Farkas and Bohnsack describe advances in understanding targeting of tail-anchored (TA) proteins, including their insertion into the ER, how nascent TA proteins are selectively captured at the ribosome, and recently discovered quality control pathways.
Abstract
Tail-anchored (TA) proteins fulfill diverse cellular functions within different organellar membranes. Their characteristic C-terminal transmembrane segment renders TA proteins inherently prone to aggregation and necessitates their posttranslational targeting. The guided entry of TA proteins (GET in yeast)/transmembrane recognition complex (TRC in humans) pathway represents a major route for TA proteins to the endoplasmic reticulum (ER). Here, we review important new insights into the capture of nascent TA proteins at the ribosome by the GET pathway pretargeting complex and the mechanism of their delivery into the ER membrane by the GET receptor insertase. Interestingly, several alternative routes by which TA proteins can be targeted to the ER have emerged, raising intriguing questions about how selectivity is achieved during TA protein capture. Furthermore, mistargeting of TA proteins is a fundamental cellular problem, and we discuss the recently discovered quality control machineries in the ER and outer mitochondrial membrane for displacing mislocalized TA proteins.
Introduction
The protein components of biological membranes expand their functionality beyond physical barriers by acting as gateways, allowing intercompartment communication as well as facilitating transport and other membrane-associated processes. Membrane proteins collectively constitute ∼30% of the proteome of most organisms (Krogh et al., 2001), and their biogenesis represents a major challenge for cells. Their hydrophobic transmembrane domains (TMDs), essential for integration into the lipid bilayer and functionality, render such proteins inherently prone to aggregation in the aqueous cytosolic environment. Dedicated targeting strategies for chaperoning such proteins to their target membranes are therefore necessary. Proteins destined for the ER that carry short signal sequences at their N-terminal end and/or internal TMDs are typically recognized cotranslationally by the signal recognition particle (SRP). This arrests translation and induces relocalization of the ribosome nascent chain complex (RNC) to the ER-bound Sec61 translocon, where the newly synthesized protein is channeled directly into the ER lumen and/or membrane (reviewed in Akopian et al., 2013; Rapoport et al., 2017). In both yeast and mammals, a macromolecular ER membrane protein complex (EMC) cooperates with the translocon by assisting the cotranslational folding and biogenesis of polytopic membrane proteins in the ER as well as itself acting as a membrane insertase, mediating the correct topological insertion of the first TMD of specific G-coupled receptors (Bai et al., 2020; Chitwood et al., 2018; Shurtleff et al., 2018). Furthermore, an SRP-independent ER targeting pathway (SND) has recently been revealed in yeast, where proteins containing TMDs in their central regions are captured by Snd1 and directed toward a Sec61 translocon associated with Snd2 and Snd3 (Aviram et al., 2016). This pathway appears to have a broad client spectrum and has been suggested to functionally compensate when other ER targeting pathways are impaired.
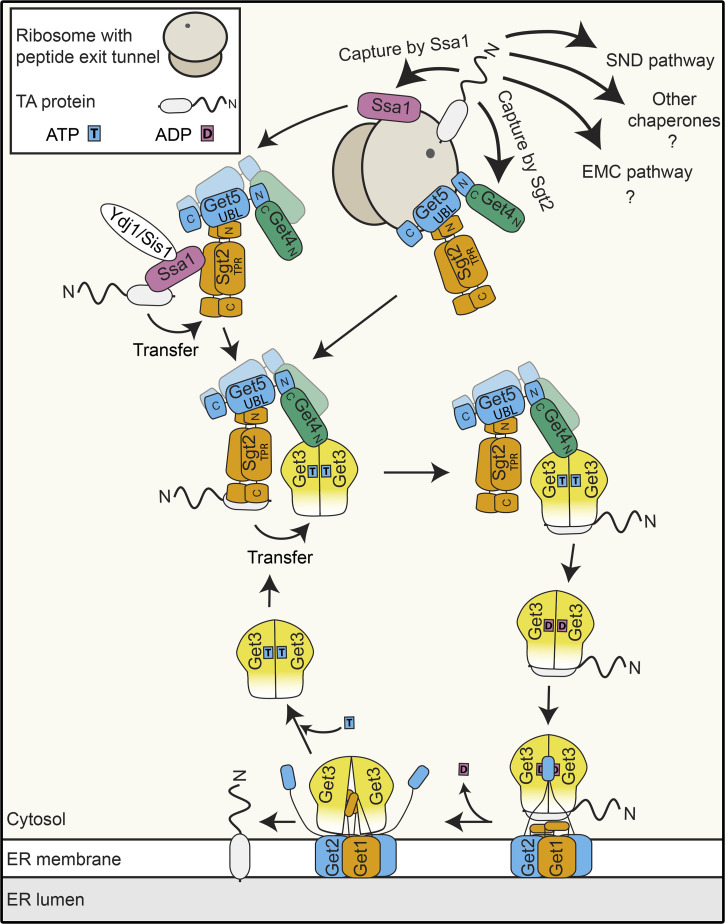
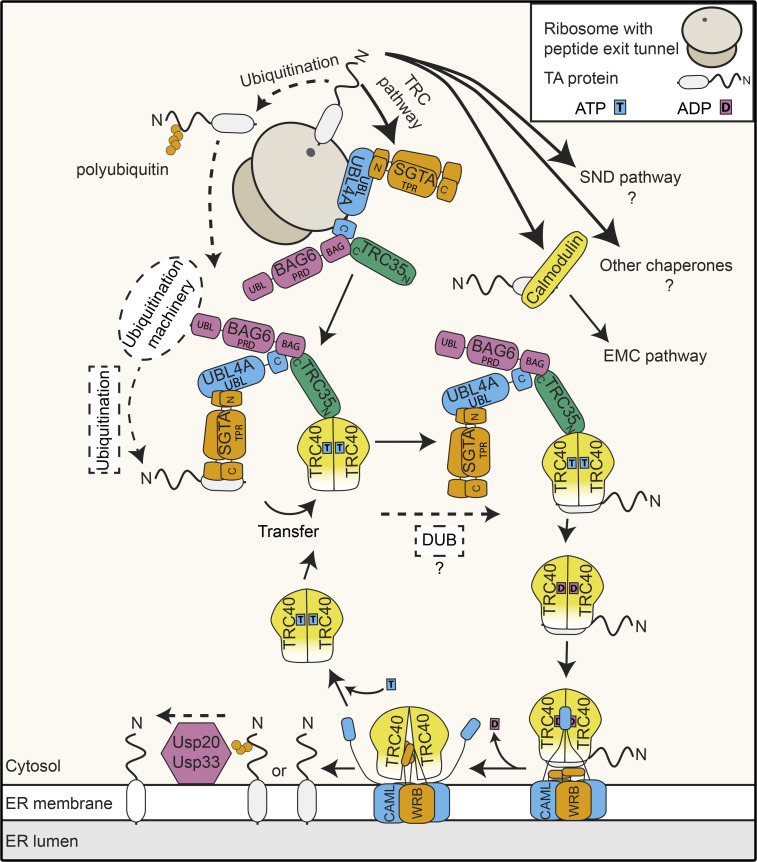
Tail-anchored (TA) proteins represent a specific class of membrane proteins characterized by a single TMD close to their C-terminus (reviewed in Kutay et al., 1993). The TA protein family contains >50 members in yeast (Beilharz et al., 2003), >500 in plants (Kriechbaumer et al., 2009), and >300 in humans (Kalbfleisch et al., 2007), which populate different membranes (ER, Golgi, and inner nuclear, outer mitochondrial, and peroxisomal membranes). These proteins fulfill diverse membrane-associated functions ranging from regulating intracellular vesicular trafficking (SNARE proteins) to apoptosis, autophagy, lipid biosynthesis, and protein degradation. The topology of TA proteins dictates their posttranslational targeting, as translation termination occurs concurrent with emergence of the TMD from the polypeptide exit tunnel of the ribosome. A major route to the ER for TA proteins is the evolutionarily conserved guided entry of TA proteins (GET) pathway in yeast and the homologous transmembrane recognition complex (TRC) pathway in mammals (Figs. 1 and 2; Borgese et al., 2019; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). The established view of the GET/TRC pathway (reviewed in Borgese et al., 2019; Chio et al., 2017) involves the posttranslational capture of TA proteins by a pretargeting complex composed of the homodimeric, cytosolic chaperone Sgt2 (yeast)/SGTA (mammals) and the Get4-Get5 heterodimer (yeast)/TRC35-UBL4A-BAG6 complex (mammals; Jonikas et al., 2009; Mariappan et al., 2011; Wang et al., 2010). Interaction of the pretargeting complex with an ATP-bound form of the ATPase Get3 (yeast)/TRC40 (mammals) results in transfer of the TA protein from Sgt2/SGTA to Get3/TRC40 such that the TMD is shielded within a hydrophobic pocket of Get3/TRC40 (Bozkurt et al., 2009; Mateja et al., 2009; Stefanovic and Hegde, 2007; Suloway et al., 2009). ATP hydrolysis triggered by interaction of Get3/TRC40 with the TA protein, coupled with conformational changes in Get3/TRC40 induced by interaction with Get4-Get5/TRC35-UBL4A-BAG6, drives dissociation of TA protein-bound Get3/TRC40 from the pretargeting complex, allowing delivery to the ER-bound GET receptor composed of Get1 and Get2 (yeast)/tryptophan-rich basis protein (WRB) and calcium-modulating cyclophilin ligand (CAML; mammals; Mariappan et al., 2011; Mateja et al., 2015; Stefer et al., 2011). Receptor binding triggers ADP release and conformational rearrangement of Get3/TRC40, allowing TA protein insertion into the membrane and recycling of Get3/TRC40 (reviewed in Mateja and Keenan, 2018). Although much less is currently known about the GET pathway in plants, homologues of the GET pathway components have been identified or are predicted (Xing et al., 2017; Srivastava et al., 2017; Asseck et al., 2021), and two TA SNARE proteins have been shown to be affected by lack of Arabidopsis thaliana (At)GET3 (Xing et al., 2017). A growing body of evidence indicates functional redundancy between different pathways for targeting TA proteins to the ER (Figs. 1 and 2; Casson et al., 2017). Although some TA proteins of the secretory pathway fulfill essential cellular functions, deletion of GET pathway components is not lethal in yeast or plants, and for many TA proteins, lack of GET pathway components reduces but does not abolish ER targeting. Consistent with this notion, both the EMC insertase (Guna et al., 2018) and the SND pathway (Aviram et al., 2016) have been shown to support the ER targeting of TA proteins.
Figure 1.
TA protein targeting to the ER in yeast. Nascent TA proteins emerging from the ribosome can be captured by alternative ER-targeting machineries. A major route to the ER is via the GET pathway, involving ribosome-associated capture by Sgt2, followed by Get4/Get5-mediated handover to the Get3 ATPase and insertion into the ER membrane by a heterotetrameric GET receptor complex composed of Get1 and Get2.
Figure 2.
TA protein targeting to the ER in mammals. Mammalian TA proteins are predominantly targeted to the ER by the TRC pathway. After capture by SGTA, together with the BAG6 complex (BAG6, UBL4A, and TRC35), the TA protein is passed to the TRC40 chaperone for delivery to the ER-bound receptor complex formed by WRB and CAML. BAG6 has dual functions bridging ER targeting and ubiquitination of TA proteins and can be antagonized by SGTA. Ubiquitinated TA proteins can be deubiquitinated by ER-associated UPS20/UPS33.
Despite a wealth of knowledge on some aspects of TA protein targeting to the ER by the GET/TRC pathway, other features and mechanistic details remain enigmatic. How TA proteins can be captured posttranslationally but also reliably avoid aggregation during handover from the ribosome to the pretargeting complex or other chaperones is an inherent conundrum of their biogenesis. Knowledge on the architecture of the GET/TRC receptor complex and mechanistic understanding of how the TA protein, delivered to the receptor by Get3/TRC40, is inserted into the ER membrane, have been limited by the technically challenging nature of structural analyses of membrane-bound complexes. Furthermore, how the fidelity of TA protein targeting to different membranes is ensured is poorly understood, and little is currently known about how the actions of TA protein targeting and quality control are coordinated. In this review, we describe recent advances that address these key aspects of the GET/TRC pathway and TA protein biogenesis.
Capture of nascent TA proteins
In contrast to cotranslational protein targeting, where capture of client proteins and their delivery to the ER-bound receptor complex is performed by the SRP complex, posttranslational targeting of TA proteins to the ER by the GET/TRC pathway is a more stepwise process, involving a dynamically assembling pretargeting complex that mediates initial capture but then hands over substrates to another chaperone for delivery to the membrane-bound receptor. The existence of a modular pretargeting complex appears to be a feature of the GET/TRC pathway conserved throughout eukaryotes, but compositional and structural differences between species are apparent. In yeast, Get4-Get5 form an obligate heterodimer, whereas the homologous TRC35 and UBL4A interact via an additional component BAG6 (Chang et al., 2010; Chartron et al., 2010; Mariappan et al., 2010; Mock et al., 2015). Biochemical evidence shows that a Get4/TRC35 homologue exists in plants, and homologues of Get5/UBL4A and Sgt2/SGTA are predicted from in silico analyses (Srivastava et al., 2017; Xing et al., 2017). A BAG6 homologue has also been identified in plants (https://www.arabidopsis.org/servlets/TairObject?id=35038&type=locus); however, it remains unknown if this protein associates with components of the GET pathway and/or contributes to TA protein targeting. These differences likely reflect subtle variations in the mechanism of TA protein capture between species and/or the greater need for regulation and surveillance in multicellular organisms.
Ribosome binding of pretargeting complex components
Due to the position of the TMD at the C-terminus of TA proteins, the GET/TRC pathway must capture clients posttranslationally. However, posttranslational capture has the inherent caveat of protein aggregation in the narrow window between emergence of the TMD from the ribosome exit tunnel and protein capture by a chaperoning factor. The first hint how this issue may be overcome came with the intriguing discovery of Get5 in a high-throughput screen for ribosome-associated proteins in yeast (Fleischer et al., 2006). This observation raised the possibility of a physical connection between the upstream components of the GET pathway and the translation machinery, despite the posttranslational nature of the final capture event. Detection of the Get4-Get5 heterodimer associated with polysomes importantly confirmed recruitment of these proteins to actively translating ribosomes (Zhang et al., 2016), further supporting that the GET pathway pretargeting complex might be poised on the ribosomes ready to shield nascent TA proteins directly as they emerge from the exit tunnel. In vitro reconstitution confirmed a high-affinity interaction between Get4-Get5 and ribosomes, and protein–protein and protein–RNA cross-linking analyses pinpointed the polypeptide exit tunnel of the ribosome as the Get4-Get5 binding site (Fig. 1; Zhang et al., 2021). Get4-Get5 bridge interactions between Sgt2 and Get3 to facilitate handover of the TA protein to the downstream chaperone, and therefore functional significance of Get4-Get5 ribosome binding must be coupled to TA protein capture by Sgt2. Indeed, it was recently shown that Get4-Get5 act as a binding platform for recruitment of Sgt2 to ribosomes and that the presence of Get4-Get5 on ribosomes enhances TA protein capture by Sgt2 (Zhang et al., 2021). Crystal structures of GET pathway subcomplexes demonstrate that the N-terminal domains of the Sgt2 homodimer interact with the central UBL domain of Get5, while the N-terminal region of Get5 mediates interaction with C-terminal region of Get4 (Chang et al., 2010; Chartron et al., 2012a; Simon et al., 2013). As Get5 appears to simultaneously interact with Get4, Sgt2, and ribosomes, it is tempting to speculate that ribosome binding of the pretargeting complex may be mediated by the C-terminal region of Get5. However, due to the multimeric nature of the pretargeting complex and evidence that the Get5 C-terminal region also mediates homodimerization (Chartron et al., 2012b), structural analyses of Get4-Get5-Sgt2–bound ribosomes will be necessary to resolve in detail the architecture of GET pathway pretargeting complex–bound ribosomes.
The mammalian pretargeting complex components UBL4A, TRC35, BAG6, and SGTA also associate with RNC complexes (Fig. 2; Leznicki and High, 2020; Mariappan et al., 2010), implying that the mechanism of ribosome-associated capture of TA proteins is also employed in mammalian cells to circumvent protein aggregation upon the TMD encountering the aqueous cytosol during targeting. However, the mammalian-specific pretargeting complex component BAG6, rather than the Get5-homologous UBL4A, appears to act as the key ribosome adaptor, and SGTA can also interact with RNC complexes independently of TRC35-UBL4A (Leznicki and High, 2020; Mariappan et al., 2010).
Notably, Get3/TRC40, which receive the TA protein from the pretargeting complex, are not ribosome associated (Mariappan et al., 2010; Zhang et al., 2016). This implies that following Get4-Get5–facilitated capture of the TA protein by ribosome-associated Sgt2, the pretargeting complex should dissociate from the ribosome to encounter Get3. It is possible that a conformational change in the pretargeting complex, induced by TA protein binding, triggers release from the ribosome.
Recognition of ribosomes synthesizing TA proteins
The discovery of ribosome-associated populations of the GET pretargeting complex and the BAG6 complex and SGTA in yeast and mammals, respectively, raises the question of how these complexes, which are significantly less abundant than cytosolic ribosomes, are able to identify ribosomes synthesizing TA proteins. Analogous to SRP, which probes the translating ribosome population, preferentially binding ribosomes translating proteins with a signal sequence (Berndt et al., 2009; Holtkamp et al., 2012; Ogg and Walter, 1995; Voorhees and Hegde, 2015), yeast Get4-Get5 show increased association with RNCs containing a TA/TMD in the exit tunnel (Zhang et al., 2021). Mammalian SGTA is likewise selectively recruited to ribosomes synthesizing membrane proteins (Leznicki and High, 2020). This implies that a similar substrate-scanning mechanism, involving transient association events followed by high-affinity docking only onto appropriate ribosomes, is employed in both the co- and posttargeting pathways. The underlying mechanism of how the GET/TRC pretargeting components sense the presence of the TA/TMD in the exit tunnel still remains to be elucidated. Intriguingly, the newly identified ribosome-binding site of the GET pathway pretargeting complex overlaps with that of SRP, implying mutually exclusive ribosome occupancy. Consistent with this, the presence of Get4-Get5 on RNCs with a TMD in the exit tunnel reduces the amount of SRP bound, and strong binding of SRP to an exposed TMD leads to displacement of Get4-Get5 (Zhang et al., 2021). This suggests a compelling model where the GET/TRC pretargeting complex, SRP, and potentially ribosome-bound Snd1 (Fleischer et al., 2006) constantly screen the translating pool of ribosomes and outcompete each other for ribosome binding upon encountering one synthesizing an optimal substrate.
Additional players in initial TA protein capture
It is well established both in vitro and in vivo that TA protein targeting by the GET/TRC pathways involves a chaperone exchange in which the TMD, initially shielded by a hydrophobic patch within the C-terminal region of Sgt2, is handed over to Get3, where it is protected within a dedicated hydrophobic groove formed in the ATP-bound state. While Sgt2 has long been considered the upstream component of the GET pathway, it has recently emerged that the abundant, Hsp70-like chaperone Ssa1 can also act as a highly efficient nascent TA protein chaperone and that its effectiveness in TA protein trapping is enhanced by the J domain–containing cochaperone protein Ydj1 (Fig. 1; Cho et al., 2021; Cho and Shan, 2018). Ssa1 and numerous other chaperones physically interact with Sgt2 via its tetratricopeptide repeat domain (Cho and Shan, 2018; Krysztofinska et al., 2017). As this domain of Sgt2 is important for ER targeting of TA proteins in vivo, this supports a potential role of other chaperones alongside Sgt2. Transfer of TA protein cargoes from Ssa1 to Sgt2 is energetically favorable and stimulated by the J domain proteins Ydj1 and Sis1. Interestingly, Ydj1 and Sis1 appear to function redundantly in targeting of a reporter TA protein to the ER (Cho et al., 2021), perhaps suggesting that the chaperone cascade protecting TA proteins during their biogenesis can be more extensive than initially anticipated. However, it still remains to be determined how much of a contribution these proteins make to endogenous TA protein targeting within the native cellular environment. Interestingly, human cells lacking SGTA or BAG6 are viable, and TA protein targeting to the ER can still be accomplished in the absence of these factors (Culver and Mariappan, 2021). It is possible, therefore, for capture by the pretargeting complex to be bypassed, and perhaps in this context, other chaperones, analogous to those characterized in yeast, contribute to the initial protection of nascent TA proteins before their association with TRC40. Along this line, TA proteins with less hydrophobic TMDs, which use the EMC rather than the TRC pathway, have been shown to be chaperoned through the cytosol by calmodulin (Fig. 2; Guna et al., 2018). The existence of alternative, partially redundant, targeting pathways, supported by the nonlethality of yeast, plant, and human GET pretargeting factor knockouts (Stefanovic and Hegde, 2007; Schuldiner et al., 2008; Jonikas et al., 2009; Srivastava et al., 2017; Xing et al., 2017; Culver and Mariappan, 2021), likely helps ensure high-fidelity and robust targeting of TA proteins in vivo.
Selectivity of TA protein capture
It has become increasingly clear that nascent TA proteins emerge into a crowded environment where encounters with different machineries can direct them toward different fates, such as targeting to the ER via different routes, targeting to different organelles, or potentially, degradation. These observations suggest a finely tuned process of nascent TA protein capture to direct different proteins to the appropriate destinations and highlight the question of how TA proteins are selectively captured. While all TA proteins share the common characteristic of a single TMD close to the C-terminus that can serve as a targeting signal, the length and hydrophobicity of this TMD, as well as the net charge of the downstream sequence (also termed C-terminal element [CTE]), vary significantly, and these features are important determinants of the ultimate destination of the protein (Beilharz et al., 2003; Borgese et al., 2019). TA proteins of the outer mitochondrial membrane (OMM) and peroxisomes are typified by short, less hydrophobic TMDs, and positively charged CTEs are characteristic features of peroxisomal TA proteins (Horie et al., 2002). In contrast, ER and Golgi TA proteins generally have relatively long, hydrophobic TMDs, and the charge of their CTEs varies considerably (Rao et al., 2016; Borgese et al., 2019). The physiochemical properties of ER TMDs favor capture by Sgt2/SGTA and binding by Get3/TRC40, whereas TA proteins with less hydrophobic TMDs are poor substrates (Coy-Vergara et al., 2019; Guna et al., 2018). This implies that an important layer of capture selectivity is already encoded within the proteins themselves. Strict categorization of different organellar TA proteins based on physiochemical properties is not possible, however, as differently localized TA proteins have partially overlapping properties, indicating that this criterion alone is insufficient to ensure correct targeting.
TA proteins not only need to be directed to different target membranes, but they also need to be recognized as bona fide membrane proteins. The hydrophobic sequences that are integral features of membrane proteins must be distinguished from exposed hydrophobic patches of misfolded, nonmembrane proteins that serve as signals for recruitment of the protein quality control machinery. Interestingly, the BAG6 component of the mammalian pretargeting complex sits at the nexus between the alternative fates of targeting and degradation; a minimal C-terminal BAG domain in BAG6 scaffolds interactions between TRC35 and UBL4A and is sufficient for TA protein targeting to the ER (Mock et al., 2015), while the N-terminal UBL domain of the protein promotes recruitment of the ubiquitination machinery to mediate quality control of aberrant proteins (Fig. 2; Rodrigo-Brenni et al., 2014). In this context, BAG6 has been implicated in rerouting SGTA-bound TA proteins that are not efficiently relayed to TRC40 toward the degradation pathway (Shao et al., 2017). SGTA has emerged as another central player in determining the fate of TA proteins, as it is able to antagonize BAG6-facilitated protein ubiquitination. Interestingly, SGTA not only reduces the likelihood of protein ubiquitination by shielding the hydrophobic TMD but also promotes active deubiquitination of BAG6 complex–associated proteins (Leznicki and High, 2012). In this way, SGTA could contribute to the recovery of nascent TA proteins aberrantly marked for degradation by BAG6-associated ubiquitin ligases. Notably, the interplay between the BAG complex and SGTA in determining protein fate extends beyond TA proteins to other membrane proteins targeted cotranslationally (Leznicki and High, 2020). Intriguingly, it was recently shown that ubiquitination of TA proteins can occur independently of BAG6, and that ubiquitinated TA proteins can still be handled by TRC40 and directed to the ER, where they are fully deubiquitinated by USP20/USP33 before or after membrane insertion (Fig. 2; Culver and Mariappan, 2021). It remains unclear mechanistically how exactly ubiquitinated TA proteins evade proteosome-mediated degradation in the cytosol, but it is possible that either the nature of the ubiquitination and/or rapid capture by TRC40 enables them to efficiently reach their destination and be deubiquitinated. It will be interesting to determine if this ubiquitination-deubiquitination cycle simply represents a futile mislabeling and recovery process or whether it fulfils a specific function during TA protein biogenesis.
Delivery of TA proteins into the ER
Both the GET/TRC receptor and the EMC complex have emerged as gateways into the ER for TA proteins (Guna et al., 2018; Schuldiner et al., 2008; Vilardi et al., 2011; Yamamoto and Sakisaka, 2012). TA proteins destined for ER insertion via the GET/TRC receptor converge on the homodimeric cytosolic chaperone Get3/TRC40, which escorts them to the ER-bound GET/TRC insertase (McDowell et al., 2020; Wang et al., 2014). Docking of ADP and TA protein–bound Get3/TRC40 onto the GET receptor allows transfer of the TA protein to the receptor, which subsequently mediates their insertion into the membrane (Wang et al., 2014). Then, upon ADP release, Get3/TRC40 is recycled for another round of ATP binding and TA protein targeting (reviewed in detail in Chio et al., 2017).
Evolutionary conservation of the GET receptor complex
The Get1 component of the GET receptor is a member of the Oxa1 superfamily of insertase proteins, which includes bacterial YidC and eukaryotic EMC3 (Anghel et al., 2017; McDowell et al., 2021), and it shares its three-TMD topology with other members of this family. Get1 sequence conservation among different phyla readily facilitated the identification of homologues in mammals (WRB) and plants (AtGet1; Srivastava et al., 2017; Xing et al., 2017). In contrast, Get2 homologues are more divergent, and sequence conservation is limited to the functionally essential N-terminal Get3-binding sequence and, to a lesser extent, the three TMDs (Borgese, 2020). CAML has nevertheless been identified as a functional and structural homologue of Get2 in mammals (Yamamoto and Sakisaka, 2012), and demonstrated to complement phenotypes associated with loss of Get1/Get2 when coexpressed with WRB in budding yeast (Vilardi et al., 2014). The existence of Get2 orthologues or functional homologues in other phyla were uncertain for a long time. The recent identification of the archaeplastidic Get2 homologue AtGet2(Asseck et al., 2021) and in silico prediction of Get2 homologues in mollusks and arthropods (Borgese, 2020), however, now strongly support that not only Get1/WRB, but the GET receptor complex as a whole is conserved among eukaryotes.
Architecture and stoichiometry of the GET receptor
The interaction between Get1/WRB and Get2/CAML is mediated by their TMDs, which are also necessary for the insertase function of the complex (Vilardi et al., 2014; Wang et al., 2014). Moreover, mammalian WRB and CAML, and likely other homologues as well, are thought to exist as an obligate complex, mutually stabilizing each other. Indeed, the expression levels of WRB or CAML decrease in the absence of the other subunit, likely because of destabilization of the remaining subunit (Colombo et al., 2016; Rivera-Monroy et al., 2016). However, the effects of the individual components seem to be asymmetric on each other. Namely, WRB can insert into the ER membrane correctly in the absence of CAML and is later degraded as an orphan subunit, whereas WRB is required for CAML to assume a correct topology in the first place (Carvalho et al., 2019; Inglis et al., 2020). More specifically, the second TMD of CAML remains exposed to the ER lumen in the absence of WRB, where it acts as a degron, triggering protein degradation. Interestingly, its topology can be corrected posttranslationally, and its degradation prevented when WRB is available in the membrane (Inglis et al., 2020). It remains to be seen whether a similar inter-subunit interplay also occurs in other species; however, results from A. thaliana imply that correct assembly of the GET receptor may be differently controlled in plants, as ectopically expressed AtGet2 remains stable in the absence of AtGet1 (Asseck et al., 2021).
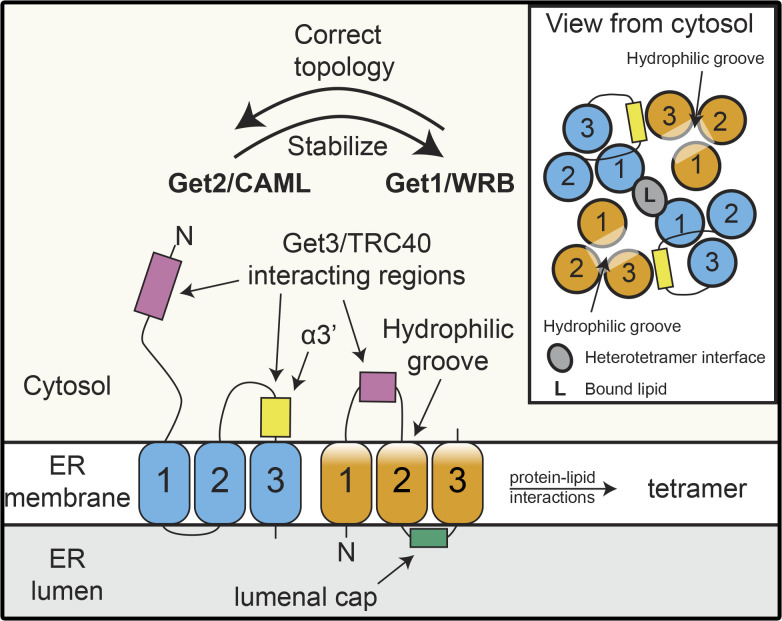
Although formation of a Get1/Get2 heterodimer is recognized as a prerequisite for a minimal functional receptor complex, a higher-order stoichiometry of the insertion-competent GET receptor complex has long been actively discussed. Due to the symmetric, homodimeric nature of Get3 in the TA protein–loaded complex, two analogous binding sites for both Get1 and Get2 are offered (Stefer et al., 2011). Despite a partial overlap of the Get1 and Get2 interaction sites on Get3, simultaneous binding of Get3 by Get1 and Get2 has been observed in crystal structures of Get3, together with the cytosolic domains of Get1 and Get2 (Stefer et al., 2011). This gave rise to the notion of a heterotetrameric structure of the GET receptor composed of two Get1 and two Get2 subunits, which could bind a single Get3 dimer with high affinity (Mariappan et al., 2011; Stefer et al., 2011). However, results obtained with in vitro reconstituted proteoliposomes demonstrated that a single dimer of Get1/Get2 can be sufficient for insertion of TA substrates into the membrane (Zalisko et al., 2017). Nevertheless, the heterotetrameric arrangement is supported by recent high-resolution cryo-EM structures of Get3/TRC40 homodimers docked onto the yeast and mammalian GET receptors, which reveal the formation of a heterotetrameric receptor complex upon Get3/TRC40 binding (Figs. 1, 2, and 3; McDowell et al., 2020). These new structures further consolidate the previously proposed model (Mariappan et al., 2011; Stefer et al., 2011) that Get3/TRC40 is initially captured by the extended cytosolic domains of Get2/CAML before contacting Get1/WRB (Figs. 1, 2, and 3). This arrangement, coupled with the finding that Get1-Get2 can simultaneously bind two Get3 molecules (McDowell et al., 2020), opens the possibility for a relay system where translocation of a first Get3/TRC40-TA protein complex to Get1 immediately allows capture of a second substrate complex by Get2, potentially increasing the efficiency of receptor complex loading and minimizing the risk of mistargeting. Interestingly, the GET/TRC receptor seems to be stabilized at the heterotetramer interface by not only protein–protein but also protein–lipid interactions, indicating that the lipid environment of the ER membrane may also influence the oligomeric state of the receptor. Importantly, complementation experiments in yeast demonstrate that disrupting lipid binding and thus the formation of the heterotetramer leads to in vivo loss of function of the receptor manifesting in TA protein mislocalization (McDowell et al., 2020). Therefore, although a Get1/Get2 dimer appears to be sufficient for the insertase function of the receptor in vitro, it is highly likely that a tetrameric complex is required to ensure efficient and accurate TA protein targeting within the cellular environment.
Figure 3.
Architecture of the Get1/WRB and Get2/CAML in the GET/TRC receptor complex. Get1/WRB and Get2/CAML both possess three TMDs (labeled 1–3) and rely on each other for stability and correct assembly within the ER membrane. The cytosolic regions of Get2/CAML (N-terminus and a loop between TMD 2 and 3) and a cytosolic region of Get1/WRB between TMDs 1 and 2 are docking sites for Get3/TRC40 carrying a TA protein cargo. A hydrophilic groove formed by the Get1/WRB TMDs and Get2/CAML TMD3 is proposed to serve as an insertion route for TA proteins to enter the membrane. Assembly of the heterodimer shown here into the final heterotetrameric structure of the receptor upon Get3/TRC40 binding involves protein–lipid interactions.
Mechanistic view of TA protein insertion into the lipid bilayer
Recent structural advances provide exciting mechanistic insights into the details of TA insertion, both by the GET/TRC receptor (McDowell et al., 2020) and the EMC complex (Bai et al., 2020; Miller-Vedam et al., 2020; O’Donnell et al., 2020; Pleiner et al., 2020). In the case of the GET/TRC receptor, within the membrane, the TMDs of WRB, together with TMD3 of CAML are arranged such that a hydrophilic groove, sealed at the luminal face but accessible from the cytosol, is assembled (Fig. 3). This likely serves as a substrate entry point with the charged, extreme C-terminus of the TA protein drawn in by interactions with the numerous hydrophilic residues of the receptor channel, thus bringing the TMD in close proximity to the destabilized bilayer, allowing insertion. This corroborates previous results obtained by cross-linking nascent TA substrates to the receptor in vitro, which pinpointed the region around the hydrophilic groove as the entry point for TA proteins into the membrane (Wang et al., 2014). Interestingly, assembly of the GET receptor as a heterotetrameric complex means that tandem hydrophilic grooves generated by each Get1/WRB-Get2/CAML pair represent two alternative routes into the membrane. The asymmetric binding of the TA protein within the Get3 dimer (Mateja et al., 2015), means that depending on the orientation of the docking, insertion via a particular channel will be favored. Dynamic modeling of interactions between the receptor and the TRC40 dimer in different conformations/nucleotide-bound states (Mateja et al., 2015; Stefer et al., 2011) suggest that transition of Get3/TRC40 to the open conformation leads to rearrangement of complex such that the C-terminus of the released TA protein is oriented toward the hydrophilic groove (McDowell et al., 2020). Complementary structural analyses of the EMC complexes (Bai et al., 2020; Miller-Vedam et al., 2020; O’Donnell et al., 2020; Pleiner et al., 2020) reveal analogous hydrophilic groove features, indicating that a common insertion mechanism is used by evolutionarily distant membrane receptors to accomplish insertion of a diverse set of membrane proteins (Bai and Li, 2021; McDowell et al., 2021). For the GET/TRC receptor, it is not yet clear how exactly the TA protein transits from this hydrophilic groove to become fully immersed in the membrane, but an amphipathic helix of Get1/WRB that lies close to the membrane has been suggested to cause membrane distortions that could facilitate TA protein integration.
Another key feature of the GET receptor revealed by the new structures is a short helix, α3′, present in the cytosolic domains of both yeast Get2 and human CAML, which binds the TA-binding domain of Get3/TRC40 and serves a gating function for the hydrophilic groove (Fig. 3; McDowell et al., 2020). Contact between the Get2/CAML helix and the Get3/TRC40 TA-binding domain induce conformational rearrangements crucial to the release and insertion of TA substrates in vivo, further underlining the role the GET receptor plays in stimulating substrate release from Get3. It remains to be determined whether, mechanistically, this helix facilitates TA protein insertion by preventing backsliding of the TMD out of the hydrophilic groove or actively driving the TA protein into the channel.
Quality control and rescue of TA protein targeting
The multiple possible destinations for TA proteins, together with the broad spectrum of physical properties of secretory pathway TA proteins, the existence of alternative ER targeting and insertion strategies, and the complexity of ensuring optimal capture, renders targeting of TA proteins to the ER inherently prone to errors. Defects in this process can manifest as either redirection of ER TA proteins to other membranes or the spurious ER insertion of non-ER TA proteins. Given the importance of TA protein functions and the toxic effects of cytosolic protein aggregates, either of these scenarios can have seriously detrimental effects on cells, necessitating robust surveillance, recovery, and degradation pathways (Jiang, 2021).
Removal of TA proteins misdirected to the OMM
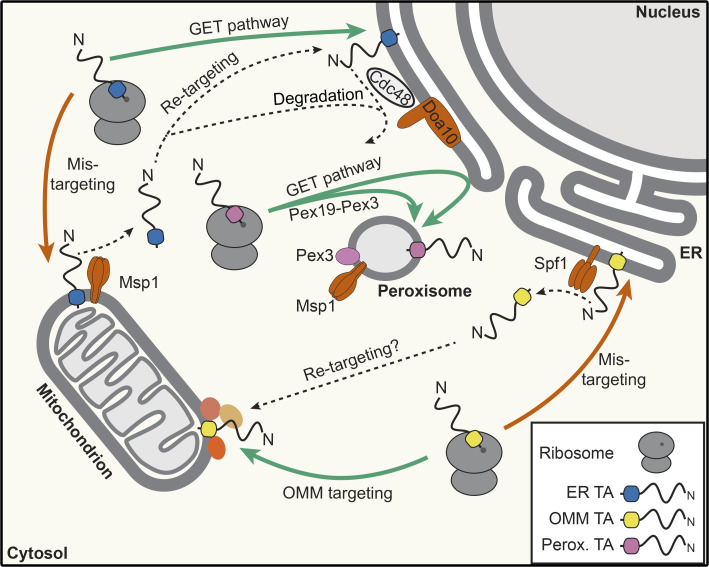
In contrast to ER TA proteins, some TA proteins of the OMM have been proposed to be inserted directly, potentially due to the specific lipid composition of the OMM (Figueiredo Costa et al., 2018). The hydrophobicity of their TMDs are lower than that of secretory pathway TA proteins (Borgese et al., 2001; Beilharz et al., 2003), which, considering that high TMD hydrophobicity is necessary for recognition by the GET pathway (Guna et al., 2018), helps explain how they avoid capture and subsequent delivery to the ER by Get3. In contrast, peroxisomal TA proteins can either use the Pex19-Pex3 machinery to directly reach peroxisomes (reviewed in Mayerhofer, 2016) or be first targeted to the ER by the GET pathway (Schuldiner et al., 2008) before reaching peroxisomes (Fig. 4).
Figure 4.
Quality control machineries regulating distribution of mislocalized TA proteins between the ER and other organelles. ER-destined and peroxisome (Perox.)-destined TA proteins can mislocalize to the OMM, and OMM TA proteins can mislocalize to the ER. ATP-dependent machines in these membranes can recognize and displace mistargeted protein while correctly localized proteins are retained, for example by interactions with binding partners (indicated by unnamed, colored circles on the OMM). Nomenclature is as in yeast.
It has been observed that ER-inserted TA proteins can mislocalize to mitochondria when the functionality of the GET pathway is impaired (Schuldiner et al., 2008). A salient example is yeast Pex15, which is a peroxisome-destined TA protein first inserted into the ER but that mislocalizes to mitochondria not only when the C-terminal 30 amino acids or Pex19 are lacking, but also in the absence of Get3 (Schuldiner et al., 2008; Okreglak and Walter, 2014; Li et al., 2019). Furthermore, a basal level of mistargeting of secretory pathway TA proteins, such as yeast Gos1, to the OMM is observed even in the presence of a functional GET/TRC pathway, implying that some level of mistargeting is unavoidable (Chen et al., 2014). This therefore raises the question of how mistargeted TA proteins are recognized and cleared while the appropriately localized proteins are retained.
A mechanism by which TA proteins incorrectly inserted into the OMM can be extracted has recently been described; the highly evolutionarily conserved mitochondrial and peroxisomal AAA-ATPase Msp1 (ATAD1 in mammals) has been shown to be essential for the removal of mitochondrially mislocalized Pex15 and is thought to act as a general dislocase of TA proteins mislocalized to the mitochondria (Fig. 4; Chen et al., 2014; Okreglak and Walter, 2014). This explains why double-mutant yeast strains lacking both GET pathway components and Msp1 mislocalize TA proteins noticeably to mitochondria (Li et al., 2019). Accordingly, GET pathway components and Msp1 show a synthetic negative genetic effect (Chen et al., 2014; Okreglak and Walter, 2014), emphasizing the importance of high-fidelity protein targeting and quality control. Msp1 forms hexamers in the OMM, and in vitro, its ATPase activity is sufficient to drive the removal of TA proteins from proteoliposomes (Wohlever et al., 2017), demonstrating that Msp1 alone can both recognize its substrates and drive their subsequent extraction from the membrane. Recent visualizations of Msp1-substrate complexes provide insights into the mechanism of membrane extraction; the TA protein substrate enters via a single, hydrophobic site and is then anchored within the hydrophobic pore by a network of aromatic amino acids (Wang et al., 2020). ATP hydrolysis, the driving force for membrane extraction, is coordinated with subunit positioning within the complex via specific elements at the subunit interfaces (Wang et al., 2020). Interestingly, based on in vitro experiments with MBP-tagged Pex3, it has been suggested that the unfoldase activity of Msp1 may be regulated by Pex3 on peroxisomes (Castanzo et al., 2020), but it remains to be determined whether a similar regulatory mechanism exists in the OMM as well.
The stringency of recognition of mitochondrially mislocalized TA proteins relies on at least a twofold recognition mechanism by Msp1. First, basic residues typical of the luminal tails of peroxisomal TAs are recognized when exposed in the mitochondrial intermembrane space (Li et al., 2019). Second, exposed hydrophobic amino acids close to the membrane, present in various secretory pathway and peroxisomal TA proteins, also act as a recognition signal for Msp1 (Li et al., 2019). Furthermore, there is evidence indicating that orphan TA proteins lacking binding partners within the OMM are more readily recognized and displaced by Msp1 (Weir et al., 2017; Dederer et al., 2019). This indicates that besides the biophysical properties of Msp1 substrates, their ability to form functional protein–protein interactions with other membrane proteins is an important discriminating factor that determines whether a TA protein is retained in or removed from the OMM. Mechanistically, lack of a binding partner may render mislocalized TA proteins less stably anchored within the membrane, or, when separated from their normal interaction partners, mistargeted proteins may be more likely to expose hydrophobic patches to the cytosol, both of which would increase the chance of expulsion by Msp1.
It is possible that after removal from the OMM by Msp1, secretory pathway TA proteins returned to the cytosol could be retargeted if they are captured by Get3/TRC40 or other chaperones capable of directing them to the ER-bound insertase machineries. However, a cellular machinery also needs to exist that degrades excess TA proteins in the cytosol or at the ER to ensure protein homeostasis. Indeed, the E3 ubiquitin ligase Doa10, an important player in ER-associated degradation, has recently emerged as a quality control factor responsible for sensing and ubiquitination of spurious and excess TA proteins ejected from the OMM. It has been suggested that Doa10-mediated ubiquitination may take place either in the cytosol (Dederer et al., 2019) or after retargeting to the ER, where their extraction and degradation is facilitated by the AAA-ATPase Cdc48 (Matsumoto et al., 2019; Fig. 4).
Mistargeting of mitochondrial TA proteins to the ER
It is not only the case that nonmitochondrial TA protein are misdirected to the OMM; also, reciprocal events can occur as OMM TMD-containing proteins are observed in the ER, especially when mitochondrial targeting signals are masked, when the mitochondrial import machineries are overloaded, or upon mitochondrial dysfunction (Hansen et al., 2018; Xiao et al., 2021; Vitali et al., 2018). More specifically, the GET pathway itself has been shown to contribute to the ER mislocalization of OMM TA proteins (Vitali et al., 2018; Xiao et al., 2021). While the high efficiency of mitochondrial targeting appears to keep such mistargeting to a minimum, perturbation of the equilibrium between OMM targeting and ER mistargeting by the GET pathway can lead to aberrant accumulation of clients in the wrong membrane. Similar to nonmitochondrial TA proteins ejected from the OMM by Msp1, a parallel ATP-dependent mechanism for displacing TA proteins incorrectly introduced into the ER has recently been discovered (Fig. 4; McKenna et al., 2020; Qin et al., 2020). Structural and biochemical analyses of the ER-bound P5A-type ATPase Spf1 (yeast)/ATP13A1 (human)/CATP-8 (Caenorhabditis elegans) have identified it as a major quality control factor in the ER. Spf1 contains a substrate- binding pocket, laterally accessible from the membrane, that has been proposed to flip ER-associated proteins, promoting their release back into the cytosol or their topological rearrangement. Precisely how specificity for mislocalized/misinserted proteins is achieved remains unclear, but it is suggested that the lower hydrophobicity of non-ER TMDs may favor their dislocation. Furthermore, in cells lacking Spf1, the ergosterol content of the ER is altered to more closely resemble the OMM, implying that membrane composition may contribute to TA protein distribution and that Spf1 could also influence TA protein mislocalization by regulating the lipid composition of membranes (Krumpe et al., 2012). In yet another analogy to Msp1-mediated rejection of mislocalized TA proteins from the OMM, it is likely that mitochondrial proteins extracted from the ER may then be successfully retargeted to their desired destination. Indeed, the recently described ER–surface-mediated protein targeting pathway sets a precedent for such a route (Hansen et al., 2018).
Concluding remarks
The initial momentum of the TA protein–targeting field following the discovery of the GET/TRC pathway has not abated. Recent years have seen not only a deepening mechanistic understanding of the intricacies of this pathway but also growing knowledge on its interplay with other targeting pathways and the safety nets present to ensure the fidelity of TA protein targeting. Building on the early structural analyses of individual proteins and protein domain complexes, recent advances in cryo-EM have now enabled visualization of larger complexes, such as the targeting factor–bound membrane insertase, allowing the route of TA proteins from the cytosol into the ER membrane to be anticipated. Alongside high-throughput microscopy–based screens, further analyses of TA protein targeting in cells have uncovered alternative routes that TA protein can take to reach the ER and have highlighted functional redundancies. The existence of different pathways for TA protein targeting and insertion into the ER, on the one hand, suggest a robust system with backup strategies in place in case a client protein escapes its normal targeting route, but on the other hand, emphasize the complexity of the capture process and selection of the optimal targeting pathway. In turn, these additional layers of complexity underscore the need for quality control pathways. The discovery of active removal of mislocalized TA proteins not only provides a mechanism for such quality control but perhaps also suggests a dynamic aspect to TA protein targeting wherein TA proteins within membranes can be removed and then either retargeted to their correct destination or reinserted if appropriate. Altogether, a picture emerges of an array of capture, insertion, and ejection machineries that are finely balanced in terms of substrate preferences to optimize TA protein localization within the cell.
Acknowledgments
We thank Blanche Schwappach for her support and helpful comments on the manuscript.
Work in the Bohnsack laboratory on the GET pathway is funded by the Deutsche Forschungsgemeinschaft (SFB1190 to K.E. Bohnsack).
The authors declare no competing financial interests.
Author contributions: K.E. Bohnsack and A. Farkas wrote the manuscript and prepared the figures.
References
- Akopian, D., Shen K., Zhang X., and Shan S.O.. 2013. Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82:693–721. 10.1146/annurev-biochem-072711-164732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anghel, S.A., McGilvray P.T., Hegde R.S., and Keenan R.J.. 2017. Identification of Oxa1 Homologs Operating in the Eukaryotic Endoplasmic Reticulum. Cell Rep. 21:3708–3716. 10.1016/j.celrep.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseck, L.Y., Mehlhorn D.G., Monroy J.R., Ricardi M.M., Breuninger H., Wallmeroth N., Berendzen K.W., Nowrousian M., Xing S., Schwappach B., et al. 2021. Endoplasmic reticulum membrane receptors of the GET pathway are conserved throughout eukaryotes. Proc. Natl. Acad. Sci. USA. 118:e2017636118. 10.1073/pnas.2017636118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram, N., Ast T., Costa E.A., Arakel E.C., Chuartzman S.G., Jan C.H., Haßdenteufel S., Dudek J., Jung M., Schorr S., et al. 2016. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 540:134–138. 10.1038/nature20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, L., and Li H.. 2021. Cryo-EM structures of the endoplasmic reticulum membrane complex. FEBS J.:febs.15786. 10.1111/febs.15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, L., You Q., Feng X., Kovach A., and Li H.. 2020. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature. 584:475–478. 10.1038/s41586-020-2389-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz, T., Egan B., Silver P.A., Hofmann K., and Lithgow T.. 2003. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 278:8219–8223. 10.1074/jbc.M212725200 [DOI] [PubMed] [Google Scholar]
- Berndt, U., Oellerer S., Zhang Y., Johnson A.E., and Rospert S.. 2009. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA. 106:1398–1403. 10.1073/pnas.0808584106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese, N.2020. Searching for remote homologs of CAML among eukaryotes. Traffic. 21:647–658. 10.1111/tra.12758 [DOI] [PubMed] [Google Scholar]
- Borgese, N., Gazzoni I., Barberi M., Colombo S., and Pedrazzini E.. 2001. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Cell Biol. 12(8):2482–2496. 10.1091/mbc.12.8.2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese, N., Coy-Vergara J., Colombo S.F., and Schwappach B.. 2019. The Ways of Tails: the GET Pathway and more. Protein J. 38:289–305. 10.1007/s10930-019-09845-4 [DOI] [PubMed] [Google Scholar]
- Bozkurt, G., Stjepanovic G., Vilardi F., Amlacher S., Wild K., Bange G., Favaloro V., Rippe K., Hurt E., Dobberstein B., and Sinning I.. 2009. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. USA. 106:21131–21136. 10.1073/pnas.0910223106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, H.J.F., Del Bondio A., Maltecca F., Colombo S.F., and Borgese N.. 2019. The WRB Subunit of the Get3 Receptor is Required for the Correct Integration of its Partner CAML into the ER. Sci. Rep. 9:11887. 10.1038/s41598-019-48363-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson, J., McKenna M., Haßdenteufel S., Aviram N., Zimmerman R., and High S.. 2017. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 130:3851–3861. 10.1242/jcs.207829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanzo, D.T., LaFrance B., and Martin A.. 2020. The AAA+ ATPase Msp1 is a processive protein translocase with robust unfoldase activity. Proc. Natl. Acad. Sci. USA. 117:14970–14977. 10.1073/pnas.1920109117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.-W., Chuang Y.-C., Ho Y.-C., Cheng M.-Y., Sun Y.-J., Hsiao C.-D., and Wang C.. 2010. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 285:9962–9970. 10.1074/jbc.M109.087098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron, J.W., Suloway C.J.M., Zaslaver M., and Clemons W.M. Jr. 2010. Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. USA. 107:12127–12132. 10.1073/pnas.1006036107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron, J.W., VanderVelde D.G., and Clemons W.M. Jr. 2012a. Structures of the Sgt2/SGTA dimerization domain with the Get5/UBL4A UBL domain reveal an interaction that forms a conserved dynamic interface. Cell Rep. 2:1620–1632. 10.1016/j.celrep.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron, J.W., VanderVelde D.G., Rao M., and Clemons W.M. Jr. 2012b. Get5 carboxyl-terminal domain is a novel dimerization motif that tethers an extended Get4/Get5 complex. J. Biol. Chem. 287:8310–8317. 10.1074/jbc.M111.333252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-C., Umanah G.K.E., Dephoure N., Andrabi S.A., Gygi S.P., Dawson T.M., Dawson V.L., and Rutter J.. 2014. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 33:1548–1564. 10.15252/embj.201487943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio, U.S., Cho H., and Shan S.O.. 2017. Mechanisms of Tail-Anchored Membrane Protein Targeting and Insertion. Annu. Rev. Cell Dev. Biol. 33:417–438. 10.1146/annurev-cellbio-100616-060839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, P.J., Juszkiewicz S., Guna A., Shao S., and Hegde R.S.. 2018. EMC Is Required to Initiate Accurate Membrane Protein Topogenesis. Cell. 175:1507–1519.e16. 10.1016/j.cell.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H., and Shan S.O.. 2018. Substrate relay in an Hsp70-cochaperone cascade safeguards tail-anchored membrane protein targeting. EMBO J. 37:e99264. 10.15252/embj.201899264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H., Shim W.J., Liu Y., and Shan S.O.. 2021. J-domain proteins promote client relay from Hsp70 during tail-anchored membrane protein targeting. J. Biol. Chem. 296:100546. 10.1016/j.jbc.2021.100546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, S.F., Cardani S., Maroli A., Vitiello A., Soffientini P., Crespi A., Bram R.F., Benfante R., and Borgese N.. 2016. Tail-anchored protein biogenesis in mammals: function and reciprocal interactions of the two subunits of the TRC40 receptor. J. Biol. Chem. 291:15292–15306. 10.1074/jbc.M115.707752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy-Vergara, J., Rivera-Monroy J., Urlaub H., Lenz C., and Schwappach B.. 2019. A trap mutant reveals the physiological client spectrum of TRC40. J. Cell Sci. 132:jcs230094. 10.1242/jcs.230094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver, J.A., and Mariappan M.. 2021. Deubiquitinases USP20/33 promote the biogenesis of tail-anchored membrane proteins. J. Cell Biol. 220:e202004086. 10.1083/jcb.202004086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dederer, V., Khmelinskii A., Huhn A.G., Okreglak V., Knop M., and Lemberg M.K.. 2019. Cooperation of mitochondrial and ER factors in quality control of tail-anchored proteins. eLife. 8:e45506. 10.7554/eLife.45506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo Costa, B., Cassella P., Colombo S.F., and Borgese N.. 2018. Discrimination between the endoplasmic reticulum and mitochondria by spontaneously inserting tail-anchored proteins. Traffic. 19:182–197. 10.1111/tra.12550 [DOI] [PubMed] [Google Scholar]
- Fleischer, T.C., Weaver C.M., McAfee K.J., Jennings J.L., and Link A.J.. 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20:1294–1307. 10.1101/gad.1422006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guna, A., Volkmar N., Christianson J.C., and Hegde R.S.. 2018. The ER membrane protein complex is a transmembrane domain insertase. Science. 359:470–473. 10.1126/science.aao3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, K.G., Aviram N., Laborenz J., Bibi C., Meyer M., Spang A., Schuldiner M., and Herrmann J.M.. 2018. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science. 361:1118–1122. 10.1126/science.aar8174 [DOI] [PubMed] [Google Scholar]
- Holtkamp, W., Lee S., Bornemann T., Senyushkina T., Rodnina M.V., and Wintermeyer W.. 2012. Dynamic switch of the signal recognition particle from scanning to targeting. Nat. Struct. Mol. Biol. 19:1332–1337. 10.1038/nsmb.2421 [DOI] [PubMed] [Google Scholar]
- Horie, C., Suzuki H., Sakaguchi M., and Mihara K.. 2002. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell. 13:1615–1625. 10.1091/mbc.01-12-0570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis, A.J., Page K.R., Guna A., and Voorhees R.M.. 2020. Differential Modes of Orphan Subunit Recognition for the WRB/CAML Complex. Cell Rep. 30:3691–3698.e5. 10.1016/j.celrep.2020.02.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H.2021. Quality control pathways of tail-anchored proteins. Biochim. Biophys. Acta Mol. Cell Res. 1868:118922. 10.1016/j.bbamcr.2020.118922 [DOI] [PubMed] [Google Scholar]
- Jonikas, M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S., and Schuldiner M.. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 323:1693–1697. 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch, T., Cambon A., and Wattenberg B.W.. 2007. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic. 8:1687–1694. 10.1111/j.1600-0854.2007.00661.x [DOI] [PubMed] [Google Scholar]
- Kriechbaumer, V., Shaw R., Mukherjee J., Bowsher C.G., Harrison A.M., and Abell B.M.. 2009. Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic. 10:1753–1764. 10.1111/j.1600-0854.2009.00991.x [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson B., von Heijne G., and Sonnhammer E.L.L.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Krumpe, K., Frumkin I., Herzig Y., Rimon N., Özbalci C., Brügger B., Rapaport D., and Schuldiner M.. 2012. Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol. Biol. Cell. 23:3927–3935. 10.1091/mbc.e11-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysztofinska, E.M., Evans N.J., Thapaliya A., Murray J.W., Morgan R.M.L., Martinez-Lumbreras S., and Isaacson R.L.. 2017. Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp. Front. Mol. Biosci. 4:68. 10.3389/fmolb.2017.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay, U., Hartmann E., and Rapoport T.A.. 1993. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3:72–75. 10.1016/0962-8924(93)90066-A [DOI] [PubMed] [Google Scholar]
- Leznicki, P., and High S.. 2012. SGTA antagonizes BAG6-mediated protein triage. Proc. Natl. Acad. Sci. USA. 109:19214–19219. 10.1073/pnas.1209997109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki, P., and High S.. 2020. SGTA associates with nascent membrane protein precursors. EMBO Rep. 21:e48835. 10.15252/embr.201948835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zheng J., Wu X., and Jiang H.. 2019. Mitochondrial AAA-ATPase Msp1 detects mislocalized tail-anchored proteins through a dual-recognition mechanism. EMBO Rep. 20:e46989. 10.15252/embr.201846989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan, M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R.J., and Hegde R.S.. 2010. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 466:1120–1124. 10.1038/nature09296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan, M., Mateja A., Dobosz M., Bove E., Hegde R.S., and Keenan R.J.. 2011. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 477:61–66. 10.1038/nature10362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja, A., and Keenan R.J.. 2018. A structural perspective on tail-anchored protein biogenesis by the GET pathway. Curr. Opin. Struct. Biol. 51:195–202. 10.1016/j.sbi.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja, A., Paduch M., Chang H.-Y., Szydlowska A., Kossiakoff A.A., Hegde R.S., and Keenan R.J.. 2015. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 347:1152–1155. 10.1126/science.1261671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja, A., Szlachcic A., Downing M.E., Dobosz M., Mariappan M., Hegde R.S., and Keenan R.J.. 2009. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 461:361–366. 10.1038/nature08319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, S., Nakatsukasa K., Kakuta C., Tamura Y., Esaki M., and Endo T.. 2019. Msp1 Clears Mistargeted Proteins by Facilitating Their Transfer from Mitochondria to the ER. Mol. Cell. 76:191–205.e10. 10.1016/j.molcel.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Mayerhofer, P.U.2016. Targeting and insertion of peroxisomal membrane proteins: ER trafficking versus direct delivery to peroxisomes. Biochim. Biophys. Acta. 1863:870–880. 10.1016/j.bbamcr.2015.09.021 [DOI] [PubMed] [Google Scholar]
- McDowell, M.A., Heimes M., Fiorentino F., Mehmood S., Farkas Á., Coy-Vergara J., Wu D., Bolla J.R., Schmid V., Heinze R., et al. 2020. Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex. Mol. Cell. 80:72–86.e7. 10.1016/j.molcel.2020.08.012 [DOI] [PubMed] [Google Scholar]
- McDowell, M.A., Heimes M., and Sinning I.. 2021. Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat. Struct. Mol. Biol. 28:234–239. 10.1038/s41594-021-00567-9 [DOI] [PubMed] [Google Scholar]
- McKenna, M.J., Sim S.I., Ordureau A., Wei L., Harper J.W., Shao S., and Park E.. 2020. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science. 369:eabc5809. 10.1126/science.abc5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Vedam, L.E., Bräuning B., Popova K.D., Schirle Oakdale N.T., Bonnar J.L., Prabu J.R., Boydston E.A., Sevillano N., Shurtleff M.J., Stroud R.M., et al. 2020. Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. eLife. 9:e62611. 10.7554/eLife.62611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock, J.-Y., Chartron J.W., Zaslaver M., Xu Y., Ye Y., and Clemons W.M. Jr. 2015. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proc. Natl. Acad. Sci. USA. 112:106–111. 10.1073/pnas.1402745112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell, J.P., Phillips B.P., Yagita Y., Juszkiewicz S., Wagner A., Malinverni D., Keenan R.J., Miller E.A., and Hegde R.S.. 2020. The architecture of EMC reveals a path for membrane protein insertion. eLife. 9:e57887. 10.7554/eLife.57887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg, S.C., and Walter P.. 1995. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell. 81:1075–1084. 10.1016/S0092-8674(05)80012-1 [DOI] [PubMed] [Google Scholar]
- Okreglak, V., and Walter P.. 2014. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. USA. 111:8019–8024. 10.1073/pnas.1405755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiner, T., Tomaleri G.P., Januszyk K., Inglis A.J., Hazu M., and Voorhees R.M.. 2020. Structural basis for membrane insertion by the human ER membrane protein complex. Science. 369:433–436. 10.1126/science.abb5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Q., Zhao T., Zou W., Shen K., and Wang X.. 2020. An Endoplasmic Reticulum ATPase Safeguards Endoplasmic Reticulum Identity by Removing Ectopically Localized Mitochondrial Proteins. Cell Rep. 33:108363. 10.1016/j.celrep.2020.108363 [DOI] [PubMed] [Google Scholar]
- Rao, M., Okreglak V., Chio U.S., Cho H., Walter P., and Shan S.O.. 2016. Multiple selection filters ensure accurate tail-anchored membrane protein targeting. eLife. 5:e21301. 10.7554/eLife.21301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport, T.A., Li L., and Park E.. 2017. Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 33:369–390. 10.1146/annurev-cellbio-100616-060439 [DOI] [PubMed] [Google Scholar]
- Rivera-Monroy, J., Musiol L., Unthan-Fechner K., Farkas Á., Clancy A., Coy-Vergara J., Weill U., Gockel S., Lin S.-Y., Corey D.P., et al. 2016. Mice lacking WRB reveal differential biogenesis requirements of tail-anchored proteins in vivo. Sci. Rep. 6:39464. 10.1038/srep39464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni, M.C., Gutierrez E., and Hegde R.S.. 2014. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol. Cell. 55:227–237. 10.1016/j.molcel.2014.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner, M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H.D., Schwappach B., and Weissman J.S.. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 134:634–645. 10.1016/j.cell.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, S., Rodrigo-Brenni M.C., Kivlen M.H., and Hegde R.S.. 2017. Mechanistic basis for a molecular triage reaction. Science. 355:298–302. 10.1126/science.aah6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff, M.J., Itzhak D.N., Hussmann J.A., Schirle Oakdale N.T., Costa E.A., Jonikas M., Weibezahn J., Popova K.D., Jan C.H., Sinitcyn P., et al. 2018. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. eLife. 7:e37018. 10.7554/eLife.37018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, A.C., Simpson P.J., Goldstone R.M., Krysztofinska E.M., Murray J.W., High S., and Isaacson R.L.. 2013. Structure of the Sgt2/Get5 complex provides insights into GET-mediated targeting of tail-anchored membrane proteins. Proc. Natl. Acad. Sci. USA. 110:1327–1332. 10.1073/pnas.1207518110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, R., Zalisko B.E., Keenan R.J., and Howell S.H.. 2017. The GET System Inserts the Tail-Anchored Protein, SYP72, into Endoplasmic Reticulum Membranes. Plant Physiol. 173:1137–1145. 10.1104/pp.16.00928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic, S., and Hegde R.S.. 2007. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 128:1147–1159. 10.1016/j.cell.2007.01.036 [DOI] [PubMed] [Google Scholar]
- Stefer, S., Reitz S., Wang F., Wild K., Pang Y.-Y., Schwarz D., Bomke J., Hein C., Löhr F., Bernhard F., et al. 2011. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 333:758–762. 10.1126/science.1207125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway, C.J.M., Chartron J.W., Zaslaver M., and Clemons W.M. Jr. 2009. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. USA. 106:14849–14854. 10.1073/pnas.0907522106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi, F., Lorenz H., and Dobberstein B.. 2011. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J. Cell Sci. 124:1301–1307. 10.1242/jcs.084277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi, F., Stephan M., Clancy A., Janshoff A., and Schwappach B.. 2014. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS One. 9:e85033. 10.1371/journal.pone.0085033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali, D.G., Sinzel M., Bulthuis E.P., Kolb A., Zabel S., Mehlhorn D.G., Figueiredo Costa B., Farkas Á., Clancy A., Schuldiner M., et al. 2018. The GET pathway can increase the risk of mitochondrial outer membrane proteins to be mistargeted to the ER. J. Cell Sci. 131:jcs211110. 10.1242/jcs.211110 [DOI] [PubMed] [Google Scholar]
- Voorhees, R.M., and Hegde R.S.. 2015. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife. 4:e07975. 10.7554/eLife.07975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Brown E.C., Mak G., Zhuang J., and Denic V.. 2010. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell. 40:159–171. 10.1016/j.molcel.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Chan C., Weir N.R., and Denic V.. 2014. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature. 512:441–444. 10.1038/nature13471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Myasnikov A., Pan X., and Walter P.. 2020. Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction. eLife. 9:e54031. 10.7554/eLife.54031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, N.R., Kamber R.A., Martenson J.S., and Denic V.. 2017. The AAA protein Msp1 mediates clearance of excess tail-anchored proteins from the peroxisomal membrane. Elife. 6. e28507. 10.7554/eLife.28507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlever, M.L., Mateja A., McGilvray P.T., Day K.J., and Keenan R.J.. 2017. Msp1 Is a Membrane Protein Dislocase for Tail-Anchored Proteins. Mol. Cell. 67:194–202.e6. 10.1016/j.molcel.2017.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, T., Shakya V.P., and Hughes A.L.. 2021. ER targeting of non-imported mitochondrial carrier proteins is dependent on the GET pathway. Life Sci. Alliance. 4:e202000918. 10.26508/lsa.202000918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, S., Mehlhorn D.G., Wallmeroth N., Asseck L.Y., Kar R., Voss A., Denninger P., Schmidt V.A.F., Schwarzländer M., Stierhof Y.-D., et al. 2017. Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc. Natl. Acad. Sci. USA. 114:E1544–E1553. 10.1073/pnas.1619525114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y., and Sakisaka T.. 2012. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol. Cell. 48:387–397. 10.1016/j.molcel.2012.08.028 [DOI] [PubMed] [Google Scholar]
- Zalisko, B.E., Chan C., Denic V., Rock R.S., and Keenan R.J.. 2017. Tail-Anchored Protein Insertion by a Single Get1/2 Heterodimer. Cell Rep. 20:2287–2293. 10.1016/j.celrep.2017.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., De Laurentiis E., Bohnsack K.E., Wahlig M., Ranjan N., Gruseck S., Hackert P., Wölfle T., Rodnina M.V., Schwappach B., and Rospert S.. 2021. Ribosome-bound Get4/5 facilitates the capture of tail-anchored proteins by Sgt2 in yeast. Nat. Commun. 12:782. 10.1038/s41467-021-20981-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Schäffer T., Wölfle T., Fitzke E., Thiel G., and Rospert S.. 2016. Cotranslational Intersection between the SRP and GET Targeting Pathways to the Endoplasmic Reticulum of Saccharomyces cerevisiae. Mol. Cell. Biol. 36:2374–2383. 10.1128/MCB.00131-16 [DOI] [PMC free article] [PubMed] [Google Scholar]