Abstract
Background.
The effect of stress on alcohol consumption in humans is highly variable and underlying processes are not yet understood. Attempts to model a positive relationship between stress and increased ethanol consumption in animals have been only modestly successful. Our hypothesis is that individual differences in stress effects on ethanol consumption are mediated by genetics.
Methods.
We measured alcohol consumption, using the drinking-in-the-dark (DID) paradigm in females from two inbred mouse strains, C57BL/6J (B6) and DBA/2J (D2) and 35 of their inbred progeny (the BXD family). A control group was maintained in normal housing and a stress group was exposed to chronic mild stress (CMS), consisting of unpredictable stressors over seven weeks. These included predator, social, and environmental perturbations. Alcohol intake was measured over sixteen weeks in both groups during Baseline (preceding 5-week period), CMS (intervening 7-week period), and post-stress (final 4-week period).
Results.
We detected a strong effect of CMS on alcohol intake. A few strains demonstrated CMS-related increased alcohol consumption; however, most showed decreased intake. We identified one nearly significant quantitative trait locus on chromosome 5 that contains the neuronal nitric oxide synthase gene (Nos1). The expression of Nos1 is frequently changed following alcohol exposure and variants in this gene segregating among the BXD population may modulate alcohol intake in response to stress.
Conclusions.
The results we present here represent the first study to combine chronic stress and alcohol consumption in a genetic reference population of mice. Differences in susceptibility to the effects of stressful environments vis a vis alcohol use disorders would suggest that the differences have at least some basis in genetic constitution. We have also nominated a likely candidate gene underlying the large individual differences in effects of stress on alcohol consumption.
Keywords: forward genetic analysis, quantitative trait loci analysis, ethanol chronic mild stress, Nos1
INTRODUCTION
A seminal paper by Cloninger (1987) developed the idea that alcohol use disorders can be usefully divided into at least two types. The abusive-early onset type is defined as secondary to anti-social personality disorder, whereas the dependent type is defined as being associated with relief from or response to stressful situations. The adult onset type includes harm-avoidance and susceptibility to stress. Attempts to model stress-related alcohol use disorder in animals have been challenging. Researchers have employed acute and repeated stressors, including foot shock, cold water immersion, restraint, and social defeat, and some have introduced genetics as a key variable in the research. The results have been mixed and no consistent pattern has emerged. For example, in the high alcohol consuming mouse strain, C57BL/6J (B6), acute foot shock and repeated restraint increase alcohol consumption (Matthews et al., 2008; Farook et al., 2009), or has no effect (restraint) (Tambour et al., 2008). Alternatively, repeated cold-water immersion decreases alcohol consumption in B6 mice (Boyce-Rustay et al., 2008). Yang and colleagues (2008) showed that repeated restraint increased alcohol consumption in 129/SvEv but not in the B6 strain. In lines selected for high alcohol preference, repeated foot shock increases alcohol consumption in adolescent but not adult animals (Chester et al., 2008).
We propose that the crux of stress-related alcohol consumption requires understanding the genetic basis for individual differences in at least two domains. One domain has to do with appetite for alcohol. Another domain has to do with response to stress or reactivity. We propose that the genetic basis for individual differences in one domain may only partially overlap with the other. This hypothesis may at least partially explain inconsistencies in mice, and even in humans (Hotopf et al., 2003). In most cases, the models address only one domain while either ignoring or inadequately addressing the other and often without accounting for possibly important genetic factors. A more comprehensive model must combine: 1) a method of stress that more closely models the kinds of stressors humans experience (i.e. unpredictable, chronic); 2) a method of alcohol consumption that can capture a wide range of amounts consumed and that achieves intoxicating levels; 3) physiological sequelae of the stress; and 4) a population that can be used to dissect genetic factors modulating phenotypic response to stress, alcohol consumption and their interaction. We have developed such a model that captures the genetic influence at the intersection of intoxicating alcohol consumption and stress through incorporation of a drinking in the dark (DID) alcohol intake paradigm combined with the application of chronic mild stress (CMS) in the well characterized BXD genetic reference population.
The BXD genetic reference population is currently the largest and best characterized recombinant inbred panel available. This population is segregating for 5 million gene variants that distinguish the parental B6 and D2 strains, has been densely genotyped, and includes a sufficient number of strains (~160) to power systems genetics, traditional forward genetics, and gene by environment (GXE) approaches. The BXDs are particularly amenable for GXE studies because individual genotypes (strains) can be resampled under many different conditions and the analysis of trait heritability, and subsequent quantitative trait locus (QTL) mapping, can be improved by increasing both the number of genotypes and individuals of each genotype that are sampled. The BXD family has also been densely profiled for molecular, pharmacological, behavioral, and physiological traits, including alcohol intake (Phillips et al., 1994; Rodriguez et al., 1994; Gill et al., 1996; Fernandez et al., 1999), response to stress (Jung et al., 2017; Shea et al., 2015; Carhuantanta et al., 2014), and gene expression in numerous tissues, including brain regions. However, the genetics of alcohol intake in response to chronic stress has never been profiled in this, or any, model population. Thus, using the BXD platform, we can nominate candidate genes that modulate stress-induced changes in voluntary alcohol intake and compare our results against the nearly 6,000 phenotypes and millions of gene expression profiles in numerous tissues that have been generated for the BXD panel.
We have selected the DID paradigm to measure voluntary alcohol consumption in the BXD population. Some methods of measuring alcohol intake (e.g. 2-bottle choice, 24 h acceptance) have been criticized because it can be difficult to ascertain whether the animals may be drinking (or not) because of taste and thus the validity of the measure as pharmacologically relevant is questionable. DID is a modified acceptance procedure, that takes advantage of normal circadian cycles of feeding to elicit high voluntary alcohol intake (Rhodes et al., 2005). Thus, DID allows for binge drinking in animals so inclined.This method has the advantage of correlating highly with other ethanol intake measures across strains, and some animals achieve intoxicating blood ethanol concentrations (Rhodes et al., 2007). We previously reported that DID alcohol intake in the BXD family is heritable and variable (Mulligan et al., 2018).
Finally, we have chosen to apply CMS as the environmental factor to study the influence of stress on alcohol intake. CMS is the application of a variety of unpredictable perturbations to the animals (Willner et al., 1987). There are usually four classes of perturbations; unpredictable changes in light-dark cycle, environmental challenge, social, and predator stress. Physiological and behavioral responses to chronic stress as applied in the CMS paradigm are strongly dependent on genetic background (Ibarguen-Vargas et al., 2008), and behavioral changes produced by CMS can be reversed by antidepressants (for review, see Willner, 2005). Furthermore, variation in cognitive and emotional behavior in response to chronic variable stress is heritable among the BXDs (Jung et al., 2017; Shea et al., 2015; Carhuantanta, et al., 2014). Here, we report changes in voluntary DID alcohol intake during and after chronic stress exposure. We demonstrate that these changes are heritable in the BXD population and identify genetic loci and candidate genes that moderate individual differences in the effect of stress on alcohol consumption.
MATERIALS AND METHODS
Animals.
The study included females (aged 50 to 90 ±2 days at the start of the experiment) from B6, D2, and 34 advanced intercross BXD strains (n = 1 to 6 per strain and condition, Stable 1). Mice were singly housed in shoebox cages with food and water provided ad libitum except when the water was replaced by 20% (v/v) ethanol (see below). The ambient temperature was 21 ± 2 ºC and the light cycle was 23:00/11:00 lights on/off. The animals were weighed weekly to assess health and to provide the basis for calculating the amount of ethanol consumed in g/kg. All procedures were approved by the UTHSC Institutional Animal Care and Use Committee.
Drinking in the Dark (DID).
DID consisted of four consecutive days of having the drinking water replaced by 20% (v/v) ethanol prepared from 95% USP neutral grain spirits for 2 or 4h. For the first 3 days, the water bottles are removed from home cages 3 h after lights out (i.e. 14:00 hours) and replaced by 15 ml conical centrifuge tubes containing 20% ethanol fitted with sipper tubes for 2h. On the fourth day, the same procedure was followed with the exception that the ethanol was left on the home cages for 4h. DID was measured Tuesday through Friday for all 16 weeks. The centrifuge tubes were refilled daily and weighed immediately before being placed on the cages. At the end of 2 or 4h, the tubes were removed from the cages and weighed to yield the amount of fluid consumed. The ethanol content in the fluid was assessed and divided by body weight to give g/kg consumed. Detailed description of methods can be found in Mulligan et al. (2018). We averaged the 2h per week and 4h across all weeks, so there are 3X5 week data for 2h and 1X5 for 4h in baseline, 3X7 and 1X7 for 2h and 4h respectively for the CMS period and 3X4 and 1X4 for post stress, respectively.
Chronic Mild Stress (CMS).
During a period of seven weeks, mice received two disturbances per day (see Supplemental Figure 1). These consisted of being exposed to wet bedding for 1h, 1cm of water in the bottom of the cage with no bedding for 1h, no bedding for 1h, confinement in a 3X3X3 cm plastic box for 15 min, tilted cage at 45º for 1h, exposure to foreign mouse or fox urine odor for 1h. The other stressor was a complete phase shift in the light: dark cycle over each weekend. Disruption of the light cycle (lights constantly on) occurred over the weekend (starting Friday 11:00 AM) with the light cycle resuming at 11 A.M. on Monday (lights off). The schedule was repeated weekly over the duration of the study.
Treatment Periods.
The entire protocol took 16 weeks to complete and was subdivided into three periods: Baseline (initial 5 weeks), CMS (next 7 weeks), and post-stress (final 4 weeks). Alcohol was offered as described above for all periods and experimental groups. During the intervening 7-week CMS period, control animals were subjected to normal housing and experimental (stress) animals were subjected to CMS.
Data Analysis.
Mean 2 or 4 h intake for each period (Baseline, CMS, post-stress) was computed for each subject. Difference scores for CMS minus Baseline (contrast 1) and post-stress minus Baseline (contrast 2) were first computed at the individual subject level. The resulting difference scores for each contrast were then averaged by strain and treatment (stress vs. control) group. The effect of stress on ethanol intake was evaluated by analysis of variance for mean intake difference scores (CMS minus Baseline and post-stress minus Baseline) for two between-subjects variables (Strain, Stress) experiment. However, to illustrate the interaction effect of stress and period on alcohol consumption, we present strain effects for the difference scores between the two treatment groups for each period comparison as described below and in Figure 1. These effects are then used for genetic mapping of the effect of stress on alcohol consumption.
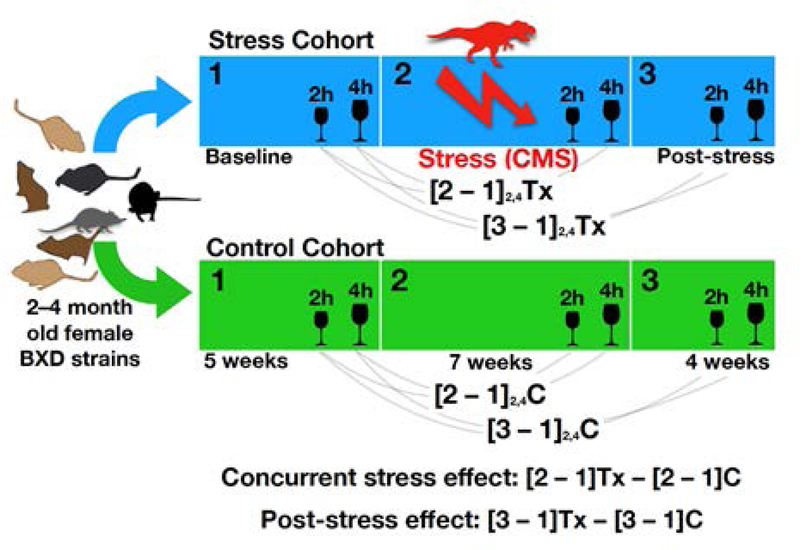
Figure 1. Study design.
C designates control group and Tx designates stress group.
Comparison 1 Effect of simultaneous CMS on alcohol intake:
For each strain, the mean difference values for contrast 1 were subtracted between treatment groups (stress – control). Positive values indicate higher alcohol consumption in the stress group relative to the control group at the end of the CMS period relative to Baseline and negative values indicate relatively lower consumption.
Simplified: Mean Stress [CMS-Baseline] - Mean Control [Stress-Baseline]
Comparison 2 Trace effect of CMS on alcohol intake:
For each strain, the mean difference values for contrast 2 were subtracted between treatment groups (stress – control). Positive values indicate higher alcohol consumption in the stress group relative to the control group at the end of the post-stress period relative to the Baseline period and negative values indicate lower relative consumption.
Simplified: Mean Stress [post-CMS - Baseline] - Mean Control [post-CMS - Baseline]
Heritability.
Broad sense heritability was calculated from the ANOVA results table as 0.5VG(enetic)/(0.5VG + VE(nvironment), where the variance between strain means (ANOVA strain mean square) is equivalent to VG and the variance within strains (ANOVA mean square error or Residual Mean Sq) is equivalent to VE (Hegmann and Possidente, 1981; Belknap, 1998; Petretto et al., 2006).
QTL Mapping.
Traditional QTL mapping was performed in GeneNetwork (GN, www.GeneNetwork.org) as described in Mulligan et al. (2018). In brief, a simple regression method (Haley-Knott) was used to compute QTL probability given BXD strain genotypes and alcohol intake traits averaged by strain (Chesler et al., 2005; Mulligan et al., 2017). Genome-wide suggestive (adjusted p < 0.63) and significant (adjusted p < 0.05) thresholds were determined based on 1,000 permutations of the trait data for each phenotype (GN default). A 1.5 LOD drop from the top marker was used to define QTL confidence interval regions. Data used for QTL mapping have been deposited to GN (Mouse BXD Phenotypes data set) as trait IDs 21307 through 21318.The HQF BXD Neocortex ILM6v1.1 (Dec10v2) RankInv data found on GeneNetwork.org was used for cisQTL mapping of the phenotypes to identify candidate genes.
Candidate Gene Prioritization.
Lists of polymorphic genes were obtained using public D2 strain sequence data from the Mouse Genomes Project (Keane et al., 2011) accessible at www.sanger.ac.uk/sanger/Mouse_SnpViewer. Brain gene expression was determined by querying GN brain microarray data sets and brain and peripheral microarray data sets publically available at biogs.org (Wu et al., 2009). Biological function related to stress or alcohol was determined using the Chilibot (www.chilibot.net) text-mining application (Chen and Sharp, 2004).
RESULTS
Significant effect of strain and stress on alcohol intake.
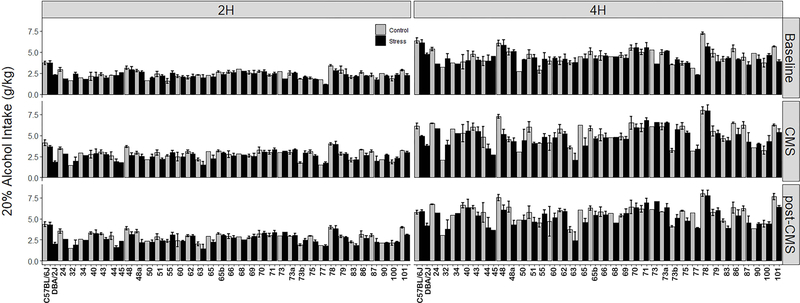
Strain averaged 2 or 4 h DID alcohol intake varied among strains at baseline and during and after stress (Figure 2). For the 2 h and 4 h intake during the Baseline period, ANOVA revealed a significant strain effect (F36,154=7.02 and 5.62 respectively, p<0.001 for both). Heritability estimates are 0.78 and 0.73 for 2 h and 4 h intake respectively. During the Baseline period, alcohol intake is increased on average over the five weeks of measurement, although there is considerable and heritable genetic variation with some strains demonstrating decreased alcohol intake. Variation among strains in alcohol intake during the Baseline period has been linked to regions on Chrs 2, 4, 6, 8, and 12 and several high priority candidate genes have been nominated. This work has been published (Mulligan et al., 2018).
Figure 2. Average Strain Alcohol Intake Over All Periods.
Left and right panel display 2h and 4h intake, respectively. Intake for each period is shown in rows. Alcohol (20%) intake in grams of alcohol per kilogram body weight is shown on the y-axis. Strains are shown on the x-axis. Intake for control and stress group are displayed for each strain.
During the stress period, ANOVA showed significant strain effects for 2 h and 4 h intake (F36,154=5.74 and 5.77 respectively, p<0.001 for both). The effects of treatment for both were also significant (F1,154=12.55 and 11.47 respectively, p<0.001 for both). The treatment by strain interactions was not significant (F34,154 = 1.28, p = 0.16) for the 2 h access period and significant (F34,154 = 1.61, p < 0.05) for the 4h access period.To estimate heritabilities, we analyzed the results for the stress and control groups separately. For the stress group, the heritability estimate for 2h intake was 0.63 and for the controls, 0.65. For the 4 h intake, the heritability estimate for the stress group was 0.64 and for the controls, 0.66.
For 2 h and 4 h intake during the post-stress period, ANOVA showed significant strain effects (F36,154=8.68 and 5.06 respectively, p<0.001 for both). The effect of treatment group was also significant for both (F1,154=22.01 and 14.46 respectively, p<0.001 for both). There was a suggestive interaction effect for the 2 h access period (F34,154=1.39, p = 0.09), however, the strain by treatment interaction was not significant for the 4 h access period (F<1). For the previously stressed group, the heritability estimate for 2 h intake was 0.74 and for the controls, 0.69. For the 4 h intake, the heritability estimate for the previously stressed group was 0.63 and for the controls, 0.58.
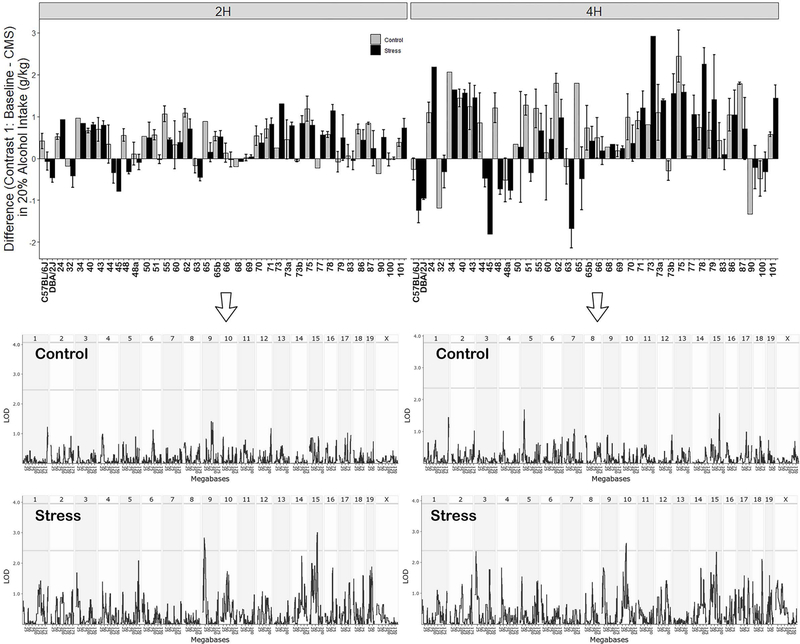
To better evaluate the effect of treatment on alcohol intake in the control and stress groups, we first calculated a difference score for each subject and then averaged by strain, treatment group and period. In the first contrast (see Figure 1) we evaluated the average strain difference in intake for the control and stress groups between the Baseline and CMS periods (Figure 3). There was a significant strain effect for both the 2 h (F36,154 = 4.85, p < 0.001) and 4 h (F36,154 = 5.2, p < 0.001) access periods. There was also a signifiant treatment effect for the 4 h (F1,154 = 4.29, p < 0.05), but not for the 2 h access period (F1,154 = 2.66, p = 0.10). There was a significant interaction effect for both the 2 h (F34,154 = 1.62, p < 0.05) and 4 h (F34,154 = 1.80, p < 0.01) access periods indicating a differential effect of stress on alcohol intake dependent on strain (genetic background). Consistent with relatively stable drinking across periods for the control group, suggestive QTL for the difference in alcohol intake between the Baseline and CMS periods were only identified for the stress group (Figure 3). Suggestive QTLs included loci on Chrs 9 and 15 for the 2 h access period and a locus on Chr 10 for the 4 h access period (Figure 3).
Figure 3. Average Strain Difference in Intake for Contrast 1 (Baseline – CMS Periods).
Left and right panel display 2h and 4h intake, respectively.. Alcohol (20%) intake in grams of alcohol per kilogram body weight is shown on the y-axis. Strains are shown on the x-axis. Intake for control and stress group are displayed for each strain. Below each bar graph is the corresponding interval map for the Control and Stress groups. The strength of the association (logarithm of the odds or LOD score) is plotted on the y-axis across the genome (x-axis). The threshold for genom-wide signifcance based on permutation is shown by the horizontal line.
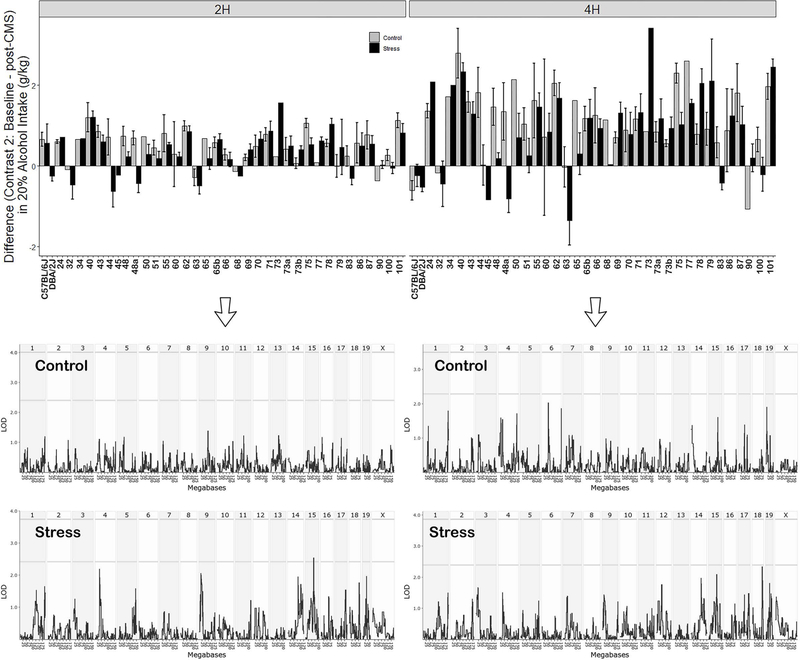
In the second contrast (see Figure 1) we evaluated the average strain difference in intake for the control and stress groups between the Baseline and post-CMS periods (Figure 4). There was a significant strain effect for both the 2 h (F36,154 = 3.87, p < 0.001) and 4 h (F36,154 = 4.09, p < 0.001) access periods. There was also a signifiant treatment effect for both access periods (F1,154 = 4.91 for 2 h and F1,154 = 5.24 for 4 h, p < 0.05). The effect of the interaction was not significant for either access period. The lack of a significant interaction effect for this contrast may indicate that the longer lasting effects of stress on intake are primarily additive instead of interactive with genotype.
Figure 4. Average Strain Difference in Intake for Contrast 2 (Baseline – post-CMS Periods).
Left and right panel display 2h and 4h intake, respectively.. Alcohol (20%) intake in grams of alcohol per kilogram body weight is shown on the y-axis. Strains are shown on the x-axis. Intake for control and stress group are displayed for each strain. Below each bar graph is the corresponding interval map for the Control and Stress groups. The strength of the association (logarithm of the odds or LOD score) is plotted on the y-axis across the genome (x-axis). The threshold for genome-wide significance or suggestive loci based on permutation is shown by the upper and lower horizontal lines, respectively.
Because of the experimental design, we can present the effects of stress on alcohol consumption in two ways. First, we can analyze the contemporaneous effects of stress on alcohol intake by comparing intake in the stress and control group measured during CMS with intake measured during Baseline (Comparison 1, Figure 1). Second, we can analyze the trace effects of stress by comparing post-stress intake in the stress and control group with Baseline intake (Comparison 2, Figure 1). Furthermore, we present the average difference in intake due to period and treatment for both the 2 h and 4 h access periods for each comparison (Figure 5).
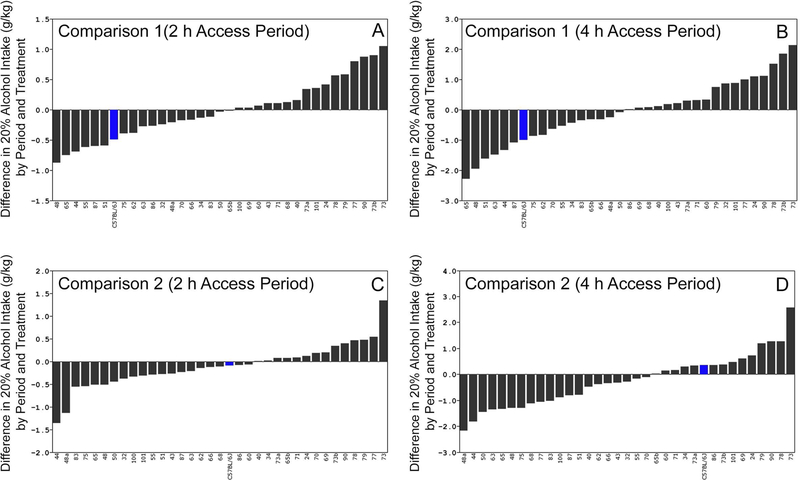
Figure 5. Strain Distribution of Differences for Comparisons 1 and 2.
Consumption (g/kg) represented as mean difference score on the y-axis and strain shown on the x-axis. B6 and D2 filled in blue and red, respectively. Strain distributions organized by rank for each trait. Error bars not shown for difference of differences. Difference in consumption due to chronic stress (Comparison 1) shown in A for 2h intake and B for 4h intake. Difference in consumption following chronic stress shown in C for 2h intake and D for 4h intake. (A) Comparison 1 for 2h intake, CMS group minus control and CMS period (mean of 7 weeks) minus Baseline (mean of 5 weeks). (B) Differences in 4h DID, stress group minus control and CMS period minus Baseline period. (C) Comparison 2, strain distribution of differences in 2h DID, CMS group minus control and post-CMS period minus Baseline. (D) Strain distribution of differences in 4h DID, CMS group minus control and post-CMS period minus Baseline. For Comparison 2, most strains demonstrate a decrease in intake associated with chronic stress relative to baseline (negative value). Some strains, notably BXD73, demonstrate increased voluntary alcohol intake during CMS (Comparison 1) and following CMS (Comparison 2).
Comparison 1, 2 and 4h alcohol intake.
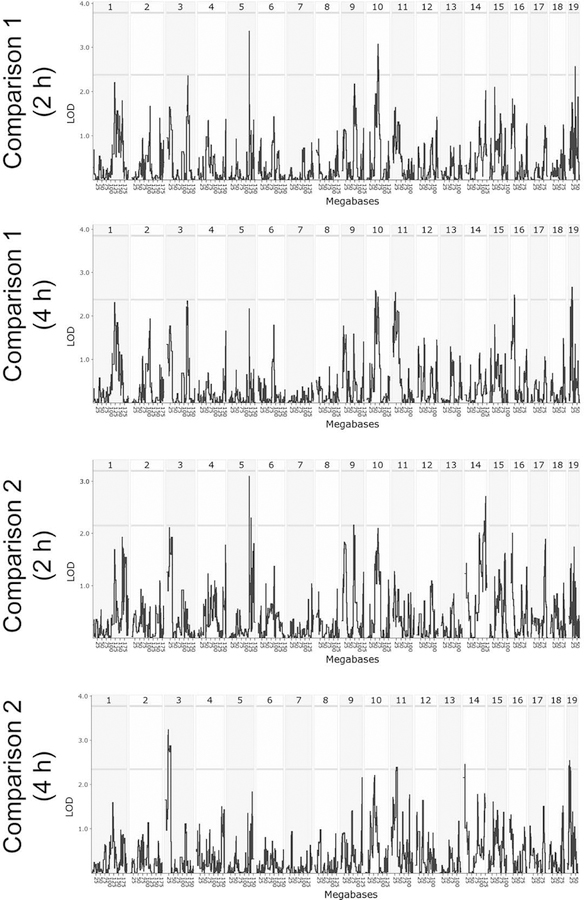
CMS produced a contemporaneous effect on alcohol intake (operationally defined as a 0.5 or 1 g/kg respective change in intake for the 2h and 4 h access periods) that was dependent on strain (Figure 5). CMS decreased 2 h alcohol intake relative to Baseline in some strains (B6 and BXD 51, 87, 55, 44, 65, and 48) and increased alcohol intake in other strains (BXDs 78, 79, 77, 90, 73b, and 73; Figure 5A). Similar results were observed for the 4 h access period. CMS decreased 4 h alcohol intake relative to Baseline in some strains (B6 and BXDs 87, 44, 63, 51, 48, and 65) and increased intake in other strains (BXDs 24, 90, 78, 73b, and 73; Figure 5B). Suggestive loci for the contemporaneous effect of stress (Figure 6) were identified on Chrs 5 (2 h), 10 (2 and 4 h), 11 (4 h), 16 (4 h), and 19 (2 and 4 h).
Figure 6. Suggestive loci for the contemporaneous and trace effects of stress.
QTL maps are provided for 2 h and 4 h access periods for contemporaneous (Comparison 1) and trace (Comparison 2) effects of stress. The strength of the association (logarithm of the odds or LOD score) is plotted on the y-axis across the genome (x-axis). The threshold for genome-wide significance or suggestive loci based on permutation is shown by the top and bottom horizontal lines, respectively. Suggestive loci for the contemporaneous effect of stress were identified on Chrs 5 (2 h), 10 (2 and 4 h), 11 (4 h), 16 (4 h), and 19 (2 and 4 h). Suggestive loci for the trace effect of stress were identified on Chrs 3 (4 h), 5 (2 h), 11 (4 h), 14 (2 h and 4 h), and 19 (4 h).
Comparison 2, 2 and 4h alcohol intake.
CMS produced a trace effect on alcohol intake (operationally defined as a 0.5 or 1 g/kg respective change in intake for the 2h and 4 h access periods) that was dependent on strain (Figure 5). Intake (2 h) during the post-CMS period relative to Baseline was decreased by stress in some strains (BXDs 48a and 44) and dincreased in other strains (BXDs 73 and 77; Figure 5C). Intake during the 4 h access period was also decreased by stress in some strains (BXDs 83, 77, 68, 75, 48, 65, 63, 50, 44, and 48a) and increased in others (BXDs 79, 90, 78, and 73, Figure 5D). Of note, the strain distribution for alcohol intake for the effect of contemporaneous stress (Comparison 1) is fairly evenly distributed. However, the distribution for trace effects (Comparison 2) is left shifted towards decreased consumption. In both comparisons, BXD 73 is a high response strain (increased intake). In contrast, BXDs 48a and 44 are low responder strains only when considering trace effects of stress on alcohol intake in Comparison 2. Suggestive loci for the trace effect of stress (Figure 6) were identified on Chrs 3 (4 h), 5 (2 h), 11 (4 h), 14 (2 h and 4 h), and 19 (4 h).
Evaluation of candidate genes.
A nearly significant (LOD > 3) QTL on Chr 5 is located at 117.790 Mb (peak marker at rs36606069) for the 2 h access period for both contemporaneous and trace effects of CMS on alcohol intake. B6 alleles drive higher alcohol intake for both effects. The 1.5 LOD confidence interval for this QTL is between 117.783 and 117.811 Mb. There is only a single polymorphic gene located near this interval, Nos1 (neuronal nitric oxide synthase).
Additionally, suggestive (LOD = 3) loci on Chr 10 located at 64.7 Mb (peak marker at rs13480625) for 2 and 4 h contemporaneous effects of CMS on alcohol intake and on Chr 3 located at 18.711Mb (peak marker at rs230252925) for trace effects of CMS on alcohol intake were also identified. The confidence interval for the Chr 10 contemporaneous is 10 Mb (between 58.402 to 68.341 Mb) and includes 47 polymorphic genes (STable 2). The confidence interval is slightly larger for the Chr 3 locus (between 16.490Mb to 33.754 Mb) and includes 68 polymorphic genes (STable 2). Brain gene expression in relevent brain regions (amygdala, hippocampus, hypothalamus, and nucleus accumbens) and biological function were used to prioritize candidate genes from each interval that might be involved in mediating the contemporaneous or trace effects of stress on alcohol intake.High priority candidates for the Chr 10 region and contemporaneous stress effects include: P4ha1 (alcohol), Tet1 (alcohol and stress), and Ascc1, Ccar1, Hk1, Jmjd1c, Lims1, Npffr1, Nrbf2, Pcbd1, and Srgn (stress). All genes are polymorphic and expressed at moderate to high levels in relevant brain regions. High priority candidates for the Chr 3 region and trace stress effects include: Actl6a and Slc2a2 (alcohol), Cyp7b1, Hltf, Sec 62, and Tnik (stress), and Cp, Crh, Eif5a2, Mfn1, Nrlgn1, Pik3ca, and Pld1 (stress and alcohol).
Discussion.
We have demonstrated once again that ethanol consumption among BXD strains is highly variable. Moreover, we have also demonstrated that ethanol consumption in response to chronic stress is variable, heritable, and modulated by genes from several different genomic loci. Of great interest is our observation that most of the BXD strains showed decreased ethanol consumption in response to CMS. This result is striking as we previously reported that most BXD strains increase DID ethanol consumption over a 5-week period in the absence of stress (Mulligan et al. 2018). These results support the hypothesis that chronic stress tends to reduce voluntary alcohol consumption in non-dependent individuals. In further support of this hypothesis, a recent study by Anderson and colleagues (2016) revealed that forced swim stress increased alcohol intake in the B6 strain only when stress preceded a model of dependence-like drinking (chronic intermittent ethanol or CIE paradigm; Lopez et al 2017). There was no effect of stress on DID alcohol intake or intake using an intermittent alcohol access paradigm and stress decreased alcohol intake in a continuous access model. Dependence-like drinking in the CIE model has also been measured among BXD strains. However, when comparing our 2 h drinking values (Figure 5, GN trait 20407) for comparisons 1 (contemporaneous effects of stress on alcohol intake) or 2 (trace effects of stress on alcohol intake) with alcohol intake measures (2 h access of 15% ethanol following CIE quantified as the percent change from baseline) in the CIE model (GN trait 12741) the measures are uncorrelated (r15df = −0.1, p > 0.7 for comparison 1 and CIE and r15df = −0.3, p > 0.3 for comparison 2 and CIE). These findings suggest that different genetic mechanisms drive stress-induced alcohol intake in voluntary alcohol intake methods (i.e. DID) compared to those that model dependence-induced consumption (i.e. CIE). Here we demonstrate that stress can alter the trajectory of innate high or low alcohol intake, and in most BXD strains stress was associated with decreased intake. In the design of this research, we chose to use the chronic mild stress model of Willner (1987;2005;2016). In this paradigm, the composite of all perturbations is considered as a whole, and no attempts are made to separate contributions of the various components to the criteria measures. Consequently, we have not attempted to differentiate among the perturbations, including altered light cycle. Whether the changes in DID were affected by light cycle perturbations independently from the others is interesting and the topic for another study.
Despite the general observation that stress decreased DID alcohol intake in the majority of strains, we also observed stress-related increases of at least 0.5 g/kg over 2h access in 4 strains with two related strains (BXD 73 and 73b) showing the greatest increase in the contemporaneous comparison. more than 2 g/kg in 4h access and more than 1 g/kg increase during 2h access. This result is particularly intriguing because BXD 73 is one of several BXD strains that are highly related ~90% genetic background identity). This includes the 48 family (48 and 48a) and the 65 family (65 and 65b) in addition to the 73 family (73, 73a, and 73b). Each family member should demonstrate highly similar patterns of alcohol intake throughout the study owing to their shared genetic background. However, this assumption is not met among the 73 and 65 families (Figure 5) in which at least one family member shows distinctly different patterns of intake (Figures 2,3,4&5). Owing to the reduced background complexity of these strains, identification of the causal variants that produce large phenotypic divergence in stress-induced alcohol intake can be expedited through sequencing, generation of reduced complexity crosses, and fine mapping of candidate loci (Ashbrook et al., 2017).
The BXD panel has been used to identify QTLs that modulate alcohol intake and stress response independently. These previous studies in the BXD panel revealed heritable differences in alcohol intake, endocrine function, and stress response and suggest latent relationships between traits. For example, variation in basal levels of the neurosteroid precursor to corticosterone were correlated with several alcohol related traits in the BXDs (Porcu et al., 2011), suggesting a link between endocrine function, response to stress, and alcohol response. In addition, chronic stress resulted in heritable strain variation in cognitive performance (Jung et al., 2017; Shea et al., 2015) and emotional behavior (Carhuantanta et al., 2014), and these traits were modulated by distinct genetic loci. Remarkably, the QTLs identified in this study do not overlap with the stress- or alcohol-related QTLs associated with the above studies suggesting that they are novel loci modulating voluntary intake following stress. All of these loci are suggestive and the further testing of additional BXD strains will be needed to reproduce and validate these loci. However, one locus on chromosome 5 was associated with variation in both stress-induced contemporaneous and trace alcohol intake and was very close to reaching genome-wide significance (Figure 6). The only candidate gene within this narrow confidence interval is the neuronal nitric oxide synthase gene (Nos1). Rodent Nos1 expression (mRNA and/or protein level) is frequently changed in numerous tissue following alcohol exposure (Finn et al., 2018; Situmorang JG et al., 2018; Silva et al., 2016; Karadayian et al., 2014; Yazir et al., 2012; Bagyánszki et al., 2011; Naassila et al., 2003; Spanagel et al., 2002) and it may exhibit sexually dimorphic expression following binge alcohol consumption in the B6 strain (Finn et al., 2018). The Nos1 locus is polymorphic between the B6 and D2 strains (235 SNPs and 88 small InDels), including a SNP in a putative splice region (rs108543252). All Nos1 variants are non-coding, and the causal variant and genetic mechanism by which variation at this locus may influence behavior has yet to be determined. However, the results of our genetic mapping study combined with evidence for alcohol-induced variation across a number of model systems strongly implicates Nos1 as a genetic mediator of stress-induced alcohol intake.
In addition to Nos 1, we also identified several salient candidate genes for the contemporaneous and trace effects of stress on alcohol intake in the BXD population on Chrs 10 and 3 respectively (STable 1). Several polymorphic genes in the Chr 3 region are especially salient based on their known biological function and their moderate to high expression in brain regions implicated in alcohol intake, addiction, and response to stress (amygdala, hippocampus, hypothalamus, nucleus accumbens). These include Crh, Nlgn1, and Pik3ca. All of these candidates jave a known role in both alcohol and stress responses. Corticotropin-releasing hormone (Crh) is one of the main peptide hormones responsible for transduction of stress responses and HPA axis activation. Alterations in Crh signaling due to stress exposure or genetic modification are associated with changes in alcohol consumption or seeking behavior in rodent models (Yang et al., 2018; Broccoli et al., 2018). Neuroligins are synaptic cell adhesion proteins important for the organization of excitatory and inhibitory synapses. Nlgn1 has been associated with the risk of alcohol dependence in human populations (Zuo et al., 2013) and is thought to contribute to synaptic plasticity associated with fear learning (Stork et al., 2001). The Pik3ca gene encodes the important intracellular signaling enzyme phosphatidylinositol 3-kinase (PI3K). Chronic social defeat stress (Deng et al., 2019) and withdrawal from alcohol (Qiao et al., 2018) both lead to alterations in intracellular signaling cascades mediated in part through changes in the lavel or activity of PI3K activity, suggesting that this gene may mediate the effects of both stress and alcohol. Several candidate genes were also identified in the Chr 10 region, including the ten-eleven translocation protein Tet1 responsible for the important DNA modification, 5-hydroxymethylcytosine. Genetic deletion of Tet1 in mice resulted in lower sensitivity to the effects of chronic restraint stress (Cheng et al., 2018). In addition, in humans, TET1 mRNA tended to be altered in psychotic patients with comorbid chronic alcohol abuse (Guidotti et al., 2013). Although we have identified several salient candidate genes that may mediate the contemporaneous or trace effects of stress on alcohol intake, a caveat of this study is that we were only able to identify suggestive QTL, and with the exception of Nos1, there are several salient candidates within each locus. Ultimately, quantification of gene expression profiles in multiple brain regions from the subjects of this study will lead to identification of the main molecular drivers of stress-induced alterations in alcohol intake in the BXD population. .
The two types of alcohol use disorders concept advanced by Cloninger (1987) and Sigvardsson et al. (1996) has received some criticism as being too simplistic (e.g., Hall and Sannibale, 1996), Indeed, many individuals with the disorder display behaviors of both types. While the typology approach to understanding the disorder may have some heuristic value, it might be better to think of alcohol use disorder etiology as continua of complementary genetic and environmental underpinnings. So, individuals displaying predominantly Type 2 characteristics would be influenced primarily by genetics and those displaying Type 1 characteristics would be under the influence of both genetics and environment and their interactions. While determining familial or genetic influence in Type 2 may be somewhat straightforward, determining such influence in Type 1 is more problematic as the right genotype needs to be exposed to the precipitating environmental conditions. Of course, conducting the relevant studies in humans would be fraught with ethical concerns. This is where a carefully designed animal study would help. By subjecting a genetic reference population of mice to chronic stressors, we have shown that the genetic contribution to chronic stress-related alcohol consumption via heritability estimates is importantly greater than alcohol consumption in the absence of the stress. Moreover, we have been able to nominate several candidate genes that may contribute to the stress-related change in alcohol consumption.
Conclusion.
Differences in susceptibility to the effects of stressful environments vis a vis alcohol use disorders would suggest that the differences have at least some basis in genetic constitution. The results that we present here in a genetic reference population of mice lend initial support for the hypothesis and argue for more extensive study to identify genes and biomarkers that confer differential risk.
Supplementary Material
Table 1.
Correlation matrix (pairwise Pearson product-moment correlations) of strain means of DID differences comparing CMS minus Baseline (Comparison 1) and Baseline post-stress minus Baseline (Comparison 2) periods.
| Phenotype | CMS-Baseline 2h | CMS-Baseline 4h | PostCMS-Baseline 2h |
|---|---|---|---|
| CMS-Baseline, 4h | 0.91 | ||
| Post-stress-Baseline 2h | 0.74 | 0.65 | |
| Post-stress-Baseline 4h | 0.65 | 0.68 | 0.83 |
p<0.001 for all, df=33
Acknowledgments
The authors thank Daming Zhuang and Shuanying Leng for excellent technical assistance. Partial support provided by USPHS Grants R01AA021951 and U01AA013499 Support was also provided by the State of Tennessee, Center for Integrative and Translational Genetics program.
Footnotes
The authors declare no conflicts of interest.
References
- Anderson RI, Lopez MF, Becker HC (2016) Forced swim stress increases ethanol consumption in C57BL/6J mice with a history of chronic intermittent ethanol exposure. Psychopharmacology (Berl) 233:2035–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbrook DG, Mulligan MK, Williams RW. (2018) Post-genomic behavioral genetics: From revolution to routine. Genes Brain Behav 17:e1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagyánszki M, Torfs P, Krecsmarik M, Fekete E, Adriaensen D, Van Nassauw L, Timmermans JP, Kroese AB (2011). Chronic alcohol consumption induces an overproduction of NO by nNOS- and iNOS-expressing myenteric neurons in the murine small intestine. Neurogastroenterol Motil 23(6):e237–48. [DOI] [PubMed] [Google Scholar]
- Belknap JK (1998). Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet 28:29–38. [DOI] [PubMed] [Google Scholar]
- Boyce-Rustay JM, Janos AL, Holmes A. (2008) Effects of chronic swim stress on EtOH-related behaviors in C57BL/6J, DBA/2J and BALB/cByJ mice. Behav Brain Res 186:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli L, Uhrig S, von Jonquieres G, Schönig K, Bartsch D, Justice NJ, Spanagel R, Sommer WH1, Klugmann M, Hansson AC (2018). Targeted overexpression of CRH receptor subtype 1 in central amygdala neurons: effect on alcohol-seeking behavior. Psychopharmacology 236(6):1821–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carhuatanta KA, Shea CJ, Herman JP, Jankord R (2014) Unique genetic loci identified for emotional behavior in control and chronic stress conditions. Front Behav Neurosci 8:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sharp BM (2004). Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics 5:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Sun M, Chen L, Li Y, Lin L, Yao B, Li Z, Wang Z, Chen J, Miao Z, Xin N, Huang L, Allen EG, Wu H, Xu X, Jin P (2018). Ten-Eleven Translocation Proteins Modulate the Response to Environmental Stress in Mice. Cell Rep 25(11) :3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. (2005). Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet 37:233–242. [DOI] [PubMed] [Google Scholar]
- Chester JA, Barrenha GD, Hughes ML, Keuneke KJ. (2008) Age-and sex-dependent effects of footshock stress on subsequent alcohol drinking and acoustic startle behavior in mice selectively bred for high alcohol preference. Alcohol Clin Exper Res 32:1782–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR, Bohman M, Sigvardsson S. (1981) Inheritance of alcohol abuse: cross fostering analysis of adoptive men. Arch Gen Psychiatry 38:861–868 [DOI] [PubMed] [Google Scholar]
- Cloninger CR. (1987). Neurogenetic adaptive mechanisms in alcoholism. Science 236: 410–416. [DOI] [PubMed] [Google Scholar]
- Deng Z, Yuan C, Yang J, Peng Y, Wang W, Wang Y, Gao W (2019). Behavioral defects induced by chronic social defeat stress are protected by Momordica charantia polysaccharides via attenuation of JNK3/PI3K/AKT neuroinflammatory pathway. Ann Transl Med 7(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. (1998) Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc Natl Acad Sci (USA), 85:5274–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farook JM, Lewis B, Littleton JM, Barron S. (2009) Topiramate attenuates the stress-induced increase in alcohol consumption and preference in male C57BL/6J mice. Physiol Behav 96:189–193. [DOI] [PubMed] [Google Scholar]
- Fernandez JR, Vogler GP, Tarantino LM, Vignetti S, Plomin R, McClearn GE (1999). Sex-exclusive quantitative trait loci influences in alcohol-related phenotypes. Am J Med Genet 88:647–652. [DOI] [PubMed] [Google Scholar]
- Finn DA, Hashimoto JG, Cozzoli DK, Helms ML, Nipper MA, Kaufman MN, Wiren KM, Guizzetti M (2018). Binge Ethanol Drinking Produces Sexually Divergent and Distinct Changes in Nucleus Accumbens Signaling Cascades and Pathways in Adult C57BL/6J Mice. Front Genet 9:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill K, Liu Y, Deitrich RA (1996). Voluntary alcohol consumption in BXD recombinant inbred mice: relationship to alcohol metabolism. Alcohol Clin Exp Res 20:185–190. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR (2013). DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcohol Clin Exp Res 37(3):417–24. [DOI] [PubMed] [Google Scholar]
- Hall W, Sannibale C. (1996) Are there two types of alcoholism? The Lancet 348:1258. [DOI] [PubMed] [Google Scholar]
- Hotopf M, David AS, Hull L, Nikalaou V, Unwin C, Wessely S (2003). Gulf war illness--better, worse, or just the same? A cohort study. BMJ 327(7428):1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegmann JP, Possidente B. (1981) Estimating genetic correlations from inbred strains. Behavior Genetics 11(2):103–114. [DOI] [PubMed] [Google Scholar]
- Ibarguen-Vargas Y, Surget A, Touma C, Palme R, Belzung C. (2008) Multifaceted strain-specific effects in a mouse model of depression and of antidepressant reversal. Psychoneuroendocrinology 33:1357–1368. [DOI] [PubMed] [Google Scholar]
- Jung SH, Brownlow ML, Pellegrini M, Jankord R. (2017) Divergence in Morris water maze-based cognitive performance under chronic stress is associated with the hippocampal whole transcriptomic modification in mice. Front Mol Neurosci 10:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellåker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assunção JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. (2011). Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadayian AG, Bustamante J, Czerniczyniec A, Cutrera R, Lores-Arnaiz S (2014). Effect of melatonin on motor performance and brain cortex mitochondrial function during ethanol hangover. Neuroscience 269:281–9. [DOI] [PubMed] [Google Scholar]
- Lopez MF, Miles MF, Williams RW, Becker HC. (2017) Variable effects of chronic intermittent ethanol exposure on ethanol drinking in a genetically diverse mouse cohort. Alcohol 58:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DB, Morrow AL, O’Buckley T, Flanigan TJ, Berry RB, Cook MN, Mittleman G, Goldowitz D, Tokunaga S, Silvers JM. (2008) Acute mild footshock alters ethanol drinking and plasma corticosterone levels in C57BL/6J male mice, but not DBA/2J or A/J male mice. Alcohol 42:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Zhao W, Dickerson M, Arends D, Prins P, Cavigelli SA, Terenina E, Mormède P, Lu L, Jones BC. (2018) Genetic contribution to initial and progressive alcohol intake among recombinant inbred strains of mice. Front Genet 9:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Mozhui K, Prins P, Williams RW (2017). GeneNetwork: A Toolbox for Systems Genetics. Methods Mol Biol 1488:75–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naassila M, Pierrefiche O, Beaugé FJ, Sébire N, Daoust M (2003). Chronic ethanol exposure differentially regulates NOS1 mRNA levels depending on rat brain area. Neurosci Lett 338(3):221–4. [DOI] [PubMed] [Google Scholar]
- Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, Fischer J, Maatz H, Kren V, Pravenec M, Hubner N, Aitman TJ. (2006). Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet 2(10):e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK (1994). Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res 18:931–941. [DOI] [PubMed] [Google Scholar]
- Porcu P, O’Buckley TK, Song SC, Harenza JL, Lu L, Wang X, Williams RW, Miles MF, Morrow AL (2011). Genetic analysis of the neurosteroid deoxycorticosterone and its relation to alcohol phenotypes: identification of QTLs and downstream gene regulation. PLos One 6(4):e18405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Gai H, Su R, Deji C, Cui J, Lai J, Zhu Y (2018). PI3K-AKT-GSK3β-CREB signaling pathway regulates anxiety-like behavior in rats following alcohol withdrawal. J Affect Disord 235:96:104. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC (2005). Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav 84: 53–63. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Ford MM. Yu CH, Brown LL, Finn DA, Garland T Jr, Crabbe JC. (2007). Mouse inbred strain differences in ethanol drinking to intoxication. Genes Brain Behav 6:1–18. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. (1994). Alcohol acceptance, preference, and sensitivity in mice. I. Quantitative genetic analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res 18:1416–1422. [DOI] [PubMed] [Google Scholar]
- Shea CJ Carhuatanta KA, Wagner J, Bechmann N, Moore R, Herman JP, Jankord R (2015) Variable impact of chronic stress on spatial learning and memory in BXD mice. Physiol Behav 150:69–77. [DOI] [PubMed] [Google Scholar]
- Sigvardsson S, Bohman M, Cloninger CR (1996) Replication of the Stockholm Adoption Study of Alcoholism. Arch Gen. Psychiatry 53:681–687. [DOI] [PubMed] [Google Scholar]
- Silva SM, Silva S, Meireles M, Leal S (2015). nNOS is involved in cardiac remodeling induced by chronic ethanol consumption. Toxicology 329:98–105. [DOI] [PubMed] [Google Scholar]
- Situmorang JH, Lin HH, Lo H, Lai CC (2018). Role of neuronal nitric oxide synthase (nNOS) at medulla in tachycardia induced by repeated administration of ethanol in conscious rats. J Biomen Sci 25(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R, Siegmund S, Cowen M, Schroff KC, Schumann G, Fiserova M, Sillaber I, Wellek S, Singer M, Putzke J (2002). The neuronal nitric oxide synthase gene is critically involved in neurobehavioral effects of alcohol. J Neurosci 22(19):8876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork O, Stork S, Pape HC, Obata K (2001). Identification of genes expressed in the amygdala during the formation of fear memory. Learn Mem 8(4):209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambour S, Brown LL, Crabbe JC. (2008) Gender and age at drinking onset affect voluntary alcohol consumption but neither the alcohol deprivation effect nor the response to stress in mice. Alcohol Clin Exp 32:2100–2106. [DOI] [PubMed] [Google Scholar]
- Willner P, Towell A, Sampson D, Sophokleous S, Muscat R. (1987) Reduction of sucrose preference by chronic, unpredictable mild stress and its restoration by a tricyclic antidepressant. Psychopharmacology (Berl) 93:358–364. [DOI] [PubMed] [Google Scholar]
- Willner P. (2005) Chronic mild stress (CMS) revisited: consistency and behavioural-neurobiological concordance in the effects of CMS. Neuropsychobiology 52:90–110. [DOI] [PubMed] [Google Scholar]
- Willner P. (2016) The chronic mild stress (CMS) model of depression: History, evaluation and usage. Neurobiology of Stress 6:78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd, Su AI (2009). BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10(11):R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ju L, Yang C, Xue J, Setlow B, Morey TE, Gravenstein N, Seubert CN, Vasilopoulos T, Martynyuk AE (2018). Effects of combined brief etomidate anesthesia and postnatal stress on amygdala expression of Cl- cotransporters and corticotropin-releasing hormone and alcohol intake in adult rats. Neurosci Lett 685:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang S, Rice KC, Munro CA, Wand GS. (2008) Restraint stress and ethanol consumption in two mouse strains. Alcohol Clin Exp Res 32:840–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazir Y, Tugay M, Utkan Z, Utkan T (2012). Effects of chronic ethanol consumption on rat upper gastrointestinal system: functional and histologic findings. Alcohol 46(7):649–55. [DOI] [PubMed] [Google Scholar]
- Zuo L, Wang K, Zhang X, Pan X, Wang G, Krystal JH, Zhang H, Luo X (2013). Sex chromosome-wide association analysis suggested male-specific risk genes for alcohol dependence. Psychiatr Genet 23(6):233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.