Abstract
Background
Lung cancer mainly includes non‐small cell lung cancer (NSCLC). Lung adenocarcinoma (LUAD) is the main subtype of NSCLC. Long non‐coding RNAs (LncRNAs) had been found to exert numerous functions on the progressions of cancers. MicroRNAs often exist as the target of LncRNAs to regulate a series of signaling pathways in human. We explored the effects and molecular mechanism of LncRNA SGMS1‐AS1 on the procedures of LUAD cells.
Methods
The ENCORI and GEPIA databases were used to analyze the differences in SGMS1, miR‐106a‐5p, and MYLIP between LUAD and normal tissue. Their expression levels were examined by RT‐PCR. CCK8, colony formation, migration, and invasion assay were conducted in LUAD cells which had silenced SGMS1‐AS1. To verify the relationship between SGMS1‐AS1, miR‐106a‐5p, and MYLIP, we overexpressed miR‐106a‐5p inhibitor or MYLIP in LUAD cells after decreasing SGMS1‐AS1 and repeated the above assays.
Results
SGMS1‐AS1 was downregulated in LUAD tissue as well as cells, which was related to good prognosis of patients with lung adenocarcinoma. Additionally, knockdown of SGMS1‐AS1 promoted proliferation, migration, invasion, and epithelial mesenchymal transition (EMT) progression of LUAD cells, which meant that SGMS1‐AS1 inhibited the progression of LUAD cells. Furthermore, miR‐106a‐5p was the direct target of SGMS1‐AS1 and transfecting miR‐106a‐5p inhibitor could reversed the impact induced by knockdown of SGMS1‐AS1. Subsequently, we found that MYLIP was the target of miR‐106a‐5p, which was negatively correlated with miR‐106a‐5p, but had high positive correlation with SGMS1‐AS1. Consistently, overexpression MYLIP partly eliminated the effects on A549 cells induced by silencing of SGMS1‐AS1.
Conclusion
LncRNA SGMS1‐AS1 inhibits the proliferation, invasion, migration and EMT progression of LUAD cells via targeting miR‐106a‐5p/MYLIP axis.
Keywords: LncRNA‐SGMS1‐AS1, LUAD, miR‐106A‐5p, MYLIP
SGMS1‐AS1 is downregulated in LUAD and is correlated to good prognosis of patients. (a) The GEPIA and ENCORI databases illustrate that the expression of SGMS1‐AS1 is abnormal in multiple cancers and lower in LUAD tissue compared to normal tissue. (b) The overall survival rate of LUAD patients with a low level of SGMS1‐AS1 is poor according to the ENCORI website. (c) Quantitative reverse transcription PCR (qRT‐PCR) was conducted to compare the expression level of SGMS1‐AS1 between LUAD and adjacent tissues of 18 patients. (d) The result of qRT‐PCR revealed that the mRNA level of SGMS1‐AS1 in LUAD cell lines (A549, H1975, and PC9) was consistently lower than that in normal cell lines human bronchial epithelioid cells (HBE).

INTRODUCTION
Lung cancer is the most deadly cancer in the world, 1 and kills more people than breast and prostate cancer combined. 2 Lung cancer consists of small cell lung cancer (SCLC) and non‐small cell lung cancer (NSCLC), with NSCLC accounting for approximately 85% of the total. 3 Lung adenocarcinoma (LUAD), a subtype of NSCLC, occurs in the highest proportion in lung cancer (about 40%). 4 Over recent decades, emerging treatment technologies have been employed to prolonged the survival time of patients with LUAD. Tyrosine kinase inhibitors (TKIs), 5 one of the molecular targeted therapy medicines for LUAD sufferers, had reached to its peak of improving life span of patients. 6 , 7 Thus there is an urgent need to study the pathogenesis of LUAD and find new, accurate, and effective therapeutic targets.
Genome sequencing projects have found that non‐coding RNA accounts for approximately 98% of transcriptome in humans, of which long non‐coding RNA (LncRNA) is a crucial part. 8 LncRNA, longer than 200 bases in non‐coding RNA, is expressed in various cells as well as species. 9 , 10 Scientists have verified that LncRNA exerts a vital role in diverse biological procedures as one type of transcription factor. 11 , 12 , 13 In addition, increasing LncRNAs were confirmed to have an effect on the progression of cancers as tumor suppressors or oncogenes. 14 , 15 SGMS1‐AS1 is a type of newly discovered LncRNA that transcripts ubiquitously in testis, thyroid, and 25 other tissues. SGMS1‐AS1 is expressed aberrantly in multiple cancers according to the GEPIA database, but its function as well as molecular mechanism on tumors are unclear. Interestingly, we found SGMS1‐AS1 decreased dramatically in LUAD through bioinformatic analysis, and the survival time of patients with high expression of SGMS1‐AS1 was longer than that of patients with low expression. It is therefore reasonable to assume that SGMS1‐AS1 could inhibit the development of LUAD.
MicroRNAs (miRNAs), endogenous 17–25 nt small RNAs, can regulate post‐transcriptional suppression or post‐translational repression by binding to the 3′‐untranslated regions (UTR) of target genes. 16 The ENCORI database predicted that miR‐106a‐5p is one of the target genes of lncRNA SGMS1‐AS1. It has a sensitized impact on the development as well as metastasis of NSCLC according to previous reports. 17 Additionally, upregulation of miR‐106a‐5p could give rise to poor prognosis of NSCLC and its expression is decreased significantly after chemotherapy. 18
Myosin regulatory light‐chain interacting protein (MYLIP), one of the ubiquitin ligases, has a crucial effect on various cancers such as prostate cancer cells, breast cancer, and cervical cancer. 19 , 20 , 21 In 2020, MYLIP was found to be correlated with poor prognosis when it downregulated in NSCLC. Nevertheless, the mechanism of the above functions has rarely been studied. 22 In addition, bio‐analysis showed that the mRNA level of MYLIP in LUAD was dramatically decreased. We therefore hypothesized that MYLIP could inhibit the process of LUAD.
The main objective of our research was to elucidate the effect of SGMS1‐AS1 on the proliferation, invasion, migration, and epithelial mesenchymal transition (EMT) procedure of LUAD cells and its molecular mechanism via the miR‐106a‐5p/MYLIP axis.
METHODS
Ethics statement
The research was permitted by the Institutional Review Board of the Cancer Hospital of the China Medical University and abided by the principle of the Declaration of Helsinki. All patients mentioned in this text volunteered to participate the study and signed the consent form.
Patients and cell culture
All LUAD tissues and normal tissues were obtained from 18 patients who had been diagnosed with lung adenocarcinoma in Qi Lu hospital. In addition, all the tissue samples were acquired from surgical excision and conserved at −80°C in a freezer. Human bronchial epithelial cell (HBE) and all LUAD cell lines (A549, PC9, H1975) were bought from the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI‐1640 medium supplemented with 10% fetal bovine serum, 1% streptomycin, and 1% penicillin at 37°C with 95% air and 5% CO2.
Dual‐luciferase reporter assay
The MYLIP assumed binding sites of miR‐106a‐5p were predicted and 3′‐UTR fragments carrying wildtype (WT) or mutant type (MUT) presumptive nucleotide sequences were cloned into pGL3 vector (Promega). The LUAD cells were seeded in 24‐well plates and transfected with MYLIP 3′‐UTR‐WT/MUT and miR‐106a‐5p inhibitor together. After 24 h, a dual specific luciferase assay kit (Promega) was used for the reporter assays.
RT‐PCR
All cells were lysed with TRIzol reagent after washing with phosphate buffer saline (PBS), then RNA was isolated according the standard protocol offered by the reagent. The extracted total RNA was transcribed to cDNA following the manufacturer's instructions (Fermentas) for detection. Quantitative PCR was used to measure the mRNA level and the values obtained were normalized to the glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) load. The primers used are presented in Table 1.
TABLE 1.
Primer sequences
| Gene | RT‐PCT primer |
|---|---|
| MiR‐106a‐5p |
Forward: 5′‐GATGCTCAAAAAGTGCTTACAGTGCA‐3′ Reverse: 5′‐TATGGTTGTTCTGCTCTCTGTCTC‐3′ |
| SGMS1‐AS1 |
Forward: 5′‐GGCTGTGTTTCTGCATACTCC‐3′ Reverse: 5′‐ACGACCCAGCATACATACTGA‐3′ |
| MYLIP |
Forward: 5′‐ATAACAGAGACGCACGCATT‐3′ Reverse: 5′‐TCCTGGCATGGTCATACACCT‐3′ |
Cell viability assay
LUAD cells were seeded in 96‐well plates at a density of 1 × 104 per well and cultured for 24 h. Cell viability was assayed using a Cell Counting Kit‐8 (CCK‐8, Dojindo Laboratories) following the manufacturer's protocol. The concentration required to prohibit cell growth by 50% (IC50) was calculated from viability curves as previously reported.
Colony formation assay
LUAD cells were counted and seeded in 6‐well plates for about 14 days. The medium was changed or replenished every 3–4 days. The cells were washed with PBS and fixed with 4% paraformaldehyde for 15–30 min. After fixing, the colonies were dyed with 0.1% crystal violet for 10 min and then washed with PBS or liquid water several times. Finally, the cells were photographed and the amounts of colony were counted using image J software.
Immunoblotting analysis
Cells were transfected with the indicated vectors and harvested after 24 h. SDS (1%) was used to lysate cells and extract total protein, and the concentration of protein was measured by Bradford Protein Assay (Thermo). Then 20 μg of protein sample was loaded into SDS‐PHAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was blanked by tris‐hcl buffer solusion plus tween containing 3–5% milk. The PDFV membrane was incubated with specific primary antibodies GAPDH (Rabbit mAb #2118, Cell Signaling Technology), E‐cadherin (Rabbit mAb #3195, Cell Signaling Technology), N‐cadherin (20874‐1‐AP, Proteintech), and Vimentin (ab92547, ABcam) overnight at 4°C, followed by incubation with secondary antibodies at room temperature for 1–2 h. Finally, the proteins were visualized with an Immobilon Western Chemiluminescent HRP Substrate kit (Merck Millipore).
Trans well migration and invasion assay
A total of 5 × 104 LUAD cells was seeded into the upper chamber (Corning) with serum‐free medium, then 600 μl of medium with 10% fetal bovine serum was put into the lower chamber. The cells were fixed and stained with 4% paraformaldehyde and 0.1% crystal violet (Beyotime) after incubating for 24 h. The cells which were still in the upper chamber were also removed. Finally, the number of cells that migrated to the lower chamber was recorded and counted with a microscope (Nikon) as the migration result. For the invasion assay the same protocol was employed apart from using Matrigel to coat the upper chamber membrane and seeding LUAD cells into camber in addition to incubating cells for 48–72 h.
Statistical analysis
The data are presented as means ± SD. The significant differences between compared groups were determined with Student's t‐test or one‐way ANOVA test. The differences were considered significant at *p < 0.05 and **p < 0.01.
RESULTS
LncRNA SGMS1‐AS1 expression is downregulated in LUAD tissues and is correlated with good patient prognosis
The expression data of LncRNA SGMS1‐AS1 in cancer tissues were obtained from the GEPIA and ENCORI databases, and showed that the expression level of SGMS1‐AS1 was abnormal in various cancers and dramatically downregulated in LUAD tissues compared to normal tissues (Figure 1(a)). Accordingly, survival analysis demonstrated that a higher SGMS1‐AS1 level contributed to better patient prognosis (Figure 1(b)). To confirm this data, we performed RT‐PCR to estimate mRNA load in LUAD as well as adjacent samples 18 cases of patients who were diagnosed with LUAD (Figure 1(c)). Moreover, the mRNA level of SGMS1‐AS1 measured was consistently lower in LUAD cell lines (A549, H1975, PC9) compared to the normal cell line HBE (p < 0.01) (Figure 1(d)). The SGMS1‐AS1 expressions in A549 and PC9 were similar so A549 and PC9 were chosen to perform subsequent experiments. All the data show that LncRNA SGMS1‐AS1 may be closely related to the progression of LUAD.
FIGURE 1.
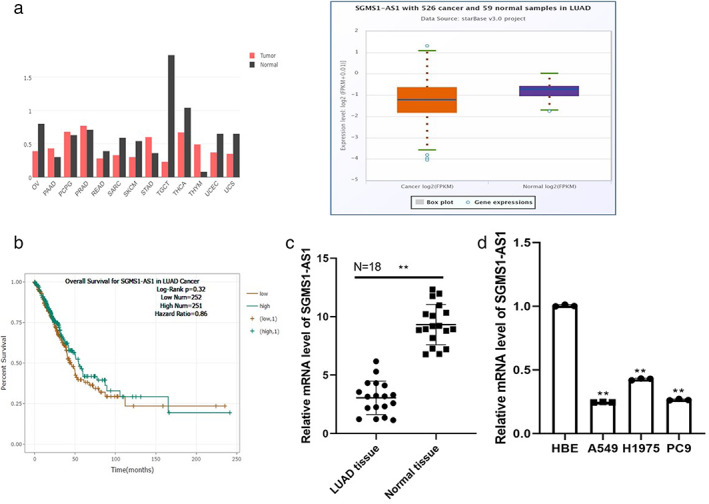
SGMS1‐AS1 is downregulated in LUAD and correlated to good prognosis for patients. (a) The GEPIA and ENCORI databases illustrate the expression of SGMS1‐AS1 is abnormal in multiple cancers and lower in LUAD tissue compared to normal tissue. (b) The overall survival rate of LUAD patients with a low level of SGMS1‐AS1 is poor according to the ENCORI website. (c) Quantitative reverse transcription PCR (qRT‐PCR) was conducted to compare the expression level of SGMS1‐AS1 between LUAD and para‐carcinoma tissues of 18 patients. (d) The qRT‐PCR revealed that the mRNA level of SGMS1‐AS1 in LUAD cell lines (A549, H1975, and PC9) was consistently lower than that in normal cell lines (HBE)
SGMS1‐AS1 inhibits LUAD cells proliferating, migrating, and invading, and the process of EMT
To verify the function of SGMS1‐AS1 in the procedure of LUAD cell lines, SGMS1‐AS1 siRNA was transfected into A549 and PC9 cells to reduce the SGMS1‐AS1 level. The mRNA load was obviously depressed in the two siRNA samples compared with the control group (p < 0.01) (Figure 2(a)). Next, CCK8 assay and colony formation assay were employed to verify the effect of SGMS‐AS1 on A549 and PC9 cells. We found that the ability of cells to proliferate was dramatically increased after silencing SGMS1‐AS1 by siRNA (p < 0.05; Figure 2(b,c)). Consistently, transwell migration and invasion assay confirmed the inhibiting impact of SGMS1‐AS1 on A549 and PC9, namely, knockdown of SGM1S1‐AS1 could forcefully promote cells to migrate and invade (p < 0.01; Figure 2(d,e)). Western blot was used to examine the protein level involved in the EMT of LUAD cells. The results showed that in A549 cells SGMS1‐AS1 inhibition clearly reduced the protein level of E‐cadherin, but increased the expression of N‐cadherin, which confirms that SGMS1‐AS1 inhibits the EMT of LUAD cells (Figure 2(f)).
FIGURE 2.
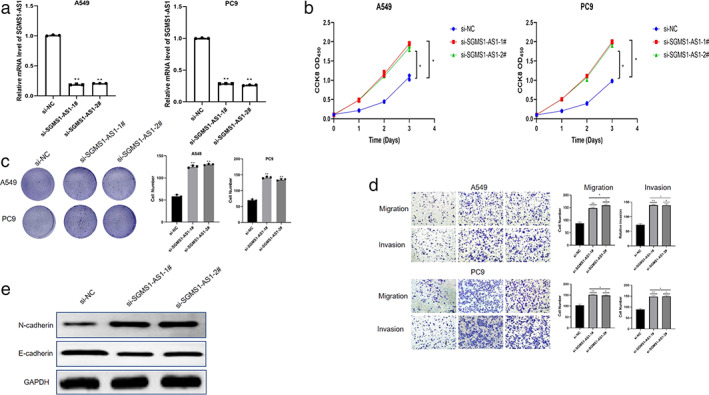
SGMS1‐AS1 inhibits the proliferation, migration, invasion, and EMT progression of LUAD cells. (a) A549 and PC9 were transfected with two siRNAs and qRT‐PCR was applied to examine the mRNA level of SGMS1‐AS1. (b, c) The ability of LUAD cells was determined by CCK8 assay and colony formation assay respectively. (d, e) Treanswell assays were conducted to measure the effect of knockdown of SGMS1‐AS1 on A549 and PC9. (f) The relative protein levels were examined by western blotting analysis
MiR‐106a‐5p is the direct target of SGMS1‐AS1 and is negatively correlated with SGMS1‐AS1
We consulted a reliable website to require the downstream of SGMS1‐AS1 (http://starbase.sysu.edu.cn/panCancer.php) to elucidate the function mechanism of SGMS1‐AS1 on LUAD. MiR‐106a‐5p was stated to be the direct target of SGMS1‐AS1 by the ENCORI database (Figure 3(a)). This database also showed that the expression of SGMS1‐AS1 was negatively related with that of miR‐106a‐5p (Figure 3(b)). To verify the relationship between the two genes, we employed the luciferase reporter assay and the data illustrated that heterotopic expression of miR‐106A‐5p had no obvious effect on the luciferase activity of mutant SGMS1‐AS1 but significantly depressed that of wild‐type SGMS1‐AS1 (p < 0.01; Figure 3(c)). Subsequently, the mRNA level of miR‐106a‐5p increased dramatically in A549 and PC9 after transfecting with siSGMS1‐AS1. In addition, silencing miR‐106a‐5p increased the expression of SGMSI‐AS1 (p < 0.01; Figure 3(d,e)). The expression load of miR‐106a‐5p was increased in LUAD cells (A549, H1975, PC9) compared to normal cells (HBE) (p < 0.01; Figure 3(f)). All the findings showed that SGMS1‐AS1 is likely to inhibit the progression of LUAD cells by regulating the miR‐106a‐5p.
FIGURE 3.
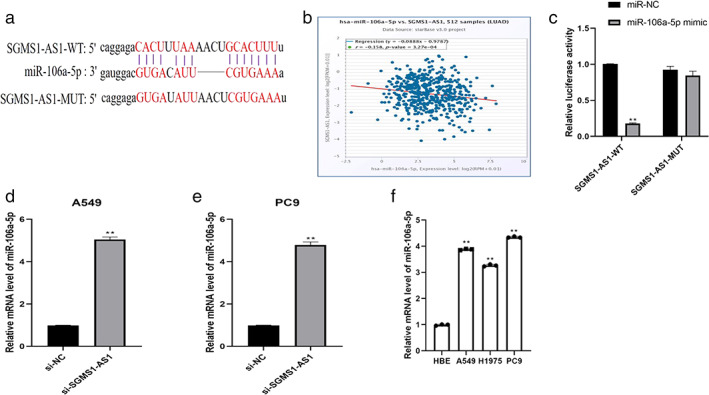
MiR‐106a‐5p is the direct target of SGMS1‐AS1 and is negatively correlated with SGMS1‐AS1. (a) The ENCORI database was used to predict that SGMS1‐AS1 could bind to miR‐106a‐5p. (b) The expression of SGMS1‐AS1 was negatively related with miR‐106a‐5p by the ENCORI database. (c) MiR‐106‐5p mimics decreased the expression level of SGMS1‐AS1 3′‐UTR‐WT luciferase activity significantly whereas it had no obvious effect on 3′‐UTR‐MUT. (d, e) qRT‐PCR experiments verified the relationship between SGMS1‐AS1 and miR‐106a‐5p. (f) The mRNA level of miR‐106a‐5p was upregulated dramatically in LUAD cells compared with normal cells
Inhibiting miR‐106a‐5p can counteract the regulatory effect of SGMS1‐AS1 on LUAD cells
To confirmed that SGMS1‐AS1 exerted an inhibitory impact on A549, we transfected miR‐106a‐5p inhibitor into A549 after knockdown of SGMS1‐AS1. The results demonstrated that decreasing miR‐106a‐5p clearly depressed the proliferative ability of cells induced by siSGMS1‐AS1 (p < 0.05; Figure 4(a,b)). Consistently, miR‐106a‐5p partially eliminated a series of effects of SGMS1 on A549 cells (p < 0.05; Figure 4(c,d)). Furthermore, downregulation of miR‐106a‐5p changed the siSGMS1‐AS1‐caused regulatory proteins related to the EMT procedure (Figure 4(e)). In conclusion, miR‐106a‐5p changed the influence of SGMS1‐AS1 on LUAD cells.
FIGURE 4.
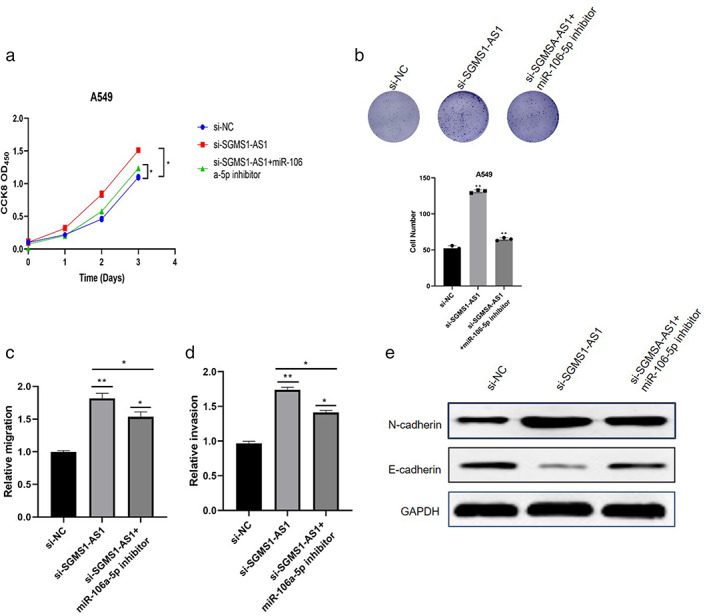
Inhibiting miR‐106a‐5p can counteract the regulatory effect of SGMS1‐AS1 on LUAD cells. (a, b) CCK8 assay and colony formation assay were conducted to measure the proliferative ability of A549 after transfecting the indicted vector. (c, d) Transmembrane cells were counted to compare the migration and invasion ability. (e) The protein levels related to EMT were detected after treatment with miR‐106a‐5p inhibitor in the indicated cells
MYLIP is the target of miR‐106a‐5p and is regulated by SGMS1‐AS1
We found that MYLIP is the direct target of miR‐106a‐5p by browsing the ENCORI website (Figure 5(a)). Luciferase reporter assay was conducted to verify that miR‐106a‐5p mimic notably downregulated the mRNA level of wild‐type MYLIP rather than the mutant one (p < 0.01; Figure 5(b)). Consistently, decreasing miR‐106a‐5p upregulated the expression of MYLIP in cells (p < 0.01; Figure 5(c)). Subsequently, we discovered a positive relationship between MYLIP and SGMS1‐AS1 from the ENCORI database (Figure 5(d)). As presented in Figure 5(e), the mRNA level of MYLIP was depressed significantly after silencing SGMS1‐AS1 in A549 (p < 0.01; Figure 5(e)). Interestingly, MYLIP was found to be less expressed in LUAD than in normal tissue, and low expression of MYLIP was associated with poor prognosis of patients diagnosed with LUAD according to GEPIA database (Figure 5(f,g)). From all the results, we concluded that SGMS1‐AS1 could regulate MYLIP by miR‐106‐5p.
FIGURE 5.
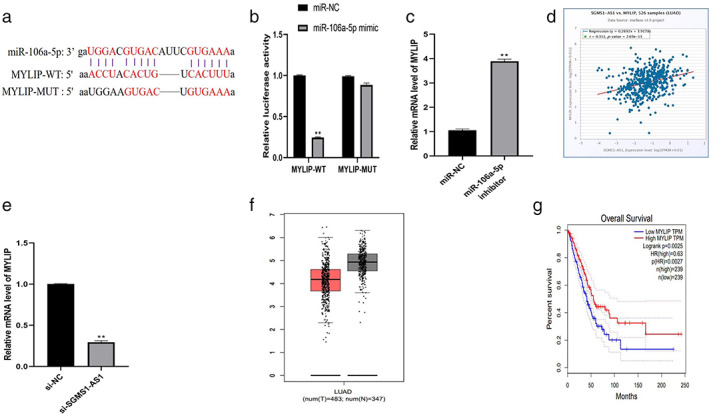
MYLIP is the target of miR‐106a‐5p and is regulated by SGMS1‐AS1. (a) The ENCORI website predicted that MYLIP is the direct target of miR‐106a‐5p. (b) Luciferase reporter assay showed that miR‐106a‐5p mimic suppressed the level of MYLIP‐WT ability but not MYLIP‐MUT ability. (c) Inhibiting miR‐106a‐5p depressed the expression level of MYLIP. (d) The ENCORI database demonstrated that MYLIP is positively correlated with SGMS1‐AS1. (e) Decreasing SGMS1‐AS1 could downregulate the mRNA level of MYLIP in A549. (f, g) The GEPIA database showed that MYLIP expression was lower in LUAD tissue than in normal tissue and a high level of MYLIP had an obvious relationship with poor prognosis of patients
SGMS1‐AS1 regulates a series of progressions of LUAD cells via the miR‐106a‐5p/MYLIP axis
We overexpressed MYLIP into cells which silenced SGMS1‐AS1 to verify the function of the SGMSA‐AS1/miR‐106a‐5p/MYLIP axis. The results illustrate that the proliferation, migration, and invasion ability was restored by clearly overexpressing MYLIP (p < 0.05; Figure 6(a–c)). Furthermore, ectopic expression of MYLIP upregulated the level of E‐cadherin but downregulated that of N‐cadherin, which was changed by siSGMS1‐AS before (Figure 6(d)). In summary, SGMS1‐AS1 inhibited LUAD progression by targeting the miR‐106a‐5p/MYLIP axis.
FIGURE 6.
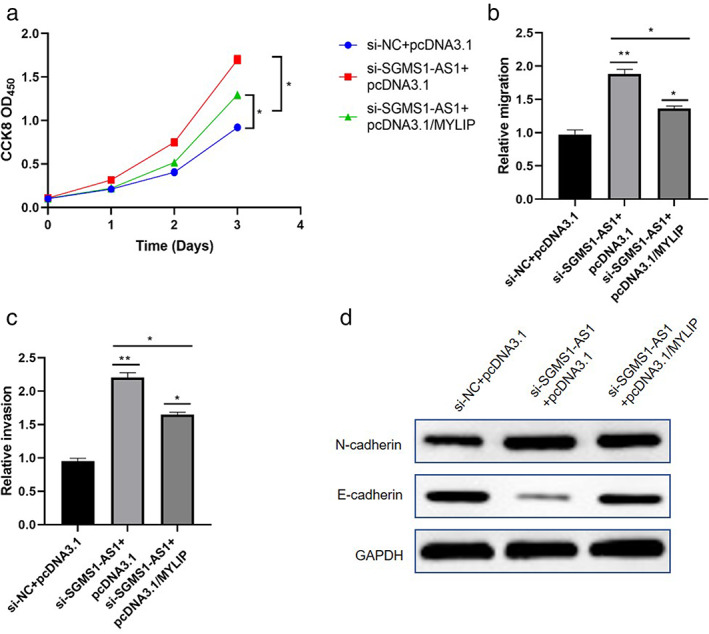
SGMS1‐AS1 regulated the progression of LUAD cells via the miR‐106a‐5p/MYLIP axis. (a–c) Overexpressed MYLIP could reverse the effect of SGMS1‐AS1 on migration and invasion of LUAD cells. (d) Overexpressed MYLIP could restore the level of proteins related to EMT procedure was affected by knockdown of SGMS1‐AS1
DISCUSSION
Lung cancer is the leading cause of cancer‐related deaths worldwide, resulting in about 142 607 deaths in the United States in 2019. The overwhelming majority of patients with lung cancer suffer from NSCLC, the most common type of carcinoma of the lung. 23 NSCLC contains a variety of cancer subtypes, such as LUAD, lung squamous carcinoma (LUSC), and large cell cancers. Approximately 30% of patients have LUSC, most of patients are diagnosed with LUAD, which stems from Clara cells or type II pneumocytes. Even though emerging therapies have been employed, the overall survival rate has not improved in recent decades, which emphasizes the significance of new therapeutic treatments for LUAD. 24 , 25
LncRNAs (long non‐coding RNAs) represent low‐grade coding ability but are defined as RNA transcripts. In‐depth research has indicated that LncRNAs are used in various biological mechanisms, such as controlling cell growth, apoptosis, signal transduction, and other pathologic functions. Recent studies revealed that ectopic expression of LncRNA had a considerable role in both the progression and metastasis of cancers. 26 We discovered that SGMS1‐AS1 was aberrantly expressed in LUAD tissues and was correlated with prognosis of patients. However, its impacts as well as functional mechanisms for LUAD have not yet been clarified. We explored the effects of SGMS1‐AS1 on progression, such as the proliferation, migration, invasion, and EMT of lung adenocarcinoma cells. We concluded that SGMS1‐AS1 could inhibit developments of LUAD cells to a certain degree.
Novel technologies have demonstrated that the majority of the genome is expressed into RNA, whereas only a few human genes translate to proteins, around 1–2%. Those without coding potential are called non‐coding RNAs, such as microRNA and LncRNA. 27 MicroRNA, ~25 nt nucleotides, usually generates a complex with GW182 and AGO2 (two kind of proteins) called miRNA‐induced silencing complex (miRISC). miRNA then binds to the 3′‐UTR of the target mRNA to prevent its translation into protein. Meanwhile, LncRNA combines with microRNA in a similar way to regulate the downstream gene of miRNA. 28 MiR‐106a‐5p, the direct target of SGMS1‐AS1, has been reported to promote the development of NSCLC, but the detailed molecular mechanism is unclear. Our studies showed that miRNA‐106a has a negative correlation with SGMS1‐AS1 and reversed the effect of SGMS1‐AS1 on LUAD cells. MYLIP is a crucial E3 ubiquitin ligase that has been reported to exert a vital role in various cancers. Notably, MYLIP is the target of miR‐106a‐5p and is positively related to LncRNA SGMS1‐AS1. Overexpressing MYLIP in cells could renovate the function induced by knockdown of SGMS1, which indicates that SGMS1 regulates the progression of LUAD cells by the miR‐106a‐5p/MYLIP axis.
In conclusion, our research verified that LncRNA SGMS1‐AS1 inhibits the proliferation, migration, invasion, and EMT of LUAD cells via the miR‐106a‐5p/MYLIP axis, which provides a potential target for treating LUAD.
CONFLICT OF INTEREST
This article did not contain any conflict of interest.
Liu T, Yang C, Wang W, Liu C. LncRNA SGMS1‐AS1 regulates lung adenocarcinoma cell proliferation, migration, invasion, and EMT progression via miR‐106a‐5p/MYLI9 axis. Thorac Cancer. 2021;12:2104–2112. 10.1111/1759-7714.14043
REFERENCES
- 1. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103:463–73. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 3. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446–54. [DOI] [PubMed] [Google Scholar]
- 4. Zhang C, Zhang Z, Zhang G, Zhang Z, Luo Y, Wang F, et al. Clinical significance and inflammatory landscapes of a novel recurrence‐associated immune signature in early‐stage lung adenocarcinoma. Cancer Lett. 2020;479:31–41. [DOI] [PubMed] [Google Scholar]
- 5. Pakkala S, Ramalingam SS. Personalized therapy for lung cancer: striking a moving target. JCI Insight. 2018;3. 10.1172/jci.insight.120858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Auperin A, Le Pechoux C, Rolland E, Curran WJ, Furuse K, Fournel P, et al. Meta‐analysis of concomitant versus sequential radiochemotherapy in locally advanced non‐small‐cell lung cancer. J Clin Oncol. 2010;28:2181–90. [DOI] [PubMed] [Google Scholar]
- 7. Travis WD, Brambilla E, Riely GJ. New pathologic classification of lung cancer: relevance for clinical practice and clinical trials. J Clin Oncol. 2013;31:992–1001. [DOI] [PubMed] [Google Scholar]
- 8.John SM. Non‐coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017;33:540–52. [DOI] [PubMed] [Google Scholar]
- 11. Boon RA, Jae N, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67:1214–26. [DOI] [PubMed] [Google Scholar]
- 12. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33:665–76. [DOI] [PubMed] [Google Scholar]
- 14. Tang Y, Cheung BB, Atmadibrata B, Marshall GM, Dinger ME, Liu PY, et al. The regulatory role of long noncoding RNAs in cancer. Cancer Lett. 2017;391:12–9. [DOI] [PubMed] [Google Scholar]
- 15. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu YF, Yu JR, Yang Z, Zhu GX, Gao P, Wang H, et al. Promoter hypomethylation mediated upregulation of MicroRNA‐10b‐3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC). J Exp Clin Cancer Res. 2018;37:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie X, Liu H‐T, Mei J, Ding F‐B, Xiao H‐B, Hu F‐Q, et al. miR‐106a promotes growth and metastasis of non‐small cell lung cancer by targeting PTEN. Int J Clin Exp Pathol. 2015;8(4):3827–34. [PMC free article] [PubMed] [Google Scholar]
- 18. Tian Y, Sun C, Zhang L, Pan Y. Clinical significance of miRNA ‐ 106a in non‐small cell lung cancer patients who received cisplatin combined with gemcitabine chemotherapy. Cancer Biol Med. 2018;15:157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito S, Ueno A, Ueda T, Nakagawa H, Taniguchi H, Kayukawa N, et al. CNPY2 inhibits MYLIP‐mediated AR protein degradation in prostate cancer cells. Oncotarget. 2018;9(25):17645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao L, Zhao Y, He Y, Mao Y. miR‐19b promotes breast cancer metastasis through targeting MYLIP and its related cell adhesion molecules. Oncotarget. 2017;8(38):64330–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ni M, Yan Q, Xue H, Du Y, Zhao S, Zhao Z. Identification of MYLIP gene and miRNA‐802 involved in the growth and metastasis of cervical cancer cells. Cancer Biomark. 2021;30:287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun Q, Li X, Xu M, Zhang L, Zuo H, Xin Y, et al. Differential expression and bioinformatics analysis of circRNA in non‐small cell lung cancer. Front Genet. 2020;11:586814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non‐small cell lung cancer: a review. JAMA. 2019;322:764–74. [DOI] [PubMed] [Google Scholar]
- 24. Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non‐small cell lung cancer. Trends Mol Med. 2019;25:585–94. [DOI] [PubMed] [Google Scholar]
- 25. Kleczko EK, Kwak JW, Schenk EL, Nemenoff RA. Targeting the complement pathway as a therapeutic strategy in lung cancer. Front Immunol. 2019;10:954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu Q, Egranov SD, Lin C, Yang L. Long noncoding RNA loss in immune suppression in cancer. Pharmacol Ther. 2020;213:107591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beermann J, Piccoli MT, Viereck J, Thum T. Non‐coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–325. [DOI] [PubMed] [Google Scholar]
- 28. Muller V, Oliveira‐Ferrer L, Steinbach B, Pantel K, Schwarzenbach H. Interplay of lncRNA H19/miR‐675 and lncRNA NEAT1/miR‐204 in breast cancer. Mol Oncol. 2019;13:1137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]


