Abstract
Chronic pain is the leading cause of disability worldwide1 and commonly associated with comorbid disorders2. However, the role of diet in chronic pain is poorly understood. Of particular interest is the Western-style diet, enriched with ω-6 polyunsaturated fatty acids (PUFAs) that accumulate in membrane phospholipids and oxidize into pronociceptive oxylipins3,4. Here we report that mice administered a ω-6 PUFA-enriched diet develop persistent nociceptive hypersensitivities, spontaneously-active and hyper-responsive glabrous afferent fibers, and histologic markers of peripheral nerve damage reminiscent of a peripheral neuropathy. Linoleic and arachidonic acids accumulate in lumbar dorsal root ganglia, with increased liberation via elevated PLA2 activity. Pharmacological and molecular inhibition of PLA2g7 or diet reversal with high ω-3 PUFAs attenuate nociceptive behaviors, neurophysiologic abnormalities, and afferent histopathology induced by high ω-6 intake. Additionally, ω-6 PUFA accumulation exacerbates allodynia observed in preclinical inflammatory and neuropathic pain models, and is strongly correlated with multiple pain indices of clinical diabetic neuropathy. Collectively, these data reveal dietary enrichment with ω-6 PUFAs as a novel etiology of peripheral neuropathy and risk factor for chronic pain, and implicate multiple therapeutic considerations for clinical pain management.
Although medical recommendations about diet are made for cardiovascular disease5, diabetes6, and autoimmune diseases7, this is not the case for most pain disorders. Poor nutrition certainly could be a risk factor for chronic pain conditions, especially with excess intake of ω-6 PUFAs, including linoleic acid (LA) and arachidonic acid (AA). Cellular membrane levels of these essential fatty acids are regulated by dietary intake and necessitate ~1% of total calories. The average daily Western diet however contains 10–20-fold greater ω-6 levels8,9. This discrepancy may have considerable clinical significance since ω-6 PUFAs undergo oxidation into pronociceptive oxylipins10–14. Elevated ω-6 levels are associated with pain conditions such as irritable bowel syndrome15, rheumatoid arthritis16, and headache17. Therefore, we evaluated the role of dietary ω-6 PUFAs in the development of persistent pain.
To determine whether elevated dietary ω-6 PUFAs affect nociceptive thresholds, we administered either a high ω-6 diet (H6D) composed of 11.8% kcal/kg ω-6 PUFAs or an isocaloric low ω-6 diet (L6D) composed of 0.4% kcal/kg ω-6 PUFAs to male and female mice for 24 weeks (Supplementary Table 1). Strikingly, both males and females on the H6D developed persistent hypersensitivity to mechanical and heat stimulation, which peaked at 8 weeks (Extended Data Fig. 1). Additional testing at 8 weeks revealed that H6D mice exhibit mechanical hypersensitivity across a range of stimulus intensities, including dynamic brush-evoked stimulation (Fig. 1a,b). H6D mice also demonstrated hypersensitivity to noxious cold and heat (Fig. 1c,d). Saturated fatty acid (SFA) content, but not monounsaturated or ω-3 PUFA levels, was modified to compensate for the different ω-6 PUFAs levels in each diet (Supplementary Table 1). Since SFAs can affect the physical properties of cell membranes, it is possible that the changes in dietary content could contribute to the observed phenotype. However, before receiving the L6D or H6D, the mice were maintained on a standard chow diet (2.9% ω-6 levels, 0.8% SFA levels), and exhibited behavioral responses that were indistinguishable from the L6D responses (Extended Data Fig. 1).
Figure 1.
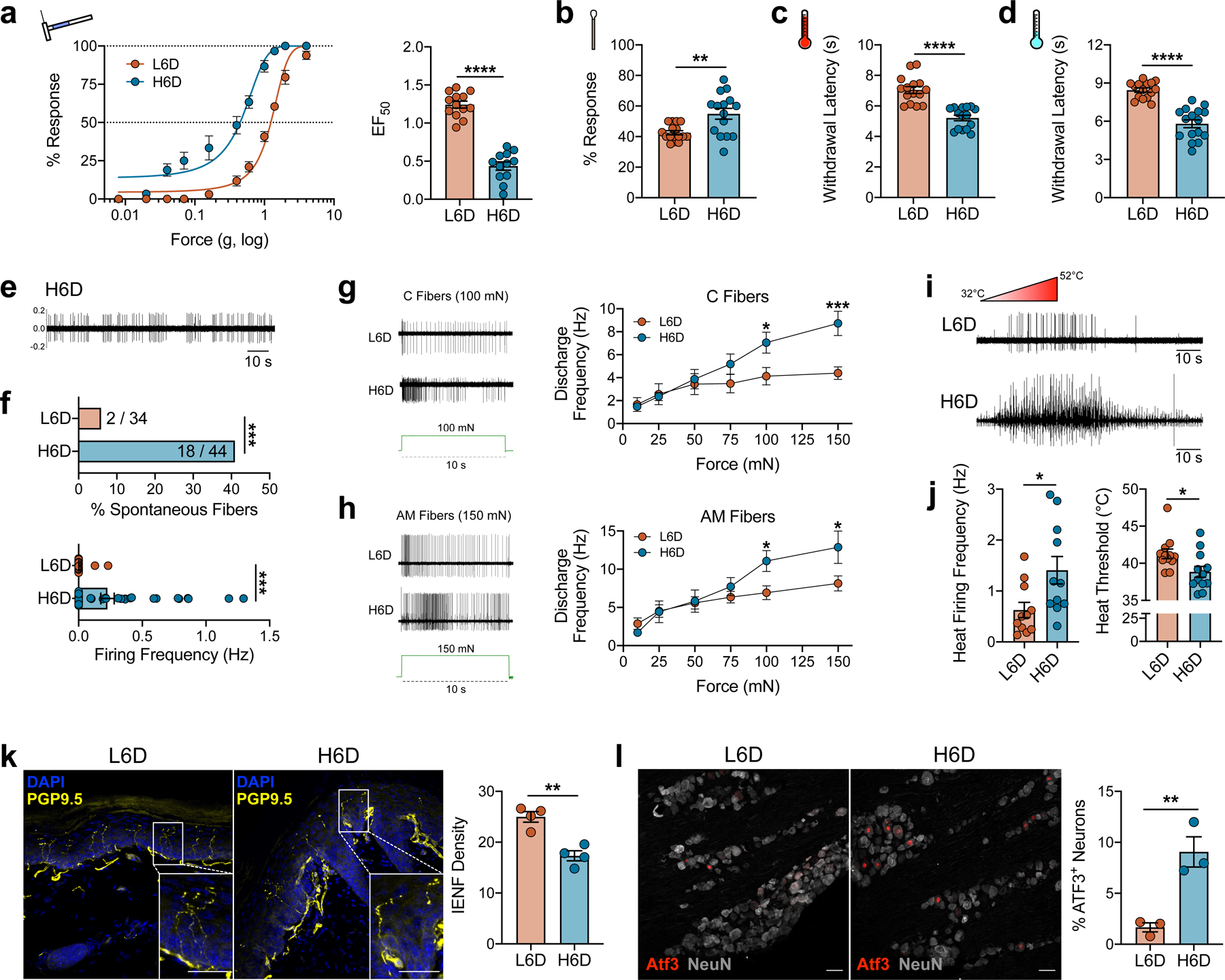
An ω-6 fatty acid-enriched diet induces a peripheral neuropathy-like phenotype in mice. a–d, Effects of a high 11.8% ω-6 fatty acid diet (H6D) or low 0.4% ω-6 fatty acid diet (L6D) on nociceptive thresholds after 8 weeks. a, Mechanical force-response curves (left) and EF50 values (right) as determined with nonlinear regression (L6D mice, n=13; H6D mice, n=12). ****P<0.0001. b, Responsiveness to dynamic brush stimulation (L6D, n=16; H6D, n=15). **P=0.0022. c,d, Paw withdrawal latencies (s) to radiant heat (c) and cold (d) stimulation (L6D, n=15; H6D, n=16). ****P<0.0001 (c,d). e-j, Effects of diet on the neurophysiologic firing properties of mouse glabrous afferent fibers. e, Representative recording of spontaneous firing activity from a H6D mouse after 8 weeks. f, Percentage of spontaneously-active fibers with discharge frequencies (L6D, n=34; H6D, n=44). ***P=0.0005 (top), ***P=0.0006 (bottom). g,h, Representative firing activities of C fibers (g) and AM fibers (h) responding to 100-mN and 150-mN square force stimulation for 10 s (in green), respectively. Discharge frequencies for each force tested are to the right for each fiber subtype (L6D-C, n=14; H6D-C, n=13; L6D-AM, n=23; H6D-AM, n=12). ***P=0.0007 (g), *P=0.0432 (g), *P=0.0387 (h-100), 0.0288 (h-150). i, Representative firing activities during a heat ramp. j, Discharge frequencies (left) and heat thresholds (right) of heat-responsive fibers (L6D, n=12; H6D, n=12). *P=0.0204 (left), *P=0.0213 (right). k, Representative immunofluorescent staining of PGP9.5 (yellow) with DAPI (blue) to visualize intraepidermal nerve fibers (IENFs) in glabrous hindpaw skin, scale bar: 50 μm. Insets: Designated boxed regions magnified 2.5-fold, scale bar: 25 μm. IENF densities (right, L6D, n=4; H6D, n=4). **P=0.0018. l, Representative immunofluorescent staining of activating transcription factor 3 (ATF3, red) expression in NeuN+ lumbar DRG perikarya (gray), scale bar: 50 μm. Percentage ATF3+ neurons (right, L6D, n=3; H6D, n=3). **P=0.0087. Data were collected from mice after 8 weeks on the respective diet. All data are mean ± SEM. Error bars may be within the size of the symbol. Statistical tests used were unpaired two-tailed Student’s t test (a-d, fbottom, j-l), two-sided Fisher’s exact test (ftop), and two-way ANOVA with Sidak’s post-hoc test (g,h).
We performed single-fiber electrophysiologic recordings from ex vivo glabrous skin-tibial nerve preparations to characterize the effect of the H6D after 8 weeks on the detection and firing properties of peripheral afferent fibers. Interestingly, we found that 40.9% of recorded fibers from H6D mice exhibit spontaneous firing as compared to just 5.8% in L6D mice (Fig. 1e,f). The H6D increased mechanical-evoked activity in C- and A-fibers as well as post-stimulus afterdischarge (Fig. 1g,h and Extended Data Fig. 2a,b). Moreover, using a Peltier thermal delivery system, we determined that heat-activated fibers from H6D mice had reduced activation thresholds, increased firing frequency, and prolonged post-stimulus activity (Fig. 1i,j and Extended Data Fig. 2c). Conduction velocities for both C and AM fibers were unchanged between diet groups (Extended Data Fig. 2d). Overall, the H6D-induced hyper-responsiveness of afferent fibers to mechanical and heat stimuli parallels the mechanical- and heat-evoked hypersensitivities observed behaviorally in the same plantar hindpaw tissue.
We subsequently evaluated histologic markers of peripheral nerve damage to determine the presence of diet-induced neuronal damage. H6D mice exhibited a significant reduction in intraepidermal nerve fiber (IENF) density, an established marker of preclinical and clinical peripheral neuropathy18,19, in glabrous skin after 8 weeks on diet (Fig. 1k and Extended Data Fig. 2e). More perikarya from DRG as well as the trigeminal ganglia (TG) from H6D mice expressed the neuronal stress marker, activating transcription factor 3 (ATF3)20,21, compared to L6D mice on diet for at least 8 weeks (Fig. 1l and Extended Data Fig. 2f,g). In contrast, immunolabeling of the lumbar spinal cord with Iba1 and c-fos, markers for microglial and spinal neuron activation, respectively, was not different between groups (Extended Data Fig. 2h,i). Together, the reduction in glabrous IENF density and up-regulation of ATF3 in DRG and TG demonstrate the onset of peripheral nerve damage in mice within 8 weeks on the H6D. In total, the behavioral, neurophysiologic, and pathohistologic data indicate that mice quickly develop a peripheral neuropathy-like phenotype when fed a ω-6 PUFA-enriched western-style diet.
Given that high-fat diet consumption is commonly associated with dyslipidemia, insulin insensitivity, and glucose dysregulation22–26, we tested whether the H6D triggered onset of diabetes and a subsequent neuropathy. Blood glucose and HbA1c levels from H6D mice were comparable to mice given the L6D or normal chow, while db/db diabetic mice exhibited elevated levels of both (Extended Data Fig. 3a,b). Additionally, weekly food consumption and body weights in H6D mice were no different than those observed in L6D mice (Extended Data Fig. 3c,d). Therefore, the observed neuropathy-like phenotype does not result from the induction of diabetes.
Rats previously given a H6D showed LA and AA accumulation in multiple tissues, including brain27. Since diet-induced nociceptive behaviors can be peripherally- and/or centrally-mediated13,28–31, we next evaluated changes to lipid composition for lumbar DRG and spinal cord of H6D mice after 8 weeks using unbiased shotgun lipidomics. Striking changes were observed across lipid classes in lumbar DRG, but not spinal cord, for both H6D male and female mice compared to L6D mice (Extended Data Fig. 4a,b). We quantified total ω-6 lipids in each tissue, and found that both LA and AA levels were elevated, but only in H6D DRG (Fig. 2a,b). Sub-profiling revealed non-uniform accumulation of LA and AA across lipid classes, with robust increases amongst membrane-associated lipids, whereas ω-3 PUFAs remained largely unchanged (Extended Data Fig. 4c,d). Interestingly, we observed marked increases in lysophospholipid levels in DRG of H6D mice (Fig. 2c). Lysophospholipids arise in abundance following enzymatic cleavage of sn-2 fatty acids by phospholipase A2 (PLA2) enzymes, and are known to be elevated in diabetes, coronary heart disease,32 and even neuropathic pain33. The release of sn-2-localized LA and AA from membrane phospholipids by PLA2 initiates their conversion into oxidized, pronociceptive metabolites34. Thus, we hypothesized that increased release of ω-6 fatty acids by PLA2 in DRG neurons elevates production of pronociceptive metabolites that underlie the H6D-associated neuropathic phenotype.
Figure 2.
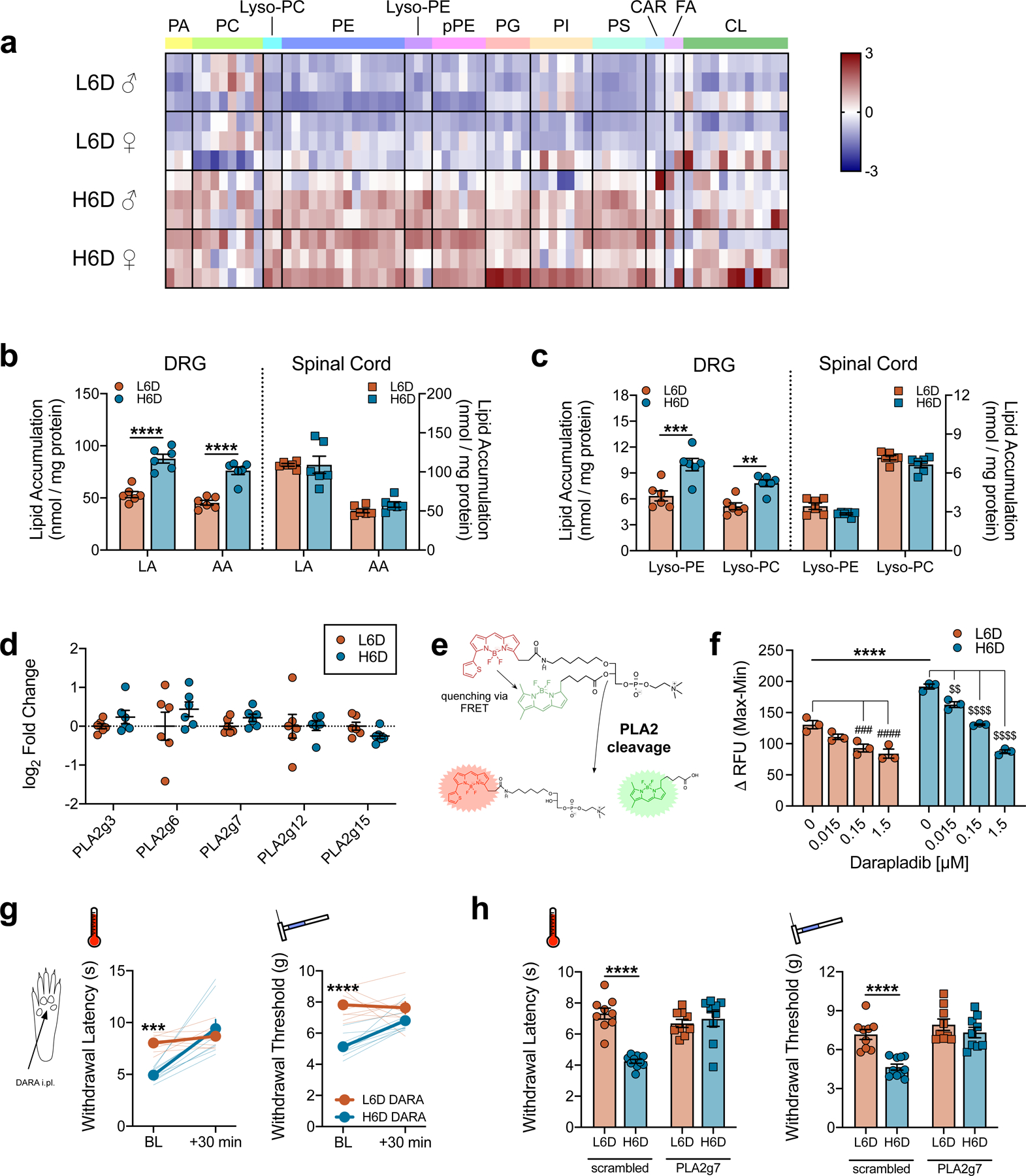
The H6D increases membrane loading of ω-6 PUFAs and stimulates PLA2 activity in peripheral afferent neurons. a, Heatmap of linoleic acid (LA) and arachidonic acid (AA)-esterified lipid species in lumbar DRG from male (♂) and female (♀) mice on either diet. Lipid classes are designated atop the heatmap. Scale bar represents z-score transformations for each lipid species (n=3 mice/sex/diet). b,c, Total accumulation of LA/AA (b) and lysophospholipids (c) in lumbar DRG and spinal cord of mice on either diet (n=6/group). ****P<0.0001 (b), ***P=0.0002 (c-lyso-PE), **P=0.0044 (c-lyso-PC). d, PLA2 isozyme transcript expression in lumbar DRG (n=6 mice/group). e, Schematic detailing changes to a fluorogenic BODIPY substrate following sn-2 bond cleavage by PLA2 enzymes. f, PLA2 activity in DRG homogenates in response to pretreatment with vehicle (DMSO) or the PLA2g7-selective inhibitor, darapladib (n=3 biologically independent mouse samples/group). ****P<0.0001 H6D-0 vs L6D-0, ####P<0.0001 L6D-1.5 vs L6D-0, ###P=0.0005 L6D-0.15 vs L6D-0, $ $ $ $P<0.0001 H6D-1.5/0.15 vs H6D-0, $ $P=0.0059 H6D-0.015 vs H6D-0. g, Acute effects (30 min) of intraplantarly-injected darapladib (DARA) on heat- and mechanical-evoked nociception. Bold lines represent means ± SEM determined from individual animal responses (faint lines) before and after treatment (L6D, n=9; H6D, n=8). ***P=0.0003 (left), ****P<0.0001 (right). h, Withdrawal responses to heat and mechanical stimulation following daily intrathecal (i.t., q.d. x 3 days) injection with either scrambled or PLA2g7-directed siRNA (L6D-scr, n=9; H6D-scr, n=10; L6D-g7, n=9; H6D-g7, n=9). ****P<0.0001 (scrambled). All data are mean ± SEM. Error bars may be within the size of the symbol. Statistical tests used were two-way ANOVA (b,c), mixed-effects ANOVA (g), and one-way ANOVA (f,h), all with Sidak’s post-hoc test.
The expression and activity of PLA2 isozymes govern the release of membrane phospholipid-bound LA and AA35. We first assessed whether the H6D altered PLA2 isozyme expression in lumbar DRG. Previous single-cell RNA-sequencing identified PLA2g7 as the most prominent isoform in lumbar DRG neurons36, accounting for 70–90% of PLA2 transcripts across all sensory neuron subclasses (Extended Data Fig. 5a). We replicated these findings with qPCR using whole DRG RNA extracts as well as by immunolabeling PLA2g7 expression across established afferent neuron subclasses (Extended Data Fig. 5b-e). Surprisingly, expression of PLA2g7 and other prominent isozymes were unchanged in DRG from H6D-fed mice (Fig. 2d). Circulating PLA2g7 levels also were not different between L6D and H6D mice, despite increased LA in H6D plasma (Extended Data Fig. 5f,g). Since the H6D did not alter PLA2 expression, we proceeded to evaluate changes in activity.
A recent clinical trial reported increased plasma PLA2G7 activity in patients on a 8-week LA-rich diet37. Thus, we investigated changes in PLA2 activity from H6D afferent neurons by exposing purified DRG homogenates to a phospholipid reporter containing dual fluorogenic boron dipyrromethene (BODIPY) acyl chains (Fig. 2e). H6D DRG homogenates exhibited greater fluorescent output compared to L6D, indicating elevated PLA2 activity (Fig. 2f and Extended Data Fig. 6a). Expression of annexins, established repressors of PLA2 activity, in H6D DRG was unchanged (Extended Data Fig. 6b). However, there was an accumulation of PLA2g7 protein in the cytosolic fraction of H6D DRG homogenates, implicating a H6D-induced change in intracellular PLA2g7 turnover (Extended Data Fig. 6c).
To assess the contribution of PLA2g7, DRG lysates were pre-incubated with the selective inhibitor, darapladib38, before BODIPY exposure. PLA2 activity was reduced in a concentration-dependent manner, with 80% maximal inhibition, suggesting that PLA2g7 mediates the majority of PLA2 activity in DRG (Fig. 2f and Extended Data Fig. 6d,e). Lipidomics determined that ω-6 lipids accumulate in the glabrous skin of H6D mice, like the DRG (Extended Data Fig. 6f). Therefore, we next tested whether inhibition of PLA2g7 in glabrous skin attenuates H6D-induced nociceptive behaviors. Administration of a local, intraplantar injection of darapladib dose-dependently reversed both heat and mechanical hypersensitivity in H6D mice (Fig. 2g and Extended Data Fig. 6g). To validate selective inhibition of PLA2g7 with darapladib, we administered intrathecally PLA2g7-directed siRNA, which reduced PLA2g7 expression in DRG and spinal cord (Extended Data Fig. 6h,i). As with darapladib, PLA2g7 knockdown reversed the H6D-induced heat and mechanical hypersensitivities, whereas no effect was seen with scrambled siRNA (Fig. 2h). Glabrous IENF density was unaffected by siRNA treatment (Extended Data Fig. 6j). PLA2g7 siRNA reduced transcript expression in L6D mice, but no effect was observed behaviorally. To further link elevated ω-6 PUFA release to nociceptive hypersensitivity, darapladib and PLA2g7 siRNA treatments were tested on db/db mice, and both attenuating the observed mechanical allodynia (Extended Data Fig. 6k-n). These collective data indicate that blocking oxylipin generation through inhibition of the predominate PLA2 isozyme in lumbar DRG is sufficient to reverse H6D-induced hypersensitivity.
Since blocking PLA2-mediated lipid release reversed the hypersensitivity, we next tested whether balancing lipid membrane content through dietary intervention would also reverse the phenotype. It is well-documented that ω-3 PUFA oxylipins exhibit anti-inflammatory and anti-nociceptive effects, which directly counter the pro-inflammatory, pronociceptive effects caused by oxidized ω-6 metabolites39–41. It is unclear however, if re-establishing ω-6/ω-3 balance with diet could reverse the H6D-induced neuropathy phenotype. To test this, mice were fed the H6D for 8 weeks, at which point they were switched to a high ω-3 diet (H3D), the L6D, or continued on the H6D for another 8 weeks (Fig. 3a and Supplementary Table 1). Mice placed on the L6D demonstrated marginally improved nociceptive thresholds compared to H6D mice, whereas switching to the H3D completely rescued behavioral hypersensitivities (Fig. 3b,c). H3D mice exhibited fewer spontaneously-active glabrous afferent fibers, and both C- and A-fibers demonstrated recoveries in mechanical responsiveness, post-stimulus afterdischarge, and heat thresholds (Fig. 3d-g and Extended Data Fig. 7a-b). H3D IENF densities and DRG ATF3 expression recovered relative to historical L6D levels (Fig. 3h,i and Extended Data Fig. 7c,d). Furthermore, LA levels and PLA2 activity in DRG of H3D mice were reduced (Fig. 3j,k). Combined, the H3D switch rescued the behavioral, electrophysiologic, pathohistologic, and metabolic alterations associated with the H6D-induced neuropathy phenotype.
Figure 3.
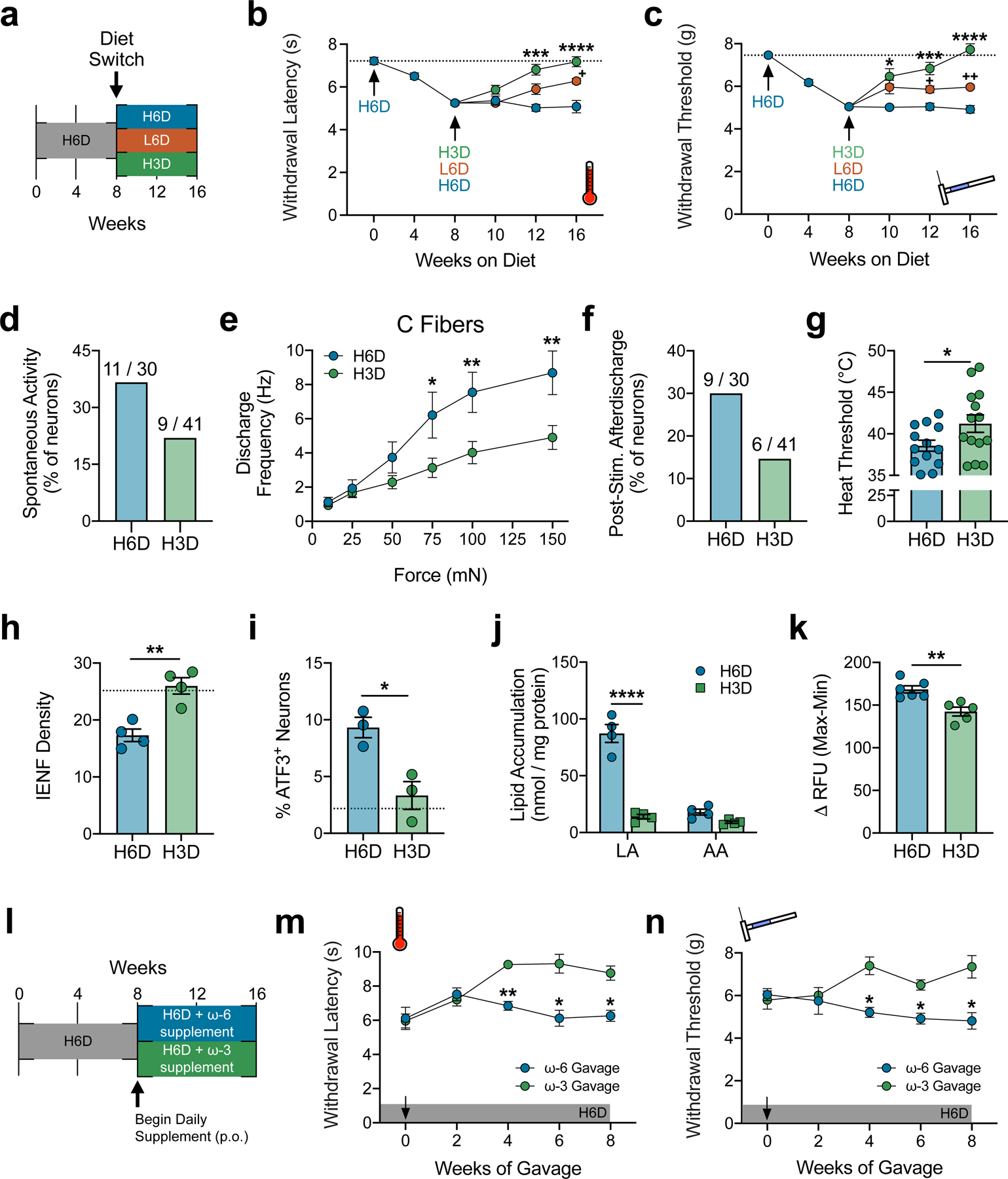
An ω-3 fatty acid-enriched diet rescues the H6D-induced neuropathy-like phenotype. a, Schematic of the diet reversal paradigm, showing mice after 8 weeks on the H6D either continue on the H6D, receive the L6D, or receive a high 7.3% ω-3 diet (H3D) for 8 additional weeks. b,c, Effects of the H3D reversal on H6D-induced heat (b) and mechanical (c) hypersensitivities (H6D, n=11; L6D, n=12; H3D, n=12). ****P<0.0001 H3D-16 vs H6D-16 (b,c), ***P=0.0002 H3D-12 (b), 0.0008 H3D-12 (c) vs H6D-12, *P=0.0363 H3D-10 vs H6D-10 (c), ++P=0.0014 L6D-16 vs H6D-16 (c), +P=0.0352 L6D-16 vs H6D-16 (b), 0.0285 L6D-12 vs H6D-12 (c). d, Percentage of spontaneously-active fibers (H6D, n=30; H3D, n=41). e, Discharge frequencies of mechanical-responsive C fibers during stimulation (H6D, n=11; H3D, n=12). **P=0.0072 (H3D-150), 0.0094 (H3D-100), *P=0.0285 (H3D-75). f, Percentage of fibers exhibiting post-stimulus afterdischarge following mechanical force application (H6D, n=30; H3D, n=41). g, Heat thresholds of heat-responsive fibers (H6D, n=13; H3D, n=14). *P=0.0466. h, IENF densities (H6D, n=4; H3D, n=4). **P=0.003. i, Percentage of ATF3+ neurons in lumbar DRG (H6D, n=3; H3D, n=3). *P=0.0169. The dotted line (h,i) reflects historical mean IENF density and %ATF3+ neurons, respectively, from C57BL6/J mice on the L6D. j, Total accumulation of LA and AA in mouse lumbar DRG (H6D, n=4; H3D, n=4). ****P<0.0001 (LA). k, PLA2 activity in DRG homogenates (H6D, n=6; H3D, n=5). **P=0.0043. l, Schematic of the 16-week H6D with supplementation paradigm, showing that after 8 weeks on the H6D, mice begin to receive either an additional ω-6 or ω-3 supplement daily via gavage in addition to the H6D. m,n, Effects of the ω-6 and ω-3 supplement on H6D-induced heat (m) and mechanical (n) hypersensitivities (ω-6, n=4; ω-3, n=4). **P=0.0023 (m-4wk), *P=0.0243 (m-6wk), *P=0.0194 (m-8wk), *P=0.0333 (n-4wk), *P=0.0205 (n-6wk), *P=0.0492 (n-8wk). All data are mean ± SEM. Statistical tests used were mixed-effects ANOVA with Geisser-Greenhouse’s correction and Tukey’s post-hoc test (b,c), two-way ANOVA with Sidak’s post-hoc test (e,j,m,n), and unpaired two-tailed Student’s t test with Welch’s correction (g-k).
Despite the translational potential here, patients struggle to implement dietary and behavioral changes into their daily routines42. Nutritional supplementation may represent a more practical alternative to daily regulation of dietary PUFAs. To investigate this, mice were placed on the H6D for 8 weeks then began receiving a daily ω-6 or ω-3 supplement via gavage while also continuing the H6D (Fig. 3l). Interestingly, after 4 weeks of gavage, mice receiving the ω-3 supplement exhibited partial recoveries of their pre-H6D nociceptive thresholds (Fig. 3m,n). Continued supplementation maintained these recoveries for the final 4 weeks of testing. These ω-3 supplementation data, along with the H3D reversal data, indicate that both increasing ω-3 levels and decreasing ω-6 levels are required to reverse the H6D phenotype.
Clinically, the development of one pain condition markedly increases the risk of developing additional pain comorbidities43. Thus, we next questioned whether the H6D would exacerbate and/or prolong nociceptive hypersensitivity under conditions of inflammatory or neuropathic injury. To model persistent inflammatory pain44, L6D, H6D, and H3D mice were injected with Complete Freund’s Adjuvant (CFA). The H6D prolonged CFA-induced heat and mechanical hypersensitivity 3-fold compared to L6D and H3D mice (Fig. 4a,b). We again utilized db/db mice to model type 2 diabetes-associated peripheral neuropathy45,46. Diabetic mice on the H6D for just 6 weeks developed a greater mechanical allodynia compared to db/db mice on the L6D, whereas those on the H3D did not exhibit mechanical allodynia but rather mechanical responses commensurate with those from historical C57BL6/J control mice on normal chow (Fig. 4c). Interestingly, db/db mice given either the L6D or H3D exhibited normal responses to noxious heat stimulation, contrasting the hypersensitivity induced by the H6D (Fig. 4d). Together, these data suggest that a H6D risks prolonged and/or exacerbated pain when coupled with persistent inflammation or neuropathic injury, but also can be ameliorated with a H3D supplement.
Figure 4.
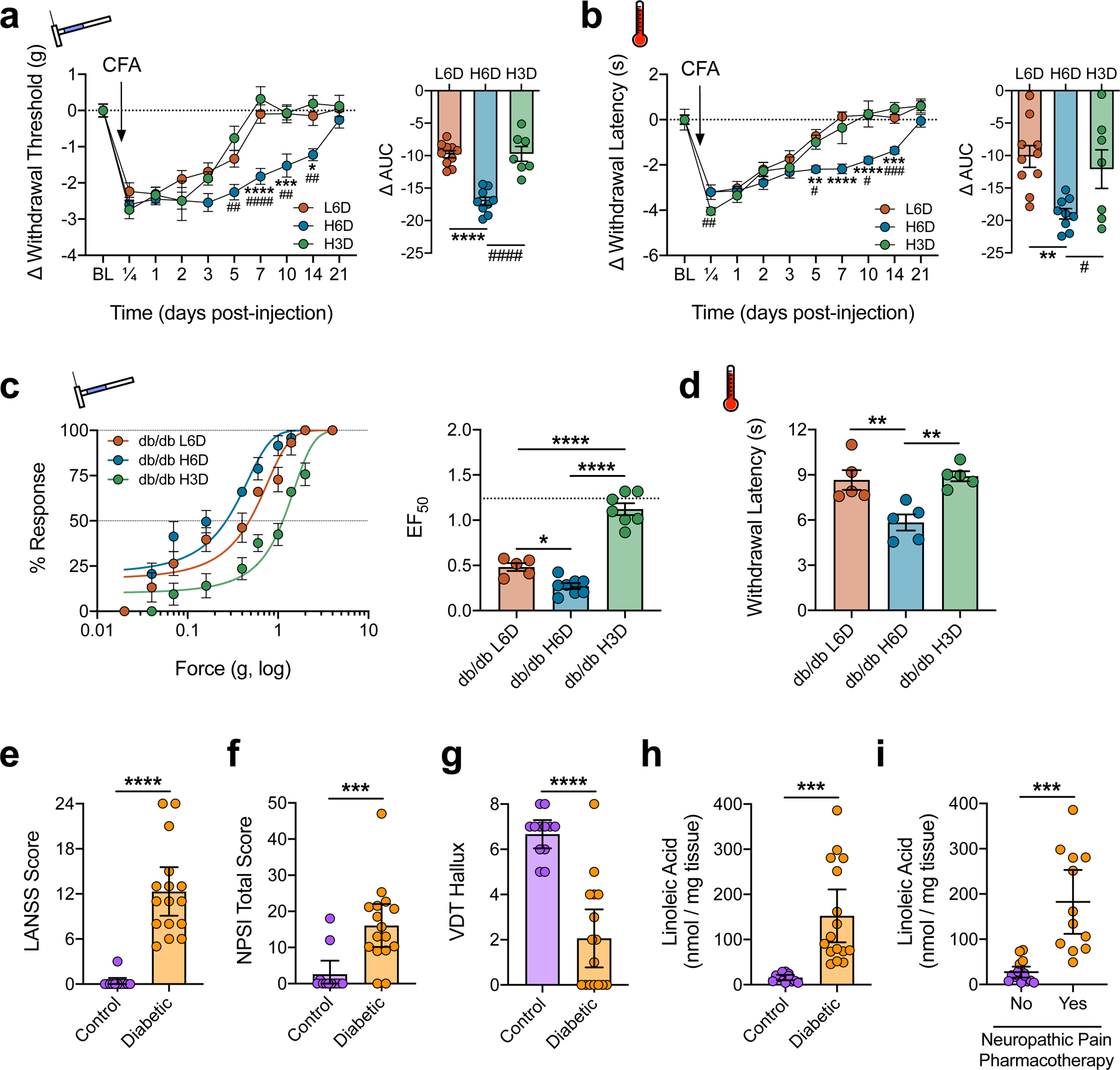
Diet-specific modulation of nociceptive behaviors associated with inflammatory and neuropathic pain. a,b, Time course of L6D-, H6D-, and H3D-specific changes to withdrawal responses from mechanical (a) and heat (b) stimulation following administration of Complete Freund’s Adjuvant (CFA). Change from baseline was calculated for each mouse since mean pre-CFA baselines differed across groups. Right (a,b): Areas under the curve (L6D, n=10; H6D, n=9; H3D, n=7). Time courses (comparisons to H6D): ####P<0.0001 H3D-7 (a), ##P=0.0013 H3D-5 (a), ##P=0.0021 H3D-10 (a), ##P=0.0032 H3D-14 (a), ****P<0.0001 L6D-7 (a), ***P=0.0005 L6D-10 (a), *P=0.0334 L6D-14 (a), ###P=0.0003 H3D-14 (b), #P=0.0174 H3D-5 (b), #P=0.0266 H3D-10 (b), ****P<0.0001 L6D-7/10 (b), ***P=0.0013 L6D-14 (b), **P=0.0021 L6D-15 (b). AUCs (comparisons to H6D): ####P<0.0001 H3D (a), ****P<0.0001 L6D (a), #P=0.0472 H3D (b), **P=0.0045 L6D (b). c, Mechanical force-response curves (left) with EF50 values (right) determined with nonlinear regression for 12-week-old db/db mice fed either the L6D, H6D, or H3D for 6 weeks (L6D, n=5; H6D, n=8; H3D, n=7). The dotted line (EF50 plot) reflects the historical mean EF50 value for C57BL6/J mice on the L6D. ****P<0.0001 L6D/H6D vs H3D, *P=0.0244 L6D vs H6D. d, Paw withdrawal latencies to radiant heat stimulation. **P=0.0064 L6D vs H6D, 0.0035 H3D vs H6D. e-g, Scatter plots of LANSS Pain Scale scores (e), NPSI total scores (f), and hallux vibration detection thresholds (g, a.u.) for control (purple) and diabetic neuropathy (orange) subjects (control, n=12; diabetic, n=16). ****P<0.0001 (e,g), ***P=0.0005 (f). h,i, Total LA content in skin biopsies from the lateral malleolus. Subjects are grouped based on diagnosis (h) or whether they are actively prescribed a neuropathic pain pharmacotherapy regimen (i). ***P=0.0002 (h), 0.0005 (i). Data are mean ± SEM (a-d) or mean ± 95% confidence intervals (e-i). Error bars for some data points are within the size of the symbol. Statistical tests used were two-way ANOVA with Geisser-Greenhouse’s correction and Tukey’s post-hoc test (atime,btime), one-way ANOVA with Tukey’s post-hoc test (aAUC,bAUC,c,d), unpaired two-tailed Mann-Whitney (e-g), and unpaired two-tailed Student’s t test (h,i).
These findings could have far-reaching clinical significance. In consideration of the clinical link between dietary LA intake and diabetes47 along with our findings in diabetic mice, we next tested for associations between skin ω-6 levels and pain symptoms reported by diabetic subjects with painful neuropathy (Supplementary Table 2). Compared to age-matched non-diabetic controls, subjects with diabetic neuropathy reported more neuropathic pain-related symptoms, based on scores derived from the LANSS Pain Scale48 and the NPSI questionnaire49, and exhibited diminished vibration detection thresholds of the halluces (Fig. 4e-g). Moreover, total LA content was substantially elevated in ankle skin biopsies from diabetic subjects (Fig. 4h and Extended Data Fig. 8a). Despite the small sample size (n=28 total participants), robust correlations were observed between skin LA levels and all 3 clinical indices (Extended Data Fig. 8b-d). We also grouped subjects based on active prescriptions for medications commonly used to manage neuropathic pain (e.g., gabapentinoids, SSRIs, SNRIs). Remarkably, 75% (12/16) of diabetic subjects with elevated LA levels required neuropathic pain pharmacotherapy (Fig. 4i). The 4 diabetic subjects not receiving treatment showed LA levels below 100 nmol/mg tissue. Four diabetics prescribed neuropathic pharmacotherapy also had low skin LA levels. Interestingly however, they were either on a starting dose of the medication, rated pain below 5 (0–10 scale), or reported >90% pain relief (Supplementary Table 2). These data suggest that patients with low skin LA levels may be best managed conservatively with either symptomatic analgesics or low-dose neuropathic pharmacotherapy, while patients with LA levels >100 nmol/mg require more robust intervention with neuropathic pharmacotherapy. We believe these data warrant continued investigation of peripheral fatty acid and metabolite levels as potential pain biomarkers. Such biomarkers could provide clinicians with reliable objective endpoints to guide diagnoses as well as decision-making on treatment regimens, including therapeutic diets.
In summary, we demonstrated that an ω-6 PUFA-enriched diet loads DRG and skin with LA and AA within 8 weeks, leading to elevated PLA2-mediated lipid release, the onset of peripheral nerve damage, and the development of nociceptive hypersensitivity. Our findings support the inclusion of pain disorders with other prominent diseases that necessitate medical oversight of patients’ diet. We also propose diet as a pain risk factor, based on observations that elevated ω-6 PUFAs exacerbate nociceptive hypersensitivities in preclinical models of inflammatory and neuropathic pain, as well as the robust clinical associations determined between skin LA content and multiple pain indices in diabetic neuropathy. These are consistent with high-BMI patients being increasingly susceptible to chronic pain conditions50,51. We further show that darapladib, a PLA2G7-selective inhibitor under clinical investigation for non-pain conditions52,53, attenuates H6D-induced hypersensitivity, while dietary replacement with ω-3 PUFAs completely reverts the phenotype. While they both may provide novel approaches for management of chronic pain disorders, the role of PLA2g7 is particularly intriguing, considering the recent association of a loss-of-function polymorphism with reduced migraine risk54 as well as the identification of several expression quantitative trait loci in human DRG tissue55. And while it is unknown whether a PLA2g7 inhibitor would diminish concomitant diet therapy, for example, future exploration is still needed given the lack for effective analgesics for so many prevalent pain conditions.
Methods
Animals.
All animal experiments conformed to the Guidelines for the Use of Animals in Research as put forward by the International Association for the Study of Pain, and to protocols approved by the University Texas Health Science Center at San Antonio (UTHSCSA) Animal Care and Use Committee. Mouse experiments were initiated at 8–10-weeks of age in both male and female C57BL/6J mice (#000664, The Jackson Laboratory). Male BKS.Cg-Dock7m+/+Leprdb/J (db/db56, #000642, The Jackson Laboratory) mice 16-weeks of age were used as positive controls to model a diabetic neuropathy phenotype. Mice were housed in groups of 4–5, maintained on a 12-hour light–dark cycle with ambient temperatures between 20 and 22 °C, and with free access to food and water.
Diets.
Randomized groups of mice were fed isocaloric diets containing 10% g/kg total fat with an energy density of 22.9% kcal/kg for at least 8 weeks to maintain equivalency of nutrient and calorie intake. L6D, H6D, and H3D were formulated by Dyets Inc. based on the AIN-93G diet57, and modified to control the amount of ω-6 or ω-3 PUFAs (Supplementary Table 1). As described previously27, the H6D consisted of 11.8% kcal/kg ω-6 PUFAs, 1.0% kcal/kg ω-3 PUFAs, 2.2% kcal/kg MUFAs, and 7.9% kcal/kg SFA (Dyet #181189) and the L6D consisted of 0.4% kcal/kg ω-6 PUFAs, 1.0% kcal/kg ω-3 PUFAs, 1.2% kcal/kg MUFAs, and 20.3% kcal/kg SFA (Dyet #180784). H3D consisted of 0.9% kcal/kg ω-6 PUFAs, 8.6% kcal/kg ω-3 PUFAs, 6.0% kcal/kg MUFAs, and 7.0% kcal/kg SFA (Dyet #104593). For rescue experiments, mice were placed on H6D for 8 weeks, then switched to either H6D or H3D for at least another 8 weeks. All diets were stored at −20°C and used for no longer than 6 months. Food was replaced weekly. Animals and food were weighed weekly to monitor changes in body weights and food consumption. Weekly food intake (per cage of 5 mice) was calculated by the change in food weight measured at the beginning and end of each week. Normal chow consisting of 2.6% kcal/kg ω-6 PUFAs, 0.3% kcal/kg ω-3 PUFAs, 1.3% kcal/kg MUFAs, and 0.8% kcal/kg SFA (Teklad LM-485; Envigo) was given to control and db/db mice for metabolic comparison.
For the oral gavage experiment, following 8 weeks on the H6D, mice received orally 200 ul (q.d., p.o.) of either high LA safflower oil (BodyBio, SP125, ~424 mg LA daily) or an over-the-counter fish oil (Carlson, The Very Finest Fish Oil, Norwegian, ~64 mg ω-3 daily, 32 mg EPA, 20 mg DHA) on Mondays through Fridays in addition to the H6D ad libitum for 8 weeks. A recovery period was allowed on Saturdays and Sundays to not overly-stress the mice from gavage volume.
Behavioral testing.
All experiments were performed by blinded observers and the assay order was randomized on each testing day.
Mechanical stimulation assays.
Paw withdrawal thresholds (PWT) to noxious mechanical stimulation were evaluated using Ugo Basile dynamic plantar aesthesiometer equipped with an 0.8-mm rigid von Frey filament as previously described58. Briefly, animals were randomized into plastic observation boxes on an acrylic grid platform and acclimated for 60 min. The aesthesiometer was positioned under the mouse to stimulate the mid-plantar area of the hindpaw with a force ramp up to 15 g over a 7 sec period. The force at which withdrawal occurred was recorded. All animals were tested a minimum of three times and the average was used for statistical analysis. In order to more thoroughly characterize mechanical hypersensitivity, a range of von Frey fibers from 0.008–4 g (11 fibers) were used as described previously59. Briefly, mice were acclimated in observation boxes on a wire mesh floor for 60 min. Von Frey fibers contacted the midplantar surface of the hindpaw until slight buckling of the filament was observed, then held for 2 sec. Each fiber was probed 5 times per mouse with at least 30 sec between applications. Withdrawal of the paw was noted as a positive response and no movement was a negative. Values were recorded as percent response for each fiber. Force-response curves were generated using nonlinear least-squares regression of the mean and used to calculate the half maximal effective force (EF50)60.
Brush test.
Dynamic tactile response was assessed using a cotton swab brushed quickly across the plantar surface of the hindpaw as described previously61. Animals were acclimated for 30–60 minutes in boxes atop mesh grid flooring. The “puffed out” cotton swab was used to brush the length of the ventral hindpaw in a continuous motion. Positive responses were defined as withdrawal or rapid shaking of the paw. Each animal was tested 5 times on each paw with a minimum of 30 sec between tests.
Heat stimulation assay.
Paw withdrawal latency (sec) to heat stimulation was assessed using the radiant heat test62. Animals were acclimated in plastic observation boxes for 30–60 min prior to testing. The mid-plantar surface of the mouse hindpaw was exposed to a radiant heat source through a glass floor until paw withdrawal. The intensity of the heat source was adjusted to produce a standard baseline of ~8 sec in naïve wild type mice, with a maximum cutoff of 20 sec. All animals were tested a minimum of three times with at least 60 sec between recordings and the average of all recordings was used for statistical analysis. Mechanical thresholds were determined from the same animals on the same days.
Cold stimulation assay.
Response to cold stimulus was measured according to an adaptation of a previously described protocol63. Mice were placed in plastic observation boxes on top of a 3/16” tempered glass flooring and allowed to acclimate for at least 30 min. A 10-mL syringe was sectioned above the Leur-lock and tightly packed with finely crushed dry ice. The syringe was pressed firmly on the bottom of the tempered glass directly below the hindpaw while measuring paw withdrawal latency (sec) with a stop-watch. Thickness of tempered glass and syringe size were selected in order to establish baseline measurements of ~8 sec. Each animal was tested 3 times on each paw with a minimum of 1 min between tests.
Complete Freund’s Adjuvant (CFA) pain model.
Complete Freund’s adjuvant (CFA; Sigma) was diluted 1:1 with saline and injected intraplantarly (i.pl.) into the right hindpaw of mice using a 30G insulin syringe filled to 20 µl. Thermal and mechanical readings were taken prior to the injection and then at 0.25, 1, 2, 3, 5, 7, 10, 14, and 21 days after injection. Animals were followed until they returned to the original baseline thresholds. Since pre-CFA baselines were different between diet cohorts, data were normalized as the change from baseline withdrawal response.
Blood sampling.
HbA1c and fasting blood glucose levels were measured in mice after 8 weeks on either H6D, L6D, or H3D. Whole blood was collected from the submandibular branch of the jugular vein into EDTA-coated Microvette CB300 collection tube (Kent Scientific). HbA1c levels were determined using a mouse whole blood assay kit according to the manufacturer’s instructions (#83010, Crystal Chem). Briefly, whole blood was mixed with lysis buffer for 10 minutes then mixed with protease buffers and incubated at 37°C for 5 minutes. Absorbance was then measured at 700-nm on a VersaMax microplate reader (SoftMax Pro 7.1, Molecular Devices). To measure fasting blood glucose, mice first were fasted for 5 hours to optimize physiological context64,65. Then, a single drop of blood was collected from the submandibular branch of the jugular vein on a blood glucose test strip and analyzed with the AlphaTrak2 Blood Glucose Monitoring System. Blood from WT and db/db mice on normal chow served as a negative and positive controls, respectively.
Shotgun lipidomics.
DRG and spinal cord tissues were homogenized in 0.5 mL 10x diluted PBS in 2.0 ml cryogenic vials (Corning) by using the Precellys® Evolution (Bertin). Protein assay on the homogenates was performed by using a bicinchoninic acid protein assay kit (Thermo Scientific) with bovine serum albumin as standards. The rest of the homogenate was accurately transferred into a disposable glass culture test tube, and a mixture of lipid internal standards was added prior to lipid extraction for quantification of all reported lipid species. Lipid extraction was performed by using a modified Bligh and Dyer procedure as described previously66. Individual lipid extracts were resuspended into a volume of 400 μl of chloroform/methanol (1:1, v/v) per mg of protein and flushed with nitrogen, capped, and stored at −20°C for lipid analysis. For shotgun lipidomics, lipid extracts were further diluted to a final concentration of ~500 fmol/µL, and the mass spectrometric analysis was performed on a QQQ mass spectrometer (Thermo TSQ Quantiva) equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience) as previously described67. Identification and quantification of lipid molecular species were performed using an automated software program (Xcalibur)68,69. Data were normalized to protein (per mg). All membrane-bound lipids (defined as lipids attached to membrane-anchoring headgroups) were then grouped together and the total concentration of the ω-6 fatty acids, linoleic acid (18:2) and arachidonic acid (20:4), were determined between L6D and H6D and used for statistical analysis.
Quantification of total tissue lipids.
The lipid extraction protocol was adapted from the Bligh-Dyer method70. Briefly, frozen tissue samples were weighed, then sectioned on a cryostat at 16 µm and collected into a cold-acclimated glass test tube. Each tube then was placed on ice, received 2 ml extraction buffer (50% methanol, 25% chloroform, 20% ddH2O, 0.02% butylated hydroxytoluene), was spiked with an internal standard (5 µl/ml LA-d4, 1 µl/ml AA-d8, 5 µl/ml, 1 µl/ml EPA-d5, 1 µl/ml DHA-d5, and 5 µg/ml α-linolenic acid-d16), and was vortexed for 30 sec every 5 min for 15 min. Samples were centrifuged at 5000 x g at 4°C for 10 min, and the bottom organic phase was collected in a separate glass tube. The aqueous phase was re-extracted with 1 ml extraction buffer, and following collection of the second bottom phase, samples were dried down under a steady stream of nitrogen. To analyze total lipid pools, samples next underwent base-catalyzed saponification as described previously71,72. Briefly, dried samples were re-suspended with 850 µl methanol/chloroform solution (8:1) and 150 µl 40% potassium hydroxide solution, placed under nitrogen, then heated to 37°C for 60 min. Following, samples received 700 µl 0.05M phosphate buffer and 300 µl 1M HCl (pH<5) then were extracted twice with 2 ml hexane. Both upper phases were combined then dried down under nitrogen. Samples were stored at −80°C until processing with LC-MS/MS.
Dried samples were re-constituted with 150 µl ethanol and transferred to 1 ml autosampler vials for LC-MS/MS. A Waters Acquity UPLC system was used to perform reversed-phase separation of the free fatty acids. A Thermo Electron Corporation BDS Hypersil C18 column (50 mm x 2.1 mm i.d.) with a 5-μm particle size was held at 45°C. Solvent A was 5 mM ammonium acetate in water and solvent B was acetonitrile with 10% 2-propanol and 0.2% acetic acid. All solvents were Fisher Scientific Optima LC/MS grade. The gradient was set-up as follows: 0 min – 40% A:60% B, 4 min – 5% A:95% B, 4.5 min – 5% A:95% B, 4.6 min – 40% A:60% B, 6.5 min – 40% A:60% B. The autosampler was held at 5 °C. The injection volume was 5 μl and the flow rate was constant 0.5 mL/min. A TQD tandem quadrupole mass spectrometer (Waters) was utilized for free fatty acid concentration determination. An ESI source in negative ion mode was used with the capillary voltage set to 1.5 kV. The source temperature was set at 150°C with a desolvation temperature of 400°C. Argon was used as the collision gas for CID at 0.10 mL/min and nitrogen was used for the cone gas flow at 12 L/hr and the desolvation gas flow at 600 L/hr. Multi-reaction monitoring (MRM) channels were used for both LA and AA, but since LA does not produce detectable fragment ions, the channel utilized the parent ion mass of 279.2 for the fragment ion as well. This will exclude species of the same mass that do produce fragment ions. AA MRM transitions were 303.2→205 and 303.2→259. The collision energy for LA and AA was 10 eV. Waters Corporation software TargetLynx was used to perform channel integration and smoothing. Analytical standards of LA and AA (Cayman Chemical) were used to establish calibration curves. The calibration ranged from 10 ppm to 10 ppb for each standard in ethanol.
Single-fiber recordings of glabrous skin-tibial nerve preparations.
To provide a neurophysiological correlate to H6D-induced behaviors, we utilized ex vivo glabrous skin-tibial/sural nerve preparations as described previously73–77. Tibial and sural nerves were selected based on anatomical distribution of terminal nerve endings within the glabrous skin of the mouse hindpaw78, using the same region tested for evoked behavioral assays described above. Mice were briefly anesthetized with isofluorane and then sacrificed via cervical dislocation. The right leg was shaved followed by careful dissection of the skin-nerve preparation, which included the glabrous skin and attached tibial/sural nerves. The preparation was transferred to the organ bath chamber of a 3D-printed stage (courtesy of Peter Reeh) with oxygenated standard interstitial fluid (SIF, pH 7.4) consisting of: 123 mM NaCl, 3.5 mM KCl, 2.0 mM CaCl2, 0.7 mM MgSO4, 1.7 mM NaH2PO4, 9.5 mM NaC6H11O7, 5.5 mM glucose, 7.5 mM sucrose, 10 mM HEPES, and perfusing at 32 ± 0.7°C (Eco Silver, LAUDA-Brinkmann) at a flow rate of 15–16 ml/min (Masterflex L/S, Cole-Parmer). The skin was positioned corium-side up and pinned down with insect needles to a silicon rubber base (Sylgard 184, Dow Corning) in the organ chamber. The nerve was threaded through a 1-mm hole into the adjacent recording chamber and placed atop a mirror plate. Low viscosity mineral oil (M5904, Sigma) was applied onto the nerve on the mirror plate during teasing and recording to electrically isolate it from the rest of the perfusion chamber. Using a stereomicroscope (SMZ 745T, Nikon) positioned over the recording chamber, the nerve was de-sheathed and single filaments were carefully teased apart for recording. Teased filaments were wrapped around a 0.25 mm silver wire electrode (AGW1010, World Precision Instruments) to record activity. Both the reference (grounded extracellular milieu) and recording (nerve) electrodes in the recording chamber fed into a low-noise headstage probe (DAM80p, World Precision Instruments) which relayed the signal to a DAM80 differential amplifier (World Precision Instruments). Incoming signals were amplified 1000-fold and bandpass-filtered between 300 and 1000 Hz. Amplified signals were filtered through a Hum Bug 50/60 Hz Noise Eliminator (Quest Scientific) then passed to a TBS1000B digital oscilloscope (Tektronix), a Model 330 audio monitor (A-M Systems), and a Micro1401–3 digital data acquisition system (Cambridge Electronic Design). Data were recorded with Spike2 (Cambridge Electronic Design).
Sensory profiling of teased fibers.
Receptive fields (RF) were initially identified based on mechanical responsiveness to a blunt glass rod. Upon positive identification of a RF, the following steps were utilized to characterize the sensory profile of the teased fiber: (1) spontaneous activity – The presence of spontaneous activity was assessed for the first 2 min of recording, (2) mechanical stimulation – A precision force-controlled mechanical stimulator (Series 300C-I Dual Mode Servo System, Aurora Scientific) was used to evaluate fiber mechano-sensitivity. The mechanical cylinder (0.7 mm tip diameter) was positioned over the RF of interest, followed by computer-controlled application of square force stimuli (force range of 5 – 200 mN with 10 sec duration). To prevent sensitization/desensitization of the recorded fiber, 60 sec intervals were given between force applications, (3) heat stimulation – A Peltier-based thermal stimulus delivery system (CS1, Cool Solutions) was used to assess fiber responsiveness to controlled heat application79. A custom-designed cylindrical ring (OnShape) that was 3D-printed on a Form 2 using Tough Resin (FormLabs) was used to isolate the RF of interest. Vacuum grease (Dow Corning) tightly sealed the ring in position. An insulated afferent tube connected to the Peltier device, an efferent tube, and a thermocouple were then positioned within the ring. A dispensing pump (IPC24, Ismatec) drove the push-pull superfusion (60 ul/sec) of SIF over the RF within the ring (Extended Data Fig. 2c). The SIF temperature increased from 30℃ to 52.0–60.5℃ during heat ramps (30 sec). After 30 sec of heating, the Peltier was stopped and temperature returned to baseline within 90 sec. At least 5 min intervals were given between heating to prevent sensitization/desensitization of the isolated RF, (4) conduction velocity – A 2.0 MΩ Parylene-coated tungsten metal stimulating electrode (TM33B20, World Precision Instruments) was positioned within the RF of interest. A stimulus isolator (A365, World Precision Instruments) and pulse generator (A310, World Precision Instruments) were used to deliver electrical pulses to the RF. Upon electrical stimulation of the fiber, digital calipers were used to measure the conduction distance (in mm) between the RF and the recording electrode. Conduction velocity (CV) was calculated by dividing this distance by the latency of the firing fiber from the stimulating artefact and represented as meters per second (m/s).
Spike detection and analysis.
Recordings were analyzed with off-line template-matching in Spike2 version 8.14 (Cambridge Electronic Design). Settings for spike template matching required a minimum 70% of sampling points within a template and 25% maximum amplitude change. Waveform data was interpolated linearly. High-pass filter time constant was set to 6.4 ms. Triggers were set at least 3 times the baseline noise level. Conduction velocity (CV), calculated by dividing the conduction distance over the spike electrical latency, was used to classify fiber type according to the following cut-offs: C fibers <0.8 m/s; AM fibers >1.2 m/s80. Spontaneously active fibers were defined with a minimum unprovoked discharge frequency of 0.1 Hz81. Mechanical discharge frequencies were determined for each force application as the number of firings within the 10 sec ramp period. Post-stimulus after-discharge was identified as persistent activity occurring after completion of any force ramp. For heat responses, total afferent activity was used instead of discharge frequency as recordings often included individual units with variable amplitudes that could not always be discriminated74. Heat threshold temperatures were identified as the temperature that initiated the first afferent activity during a heat ramp. Post-stimulation activity was calculated as the number of action potentials during the time period defined as the point when the recovering temperature reaches a fiber’s heat threshold through to baseline.
Immunohistochemistry and imaging.
L3-L5 dorsal root ganglia (DRG) and 3 mm biopsies of glabrous hindpaw skin were immediately dissected after sacrifice. Tissues were immersion-fixed in 4% paraformaldehyde in 0.1M phosphate buffer (PB) for 2 hr. Tissue samples were washed 3 x 15 min in PB, immersed in 10% sucrose at 4°C overnight, transferred to 30% sucrose at 4°C overnight and then stored at −20°C. For cryo-sectioning, tissues were thawed and placed in OCT (Tissue-Tek) prior to freezing on dry ice. Sections of DRG (14 μm) and paw tissue (20 μm) were cut with a cryostat (Microm HM550, ThermoFisher Scientific) and thaw-mounted onto Superfrost Plus slides (Fisherbrand). Slides were air-dried at RT and stored at −20°C. Staining was performed using MAXpack Immunostaining Media Kit (Active Motif). Tissue sections were blocked with MAXblock Blocking Medium for 1 h at room temperature, followed by incubation with primary antibodies diluted in MAXbind Staining Medium at 4°C overnight. Primary antibodies included: PGP9.5 (1:1000; AB1761-I, Millipore), ATF3 (1:400; ab207434, Abcam), NeuN (1:500; ABN91, Millipore), NFH (1:5000; 822601, Biolegend), TRPV1 (1:700; GP14100, Neuromics), PLA2g7 (1:500; 15526–1-AP, ProteinTech), and GFRA2 (1:500; AF429, R&D Systems). Isolectin GS-IB4 Alexa Fluor® 488 Conjugate (1:800 from stock; 121411, ThermoFisher) was utilized during primary incubation as well. Sections were then washed 3 x 10 min in MAXwash solution and incubated with secondary antibodies for 1 hr at RT. The following secondary antibodies (Jackson ImmunoResearch) were used: Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L), Alexa Fluor® 488 AffiniPure Donkey Anti-Rabbit IgG (H+L), Alexa Fluor® 568 Donkey anti-Rabbit IgG (H+L), Alexa Fluor® 647 AffiniPure Donkey Anti-Guinea Pig IgG (H+L), Rhodamine Red-X AffiniPure Donkey Anti-Goat IgG (H+L) all at 1:500 dilution from stock. After washing 3 x 10 min with MAXwash, DAPI (0.02 μg/ml, Sigma; D9542) was applied for 10 min at RT, followed by 2 x 10 min washes with MAXwash and then 2 x 5 min in ddH2O. Slides air-dried in the dark for 20 min then were mounted with ProLong Diamond (ThermoFisher Scientific) and no. 1.5 high precision cover slips (Zeiss) for imaging.
Confocal Microscopy.
Images were obtained with a Nikon C1si laser scanning confocal microscope equipped with: 402-nm diode, 488.1-nm solid state, 561.4-nm diode-pumped solid state, and 639-nm diode lasers. Objectives (Nikon) used include: a 10X 0.4 NA Plan Apo air, a 20X 0.75 NA Plan Apo DIC air, and a 40X 0.95 NA collar-corrected Plan Apo air. Image acquisition settings were 1024 x 1024 resolution, 12-bit image depth, 2.0 μs/pixel scan speed, 30 μm confocal aperture (pinhole diameter), and sequential channel scan. Z-stacks were taken at 0.3–0.6 μm optical steps (20X, 40X objectives). All images for each tissue type were taken at identical gain settings. Laser power, HV, and offset were adjusted to maximize dynamic range while avoiding pixel saturation (EZ-C1 v3.8, Nikon). Adjustments of brightness/contrast, look-up tables, and z-stack reconstructions were performed in FIJI82–85.
Intraepidermal Nerve Fiber (IENF) Density Quantification.
To evaluate changes in epidermal innervation density, immunolabeled IENFs projected across the dermal-epidermal junction to the epidermis were counted in randomly selected fields of view from each section. The following exclusion criteria were used when counting IENFs: (1) fragments of nerve fibers in the epidermis that did not clearly cross the dermal-epidermal junction were not counted and (2) fibers that approached but did not cross the dermal-epidermal junction, as determined by basal epidermal autofluorescence, were not counted86,87. To reduce bias, counting was conducted by 2 blinded observers on 3–5 non-consecutive sections per animal with 4 animals per group. The averaged fiber count for each section was divided by the length of the dermal-epidermal junction to calculate the IENF density (IENFs/mm of tissue). Mean IENF density ± SEM for each group was used to determine statistical significance.
ATF3 Quantification.
To measure changes in the expression of the transcription factor, ATF3, as a marker of neuronal injury20, the number of ATF3+ neurons in immunolabeled sections from L3-L5 DRG were counted. Images underwent thresholding before manual counting of the stained nuclei. For each image, the percentage of ATF3-labeled neurons was calculated by dividing total ATF3+ neurons by total NeuN+ neurons × 100. Using FIJI, quantifications were performed on 3–5 non-overlapping sections per mouse to determine individual means. Group means ± SEM were calculated using individual means from 3–4 mice per group.
PLA2 activity assay.
DRGs from H6D, L6D, and H3D animals were dissected, placed into cold lysis buffer (50 mM Tris HCl, pH 8, 2 mM DTT, 1 mM MgCl2, complete Mini protease inhibitor) and incubated on ice for 15 minutes. DRGs were homogenized using a 2-ml Dounce homogenizer, transferred to 1.5 ml tubes and centrifuged for 20 min at 15,000g and 4°C. Supernatants were collected into new 2-ml tubes. Total protein concentration was determined using the Bio-Rad Bradford protein assay kit according to the manufacturer’s instructions (ThermoFisher Scientific, #23225). An adapted BODIPY (Invitrogen, #A10072) assay was used for continuous monitoring of phospholipase A2 (PLA2) activity from homogenates based on the manufacturer’s instructions and previous publication. Briefly, 1 mM stock of Red/Green BODIPY PC-A2 was prepared in DMSO. To prepare the liposome mixture, equal volumes of 10 mM DOPC, 10 mM DOPG and 1 mM BODIPY dye were mixed in a microcentrifuge tube and slowly injected into assay buffer while rapidly stirring. DRG lysates were added to microplate wells in appropriate concentrations. A buffer-only well was use as a negative control and bee venom (PLA2g3) was used as a positive control. The liposome mixture was then added 1:1 to the microplate wells. In PLA2 inhibitor experiments, the drug was incubated with the DRG lysate for 15 minutes prior to addition of the liposomal substrate. FlexStation 3 with SoftMax Pro 4.3 (Molecular Devices) was set to 37°C and used for kinetic recording of 520 nm and 570 nm emission for 30 minutes. Peak maximum minus minimum fluorescent signals for each wavelength (a.u.) were used for statistical analysis for each sample.
In vivo treatment.
All drugs were prepared in coded syringes on the day of injection by an individual not performing behavioral testing in order to blind the experimenter. Animals were initially acclimated to observation boxes atop thermal or mechanical platforms for a minimum of 30 min prior to collection of baseline recordings. After baselines, animals were administered vehicle or testing compound. Darapladib (Selleckchem; SB-480848) was dissolved in dimethyl sulfoxide (DMSO) and Tween-20 then diluted in phosphate buffered saline (PBS) to 2% of each for final concentrations of 300 nmol and 30 nmol. Control animals received equivalent volume of the darapladib vehicle.
Knockdown of PLA2g7 was performed using Dharmacon SMARTpool small interfering RNA (siRNA) against four separate PLA2g7 target sequences. Control groups received non-targeting scrambled (scr) siRNA (Dharmacon). The PLA2g7 or scrambled siRNA was injected intrathecally (i.t., 2 µg) for three consecutive days for a cumulative dose of 6 µg. Behavior was tested on 2 days after final injection by blinded observers, followed by collection of DRGs for protein and transcript quantification.
qPCR.
L3–5 DRG were harvested and immediately frozen on dry ice then stored at −80°C. DRG tissue was homogenized as described previously88 in 900 μl QIAzol lysis reagent (QIAGEN) and placed on ice for 5 min. RNA was isolated according to the manufacturer’s instructions (Qiagen; RNeasy Plus Universal Mini Kit, #73404). Briefly, 100 µl gDNA eliminator and 180 µl chloroform were added to each tube and shaken for 15 s. Tubes were spun at 12,000 g for 15 min and the top aqueous phase was removed for further purification with ethanol and wash buffers and elution with RNase-free water (NanoDrop, Thermo). cDNA was then synthesized from 1 μg RNA using Superscript III First Strand Synthesis kit (Invitrogen; #18080051). Amplification of target sequences was performed using TaqMan Fast Advanced Master Mix (Applied Biosystems; #4444557) according to the manufacturer’s instructions plus selected primers for Rn18S (Mm03928990_g1, ThermoFisher), PLA2g3 (Mm00555594_m1, ThermoFisher), PLA2g6 (Mm00479527_m1, ThermoFisher), PLA2g7 (Mm00479105_m1, ThermoFisher), PLA2g12 (Mm01316982_m1, ThermoFisher), PLA2g15 (Mm00505425_m1, ThermoFisher), ANXA1 (Mm00440225_m1, ThermoFisher), ANXA2 (Mm01150673_m1, ThermoFisher), ANXA3 (Mm00442685_m1, ThermoFisher), ANXA5 (Mm01293059_m1, ThermoFisher), and ANXA6 (Mm00478966_m1, ThermoFisher). qPCR was performed using the StepOnePlus Real-time PCR System (Applied Biosystems, #4376600). A no-template control (NTC) was used as a negative control. Cycles of thresholds (Ct) were collected and normalized to the internal control, Rn18S. Normalized ∆∆Ct to Rn18S reference were used for statistical analysis89.
Western Blot.
Reinforced screw cap microtubes (2-ml) pre-filled with 2.8 mm ceramic beads (19–628, Omni International) were filled with 900 μl RIPA buffer (ThermoFisher, #89900) with cOmplete mini protease inhibitors (Roche; #11836145001). Thawed DRG tissue samples were then transferred to bead tubes and placed on ice. The processing chamber of a Bead Ruptor 24 (Omni International; #19–040E) was pre-cooled to 0°C with an attached BR-Cryo Cooling Unit (Omni International; #19–8005). Samples were placed in the tube carriage, homogenized for 30 sec at 7.10 m/s, then placed on ice for 5 min. Samples were transferred to fresh tubes and centrifuged for 3 min at 8,000 x g and 4°C. Supernatants were collected and stored at −80°C until use. After total protein determination, supernatants were prepared for SDS-PAGE on 4–12% gradient Bis-Tris gels according to the NuPAGE protocol (Novex). Proteins were transferred to PVDF membranes via iBlot2 (ThermoFisher). Immunoblots were probed with polyclonal rabbit anti-PLA2G7 antibody (1:500, 15526–1-AP, ProteinTech). GAPDH was measured as a loading control with a monoclonal mouse anti-GAPDH antibody (1:1000, sc-32233, Santa Cruz). Donkey anti-rabbit IR800 and donkey anti-mouse IR680 secondary antibodies (1:10,000, LI-COR Biosciences) were used to detect PLA2G7 and GAPDH, respectively. Immunoblots were imaged using a LI-COR Odyssey infrared imager and relative band intensities were quantified using Image Studio (LI-COR Biosciences).
Human study and sample collection.
Clinical sensory data and skin biopsies collected from 12 control subjects and 16 subjects with type 2 diabetes and diabetic neuropathy (all ages >50 years) were included as part of an ongoing observational trial at UTHSCSA. Regulatory approval was obtained from the Human Subjects Institutional Review Board at the University of Texas Health Science Center at San Antonio (#20160027HU). Subject information is available in Supplementary Table 2. Informed consent was obtained from subjects according to protocol guidelines set by the Institutional Review Board. Subjects completed the self-reported pain questionnaires for the Leeds Assessment of Neuropathic Symptoms and Signs (LANSS) and Neuropathic Pain Symptom Inventory (NPSI), followed by sensory testing as described48. Vibration detection thresholds then were determined for both halluces for each patient, followed by collection of a skin punch biopsy. Study data were collected and managed using REDCap electronic data capture tools hosted at UTHSCSA90.
For vibration testing, a Rydel-Seiffer tuning fork (US Neurological) was used on the left and right hallux (i.e., great toe). The principal neurologist placed the base of the tuning fork, with the dampers facing the researcher, on the bony prominences. To create a vibration, the tynes were pressed together between the thumb and index finger, then released such that the illusion of two triangles is visible on each damper marked on a scale from 0 to 8. As the intensity of the vibration diminishes, the two triangles move closer together and the point of intersection moves slowly upward. Each patient vocalizes once the vibration cannot be detected and the intensity score was recorded. This procedure was repeated four times per hallux, with the first reading being discarded and the last three being recorded.
Lastly, following injection with 2% lidocaine/epinephrine (Hospira) solution to anesthetize the skin, punch biopsies were taken 10 cm proximal to the lateral malleolus of the ankle to a depth of 4 mm with a sterile 4-mm circular punch. Biopsies were collected in sterile 2-ml cryotubes and immediately snap-frozen on liquid nitrogen. Samples were stored at −80°C until processing for total lipid analysis.
Statistical analysis and reproducibility.
Prism 9.0 (GraphPad) was used for statistical analysis. Statistical tests utilized are specified at the end of each figure legend. Generally, unpaired, two-tailed Student’s t tests were used for analyses between two groups, whereas one-way or two-way analysis of variance (ANOVA) coupled with a specific multiple comparisons test were used for multiple groups and/or conditions. Sample sizes were designed to generate 80% power at a two-sided P<0.05. Unless specified, data are presented as mean ± SEM, with values derived from independent biological replicates. Information pertaining to use of technical replicates can be found within the methods (e.g., qPCR, HPLC-MS/MS, IHC). All experiments that utilized male and females were tested for possible sex differences and if none were observed, the animals were combined for further analysis91. OmniGraffle version 7.18.1 (The Omni Group) was used for illustrations and figure organization.
Extended Data
Extended Data Fig. 1.
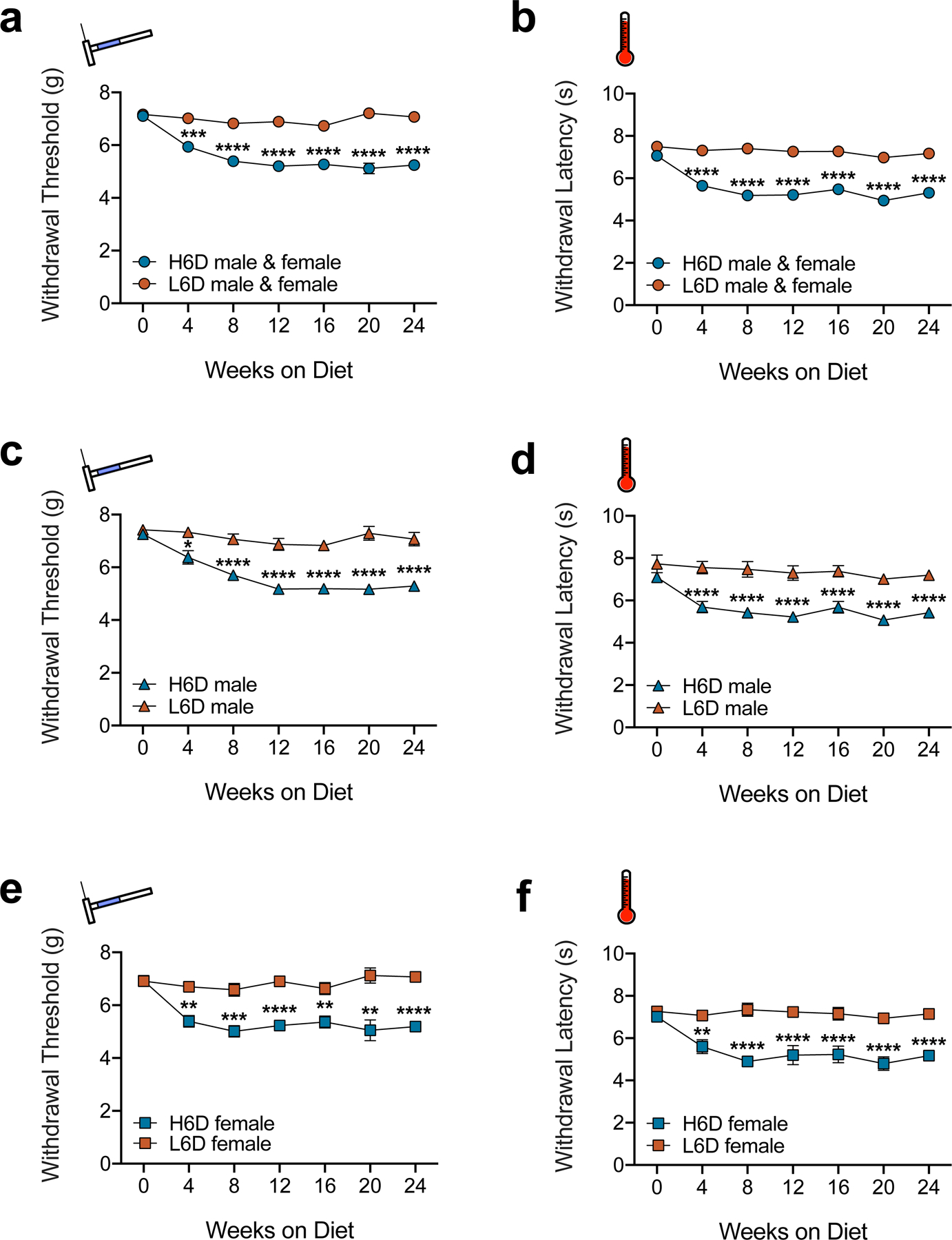
H6D induces persistent nociceptive hypersensitivities in both male and female mice. a-f, Time courses of changes in mechanical withdrawal threshold (a,c,e) and heat withdrawal latency (b,d,f) for male and female mice on the H6D and L6D. The top two plots (a,b) represent compiled male and female responses (L6D, n=24; H6D, n=29). a, ***P=0.0003 week 4, ****P<0.0001 weeks 8–24. b, ****P<0.0001 weeks 4–24. The middle plots (c,d) are male-only responses (L6D, n=12; H6D, n=16). c, *P=0.0401 week 4, ****P<0.0001 weeks 8–24. d, ****P<0.0001 weeks 4–24. The bottom plots (e,f) are female-only responses (L6D, n=12; H6D, n=13). e, **P=0.0055 week 16, 0.0023 week 20, ***P=0.0009 week 4, 0.0005 week 8, ****P<0.0001 weeks 12,24. f, **P=0.0022 week 4, ****P<0.0001 weeks 8–24. Data are mean ± SEM. Error bars for some data points are within the size of the symbol. The statistical test used was two-way repeated-measures ANOVA with Sidak’s post-hoc test (a-f).
Extended Data Fig. 2. The H6D sensitizes afferent fibers to mechanical and heat stimuli.
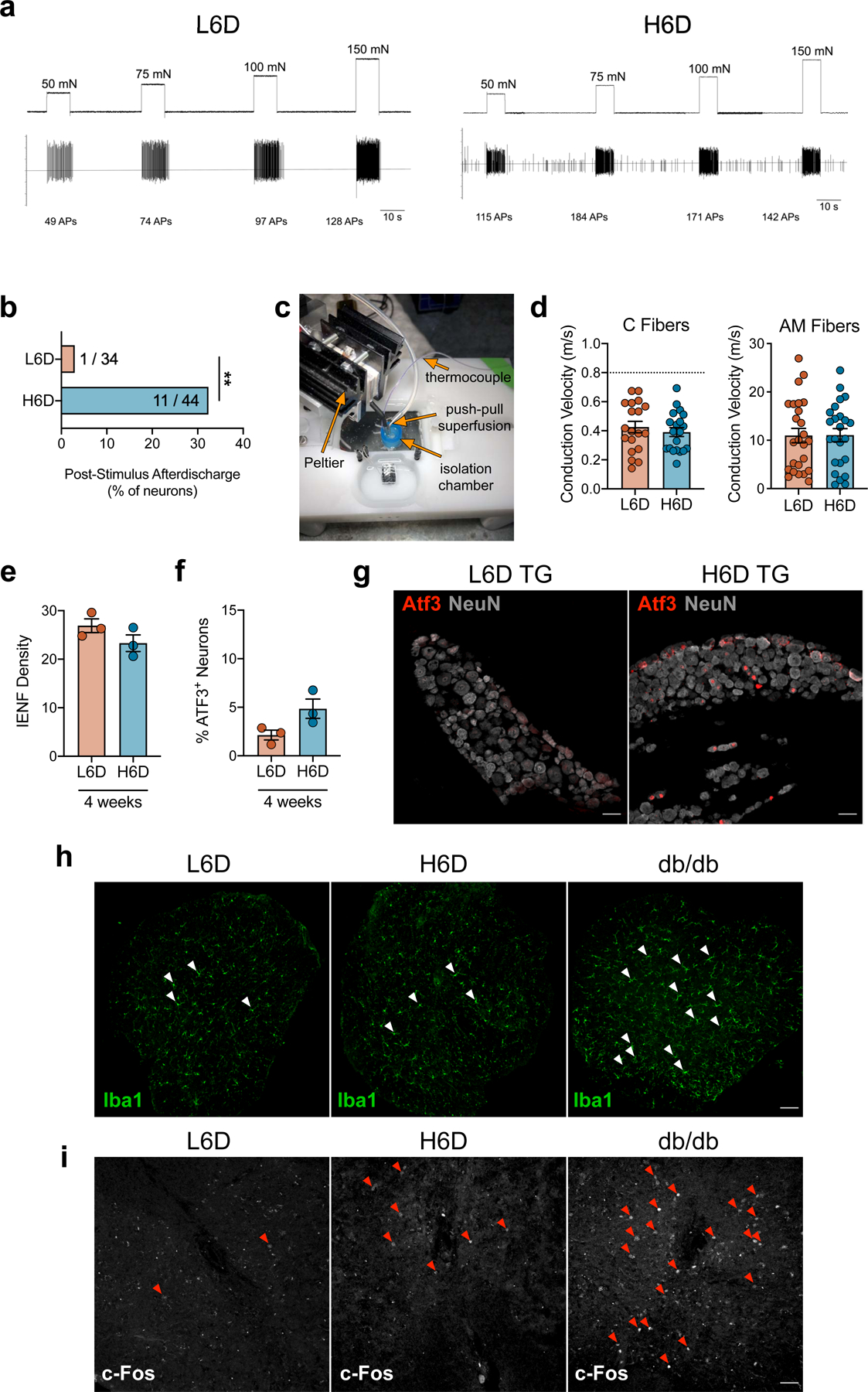
a, Representative recording wavemarks from L6D and H6D mice during mechanical force application. The number of action potentials are denoted beneath each stimulation for each recording. b, Percentage of fibers exhibiting post- stimulus afterdischarge following mechanical force application. Values represent the number of fibers exhibiting afterdischarge over the total recorded fibers for each group (L6D, n=34; H6D, n=44). **P=0.0096. c, The Peltier-based heat delivery system setup. d, Conduction velocities determined for recorded C (left) and AM (right) fibers from L6D and H6D mice (L6D-C, n=19; H6D-C, n=20; L6D-AM, n=25; H6D-AM, n=24). Dotted line represents cut-off value for C fiber classification. e,f, Glabrous IENF densities (e) and percentage of ATF3+ neurons in lumbar DRG (f) after 4 weeks on the L6D or H6D (L6D, n=3; H6D, n=3). g, Representative immunofluorescent staining of ATF3 expression in trigeminal ganglia (TG) from L6D and H6D mice co-localized with NeuN, scale bars: 50 μm (n=2 mice/group). h,i, Immunofluorescent staining of Iba1 (h) and c-Fos (i) expression in the lumbar spinal cord of L6D and H6D mice. Positive control tissue was utilized from db/db mice. White arrowheads designate microglia (h), 10X magnification, scale bar: 50 μm (n=2 mice/group). Red arrowheads designate c-Fos+ nuclei (i), 20X magnification, scale bar: 50 μm (n=2 mice/group). All data are mean ± SEM. The statistical test used was a two-sided Fisher’s exact test (b).
Extended Data Fig. 3. The H6D does not induce a diabetic phenotype.
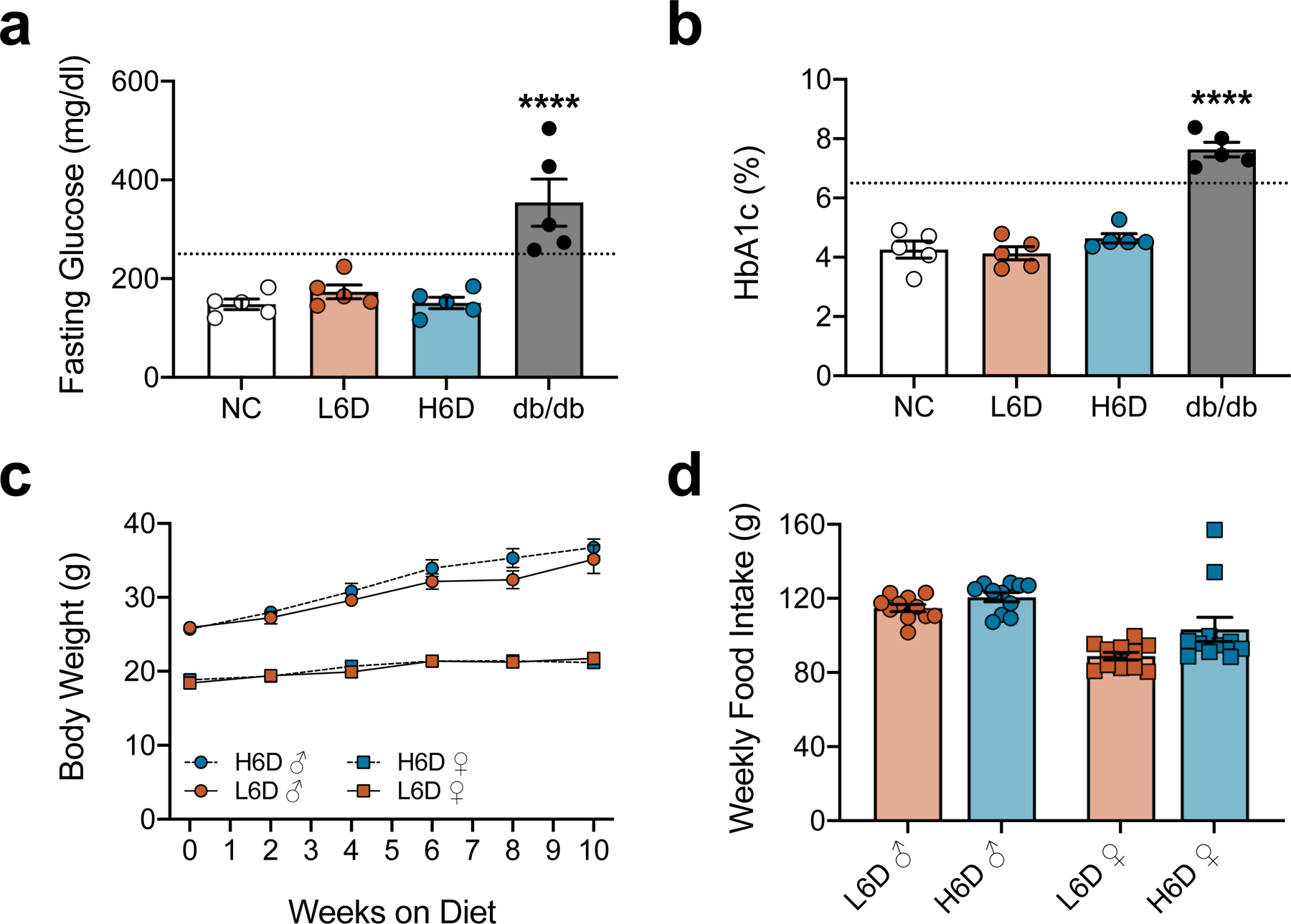
a,b, Scatter plots of fasting blood glucose levels (a) and HbA1c levels (b) from mice on L6D and H6D for 8 weeks. Mice on normal chow (NC) and 16-week-old db/db mice served as negative and positive controls, respectively (NC, n=5; L6D, n=5; H6D, n=5; db/db, n=5). Dotted lines in each figure represent established cut-offs for type 2 diabetes. ****P<0.0001 (db/db vs NC). c,d, Weekly monitoring of body weights (c) and food intake (d) for both male and female mice on either L6D or H6D. Data are mean ± SEM. Error bars for some data points are within the size of the symbol. Statistical test used was one-way ANOVA with Tukey’s post-hoc test (a,b).
Extended Data Fig. 4. The H6D alters lipid composition in DRG, but not spinal cord.
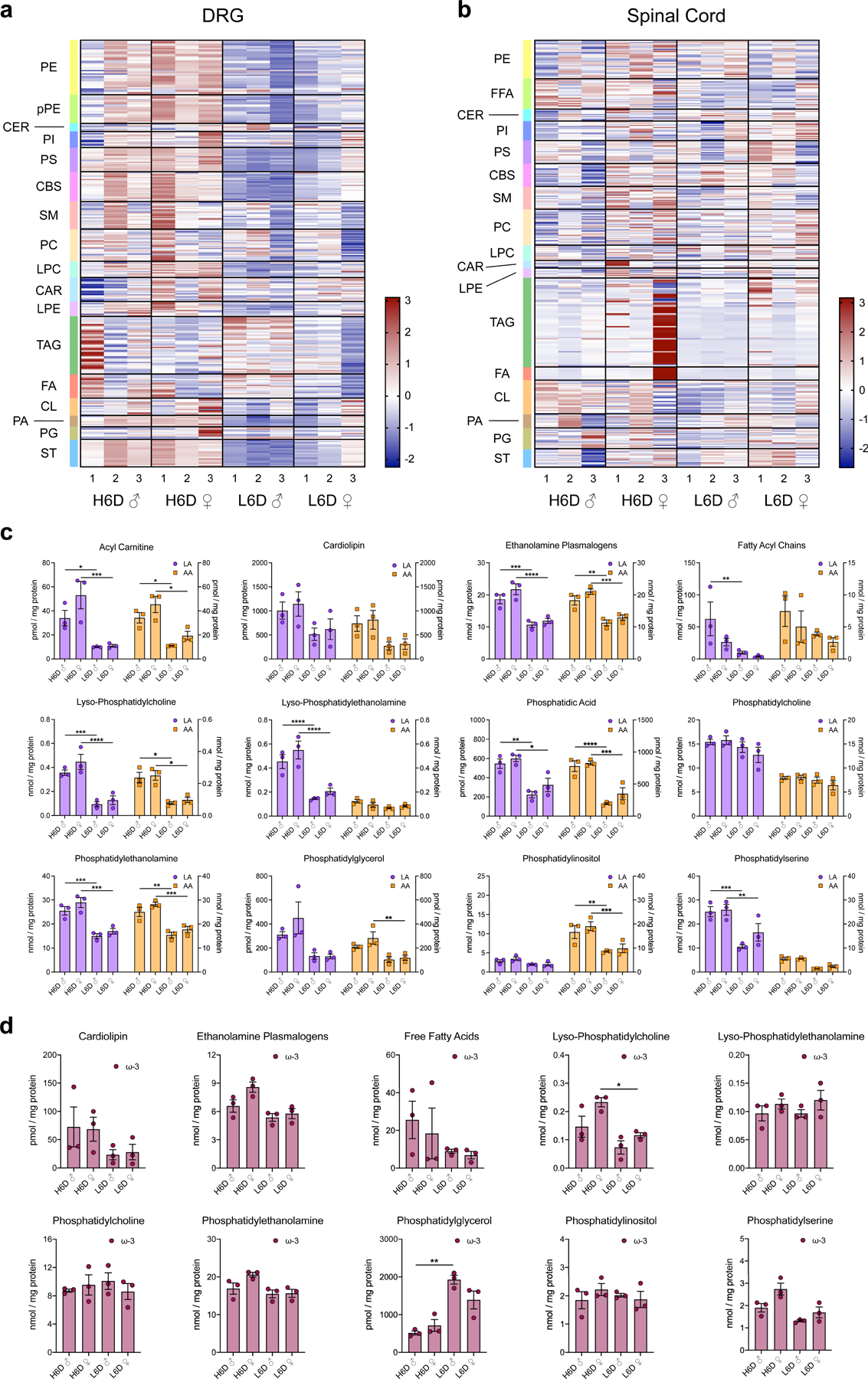
a,b, Heatmaps of lipid species from lumbar DRG (a) and spinal cord (b) from male (♂) and female (♀) mice on either the H6D or L6D. Lipid classes are designated to the left of each heatmap. Scale bar represents z-score transformations for each lipid species. c,d, Scatter plots of LA- and AA-esterified lipids (c) as well as ω-3 content (d, EPA, DHA levels) in DRG sub-profiled by lipid class for male and female mice on either diet (n=3 mice/group/sex). c, Acyl carnitine (LA): *P=0.0346, ***P=0.0004. Acyl carnitine (AA): *P=0.0417 (♂), *P=0.0214 (♀). Ethanolamine plasmalogens (LA): ***P=0.0008, ****P<0.0001. Ethanolamine plasmalogens (AA): **P=0.0027, ***P=0.0007. Fatty acyl chains (LA): **P=0.0069. Lyso- phosphatidylcholine (LA): ***P=0.0002, ****P<0.0001. Lyso- phosphatidylcholine (AA): *P=0.0161, *P=0.0215. Lyso- phosphatidylethanolamine (LA): ****P<0.0001. Phosphatidic acid (LA): **P=0.005, *P=0.0166. Phosphatidic acid (AA): ****P<0.0001, ***P=0.0001. Phosphatidylethanolamine (LA): ***P=0.0006, ***P=0.0002. Phosphatidylethanolamine (AA): **P=0.0015, ***P=0.0007. Phosphatidylglycerol (AA): **P=0.0033. Phosphatidylinositol (AA): **P=0.003, ***P=0.001. Phosphatidylserine (LA): ***P=0.0001, **P=0.0072. d, Lyso-phosphatidylcholine: *P=0.0316. Phosphatidylglycerol: **P=0.0062. Data are mean ± SEM. Statistical tests used were two-way ANOVA with Tukey’s post- hoc test (c) and one-way ANOVA with Tukey’s post-hoc test (d).
Extended Data Fig. 5. PLA2g7 expression predominates in neuronal subpopulations of the lumbar DRG.
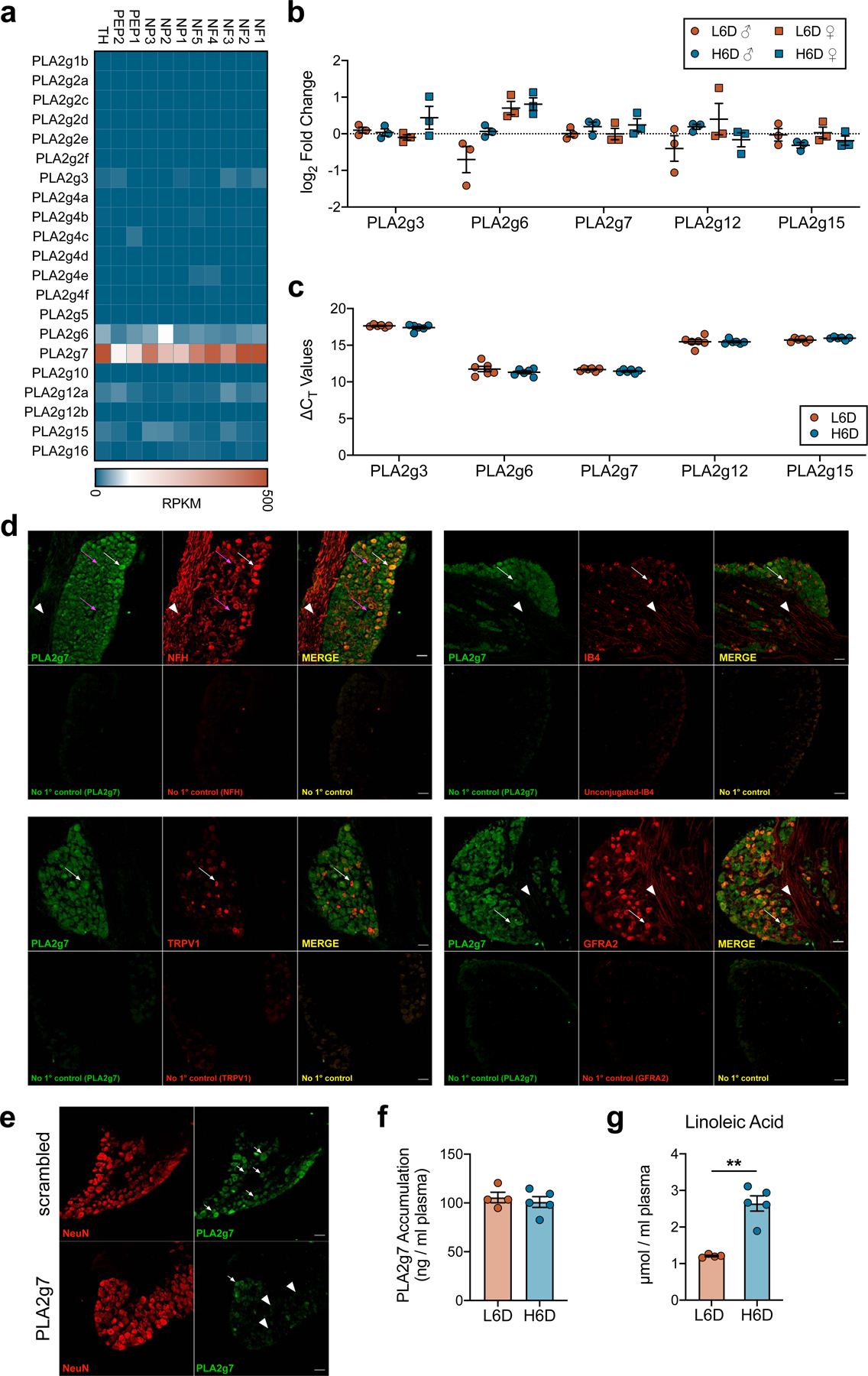
a, Heatmap indicating PLA2 isozyme expression across established neuronal subpopulations of the mouse lumbar DRG. Single-cell RNA-seq data were reproduced with permission36. b,c, qPCR data showing PLA2 isozyme expression and change in cycle threshold values relative to 18S rRNA in lumbar DRG from H6D and L6D mice (b: n=3/group. c: L6D, n=6; H6D, n=6). d, Representative immunofluorescent staining of PLA2g7 expression in mouse lumbar DRG and co- localization with neuronal subtype-specific markers. No primary controls are included for each marker as designated. The magenta arrows highlight two small diameter neurons, one with high PLA2g7 expression and one with low to moderate expression, that are negative for NFH. White arrows designate cell bodies with PLA2g7+ staining and their co- localization with the respective subtype marker. White arrowheads highlight axons projecting through the ganglia that exhibit virtually no PLA2g7 expression compared to cell bodies. Scale bars: 50 μm (n=2 mice). e, Representative immunofluorescent staining of lumbar DRG from naïve control mice that received either scrambled or PLA2g7-directed siRNA intrathecally for the purpose of PLA2g7 antibody validation. White arrows highlight PLA2g7+ staining of neuronal cell bodies, whereas the white arrowheads designate cells exhibiting loss of PLA2g7 immunoreactivity. Scale bars: 50 μm, (n=2 mice/group). f,g, Circulating plasma PLA2g7 levels (f) and plasma LA accumulation (g) from L6D and H6D mice after 8 weeks (L6D, n=4; H6D, n=5). **P<0.0022. Data are mean ± SEM. Error bars for some data points are within the size of the symbol. Statistical test used was unpaired two-tailed Student’s t test (g).
Extended Data Fig. 6. Pharmacological inhibition and genetic knockdown of PLA2g7 in DRG neurons reduces PLA2 activity and attenuates nociceptive hypersensitivities.
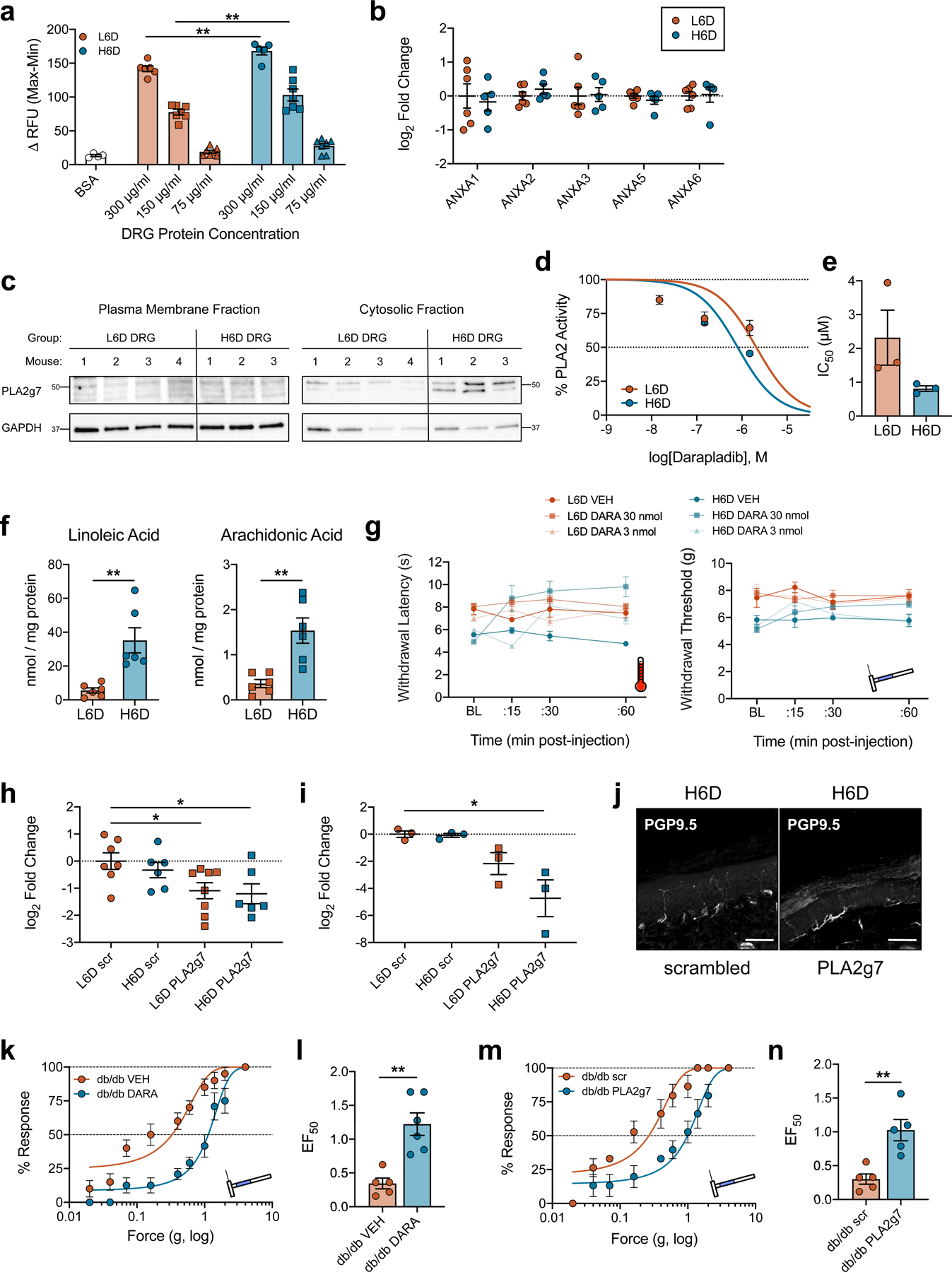
a, Optimization of DRG protein concentration used with the PLA2 BODIPY activity assay. H6D DRG homogenates demonstrate increased activity at multiple concentrations compared to L6D (BSA, n=4; L6D-300, n=6; L6D-150, n=7; L6D- 75, n=7; H6D-300, n=5; H6D-150, n=7; H6D-75, n=7 DRG replicates/group). **P=0.0055 (300), 0.0023 (150). b, qPCR data showing annexin isozyme expression in L6D and H6D DRG (L6D, n=6; H6D, n=5). c, Immunoblots of PLA2g7 and GAPDH protein expression in membrane and cytosolic fractions from homogenized DRG (L6D, n=4 mice; H6D, n=3 mice). Molecular weight markers (kDa) are adjacent to each target. d, Concentration-response curves for darapladib-mediated inhibition of PLA2 activity for DRG (n=3/group). e, Darapladib half-maximal inhibitory concentrations (IC50) as determined by nonlinear regression (n=3/group). f, Total LA and AA levels determined from glabrous hindpaw skin punches (L6D, n=6; H6D, n=6). **P=0.0098 (LA), 0.0071 (AA). g, Dose-response timecourses for i.pl. darapladib on heat- and mechanical- evoked nociception (L6D-VEH, n=6; L6D-3, n=4; L6D-30, n=9; H6D-VEH, n=6; H6D-3, n=5; H6D-30, n=9). h,i, PLA2g7 qPCR data for lumbar DRG (h) and spinal cord (i) following intrathecal siRNA treatment (q.d. x 3d) (h: L6D-scr, n=7; H6D- scr, n=6; L6D-PLA2g7, n=8; H6D-PLA2g7, n=6. i: n=3/group). h, *P=0.0429 (L6D-PLA2g7), 0.0371 (H6D-PLA2g7). i, *P=0.0333. j, Immunofluorescent images of glabrous IENFs from siRNA- treated mice, scale bar: 50 μm (n=2 mice/group). k,l, Mechanical force-response curves (k) and EF50 values (l) for 16-week db/db mice injected i.pl. with either vehicle or darapladib (db/db-veh, n=5; db/db-DARA, n=6). l, **P=0.0020. m,n, Mechanical force-response curves (m) and EF50 values (n) for a different cohort of 16-week db/db mice following i.t. siRNA injections (db/db-veh, n=5; db/db-darapladib, n=5). n, **P=0.0066. All data are mean ± SEM. Error bars for some data points are within the size of the symbol. Statistical tests used were one-way ANOVA with Sidak’s post-hoc test (a), Tukey’s post-hoc test (d), or Dunnett’s post-hoc test (h,i), two- way ANOVA with Tukey’s post-hoc test (c), and unpaired two-tailed Student’s t test (f,l,n).
Extended Data Fig. 7. H3D reverses H6D- induced changes in afferent fibers.
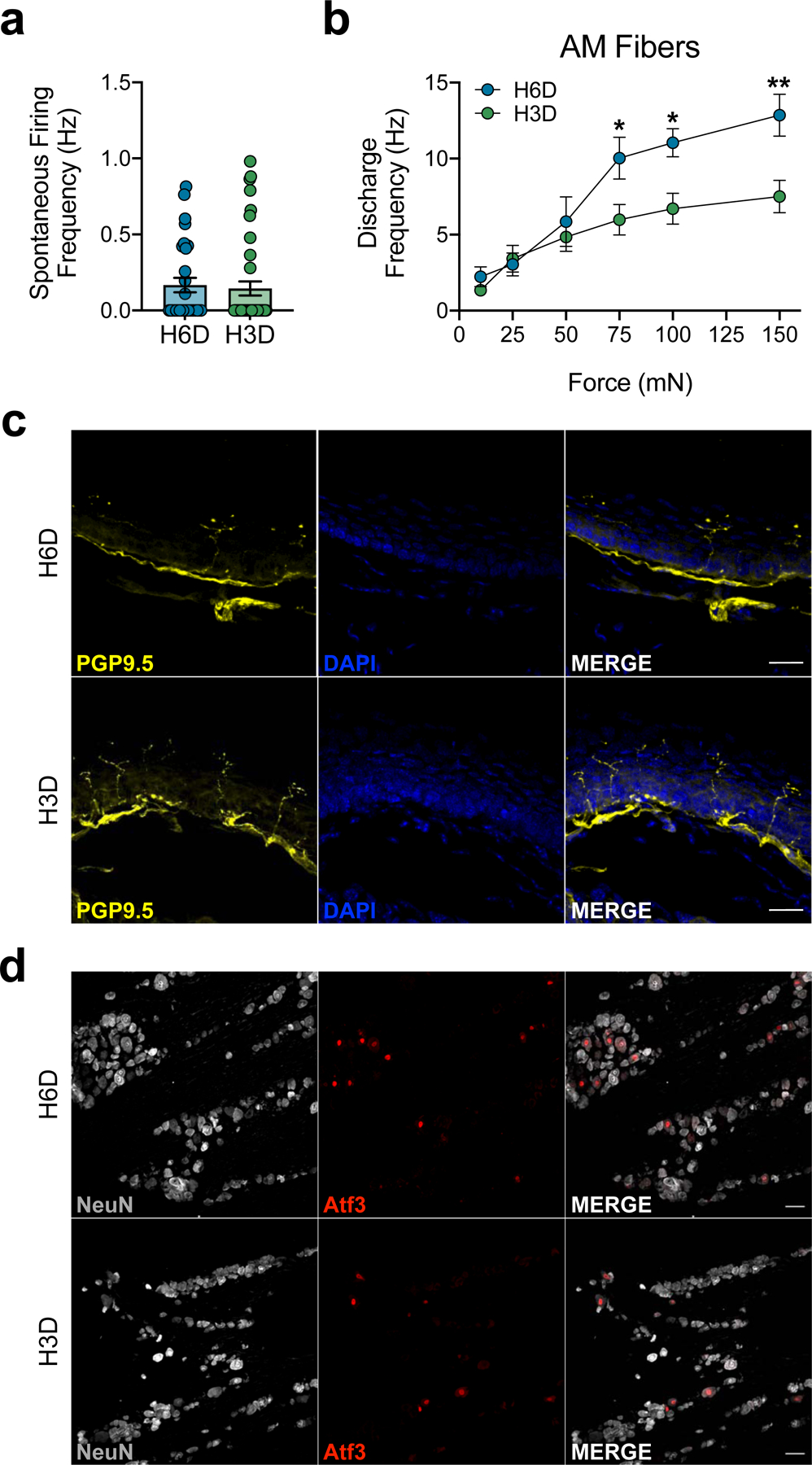
a, Discharge frequencies of spontaneously-active fibers (H6D, n=30; H3D, n=41). b, Discharge frequencies of AM fibers (H6D, n=13; H3D, n=21). *P=0.0353 (75), 0.0335 (100), **P=0.0047 (150). c,d, Representative immunofluorescence staining of glabrous hindpaw skin IENFs (c) and ATF3 expression in lumbar DRG neurons (d) in H6D and H3D mice. Scale bars: 25 μm (c; n=4 mice/group), 50 μm (d; n=3 mice/group). The representative images supplement Figures 3h and 3i, respectively. Data are mean ± SEM. Error bars for some data points are within the size of the symbol. Statistical test used was two-way ANOVA with Sidak’s post-hoc test (b).
Extended Data Fig. 8. Increased LA content in skin of diabetic subjects with painful neuropathy.
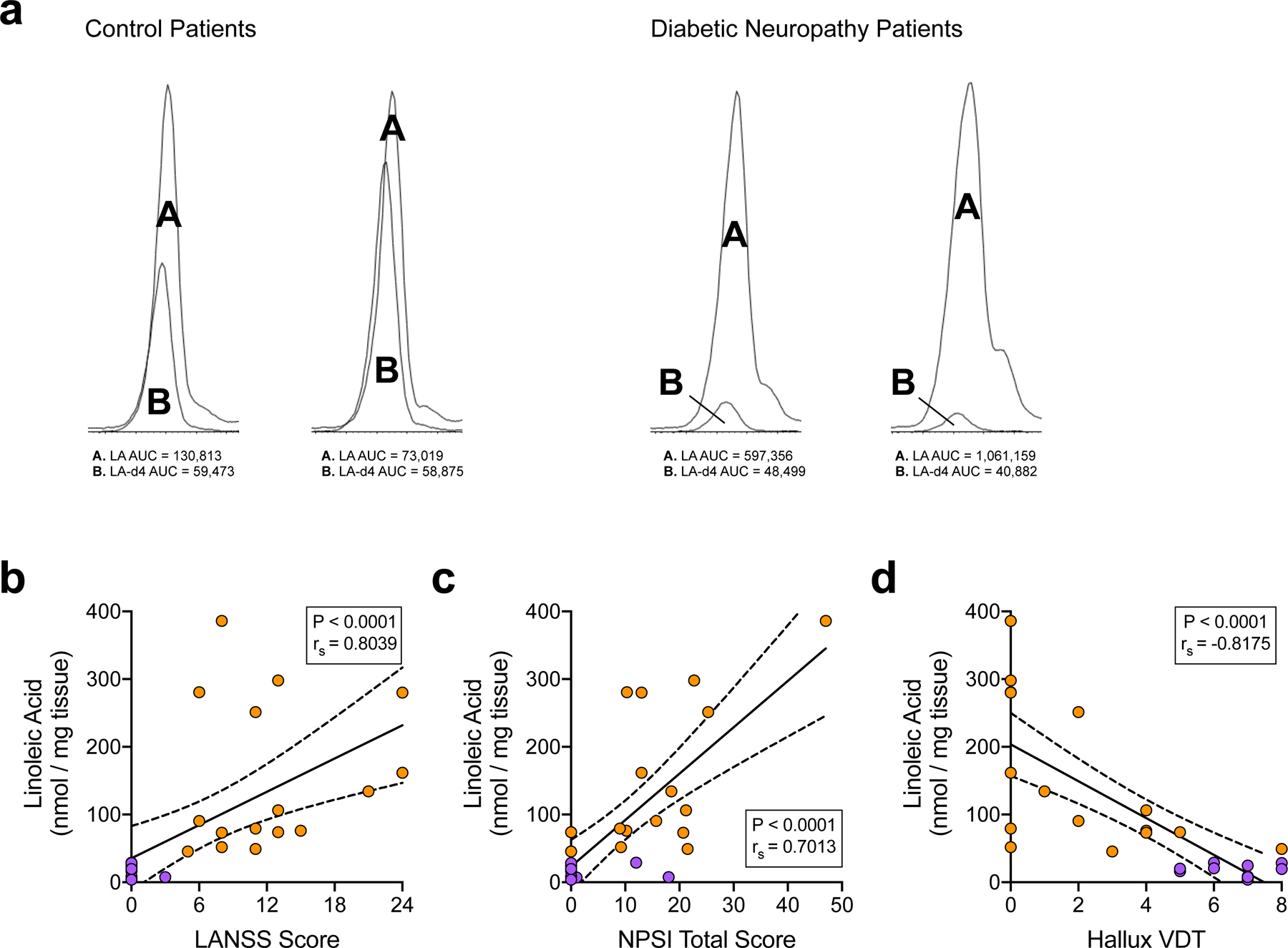
a, Chromatogram snapshots of the endogenous LA peak (labeled A) overlaid with the LA-d4 internal control peak (labeled B) for skin biopsy extracts from diabetic and control subjects. Integrated AUC values (a.u.) for each peak are beneath each chromatogram. b-d, Correlation analyses between subject skin LA levels and their respective LANSS scores (b), NPSI scores (c), and hallux vibration detection thresholds (d) (control, n=12; diabetic, n=16). Linear regression identified the best-fit line (solid line) with 95% confidence intervals (dotted lines). Inset boxes contain Spearman coefficients (rs) and corresponding P-values. Statistical test used was two-tailed Spearman correlation.
Supplementary Material
Acknowledgments
The project described was supported in part by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1TR002645 (K.M.H). Additional support by the National Institutes of Health includes grants: R01NS110948 (K.M.H.), T32DE14318 (P.M.L., A.R.F., K.M.H.), T32GM113896 (J.T.B.), F30AT009949 (J.T.B.), F32DK113841 (P.M.L.), F30DE028486 (A.R.F.), and a grant from the Ella and Williams Owen’s Foundation (K.M.H.). Clinical data were managed using REDCap software supported by UL1RR024982. Certain mass spectrometric analyses were carried out on equipment supported by the U.S. Department of Agriculture, Agricultural Research Service, under agreement No. 58–3094-8–012. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or U.S. Department of Agriculture. We thank Xianlin Han and lab for expertise and guidance on the shotgun lipidomics. We thank Mayur Patil and Ping Wu for technical assistance as well as Anibal Diogenes, Nikita Ruparel, Asma Khan, and Armen Akopian for fruitful discussions.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Data Availability
Datasets generated during this study are included as source data files under Supplementary Information. Any other data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Vos T, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills SEE, Nicolson KP & Smith BH Chronic pain: a review of its epidemiology and associated factors in population-based studies. Br. J. Anaesth 123, e273–e283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabbs M, Leng S, Devassy JG, Monirujjaman M & Aukema HM Advances in our understanding of oxylipins derived from dietary PUFAs. Adv. Nutr 6, 513–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearer GC & Walker RE An overview of the biologic effects of omega-6 oxylipins in humans. Prostaglandins Leukot. Essent. Fatty Acids 137, 26–38 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Simopoulos AP Evolutionary aspects of diet, the omega-6/omega-3 ratio and genetic variation: nutritional implications for chronic diseases. Biomed. Pharmacother 60, 502–507 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Mann JI Diet and risk of coronary heart disease and type 2 diabetes. Lancet 360, 783–789 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Manzel A, et al. Role of “western diet” in inflammatory autoimmune diseases. Curr. Allergy Asthma Rep 14, 404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasbalg TL, Hibbeln JR, Ramsden CE, Majchrzak SF & Rawlings RR Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am. J. Clin. Nutr 93, 950–962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kris-Etherton PM, et al. Polyunsaturated fatty acids in the food chain in the United States. Am. J. Clin. Nutr 71, 179S–188S (2000). [DOI] [PubMed] [Google Scholar]
- 10.Patwardhan AM, et al. Heat generates oxidized linoleic acid metabolites that activate TRPV1 and produce pain in rodents. J. Clin. Invest 120, 1617–1626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eskander MA, et al. Persistent nociception triggered by nerve growth factor (NGF) is mediated by TRPV1 and oxidative mechanisms. J. Neurosci 35, 8593–8603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green DP, Ruparel S, Roman L, Henry MA & Hargreaves KM Role of endogenous TRPV1 agonists in a postburn pain model of partial-thickness injury. Pain 154, 2512–2520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsden CE, et al. A systems approach for discovering linoleic acid derivatives that potentially mediate pain and itch. Sci Signal 10(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sisignano M, et al. Targeting CYP2J to reduce paclitaxel-induced peripheral neuropathic pain. Proc. Natl. Acad. Sci. U. S. A 113, 12544–12549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke G, et al. Marked elevations in pro-inflammatory polyunsaturated fatty acid metabolites in females with irritable bowel syndrome. J. Lipid Res 51, 1186–1192 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adam O, et al. Anti-inflammatory effects of a low arachidonic acid diet and fish oil in patients with rheumatoid arthritis. Rheumatol. Int 23, 27–36 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Calder PC Session 3: Joint Nutrition Society and Irish Nutrition and Dietetic Institute Symposium on ‘Nutrition and autoimmune disease’ PUFA, inflammatory processes and rheumatoid arthritis. Proc. Nutr. Soc 67, 409–418 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Devigili G, et al. The diagnostic criteria for small fibre neuropathy: from symptoms to neuropathology. Brain 131, 1912–1925 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauria G, et al. European Federation of Neurological Societies/Peripheral Nerve Society Guideline on the use of skin biopsy in the diagnosis of small fiber neuropathy. Report of a joint task force of the European Federation of Neurological Societies and the Peripheral Nerve Society. Eur. J. Neurol 17, 903–912, e944–909 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Hunt D, Raivich G & Anderson PN Activating transcription factor 3 and the nervous system. Front. Mol. Neurosci 5, 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsujino H, et al. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol. Cell. Neurosci 15, 170–182 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Bray GA & Popkin BM Dietary fat intake does affect obesity! Am. J. Clin. Nutr 68, 1157–1173 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Forouhi NG, et al. Association of plasma phospholipid n-3 and n-6 polyunsaturated fatty acids with type 2 diabetes: The EPIC-InterAct case-cohort study. PLoS Med 13, e1002094 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fung TT, Schulze M, Manson JE, Willett WC & Hu FB Dietary patterns, meat intake, and the risk of type 2 diabetes in women. Arch. Intern. Med 164, 2235–2240 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Mente A, et al. Association of dietary nutrients with blood lipids and blood pressure in 18 countries: a cross-sectional analysis from the PURE study. Lancet Diabetes Endocrinol 5, 774–787 (2017). [DOI] [PubMed] [Google Scholar]
- 26.van Dam RM, Willett WC, Rimm EB, Stampfer MJ & Hu FB Dietary fat and meat intake in relation to risk of type 2 diabetes in men. Diabetes Care 25, 417–424 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Ramsden CE, et al. Dietary linoleic acid-induced alterations in pro- and anti-nociceptive lipid autacoids: Implications for idiopathic pain syndromes? Mol. Pain 12(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patwardhan AM, Scotland PE, Akopian AN & Hargreaves KM Activation of TRPV1 in the spinal cord by oxidized linoleic acid metabolites contributes to inflammatory hyperalgesia. Proc. Natl. Acad. Sci. U. S. A 106, 18820–18824 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffin TM, et al. Diet-induced obesity differentially regulates behavioral, biomechanical, and molecular risk factors for osteoarthritis in mice. Arthritis Res. Ther 12, R130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee E, et al. Transient receptor potential vanilloid type-1 channel regulates diet-induced obesity, insulin resistance, and leptin resistance. FASEB J 29, 3182–3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tramullas M, Finger BC, Dinan TG & Cryan JF Obesity takes its toll on visceral pain: High-fat diet induces Toll-like receptor 4-dependent visceral hypersensitivity. PLoS One 11, e0155367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilensky RL, et al. Inhibition of lipoprotein-associated phospholipase A2 reduces complex coronary atherosclerotic plaque development. Nat. Med 14, 1059–1066 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace VC, Cottrell DF, Brophy PJ & Fleetwood-Walker SM Focal lysolecithin-induced demyelination of peripheral afferents results in neuropathic pain behavior that is attenuated by cannabinoids. J. Neurosci 23, 3221–3233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piomelli D & Sasso O Peripheral gating of pain signals by endogenous lipid mediators. Nat. Neurosci 17, 164–174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis EA, Cao J, Hsu YH, Magrioti V & Kokotos G Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev 111, 6130–6185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Kim M, et al. Impact of 8-week linoleic acid intake in soy oil on Lp-PLA2 activity in healthy adults. Nutr. Metab. (Lond.) 14, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackie JA, et al. The identification of clinical candidate SB-480848: a potent inhibitor of lipoprotein-associated phospholipase A2. Bioorg. Med. Chem. Lett 13, 1067–1070 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Goldberg RJ & Katz J A meta-analysis of the analgesic effects of omega-3 polyunsaturated fatty acid supplementation for inflammatory joint pain. Pain 129, 210–223 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Laye S, Nadjar A, Joffre C & Bazinet RP Anti-inflammatory effects of omega-3 fatty acids in the brain: Physiological mechanisms and relevance to pharmacology. Pharmacol. Rev 70, 12–38 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Xu ZZ, et al. Resolvins RvE1 and RvD1 attenuate inflammatory pain via central and peripheral actions. Nat. Med 16, 592–597, 591p following 597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bazata DD, Robinson JG, Fox KM, Grandy S & Group SS Affecting behavior change in individuals with diabetes: findings from the Study to Help Improve Early Evaluation and Management of Risk Factors Leading to Diabetes (SHIELD). Diabetes Educ 34, 1025–1036 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Davis JA, Robinson RL, Le TK & Xie J Incidence and impact of pain conditions and comorbid illnesses. J. Pain Res 4, 331–345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fehrenbacher JC, Vasko MR & Duarte DB Models of inflammation: Carrageenan- or complete Freund’s adjuvant (CFA)-induced edema and hypersensitivity in the rat. Curr. Protoc. Pharmacol Chapter 5, Unit5 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi K, et al. The db/db mouse, a model for diabetic dyslipidemia: molecular characterization and effects of Western diet feeding. Metabolism 49, 22–31 (2000). [DOI] [PubMed] [Google Scholar]
- 46.O’Brien PD, Sakowski SA & Feldman EL Mouse models of diabetic neuropathy. ILAR J 54, 259–272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alvheim AR, et al. Dietary linoleic acid elevates endogenous 2-AG and anandamide and induces obesity. Obesity (Silver Spring) 20, 1984–1994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett M The LANSS Pain Scale: the Leeds assessment of neuropathic symptoms and signs. Pain 92, 147–157 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Bouhassira D, et al. Development and validation of the Neuropathic Pain Symptom Inventory. Pain 108, 248–257 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Mauck MC, et al. Obesity increases the risk of chronic pain development after motor vehicle collision. Pain 160, 670–675 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Okifuji A & Hare BD The association between chronic pain and obesity. J. Pain Res 8, 399–408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White HD, et al. Darapladib for preventing ischemic events in stable coronary heart disease. N. Engl. J. Med 370, 1702–1711 (2014). [DOI] [PubMed] [Google Scholar]
- 53.O’Donoghue ML, et al. Effect of darapladib on major coronary events after an acute coronary syndrome: the SOLID-TIMI 52 randomized clinical trial. JAMA 312, 1006–1015 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Haghdoost F, et al. Association between Ala379Val polymorphism of lipoprotein-associated phospholipase A2 and migraine without aura in Iranian population. Iran J Neurol 15, 80–84 (2016). [PMC free article] [PubMed] [Google Scholar]
- 55.Parisien M, et al. Effect of human genetic variability on gene expression in dorsal root ganglia and association with pain phenotypes. Cell Rep 19, 1940–1952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hummel KP, Dickie MM & Coleman DL Diabetes, a new mutation in the mouse. Science 153, 1127–1128 (1966). [DOI] [PubMed] [Google Scholar]
- 57.Reeves PG, Nielsen FH & Fahey GC Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr 123, 1939–1951 (1993). [DOI] [PubMed] [Google Scholar]
- 58.Gibbs JL, Flores CM & Hargreaves KM Attenuation of capsaicin-evoked mechanical allodynia by peripheral neuropeptide Y Y1 receptors. Pain 124, 167–174 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Chaplan SR, Bach FW, Pogrel JW, Chung JM & Yaksh TL Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63 (1994). [DOI] [PubMed] [Google Scholar]
- 60.White S, Marquez de Prado B, Russo AF & Hammond DL Heat hyperalgesia and mechanical hypersensitivity induced by calcitonin gene-related peptide in a mouse model of neurofibromatosis. PLoS One 9, e106767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garrison SR, Dietrich A & Stucky CL TRPC1 contributes to light-touch sensation and mechanical responses in low-threshold cutaneous sensory neurons. J. Neurophysiol 107, 913–922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hargreaves K, Dubner R, Brown F, Flores C & Joris J A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain 32, 77–88 (1988). [DOI] [PubMed] [Google Scholar]
- 63.Brenner DS, Golden JP & Gereau R.W.t. A novel behavioral assay for measuring cold sensation in mice. PLoS One 7, e39765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayala JE, et al. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis. Model. Mech 3, 525–534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han BG, et al. Markers of glycemic control in the mouse: comparisons of 6-h- and overnight-fasted blood glucoses to Hb A1c. Am. J. Physiol. Endocrinol. Metab 295, E981–986 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang M & Han X Multidimensional mass spectrometry-based shotgun lipidomics. Methods Mol. Biol 1198, 203–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han X, Yang K & Gross RW Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom 22, 2115–2124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang M, Wang C, Han RH & Han X Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res 61, 83–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang K, Cheng H, Gross RW & Han X Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem 81, 4356–4368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bligh EG & Dyer WJ A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 71.Pettinella C, Lee SH, Cipollone F & Blair IA Targeted quantitative analysis of fatty acids in atherosclerotic plaques by high sensitivity liquid chromatography/tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 850, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Quehenberger O, Armando AM & Dennis EA High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta 1811, 648–656 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reeh PW Sensory receptors in a mammalian skin-nerve in vitro preparation. Prog. Brain Res 74, 271–276 (1988). [DOI] [PubMed] [Google Scholar]
- 74.Banik RK & Brennan TJ Sensitization of primary afferents to mechanical and heat stimuli after incision in a novel in vitro mouse glabrous skin-nerve preparation. Pain 138, 380–391 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zimmermann K, et al. Phenotyping sensory nerve endings in vitro in the mouse. Nat. Protoc 4, 174–196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hogan D, Baker AL, Moron JA & Carlton SM Systemic morphine treatment induces changes in firing patterns and responses of nociceptive afferent fibers in mouse glabrous skin. Pain 154, 2297–2309 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moehring F, et al. Keratinocytes mediate innocuous and noxious touch via ATP-P2X4 signaling. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duraku LS, et al. Spatiotemporal dynamics of re-innervation and hyperinnervation patterns by uninjured CGRP fibers in the rat foot sole epidermis after nerve injury. Mol. Pain 8, 61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reid G, Amuzescu B, Zech E & Flonta ML A system for applying rapid warming or cooling stimuli to cells during patch clamp recording or ion imaging. J. Neurosci. Methods 111, 1–8 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Koltzenburg M, Stucky CL & Lewin GR Receptive properties of mouse sensory neurons innervating hairy skin. J. Neurophysiol 78, 1841–1850 (1997). [DOI] [PubMed] [Google Scholar]
- 81.Banik RK & Brennan TJ Spontaneous discharge and increased heat sensitivity of rat C-fiber nociceptors are present in vitro after plantar incision. Pain 112, 204–213 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Collins TJ ImageJ for microscopy. Biotechniques 43, 25–30 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Linkert M, et al. Metadata matters: access to image data in the real world. J. Cell Biol 189, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.LoCoco PM, et al. Pharmacological augmentation of nicotinamide phosphoribosyltransferase (NAMPT) protects against paclitaxel-induced peripheral neuropathy. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beiswenger KK, Calcutt NA & Mizisin AP Epidermal nerve fiber quantification in the assessment of diabetic neuropathy. Acta Histochem 110, 351–362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.LoCoco PM, et al. Reliable approaches to extract high-integrity RNA from skin and other pertinent tissues used in pain research. PAIN Reports e818, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livak KJ & Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 90.Harris PA, et al. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform 42, 377–381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beery AK Inclusion of females does not increase variability in rodent research studies. Curr Opin Behav Sci 23, 143–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets generated during this study are included as source data files under Supplementary Information. Any other data that support the findings of this study are available from the corresponding author upon request.


