Abstract
Interferon (IFN)-induced guanosine triphosphate hydrolysing enzymes (GTPases) have been identified as cornerstones of IFN-mediated cell-autonomous defence. Upon IFN stimulation, these GTPases are highly expressed in various host cells, where they orchestrate anti-microbial activities against a diverse range of pathogens such as bacteria, protozoan and viruses. IFN-induced GTPases have been shown to interact with various host pathways and proteins mediating pathogen control via inflammasome activation, destabilising pathogen compartments and membranes, orchestrating destruction via autophagy and the production of reactive oxygen species as well as inhibiting pathogen mobility. In this mini-review, we provide an update on how the IFN-induced GTPases target pathogens and mediate host defence, emphasising findings on protection against bacterial pathogens.
Keywords: bacterial defence, cell-autonomous defence, GTPase, interferons
IFN signalling and induction of IFN-stimulated genes
Exposure of cells to interferons (IFN) results in the induction of a network of genes that combat infections, leading to so-called IFN-mediated cell-autonomous defence [1–5]. This network is a finely tuned mechanism to balance launching an efficient pathogen control while preventing collateral tissue damage. In the last two decades, IFN-induced GTPases have become a focus of attention as key mediators of IFN-mediated host defence.
There is abundant evidence for the vital role of IFN in combating an array of pathogens, including key roles in defence against bacteria [2,6–22]. Ten mammalian IFNs are known, with seven found in humans [23,24]. Based on genetic loci, homology in amino acid sequence and receptor binding, IFNs are currently divided into three groups, namely type I, II and III [1,25].
Upon binding their specific receptors, IFNs activate signal transduction via the JAK/STAT pathway which leads to the formation of the transcription factor complex IFN-stimulated gene factor 3 (ISGF3), consisting of phosphorylated STAT1/STAT2 and IRF9, for type I and type III IFNs and the transcription factor gamma-activated factor (GAF), a homodimer of phosphorylated STAT1, for type 2 IFN-specific signalling [26–28]. These activated transcription factors translocate into the nucleus and bind to their specific promotor elements, IFN-stimulated response element (ISRE) and gamma-activated sequence (GAS) for type I/III and type II, respectively [26–28]. The binding of these transcription factors can modulate the transcription of up to 2000 IFN-stimulated genes (ISGs) [1–3], resulting in immunomodulatory, anti-proliferative and anti-pathogenic consequences [2,29]. Even though these IFNs possess distinct receptors, transcription factors and promotor binding sites, the activation of ISGs via IFNs is complex. All types of IFN show non-canonical signalling, some ISGs are also controlled by IFN regulatory factors (IRFs); which in turn are also ISGs; other ISGs are constitutively expressed at low levels in addition to being IFN-inducible and another portion ISGs are also induced by NF-κB signalling [28,30–35].
Families of IFN-induced GTPases
GTPases induced by IFN have been identified as crucial effectors in IFN-mediated pathogen control [36–56]. These large GTPases can be divided into four subfamilies based on their paralogy and molecular mass [57]. The four subfamilies are the 21–47 kDa immunity-related GTPases (IRGs), the 65–73 kDa guanylate-binding proteins (GBPs), the 72–82 kDa myxoma (MX) resistance proteins and the 200–285 kDa very large inducible GTPases (VLIGs/GVINs) [58–60]. In the following, we will mainly focus on IRG and GBP GTPases and their functions in cell-autonomous defence against bacteria.
Mice have 23 IRGs and this family of genes has been mostly lost in humans, apart from IRGM1 and IRGC [61,62]. The IRGs can be divided into two classes; the primarily cytosolic ‘GKS’ IRGs which possess a conserved canonical GX4GKS sequence in the first nucleotide-binding motif (G1) and the predominantly membrane-bound ‘GMS’ IRGs which possess the non-canonical GX4GMS sequence in their G1 nucleotide-binding motif [60,61]. The ‘GMS’ IRGs control the activity of ‘GKS’ IRGs by controlling the GDP to GTP switch, thus acting as guanosine dissociation inhibitors (GDIs) [59,63]. In the absence of ‘GMS’ IRGs, ‘GKS’ IRGs are constitutively active, form cytoplasmic aggregates and fail to localise to their respective cellular compartment, Toxoplasma gondii parasitophorous vacuole and Chlamydia trachomatis inclusions [59,63,64].
Thus far, 7 human GBP (hGBP) genes (GBP1-GBP7) located on chromosome 1 and 11 mouse GBPs (mGBPs) (Gbp2b-Gbp11) have been identified [49,65–67]. The mGBPs are organised in clusters on chromosome 3 (Gbp2b, Gbp2, Gbp3, Gbp5, Gbp7 and a pseudomGbp2b) and chromosome 5 (Gbp4, Gbp6, Gbp8, Gbp9, Gbp10, Gbp11 and a pseudomGbp2) [68]. Transcription of human and mGBPs can be triggered by type 1 and 2 IFN as well as other inflammatory cytokines and TLR ligands, although the quantitative responses vary substantially between the different GBPs and cytokines [49,69,70].
IFN-induced GTPases belong to the dynamin-protein family as judged by structural similarities and shared biochemical characteristics [57,71,72]. As members of the dynamin protein family, they possess a large GTPase domain (∼300 amino acids), a middle domain and a GTPase effector domain (GED) [73]. In addition to these three domains, many IFN-induced GTPases also possess other domains and motifs for protein–protein and protein–membrane interactions [44,73–75]. In contrast with dynamin, at least some IFN-induced GBPs can hydrolyse GTP to GDP and GDP to GMP, though their GTPase activation is still dependent on oligomerisation [73,76,77]. These dynamin-related characteristics enable IFN-induced GTPases to operate either as mechanoenzymes or as an assembly platform to co-ordinate diverse functions [57]. For instance, they govern vesicular trafficking and the coordination of protein complex assembly to stimulate autophagic, membranolytic, oxidative and inflammasome-related anti-microbial activities upon cytosolic bacteria as well as on pathogen containing vacuoles [2,57,78–81].
Mechanisms of host defence by IFN-induced GTPases
Targeting of specific pathogens by GBPs and IRGs
To execute anti-microbial functions, GBPs and IRGs co-localise with pathogens invading the host cell. GBPs and IRGs are typically found in the cytosol, in vesicle-like structures and on endomembranes, but translocate to pathogen compartments and cytosolic bacteria which have escaped from the phagosome (Figure 1) [70,75,82,83]. Bacteria shown to interact with GBPs and IRGs include Listeria monocytogenes, Legionella pneumophila, Shigella flexneri, Mycobacterium bovis BCG, Chlamydia trachomatis, Francisella novicida, Salmonella typhimurium, Brucella abortus, Yersinia pseudotuberculosis and Burkholderia thailandensis [2,38–40,42–44,47,56,65,69,84–96].
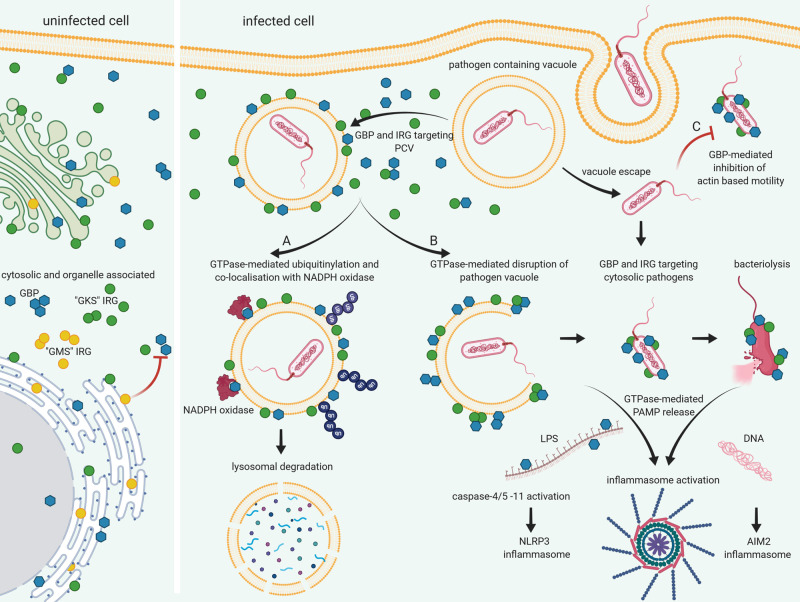
Figure 1. Mechanisms of pathogen clearance by IFN-induced GTPases.
In uninfected cells, GBPs and IRGs are found in the cytosol, in vesicle-like clusters, associated with endomembranes and the nucleus. ‘GMS’ IRGs control the activation of GBPs and host membrane located ‘GKS’ IRGs act as guanosine dissociation inhibitors. During infection GBPs and IRGs co-localise with pathogen containing vacuoles (PCV) and cytosolic pathogens within minutes of pathogen entry. (A) GBP association with PCV may lead to the accumulation of ubiquitin and subsequent destruction of the invading pathogen. GBP7 is essential for NADPH oxidase holoenzyme assembly on PCV. (B) GBP and IRG co-localisation with the PCV and cytosolic bacteria leads to the disruption of vacuole and membrane integrity, releasing PAMPS into the cytosol. GBPchrom3, GBP1, GBP2, GBP3, GBP4 mediate activation of caspase4/5 or 11 during Salmonella, Legionella or Chlamydia infection by cytosolic LPS release leading to pyroptosis. Association with GBPs and IRGB10 leads to loss of membrane integrity and bacteriolysis with subsequent AIM2 activation during Francisella infection. (C) In addition to disrupting PCV and bacterial membrane integrity, GBPs also mediate host defence via the inhibition of actin-based motility of Burkholderia and Shigella pathogens. Created with BioRender.com.
Even though the exact molecular mechanism which enables them to target and destroy pathogens is not fully understood, it has been shown that human and mGBPs form homo- and hetero- and polymers to fulfil their anti-microbial function [50,97]. Kravets et al. [50] showed that mGBPs accumulate on T. gondii vacuoles in densely packed multimers consisting of several thousand monomers. Furthermore, these proteins seem to locate to the pathogens and associated membranes in a hierarchical manner, with GBP1, GBP2 and GBP5 leading the way due to a CaaX prenylation motif at the C-terminus of the protein, which enables them to bind to membranes and to recruit non-prenylated GBPs to their location [50,82]. In addition to targeting various pathogens and their vacuoles directly, in mouse cells ‘GMS’ IRGs have been suggested to influence the localisation and activation of GBPs and ‘GKS’ IRGs on target membranes via a ‘missing-self’ signal [64]. This control of GBP and IRG activation and aggregation on host membranes via the ‘GMS’ IRG family proteins (IRGM), is further supported by the targeting of GBPs and IRGs onto lipid droplets from which IRGM1 and IRGM3 have been removed independently of infection [64]. Based on this observation, it was suggested that a lack of IRGM proteins and therefore the mistargeting of self-membranes through activated GBPs and IRGs as well as the formation of cytosolic clusters leads to a diminished pool of available GBPs and IRGs which could effectively target C. trachomatis and T. gondii [64]. It should be noted that there is some data that is not consistent with the ‘missing-self’ hypothesis [37,44,67], although some of this was refuted in later publications [98,99].
Park et al. [100] proposed a ‘triple check’ model for targeting of mGBPs and IRGs to pathogen vacuoles. This model suggests that pathogen vacuoles are targeted by the autophagy conjugation system by depositing microtubule-associated protein light chain 3 (LC3) and its homologues on the pathogen vacuole. IFN-γ stimulation would ‘trigger’ LC3 on these membranes, either via posttranslational modifications or via the addition of factors such as ubiquitin, to act as a guanine nucleotide exchange factor (GEF) for GBPs and IRGs and activate them. Misguiding of GBPs to endomembranes would be avoided through the protective function of IRGM proteins, which act as GDIs for GBPs and IRGs [63,101]. How the LC3 conjugation system recognises pathogen vacuoles remains unknown. However, Brown et al. [102] have suggested that the autophagy conjugation complex or some upstream sensor of this complex recognises changes to the membranes occupied by pathogens, such as missing-self (e.g. lack of IRGM proteins), changed-self (e.g. rearranged protein and lipid composition) and non-self (e.g. pathogen effectors and secretion systems). It was also suggested that the binding of this complex to membranes might be facilitated via autophagy related-protein ATG5 [103], as ATG5 from the autophagy conjugation complex can bind membranes via an unknown lipid moiety [104]. This model is supported by the observations that ATG5 and LC3 are found on murine norovirus (MNV) membranous replication complexes [105] and also T. gondii vacuoles without prior IFN-γ stimulation in mouse macrophages [100,106]. Furthermore, GBPs and IRGs are unable to target pathogen containing vacuoles and aggregate in the cytosol in cells lacking all ATG5 or all LC3 homologues [102,106,107]. Whether this model applies to other pathogens and host species, especially with humans which lack most IRGs, remains to be investigated.
To what extent IRGs and GBPs co-operate in targeted co-localisation to pathogens or pathogen vacuoles remains unclear, as reciprocal dependence has been observed. For example, IRGM1 and 3 are needed for targeting of mGBPs to T. gondii vacuoles and pathogen control in MEFs, whereas they are dispensable for Leishmania donovani control [41,52]. On the other hand, the localisation of IRGs can also be dependent on GBPs, as the targeting of IRGB10 and IRGB6, to F. novicida, T. gondii and E. coli are dependent on GBPs from chromosome 3, as these IRGs failed to co-localise with pathogen inclusions in cells lacking GBPs on mouse chromosome 3 [39,108].
Different GBPs and IRGs have been shown to target specific pathogens, though the underlying mechanisms for this specificity is only now being uncovered [42,52,65,94,109]. Kohler et al. [89] have suggested that changes in the C-terminal polybasic motif (PBM) in primate GBP1s are responsible for the pathogen specificity towards S. flexneri. In line with this, it was shown that the unique triple-arginine cassette in the PBM of hGBP1 is responsible for targeting S. flexneri [43]. The highly divergent C-terminal amino acid sequence in mGBPs might also indicate a non-redundant function in determining pathogen specificity [49]. In addition, alternative splicing variants of GBPs might play a role in specific pathogen targeting, since a splicing variant of mGBP5, mGBP5a, was present in L. monocytogenes infected mouse liver but absent from T. gondii infected liver [49]. Besides the co-localisation of GBPs and IRGs with particular pathogens, differences in GBP targeting of the same pathogens have also been observed in distinct cell types of the same host species. For example, hGBP1 co-localises with T. gondii in mesenchymal stromal cells and THP1 but not A549 cells [69,96,110,111]. This remarkable diversity of targeting strategies for specific pathogens might be due to the diverse genetic backgrounds and proteomes of different host species and cell types as well as pathogen-specific virulence factors and intracellular life cycles.
Mechanisms of pathogen clearance by IFN-induced GTPases
The anti-microbial mechanisms of IFN-induced GTPases that are discussed below are represented in Figure 1.
Ubiquitination and lysosomal destruction mediated by GBPs and IRGs
GBPs and IRGs can mediate pathogen control by induction of autophagy and ubiquitin-mediated destruction of pathogen vacuoles [112–114]. mGBP7 interacts with and recruits the autophagy protein ATG4B to Mycobacterium-containing vacuoles [65], which promotes the expansion of autophagic membranes around the bacteria and damaged bacterial compartments [2,57], leading to degradation of the pathogen via lysosome fusion [65]. Haldar et al. [41] demonstrated that IFN-γ-induced IRGM1 and IRGM3 control the recruitment of the E3 ligase tumour necrosis factor receptor-associated factor 6 (TRAF6) and subsequent ubiquitination of vacuoles of T. gondii and C. trachomatis. Following ubiquitination, GBPs co-localise with vacuoles in a sequestosome 1 (SQSTM1/p62)-dependent manner and mark these vacuoles for destruction [41]. IRGM-dependent autophagy was also shown for Mycobacterium infections though the exact mechanism remains unclear [45,114]. It seems likely that ‘GMS’ proteins IRGM1 and IRGM3 co-ordinate the localisation of other GKS IRGs to pathogen vacuoles, as virulent T. gondii strains and C. muridarum inhibit ‘GKS’ IRG activity and vacuole co-localisation of these IRG proteins thereby avoiding ubiquitination of the replicative niche [41,115-117].
In addition to mediating the ubiquitination of pathogen compartments and the subsequent lysosomal destruction via controlling the ‘GKS’ IRGs activity, IRGM1 has been shown to target M. tuberculosis vacuoles directly [37,65]. The recruitment of IRGM1 to pathogen containing vacuoles appears to facilitate fusion with lysosomes, as lysosomal fusion of M. tuberculosis vacuoles is impaired in Irgm1-deficient mutants [37]. The C-terminal amphipathic helix (αK) of IRGM1 binds to Mycobacterium vacuoles by interaction with phosphoinositide-3,4-bisphosphate (PtdIns[3,4]P2) and PtdIns[3,4,5]P3[44].
For protection against the lung pathogen L. pneumophila, IRG-dependent as well as IRG-independent pathways have been described. Both IRGM1 and IRGM3, have been implicated in IFN-mediated control of L. pneumophila [9,36]. The binding of IRGM1 to the intracellular replicative niche of L. pneumophila, the Legionella-containing vacuole (LCV), results in the co-localisation of other IRG proteins and subsequent ubiquitination of the LCV, thereby leading to LCV degradation through autophagy [4]. GBP1 and GBP2 are involved in an IRGM-independent resistance against L. pneumophila, as the bacterial protein secretion system on the LCV is recognised as a PAMP, leading to binding of the cytosolic carbohydrate-binding protein galectin-3. The binding of galectin-3 to the LCV recruits GBP1 and GBP2 to the LCV, as well as subsequent ubiquitination and targeting by p62, which leads to the degradation of the bacteria via autophagy [93]. This IRGM-independent and GBP-dependent ubiquitination during Legionella infection is in contrast with the previously mentioned IRGM-dependent and GBP-independent ubiquitination of T. gondii vacuoles as well as C. trachomatis inclusions [41].
In most cases, the ubiquitination of pathogens and their compartments is a host-derived response which favours host survival and promotes pathogen control. In contrast with this, it was shown that the hGBP1-mediated, poly-ubiquitin coat on S. flexneri is not host-derived but mediated by a bacterial-derived E3 ubiquitin ligase IpaH9.8, which recognises, binds and ubiquitinates GBP1, GBP2 and GBP4 but not GBP3 and labels them for proteasome-mediated degradation [42,43,90]. This poly-ubiquitination reverses the GBP-mediated restriction and enables the bacteria to form actin tails and spread efficiently from cell to cell [42,43,90].
GBP-mediated production of reactive oxygen species (ROS)
Another host resistance pathway that mediates IFN-induced pathogen control is the production of ROS. NOX2 is an NADPH oxidase that is able to generate superoxide, which has microbicidal properties [118]. During L. monocytogenes and M. bovis BCG infection, mGBP7 binds the membrane-bound heterodimer gp91phox-p22phox (cytochrome b558) and cytosolic p67phox [65]. Thus, GBP7 acts as a linker between membrane-bound and cytosolic NOX2 components to assemble and activate the NOX2 holoenzyme on pathogen compartments after IFN-γ stimulation [65].
GBP-mediated inflammasome activation
Recent work has linked IFN-induced GTPases with inflammasome activation in various host cells and in response to a diverse range of pathogens. IFN-induced GTPases appear to influence inflammasome activation by promoting inflammasome complex assembly and targeting pathogens and their compartments to increase the access of PAMPs to cytosolic inflammasome components. These two mechanisms of inflammasome activation can work in concert to achieve adequate inflammasome activation and thus host defence.
GBP5 is involved in the assembly of the NOD-, LRR- and pyrin domain-containing 3 (NLRP3) inflammasome during Listeria or Salmonella spp. infections via tetramerisation of GBP5 [119-121]. Assembly of the NLRP3 inflammasome leads to the induction of pyroptosis in order to control bacterial infections. Deletion of single mGBPs from chromosome 3 revealed unique functions for GBPs; namely, GBP5 binding to the pyrin domain of NLRP3 and GBP2 binding of apoptosis-associated speck-like protein containing a CARD (ASC) [121]. Due to their ability to form heterodimers, GBP2 and GBP5 thus facilitate the assembly of the NLRP3 inflammasome to activate caspase-1 [121].
Several observations have shown that GBPs activate inflammasomes by either directly sensing bacterial products or facilitating access to bacterial PAMPs. The induction of inflammasomes via GBPs and IRGs can result in canonical (caspase-1) or non-canonical (caspase-11, human caspase-4/5) mediated pyroptosis. It was shown that GBPs from mouse chromosome 3, namely GBP2, GBP5, as well as IRGB10 are essential for the activation of the AIM2 inflammasome in F. novicida infected macrophages, as cells lacking these IFN-induced GTPases showed decreased inflammasome activation [39,87,88]. GBPs from chromosome 3 also control the non-canonical activation of caspase-11 in response to L. pneumophila as well as pathogenic and non-pathogenic E. coli outer membrane vesicles and free LPS injected into the cytosol [36,122,123]. GBPs from chromosome 3 were also essential for caspase-1 activation and IL1-α as well as IL-1ß release in response to B. abortus and Y. pseudotuberculosis infections [47,92]. It has been suggested that GBPs might be influencing the membrane dynamics of outer membrane vesicles and the integrity of pathogen membranes due to their dynamin-like activities thus exposing lipid A of LPS and other PAMPS to the cytosol, hence making them accessible for inflammasome activation [39,122,124].
In line with these previous observations suggesting GBPs mediate LPS release and/or recognition by the inflammasome, hGBP1 was recently identified as a novel cytosolic LPS sensor [69,94-96]. hGBP1 binds to LPS via electrostatic interactions between the negatively charged LPS and positively charged amino acid residues of hGBP1 [94,97]. Detection of LPS via hGBP1 results in the recruitment of hGBP2-4 to cytosolic Salmonella, this GBP coat in turn recruits and activates caspase-4 [69,94,95]. Based on these observations and their own, Kutsch et al. [97] presented a model of hGBP1 acting as a detergent on the bacterial LPS layer. hGBP1 was identified as a LPS sensing and binding protein, which disrupts the O-antigen barrier of Gram-negative bacteria through the insertion of the farnesyl tail of hGBP1 molecules into this layer, thereby disrupting the interactions between LPS molecules mediated by the O-antigens [97]. A triple-arginine motif in the C-terminal end of GBP1 mediates the binding of hGBP1 to the pathogen LPS O-antigen [43]. The insertion of hGBP1 into the LPS layer seemingly changes the membrane stiffness and fluidity, thus making the bacteria more accessible to caspase-4 activation and more susceptible to the anti-microbial activity of polymyxin B, as well as potentially influencing the function of other pathogen proteins inserted into the outer membrane such as Shigella IcsA [97].
Using different GBP1 catalytic mutants, Xavier et al. [125] identified a novel pathway of NLRP3 activation mediated by hGBP1. This group discovered that hGBP1 recruitment to C. trachomatis inclusions activates GTP hydrolysis to GMP and the subsequent generation of uric acid activates the NLRP3 inflammasome [125]. This novel pathway suggests that, in contrast with previous findings [69,94–97], inflammasome activation can be independent of PAMP release in human cells, relying only on the hydrolytic activity of hGBP1 [125]. Whether this activation is unique to the Chlamydia inclusion or represents a more general response towards other pathogens, remains to be investigated.
IFN-induced GTPases and actin-based motility
Recent findings have demonstrated that IFN-induced GBPs can inhibit the actin-based motility of intracellular bacteria. hGBPs target cytosolic S. flexneri after IFN-γ exposure and interfere with actin tail formation, which is required for cytosolic mobility and cell to cell spread [42,43]. GBP1 is essential for IFN-γ mediated inhibition of actin tail formation as well as recruitment of GBP2, 3 and 4 to the pathogen [42]. GBP-mediated inhibition of actin tails hindered the bacteria from spreading efficiently from cell to cell and resulted in large microcolonies forming in infected cells but significantly fewer cells becoming infected [42]. In addition to S. flexneri, hGBP1 also targets B. thailandensis through a C-terminal triple-arginine motif that binds O-antigen [43].
mGBPs also inhibit the formation of actin tails and the formation of multinucleated giant cells (MNGCs) during B. thailandensis infection by interfering with Arp2/3-mediated actin nucleation and cytoskeletal remodelling [86]. Cells lacking multiple GBPs from chromosome 3 as well as Gbp2−/− and Gbp5−/− cells showed an increased number of MNGCs and increased bacterial load [86].
Perspectives
Importance of the field: IFN-induced GTPases play a significant role in cell-autonomous defence against a wide variety of pathogens. They initiate and regulate a diverse range of host defence pathways and an appreciation of the roles of IFN-induced GTPases in host defence could lead to more effective anti-microbial treatments.
Current thinking: Individual IFN-induced GTPases possess unique functions that tailor the response to different pathogens and mediate their anti-microbial function by compromising the integrity of pathogen-related membranes, releasing PAMPS into the cytosol, inducing bactericidal small molecules, marking pathogens for destruction or inhibiting pathogen mobility.
Future directions: Identifying GTPase binding partners that mediate their specific function and regulate their activities, will be crucial in enhancing our understanding of how these GTPases mediate IFN-induced cell-autonomous defence against various pathogens.
Abbreviations
- GBPs
guanylate-binding proteins
- GDIs
guanosine dissociation inhibitors
- hGBP
human GBP
- IFN
interferon
- IRGs
immunity-related GTPases
- ISGs
IFN-stimulated genes
- LC3
microtubule-associated protein light chain 3
- LCV
Legionella-containing vacuole
- mGBPs
mouse GBP
- MNGCs
multinucleated giant cells
- PBM
polybasic motif
- ROS
reactive oxygen species
Conflicts of interest
The authors have no conflicts of interest.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
ELH and IVD are supported by the National Health and Medical Research Council of Australia APP1145244. This work was supported by DFG IRTG 2168. WK is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy EXC2151 – 390873048.
Open Access
Open access for this article was enabled by the participation of the University of Melbourne in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Author Contributions
Conception: H.L.R., I.V.D., E.L.H. Drafting: H.L.R., I.V.D., E.L.H., Revising and critiquing: H.L.R., W.K., I.V.D., E.L.H. Funding: W.K., I.V.D., E.L.H. All authors give final approval of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- 1.Rusinova, I., Forster, S., Yu, S., Kannan, A., Masse, M., Cumming, H.et al. (2013) INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41, D1040–D1046 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacMicking, J.D. (2012) Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12, 367–382 10.1038/nri3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw, A.E., Hughes, J., Gu, Q., Behdenna, A., Singer, J.B., Dennis, T.et al. (2017) Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 15, e2004086 10.1371/journal.pbio.2004086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilla-Moffett, D., Barber, M.F., Taylor, G.A. and Coers, J. (2016) Interferon-inducible GTPases in host resistance, inflammation and disease. J. Mol. Biol. 428, 3495–3513 10.1016/j.jmb.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang, S., Meng, Q., Maminska, A. and MacMicking, J.D. (2019) Cell-autonomous immunity by IFN-induced GBPs in animals and plants. Curr. Opin. Immunol. 60, 71–80 10.1016/j.coi.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu, B., Ebensperger, C., Dembic, Z., Wang, Y., Kvatyuk, M., Lu, T.et al. (1998) Targeted disruption of the interferon-gamma receptor 2 gene results in severe immune defects in mice. Proc. Natl Acad. Sci. U.S.A. 95, 8233–8238 10.1073/pnas.95.14.8233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrero, J.A. (2013) Confounding roles for type I interferons during bacterial and viral pathogenesis. Int. Immunol. 25, 663–669 10.1093/intimm/dxt050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banks, D.A., Ahlbrand, S.E., Hughitt, V.K., Shah, S., Mayer-Barber, K.D., Vogel, S.N.et al. (2019) Mycobacterium tuberculosis inhibits autocrine type I IFN signaling to increase intracellular survival. J. Immunol. 202, 2348–2359 10.4049/jimmunol.1801303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippmann, J., Müller, H.C., Naujoks, J., Tabeling, C., Shin, S., Witzenrath, M.et al. (2011) Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell. Microbiol. 13, 1668–1682 10.1111/j.1462-5822.2011.01646.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Opitz, B., Vinzing, M., Van Laak, V., Schmeck, B., Heine, G., Günther, S.et al. (2006) Legionellapneumophila induces IFNβ in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J. Biol. Chem. 281, 36173–36179 10.1074/jbc.M604638200 [DOI] [PubMed] [Google Scholar]
- 11.Flynn, J.L., Chan, J., Triebold, K.J., Dalton, D.K., Stewart, T.A. and Bloom, B.R. (1993) An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 178, 2249–2254 10.1084/jem.178.6.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, S., Hendriks, W., Althage, A., Hemmi, S., Bluethmann, H., Kamijo, R.et al. (1993) Immune response in mice that lack the interferon-gamma receptor. Science 259, 1742–1745 10.1126/science.8456301 [DOI] [PubMed] [Google Scholar]
- 13.Dussurget, O., Bierne, H. and Cossart, P. (2014) The bacterial pathogen Listeria monocytogenes and the interferon family: type I, type II and type III interferons. Front. Cell Infect. Microbiol. 4, 50 10.3389/fcimb.2014.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norazmi, M.N. (2017) Interferon-β controls non-tuberculous mycobacterial infection in mice. Virulence 8, 1085–1087 10.1080/21505594.2017.1341035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shtrichman, R. and Samuel, C.E. (2001) Review: the role of gamma interferon in antimicrobial immunity. Curr. Opin. Microbiol. 4, 251–259 10.1016/S1369-5274(00)00199-5 [DOI] [PubMed] [Google Scholar]
- 16.Wei, J., Ma, Y., Wang, L., Chi, X., Yan, R., Wang, S.et al. (2017) Alpha/beta interferon receptor deficiency in mice significantly enhances susceptibility of the animals to pseudorabies virus infection. Vet. Microbiol. 203, 234–244 10.1016/j.vetmic.2017.03.022 [DOI] [PubMed] [Google Scholar]
- 17.Deckert-Schlüter, M., Rang, A., Weiner, D., Huang, S., Wiestler, O.D., Hof, H.et al. (1996) Interferon-gamma receptor-deficiency renders mice highly susceptible to toxoplasmosis by decreased macrophage activation. Lab. Invest. 75, 827–841 PMID: [PubMed] [Google Scholar]
- 18.Mastroeni, P., Clare, S., Khan, S., Harrison, J.A., Hormaeche, C.E., Okamura, H.et al. (1999) Interleukin 18 contributes to host resistance and gamma interferon production in mice infected with virulent Salmonella typhimurium. Infect. Immun. 67, 478–483 10.1128/IAI.67.2.478-483.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swihart, K., Fruth, U., Messmer, N., Hug, K., Behin, R., Huang, S.et al. (1995) Mice from a genetically resistant background lacking the interferon gamma receptor are susceptible to infection with Leishmania major but mount a polarized T helper cell 1-type CD4+ T cell response. J. Exp. Med. 181, 961–971 10.1084/jem.181.3.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantin, E., Tanamachi, B., Openshaw, H., Mann, J. and Clarke, K. (1999) Gamma interferon (IFN-gamma) receptor null-mutant mice are more susceptible to herpes simplex virus type 1 infection than IFN-gamma ligand null-mutant mice. J. Virol. 73, 5196–5200 10.1128/JVI.73.6.5196-5200.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaacs, A. and Lindenmann, J. (1957) Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147, 258–267 10.1098/rspb.1957.0048 [DOI] [PubMed] [Google Scholar]
- 22.Findlay, G.M. and MacCallum, F.O. (1937) An interference phenomenon in relation to yellow fever and other viruses. J. Pathol. Bacteriol. 44, 405–424 10.1002/path.1700440216 [DOI] [Google Scholar]
- 23.Pestka, S. (2007) The interferons: 50 years after their discovery, there is much more to learn. J. Biol. Chem. 282, 20047–20051 10.1074/jbc.R700004200 [DOI] [PubMed] [Google Scholar]
- 24.Donnelly, R.P. and Kotenko, S.V. (2010) Interferon-lambda: a new addition to an old family. J. Interferon Cytokine Res. 30, 555–564 10.1089/jir.2010.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestka, S., Krause, C.D. and Walter, M.R. (2004) Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32 10.1111/j.0105-2896.2004.00204.x [DOI] [PubMed] [Google Scholar]
- 26.Schroder, K., Hertzog, P.J., Ravasi, T. and Hume, D.A. (2004) Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189 10.1189/jlb.0603252 [DOI] [PubMed] [Google Scholar]
- 27.Saha, B., Jyothi Prasanna, S., Chandrasekar, B. and Nandi, D. (2010) Review article: gene modulation and immunoregulatory roles of interferonγ. Cytokine 50, 1–14 10.1016/j.cyto.2009.11.021 [DOI] [PubMed] [Google Scholar]
- 28.Platanias, L.C. (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- 29.De Andrea, M., Ravera, R., Gioia, D., Gariglio, M. and Landolfo, S. (2002) Review article: the interferon system: an overview. Eur. J. Paediatr. Neurol. 6, A41–A46 10.1053/ejpn.2002.0573 [DOI] [PubMed] [Google Scholar]
- 30.Wang, W., Xu, L., Su, J., Peppelenbosch, M.P. and Pan, Q. (2017) Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 25, 573–584 10.1016/j.tim.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platanitis, E. and Decker, T. (2018) Regulatory networks involving STATs, IRFs, and NFκB in inflammation. Front. Immunol. 9, 2542 10.3389/fimmu.2018.02542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green, R., Ireton, R.C. and Gale, Jr, M. (2018) Interferon-stimulated genes: new platforms and computational approaches. Mamm. Genome 29, 593–602 10.1007/s00335-018-9755-6 [DOI] [PubMed] [Google Scholar]
- 33.Mostafavi, S., Yoshida, H., Moodley, D., LeBoité, H., Rothamel, K., Raj, T.et al. (2016) Parsing the interferon transcriptional network and its disease associations. Cell 164, 564–578 10.1016/j.cell.2015.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kane, M., Zang, T.M., Rihn, S.J., Zhang, F., Kueck, T., Alim, M.et al. (2016) Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe 20, 392–405 10.1016/j.chom.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoggins, J.W. (2019) Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 6, 567–584 10.1146/annurev-virology-092818-015756 [DOI] [PubMed] [Google Scholar]
- 36.Pilla, D.M., Hagar, J.A., Haldar, A.K., Mason, A.K., Degrandi, D., Pfeffer, K.et al. (2014) Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl Acad. Sci. U.S.A. 111, 6046–6051 10.1073/pnas.1321700111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacMicking, J.D., Taylor, G.A. and McKinney, J.D. (2003) Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science 302, 654–659 10.1126/science.1088063 [DOI] [PubMed] [Google Scholar]
- 38.Man, S.M., Karki, R., Malireddi, R.K., Neale, G., Vogel, P., Yamamoto, M.et al. (2015) The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467–475 10.1038/ni.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Man, S.M., Karki, R., Sasai, M., Place, D.E., Kesavardhana, S., Temirov, J.et al. (2016) IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell 167, 382–396.e17 10.1016/j.cell.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Zeer, M.A., Al-Younes, H.M., Braun, P.R., Zerrahn, J. and Meyer, T.F. (2009) IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS One 4, e4588 10.1371/journal.pone.0004588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haldar, A.K., Foltz, C., Finethy, R., Piro, A.S., Feeley, E.M., Pilla-Moffett, D.M.et al. (2015) Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc. Natl Acad. Sci. U.S.A. 112, E5628–E5637 10.1073/pnas.1515966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandel, M.P., Pathe, C., Werner, E.I., Ellison, C.J., Boyle, K.B., von der Malsburg, A.et al. (2017) GBPs inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe 22, 507–518.e5 10.1016/j.chom.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piro, A.S., Hernandez, D., Luoma, S., Feeley, E.M., Finethy, R., Yirga, A.et al. (2017) Detection of cytosolic Shigella flexneri via a C-terminal triple-arginine motif of GBP1 inhibits actin-based motility. mBio 8, e01979-17 10.1128/mBio.01979-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiwari, S., Choi, H.P., Matsuzawa, T., Pypaert, M. and MacMicking, J.D. (2009) Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat. Immunol. 10, 907–917 10.1038/ni.1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh, S.B., Davis, A.S., Taylor, G.A. and Deretic, V. (2006) Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441 10.1126/science.1129577 [DOI] [PubMed] [Google Scholar]
- 46.Meunier, E., Dick, M.S., Dreier, R.F., Schürmann, N., Kenzelmann Broz, D., Warming, S.et al. (2014) Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 509, 366–370 10.1038/nature13157 [DOI] [PubMed] [Google Scholar]
- 47.Costa Franco, M.M., Marim, F., Guimarães, E.S., Assis, N.R.G., Cerqueira, D.M., Alves-Silva, J.et al. (2018) Brucella abortus triggers a cGAS-independent STING pathway to induce host protection that involves guanylate-binding proteins and inflammasome activation. J. Immunol. 200, 607–622 10.4049/jimmunol.1700725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marinho, F.V., Fahel, J.S., de Araujo, A., Diniz, L.T.S., Gomes, M.T.R., Resende, D.P.et al. (2020) Guanylate binding proteins contained in the murine chromosome 3 are important to control mycobacterial infection. J. Leukoc. Biol. 108, 1279–1291 10.1002/JLB.4MA0620-526RR [DOI] [PubMed] [Google Scholar]
- 49.Degrandi, D., Konermann, C., Beuter-Gunia, C., Kresse, A., Wurthner, J., Kurig, S.et al. (2007) Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 179, 7729–7740 10.4049/jimmunol.179.11.7729 [DOI] [PubMed] [Google Scholar]
- 50.Kravets, E., Degrandi, D., Ma, Q., Peulen, T.O., Klumpers, V., Felekyan, S.et al. (2016) Guanylate binding proteins directly attack Toxoplasma gondii via supramolecular complexes. Elife 5, e11479 10.7554/eLife.11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steffens, N., Beuter-Gunia, C., Kravets, E., Reich, A., Legewie, L., Pfeffer, K.et al. (2020) Essential role of mGBP7 for survival of Toxoplasma gondii infection. mBio 11, e02993-19 10.1128/mBio.02993-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haldar, A.K., Nigam, U., Yamamoto, M., Coers, J. and Goyal, N. (2020) Guanylate binding proteins restrict Leishmania donovani growth in nonphagocytic cells independent of parasitophorous vacuolar targeting. mBio 11, e01464-20 10.1128/mBio.01464-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun, E., Hotter, D., Koepke, L., Zech, F., Groß, R., Sparrer, K.M.J.et al. (2019) Guanylate-binding proteins 2 and 5 exert broad antiviral activity by inhibiting furin-mediated processing of viral envelope proteins. Cell Rep. 27, 2092–2104.e10 10.1016/j.celrep.2019.04.063 [DOI] [PubMed] [Google Scholar]
- 54.Itsui, Y., Sakamoto, N., Kakinuma, S., Nakagawa, M., Sekine-Osajima, Y., Tasaka-Fujita, M.et al. (2009) Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50, 1727–1737 10.1002/hep.23195 [DOI] [PubMed] [Google Scholar]
- 55.Nordmann, A., Wixler, L., Boergeling, Y., Wixler, V. and Ludwig, S. (2012) A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication. FASEB J. 26, 1290–1300 10.1096/fj.11-189886 [DOI] [PubMed] [Google Scholar]
- 56.Lindenberg, V., Molleken, K., Kravets, E., Stallmann, S., Hegemann, J.H., Degrandi, D.et al. (2017) Broad recruitment of mGBP family members to Chlamydia trachomatis inclusions. PLoS One 12, e0185273 10.1371/journal.pone.0185273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim, B.-H., Shenoy, A.R., Kumar, P., Bradfield, C.J. and MacMicking, J.D. (2012) IFN-inducible GTPases in host cell defense. Cell Host Microbe 12, 432–444 10.1016/j.chom.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacMicking, J.D. (2004) IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25, 601–609 10.1016/j.it.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 59.Martens, S. and Howard, J. (2006) The interferon-inducible GTPases. Annu. Rev. Cell Dev. Biol. 22, 559–589 10.1146/annurev.cellbio.22.010305.104619 [DOI] [PubMed] [Google Scholar]
- 60.Boehm, U., Guethlein, L., Klamp, T., Ozbek, K., Schaub, A., Fütterer, A.et al. (1998) Two families of GTPases dominate the complex cellular response to IFN-gamma. J. Immunol. 161, 6715–6723 PMID: [PubMed] [Google Scholar]
- 61.Bekpen, C., Hunn, J.P., Rohde, C., Parvanova, I., Guethlein, L., Dunn, D.M.et al. (2005) The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 6, R92–R92 10.1186/gb-2005-6-11-r92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bekpen, C., Xavier, R.J. and Eichler, E.E. (2010) Human IRGM gene ‘to be or not to be’. Semin. Immunopathol. 32, 437–444 10.1007/s00281-010-0224-x [DOI] [PubMed] [Google Scholar]
- 63.Hunn, J.P., Koenen-Waisman, S., Papic, N., Schroeder, N., Pawlowski, N., Lange, R.et al. (2008) Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 27, 2495–2509 10.1038/emboj.2008.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haldar, A.K., Saka, H.A., Piro, A.S., Dunn, J.D., Henry, S.C., Taylor, G.A.et al. (2013) IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog. 9, e1003414 10.1371/journal.ppat.1003414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim, B.H., Shenoy, A.R., Kumar, P., Das, R., Tiwari, S. and MacMicking, J.D. (2011) A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 332, 717–721 10.1126/science.1201711 [DOI] [PubMed] [Google Scholar]
- 66.Olszewski, M.A., Gray, J. and Vestal, D.J. (2006) In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interferon Cytokine Res. 26, 328–352 10.1089/jir.2006.26.328 [DOI] [PubMed] [Google Scholar]
- 67.Shenoy, A.R., Kim, B.H., Choi, H.P., Matsuzawa, T., Tiwari, S. and MacMicking, J.D. (2007) Emerging themes in IFN-gamma-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology 212, 771–784 10.1016/j.imbio.2007.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kresse, A., Konermann, C., Degrandi, D., Beuter-Gunia, C., Wuerthner, J., Pfeffer, K.et al. (2008) Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC Genom. 9, 158 10.1186/1471-2164-9-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fisch, D., Bando, H., Clough, B., Hornung, V., Yamamoto, M., Shenoy, A.R.et al. (2019) Human GBP1 is a microbe-specific gatekeeper of macrophage apoptosis and pyroptosis. EMBO J. 38, e100926 10.15252/embj.2018100926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tripal, P., Bauer, M., Naschberger, E., Mörtinger, T., Hohenadl, C., Cornali, E.et al. (2007) Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 27, 44–52 10.1089/jir.2007.0086 [DOI] [PubMed] [Google Scholar]
- 71.Ghosh, A., Uthaiah, R., Howard, J., Herrmann, C. and Wolf, E. (2004) Crystal structure of IIGP1: a paradigm for interferon-inducible p47 resistance GTPases. Mol. Cell 15, 727–739 10.1016/j.molcel.2004.07.017 [DOI] [PubMed] [Google Scholar]
- 72.Prakash, B., Praefcke, G.J., Renault, L., Wittinghofer, A. and Herrmann, C. (2000) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403, 567–571 10.1038/35000617 [DOI] [PubMed] [Google Scholar]
- 73.Praefcke, G.J. and McMahon, H.T. (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- 74.Martens, S., Sabel, K., Lange, R., Uthaiah, R., Wolf, E. and Howard, J.C. (2004) Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: retargeting to plasma membrane induced by phagocytosis. J. Immunol. 173, 2594–2606 10.4049/jimmunol.173.4.2594 [DOI] [PubMed] [Google Scholar]
- 75.Modiano, N., Lu, Y.E. and Cresswell, P. (2005) Golgi targeting of human guanylate-binding protein-1 requires nucleotide binding, isoprenylation, and an IFN-gamma-inducible cofactor. Proc. Natl Acad. Sci. U.S.A. 102, 8680–8685 10.1073/pnas.0503227102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neun, R., Richter, M.F., Staeheli, P. and Schwemmle, M. (1996) GTPase properties of the interferon-induced human guanylate-binding protein 2. FEBS Lett. 390, 69–72 10.1016/0014-5793(96)00628-X [DOI] [PubMed] [Google Scholar]
- 77.Schwemmle, M. and Staeheli, P. (1994) The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem. 269, 11299–11305 10.1016/S0021-9258(19)78125-3 [DOI] [PubMed] [Google Scholar]
- 78.Man, S.M., Place, D.E., Kuriakose, T. and Kanneganti, T.D. (2017) Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. J. Leukoc. Biol. 101, 143–150 10.1189/jlb.4MR0516-223R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomes, M.T.R., Cerqueira, D.M., Guimarães, E.S., Campos, P.C. and Oliveira, S.C. (2019) Guanylate-binding proteins at the crossroad of noncanonical inflammasome activation during bacterial infections. J. Leukoc. Biol. 106, 553–562 10.1002/JLB.4MR0119-013R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Santos, J.C. and Broz, P. (2018) Sensing of invading pathogens by GBPs: at the crossroads between cell-autonomous and innate immunity. J. Leukoc. Biol. 104, 729–735 10.1002/JLB.4MR0118-038R [DOI] [PubMed] [Google Scholar]
- 81.Tretina, K., Park, E.S., Maminska, A. and MacMicking, J.D. (2019) Interferon-induced guanylate-binding proteins: guardians of host defense in health and disease. J. Exp. Med. 216, 482–500 10.1084/jem.20182031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Britzen-Laurent, N., Bauer, M., Berton, V., Fischer, N., Syguda, A., Reipschlager, S.et al. (2010) Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One 5, e14246 10.1371/journal.pone.0014246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vestal, D.J., Gorbacheva, V.Y. and Sen, G.C. (2000) Different subcellular localizations for the related interferon-induced GTPases, MuGBP-1 and MuGBP-2: implications for different functions? J. Interferon Cytokine Res. 20, 991–1000 10.1089/10799900050198435 [DOI] [PubMed] [Google Scholar]
- 84.Tietzel, I., El-Haibi, C. and Carabeo, R.A. (2009) Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One 4, e6499 10.1371/journal.pone.0006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rupper, A.C. and Cardelli, J.A. (2008) Induction of guanylate binding protein 5 by gamma interferon increases susceptibility to Salmonella enterica serovar Typhimurium-induced pyroptosis in RAW 264.7 cells. Infect. Immun. 76, 2304–23015 10.1128/IAI.01437-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Place, D.E., Briard, B., Samir, P., Karki, R., Bhattacharya, A., Guy, C.S.et al. (2020) Interferon inducible GBPs restrict Burkholderia thailandensis motility induced cell-cell fusion. PLoS Pathog. 16, e1008364 10.1371/journal.ppat.1008364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meunier, E., Wallet, P., Dreier, R.F., Costanzo, S., Anton, L., Ruhl, S.et al. (2015) Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16, 476–484 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wallet, P., Benaoudia, S., Mosnier, A., Lagrange, B., Martin, A., Lindgren, H.et al. (2017) IFN-γ extends the immune functions of Guanylate Binding Proteins to inflammasome-independent antibacterial activities during Francisella novicida infection. PLoS Pathog. 13, e1006630 10.1371/journal.ppat.1006630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kohler, K.M., Kutsch, M., Piro, A.S., Wallace, G.D., Coers, J. and Barber, M.F. (2020) A rapidly evolving polybasic motif modulates bacterial detection by guanylate binding proteins. mBio 11, e00340-20 10.1128/mBio.00340-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li, P., Jiang, W., Yu, Q., Liu, W., Zhou, P., Li, J.et al. (2017) Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378–383 10.1038/nature24467 [DOI] [PubMed] [Google Scholar]
- 91.Coers, J. and Haldar, A.K. (2015) Ubiquitination of pathogen-containing vacuoles promotes host defense to Chlamydia trachomatis and Toxoplasma gondii. Commun. Integr. Biol. 8, e1115163 10.1080/19420889.2015.1115163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zwack, E.E., Feeley, E.M., Burton, A.R., Hu, B., Yamamoto, M., Kanneganti, T.D.et al. (2017) Guanylate binding proteins regulate inflammasome activation in response to hyperinjected yersinia translocon components. Infect. Immun. 85, e00778-16 10.1128/IAI.00778-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feeley, E.M., Pilla-Moffett, D.M., Zwack, E.E., Piro, A.S., Finethy, R., Kolb, J.P.et al. (2017) Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc. Natl Acad. Sci. U.S.A. 114, E1698–E1706 10.1073/pnas.1615771114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santos, J.C., Boucher, D., Schneider, L.K., Demarco, B., Dilucca, M., Shkarina, K.et al. (2020) Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria. Nat. Commun. 11, 3276 10.1038/s41467-020-16889-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wandel, M.P., Kim, B.H., Park, E.S., Boyle, K.B., Nayak, K., Lagrange, B.et al. (2020) Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol. 21, 880–891 10.1038/s41590-020-0697-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fisch, D., Clough, B., Domart, M.C., Encheva, V., Bando, H., Snijders, A.P.et al. (2020) Human GBP1 differentially targets salmonella and toxoplasma to license recognition of microbial ligands and caspase-mediated death. Cell Rep. 32, 108008 10.1016/j.celrep.2020.108008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kutsch, M., Sistemich, L., Lesser, C.F., Goldberg, M.B., Herrmann, C. and Coers, J. (2020) Direct binding of polymeric GBP1 to LPS disrupts bacterial cell envelope functions. EMBO J. 39, e104926 10.15252/embj.2020104926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Springer, H.M., Schramm, M., Taylor, G.A. and Howard, J.C. (2013) Irgm1 (LRG-47), a regulator of cell-autonomous immunity, does not localize to mycobacterial or listerial phagosomes in IFN-γ-induced mouse cells. J. Immunol. 191, 1765–1774 10.4049/jimmunol.1300641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maric-Biresev, J., Hunn, J.P., Krut, O., Helms, J.B., Martens, S. and Howard, J.C. (2016) Loss of the interferon-γ-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC Biol 14, 33 10.1186/s12915-016-0255-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park, S., Choi, J., Biering, S.B., Dominici, E., Williams, L.E. and Hwang, S. (2016) Targeting by AutophaGy proteins (TAG): targeting of IFNG-inducible GTPases to membranes by the LC3 conjugation system of autophagy. Autophagy 12, 1153–1167 10.1080/15548627.2016.1178447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Choi, J., Biering, S.B. and Hwang, S. (2017) Quo vadis? Interferon-inducible GTPases go to their target membranes via the LC3-conjugation system of autophagy. Small GTPases 8, 199–207 10.1080/21541248.2016.1213090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brown, H.M., Biering, S.B., Zhu, A., Choi, J. and Hwang, S. (2018) Demarcation of viral shelters results in destruction by membranolytic GTPases: antiviral function of autophagy proteins and interferon-inducible GTPases. Bioessays 40, e1700231 10.1002/bies.201700231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ang, D.K.Y., Ong, S.Y., Brown, A.S., Hartland, E.L. and van Driel, I.R. (2012) A method for quantifying pulmonary Legionella pneumophila infection in mouse lungs by flow cytometry. BMC Res. Notes 5, 448 10.1186/1756-0500-5-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Romanov, J., Walczak, M., Ibiricu, I., Schüchner, S., Ogris, E., Kraft, C.et al. (2012) Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 31, 4304–4317 10.1038/emboj.2012.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hwang, S., Maloney, N.S., Bruinsma, M.W., Goel, G., Duan, E., Zhang, L.et al. (2012) Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11, 397–409 10.1016/j.chom.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Choi, J., Park, S., Biering, S.B., Selleck, E., Liu, C.Y., Zhang, X.et al. (2014) The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 40, 924–935 10.1016/j.immuni.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao, Z., Fux, B., Goodwin, M., Dunay, I.R., Strong, D., Miller, B.C.et al. (2008) Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 10.1016/j.chom.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamamoto, M., Okuyama, M., Ma, J.S., Kimura, T., Kamiyama, N., Saiga, H.et al. (2012) A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 37, 302–313 10.1016/j.immuni.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 109.Jayakumar, A., Donovan, M.J., Tripathi, V., Ramalho-Ortigao, M. and McDowell, M.A. (2008) Leishmania major infection activates NF-kappaB and interferon regulatory factors 1 and 8 in human dendritic cells. Infect. Immun. 76, 2138–2148 10.1128/IAI.01252-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qin, A., Lai, D.H., Liu, Q., Huang, W., Wu, Y.P., Chen, X.et al. (2017) Guanylate-binding protein 1 (GBP1) contributes to the immunity of human mesenchymal stromal cells against Toxoplasma gondii. Proc. Natl Acad. Sci. U.S.A. 114, 1365–1370 10.1073/pnas.1619665114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnston, A.C., Piro, A., Clough, B., Siew, M., Virreira Winter, S., Coers, J.et al. (2016) Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii. Cell Microbiol. 18, 1056–1064 10.1111/cmi.12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alonso, S., Pethe, K., Russell, D.G. and Purdy, G.E. (2007) Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc. Natl Acad. Sci. U.S.A. 104, 6031–6036 10.1073/pnas.0700036104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ponpuak, M., Davis, A.S., Roberts, E.A., Delgado, M.A., Dinkins, C., Zhao, Z.et al. (2010) Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32, 329–341 10.1016/j.immuni.2010.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gutierrez, M.G., Master, S.S., Singh, S.B., Taylor, G.A., Colombo, M.I. and Deretic, V. (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- 115.Coers, J., Bernstein-Hanley, I., Grotsky, D., Parvanova, I., Howard, J.C., Taylor, G.A.et al. (2008) Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J. Immunol 180, 6237–6245 10.4049/jimmunol.180.9.6237 [DOI] [PubMed] [Google Scholar]
- 116.Zhao, Y., Ferguson, D.J., Wilson, D.C., Howard, J.C., Sibley, L.D. and Yap, G.S. (2009) Virulent Toxoplasma gondii evade immunity-related GTPase-mediated parasite vacuole disruption within primed macrophages. J. Immunol. 182, 3775–3781 10.4049/jimmunol.0804190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haldar, A.K., Piro, A.S., Finethy, R., Espenschied, S.T., Brown, H.E., Giebel, A.M.et al. (2016) Chlamydia trachomatis is resistant to inclusion ubiquitination and associated host defense in gamma interferon-primed human epithelial cells. mBio 7, e01417-16 10.1128/mBio.01417-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang, Y., Bazhin, A.V., Werner, J. and Karakhanova, S. (2013) Reactive oxygen species in the immune system. Int. Rev. Immunol. 32, 249–270 10.3109/08830185.2012.755176 [DOI] [PubMed] [Google Scholar]
- 119.Shenoy, A.R., Wellington, D.A., Kumar, P., Kassa, H., Booth, C.J., Cresswell, P.et al. (2012) GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science 336, 481–485 10.1126/science.1217141 [DOI] [PubMed] [Google Scholar]
- 120.Caffrey, D.R. and Fitzgerald, K.A. (2012) Immunology. Select inflammasome assembly. Science 336, 420–421 10.1126/science.1222362 [DOI] [PubMed] [Google Scholar]
- 121.Kim, B.H., Chee, J.D., Bradfield, C.J., Park, E.S., Kumar, P. and MacMicking, J.D. (2016) Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nat. Immunol. 17, 481–489 10.1038/ni.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Finethy, R., Luoma, S., Orench-Rivera, N., Feeley, E.M., Haldar, A.K., Yamamoto, M.et al. (2017) Inflammasome activation by bacterial outer membrane vesicles requires guanylate binding proteins. mBio 8, e01188-17 10.1128/mBio.01188-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Santos, J.C., Dick, M.S., Lagrange, B., Degrandi, D., Pfeffer, K., Yamamoto, M.et al. (2018) LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. 37, e98089 10.15252/embj.201798089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu, B.C., Sarhan, J., Panda, A., Muendlein, H.I., Ilyukha, V., Coers, J.et al. (2018) Constitutive interferon maintains GBP expression required for release of bacterial components upstream of pyroptosis and anti-DNA responses. Cell Rep. 24, 155–168.e5 10.1016/j.celrep.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xavier, A., Al-Zeer, M.A., Meyer, T.F. and Daumke, O. (2020) hGBP1 coordinates chlamydia restriction and inflammasome activation through sequential GTP hydrolysis. Cell Rep. 31, 107667 10.1016/j.celrep.2020.107667 [DOI] [PubMed] [Google Scholar]