Abstract
SARS-CoV-2 viral contagion has given rise to a worldwide pandemic. Although most children experience minor symptoms from SARS-CoV-2 infection, some have severe complications including Multisystem Inflammatory Syndrome in Children. Neuroblastoma patients may be at higher risk of severe infection as treatment requires immunocompromising chemotherapy and SARS-CoV-2 has demonstrated tropism for nervous cells. To date, there is no sufficient epidemiological data on neuroblastoma patients with SARS-CoV-2. Therefore, we evaluated datasets of non-SARS-CoV-2 infected neuroblastoma patients to assess for key genes involved with SARS-CoV-2 infection as possible neuroblastoma prognostic and infection biomarkers. We hypothesized that ACE2, CD147, PPIA and PPIB, which are associated with viral-cell entry, are potential biomarkers for poor prognosis neuroblastoma and SARS-CoV-2 infection.
We have analysed three publicly available neuroblastoma gene expression datasets to understand the specific molecular susceptibilities that high-risk neuroblastoma patients have to the virus. Gene Expression Omnibus (GEO) GSE49711 and GEO GSE62564 are the microarray and RNA-Seq data, respectively, from 498 neuroblastoma samples published as part of the Sequencing Quality Control initiative. TARGET, contains microarray data from 249 samples and is part of the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative. ACE2, CD147, PPIA and PPIB were identified through their involvement in both SARS-CoV-2 infection and cancer pathogenesis.
In-depth statistical analysis using Kaplan-Meier, differential gene expression, and Cox multivariate regression analysis, demonstrated that overexpression of ACE2, CD147, PPIA and PPIB is significantly associated with poor-prognosis neuroblastoma samples. These results were seen in the presence of amplified MYCN, unfavourable tumour histology and in patients older than 18 months of age. Previously, we have shown that high levels of the nerve growth factor receptor NTRK1 together with low levels of the phosphatase PTPN6 and TP53 are associated with increased relapse-free survival of neuroblastoma patients. Interestingly, low levels of expression of ACE2, CD147, PPIA and PPIB are associated with this NTRK1-PTPN6-TP53 module, suggesting that low expression levels of these genes are associated with good prognosis. These findings have implications for clinical care and therapeutic treatment. The upregulation of ACE2, CD147, PPIA and PPIB in poor-prognosis neuroblastoma samples suggests that these patients may be at higher risk of severe SARS-CoV-2 infection. Importantly, our findings reveal ACE2, CD147, PPIA and PPIB as potential biomarkers and therapeutic targets for neuroblastoma.
Keywords: Neuroblastoma, SARS-CoV-2, Biomarkers, ACE2, CD147, PPIA, PPIB, NTRK1, TP53, PTPN6
Highlights
-
•
ACE2, CD147, PPIA, and PPIB are involved in SARS-CoV-2 viral cell-entry.
-
•
We determined whether ACE2, CD147, PPIA, and PPIB are potential neuroblastoma biomarkers.
-
•
High ACE2, CD147, PPIA, and PPIB expression is associated with poor prognosis neuroblastomas increasing the risk of SARS-COV-2 infection.
-
•
ACE2, CD147, PPIA, and PPIB are potential biomarkers for neuroblastoma and SARS-CoV-2 infection.
1. Introduction
High infection rate of SARS-CoV-2 virus has resulted in a global pandemic [1]. Severe infection elicits a hyper inflammatory response [2] and pose a risk to immunocompromised patients with pre-existing medical conditions [2]. Therefore, there is a need to understand the detrimental effects of SARS-CoV-2 infection on paediatric cancer patients, such as those with neuroblastoma.
Although mortality due to SARS-CoV-2 infection in children is low and most children experience minor symptoms from infection [3], some develop neurological complications including encephalopathy, seizure and peripheral nerve palsies [4], or the life threatening Multisystem Inflammatory Syndrome in Children [3]. Since close to 90% of neuroblastoma diagnosis is primarily in the first 12–24 months of age [5], patients could be at a greater risk of SARS-CoV-2 infection, as children under 5-years of age are more likely to develop severe symptoms [3,6]. Moreover, patients may be at higher-risk of infection due to their immunocompromised status as a result of chemotherapy treatment and specific molecular susceptibilities to the virus caused by cancer [6]. Since SARS-CoV-2 contagion will, most likely, continue in the future, it is critical to understand whether neuroblastoma patients are at higher risk of severe infection.
Neuroblastomas are tumours of sympathoadrenergic origin and the most frequent neoplasms of infancy [5]. Long-term survival with high-risk type of neuroblastoma is poor. Prognosis depends upon patient age and tumour biology [5]. Good-prognosis neuroblastomas which are differentiated and have favourable cytogenetics [5,7], express high levels of the nerve growth factor (NGF) receptor tyrosine-kinase NTRK1 [7,8], whereas poor-prognosis tumours which are poorly differentiated, contain segmental chromosomal-abnormalities (SCA) such as 17p gain, 1q and 11q loss, and have MYCN-amplification [7,8], express the neurotrophin receptor tyrosine-kinase NTRK2 [7,8]. On NGF-stimulation, NTRK1 is activated by tyrosine phosphorylation at positions 490, 670, 674, 675, and 785 [8]. This induces signalling promoting differentiation of neuroblastoma cells [8]. Neuroblastomas are characterised by displaying high cell heterogeneity, which is reflected in their clinical presentation and poor outcomes [5,7,8]. The varied location of the tumours together with complex histopathology and biological characteristics makes neuroblastoma difficult to treat with high-risk patients having poor survival outcomes [5].
We have demonstrated that the tumour suppressor TP53 represses expression of the phosphatase PTPN6, resulting in activation of NTRK1 [9]. Furthermore, we have shown that this type of NTRK1-activation is independently and significantly associated with 5-year relapse free survival (RFS) of children with neuroblastoma including patients with high-risk type tumours [10]. This module is independently associated with RFS in the presence of MYCN-amplification, SCA and histology status. Therefore, suggesting that together NTRK1, wild-type TP53 and low levels of PTPN6 expression could be a predictive biomarker of good prognosis for neuroblastoma [10].
ACE2, an enzyme belonging to the renin-angiotensin-system that cleaves Angiotensin I to Angiotensin II, behaves as the major cell-entry receptor for SARS-CoV-2 [11]. ACE2 binds the SARS-CoV-2 S1 subunit of the Spike (S) protein, facilitating cell-entry [11]. ACE2 expression is decreased in breast-cancer and hepatocellular carcinoma [12,13]. Upregulation of the ACE2/Ang-(1–7)/Mas axis inhibits breast-cancer cell migration and invasion [14]. However, ACE2 upregulation is associated with poor-survival in lung adenocarcinoma [15,16] and the ACE2/Ang-(1–7)/Mas axis promotes migration and invasion of renal carcinoma cells [17]. These findings suggest that ACE2 plays a cancer specific role.
CD147, a cell-surface receptor and interleukin signalling mediator [18], interacts with SARS-CoV-2 S protein to mediate viral infection [18]. Loss of CD147 inhibits increased SARS-CoV-2 viral load, and expression of CD147, in non-susceptible cells, facilitates viral-entry [19]. In breast- and colorectal-cancer high CD147 expression is associated with poor disease free survival (DFS) [19]. CD147 is expressed in neuroblastoma exosomes [20] and high expression is associated with undifferentiated tumours [21].
PPIA and PPIB which are peptidyl-prolyl cis-trans isomerases involved in inflammation [22], facilitate viral-entry of SARS-CoV by interacting with CD147 [23]. They interact with nsp-1of SARS-CoV and are incorporated into the viral capsid and then released, enabling further virus binding to CD147 and subsequent infection of CD147-expressing cells [23]. They are overexpressed in glioblastoma multiforme and non-small-cell lung carcinoma [24]. PPIA correlates with poor outcome in gastric-cancer [24] and PPIB promotes cell proliferation and invasion in gastric and non-small cell lung cancer [25].
Given the heterogeneous histological nature of neuroblastoma, the identification of neuroblastoma biomarkers is of key significance for patient outcome. Our aim is to identify potential biomarkers that could be used as prognostic indicators and potential treatment targets for neuroblastoma. We have hypothesized that ACE2, CD147, PPIA, and PPIB which are involved in SARS-CoV-2 viral cell-entry are potential biomarkers for neuroblastoma and SARS-CoV-2 infection.
2. Methods
2.1. Datasets
Three neuroblastoma gene expression datasets of non-SARS-Cov2 infected patients were assessed. Gene Expression Omnibus (GEO) database, accession number GSE49711, 498 samples of Agilent customized 4 × 44K oligonucleotide microarray data were downloaded, of which 492 have event free survival (EFS) data and MYCN amplification status, were analysed. For validation, Illumina HiSeq 2000 RNA-Seq data from the same samples was downloaded from GEO GSE62564 and analysed in the same fashion. Microarray data was measured as the log-base-2 of the number of bases aligned in the gene, and the RNA-Seq data was measured in log2RPM. (GEO, https://www.ncbi.nlm.nih.gov/geo/). The third dataset contained Affymetrix Exon-ST microarray data from 249 samples analysed as part of the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative and was downloaded from cBioportal as z-scores (https://software.broadinstitute.org/cancer/cga/target) In this dataset, 243 samples had event free survival (EFS) data and MYCN amplification status. All datasets contained tumour histology and age at diagnosis information. Clinical characteristics of each dataset can be seen in Supplementary Table 1.
GSE49711 and GSE62564 datasets were non-normally distributed due to a peak of values at 0 for the microarray data (GSE49711) and at less than −6 for the RNA-Seq data (GSE62564). These non-normal peaks are most likely due to undetected reads. Since z-scores must be calculated from normally distributed data, the non-normal scores were removed. Z-scores were calculated by comparing the expression of each gene in each sample to the expression of the gene across all samples.
Gene expression in samples stratified by MYCN amplification, age at diagnosis, tumour histology, and NTRK1-TP53-PTPN6 module presence, was assessed by comparing mean z-scores using the independent t-test. For the NTRK1-TP53-PTPN6 module, samples with TP53 and PTPN6 z-scores < 0 and NTRK1 z-scores > 0 were considered as having the presence of the module whereas samples with TP53 and PTPN6 z-scores > 0 and NTRK1 z-scores < 0 were considered as having absence of the module. This approach is consistent with published studies characterising this module [10].
2.2. Statistical analysis
Z-scores were used to categorise gene expression into low, moderate or high. A z-score of less than −0.5 indicated low, −0.5 to 0.5 indicated moderate and greater than 0.5 indicated high expression, respectively. The Kaplan-Meier method was used to calculate estimates of cumulative survival probabilities (survival function). Plots were generated and the log-rank statistic was used to determine the statistical significance of the differences between the survival functions. EFS was defined as the time, in days/years, between treatment or the start of a study, and the event was defined as relapse, progression, death or study end. A Cox regression hazard model was generated and multivariate analysis was performed. Stratification by MYCN amplification, tumour histology, age at diagnosis and presence of the NTRK1-PTPN6-TP53 module was undertaken. The statistical significance threshold was taken at 95% confidence interval and a P = 0.05 or less.
3. Results
3.1. ACE2, CD147, PPIA and PPIB expression and association with event free survival
To assess whether expression of ACE2, CD147, PPIA, or PPIB could have prognostic value in neuroblastoma, multivariate analysis was undertaken in 492 samples obtained from the GSE49711 dataset. Although GSE49711 was used for the main analysis, results were confirmed with the GSE62564 and TARGET datasets.
Multivariate analysis (Table 1) showed that high and moderate ACE2 (hazard ratio (HR) = 1.48, P = 0.05 and HR = 1.58, P = 0.03, respectively), high CD147 (HR = 2.13, P = < 0.005) and high and moderate PPIA expression (HR = 2.74, P < 0.005 and HR = 2.01, P < 0.005, respectively) were independently associated with poor-neuroblastoma survival. High PPIB expression trended towards poor-survival (HR = 1.29, P = 0.28). Interestingly, removal of PPIA from the model demonstrated that high PPIB expression was significantly associated with poor-survival (HR = 1.68, P = 0.02) (Supplementary Table 2) suggesting that the prognostic effects of PPIB relies on its correlation with PPIA.
Table 1.
Multivariate analysis a of 492 neuroblastoma samples expressing ACE2, CD147, PPIA and PPIB (GSE49711).
| Variable | Hazard ratio | Pb |
|---|---|---|
| ACE2 | ||
| Low | 1 | |
| Moderate | 1.58 | .03 |
| High | 1.48 | .05 |
| CD147 | ||
| Low | 1 | |
| Moderate | 1.46 | .10 |
| High | 2.13 | <.005 |
| PPIA | ||
| Low | 1 | |
| Moderate | 2.01 | <.005 |
| High | 2.74 | <.005 |
| PPIB | ||
| Low | 1 | |
| Moderate | 0.85 | .46 |
| High | 1.29 | .28 |
Multivariate analysis Cox regression model.
P values lower than 0.05 were deemed significant.
Multivariate analysis was undertaken in samples stratified by MYCN amplification, age at diagnosis, tumour histology and the NTRK1-PTPN6-TP53 module. Analysis adjusted for MYCN amplification, a key poor prognosis marker [7], showed moderate ACE2 (HR = 1.51, P = 0.04), high CD147 (HR = 1.97, P = 0.01) and high and moderate PPIA expression (HR = 2.30, P < 0.005 and HR = 1.85, P = 0.01, respectively) significantly associated with poor prognosis independent of MYCN amplification (Supplementary Table 3). Neuroblastomas are classified as differentiating (with ganglionic characteristics) or undifferentiated (with neuroblastic characteristics) [5]. Adjustment by tumour histology showed that only moderate and high PPIA expression was significantly associated with poor-survival independent of tumour histology (HR = 1.65, P = 0.04 and HR = 2.33, P < 0.005, respectively) (Supplementary Table 4). Since age at diagnosis affects patient outcome [5], expression was analysed in patients younger and older than 18 months of age. Moderate ACE2 (HR = 1.56, P = 0.03), high CD147 (HR = 1.99, P = 0.01), and moderate and high PPIA expression (HR = 1.80, P = 0.01 and HR = 2.27, P < 0.005, respectively) was significantly associated with poor-survival independent of age at diagnosis (Supplementary Table 5).
These results were supported by Kaplan-Meier analysis which showed significant separation in survival outcome between high, moderate and low expression of ACE2, CD147, PPIA or PPIB (Fig. 1A–D). It was estimated that patients with high ACE2 expression had a 59.4% probability of 5-year event free survival (EFS) with a median survival-time of over 5-years, whereas low expression had a 73.3% probability of 5-year EFS with a median survival-time of over 5-years. For CD147 high levels had a 38.1% probability of 5-year EFS with a median survival-time of 1.83 years, whereas with low expression the probability of survival increased to 81.6% with a median survival-time of over 5-years. For PPIA and PPIB patients with high expression had a 36.9% and 37.7%, respectively, probability of survival with a median survival-time of 1.8 and 2.23 years, respectively, whereas with low expression survival increased to 82.6% and 75.1%, with a median survival-time of over 5-years, respectively.
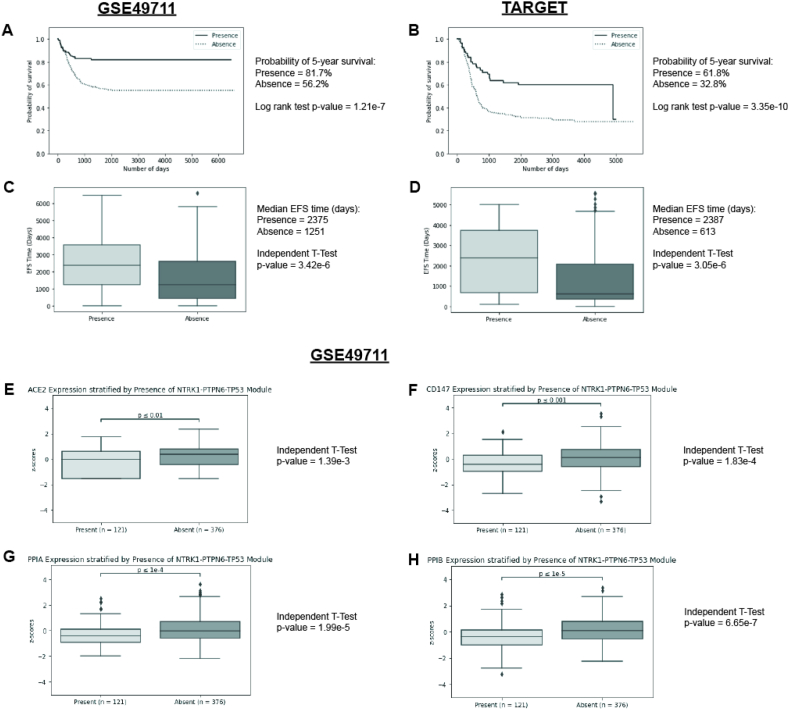
Fig. 1.
Kaplan-Meier survival curves and Box plots (GSE49711) for 492 neuroblastoma samples expressing ACE2, CD147, PPIA, and PPIB. For each gene analysed, samples were stratified into low, medium and high expression groups.
Kaplan-Meier curves were plotted for each group, and survival distributions curves were compared with the log-rank test. Samples with high expression show significantly decreased EFS when compared to samples with low expression A) ACE2, B) CD147, C) PPIA and D) PPIB
Samples were stratified into groups by MYCN amplification and Box plots were plotted using EFS data. Independent t-tests and log-rank tests were used to determine significance E) ACE2, F) CD147, G) PPIA and H) PPIB.
Gene expression was analysed for association with MYCN amplification, age at diagnosis and tumour histology. Samples were stratified based on the presence of each prognostic marker, and expression of each gene was compared between the stratified groups using the independent t-test (Fig. 1E–H). MYCN amplification was significantly associated with upregulation of ACE2, CD147, PPIA and PPIB in GSE49711. Similar results were obtained with GSE62564 dataset. PPIA was seen significantly associated in TARGET (Supplementary Table 6). Unfavourable tumour histology was associated with upregulation of CD147, PPIA, and PPIB (Supplementary Fig. 1 A-D) in GSE49711 and GSE62564 (Supplementary Table 7). Significant association remained for PPIA and PPIB in TARGET (Supplementary Table 7). Samples from patients older than 18 months of age were associated with upregulation of all four genes (Supplementary Fig. 1 E-H) in GSE49711. Similar results were observed with GSE62564 (Supplementary Table 8). In TARGET, results remained significant for PPIB (Supplementary Table 8).
3.2. ACE2, CD147, PPIA and PPIB expression and association with NTRK1-PTPN6-TP53
To verify that samples expressing high levels of NTRK1 together with low levels of PTPN6 and TP53 are indicative of good prognosis as previously demonstrated by our laboratory [10], EFS was analysed using the Kaplan Meier method and an independent t-test. It was estimated that patients with NTRK1-PTPN6-TP53 expression had an 81.7% probability of 5-year EFS with a median survival-time of over 5-years, whereas lack of expression had a 56.2% probability of 5-year EFS with a median survival-time of 3.4 years in GSE49711 (Fig. 2 A). Similar results were obtained with TARGET (Fig. 2 B). It was estimated that patients with NTRK1-PTPN6-TP53 expression had a 61.8% probability of 5-year EFS with a median survival-time of over 5-years, whereas lack of expression had a 32.8% probability of 5-year EFS with a median survival-time of 1.7 years. Therefore, verifying association of the module with prolonged EFS [10].
Fig. 2.
Kaplan-Meir survival curves for 492 neuroblastoma samples expressing high levels of NTRK1 together with low levels of PTPN6 and TP53 (presence of module) or low levels of NTRK1 together with high levels of PTPN6 and TP53 (absence of module). Survival distributions curves were compared with the log-rank test A) GEO GSE49711 dataset and B) TARGET dataset.
Box plots were plotted for both datasets using EFS data. Independent t-tests and log-rank tests were used to determine significance C) GEO GSE49711 dataset and D) TARGET dataset.
Samples were stratified by presence of the NTRK1-PTPN6-TP53 module (GEO GSE49711 dataset). Box plots were plotted using EFS data. Independent t-tests and log-rank tests were used to determine significance E) ACE2, F) CD147, G) PPIA and H) PPIB. In samples with the NTRK1-PTPN6-TP53 module, low expression of ACE2, CD147, PPIA or PPIB show significantly increased EFS.
Multivariate analysis of ACE2, CD147, PPIA and PPIB in samples expressing NTRK1-PTPN6-TP53 showed this module significantly associated with good-prognosis independent of ACE2, CD147, PPIA and PPIB expression (HR = 0.47, P = < 0.005) (Table 2). Furthermore, in GSE49711, NTRK1-PTPN6-TP53 was significantly associated with downregulation of ACE2, CD147, PPIA and PPIB (Fig. 2E–H). These results remained significant for GSE62564 and in the case of the TARGET dataset for PPIA and PPIB (Supplementary Table 9).
Table 2.
Multivariate analysis a of 492 neuroblastoma samples expressing ACE2, CD147, PPIA and PPIB adjusted for the NTRK1-PTPN6-TP53 module (GSE49711).
| Variable | Hazard ratio | Pb |
|---|---|---|
| NTRK1-PTPN6-TP53 module | ||
| Absent | 1 | |
| Present | 0.47 | <.005 |
| ACE2 | ||
| Low | 1 | |
| Moderate | 1.50 | .05 |
| High | 1.36 | .13 |
| CD147 | ||
| Low | 1 | |
| Moderate | 1.41 | .14 |
| High | 2.17 | <.005 |
| PPIA | ||
| Low | 1 | |
| Moderate | 1.98 | <.005 |
| High | 2.61 | <.005 |
| PPIB | ||
| Low | 1 | |
| Moderate | 0.81 | .31 |
| High | 1.14 | .57 |
Multivariate analysis Cox regression model.
P values lower than 0.05 were deemed significant.
It is important to indicate that stratification by SCA was not undertaken. From the three datasets, only TARGET had information regarding SCA in the form of 59 samples. This low sample number was not statistically powerful to carry out the analysis.
4. Discussion
Together, these results show that ACE2, CD147, PPIA and PPIB which are genes involved in viral-entry of SARS-CoV-2, are upregulated and significantly associated with poor-prognosis neuroblastomas with MYCN amplification, unfavourable histology and in patients older than 18 months of age. Moreover, their expression is downregulated in tumours expressing NTRK1-PTPN6-TP53. This strongly suggests that poor-prognosis patients may be at a higher risk of SARS-CoV-2 infection when compared to those expressing NTRK1-PTPN6-TP53. Furthermore, the upregulation of ACE2, CD147, PPIA and PPIB may not only render poor-prognosis neuroblastoma patients more vulnerable to infection, but may also lead to increased viral load and more severe infection.
These findings are supported by our preliminary identification of enriched-gene-clusters and enriched-pathways in prognosis-related clusters of neuroblastoma samples (using the TARGET dataset), with potential to interact with SARS-CoV-2 proteins. Results showed that in potential poor-prognosis clusters PI3K-AKT-mTOR signalling and CD147, PPIA, PPIB and NTRK1 could be implicated in interactions with the SARS-CoV NSP3 protein. NSP3 is of relevance as SARS-CoV NSP3 shares 94% sequence homology with SARS-CoV-2 NSP3 [26]. The NSP3 protein has immuno-evasive properties by inhibiting interferon signalling and preventing attack from the host's immune system [27]. Interestingly, interferon signalling was also prevalent in interactions investigated in poor prognosis S-type and Mesenchymal neuroblastoma cell lineages. However, these results need further investigation due to the complex and heterogeneous nature of neuroblastoma.
Since SARS-CoV-2 infection is likely to persist in the future, these findings may have implications for patient care. CD147 participates in neuroblastoma tumour growth and metastasis [28]. Inhibition of CD147 is, therefore, a potential treatment strategy for slowing tumour growth. Evidence demonstrates that inhibition of CD147 in glioblastoma can reduce tumour cell invasion [29]. Meplazumab, a biological drug which blocks CD147 on the surface of Vero E6 cells and inhibits SARS-CoV-2 replication [30], could potentially treat SARS-CoV-2 infection and neuroblastoma.
Although ACE2 has been associated with SARS-CoV cell-entry [31], and CD147 and PPIA facilitate infection of Human Immunodeficiency Virus (HIV-1) and genome replication of Hepatitis C Virus (HCV) [32,33], it is the statistically significant relationship between ACE2, CD147, PPIA and PPIB as a result of the analysis of the neuroblastoma datasets that strongly emphasises their importance as potential neuroblastoma prognostic markers.
Together these results enhance the knowledge regarding neuroblastoma patient risk to severe SARS-CoV-2 infection. Although studies have assessed SARS-CoV-2 infection rates in paediatric cancer patients [6], to date, few have specifically focused on neuroblastoma. Given that treatment of high-risk neuroblastoma needs further development, our findings of ACE2, CD147 and PPIA as significantly independent prognostic markers should be considered as potential treatment targets. Furthermore, drugs developed to target ACE2, CD147 and PPIA could be repositioned for neuroblastoma treatment.
Funding
This investigation was not supported by any funding.
Declaration of competing interest
The authors declare that they do not have conflicts of interest that could influence the research described in this paper.
Acknowledgments
Authors are thankful to David Bacon for technical help with graphics and tables.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101081.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derosa L., Melenotte C., Griscelli F., Gachot B., Marabelle A., Kroemer G., Zitvogel L. The immuno-oncological challenge of COVID-19. Nat. Can. (Que.) 2020;1:946–964. doi: 10.1038/s43018-020-00122-3. [DOI] [PubMed] [Google Scholar]
- 3.Zare-Zardini H., Soltaninejad H., Ferdosian F., Hamidieh A.A., Memarpoor-Yazdi M. Coronavirus disease 2019 (COVID-19) in children: prevalence, diagnosis, clinical symptoms, and treatment. Int. J. Gen. Med. 2020;13:477–482. doi: 10.2147/IJGM.S262098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singer T.G., Evankovich K.D., Fisher K., Demmler-Harrison G.J., Risen S.R. Coronavirus infections in the nervous system of children: a scoping review making the case for long-term neurodevelopmental surveillance. Pediatr. Neurol. 2021;117:47–63. doi: 10.1016/j.pediatrneurol.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irwin M.S., Park J.R. Neuroblastoma: paradigm for precision medicine Pediatr. Clin. North Am. 2015;62(1):225–256. doi: 10.1016/j.pcl.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Kotecha R.S. Challenges posed by COVID-19 to children with cancer. Lancet Oncol. 2020;21:e235. doi: 10.1016/S1470-2045(20)30205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakagawara A., Li Y., Izumi H., Muramori K., Inada H., Nishi M. Neuroblastoma. Jpn. J. Clin. Oncol. 2018;48:214–241. doi: 10.1093/jjco/hyx176. [DOI] [PubMed] [Google Scholar]
- 8.Ratner N., Brodeur G.M., Dale R.C., Schor N.F. The "neuro" of neuroblastoma: neuroblastoma as a neurodevelopmental disorder. Ann. Neurol. 2016;80:13–23. doi: 10.1002/ana.24659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montano X. Repression of SHP-1 expression by p53 leads to trkA tyrosine phosphorylation and suppression of breast cancer cell proliferation. Oncogene. 2009;28:3787–3800. doi: 10.1038/onc.2009.143. [DOI] [PubMed] [Google Scholar]
- 10.Youssef G., Gillett C., Rampling D., Chagtai T., Virasami A., Barton J., Edwards D., Sebire N., Anderson J., Montano X. The presence of Y674/Y675 phosphorylated NTRK1 via TP53 repression of PTPN6 expression as a potential prognostic marker in neuroblastoma. Hum. Pathol. 2019;86:182–192. doi: 10.1016/j.humpath.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q., Lu S., Li T., Yu L., Zhang Y., Zeng H., Qian X., Bi J., Lin Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Canc. Res. 2019;38:173. doi: 10.1186/s13046-019-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye G., Qin Y., Lu X., Xu X., Xu S., Wu C., Wang X., Wang S., Pan D. The association of renin-angiotensin system genes with the progression of hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2015;459:18–23. doi: 10.1016/j.bbrc.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 14.Yu C., Tang W., Wang Y., Shen Q., Wang B., Cai C., Meng X., Zou F. Downregulation of ACE2/Ang-(1–7)/Mas axis promotes breast cancer metastasis by enhancing store-operated calcium entry. Canc. Lett. 2016;376:268–277. doi: 10.1016/j.canlet.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y., Ni L., Wan H., Fan L., Fei X., Ma Q., Gao B., Xiang Y., Che J., Li Q. Overexpression of ACE2 produces antitumour effects via inhibition of angiogenesis and tumour cell invasion in vivo and in vitro. Oncol. Rep. 2011;26:1157–1164. doi: 10.3892/or.2011.1394. [DOI] [PubMed] [Google Scholar]
- 16.Samad A., Jafar T., Rafi J.H. Identification of angiotensin-converting enzyme 2 (ACE2) protein as the potential biomarker in SARS-CoV-2 infection-related lung cancer using computational analyses. Genomics. 2020;112:4912–4923. doi: 10.1016/j.ygeno.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J., Fan J., Wu F., Huang Q., Guo M., Lv Z., Han J., Duan L., Hu G., Chen L., Liao T., Ma W., Tao X., Jin Y. The ACE2/angiotensin-(1–7)/mas receptor Axis: pleiotropic roles in cancer. Front. Physiol. 2017;8 doi: 10.3389/fphys.2017.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Chen W., Zhang Z., Deng Y. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020;5:1–10. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bovenzi C.D., Hamilton J., Tassone P., Johnson J., Cognetti D.M., Luginbuhl A., Keane W.M., Zhan T., Tuluc M., Bar-Ad V., Martinez-Outschoorn U., Curry J.M. Prognostic indications of elevated MCT4 and CD147 across cancer types: a meta-analysis. BioMed Res. Int. 2015 doi: 10.1155/2015/242437. https://www.hindawi.com/journals/bmri/2015/242437/ e242437. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marimpietri D., Petretto A., Raffaghello L., Pezzolo A., Gagliani C., Tacchetti C., Mauri P., Melioli G., Pistoia V. Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumour progression. PloS One. 2013;8 doi: 10.1371/journal.pone.0075054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia J., Faca V., Jarzembowski J., Zhang Q., Park J., Hanash S. Comprehensive profiling of the cell surface proteome of Sy5Y neuroblastoma cells yields a subset of proteins associated with tumour differentiation. J. Proteome Res. 2009;8:3791–3796. doi: 10.1021/pr800964v. [DOI] [PubMed] [Google Scholar]
- 22.Lavin P.T.M., Mc Gee M.M. Cyclophilin function in Cancer; lessons from virus replication. Curr. Mol. Pharmacol. 2016;9:148–164. doi: 10.2174/1874467208666150519115443. 2015. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z., Mi L., Xu J., Yu J., Wang X., Jiang J., Xing J., Shang P., Qian A., Li Y., Shaw P.X., Wang J., Duan S., Ding J., Fan C., Zhang Y., Yang Y., Yu X., Feng Q., Li B., Yao X., Zhang Z., Li L., Xue X., Zhu P. Function of HAb18G/cd147 in invasion of host cells by severe acute respiratory Syndrome coronavirus. J. Infect. Dis. 2005;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radzikowska U., Ding M., Tan G., Zhakparov D., Peng Y., Wawrzyniak P., Wang M., Li S., Morita H., Altunbulakli C., Reiger M., U Neumann A., Lunjani N., Traidl-Hoffmann C., C Nadeau K., O'Mahony L., Akdis C., Sokolowska M. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy. 2020;75:2829–2845. doi: 10.1111/all.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng M.-R., Huang J.-A., Zhu Z.-T., Li H., Shen J.-F., Chen Q. Cyclophilin B promotes cell proliferation, migration, invasion and angiogenesis via regulating the STAT3 pathway in non-small cell lung cancer. Pathol. Res. Pract. 2019;215:152417. doi: 10.1016/j.prp.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Yoshimoto F.K. The proteins of severe acute respiratory Syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Astuti I. Ysrafil, severe acute respiratory Syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Xu J., Chen L., Zhong W.-D., Zhang Z., Mi L., Zhang Y., Gong Liao C., Bian H.-J., Jiang J.-L., Yang X.-M., Li X.-Y., Fan C.-M., Zhu P., Fu L., Chen Z.-N. HAb18G (CD147), a cancer-associated biomarker and its role in cancer detection. Histopathology. 2009;54:677–687. doi: 10.1111/j.1365-2559.2009.03280.x. [DOI] [PubMed] [Google Scholar]
- 29.Liang Q., Xiong H., Gao G., Xiong K., Wang X., Zhao Z., Zhang H., Li Y. Inhibition of basigin expression in glioblastoma cell line via antisense RNA reduces tumour cell invasion and angiogenesis. Canc. Biol. Ther. 2005;4:759–762. doi: 10.4161/cbt.4.7.1828. [DOI] [PubMed] [Google Scholar]
- 30.Bian H., Zheng Z.-H., Wei D., Zhang Z., Kang W.-Z., Hao C.-Q., Dong K., Kang W., Xia J.-L., Miao J.-L., Xie R.-H., Wang B., Sun X.-X., Yang X.-M., Lin P., Geng J.-J., Wang K., Cui H.-Y., Zhang K., Chen X.-C., Tang H., Du H., Yao N., Liu S.-S., Liu L.-N., Zhang Z., Gao Z.-W., Nan G., Wang Q.-Y., Lian J.-Q., Chen Z.-N., Zhu P. Meplazumab treats COVID-19 pneumonia: an open-labelled, concurrent controlled add-on clinical trial. medRxiv. 2020 doi: 10.1101/2020.03.21.20040691. [DOI] [Google Scholar]
- 31.Davidson A.M., Wysocki J., Batlle D. Interaction of SARS-CoV-2 and other coronavirus with ACE (Angiotensin-Converting enzyme)-2 as their main receptor. Hypertension. 2020;76:1339–1349. doi: 10.1161/HYPERTENSIONAHA.120.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pushkarsky T., Zybarth G., Dubrovsky L., Yurchenko V., Tang H., Guo H., Toole B., Sherry B., Bukrinsky M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. Unit. States Am. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watashi K., Ishii N., Hijikata M., Inoue D., Murata T., Miyanari Y., Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.