Abstract
The lack of rapid, sensitive, and deployable tuberculosis diagnostic tools is hampering the early diagnosis of tuberculosis and early detection of treatment failures. The conventional sputum smear microscopy or Xpert MTB/RIF assay cannot distinguish between alive and dead bacilli and the culture method delays providing results. Tuberculosis molecular bacterial load assay is a reverse transcriptase real-time quantitative polymerase chain reaction that quantifies viable tuberculosis bacillary load as a marker of treatment response for patients on anti-tuberculosis therapy. However, results are not synthesized enough to inform its comparative advantage to tuberculosis culture technique which is yet the gold standard of care. With this review, we searched electronic databases, including PubMed, Embase, and Web of Science, from March 2011 up to February 2021 for clinical trials or prospective cohort studies that compared tuberculosis molecular bacterial load assay with tuberculosis culture in adults. We included eight studies that meet the inclusion criteria. Tuberculosis molecular bacterial load assay surpasses culture in monitoring patients with tuberculosis during the first few weeks of anti-tuberculosis treatment. It is more desirable over culture for its shorter time to results, almost zero rates of contamination, need for less expertise on the method, early rate of decline, lower running cost, and reproducibility. Its rapid and specific tuberculosis treatment monitoring competency benefits patients and healthcare providers to monitor changes of bacillary load among isolates with drug-susceptible or resistance to anti-tuberculosis regimens. Despite of the high installing cost of the tuberculosis molecular bacterial load assay method, molecular expertise, and a well-equipped laboratory, tuberculosis molecular bacterial load assay is a cost-effective method with comparison to culture in operational running. To achieve maximum utility in high tuberculosis burden settings, an intensive initial investment in nucleic acid extraction and polymerase chain reaction equipment, training in procedures, and streamlining laboratory supply procurement systems are crucial. More evidence is needed to demonstrate the potential large-scale and sustainable use of tuberculosis molecular bacterial load assay over culture in resource-constrained settings.
Keywords: Tuberculosis, tuberculosis molecular bacterial load assay, culture, treatment monitoring
Introduction
Tuberculosis (TB) remains a worldwide threatening and one of the top 10 causes of death, with an estimated 10 million people fell ill with TB annually. The lack of rapid, sensitive, and deployable TB diagnostic tools is hampering the early diagnosis of the disease and early detection of treatment failures. The accuracy of diagnostic tests and the time taken to provide results were proven to impact TB treatment outcomes.1–4 As a result, novel diagnostic tools for monitoring treatment response and early identifying treatment failure are desperately needed.5–7 Methods of monitoring response to anti-TB treatment would be desirable during treatment especially in identifying cases failing therapy and those at risk of relapse. There is difficulty in TB treatment, regardless of the type of TB, drug-susceptible or drug-resistant, as the treatments require 6–12 months or more time, with four or more drug-combination to provide the desired outcome.
A reduction in bacterial load is the most vital currently available marker for TB treatment response. 8 For different disease conditions, pharmacodynamic biomarkers are objectively measured and assessed as a sign of pharmacologic responses to therapeutic interventions. 9 TB biomarkers can either be in a two-dimensional matrix, according to the clinical outcome (failure vs relapse) and level of surrogacy (patient vs trial),10,11 while other promising TB biomarkers are emerging. Such biomarkers, including time-to-positivity (TTP), sputum culture conversion, smear conversion, therapeutic drug monitoring (TDM), pharmacokinetics (PK), minimum inhibitory concentration (MIC), and whole blood bactericidal assay (WBA) could facilitate the development of alternative treatment strategies.
So far, no specific molecular method has been superiorly recommended as a biomarker for monitoring TB treatment response, necessitating the continued use of phenotypic methods.12,13 The conventional sputum smear microscopy or Xpert MTB/RIF assay cannot distinguish between alive and dead bacilli and the culture method delays providing results. Sputum smear microscopy as one of the phenotypic methods remains the most commonly used test for diagnosis and monitoring of treatment, despite being less sensitive and non-specific for Mycobacterium tuberculosis (M. tb), while mycobacterial culture, being identified and applied as the gold standard for TB diagnosis, has the disadvantage of providing results after a substantial period (3–4 weeks).14–17
The tuberculosis molecular bacterial load assay (TB-MBLA) is a reverse transcriptase quantitative real-time polymerase chain reaction (RT-qPCR) of 16S rRNA detection test that quantifies TB bacillary load and is used as a marker of treatment response for patients on anti-TB therapy. 18 The assay is rapid, free of contamination, and can inform the elimination rate of M. tb during treatment.8,18,19 Comparing to culture principles, which usually involves culturing mycobacteria species on either solid-based Lowenstein–Jensen medium (LJ) or liquid mycobacterial growth indicator tube (MGIT), it yields timely results. 20 However, results are not synthesized enough to inform its comparative advantage to the conventional culture methods for monitoring response to anti-TB treatment, including failures to anti-TB treatment.
We aimed to contemplate the potential of TB-MBLA over solid and liquid culture as biomarkers for monitoring treatment response. And in this review, we searched electronic databases, including PubMed, Embase, and Web of Science, from March 2010 up to February 2021 for clinical trials or prospective cohort studies that compared TB-MBLA with TB culture in adults. The search included a combination of the terms, “Tuberculosis,” “biomarkers,” “molecular bacterial load assay,” “outcome,” “treatment monitoring,” “culture,” “sputum smear microscopy” and “tuberculosis molecular methods.” We also manually searched the references of the included studies. Table 1 summarizes the glossary of research terms that we considered in the review.
Table 1.
Glossary of research terms in the review.
| Glossary | Description |
|---|---|
| Biomarker | A measurable characteristic of the organism state during treatment 18 |
| Treatment completed | Treatment completed as recommended by the national policy 8 |
| Predictive biomarkers | Biomarkers which allow the prediction of the treatment outcome 10 |
| Bacterial loads | A measure of M. tb in original sputum samples and quantified as estimated colony-forming unit in 1 mL of sputum sample (eCFU/mL)18,19 |
| Xpert MTB/RIF | Molecular method for diagnosis of TB and can provide resistance strain on rifampicin14,15 |
TB: tuberculosis.
Results
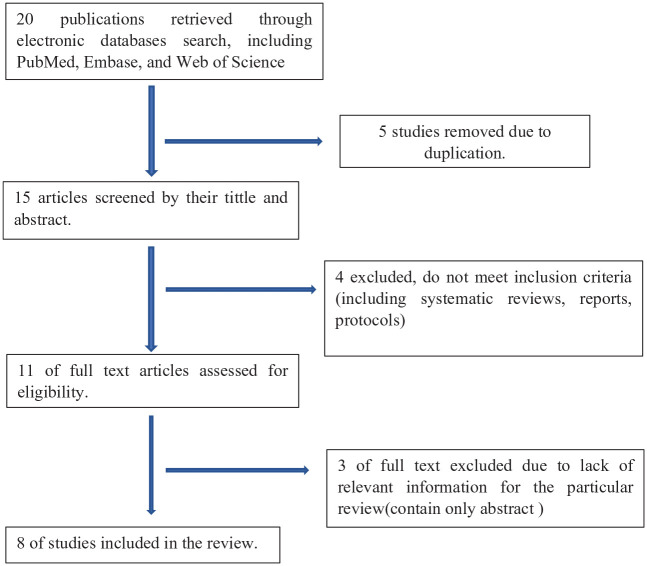
With this review, we found eight eligible clinical trial and prospective cohort studies that compared TB-MBLA with liquid or solid TB culture as a biomarker for monitoring treatment of patients with TB. Figure 1 summarizes the flow diagram of the study.
Figure 1.
Study flow diagram.
Table 2 summarizes the overall characteristics of the eight studies selected for the review and their major findings.
Table 2.
Characteristics of included studies (n = 8).
| Characteristics | Description of preferred methods | Studies |
|---|---|---|
| Shorter time to results | TB-MBLA gives rapid bacillary load count and takes a short time to results, turnaround time for TB-MBLA is early as within 2 days compared to 2–8 weeks of Culture | 8,21,22 |
| Rate of contamination | TB-MBLA is not affected by contamination and does not need a decontamination process when conducting while culture has about 10% rate of contamination | 18,19,22,23 |
| Need for less expertise on the method | Culture is more preferable while TB-MBLA needs training on how to perform the test, comparing to a culture where it learned from normal formal skills. | 22,24 |
| Early rate of decline | TB-MBLA shows precise evidence on the decline of bacterial load in response to antimicrobial treatment | 18,24–26 |
| Lower running cost | TB-MBLA is a preferred cost-effective method in terms of cost reduction in TB trials by speeding up drug development considering having higher utility for making clinical decisions with comparison to culture operational cost as it uses high-level biosafety containment laboratories despite both using Biosafety Laboratory level 3 | 22,27 |
| Reproducibility | TB-MBLA shows the degree of agreement when the experiment is repeated in different laboratory settings | 21,8 |
| Sensitivity | TB-MBLA is more sensitive than culture in picking bacilli, even small bacilli amount | 19 |
TB-MBLA: tuberculosis molecular bacterial load assay.
Table 3 summarizes the technical and operational pros and cons of TB-MBLA versus TB culture based on the evidence compiled from the studies included in the review.
Table 3.
Summarizing technical and operational pros and cons of TB-MBLA versus culture.
| Pros | Cons | Reference | |
|---|---|---|---|
| Technically | Culture: available and used in clinical settings compared to TB-MBLA with regular formal skills | TB-MBLA: mostly used in clinical trial settings, and it requires more expertise with training to conduct the test. | 8,19 |
| Operationally | TB-MBLA: rapidly quantifies viable M. tb, reproducible and may be appropriate in treatment monitoring and drug efficacy |
Culture: methods are time-exhausting and
usually have a risk of
contamination TB-MBLA: has a higher operational cost |
19,22,23,26,27 |
TB-MBLA: tuberculosis molecular bacterial load assay.
Discussion
TB-MBLA is among new technological advances in diagnostics and there are many opportunities for TB-MBLA to function preferably as a biomarker for monitoring TB treatment response. The TB-MBLA functions are based on 16S rRNA and the RT-qPCR technology, with the potential to quickly quantify viable bacilli and detect potential failures in anti-TB treatment in contrast to DNA-based techniques like Xpert MTB/RIF which is also a quick method of identifying bacilli that do not offer information on viable bacterial load. 13 TB-MBLA is capable of detecting a 16S rRNA of dormant and replicating TB cells while continuously measuring and quantifying bacterial load in the sputum of a patient. TB-MBLA process in a protocol has a three-step consisting of (1) extraction of total RNA, (2) enzymatic genomic DNA removal, and (3) RT-qPCR where cycle threshold is transformed to bacterial load.24,28 When mycobacterial cells are killed by anti-TB drugs, there is a decrease in rRNA amount and thus easily estimates the number of viable cells in a patient’s sputum sample. A decline in rRNA has been defined as a surrogate biomarker of microbial viability and bactericidal activity for anti-TB regimen, due to a cellular abundance of 16S rRNA and half-life being shorter than that of DNA. This 16S rRNA measurement has been used in the quantification of bacterial load.
TB-MBLA surpasses and has clinical importance over culture in monitoring patients with TB during the first few weeks of anti-TB treatment. It is more desirable over the culture for its shorter time to results, and it has been reported to have a superior advantage that regardless of bacterial load present in sputum, one can obtain results within 24 h after sputum expectoration. 8 Solid or liquid TB culture methods are time-consuming and are susceptible to contaminations that compromise their potential use for monitoring ant-TB treatment. The currently available culture-based methods require a delayed turnaround time of laboratory results for low-burden samples compared to TB-MBLA that rapidly give M. tb load count in a consistent pattern as shown in a model presented by Svensson et.al. 23
In the first week and the first month of treatment, TB-MBLA has demonstrated zero rates of contamination, early and rapid rate of decline of M. tb bacilli, and suitable outcomes to adjust the anti-TB regimen.21,26 Reproducibility factors of the TB-MBLA test make it robust and thus applicable in different laboratory settings.8,18 While solid or liquid TB culture methods remain time-consuming and susceptible to contaminations that compromise their potential use for monitoring ant-TB treatment, TB-MBLA has a unique potential to monitor changes in bacterial load and response to TB therapy, with the ability to show the early rate of decline of the viable M. tb count in low-burden samples.24,25 In response to therapy, TB-MBLA can rapidly give M. tb load count in a consistent pattern when compared to the culture which takes a prolonged time to provide results.
Studies demonstrated that TB-MBLA can deliver data on the number of viable bacteria as little as 4 h, and this can be used to evaluate disease severity at the initial anti-TB treatment. It yields reproducibility and robustness with regard to bacilli quantification, which would be of great help in measuring response to treatment continuously. Culture with drug susceptibility testing is considered the gold standard of care for the diagnosis of TB and its drug-resistance strains; however, it is time-taking, less precise, and is exposed to missing data that hinder its potential use for monitoring treatment as compared to TB-MBLA.
Recently, the World Health Organization (WHO) attests to the potential use of TB-MBLA for monitoring ant-TB treatment response and its potential substituting culture that has suffered from some practical limitations. 29 However, TB-MBLA is yet mostly applied in research settings because its implementation needs more training and availability and of some important equipment with needs of intensive investments for maximum implementation in resource-constrained high-burden countries. TB-MBLA is still under evaluation in some high-burden countries and the results need to be synthesized with existing literature to provide broader evidence that can be done in normal clinical areas since most of these studies in this review have been done under clinical trial settings.
Limitations
There are some limitations to this review. Most of the included studies have been done in Eastern Africa, providing a few such studies comparing TB-MBLA against culture. This was mainly because there were a few studies conducted on this particular subject. Despite this, available studies are sourced and discussed utmost with included reviews.
Conclusion
TB-MBLA surpasses culture in monitoring patients with TB during the first few weeks of anti-TB treatment. It is more desirable over the culture for its shorter time to results by providing early information on the rate of decline in bacterial load. TB-MBLA still requires molecular expertise and a well-equipped laboratory to perform. To achieve maximum utility in high TB burden settings, an intensive initial investment in nucleic acid extraction and PCR equipment, training in procedures, and streamlining laboratory supply procurement system are crucial. More evidence is needed to demonstrate the potential large-scale and sustainable use of TB-MBLA over culture in resource-constrained settings.
Acknowledgments
The authors thank the Center for Innovative Drug Development and Therapeutic Trials for Africa (CDT-Africa), College of Health Sciences, Addis Ababa University for the support rendered.
Footnotes
Author contributions: B.S. designed and made a significant contribution to this review. B.S. and T.M. conducted the literature search. B.S., L.C, E.G, C.L.W., and M.A, interpreted the data. B.S. prepared the manuscript. All the authors have reviewed and gone over the sequential amended manuscript. The last draft was read and agreed upon by all the authors.
Data availability and materials: All applicable data are within the manuscript and its supporting information files.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: No specific funding was received for this review. T.M. was supported in part by the Fogarty International Center and National Institute of Allergy and Infectious Diseases of the US National Institutes of Health under award no. D43TW009127. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
ORCID iDs: Bibie Said  https://orcid.org/0000-0002-3687-6827
https://orcid.org/0000-0002-3687-6827
Tsegahun Manyazewal  https://orcid.org/0000-0002-8360-7574
https://orcid.org/0000-0002-8360-7574
References
- 1. Kloprogge F, Mwandumba HC, Banda G, et al. Longitudinal pharmacokinetic-pharmacodynamic biomarkers correlate with treatment outcome in drug-sensitive pulmonary tuberculosis: a population pharmacokinetic-pharmacodynamic analysis. Open Forum Infect Dis 2020; 7(7): ofaa218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Visca D, Centis R, Munoz-Torrico M, et al. Post-tuberculosis sequelae: the need to look beyond treatment outcome. Int J Tuberc Lung Dis 2020; 24(8): 761–762. [DOI] [PubMed] [Google Scholar]
- 3. Alemu A, Bitew ZW, Worku T. Poor treatment outcome and its predictors among drug-resistant tuberculosis patients in Ethiopia: a systematic review and meta-analysis. Int J Infect Dis 2020; 98: 420–439. [DOI] [PubMed] [Google Scholar]
- 4. Chedid C, Kokhreidze E, Tukvadze N, et al. Association of baseline white blood cell counts with tuberculosis treatment outcome: a prospective multicentered cohort study. Int J Infect Dis 2020; 100: 199–206. [DOI] [PubMed] [Google Scholar]
- 5. Manyazewal T, Marinucci F, Belay G, et al. Implementation and evaluation of a blended learning course on tuberculosis for front-line health care professionals. Am J Clin Pathol 2017; 147(3): 285–291. [DOI] [PubMed] [Google Scholar]
- 6. Mohammed H, Oljira L, Roba KT, et al. Burden of tuberculosis and challenges related to screening and diagnosis in Ethiopia. J Clin Tuberc Other Mycobact Dis 2020; 19: 100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arinaminpathy N, Dowdy D. Understanding the incremental value of novel diagnostic tests for tuberculosis. Nature 2015; 528(7580): S60–S67. [DOI] [PubMed] [Google Scholar]
- 8. Sabiiti W, Azam K, Farmer ECW, et al. Tuberculosis bacillary load, an early marker of disease severity: the utility of tuberculosis molecular bacterial load assay. Thorax 2020; 75(7): 606–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Florian J, Campbell E, et al. Advancing biosimilar development using pharmacodynamic biomarkers in clinical pharmacology studies. Clin Pharmacol Ther 2020; 107(1): 40–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gatti A, Ceriani C, De Paschale M, et al. Quantification of neutrophil and monocyte CD64 expression: a predictive biomarker for active tuberculosis. Int J Tuberc Lung Dis 2020; 24(2): 196–201. [DOI] [PubMed] [Google Scholar]
- 11. Wallis RS, Peppard T. Early biomarkers and regulatory innovation in multidrug-resistant tuberculosis. Clin Infect Dis 2015; 61(Suppl. 3): S160–S163. [DOI] [PubMed] [Google Scholar]
- 12. Mbelele PM, Mohamed SY, Sauli E, et al. Meta-narrative review of molecular methods for diagnosis and monitoring of multidrug-resistant tuberculosis treatment in adults. Int J Mycobacteriol 2018; 7(4): 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goletti D, Petruccioli E, Joosten SA, et al. Tuberculosis biomarkers: from diagnosis to protection. Infect Dis Report 2016; 8(2): 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. du Preez K, Schaaf HS, Dunbar R, et al. Closing the reporting gap for childhood tuberculosis in South Africa: improving hospital referrals and linkages. Public Health Action 2020; 10(1): 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manyazewal T, Woldeamanuel Y, Holland DP, et al. Electronic pillbox-enabled self-administered therapy versus standard directly observed therapy for tuberculosis medication adherence and treatment outcomes in Ethiopia (SELFTB): protocol for a multicenter randomized controlled trial. Trials 2020; 21(1): 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chemeda A, Abebe T, Ameni G, et al. Utility of urine as a clinical specimen for the diagnosis of pulmonary tuberculosis in people living with HIV in Addis Ababa, Ethiopia. J Clin Tuberc Other Mycobact Dis 2019; 17: 100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nadiah Z, Koesoemadinata RC, McAllister SM, et al. Early chest X-ray in persons with presumptive tuberculosis increases Xpert® MTB/RIF diagnostic yield and efficiency. Public Health Action 2020; 10(1): 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Honeyborne I, McHugh TD, Phillips PP, et al. Molecular bacterial load assay, a culture-free biomarker for rapid and accurate quantification of sputum Mycobacterium tuberculosis bacillary load during treatment. J Clin Microbiol 2011; 49(11): 3905–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mtafya B, Sabiiti W, Sabi I, et al. Molecular bacterial load assay concurs with culture on NaOH-induced loss of mycobacterium tuberculosis viability. J Clin Microbiol 2019; 57(7): 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nair S, Oommen S, Pai V. Evaluation of culture methods for isolation of mycobacterium tuberculosis complex and its resistance among pulmonary tuberculosis cases in a tertiary care setting, Kerala: a pilot study. Int J Curr Microbiol App Sci 2018; 7(11): 452–460. [Google Scholar]
- 21. Honeyborne I, Mtafya B, Phillips PP, et al. The molecular bacterial load assay replaces solid culture for measuring early bactericidal response to antituberculosis treatment. J Clin Microbiol 2014; 52(8): 3064–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sabiiti W, Azam K, Kuchaka D, et al. Improving diagnosis and monitoring of treatment response in pulmonary tuberculosis using the molecular bacterial load assay (MBLA). Biorxiv 2019: 555995. [Google Scholar]
- 23. Svensson RJ, Sabiiti W, Kibiki GS, et al. Model-based relationship between the molecular bacterial load assay and time to positivity in liquid culture. Antimicrob Agent Chemother 2019; 63(10): e00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mbelele PM, Mpolya EA, Sauli E, et al. Mycobactericidal effects of different regimens measured by molecular bacterial load assay among people treated for multidrug-resistant tuberculosis in Tanzania. J Clin Microbiol 2021; 59(4): e02927–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sabiiti W, Ntinginya N, Kuchaka D, et al. Molecular bacterial load assay: a fast and accurate means for monitoring tuberculosis treatment response. BMJ Global Health 2017; 2(2): A8. [Google Scholar]
- 26. Hai HT, Vinh DN, Thu DDA, et al. Comparison of the Mycobacterium tuberculosis molecular bacterial load assay, microscopy and GeneXpert versus liquid culture for viable bacterial load quantification before and after starting pulmonary tuberculosis treatment. Tuberculosis 2019; 119: 101864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ribón W. Mycobacterium tuberculosis: biorisk, biosafety and biocontainment. In:Cardona PJ. (Ed.) Understanding Tuberculosis: Global Experiences and Innovative Approaches to the Diagnosis. London: IntechOpen, 2012, p. 203. [Google Scholar]
- 28. Sabiiti W, Mtafya B, De Lima DA, et al. A tuberculosis molecular bacterial load assay (TB-MBLA). J Vis Exp 2020(158): 10379160460. [DOI] [PubMed] [Google Scholar]
- 29. World Health Organization. WHO Global Tuberculosis report 2018. Geneva: WHO, 2018, https://apps.who.int/iris/handle/10665/274453 [Google Scholar]