Abstract
Purpose
The purpose of this study was to perform genetic linkage analysis and association analysis on exome genotyping from highly aggregated African American families with nonpathogenic myopia. African Americans are a particularly understudied population with respect to myopia.
Methods
One hundred six African American families from the Philadelphia area with a family history of myopia were genotyped using an Illumina ExomePlus array and merged with previous microsatellite data. Myopia was initially measured in mean spherical equivalent (MSE) and converted to a binary phenotype where individuals were identified as affected, unaffected, or unknown. Parametric linkage analysis was performed on both individual variants (single-nucleotide polymorphisms [SNPs] and microsatellites) as well as gene-based markers. Family-based association analysis and transmission disequilibrium test (TDT) analysis modified for rare variants was also performed.
Results
Genetic linkage analysis identified 2 genomewide significant variants at 7p15.2 and 7p14.2 (in the intergenic region between MIR148A and NFE2L3 and in the noncoding RNA LOC401324) and 2 genomewide significant genes (CRHR2 and AVL9) both at 7p14.3. No genomewide results were found in the association analyses.
Conclusions
This study identified a significant linkage peak in African American families for myopia at 7p15.2 to 7p14.2, the first potential risk locus for myopia in African Americans. Interesting candidate genes are located in the region, including PDE1C, which is highly expressed in the eyes, and known to be involved in retinal development. Further identification of the causal variants at this linkage peak will help elucidate the genetics of myopia in this understudied population.
Keywords: genetics, linkage analysis, family studies, genetic association
More people in the world are afflicted with myopia than any other eye disorder. The World Health Organization defines uncorrected refractive errors like myopia as visual impairments and estimates that about 153 million people are living worldwide with uncorrected refractive errors.1 One quarter of the American population is myopic, and prevalence is rising.2 Lower-income and disadvantaged populations are particularly at risk because they lack the finances to correct the impairment and thus suffer greater than more affluent populations.
Myopia is a complex disease caused by both genetic and environmental factors, making analysis of the phenotype challenging.3,4 Multiple environmental factors have been identified, including education level and time outside.3
Multiple genetic factors have also been identified to contribute to myopia risk; genetic studies of myopia consist of both family-based linkage studies and population-based association studies. Each method has its advantages/disadvantages. Population-based association studies, specifically genomewide association studies (GWAS), are more effective at identifying common variants with a small to moderate effect on the trait. Many GWAS that have found genomewide significant variants associated with myopia and its quantitative phenotype refractive error.5–13
Family-based linkage studies are effective at finding rare, highly penetrant variants. Variants that are rare in the population at large may be common within an individual family. Family-based studies can also offer better coverage of the genome via longer haplotypes. Haplotypes in population-based studies have been broken apart by generations of recombination so only a small number of variants are in linkage disequilibrium (LD). By contrast, the number of meioses that can occur within a given family is quite small, creating longer haplotypes within a family that can be used to tag rare or ungenotyped causal variants in LD with the linked variants. The drawback is that additional variants along the linked haplotype means identifying the actual causal variant is more difficult. Genomewide significant linked loci have been identified for both refractive error and myopia.14–26 Multiple studies have reported linkage using common myopia.18–26
African Americans have been particularly understudied for myopia. Initial studies showed that African Americans had a lower prevalence of myopia than Caucasians27; more recent studies show myopia prevalence in African American children is approximately equal to Caucasians.28 African American children have been shown to have both a higher percentage of new myopia cases29 and a higher odds ratio for myopia risk than Caucasian children.30 Other African groups show various prevalences in children – 3% in Ghana31 and South Africa,32 and 10% for African Caribbean children in England.23 A more recent study by Jiang et al. showed that a parent with myopia was associated with an increase of myopia risk in children of multiple populations, including African Americans.33 A 2020 review by Grzybowski et al., concluded that myopia prevalences in children are rising in Asia, Europe, and North America but are under 10% in South America and Africa.34
Despite ample evidence for myopia prevalence, there have been a paucity of genetic studies with African samples, with zero GWAS and only a handful of microsatellite-based linkage studies.35,36 Our study is the first genetic analysis using SNP genotypes in African American families with a history of myopia. We used an exome-based microarray for increased coverage of rare variants. A subset of the families used in this study were part of a previous study that found significant linkage to 7p15 with refractive error36 using microsatellites. Using myopia affection as the phenotype did not result in replication of the signal35 nor was the signal replicated in meta-analysis with other populations.16
Methods
Patient Recruitment
We collected data from 517 individuals from 106 African American families in the Philadelphia metropolitan area as part of the Family Myopia Study. Prospective participants were identified through database review, mailings, clinical visits, interviews, and referrals from private doctors. Eligible families were required to have at least three participants, including at least one parent with myopia, and one myopic sibling. All study participants provided informed consent and protocols adhered to the tenets of the Declaration of Helsinki. The study was approved by the institutional review boards of the University of Pennsylvania and the National Human Genome Research Institute.
All participants received a comprehensive eye examination, including visual acuity, slit lamp biomicroscopy, dilated fundus examination, and manifest refraction. Patients older than 41 years had their refraction measured with manifest refraction, whereas patients 41 years and younger had their refraction measured using cycloplegic refraction. The measurement used in this study was mean spherical equivalent (MSE), measured in diopters (D), which is calculated by adding the spherical component to one-half the cylindrical component and averaging for both eyes.
Genotyping and Quality Control
Five hundred seventeen subject DNA samples were genotyped using an Illumina ExomePlus array at the Center for Inherited Disease Research (CIDR) at Johns Hopkins University (Baltimore, MD, USA). Variants were filtered to a mean call rate of 99%, and any variant with a quality score of 0.15 or less was set to missing. Monomorphic markers were removed using PLINK.37 Sib-pair38 was used to identify Mendelian inconsistencies. Markers with a Mendelian inconsistency in a single family were removed from that family; markers with multiple Mendelian error were removed from all families. PLINK and Prest-Plus39 were used to verify familial relationships by calculating identity by descent (IBD) values.
Ten ungenotyped individuals were added to the data set to connect disjointed pedigrees. The existence of these individuals was confirmed by family history, but they were either unwilling or unavailable to participate. Their phenotypes and genotypes were coded as unknown.
The single-nucleotide polymorphism (SNP) data was merged with a previous set of 367 microsatellite genotypes; 493 individuals from the exome-based array had microsatellite data. The final data set consisted of 527 individuals with 98,631 markers.
Myopia Affection Classification
Subjects were classified as either affected or unaffected with myopia. Individuals with MSE ≤ −1.0 D were coded as affected. Adult participants (21 and over) with an MSE of ≥ 0.0 D were coded as unaffected or unknown if the MSE was between −1.0 and 0.0 D in order to avoid potential misclassification errors. We used extra caution when coding children as unaffected, because normal childhood developmental changes can result in misclassification. Children ages 5 to 10 years were coded as unaffected only if their MSE was ≥ 2.0 D and as unknown between −1.0 and 2.0 D. Children aged 11 to 20 years were coded as unaffected if their MSE was ≥ 1.5 D and as unknown between −1.0 and 1.5 D. All children were affected if their MSE was ≤ −1.0 D. These thresholds were based on ophthalmological guidelines as to what levels of childhood refraction are most indicative of myopia in adulthood and the large number of children deemed to be unknown is designed to allow for uncertainty in these projections. The final data set contained a total of 527 individuals (295 as affected, 100 as unaffected, and 132 as unknown/missing). The data were 58.06% female individuals (306 female individuals to 221 male individuals). The average MSE was −2.78 D with a standard deviation of 3.60. The mean age of the entire data set was 40.37 with a standard deviation of 19. The mean age of the adults (21 years and above) was 47.42 (standard deviation of 14.83) and the mean age of the children (20 years and below) was 14.15 (standard deviation of 3.62).
Allele Frequency Estimation
Allele frequencies for the data set were calculated in Sib-pair.38 Estimating allele frequencies directly from an ethnically homogeneous data set properly controls type I error rates in parametric linkage studies.40–42
Parametric Linkage Analysis
We performed both variant-based and gene-based parametric linkage analyses. In linkage analyses, all individuals (including those with missing/unknown phenotypes) are included in the analyses. This is because individuals with missing/unknown phenotypes with genotype information will provide information about allele transmission (thus contributing to the overall LOD score of the variant) and even individuals with no phenotype or genotype information may be needed to provide relationship information to connect relatives with data and avoid disjointed pedigrees. Analyses assumed an autosomal dominant mode of inheritance, with a disease allele frequency of 0.01 and a penetrance of 0.9 for disease allele carriers and 0.1 for noncarriers. The 0.1 penetrance for noncarriers (the phenocopy rate) allow for a 10% chance that an individual that has myopia for reasons other than a high-risk variant (e.g. environmental factors or polygenic inheritance).
Variant-based analyses were two-point linkage analyses performed between the phenotype and each individual SNP, using TwoPointLods.43 Gene-based analyses used the collapsed haplotype pattern (CHP) method in SEQLinkage44 to build multi-allelic pseudomarkers, which corresponded to a gene. Two-point linkage analysis was then performed on the pseudomarkers using Merlin.45 We performed two sets of gene-based analysis: one analysis using rare variants only (MAF ≤ 0.05) and one analysis using all variants.
Family-Based Association Analysis
We used the family-based association test (FBAT)46 to perform variant-based and gene-based analysis association analyses. We also used the rare variant transmission disequilibrium test (RV-TDT)47 by choosing the most informative trio out of the extended families - two parents (one affected and one unaffected) and affected child trio with the highest genotyping rate.
Functional Annotation
All variants were annotated using wANNOVAR48,49 and CRAVAT,50,51 which provide information about SNP location, function, and frequency across multiple populations. They also provide protein predictions from multiple programs such as SIFT,52–56 PolyPhen2,57 CADD,58,59 and REVEL.60
Gene Expression in Human Ocular Tissues
To identify high-priority candidate genes, we examined ocular tissue expression of the significant/suggestive genes from our analyses. Gene expression in human ocular tissues was inspected in the publicly available web resources eyeIntegration61 and The Ocular Tissue Databases.62 The eyeIntegration database provides the largest RNA-seq based transcriptome database of healthy human eye tissues and hundreds of Genotype-Tissue Expression (GTEx) tissue samples.63,64 The Ocular Tissue Database contains microarray expression data of 10 normal human ocular tissues.65 We compared the expression of our significant and suggestive genes in the eyes against two reference tissues (whole blood and pan-body synthetic subtissues). The pan-body synthetic set was comprised of a stratified sample of 54 tissues in the GTEx data set.
Results
Variant-Based Linkage Results
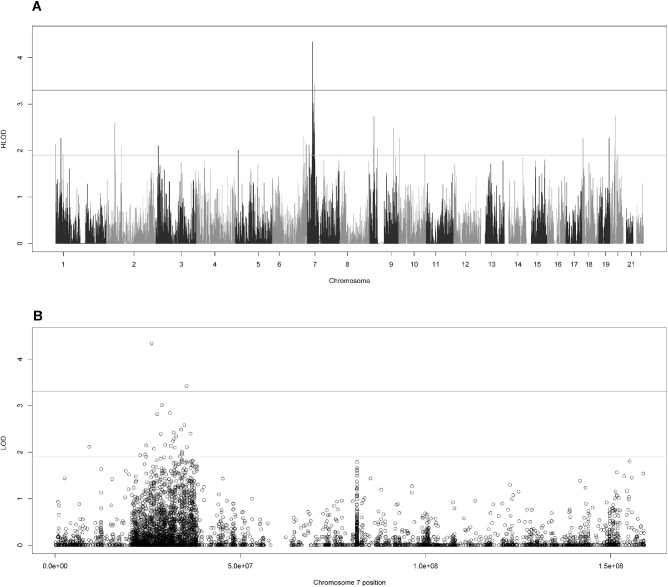
Two variants exhibited genomewide significant linkage to myopia using a definition of genomewide significant as (H)LOD ≥ 3.3 and genomewide suggestive as (H)LOD ≥ 1.9 in accordance with the recommendations of Lander and Kruglyak.66 The rs4719841 is in the intergenic region between MIR148A and NFE2L3 at 7p15.2 (HLOD = 4.34), and rs235397 is located in the noncoding RNA LOC401324 at 7p14.2 (HLOD = 3.43; Fig. 1A). Twenty-five suggestively linked SNPs were found at 7p15.2-14.2 (Fig. 1B). Thirty-three suggestive SNPs were found on other chromosomes, with the largest concentration at 1p36.1-36.11 (Table 1).
Figure 1.
HLOD scores for variant-based two-point linkage analysis. (A) The genomewide HLOD scores (B) the HLOD scores for chromosome 7. In both, the lines at 3.3 and 1.9 represent the respective significant and suggestive thresholds as suggested by Lander and Kruglyak.
Table 1.
All Significant and Suggestive HLOD Scores From Variant Based Linkage
| CHR | rsID | POS | HLOD | GENE | FUNC | EXON | FREQ | SIFT | POLYPH | FATHMM | CADD | REVEL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | rs4719841 | 25997536 | 4.34 | MIR148A; NFE2L3 | Intergenic | . | 0.27 | . | . | . | . | . |
| 7 | rs235397 | 35372749 | 3.42 | LOC401324 | ncRNA | . | 0.20 | . | . | . | . | . |
| 7 | rs6462100 | 28754095 | 3.01 | CREB5 | Intronic | . | 0.40 | . | . | . | . | . |
| 7 | rs7797330 | 30895010 | 2.84 | INMT-MINDY4 | ncRNA | . | 0.38 | . | . | . | . | . |
| 7 | rs7779240 | 27562660 | 2.82 | EVX1; HIBADH | Intergenic | . | 0.15 | . | . | . | . | . |
| 20 | rs3746736 | 23424613 | 2.75 | CSTL1 | Exonic | nonsyn | 0.20 | T | B | T | 0.003 | 0.086 |
| 9 | rs10757225 | 21555445 | 2.73 | MIR31HG | ncRNA | . | 0.18 | . | . | . | . | . |
| 2 | rs1920511 | 41792845 | 2.59 | SLC8A1; LINC01913 | Intergenic | . | 0.33 | . | . | . | . | . |
| 7 | rs10270663 | 34786398 | 2.58 | NPSR1-AS1 | ncRNA | . | 0.20 | . | . | . | . | . |
| 7 | rs1427483 | 33959239 | 2.49 | BMPER | Intronic | . | 0.29 | . | . | . | . | . |
| 9 | rs61757558 | 117118379 | 2.48 | AKNA | Exonic | nonsyn | 0.06 | D | B | T | 22.6 | 0.019 |
| 7 | rs2270219 | 31877261 | 2.42 | PDE1C | Intronic | . | 0.23 | . | . | . | . | . |
| 7 | rs3735400 | 36438709 | 2.40 | ANLN | Exonic | nonsyn | 0.12 | D | D | T | 29.7 | 0.204 |
| 7 | rs6462088 | 28504566 | 2.40 | CREB5 | Intronic | . | 0.24 | . | . | . | . | . |
| 7 | rs2011974 | 32611392 | 2.35 | AVL9 | Intronic | . | 0.34 | . | . | . | . | . |
| 6 | rs214950 | 152708310 | 2.29 | SYNE1 | Exonic | nonsyn | 0.15 | D | B | T | 7.324 | 0.104 |
| 7 | rs10266620 | 31957550 | 2.29 | PDE1C | Intronic | . | 0.26 | . | . | . | . | . |
| 1 | rs7550997 | 26596080 | 2.27 | CEP85 | Exonic | nonsyn | 0.18 | T | B | T | 15.09 | 0.043 |
| 1 | rs8564 | 26605069 | 2.27 | CEP85 | UTR3 | . | 0.18 | . | . | . | . | . |
| 1 | rs7544 | 26607726 | 2.27 | SH3BGRL3 | UTR3 | . | 0.18 | . | . | . | . | . |
| 1 | rs10493030 | 26561856 | 2.27 | CEP85 | Intronic | . | 0.18 | . | . | . | . | . |
| 1 | rs10902732 | 26606174 | 2.27 | SH3BGRL3; CEP85 | Intergenic | 0.18 | . | . | . | . | . | |
| 1 | rs11247900 | 26612460 | 2.27 | UBXN11 | Exonic | syn | 0.18 | . | . | . | . | . |
| 1 | rs11577318 | 26601570 | 2.27 | CEP85 | Intronic | . | 0.18 | . | . | . | . | . |
| 1 | rs17163746 | 26564230 | 2.27 | CEP85 | Intronic | . | 0.18 | . | . | . | . | . |
| 1 | rs17163749 | 26568165 | 2.26 | CEP85 | Intronic | . | 0.18 | . | . | . | . | . |
| 10 | rs61729846 | 5920244 | 2.26 | ANKRD16 | Exonic | nonsyn | 0.20 | D | D | T | 26.1 | 0.690 |
| 18 | rs387462 | 3458997 | 2.26 | TGIF1 | downstream | . | 0.36 | . | . | . | . | . |
| 7 | rs6952967 | 31795856 | 2.24 | PDE1C | Intronic | . | 0.47 | . | . | . | . | . |
| 6 | rs791183 | 160610124 | 2.23 | SLC22A1; SLC22A2 | Intergenic | . | 0.40 | . | . | . | . | . |
| 7 | rs1420123 | 29647662 | 2.16 | PRR15; LOC646762 | Intergenic | . | 0.27 | . | . | . | . | . |
| 7 | rs1029602 | 24571485 | 2.15 | NPY; MPP6 | Intergenic | . | 0.38 | . | . | . | . | . |
| 7 | rs4291168 | 31178749 | 2.14 | ADCYAP1R1; NEUROD6 | Intergenic | . | 0.18 | . | . | . | . | . |
| 20 | rs6036107 | 22403287 | 2.13 | LOC284788; LINC00261 | Intergenic | . | 0.33 | . | . | . | . | . |
| 1 | rs2228579 | 1223385 | 2.13 | SCNN1D | Exonic | nonsyn | 0.33 | T | B | T | 0.003 | 0.036 |
| 6 | rs34544438 | 167438292 | 2.12 | FGFR1OP | Exonic | nonsyn | 0.07 | T | B | T | 0.268 | 0.119 |
| 7 | rs1285407 | 9266388 | 2.11 | NXPH1; PER4 | Intergenic | . | 0.12 | . | . | . | . | . |
| 7 | rs12113424 | 35423720 | 2.11 | LOC401324; HERPUD2 | Intergenic | . | 0.19 | . | . | . | . | . |
| 2 | rs13424561 | 73868446 | 2.11 | NAT8 | Exonic | nonsyn | 0.11 | T | B | T | 0.166 | 0.01 |
| 3 | rs36117895 | 11400019 | 2.10 | ATG7 | Exonic | nonsyn | 0.12 | D | P | T | 25.3 | 0.227 |
| 9 | rs10511687 | 20764870 | 2.10 | FOCAD | Exonic | nonsyn | 0.32 | T | B | T | 14.75 | 0.069 |
| 7 | rs6415258 | 32192596 | 2.10 | PDE1C | Intronic | . | 0.27 | . | . | . | . | . |
| 7 | rs212837 | 26695215 | 2.08 | C7orf71; SKAP2 | Intergenic | . | 0.35 | . | . | . | . | . |
| 1 | rs12138111 | 26590432 | 2.07 | CEP85 | Intronic | . | 0.19 | . | . | . | . | . |
| 18 | rs77600482 | 3460731 | 2.06 | TGIF1; GAPLINC | Intergenic | . | 0.15 | . | . | . | . | . |
| 9 | rs10973446 | 37638744 | 2.05 | TOMM5; FRMPD1 | Intergenic | . | 0.45 | . | . | . | . | . |
| 18 | rs381234 | 3464650 | 2.02 | TGIF1; GAPLINC | Intergenic | . | 0.38 | . | . | . | . | . |
| 7 | rs731844 | 34150264 | 2.02 | BMPER | Intronic | . | 0.39 | . | . | . | . | . |
| 5 | rs7715811 | 13769974 | 2.01 | DNAH5 | Intronic | . | 0.39 | . | . | . | . | . |
| 5 | rs1502050 | 13779743 | 2.01 | DNAH5 | Intronic | . | 0.38 | . | . | . | . | . |
| 7 | rs10224983 | 34180326 | 1.98 | BMPER | Intronic | . | 0.15 | . | . | . | . | . |
| 7 | rs961652 | 34111660 | 1.95 | BMPER | Intronic | . | 0.30 | . | . | . | . | . |
| 7 | rs16480 | 24311069 | 1.95 | STK31; NPY | Intergenic | . | 0.38 | . | . | . | . | . |
| 7 | rs2033670 | 22929061 | 1.94 | SNHG26; FAM126A | Intergenic | . | 0.33 | . | . | . | . | . |
| 10 | rs7071768 | 129903016 | 1.93 | MKI67 | Exonic | nonsyn | 0.47 | T | B | T | 0.001 | 0.022 |
| 1 | rs10908292 | 36764770 | 1.92 | THRAP3 | Intronic | . | 0.39 | . | . | . | . | . |
| 20 | rs5741809 | 36956026 | 1.92 | BPI | Exonic | nonsyn | 0.14 | T | B | T | 0.001 | 0.011 |
| 7 | rs2392246 | 33571828 | 1.91 | BBS9 | Intronic | . | 0.12 | . | . | . | . | . |
| 7 | rs976681 | 24530016 | 1.91 | NPY; MPP6 | Intergenic | . | 0.36 | . | . | . | . | . |
The list of all significant and suggestive variants from the variant-based linkage analyses, as sorted by HLOD. Here, the headers represent: CHR = chromosome, rsID = rsID of the SNP, POS = physical position in base pairs of the SNP, HLOD = heterogeneity LOD score across all 106 families, GENE = Gene location of the SNP (if intergenic then the two closest genes), FUNC = function of the SNP (e.g. exonic, intronic), EXON = if exonic, the exonic function of the SNP (nonsyn = nonsynonymous, syn = synonymous), FREQ = frequency of the variant in gnomAD Africans, SIFT = SIFT prediction (T = tolerated, D = damaging), POLY = PolyPhen2 prediction score (B = benign, P = possibly damaging, D = damaging), FATHMM = FATHMM prediction (T = tolerated), CADD = CADD phred score ≥ 10 corresponds to 10% most deleterious substitutions in genome, ≥ 20 corresponds to 1% most deleterious substitutions in the genome, etc.), REVEL = REVEL score (corresponds to proportion of trees in random forest algorithm that classified variant as pathogenic).
Functional Annotation of Variants
The rs4719841 is intergenic and has an MAF in the gnomAD database of 0.2656 and 0.26 in our data set. The rs235397, in the noncoding RNA LOC401324 at 7p14.2, has an MAF of 0.2 both in gnomAD and our data set. The only exonic variant in 7p15.2-14.2 was rs3735400 (HLOD = 2.22). The rs3735400 is in ANLN (7p14.2), is a nonsynonymous exonic variant, has a Combined Annotation Dependent Depletion (CADD) score over 29 and predicted damaging by SIFT and PolyPhen2, and has an MAF of 0.1201 in the gnomAD 0.13 in our data set.
Gene-Based Linkage Results
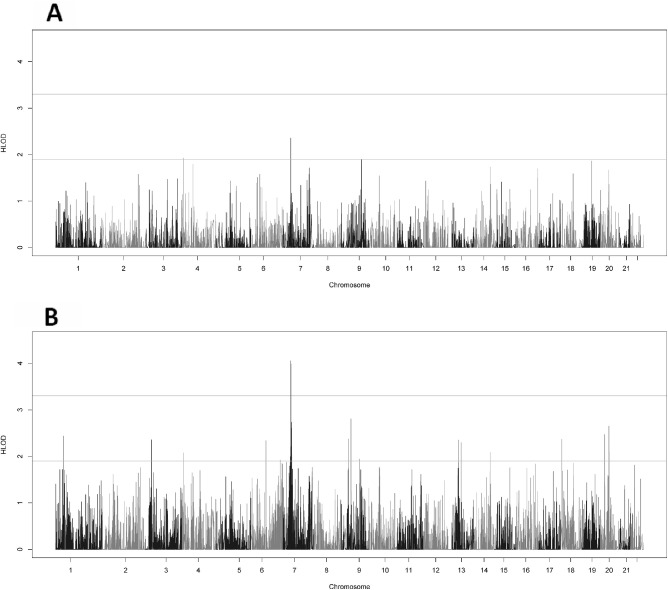
The gene-based analysis using rare variants (MAF ≤ 0.05) produced no genomewide significant results and four genome-wide suggestive genes (Fig. 2A). The two top genes were located at 7p14.3 - INMT-MINDY4 (HLOD = 2.35) and MINDY4 (HLOD = 1.99).
Figure 2.
Genome wide HLODs scores for gene-based two-point linkage analysis. (A) The gene-based HLOD scores using only the rare variants (MAF ≤ 0.05) and (B) the gene-based HLOD scores using all variants. The lines at 3.3 and 1.9 represent the respective significant and suggestive thresholds as suggested by Lander and Kruglyak.
Using all variants, the genomewide significant signal is recovered at the 7p14.3 band (Fig. 2B). This makes sense, as the variants identified in the variant-based analysis were common (MAF > 0.05). The genomewide significant genes were CRHR2 (HLOD = 4.06) and AVL9 (HLOD = 3.99). There were an additional 23 suggestive genes, with 7 genes in the 7p14.3-14.2 region (Table 2, Supplementary Fig. S1).
Table 2.
All Significant and Suggestive Genes From the Gene-based Linkage Analysis
| CHR | POS | GENE | CUMUL LOD | HLOD | VARIANTS |
|---|---|---|---|---|---|
| 7 | 48.34 | CRHR2 | 3.98 | 4.06 | All |
| 7 | 50.78 | AVL9 | 3.99 | 3.99 | All |
| 9 | 59.79 | DNAI1 | 2.81 | 2.81 | All |
| 7 | 52.59 | NPSR1-AS1 | 2.74 | 2.74 | All |
| 7 | 52.05 | BMPER | 1.86 | 2.73 | All |
| 20 | 54.18 | BPIFA2 | 2.64 | 2.65 | All |
| 7 | 54.00 | SEPT7 | 2.34 | 2.61 | All |
| 20 | 28.26 | PAK7 | 1.75 | 2.47 | All |
| 1 | 49.16 | EPHB2 | 2.44 | 2.44 | All |
| 9 | 43.87 | FOCAD | 2.14 | 2.37 | All |
| 18 | 8.08 | SMCHD1 | 2.37 | 2.37 | All |
| 3 | 39.61 | EFHB | 1.77 | 2.36 | All |
| 7 | 48.59 | INMT-FAM188B | 2.35 | 2.35 | Rare |
| 13 | 36.59 | CSNK1A1L | 2.15 | 2.35 | All |
| 6 | 92.24 | COL12A1 | 2.34 | 2.34 | All |
| 13 | 52.86 | SETDB2 | 1.94 | 2.30 | All |
| 7 | 50.30 | PDE1C | 2.21 | 2.21 | All |
| 20 | 55.10 | CEP250 | 1.93 | 2.10 | All |
| 14 | 97.59 | SERPINA9 | 1.35 | 2.09 | All |
| 4 | 9.39 | EVC | 0.92 | 2.08 | All |
| 7 | 54.69 | KIAA0895 | 1.34 | 2.04 | All |
| 7 | 46.21 | LOC646762 | 1.49 | 1.99 | All |
| 7 | 48.61 | FAM188B | 1.99 | 1.99 | Rare |
| 9 | 111.62 | ZNF462 | 1.95 | 1.95 | All |
| 4 | 9.39 | EVC | 1.93 | 1.93 | Rare |
| 6 | 178.62 | PACRG | 0.72 | 1.92 | All |
| 1 | 49.16 | EPHB2 | 1.92 | 1.92 | All |
| 7 | 50.30 | PDE1C | 0.31 | 1.91 | All |
| 9 | 123.13 | AKNA | 1.90 | 1.90 | Rare |
The list of all significant and suggestive genes from the gene-based linkage analyses, as sorted by HLOD. Here the headers represent: CHR = chromosome, POS = genetic position in cM of the gene, GENE = gene, CUMUL LOD = cumulative LOD score for the gene across all 106 families, HLOD = heterogeneity LOD score for the gene across all 106 families, VARIANTS = type of variants used in this test (All = all variants were used, Rare = only rare variants (MAF ≤ 0.05) were used).
Association Results
No genomewide significant (5 × 10−8) results were found in the association analyses. The most significant FBAT variant P value was rs887468 (1.2 × 10−5) in PSORS1C3 at 6q21.33. The most significant FBAT gene-based P value was KCNS1 (4.97 × 10−5) at 20q12. The most significant RV-TDT P value was the intergenic SNP rs11929331 (2.5 × 10−4) between LINC01994 and ATP11B at 3q26.44.
Gene Expression in Human Ocular Tissue
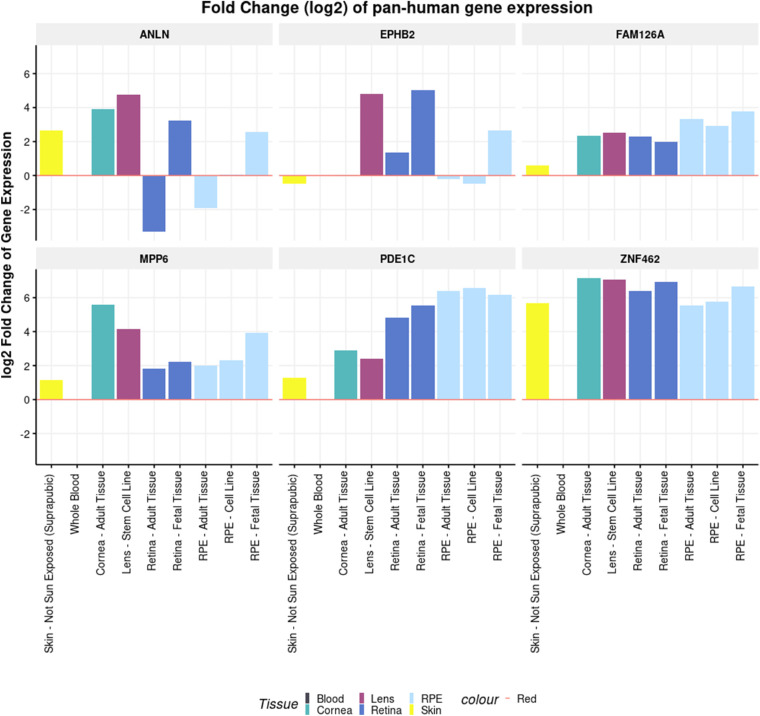
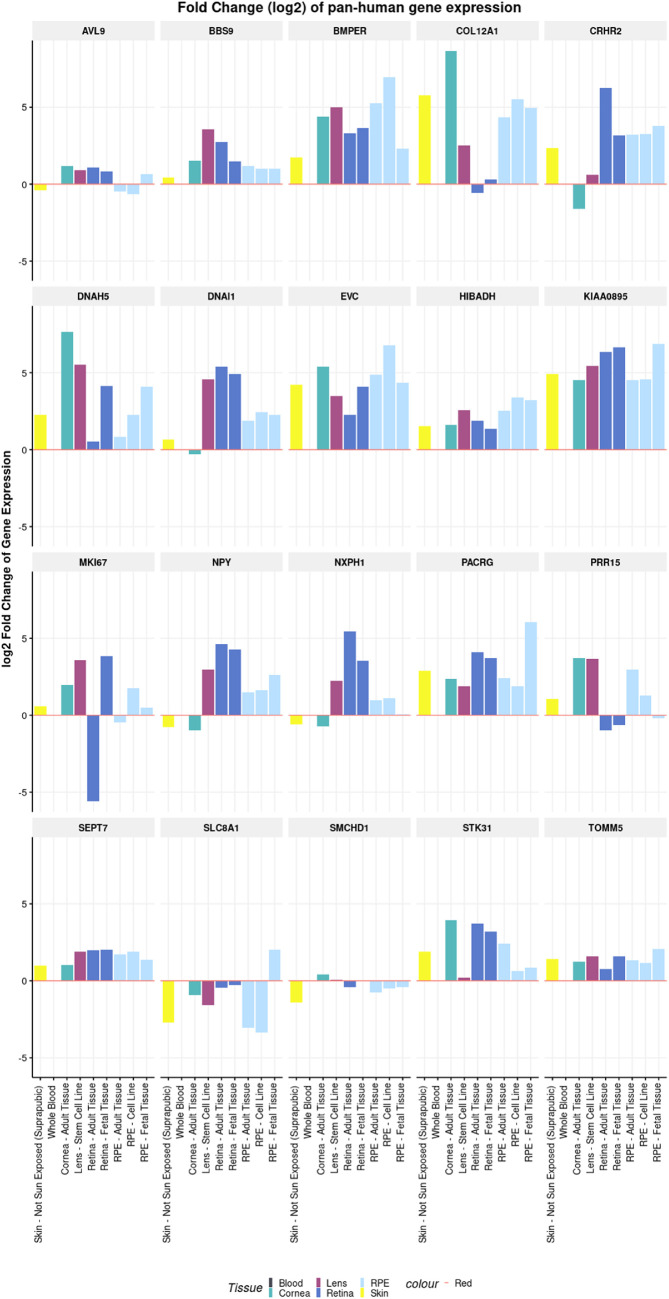
The eyeIntegration and Ocular Tissue databases revealed that a vast majority of the significant/suggestively genes were expressed in ocular tissues (Supplementary Table S1, Supplementary Figs. S2, S3). The suggestive genes (including ANLN and PDE1C) from the variant-based and gene-based analyses were found to have higher expression in most tissues of the eyes compared to whole blood and pan-body synthetic subtissues (Fig. 3, Supplementary Table S1). Other significant and suggestive genes (including AVL9, BBS9, and BMPER) have a higher expression in most of the ocular tissues compared to at least one reference tissue group (Fig. 4, Supplementary Table S1). The Ocular Tissue Database showed that EPHB2, PDE1C, NPY, EVC, and COL12A1 have good expression in the adult human sclera, which may play a role in myopia pathogenesis (see Supplementary Figs. S2, S3). Both databases are primarily derived from European ancestry individuals, so expression may vary in African Americans, but there are no known databases with African American eye expression tissue data.
Figure 3.
Pan-human tissue differential expression of ANLN, FAM126A, MPP6, PDE1C, EPHB2, and ZNF462. The x-axis shows the different types of tissues used in the test. The y-axis shows the log2 fold change of gene expression. The differential expression is being shown relative to the reference tissue (whole blood).
Figure 4.
Pan-human tissue differential expression of additional significant and suggestive genes in the two-point linkage analysis. The x-axis shows the different types of tissues used in the test. The y-axis shows the log2 fold change of gene expression. The differential expression is being shown relative to the reference tissue (whole blood).
Discussion
We have identified a genomewide significant linkage between four markers (2 SNPs and 2 genes) at 7p15.2-14.2 and myopia in African American families. This is the largest genetic analysis of myopia in the African American population and the first using SNP genotypes. This is the first study to report a myopia risk locus in African Americans. African Americans have been severely understudied with respect to myopia and refractive error. We note that the 7p15 linkage signal is not entirely novel, as we have previously identified a genomewide significant linkage to refractive error at 7p15 in a subset of these African American families36 and replicated it in a Caucasian data set.67 Both those studies used a different phenotype (quantitative refractive error scores) and the African American study used only microsatellite data and pointedly did not find significant linkage anywhere when myopia affection (the trait used in this study) was the analyzed trait.35 The risk locus identified at 7p14.2 is entirely novel; it was not identified in the microsatellite study. Further, the increased granularity of the SNP data (especially with the addition of the rare exonic variants), allowed for the location of regions and genes where the linked variants are accumulating, which cannot be done in a study with sparse microsatellite data only. This allowed us to go beyond defining a single linked region and offer specific genes within that linked region that might be causal, which is discussed in detail in the following paragraphs.
The risk locus identified at 7p15 is known as MYP17, and, despite multiple replications, the causal gene remains unknown. Our significant linkage signals in the variant-based analysis are in an intergenic region between the transcription factor NFE2L3 and the noncoding RNAs MIR148A at 7p15.2 and LOC401324 at 7p14.2. None of these genes have any previously known connection to eye disorders. The significant genes found by the gene-based analysis (CRHR2 and AVL9) were located slightly downstream at 7p14.3 and have no known association with eye disease. AVL9 and CRHR2 expressions were enriched in most ocular tissues versus reference tissue groups.
Linkage peaks are often broad and locus heterogeneity and other factors add to the uncertainty. Thus, the true causal variant(s) may lie anywhere within the linked region. Thus, it is important to examine the entire linked 7p region for candidate genes, which does identify multiple good candidates.
The rs3735400 (HLOD = 2.4) in the anillin (ANLN) gene were predicted damaging by both SIFT and PolyPhen2 and had a CADD score over 29. Thus, the variant is likely to have a deleterious effect on gene/protein function. Anillin is an actin binding protein that is involved in cell migration, cell growth, and cytokinesis; it regulates actin cytoskeletal dynamics and has been implicated in multiple cancers.68 Its expression was shown to be elevated in eye tissue.
PDE1C (7p14.3) exhibited suggestive evidence of linkage in the gene-based test (HLOD = 2.2) and had four suggestive intronic variants. PDE1C is a phosphodiesterase that has been reported to be involved in avian and rodent retinal development.69,70 PDE1C is expressed significantly higher in both adult and fetal retina tissue and adult and cell line RPE compared to whole blood and pan-body tissues. BMPER (7p14.3) was found to be suggestively linked in both the variant-based and gene-based analyses. BMPER has been shown to be functionally involved in eye organogenesis in mice.71 BBS9, which was found to have a genomewide suggestive intronic variant (HLOD = 1.9), is known to cause Bardet-Biedl syndrome, a symptom of which includes retinal degeneration.72,73 Both genes were expressed higher in ocular tissue than the reference set.
In addition to the significantly linked region on 7p, we also identified several genomewide suggestive linkage signals, the most interesting being located at 1p36. This suggestive signal is a replication of a well-documented myopia/refractive error risk locus, MYP14, at 1p36.15,67,74,75 This is the first time that this linkage to myopia has been documented in African Americans. The causal gene at MYP14 remains elusive. Six of the 9 suggestive SNPs at 1p36 were intronic SNPs located in CEP85. CEP85 is involved in centromere disjunction and has no known connection to eye disease.76 Other centromere proteins in the CEP family have been found implicated in eye disease, like CEP250 in Usher syndrome (which has retinitis pigmentosa as one of its symptoms).77
The association analyses did not identify any genomewide significant signals. The top variant was in an intron of the psoriasis gene PSORS1C3. The top gene was KCNS1 at 20q12, a voltage gated potassium channel protein that is functional in lens epithelia.78 The top SNP in the RV-TDT analysis in an intergenic region between LINC01994 and ATP11B at 3q26.44. The 3q26 is the site of the known myopia locus MYP879 and ATP11B has been shown to be present in mouse retina.80
It is not surprising that the linkage and association results tended to disagree, because they are testing for different things. The family-based association analysis relies on the risk allele being shared across families, either identical by state (IBS) or IBD. Linkage tracks co-segregation of haplotypes and the phenotype within a family but does not require that these haplotypes contain alleles IBS across different families.
Although we have given evidence for some interesting candidate genes for the 7p and 1p signals any implication of causality is speculative at this point. Several genes, including the eye development genes PDE1C and BMPER, are good candidates for future follow-up studies, but additional studies will be needed to determine which variants are responsible for the observed linkage peaks. We also note that any novel linkage findings will need further replication in independent data sets. This study's major limitation was the coverage of the exome-enriched microarray. The causal variant may not have been identified; it is more likely that ungenotyped variants are segregating on the linked haplotypes in each family in LD with the more common variants identified here. This haplotype tagging ability is one of the advantages of family-based linkage studies. This explains why most of the linked variants on the 7p haplotype are common; they are most likely tagging rare variants that were not genotyped on this limited microarray. This also explains why we only identified a genomewide suggestive signal at 7p when using only rare variants in the gene-based tests and why the genomewide significant signal was recovered upon returning the common variants to the analysis. Thus, targeted sequencing of the linked regions, particularly on 7p and 1p, is a necessary future step in determining the causal variant(s).
The expression databases helped to narrow down our candidate genes. We should exercise caution in the interpretation of these findings because the samples from the transcriptome databases were from healthy human eye tissues and their expression profile may differ from the diseased eye. We also do not have a refractive error phenotype on the eyes used in these databases. Further, the tissue used in these databases were primarily from European ancestry individuals, so any expression results must be evaluated within that context.
We have identified a genomewide significant linkage of myopia to the 7p15.2-14.2 region in African Americans. This is the first significant myopia risk locus found in African Americans. This supports a previous study which has linked 7p15 with refractive error.36,67 We also identified several genomewide suggestive signals, including replication of the MYP14 locus at 1p36 for the first time in African Americans. We note that linkage analyses like this one identifies large linked regions and this study aimed to identify good candidate genes/variants and which families are most informative. We plan further whole genome sequencing of our most informative families, which will give us increased coverage of the genome, particularly intronic regions. Deep coverage of these linked regions may elucidate the causal genes/variants.
Supplementary Material
Acknowledgments
The authors thank the study participants and their families.
Funded in part by the National Eye Institute Grant R01 EY020483 as well as the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Disclosure: C.L. Simpson, None; A.M. Musolf, None; R.Y. Cordero, None; J.B. Cordero, None; L. Portas, None; F. Murgia, None; D.D. Lewis, None; C.D. Middlebrooks, None; E.B. Ciner, None; J.E. Bailey-Wilson, None; D. Stambolian, None
References
- 1. Resnikoff S, Pascolini D, Mariotti SP, Pokharel GP.. Global magnitude of visual impairment caused by uncorrected refractive errors in 2004. Bull World Health Organ. 2008; 86: 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vitale S, Sperduto RD, Ferris FL 3rd. Increased prevalence of myopia in the United States between 1971-1972 and 1999-2004. Arch Ophthalmol. 2009; 127: 1632–1639. [DOI] [PubMed] [Google Scholar]
- 3. Stambolian D. Genetic susceptibility and mechanisms for refractive error. Clin Genet. 2013; 84: 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wojciechowski R, Hysi PG.. Focusing in on the complex genetics of myopia. PLoS Genet. 2013; 9: e1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fan Q, Verhoeven VJ, Wojciechowski R, et al.. Meta-analysis of gene-environment-wide association scans accounting for education level identifies additional loci for refractive error. Nat Commun. 2016; 7: 11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simpson CL, Wojciechowski R, Oexle K, et al.. Genome-wide meta-analysis of myopia and hyperopia provides evidence for replication of 11 loci. PLoS One. 2014; 9: e107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stambolian D, Wojciechowski R, Oexle K, et al.. Meta-analysis of genome-wide association studies in five cohorts reveals common variants in RBFOX1, a regulator of tissue-specific splicing, associated with refractive error. Hum Mol Genet. 2013; 22: 2754–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tedja MS, Wojciechowski R, Hysi PG, et al.. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat Genet. 2018; 50: 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verhoeven VJ, Hysi PG, Saw SM, et al.. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012; 131: 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verhoeven VJ, Hysi PG, Wojciechowski R, et al.. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013; 45: 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kiefer AK, Tung JY, Do CB, et al.. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 2013; 9: e1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wong YL, Hysi P, Cheung G, et al.. Genetic variants linked to myopic macular degeneration in persons with high myopia: CREAM Consortium. PLoS One. 2019; 14: e0220143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hysi PG, Young TL, Mackey DA, et al.. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010; 42: 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wojciechowski R, Bailey-Wilson JE, Stambolian D.. Fine-mapping of candidate region in Amish and Ashkenazi families confirms linkage of refractive error to a QTL on 1p34-p36. Mol Vis. 2009; 15: 1398–1406. [PMC free article] [PubMed] [Google Scholar]
- 15. Wojciechowski R, Moy C, Ciner E, et al.. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006; 119: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wojciechowski R, Stambolian D, Ciner E, Ibay G, Holmes TN, Bailey-Wilson JE.. Genomewide linkage scans for ocular refraction and meta-analysis of four populations in the Myopia Family Study. Invest Ophthalmol Vis Sci. 2009; 50: 2024–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ibay G, Doan B, Reider L, et al.. Candidate high myopia loci on chromosomes 18p and 12q do not play a major role in susceptibility to common myopia. BMC Med Genet. 2004; 5: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stambolian D, Ibay G, Reider L, et al.. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet. 2004; 75: 448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Musolf AM, Simpson CL, Moiz BA, et al.. Caucasian families exhibit significant linkage of myopia to chromosome 11p. Invest Ophthalmol Vis Sci. 2017; 58: 3547–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Musolf AM, Simpson CL, Long KA, et al.. Myopia in Chinese families shows linkage to 10q26.13. Mol Vis. 2018; 24: 29–42. [PMC free article] [PubMed] [Google Scholar]
- 21. Musolf AM, Simpson CL, Alexander TA, et al.. Genome-wide scans of myopia in Pennsylvania Amish families reveal significant linkage to 12q15, 8q21.3 and 5p15.33. Hum Genet. 2019; 138: 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simpson CL, Musolf AM, Li Q, et al.. Exome genotyping and linkage analysis identifies two novel linked regions and replicates two others for myopia in Ashkenazi Jewish families. BMC Med Genet. 2019; 20: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rudnicka AR, Owen CG, Nightingale CM, Cook DG, Whincup PH.. Ethnic differences in the prevalence of myopia and ocular biometry in 10- and 11-year-old children: the Child Heart and Health Study in England (CHASE). Invest Ophthalmol Vis Sci. 2010; 51: 6270–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammond CJ, Andrew T, Mak YT, Spector TD.. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004; 75: 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen CY, Stankovich J, Scurrah KJ, et al.. Linkage replication of the MYP12 locus in common myopia. Invest Ophthalmol Vis Sci. 2007; 48: 4433–4439. [DOI] [PubMed] [Google Scholar]
- 26. Schäche M, Chen CY, Pertile KK, et al.. Fine mapping linkage analysis identifies a novel susceptibility locus for myopia on chromosome 2q37 adjacent to but not overlapping MYP12. Mol Vis. 2009; 15: 722–730. [PMC free article] [PubMed] [Google Scholar]
- 27. Katz J, Tielsch JM, Sommer A.. Prevalence and risk factors for refractive errors in an adult inner city population. Invest Ophthalmol Vis Sci. 1997; 38: 334–340. [PubMed] [Google Scholar]
- 28. Kleinstein RN, Jones LA, Hullett S, et al.. Refractive error and ethnicity in children. Arch Ophthalmol. 2003; 121: 1141–1147. [DOI] [PubMed] [Google Scholar]
- 29. Kleinstein RN, Sinnott LT, Jones-Jordan LA, et al.. New cases of myopia in children. Arch Ophthalmol. 2012; 130: 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Borchert MS, Varma R, Cotter SA, et al.. Risk factors for hyperopia and myopia in preschool children the multi-ethnic pediatric eye disease and Baltimore pediatric eye disease studies. Ophthalmology. 2011; 118: 1966–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumah BD, Ebri A, Abdul-Kabir M, et al.. Refractive error and visual impairment in private school children in Ghana. Optom Vis Sci. 2013; 90: 1456–1461. [DOI] [PubMed] [Google Scholar]
- 32. Naidoo KS, Raghunandan A, Mashige KP, et al.. Refractive error and visual impairment in African children in South Africa. Invest Ophthalmol Vis Sci. 2003; 44: 3764–3770. [DOI] [PubMed] [Google Scholar]
- 33. Jiang X, Tarczy-Hornoch K, Cotter SA, et al.. Association of parental myopia with higher risk of myopia among multiethnic children before school age. JAMA Ophthalmol. 2020; 138: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grzybowski A, Kanclerz P, Tsubota K, Lanca C, Saw SM.. A review on the epidemiology of myopia in school children worldwide. BMC Ophthalmol. 2020; 20: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ciner E, Ibay G, Wojciechowski R, et al.. Genome-wide scan of African-American and white families for linkage to myopia. Am J Ophthalmol. 2009; 147: 512-517 e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D.. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008; 32: 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Purcell S, Neale B, Todd-Brown K, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duffy D. SIB-PAIR: A program for simple genetic analysis v1.00.beta. Queensland, Australia: Queensland Institute of Medical Research; 2008. [Google Scholar]
- 39. Sun L. Detecting pedigree relationship errors. Methods Mol Biol. 2012; 850: 25–46. [DOI] [PubMed] [Google Scholar]
- 40. Mandal DM, Sorant AJ, Atwood LD, Wilson AF, Bailey-Wilson JE.. Allele frequency misspecification: effect on power and Type I error of model-dependent linkage analysis of quantitative traits under random ascertainment. BMC Genet. 2006; 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mandal DM, Wilson AF, Bailey-Wilson JE.. Effects of misspecification of allele frequencies on the power of Haseman-Elston sib-pair linkage method for quantitative traits. Am J Med Genet. 2001; 103: 308–313. [PubMed] [Google Scholar]
- 42. Mandal DM, Wilson AF, Elston RC, Weissbecker K, Keats BJ, Bailey-Wilson JE.. Effects of misspecification of allele frequencies on the type I error rate of model-free linkage analysis. Hum Hered. 2000; 50: 126–132. [DOI] [PubMed] [Google Scholar]
- 43. Thomas A. TwoPointsLods. TwoPointLods: http://www-genepi.med.utah.edu/∼alun/software/.
- 44. Wang GT, Zhang D, Li B, Dai H, Leal SM.. Collapsed haplotype pattern method for linkage analysis of next-generation sequence data. Eur J Human Genetics EJHG. 2015; 23: 1739–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abecasis GR, Cherny SS, Cookson WO, Cardon LR.. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002; 30: 97–101. [DOI] [PubMed] [Google Scholar]
- 46. Laird NM, Horvath S, Xu X.. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000; 19(Suppl 1): S36–42. [DOI] [PubMed] [Google Scholar]
- 47. He Z, O'Roak BJ, Smith JD, et al.. Rare-variant extensions of the transmission disequilibrium test: application to autism exome sequence data. Am J Hum Genet. 2014; 94: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chang X, Wang K.. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012; 49: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Douville C, Carter H, Kim R, et al.. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics. 2013; 29: 647–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masica DL, Douville C, Tokheim C, et al.. CRAVAT 4: Cancer-Related Analysis of Variants Toolkit. Cancer Res. 2017; 77: e35–e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Z, Shen D, Parsons DW, et al.. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004; 304: 1164–1166. [DOI] [PubMed] [Google Scholar]
- 53. Zhao Y, Zhao F, Zong L, et al.. Exome sequencing and linkage analysis identified tenascin-C (TNC) as a novel causative gene in nonsyndromic hearing loss. PLoS One. 2013; 8: e69549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumar P, Henikoff S, Ng PC.. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 55. Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC.. SIFT missense predictions for genomes. Nat Protoc. 2016; 11: 1–9. [DOI] [PubMed] [Google Scholar]
- 57. Adzhubei I, Jordan DM, Sunyaev SR.. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics. 2013; Chapter 7: Unit7.20. [DOI] [PMC free article] [PubMed]
- 58. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014; 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M.. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019; 47: D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ioannidis NM, Rothstein JH, Pejaver V, et al.. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016; 99: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bryan JM, Fufa TD, Bharti K, Brooks BP, Hufnagel RB, McGaughey DM.. Identifying core biological processes distinguishing human eye tissues with precise systems-level gene expression analyses and weighted correlation networks. Hum Mol Genet. 2018; 27: 3325–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. The Ocular Tissue Database: https://genome.uiowa.edu/otdb/ .
- 63. Bryan JM, Fufa TD, Bharti K, Brooks BP, Hufnagel RB, McGaughey DM.. Identifying core biological processes distinguishing human eye tissues with precise systems-level gene expression analyses and weighted correlation networks. Hum Mol Genet. 2018; 27: 3325–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Swamy V, McGaughey D.. Eye in a disk: eyeIntegration human pan-eye and body transcriptome database version 1.0. Invest Ophthalmol Vis Sci. 2019; 60: 3236–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wagner AH, Anand VN, Wang W-H, et al.. Exon-level expression profiling of ocular tissues. Exp Eye Res. 2013; 111: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lander E, Kruglyak L.. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995; 11: 241–247. [DOI] [PubMed] [Google Scholar]
- 67. Klein AP, Duggal P, Lee KE, et al.. Linkage analysis of quantitative refraction and refractive errors in the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2011; 52: 5220–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lian YF, Huang YL, Wang JL, et al.. Anillin is required for tumor growth and regulated by miR-15a/miR-16-1 in HBV-related hepatocellular carcinoma. Aging. 2018; 10: 1884–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Deplano S, Giorgi M, Maccarone R, et al.. Gene expression and protein localization of calmodulin-dependent phosphodiesterase during ontogenesis of chick retina. J Neurosci Res. 2008; 86: 1017–1023. [DOI] [PubMed] [Google Scholar]
- 70. Santone R, Giorgi M, Maccarone R, Basso M, Deplano S, Bisti S.. Gene expression and protein localization of calmodulin-dependent phosphodiesterase in adult rat retina. J Neurosci Res. 2006; 84: 1020–1026. [DOI] [PubMed] [Google Scholar]
- 71. Ikeya M, Kawada M, Kiyonari H, et al.. Essential pro-Bmp roles of crossveinless 2 in mouse organogenesis. Development. 2006; 133: 4463–4473. [DOI] [PubMed] [Google Scholar]
- 72. Jiang J, Promchan K, Jiang H, et al.. Depletion of BBS Protein LZTFL1 Affects Growth and Causes Retinal Degeneration in Mice. J Genetics Genomics = Yi chuan xue bao 2016; 43: 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Veleri S, Bishop K, Dalle Nogare DE, et al.. Knockdown of Bardet-Biedl syndrome gene BBS9/PTHB1 leads to cilia defects. PLoS One 2012; 7: e34389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li YJ, Guggenheim JA, Bulusu A, et al.. An international collaborative family-based whole-genome linkage scan for high-grade myopia. Invest Ophthalmol Vis Sci. 2009; 50: 3116–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Abbott D, Li YJ, Guggenheim JA, et al.. An international collaborative family-based whole genome quantitative trait linkage scan for myopic refractive error. Mol Vis. 2012; 18: 720–729. [PMC free article] [PubMed] [Google Scholar]
- 76. Chen C, Tian F, Lu L, et al.. Characterization of Cep85 - a new antagonist of Nek2A that is involved in the regulation of centrosome disjunction. J Cell Sci. 2015; 128: 3290–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Khateb S, Zelinger L, Mizrahi-Meissonnier L, et al.. A homozygous nonsense CEP250 mutation combined with a heterozygous nonsense C2orf71 mutation is associated with atypical Usher syndrome. J Med Genet. 2014; 51: 460–469. [DOI] [PubMed] [Google Scholar]
- 78. Shepard AR, Rae JL.. Electrically silent potassium channel subunits from human lens epithelium. Am J Physiol. 1999; 277: C412–424. [DOI] [PubMed] [Google Scholar]
- 79. Andrew T, Maniatis N, Carbonaro F, et al.. Identification and replication of three novel myopia common susceptibility gene loci on chromosome 3q26 using linkage and linkage disequilibrium mapping. PLoS Genet. 2008; 4: e1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang J, Molday LL, Hii T, et al.. Proteomic analysis and functional characterization of P4-ATPase phospholipid flippases from murine tissues. Sci Rep. 2018; 8: 10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.