Abstract
Immunoglobulin class switch recombination (CSR) occurs in activated B cells with increased mitochondrial mass and membrane potential. Transcription factor Yin Yang 1 (YY1) is critical for CSR and for formation of the DNA loops involved in this process. We therefore sought to determine if YY1 knockout impacts mitochondrial gene expression and mitochondrial function in murine splenic B cells, providing a potential mechanism for regulating CSR. We identified numerous genes in splenic B cells differentially regulated when cells are induced to undergo CSR. YY1 conditional knockout caused differential expression of 1129 genes, with 59 being mitochondrial-related genes. ChIP-seq analyses showed YY1 was directly bound to nearly half of these mitochondrial-related genes. Surprisingly, at the time when YY1 knockout dramatically reduces DNA loop formation and CSR, mitochondrial mass and membrane potential were not significantly impacted, nor was there a significant change in mitochondrial oxygen consumption, extracellular acidification rate, or mitochondrial complex I or IV activities. Our results indicate that YY1 regulates numerous mitochondrial-related genes in splenic B cells, but this does not account for the impact of YY1 on CSR or long-distance DNA loop formation.
Keywords: Class switch recombination, Immunoglobulin, Mitochondria, Transcription, YY1
Introduction
Yin Yang 1 (YY1) is a ubiquitously expressed zinc finger transcription factor that plays fundamental roles in gene expression, proliferation, differentiation, embryogenesis, replication, and apoptosis [1–35]. YY1 was first identified as a key regulator of viral genes, ribosomal protein genes, and the immunoglobulin kappa (Igκ) 3′ enhancer [28, 35, 36], and regulates a huge variety of genes. YY1 can also mediate mammalian Polycomb Group function by recruiting components of the Polycomb Group complexes to DNA [3, 24, 25, 32, 37, 38]. Consistent with its diverse functionality, genetic ablation of the yy1 gene in mice results in peri-implantation lethality [39]. Conditional deletion in the B-cell lineage (via mb1-CRE expression) results in developmental arrest at the pro-B cell stage [40], and other CRE drivers have shown that YY1 is crucial for multiple B-cell stages, including germinal center B-cell development [4, 20, 30].
YY1 is a key factor for controlling long-distance DNA loops between regulatory DNA sequences in a variety of contexts including the Vh, Vκ, and Th2 cytokine loci, as well as regulatory sequences within numerous cell types [17, 31, 40–43]. The Ig genes are prime examples of the importance of long-distance DNA interactions as their somatic rearrangement requires YY1-dependent linkage of DNA sequences separated by as much as three megabases [9, 25, 37, 40, 41, 44]. Similarly, Ig class switch recombination (CSR) requires formation of cytokine-inducible loops between switch region DNA sequences and a large 220 kb DNA loop synapse between the IgH Eμ and 3’RR enhancers [45]. Interestingly, YY1 conditional knockout dramatically reduces CSR and long-distance DNA loops required for CSR [24, 33].
Several studies have implicated YY1 as a regulator of mitochondrial-related gene expression [4, 20, 46]. Skeletal-muscle-specific YY1 knockout (YY1mKO) mice show severely defective mitochondrial morphology, decreased oxidative function associated with exercise intolerance, signs of mitochondrial myopathy, and short stature. These muscle-specific effects are believed to be due to regulation by YY1-PGC-1α transcriptional complexes via mTOR signaling [46]. RNA-seq data indicate that YY1 controls genes are involved in mitochondrial bioenergetics in addition to many other genes [20].
Mitochondria are essential hubs of metabolic activity, regulating various cellular signaling pathways. Many studies have shown links between immune function and mitochondrial processes [47–55]. Recently, a strong association between individual B-cell fates and mitochondrial functions was shown by Jang et al. [56]. This work showed that CSR occurs specifically in activated B cells with increased mitochondrial mass and membrane potential, whereas plasma cell differentiation occurs in cells with decreased mitochondrial mass and potential [56]. Other studies showed lowered glucose or inhibitors of oxidative phosphorylation impacts CSR [57]. We previously showed that although YY1 is critical for CSR and the DNA loops involved in this process, deletion of YY1 does not alter expression of genes directly involved in CSR [4, 24, 33]. In light of the importance of mitochondrial function for lymphocyte activities, and the increased mitochondrial mass and potential associated with CSR, we sought to determine if YY1 knockout would impact mitochondrial gene expression and function in splenic B cells, providing a potential mechanism for regulating CSR.
Using RNA-seq approaches, we identified many genes in splenic B cells that are differentially regulated when cells are induced to undergo CSR. We found that conditional YY1 knockout impacted expression of numerous genes, and 59 of these genes are involved in mitochondrial function, including lipid metabolism, amino acid metabolism, and oxidative phosphorylation. Surprisingly, we found that at the time when YY1 knockout dramatically reduces DNA loop formation and CSR, mitochondrial mass and membrane potential are not significantly impacted, nor is there a significant change in mitochondrial oxygen consumption, extracellular acidification rate (ECAR), or function of mitochondria complex I or IV, although there is a small shift toward glycolysis. Our results indicate that YY1 regulates numerous mitochondrial-related genes in splenic B cells, but this does not account for the impact of YY1 on CSR, or long-distance DNA loop formation.
Results
YY1 controls numerous mitochondrial-related genes in splenic B cells
Deletion of YY1 in splenic B cells results in a dramatic reduction in Eμ-3’RR enhancer long-distance DNA loops, nuclear activation-induced cytidine deaminase (AID) levels, as well as immunoglobulin heavy chain (IgH) CSR [24, 33, 34]. To determine if YY1 might regulate CSR through regulation of mitochondrial-related genes, we performed RNA-seq analyses comparing naïve splenic B cells with those induced to undergo CSR by treatment with bacterial lipopolysaccharide (LPS) plus IL4 before and after conditional knockout of YY1.
A total of 5540 genes were differentially regulated upon LPS plus IL4 stimulation of splenic B cells (which induces CSR to IgG1), with 2906 genes increasing expression and 2634 genes decreasing expression (Fig. 1, bottom bracket heatmap panels; Supporting Information Table 1). Gene ontology (GO) analysis identified terms of differentially regulated gene clusters as those involved in cell cycle, mitotic chromosome condensation, and cholesterol biosynthetic processes (Table 1). To determine genes regulated by YY1, we used an ex vivo deletion system that deletes the yy1 gene through action of recombinant TAT-CRE protein [4, 24, 33]. Using yy1f/f mice, which contain the first yy1 exon flanked by loxP sites [33, 34, 39, 40], splenic B cells were isolated, and cells were either mock treated or treated with recombinant TAT-CRE protein to delete the yy1 gene [33], and then cultured in LPS plus IL4 for 3 days to induce CSR. During this time, YY1 protein decays to nearly undetectable levels and CSR drops dramatically [33] (see also Supporting Information Fig. 1 and 2B), coincident with significant drops in long-distance DNA loops between the Eμ and 3’RR enhancers [24, 33, 34]. RNA was prepared from these samples for RNA-seq analyses.
Figure 1.
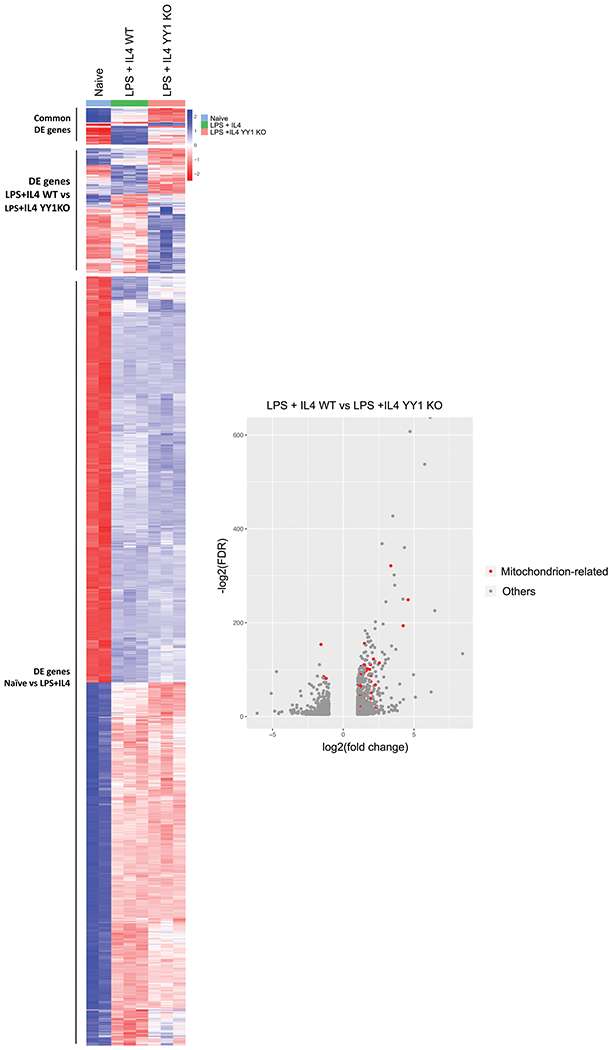
Differential expression in murine splenic B cells of genes induced by LPS plus IL4 treatment and by YY1 knockout. RNA was isolated from naive yy1f/f splenic B cells, or cells stimulated with LPS plus IL4 for 3 days and prepared for RNA-seq analyses. Genes that were differentially regulated twofold or greater with a FDR of <0.05 are shown in the heatmap and in Supporting information Table 1. In addition, isolated splenic B cells from yy1f/f mice were either mock treated or treated with recombinant TAT-CRE protein to delete the yy1 gene. Cells were stimulated with LPS plus IL4 for 3 days and RNA was isolated for RNA-seq analyses. Differentially regulated genes are shown in the heatmap and in Supporting Information Table 2. Volcano plots show genes that increase (left) or decrease (right) after YY1 knockout. Red dots represent mitochondrial-related genes. RNA-seq was performed in triplicate from three independent experiments.
Table 1.
GO analysis of Differentially Regulated Genes in Naïve vs LPS + IL4 Splenic B Cells
| Annotation cluster | GO terms | Gene number | P value |
|---|---|---|---|
| Annotation cluster 1 | Cell cycle | 203 | 1.88E-39 |
| Cell division | 143 | 1.58E-35 | |
| Mitotic nuclear division | 118 | 1.45E-34 | |
| Annotation cluster 2 | Mitotic chromosome condensation | 9 | 5.26E-05 |
| Chromosome condensation | 8 | 2.30E-03 | |
| Meiotic chromosome condensation | 4 | 1.74E-02 | |
| Annotation cluster 3 | Cholesterol biosynthetic process | 13 | 3.76E-04 |
| Sterol biosynthetic process | 11 | 1.29E-03 |
After YY1 deletion, a total of 1129 genes were differentially regulated with expression of 539 genes increasing and 590 genes decreasing (Fig. 1, middle bracket of heatmap, Supporting Information Table 2). GO analysis of differentially expressed genes identified top functions as mitochondrion, metal binding, and tumor necrosis factor (Table 2). Fifty-nine differentially regulated genes after YY1 knockout were mitochondrial related, and expression of 51 genes decreased after YY1 deletion, and eight increased (Fig. 1, Volcano plot, and Table 3). Thus, most of these mitochondrial-related genes are positively regulated by YY1 in splenic B cells after induction of CSR. Of the genes that change expression upon YY1 knockout, 10 are involved in sugar and fatty acid metabolism, 11 in amino acid metabolism, eight in oxidative phosphorylation, nine in mitochondrial DNA or nucleotide metabolism, five in mitochondrial membranes, one in mitochondrial potential maintenance, and one in mitochondrial fusion, with other functions scattered among ion transport, cell signaling, or tRNA metabolism (11 genes) (Table 3). Fatty acid beta-oxidation genes were among the most commonly deregulated genes after YY1 knockout (Eci1, Hsd17b10, Ech1, EtfB, Echs1, Decr1), as were genes involved in oxidative phosphorylation electron transport (Ndufaf6, Coq4, Atp5s1, Ndufc1, and Cox20). Genes involved in amino acid metabolism were the second highest number of deregulated genes. Our RNA-seq data were verified by qPCR for a subset of transcripts that are differentially regulated after YY1 knockout, and this was further confirmed by western blots (Supporting Information Fig. 2A and B). In addition, we evaluated transcripts by preparing RNA from IgG1 switched verses unswitched cells. There was little difference in expression of selected genes between these two groups of cells, indicating that YY1 loss impacted gene expression changes in both switched and unswitched cells (Supporting Information Fig. 2C). YY1 and switch γ1 transcripts served as controls.
Table 2.
GO Analysis of Genes Regulated by YY1 Knockout in Splenic B Cells
| Annotation cluster | GO terms | Gene number | P value |
|---|---|---|---|
| Annotation cluster 1 | Mitochondrion | 59 | 2.32E-05 |
| Transit peptide | 22 | 1.41E-04 | |
| Transit peptide:Mitochondrion | 21 | 1.01E-03 | |
| Annotation cluster 2 | Metal-binding | 78 | 2.44E-03 |
| Zinc-finger | 42 | 2.70E-03 | |
| Zinc | 48 | 2.21E-02 | |
| Metal ion binding | 77 | 2.49E-02 | |
| Annotation cluster 3 | Tumour necrosis factor | 4 | 7.08E-04 |
| Tumor necrosis factor receptor binding | 4 | 2.05E-02 | |
| Defense response to Gram-positive bacterium | 6 | 2.50E-02 |
Table 3.
Mitochondrial-related genes regulated by YY1 arranged by function
| Name | Log Fold Change | False Discovery Rate | Promoter Binding | Mitochondrial Function |
|---|---|---|---|---|
| Fatty Acid Metabolism (70% direct YY1 targets) | ||||
| Eci1 | 4.58 | 1.17E-75 | +++ | Fatty acid beta oxidation |
| Hsd17b10 | 3.36 | 1.89E-97 | + | Fatty acid and steroid oxidation |
| Ech1 | 2.55 | 3.74E-35 | ++++ | Fatty acid beta oxidation |
| Hint2 | 2.32 | 1.29E-06 | + | Steroid biosynthesis |
| Pccb | 1.97 | 5.68E-27 | ++++ | Catabolism of propionyl CoA |
| Acot2 | 1.95 | 3.17E-12 | + | Reg levels of Acyl-CoA |
| Echs1 | 1.92 | 4.79E-28 | Fatty acid beta-oxidation | |
| Etfb | 1.84 | 0.00041 | Fatty Acid and AA catabolism | |
| Mpdu1 | 1.57 | 5.59E-37 | ++++ | Synth of Lipid linked Oligosacc |
| Decr1 | 1.18 | 1.06E-18 | Fatty acid beta-oxidation | |
| Amino Acid Metabolism (27% direct YY1 targets) | ||||
| Ivd | 1.92 | 4.67E-34 | Leucine catabolism | |
| Dbt | 1.72 | 1.26E-16 | + | Breakdown of branched AAs |
| Phykpl | 1.61 | 3.60E-30 | Lysine metabolism | |
| Hmgcl | 1.45 | 4.27E-13 | + | Leucine degradation |
| Mapk3 | 1.23 | 1.64E-14 | Cell signaling | |
| Kmo | 1.23 | 1.02E-18 | Tryptophan metabolism | |
| Bcat1 | 1.21 | 3.00E-20 | Amino acid metabolism | |
| Mpst | 1.16 | 8.02E-08 | ++ | Cysteine metabolism |
| Bckdha | 1.11 | 3.88E-05 | Branched AA metabolishm | |
| 9030617O03Rik | −1.57 | 5.70E-47 | Glutamine metabolism | |
| Slc25a29 | −2.7 | 0.000763 | Mito transporter of basic AA | |
| Nucleic Acid Related (33% direct YY1 targets) | ||||
| Clybl | 2.14 | 1.23E-37 | Vitamin B12 metabolism | |
| Qtrt1 | 1.50 | 3.66E-15 | tRNA synthesis | |
| Mrps18b | 1.48 | 1.24E-29 | ++++ | Ribosomal protein |
| Wars2 | 1.14 | 8.76E-14 | tRNA synthesis | |
| Clpb | 1.12 | 2.80E-13 | ATPase | |
| Dcps | 1.10 | 2.40E-21 | mRNA capping | |
| Dhodh | 1.08 | 3.77E-08 | + | Pyrimidine metabolism |
| Slc25a24 | −1.11 | 1.09E-05 | ATP-Mg membrane transporter | |
| Qrsl1 | −1.38 | 2.52E-26 | ++++ | Gln-tRNA formation |
| Oxidative Phosphorylation/Energy Production (63% direct YY1 targets) | ||||
| Ndufaf6 | 2.27 | 4.85E-21 | ++++ | Ox-Phos. Assembly Complex 1 |
| Coq4 | 1.71 | 2.51E-09 | CoQ biosynth, Electron transp. | |
| Suclg2 | 1.49 | 5.51E-34 | TCA cycle | |
| Prdx3 | 1.26 | 6.45E-33 | + | Protects from oxidative stress, peroxide reductase |
| Ckb | 1.24 | 3.65E-06 | Energy homeostasis | |
| Atp5sl | 1.21 | 1.51E-20 | +++ | Ox Phos Complex 1 |
| Ndufc1 | 1.20 | 1.46E-08 | ++++ | Complex 1 subunit |
| Cox20 | 1.16 | 2.20E-11 | ++++ | Ox Phos Cytochrome C oxidase |
| Mitochondrial Membranes (43% direct YY1 targets) | ||||
| Nipsnap1 | 4.23 | 5.75E-59 | Mito DNA maint, vesic transp. | |
| Chchd10 | 1.87 | 5.29E-31 | Cristae morphology | |
| Alkbh7 | 1.77 | 0.027002 | + | Controls mito memb potential |
| Miga1 | 1.70 | 2.58E-31 | ++++ | Controls Mitochondrial fusion |
| Vat1 | 1.63 | 6.91E-10 | + | Vesicle transport |
| Letmd1 | 1.07 | 2.06E-18 | Mito outer membrane protein | |
| Tspoap1 | −1.19 | 4.41E-14 | Vesicle release | |
| Signal Transduction/Cell Cycle (67% direct YY1 targets) | ||||
| Pebp1 | 1.33 | 8.40E-13 | + | Signal transduction: inhib Raf |
| Raf1 | 1.08 | 1.02E-18 | ++++ | Cell signaling |
| Cdkn2a | −1.21 | 3.49E-25 | Cell cycle regulation | |
| Other Functions (18% direct YY1 targets) | ||||
| As3mt | 1.99 | 6.46E-16 | Arsenite methyltransferase | |
| Nefh | 1.95 | 3.70E-23 | Neurofilament | |
| Sfxn1 | 1.50 | 1.21E-47 | +++ | Iron transport |
| Cd36 | 1.27 | 4.35E-16 | Cell adhesion | |
| Spryd4 | 1.26 | 8.62E-08 | ++ | Unknown |
| Fam136a | 1.24 | 5.39E-28 | Unknown | |
| Thra | 1.17 | 0.000561 | Nuclear hormone receptor | |
| Pyurf | 1.09 | 4.83E-17 | Unknown | |
| Dnajc30 | 1.05 | 0.000516 | Chaperone | |
| Lrrc24 | −1.03 | 0.002475 | Protein modification | |
| Sox4 | −1.32 | 0.002928 | Transcription factor | |
Direct and indirect regulation of mitochondria-related genes
The very strong representation of mitochondrial-related genes differentially expressed after YY1 knockout suggested that YY1 directly binds to the promoters of a number of these genes. To test this, we performed ChIP-seq experiments on chromatin isolated from splenic B cells. YY1 ChIP-seq analyses showed that YY1 bound to 3069 locations in the genome (Supporting Information Table 3). YY1 bound to a larger fraction of genes that were upregulated after YY1 knockout (i.e., repressed by YY1) than were activated by YY1 (Fig. 2A, gray shading). The largest fraction of YY1 binding sites in splenic B cells was in promoter regions (Fig. 2B and C). YY1 bound to 2179 promoters, and the top-enriched GO term of these 2179 genes was “Mitochondrion” (371 genes belonged to this term, Table 4). Other GO terms of genes that bind to YY1 were ribosome, transcription, RNA processing, DNA repair, cell cycle, helicase activity, and mitochondrial respiratory chain complex 1 (Table 4). A large fraction of YY1 binding sites co-localized with repressive histone mark H3K9me3 (18.1%, Supporting Information Table 3). YY1 also overlapped with repressive mark H3K27me3 (3.6%) and H3K27me2 (6.3%) (Supporting Information Table 3). In addition, YY1 overlapped many sites with histone modifications indicative of transcriptional activity including H3K9ac (10.5%), H3K4me3 (7.6%), H3K4ac (5.5%), and H3K27ac (7.9%) (Supporting Information Table 3). These patterns are consistent with the role of YY1 as both a transcriptional repressor and a transcriptional activator [1, 2, 28, 35, 36, 58, 59]
Figure 2.
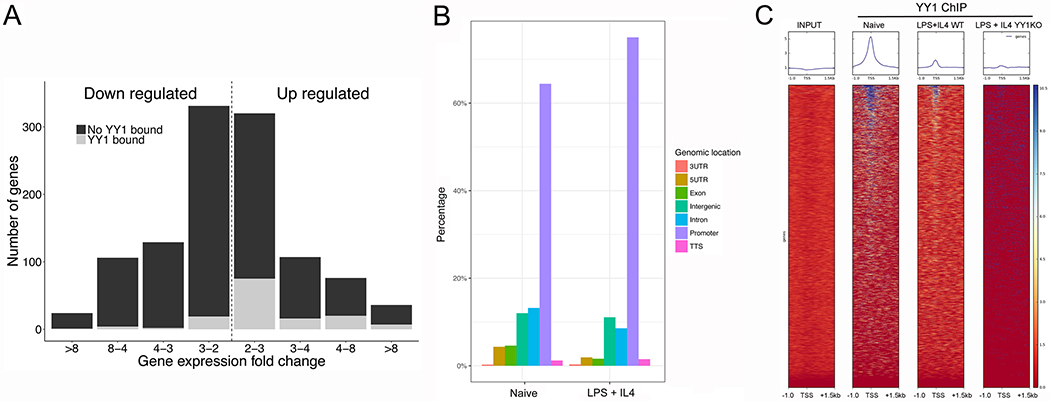
YY1 predominately binds to promoters in murine splenic B cells. (A) YY1 binding is plotted in relation to change in expression levels after YY1 knockout and whether YY1 bound to promoter regions (gray shading) or had no YY1 binding (black shading). (B) YY1 Chip-seq data are plotted according to binding site locations within the genome. (C). YY1 localization is plotted in the 2.5 kb region surrounding known promoters. Data are representative of three independent ChIP-seq experiments. The input lane is DNA from naïve splenic B cells.
Table 4.
GO Analysis of Genes That Bind to YY1
| Annotation cluster | GO terms | Gene number | P value |
|---|---|---|---|
| Annotation cluster 1 | Mitochondrion | 371 | 1.15E-34 |
| Transit peptide | 152 | 1.20E-31 | |
| Annotation cluster 2 | Ribonucleotprotein complex | 119 | 3.15E-32 |
| Ribosome | 83 | 1.30E-28 | |
| Annotation cluster 3 | Transcription | 350 | 2.30E-18 |
| Regulation of Transcription | 394 | 4.70E-15 | |
| DNA binding | 277 | 1.03E-05 | |
| Annotation cluster 4 | RNA processing | 73 | 5.05E-17 |
| Spliceosomal complex | 47 | 4.53E-09 | |
| Catalytic step 2 spliceosome | 34 | 4.53E-09 | |
| mRNA splicing, via spliceosome | 30 | 4.48E-05 | |
| Annotation Cluster 5 | Metal ion binding | 495 | 7.10E-09 |
| Zinc ion binding | 169 | 6.56E-05 | |
| Annotation Cluster 6 | Nucleotide binding | 361 | 6.45E-21 |
| ATP binding | 247 | 3.20E-08 | |
| Annotation Cluster 7 | Cellular response to DNA damage stimulus | 102 | 4.23E-12 |
| DNA repair | 79 | 4.43E-10 | |
| Annotation Cluster 8 | Cell cycle | 133 | 1.29E-11 |
| Mitotic nuclear division | 67 | 3.23E-08 | |
| Cell division | 83 | 4.18E-08 | |
| Annotation Cluster 9 | Protein transport | 137 | 3.39E-14 |
| Annotation Cluster 10 | Helicase activity | 40 | 1.58E-08 |
| ATP-dependent RNA helicase activity | 23 | 1.45E-06 | |
| RNA secondary structure unwinding | 16 | 1.57E-04 | |
| Annotation Cluster 11 | NADH dehydrogenase activity | 10 | 2.00E-06 |
| Respiratory chain | 21 | 4.59E-06 | |
| Mitochondrial respiratory chain complex I | 18 | 1.14E-05 | |
| NADH dehydrogenase (ubiquinone) activity | 13 | 4.33E-04 | |
| Oxidoreductase activity, acting on NAD(P)H | 6 | 0.008826 |
We evaluated YY1 binding to the 59 mitochondrial-related genes that are differentially regulated after YY1 knockout. YY1 directly bound to the promoters of 25 of these genes, with binding to 15 being particularly strong, and binding of the remaining 10 genes being somewhat weaker (Fig. 3; Table 3). The pattern of binding over genes involved in fatty acid metabolism was particularly strong (70% of differentially regulated genes in this category directly bound to YY1) (Fig. 3; Table 3). In addition, 63% of differentially regulated genes involved in oxidative phosphorylation directly bound YY1. Thus, our ChIP-seq data indicate that YY1 directly binds to the promoters of 42% of mitochondrial-related genes regulated by YY1 in splenic B cells. The remaining 34 mitochondrial-related genes regulated by YY1 must be controlled by either indirect mechanisms or by binding to sites distal to the 2500 bp surrounding the promoters we evaluated.
Figure 3.
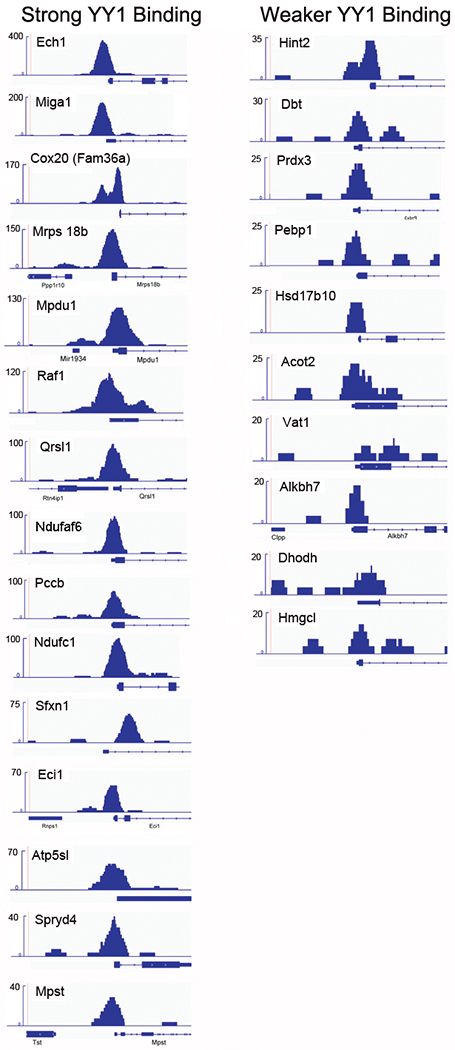
YY1 binds to mitochondrial-related promoters. ChIP-seq data from naïve splenic B cells were evaluated over the promoters of the 59 mitochondrial-related genes that show differential gene expression after YY1 knockout. Fifteen promoters showed particularly strong YY1 binding defined as 40 or higher on the Y-axis scale (left). Ten promoters bound YY1 more weakly, defined as between 20 and 35 on the Y-axis scale (right). The ChIP-seq data are representative of two independent ChIP-seq experiments.
Mitochondrial functions after YY1 knockout in splenic B cells
We set out to determine the impact of YY1 knockout on mitochondrial function in B cells induced to undergo CSR. Murine CD23+ splenic B cells were either mock treated or treated with recombinant TAT-CRE protein to delete the endogenous yy1 gene ex vivo, and then stimulated with LPS plus IL4 to induce CSR. Consistent with our published work [33], YY1 protein levels were greatly reduced 3 days after TAT-CRE treatment, but there was no impact on cell proliferation (Supporting Information Figs. 2B and 3A). While CSR dropped dramatically, there was no change in cell viability, or levels of B-cell activation markers CD69 or CD95 (Fig. 4A). There was, however, a small increase in the level of annexin V staining (Supporting Information Fig. 3B). Mitochondrial mass and potential were quantitated using MitoTracker Green and DeepRed dyes, respectively. We found no significant difference between mock (labeled as control) and TAT-CRE (labeled as YY1 KO) treated cells (Fig. 4B). We also evaluated IgG1 switched compared to unswitched cells, as well as differentiation to CD138 high plasmablasts/plasma cells. Again, while CSR dropped dramatically in TAT-CRE-treated samples (Fig. 4C), there was no difference in mitochondrial mass or potential comparing switched to unswitched cells (Fig. 4C and D; IgG1−CD138− vs. IgG1+CD138−). Differentiation to CD138 high plasmablasts/plasma cells was reduced after YY1 knockout (Fig. 4C, IgG1−, CD138+ cells), and these cells showed a small but significant increase in mitochondrial mass (Fig. 4D).
Figure 4.
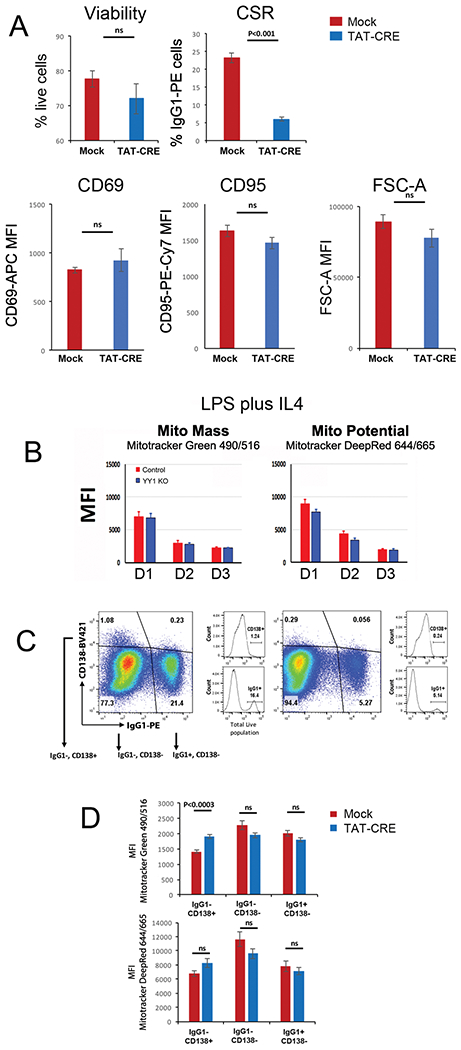
YY1 knockout does not impact mitochondrial mass or potential at day 3. Splenic B cells from yy1f/f mice were either mock treated (red bars) or treated with recombinant TAT-CRE protein to delete the yy1 gene (blue bars). Cells were stimulated with LPS plus IL4 for 3 days. (A) Percentage of live cells was evaluated by Zombie Aqua™ dye or DAPI, and percent CSR to IgG1 was determined by flow cytometry with anti-IgG1 antibody. Experiments are from 15 independent experiments with one mouse per experiment and statistical significance was determined by a two-tailed Student’s t-test. Levels of CD69 and CD95 were determined by flow cytometry with anti-CD69 and anti-CD95 antibodies. Experiments are from three experiments and statistical significance was determined by a two-tailed Student’s t-test. (B) Cells were evaluated for mitochondrial mass or potential by flow cytometry analysis with MitoTracker red or green, respectively, on days 1, 2, or 3. None of the changes reached statistical significance. Data are from four independent experiments with two animals per experiment. (C and D) On day 3, cells were evaluated by flow cytometry for IgG1 and CD138 expression, and cells in IgG1− CD138+, IgG1− CD138−, or IgG1+ CD138− fractions were evaluated for mitochondrial mass and potential by flow cytometry using MitoTracker green or red. The CD138− populations in (C) and (D) are a combination of CD138 intermediate and low cells. Thus, IgG1− CD138+ cells in the upper left quadrant of panel (C) are analyzed in (D); the IgG1− and CD138− cells in the lower left quadrant of (C) are analyzed in (D), and the IgG1+ CD138− cells in the lower right quadrant in (C) are analyzed in (D). Mitotracker DeepRed falls under the APC channel (644/665 exc/emi) and Mitotracker Green falls under the FITC channel (490/516; exc/emi). Results are from three independent experiments with two mice per experiment. Shown are the mean and standard deviation. Statistical significance was determined by a two-tailed Student’s t-test.
Mitochondrial function was also assessed through extracellular flux analysis. The oxygen consumption rate (OCR; a measure of oxidative phosphorylation) and ECAR (a measure of aerobic glycolysis) profiles were determined in LPS-stimulated splenic B cells before and after YY1 knockout. Basal oxygen consumption was unaffected by YY1 knockout (Fig. 5A). Similarly, drops in ECAR were not significant (Fig 5B), nor was there a drop in basal respiration or protein leak (Fig. 5C). Derived measures calculated after adding inhibitors showed modest drops in maximal and spare respiratory capacity after YY1 knockout, although these drops did achieve statistical significance (Fig. 5C).
Figure 5.
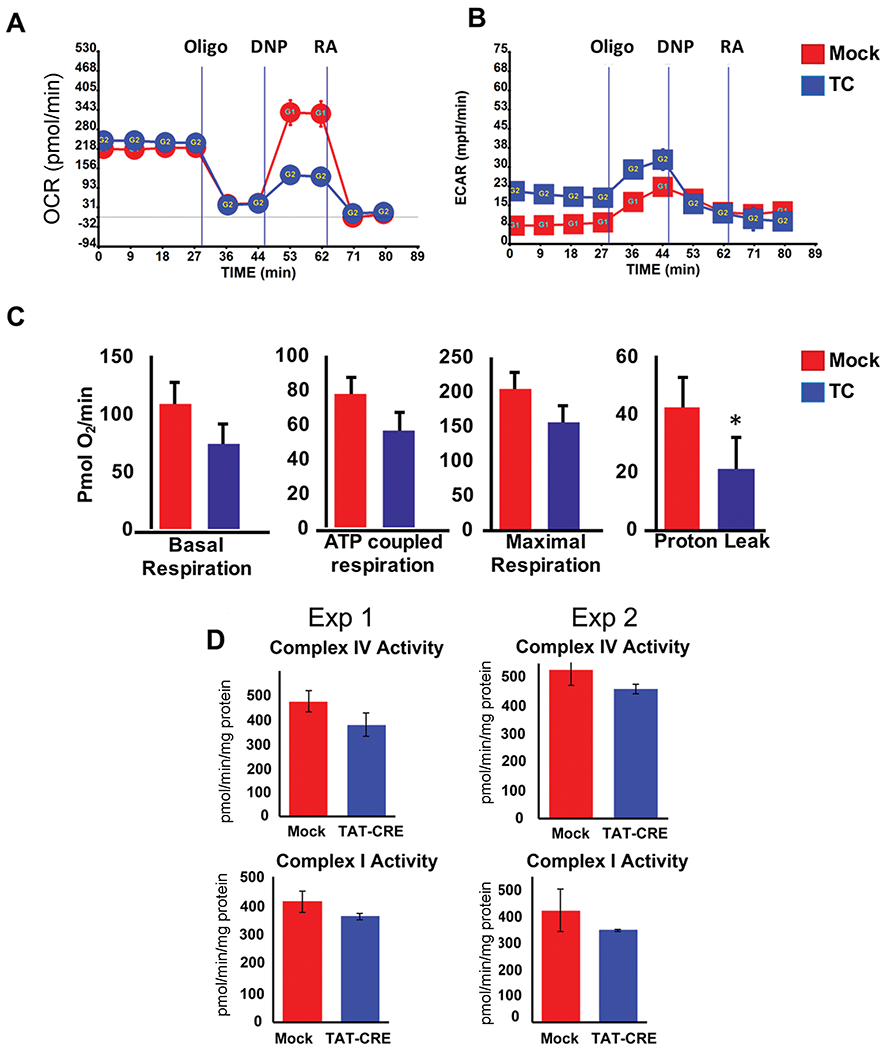
YY1 knockout ex vivo minimally impacts mitochondrial respiration during the first three days. Splenic B cells from yy1f/f mice were either mock treated or treated with recombinant TAT-CRE protein to delete the yy1 gene. Cells were stimulated with LPS for 3 days then evaluated for (A) oxygen consumption rate, (B) extracellular acidification rate, (C) basal respiration, ATP coupled respiration, maximal respiration, or proton leak Seahorse Extracellular Flux (XF) Analyzer (Seahorse Bioscience, Billerica, MA, USA). Data are expressed as mean ± SEM of four values, and representative of three independent experiments with two mice per experiment. (D) Mitochondrial complex activities (complexes I and IV) were measured using isolated mitochondria from mock and TAT-CRE-treated B cells. Assays were performed in triplicate and repeated twice. Differences in each experiment did not reach statistical significance.
To measure mitochondrial electron transport chain complex activities, mitochondria were isolated from LPS plus IL4-stimulated splenic B cells and activity of electron transport chain complexes I and IV was performed. We observed only a modest decrease of the activity of each complex following YY1 knockout (Fig. 5D). Further, we performed western blot analyses of various oxidative phosphorylation complex subunits and observed no significant difference of proteins in mock as compared to TAT-CRE-treated cells (Supporting Information Fig. 2D). Similar to published work [57], treatment of splenic B cells with oligomycin A, a mitochondrial poison, resulted in reduced CSR in both mock and TAT-CRE-treated cells (Supporting Information Fig. 4), indicating a likely impact of mitochondria on CSR. Therefore, although YY1 knockout affected numerous mitochondrial genes, in the time-frame studied here, our results showed little influence of YY1 knockout on oxidative phosphorylation, electron transport complex activity, or protein levels. To explore possible changes at later time points, we provided fresh media every other day, which extends the life of our cultures. While there was no significant difference in viability at day 3 after TAT-CRE treatment, we found a rapid loss of viability of YY1 knockout (TAT-CRE treated) compared to mock treated samples at later time points (Fig. 6A). Interestingly, surviving cells that grew out on days 5 and 7 showed increasing percentages of cells that had not deleted YY1 resulting in an increase in CSR through time (Fig. 6B). This corresponded with increased mitochondrial potential on day 5 as well (Fig. 6C). Thus, the loss of YY1 results in considerable cell death at later time points (days 5 and 7). As the impact of YY1 knockout on CSR is demonstrable by day 3 when YY1 knockout cells are viable, proliferating normally, and show no change in mitochondrial functions, loss of CSR at this time is not related to YY1 impact on mitochondrial function.
Figure 6.
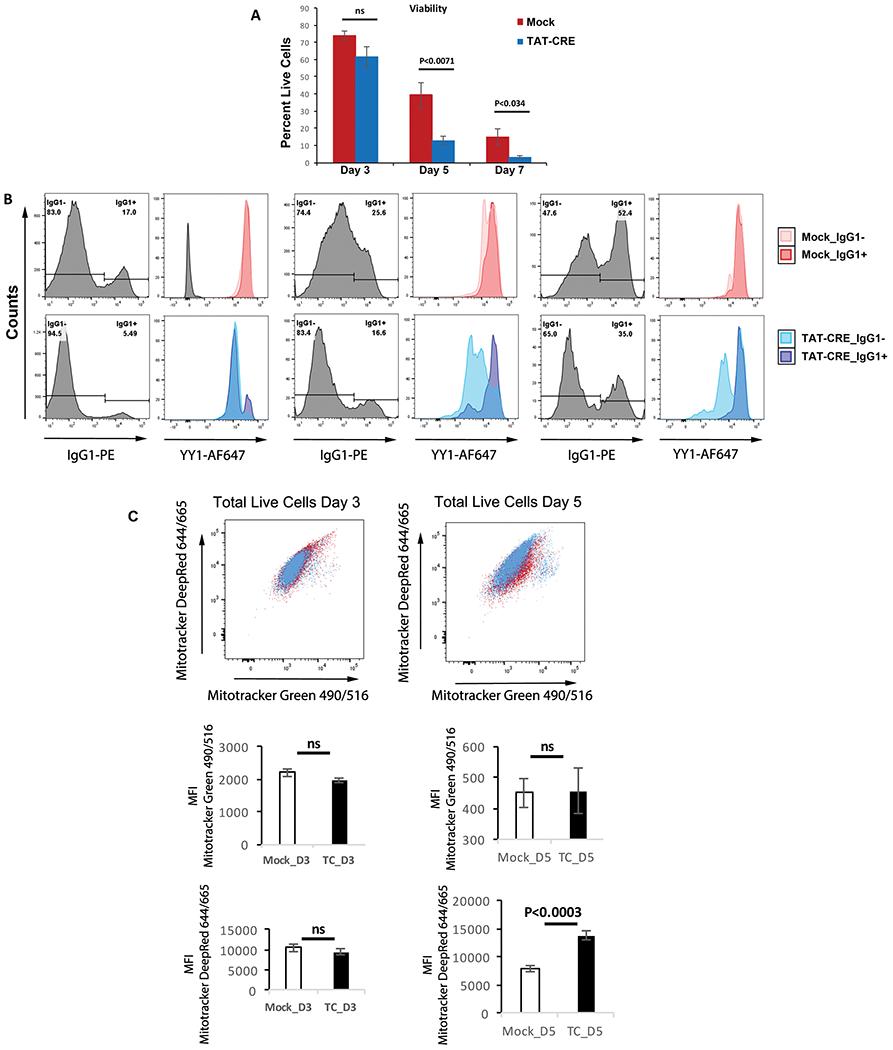
Impact of YY1 knockout on viability, CSR, and mitochondrial function. Splenic B cells from yy1f/f mice were wither mock treated or treated with TAT-CRE followed by stimulation with LPS + IL4 for either 3, 5, or 7 days. (A) Cells were evaluated for viability by flow cytometry with Zombie Aqua™ dye. (B) Levels of CSR or YY1 protein were determined by flow cytometry with either anti-IgG1 or anti-YY1 antibodies. (C) Mitochondrial function was evaluated by staining cells with MitoTracker deep red or MitoTracker green. MitoTracker DeepRed falls under the APC channel (644/665 exc/emi) and MitoTracker Green falls under the FITC channel (490/516; exc/emi). MFI is plotted in the lower panels. Experiments were performed three times with two biological replicates each time, and statistical significance was determined by a two-tailed Students’ t-test.
Various studies have shown that compared to naïve B cells, glucose uptake increases upon B-cell activation with various stimuli [60, 61], and glucose levels can impact CSR [57]. Glucose uptake experiments can measure potential changes in metabolic activity. As our mitochondrial complex activities showed only a modest change, we explored the uptake of the fluorescent glucose analog, 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2-NBDG), in unstimulated and LPS-stimulated cells before and after knockout of YY1. As expected, LPS caused an increase in glucose uptake (Fig. 7A, compare unstimulated mock to LPS mock samples). TAT-CRE-treated unstimulated cells showed a significant increase in uptake compared to mock-treated unstimulated cells (Fig. 7A, two left samples). Comparing LPS-stimulated mock to TAT-CRE samples, there was a 14% drop in glucose uptake suggesting YY1 knockout reduces glucose uptake (Fig. 7A, two right samples, and Fig. 7B, top two flow cytometry panels). Although the drop in glucose uptake was small after YY1 knockout (14%), it did reach statistical significance (p < 0.05) (Fig. 7A).
Figure 7.
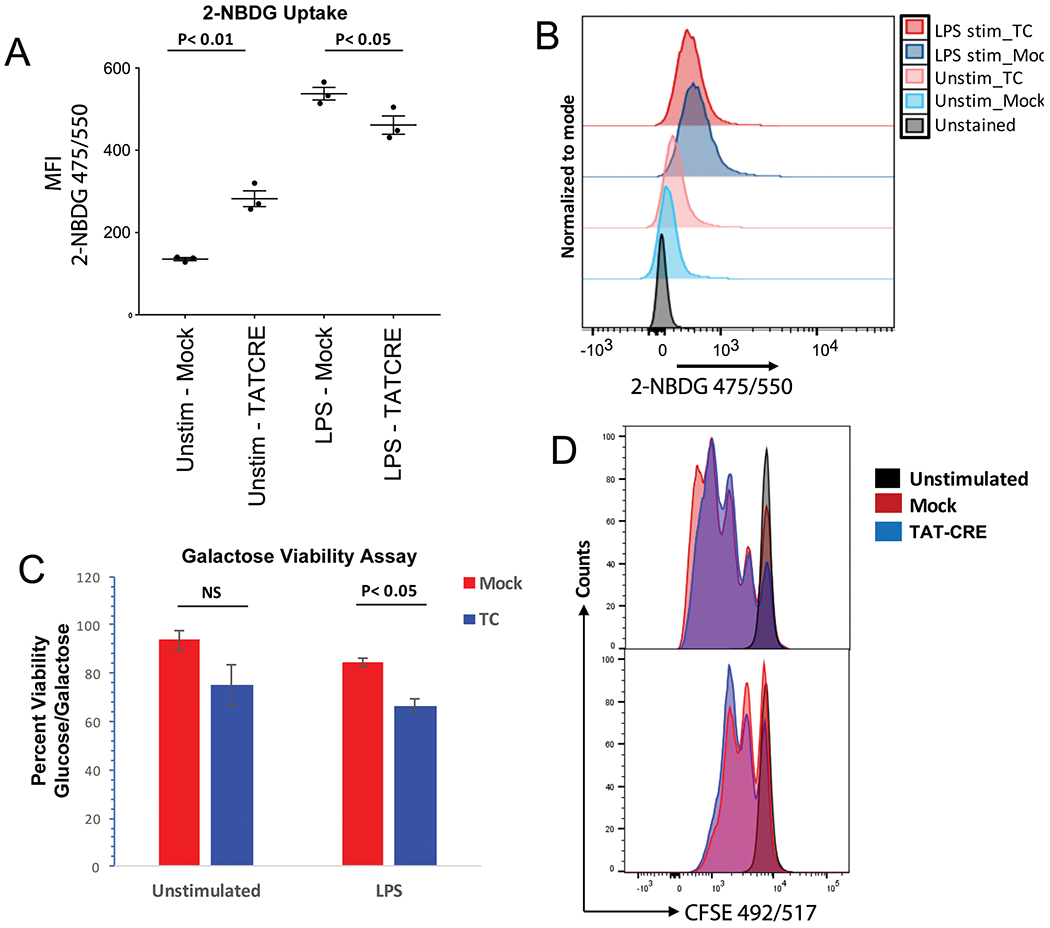
YY1 knockout cells increase their dependence on glycolysis. (A) Splenic B cells from yy1f/f mice were either mock treated or treated with recombinant TAT-CRE protein to delete the yy1 gene. Cells were grown in media containing the glucose analog 2-NBDG and left either unstimulated or stimulated with LPS for 2 days followed by evaluation of glucose uptake by flow cytometry. Data are shown from three independent experiments with three mice per experiment. (B) A representative flow cytometry plot from three independent experiments is shown for 2-NBDG uptake in unstimulated mock and TAT-CRE samples, or LPS-stimulated mock and TAT-CRE samples. (C) Cell viability was measured in the presence of either glucose- or galactose-containing media in mock (red bars) or TAT-CRE-treated cells (blue bars) from either unstimulated, or LPS-treated splenic B cells. Statistical significance (p < 0.05) was observed in the LPS-treated TAT-CRE samples compared to mock samples. Data are taken from three independent experiments and statistical significance was determined by a two-tailed Students’ t-test. (D) CFSE staining and evaluation by flow cytometry of cells in glucose or galactose media. Shown is a representative flow cytometry plot from five independent experiments with one mouse per experiment. 2-NBDG 475/550 (exc/emi) and CFSE (492/517; exc/emi) both fall under the FITC channel.
As there were no major effects on mitochondrial function 3 days after YY1 knockout in B cells, we explored whether YY1 knockout B cells might be more glycolytic for energy production compared to control B cells. Cells that are more dependent upon glycolysis for energy production fail to survive in galactose-containing media, as compared to glucose supplemented media. Therefore, we performed a galactose sensitivity assay to quantitate the effect of a nonmetabolizable reagent on the viability of YY1 knockout cells. Control and YY1 knockout splenic B cells were cultured in RPMI media containing either glucose (1 mg/mL) or galactose (2 mg/mL) for 48 h. We found that YY1 knockout cells were more sensitive to absence of glucose showing reduced viability (Fig. 7C). Thus, YY1 knockout caused a shift toward more glycolytic metabolism. Cells grown in galactose containing media also showed a decrease in proliferation (Fig. 7D).
Mitochondrial mass and potential after YY1 knockout in vivo
Although we observed differential regulation of numerous mitochondrial-related genes after YY1 knockout in primary splenic B cells, our results did not show significant changes in OCR, ECAR, basal respiration, ATP coupled respiration, mitochondrial oxidative complex activities, or maximal respiration. This could be due to the kinetics of our ex vivo conditional knockout system. It takes approximately 2–3 days for YY1 protein levels to decay to nearly undetectable levels after treatment with TAT-CRE [33]. YY1 must first decay to sufficiently low levels to impact gene expression (or other functions), and then expression of mitochondrial-related gene expression will consequently change with further delayed loss of the encoded proteins through protein turnover. Thus, in the time frame of 2–3 days studied here after ex vivo YY1 knockout, changed expression of mitochondrial-related genes may not sufficiently alter mitochondrial function.
To address this, we used yy1f/f CD20TamCRE mice, which can be induced to delete the endogenous yy1 gene in vivo by tamoxifen treatment. These mice, and yy1f/f controls, were injected with tamoxifen over three consecutive days to delete the yy1 gene in vivo in the yy1f/f CD20TamCRE mice, and then splenic B cells were isolated for evaluation of mitochondrial function. As anticipated, naïve B cells from yy1f/f CD20TamCRE mice that did not receive tamoxifen injection, as well as mice injected with the immunogen NP-OVA retained YY1 expression (Supporting Information Fig. 5, far right of top and middle panels). However, consistent with published work [20], injection with tamoxifen for three consecutive days resulted in YY1 deletion and dramatically reduced numbers of germinal center cells in yy1f/f CD20TamCRE mice (Supporting Information Fig. 5, bottom panel). We previously also saw a very dramatic loss of germinal center cells and absence of germinal centers when YY1 was knocked out using gamma1-CRE [4]. Isolated splenic B cells from yy1f/f mice injected with tamoxifen were stimulated with LPS plus IL4, and as this line lacked the TAM-CRE gene, these cells retained YY1 and underwent robust CSR (Supporting Information). On the contrary, splenic B cells from tamoxifen-treated yy1f/f CD20TamCRE mice failed to undergo CSR and lost YY1 expression (Supporting Information).
Unstimulated naïve B cells from tamoxifen-treated yy1f/f CD20TamCRE mice showed a significant drop in mitochondrial mass and potential compared to yy1f/f mice, indicating the YY1 knockout did impact mitochondrial function (Fig. 8). However, similar to our ex vivo YY1 deletion studies, in the context of LPS, LPS plus IL4, or anti-IgM stimulation, there was no significant drop in mitochondrial mass or potential. Thus, these inducers are able to overcome, at least in part, the impact of YY1 knockout on mitochondrial function.
Figure 8.
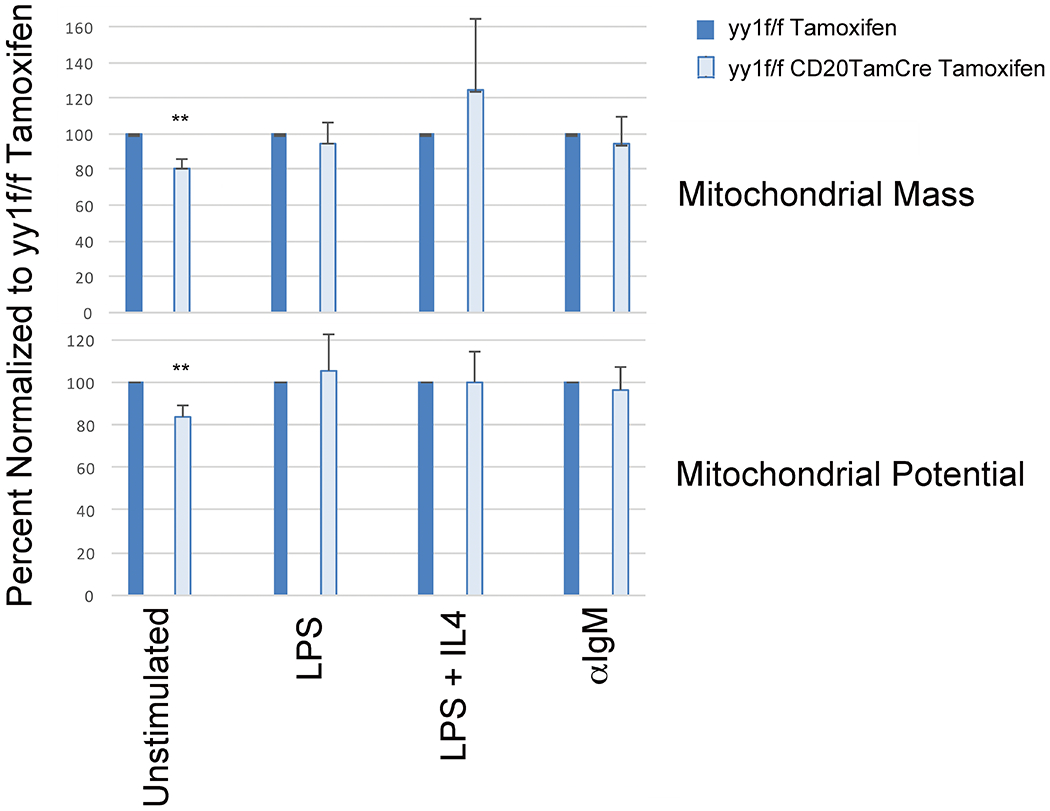
YY1 knockout in vivo impacts mitochondrial mass and potential in naïve splenic B cells, but not in cells stimulated with LPS, LPS plus IL4, or anti-IgM. Mice (yy1f/f or yy1f/f CD20TamCre) were injected for three consecutive days with tamoxifen to induce deletion of the yy1 gene (deletion occurs in yy1f/f CD20TamCre mice, not in yy1f/f mice). Subsequently, splenic B cells were isolated and either left unstimulated (naïve) or stimulated with LPS, LPS plus IL4, or anti-IgM, then evaluated for mitochondrial mass and potential by flow cytometry. Data are mean and with standard deviation taken from three independent experiments with two mice per experiment, and statistical significance was determined by a two-tailed Students’ t-test.
We conclude that YY1 knockout does impact mitochondrial-related gene expression and mitochondrial function in naïve, unstimulated splenic B cells, but in cells induced to undergo CSR, mitochondrial function is minimally affected, and this cannot explain the impact of YY1 knockout on CSR [33], or the formation of long-distance DNA loops [24].
Discussion
We found that induction of splenic B cells to undergo CSR by LPS plus IL4 treatment resulted in differential expression of numerous genes, with 1129 genes differentially regulated by YY1 knockout. Fifty-nine of these genes are related to mitochondrial function, and YY1 bound directly to the promoters of 25 of these genes. YY1 impact on mitochondrial-related gene expression has been noted in other systems. Top pathways in skeletal muscle that are downregulated by muscle-specific YY1 conditional knockout include metabolic and regulatory mitochondrial-related genes [62]. In intestinal stem cells, conditional YY1 knockout results in reduced mitochondrial complex I gene expression and altered energy metabolism [27]. Mitochondrial-related genes were also among the top categories of dysregulated genes after conditional YY1 knockout in pro-B cells, and ChIP-seq data showed enriched YY1 binding at mitochondrial-related promoters [20]. Thus, similar to our work, downregulated genes after YY1 knockout in multiple systems impacted mitochondrial-related genes, and those involved in generation of precursor metabolites, oxidation, and metabolic processes. Our RNA-seq experiments in splenic B cells also showed reduced expression of a number of genes involved in oxidative phosphorylation including those involved with complexes 1 and IV assembly and function.
Our in vivo YY1 deletion studies showed that YY1 knockout causes a significant drop in mitochondrial mass and potential in naïve unstimulated splenic B cells. However, in the context of LPS plus IL4 induction we observed only modest effects on mitochondrial mass and potential, and our ex vivo studies showed no differences in oxygen consumption and extracellular acidification. Although we observed modest decreases in maximal and ATP coupled respiration during flux analysis, mitochondrial complex activities and levels were not significantly altered. Therefore, mitochondria remain active 3 days after YY1 knockout. This was further supported by a drop in CSR following oligomycin A treatment in mock as well as TAT-CRE-treated cells. Thus, mitochondria are not compromised 3 days after YY1 knockout in splenic B cells.
Glucose levels can impact CSR [57], but our glucose uptake experiments showed only a small drop in uptake after YY1 deletion (14%), though this small drop did achieve statistical significance. This small change is in contrast to the dramatic effect 3 days after YY1 knockout on CSR and long-distance DNA loop formation, when Ig CSR and long-distance Eμ-3’RR DNA loops drop approximately fivefold [24, 33]. These results strongly argue that the impact of YY1 on CSR is not due to changes in gene expression that impact mitochondrial function or glucose uptake. We also previously failed to detect changes in gene expression of CSR-related genes after YY1 knockout [24].
Instead, our data argue that the YY1 impact on CSR and long-distance DNA loops is not due to changes in mitochondrial or glycolytic energy production, or to changes in gene expression, but is perhaps due to YY1 playing a structural role in DNA loop formation or in regulation of nuclear AID levels [33, 34]. YY1-related long-distance DNA looping systems involving promoter–enhancer, enhancer–enhancer, and promoter–promoter interactions have been observed in many cell types and it has been argued that YY1 plays a structural role in DNA loop formation [31, 63]. In B and T cells, YY1 is involved in controlling a variety of long-distance DNA loops at the Vh, Vκ, and Th2 cytokine loci [17, 31, 37, 40–43]. Our prior data showed that YY1 is critical for the Eμ-3’RR long-distance DNA loop involved in CSR [24]. Notably, we found that a YY1 mutant lacking the YY1 transcription activation domain is fully capable of rescuing CSR and long-distance DNA loops in a yy1-null background [24]. As this mutant lacks the AID interaction domain in YY1, we propose that YY1 regulates CSR by controlling long-distance DNA loops.
Materials and methods
Mice
yy1flox/flox (yy1f/f) mb1-CRE mice on a C57BL/6 background were a gift from Y. Shi (Harvard). CD20TAM-CRE mice were obtained from D. Allman (University of Pennsylvania) and bred to yy1f/f mice to generate CD20TAM-CRE yy1f/f mice. Mice were bred and maintained under pathogen-free conditions. yy1f/f, yy1f/fCD20TAM-CRE, or C57BL/6 control mice between 8 and 12 weeks of age were used for experiments. All animal studies were performed following recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania (Protocol 803080).
Purification of splenic B cells, TAT-CRE treatment, and in vitro stimulation
Follicular B cells were purified from mouse spleen with anti-CD23-biotin (Biolegend) and streptavidin microbeads (MACS; Miltenyi Biotec). Conditional YY1 knockout in splenic B cells was performed ex vivo using TAT-CRE enzyme purified from bacteria, as previously described [33, 34]. Briefly, cells were washed three times with opti-MEM (Invitrogen) and then incubated with TAT-CRE recombinant protein for 45 min at 37°C. To inactivate TAT-CRE, fetal bovine serum was added to a final concentration of 10%. Cells were washed with splenic B medium (RPMI 1640, 10% HyClone fetal bovine serum (Thermo Scientific), sodium pyruvate, 55 μM 2-mercaptoethanol (Sigma), minimal essential medium (MEM) nonessential amino acids, 2 mM l-glutamine, 1% Penicillin-Streptomycin (Invitrogen), and then cultured at 37°C in a 5% CO2 atmosphere. Cells were activated ex vivo with 10 μg/mL bacterial LPS (Sigma) plus 20 ng/mL IL4 to stimulate proliferation and CSR.
In vivo tamoxifen treatment
yy1f/f CD20Tam-CRE or yy1f/f mice were injected intraperitoneally with 100 μL (20 mg/mL) of tamoxifen (Sigma) dissolved in corn oil daily for 3 days. Two days after the last injection, spleens were harvested and CD23+ B cells were isolated. In some experiments, NP-ova immunization was performed prior to tamoxifen injection.
Flow cytometry and cell sorting
After various days of activation, splenic B cells were washed with prewarmed MACS buffer (PBS, pH 7.2, supplemented with 0.5% BSA and 2 mM EDTA) and 1 × 106 cells were resuspended in 100 μL of staining buffer containing MitoTracker Green (20 nM; M7514; Invitrogen) and MitoTracker DeepRed (20 nM; M22426; Invitrogen), and were incubated at 37°C for 10 min. Stained cells were washed with 1 mL of prewarmed MACS buffer and kept on ice to quench further staining. Samples were analyzed by flow cytometry within 30 min. In some experiments, cells were first stained with anti-IgG1 (PE Rat Anti-Mouse IgG1, Clone A85-1, Cat # 550083; BD Bioscience) anti-CD138 (BV421 Rat Anti-Mouse CD138, Clone 281–2, Cat #562610; BD Bioscience), or anti-CD69 (APC Hamster anti-mouse CD69 clone H1.2F3; BD Bioscience) antibodies for 30 min followed by MitoTracker dyes. Other antibodies used and analyses of germinal center cells are described in our previously published work [4]. Data were collected on a BD LSR II or Fortessa flow cytometer and analyzed with FlowJo software (Tree Star). Exclusion by Zombie Aqua™ (Biolegend) or DAPI was used to identify live cells and doublets were excluded by forward and side scatter height versus width analysis.
RNA-Seq
Total RNA (1.0 μg) from each sample was used to construct RNA-Seq libraries with a unique index using the TruSeq Stranded mRNA Sample Preparation kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA) was performed for library quantification. cDNA libraries were sequenced using the Illumina NextSeq 500 system (Illumina). Sequencing was performed as 75 bp paired-end reads. RNA-Seq reads were aligned to the mouse reference genome (mm10) using Tophat 2.1.0 with default parameters. The number of reads mapped to each gene was counted by htseq-count (http://www-huber.embl.de/users/anders/HTSeq/) based on the annotation from ENSEMBL (http://uswest.ensembl.org/) mouse gene annotation v85. Identification of differentially expressed genes was performed by edgeR. Differential expression was defined as a fold change greater than 2 and false discovery rate (FDR) < 0.05. FDR was calculated based on Benjamini and Hochberg multiple testing correction. RNA-seq data are available in the Gene Expression Omnibus (GEO) database under the accession numbers GSE104097 (GSM2789711; GSM2789713; GSM2789715) and GSE145161.
ChIP-Seq
Naïve splenic B cells were cross-linked with 1% formaldehyde for 10 min at room temperature. After cell lysis, chromatin was sheared by using a Covaris S220 for 20 min with 200 cycle/burst, 10 duty factor, and 140 peak power. Note that 100 μg of sheared chromatin was used per immunoprecipitation, and 1% was reserved as input control. Sheared chromatin was incubated overnight in 4°C with preblocked Dynabeads Protein G (Invitrogen) coated with YY1 antibody (#414; Santa Cruz Biotechnology). The following day, beads were washed and DNA was purified by using the QIAquick PCR Purification Kit. Note that 6 ng from each sample was used to construct ChIP-Seq libraries with a unique index using the TruSeq ChIP Library Preparation Kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. ChIP-Seq reads were aligned to the mouse reference genome (mm10) using Bowtie 1.1.2. Duplicated reads were removed by Samtools. Significant YY1 peaks were called using MACS (v2.1.0), with a FDR ≤ 5%. Annotation of Chip-Seq data was performed by Homer. Bedtools were used to process the bed files and overlapping peaks were defined as peaks sharing more than 10% of the length of the shorter peak. ChIP-seq data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE104097.
qPCR analyses
Total RNA was isolated from LPS + IL4 stimulated splenic B cells using TRIzol™ Reagent (Thermo Fisher Scientific) and reverse transcribed with SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s protocol. qPCR was performed with SYBR green on a ViiA 7 Real-Time PCR System using primers listed in Supporting Information Table 4. Relative mRNA levels were calculated using ΔΔCt after normalizing to 18s rRNA.
Western blots
Total cell lysates were prepared with RIPA buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease and phosphatase inhibitor cocktail (Thermo Scientific) for 30 min on ice. Note that 25 μg of protein from each sample was loaded and analyzed by SDS-PAGE. Proteins were visualized using the ECL kit (Thermo Fisher Scientific) and subjected to autoradiography using ChemiDoc. Bands were quantified using ImageJ software. Antibodies used were Clybl (17314-1-AP), Ech1 (11385-1-AP), PCCB (11139-1-AP), ACOT2 (15633-1-AP), Raf1 (51140-1-AP), Mrps18b (16139-1-AP), YY1 (sc-7341), β-actin (ab8227), and β-tubulin (sc-5274) (Proteintech and Santa Cruz Biotechnology).
Isolation of mitochondria
Mitochondria from activated B cells were prepared by differential centrifugation as described previously [64]. Briefly, cells were homogenized with a Dounce glass homogenizer in H-medium (70 mM Sucrose, 220 mM Mannitol, 10 mM HEPES, pH 7.4, 1 mM EGTA-containing 1 mM PMSF, 1 μg/mL each of pepstatin, leupeptin, aprotinin, and antipain) and homogenate was centrifuged two times at 1000 RPM for 10 min to pellet nuclei and cell debris. The supernatant was centrifuged at 8000 RPM for 20 min to pellet the mitochondrial fraction. The pellet was resolved in H-medium and passed through 0.8 M sucrose to minimize microsomal contamination. Protein concentration was determined by Lowry assay.
Seahorse platform measurements
Simultaneous measurement of OCR and ECAR were measured using a Seahorse Extracellular Flux (XF) Analyzer (Seahorse Bioscience). We measured basal OCR and ECAR, along with sequential injection of various inhibitors, oligomycin (2 μg/mL), 2,4 dinitrophenol (100 μM), and rotenone (1 μM). Oligomycin is an ATP synthase inhibitor (complex V) and differentiates oxygen consumption that is used for ATP synthesis from proton leak across the inner mitochondrial membrane. DNP is an ionophore and uncouples the proton transport for ATP synthesis, resulting in the maximal OCR without ATP production, reflecting the maximal respiratory capacity of the cells. The difference between maximal OCR (after addition of DNP) and basal OCR provides the mitochondrial spare respiratory capacity. Rotenone inhibits mitochondrial complex I and interferes with the use of NADH for ATP synthesis.
Assay of electron transport chain complexes
Assays for complexes I and IV were performed as described by Birch-Machin and Turnbull [65] using a Cary 1E UV-Vis Spectrophotometer. Briefly, complex I activity (NADH: ubiquinone oxidoreductase) was measured by incubating 20 μg of mitochondria in 1 mL of assay buffer containing 25 mM potassium phosphate, pH 7.4, 5 mM MgCl2, 2 mM NaCN, 2.5 mg/mL BSA, 13 mM NADH, 65 μM ubiquinone and 2 μg/mL antimycin A, and measuring the decrease in absorbance at 340 nm due to NADH oxidation. Complex IV activity was measured by incubating 20 μg of mitochondria in 25 mM potassium phosphate, pH 7.4, 0.45 mM dodecyl maltoside, and 15 mM reduced cytochrome c and following the decrease in absorbance at 550 nm due to cytochrome c oxidation.
Galactose viability assay
Splenic B cells were seeded in glucose-free RPMI medium supplemented with either 1 mg/mL galactose or 2 mg/mL galactose, 10% dialyzed fetal bovine serum, 25 mM HEPES, 55 μM 2-mercaptoethanol, 1× MEM, nonessential amino acids, and 1% Penicillin-Streptomycin (Invitrogen). Cells grown in medium with glucose were used for comparison for each cell type. After 48 h, cells were stained with DAPI and quantified by flow cytometry to check viability.
Oligomycin A-dependent CSR kinetics
Mock and TAT-CRE-treated splenic B cells were activated ex vivo with LPS plus IL4, followed by 10 nM oligomycin A treatment for either 12 or 24 h. Cells were harvested and stained for CSR.
Statistical analysis
Statistical analysis was performed using an unpaired, two-tailed Student’s t-test unless otherwise noted.
Supplementary Material
Acknowledgements:
This work was supported by NIH R01 grants AI079002 and GM111384 to M.L.A. and GM34883 to N.G.A. We thank Corbett Berry for discussions and data interpretation, and Amit Singh for help with flow cytometry data analyses.
Abbreviations:
- AID
activation-induced cytidine deaminase
- CSR
class switch recombination
- FDR
false discovery rate
- OCR
oxygen consumption rate
- YY1
Yin Yang 1
- GO
gene ontology
Footnotes
Conflict of Interest: The authors declare no commercial or financial conflict of interest.
Additional supporting information may be found online in the Supporting Information section at the end of the article.
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.201948385
References
- 1.Gordon S, Akopyan G, Garban H and Bonavida B, Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 2006. 25: 1125–1142. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Lee J and Galvin KM, Everything you have ever wanted to know about Yin Yang1 Biochim. Biophys. Acta 1997. 1332: F49–F66. [DOI] [PubMed] [Google Scholar]
- 3.Atchison L, Ghias A, Wilkinson F, Bonini N and Atchison ML, The YY1 transcription factor functions as a PcG protein in vivo. EMBO J. 2003. 22: 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee A, Sindhava V, Vuyyuru R, Jha V, Hodawadekar S, Manser T and Atchison ML, YY1 is required for germinal center B cell development. PLoS One 2016. 11: e0155311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas MJ and Seto E, Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 1999. 236: 197–208. [DOI] [PubMed] [Google Scholar]
- 6.Bhalla SS, Robitaille L and Nemer M, Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J. Biol. Chem 2001. 276: 11439–11445. [DOI] [PubMed] [Google Scholar]
- 7.Basu A, Wilkinson FH, Colavita K, Fennelly C and Atchison ML, YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res. 2014. 42: 2208–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y et al. ,YY1 functions with INO80 to activte transcription. Nat. Struct. Molec. Biol 2007. 14: 872–874. [DOI] [PubMed] [Google Scholar]
- 9.Calame K and Atchison M, YY1 helps bring loose ends together. Genes Dev. 2007. 21: 1145–1152. [DOI] [PubMed] [Google Scholar]
- 10.Chen F, Sun H, Zhao Y and Wang H, YY1 in Cell Differentiation and Tissue Development. Crit. Rev. Oncogen 2017. 22: 131–141. [DOI] [PubMed] [Google Scholar]
- 11.Chinnappan D, Xiao D, Ratnasari A, Andry C, King TC and Weber HC, Transcription factor YY1 expression in human gastrointestinal cancer cells. Int. J. Oncol 2009. 34: 1417–1423. [PubMed] [Google Scholar]
- 12.de Nigris F, Botti C, Rossiello R, Crimi E, Sica V and Napoli C, Cooperation between Myc and YY1 provides novel silencing transcriptional targets of α3β1-integrin in tumour cells. Oncogene 2007. 26: 382–394. [DOI] [PubMed] [Google Scholar]
- 13.Donohoe ME, Zhang L-F, Xu N, Shi Y and Lee JT, Identification of a Ctcf cofactor, YY1, for the X chromosome binary switch. Mol. Cell 2007. 25: 43–56. [DOI] [PubMed] [Google Scholar]
- 14.Erkeland S, Valkhof M, Heijmans-Antonissen C, Delwel R, Valk PJM, Hermans MHA and Touw IP, The gene encoding transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood 2003. 101: 1111–1117. [DOI] [PubMed] [Google Scholar]
- 15.Green MR, Monti S, Dalla-Favera R, Pasqualucci L, Walsh NC, Schmidt-Supprian M, Kutok JL et al. , Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl Acad. Sci. USA 2011. 108: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronroos E, Terentiev AA, Punga T and Ericsson J, YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci USA 2004. 101: 12165–12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang SS, Kim YU, Lee S, Jang SW, Kim MK, Koh BH, Lee W et al. , Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc. Natl Acad. Sci. USA 2013. 110: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon Y and Lee JT, YY1 tethers Xist RNA to the inactive X nucleation center. Cell 2011. 146: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J and Kim JD, In vivo YY1 knockdown effects on genomic imprinting. Hum. Mol. Genet 2008. 17: 391–401. [DOI] [PubMed] [Google Scholar]
- 20.Kleiman E, Jia H, Loguercio S, Su AI and Feeney AJ, YY1 plays an essential role at all stages of B-cell differentiation. Proc. Natl Acad. Sci. USA 2016. 113: E3911–E3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J-S, Galvin KM, See RH, Eckner R, Livingston D, Moran E and Shi Y, Relief of YY1 transcriptional repression by adenovirus E1A is mediated by EIA-associated protein p300. Genes Dev.1995. 9: 1188–1198. [DOI] [PubMed] [Google Scholar]
- 22.Lee T-C, Zhang Y and Schwartz RJ, Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 1994. 9: 1047–1052. [PubMed] [Google Scholar]
- 23.Lorente M, Perez C, Sanchez C, Donohoe M, Shi Y and Vidal M, Homeotic transformatinos of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech. Develop 2006. 123: 312–320. [DOI] [PubMed] [Google Scholar]
- 24.Mehra P, Gerasimova T, Basu A, Jha V, Banerjee A, Sindhava V, Gray F et al. , YY1 controls Eμ-3’RR DNA loop formation and immunoglobulin heavy chain class switch recombination. Blood Adv. 2016. 1: 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan X, Papasani M, Hao Y, Calamito M, Wei F, Quinn WJI, Basu A et al. , YY1 controls Igκ repertoire and B-cell development, and localizes with condensin on the Igκ locus. EMBO J. 2013. 32: 1168–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel SR and Dressler GR, Expression of Pax2 in the intermediate mesoderm is regulated by YY1. Dev. Biol 2004. 267: 505–516. [DOI] [PubMed] [Google Scholar]
- 27.Perekatt AO, Valdez MJ, Davila M, Hoffman A, Bonder EM, Gao N and Verzi MP, YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc. Natl Acad. Sci. USA 2014. 111: 7695–7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Seto E, Chang L-S and Shenk T, Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 1991. 67: 377–388. [DOI] [PubMed] [Google Scholar]
- 29.Sucharov CC, Dockstader K and McKinsey TA,YY1 protects cardiac myocytes from pathologic hypertrophy by interacting with HDAC5. Mol. Biol. Cell 2008. 19: 4141–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trabucco SE, Gerstein RM and Zhang H, YY1 Regulates the Germinal Center Reaction by Inhibiting Apoptosis. J. Immunol 2016. 197: 1699–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ et al. , YY1 is a structural regulator of enhancer-promoter loops. Cell 2017. 171: 1573–1588.e1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson FH, Park K and Atchison ML, Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl Acad. Sci. USA 2006. 103: 19296–19301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaprazna K and Atchison ML, YY1 controls immunoglobulin class switch recombination and nuclear activation-induced deaminase levels. Mol. Cell. Biol 2012. 32: 1542–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaprazna K, Basu A, Tom N, Jha V, Hodawadekar S, Radova L, Malcikova J et al. , Transcription factor YY1 can control AID-mediated mutagenesis. Eur. J. Immunol 2017. 48: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hariharan N, Kelley DE and Perry RP, d, a trancription factor that binds to downstream elements in several polymerase II promoters, is a functionally diverse zinc finger protein. Proc. Natl. Acad. Sci. USA 1991. 88: 9799–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park K and Atchison ML, Isolation of a candidate repressor/activator, NF-E1 (YY-1, δ), that binds to the immunoglobulin κ 3’ enhancer and the immunoglobulin heavy-chain μE1 site. Proc. Natl. Acad. Sci. USA 1991. 88: 9804–9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atchison ML, Function of YY1 in long-distance DNA interactions. Frontiers Immunol. 2014. 5: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shrivastava A and Calame K, An analysis of genes regulated by the multi-functional transcriptional regulatory Yin Yang-1. Nucleic Acids Res. 1994. 22: 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E and Shi Y, Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol 1999. 19: 7237–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Schmidt-Supprain M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R et al. , Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007. 21: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, Sun Q et al. , Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity 2013. 39: 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibcus JH and Dekker J, The hierarchy of the 3D genome. Mol. Cell 2013. 49: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorkin DU, Leung D and Ren B, The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 2014. 14: 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM et al. , Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell 2011. 147: 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW et al. , S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 2007. 27: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK and Puigserver P, mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007. 450: 736–740. [DOI] [PubMed] [Google Scholar]
- 47.Sandhir R, Halder A and Sunkaria A, Mitochondria as a centrally positioned hub in innate immune response. Biochim. Biophys. Acta 2017. 1863: 1090–1097. [DOI] [PubMed] [Google Scholar]
- 48.Cherry C, Thompson B, Saptarshi N, Wu J and Hoh J, 2016: A ‘Mitochondria’ Odyssey. Trends Mol. Med. 2016. 22: 391–403. [DOI] [PubMed] [Google Scholar]
- 49.Buck MD, Sowell RT, Kaech SM and Pearce EL, Metabolic Instruction of Immunity. Cell 2017. 169: 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dufort FJ, Bleiman BF, Gumina MR, Blair D, Wagner DJ, Roberts MF, Abu-Amer Y et al. , Cutting edge: IL-4-mediated protection of primary B lymphocytes from apoptosis via Stat6-dependent regulation of glycolytic metabolism. J. Immunol 2007. 179: 4953–4957. [DOI] [PubMed] [Google Scholar]
- 51.Doughty CA, Bleiman BF, Wagner DJ, Dufort FJ, Mataraza JM, Roberts MF and Chiles TC, Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood 2006. 107: 4458–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearce EL, Poffenberger MC, Chang CH and Jones RG, Fueling immunity: insights into metabolism and lymphocyte function. Science 2013. 342: 1242454.24115444 [Google Scholar]
- 53.Mehta MM, Weinberg SE and Chandel NS, Mitochondrial control of immunity: beyond ATP. Nat. Rev. Immunol 2017. 17: 608–620. [DOI] [PubMed] [Google Scholar]
- 54.Milasta S, Dillon CP, Sturm OE, Verbist KC, Brewer TL, Quarato G, Brown SA et al. Apoptosis-inducing-factor-dependent mitochondrial function is required for T cell but not B cell function. Immunity 2016. 44: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akkaya M, Traba J, Roesler AS, Miozzo P, Akkaya B, Theall BP, Sohn H et al. , Second signals rescue B cells from activation-induced mitochondrial dysfunction and death. Nat. Immunol 2018. 19: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jang KJ, Mano H, Aoki K, Hayashi T, Muto A, Nambu Y, Takahashi K et al. , Mitochondrial function provides instructive signals for activation-induced B-cell fates. Nat. Commun 2015. 6: 6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waters LR, Ahsan FM, Wolf DM, Shirihai O and Teitell MA, Initial B cell activation induces metabolic reprogramming and mitochondrial remodeling. iScience 2018. 5: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bushmeyer S, Park K and Atchison ML, Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem 1995. 270: 30213–30220. [DOI] [PubMed] [Google Scholar]
- 59.Austen M, Luscher B and Luscher-Firzlaff JM, Characterization of the transcriptional regulator YY1. J. Biol. Chem 1997. 272: 1709–1717. [DOI] [PubMed] [Google Scholar]
- 60.Caro-Maldonado A, Wang R, Nichols AG, Kuraoka M, Milasta S, Sun LD, Gavin AL et al. , Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J. Immunol 2014. 192: 3626–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Manteiga JM, Mari S, Godejohann M, Spraul M, Napoli C, Cenci S, Musco G et al. , Metabolomics of B to plasma cell differentiation. J. Proteome Res 2011. 10: 4165–4176. [DOI] [PubMed] [Google Scholar]
- 62.Blattler SM, Verdeguer F, Liesa M, Cunningham JT, Vogel RO, Chim H, Liu H et al. , Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Mol. Cell. Biol 2012. 32: 3333–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beagan JA, Duong MT, Titus KR, Zhou L, Cao Z, Ma J, Lachanski CV et al. , YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017. 27: 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhat NK, Niranjan BG and Avadhani NG, Qualitative and comparative nature of mitochondrial translation products in mammalian cells. Biochemistry 1982. 21: 2452–2460. [DOI] [PubMed] [Google Scholar]
- 65.Birch-Machin MA and Turnbull DM, Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001. 65: 97–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


