Abstract
Protein–protein interactions (PPIs) orchestrate nearly all biological processes. They are also considered attractive drug targets for treating many human diseases, including cancers and neurodegenerative disorders. Protein-fragment complementation assays (PCAs) provide a direct and straightforward way to study PPIs in living cells or multicellular organisms. Importantly, PCAs can be used to detect the interaction of proteins expressed at endogenous levels in their native cellular environment. In this review, we present the principle of PCAs and discuss some of their advantages and limitations. We describe their application in large-scale experiments to investigate PPI networks and to screen or profile PPI targeting compounds.
Keywords: drug targets, high-throughput screening, large-scale, protein-fragment complementation assays, protein–protein interactions, proteome-wide analysis
Introduction
Protein–protein interactions (PPIs) constitute a complex cellular network and play a central role in nearly all biological processes, including DNA replication, transcription, signal transduction, enzymatic reactions, cell-to-cell communication, and membrane transport [1,2]. Alterations in protein–protein interactions (either by breakdown or the formation of novel PPIs) may lead to various diseases in humans, including cancers [3,4] or neurodegenerative disorders [5]. PPIs are attractive molecular targets with a vast therapeutic potential [6–8].
A wide range of methods are available to investigate cellular PPIs [9,10]. Biochemical methods such as immuno- and affinity purification are widely used. Coupled to mass spectrometry they enable proteome-wide analysis of PPI networks [11]. However, these techniques remove proteins from their native cellular environment, which disturbs numerous physiological interactions. In contrast, several methods allow to monitor PPIs in intact living cells and organisms, with minimal cellular perturbations. These include Förster resonance energy transfer (FRET) [12], bioluminescence resonance energy transfer (BRET) [13], fluorescence cross-correlation spectroscopy (FCCS) [14], proximity labelling [15], and protein-fragment complementation assays (PCAs) [16,17]. With the advent of gene editing and PCR tagging technologies [18–20], these methods now enable exploring the interaction network of proteins expressed at near endogenous levels in virtually any model organism or tissue culture system. Importantly, several of the aforementioned methods are suitable to probe transient PPIs and to quantify their dynamics over time. In this review, we present the principle and development of PCAs and their application in large-scale studies. The recent advances are highlighted and the main advantages and limitations of PCA-based methods are also discussed.
General principles of protein-fragment complementation assays
Development of PCA reporters
The idea of using the functional complementation of protein fragments to probe PPIs in living cells dates back to 1989, when Fields and Song described the yeast two-hybrid system (Y2H) [21]. This method relies on monitoring the interaction of two proteins of interest (a ‘bait’ and a ‘prey’) via the transcriptional activation of a reporter gene (usually encoding a selectable marker or a colorimetric enzyme). The bait and the prey are genetically fused to two fragments of the Gal4 transcription factor, the DNA binding domain and the activation domain, respectively, and expressed in yeast cells. Interaction of these ‘hybrid’ proteins restores the activity of the transcription factor and controls the expression of the reporter gene [22]. Y2H was optimised for large-scale analysis and largely contributed to the construction of proteome-wide PPI maps in multiple organisms, from yeast to humans [23–27].
Contrary to Y2H, PCAs do not rely on the cellular transcriptional machinery to detect PPIs. They make use of monomeric reporter proteins whose activity can be directly detected (enzymes or fluorescent proteins). The structure of the reporter protein is engineered to split it into two inactive but complementary fragments. When fused to interacting proteins, these fragments are brought into close proximity, which triggers the reconstitution of the reporter and the detection of its activity (Figure 1). Hence, Y2H and PCAs are conceptually different methods and have distinct applications. Johnsson and Varshavsky described a PCA precursor technique using ubiquitin as a reporter [28]. However, in this assay detection of reconstituted ubiquitin is indirect: it requires cellular proteases to cleave it and immunoblotting to reveal the cleavage. The first bona fide PCA was based on a mutated version of murine dihydrofolate reductase (DHFR) [29] (Figure 2). Since then, several other enzymes (e.g. β-lactamase, luciferases) and fluorescent proteins have been engineered into PCA reporters [30–38] allowing multiple modes of PPI detection (Figure 3). Fluorescent protein PCAs are very popular because they enable the visualisation of PPIs in living cells using fluorescence microscopy. The most frequent are based on the yellow fluorescent protein (YFP) or its variant Venus. These assays are often referred to as Bimolecular Fluorescence Complementation (BiFC). Among enzymatic reporters, DHFR, which confers resistance to the anti-folate drug methotrexate, allows probing PPIs using simple and inexpensive survival selection assays. These do not require specialised reagents or equipment and can be easily scaled up. Luciferases are enzymes that catalyse the oxidation of substrate luciferins in a reaction that emits light. Various luciferases with different properties have been used as PCA reporters, including Renilla (RLuc) [37], firefly (FLuc) [34], Gaussia (GLuc) [38], and NanoLuc [36] luciferases. Luciferase PCAs are well suited for large-scale studies because they are performed in microtiter plates and are easily scalable.
Figure 1. Principle of PCAs.
A pair of investigated proteins of interest (POI 1 & POI 2) is genetically fused to inactive fragments of a reporter protein. When POI 1 and POI 2 interact, the reporter protein fragments are brought into proximity, which enables the reconstitution of the reporter protein and the detection of a particular signal, depending on the assay type.
Figure 2. Landmark publications in PCA development towards large-scale applications.
Timeline of selected discoveries and publications. Green boxes highlight landmark development of PCA reporters, orange boxes highlight landmark publications mapping PPI networks and red boxes highlight landmark publications using PCAs to screen or profile PPI modulators.
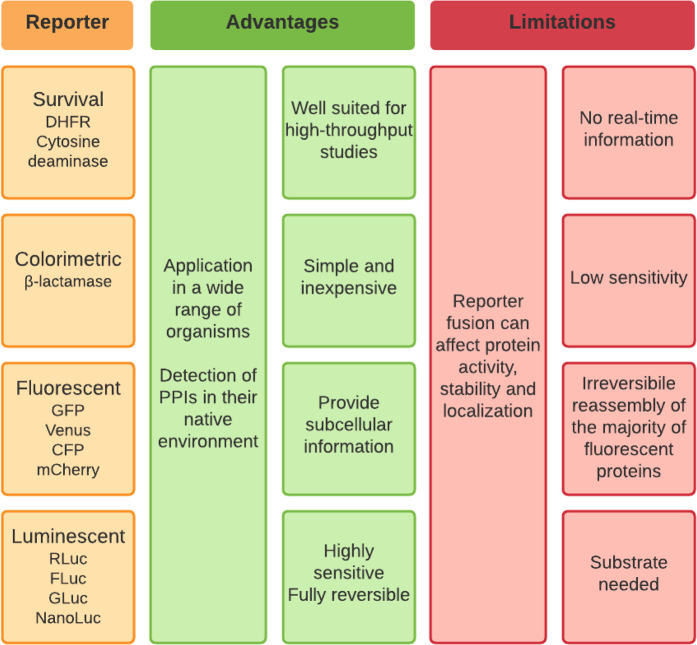
Figure 3. Main PCA reporters with their advantages and limitations.
Presentation of different types of PCAs (colorimetric, fluorescent, luminescent or survival assays) with examples of their main reporters (abbreviations: GFP — green fluorescent protein, CFP — cyan fluorescent protein, RLuc — Renilla luciferase, FLuc — firefly luciferase, GLuc — Gaussia luciferase, NanoLuc — NanoLuc® luciferase, DHFR — dihydrofolate reductase) and some of their advantages and limitations.
Properties, advantages and limitations of PCAs
PCAs constitute attractive approaches for investigating PPIs in living cells. Since they detect PPIs directly, they do not require the use of dedicated host cells (as it is the case for Y2H approaches) and can be used in any genetically amenable model system. They enable to explore the interaction of proteins in their native cellular context, expressed at physiological levels from endogenously tagged genes, and subjected to post-translational modifications and other regulatory mechanisms. Apart from widely used model organisms, such as mammalian cells, plants, fruit fly, or budding yeast, PCAs can serve to explore PPIs in pathogens, e.g. the pathogenic yeast Candida albicans [39] or the parasite Plasmodium falciparum [40].
An important feature of PCAs is that they are exquisitely sensitive to topology. The efficiency of the reconstitution of the PCA reporter depends on the spatial distance and the mobility of the individual fragments fused to the bait and prey proteins. Hence, N-terminal, C-terminal, or internal fusions are not equivalent and can yield different results. The linkers are also important and influence PCA signals. Using the DHFR PCA in yeast, it has been shown that long linkers significantly improve the detection of PPIs and allow capturing indirect interactions in multi-subunit complexes [41]. As a rule of thumb, PCAs are suitable to probe PPIs when the tagged protein extremities are <100 Å apart [41,42]. In addition to distance consideration, fusing proteins to reporter fragments can affect their function in different ways. Like any other tag, PCA fragments can create steric hindrances that impair protein activity, interaction or localisation. Moreover, certain unfolded PCA fragments may destabilise fusion proteins [43,44]. Generally, small PCA reporters that can be split in stable fragments are less likely to perturb protein activities. For example, the NanoBiT PCA reporter (engineered from the small (19 kDa) NanoLuc luciferase) has been optimised for both the size and stability of its fragments [36].
The dynamics of the reporter reconstitution is a critical parameter of PCAs. For most applications, reconstitution of the reporter should not happen spontaneously, should be reversible, and should not interfere with the binding properties of the bait and prey proteins. In other words, the PCA fragments should have low intrinsic affinity so that they only reconstitute the reporter when their ‘local’ concentration is raised due to the interaction of the bait and prey proteins. To our knowledge, the affinity of PCA fragments has been determined for FLuc (KD = 21 µM) [36], NanoBiT (KD = 190 µM) [36] and the recently developed fluorescent PCA termed splitFAST (KD ∼ 1 µM) [45]. Still, many other PCA reporters (including β-lactamase, DHFR and luciferases) have been demonstrated to be reversible [31,38,42,46], with the notable exception of GFP-family fluorescent proteins [33,47]. Irreversible PCAs may artificially trap and amplify non-physiological interactions, especially when proteins are highly expressed, locally tethered or constrained in dense compartments. They are thus best suited to probe irreversible protein interactions or for applications that benefit from molecular trapping [47]. Despite this limitation, fluorescent PCAs had a tremendous impact on the study of PPIs in living cells and new generations of reversible fluorescent reporters are actively being developed [45,48,49].
The sensitivity of PCAs depends on the detection method and equipment used to measure the activity of the reporter as well as the level of background produced by the cells. In general, enzymatic reporters are very sensitive because they enable signal amplification. The DHFR PCA has been reported to be sufficiently sensitive to detect as little as 25 complexes per cell [50]. Luciferase reporters are similarly able to detect the interaction of many endogenously expressed proteins, although their detection limit has, to our knowledge, not been rigorously determined. Importantly, luciferase PCAs produce linear luminescence signals over a large dynamic range and enable to monitor the assembly and disassembly of PPIs in near real-time [36,38,46]. They are thus especially well-suited to dissect the actions of drugs and for drug screening applications. In contrast, GFP-family reporters have a long maturation time (typically minutes to hours), which prevents time-course studies and may even misinform spatial interpretations.
PCAs applied to large-scale analysis of PPIs
Unravelling PPI networks
The elucidation of PPI networks is essential to understand the organisation and regulation of cellular processes. To investigate PPI networks using PCAs, several genome-wide reporter fragment libraries have been developed in budding yeast, mammalian cells and fruit fly (Table 1). Libraries that have been established with a single fragment enable to systematically probe the interactions of selected bait proteins with an array of preys (as exemplified in Figure 4), while two-fragment libraries enable probing any PPI combination. Examples of studies using these and other resources are presented hereafter.
Table 1. PCA resources for large-scale PPI investigation.
| Organism | PCA reporter and its fragment(s) | Details | Source |
|---|---|---|---|
| S. cerevisiae | mDHFR, 2 fragments (DHFR F[1,2], DHFR F[3]) | 4326 strains with the DHFR F[1,2] fragment 4804 strains with the DHFR F[3] fragment C-terminally tagged |
[42] Commercially available (Horizon Discovery) |
| mDHFR, 2 fragments (DHFR F[1,2], DHFR F[3]) | 1741 strains with the DHFR F[1,2] fragment 1113 strains with the DHFR F[3] fragment C-terminally tagged, barcoded in duplicate |
[55] | |
| Venus, 2 fragments (VN173, VC155) | 5809 strains with the VN173 fragment 5671 strains with the VC fragment C-terminally tagged |
[56,58] Partially commercially available (VN173, Bioneer) |
|
| NanoBiT, 2 fragments (LgBiT, SmBiT) | 5580 strains tagged with the LgBiT fragment 4981 strains tagged with the SmBiT fragment C-terminally tagged |
[69] and unpublished results | |
| D. melanogaster | Venus, 1 fragment (VN173) Cerulean, 2 fragments (CN173, CC155) |
450 fly lines focused on transcription factors N- and C-terminally tagged |
[60] |
| Mammalian cells | YFP, 1 fragment (YFPc) | 11 880 ORFs cloned in retroviral plasmids N- and C-terminally tagged |
[61] |
| Venus, 1 fragment (VC159) | cDNA library cloned in a retroviral plasmid N-terminally tagged |
[86] | |
| Gaussia luciferase, 2 fragments (hGLuc(1), hGLuc(2)) | ORFs from 2573 protein pairs cloned in mammalian expression vectors compatible with the human ORFeome (17 408 protein coding genes) N-terminally tagged |
[27] | |
| NanoBiT, 2 fragments (LgBiT, SmBiT) | ORFs from 138 protein pairs cloned in mammalian expression vectors compatible with the human ORFeome (17 408 protein coding genes) N- and C-terminally tagged |
[66] | |
| NanoLuc, 2 fragments (N2H[F1], N2H[F2]) | ORFs from 138 protein pairs cloned in mammalian expression vectors compatible with the human ORFeome (17 408 protein coding genes) N- and C-terminally tagged |
[66] |
Figure 4. Use of PCAs to investigate PPI networks.
PCAs are typically used to probe the interaction of a single (or a few) ‘bait’ protein(s) against a large number of ‘preys’ arrayed in 96, 384 or 1536 pin/well formats. Cells or strains expressing the bait protein(s) of interest tagged with a reporter fragment are distributed into appropriate plates (blue) and transformed or crossed with a library of the preys tagged with the complementary reporter fragment (red). Cells or strains expressing both bait and prey proteins are then selected and the reconstitution of the PCA reporter is measured by survival tests, colorimetric assays or fluorescence/luminescent approaches.
The first large-scale PCA experiment used the DHFR reporter. Tarassov and colleagues [42] constructed genome-wide yeast libraries of both DHFR fragments and conducted a systematic screen, which detected 2770 PPIs involving 1124 proteins. Interestingly, ∼80% of these PPIs were previously unknown. This indicates that PCAs interrogate a different space of the interactome than biochemical and Y2H assays and highlights the need of using complementary approaches for PPI mapping. This system was later applied to analyse the modulation of more than 1000 PPIs in response to DNA damage [51]. The DHFR PCA reporter was also combined with barcoding and next-generation sequencing to investigate PPIs at a high-throughput in pooled cultures [52]. This approach, now termed protein–protein interaction sequencing (PPiSeq), was used to examine the dynamics of large numbers of yeast protein pairs across multiple environmental conditions [53,54]. Recently, PPiSeq was scaled up to quantify the relative abundance of ∼9% of all possible yeast protein pairs (1.6 million). It revealed that most PPIs detected with this method are regulated and only observed in some specific growth conditions [55]. These studies demonstrate the power of using pooled rather than individual cultures for large-scale interaction studies.
BiFC is a popular approach for large-scale PPI studies. Sung and colleagues [56] used a genome-wide library of yeast strains tagged with the N-terminal fragment of the Venus fluorescent protein to probe the interactome of the small ubiquitin-like modifier (SUMO). SUMO is post-translationally conjugated to lysine residues of target proteins to regulate their activity. By comparing the signals produced by the wild type and a mutated form of SUMO that cannot be conjugated to proteins, they were able to identify 280 putative SUMO conjugates, among which 31 were validated in a band-shift assay. We utilised the same collection of yeast strains to systematically search for ubiquitin ligases interacting with the Ubc6 conjugating enzyme. This resulted in the identification of a novel protein quality control pathway involved in the maintenance of the inner nuclear membrane [57]. Recently, Kim and colleagues [58] constructed a complementary library of yeast strains, tagged with the C-terminal fragment of Venus, which enabled them to investigate protein homodimerisation. They identified 630 proteins producing a detectable fluorescent signal, yet the controls they included led them to classify ∼70% of these proteins as false-positives. We also observed a high rate of false-positives, when we investigated the interaction network of ubiquitin ligases and ubiquitin-conjugating enzymes using a sensitive microscopy setup ([44] and unpublished results). These results highlight the importance of designing appropriate controls for the analysis and interpretation of PCA experiments, especially while using irreversible reporters. Large-scale BiFC studies have also been performed in other organisms including plants [59], fruit fly [60] and mammalian cells [61]. Technological developments are essential to empower high-throughput data acquisition and analysis of large-scale BiFC experiments. To examine the interactions of six core telomeric subunits with 12 000 human proteins Lee and colleagues [61] performed fluorescence measurements with a high-throughput flow cytometer and processed them with a specially developed automated data analysis pipeline. They could identify ∼300 proteins associated with the six core telomeric proteins, including possible regulators of telomere biology, such as protein kinases and ubiquitin ligases. Like for the DHFR PCA, BiFC studies may benefit from sequencing technologies. Two preprint publications propose to combine flow cytometry, cell sorting and next-generation sequencing to analyse high-throughput BiFC experiments, which could make it possible to perform BiFC screens in pooled cultures [62,63].
Luminescence-based assays are well suited for large scale studies. The GLuc PCA is one of the most prevalent because it offers bright luminescence with a high signal to background ratio, and it is able to detect a large fraction of well-described PPIs [64]. Gilad and colleagues [65] used it for a large-scale analysis of PPIs within the programmed cell death network of mammalian cells. They used a library composed of 63 proteins from the autophagy and apoptosis machineries, as well as regulatory proteins. By screening all possible combination pairs, they identified 46 previously unknown interactions within this network. The GLuc PCA also served as an orthogonal assay to validate the human reference interactome [64]. NanoLuc PCAs show very good performances [36,66,67] and are very well suited for high-throughput studies. Moreover, contrary to GLuc, NanoLuc produces a stable luminescence signal, which enables investigating the temporal dynamics of PPIs in near real-time. This property has recently been used to profile the dissociation of a family of heterotrimeric G proteins after stimulation by various GPCR ligands [68]. Thus, NanoLuc PCAs open the perspective of revealing the dynamics of PPI networks in response to varying physiological conditions. To this end, we have constructed genome-wide libraries of yeast strains tagged with the NanoBiT PCA fragments (Table 1) ([69] and unpublished results).
PPI in diseases, modulation by chemical compounds
A large fraction of disease-associated mutations have been found to perturb PPIs [70]. PCAs can serve to investigate the functional impact of such mutations and resolve disease mechanisms as exemplified in recent studies of mutations causing mitochondrial diseases [71] and Alport Syndrome [72]. In addition, PPIs are potential drug targets for many diseases. PCAs can be applied at all stages of the drug discovery process from target identification to preclinical validation. One of the assets of PCAs is that they can be applied to probe very diverse therapeutically relevant PPIs in a single setup. For example, Li and colleagues [73] established a simple platform, named ReBiL, based on the reversible firefly luciferase PCA. They used it to probe five different PPIs (UBE2T/FANCL, p53/Mdm2, p53/Mdm4, Mdm2/Mdm4 and BRCA1/BARD1) and to elucidate the mechanism of action of small-molecule- and peptide-based compounds. Several other drug screening platforms have also been successfully established with various luciferase PCAs [74–79]. In general, luciferase-based PCAs are well-suited to perform large-scale drug screens (Figure 5) because of their sensitivity and reversibility. Sensitivity is important to miniaturise the assay and perform it in a high-throughput format, while reversibility is critical to accurately monitor the inhibitory effects of drugs on PPIs. Using such a platform, a recent study screened ∼45 000 bioactive molecules to identify compounds that directly inhibit dimerisation of the NF-κB protein p65 [80]. This enabled to identify withaferin A, a previously characterised molecule with anti-inflammatory and anticancer activities, as allosteric inhibitor of NF-κB dimerisation. In another study, ∼100 000 pure natural plant molecules were screened to identify inhibitors of Spt5, a disease-associated transcription elongation factor that directly interacts with Rpb1, the large subunit of RNA polymerase II [81]. The authors identified several compounds that interfere with Spt5/Rpb1 PCA signal and Spt5 functions in cells. Interestingly, in vitro experiments revealed that these compounds likely do not inhibit Spt5/Rpb1 interaction but rather induce conformational changes that modulate Spt5 activity. These studies illustrate that PCAs can be used to identify various classes of molecules targeting disease-related PPIs and to accelerate the development of new therapeutic strategies.
Figure 5. Use of PCAs to screen PPI modulators.
(a) Reversible PCAs are well-suited to investigate PPI modulators. (b) Screening of a library of compounds to identify molecules that target a given PPI of interest. (c) Screening of an array of PPIs to analyse how PPI networks are modulated by a given compound.
PCAs can also be used to profile the cellular response to drugs by examining their effect on a range of PPIs (Figure 5). After the development of the first PCAs, Remy and Michnick [82] examined, in a proof of concept study, the effect of two immunosuppressants, wortmannin and rapamycin, on a set of 35 PPIs involved in a signal transduction pathway controlling translation initiation. A later study [83], selected 49 PPIs involved in ten different cellular processes (e.g. cell cycle control, apoptosis, ubiquitin-mediated proteolysis) and analysed their response to 107 different drugs from six therapeutic areas (cancer, inflammation, cardiovascular disease, diabetes, neurological disorders and infectious disease). The results obtained were remarkably informative: related drugs displayed similar activity profiles and unexpected off-target effects of certain drugs could be revealed. The barcoded DHFR PCA has also been employed in pooled cultures to analyse the impact of 80 small molecules on 384 yeast PPIs, revealing an unexpected effect of the anticancer drug doxorubicin on transcription [52]. More recently, Stynen and colleagues [84] mapped the effect of two drugs, the immunosuppressant rapamycin and the type 2 diabetes drug metformin, on 3500 yeast proteins. Although metformin has no established direct target [85], they observed that it interferes with the distribution of iron in the cell. These results indicate that large-scale analysis of PPIs can enable the discovery of unpredicted effects of drugs, and thereby is useful to facilitate drug development.
Conclusion
PCAs have shown broad applicability in a wide range of PPI detection and drug screening investigations across different cells and organisms. Protocols using PCAs in living cells may not only be applied to study individual PPIs but also a high number of distinct PPIs in large-scale assays. The availability of multiple PCA reporters with complementary properties and different readouts give flexibility in the choice of the most appropriate system according to research needs. Thus PCAs are, together with orthogonal biochemical and other approaches, methods of choice to build high-quality protein interaction networks. This is important, not only to understand how proteins operate collectively to achieve cellular functions, but also to reveal how their perturbation leads to pathological states and how they can be restored by therapies. Hence, large-scale PCAs studies are helpful in deciphering genotype to phenotype relationships and resolving the molecular mechanisms underlying diseases and drug action.
Perspectives
PPIs play an ubiquitous and fundamental role in most biological processes. They contribute to the development of various diseases and are considered as possible therapeutic targets. Identifying PPIs provides information on the function and regulation of individual proteins and their integration in biological networks.
PCAs can not only detect stable but also weak and transient PPIs in living cells. They can be applied in large-scale studies to profile PPI networks and screen PPI modulators.
Future developments in PCA strategies are expected to facilitate the investigation of numerous PPIs in parallel as well as their dynamics upon physiological perturbations.
Abbreviations
- BiFC
bimolecular fluorescence complementation
- BRET
bioluminescence resonance energy transfer
- CFP
cyan fluorescent protein
- DHFR
dihydrofolate reductase
- FCCS
fluorescence cross-correlation spectroscopy
- FLuc
firefly luciferase
- FRET
Förster resonance energy transfer
- GFP
green fluorescent protein
- GLuc
Gaussia luciferase
- GPCR
G protein-coupled receptor
- LgBiT
large NanoBiT fragment
- NanoBiT
NanoLuc binary technology
- ORF
open reading frame
- PCA
protein-fragment complementation assay
- POI
protein of interest
- PPI
protein–protein interaction
- PPiSeq
protein–protein interaction sequencing
- RLuc
Renilla luciferase
- SmBiT
small NanoBiT fragment
- SUMO
small ubiquitin-like modifier
- Y2H
yeast two-hybrid
- YFP
yellow fluorescent protein
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Science Centre (NCN), Poland, under project no. 2016/21/D/NZ1/00285 to E.B. Also, E.B. acknowledges the French Government and the Embassy of France in Poland, and the Foundation for Polish Sciences (FNP). N.L. received the French Government Scholarship (BGF) to perform a joint-PhD project. G.R. was supported by the ANR grant ANR-16-CE11-0021-01 and the Institut National de la Santé et de la Recherche Médicale. A.S. was supported by an ARED grant from the Région Bretagne.
Author Contributions
All authors (E.B., N.L., A.S., R.W. and G.R.) contributed to the writing and editing of the manuscript and the figures. E.B. and G.R. conceptualised, wrote the main draft and finalised the manuscript, N.L. prepared the figures.
References
- 1.Nooren, I.M.A. and Thornton, J.M. (2003) Diversity of protein–protein interactions. EMBO J. 22, 3486–3492 10.1093/emboj/cdg359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keskin, O., Tuncbag, N. and Gursoy, A. (2016) Predicting protein–protein interactions from the molecular to the proteome level. Chem. Rev. 116, 4884–4909 10.1021/acs.chemrev.5b00683 [DOI] [PubMed] [Google Scholar]
- 3.Ivanov, A.A., Khuri, F.R. and Fu, H. (2013) Targeting protein–protein interactions as an anticancer strategy. Trends Pharmacol. Sci. 34, 393–400 10.1016/j.tips.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, Z., Ivanov, A.A., Su, R., Gonzalez-Pecchi, V., Qi, Q., Liu, S.et al. (2017) The OncoPPi network of cancer-focused protein–protein interactions to inform biological insights and therapeutic strategies. Nat. Commun. 8, 14356 10.1038/ncomms14356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freilich, R., Betegon, M., Tse, E., Mok, S.A., Julien, O., Agard, D.A.et al. (2018) Competing protein-protein interactions regulate binding of Hsp27 to its client protein tau. Nat. Commun. 9, 4563 10.1038/s41467-018-07012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells, J.A. and McClendon, C.L. (2007) Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450, 1001–1009 10.1038/nature06526 [DOI] [PubMed] [Google Scholar]
- 7.Andrei, S.A., Sijbesma, E., Hann, M., Davis, J., O'Mahony, G., Perry, M.W.D.et al. (2017) Stabilization of protein-protein interactions in drug discovery. Expert Opin. Drug Discov. 12, 925–940 10.1080/17460441.2017.1346608 [DOI] [PubMed] [Google Scholar]
- 8.Massari, S., Desantis, J., Nizi, M.G., Cecchetti, V. and Tabarrini, O. (2020) Inhibition of influenza virus polymerase by interfering with its protein-protein interactions. ACS Infect. Dis. 10.1021/acsinfecdis.0c00552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berggård, T., Linse, S. and James, P. (2007) Methods for the detection and analysis of protein-protein interactions. Proteomics 7, 2833–2842 10.1002/pmic.200700131 [DOI] [PubMed] [Google Scholar]
- 10.Titeca, K., Lemmens, I., Tavernier, J. and Eyckerman, S. (2019) Discovering cellular protein-protein interactions: technological strategies and opportunities. Mass Spectrom. Rev. 38, 79–111 10.1002/mas.21574 [DOI] [PubMed] [Google Scholar]
- 11.Smits, A.H. and Vermeulen, M. (2016) Characterizing protein-protein interactions using mass spectrometry: challenges and opportunities. Trends Biotechnol. 34, 825–834 10.1016/j.tibtech.2016.02.014 [DOI] [PubMed] [Google Scholar]
- 12.Hochreiter, B., Garcia, A.P. and Schmid, J.A. (2015) Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors 15, 26281–26314 10.3390/s151026281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfleger, K.D.G., Seeber, R.M. and Eidne, K.A. (2006) Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat. Protoc. 1, 337–345 10.1038/nprot.2006.52 [DOI] [PubMed] [Google Scholar]
- 14.Bacia, K., Kim, S.A. and Schwille, P. (2006) Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods 3, 83–89 10.1038/nmeth822 [DOI] [PubMed] [Google Scholar]
- 15.Gingrasm, A.C., Abe, K.T. and Raught, B. (2019) Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 48, 44–45 10.1016/j.cbpa.2018.10.017 [DOI] [PubMed] [Google Scholar]
- 16.Michnick, S.W., Ear, P.H., Manderson, E.N., Remy, I. and Stefan, E. (2007) Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discov. 6, 569–582 10.1038/nrd2311 [DOI] [PubMed] [Google Scholar]
- 17.Remy, I. and Michnick, S.W. (2007) Application of protein-fragment complementation assays in cell biology. Biotechniques 42, 137–141 10.2144/000112396 [DOI] [PubMed] [Google Scholar]
- 18.Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A. and Charpentier, E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doudna, J.A. and Charpentier, E. (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 20.Fueller, J., Herbst, K., Meurer, M., Gubicza, K., Kurtulmus, B., Knopf, J.D.et al. (2020) CRISPR-Cas12a–assisted PCR tagging of mammalian genes. J. Cell. Biol. 219, e201910210 10.1083/jcb.201910210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fields, S. and Song, O.K. (1989) A novel genetic system to detect protein–protein interactions. Nature 340, 245–246 10.1038/340245a0 [DOI] [PubMed] [Google Scholar]
- 22.Chien, C.T., Bartel, P.L., Sternglanz, R. and Fields, S. (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl Acad. Sci. U.S.A. 88, 9578–9582 10.1073/pnas.88.21.9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R.et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 10.1038/35001009 [DOI] [PubMed] [Google Scholar]
- 24.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. and Sakaki, Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. U.S.A. 98, 4569–4574 10.1073/pnas.061034498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Formstecher, E., Aresta, S., Collura, V., Hamburger, A., Meil, A., Trehin, A.et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 10.1101/gr.2659105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, H., Braun, P., Yildirim, M.A., Lemmens, I., Venkatesan, K., Sahalie, J.et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 10.1126/science.1158684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luck, K., Kim, D.K., Lambourne, L., Spirohn, K., Begg, B.E., Bian, W.et al. (2020) A reference map of the human binary protein interactome. Nature. 580, 402–408 10.1038/s41586-020-2188-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnsson, N. and Varshavsky, A. (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. U.S.A. 91, 10340–10344 10.1073/pnas.91.22.10340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier, J.N., Campbell-Valois, F.X. and Michnick, S.W. (1998) Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl Acad. Sci. U.S.A. 95, 12141–12146 10.1073/pnas.95.21.12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh, I., Hamilton, A.D. and Regan, L. (2000) Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J. Am. Chem. Soc. 122, 5658–5659 10.1021/ja994421w [DOI] [Google Scholar]
- 31.Galarneau, A., Primeau, M., Trudeau, L.E. and Michnick, S.W. (2002) Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein-protein interactions. Nat. Biotechnol. 20, 619–622 10.1038/nbt0602-619 [DOI] [PubMed] [Google Scholar]
- 32.Wehrman, T., Kleaveland, B., Her, J.H., Balint, R.F. and Blau, H.M. (2002) Protein–protein interactions monitored in mammalian cells via complementation of β-lactamase enzyme fragments. Proc. Natl Acad. Sci. U.S.A. 99, 3469–3474 10.1073/pnas.062043699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu, C.D., Chinenov, Y. and Kerppola, T.K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 10.1016/s1097-2765(02)00496-3 [DOI] [PubMed] [Google Scholar]
- 34.Luker, K.E., Smith, M.C.P., Luker, G.D., Gammon, S.T., Piwnica-Worms, H. and Piwnica-Worms, D. (2004) Kinetics of regulated protein–protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl Acad. Sci. U.S.A. 101, 12288–12293 10.1073/pnas.0404041101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ear, P.H. and Michnick, S.W. (2009) A general life-death selection strategy for dissecting protein functions. Nat. Methods 6, 813–816 10.1038/nmeth.1389 [DOI] [PubMed] [Google Scholar]
- 36.Dixon, A.S., Schwinn, M.K., Hall, M.P., Zimmerman, K., Otto, P., Lubben, T.H.et al. (2016) Nanoluc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 10.1021/acschembio.5b00753 [DOI] [PubMed] [Google Scholar]
- 37.Paulmurugan, R. and Gambhir, S.S. (2003) Monitoring protein-protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 75, 1584–1589 10.1021/ac020731c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remy, I. and Michnick, S.W. (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3, 977–979 10.1038/nmeth979 [DOI] [PubMed] [Google Scholar]
- 39.Lai, W.-C., Sun, H.S. and Shieh, J.-C. (2020) Establishment of tetracycline-regulated bimolecular fluorescence complementation assay to detect protein-protein interactions in Candida albicans. Sci. Rep. 10, 2936 10.1038/s41598-020-59891-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levray, Y.S., Berhe, A.D. and Osborne, A.R. (2020) Use of split-dihydrofolate reductase for the detection of protein-protein interactions and simultaneous selection of multiple plasmids in Plasmodium falciparum. Mol. Biochem. Parasitol. 238, 111292 10.1016/j.molbiopara.2020.111292 [DOI] [PubMed] [Google Scholar]
- 41.Chrétien, A.-È., Gagnon-Arsenault, I., Dubé, A.K., Barbeau, X., Després, P.C., Lamothe, C.et al. (2018) Extended linkers improve the detection of protein-protein interactions (PPIs) by dihydrofolate reductase protein-fragment complementation assay (DHFR PCA) in living cells. Mol. Cell. Proteomics 17, 373–383 10.1074/mcp.TIR117.000385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarassov, K., Messier, V., Landry, C.R., Radinovic, S., Serna Molina, M.M., Shames, I.et al. (2008) An in vivo map of the yeast protein interactome. Science 320, 1465–1470 10.1126/science.1153878 [DOI] [PubMed] [Google Scholar]
- 43.Ohmuro-Matsuyama, Y., Chung, C.-I. and Ueda, H. (2013) Demonstration of protein-fragment complementation assay using purified firefly luciferase fragments. BMC Biotechnol. 13, 31 10.1186/1472-6750-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blaszczak, E., Prigent, C. and Rabut, G. (2016) Bimolecular fluorescence complementation to assay the interactions of ubiquitylation enzymes in living yeast cells. Methods Mol. Biol. 1449, 223–241 10.1007/978-1-4939-3756-1_13 [DOI] [PubMed] [Google Scholar]
- 45.Tebo, A.G. and Gautier, A. (2019) A split fluorescent reporter with rapid and reversible complementation. Nat. Commun. 10, 2822 10.1038/s41467-019-10855-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stefan, E., Aquin, S., Berger, N., Landry, C.R., Nyfeler, B., Bouvier, M.et al. (2007) Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc. Natl Acad. Sci. U.S.A. 104, 16916–16921 10.1073/pnas.0704257104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romei, M.G. and Boxer, S.G. (2019) Split green fluorescent proteins: scope, limitations, and outlook. Annu. Rev. Biophys. 48, 19–44 10.1146/annurev-biophys-051013-022846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tchekanda, E., Sivanesan, D. and Michnick, S.W. (2014) An infrared reporter to detect spatiotemporal dynamics of protein-protein interactions. Nat. Methods 11, 641–644 10.1038/nmeth.2934 [DOI] [PubMed] [Google Scholar]
- 49.To, T.-L., Zhang, Q. and Shu, X. (2016) Structure-guided design of a reversible fluorogenic reporter of protein-protein interactions. Protein Sci. 25, 748–753 10.1002/pro.2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Remy, I. and Michnick, S.W. (1999) Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl Acad. Sci. U.S.A. 96, 5394–5399 10.1073/pnas.96.10.5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rochette, S., Gagnon-Arsenault, I., Diss, G. and Landry, C.R. (2014) Modulation of the yeast protein interactome in response to DNA damage. J. Proteomics 100, 25–36 10.1016/j.jprot.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 52.Schlecht, U., Miranda, M. and Suresh, S. (2012) Multiplex assay for condition-dependent changes in protein–protein interactions. Proc. Natl Acad. Sci. U.S.A. 23, 9213–9218 10.1073/pnas.1204952109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celaj, A., Schlecht, U., Smith, J.D., Xu, W., Suresh, S., Miranda, M.et al. (2017) Quantitative analysis of protein interaction network dynamics in yeast. Mol. Syst. Biol. 13, 934 10.15252/msb.20177532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlecht, U., Liu, Z., Blundell, J.R., St Onge, R.P. and Levy, S.F. (2017) A scalable double-barcode sequencing platform for characterization of dynamic protein-protein interactions. Nat. Commun. 8, 15586 10.1038/ncomms15586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, Z., Miller, D., Li, F., Liu, X. and Levy, S.F. (2020) A large accessory protein interactome is rewired across environments. eLife 9, e62365 10.7554/eLife.62365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung, M.-K., Lim, G., Yi, D.-G., Chang, Y.J., Yang, E.B., Lee, K.et al. (2013) Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res. 23, 736–746 10.1101/gr.148346.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khmelinskii, A., Blaszczak, E., Pantazopoulou, M., Fischer, B., Omnus, D.J., Le Dez, G.et al. (2014) Protein quality control at the inner nuclear membrane. Nature 516, 410–413 10.1038/nature14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim, Y., Jung, J.P., Pack, C.G. and Huh, W.K. (2019) Global analysis of protein homomerization in Saccharomyces cerevisiae. Genome Res. 29, 135–145 10.1101/gr.231860.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee, L.Y., Wu, F.H., Hsu, C.T., Shen, S.C., Yeh, H.Y., Liao, D.C.et al. (2012) Screening a cDNA library for protein–protein interactions directly in planta. Plant Cell 24, 1746–1759 10.1105/tpc.112.097998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bischof, J., Duffraisse, M., Furger, E., Ajuria, L., Giraud, G., Vanderperre, S.et al. (2018) Generation of a versatile BiFC ORFeome library for analyzing protein–protein interactions in live Drosophila. eLife 7, e38853 10.7554/eLife.38853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee, O.-H., Kim, H., He, Q., Baek, H.J., Yang, D., Chen, L.Y.et al. (2011) Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol. Cell Proteomics 10, M110.001628 10.1074/mcp.M110.001628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blakely, K., Mero, P., Arnold, R., Saleem, A. and Misquitta, C. (2019) Cell cycle regulation of mitochondrial protein import revealed by genome-scale pooled bimolecular fluorescence complementation screening. bioRxiv 10.1101/770669 [DOI] [Google Scholar]
- 63.Shang, L., Zhang, Y., Liu, Y., Jin, C., Yuan, Y., Tian, C.et al. (2020) A yeast BiFC-seq method for genome-wide interactome mapping. bioRxiv 10.1101/2020.06.16.154146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cassonnet, P., Rolloy, C., Neveu, G., Vidalain, P.O., Chantier, T., Pellet, J.et al. (2011) Benchmarking a luciferase complementation assay for detecting protein complexes. Nat. Methods 8, 990–992 10.1038/nmeth.1773 [DOI] [PubMed] [Google Scholar]
- 65.Gilad, Y., Shiloh, R., Ber, Y., Bialik, S. and Kimchi, A. (2014) Discovering protein-protein interactions within the programmed cell death network using a protein-fragment complementation screen. Cell Rep. 8, 909–921 10.1016/j.celrep.2014.06.049 [DOI] [PubMed] [Google Scholar]
- 66.Choi, S.G., Olivet, J., Cassonnet, P., Vidalain, P.O., Luck, K., Lambourne, L.et al. (2019) Maximizing binary interactome mapping with a minimal number of assays. Nat. Commun. 10, 3907 10.1038/s41467-019-11809-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biewenga, L., Rosier, B.J.H.M. and Merkx, M. (2020) Engineering with NanoLuc: a playground for the development of bioluminescent protein switches and sensors. Biochem. Soc. Trans. 10.1042/BST20200440 [DOI] [PubMed] [Google Scholar]
- 68.Inoue, A., Raimondi, F., Kadji, F.M.N., Singh, G., Kishi, T., Uwamizu, A.et al. (2019) Illuminating G-protein-coupling selectivity of GPCRs. Cell 177, 1933–1947 10.1016/j.cell.2019.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Boulch, M., Brossard, A., Le Dez, G., Léon, S. and Rabut, G. (2020) Sensitive detection of protein ubiquitylation using a protein fragment complementation assay. J Cell. Sci. 133, jcs240093 10.1242/jcs.240093 [DOI] [PubMed] [Google Scholar]
- 70.Sahni, N., Yi, S., Taipale, M., Fuxman Bass, J.I., Coulombe-Huntington, J., Yang, F.et al. (2015) Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 10.1016/j.cell.2015.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barshad, G., Zlotnikov-Poznianski, N., Gal, L., Schuldiner, M. and Mishmar, D. (2019) Disease-causing mutations in subunits of OXPHOS complex I affect certain physical interactions. Sci. Rep. 9, 9987 10.1038/s41598-019-46446-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Omachi, K., Kamura, M., Teramoto, K., Kojima, H., Yokota, T., Kaseda, S.et al. (2018) A split-luciferase-based trimer formation assay as a high-throughput screening platform for therapeutics in alport syndrome. Cell Chem. Biol. 25, 634–643 10.1016/j.chembiol.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 73.Li, Y.-C., Rodewald, L.W., Hoppmann, C., Wong, E.T., Lebreton, S., Safar, P.et al. (2014) A versatile platform to analyze low-affinity and transient protein-protein interactions in living cells in real time. Cell Rep. 9, 1946–1958 10.1016/j.celrep.2014.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ashkenazi, S., Plotnikov, A., Bahat, A. and Dikstein, R. (2017) Effective cell-free drug screening protocol for protein-protein interaction. Anal. Biochem. 532, 53–59 10.1016/j.ab.2017.05.030 [DOI] [PubMed] [Google Scholar]
- 75.Littmann, T., Buschauer, A. and Bernhardt, G. (2019) Split luciferase-based assay for simultaneous analyses of the ligand concentration- and time-dependent recruitment of β-arrestin2. Anal. Biochem. 573, 8–16 10.1016/j.ab.2019.02.023 [DOI] [PubMed] [Google Scholar]
- 76.Sekiba, K., Otsuka, M. and Koike, K. (2019) Identifying inhibitors of the HBx-DDB1 interaction using a split luciferase assay system. J. Vis. Exp. 154, e60652 10.3791/60652 [DOI] [PubMed] [Google Scholar]
- 77.Tsang, T.F., Qiu, Y., Lin, L., Ye, J., Ma, C. and Yang, X. (2019) Simple method for studying in vitro protein-protein interactions based on protein complementation and its application in drug screening targeting bacterial transcription. ACS Infect. Dis. 5, 521–527 10.1021/acsinfecdis.9b00020 [DOI] [PubMed] [Google Scholar]
- 78.Zhang, J., Hu, Y., Wu, N. and Wang, J. (2020) Discovery of influenza polymerase PA-PB1 interaction inhibitors using an in vitro split-luciferase complementation-based assay. ACS Chem. Biol. 15, 74–82 10.1021/acschembio.9b00552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cooley, R., Kara, N., Hui, N.S., Tart, J., Roustan, C., George, R.et al. (2020) Development of a cell-free split-luciferase biochemical assay as a tool for screening for inhibitors of challenging protein-protein interaction targets. Wellcome Open Res. 5, 20 10.12688/wellcomeopenres.15675.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ashkenazi, S., Plotnikov, A., Bahat, A., Ben-Zeev, E., Warszawski, S. and Dikstein, R. (2016) A novel allosteric mechanism of NF-κB dimerization and DNA binding targeted by an anti-inflammatory. Drug Mol. Cell Biol. 36, 1237–1247 10.1128/MCB.00895-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bahat, A., Lahav, O., Plotnikov, A., Leshkowitz, D. and Dikstein, R. (2019) Targeting Spt5-Pol II by small-molecule inhibitors uncouples distinct activities and reveals additional regulatory roles. Mol. Cell 76, 617–631 10.1016/j.molcel.2019.08.024 [DOI] [PubMed] [Google Scholar]
- 82.Remy, I. and Michnick, S.W. (2001) Visualization of biochemical networks in living cells. Proc. Natl Acad. Sci. U.S.A. 98, 7678–7683 10.1073/pnas.131216098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.MacDonald, M.L., Lamerdin, J., Owens, S., Keon, B.H., Bilter, G.K., Shang, Z.et al. (2006) Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2, 329–337 10.1038/nchembio790 [DOI] [PubMed] [Google Scholar]
- 84.Stynen, B., Abd-Rabbo, D., Kowarzyk, J., Miller-Fleming, L., Aulakh, S.K., Garneau, P.et al. (2018) Changes of cell biochemical states are revealed in protein homomeric complex dynamics. Cell 175, 1418–1429 10.1016/j.cell.2018.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spiering, M.J. (2019) The mystery of metformin. J. Biol. Chem. 294, 6689–6691 10.1074/jbc.CL119.008628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Komiya, M., Ito, A., Endo, M., Hiruma, D., Hattori, M., Saitoh, H.et al. (2017) A genetic screen to discover SUMOylated proteins in living mammalian cells. Sci. Rep. 7, 17443 10.1038/s41598-017-17450-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Remy, I. and Michnick, S.W. (2004) A cDNA library functional screening strategy based on fluorescent protein complementation assays to identify novel components of signaling pathways. Methods 32, 381–388 10.1016/j.ymeth.2003.10.011 [DOI] [PubMed] [Google Scholar]