Abstract
Direct neuronal reprogramming is an innovative new technology that involves the conversion of somatic cells to induced neurons (iNs) without passing through a pluripotent state. The capacity to make new neurons in the brain, which previously was not achievable, has created great excitement in the field as it has opened the door for the potential treatment of incurable neurodegenerative diseases and brain injuries such as stroke. These neurological disorders are associated with frank neuronal loss, and as new neurons are not made in most of the adult brain, treatment options are limited. Developmental biologists have paved the way for the field of direct neuronal reprogramming by identifying both intrinsic cues, primarily transcription factors (TFs) and miRNAs, and extrinsic cues, including growth factors and other signaling molecules, that induce neurogenesis and specify neuronal subtype identities in the embryonic brain. The striking observation that postmitotic, terminally differentiated somatic cells can be converted to iNs by mis-expression of TFs or miRNAs involved in neural lineage development, and/or by exposure to growth factors or small molecule cocktails that recapitulate the signaling environment of the developing brain, has opened the door to the rapid expansion of new neuronal reprogramming methodologies. Furthermore, the more recent applications of neuronal lineage conversion strategies that target resident glial cells in situ has expanded the clinical potential of direct neuronal reprogramming techniques. Herein, we present an overview of the history, accomplishments, and therapeutic potential of direct neuronal reprogramming as revealed over the last two decades.
Keywords: direct neuronal reprogramming, lineage conversion, transcription factors, micro-RNA, astrocytes, fibroblasts, small molecules, epigenetics
Introduction
Brain injury and neurodegenerative diseases are among the leading causes of disability-adjusted life years (DALYs) and deaths globally (GBD 2016 Neurology Collaborators, 2019). While etiologies differ, these neurological conditions share the common feature of neuronal loss in the brain and/or spinal cord, the major constituents of the central nervous system (CNS). Stroke, the most common type of brain injury results in lifelong cognitive and motor deficits, and can rob an individual of their capacity to participate productively in society and ultimately lead to early death (GBD 2016 Neurology Collaborators, 2019). Similarly, neurodegenerative disorders that impact the CNS are ultimately fatal and include Alzheimer’s disease (AD), frontotemporal dementia (FTD), other dementias (Korsnes and Winkler, 2020), Amyotrophic Lateral Sclerosis (ALS) (Xu et al., 2020), and Parkinson’s disease (PD) (GBD 2016 Parkinson’s Disease Collaborators, 2018). In all of these disorders, neuronal loss results in permanent damage as neurogenesis does not occur in most of the adult mammalian CNS. Indeed, there are only a few active neurogenic zones in the adult mammalian brain, including the ventricular-subventricular zone (V-SVZ) lining the lateral ventricles, which replenishes neurons in the olfactory bulb in mouse and the striatum in humans, and the subgranular zone (SGZ), which generates new neurons for the dentate gyrus in all mammals (Spalding et al., 2013; Ernst et al., 2014; Urban and Guillemot, 2014; Gotz et al., 2016; Ruddy and Morshead, 2017; Boldrini et al., 2018; Sorrells et al., 2018). The purpose of this review is to summarize the use of direct neuronal reprogramming strategies to convert somatic cells to induced neurons (iNs) as a novel method to create new neurons for brain repair, disease modeling and drug screening.
The concept of cellular reprogramming was first formalized in 2006, with the Nobel Prize winning discovery that terminally differentiated murine (and later human) fibroblasts could be converted to a pluripotent state by the expression of a cocktail of four transcription factors (TFs): Oct-3/4 (official gene name, Pou5f1), Sox2, Myc, and Klf4, now colloquially called the Yamanaka factors (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Figure 1). The ability to differentiate iPSCs into a variety of somatic cell types has facilitated disease modeling, drug discovery, and led to pre-clinical and clinical trials that investigate the use of iPSC cell products as therapeutic treatments (reviewed in Jha et al., 2021; Qian and Tcw, 2021). The principal advantage to iPSC-derived cell products is that they lend themselves to a personalized medicine approach, as a patient’s own cells are used as donor cells, so the non-renewability of most cell types does not act as a limitation. However, there are justified concerns with the use of iPSC-derived neurons, including the potential association of pluripotency and tumorigenicity (Lee et al., 2013).
FIGURE 1.
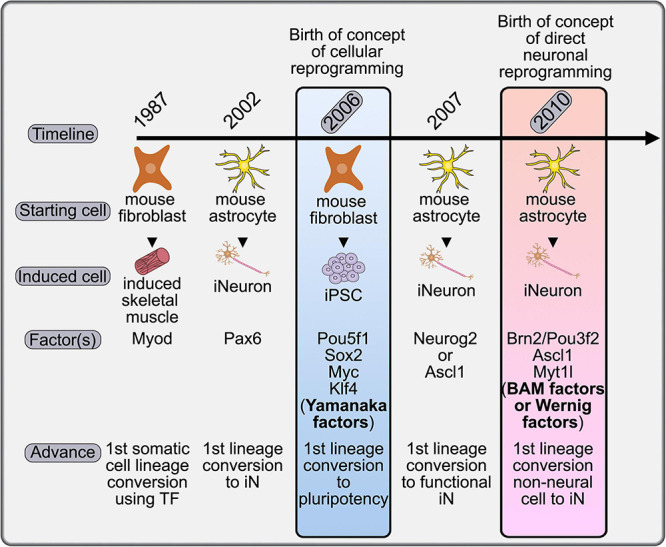
Timeline placing the concept of direct neuronal reprogramming in a historical context. The first evidence for somatic cell conversion was achieved in 1987 with the conversion of mouse fibroblasts to skeletal muscle cells. This procedure was adapted to the generation of iNeurons in 2002 and 2007 using astrocytes as a starting population, although not yet termed direct neuronal reprogramming. It was in 2006 that the field of cellular reprogramming was born with the demonstration that the Yamanaka factors could generate iPSCs. The formal launch of the direct neuronal reprogramming field came in 2010 with the identification of the BAM factors, which could convert fibroblasts, a non-neural cell type, to iNs without passing through a transient iPSC state.
Direct cellular reprogramming involves the conversion of one heterologous cell type to another without passing through a pluripotent stage, and is considered a major advance for therapeutic innovation because it removes the tumorigenic potential of iPSC-derived cells (Lee et al., 2013). Although not formally defined as “cellular reprogramming,” the transdifferentiation of one somatic cell to another preceded the Yamanaka findings by several years with the demonstration that murine C3H10T1/2 embryonic fibroblast cells could be converted to a skeletal muscle identity by the misexpression of a single TF, the basic-helix-loop-helix (bHLH) gene, Myod (Davis et al., 1987; Figure 1). In this review we summarize efforts to reprogram a variety of somatic cells to iNs using combinations of TFs, miRNAs, and small molecules, and the application of such an approach to disease modeling, drug screening, and brain repair.
Direct Neuronal Reprogramming in vitro
The conversion of somatic cells to iNs has been achieved using both human and murine cells, primarily including astrocytes and fibroblasts, but also other starting cell populations. The TFs, miRNAs, and extrinsic cues used for direct neuronal programming in vitro are summarized herein (Figure 2A). Developmental neuroscience has paved the way for these studies, with most reprogramming cocktails incorporating TFs that drive the sequential conversion of neural stem cells (NSCs) to neural progenitor cells (NPCs), to immature and then mature neurons (summarized in Table 1; Figure 2B). Future studies will be required to determine how the molecular regulators used in these cocktails interact with each other to form gene regulatory networks (GRNs), and importantly, to compare and contrast the GRNs associated with neuronal reprogramming vs. those driving neurogenesis in the embryonic brain (e.g., Han et al., 2020).
FIGURE 2.
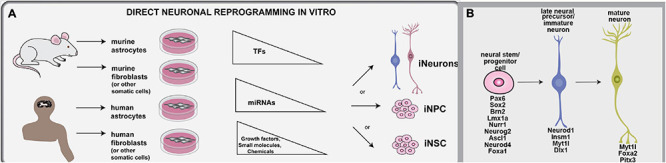
Methodologies used for direct neuronal reprogramming in vitro. (A) Direct neuronal reprogramming involves the direct conversion of somatic cells, usually astrocytes or fibroblasts, either directly into iNs or into iNSCs or iNPCs, which in turn can be differentiated into neurons. (B) Neuronal lineage conversion has been studies in murine and human cells for the most part, and involves the forced expression of lineage specifying transcription factors or miRNAs and the exposure to growth factors and other small molecules and chemicals that can make a pro-neurogenic environment.
TABLE 1.
In vitro reprogramming.
| Source cell | Factors | Subtype | Efficiency | Functional outcome | References |
| Human astrocytes | Ascl1, Nurr1, Lmx1a | DA | 18.2 ± 1.5% | Dopamine release, electrophysiologically active, generate action potentials | Addis et al., 2011 |
| Human fibroblast | Brn2, Ascl1, Myt1l (BAM) | Glutamatergic and GABAergic | 16 ± 4.3% (embryonic) 4.3 ± 1.1% (postnatal) | Electrophysiologically active, generate action potentials | Pfisterer et al., 2011a |
| BAM, Neurod1 | Glutamatergic and GABAergic | 2–4% | Electrophysiologically active, generate action potentials, form synapses | Pang et al., 2011 | |
| BAM | Glutamatergic and GABAergic | 19.5–20% | Electrophysiologically active, generate action potentials, form synapses | Vierbuchen et al., 2010; Chanda et al., 2014; Treutlein et al., 2016 | |
| BAM, Lmx1a, Foxa2 | Dopaminergic (DA) | 10% | Electrophysiologically active, generate action potentials, dopamine synthesis | Pfisterer et al., 2011a | |
| Ascl1, Nurr1, Lmx1a | DA | 6–10% | Electrophysiologically active, generate action potentials | Caiazzo et al., 2011 | |
| Ascl1, Neurog2, Nurr1, Pitx3, Sox2 | DA | 1–2% | Electrophysiologically active, generate action potentials, Transplanted to striatum in PD rat model. | Liu et al., 2011 | |
| NEUROG2, SOX11, ISL1, and LHX3 | Cholinergic motor neurons | 86–96% | Electrophysiologically active, generate action potentials | Liu et al., 2016 | |
| Brn2, Myt1l, miR-124 | Glutamatergic and GABAergic | 4–8% | Electrophysiologically active, generate action potentials, form synapses | Ambasudhan et al., 2011 | |
| miR-124, miR-9/9*with Neurod2/Ascl1 and Myt1l | Glutamatergic and GABAergic | 80% | Electrophysiologically active, generate action potentials | Yoo et al., 2011 | |
| miR-124, miR-9/9* with CTIP2, DLX1, DLX2, and MYT1L | Medium spiny neurons | 76–93%, 80% | Electrophysiologically active, generate action potentials | Victor et al., 2014; Abernathy et al., 2017 | |
| Human vascular pericytes | BAM, Tlx3 and miR-124 | Cholinergic neurons | 80% | Not tested | Liang X. G. et al., 2018 |
| Human cord blood cells | Sox2, Myc | Glutamatergic and GABAergic | 80% | Electrophysiologically active, generate action potentials | Castano et al., 2014 |
| Human pericytes | Sox2, Ascl1 | Glutamatergic and GABAergic | 48% | Electrophysiologically active, generate action potentials, form synapses | Karow et al., 2012 |
| Human umbilical cord mesenchymal stem cells | Sox2, Ascl1/Sox2, Neurog2 | Glutamatergic and GABAergic | 50% | Electrophysiologically active, generate action potentials | Araujo et al., 2018 |
| Mouse astrocytes | Neurog2/Ascl1 | Glutamatergic and GABAergic | 85% | Electrophysiologically active, generate action potentials, limited synapse formation | Berninger et al., 2007a |
| Neurog2 | Glutamatergic | 70% | Electrophysiologically active, generate action potentials | McKay et al., 2010; Heinrich et al., 2011 | |
| Ascl1 + Dlx1 | GABAergic | 70% | Electrophysiologically active, generate action potentials | McKay et al., 2010; Heinrich et al., 2011 | |
| Neurog2 | Glutamatergic | 54–73% | Electrophysiologically active, generate action potentials, transplantation to SVZ | Chouchane et al., 2017 | |
| Neurod4 + Insm1 | Glutamatergic and GABAergic | 40%; 60% | Electrophysiologically active, generate action potentials | Masserdotti et al., 2015; Rao et al., 2021 | |
| Ascl1/Neurog2 with Bcl2 | Glutamatergic and GABAergic | 60–80% | Electrophysiologically active, generate action potentials | Gascon et al., 2016 | |
| Mouse cerebellar astrocytes | Neurog2/Ascl1 | GABAergic neurons | 54–73% | Electrophysiologically active, generate action potentials, engrafted in SVZ, and migrated to olfactory bulb | Chouchane et al., 2017 |
| Astrocytes and fibroblasts | Ascl1, Phox2b, AP2a, Gata3, Hand2, Nurr1, Phox2a | Nor-adrenergic neurons | 41.8% | Electrophysiologically active, generate action potentials | Li et al., 2019 |
| Mouse fibroblasts | BAM | Glutamatergic and GABAergic | 20% | Electrophysiologically active, generate action potentials | Vierbuchen et al., 2010 |
| Mouse adult fibroblasts | BAM, Lhx3, Neurog2/Hb9, Inl1 | Motor neurons | 80.6% | Electrophysiologically active, generate action potentials, engrafted in chick neural tube | Son et al., 2011 |
| MEF, adult tail tip fibroblasts (TTF) | Ascl1, Nurr1, Lmx1a (ANL) | DA | 18.2 ± 1.5% | Electrophysiologically active, generate action potentials, dopamine release | Addis et al., 2011; Caiazzo et al., 2011 |
| Mouse TTF | ANL, Pitx3, Foxa2 En1 | DA | 5–9% | Electrophysiologically active, generate action potentials, transplantation in striatum | Kim et al., 2011b |
| MEF | Foxg1, Sox2, Ascl1, Dlx5, Lhx6 | GABAergic | 9.4% | Electrophysiologically active, generate action potentials, transplanted to hippocampus | Colasante et al., 2015 |
| Mouse adipocyte progenitor cells and hepatocytes | BAM | Glutamatergic and GABAergic | 3–6% | Electrophysiologically active, generate action potentials | Yang et al., 2013 |
| Mouse IPSC derived cardiomyocytes | BAM, Neurod1 | Glutamatergic and GABAergic | 35.5% | Electrophysiologically active, generate action potentials | Chuang et al., 2017 |
| Mouse olfactory ensheathing cells | Neurog2 | Glutamatergic | 80% | Electrophysiologically active, generate action potentials, form synapses | Sun et al., 2019 |
| Mouse-cochlear non-sensory epithelial cells, Spiral ganglion non-neuronal cells | Ascl1, Neurod1 | Primary auditory neurons | 49–55% | iNs extend projections | Noda et al., 2018 |
| Mouse microglia | Neurod1 | Glutamatergic and GABAergic | 25–35% | Electrophysiologically active, generate action potentials | Matsuda et al., 2019 |
| Rat fibroblast | ANL | DA | 7% | 90% of the DA transplanted into rat model. | Dell’Anno et al., 2014 |
| Rat fibroblast | BAM | iN | 0.4–5.9% | Transplant to striatum or hippocampus | Torper et al., 2013 |
| In vivo cell reprogramming | |||||
| Proliferating Glial cells | Neurog2, FGF, EGF | Glutamatergic | 72 ± 5% | Not tested | Grande et al., 2013 |
| Astrocytes | Neurog2, DAPT | Glutamatergic and GABAergic | 50% (cortex), 30% (cerebellum), 20% (SC) | Not tested | Hu et al., 2019 |
| Astrocytes | Neurog2, Nurr1 | Glutamatergic and GABAergic | 80% | Electrophysiologically active, generate action potentials, form synapses, long distance axonal projections | Mattugini et al., 2019 |
| Striatal astrocytes | Ascl1, Neurod1, Nurr1 | DA | 16% | Electrophysiologically active, generate action potentials, motor behavioral recovery | Rivetti di Val Cervo et al., 2017 |
| Striatal Neurons | Sox2, Nurr1, Lmx1a and Foxa1 TFs and Valproic acid, | DA | 79% | Electrophysiologically active, generate action potentials, dopamine restoration | Niu et al., 2018 |
| Cortical astrocytes | Neurod1 | Glutamatergic neurons | 92.8% | Electrophysiologically active, generate action potentials | Guo Z. et al., 2014 |
| NG2 glia | Neurod1 | Glutamatergic and GABAergic | 42.5% | Electrophysiologically active, generate action potentials | Guo Z. et al., 2014 |
| Striatal microglia | Neurod1 | Glutamatergic and GABAergic | 25–35% | Electrophysiologically active, generate action potentials, and spontaneous synaptic currents | Matsuda et al., 2019 |
| Reactive astrocytes | Neurod1 | Glutamatergic and GABAergic | 70% | Electrophysiologically active, generate action potentials, motor behavior improvement | Chen et al., 2020 |
| Striatal non-reactive astrocytes | Neurod1 | Glutamatergic neurons | 80% | Not tested | Brulet et al., 2017 |
| Astrocytes | Neurod1 | Glutamatergic | 20–27% | Improved locomotor activities and functional recovery | Livingston et al., 2020 |
| Striatal astrocytes | Neurod1, Dlx2 | GABAergic medium spiny neurons | 80% | Electrophysiologically active, generate action potentials, extension of life span, and improved motor functions | Wu et al., 2020 |
In vitro Reprogramming of Astrocytes to iNs
Transcription Factor-Based Reprogramming of Astrocytes to iNs
The first evidence for direct neuronal reprogramming came in 2002, with the conversion of murine postnatal day (P) 5-11 astrocytes to iNs by the forced expression of Pax6, a homeodomain TF with an essential role in cortical development (Heins et al., 2002; Figure 1). Pax6 repressed expression of astrocytic glial fibrillary acidic protein (GFAP) and initiated expression of the neuronal marker, β-tubulin-III (Heins et al., 2002). Even though the term “direct neuronal reprogramming” was not used in the Heins et al. (2002) study, and was only later coined in 2010 (Vierbuchen et al., 2010), Heins et al. (2002) were the first to demonstrate that a terminally differentiated somatic cell fate could be converted to a neuron, and these “reprogrammed” cells can thus be referred to as iNs.
In 2007, iNs were generated from murine P5-7 cortical astrocytes by expressing one of two proneural bHLH genes: Neurogenin 2 (Neurog2) or Ascl1 (previous gene name, Mash1), and were shown to be functional as they could fire action potentials (Berninger et al., 2007a). In the embryonic forebrain, Neurog2 and Ascl1 promote neurogenesis and specify neuronal subtype identities: Neurog2 specifies an excitatory, glutamatergic neuronal phenotype and Ascl1 specifies an inhibitory, GABAergic neuronal identity (reviewed in Oproescu et al., 2021). Neurog2 and Ascl1 also specify distinct glutamatergic and GABAergic neuronal fates, respectively, when they are overexpressed in multipotent NSCs from the adult mouse brain (Berninger et al., 2007b), although this is an example of induced differentiation and not trans-differentiation, the focus of this review.
When P5-P7 murine astrocytes were lineage converted to iNs, Neurog2-iNs expressed Tbr1, a T-box TF that is a marker of maturing glutamatergic neurons. However, Ascl1-iNs did not express markers of a GABAergic fate (Berninger et al., 2007a). Moreover, even though both Neurog2- and Ascl1-iNs fired action potentials, synapse formation was limited (Berninger et al., 2007a). These limitations were overcome by changing the mode of gene delivery to a retroviral construct that was less prone to silencing and by using a stronger CAG promoter that achieved higher, sustained levels of proneural gene expression (Heinrich et al., 2010, 2011). With these modifications, Neurog2-iNs derived from postnatal astrocytes were glutamatergic, fired action potentials and formed synapses (Heinrich et al., 2010, 2011). In addition, when Ascl1 was co-expressed with Dlx2, a homeodomain TF involved in specifying a GABAergic identity (reviewed in Wonders and Anderson, 2006), synaptically mature and electrically active iNs were generated (Heinrich et al., 2010, 2011). Interestingly, there are also regional differences in the response of astrocytic populations. For example, when Neurog2 or Ascl1 were expressed in early postnatal murine astrocytes isolated from the cerebellum, they generated inhibitory GABAergic iNs, which migrated to the olfactory bulb after transplantation into the subventricular zone (Chouchane et al., 2017). Conversely, only Neurog2, and not Ascl1, could transdifferentiate cortical astrocytes into excitatory glutamatergic neurons that integrated into the cortex (Chouchane et al., 2017). Thus, not only is it important to consider which TF to overexpress for lineage conversion, but also the starting somatic cell source, including its regional identity, as well as the mode of gene delivery and promoter choice.
To understand how Neurog2 and Ascl1 promote astrocyte-to-neuron lineage conversion, transcriptomic studies were performed after the expression of either factor in P6-P7 murine astrocytes (Masserdotti et al., 2015). While both proneural TFs rapidly induce neuronal differentiation and alter gene expression, there is only a ∼3% overlap in induced genes, consistent with the observation that these TFs specify distinct neuronal phenotypes in the embryo (Masserdotti et al., 2015). One of the common genes activated by both Neurog2 and Ascl1 is Neurod4, a proneural bHLH gene with a role in early neural lineage development, which functions downstream of Neurog2 in embryonic cortical progenitors (reviewed in Oproescu et al., 2021). Notably, misexpression of Neurod4 in P6-P7 astrocytes could similarly induce neuronal marker expression. However, in order for Neurod4-iNs to functionally mature, co-expression with Insm1, another commonly regulated TF, was required (Masserdotti et al., 2015). In support of these findings, a follow-up study using Ascl1 chromatin immunoprecipitation sequencing (ChIP-seq) and functional knockdown studies also identified Neurod4 as an essential Ascl1 transcriptional target during the conversion of murine astrocytes to iNs (Rao et al., 2021). Additional essential downstream genes identified during this study included Klf10, Chd7, and Myt1l (Rao et al., 2021). Notably, another important finding of the Masserdotti et al. (2015) study was that astrocytes lose their capacity to undergo neuronal lineage conversion over time, which is due in part to increased activity of the repressor element 1 (RE1) silencing transcription factor (REST) repressive protein complex (Masserdotti et al., 2015). Notably, REST prevents Neurog2 from activating Neurod4 expression during iN lineage conversion (Masserdotti et al., 2015).
Metabolic Reprogramming Accompanies Astrocyte to iN Lineage Conversion
A major issue with direct reprogramming of P5-P7 astrocytes to iNs by the proneural TFs is that most astrocytes die within a few days post-transduction, except those few cells that make it to the iN fate more quickly (Gascon et al., 2016). To overcome this obstacle, Gascon et al. (2016) misexpressed Ascl1 or Neurog2 along with Bcl2, an anti-apoptotic gene that facilitated a more rapid rate of neuronal conversion, which caused more iNs to survive and mature (Gascon et al., 2016). Yet unexpectedly, the effect of Bcl2 on neuronal lineage conversion was attributed to its role in reducing oxidative stress and a reduction in reactive oxygen species (ROS), and not its role in preventing apoptosis (Gascon et al., 2016). These studies raised the idea that aerobic metabolism, which produces ROS as a byproduct, may be an important consideration for reprogramming strategies.
Metabolic profiles of brain cells differ from one another—in general, NPCs in the developing brain favor glycolysis, while differentiated cells use oxidative phosphorylation (OXPHOS), which occurs in the mitochondria (Herrero-Mendez et al., 2009; Zheng et al., 2016). Mitochondria respond to stress by undergoing fusion with other mitochondria, while fission creates new mitochondria to adapt to an increase in energy demand. Interestingly, the proportion of mitochondria in each of these states differs between cortical astrocytes and neurons (Motori et al., 2013; Misgeld and Schwarz, 2017). In cortical astrocytes, mitochondria have elongated shapes consistent with mitochondrial fusion (Motori et al., 2013), while neurons exhibit smaller mitochondria, consistent with mitochondrial fission (Misgeld and Schwarz, 2017). Strikingly, Ascl1-iNs generated from murine P5 astrocytes also undergo a transition to a fission-like smaller mitochondrial morphology (Russo et al., 2020). Furthermore, LC-MS/MS of mitochondria from cortical neurons and astrocytes revealed a large number of differences, and a neuronal mitochondrial phenotype was recapitulated in Ascl1-iNs to varying degrees, depending on the extent of neuronal conversion (Russo et al., 2020). Finally, strikingly, the importance of differences in mitochondrial content was demonstrated by co-expressing Ascl1 with neuronal-enriched antioxidant genes Sod1 and Prdx2, which augmented astrocyte to iN lineage conversion (Russo et al., 2020).
Taken together, these studies demonstrate the capacity for lineage conversion between two neural lineages and highlight the striking capacity of the proneural bHLH TFs to specify neuronal subtype identities through the initiation of distinct neuronal programs.
Direct Neuronal Reprogramming of Fibroblasts to iNs
Brn2-Ascl1-Myt1l (BAM)-Mediated Direct Neuronal Reprogramming
The term “direct neuronal reprogramming” was first coined in 2010 based on the lineage conversion of murine embryonic or postnatal fibroblasts into functional iNs through misexpression of a triple TF cocktail called the BAM or Wernig factors in honor of their discoverer (Figure 1). The BAM factors are Brn2 (official gene name, Pou3f2), Ascl1 and Myt1l (Vierbuchen et al., 2010). Unlike astrocytes, which can be converted to a neuronal identity by a single TF, optimal transdifferentiation of fibroblasts to iNs required three TFs in this study. This finding suggested that the barriers to iN reprogramming were less easily surmountable in fibroblasts, likely because the embryonic lineage is not neural, in contrast to astrocytes (Vierbuchen et al., 2010). While both excitatory and inhibitory postsynaptic currents could be evoked in BAM-iNs, most currents were excitatory (Vierbuchen et al., 2010). BAM gene expression could also be triggered in mouse embryonic fibroblasts (MEFs) using deactivated Cas9 fused to a transactivation domain (dCas9-VP64). dCas9-VP64 was targeted to BAM genes to initiate transactivation using guide RNAs (gRNAs) to each BAM gene (Black et al., 2016). Notably, this approach also generated iNs that could fire action potentials (Black et al., 2016). The ability to multiplex gRNAs makes this type of strategy attractive moving forward, especially to drive neuronal subtype identities that may require even more genes, as described further below.
In the interest of translating neuronal reprogramming technology to clinical practice, investigators turned their attention to human and non-human primate fibroblasts as a source cell type for lineage conversion. BAM TFs were shown to convert human fibroblasts isolated from 5.5- to 7-week-old fetuses to functionally active iNs (Pfisterer et al., 2011a, b). However, the efficiency of iN conversion was enhanced by co-expressing BAM TFs with the bHLH TF, NEUROD1, both in postnatal day (P) 1–3- or 8–10-week-old human fetal fibroblasts (Pang et al., 2011), and in embryonic skin fibroblasts from a non-human primate (marmoset) (Zhou et al., 2014). Interestingly, BAM-induced iN conversion could also be triggered in vivo after transplanting fibroblasts carrying doxycycline-inducible constructs into the striatum or hippocampus of adult rats, leading to iN integration in vivo (Torper et al., 2013).
There are striking differences reported in the efficacy of BAM TFs for neuronal reprogramming efficiency (Table 1). Reasons may be as simple as differences in culturing conditions and media components that alter the host fibroblasts. Moreover, fibroblast sources differ between studies, including human foreskin fibroblasts (Pang et al., 2011), or fibroblasts derived from fetuses after removal of the head, vertebral column, dorsal root ganglia, and internal organs (Pfisterer et al., 2011a, b). Indeed, a recent study has found that only a subset of mouse embryonic fibroblasts (MEFs) are amenable to reprogramming to an iPSC state, with cell competition resulting in the rapid domination by “elite clones” (Shakiba et al., 2019). Transcriptomic and epigenetic differences in MEFs that respond to overexpression of the Yamanaka factors have also been cataloged in great detail, with cells that do not become bona-fide iPSCs termed “F-class cells” (reviewed in Vidal et al., 2015). Further studies will be required to determine whether there are subsets of fibroblasts that are more easily converted to iNs by the usage of different TF combinations.
After the initial demonstration that BAM TFs could convert fibroblasts to iNs, there were efforts to reduce the number of required factors. Interestingly, overexpression of ASCL1 alone can convert fibroblasts to excitatory iNs that make synapses, albeit not as efficiently, highlighting the importance of using all three BAM TFs (Vierbuchen et al., 2010; Chanda et al., 2014; Treutlein et al., 2016). Subsequent studies investigated the contributions of each of the BAM TFs to neuronal conversion. Ascl1 was found to be a pioneer factor, as it opens the chromatin surrounding neural lineage genes, enabling Brn2 binding to chromatin transactivate downstream genes in neurogenesis (Wapinski et al., 2013). The ability of Ascl1 to open the chromatin occurs rapidly, within the first few days of the reprogramming process (Wapinski et al., 2017). The following 3 weeks of neuronal differentiation involves the functions of critical downstream TFs such as Zfp238, Sox8, and Dlx3 (Wapinski et al., 2017).
In contrast to Ascl1 and Brn2, Myt1l does not play a direct role in neurogenesis, but rather serves as a multi-lineage repressor that prevents the selection of alternative, non-neural lineages (Wapinski et al., 2013; Mall et al., 2017). In this regard, it is interesting that Myod1, a bHLH gene that normally specifies a skeletal muscle identity, but which binds to the same E-boxes as Ascl1, can induce the conversion of fibroblasts to iNs when co-expressed with Myt1l (Lee Q. Y. et al., 2020). Myt1l suppresses the activation of skeletal muscle genes so that the non-neural bHLH factor Myod1 can convert fibroblasts to iNs (Lee Q. Y. et al., 2020).
The three BAM TFs thus each play critical roles in the neuronal reprogramming process. However, investigators have continued their quest to optimize reprogramming approaches, leading to additional critical insights into the underlying mechanisms of neuronal lineage conversion.
Other TF Combinations Confer Neuronal Subtype Identity Onto iNs
Dopaminergic iNs
After the seminal findings demonstrating BAM-mediated neuronal lineage conversion, several follow-up studies tested variations of the same theme. These studies used TF cocktails to generate iNs with the new goal of conferring different neuronal subtype identities. By generating iNs with specific neurotransmitter phenotypes, studies aimed to devise new strategies for neuronal repair of neurological disorders. For example, in Parkinson’s disease (PD), dopaminergic (DA) neurons in the nigrostriatal pathway are lost. Therefore, one of the first cell types targeted was DA neurons, for its therapeutic potential in the treatment of PD. A series of articles describing the generation of iDA neurons using direct neuronal reprogramming were first published in 2011. These studies described the conversion of fibroblasts to iDA neurons using a modification of the original Wernig strategy. The three BAM TFs were combined with Lmx1a and Foxa2, which specify a DA neuronal identity during development, to form a five TF cocktail that could convert human fibroblasts into induced DA neurons (iDAs) (Pfisterer et al., 2011a, b). Similarly, Ascl1 combined with Nurr1 (official gene name, Nr4a2) and Lmx1a, a triple TF cocktail termed ANL, could convert mouse embryonic or adult tail tip fibroblasts, or human adult dermal or lung fibroblasts, into iDAs that had features of mature DA neurons (Addis et al., 2011; Caiazzo et al., 2011). A follow-up study of ANL-iDAs revealed that they could integrate in the striatum in a PD rat model, and improve motor behavior after DREADD-dependent activation (Dell’Anno et al., 2014).
Next, in an independent screen of eight TFs, a new combination of five TFs was identified; Ascl1, Ngn2 (official gene name, Neurog2), Sox2, Nurr1 (official gene name, Nr4a2), and Pitx3, which could reprogram human fibroblasts to iDA neurons (Liu et al., 2011). The transplantation of these cells into the striatum of a 6-hydroxydopamine (6-OHDA lesion) PD rat model also showed improved motor behavior (Liu et al., 2011). Using a similar screening strategy of 11 TFs, Ascl1 and Pitx3 were revealed to be capable of converting murine tail tip fibroblasts to an immature iDA fate, with the added expression of Lmx1a, Nurr1 (official gene name, Nr4a2), Foxa2 and En1 leading to the expression of mature DA neuronal markers—a so-called six TF reprogramming cocktail (Kim et al., 2011b). This six TF cocktail also induced the generation of functional iDA neurons that could rescue motor behavior in 6-OHDA treated PD-like mice (Kim et al., 2011b). More recently, a three TF cocktail of Brn2, Sox2, and Foxa1 was shown to convert murine fibroblast cells into DA precursor cells that could efficiently integrate and functionally differentiate into DA neurons in a MPTP lesioned PD mouse model after transplantation (Tian et al., 2015).
GABAergic iNs
An imbalance in excitatory-inhibitory circuits is a feature of many neuropsychiatric and epileptogenic disorders. Therefore, another neuronal type targeted for their neuronal repair potential has been GABAergic inhibitory neurons. In a screen of 21 TFs with known roles in specifying a GABAergic neuronal identity during development, a combination of five TFs was identified that could trans-differentiate MEFs into induced GABAergic neurons (iGABA-iNs): Foxg1, Sox2, Ascl1, Dlx5, and Lhx6 (Colasante et al., 2015). Ascl1, Dlx5, and Lhx6 have well-documented roles in the development of GABAergic neurons during embryogenesis. Conversely, the roles of Foxg1 and Sox2 were unexpected, but in this study, both TFs were shown to be required to facilitate Ascl1 binding to the Dlx1/2 enhancer (Colasante et al., 2015). The five TF-iGABA-iNs were transplanted into the adult murine hippocampus and shown to integrate functionally by optogenetic activation (Colasante et al., 2015).
Noradrenergic iNs
Several neural disorders related to sleep, arousal, and attention -hyperactivity disorder are linked to the dysfunction in the noradrenergic (NA) neurons (Benarroch, 2009). A seven-TF cocktail of TFs, namely Ascl1, Phox2b, AP2a (official gene name Tfap2a), Gata3, Hand2, Nurr1 (official gene name Nr4a2), and Phox2a, could reprogram both murine fibroblasts and astrocytes into functional induced NA neurons (iNA) that can produce noradrenaline (Li et al., 2019). Strikingly, iNAs were electrically active, and when co-cultured with murine cardiac myocytes, they could induce myocyte beating, which provides strong support for their functionality (Li et al., 2019).
Motor Neurons
Similar to most parts of the adult mammalian brain, neurogenesis does not occur in the adult mammalian spinal cord to any significant extent. Finding new strategies to replace lost motor neurons has thus become a goal for the future of repair strategies and clinical therapeutic applications. The first demonstration that induced motor neurons (iMNs) could be generated in vitro involved the misexpression of the three BAM TFs with Lhx3 (4 TF cocktail) and either Neurog2, Hb9 (official gene name Mnx1) or all of these genes together with Isl1 (5 TF cocktails) in adult mouse tail tip fibroblasts (Son et al., 2011). The 4TF- and 5TF- iMNs fired action potentials and formed neuromuscular junctions with chick myotubes (Son et al., 2011). Mazzoni et al. (2013) further refined this process to a triple TF cocktail: Neurog2, Isl1, and Lhx3. This triple TF cocktail was applied to mouse embryonic stem cells and induced differentiation into iMNs (Mazzoni et al., 2013). In this context, the authors were able to more carefully dissect apart the transcriptional events that underlie the acquisition of a MN identity, and revealed that there are two phases: firstly, Neurog2, Isl1, and Lhx3 initially bind the same transcriptomic regions, followed by later differentiation events that are triggered by Lhx3 and Isl1 relocating to new regulatory sites in the genome (Velasco et al., 2017). To translate these findings to human fibroblasts, four TFs were shown to generate functional, cholinergic iMNs: NEUROG2, SOX11, ISL1, and LHX3 (Liu et al., 2016). Interestingly, while NEUROG2 alone combined with small molecules could induce the acquisition of a cholinergic iMN fate, mature MN markers (ISL1, LHX3) were not expressed (Liu et al., 2013). In contrast, the four TF-iMNs were electrically active and could form functional neuromuscular junctions (Liu et al., 2016).
Taken together, these studies demonstrate that a large number of different TFs can induce fibroblast to iN lineage conversion. This conclusion was further supported by a large-scale screen of 598 TFs, which identified 76 TF pairs that could generate iNs (Tsunemoto et al., 2018). Many starting paths can thus converge on the same final destination: the generation of iNs, as long as the developmental programs associated with neuronal differentiation are re-initiated. It seems fair to say from these findings, that once initiated, the neuronal differentiation program becomes self-propagating, at least to some extent. In this regard it is interesting that a CRISPR activation screen for inducers of neuronal differentiation in mouse embryonic stem cells (ESCs) identified a neuronal cell identity regulatory network that is maintained by several core factors (Liu et al., 2018). While technically not a directed neuronal differentiation study, this study supports the idea that there is not just one GRN that supports neurogenesis during reprogramming (Liu et al., 2018). However, given the evidence that the directed differentiation of ESCs into iGABA-iNs is further facilitated by the sequential expression of different TF modules (Au et al., 2013), the order in which these different network nodes are turned on is important, likely also in the reprogramming process.
miRNA Mediated Reprogramming of Fibroblasts to iNs
Additional advances in the direct neuronal reprogramming field were made with the inclusion of microRNAs (miR) into conversion cocktails (Figure 2). miRNAs are short non-coding RNAs that act as negative regulators of gene expression, either by promoting mRNA degradation or by blocking translation. Several miRNAs have been identified that promote neural lineage development by repressing the expression of genes that are not normally expressed in neural lineages (reviewed in Lang and Shi, 2012). miRNA technology can be incorporated to aid in the inhibition of alternative cell fates during cellular reprogramming. The first miRNA shown to aid direct neuronal reprogramming was miR-124, which was selected based on its abundant expression in the CNS and its role in attenuating the expression of several known negative regulators of neurogenesis, such as the poly-pyrimidine-tract-binding protein PTBP1, the neural progenitor (np) Brg/Brm-associated factors (BAF) complex, and REST (Ambasudhan et al., 2011; Xue et al., 2013). PTBP1 binds and regulates RNA biology in various ways. For instance, PTBP1 alters mRNA splicing and prevents miRNA activation through competitive binding or altering mRNA secondary structure (reviewed in Keppetipola et al., 2012). npBAF is a neural-specific SWI/SNF complex that is involved in chromatin remodeling, and overexpressing npBAF in fibroblasts can promote neuronal lineage conversion (reviewed in Staahl and Crabtree, 2013). Finally, REST forms a transcriptional repressor complex that is essential to silence neural lineage gene expression in non-neural cells (reviewed in Maksour et al., 2020). REST and miR-124 form a regulatory loop, with miR-124 repressing several genes in the REST complex, and REST repressing miR-124 expression (Xue et al., 2013).
Prior studies have revealed that overexpression of miR-124 may induce adult murine NPCs to differentiate into neurons (Cheng et al., 2009). To test whether miR-124 can aid direct neuronal reprogramming, a screen was performed by overexpressing miR-124 with different combinations of 11 TFs in human fibroblasts (Ambasudhan et al., 2011). This study identified MYT1L, BRN2 (official gene name POU3F2), and miR-124 as the most efficient triple factor combination to induce the formation of electrically active iNs (Ambasudhan et al., 2011). As ASCL1 was the only BAM component not in this reprogramming cocktail, it appears that at least some of ASCL1’s functions may be compensated for by miR-124.
In a similar study, miR-124 was combined with miR-9/9∗; the latter functions to represses BAF53a gene expression, a subunit of the npBAF complex involved in chromatin remodeling (Yoo et al., 2011). Subsequent studies identified downstream events that underlie miR-124 and mir-9/9∗ neuronal lineage conversion, and identified two sequential steps: (1) silencing of the TFs KLF4 and KLF5, to repress a fibroblast fate, and (2) upregulation of RN7SK, a small nuclear RNA required to activate neuronal lineage genes through changing chromatin accessibility (Cates et al., 2021). The importance of the epigenome in reprogramming is discussed further below.
TFs and other miRs were also combined with miR-124 and mir-9/9∗ to further improve the conversion of human fibroblasts to iNs. For example, the combined misexpression of miR-302/367, a cluster containing five miRNAs that also facilitates reprogramming to generate iPSCs, with miR-124 and miR-9/9∗, enhances neuronal lineage conversion (Zhou et al., 2015). Similarly, co-expression of miR-124 and mir-9/9∗ with ASCL1 could convert fibroblasts from other species, such as adult porcine and marmoset fibroblasts, into iNs (Habekost et al., 2020; Nemoto et al., 2020). However, the conversion of human fibroblasts from neonatal foreskin or adult dermis to iNs was most efficient when miR-124 and miR-9/9∗ were combined either with NEUROD2, or with both ASCL1 and MYT1L (Yoo et al., 2011).
Overexpression of miRs has also been incorporated into neuronal reprogramming strategies that are targeted toward neuronal subtype specification. For example, co-expression of miR-124 and miR-9/9∗ with CTIP2 (official gene name, BCL11B), DLX1, DLX2, and MYT1L (termed CDM) effectively transformed human fibroblasts into induced medium spiny neurons (iMSNs) (Victor et al., 2014). Of promise, transplantation of the iMSNs into the striatum, where these neurons are normally found, led to successful engraftment and integration into the host neural network (Richner et al., 2015). Similarly, co-expression of miR-124 and miR-9/9∗ with ISL1 and LHX3 promoted the differentiation of iMSNs that possessed subtype-specific transcriptomic and epigenomic profiles (Abernathy et al., 2017).
Given the capacity of miRNAs to promote neuronal lineage conversion, it is not surprising that an upstream regulator, PTB (official gene name, PTBP1), can also influence neuronal reprogramming (Xue et al., 2013). The knockdown of PTBP1 converted MEFs to iNs by inhibiting the ability of REST to repress neural lineage gene expression, and by regulating mRNA splicing and stability (Xue et al., 2013). Conversely, PTBP1 knockdown in human adult fibroblasts (HAFs) could not promote lineage conversion to a mature neuronal phenotype, unless it supported by the additional knockdown of another PTB paralog, nPTB (official gene name, PTBP2). In human cells, PTBP2 knockdown leads to expression of BRN2 and miRNA-9 which are both critical determinants of neuronal differentiation and neuronal reprogramming, as highlighted above (Xue et al., 2016).
As a final note, the ability of miR-124 to repress neuronal gene expression has also been exploited in direct neuronal reprogramming to deactivate the reprogramming factors after they acquire an iN identity (Lau et al., 2014). By linking four copies of the miR-124 target sequence to ASCL1, BRN2, and MYT11, the authors were able to demonstrate that upon the conversion of human fibroblasts to iNs, the expression of these conversion factors were silenced (Lau et al., 2014). This type of approach is more amenable to human therapies, because if high levels of transgene expression is not maintained, the risk of tumor formation is reduced.
Epigenetic Changes Drive Fibroblast to iN Reprogramming
Cellular reprogramming involves the shutdown of GRNs associated with the starting somatic cell type, and the activation of new gene expression profiles associated with the reprogrammed cell type. These changes are tightly linked to chromatin opening and closing so that the appropriate target genes are accessible to lineage specifying TFs. However, the process of lineage conversion faces the unsurprising challenge that some starting somatic cell populations retain their cell-type specific gene regulatory networks. For instance, iNs reprogrammed from fibroblasts continued to retain some fibroblast genes, a finding that was evident in a large-scale screen of 598 TFs that identified 76 TF pairs that could induce neuronal reprogramming (Tsunemoto et al., 2018). Understanding how chromatin dynamics contribute to incomplete lineage conversion is therefore important.
To begin with the investigation of iNs reprogrammed from fibroblasts, several bHLH TFs have been identified with pioneering activity (e.g., Ascl1), which means that they can bind to and facilitate the opening of closed chromatin so that other neurogenic TFs (e.g., Brn2) can bind target sites (Wapinski et al., 2013). Similarly, Neurod1 binds and rearranges closed chromatin in target regions during microglia to iN conversion, especially those sites with bivalent trimethylation, which is a feature of genes that are poised for transcription, as described below (Matsuda et al., 2019). In contrast, Neurog2 cannot act as a pioneer factor unless combined with two small molecules, forskolin, and dorsomorphin, which help to remove barriers to chromatin binding (Smith et al., 2016).
Unlike these pioneer TFs, most other TFs require that the chromatin surrounding their target sites are first opened before they can bind. A new concept emerging in cellular reprogramming is that epigenetic regulators act as gatekeepers that maintain terminally differentiated cell fates which in turn prevent lineage conversion (Gascon et al., 2017b). Gatekeepers control chromatin “opening” (accessible) and “closing” (inaccessible) to regulate the binding of lineage-specifying TFs that initiate differentiation programs. The status of chromatin (i.e., open or closed) is controlled by three main events: (1) histone modifications, (2) DNA methylation and (3) nucleosome remodeling, the modulation of each of which, can influence neuronal reprogramming, as discussed below.
Histone Modifications
The most extensively studied histone modifications involve the acetylation or methylation of specific lysine residues (K) in histone subunits H3 or H4. Changes are commonly found in the regulatory regions of actively transcribed genes, including H3K9 acetylation (ac), or the addition of one (me1), two (me2) or optimally three (me3) methyl groups to H3K4 (H3K4me1-3) at transcription start sites. Conversely, H3K36 (H3K36me1-3) methylation is found in the body of actively transcribed genes. The chromatin marks associated with gene silencing include H3K9me3, H3K27me3, and monoubiquitinylation of H2AK119ub. Lineage gatekeepers primarily act via the polycomb repressive complexes 1/2 (PRC1/2), which compact chromatin by catalyzing the repressive H3K27me3 and H2AK119ub modifications (Tursun et al., 2011; Cheloufi et al., 2015; Gascon et al., 2017a).
Chromatin changes have been most extensively studied during reprogramming to pluripotency to generate iPSCs. H3K9me3 acts as a barrier to iPSC reprogramming, such that the overexpression of histone demethylases (e.g., Jmjd2c) or the knockdown of histone methylases (e.g., G9a, Setdb1) specific to H3K9me3 aids reprogramming (reviewed in Stricker and Gotz, 2021). Similarly, H3K27me3 marks on neuronal lineage genes are among the earliest changes to occur when microglia are reprogrammed to iNs by Neurod1 (Matsuda et al., 2019). A loss of H3K4 methylation occurs in genes that maintain the starting somatic cell state early on during the iPSC reprogramming process (reviewed in Stricker and Gotz, 2021).
In some instances, both activating (H3K4me3) and repressive (H3K27me3) chromatin modifications can co-occur in the regulatory regions of genes that are “poised” for transcriptional activation (Bernstein et al., 2006). To facilitate the activation of adult NSCs, Sox2 binds to “poised” genes, limiting PRC2 activity at these bivalent sites to reduce H3K27me3 modifications and increase H3K9ac marks at critical neurogenic loci, such as at Neurog2 and Neurod1 (Amador-Arjona et al., 2015). It is intriguing to speculate that a similar role for SOX2 may be found during the reprogramming of fibroblasts to iNs. Indeed, SOX2 augments Neurog2- or Ascl1-mediated reprogramming of human umbilical cord mesenchymal stem cells into iNs (Araujo et al., 2018), and is included in conjunction with proneural genes and other TFs to form iGABA neurons (Colasante et al., 2015) and iDA neurons (Liu et al., 2011).
Ascl1 preferentially binds a different trivalent mark during BAM-induced MEF to iN reprogramming; H3K27a, H3K4me1, and H3K9me3 (Wapinski et al., 2013). It is surprising that the H3K9me3 mark, which normally represses iPSC reprogramming, is required for Ascl1 binding and iN lineage conversion, since overexpression of Jmjd2c, which removes this mark, reduces neuronal conversion efficiency (Wapinski et al., 2013).
While there remains much to be sorted out in terms of understanding the contribution of histone modification to neuronal lineage conversion, it is clear that these changes play a fundamental role in cellular reprogramming.
DNA Methylation
DNA is mainly methylated on cytosines by DNA methyltransferases (DNMTs), primarily in CpG dinucleotides that are located in regulatory regions of the genome. To a smaller extent DNA can also be methylated on non-CpG sequences (termed CpH), which are often found in non-regulatory regions of the genome. Strikingly, CpH methylation is most abundant in mature mammalian neurons and not frequently found in other cell types (Guo J. U. et al., 2014). Methylated cytosines repress transcription by closing chromatin through the recruitment of methylated-DNA binding domain (MBD) proteins, that recruit additional factors to confer a repressive chromatin structure. The removal of the repressive DNA methyl groups is achieved by ten-eleven translocation (TET) methylcytosine dioxygenases. Recent studies have revealed that the DNA methylome is dynamically regulated during BAM-induced MEF to iN reprogramming (Luo et al., 2019). Strikingly, Ascl1 also displays pioneer functions in this regard, through inducing CpH methylation in a neuron-like signature in iNs, which is a DNMT3A dependent process (Luo et al., 2019).
Chromatin Remodeling
Nucleosome remodeling involves ATP-dependent nucleosome movements that are facilitated by altering how DNA interacts with histones, which can result in opening of the chromatin to make it accessible for TF binding. The main regulators of chromatin remodeling are different BAF (Brg1/Brm-associated factor) complexes, also known as a SWI/SNF complexes, which disassemble nucleosomes to convert heterochromatin to euchromatin. Surprisingly, a role for chromatin remodeling in neuronal reprogramming was revealed in β-actin (official gene name, Actb) KO mice (Xie et al., 2018). Actb KO MEFs have a reduced capacity to undergo neuronal lineage conversion because Brg1, a BAF subunit, cannot bind chromatin, which results in high levels of sustained H3K9 and H3K27 methylation and consequent chromatin compaction (Xie et al., 2018).
Small Molecules Aid in vitro Neuronal Reprogramming
As expected, media composition is critically important for all in vitro neuronal reprogramming strategies and has roles that include supporting iN survival and augmenting the effects of overexpressing neural TFs and miRNAs. As such, in parallel to tests of intrinsic factors, investigators have been working on generating defined media to promote neuronal lineage conversion without the requirement to express exogenous genes or miRNAs. Critical studies that highlight the impact of media components on neuronal reprogramming are summarized in Table 2 (for murine studies) and Table 3 (for human studies). Here we simply highlight the rationale for choosing various factors.
TABLE 2.
Small molecules used in iN lineage conversion strategies in rodent cells.
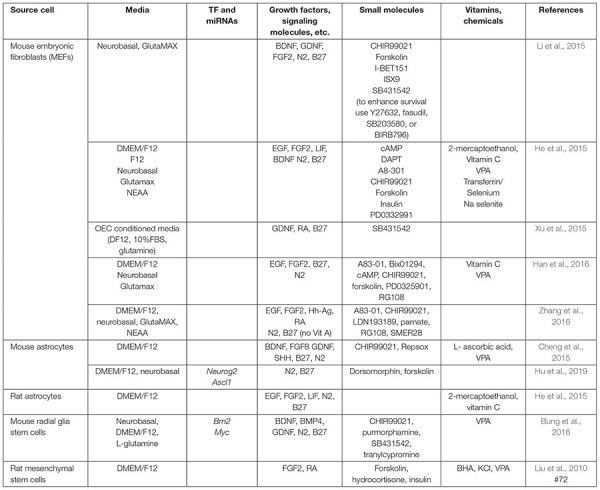 |
TABLE 3.
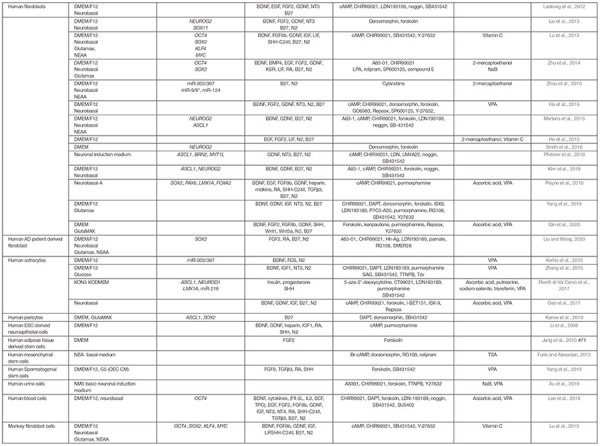
Small molecules used in iN lineage conversion strategies in human cells.
 |
As with the direct manipulation of gene expression, media design makes the same considerations of which pathways to activate to promote neuronal differentiation, and to inactivate or prevent the acquisition of alternative cell fates. Factors added to culture media can be roughly considered in three groups: (1) growth factors, supplements, cytokines, and other signaling molecules, (2) small molecule agonists or antagonists of signaling molecules, and (3) other vitamins or chemicals that impact cellular status (pH, reducing capacity, etc.). In general, these molecules/chemicals either turn on (summarized in Figure 3) or turn off (summarized in Figure 4) pathways to promote neuronal lineage conversion.
FIGURE 3.
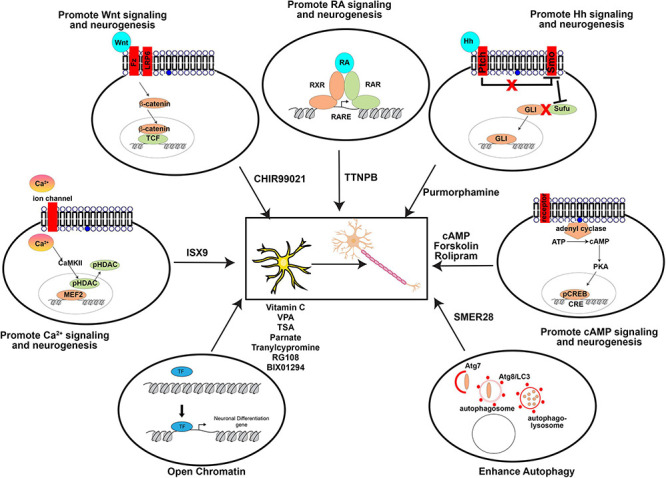
Pathways activated by small molecules to aid direct neuronal reprogramming. A summary of the agonists/antagonists used in neuronal reprogramming cocktails that activate critical signaling pathways or cellular processes.
FIGURE 4.
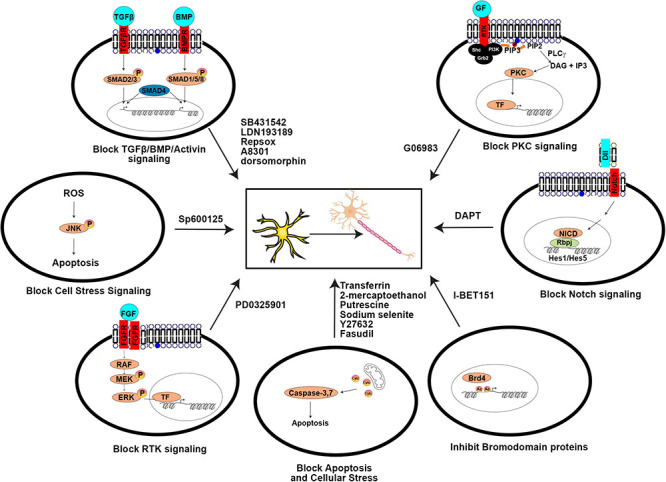
Signaling pathways blocked by small molecules to aid direct neuronal reprogramming. A summary of the agonists/antagonists used in neuronal reprogramming cocktails that inhibit critical signaling pathways or cellular processes.
Growth Factors, Supplements, Cytokines, and Other Signaling Molecules
The most common growth factor added to cell culture media are fibroblast growth factors (FGF2/4/8), epidermal growth factor (EGF) or insulin growth factor (IGF), which support general cell proliferation, along with N2 and/or B27 defined growth supplements that supports neuronal growth and survival. Other growth factors are added because they promote neuronal differentiation, such as Wnts, retinoic acid, sonic hedgehog (SHH), and the sex steroid, progesterone (reviewed in Rivetti di Val Cervo et al., 2017; Oproescu et al., 2021).
Small Molecule Antagonists
Several signaling pathways are targeted for shutdown by small molecule antagonists for neuronal reprogramming, as described below:
Activin/BMP/TGFβ/ALK Signaling
Signaling through these TGFβ family pathways specifies mesoderm and endoderm lineages and must be inhibited to allow neural fate specification (Chambers et al., 2009). Notably, BMP signaling also promotes Ascl1 degradation and therefore promotes gliogenesis and inhibits neurogenesis (Vinals et al., 2004). Inhibitors of these pathways include SB431542, LDN193189, dorsomorphin, RepSox, and A8301.
Notch Signaling
Notch binds Jag/Dll ligands on adjacent cells, ultimately leading to the downstream transcription of Hes1/Hes5, bHLH TFs that inhibit proneural gene expression and prevent neuronal differentiation. Repression of Notch signaling is achieved using DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester), a gamma secretase inhibitor that prevents cleavage of the Notch receptor so that the Notch intracellular domain (NICD) does not translocate to the nucleus to assist in the transcriptional activation of the downstream Hes1/Hes5 bHLH effectors (reviewed in Oproescu et al., 2021).
Bromodomain Binding Proteins
Bromodomain proteins bind acetylated proteins, such as histones. I-BET151 is a BET bromodomain inhibitor that prevents these proteins from binding their targets, with diverse effects, including cytokine-induction of downstream STATs.
Cell Death Pathways
Antagonists such as ROCK inhibitors (e.g., Y27632, fasudil; Li et al., 2015) are also used to promote cell survival during neuronal lineage conversion. Furthermore, using a P7C3 or a P7C3-A20 derivative may be helpful as they are nicotinamide phosphoribosyl transferase (NAMPT) that have neuroprotective effects promoting survival (Yang et al., 2019).
Other Signaling Pathways
Inhibition of other signaling pathways has also been found to improve the conversion of fibroblasts to iNs, including: PKC signaling, which is blocked with G06983, JNK signaling, which is blocked with SP600125, p38MAPK signaling, which is blocked with SB203580 or BIRB796, and MEK signaling, which is blocked with PD0325901 (Han et al., 2012; Hu et al., 2015; Li et al., 2015).
Epigenetic Modifiers
Several factors that open chromatin have been used in neuronal reprogramming, with the rationale that they will facilitate binding of lineage-specifying TFs to their target sites. Included are inhibitors of DNA methylation, such as 5-azacytidine or 5-aza-2’-deoxycytidine. Similarly, inhibitors of DNA methyltransferase (DNMT), namely RG108 or BIX01294, can prevent DNA methylation, which also opens up the chromatin for TF binding. Tranylcypromine is an inhibitor of lysine-specific demethylase 1 (LSD1), which converts H3K4me2 to H3K4me1 or H3K4me0 by removing a chromatin mark associated with open chromatin used in neuronal reprogramming (Bung et al., 2016). Similarly, parnate is also a histone demethylase inhibitor used in neuronal reprogramming (Zhang et al., 2016).
Small Molecule Agonists
Rather than adding signaling molecules directly, small molecules can also be used to activate pathways that promote neuronal differentiation. Neuronal differentiation pathways that have been targeted with small molecules include:
Wnt Signaling
Wnts promote embryonic neurogenesis (Li et al., 2012), the activation of which is achieved using a small molecule that inhibits GSK3 activity (e.g., CHIR99021), as GSK3 is a downstream negative regulator of WNT signaling.
Cyclic-AMP (cAMP) Signaling
Activation of adenyl cyclase converts ATP to cAMP, which activates PKA, leading to phosphorylation of cAMP response element binding (CREB) protein (Montminy and Bilezikjian, 1987), which then translocates to the nucleus to initiate transcription and neuronal differentiation (Shan et al., 2008). This pathway can be achieved by adding cAMP directly, or using forskolin (Jang et al., 2010). In addition, rolipram can activate cAMP by inhibiting type 4 cyclic nucleotide phosphodiesterases (PDE4) (Sasaki et al., 2007).
Retinoic Acid (RA) Signaling
RA is a potent activator of neurogenesis, and acts as a ligand for nuclear receptors of the RXR and RAR TF family, which dimerize to bind RARE elements in the genome (Diez del Corral et al., 2003). RA signaling can be activated in vitro using TTNPB (Maloney et al., 2000).
Hedgehog (Hh) Signaling
Hh binds the Ptch receptor, which then releases its inhibitory control of Smo. Once Smo is activated, it represses Sufu, releasing Sufu inhibition of the Gli TFs. Thus, as a consequence of Hh activation of Ptch, Gli TFs can translocate to the nucleus and activate transcription. Hh signaling is activated by purmorphamine, which binds the Smo receptor to facilitate neuronal differentiation and reprogramming (Li et al., 2008).
Calcium-Dependent Signaling
Ca2+ enters the cell through ion channels, and once intracellular Ca2+ concentrations increase, CaMKII is activated. CaMKII can phosphorylate HDACs, leading to their translocation out of the nucleus, to allow different TFs to promote neurogenesis, such as MEF2. Once HDAC interactions are removed, these TFs then transactivate neurogenic genes. ISX9 induces neural lineage development by activating calcium dependent signaling (Petrik et al., 2012).
Autophagy
Autophagy sustains cellular homeostasis by removing damaged organelles so that they can be replaced by new healthy organelles. Autophagy begins with Atg7, which acts like an E1 activator protein for ubiquitin ligases, which helps to “mark” damaged organelles, and activates Atg8/LC3. Atg8 is then required followed by the sequential formation of autophagosomes and then autophagolysosomes. Autophagy can be induced in an mTOR-independent fashion using SMER28, which enhances neuronal reprogramming (Zhang et al., 2016).
Other Vitamins or Chemicals
Vitamin C (Ascorbic Acid)
Vitamin C is added to reprogramming cocktails not only for its role as an antioxidant, but also for its activity as a modulator of the epigenome (Lee Chong et al., 2019). Vitamin C increases the activity of the Jumonji-C domain histone demethylases (JHDMs), which remove repressive H3K9 and H3K27 marks from chromatin. Vitamin C also increases the activity of ten-eleven translocation (TET) family DNA hydroxylases which “erase” DNA methylation (Lee Chong et al., 2019). Thus, vitamin C facilitates somatic cell reprogramming, both for iPSCs and for iNs, by opening up the chromatin to make it accessible for TF lineage determinant (Esteban and Pei, 2012; Lee Chong et al., 2019).
HDAC Inhibitors
HDAC inhibitors include valproic acid (VPA), sodium butyrate (NaB) and trichostatin A (TSA), prevent the removal of acetyl groups from histones. This therefore confers a generally open chromatin structure due to electrostatic repulsion between negatively charged DNA and acetylated histone groups. For example, NaB facilitates somatic cell reprogramming to an iPSC fate through preventing Oct4 reprogramming TF from associating with HDACs (Zhang et al., 2014). Consequently, Oct4 can elevate expression of the miR-202/367 cluster (Zhang et al., 2014) and is also important for neuronal lineage conversion (Zhou et al., 2015).
Other Chemical Agonists
Putrescine is a diamine that controls cellular stress response, regulates transcription, promotes stem cell self-renewal (Zhao et al., 2012), and enhances neuronal reprogramming (Rivetti di Val Cervo et al., 2017). 2-mercaptoethanol, which is a reducing agent, is also added to media during reprogramming as a potent reducing agent that removes free radicals, thereby preventing cell death. Transferrin binds iron, which reduces iron-induced cell death of iNs (Cozzi et al., 2019). Sodium selenite inhibits ROS-mediated apoptosis of neural cells (Yeo and Kang, 2007).
Neuronal Reprogramming Involving a Neural Precursor Cell or Neural Stem Cell Intermediate
Two major neural trans-differentiation approaches are being used: direct transformation of source cells into iNs, as described above, or the prior conversion of somatic cells to an induced neural progenitor cell (iNPC) or induced NSC (iNSC) state before the induction of neuronal differentiation. The advantage of first generating iNPCs or iNSCs is that these cells are proliferative and can be expanded before terminal differentiation to non-expandable iNs.
The first protocol developed to generate iNPCs from MEFs involved the transient expression of the four Yamanaka factors (Oct4, Sox2, Klf4, Myc) with the addition of FGF2/4 and EGF to support iNPC proliferation (Kim et al., 2011a). Culturing iNPCs in media containing an N2 supplement promoted neuronal differentiation, yielding functional iNs that invoke action potentials (Kim et al., 2011a). Importantly, with this paradigm, the MEFs did not pass through a pluripotent iPSC state, which reduces the risk of tumorigenicity (Kim et al., 2011a). Since then, efforts were also made to introduce the four Yamanaka TFs in an integration free manner into human and monkey fibroblasts using a Sendai virus that could be inactivated by heat (39°C) (Lu et al., 2014). In this study, small molecules such as LIF, SB431542 (to block TGFb) and CHIR99021 (to block GSK3 and activate Wnt) were also included (Lu et al., 2014). Human neonatal dermis-derived fibroblasts or adult adipose-derived stem cells have also been converted to an iNSC identity by misexpressing the four Yamanaka TFs and growing on a MEF feeder layer (Cairns et al., 2016). When the MEF feeder layer was later removed this allowed for the differentiation of human iNSCs into neuronal and glial cells that were successfully engrafted into either an embryonic chick brain and in a 3D human brain tissue model (Cairns et al., 2016). Finally, misexpression of the BAM factors together with Bcl-xl in murine embryonic fibroblasts or adult tail tip fibroblasts converts fibroblasts to an intermediate iNPC identity (Lim et al., 2015). Neuronal differentiation can then be achieved in a second step through the misexpression of TFs such as Nurr1 and Foxa1, which confer a dopaminergic neuronal identity (Lim et al., 2015).
Several groups have also subtracted some of the Yamanaka TFs and were still able to generate iNPCs/iNSCs. For example, by expressing the four Yamanaka TFs but restricting Oct4 expression to the initial reprogramming phase, mouse fibroblasts could form iNSCs that had the capacity to self-renew for >50 passages, a hallmark feature of stem cells (Thier et al., 2012). Similarly, the expression of OCT4, NANOG, or SOX2 in human astrocytes generated iNPCs, that could be differentiated into iNs in vivo through the additional expression of ASCL1 (Corti et al., 2012). The expression of only Sox2, Klf4, and Myc together with Brn4 (official gene name, Pou3f4) and Tcf3 could also induce mouse fibroblasts to a self-renewing iNSC identity (Han et al., 2012). In another study, self-renewing iNSCs were generated by the misexpression of a single Yamanaka factor, Sox2, in murine and human fibroblasts (Ring et al., 2012). Similarly, misexpression of OCT4 alone could induce human fibroblasts to transdifferentiate into iNPCs with trilineage potential (Mitchell et al., 2014). In a similar study, OCT4 overexpression was combined with several small molecules (CHIR99021, A83-01, sodium butyrate, LPA, rolipram, SP600125), termed cell activation and signaling-directed (CASD), to improve the conversion of human fibroblasts to iNSCs (Zhu et al., 2014). However, there is some concern for all the Yamanaka factors in the use of neuronal reprogramming due to their tumorigenic potential. For example, Oct4 misexpression can lead to the formation of dysplastic lesions (Hochedlinger et al., 2005), suggesting that this type of approach may not be suitable for clinical applications.
To overcome the low reprogramming efficiency from fibroblast into iNSCs, Sox2 can be used in addition to Klf4, Myc and Brn4, as these factors are known to reprogram fetal fibroblast into neural restricted precursor cells (Kim et al., 2014; Zou et al., 2014). In a subsequent screen of 11 neural TFs, the combination of Sox2, Brn2, and Foxg1 was found to be sufficient to convert murine fibroblasts to iNPCs with trilineage potential (Lujan et al., 2012). A screen of 14 TFs identified a panel of eight TFs (Brn2, Hes1, Hes3, Klf4, Myc, NICD, Plagl1, Rfx4) that can convert not only MEFs, but also adult liver cells and B lymphocytes into iNSCs (Cassady et al., 2014). In another expanded screen of 25 TFs, both 6 TF and 7 TF combinations were identified that could convert human fibroblasts into iNPs (Hou et al., 2017). Thus, as with programs to generate iNs directly, there are a variety of different molecular cocktails that can be used to generate iNSCs or iNPCs.
Direct Neuronal Reprogramming of Other Somatic Cell Types to iNs
While initial studies on direct neuronal reprogramming used either astrocytes or fibroblasts as starting cell types, a number of groups have begun to explore the possibility of converting other somatic cell types to iNs, including brain pericytes, olfactory ensheathing cells (OECs), adipocyte progenitor cells, hepatocytes, bone marrow derived mesenchymal stem cells, microglia, and others. Technically, some of these protocols should be classified as induced differentiation rather than lineage conversion strategies as they began with cells in a progenitor cell state, but as they involve the interconversion of one cell lineage to another, they are described herein.
The use of the BAM TFs, either alone or in combination with other factors, has been applied to the reprogramming of other somatic cell types in addition to fibroblasts with great success. Subsequent studies confirmed that BAM TFs could also trans-differentiate other murine somatic cell sources into iNs, including adipocyte progenitor cells (Yang et al., 2013) and hepatocytes, both from early postnatal mice, with the latter study further confirming silencing of the hepatocyte transcriptome (Marro et al., 2011). In another study, BAM TFs were combined with Neurod1 and were shown to convert murine and human iPSC-derived cardiomyocytes, which are a mesodermal lineage, to iNs that could fire action potentials (Chuang et al., 2017). In a similar study, vascular pericytes isolated from the human brain were transdifferentiated into iNs using BAM TFs along with Tlx3 and miR-124 (Liang X. G. et al., 2018). Interestingly, in pericytes, Myt1l, one of the BAM components, was sufficient on its own to promote pericyte to iN conversion, and it did so in part by elevating the expression levels of Ascl1, Brn2, and Neurog2 (Liang X. G. et al., 2018). Given that Myt1l’s role in neuronal lineage conversion has been attributed to its role in turning off the expression of non-neural genes, this finding raises the intriguing possibility that neural lineages are somehow a default fate of brain pericytes. Taken together, these studies support the robustness of the BAM cocktail for neuronal lineage conversion of a vast array of cell types.
Neuronal reprogramming of other somatic cell lineages has also been achieved using some of the original Yamanaka factors. For instance, Sox2, a SoxB1 TF that is both used to generate iPSCs and is a critical determinant required for specifying and maintaining a NPC identity during development has been primarily used by investigators (Zhang and Cui, 2014). In one study, SOX2 was misexpressed along with MYC, also one of the original Yamanaka factors, in CD133 positive human cord blood cells, resulting in the formation of iNs that were functionally active (Castano et al., 2014). Notably, the same conversion strategy was less efficient at converting peripheral blood mononuclear cells into functionally active iNs, highlighting intrinsic differences in the responses of different host cells to reprogramming factors (Castano et al., 2014). Similarly, Oct4 was used to transform cord blood and adult peripheral blood cells to iNPCs but required the addition of small molecules inhibitors of SMAD and GSK3 signaling (Lee et al., 2015), contrasting to fibroblasts in which Oct4 alone was sufficient for neuronal lineage conversion (Mitchell et al., 2014). Notably, blood-derived iNPCs differentiated into astrocytes or glutamatergic excitatory iNs (and not oligodendrocytes) after xenografting into the murine brain and could also be directed to a dopaminergic identity if pre-treated with Shh and FGF8b (Lee et al., 2015).
SOX2 was also combined with the proneural gene ASCL1 to transdifferentiate pericytes isolated from the human brain into functionally active iNs (Karow et al., 2012). However, in a follow-up study using scRNAseq to study lineage trajectories, the transdifferentiation of human pericytes to iNs was shown to traverse a NSC-like intermediate state, arguing that this procedure is not technically “direct” neuronal reprogramming (Karow et al., 2018). Similarly, Sox2/Ascl1 and Sox2/Neurog2 combinations were demonstrated to convert human umbilical cord mesenchymal stem cells (UMSC) to electrically active iNs (Araujo et al., 2018). Interestingly, when Neurog2 was expressed in rat mesenchymal stem cells (MSCs) that were transplanted into the striatum 1 day post-middle cerebral artery occlusion (MCAO) stroke in rats, the MSCs transformed into a NPC-like intermediate, and then transdifferentiated into electrically active iNs that could facilitate behavioral recovery (Cheng et al., 2014). As described further below, as the MSCs went through a NPC intermediate, it is not technically direct reprogramming, suggesting that Neurog2 alone is not sufficient to directly promote lineage conversion, at least in this non-neural starting cell type, without a progenitor intermediate (Cheng et al., 2014).
Another source of somatic cells that are “readily available” for reprogramming are olfactory ensheathing cells (OECs), a glial cell type in the olfactory epithelium (Sun et al., 2019). Interestingly, OECs can be converted to iNs by the expression of Neurog2 alone, which yields iNs that generate action potentials, form synapses, and integrate into the adult spinal cord (Sun et al., 2019). Given that glial cells are also neural, it is perhaps not surprising that OECs can be converted to iNs with one proneural TF. Indeed, conditioned media from OECs can induce the differentiation of spermatogonial stem cells into dopaminergic neurons when combined with other small molecules in a defined media (Yang et al., 2014). Early postnatal murine Sertoli cells could also be transdifferentiated into iNSCs using a cocktail of nine TFs (Ascl1, Neurog2, Hes1, Id1, Pax6, Brn2, Sox2, Myc, Klf4), which display trilineage potential, generating astrocytes, oligodendrocytes and neurons that are dopaminergic, GABAergic, or cholinergic (Sheng et al., 2011).
Neuronal reprogramming has also been applied to the inner ear, with the goal of replacing primary auditory neurons, which is achieved by the expression of Ascl1, or Ascl1 together with Neurod1 in cochlear non-sensory epithelial cells to form iNs that have the phenotype of auditory neurons (Nishimura et al., 2014). Notably, Neurod1 and Ascl1 were also co-expressed in spiral ganglion non-neuronal cells to generate iNs that had a transcriptomic profile that resembled primary auditory neurons, and which could project to cochlear hair cells in co-cultures in vitro (Noda et al., 2018).
Strikingly, in a screen of eight TFs, including all of the BAM TFs, Sox2, Neurog2, and Neurod1, as well as miR-124 and MBD3, an epigenetic modifier, misexpression of Neurod1 alone was found to be sufficient to convert murine microglial to iNs (Matsuda et al., 2019). Notably, in this study, Neurod1 acts as a pioneer factor, binding to and opening closed regions of the chromatin, to facilitate neuronal lineage conversion (Matsuda et al., 2019).
In summary, a variety of different somatic cell types can be converted to iNs, with different TF combinations active depending on the starting cell type. One of the important revelations from these studies was that in some instances, somatic cells transit through a NPC or NSC intermediate stage.
Applications for Induced Neurons Generated in vitro via Direct Neuronal Reprogramming
Important Considerations for Disease Modeling Using Patient-Derived iNs
One of the main applications for patient-derived iNs is modeling of neurological disease and neurodevelopmental disorders, including those with genetic or environmental causes. Several considerations must be taken into account to optimize lineage conversion for disease modeling, especially when somatic cells are reprogrammed from elderly patients (Figure 5):
FIGURE 5.
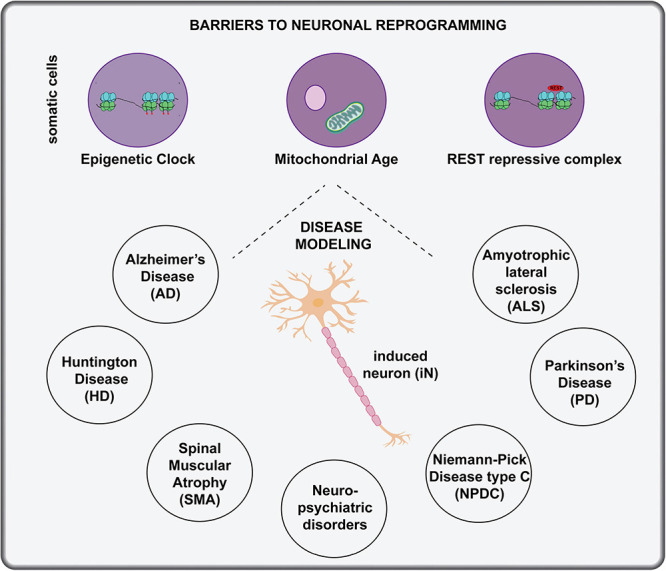
Applications of in vitro direct neuronal reprogramming. The generation of iNs in a dish has great potential for disease modeling, but several considerations must be made when using host cells from aged individuals, including changes to the epigenetic clock, mitochondrial age and activity of the REST repressive complex. Diseases that have been modeled with in vitro neuronal lineage conversion beginning with patient cells include AD, ALS, PD, HD, Niemann-Pick disease type C, and neuro-psychiatric disorders.
Epigenetic Clock
Direct reprogramming to generate iNs has clear advantages for studying the pathology of age-related neurological disorders because iPSC-derived iNs reset their epigenetic clocks and therefore may require long-term culturing to mimic aging and re-initiate disease (reviewed in Stricker and Gotz, 2021). Indeed, a direct comparison of iNs derived from iPSCs or tail tip fibroblasts revealed that the latter retained epigenetic signatures of the starting fibroblasts and expressed higher levels of genes associated with DNA damage and oxidative stress compared to iPSC-iNs (Yang et al., 2015). These findings are consistent with the idea that DNA methylation patterns change with biological age (or with cell passages in a dish), establishing a molecular clock that is essentially reset to the beginning after a somatic cell is converted to a pluripotent iPSC state (Horvath, 2013; Bell et al., 2019). In contrast, two studies revealed that the epigenetic signatures of the starting cell are maintained during direct neuronal reprogramming using miR-124 and miR-9/9∗ combined with CDM TFs (Huh et al., 2016), or combining the TFs Neurog2 and Ascl1 with small molecules (Mertens et al., 2015). Together these studies suggest that direct neuronal reprogramming is better than using iPSC-iNs for modeling diseases, especially those with an aging link.
Mitochondrial Age
Mitochondrial function decreases in neurons as they age, and importantly, the reduced OXPHOS capacity is recapitulated in iNs derived from “old” donor fibroblasts that are isolated from individuals 40 years of age and above (Kim et al., 2018). This finding demonstrates that iNs derived through direct neuronal reprogramming, in this case using Neurog2, Ascl1, and small molecules, can be used to model age related pathologies, including many neurodegenerative diseases (Kim et al., 2018).
REST Repressive Complex
Neuronal lineage conversion is less efficient when starting with cells obtained from older vs. younger individuals (Drouin-Ouellet et al., 2017). One study attributed these differences to elevated activity of the REST repressive complex in aged fibroblasts, which prevents the activation of neural-specific mRNAs and miRNAs, including miR-124 and miR-9/9∗, highlighted above as critical reprogramming factors (Drouin-Ouellet et al., 2017). The knockdown of REST activity thus offers a path forward for investigators experiencing difficulty in reprogramming fibroblasts from elderly patients.
With these considerations in mind, a snapshot of current efforts to use iNs derived from direct neuronal reprogramming to model neurological disorders is provided in the next section and summarized in Figure 5.
Disease Modeling Using Patient-Derived iNs
Alzheimer’s Disease (AD)
More than 90% of patients with AD exhibit late-onset, sporadic disease with no family history. In comparison, early-onset familial dementia is associated with known mutations in genes such as PSEN1, PSEN2, and APP (reviewed in Nardini et al., 2021). Several groups have now modeled neuronal pathology using iNs derived from sporadic AD patients. The conversion of human fibroblasts to iNs was achieved by expression of the BAM TFs together with NEUROD1, with neuronal conversion rates further enhanced by growing cells on nanopatterned substrates (Kim et al., 2017). This conversion strategy was applied either to human fibroblasts with induced expression of a mutant APP or derived from sporadic AD patient fibroblasts with an APOE ε3/4 allele that is an AD risk factor (Kim et al., 2017). Derivative iNs from AD patients exhibited increased amyloid-β42 accumulation and tau hyperphosphorylation, characteristic features of AD, and also displayed an increased vulnerability to peroxide-mediated cell death (Kim et al., 2017). Strikingly, using transcriptomic studies, this study identified DSG2 as an associated risk factor, the knockdown of which could rescue APP processing defects in iNs derived from patient fibroblasts with the APOE ε3/4 allele (Kim et al., 2017).
The generation of iNPCs has also been applied to fibroblasts from patients with sporadic and familial AD, using the six TF and seven TF reprogramming strategies described above (Hou et al., 2017). When the AD-iNPCs were converted to neurons, these neurons also accumulated amyloid-β42 and displayed tau hyperphosphorylation (Hou et al., 2017). Similarly, another group generated iNSCs from fibroblasts isolated from elderly patients with sporadic AD by expressing SOX2 and culturing the cells in media that contains a variety of small molecules (summarized in Tables 2, 3; Liu and Wang, 2020). The AD patient derived iNSCs could then be differentiated into iNs in vitro, and as in the above studies accumulated amyloid-β and high levels of phospho-tau (Liu and Wang, 2020). Future studies could consider the use of these cells for drug screening and additional modeling of disease pathophysiology.
Amyotrophic Lateral Sclerosis (ALS)
ALS is a terminal neurodegenerative disease that results in a loss of motor neurons (MNs) in the brain and spinal cord and leads to a fast deterioration of motor function, culminating in death between 3 and 5 years post diagnosis. Upper MNs in layer V of the neocortex degenerate and evidence in animal models and humans support a “dying forward model” of ALS, in which upper MN pathology precedes lower MN loss (Vucic and Kiernan, 2006; Ozdinler et al., 2011; Menon et al., 2015; Thomsen et al., 2018). Most ALS diagnoses are sporadic, with only 5–10% of cases inherited (Pasinelli and Brown, 2006). Of the familial cases, several mutations have been identified in SOD1, which encodes for a Cu-Zn superoxide dismutase (Bendotti and Carri, 2004). Hexa-repeat expansion mutations in chromosome 9 open reading frame 72 (C9ORF72) also occurs in ∼50% of familial ALS patients (Choi et al., 2019). Finally, mutations in TAR DNA binding protein 43 (TDP-43) and fused in sarcoma (FUS) (Lai et al., 2011) are also found in patients with ALS.
To model ALS, iNPCs were generated by misexpressing Yamanaka factors in fibroblasts isolated from patients with a SOD1A4V mutation, C9orf72 expansions, or sporadic disease (Meyer et al., 2013). ALS and control iNPCs were then converted to iAstrocytes, which were co-cultured with motor neurons, which revealed that ALS-derived iAstrocytes, but not glial cells derived from control individuals, induced motor neuron death (Meyer et al., 2013). In a subsequent study, fibroblasts from ALS patients bearing FUS mutations were converted to cholinergic iMNs by overexpressing NEUROG2, SOX11, LHX3, and ISL1 (Liu et al., 2016). While the ALS iMNs had typical neuronal morphologies and the same marker expression as iMNs derived from healthy individuals, ALS iMNs had an increased propensity to undergo cell death, and displayed electrophysiological deficits (Liu et al., 2016). Finally, as the ALS iMNs could accurately model the pathophysiology of disease, a small molecule screen was also performed, which identified kenpaullone as a factor that increases iMN survival, a promising finding for the future development of new therapeutics (Liu et al., 2016).
Huntington Disease (HD)
HD is associated with a poly-glutamine expansion in the Htt protein, which leads to the formation of Htt protein aggregates that are associated with neuronal death (Li and Li, 2004). Expansions of 36 or more CAG repeats, encoding glutamine (Q), in Htt are considered pathogenic (Tabrizi et al., 2020). The first study to apply direct neuronal reprogramming to HD knocked down PTBP1 in fibroblasts from individuals with 16Q (non-pathogenic), 68Q (pathogenic), and 86Q repeats (pathogenic) (Liu et al., 2014). Notably, the iNs derived from individuals that harbor pathogenic Htt polyQ expansions formed Htt aggregates, and displayed abnormally thin and short neurites (Liu et al., 2014). HD patient fibroblast cells that were trans-differentiated to iNPCs using the 6-TF and 7-TF cocktails above were then differentiated into iNs, which displayed pathological hallmarks of disease, including an increase in DNA damage (Hou et al., 2017). Similarly, although not technically direct neuronal reprogramming, an increase in the DNA damage response and higher oxidative stress was observed in iNs derived from HD patient-derived iPSCs (Chiu et al., 2015). Taken together, these studies demonstrate that some features of HD pathophysiology may be accurately modeled by iNs.
Parkinson’s Disease (PD)
As with the other neurodegenerative diseases, there are both sporadic and genetic forms of PD (Torrent et al., 2015). The death of dopaminergic neurons in familial PD is associated with oxidative stress and has been linked to several potentially pathogenic mutations in genes involved in mitochondrial homeostasis, such as PARK2, PINK1, PARK7, SNCA, GBA1, and LRRK2 (Garcia and Sidransky, 2021). A large number of studies have used iPSC-derived iNs to study PD pathophysiology, with difficulties related to the epigenetic age of these cells (reviewed in Torrent et al., 2015). Direct neuronal reprogramming may circumvent these issues, as highlighted above, and was first applied using the misexpression of Yamanaka factors to generate iNPCs from sporadic and familial PD with a LRRK2 mutation (Lee et al., 2019). In a follow-up study, PD patient-derived iNPCs displayed a higher rate of apoptosis, which was reduced by treatment with cryptotanshinone (CTN), where CTN modulates mitochondrial ROS and membrane potential to reduce cellular apoptosis (Lee J. E. et al., 2020). Follow-up studies are required to determine whether iNs derived from the PD patient iNPCs display pathological features.
Spinal Muscular Atrophy (SMA)
SMA is an autosomal recessive disorder associated with a homozygous mutation of SMN1, and results in spinal motor neuron death. The pathophysiology of SMA has recently been characterized by generating iMNs from patient and control fibroblasts through the expression of a cocktail of eight TFs: ASCL1, ISL1, NEUROD1, BRN2, HB9, LHX3, MYT1L, and NEUROG2 (Zhang et al., 2017). Notably, SMA patient-derived iMNs had a reduced rate of neurite outgrowth, and displayed signs of neurite degeneration in long-term culture. These features highlight the potential of this model to further understand disease biology (Zhang et al., 2017).
Niemann-Pick Disease Type C (NPDC)
NPDC is an autosomal recessive neurodegenerative disease associated with homozygous mutations in NPC1 or NPC2 (Carstea et al., 1997). NPDC is a liposomal storage disorder, and manifests early in life with dementia, cerebellar ataxia and other symptoms that progresses ultimately to death. Modeling NPDC has been achieved using the direct conversion of patient fibroblasts to iNPCs by misexpressing SOX2 and the high mobility group A2 (HMGA2) protein, which alters chromatin structure by binding to AT rich regions of the genome (Sung et al., 2017). Prior studies have demonstrated that HMGA2 augments SOX2’s ability to reprogram somatic cells to iNSCs (Chang et al., 2005). In this study, iNPCs derived from NPDC patients were shown to have defects in self-renewal, neurogenesis, and cholesterol homeostasis (Sung et al., 2017). As the patient-derived iNSCs phenocopy several aspects of NPDC, future studies can examine their utility for drug screening.
Neuropsychiatric Disorders
Neuropsychiatric disorders are diverse in origin and in manifestation, and lead to a variety of defects, such as in cognition, psychoses, neuroses, and mental impairment. In many cases, the etiology of these disorders remains unknown or poorly characterized. A large number of studies have linked miRNA disruptions to schizophrenia, autism spectrum disorder, depression, and other neuropsychiatric disorders (reviewed in Bavamian et al., 2015). One example is bipolar disorder (BD), in which patient may exhibit cycles of mania followed by depressive episodes. The generation of iNs from BD patients and healthy controls was achieved by misexpressing miR-9, miR-124, NEUROD2, ASCL1, and MYT1L (Bavamian et al., 2015). The iNs from BD patients expressed elevated levels of miR-34a, which was also increased in autopsy specimens (Bavamian et al., 2015). Consequently, miR-34a target genes, such as ANK3 and CACNB3, were elevated in BD patient-derived iNs. This study highlights the potential of utilizing neuronal reprogramming to identify the genetic contributors of a neuropsychiatric disorder.
In vivo Neuronal Reprogramming of Endogenous Glial Cells
Predominant Use of bHLH Transcription Factors for in vivo Reprogramming
The first evidence that endogenous glial cells could be converted to iNs by TF misexpression was obtained in a stab wound injury model in the adult mouse brain, with Pax6 driving the neuronal conversion of resident glial cells (Buffo et al., 2005). Since then, in vivo reprogramming of reactive glia, including NG2 glia and astrocytes, to iNs was achieved by expressing a variety of TFs alone or in combination with other molecules, but the factors used most often are bHLH TFs. Critical studies on these bHLH TFs are summarized herein and in Figure 6.
FIGURE 6.
Applications of in vivo direct neuronal reprogramming to treat injury and/or neurodegenerative disease. Direct neuronal reprogramming has therapeutic potential for brain injury (e.g., glial scars, stroke) and neurodegenerative diseases, such as PD, HD, and ALS. The advantage of this approach is that endogenous glial cells can be directly reprogrammed to replace lost neurons at the site of injury or trauma.
Employing Neurog2 for in vivo Neuronal Reprogramming
Neurog2 alone is inherently inefficient at inducing neuronal lineage conversion, especially in vivo, but several groups have enhanced reprogramming by combining Neurog2 with various other factors. Specifically, in vivo neuronal reprogramming has been enhanced by expressing Neurog2 in glial cells together with the co-injection of growth factors (FGF2, EGF) (Grande et al., 2013), co-injection of DAPT to block Notch signaling (Hu et al., 2019), co-expression with Bcl2 and injection of antioxidants (Gascon et al., 2016), or through co-expression with Nurr1 (Mattugini et al., 2019). Notably, a study of focal ischemia, a well-characterized stroke model, further enhanced the ability of Neurog2 to drive glial conversion to mature DCX-/NEUN+ iNs compared to the immature DCX+ iNs that predominated in the uninjured brain (Grande et al., 2013). In contrast, in a conflicting report, the misexpression of Neurog2 and Bcl2 in reactive astrocytes post ischemia led to very low neuronal conversion rates, both in young and old mice (Gresita et al., 2019).
These studies suggest that the astrocyte changes that accompany injury, such as reactive gliosis, may impact glial cell plasticity and hence influence neuronal reprogramming. Moreover, astrocytes in different brain regions have a differential capacity to undergo neuronal lineage conversion, with Neurog2 triggering the conversion of GFAP+ glial cells to iNs more effectively in the cortex > cerebellum > spinal cord (Hu et al., 2019). These studies highlight the sensitivity of reprogramming TFs to environmental context, and suggest that their activities are tightly regulated. Such a tight regulation fits with the regulatory controls that govern neural-specific bHLH function during normal embryonic and adult neurogenesis (reviewed in Oproescu et al., 2021).
Use of Ascl1 for in vivo Neuronal Reprogramming
In a 2014 study, resident NG2 glia were induced to differentiate into neurons either by the overexpression of Ascl1 with Sox2, or Sox2 alone, triggering the generation of both immature DCX+ or mature DCX-/NEUN+ cells that were electrically active based on patch clamp recordings (Heinrich et al., 2014). Ascl1 has also been combined with Lmx1a and Nurr1 (ALN) to convert endogenous NG2 glia to inhibitory iNs in a dopamine-depleted (6-OHDA treated) mouse model of Parkinson’s disease (Pereira et al., 2017). The ALN-iNs integrated into the existing neuronal circuitry in the striatum and midbrain of 6-OHDA treated mice (Pereira et al., 2017). Interestingly, although this same combination of factors generated dopaminergic iNs in vitro (Torper et al., 2013), it could not activate tyrosine hydroxylase (TH) expression in iNs generated in vivo (Pereira et al., 2017). In contrast, the combination of a slightly different combination of TFs (Ascl1, NeuroD1, Lmx1a) and miR-218 efficiently programmed striatal astrocytes into functional induced dopaminergic neurons (iDANs), also in a 6-OHDA model of Parkinson’s disease (Rivetti di Val Cervo et al., 2017). Not only were the iDANs able to fire action potentials, but there was also an associated behavioral recovery based on improved performance on motor tasks (Rivetti di Val Cervo et al., 2017). However, one confounding factor is that this study did not use a genetic reporter for the reprogrammed iNs, so some concern has been expressed that the identified TH+ cells arose in situ from the 6-OHDA lesion itself (Pereira et al., 2017).
Nevertheless, these studies hold promise that a neurodegenerative disease such as Parkinson’s disease may be treated by direct neuronal reprogramming in the future. Indeed, improvements have been made in the TF cocktails used for iDAN reprogramming. For example, by misexpressing Sox2, Nurr1, Lmx1a, and Foxa1 TFs and injecting Valproic acid, striatal glia could be converted to iDANs in vivo (Niu et al., 2018). It thus seems only a matter of time until an effective TF combination is found to generate iDANs in vivo for the treatment of PD.
Use of Neurod1 for in vivo Neuronal Reprogramming
Neurod1 has proven to be a potent reprogramming vehicle in vivo, driving the conversion of activated astrocytes or NG2 glia into functional iNs in a stab wound brain injury model and in Alzheimer’s disease model mice (Guo Z. et al., 2014). While Neurod1 effectively converts cortical astrocytes to glutamatergic iNs, consistent with its subtype differentiation properties in development, Neurod1 converted NG2 glia to glutamatergic and GABAergic iNs (Guo Z. et al., 2014). Notably, Neurod1 could also convert striatal microglia to functional neurons in vivo, even without intended injury, although injecting the viral construct may itself trigger a stab wound response (Matsuda et al., 2019). To avoid this confounding factor, Neurod1 was delivered intravascularly to the brain by packaging in adeno-associated virus (AAV) 9, which crosses the blood-brain-barrier without the need for injections into the brain. With this mode of delivery, non-reactive astrocytes in the striatum were reprogrammed to iNs (Brulet et al., 2017). Surprisingly, however, cortical astrocytes were not converted to iNs with the intravascular AAV9 delivery approach (Brulet et al., 2017). These findings are consistent with the demonstration that astrocytes show region-specific responses to neuronal reprogramming factors (Hu et al., 2019).
Glial scars form after injury to form a functional barrier between the injured and healthy brain. While they serve an important role in the injury response, they also permanently remodel the brain and can be detrimental in the long-term (Sofroniew, 2015). Interestingly, the overexpression of Neurod1 in astrocytes generated functional iNs and reduced glial toxicity after a stab wound brain injury (Zhang et al., 2020). Moreover, the use of the GFAP promoter in this study appeared to more selectively target the conversion of A1 astrocytes, which secrete inflammatory cytokines, vs. the A2 astrocytes, which are neuroprotective (Zhang et al., 2020). There was also significant tissue repair in the wounded region of the brain after Neurod1 expression, including rebalancing of the neuron: astrocyte ratio, improvement of the blood-brain-barrier, and a decrease in both reactive astrocytes and microglia (Zhang et al., 2020).
Neurod1-induced lineage conversion of reactive astrocytes has also been applied to the treatment of a focal stroke model in mice induced using endothelin-1, a vasoconstrictor (Chen et al., 2020; Livingston et al., 2020). In both of these studies, Neurod1-induced the effective transdifferentiation of astrocytes to functional iNs in vivo, and provided a significant enhancement of motor behavior (Chen et al., 2020; Livingston et al., 2020). These studies provide important proof-of-concept evidence that a neuronal reprogramming strategy could be applied to the treatment of brain injury.
Finally, Neurod1-induced neuronal reprogramming has also been applied to the treatment of a neurodegenerative disease. When Neurod1 was co-expressed with Dlx2, it could drive the efficient conversion of striatal astrocytes to GABAergic, induced medium spiny neurons (iMSNs) in a mouse model of Huntington Disease (R6/2 mice) (Wu et al., 2020). Not only were the iMSNs functional, but this gene therapy also extended the lifespan of R6/2 mice, delayed their onset of motor deficits, and caused reduced neuronal pathology, which provides encouraging support for the potential of neuronal reprogramming to treat neurodegeneration (Wu et al., 2020).
Therapeutic Potential of in vivo Reprogramming in Comparison to the Current Strategies
Comparing the Potential of Exogenous vs. Endogenous Neuronal Repair
In neurodegenerative diseases, dying neurons are not replaced, so new therapeutic options are required. Similarly, even though some neurogenesis occurs in the infarct region post-stroke, due to NSC migration from neurogenic zones in the SGZ and V-SVZ, very few new neurons are made and functional recovery is not complete (Rahman et al., 2021). New strategies that drive neuronal replacement are thus required. When devising neuronal replacement strategies for brain injury or neurodegenerative diseases, there are two potential approaches—endogenous or exogenous repair. In this review, we focus on the potential of direct neuronal reprogramming, which could be used to target endogenous glial cells, or to generate iNs for transplant and exogenous repair.
Potential of Exogenous Repair
Cell transplantation in the CNS has long been considered as a potential therapeutic approach, and could in theory be applied to iNs. Indeed, stem cell transplantations into the brain have been investigated since the 1980s, with several human clinical trials initiated (reviewed in Fan et al., 2020; Osborn et al., 2020; Stoddard-Bennett and Pera, 2020). For instance, a search of https://clinicaltrials.gov/ for “stem cell” and various neurological disorders revealed that 31 clinical trials have been initiated for PD, 60 for ALS, and 93 for stroke. Yet, despite these many trials, stem cell transplants are not yet standard-of-care treatments for any of these disorders. Many noteworthy challenges exist, including cell source and purity, potential tumorigenicity, and immune tolerance (Michelsen et al., 2015; Doerr et al., 2017). Cell purity is of particular concern when transplanting neurons derived from pluripotent cells, which are potentially tumorigenic, so the development of robust strategies to remove undifferentiated cells is critical. Interestingly, by linking a cell death gene (Tk, thymidine kinase) to the promoter of a cell division gene (Cdk1), one group has devised a strategy that can kill any proliferating cells that may have escaped cell purification steps (Liang Q. et al., 2018).
While immune rejection could be prevented by using autologous cell transplantations, for instance of iPSC-derived cells (reviewed in Osborn et al., 2020), the hope would be that a universal supply of iNs for transplant could be developed. To avoid lifelong immunosuppression, one approach would be to generate a panel of human leukocyte antigen (HLA)-typed iPSCs that could be matched to patients (Doi et al., 2020; Piao et al., 2021). Another innovative approach to tackle the issue of host-immunoreactivity and graft rejection has involved the development of a “cell cloaking” strategy, which involves the expression of eight immunomodulatory genes, allowing transplanted cells to escape rejection in allogeneic hosts (Harding et al., 2019; Lanza et al., 2019). Future extensive studies will be required to develop these approaches for clinical application.
Potential of Endogenous Repair
The main advantage of neuronal reprogramming in situ is the possibility that resident glial cells in the brain can be targeted, so that brain rejection is not an issue. The ability to move forward with an endogenous repair approach is supported by major advances made in AAV gene therapy, including many examples now of AAVs moving to the clinic, especially to treat monogenic diseases. Some examples for CNS disorders include an AAV2 gene therapy designed to express RPE65 in the eye to treat Leber Congenital Amaurosis (Luxturna®) and an AAV9 vector that expresses SMN1 in motor neurons to treat Spinal Muscular Atrophy (Spinaraza®).
Future Challenges and Safety Considerations for Endogenous Neuronal Repair
Effects of Astrocyte Loss/Neuronal Gain
The main approach for neuronal reprogramming is to convert astrocytes to iNs, but an important consideration is the effect of the consequent astrocyte loss on brain function. Indeed, there is some evidence that the loss of local astrocytes affects brain homeostasis in PD patients (reviewed in Khakh and Deneen, 2019). Future studies will be required to investigate the consequences of glial loss in detail. It is also important to consider that iNs may form aberrant connections that are counterproductive, or even detrimental, triggering epileptic foci, for example.
Toxicity of Overexpressed TFs
An early study used retroviruses to overexpress several neural bHLH TFs, including Neurog2, Ascl1, and Neurod1, in the P0/P1 cortical subventricular zone, and while these genes efficiently induced neurogenesis, the newly derived neurons ultimately underwent apoptosis (Cai et al., 2000). This study suggests that the long-term expression of developmental bHLH genes in mature neurons is ultimately toxic. It is therefore important to devise reprogramming strategies that prevent sustained expression of bHLH genes. While most studies use a GFAP astrocytic promoter to drive bHLH gene expression, which theoretically should not be active in the new iNs, detailed studies are required to test this assumption.
Cellular Context
An important consideration for moving neuronal reprogramming into the clinic is that the TFs and miRNAs that trigger neuronal lineage conversion all function in a context-dependent fashion. Understanding how the diseased brain environment affects their functions is thus critical. For instance, Ascl1 is a potent neuronal reprogramming factor in fibroblasts (Vierbuchen et al., 2010; Caiazzo et al., 2011; Kim et al., 2011b; Pang et al., 2011; Pfisterer et al., 2011a; Son et al., 2011), hepatocytes (Marro et al., 2011), cardiomyocytes (Chuang et al., 2017), astrocytes (Rivetti di Val Cervo et al., 2017), and pluripotent cells (Yang et al., 2017), but not in the adult neocortex (Grande et al., 2013), hippocampus and spinal cord (Ohori et al., 2006; Jessberger et al., 2008). Similarly, Neurog2 is used less often for neuronal reprogramming because it must be combined with other signals to become a potent lineage converter (Gascon et al., 2016). It currently remains to be determined how the diseased brain affects neuronal reprogramming efficiency. When considering how TFs/miRNAs operate in a diseased brain, especially in vivo, one must consider that cell fate is ultimately dictated by the balance of inducers and repressors (Yamashita et al., 2019). The impact of several negative regulators of neurogenesis (e.g., p53-p21 pathway, REST repressor complex, oxidative stress) that may suppress reprogramming processes in a diseased brain remain to be fully elucidated (Cheloufi et al., 2015; Jiang et al., 2015; Masserdotti et al., 2015; Gascon et al., 2016). Addressing the “real life” hurdles of the compromised in vivo environment of the diseased/damaged brain will be essential to successfully using neuronal reprogramming for neural repair in the clinic.
Long-Term Toxicity of AAVs
AAVs delivered to the brain may trigger an immune reaction in situ, or if the blood-brain-barrier is compromised, which generally occurs in neurological diseases, escape into the CSF, blood or draining lymph nodes to infect peripheral organs. Potential AAV toxicity must, therefore be tested extensively before moving to the clinic.
Delivery Challenges
Our ability to deliver therapeutics to the brain is limited by the blood-brain-barrier (BBB), which blocks all but the smallest molecules from entering via the bloodstream. Intracranial injections are thus currently the method of choice for viral delivery to the brain in animal models, and while this approach can be used in humans, it is surgically invasive. Magnetic resonance imaging (MRI)-guided focused ultrasound (FUS) is a new, minimally invasive approach that transiently increases the permeability of the BBB to provide therapeutic access to the brain. FUS has been used for delivery of antibodies (Jordao et al., 2010), genes (Thevenot et al., 2012; Weber-Adrian et al., 2015), stem cells (Burgess et al., 2011), and immunoglobulins (Dubey et al., 2020) to the brain and spinal cord in mouse models. Moreover, FUS has been used to transiently permeabilize the BBB in human patients with ALS (Abrahao et al., 2019) or AD (Lipsman et al., 2018; Meng et al., 2019). Future investigations using FUS-mediated AAV gene delivery, or other minimally invasive techniques, will improve the feasibility of using gene therapies for direct neuronal reprogramming.
Conclusion
The development of experimental paradigms for direct somatic cell reprogramming into any desired lineage, without a transient pluripotent state, was a major breakthrough in the field of lineage conversion. Remarkable achievements have been made in identifying TFs, miRNAs, small molecules and other factors that drive direct neuronal reprogramming, both in vitro and in vivo. The potential application of this technology to therapeutic scenarios is just coming to the forefront, and provides new promise for the large number of individuals afflicted by neurodegenerative disease or brain injury.
Author Contributions
LV and CS: conceptualization. LV, EP, LD, and CS: writing—review and editing. TF: artwork. CS: funding acquisition. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- AD
Alzheimer’s disease
- ALS
Amyotrophic lateral sclerosis
- ANL
Ascl1-Nurr1-Lmx1a
- BAF
Brg/Brm-associated factors
- BAM
Brn2-Ascl1-Myt1l
- BDNF
Brain derived neurotrophic factor
- bHLH
basic-helix-loop-helix
- BMP
Bone morphogenic protein
- CNS
central nervous system
- CREB
cyclic-AMP response element binding protein
- CTN
Cryptotanshinone
- DALY
disability-adjusted life years
- DNMT
DNA methyltransferase
- DREADD
Designer Receptors Exclusively Activated by Designer Drugs
- ESC
embryonic stem cells
- FGF
fibroblast growth factor
- FTD
frontotemporal dementia
- GDNF
Glial cell derived neurotrophic factor
- GFAP
glial fibrillary acidic protein
- HAFs
human adult fibroblasts
- HD
Huntington’s disease
- HDAC
histone deacetylase
- hESC
human embryonic stem cell
- iDA
induced dopaminergic neurons
- iMNs
induced motor neurons
- iN
induced neurons
- iPSC
induced pluripotent stem cell
- JHDM
Jumonji-C domain histone demethylases
- LC-MS
liquid chromatography- mass spectroscopy
- LIF
Leukemia inhibitory factor
- lncRNA
long non-coding RNA
- MCAO
middle cerebral artery occlusion
- MEF
mouse embryonic fibroblasts
- miR
microRNA
- Mll1
Mixed lineage leukemia-1
- MRIgFUS
magnetic resonance guided focused ultrasound
- MSN
medium spiny neurons
- NaB
sodium butyrate
- NAMPT
nicotinamide phosphoribosyl transferaseNeurog2, Neurogenin 2
- NICD
Notch intracellular domain
- NPC
Neural progenitor cell
- NPDC
Niemann-Pick disease type C
- NSC
Neural stem cell
- OEC
Olfactory Ensheathing cells
- OHDA
hydroxydopamine
- OPC
oligodendrocyte precursor cells
- OXPHOS
oxidative phosphorylation
- P
postnatal
- PD
Parkinson’s disease
- PNS
peripheral nervous system
- PTBP
poly-pyrimidine-tract-binding protein
- RA
Retinoic acid
- REST
(RE1) silencing transcription factor
- ROS
reactive oxygen species
- SGZ
subgranular zone
- SHH
sonic hedgehog
- SMA
spinal muscular atrophy
- TET
ten-eleven translocation
- Tet3
TET methylcytosine dioxygenase3
- TF
transcription factor
- TGFb
Transforming growth factor b
- TIGAR
TP53 inducible glycolysis and apoptosis regulator
- UMSC
Human umbilical cord mesenchymal stem cells
- VPA
valproic acid
- V-SVZ
ventricular-subventricular zone
- YAP
Yes-associated protein.
Footnotes
Funding. This work was supported by operating grants to CS from the Canadian Institutes of Health Research (CIHR PJT–162108) and the University of Toronto’s Medicine by Design initiative, which receives funding from the Canada First Research Excellence Fund (CFREF). LV was supported by a University of Toronto Department of Laboratory Medicine and Pathobiology Admission Award. EP was supported by a University of Toronto Department of Biochemistry Fellowship and an Ontario Graduate Scholarship. LD was supported by a University of Toronto Department of Laboratory Medicine and Pathobiology fellowship, CIHR Frederick Banting and Charles Best Masters award, and Vision Science Research Program awards. CS holds the Dixon Family Chair in Ophthalmology Research.
References
- Abernathy D. G., Kim W. K., McCoy M. J., Lake A. M., Ouwenga R., Lee S. W., et al. (2017). MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell 21 332.e9–348.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahao A., Meng Y., Llinas M., Huang Y., Hamani C., Mainprize T., et al. (2019). First-in-human trial of blood-brain barrier opening in amyotrophic lateral sclerosis using MR-guided focused ultrasound. Nat. Commun. 10:4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis R. C., Hsu F. C., Wright R. L., Dichter M. A., Coulter D. A., Gearhart J. D. (2011). Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One 6:e28719. 10.1371/journal.pone.0028719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Arjona A., Cimadamore F., Huang C. T., Wright R., Lewis S., Gage F. H., et al. (2015). SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 112 E1936–E1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S. A., et al. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9 113–118. 10.1016/j.stem.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo J. A. M., Hilscher M. M., Marques-Coelho D., Golbert D. C. F., Cornelio D. A., Batistuzzo de Medeiros S. R., et al. (2018). Direct reprogramming of adult human somatic stem cells into functional neurons using Sox2, Ascl1, and Neurog2. Front. Cell Neurosci. 12:155. 10.3389/fncel.2018.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au E., Ahmed T., Karayannis T., Biswas S., Gan L., Fishell G. (2013). A modular gain-of-function approach to generate cortical interneuron subtypes from ES cells. Neuron 80 1145–1158. 10.1016/j.neuron.2013.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavamian S., Mellios N., Lalonde J., Fass D. M., Wang J., Sheridan S. D., et al. (2015). Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol. Psychiatry 20 573–584. 10.1038/mp.2014.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C. G., Lowe R., Adams P. D., Baccarelli A. A., Beck S., Bell J. T., et al. (2019). DNA methylation aging clocks: challenges and recommendations. Genome Biol. 20:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch E. E. (2009). The locus ceruleus norepinephrine system: functional organization and potential clinical significance. Neurology 73 1699–1704. 10.1212/wnl.0b013e3181c2937c [DOI] [PubMed] [Google Scholar]
- Bendotti C., Carri M. T. (2004). Lessons from models of SOD1-linked familial ALS. Trends Mol. Med. 10 393–400. 10.1016/j.molmed.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Berninger B., Costa M. R., Koch U., Schroeder T., Sutor B., Grothe B., et al. (2007a). Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 27 8654–8664. 10.1523/jneurosci.1615-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger B., Guillemot F., Gotz M. (2007b). Directing neurotransmitter identity of neurones derived from expanded adult neural stem cells. Eur. J. Neurosci. 25 2581–2590. 10.1111/j.1460-9568.2007.05509.x [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Black J. B., Adler A. F., Wang H.-G., D’Ippolito A. M., Hutchinson H. A., Reddy T. E., et al. (2016). Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell 19 406–414. 10.1016/j.stem.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M., Fulmore C. A., Tartt A. N., Simeon L. R., Pavlova I., Poposka V., et al. (2018). Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell 22 589.e5–599.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulet R., Matsuda T., Zhang L., Miranda C., Giacca M., Kaspar B. K., et al. (2017). NEUROD1 instructs neuronal conversion in non-reactive astrocytes. Stem Cell Rep. 8 1506–1515. 10.1016/j.stemcr.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo A., Vosko M. R., Erturk D., Hamann G. F., Jucker M., Rowitch D., et al. (2005). Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. Proc. Natl. Acad. Sci. U.S.A. 102 18183–18188. 10.1073/pnas.0506535102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bung R., Worsdorfer P., Thier M. C., Lemke K., Gebhardt M., Edenhofer F. (2016). Partial dedifferentiation of murine radial glia-type neural stem cells by Brn2 and c-Myc yields early neuroepithelial progenitors. J. Mol. Biol. 428 1476–1483. 10.1016/j.jmb.2015.10.028 [DOI] [PubMed] [Google Scholar]
- Burgess A., Ayala-Grosso C. A., Ganguly M., Jordao J. F., Aubert I., Hynynen K. (2011). Targeted delivery of neural stem cells to the brain using MRI-guided focused ultrasound to disrupt the blood-brain barrier. PLoS One 6:e27877. 10.1371/journal.pone.0027877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Morrow E. M., Cepko C. L. (2000). Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development 127 3021–3030. 10.1242/dev.127.14.3021 [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Dell’Anno M. T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476 224–227. 10.1038/nature10284 [DOI] [PubMed] [Google Scholar]
- Cairns D. M., Chwalek K., Moore Y. E., Kelley M. R., Abbott R. D., Moss S., et al. (2016). Expandable and rapidly differentiating human induced neural stem cell lines for multiple tissue engineering applications. Stem Cell Rep. 7 557–570. 10.1016/j.stemcr.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea E. D., Morris J. A., Coleman K. G., Loftus S. K., Zhang D., Cummings C., et al. (1997). Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277 228–231. 10.1126/science.277.5323.228 [DOI] [PubMed] [Google Scholar]
- Cassady J. P., D’Alessio A. C., Sarkar S., Dani V. S., Fan Zi P., Ganz K., et al. (2014). Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells. Stem Cell Rep. 3 948–956. 10.1016/j.stemcr.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano J., Menendez P., Bruzos-Cidon C., Straccia M., Sousa A., Zabaleta L., et al. (2014). Fast and efficient neural conversion of human hematopoietic cells. Stem Cell Rep. 3 1118–1131. 10.1016/j.stemcr.2014.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates K., McCoy M. J., Kwon J. S., Liu Y., Abernathy D. G., Zhang B., et al. (2021). Deconstructing stepwise fate conversion of human fibroblasts to neurons by MicroRNAs. Cell Stem Cell 28 127.e9–140.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., Studer L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27 275–280. 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S., Ang C. E., Davila J., Pak C., Mall M., Lee Q. Y., et al. (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3 282–296. 10.1016/j.stemcr.2014.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. Y., Reid P. C., Sugii S., Ohgami N., Cruz J. C., Chang C. C. (2005). Niemann-Pick type C disease and intracellular cholesterol trafficking. J. Biol. Chem. 280 20917–20920. [DOI] [PubMed] [Google Scholar]
- Cheloufi S., Elling U., Hopfgartner B., Jung Y. L., Murn J., Ninova M., et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528 218–224. 10.1038/nature15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C., Ma N. X., Pei Z. F., Wu Z., Do-Monte F. H., Keefe S., et al. (2020). A NeuroD1 AAV-based gene therapy for functional brain repair after ischemic injury through in vivo astrocyte-to-neuron conversion. Mol. Ther. 28 217–234. 10.1016/j.ymthe.2019.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Lu X. C., Hao H. Y., Dai X. L., Qian T. D., Huang B. S., et al. (2014). Neurogenin 2 converts mesenchymal stem cells into a neural precursor fate and improves functional recovery after experimental stroke. Cell. Physiol. Biochem. 33 847–858. 10.1159/000358657 [DOI] [PubMed] [Google Scholar]
- Cheng L., Gao L., Guan W., Mao J., Hu W., Qiu B., et al. (2015). Direct conversion of astrocytes into neuronal cells by drug cocktail. Cell Res. 25 1269–1272. 10.1038/cr.2015.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. C., Pastrana E., Tavazoie M., Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12 399–408. 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu F. L., Lin J. T., Chuang C. Y., Chien T., Chen C. M., Chen K. H., et al. (2015). Elucidating the role of the A2A adenosine receptor in neurodegeneration using neurons derived from Huntington’s disease iPSCs. Hum. Mol. Genet. 24 6066–6079. 10.1093/hmg/ddv318 [DOI] [PubMed] [Google Scholar]
- Choi S. Y., Lopez-Gonzalez R., Krishnan G., Phillips H. L., Li A. N., Seeley W. W., et al. (2019). C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat. Neurosci. 22 851–862. 10.1038/s41593-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchane M., Melo de Farias A. R., Moura D. M. D. S., Hilscher M. M., Schroeder T., Leão R. N., et al. (2017). Lineage reprogramming of astroglial cells from different origins into distinct neuronal subtypes. Stem Cell Rep. 9 162–176. 10.1016/j.stemcr.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang W., Sharma A., Shukla P., Li G., Mall M., Rajarajan K., et al. (2017). Partial reprogramming of pluripotent stem cell-derived cardiomyocytes into neurons. Sci. Rep. 7:44840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G., Lignani G., Rubio A., Medrihan L., Yekhlef L., Sessa A., et al. (2015). Rapid conversion of fibroblasts into functional forebrain GABAergic interneurons by direct genetic reprogramming. Cell Stem Cell 17 719–734. 10.1016/j.stem.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Corti S., Nizzardo M., Simone C., Falcone M., Donadoni C., Salani S., et al. (2012). Direct reprogramming of human astrocytes into neural stem cells and neurons. Exp. Cell Res. 318 1528–1541. 10.1016/j.yexcr.2012.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi A., Orellana D. I., Santambrogio P., Rubio A., Cancellieri C., Giannelli S., et al. (2019). Stem cell modeling of neuroferritinopathy reveals iron as a determinant of senescence and ferroptosis during neuronal aging. Stem Cell Rep. 13 832–846. 10.1016/j.stemcr.2019.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51 987–1000. 10.1016/0092-8674(87)90585-x [DOI] [PubMed] [Google Scholar]
- Dell’Anno M. T., Caiazzo M., Leo D., Dvoretskova E., Medrihan L., Colasante G., et al. (2014). Remote control of induced dopaminergic neurons in parkinsonian rats. J. Clin. Invest. 124 3215–3229. 10.1172/jci74664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez del Corral R., Olivera-Martinez I., Goriely A., Gale E., Maden M., Storey K. (2003). Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40 65–79. 10.1016/s0896-6273(03)00565-8 [DOI] [PubMed] [Google Scholar]
- Doerr J., Schwarz M. K., Wiedermann D., Leinhaas A., Jakobs A., Schloen F., et al. (2017). Whole-brain 3D mapping of human neural transplant innervation. Nat. Commun. 8:14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi D., Magotani H., Kikuchi T., Ikeda M., Hiramatsu S., Yoshida K., et al. (2020). Pre-clinical study of induced pluripotent stem cell-derived dopaminergic progenitor cells for Parkinson’s disease. Nat. Commun. 11:3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin-Ouellet J., Lau S., Brattas P. L., Rylander Ottosson D., Pircs K., Grassi D. A., et al. (2017). REST suppression mediates neural conversion of adult human fibroblasts via microRNA-dependent and -independent pathways. EMBO Mol. Med. 9 1117–1131. 10.15252/emmm.201607471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey S., Heinen S., Krantic S., McLaurin J., Branch D. R., Hynynen K., et al. (2020). Clinically approved IVIg delivered to the hippocampus with focused ultrasound promotes neurogenesis in a model of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 117 32691–32700. 10.1073/pnas.1908658117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A., Alkass K., Bernard S., Salehpour M., Perl S., Tisdale J., et al. (2014). Neurogenesis in the striatum of the adult human brain. Cell 156 1072–1083. 10.1016/j.cell.2014.01.044 [DOI] [PubMed] [Google Scholar]
- Esteban M. A., Pei D. (2012). Vitamin C improves the quality of somatic cell reprogramming. Nat. Genet. 44 366–367. 10.1038/ng.2222 [DOI] [PubMed] [Google Scholar]
- Fan Y., Winanto, Ng S. Y. (2020). Replacing what’s lost: a new era of stem cell therapy for Parkinson’s disease. Transl. Neurodegeneration 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk R. T., Alexanian A. R. (2013). Enhanced dopamine release by mesenchymal stem cells reprogrammed neuronally by the modulators of SMAD signaling, chromatin modifying enzymes, and cyclic adenosine monophosphate levels. Transl. Res. 162 317–323. 10.1016/j.trsl.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Gao L., Guan W., Wang M., Wang H., Yu J., Liu Q., et al. (2017). Direct generation of human neuronal cells from adult astrocytes by small molecules. Stem Cell Rep. 8 538–547. 10.1016/j.stemcr.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E. J., Sidransky E. (2021). Genetics provides new individualized therapeutic targets for Parkinson’s disease. Neural Regeneration Res. 16 994–995. 10.4103/1673-5374.297076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon S., Masserdotti G., Russo G. L., Gotz M. (2017a). Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell 21 18–34. 10.1016/j.stem.2017.06.011 [DOI] [PubMed] [Google Scholar]
- Gascon S., Murenu E., Masserdotti G., Ortega F., Russo G. L., Petrik D., et al. (2016). Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell 18 396–409. 10.1016/j.stem.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Gascon S., Ortega F., Gotz M. (2017b). Transient CREB-mediated transcription is key in direct neuronal reprogramming. Neurogenesis 4:e1285383. 10.1080/23262133.2017.1285383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2016 Neurology Collaborators (2019). Global, regional, and national burden of neurological disorders, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 18 459–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2016 Parkinson’s Disease Collaborators (2018). Global, regional, and national burden of Parkinson’s disease, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 17 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Nakafuku M., Petrik D. (2016). Neurogenesis in the developing and adult brain-similarities and key differences. Cold Spring Harb. Perspect. Biol. 8:a018853. 10.1101/cshperspect.a018853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande A., Sumiyoshi K., ópez-Juárez A. L., Howard J., Sakthivel B., Aronow B., et al. (2013). Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat. Commun. 4:2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresita A., Glavan D., Udristoiu I., Catalin B., Hermann D. M., Popa-Wagner A. (2019). Very low efficiency of direct reprogramming of astrocytes into neurons in the brains of young and aged mice after cerebral ischemia. Front. Aging Neurosci. 11:334. 10.3389/fnagi.2019.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. U., Su Y., Shin J. H., Shin J., Li H., Xie B., et al. (2014). Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17 215–222. 10.1038/nn.3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. (2014). In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 14 188–202. 10.1016/j.stem.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habekost M., Jorgensen A. L., Qvist P., Denham M. (2020). MicroRNAs and Ascl1 facilitate direct conversion of porcine fibroblasts into induced neurons. Stem Cell Res. 48:101984. 10.1016/j.scr.2020.101984 [DOI] [PubMed] [Google Scholar]
- Han D. W., Tapia N., Hermann A., Hemmer K., Hoing S., Arauzo-Bravo M. J., et al. (2012). Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10 465–472. 10.1016/j.stem.2012.02.021 [DOI] [PubMed] [Google Scholar]
- Han S., Wilkinson G. A., Okawa S., Adnani L., Dixit R., Faisal I., et al. (2020). Proneural genes define ground state rules to regulate neurogenic patterning and cortical folding. bioRxiv [Preprint]. 10.1101/2020.09.22.307058v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-C., Lim Y., Duffieldl M. D., Li H., Liu J., Abdul Manaph N. P., et al. (2016). Direct reprogramming of mouse fibroblasts to neural stem cells by small molecules. Stem Cells Int. 2016 1–11. 10.1155/2016/4304916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding J., Vintersten-Nagy K., Shutova M., Yang H., Tang J. K., Massumi M., et al. (2019). Induction of long-term allogeneic cell acceptance and formation of immune privileged tissue in immunocompetent hosts. bioRxiv [Preprint]. 10.1101/716571v1 [DOI] [Google Scholar]
- He S., Guo Y., Zhang Y., Li Y., Feng C., Li X., et al. (2015). Reprogramming somatic cells to cells with neuronal characteristics by defined medium both in vitro and in vivo. Cell Regen (Lond.) 4:12. 10.1186/s13619-015-0027-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Bergami M., Gascon S., Lepier A., Vigano F., Dimou L., et al. (2014). Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Rep. 3 1000–1014. 10.1016/j.stemcr.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Blum R., Gascon S., Masserdotti G., Tripathi P., Sanchez R., et al. (2010). Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 8:e1000373. 10.1371/journal.pbio.1000373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Gascon S., Masserdotti G., Lepier A., Sanchez R., Simon-Ebert T., et al. (2011). Generation of subtype-specific neurons from postnatal astroglia of the mouse cerebral cortex. Nat. Protoc. 6 214–228. 10.1038/nprot.2010.188 [DOI] [PubMed] [Google Scholar]
- Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K. L., Hack M. A., et al. (2002). Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 5 308–315. 10.1038/nn828 [DOI] [PubMed] [Google Scholar]
- Herrero-Mendez A., Almeida A., Fernandez E., Maestre C., Moncada S., Bolanos J. P. (2009). The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat. Cell Biol. 11 747–752. 10.1038/ncb1881 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., Yamada Y., Beard C., Jaenisch R. (2005). Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121 465–477. 10.1016/j.cell.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Horvath S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P. S., Chuang C. Y., Yeh C. H., Chiang W., Liu H. J., Lin T. N., et al. (2017). Direct conversion of human fibroblasts into neural progenitors using transcription factors enriched in human ESC-derived neural progenitors. Stem Cell Rep. 8 54–68. 10.1016/j.stemcr.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Qiu B., Guan W., Wang Q., Wang M., Li W., et al. (2015). Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17 204–212. 10.1016/j.stem.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Hu X., Qin S., Huang X., Yuan Y., Tan Z., Gu Y., et al. (2019). Region-restrict astrocytes exhibit heterogeneous susceptibility to neuronal reprogramming. Stem Cell Rep. 12 290–304. 10.1016/j.stemcr.2018.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh C. J., Zhang B., Victor M. B., Dahiya S., Batista L. F., Horvath S., et al. (2016). Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. eLife 5:e18648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Cho H. H., Cho Y. B., Park J. S., Jeong H. S. (2010). Functional neural differentiation of human adipose tissue-derived stem cells using bFGF and forskolin. BMC Cell Biol. 11:25. 10.1186/1471-2121-11-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger S., Toni N., Clemenson G. D., Jr., Ray J., Gage F. H. (2008). Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat. Neurosci. 11 888–893. 10.1038/nn.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha B. S., Farnoodian M., Bharti K. (2021). Regulatory considerations for developing a phase I investigational new drug application for autologous induced pluripotent stem cells-based therapy product. Stem Cells Transl. Med. 10 198–208. 10.1002/sctm.20-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Xu Z., Zhong P., Ren Y., Liang G., Schilling H. A., et al. (2015). Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat. Commun. 6:10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordao J. F., Ayala-Grosso C. A., Markham K., Huang Y., Chopra R., McLaurin J., et al. (2010). Antibodies targeted to the brain with image-guided focused ultrasound reduces amyloid-beta plaque load in the TgCRND8 mouse model of Alzheimer’s disease. PLoS One 5:e10549. 10.1371/journal.pone.0010549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow M., Camp J. G., Falk S., Gerber T., Pataskar A., Gac-Santel M., et al. (2018). Direct pericyte-to-neuron reprogramming via unfolding of a neural stem cell-like program. Nat. Neurosci. 21 932–940. 10.1038/s41593-018-0168-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow M., Sanchez R., Schichor C., Masserdotti G., Ortega F., Heinrich C., et al. (2012). Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 11 471–476. 10.1016/j.stem.2012.07.007 [DOI] [PubMed] [Google Scholar]
- Keppetipola N., Sharma S., Li Q., Black D. L. (2012). Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 47 360–378. 10.3109/10409238.2012.691456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkis I., Ghasemi-Kasman M., Hajikaram M., Baharvand H., Javan M. (2015). MicroRNA-mediated in vitro and in vivo direct conversion of astrocytes to neuroblasts. PLoS One 10:e0127878. 10.1371/journal.pone.0127878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B. S., Deneen B. (2019). The emerging nature of astrocyte diversity. Annu. Rev. Neurosci. 42 187–207. 10.1146/annurev-neuro-070918-050443 [DOI] [PubMed] [Google Scholar]
- Kim H., Yoo J., Shin J., Chang Y., Jung J., Jo D. G., et al. (2017). Modelling APOE varepsilon3/4 allele-associated sporadic Alzheimer’s disease in an induced neuron. Brain 140 2193–2209. 10.1093/brain/awx144 [DOI] [PubMed] [Google Scholar]
- Kim J., Efe J. A., Zhu S., Talantova M., Yuan X., Wang S., et al. (2011a). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108 7838–7843. 10.1073/pnas.1103113108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Su S. C., Wang H., Cheng A. W., Cassady J. P., Lodato M. A., et al. (2011b). Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell 9 413–419. 10.1016/j.stem.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. M., Flaßkamp H., Hermann A., Araúzo-Bravo M. J., Lee S. C., Lee S. H., et al. (2014). Direct conversion of mouse fibroblasts into induced neural stem cells. Nat. Protoc. 9 871–881. [DOI] [PubMed] [Google Scholar]
- Kim Y., Zheng X., Ansari Z., Bunnell M. C., Herdy J. R., Traxler L., et al. (2018). Mitochondrial aging defects emerge in directly reprogrammed human neurons due to their metabolic profile. Cell Rep. 23 2550–2558. 10.1016/j.celrep.2018.04.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsnes M. S., Winkler A. S. (2020). Global, regional, and national burden of dementia, 1990-2016: predictions need local calibration. Neurology 94 718–719. 10.1212/wnl.0000000000009301 [DOI] [PubMed] [Google Scholar]
- Ladewig J., Mertens J., Kesavan J., Doerr J., Poppe D., Glaue F., et al. (2012). Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 9 575–578. 10.1038/nmeth.1972 [DOI] [PubMed] [Google Scholar]
- Lai S. L., Abramzon Y., Schymick J. C., Stephan D. A., Dunckley T., Dillman A., et al. (2011). FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 32 550.e1–550.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M. F., Shi Y. (2012). Dynamic roles of microRNAs in neurogenesis. Front. Neurosci. 6:71. 10.3389/fnins.2012.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza R., Russell D. W., Nagy A. (2019). Engineering universal cells that evade immune detection. Nat. Rev. Immunol. 19 723–733. 10.1038/s41577-019-0200-1 [DOI] [PubMed] [Google Scholar]
- Lau S., Rylander Ottosson D., Jakobsson J., Parmar M. (2014). Direct neural conversion from human fibroblasts using self-regulating and nonintegrating viral vectors. Cell Rep. 9 1673–1680. 10.1016/j.celrep.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Lee A. S., Tang C., Rao M. S., Weissman I. L., Wu J. C. (2013). Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 19 998–1004. 10.1038/nm.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Sim H., Yoo H. M., Lee M., Baek A., Jeon Y. J., et al. (2020). Neuroprotective effects of cryptotanshinone in a direct reprogramming model of Parkinson’s disease. Molecules 25:3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Mitchell R. R., McNicol J. D., Shapovalova Z., Laronde S., Tanasijevic B., et al. (2015). Single transcription factor conversion of human blood fate to NPCs with CNS and PNS developmental capacity. Cell Rep. 11 1367–1376. 10.1016/j.celrep.2015.04.056 [DOI] [PubMed] [Google Scholar]
- Lee M., Sim H., Ahn H., Ha J., Baek A., Jeon Y. J., et al. (2019). Direct reprogramming to human induced neuronal progenitors from fibroblasts of familial and sporadic parkinson’s disease patients. Int. J. Stem Cells 12 474–483. 10.15283/ijsc19075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Q. Y., Mall M., Chanda S., Zhou B., Sharma K. S., Schaukowitch K., et al. (2020). Pro-neuronal activity of Myod1 due to promiscuous binding to neuronal genes. Nat. Cell Biol. 22 401–411. 10.1038/s41556-020-0490-3 [DOI] [PubMed] [Google Scholar]
- Lee Chong T., Ahearn E. L., Cimmino L. (2019). Reprogramming the epigenome with vitamin C. Front. Cell Dev. Biol. 7:128. 10.3389/fcell.2019.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mattar P., Zinyk D., Singh K., Chaturvedi C. P., Kovach C., et al. (2012). GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 32 7791–7805. 10.1523/jneurosci.1309-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Shi Y., Yao X., Wang X., Shen L., Rao Z., et al. (2019). Conversion of astrocytes and fibroblasts into functional noradrenergic neurons. Cell Rep. 28 682.e7–697.e7. [DOI] [PubMed] [Google Scholar]
- Li S. H., Li X. J. (2004). Huntingtin and its role in neuronal degeneration. Neuroscientist 10 467–475. 10.1177/1073858404266777 [DOI] [PubMed] [Google Scholar]
- Li X., Zuo X., Jing J., Ma Y., Wang J., Liu D., et al. (2015). Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17 195–203. 10.1016/j.stem.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Li X. J., Hu B. Y., Jones S. A., Zhang Y. S., Lavaute T., Du Z. W., et al. (2008). Directed differentiation of ventral spinal progenitors and motor neurons from human embryonic stem cells by small molecules. Stem Cells 26 886–893. 10.1634/stemcells.2007-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Monetti C., Shutova M. V., Neely E. J., Hacibekiroglu S., Yang H., et al. (2018). Linking a cell-division gene and a suicide gene to define and improve cell therapy safety. Nature 563 701–704. 10.1038/s41586-018-0733-7 [DOI] [PubMed] [Google Scholar]
- Liang X. G., Tan C., Wang C. K., Tao R. R., Huang Y. J., Ma K. F., et al. (2018). Myt1l induced direct reprogramming of pericytes into cholinergic neurons. CNS Neurosci. Ther. 24 801–809. 10.1111/cns.12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.-S., Chang M.-Y., Kim S.-M., Yi S.-H., Suh-Kim H., Jung S. J., et al. (2015). Generation of dopamine neurons from rodent fibroblasts through the expandable neural precursor cell stage. J. Biol. Chem. 290 17401–17414. 10.1074/jbc.m114.629808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsman N., Meng Y., Bethune A. J., Huang Y., Lam B., Masellis M., et al. (2018). Blood-brain barrier opening in Alzheimer’s disease using MR-guided focused ultrasound. Nat. Commun. 9:2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. L., Zang T., Zhang C. L. (2016). Direct lineage reprogramming reveals disease-specific phenotypes of motor neurons from human ALS patients. Cell Rep. 14 115–128. 10.1016/j.celrep.2015.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-L., Zang T., Zou Y., Chang J. C., Gibson J. R., Huber K. M., et al. (2013). Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4:2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li F., Stubblefield E. A., Blanchard B., Richards T. L., Larson G. A., et al. (2011). Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 22 321–332. 10.1038/cr.2011.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jiang X., Yu M. K., Dong J., Zhang X., Tsang L. L., et al. (2010). Switching from bone marrow-derived neurons to epithelial cells through dedifferentiation and translineage redifferentiation. Cell Biol. Int. 34 1075–1083. 10.1042/CBI20100516 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang H. (2020). Modeling sporadic Alzheimer’s disease by efficient direct reprogramming of the elderly derived disease dermal fibroblasts into neural stem cells. J. Alzheimers Dis. 73 919–933. 10.3233/jad-190614 [DOI] [PubMed] [Google Scholar]
- Liu Y., Xue Y., Ridley S., Zhang D., Rezvani K., Fu X. D., et al. (2014). Direct reprogramming of Huntington’s disease patient fibroblasts into neuron-like cells leads to abnormal neurite outgrowth, increased cell death, and aggregate formation. PLoS One 9:e109621. 10.1371/journal.pone.0109621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yu C., Daley T. P., Wang F., Cao W. S., Bhate S., et al. (2018). CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 23 758.e8–771.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston J., Lee T., Daniele E., Phillips C., Krassikova A., Enbar T., et al. (2020). Direct reprogramming of astrocytes to neurons leads to functional recovery after stroke. bioRxiv [Preprint]. 10.1101/2020.02.02.929091. [DOI] [Google Scholar]
- Lu H. F., Chai C., Lim T. C., Leong M. F., Lim J. K., Gao S., et al. (2014). xeno-free and feeder-free culture system for the derivation, expansion and direct differentiation of transgene-free patient-specific induced pluripotent stem cells. Biomaterials 35 2816–2826. 10.1016/j.biomaterials.2013.12.050 [DOI] [PubMed] [Google Scholar]
- Lu J., Liu H., Huang C. T., Chen H., Du Z., Liu Y., et al. (2013). Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Rep. 3 1580–1591. 10.1016/j.celrep.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Chanda S., Ahlenius H., Sudhof T. C., Wernig M. (2012). Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. U.S.A. 109 2527–2532. 10.1073/pnas.1121003109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Lee Q. Y., Wapinski O., Castanon R., Nery J. R., Mall M., et al. (2019). Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. eLife 8:e40197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksour S., Ooi L., Dottori M. (2020). More than a corepressor: the role of CoREST proteins in neurodevelopment. eNeuro 7:ENEURO.0337-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall M., Kareta M. S., Chanda S., Ahlenius H., Perotti N., Zhou B., et al. (2017). Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 544 245–249. 10.1038/nature21722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney P. R., Parks D. J., Haffner C. D., Fivush A. M., Chandra G., Plunket K. D., et al. (2000). Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 43 2971–2974. 10.1021/jm0002127 [DOI] [PubMed] [Google Scholar]
- Marro S., Pang Z. P., Yang N., Tsai M.-C., Qu K., Chang H. Y., et al. (2011). Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9 374–382. 10.1016/j.stem.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Gillotin S., Sutor B., Drechsel D., Irmler M., Jørgensen H. F., et al. (2015). Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell 17 74–88. 10.1016/j.stem.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Irie T., Katsurabayashi S., Hayashi Y., Nagai T., Hamazaki N., et al. (2019). Pioneer factor neurod1 rearranges transcriptional and epigenetic profiles to execute microglia-neuron conversion. Neuron 101 472.e7–485.e7. [DOI] [PubMed] [Google Scholar]
- Mattugini N., Bocchi R., Scheuss V., Russo G. L., Torper O., Lao C. L., et al. (2019). Inducing different neuronal subtypes from astrocytes in the injured mouse cerebral cortex. Neuron 103 1086.e5–1095.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni E. O., Mahony S., Closser M., Morrison C. A., Nedelec S., Williams D. J., et al. (2013). Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 16 1219–1227. 10.1038/nn.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R. D. G., Heinrich C., Blum R., Gascón S., Masserdotti G., Tripathi P., et al. (2010). Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 8:e1000373. 10.1371/journal.pbio.1000373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., MacIntosh B. J., Shirzadi Z., Kiss A., Bethune A., Heyn C., et al. (2019). Resting state functional connectivity changes after MR-guided focused ultrasound mediated blood-brain barrier opening in patients with Alzheimer’s disease. Neuroimage 200 275–280. 10.1016/j.neuroimage.2019.06.060 [DOI] [PubMed] [Google Scholar]
- Menon P., Kiernan M. C., Vucic S. (2015). Cortical hyperexcitability precedes lower motor neuron dysfunction in ALS. Clin. Neurophysiol. 126 803–809. 10.1016/j.clinph.2014.04.023 [DOI] [PubMed] [Google Scholar]
- Mertens J., Paquola A. C. M., Ku M., Hatch E., Böhnke L., Ladjevardi S., et al. (2015). Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell 17 705–718. 10.1016/j.stem.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Ferraiuolo L., Miranda C. J., Likhite S., McElroy S., Renusch S., et al. (2013). Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. Proc. Natl. Acad. Sci. U.S.A. 111 829–832. 10.1073/pnas.1314085111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen K. A., Acosta-Verdugo S., Benoit-Marand M., Espuny-Camacho I., Gaspard N., Saha B., et al. (2015). Area-specific reestablishment of damaged circuits in the adult cerebral cortex by cortical neurons derived from mouse embryonic stem cells. Neuron 85 982–997. 10.1016/j.neuron.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Misgeld T., Schwarz T. L. (2017). Mitostasis in neurons: maintaining mitochondria in an extended cellular architecture. Neuron 96 651–666. 10.1016/j.neuron.2017.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R. R., Szabo E., Benoit Y. D., Case D. T., Mechael R., Alamilla J., et al. (2014). Activation of neural cell fate programs toward direct conversion of adult human fibroblasts into tri-potent neural progenitors using OCT-4. Stem Cells Dev. 23 1937–1946. 10.1089/scd.2014.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy M. R., Bilezikjian L. M. (1987). Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 328 175–178. 10.1038/328175a0 [DOI] [PubMed] [Google Scholar]
- Motori E., Puyal J., Toni N., Ghanem A., Angeloni C., Malaguti M., et al. (2013). Inflammation-induced alteration of astrocyte mitochondrial dynamics requires autophagy for mitochondrial network maintenance. Cell Metab. 18 844–859. 10.1016/j.cmet.2013.11.005 [DOI] [PubMed] [Google Scholar]
- Nardini E., Hogan R., Flamier A., Bernier G. (2021). Alzheimer’s disease: a tale of two diseases? Neural Regeneration Res. 16 1958–1964. 10.4103/1673-5374.308070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto A., Kobayashi R., Yoshimatsu S., Sato Y., Kondo T., Yoo A. S., et al. (2020). Direct neuronal reprogramming of common marmoset fibroblasts by ASCL1, microRNA-9/9∗, and microRNA-124 overexpression. Cells 10:6. 10.3390/cells10010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Weichert R. M., Liu W., Davis R. L., Dabdoub A. (2014). Generation of induced neurons by direct reprogramming in the mammalian cochlea. Neuroscience 275 125–135. 10.1016/j.neuroscience.2014.05.067 [DOI] [PubMed] [Google Scholar]
- Niu W., Zang T., Wang L. L., Zou Y., Zhang C. L. (2018). Phenotypic reprogramming of striatal neurons into dopaminergic neuron-like cells in the adult mouse brain. Stem Cell Rep. 11 1156–1170. 10.1016/j.stemcr.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Meas S. J., Nogami J., Amemiya Y., Uchi R., Ohkawa Y., et al. (2018). Direct reprogramming of spiral ganglion non-neuronal cells into neurons: toward ameliorating sensorineural hearing loss by gene therapy. Front. Cell Dev. Biol. 6:16. 10.3389/fcell.2018.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohori Y., Yamamoto S., Nagao M., Sugimori M., Yamamoto N., Nakamura K., et al. (2006). Growth factor treatment and genetic manipulation stimulate neurogenesis and oligodendrogenesis by endogenous neural progenitors in the injured adult spinal cord. J. Neurosci. 26 11948–11960. 10.1523/jneurosci.3127-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oproescu A. M., Han S., Schuurmans C. (2021). New insights into the intricacies of proneural gene regulation in the embryonic and adult cerebral cortex. Front. Mol. Neurosci. 14:642016. 10.3389/fnmol.2021.642016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn T. M., Hallett P. J., Schumacher J. M., Isacson O. (2020). Advantages and recent developments of autologous cell therapy for Parkinson’s disease patients. Front. Cell Neurosci. 14:58. 10.3389/fncel.2020.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdinler P. H., Benn S., Yamamoto T. H., Guzel M., Brown R. H., Jr., Macklis J. D. (2011). Corticospinal motor neurons and related subcerebral projection neurons undergo early and specific neurodegeneration in hSOD1G(9)(3)A transgenic ALS mice. J. Neurosci. 31 4166–4177. 10.1523/jneurosci.4184-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., et al. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476 220–223. 10.1038/nature10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P., Brown R. H. (2006). Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 7 710–723. 10.1038/nrn1971 [DOI] [PubMed] [Google Scholar]
- Pereira M., Birtele M., Shrigley S., Benitez J. A., Hedlund E., Parmar M., et al. (2017). Direct reprogramming of resident NG2 glia into neurons with properties of fast-spiking parvalbumin-containing interneurons. Stem Cell Rep. 9 742–751. 10.1016/j.stemcr.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik D., Jiang Y., Birnbaum S. G., Powell C. M., Kim M. S., Hsieh J., et al. (2012). Functional and mechanistic exploration of an adult neurogenesis-promoting small molecule. FASEB J. 26 3148–3162. 10.1096/fj.11-201426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Ek F., Lang S., Soneji S., Olsson R., Parmar M. (2016). Small molecules increase direct neural conversion of human fibroblasts. Sci. Rep. 6:38290. 10.1038/srep38290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., et al. (2011a). Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 108 10343–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Wood J., Nihlberg K., Hallgren O., Bjermer L., Westergren-Thorsson G., et al. (2011b). Efficient induction of functional neurons from adult human fibroblasts. Cell Cycle 10 3311–3316. 10.4161/cc.10.19.17584 [DOI] [PubMed] [Google Scholar]
- Piao J., Zabierowski S., Dubose B. N., Hill E. J., Navare M., Claros N., et al. (2021). Preclinical efficacy and safety of a human embryonic stem cell-derived midbrain dopamine progenitor product, MSK-DA01. Cell Stem Cell 28 217.e7–229.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Playne R., Jones K., Connor B. (2018). Generation of dopamine neuronal-like cells from induced neural precursors derived from adult human cells by non-viral expression of lineage factors. J. Stem Cells Regen. Med. 14 34–44. 10.46582/jsrm.1401005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Tcw J. (2021). Human iPSC-based modeling of central nerve system disorders for drug discovery. Int. J. Mol. Sci. 22:1203. 10.3390/ijms22031203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Zhao A. D., Sun M. L., Ma K., Fu X. B. (2020). Direct conversion of human fibroblasts into dopaminergic neuron-like cells using small molecules and protein factors. Mil. Med. Res. 7:52. 10.1186/s40779-020-00284-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A. A., Amruta N., Pinteaux E., Bix G. J. (2021). Neurogenesis after stroke: a therapeutic perspective. Transl. Stroke Res. 12 1–14. 10.1007/s12975-020-00841-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Z., Wang R., Li S., Shi Y., Mo L., Han S., et al. (2021). Molecular mechanisms underlying ascl1-mediated astrocyte-to-neuron conversion. Stem Cell Rep. 16 534–547. 10.1016/j.stemcr.2021.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner M., Victor M. B., Liu Y., Abernathy D., Yoo A. S. (2015). MicroRNA-based conversion of human fibroblasts into striatal medium spiny neurons. Nat. Protoc. 10 1543–1555. 10.1038/nprot.2015.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring K. L., Tong L. M., Balestra M. E., Javier R., Andrews-Zwilling Y., Li G., et al. (2012). Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11 100–109. 10.1016/j.stem.2012.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P., Romanov R. A., Spigolon G., Masini D., Martín-Montañez E., Toledo E. M., et al. (2017). Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 35 444–452. 10.1038/nbt.3835 [DOI] [PubMed] [Google Scholar]
- Ruddy R. M., Morshead C. M. (2017). Home sweet home: the neural stem cell niche throughout development and after injury. Cell Tissue Res. 371 125–141. 10.1007/s00441-017-2658-0 [DOI] [PubMed] [Google Scholar]
- Russo G. L., Sonsalla G., Natarajan P., Breunig C. T., Bulli G., Merl-Pham J., et al. (2020). CRISPR-mediated induction of neuron-enriched mitochondrial proteins boosts direct glia-to-neuron conversion. Cell Stem Cell 5 524.e7–534.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Kitagawa K., Omura-Matsuoka E., Todo K., Terasaki Y., Sugiura S., et al. (2007). The phosphodiesterase inhibitor rolipram promotes survival of newborn hippocampal neurons after ischemia. Stroke 38 1597–1605. 10.1161/strokeaha.106.476754 [DOI] [PubMed] [Google Scholar]
- Shakiba N., Fahmy A., Jayakumaran G., McGibbon S., David L., Trcka D., et al. (2019). Cell competition during reprogramming gives rise to dominant clones. Science 364:eaan0925. 10.1126/science.aan0925 [DOI] [PubMed] [Google Scholar]
- Shan Z. Y., Shen J. L., Li Q. M., Wang Y., Huang X. Y., Guo T. Y., et al. (2008). pCREB is involved in neural induction of mouse embryonic stem cells by RA. Anat. Rec. 291 519–526. 10.1002/ar.20686 [DOI] [PubMed] [Google Scholar]
- Sheng C., Zheng Q., Wu J., Xu Z., Wang L., Li W., et al. (2011). Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 22 208–218. 10.1038/cr.2011.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. K., Yang J., Liu M. L., Zhang C. L. (2016). Small molecules modulate chromatin accessibility to promote neurog2-mediated fibroblast-to-neuron reprogramming. Stem Cell Rep. 7 955–969. 10.1016/j.stemcr.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M. V. (2015). Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 16 249–263. 10.1038/nrn3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E. Y., Ichida J. K., Wainger B. J., Toma J. S., Rafuse V. F., Woolf C. J., et al. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9 205–218. 10.1016/j.stem.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells S. F., Paredes M. F., Cebrian-Silla A., Sandoval K., Qi D., Kelley K. W., et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555 377–381. 10.1038/nature25975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding K. L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H. B., et al. (2013). Dynamics of hippocampal neurogenesis in adult humans. Cell 153 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl B. T., Crabtree G. R. (2013). Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr. Opin. Neurobiol. 23 903–913. 10.1016/j.conb.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard-Bennett T., Pera R. R. (2020). Stem cell therapy for Parkinson’s disease: safety and modeling. Neural Regeneration Res. 15 36–40. 10.4103/1673-5374.264446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker S. H., Gotz M. (2021). Epigenetic regulation of neural lineage elaboration: implications for therapeutic reprogramming. Neurobiol. Dis. 148:105174. 10.1016/j.nbd.2020.105174 [DOI] [PubMed] [Google Scholar]
- Sun X., Tan Z., Huang X., Cheng X., Yuan Y., Qin S., et al. (2019). Direct neuronal reprogramming of olfactory ensheathing cells for CNS repair. Cell Death Dis. 10:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung E. A., Yu K. R., Shin J. H., Seo Y., Kim H. S., Koog M. G., et al. (2017). Generation of patient specific human neural stem cells from Niemann-Pick disease type C patient-derived fibroblasts. Oncotarget 8 85428–85441. 10.18632/oncotarget.19976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi S. J., Flower M. D., Ross C. A., Wild E. J. (2020). Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 16 529–546. 10.1038/s41582-020-0389-4 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Thevenot E., Jordao J. F., O’Reilly M. A., Markham K., Weng Y. Q., Foust K. D., et al. (2012). Targeted delivery of self-complementary adeno-associated virus serotype 9 to the brain, using magnetic resonance imaging-guided focused ultrasound. Hum. Gene Ther. 23 1144–1155. 10.1089/hum.2012.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M., Wörsdörfer P., Lakes Y., Gorris R., Herms S., Opitz T., et al. (2012). Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10 473–479. 10.1016/j.stem.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Thomsen G. M., Avalos P., Ma A. A., Alkaslasi M., Cho N., Wyss L., et al. (2018). Transplantation of neural progenitor cells expressing glial cell line-derived neurotrophic factor into the motor cortex as a strategy to treat amyotrophic lateral sclerosis. Stem Cells 36 1122–1131. 10.1002/stem.2825 [DOI] [PubMed] [Google Scholar]
- Tian C., Li Y., Huang Y., Wang Y., Chen D., Liu J., et al. (2015). Selective generation of dopaminergic precursors from mouse fibroblasts by direct lineage conversion. Sci. Rep. 5:12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O., Pfisterer U., Wolf D. A., Pereira M., Lau S., Jakobsson J., et al. (2013). Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. U.S.A. 110 7038–7043. 10.1073/pnas.1303829110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent R., De Angelis Rigotti F., Dell’Era P., Memo M., Raya A., Consiglio A. (2015). Using iPS cells toward the understanding of Parkinson’s disease. J. Clin. Med. 4 548–566. 10.3390/jcm4040548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Lee Q. Y., Camp J. G., Mall M., Koh W., Shariati S. A. M., et al. (2016). Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534 391–395. 10.1038/nature18323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemoto R., Lee S., Szucs A., Chubukov P., Sokolova I., Blanchard J. W., et al. (2018). Diverse reprogramming codes for neuronal identity. Nature 557 375–380. 10.1038/s41586-018-0103-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B., Patel T., Kratsios P., Hobert O. (2011). Direct conversion of C. elegans germ cells into specific neuron types. Science 331 304–308. 10.1126/science.1199082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban N., Guillemot F. (2014). Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell Neurosci. 8:396. 10.3389/fncel.2014.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Ibrahim M. M., Kakumanu A., Garipler G., Aydin B., Al-Sayegh M. A., et al. (2017). A multi-step transcriptional and chromatin state cascade underlies motor neuron programming from embryonic stem cells. Cell Stem Cell 20 205.e8–217.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor M. B., Richner M., Hermanstyne T. O., Ransdell J. L., Sobieski C., Deng P. Y., et al. (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84 311–323. 10.1016/j.neuron.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S. E., Stadtfeld M., Apostolou E. (2015). F-class cells: new routes and destinations for induced pluripotency. Cell Stem Cell 16 9–10. 10.1016/j.stem.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., Wernig M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463 1035–1041. 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinals F., Reiriz J., Ambrosio S., Bartrons R., Rosa J. L., Ventura F. (2004). BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J. 23 3527–3537. 10.1038/sj.emboj.7600360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic S., Kiernan M. C. (2006). Novel threshold tracking techniques suggest that cortical hyperexcitability is an early feature of motor neuron disease. Brain 129 2436–2446. 10.1093/brain/awl172 [DOI] [PubMed] [Google Scholar]
- Wapinski O. L., Lee Q. Y., Chen A. C., Li R., Corces M. R., Ang C. E., et al. (2017). Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 20 3236–3247. 10.1016/j.celrep.2017.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O. L., Vierbuchen T., Qu K., Lee Qian Y., Chanda S., Fuentes D. R., et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155 621–635. 10.1016/j.cell.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Adrian D., Thevenot E., O’Reilly M. A., Oakden W., Akens M. K., Ellens N., et al. (2015). Gene delivery to the spinal cord using MRI-guided focused ultrasound. Gene Ther. 22 568–577. 10.1038/gt.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders C. P., Anderson S. A. (2006). The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 7 687–696. 10.1038/nrn1954 [DOI] [PubMed] [Google Scholar]
- Wu Z., Parry M., Hou X. Y., Liu M. H., Wang H., Cain R., et al. (2020). Gene therapy conversion of striatal astrocytes into GABAergic neurons in mouse models of Huntington’s disease. Nat. Commun. 11:1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Jankauskas R., Mazari A. M. A., Drou N., Percipalle P. (2018). β-actin regulates a heterochromatin landscape essential for optimal induction of neuronal programs during direct reprograming. PLoS Genet. 14:e1007846. 10.1371/journal.pgen.1007846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Wu F., Gu X., Zhang J., You K., Chen Y., et al. (2019). Direct conversion of human urine cells to neurons by small molecules. Sci. Rep. 9:16707. 10.1038/s41598-019-53007-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang Y., He Z., Yang H., Gao W.-Q. (2015). Direct conversion of mouse fibroblasts to GABAergic neurons with combined medium without the introduction of transcription factors or miRNAs. Cell Cycle 14 2451–2460. 10.1080/15384101.2015.1060382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Liu T., Liu L., Yao X., Chen L., Fan D., et al. (2020). Global variation in prevalence and incidence of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J. Neurol. 267 944–953. 10.1007/s00415-019-09652-y [DOI] [PubMed] [Google Scholar]
- Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., et al. (2013). Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated MicroRNA circuits. Cell 152 82–96. 10.1016/j.cell.2012.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Qian H., Hu J., Zhou B., Zhou Y., Hu X., et al. (2016). Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 19 807–815. 10.1038/nn.4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Shang J., Nakano Y., Morihara R., Sato K., Takemoto M., et al. (2019). In vivo direct reprogramming of glial linage to mature neurons after cerebral ischemia. Sci. Rep. 9:10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu Y., Hai Y., Guo Y., Yang S., Li Z., et al. (2014). Efficient conversion of spermatogonial stem cells to phenotypic and functional dopaminergic neurons via the PI3K/Akt and P21/Smurf2/Nolz1 pathway. Mol. Neurobiol. 52 1654–1669. 10.1007/s12035-014-8966-4 [DOI] [PubMed] [Google Scholar]
- Yang N., Chanda S., Marro S., Ng Y. H., Janas J. A., Haag D., et al. (2017). Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 14 621–628. 10.1038/nmeth.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Chen R., Wu X., Zhao Y., Fan Y., Xiao Z., et al. (2019). Rapid and efficient conversion of human fibroblasts into functional neurons by small molecules. Stem Cell Rep. 13 862–876. 10.1016/j.stemcr.2019.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Jiao J., Gao R., Le R., Kou X., Zhao Y., et al. (2015). Enhanced rejuvenation in induced pluripotent stem cell-derived neurons compared with directly converted neurons from an aged mouse. Stem Cells Dev. 24 2767–2777. 10.1089/scd.2015.0137 [DOI] [PubMed] [Google Scholar]
- Yang Y., Jiao J., Gao R., Yao H., Sun X.-F., Gao S. (2013). Direct conversion of adipocyte progenitors into functional neurons. Cell. Reprogramm. 15 484–489. 10.1089/cell.2013.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo J. E., Kang S. K. (2007). Selenium effectively inhibits ROS-mediated apoptotic neural precursor cell death in vitro and in vivo in traumatic brain injury. Biochim. Biophys. Acta 1772 1199–1210. 10.1016/j.bbadis.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lei Z., Guo Z., Pei Z., Chen Y., Zhang F., et al. (2020). Development of neuroregenerative gene therapy to reverse glial scar tissue back to neuron-enriched tissue. Front. Cell Neurosci. 14:594170. 10.3389/fncel.2020.594170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yin J. C., Yeh H., Ma N. X., Lee G., Chen X. A., et al. (2015). Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell 17 735–747. 10.1016/j.stem.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Lin Y.-H., Sun Yujiao J., Zhu S., Zheng J., Liu K., et al. (2016). Pharmacological reprogramming of fibroblasts into neural stem cells by signaling-directed transcriptional activation. Cell Stem Cell 18 653–667. 10.1016/j.stem.2016.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. J., Li J. J., Lin X., Lu Y. Q., Guo X. X., Dong E. L., et al. (2017). Modeling the phenotype of spinal muscular atrophy by the direct conversion of human fibroblasts to motor neurons. Oncotarget 8 10945–10953. 10.18632/oncotarget.14641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Cui W. (2014). Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells 6 305–311. 10.4252/wjsc.v6.i3.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xiang D., Wu W. S. (2014). Sodium butyrate facilitates reprogramming by derepressing OCT4 transactivity at the promoter of embryonic stem cell-specific miR-302/367 cluster. Cell Reprogram. 16 130–139. 10.1089/cell.2013.0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Goh K. J., Ng H. H., Vardy L. A. (2012). A role for polyamine regulators in ESC self-renewal. Cell Cycle 11 4517–4523. 10.4161/cc.22772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., et al. (2016). Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 5:e13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Gu H., Fan R., Wang B., Lou J. (2015). MicroRNA 302/367 cluster effectively facilitates direct reprogramming from human fibroblasts into functional neurons. Stem Cells Dev. 24 2746–2755. 10.1089/scd.2015.0123 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Kohda K., Ibata K., Kohyama J., Akamatsu W., Yuzaki M., et al. (2014). Reprogramming non-human primate somatic cells into functional neuronal cells by defined factors. Mol. Brain 7:24. 10.1186/1756-6606-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Ambasudhan R., Sun W., Kim H. J., Talantova M., Wang X., et al. (2014). Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res. 24 126–129. 10.1038/cr.2013.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Yan Q., Zhong J., Wang K., Sun H., Yi X., et al. (2014). Direct conversion of human fibroblasts into neuronal restricted progenitors. J. Biol. Chem. 289 5250–5260. 10.1074/jbc.m113.516112 [DOI] [PMC free article] [PubMed] [Google Scholar]


