Abstract
Plasmodium falciparum resistance to artemisinin-based combination therapy (ACT) is a global threat to malaria control and elimination efforts. Mutations in the P. falciparum kelch13 gene (Pfk13) that are associated with delayed parasite clearance have emerged on the Thai-Cambodian border since 2008. There is growing evidence of widespread Pfk13 mutations throughout South-East Asia and they have independently emerged in other endemic regions. In Papua New Guinea (PNG), Pfk13 “C580Y” mutant parasites with reduced in vitro sensitivity to artemisinin have been isolated in Wewak, a port town in East Sepik Province. However, the extent of any local spread of these mutant parasites in other parts of PNG is unknown. We investigated the prevalence of Pfk13 mutations in multiple malaria-endemic regions of PNG. P. falciparum isolates (n = 1152) collected between 2016 and 2018 and assessed for Pfk13 variation by sequencing. Of 663 high quality Pfk13 sequences a total of five variants were identified. They included C580Y, a mutation at a previously documented polymorphic locus: N499K, and three previously undescribed mutations: R471C, K586E and Y635C. All variants were found in single isolates, indicating that these Pfk13 mutations were rare in the areas surveyed. Notably, C580Y was absent from Maprik district, which neighbours Wewak where C580Y mutant parasites were previously identified. The single C580Y isolate was found in the port town of Lae, Morobe Province, a potential entry site for the importation of drug resistant parasites into PNG. Although sample size in this location was small (n = 5), our identification of a C580Y mutant in this second location is concerning, highlighting the urgent need for further surveillance in Lae. Other Pfk13 mutants were rare in PNG between 2016 and 2018. Continued surveillance for molecular markers of drug resistance is critically important to inform malaria control in PNG.
Keywords: Plasmodium falciparum, Artemisinin resistance, Antimalarial drug resistance, kelch13 mutations, Malaria control, Surveillance
1. Introduction
Resistance of the human malaria parasite, Plasmodium falciparum to antimalarial drugs is a public health concern for malaria endemic countries. The parasite's ability to develop resistance to antimalarial drugs presents significant challenges for malaria treatment, control and elimination (World Health Organisation, 2019). Historically, South-East Asia has been the epicentre of single and multi-drug resistant malaria and remains a hotspot for emerging antimalarial resistance (Ashley et al., 2014; Mita et al., 2009). Currently, the World Health Organisation (WHO) recommends the use of artemisinin-based combination therapy (ACT) as first-line treatment for uncomplicated malaria (World Health Organisation, 2015). While it remains the most effective and widely used treatment in many malaria endemic countries, the growing number of countries of emerging artemisinin resistant parasites is a great concern (Leang et al., 2015; Mathieu et al., 2020; Menard et al., 2016; Uwimana et al., 2020).
Artemisinin resistance (ART-R) is characterised clinically by a delay in parasite clearance following an artemisinin-based therapy (Dondorp et al., 2009). This phenotype was first observed on the Thai-Cambodian border between 2006 and 2008 following both artemisinin mono- and combination therapy and has been linked to non-synonymous single nucleotide polymorphisms (NS-SNPs) found in the P. falciparum chromosome 13 kelch gene (Pfk13) (Ariey et al., 2014; Dondorp et al., 2009; Noedl et al., 2008; Straimer et al., 2015). Genetic analysis of P. falciparum isolates shows that Pfk13 mutations Y493H, R539T, I543T and C580Y were associated with increased ring-stage parasite survival rates in in vitro drug resistance assays and prolonged parasite clearance times in vivo (Ariey et al., 2014; Witkowski et al., 2013). Several Pfk13 mutations, some of which are associated with ART-R, have emerged independently in multiple regions along the Thai-Cambodian and Thai-Myanmar border and have spread throughout the Greater Mekong Region (Miotto et al., 2013; Takala-Harrison et al., 2015; Talundzic et al., 2015). Initially, a soft-selective sweep influenced the emergence and spread of these Pfk13 mutant alleles (Miotto et al., 2015; Takala-Harrison et al., 2015), however this evolved to a hard-sweep in which the kelch13 Haplotype Group 1 (KEL1) lineage, within which C580Y is commonly found, became the dominant lineage in artemisinin-resistant parasite populations (Amato et al., 2018). Consequently, C580Y appears to be outcompeting other mutants and reaching near-fixation in the South-East Asian population (Imwong et al., 2017). The continued use of ACTs in South-East Asia and its cumulative drug pressure on these artemisinin-resistant parasite populations has also resulted in parasite resistance to partner drugs.
PNG carries the highest burden of malaria in the WHO Western Pacific region, accounting for more than 70% of clinical cases (World Health Organisation Regional Office for the Western Pacific, 2017). In 2011, the PNG National Department of Health implemented ACTs into its treatment regimen with artemether-lumefantrine (AL) and DHA-PPQ as first- and second-line treatments respectively, for uncomplicated malaria infections (Papua New Guinea National Department of Health: National malaria treatment protocol, 2009). This was a result of high treatment failure rates associated with a high prevalence of drug resistance mutations limiting the efficacy of the previous treatment regimen of chloroquine plus SP (Casey et al., 2004; Genton et al., 2005; Karunajeewa et al., 2008; Marfurt et al., 2008, 2010). The existing ACT regimen has so far remained efficacious (Tavul et al., 2018), with no evidence of treatment failures in ongoing therapeutic efficacy studies by the PNG National Department of Health and PNG Institute of Medical Research (PNGIMR) (World Health Organisation, 2020). However, two recent reports have identified the Pfk13 C580Y mutation in a traveler returning to Australia from West New Britain Province, PNG in 2012 (Prosser et al., 2018), and in three malaria patients in presenting to a clinic in East Sepik Province in 2017 (Miotto et al., 2020). Whole genome sequence analyses showed haplotype sharing with both Indonesian and PNG parasite populations, thus indicating continuous recombination events between imported and local parasites (Miotto et al., 2020). This raises important questions regarding the presence of artemisinin resistance associated Pfk13 mutations in the broader PNG parasite population, their prevalence and implications for the existing ACT first line treatment regimen.
The PNG Institute of Medical Research together with the National Department of Health has conducted Pfk13 surveillance in PNG for many years. Previous analysis of 43 P. falciparum isolates collected in 2013 and 2014 from 12 locations across the country did not identify any Pfk13 mutations (Menard et al., 2016). In addition, genotyping of clinical infections from therapeutic efficacy studies conducted in the Milne Bay (Alotau) and East Sepik (Maprik) Provinces in 2011–2014 identified only wild type PfK13 alleles (unpublished data). Here we have investigated the presence of Pfk13 mutations in PNG by screening a large number of previously collected P. falciparum samples from cross-sectional and nationwide surveys conducted between 2016 and 2018. We establish the prevalence of Pfk13 mutations in a number of locations throughout PNG during this time frame and screen for the presence of Pfk13 mutations that may reduce artemisinin efficacy.
2. Materials and methods
2.1. Ethics and informed consent
Approval for the use of human blood samples for investigating molecular markers of antimalarial drug resistance was provided by the PNG Medical Research Advisory Committee (MRAC) and the PNGIMR Institutional Review Board (IRB) (2016-17 MIS: MRAC 15.21, IRB 1512, International Centre for Excellence in Malaria Research (ICEMR MRAC:11.12, IRB:1116), WHO Tropical Disease Research Residual Malaria Call 2015 (TDR MRAC:16.08, IRB:1517), Australia-PNG-Trilateral Malaria Project (TMP MRAC:17.11, IRB:1711)) and by the Walter and Eliza Hall Human Research and Ethics Committee (HREC: 13.14 and 12.10). All samples were collected by voluntary consent from participants or their parents or guardians.
2.2. Study sites and samples
Samples were compiled from four studies (Table 1) conducted in collaboration between the PNGIMR and the National Department of Health (NDOH) in malaria-endemic provinces across PNG. These included: (i) Malaria positive RDT (mRDT) from the national malaria indicator survey (MIS) in 2016/17, (ii) Dried blood spots (DBS) from consenting febrile and mRDT-positive individuals attending four health facilities in malaria sentinel site surveillance from 2017 to 2018, Australia-China-PNG Trilateral Malaria Project), (iii) Capillary blood (200–350 μL) from individuals > 6months in two cross-sectional surveys in 2016 in the highly malaria endemic East Sepik (ICEMR) and Madang (TDR) provinces. Due to low DNA concentration of extracted samples from RDTs (MIS), 8 of 13 provinces in the malaria indicator survey could be assessed.
Table 1.
Samples genotyped for Pfk13 mutations.
| Study1/Year of collection | Survey Type/Population | Sample Type | Pf qPCR positive | Pfk13 PCR positive | Sequencing (% of Pf isolates screened) |
|---|---|---|---|---|---|
| MIS 2016/17 | Community Survey/General population) | RDT Strips | 469 | 58 | 38 (8.1) |
| ICEMR Cross sectional 2016 | Clinical (>6 months) | Capillary blood/Microtainer | 192 | 181 | 178 (92.7) |
| TDR Cross sectional 2016 | Cross-Sectional Survey (>6months) | Capillary blood/Microtainer | 384 | 360 | 353 (91.9) |
| TMP 2017/18 | 4 Sentinel Sites, Clinical (General population) | Dried Blood spot/Filter paper | 107 | 107 | 94 (87.8) |
| Total | 1152 | 706 | 663 (57.5) |
- 1National malaria indicator survey (MIS 2016/18) from 13/22 province. Malaria RDT (available from 13/22 provinces; general population sample, all ages.
- ICEMR East Sepik cross-sectional survey 2016.
- Tropical Disease Research (TDR) Grant cross-sectional survey in Madang Province in 2016.
- Australia-China-PNG Trilateral Malaria Project (TMP): Sentinel site surveillance 2017/2018.
2.3. Laboratory procedures
2.3.1. Genomic DNA extraction
Genomic DNA was extracted from the DBS and red cell pellets samples using the Favorgen 96-Well Genomic DNA Kit (Favrogen Biotech Corp. Taiwan) as per the manufacturer's instructions. With the RDT samples, each RDT cassette was disassembled to access the internal cellulose membrane strip where two 3-4 mm-wide pieces were cuts from the sample pad area. Genomic DNA was extracted from these pieces using the Chellex-Saponin method (Plowe et al., 1995) as well as the QIAamp DNA Mini Kit (QIAGEN N.V) according to the manufacturer's instructions.
2.3.2. Identification and quantification of Plasmodium parasite DNA
Parasite DNA detection and quantification was determined using a previously described Plasmodium species-specific real-time PCR TaqMan assay where parasite density within the sample was determined by using a DNA template of known starting concentration (copy number) serially diluted from 1 × 105 down to 5 copies/μL and assayed in the same run. A standard curve in linear regression was generated using the log of the starting copy number and unknown sample densities were interpolated from the standard curve (Rosanas-Urgell et al., 2010).
2.3.3. Amplification and sequencing of Pfk13 gene
DNA samples positive for P. falciparum by quantitative PCR were used as template in a nested PCR reaction to amplify the BTB-POZ domain of the Pfk13 gene known to be associated with artemisinin resistance. The protocol was adapted from a previously published protocol (Menard et al., 2016) with slight modifications with the reaction conditions. In brief, 2–4 μL of genomic DNA was combined with 2x Reaction Buffer (Solis Bio Dyne), 0.5 μM of each primer, 2.5 mM MgCl2, 0.25 mM dNTP and 1.25 U Hot Start Taq Polymerase (Solis Bio Dyne, Denmark) in a 20 μL volume for both primary: PF 5′-CGGAGTGACCAAATCTGGGA-3′ and PR 5′-GGAATCTGGTGGTAACAGC-3′) and nested reactions: NF 5′-GCCAAGCTGCCATTCATTTG and NR 5′-GCCTTGTTGAAAGAAGCAGA-3′). Primary reaction thermocycling conditions included a hot start of 95°C (to activate the Taq Polymerase) for 15 minutes, followed by 30 cycles at 95°C for 30 seconds, 58°C for 1 minute, 65°C for 1 minute 40 seconds, and final extension for 10 minutes at 65°C. Secondary cycling conditions were similar to that of primary reaction except the annealing and extension temperatures were 60°C for 30 seconds and 1 minute respectively for 35 cycles. Final PCR reactions were assessed for Pfk13 amplification by gel-electrophoresis in a 1% Agarose gel stained with 1x ethidium bromide DNA staining dye and visualised using the Bio-Rad XR + Gel Electrophoresis Documentation System. Reactions showing successful DNA amplification of 850–900 bp fragments were sent to Macrogen Inc. (Korea) for Sanger sequencing. Sequence electropherograms were trimmed and aligned to the Pfk13 3D7 clone reference PF3D7_1343700 (PlasmoDB) using Geneious Version 6.1.8 software (Biomatters LTD, New Zealand).
3. Results
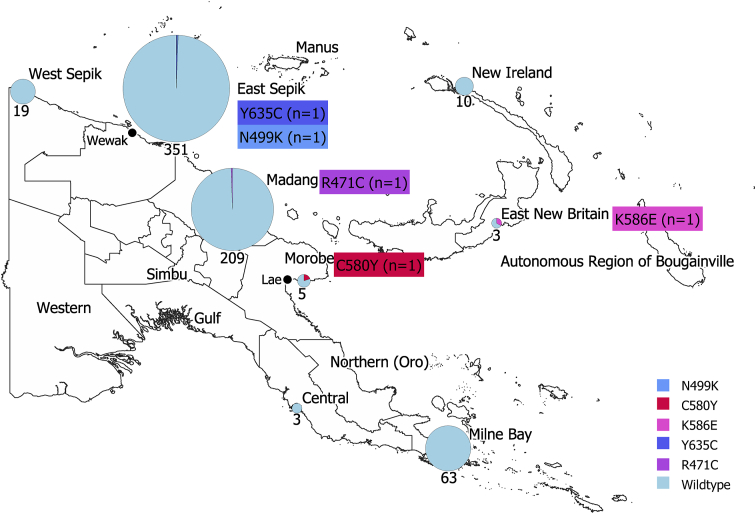
A total of 1152 P. falciparum positive samples were assessed from eight of the thirteen locations (Table 1). The five locations not assessed were included in the RDT sample set that overall performed poorly, due to low DNA quality. P. falciparum positive samples were screened by PCR to amplify an 850 (Talundzic et al., 2015) base pair fragment of Pfk13. From this, 706 (61%) samples that were successfully amplified were subjected to traditional Sanger sequencing. Sequencing returned 663 high quality sequences and when aligned to the PF3D7_1343700 reference revealed six NS Pfk13 SNPs resulting in variations in the protein product. These included C580Y and in a previously described mutant position N499D (Ouattara et al., 2015) we identified a novel allele: N499K. Three additional newly identified mutants were found: R471C, K586E and Y635C (Fig. 1). In addition, three synonymous substitutions were found at codons numbers 491, 536 and 628 (Table 2). All NS mutations were identified in single isolates. This was also the case for synonymous mutations, except for the substitution in codon 491, which was found in two isolates from Madang and Milne Bay (Table 2).
Fig. 1.
Map showing the prevalence of the Pfk13 mutations in samples that were successfully sequenced (n = 633). The size of the pie graph represents the number of samples successfully sequenced per locations. Five locations, Western, Northern, Simbu, Manus and Bougainville had RDT samples that performed poorly therefore were not assessed.
Table 2.
Pfk13 mutations identified by year and location. Mutations showing amino acid changes and proportion of mutations in the 663 samples that were successfully sequenced (n; number of mutants, N; samples successfully sequenced per site).
| Location | Year sample collected |
Nucleotide coordinate | Frequency (n/N) |
||
|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | |||
| Non-synonymous mutation (amino acid change) | |||||
| Madang | R471C | 1411_C/T | 0.48 (1/209) | ||
| East Sepik | N499K* | 1497_C/A | 0.28 (1/351) | ||
| Morobe | C580Y* | 1739_G/T | 20 (1/5) | ||
| East New Britain | K586E | 1756_G/A | 33.3 (1/3) | ||
| East Sepik | Y635C | 1904_A/T | 0.28 (1/351) | ||
| Synonymous mutations (nucleotide change) | |||||
| Milne Bay | † | 1473_T/C | 0.16 (1/63) | ||
| Madang | † | 1608_A/G | 0.28 (1/209) | ||
| Madang | † | † | 1473_T/C | 0.48 (1/209) | |
| East Sepik | † | 1884_T/C | 0.28 (1/351) | ||
*Previously described Pfk13 mutations.
†Indicating the synonymous mutation at that particular year.
4. Discussion
The decreasing sensitivity of P. falciparum to first-line malaria treatments is a continuing global health problem with the emergence and expansion of an artemisinin resistant C580Y population (KEL/PLA1) originating from Western Cambodia in 2008 (Amato et al., 2017, 2018; Dondorp et al., 2009; Noedl et al., 2008). The discovery of C580Y mutant parasites in PNG is a major concern to the national malaria control programme since AL is first line treatment (Pulford et al., 2013). By screening a large number of archived P. falciparum samples from epidemiological surveys and sentinel surveillance conducted in PNG between 2016 and 2018, we were able to examine the prevalence of Pfk13 mutations in eight endemic regions of the country. The results suggest that between 2016 and 2018 there were Pfk13 mutations in geographically distinct endemic regions of the country, including one isolate in Lae, Morobe Province found to be carrying the C580Y mutation.
Whilst all Pfk13 mutations were found to be present in only one isolate each and therefore considered rare, the C580Y mutation was detected in a population with a very small sample (n = 5). Due to the low quality of the DNA extracted from stored RDTs for Morobe, only a small number of samples were successfully genotyped. The prevalence of this mutation therefore may not be accurate, with further sampling at this site an urgent priority. Nevertheless, the presence at all of the C580Y mutation in isolates in Morobe in 2018 (this study) is concerning, with previous studies identifying the mutation several hundred kilometres away in Wewak, East Sepik Province in 2017 (Miotto et al., 2020). Firstly, due to its known phenotypic association with in vitro artemisinin resistance and secondly, its identification in a densely populated urban port town Lae (Morobe), that is a staging point for transport into other provinces, a hub for frequent human movement. Of note, Lae's locality is highly favourable for parasite importation either from the north coast region (Wewak) or West New Britain where one other C580Y mutant is believed to have originated (Prosser et al., 2018). In terms of cross-border (PNG-Indonesia) importation, there is no evidence to date of C580Y mutations in Indonesian parasites, suggesting this is unlikely. However, samples have so far only been surveyed in a single location (Timika) on the Indonesian side of New Guinea (Miotto et al., 2020).
Our discovery of a C580Y mutant in Morobe, but lack of C580Y mutants in other sites, including the area surrounding Maprik, only 50 km inland from Wewak where C580Y mutants have been detected (Miotto et al., 2020), suggests that ports may represent an important entry point into PNG for imported drug resistant parasites. It is also important to factor in other influences that may contribute to the emergence and potential spread of C580Y mutants in PNG. One such factor could be the influence of low malaria transmission on emergence of drug resistance. Malaria transmission in PNG had dropped significantly after the nationwide distribution of long-lasting insecticide treated mosquito nets (LLIN) and later implementation of ACT (Hetzel et al., 2015, 2017) but has resurged in recent years (PNGIMR, 2018). During low transmission periods, polyclonal infections are less frequent in PNG (Koepfli et al., 2017) and linkage disequilibrium between distant loci increases (Anderson et al., 2000; Kattenberg et al., 2019). Immunity also decreases, whereby inadequate immunity prevents the elimination of parasite strains that may have survived drug treatment and therefore increases their chance for selection (Ataide et al., 2017). This may provide ideal conditions for the emergence of Pfk13 C580Y mutant parasites in PNG. Whatever the case, it will be important to characterise any new C580Y mutants that are found through surveillance programs in terms of their genomic relationships with previously identified mutants, to establish whether these mutations are spreading or emerging independently in different locations.
Several other Pfk13 mutations were identified in the genetic surveillance conducted in this study. The N499K mutation is located at a documented polymorphic position found in Pfk13 in African parasites, where an alternative allele, namely N499D, has been observed (Fairhurst, 2015; Taylor et al., 2015), although there is no evidence to date of its association with artemisinin resistance. The remaining Pfk13 mutations, R471C, K586E and Y635C are novel mutations and polymorphic positions that have not been described elsewhere, to our knowledge. While all Pfk13 mutations observed in these 2016-18 samples were rare, identified in only one sample each, they are indicative of the selective pressure occurring within the local parasite population. Evaluation of any mutants for their association with phenotypic resistance will be important to establish whether they pose any risk to artemisinin effectiveness in PNG.
To date, there is no clinical evidence of resistance to ACTs in PNG, since ACTs remain efficacious in therapeutic efficacy studies (World Health Organisation, 2019). However, the occurrence of Pfk13 mutations in PNG's parasite population indicates underlying genetic changes, potentially as a result of ACT exposure. Continued drug exposure in malaria parasites has been known to induce genomic variations in the organism that enable drug tolerance and survival (Hamilton et al., 2019). However, not all Pfk13 mutations will be associated with delayed parasite clearance and resistance to artemisinin. For example, in Africa the most common allele A578S, which occurs in low frequency in the African parasite populations and also present in South-East Asian artemisinin resistant parasites, shows no association with delayed parasite clearance or artemisinin resistance in African parasites (Amaratunga et al., 2019; Kamau et al., 2015; MalariaGEN, 2016). While this may be the case for the rare novel mutations found in this study, C580Y is a well-validated and extremely common artemisinin resistance allele throughout South-East Asia (Ariey et al., 2014) and has now emerged independently in multiple locations (Mathieu et al., 2020; Miotto et al., 2020).
The findings from this study are critical for the control of malaria and demonstrate the need for ongoing genetic surveillance to determine the emergence and spread of drug resistant parasites and rigorous therapeutic efficacy studies (TES) to establish possible impacts on treatment efficacy. These studies are particularly urgent in and around regions where C580Y mutants circulate as well as among populations of frequent immigration, while parasite genotyping for drug resistance mutations will require a robust and highly sensitive methods that can be routinely conducted in PNG.
Funding
Collection of samples for this work was supported by the Global Fund to Fight AIDS, Tuberculosis and Malaria (MIS 2016/17), a WHO Tropical Disease Research Grant (WCCPRD4426109 2016/639607), a PNG-China-Australia Trilateral Research Co-operation funded by Australian Department of Foreign Affairs and Trade (DFAT), a National Institutes of Health International Centre of Excellence in Malaria Research for the South West Pacific, USA (DMID Protocol # 10–0035). The generation of molecular data was supported by STRIVE PNG, an Australian DFAT Centre for Health Security Stronger Health Systems Grant. DLG was supported by STRIVE PNG and the Australian Centre for Research Excellence in Malaria Elimination (ACREME) funded by the National Health and Medical Research Council (NHMRC) of Australia (APP1134989). LJR and IM were supported by NHMRC Research Fellowships (GNT1161627, GNT1155075). This work was also supported by the Victorian Operational Infrastructure Support Program.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank all study participants who took part in the various studies and their local leaders and authorities including Provincial Health Offices of East New Britain, East Sepik, Milne Bay, Morobe, Madang, Western Highlands, Simbu and Sandaun Provinces and the Evangelical Brotherhood Church Health Services. We acknowledge the support of the PNG National Department of Health and are grateful for the efforts of our field and laboratory staff at the PNGIMR.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2021.06.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Amaratunga C., Andrianaranjaka V.H., Ashley E., Bethell D., Björkman A., Bonnington C.A., Cooper R.A., Dhorda M., Dondorp A., Erhart A., Fairhurst R.M., Faiz A., Fanello C., Fukuda M.M., Guérin P., van Huijsduijnen R.H., Hien T.T., Hong N.V., Htut Y., Huang F., Humphreys G., Imwong M., Kennon K., Lim P., Lin K., Lon C., Mårtensson A., Mayxay M., Mokuolu O., Morris U., Ngasala B.E., Amambua-Ngwa A., Noedl H., Nosten F., Onyamboko M., Phyo A.P., Plowe C.V., Pukrittayakamee S., Randrianarivelojosia M., Rosenthal P.J., Saunders D.L., Sibley C.H., Smithuis F., Spring M.D., Sondo P., Sreng S., Starzengruber P., Stepniewska K., Suon S., Takala-Harrison S., Thriemer K., Thuy-Nhien N., Tun K.M., White N.J., Woodrow C., Group W.K.G.-P.S. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments—a WWARN individual patient data meta-analysis. BMC Med. 2019;17:1. doi: 10.1186/s12916-018-1207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato R., Lim P., Miotto O., Amaratunga C., Dek D., Pearson R.D., Almagro-Garcia J., Neal A.T., Sreng S., Suon S., Drury E., Jyothi D., Stalker J., Kwiatkowski D.P., Fairhurst R.M. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect. Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato R., Pearson R.D., Almagro-Garcia J., Amaratunga C., Lim P., Suon S., Sreng S., Drury E., Stalker J., Miotto O., Fairhurst R.M., Kwiatkowski D.P. Origins of the current outbreak of multidrug-resistant malaria in southeast Asia: a retrospective genetic study. Lancet Infect. Dis. 2018;18:337–345. doi: 10.1016/S1473-3099(18)30068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T.J., Haubold B., Williams J.T., Estrada-Franco J.G., Richardson L., Mollinedo R., Bockarie M., Mokili J., Mharakurwa S., French N., Whitworth J., Velez I.D., Brockman A.H., Nosten F., Ferreira M.U., Day K.P. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.-C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Ménard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.-C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., Sopha C., Chuor C.M., Nguon C., Sovannaroth S., Pukrittayakamee S., Jittamala P., Chotivanich K., Chutasmit K., Suchatsoonthorn C., Runcharoen R., Hien T.T., Thuy-Nhien N.T., Thanh N.V., Phu N.H., Htut Y., Han K.T., Aye K.H., Mokuolu O.A., Olaosebikan R.R., Folaranmi O.O., Mayxay M., Khanthavong M., Hongvanthong B., Newton P.N., Onyamboko M.A., Fanello C.I., Tshefu A.K., Mishra N., Valecha N., Phyo A.P., Nosten F., Yi P., Tripura R., Borrmann S., Bashraheil M., Peshu J., Faiz M.A., Ghose A., Hossain M.A., Samad R., Rahman M.R., Hasan M.M., Islam A., Miotto O., Amato R., MacInnis B., Stalker J., Kwiatkowski D.P., Bozdech Z., Jeeyapant A., Cheah P.Y., Sakulthaew T., Chalk J., Intharabut B., Silamut K., Lee S.J., Vihokhern B., Kunasol C., Imwong M., Tarning J., Taylor W.J., Yeung S., Woodrow C.J., Flegg J.A., Das D., Smith J., Venkatesan M., Plowe C.V., Stepniewska K., Guerin P.J., Dondorp A.M., Day N.P., White N.J., Tracking Resistance to Artemisinin, C. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide R., Ashley E.A., Powell R., Chan J.A., Malloy M.J., O'Flaherty K., Takashima E., Langer C., Tsuboi T., Dondorp A.M., Day N.P., Dhorda M., Fairhurst R.M., Lim P., Amaratunga C., Pukrittayakamee S., Hien T.T., Htut Y., Mayxay M., Faiz M.A., Beeson J.G., Nosten F., Simpson J.A., White N.J., Fowkes F.J. Host immunity to Plasmodium falciparum and the assessment of emerging artemisinin resistance in a multinational cohort. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3515–3520. doi: 10.1073/pnas.1615875114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey G.J., Ginny M., Uranoli M., Mueller I., Reeder J.C., Genton B., Cowman A.F. Molecular analysis of Plasmodium falciparum from drug treatment failure patients in Papua New Guinea. Am. J. Trop. Med. Hyg. 2004;70:251–255. [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst R.M. Understanding artemisinin-resistant malaria: what a difference a year makes. Curr. Opin. Infect. Dis. 2015;28:417–425. doi: 10.1097/QCO.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genton B., Baea K., Lorry K., Ginny M., Wines B., Alpers M.P. Parasitological and clinical efficacy of standard treatment regimens against Plasmodium falciparum, P. vivax and P. malariae in Papua New Guinea. Papua New Guinea Med. J. 2005;48:141–150. [PubMed] [Google Scholar]
- Hamilton W.L., Amato R., van der Pluijm R.W., Jacob C.G., Quang H.H., Thuy-Nhien N.T., Hien T.T., Hongvanthong B., Chindavongsa K., Mayxay M., Huy R., Leang R., Huch C., Dysoley L., Amaratunga C., Suon S., Fairhurst R.M., Tripura R., Peto T.J., Sovann Y., Jittamala P., Hanboonkunupakarn B., Pukrittayakamee S., Chau N.H., Imwong M., Dhorda M., Vongpromek R., Chan X.H.S., Maude R.J., Pearson R.D., Nguyen T., Rockett K., Drury E., Gonçalves S., White N.J., Day N.P., Kwiatkowski D.P., Dondorp A.M., Miotto O. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect. Dis. 2019;19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel M.W., Morris H., Tarongka N., Barnadas C., Pulford J., Makita L., Siba P.M., Mueller I. Prevalence of malaria across Papua New Guinea after initial roll-out of insecticide-treated mosquito nets. Trop. Med. Int. Health. 2015;20:1745–1755. doi: 10.1111/tmi.12616. [DOI] [PubMed] [Google Scholar]
- Hetzel M.W., Pulford J., Ura Y., Jamea-Maiasa S., Tandrapah A., Tarongka N., Lorry L., Robinson L.J., Lilley K., Makita L., Siba P.M., Mueller I. Insecticide-treated nets and malaria prevalence, Papua New Guinea, 2008-2014. Bull. World Health Organ. 2017;95:695–705B. doi: 10.2471/BLT.16.189902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Suwannasin K., Kunasol C., Sutawong K., Mayxay M., Rekol H., Smithuis F.M., Hlaing T.M., Tun K.M., van der Pluijm R.W., Tripura R., Miotto O., Menard D., Dhorda M., Day N.P.J., White N.J., Dondorp A.M. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect. Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau E., Campino S., Amenga-Etego L., Drury E., Ishengoma D., Johnson K., Mumba D., Kekre M., Yavo W., Mead D., Bouyou-Akotet M., Apinjoh T., Golassa L., Randrianarivelojosia M., Andagalu B., Maiga-Ascofare O., Amambua-Ngwa A., Tindana P., Ghansah A., MacInnis B., Kwiatkowski D., Djimde A.A. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J. Infect. Dis. 2015;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunajeewa H., Mueller I., Senn Michelle, Enmoore L., Irwin L., Servina G., O’live Oa, Griffin Suzanne, Kay Kotabe, Suano P., Taronka Nandao, Alice Ura, Lautu Dulcie, Page-Sharp Madhu, Wong Rina, Salman S., Peter Siba, Illet Kenneth F., Davis T.M.E. A combination of antimalarial combination therapy in Papua New Guinean children. N. Engl. J. Med. 2008;359:2245–2557. doi: 10.1056/NEJMoa0804915. [DOI] [PubMed] [Google Scholar]
- Kattenberg J.H., Razook Z., Keo R., Koepfli C., Jennison C., Lautu Gumal D., Fola A.A., Ome-Kaius M., Barnadas C., Siba P., Felger I., Kazura J., Mueller I., Robinson L.J., Barry A.E. Monitoring of Plasmodium falciparum and Plasmodium vivax using microsatellite markers indicates limited changes in population structure after substantial transmission decline in Papua New Guinea. bioRxiv. 2019 doi: 10.1111/mec.15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli C., Ome-Kaius M., Jally S., Malau E., Maripal S., Ginny J., Timinao L., Kattenberg J.H., Obadia T., White M., Rarau P., Senn N., Barry A.E., Kazura J.W., Mueller I., Robinson L.J. Sustained malaria control over an 8-year period in Papua New Guinea: the challenge of low-density asymptomatic Plasmodium infections. J. Infect. Dis. 2017;216:1434–1443. doi: 10.1093/infdis/jix507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang R., Taylor W.R.J., Bouth D.M., Song L., Tarning J., Char M.C., Kim S., Witkowski B., Duru V., Domergue A., Khim N., Ringwald P., Menard D. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob. Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MalariaGEN P.f.C.P. Genomic epidemiology of artemisinin resistant malaria. Elife. 2016;5 doi: 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfurt J., de Monbrison F., Brega S., Barbollat L., Muller I., Sie A., Goroti M., Reeder J.C., Beck H.P., Picot S., Genton B. Molecular markers of in vivo Plasmodium vivax resistance to amodiaquine plus sulfadoxine-pyrimethamine: mutations in pvdhfr and pvmdr1. J. Infect. Dis. 2008;198:409–417. doi: 10.1086/589882. [DOI] [PubMed] [Google Scholar]
- Marfurt J., Smith T.A., Hastings I.M., Muller I., Sie A., Oa O., Baisor M., Reeder J.C., Beck H.P., Genton B. Plasmodium falciparum resistance to anti-malarial drugs in Papua New Guinea: evaluation of a community-based approach for the molecular monitoring of resistance. Malar. J. 2010;9:8. doi: 10.1186/1475-2875-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu L.C., Cox H., Early A.M., Mok S., Lazrek Y., Paquet J.-C., Ade M.-P., Lucchi N.W., Grant Q., Udhayakumar V., Alexandre J.S., Demar M., Ringwald P., Neafsey D.E., Fidock D.A., Musset L. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. eLife. 2020;9 doi: 10.7554/eLife.51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D., Khim N., Beghain J., Adegnika A.A., Shafiul-Alam M., Amodu O., Rahim-Awab G., Barnadas C., Berry A., Boum Y., Bustos M.D., Cao J., Chen J.H., Collet L., Cui L., Thakur G.D., Dieye A., Djalle D., Dorkenoo M.A., Eboumbou-Moukoko C.E., Espino F.E., Fandeur T., Ferreira-da-Cruz M.F., Fola A.A., Fuehrer H.P., Hassan A.M., Herrera S., Hongvanthong B., Houze S., Ibrahim M.L., Jahirul-Karim M., Jiang L., Kano S., Ali-Khan W., Khanthavong M., Kremsner P.G., Lacerda M., Leang R., Leelawong M., Li M., Lin K., Mazarati J.B., Menard S., Morlais I., Muhindo-Mavoko H., Musset L., Na-Bangchang K., Nambozi M., Niare K., Noedl H., Ouedraogo J.B., Pillai D.R., Pradines B., Quang-Phuc B., Ramharter M., Randrianarivelojosia M., Sattabongkot J., Sheikh-Omar A., Silue K.D., Sirima S.B., Sutherland C., Syafruddin D., Tahar R., Tang L.H., Toure O.A., Tshibangu-wa-Tshibangu P., Vigan-Womas I., Warsame M., Wini L., Zakeri S., Kim S., Eam R., Berne L., Khean C., Chy S., Ken M., Loch K., Canier L., Duru V., Legrand E., Barale J.C., Stokes B., Straimer J., Witkowski B., Fidock D.A., Rogier C., Ringwald P., Ariey F., Mercereau-Puijalon O., Consortium K. A worldwide Map of Plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Almagro-Garcia J., Manske M., MacInnis B., Campino S., Rockett K.A., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Duong S., Nguon C., Chuor C.M., Saunders D., Se Y., Lon C., Fukuda M.M., Amenga-Etego L., Hodgson A.V.O., Asoala V., Imwong M., Takala-Harrison S., Nosten F., Su X.-z., Ringwald P., Ariey F., Dolecek C., Hien T.T., Boni M.F., Thai C.Q., Amambua-Ngwa A., Conway D.J., Djimdé A.A., Doumbo O.K., Zongo I., Ouedraogo J.-B., Alcock D., Drury E., Auburn S., Koch O., Sanders M., Hubbart C., Maslen G., Ruano-Rubio V., Jyothi D., Miles A., O'Brien J., Gamble C., Oyola S.O., Rayner J.C., Newbold C.I., Berriman M., Spencer C.C.A., McVean G., Day N.P., White N.J., Bethell D., Dondorp A.M., Plowe C.V., Fairhurst R.M., Kwiatkowski D.P. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S.O., Dhorda M., Imwong M., Woodrow C., Manske M., Stalker J., Drury E., Campino S., Amenga-Etego L., Thanh T.N., Tran H.T., Ringwald P., Bethell D., Nosten F., Phyo A.P., Pukrittayakamee S., Chotivanich K., Chuor C.M., Nguon C., Suon S., Sreng S., Newton P.N., Mayxay M., Khanthavong M., Hongvanthong B., Htut Y., Han K.T., Kyaw M.P., Faiz M.A., Fanello C.I., Onyamboko M., Mokuolu O.A., Jacob C.G., Takala-Harrison S., Plowe C.V., Day N.P., Dondorp A.M., Spencer C.C., McVean G., Fairhurst R.M., White N.J., Kwiatkowski D.P. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Sekihara M., Tachibana S.-I., Yamauchi M., Pearson R.D., Amato R., Gonçalves S., Mehra S., Noviyanti R., Marfurt J., Auburn S., Price R.N., Mueller I., Ikeda M., Mori T., Hirai M., Tavul L., Hetzel M.W., Laman M., Barry A.E., Ringwald P., Ohashi J., Hombhanje F., Kwiatkowski D.P., Mita T. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T., Tanabe K., Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol. Int. 2009;58:201–209. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Ouattara A., Kone A., Adams M., Fofana B., Maiga A.W., Hampton S., Coulibaly D., Thera M.A., Diallo N., Dara A., Sagara I., Gil J.P., Bjorkman A., Takala-Harrison S., Doumbo O.K., Plowe C.V., Djimde A.A. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am. J. Trop. Med. Hyg. 2015;92:1202–1206. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papua New Guinea National Department of Health . Dept. of Health; 2009. National Malaria Treatment Protocol. [Google Scholar]
- Plowe C.V., Djimde A., Bouare M., Doumbo O., Wellems T.E. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: Polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- PNGIMR . Papua New Guinea Institute of Medical Research; 2018. Papua New Guinea Malaria Indicator Survey 2016-2017: Malaria Prevention, Infection and Treatement. [Google Scholar]
- Prosser C., Meyer W., Ellis J., Lee R. Resistance screening and trend analysis of imported falciparum malaria in NSW, Australia (2010 to 2016) PloS One. 2018;13 doi: 10.1371/journal.pone.0197369. e0197369-e0197369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulford J., Kurumop S.F., Ura Y., Siba P.M., Mueller I., Hetzel M.W. Malaria case management in Papua New Guinea following the introduction of a revised treatment protocol. Malar. J. 2013;12 doi: 10.1186/1475-2875-12-433. 433-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosanas-Urgell A., Mueller D., Betuela I., Barnadas C., Iga J., Zimmerman P.A., del Portillo H.A., Siba P., Mueller I., Felger I. Comparison of diagnostic methods for the detection and quantification of the four sympatric Plasmodium species in field samples from Papua New Guinea. Malar. J. 2010;9:361. doi: 10.1186/1475-2875-9-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J., Gnadig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S., Gregory P.D., Urnov F.D., Mercereau-Puijalon O., Benoit-Vical F., Fairhurst R.M., Menard D., Fidock D.A. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Jacob C.G., Arze C., Cummings M.P., Silva J.C., Dondorp A.M., Fukuda M.M., Hien T.T., Mayxay M., Noedl H., Nosten F., Kyaw M.P., Nhien N.T., Imwong M., Bethell D., Se Y., Lon C., Tyner S.D., Saunders D.L., Ariey F., Mercereau-Puijalon O., Menard D., Newton P.N., Khanthavong M., Hongvanthong B., Starzengruber P., Fuehrer H.P., Swoboda P., Khan W.A., Phyo A.P., Nyunt M.M., Nyunt M.H., Brown T.S., Adams M., Pepin C.S., Bailey J., Tan J.C., Ferdig M.T., Clark T.G., Miotto O., MacInnis B., Kwiatkowski D.P., White N.J., Ringwald P., Plowe C.V. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talundzic E., Chenet S.M., Goldman I.F., Patel D.S., Nelson J.A., Plucinski M.M., Barnwell J.W., Udhayakumar V. Genetic analysis and species specific amplification of the artemisinin resistance-associated kelch propeller domain in P. Falciparum and P. Vivax. PloS One. 2015;10 doi: 10.1371/journal.pone.0136099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavul L., Hetzel M.W., Teliki A., Walsh D., Kiniboro B., Rare L., Pulford J., Siba P.M., Karl S., Makita L., Robinson L., Kattenberg J.H., Laman M., Oswyn G., Mueller I. Efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for the treatment of uncomplicated malaria in Papua New Guinea. Malar. J. 2018;17:350. doi: 10.1186/s12936-018-2494-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.M., Parobek C.M., DeConti D.K., Kayentao K., Coulibaly S.O., Greenwood B.M., Tagbor H., Williams J., Bojang K., Njie F., Desai M., Kariuki S., Gutman J., Mathanga D.P., Martensson A., Ngasala B., Conrad M.D., Rosenthal P.J., Tshefu A.K., Moormann A.M., Vulule J.M., Doumbo O.K., Ter Kuile F.O., Meshnick S.R., Bailey J.A., Juliano J.J. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J. Infect. Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwimana A., Legrand E., Stokes B.H., Ndikumana J.-L.M., Warsame M., Umulisa N., Ngamije D., Munyaneza T., Mazarati J.-B., Munguti K., Campagne P., Criscuolo A., Ariey F., Murindahabi M., Ringwald P., Fidock D.A., Mbituyumuremyi A., Menard D. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020;26:1602–1608. doi: 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Khim N., Chim P., Kim S., Ke S., Kloeung N., Chy S., Duong S., Leang R., Ringwald P., Dondorp A.M., Tripura R., Benoit-Vical F., Berry A., Gorgette O., Ariey F., Barale J.C., Mercereau-Puijalon O., Menard D. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob. Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation . third ed. WHO; 2015. Guidelines for the Treatment of Malaria; p. 315. [Google Scholar]
- World Health Organisation . vol. 1. 2019. World Malaria Report; p. 232. [Google Scholar]
- World Health Organisation . 2020. Report on Antimalarial Drug Efficacy, Resistance and Response; p. 64. [Google Scholar]
- World Health Organisation Regional Office for the Western Pacific . 2017. Regional Action Frameworkd for Malaria Control and Elimination in Western Pacific: 2016-2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.