87] |
| Bid | P55957 | Ile/Glu/ala/Asp(75)/Ser | Apoptotic pathway | 1 | G | D/M/H | [107,114] |
| BAP31 | P51572 | Ala/Ala/Val/Asp(231)/Gly | Apoptotic pathway | 1 | G | D/H | [115] |
| PARP1 | P09874 | Asp/Glu/Val/Asp(214)/Gly | Apoptotic pathway | 1 | L | D/M/H | [116] |
| SYAP1 | Q96A49 | Phe/Val/Ser/Asp(278)/Ala | Signal Transduction | 1/5 | UN | D/M/H | [89] |
| GSDMD | P57764 | Phe/Leu/Thr/Asp(275)/Gly | Pyroptosis | 1/4/5 | G | D/M/H | [89,97,99] |
| Actin | P60709 | Leu/Val/Val/Asp(11)/Asn | Cell structure | 1 | UN | D/M/H | [89] |
| Actin | P60709 | Gly/Gln/Lys/Asp(51)/Ser | Cell structure | 4 | UN | D/M/H | [89] |
| Actin | P60709 | Asp/Ser/Gly/Asp(157)/Gly | Cell structure | 1 | UN | D/M/H | [89] |
| Gelsolin | P06396 | Asp/Glu/Thr/Asp(403)/Gly | Cell structure | 1 | G | M/H | [89] |
| Spectrin | Q13813 | Asp/Glut/Thr/Asp(1185)/Ser | Cell structure | 4 | UN | D/M/H | [117] |
| ARPC5 | O15511 | Asp/Glu/Glu/Asp(29)/Gly | Cell structure | 1 | UN | D/M/H | [89] |
| RCSD1 | Q6JBY9 | Glu/Glu/Val/Asp(272)/Gly | Cell structure | 1 | UN | H | [89] |
| IQGAP1 | IQGAP1 | Asp/Glu/Val/Asp(8)/Gly | Cell structure | 1 | UN | D/M/H | [89] |
| GAPDH | P04075 | Lys/Thr/Val/Asp(189)/Gly | Metabolism | 1 | L | D/M/H | [88] |
| ENO1 | P06733 | ? | Metabolism | 1 | UN | H | [88] |
| TPI1 | P60174 | ? | Metabolism | 1 | UN | H | [88] |
| PIP4K2B | Q9UBF8 | Phe/Ser/Val/Asp(488)/Ser | Kinase | 1 | UN | D/M/H | [89] |
| BASP1 | P80723 | Thr/Lys/Ser/Asp(165)/Gly | Channel | 1 | UN | H | [89] |
| CALU | O43852 | Tyr/Ile/Gly/Asp(216)/Met | Calcium-binding | 1 | UN | D/M/H | [89] |
| USP10 | Q14694 | Leu/Glu/Asn/Asp(138)/Gly | Ubiquitin protease | 1 | UN | D/M/H | [89] |
| HOIP | Q96EP0 | Leu/Glu/Pro/Asp(348)/Leu | Ubiquitin ligase | 1 | L | M/H | [118] |
| HOIP | Q96EP0 | Leu/Val/Val/Val/Asp(387)/Ser | Ubiquitin ligase | 1 | L | D/M/H | [118] |
| TFAP2A | P05549 | Asp/Arg/His/Asp(19)/Gly | Transcription | 1 | UN | M/H | [119] |
| Max | P61244 | Ile/Glu/Val/Glu(10)/Ser | Transcription | 5 | UN | M/H | [110] |
Cytokines
Caspase-1 was originally identified as an Interleukine-1β (IL-1β)-converting enzyme and was originally characterised to cleave and mature this cytokine. Caspase-1 also processes the IL-18 pro-form into its mature form. The cytokines IL-1β and IL-18 are unconventionally secreted through GSDMD pores or following cell lysis [90,91]. Caspase-4 also cleave these cytokines, although much less efficiently than caspase-1 [75]. Cleavage of IL-1β and IL-18 leads to the recruitment of additional phagocytes and contributes to the generation of a fever. Caspase-8 also cleaves IL-1β and IL-18 during multiple situations [92,93].
Caspase-1 also cleaves IL-37, an anti-inflammatory cytokine, to promote IL-37 nuclear translocation and genetic repression of anti-inflammatory cytokines [94].
Caspase-5 has been reported to cleave IL-1α in senescent cells, a process that may contribute to aging-associated inflammation [95].
Gasdermin D
Gasdermin D (GSDMD) is a central executor of pyroptosis (Figure 3). Following cleavage by caspase-1, -4 (-11), -5 and -8, the GSDMD N-terminal fragment is freed from its inhibitory counterpart (C-terminal fragment) and is able to form pores at the plasma membrane and into different organelles [96–99]. GSDMD pores allow secretion of pro-inflammatory cytokines (IL-1β, IL-18) and the release of a myriad of DAMPS (e.g. ATP, Galectin-1) [100,101]. Additionally, GSDMD pores generate potassium efflux to allow caspase-1 activation through the NLRP3 inflammasome, downstream of the non-canonical inflammasome [102,103]. Terminal membrane rupture downstream of GSDMD pores has been shown to be mediated by the membrane protein NINJ1 [104]. In neutrophils, GSDMD cleavage by non-canonical inflammatory caspases allows the generation of neutrophil extracellular traps [105].
Other substrates
Caspase-1 can cleave proapoptotic proteins like Bid and caspase-3 and -7. It has been suggested to control infection by specific pathogens and stands as a backup cell death mechanism if pyroptosis is counteracted by pathogens [106–108].
Caspase-1, -4, -5, and -11 can cleave and inactivate cGAS to control type I IFN response and modulate antiviral responses [109].
Caspase-1 has been shown to cleave other substrates however, the functional relevance of these substrates remains unclear. For example, caspase-1 may contribute to cell demise by cleaving many structural proteins (e.g. vimentin, actin, gelsolin, IQGAP1 and others (Table 3)). Caspase-1 may also regulate RNA-mediated processes and metabolism by cleaving ribonucleoproteins [87,89] and glycolytic enzymes [88] (Table 3)).
Known caspase-4 and -5 substrates are minimal, and efforts to identify them thus far have been limited. Outside the substrates mentioned above, caspase-5 cleaves the transcription factor Max after glutamic acid [110], and SYAP1 at an unknown site [89].
Caspases and their substrates during evolution
Numerous caspase substrates are conserved throughout evolution. Inflammatory caspases are present in vertebrates, from the zebrafish [120,121] to higher primates [20] (Table 3; D (Danio rerio), M (Mus musculus), H (Homo sapiens)). Human inflammatory caspases share high similarities with higher primates caspases.
Multiple substrates seem to be conserved during evolution and many of their cleavage site position (Table 3) is highly conserved, suggesting a role for various caspases substrates throughout evolution. Ancestral reconstitutions of caspases support the co-evolution of caspases and their substrates [71,122].
Caspase inhibitors
Whereas apoptotic caspase activity is controlled by the inhibitors of apoptosis proteins (IAP) [123], endogenous inhibitors controlling inflammatory caspase activity are poorly characterised.
SerpinB1 was suggested to be an endogenous inhibitor of inflammatory caspases in some cell types, such as neutrophils [124].
The CARD-only proteins (COPs) and PYD-only proteins (POPs) can inhibit inflammatory caspases indirectly by regulating inflammasome formation and/or caspase recruitment to their activating platform [125].
The involvement of inflammatory caspases in a range of diseases containing an inflammatory component (from obesity to cancer and sepsis [126,127]) supports the urgency to develop specific inhibitors that can target one or multiple inflammatory caspases.
Vertex developed a caspase-1/4 inhibitor, VX765, that was taken to clinical trials [128]. However, the studies were halted in stage 2 due to liver toxicity [129].
The development of caspase inhibitors that target exosites highlights the possibility of more specific inhibitors.
In addition, the development of exosite inhibitors has the potential to generate inhibitors that target deleterious functions of caspases, without affecting the beneficial ones, by modulating the cleavage of selected substrates.
Perspectives
Importance in the field: Inflammatory caspases are crucial for regulated immune responses and linked to diverse pathologies from sepsis to cancer.
Summary of the current thinking: Inflammatory caspases are activated either by canonical or non-canonical inflammasomes. GBPs are novel innate immune sensors that form a non-canonical inflammasome and facilitate LPS presentation to caspase-4 and -11. Caspases recognise their substrates through substrate binding-pockets and use exosites to increase substrate selectivity.
Future directions: Research to identify and characterise novel caspase substrates will expand our understanding of inflammatory caspases in health and disease. Future research will address how inflammatory caspases activity is controlled by endogenous mechanisms and inhibitors. Targeting caspase exosites may allow for the development of more specific pharmacological inhibitors.
Acknowledgements
The figures were generated using Biorender (biorender.com). We apologized to colleagues we did not cited due to limited space.
Abbreviations
- ASC
apoptosis-associated speck-like protein
- CARD
Caspase recruitment and activation domain
- COP
CARD-only protein
- DAMP
danger-associated molecular patterns
- IDL
interdomain linker
- LTA
lipoteichoic acid
- PAMP
pathogen-associated molecular patterns
- POP
PYD-only protein
- PRR
pattern recognition receptor
- PYD
pyrin domain
- RDL
recruitment domain linker
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The Boucher lab is supported by a Springboard award (SBF006\1025) from the Academy for Medical Science and institutional funding from the University of York.
Open Access
Open access for this article was enabled by the participation of University of York in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Author Contributions
G.B. and B.H. contributed equally to this work. G.B., B.H. and R.K. wrote the review. D.B. wrote the review and supervised.
References
- 1.López-Otín, C. and Bond, J.S. (2008) Proteases: multifunctional enzymes in life and disease. J. Biol. Chem. 283, 30433–7 10.1074/jbc.R800035200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuentes-Prior, P. and Salvesen, G.S. (2004) The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384, 201–232 10.1042/BJ20041142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis, H.M. and Horvitz, H.R. (1986) Genetic control of programmed cell death in the nematode C. elegans. Cell 44, 817–829 10.1016/0092-8674(86)90004-8 [DOI] [PubMed] [Google Scholar]
- 4.Thornberry, N.A., Bull, H.G., Calaycay, J.R., Chapman, K.T., Howard, A.D., Kostura, M.J.et al. (1992) A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356, 768–774 10.1038/356768a0 [DOI] [PubMed] [Google Scholar]
- 5.Yuan, J., Shaham, S., Ledoux, S., Ellis, H.M. and Horvitz, H.R. (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75, 641–652 10.1016/0092-8674(93)90485-9 [DOI] [PubMed] [Google Scholar]
- 6.Martinon, F., Burns, K. and Tschopp, J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- 7.Schroder, K. and Tschopp, J. (2010) The inflammasomes. Cell 140, 821–832 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- 8.Broz, P. and Dixit, V.M. (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- 9.Minina, E.A., Staal, J., Alvarez, V.E., Berges, J.A., Berman-Frank, I., Beyaert, R.et al. (2020) Classification and nomenclature of metacaspases and paracaspases: no more confusion with caspases. Mol. Cell 77, 927–929 10.1016/j.molcel.2019.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baena-Lopez, L.A., Arthurton, L., Xu, D.C. and Galasso, A. (2018) Non-apoptotic caspase regulation of stem cell properties. Semin. Cell Dev. Biol. 82, 118–126 10.1016/j.semcdb.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solier, S., Fontenay, M., Vainchenker, W., Droin, N. and Solary, E. (2017) Non-apoptotic functions of caspases in myeloid cell differentiation. Cell Death Differ. 24, 1337–1347 10.1038/cdd.2017.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McArthur, K. and Kile, B.T. (2018) Apoptotic caspases: multiple or mistaken identities? Trends Cell Biol. 28, 475–493 10.1016/j.tcb.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee, A. and Williams, D.W. (2017) More alive than dead: non-apoptotic roles for caspases in neuronal development, plasticity and disease. Cell Death Differ. 24, 1411–1421 10.1038/cdd.2017.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kucka, K. and Wajant, H. (2020) Receptor oligomerization and its relevance for signaling by receptors of the tumor necrosis factor receptor superfamily. Front. Cell Dev. Biol. 8, 615141 10.3389/fcell.2020.615141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tummers, B. and Green, D.R. (2017) Caspase-8: regulating life and death. Immunol. Rev. 277, 76–89 10.1111/imr.12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schafer, Z.T. and Kornbluth, S. (2006) The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell 10, 549–561 10.1016/j.devcel.2006.04.008 [DOI] [PubMed] [Google Scholar]
- 17.Dorstyn, L., Akey, C.W. and Kumar, S. (2018) New insights into apoptosome structure and function. Cell Death Differ. 25, 1194–1208 10.1038/s41418-017-0025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmore, S. (2007) Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pop, C. and Salvesen, G.S. (2009) Human caspases: activation, specificity, and regulation. J. Biol. Chem. 284, 21777–21781 10.1074/jbc.R800084200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinon, F. and Tschopp, J. (2007) Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 14, 10–22 10.1038/sj.cdd.4402038 [DOI] [PubMed] [Google Scholar]
- 21.Saleh, M., Vaillancourt, J.P., Graham, R.K., Huyck, M., Srinivasula, S.M., Alnemri, E.S.et al. (2004) Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature 429, 75–79 10.1038/nature02451 [DOI] [PubMed] [Google Scholar]
- 22.Vande Walle, L., Jiménez Fernández, D., Demon, D., Van Laethem, N., Van Hauwermeiren, F., Van Gorp, H.et al. (2016) Does caspase-12 suppress inflammasome activation? Nature 534, E1–E4 10.1038/nature17649 [DOI] [PubMed] [Google Scholar]
- 23.Skeldon, A.M., Morizot, A., Douglas, T., Santoro, N., Kursawe, R., Kozlitina, J.et al. (2016) Caspase-12, but not caspase-11, inhibits obesity and insulin resistance. J. Immunol. 196, 437–447 10.4049/jimmunol.1501529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer, H., Koenig, U., Eckhart, L. and Tschachler, E. (2002) Human caspase 12 has acquired deleterious mutations. Biochem. Biophys. Res. Commun. 293, 722–726 10.1016/S0006-291X(02)00289-9 [DOI] [PubMed] [Google Scholar]
- 25.Howard, A.D., Kostura, M.J., Thornberry, N., Ding, G.J., Limjuco, G., Weidner, J.et al. (1991) IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1 beta precursor at two distinct sites and does not cleave 31-kDa IL-1 alpha. J. Immunol. 147, 2964–2969 PMID: [PubMed] [Google Scholar]
- 26.Cerretti, D.P., Kozlosky, C.J., Mosley, B., Nelson, N., Van Ness, K., Greenstreet, T.A.et al. (1992) Molecular cloning of the interleukin-1 beta converting enzyme. Science 256, 97–100 10.1126/science.1373520 [DOI] [PubMed] [Google Scholar]
- 27.Kostura, M.J., Tocci, M.J., Limjuco, G., Chin, J., Cameron, P., Hillman, A.G.et al. (1989) Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc. Natl Acad. Sci. U.S.A. 86, 5227–5231 10.1073/pnas.86.14.5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, S., Miura, M., Jung, Y.K., Zhu, H., Li, E. and Yuan, J. (1998) Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell 92, 501–509 10.1016/S0092-8674(00)80943-5 [DOI] [PubMed] [Google Scholar]
- 29.Lin, X.Y., Choi, M.S. and Porter, A.G. (2000) Expression analysis of the human caspase-1 subfamily reveals specific regulation of the CASP5 gene by lipopolysaccharide and interferon-gamma. J. Biol. Chem. 275, 39920–6 10.1074/jbc.M007255200 [DOI] [PubMed] [Google Scholar]
- 30.Göös, H., Fogarty, C.L., Sahu, B., Plagnol, V., Rajamäki, K., Nurmi, K.et al. (2019) Gain-of-function CEBPE mutation causes noncanonical autoinflammatory inflammasomopathy. J. Allergy Clin. Immunol. 144, 1364–1376 10.1016/j.jaci.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denecker, G., Ovaere, P., Vandenabeele, P. and Declercq, W. (2008) Caspase-14 reveals its secrets. J. Cell Biol. 180, 451–458 10.1083/jcb.200709098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown-Suedel, A.N. and Bouchier-Hayes, L. (2020) Caspase-2 substrates: to apoptosis, cell cycle control, and beyond. Front. Cell Dev. Biol. 8, 610022 10.3389/fcell.2020.610022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kersse, K., Verspurten, J., Vanden Berghe, T. and Vandenabeele, P. (2011) The death-fold superfamily of homotypic interaction motifs. Trends Biochem. Sci. 36, 541–552 10.1016/j.tibs.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 34.Dick, M.S., Sborgi, L., Rühl, S., Hiller, S. and Broz, P. (2016) ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat. Commun. 7, 11929 10.1038/ncomms11929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, A., Magupalli, V.G., Ruan, J., Yin, Q., Atianand, M.K., Vos, M.R.et al. (2014) Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boucher, D., Monteleone, M., Coll, R.C., Chen, K.W., Ross, C.M., Teo, J.L.et al. (2018) Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med. 215, 827–840 10.1084/jem.20172222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broz, P., von Moltke, J., Jones, J.W., Vance, R.E. and Monack, D.M. (2010) Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sagulenko, V., Thygesen, S.J., Sester, D.P., Idris, A., Cridland, J.A., Vajjhala, P.R.et al. (2013) AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 20, 1149–1160 10.1038/cdd.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levinsohn, J.L., Newman, Z.L., Hellmich, K.A., Fattah, R., Getz, M.A., Liu, S.et al. (2012) Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 8, e1002638 10.1371/journal.ppat.1002638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, K.S., Teo, D.E.T., Tan, K.S., Toh, G.A., Ong, H.H., Lim, C.K.et al. (2020) Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370, eaay2002 10.1126/science.aay2002 [DOI] [PubMed] [Google Scholar]
- 41.Bauernfried, S., Scherr, M.J., Pichlmair, A., Duderstadt, K.E. and Hornung, V. (2021) Human NLRP1 is a sensor for double-stranded RNA. Science 371, eabd0811 10.1126/science.abd0811 [DOI] [PubMed] [Google Scholar]
- 42.Ewald, S.E., Chavarria-Smith, J. and Boothroyd, J.C. (2014) NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect. Immun. 82, 460–468 10.1128/IAI.01170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasper, L., König, A., Koenig, P.-A., Gresnigt, M.S., Westman, J., Drummond, R.A.et al. (2018) The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat. Commun. 9, 4260 10.1038/s41467-018-06607-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murthy, A.M.V., Sullivan, M.J., Nhu, N.T.K., Lo, A.W., Phan, M.-D., Peters, K.M.et al. (2019) Variation in hemolysin A expression between uropathogenic Escherichia coli isolates determines NLRP3-dependent vs. -independent macrophage cell death and host colonization. FASEB J. 33, 7437–7450 10.1096/fj.201802100R [DOI] [PubMed] [Google Scholar]
- 45.Gross, O., Poeck, H., Bscheider, M., Dostert, C., Hannesschläger, N., Endres, S.et al. (2009) Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 459, 433–436 10.1038/nature07965 [DOI] [PubMed] [Google Scholar]
- 46.Mariathasan, S., Weiss, D.S., Newton, K., McBride, J., O'Rourke, K., Roose-Girma, M.et al. (2006) Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 10.1038/nature04515 [DOI] [PubMed] [Google Scholar]
- 47.Martinon, F., Pétrilli, V., Mayor, A., Tardivel, A. and Tschopp, J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 10.1038/nature04516 [DOI] [PubMed] [Google Scholar]
- 48.Hara, H., Seregin, S.S., Yang, D., Fukase, K., Chamaillard, M., Alnemri, E.S.et al. (2018) The NLRP6 inflammasome recognizes lipoteichoic acid and regulates gram-positive pathogen infection. Cell 175, 1651–1664.e14 10.1016/j.cell.2018.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khare, S., Dorfleutner, A., Bryan, N.B., Yun, C., Radian, A.D., de Almeida, L.et al. (2012) An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476 10.1016/j.immuni.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kay, C., Wang, R., Kirkby, M. and Man, S.M. (2020) Molecular mechanisms activating the NAIP-NLRC4 inflammasome: implications in infectious disease, autoinflammation, and cancer. Immunol. Rev. 297, 67–82 10.1111/imr.12906 [DOI] [PubMed] [Google Scholar]
- 51.Hornung, V., Ablasser, A., Charrel-Dennis, M., Bauernfeind, F., Horvath, G., Caffrey, D.R.et al. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, T.L., Idris, A., Dunn, J.A., Kelly, G.M., Burnton, C.M., Hodgson, S.et al. (2009) HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323, 1057–1060 10.1126/science.1169841 [DOI] [PubMed] [Google Scholar]
- 53.Xu, H., Yang, J., Gao, W., Li, L., Li, P., Zhang, L.et al. (2014) Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237–241 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- 54.Santos, J.C., Boucher, D., Schneider, L.K., Demarco, B., Dilucca, M., Shkarina, K.et al. (2020) Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria. Nat. Commun. 11, 3276 10.1038/s41467-020-16889-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wandel, M.P., Kim, B.-H., Park, E.-S., Boyle, K.B., Nayak, K., Lagrange, B.et al. (2020) Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol. 21, 880–891 10.1038/s41590-020-0697-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kutsch, M., Sistemich, L., Lesser, C.F., Goldberg, M.B., Herrmann, C. and Coers, J. (2020) Direct binding of polymeric GBP1 to LPS disrupts bacterial cell envelope functions. EMBO J. 39, e104926 10.15252/embj.2020104926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi, J., Zhao, Y., Wang, Y., Gao, W., Ding, J., Li, P.et al. (2014) Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- 58.Tretina, K., Park, E.-S., Maminska, A. and MacMicking, J.D. (2019) Interferon-induced guanylate-binding proteins: guardians of host defense in health and disease. J. Exp. Med. 216, 482–500 10.1084/jem.20182031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisch, D., Clough, B., Domart, M.-C., Encheva, V., Bando, H., Snijders, A.P.et al. (2020) Human GBP1 differentially targets Salmonella and Toxoplasma to license recognition of microbial ligands and caspase-mediated death. Cell Rep. 32, 108008 10.1016/j.celrep.2020.108008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanaja, S.K., Russo, A.J., Behl, B., Banerjee, I., Yankova, M., Deshmukh, S.D.et al. (2016) Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 165, 1106–1119 10.1016/j.cell.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santos, J.C., Dick, M.S., Lagrange, B., Degrandi, D., Pfeffer, K., Yamamoto, M.et al. (2018) LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. 37, e98089 10.15252/embj.201798089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Man, S.M., Karki, R., Sasai, M., Place, D.E., Kesavardhana, S., Temirov, J.et al. (2016) IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell.. 167, 382–396.e17 10.1016/j.cell.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meunier, E., Wallet, P., Dreier, R.F., Costanzo, S., Anton, L., Rühl, S.et al. (2015) Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16, 476–484 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casson, C.N., Yu, J., Reyes, V.M., Taschuk, F.O., Yadav, A., Copenhaver, A.M.et al. (2015) Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proc. Natl Acad. Sci. U.S.A. 112, 6688–6693 10.1073/pnas.1421699112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ross, C., Chan, A.H., Von Pein, J., Boucher, D. and Schroder, K. (2018) Dimerization and auto-processing induce caspase-11 protease activation within the non-canonical inflammasome. Life Sci. Alliance 1, e201800237 10.26508/lsa.201800237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee, B.L., Stowe, I.B., Gupta, A., Kornfeld, O.S., Roose-Girma, M., Anderson, K.et al. (2018) Caspase-11 auto-proteolysis is crucial for noncanonical inflammasome activation. J. Exp. Med. 215, 2279–2288 10.1084/jem.20180589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chu, L.H., Indramohan, M., Ratsimandresy, R.A., Gangopadhyay, A., Morris, E.P., Monack, D.M.et al. (2018) The oxidized phospholipid oxPAPC protects from septic shock by targeting the non-canonical inflammasome in macrophages. Nat. Commun. 9, 996 10.1038/s41467-018-03409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zanoni, I., Tan, Y., Di Gioia, M., Broggi, A., Ruan, J., Shi, J.et al. (2016) An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352, 1232–1236 10.1126/science.aaf3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pillon, N.J., Chan, K.L., Zhang, S., Mejdani, M., Jacobson, M.R., Ducos, A.et al. (2016) Saturated fatty acids activate caspase-4/5 in human monocytes, triggering IL-1β and IL-18 release. Am. J. Physiol. Endocrinol. Metab. 311, E825–E835 10.1152/ajpendo.00296.2016 [DOI] [PubMed] [Google Scholar]
- 70.Bitto, N.J., Baker, P.J., Dowling, J.K., Wray-McCann, G., De Paoli, A., Tran, L.S.et al. (2018) Membrane vesicles from Pseudomonas aeruginosa activate the noncanonical inflammasome through caspase-5 in human monocytes. Immunol. Cell Biol. 96, 1120–1130 10.1111/imcb.12190 [DOI] [PubMed] [Google Scholar]
- 71.Timmer, J.C., Zhu, W., Pop, C., Regan, T., Snipas, S.J., Eroshkin, A.M.et al. (2009) Structural and kinetic determinants of protease substrates. Nat. Struct. Mol. Biol. 16, 1101–1108 10.1038/nsmb.1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schechter, I. and Berger, A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 10.1016/S0006-291X(67)80055-X [DOI] [PubMed] [Google Scholar]
- 73.Thornberry, N.A., Rano, T.A., Peterson, E.P., Rasper, D.M., Timkey, T., Garcia-Calvo, M.et al. (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 10.1074/jbc.272.29.17907 [DOI] [PubMed] [Google Scholar]
- 74.Seaman, J.E., Julien, O., Lee, P.S., Rettenmaier, T.J., Thomsen, N.D. and Wells, J.A. (2016) Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues. Cell Death Differ. 23, 1717–1726 10.1038/cdd.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bibo-Verdugo, B., Snipas, S.J., Kolt, S., Poreba, M. and Salvesen, G.S. (2020) Extended subsite profiling of the pyroptosis effector protein gasdermin D reveals a region recognized by inflammatory caspase-11. J. Biol. Chem. 295, 11292–11302 10.1074/jbc.RA120.014259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramirez, M.L.G., Poreba, M., Snipas, S.J., Groborz, K., Drag, M. and Salvesen, G.S. (2018) Extensive peptide and natural protein substrate screens reveal that mouse caspase-11 has much narrower substrate specificity than caspase-1. J. Biol. Chem. 293, 7058–7067 10.1074/jbc.RA117.001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dix, M.M., Simon, G.M., Wang, C., Okerberg, E., Patricelli, M.P. and Cravatt, B.F. (2012) Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell 150, 426–440 10.1016/j.cell.2012.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McStay, G.P., Salvesen, G.S. and Green, D.R. (2008) Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 15, 322–331 10.1038/sj.cdd.4402260 [DOI] [PubMed] [Google Scholar]
- 79.Boucher, D., Blais, V. and Denault, J.-B. (2012) Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc. Natl Acad. Sci. U.S.A. 109, 5669–5674 10.1073/pnas.1200934109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desroches, A. and Denault, J.-B. (2019) Caspase-7 uses RNA to enhance proteolysis of poly(ADP-ribose) polymerase 1 and other RNA-binding proteins. Proc. Natl Acad. Sci. U.S.A. 116, 21521–8 10.1073/pnas.1909283116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacPherson, D.J., Mills, C.L., Ondrechen, M.J. and Hardy, J.A. (2019) Tri-arginine exosite patch of caspase-6 recruits substrates for hydrolysis. J. Biol. Chem. 294, 71–88 10.1074/jbc.RA118.005914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, K., Sun, Q., Zhong, X., Zeng, M., Zeng, H., Shi, X.et al. (2020) Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell 180, 941–955.e20 10.1016/j.cell.2020.02.002 [DOI] [PubMed] [Google Scholar]
- 83.Liu, Z., Wang, C., Yang, J., Chen, Y., Zhou, B., Abbott, D.W.et al. (2020) Caspase-1 engages full-length gasdermin D through two distinct interfaces that mediate caspase recruitment and substrate cleavage. Immunity 53, 106–114.e5 10.1016/j.immuni.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Devant, P., Cao, A. and Kagan, J.C. (2020) Evolution-inspired dissection of caspase activities enables the redesign of caspase-4 into an LPS sensing interleukin-1 converting enzyme. bioRxiv 10.1101/2020.12.07.413732 [DOI] [Google Scholar]
- 85.Varshavsky, A. (2019) N-degron and C-degron pathways of protein degradation. Proc. Natl Acad. Sci. U.S.A. 116, 358–366 10.1073/pnas.1816596116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weaver, B.P., Weaver, Y.M., Mitani, S. and Han, M. (2017) Coupled caspase and N-end rule ligase activities allow recognition and degradation of pluripotency factor LIN-28 during non-apoptotic development. Dev. Cell 41, 665–673.e6 10.1016/j.devcel.2017.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lamkanfi, M., Kanneganti, T.-D., Van Damme, P., Vanden Berghe, T., Vanoverberghe, I., Vandekerckhove, J.et al. (2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell Proteomics 7, 2350–2363 10.1074/mcp.M800132-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shao, W., Yeretssian, G., Doiron, K., Hussain, S.N. and Saleh, M. (2007) The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J. Biol. Chem. 282, 36321–9 10.1074/jbc.M708182200 [DOI] [PubMed] [Google Scholar]
- 89.Agard, N.J., Maltby, D. and Wells, J.A. (2010) Inflammatory stimuli regulate caspase substrate profiles. Mol. Cell Proteomics 9, 880–893 10.1074/mcp.M900528-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monteleone, M., Stanley, A.C., Chen, K.W., Brown, D.L., Bezbradica, J.S., von Pein, J.B.et al. (2018) Interleukin-1β maturation triggers its relocation to the plasma membrane for gasdermin-D-dependent and -independent secretion. Cell Rep. 24, 1425–1433 10.1016/j.celrep.2018.07.027 [DOI] [PubMed] [Google Scholar]
- 91.Chan, A.H. and Schroder, K. (2020) Inflammasome signaling and regulation of interleukin-1 family cytokines. J. Exp. Med. 217, e20190314 10.1084/jem.20190314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Man, S.M., Tourlomousis, P., Hopkins, L., Monie, T.P., Fitzgerald, K.A. and Bryant, C.E. (2013) Salmonella infection induces recruitment of caspase-8 to the inflammasome to modulate IL-1β production. J Immunol. 191, 5239–5246 10.4049/jimmunol.1301581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bossaller, L., Chiang, P.-I., Schmidt-Lauber, C., Ganesan, S., Kaiser, W.J., Rathinam, V.A.K.et al. (2012) Cutting edge: FAS (CD95) mediates noncanonical IL-1β and IL-18 maturation via caspase-8 in an RIP3-independent manner. J Immunol. 189, 5508–5512 10.4049/jimmunol.1202121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bulau, A.-M., Nold, M.F., Li, S., Nold-Petry, C.A., Fink, M., Mansell, A.et al. (2014) Role of caspase-1 in nuclear translocation of IL-37, release of the cytokine, and IL-37 inhibition of innate immune responses. Proc. Natl Acad. Sci. U.S.A. 111, 2650–2655 10.1073/pnas.1324140111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wiggins, K.A., Parry, A.J., Cassidy, L.D., Humphry, M., Webster, S.J., Goodall, J.C.et al. (2019) IL-1α cleavage by inflammatory caspases of the noncanonical inflammasome controls the senescence-associated secretory phenotype. Aging Cell 18, e12946 10.1111/acel.12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.He, W., Wan, H., Hu, L., Chen, P., Wang, X., Huang, Z.et al. (2015) Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kayagaki, N., Stowe, I.B., Lee, B.L., O'Rourke, K., Anderson, K., Warming, S.et al. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- 98.Ding, J., Wang, K., Liu, W., She, Y., Sun, Q., Shi, J.et al. (2016) Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 10.1038/nature18590 [DOI] [PubMed] [Google Scholar]
- 99.Shi, J., Zhao, Y., Wang, K., Shi, X., Wang, Y., Huang, H.et al. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 100.Phulphagar, K., Kühn, L.I., Ebner, S., Frauenstein, A., Swietlik, J.J., Rieckmann, J.et al. (2021) Proteomics reveals distinct mechanisms regulating the release of cytokines and alarmins during pyroptosis. Cell Rep. 34, 108826 10.1016/j.celrep.2021.108826 [DOI] [PubMed] [Google Scholar]
- 101.Russo, A.J., Vasudevan, S.O., Méndez-Huergo, S.P., Kumari, P., Menoret, A., Duduskar, S.et al. (2021) Intracellular immune sensing promotes inflammation via gasdermin D-driven release of a lectin alarmin. Nat. Immunol. 22, 154–165 10.1038/s41590-020-00844-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rühl, S. and Broz, P. (2015) Caspase-11 activates a canonical NLRP3 inflammasome by promoting K(+) efflux. Eur. J. Immunol. 45, 2927–2936 10.1002/eji.201545772 [DOI] [PubMed] [Google Scholar]
- 103.Baker, P.J., Boucher, D., Bierschenk, D., Tebartz, C., Whitney, P.G., D'Silva, D.B.et al. (2015) NLRP3 inflammasome activation downstream of cytoplasmic LPS recognition by both caspase-4 and caspase-5. Eur. J. Immunol. 45, 2918–2926 10.1002/eji.201545655 [DOI] [PubMed] [Google Scholar]
- 104.Kayagaki, N., Kornfeld, O.S., Lee, B.L., Stowe, I.B., O'Rourke, K., Li, Q.et al. (2021) NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 591, 131–136 10.1038/s41586-021-03218-7 [DOI] [PubMed] [Google Scholar]
- 105.Chen, K.W., Monteleone, M., Boucher, D., Sollberger, G., Ramnath, D., Condon, N.D.et al. (2018) Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps. Sci. Immunol. 3, eaar6676 10.1126/sciimmunol.aar6676 [DOI] [PubMed] [Google Scholar]
- 106.Taabazuing, C.Y., Okondo, M.C. and Bachovchin, D.A. (2017) Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 24, 507–514.e4 10.1016/j.chembiol.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heilig, R., Dilucca, M., Boucher, D., Chen, K.W., Hancz, D., Demarco, B.et al. (2020) Caspase-1 cleaves Bid to release mitochondrial SMAC and drive secondary necrosis in the absence of GSDMD. Life Sci. Alliance 3, e202000735 10.26508/lsa.202000735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsuchiya, K., Nakajima, S., Hosojima, S., Thi Nguyen, D., Hattori, T., Manh Le, T.et al. (2019) Caspase-1 initiates apoptosis in the absence of gasdermin D. Nat. Commun. 10, 2091 10.1038/s41467-019-09753-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang, Y., Ning, X., Gao, P., Wu, S., Sha, M., Lv, M.et al. (2017) Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 46, 393–404 10.1016/j.immuni.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 110.Krippner-Heidenreich, A., Talanian, R.V., Sekul, R., Kraft, R., Thole, H., Ottleben, H.et al. (2001) Targeting of the transcription factor Max during apoptosis: phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid residue in position P1. Biochem. J. 358, 705–715 10.1042/bj3580705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Akita, K., Ohtsuki, T., Nukada, Y., Tanimoto, T., Namba, M., Okura, T.et al. (1997) Involvement of caspase-1 and caspase-3 in the production and processing of mature human interleukin 18 in monocytic THP.1 cells. J. Biol. Chem. 272, 26595–26603 10.1074/jbc.272.42.26595 [DOI] [PubMed] [Google Scholar]
- 112.Kang, S.J., Wang, S., Hara, H., Peterson, E.P., Namura, S., Amin-Hanjani, S.et al. (2000) Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 149, 613–622 10.1083/jcb.149.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sagulenko, V., Vitak, N., Vajjhala, P.R., Vince, J.E. and Stacey, K.J. (2018) Caspase-1 is an apical caspase leading to caspase-3 cleavage in the AIM2 inflammasome response, independent of caspase-8. J. Mol. Biol. 430, 238–247 10.1016/j.jmb.2017.10.028 [DOI] [PubMed] [Google Scholar]
- 114.Luo, X., Budihardjo, I., Zou, H., Slaughter, C. and Wang, X. (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490 10.1016/S0092-8674(00)81589-5 [DOI] [PubMed] [Google Scholar]
- 115.Ng, F.W., Nguyen, M., Kwan, T., Branton, P.E., Nicholson, D.W., Cromlish, J.A.et al. (1997) P28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum. J. Cell Biol. 139, 327–338 10.1083/jcb.139.2.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Malireddi, R.K.S., Ippagunta, S., Lamkanfi, M. and Kanneganti, T.-D. (2010) Cutting edge: proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes. J. Immunol. 185, 3127–3130 10.4049/jimmunol.1001512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nath, R., Raser, K.J., Stafford, D., Hajimohammadreza, I., Posner, A., Allen, H.et al. (1996) Non-erythroid alpha-spectrin breakdown by calpain and interleukin 1 beta-converting-enzyme-like protease(s) in apoptotic cells: contributory roles of both protease families in neuronal apoptosis. Biochem. J. 319, 683–690 10.1042/bj3190683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Douglas, T. and Saleh, M. (2020) Cross-regulation between LUBAC and caspase-1 modulates cell death and inflammation. J. Biol. Chem. 295, 5216–5228 10.1074/jbc.RA119.011622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nyormoi, O., Wang, Z., Doan, D., Ruiz, M., McConkey, D. and Bar-Eli, M. (2001) Transcription factor AP-2alpha is preferentially cleaved by caspase 6 and degraded by proteasome during tumor necrosis factor alpha-induced apoptosis in breast cancer cells. Mol. Cell. Biol. 21, 4856–4867 10.1128/MCB.21.15.4856-4867.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang, D., Zheng, X., Chen, S., Wang, Z., Xu, W., Tan, J.et al. (2018) Sensing of cytosolic LPS through caspy2 pyrin domain mediates noncanonical inflammasome activation in zebrafish. Nat. Commun. 9, 3052 10.1038/s41467-018-04984-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li, J.-Y., Wang, Y.-Y., Shao, T., Fan, D.-D., Lin, A.-F., Xiang, L.-X.et al. (2020) The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E-mediated pyroptosis. J. Biol. Chem. 295, 1120–1141 10.1016/S0021-9258(17)49920-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grinshpon, R.D., Shrestha, S., Titus-McQuillan, J., Hamilton, P.T., Swartz, P.D. and Clark, A.C. (2019) Resurrection of ancestral effector caspases identifies novel networks for evolution of substrate specificity. Biochem. J. 476, 3475–3492 10.1042/BCJ20190625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lalaoui, N. and Vaux, D.L. (2018) Recent advances in understanding inhibitor of apoptosis proteins. F1000Res. 7, F1000 Faculty Rev-1889 10.12688/f1000research.16439.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Choi, Y.J., Kim, S., Choi, Y., Nielsen, T.B., Yan, J., Lu, A.et al. (2019) SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat. Immunol. 20, 276–287 10.1038/s41590-018-0303-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dorfleutner, A., Chu, L. and Stehlik, C. (2015) Inhibiting the inflammasome: one domain at a time. Immunol. Rev. 265, 205–216 10.1111/imr.12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cornelis, S., Kersse, K., Festjens, N., Lamkanfi, M. and Vandenabeele, P. (2007) Inflammatory caspases: targets for novel therapies. Curr. Pharm. Des. 13, 367–385 10.2174/138161207780163006 [DOI] [PubMed] [Google Scholar]
- 127.Man, S.M., Karki, R. and Kanneganti, T.-D. (2017) Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 277, 61–75 10.1111/imr.12534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wannamaker, W., Davies, R., Namchuk, M., Pollard, J., Ford, P., Ku, G.et al. (2007) (S)-1-((S)-2-{[1-(4-amino-3-chloro-phenyl)-methanoyl]-amino}-3,3-dimethyl-butanoyl)-pyrrolidine-2-carboxylic acid ((2R,3S)-2-ethoxy-5-oxo-tetrahydro-furan-3-yl)-amide (VX-765), an orally available selective interleukin (IL)-converting enzyme/caspase-1 inhibitor, exhibits potent anti-inflammatory activities by inhibiting the release of IL-1beta and IL-18. J. Pharmacol. Exp. Ther. 321, 509–516 10.1124/jpet.106.111344 [DOI] [PubMed] [Google Scholar]
- 129.MacKenzie, S.H., Schipper, J.L. and Clark, A.C. (2010) The potential for caspases in drug discovery. Curr. Opin Drug Discov. Devel. 13, 568–576 PMID: [PMC free article] [PubMed] [Google Scholar]