Abstract
Background
LAMA2-related muscular dystrophy including LAMA2-related congenital muscular dystrophy (LAMA2-CMD) and autosomal recessive limb-girdle muscular dystrophy-23 (LGMDR23) is caused by LAMA2 pathogenic variants. We aimed to describe the natural history and establish genotype–phenotype correlations in a large cohort of Chinese patients with LAMA2-related muscular dystrophy.
Methods
Clinical and genetic data of LAMA2-related muscular dystrophy patients enrolled from ten research centers between January 2003 and March 2021 were collected and analyzed.
Results
One hundred and thirty patients (116 LAMA2-CMD and 14 LGMDR23) were included. LAMA2-CMD group had earlier onset than LGMDR23 group. Head control, independent sitting and ambulation were achieved in 76.3%, 92.6% and 18.4% of LAMA2-CMD patients at median ages of 6.0 months (range 2.0–36.0 months), 11.0 months (range 6.0–36.0 months), and 27.0 months (range 18.0–84.0 months), respectively. All LGMDR23 patients achieved independent ambulation at median age of 18.0 months (range 13.0–20.0 months). Motor regression in LAMA2-CMD mainly occurred concurrently with rapid progression of contractures during 6–9 years old. Twenty-four LAMA2-related muscular dystrophy patients died, mostly due to severe pneumonia. Seizures occurred in 35.7% of LGMDR23 and 9.5% of LAMA2-CMD patients. Forty-six novel and 97 known LAMA2 disease-causing variants were identified. The top three high-frequency disease-causing variants in Han Chinese patients were c.7147C > T (p.R2383*), exon 4 deletion, and c.5156_5159del (p.K1719Rfs*5). In LAMA2-CMD, splicing variants tended to be associated with a relatively mild phenotype. Nonsense variants were more frequent in LAMA2-CMD (56.9%, 66/116) than in LGMDR23 (21.4%, 3/14), while missense disease-causing variants were more frequent in LGMDR23 (71.4%, 10/14) than in LAMA2-CMD (12.9%, 15/116). Copy number variations were identified in 26.4% of survivors and 50.0% of nonsurvivors, suggesting that copy number variations were associated with lower rate of survival (p = 0.029).
Conclusions
This study provides better understandings of natural history and genotype–phenotype correlations in LAMA2-related muscular dystrophy, and supports therapeutic targets for future researches.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-021-01950-x.
Keywords: Natural history, Genotype, LAMA2, Muscular dystrophy, Rare diseases
Background
LAMA2-related muscular dystrophy is an autosomal recessive disorder caused by pathogenic variants in LAMA2 gene (OMIM 156,225). LAMA2 is located on 6q22.33 and encodes for laminin-α2 subunit of the heterotrimeric extracellular protein laminin-α2β1γ1 [1]. The clinical spectrum ranges from a severe, early-onset LAMA2-related congenital muscular dystrophy (LAMA2-CMD, OMIM 607,855) to a mild, childhood-onset autosomal recessive limb-girdle muscular dystrophy-23 (LGMDR23, OMIM 618,138) [2, 3]. The phenotypic difference seems to be related not only to the residual amount of laminin-α2, but also to the location and type of pathogenic variants [4, 5]. LAMA2-CMD is one of the most common congenital muscular dystrophies (CMDs) in the world, accounting for 36.4%-48% of CMD patients [6–8]. However, the prevalence of LGMDR23 is not fully known. Although there are a few retrospective, cross-sectional studies exploring the natural history and genetic variations of LAMA2-related muscular dystrophy [4–7], long-term and large-scale studies of the natural history and genotype–phenotype correlations are limited. As promising therapeutic approaches (such as upregulation of LAMA1, mini-agrin, and laminin-α1 LN-domain nidogen-1 (αLNNd)) are getting closer to clinical application [9–12], the definition of natural history endpoints along with clinically relevant outcome measures is essential for both clinical care planning and clinical trial readiness [13].
In this study, we designed a large national multicenter study of LAMA2-related muscular dystrophy patients in the Chinese population. Detailed clinical and genetic data were collected for analyzing the symptom onset, survival length, motor development and regression, occurrence of disease-related complications and genetic features. Our purpose was to identify valid and reliable information of the natural history and genotype–phenotype correlations of LAMA2-related muscular dystrophy.
Results
The study cohort included 130 patients (116 LAMA2-CMD and 14 LGMDR23) (Additional files 1 and 2), 124 (95.4%) were Han Chinese, 79 were male, and 24 died (Fig. 1a). The median age at the last follow-up was 6.4 years (range 0.3–27.3 years) for LAMA2-CMD and 8.2 years (range 3.2–27.0 years) for LGMDR23 patients.
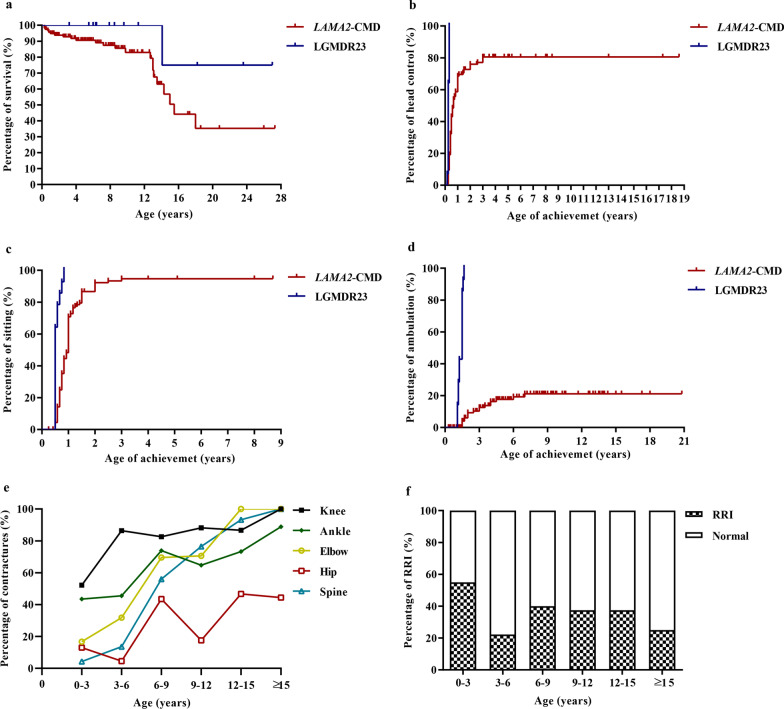
Fig. 1.
The clinical features in patients with LAMA2-related muscular dystrophy. a Kaplan–Meier survival analysis in patients with LAMA2-related muscular dystrophy. b Comparison of achievemet of head control in patients with LAMA2-related muscular dystrophy. c Comparison of achievemet of sitting in patients with LAMA2-related muscular dystrophy. d Comparison of achievemet of ambulation in patients with LAMA2-related muscular dystrophy. e Joint contractures in patients with LAMA2-CMD in different ages. f Recurrent respiratory infection in patients with LAMA2-CMD in different ages. LAMA2-CMD LAMA2-related congenital muscular dystrophy, LGMDR23 autosomal recessive limb-girdle muscular dystrophy-23, RRI recurrent respiratory infection
Clinical characteristics
Onset of symptoms
The median age of symptom onset was 0.0 months (range 0.0–6.0 months) for LAMA2-CMD and 18.0 months (range 13.0–156.0 months) for LGMDR23 (Table 1). Early symptoms appeared within the first week of life in 75.9% (88/116) of LAMA2-CMD patients. The onset symptoms in LAMA2-CMD were variable combinations of severe muscle weakness (n = 114), hypotonia (n = 114), weak cry (n = 48), neonatal feeding difficulty (n = 30), neonatal respiratory difficulty (n = 9). Four patients were treated with mechanical ventilation in the neonatal period. In LGMDR23, the onset symptoms were myopathic gait (n = 10), difficulty running and jumping (n = 3) and epilepsy (n = 1).
Table 1.
Clinical and demographic characteristics of LAMA2-related muscular dystrophy cohort
| LAMA2-CMD | LGMDR23 | |
|---|---|---|
| Age of onset, median (range), months | 0.0 (0.0–6.0) | 18.0 (13.0–156.0) |
| Age of last follow-up, median (range), years | 6.4 (0.3–27.3) | 8.2 (3.2–27.0) |
| Male, % (n) | 62.1 (72/116) | 50.0 (7/14) |
| Survivors, % (n) | 80.2 (93/116) | 92.9 (13/14) |
| Ambulation alone over 1.5 years old, % (n) | 18.4 (18/98) | 100 (14/14) |
| Regression of motor function, % (n) | 31.2 (34/109) | 7.1 (1/14) |
| Spinal deformity, % (n) | 48.6 (54/111) | 7.1 (1/14) |
| Central nervous system involvement | ||
| Cognitive impairment, % (n) | 10.3 (12/116) | 0.0 (0/14) |
| Seizure, % (n) | 9.5 (11/116) | 35.7 (5/14) |
| Typical brain white matter alterations, % (n) | 92.2 (95/103) | 69.2 (9/13) |
| Respiratory involvement | ||
| Neonatal respiratory difficulty, % (n) | 7.7 (9/116) | 0.0 (0/14) |
| Breathing difficulty during follow-up, % (n) | 26.9 (29/108) | 0.0 (0/14) |
| Feeding | ||
| Neonatal feeding difficulty, % (n) | 25.8 (30/116) | 0 (0/14) |
| Chewing difficulty over 2 years old, % (n) | 65.1 (53/90) | 0.0 (0/14) |
| Swallowing difficulty over 2 years old, % (n) | 65.1 (11/90) | 0.0 (0/14) |
LAMA2-CMD LAMA2-related congenital muscular dystrophy, LGMDR23 limb-girdle muscular dystrophy-23
Motor development and regression
Kaplan–Meier survival analysis showed differences in the motor milestones of head control, independent sitting and ambulation between LAMA2-CMD and LGMDR23 (Fig. 1b–d). Among the 114 LAMA2-CMD older than 4 months, 87 (76.3%) achieved head control (65 achieving after 4 months old) at median age of 6.0 months (range 2.0–36.0 months), but 27 never achieved; delayed head control was observed in 80.7% ((65 + 27)/114) of LAMA2-CMD. Among the 107 LAMA2-CMD older than 10 months and one nine-month-old patient with independent sitting, 100 (92.6%) achieved independent sitting (51 achieving after 10 months old) at median age of 11.0 months (range 6.0–36.0 months), 8 never achieved; delayed independent sitting was observed in 54.6% ((51 + 8)/108) of LAMA2-CMD. Among the 98 LAMA2-CMD older than 18 months, 18 (18.4%) achieved independent ambulation (14 achieving after 18 months old) at median age of 27.0 months (range 18.0–84.0 months), 80 never achieved; delayed independent ambulation was observed in 95.9% ((14 + 80)/98) of LAMA2-CMD. In LGMDR23, all patients achieved independent ambulation at median age of 18.0 months (range 13.0–20.0 months). Five LGMDR23 had difficulty in running and jumping. Motor regression assessed in 109 LAMA2-CMD was observed in head control (n = 7), rolling (n = 9), independent sitting (n = 17) and ambulation (n = 9), at median (range) ages of 9.8 (6.8–11.0), 6.0 (3.8–12.0), 8.0 (4.1–19.0) and 8.0 (1.7–11.0) years, respectively. However, only one LGMDR23 gradually lost ambulation at 5.5 years of age.
Central nervous system involvement
Seizures occurred in 9.5% (11/116) of LAMA2-CMD patients (night with epilepsy and two with febrile seizures) and 35.7% (5/14) of LGMDR23 patients (three with epilepsy and two with febrile seizures). Only one LGMDR23 patient (P123) had a family history of epilepsy. The seizure types in LAMA2-CMD were broad, including focal seizures, generalized seizures, myoclonus, typical or atypical absence seizures. Three LGMDR23 patients showed focal seizures with paroxysmal blurry vision, headache and vomiting (n = 1), clonic seizure of the limb (n = 1) and facial twitching (n = 1). The results of electroencephalography showed slow wave in occipital and posterior temporal region. The median age of the first epileptic episode was 13 years (range 0.1–15.5 years) in LAMA2-CMD, and three LGMDR23 patients developed the first epileptic episode at 11, 14 and 23 years of age, respectively. In addition, two LAMA2-CMD patients died of status epilepticus. Delayed development of cognitive function (n = 10) and regression of cognitive function following epilepsy (n = 2) were found in LAMA2-CMD, but only four of them underwent Intelligence Quotient tests.
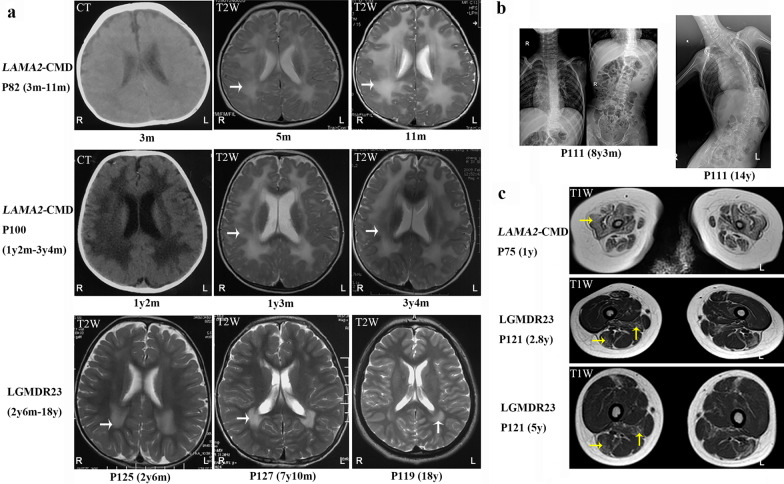
A total of 92.2% (95/103) of LAMA2-CMD patients showed widespread abnormal white matter hyperintensities on T2-magnetic resonance imaging (MRI) (Fig. 2a) as previously described [14]. In the other eight LAMA2-CMD, brain white matters on MRIs performed at ages of 5 days to 4 months were normal or focal changes, while without reexamination later. Except nine LGMDR23 patients with widespread abnormal white matter hyperintensities on T2-MRI, four LGMDR23 showed milder white matter changes predominately in the anterior and/or posterior horns of lateral ventricle (Fig. 2a). Brain MRIs of 53 LAMA2-related muscular dystrophy patients were reassessed, occipital pachygyria (18.7%, 10/53) and pontine hypoplasia (15.1%, 8/53) were found (Additional file 1). Among the ten patients with occipital pachygyria, only three had epilepsy, the correlation between occipital pachygyria and epilepsy was not observed.
Fig. 2.
The image features in patients with LAMA2-related muscular dystrophy. a Changes of brain white matter (white arrows) in LAMA2-related muscular dystrophy in different ages. b Spinal deformity (scoliosis) of patient P111 with age. c Thigh muscle MRIs. Diffuse fatty infiltration (yellow arrows) in patient P75 (1y), fatty infiltration (yellow arrows) involved in the adductor magnus and long head of biceps femoris of patient P121 (2.8y and 5y). LAMA2-CMD LAMA2-related congenital muscular dystrophy, LGMDR23 autosomal recessive limb-girdle muscular dystrophy-23
Orthopedic complications
Joint contractures assessed in 109 LAMA2-CMD patients involved the knees at first, then the ankles, elbows and hips in sequence (Fig. 1e). They progressed rapidly during 6–9 years old with the rate of 82.6%, 73.9%, 69.6% and 43.5%, respectively. Scoliosis (Fig. 2b) occurred in 40.5% (45/111) of LAMA2-CMD at median age of 6.0 years (range 0.5–12.0 years), and lordosis occurred in 8.1% (9/111) of LAMA2-CMD at median age of 3.0 years (range 2.0–7.0 years). Pectus carinatum, pectus excavatum and hip dislocation were observed in 19.0% (22/116), 12.1% (14/116) and 9.5% (11/116) of LAMA2-CMD, respectively. However, only four LGMDR23 had either one or two symptoms of tightened Achilles tendons, contractures of ankles and knees, and mild scoliosis.
Respiratory involvement
In total, 58.9% (63/107) of LAMA2-CMD and 35.7% (5/14) of LGMDR23 patients had a history of recurrent respiratory infection, mainly at ages of 0–3 years and 6–15 years (Fig. 1f). Eighteen LAMA2-CMD patients died of respiratory failure following severe pneumonia. Besides respiratory difficulty in the neonates and following severe pneumonia, 11 LAMA2-CMD developed dyspnea at median age of 11 years (range 6–24 years). Pulmonary function tests and/or polysomnographic examination were performed in 12 LAMA2-CMD at 4–18 years old, the main findings were respiratory insufficiency with restrictive ventilation dysfunction and nocturnal hypoxemia. However, only four LAMA2-CMD underwent mechanical ventilation during follow-up (at ages of 8.2, 11.2, 12.6 and 18.9 years). Although no LGMDR23 had symptoms of breathing difficulty, the pulmonary function test of P126 showed moderate to severe restrictive lung disease at 8.7 years old.
Cardiac involvement and feeding problem
Electrocardiography (ECG) and echocardiogram were performed in 63 patients (Additional file 1). ECG showed normal ECG or nonspecific changes. Echocardiogram mainly showed normal function or mild valvular regurgitation, except one decreased left ventricular ejection fraction (LVEF) of 45% in P80 at 1.8 years old. The LVEF became normal after treatment with captopril (an angiotensin converting enzyme inhibitor) for two years. In LAMA2-CMD patients over two years old, feeding problems such as chewing difficulty (58.9%, 53/90) and swallowing difficulty (12.2%, 11/90) were common. Only patients P87 and P94 underwent nasogastric feeding. P102 and P112 died of malnutrition and cachexia following swallowing difficulty at 13.5 and 18 years old, respectively.
Survival
Twenty-four patients (23 LAMA2-CMD and one LGMDR23) (Additional file 2) died at median age of 7.9 years (range 0.3–18.0 years). The remaining 106 patients were still alive at the last follow-up with median age of 6.5 years (range 0.5–27.3 years). The accumulative survival rate of LAMA2-CMD patients was approximately 50% by the age of 15 years (Fig. 1a). There were two peaks in mortality: the first year of life (13/24) and 12–15 years old (8/24). The causes of death in LAMA2-CMD included respiratory failure following severe pneumonia (18/23, 78.3%), status epilepticus (2/23, 8.7%) and malnutrition following swallowing difficulty (2/23, 8.7%). One LAMA2-CMD (P84) and one LGMDR23 (P127) died of sudden death with unknown reason, but the LAMA2-CMD patient had sustained vomiting one week before death.
Other diagnostic tests
Immunohistochemistry staining of laminin-α2 showed complete laminin-α2 deficiency in 19 LAMA2-CMD, and partial deficiency in three LAMA2-CMD and two LGMDR23 patients. Therefore, complete laminin-α2 deficiency was associated with LAMA2-CMD (p = 0.036). A total of 3/19 patients with complete laminin-α2 deficiency and 3/5 patients with partial laminin-α2 deficiency achieved ambulation (p = 0.078). The thigh muscle MRIs of 14 LAMA2-CMD showed diffuse fatty infiltration in the adductor magnus, gluteus maximus, quadriceps femoris muscle and biceps femoris (Fig. 2c). For four LGMDR23, fatty infiltration in the mid-thigh level frequently and selectivity involved in the adductor magnus and long head of biceps femoris (Fig. 2c).
Clinical characteristics correlations
Two peaks in mortality, the first year of life and 12–15 years old, were consistent with peaks of recurrent respiratory infection. In LAMA2-CMD, survival was related to head control (p = 0.022) and sitting ability (p = 0.010), and epilepsy was associated with lower rate of survival (p = 0.015) (Additional file 3). In LAMA2-CMD, spinal deformity was associated with motor regression (p < 0.001). Compared with non-ambulatory LAMA2-related muscular dystrophy patients, epilepsy was associated with the ambulatory group (p = 0.008). Seizures were more frequent in LGMDR23 (35.7%) than in LAMA2-CMD (9.5%).
Genetic characteristics
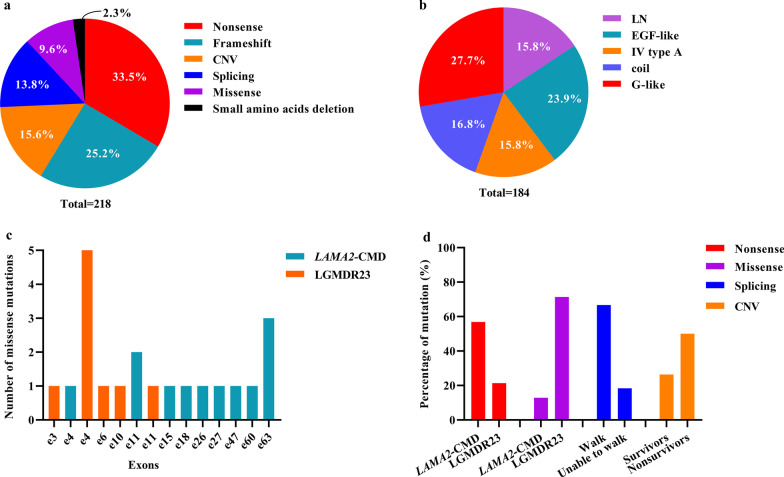
Pathogenic variants were detected in 130 patients from 111 families. Five probands’ parents were consanguineous; proband P78 and patient P19 were second cousins sharing the same great grandparent. We identified 46 novel and 97 known pathogenic variants in LAMA2 gene (Fig. 3a and Additional file 4). Among the known pathogenic variants, 54 were first reported in our previous works [8, 14–16]. The disease-causing variants in 218 (n = 111 × 2 + 1 – 5) alleles included nonsense (73/218, 33.5%), frameshift (55/218, 25.2%), CNVs (34/218, 15.6%), splicing (30/218, 13.8%), missense (21/218, 9.6%), and small amino acid deletions (5/218, 2.3%) (Fig. 4a).
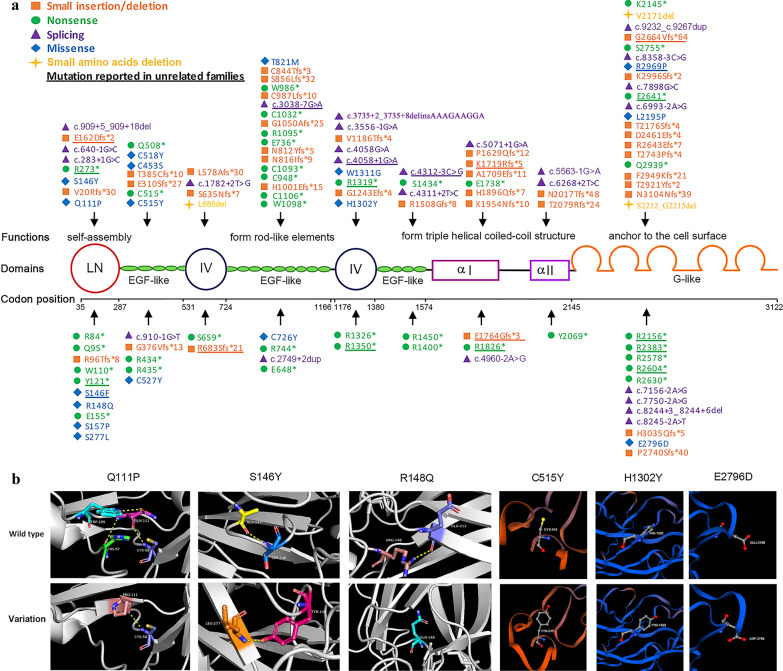
Fig. 3.
Schematic of the LAMA2 disease-causing variants. a The domain structure of laminin-α2 is illustrated. Disease-causing variants (except copy number variations) including 44 novel disease-causing variants and 36 disease-causing variants first reported in our previous works in our cohort are above the chain, and other known disease-causing variants are below. b Four novel missense variants (p.Q111P, p.S146Y, p.C515Y, p.H1302Y) and two known missense variants (p.R148Q, p.E2796D) are predicted by modeling three-dimension structure of the protein. α I laminin helical coiled-coil domain I, α II laminin helical coiled-coil domain II, IV laminin IV type A1 or A2, LN laminin N-terminal
Fig. 4.
Genetic characteristics and genotype–phenotype correlations in LAMA2-related muscular dystrophy. a Six types of disease-causing variants in 218 alleles. b Distribution of 184 allele mutations (except CNVs). c Distribution of missense disease-causing variants in LGMDR23 and LAMA2-CMD. d Nonsense in LAMA2-CMD patients, missense disease-causing variants in LGMDR23 patients, splicing disease-causing variants in ambulatory LAMA2-CMD patients and CNVs in nonsurvivors were more frequent. CNVs copy number variations; Coil laminin helical coiled-coil domain, IV type A laminin IV type A1 or A2, LN laminin N-terminal, LAMA2-CMD LAMA2-related congenital muscular dystrophy, LGMDR23 autosomal recessive limb-girdle muscular dystrophy-23, Splicing splicing disease-causing variant
It should be pointed out that based on the American College of Medical Genetics and Genomics (ACMG) variant-classification, among the four novel and 13 known missense variants, 15 were classified as variant of uncertain significance (VUS), one as likely pathogenic variant, and one as pathogenic variant (Additional file 5). Three novel variants p.Q111P, p.S146Y and p.C515Y were predicted to be damaging by four softwares, p.H1302Y was predicted to be damaging by three softwares (Additional file 5). These amino acid substitutions were on the conserved amino acid sequences, and were predicted to be damaging by modeling three-dimension structure of the protein (Fig. 3b). Among the 13 known missense variants, 12 were predicted to be damaging, but p.E2796D was predicted to be tolerated (Additional file 5). However, p.E2796D was located on the hotspot pathogenic variants region of G-like domain, the glutamic acid residue was conserved, the variant was present in population with 0.00016 (Exome Aggregation Consortium) and in accord with segregation in the family, the patient was clinically diagnosed with LAMA2-CMD without CNV. Therefore, p.E2796D was identified as a pathogenic variant. Notably, the variant c.2462C > T with a conflicting pathogenicity was classified as ACMG: 3 (VUS) in the supplementary data, while was still considered to be a disease-causing variant in this study. In our cohort, it was homozygous in two siblings with LAMA2-CMD from consanguineous parents, and was co-segregating in the family. c.2462C > T was predicted to be damaging by four tolerance predictors, was estimated with a low frequency ranging from 0.00080 (1000 Genomes Project) to 0.00238 (NHLBI Exome Sequencing Project (ESP) Exome Variant Server). Moreover, there were another three unrelated patients with LAMA2-related muscular dystrophy carrying the variant (https://www.ncbi.nlm.nih.gov/clinvar/variation/162584/). Furthermore, a missense variant at the same residue (T821P) has been reported as pathogenic/likely pathogenic without conflicts [2]. Two variants, c.4058G > A changing the last base of exon 27 and c.7898G > C changing the last base of exon 56, were predicted to produce abnormal splice sites and were classified as splicing variants.
Most of the 184 mutant alleles (except CNVs) were distributed in the laminin N-terminal (LN) (29/184, 15.8%), EGF-like domain (44/184, 23.9%), and G-like domain (51/184, 27.7%) (Fig. 4b). The 34 CNVs were mainly distributed in the LN and G-like domains. Six high-frequency pathogenic variants in Han Chinese patients were c.7147C > T (p.R2383*) (n = 11), exon 4 deletion (n = 10), c.5156_5159del (p.K1719Rfs*5) (n = 8), c.2049_2050del (p.R683Sfs*21) (n = 5), c.7921G > T (p.E2641*) (n = 5) and c.4048C > T (p.R1350*) (n = 5). Deletion of exon 4 was a founder pathogenic variant in Han Chinese patients [15].
Genotype–phenotype correlations
Genotype–phenotype correlations in patients with LAMA2-related muscular dystrophy were shown in Additional file 6 and Fig. 4c, d. Most of splicing variants (96.7%, 29/30) were found in the LAMA2-CMD patients but only one in the LGMDR23 patients. In 98 patients with LAMA2-CMD over 18 months old, splicing variants were found in 66.7% (12/18) of ambulatory patients and 17.5% (14/80) of non-ambulatory patients. Additionly, in 107 patients with LAMA2-CMD over 10 months old along with one nine-month-old patient with sitting, splicing variants were found in 12.5% (1/8) of patients never achieving sitting, 15.6% (8/51) of patients achieving sitting over than 10 months old, and 38.7% (19/49) of patients achieving sitting within 10 months old. Therefore, splicing variants tended to be associated with a relatively mild phenotype in LAMA2-CMD. Nonsense was more frequent in LAMA2-CMD patients (56.9%, 66/116) than in LGMDR23 patients (21.4%, 3/14), consequently, combinations containing nonsense (frameshift + nonsense, nonsense + nonsense, nonsense + CNV, and nonsense + splicing) were mainly found in LAMA2-CMD patients. The most high-frequency nonsense c.7147C > T was only detected in LAMA2-CMD patients. However, missense variants were more frequent in LGMDR23 patients (71.4%, 10/14) than in LAMA2-CMD patients (12.9%, 15/116), especially missense pathogenic variants in exon 4 (Fig. 4c, d). The proportion of CNV as well as frameshift was similar between LAMA2-CMD and LGMDR23. However, 26.4% of survivors and 50.0% of nonsurvivors had CNV, showing that CNV was associated with lower rate of survival (p = 0.029). On the whole, the damages of the types of disease-causing variants in LAMA2-MD were in the order: nonsense/frameshift/CNV > splicing > missense variants.
Discussion
Though muscular dystrophies due to LAMA2-related muscular dystrophy is relatively rare, they cover a distinct clinical presentations and severity with high disability and disease burden, and the further mechanism how the pathogenic variants affect the function of laminin-α2 leading to clinical heterogeneity is still unclear. Recently, two natural history studies on 46 LAMA2-related muscular dystrophy pediatric patients in the Dubowitz Neuromuscular Centre [5] and 24 LAMA2-related muscular dystrophy patients in National Institutes of Health [17] provided useful information towards trial readiness. However, international multicenter studies with detailed knowledge on long-term progression of disease as well as genotype–phenotype correlations remain challenging. Here, we report a comprehensive natural history and genetic analysis in the largest LAMA2-related muscular dystrophy cohort thus far, to address clinical and genotypic predictors of phenotypes and disease progression. This cohort represents a subgroup of particular medical needs due to severe clinical manifestations.
In the current study, we have reported that the symptom onset of LGMDR23 patients at 1–3 years old was common. This contrasted with previous studies showing the ages of symptom onset ranging from 1–3 years old to 17.0 ± 7.5 years old in LGMDR23 patients [18–20]. This might be due to the later recognization of the mild clinical presentations, limited number and different demography of the recruited patients. Therefore, for patients with symptoms of muscular dystrophy in childhood, the possibility of LGMDR23 should be considered and brain MRI scan should be further performed. Similar with the variable phenotypes in LMNA-related muscular dystrophy and POMT1-related disorders [21, 22], LAMA2-CMD and LGMDR23 could be viewed as a continuum spectrum. LAMA2-CMD patients presented with more earlier and severe muscle weakness and hypotonia at onset. Therefore, based on the age and symptoms of onset, it was relatively easy to distinguish the two groups. However, the detailed data between them were little known, especially the disease progression.
The stage of motor function directly affected patients’ quality of life. In LAMA2-CMD, head control and independent sitting were achieved in 76.3% and 92.6%, while delayed in 80.7% and 54.6%, respectively; only 18.4% of LAMA2-CMD achieved ambulation. Notably, motor regressions were frequent in LAMA2-CMD [23], and occurred concurrently with joint contractures and spinal deformity during the critical period of 6–9 years old. These suggested that most LAMA2-CMD patients could not cope with everyday activities, and needed help and care. Preventing or delaying orthopedic complications with physical therapy in the early stage could be beneficial for improving motor function [24, 25]. Orthopedic surgery might be needed for severe contractures and spinal deformities [26, 27]. More over, achievements of head control and independent sitting were predictors of longer survival in LAMA2-CMD. However, LGMDR23 showed mild and slowly progressive muscle weakness. Therefore, the evaluation of motor function is necessary for the better understanding of the two phenotypes and the better management of disease.
We only found complete laminin-α2 deficiency was associated with LAMA2-CMD, but did not observe the reported histochemical difference between ambulatory and non-ambulatory patients [4, 6, 28], this might due to the limited number of muscle biopsies available. Considering gene test becoming more and more precise, immunohistochemical staining not always corresponding to the disease severity and the limited availability of muscle biopsy on infantile patients, muscle biopsy may not be necessary for the diagnostic criteria. The pattern of muscle involvement on thigh MRI showed that fatty infiltration in the mid-thigh level predominantly and severely involved the anterior and posterior thigh muscles in LAMA2-CMD, while selectively involved the adductor magnus and long head of biceps femoris in LGMDR23. Thigh muscle MRI might be useful to predict motor function and assess disease progression, and was worthy of further investigations [29, 30].
Other disease-related complications, including respiratory involvement, seizures and feeding difficulty, not only affected the quality of life, but also affected the survival. Although early death was reported in LAMA2-CMD [4, 31, 32], long term survival has not been well documented in large cohorts. A total of 19.8% of LAMA2-CMD patients died during follow-up, the accumulative survival rate by the age of 15 years was approximately 50%. The causes of death in LAMA2-CMD were respiratory failure and pneumonia (78.3%), status epilepticus (8.7%) and malnutrition (8.7%). Respiratory insufficiency could manifest with recurrent chest infection starting from first two years of life [5], and the need for ventilation longer than 4 weeks was reported as a predictor of sudden death [33]. Therefore, LAMA2-CMD patients should receive standardized respiratory care according to current guidelines [34]. Seizures were reported in 8–50% of LAMA2-CMD [35–37], while were found in 35.7% of LGMDR23 and 9.5% of LAMA2-CMD patients in this study. Focal seizures with visual aura and autonomic signs including nausea might not be identified, and electroencephalography was not easily accessed to for the older patients, the incidence of seizures in LAMA2-CMD might be undervalued. It was worth mentioning that 5/9 LAMA2-CMD patients with epilepsy and all three LGMDR23 patients with epilepsy showed their first epileptic episode after age of 10 years, indicating that epilepsy might be more frequent in older children with LAMA2-related muscular dystrophy. Contrary to previous reports [38, 39], no correlation between occipital pachygyria and epilepsy was found in our cohort. Even though, epilepsy should be monitored, especially in older children with LAMA2-related muscular dystrophy. Malnutrition caused by feeding difficulty was also a common problem in LAMA2-CMD. In previous studies, 47.4% of LAMA2-CMD patients had feeding difficulty and required gastrostomy or nasogastric feeding before five years old [4, 40]. However, few LAMA2-CMD patients in this study underwent nasogastric feeding. Therefore, weight monitoring from early infancy and swallowing evaluation should focus on addressing feeding difficulties to minimize the failure to thrive. In a word, early prevention and interventions of complications were essential for the better quality of life and longer survival of LAMA2-CMD patients.
Splicing variant was previously thought to be more common in patients with LGMDR23 [2]. However, we found splicing variant occurred mainly in patients with LAMA2-CMD in the current study. Moreover, splicing variant tended to be associated with a relatively mild phenotype in LAMA2-CMD. Leaky splicing was reported to reduce the symptoms and self-improve clinical phenotype in some genetic diseases such as spinal muscular atrophy and neurofibromatosis type 1 [41, 42]. The impact of the different splicing variants might be related with a leaky splicing or other mechanisms which should be further studied. Recently, the adeno-associated virus carrying clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 was reported to correct a splice-site pathogenic ariant in dy2J/dy2J mouse [43]. Patients with splicing variants might benefit from the splice modulating therapy in the future. We also found that LAMA2-CMD was related to nonsense variants, LAMA2-related muscular dystrophy with nonsense variants such as c.7147C > T (p.R2383*) might benefit from the stop codon readthrough. We also confirmed that missense pathogenic variants was associated with LGMDR23, so missense pathogenic variants could be a predictor of the milder phenotype. Nearly ten percent of the pathogenic variants involved exon 4, and missense and nonsense pathogenic variants in exon 63 were only found in patients with LAMA2-CMD. Based on these findings, we speculate that exon 4 is important for laminin-211 self-assembly and exon 63 plays an important role in the function of the G-like domain. Potential therapeutic interventions including splice modulation, upregulation of the LAMA1 gene, mini-agrin and αLNNd are in development for LAMA2-related muscular dystrophy [9, 10, 38]. More knowledge of the genotype–phenotype correlations and the pathomechanism will lead to more discoveries for the therapy.
Limitations: This study has provided useful data regarding natural history and genetic features of LAMA2-related muscular dystrophy with some limitations. Even though the number of patients was substantial, most of patients included in the current study were pediatric patients with LAMA2-MD, and only limited data from adolescent and adult patients were available, which might affect the assessments especially for complications. Moreover, the number of patients with the LGMDR23 was relatively small. Considering a relative disproportion (116/14) between LAMA2-CMD and LGMDR23 subtypes, and the necessity of more LGMDR23 patients for adequate statistical power, we just did descriptive statistical analysis between the two subgroups. Finally, the number of muscle biopsies available was limited, the study of correlation between histochemical difference of laminin-α2 with the disease severity was affected.
Conclusion
In summary, in the current study, the natural history and genetic features of LAMA2-related muscular dystrophy were characterized. The study provides important information and basis for differentiating LAMA2-related muscular dystrophy subtypes and different focus of multidisciplinary management of LAMA2-related muscular dystrophy subtypes. These results should be valuable to future therapeutic trials and be crucial for future discovery of gene therapies.
Methods
Patients and study design
LAMA2-related muscular dystrophy patients were enrolled between January 2003 and March 2021 at ten research centers in China. Clinical evaluations at enrollment and annual follow-up visits were collected, the medical records were retrospectively reviewed. The inclusion criteria were a clinical and genetical diagnosis of LAMA2-related muscular dystrophy characterized by muscle weakness or hypotonia, delayed motor developmental milestones, hypercreatine kinasemia, abnormal white matter hyperintensities on T2-MRI, and pathogenic variants in LAMA2 gene. Based on the age of onset and disease severity, patients were divided into two subgroups: LAMA2-CMD and LGMDR23 [3]. Patients belonged to LAMA2-CMD if they presented with hypotonia and muscle weakness within the first six months of life, and/or delayed motor milestones in the first year of life. Patients who had mild proximal muscle weakness but with normal motor milestones in the first year of life and independent ambulation were classified as LGMDR23. Achieving or never achieving head control after 4 months old, independent sitting after 10 months old, or independent walking after 18 months old were considered as delayed motor milestones.
Clinical data and outcome measures
The primary clinical outcome measures included the onset, motor development and regression, multiple system complications and survival length. Routine laboratory data including serum creatine kinase, ECG, echocardiogram, brain MRI, thigh muscle MRI, and electromyography (EMG) were collected. Muscle biopsies were also performed, and laminin-α2 expression was detected with mAb 1922 (1:5000,100 μL; 5H2, Merck Millipore, Darmstadt, Germany). The correlations between phenotypes of the disease and clinical outcomes were analyzed.
LAMA2 pathogenic variants analysis
Genomic DNA was extracted from peripheral blood lymphocytes. Direct sequencing of LAMA2 gene before year 2013 and a custom-designed modified next-generation sequencing (NGS) starting in year 2013 were performed to identify the pathogenic variants of LAMA2 gene [14]. Multiplex ligation-dependent probe amplification (MLPA) (SALSA MLPA kit P391-A1/P392-A1, MRC-Holland, Amsterdam, The Netherlands) and custom-designed high-resolution LAMA2-targeted array-based comparative genomic hybridization (SurePrint G3 Microarray, 4 × 180 K) were performed to identify CNV [15]. The pathogenicity of identified variants was evaluated based on the ACMG guidelines. The pathogenicity of missense variants was scored using Polyphen-2, SIFT, Mutation Taster and CADD based on the recommendation of the ACMG guidelines. Moreover, the genotype characteristics and genotype–phenotype correlations (the secondary outcomes) were analyzed.
Statistical analysis
The non-normal distributed data were expressed as median (range). Pearson χ2 test, Fisher’s exact test, and Mann–Whitney U test were used wherever appropriate. GraphPad Prism 5 software (GraphPad Software, La Jolla, CA) were used to create Kaplan–Meier survival curves and graphs of clinical and genetic features. Statistical analyses were performed using SPSS (version 19.0; IBM-SPSS, Chicago, IL). Two-sided p < 0.05 was considered to be statistically significant.
Supplementary Information
Additional file 1. Clinical findings of patients with LAMA2-related muscular dystrophy.
Additional file 2. The ages of patients with LAMA2-related muscular dystrophy.
Additional file 3. Effect of motor and epilepsy on survival of LAMA2-CMD.
Additional file 4. Genetical analysis of patients with LAMA2-related muscular dystrophy.
Additional file 5. Pathogenicity analysis of 17 missense variants.
Additional file 6. Genotype-phenotype correlations in patients with LAMA2-related muscular dystrophy.
Acknowledgements
The authors are grateful to our research study participants and their families. The authors are thankful to Dr. Ching H. Wang for his critical reading and editing of the manuscript. The authors also deeply appreciate the study coordinators and supporting staff who helped acquiring clinical data and samples during the many years of this study.
Abbreviations
- αLNNd
Laminin-α1 LN-domain nidogen-1
- ACMG
American college of medical genetics and genomics
- CMD
Congenital muscular dystrophy
- CNVs
Copy number variations
- CRISPR
Clustered regularly interspaced short palindromic repeats
- ECG
Electrocardiography
- EMG
Electromyogram
- LN
Laminin N-terminal
- LAMA2-CMD
LAMA2-related congenital muscular dystrophy
- LGMDR23
Autosomal recessive limb-girdle muscular dystrophy-23
- LVEF
Left ventricular ejection fraction
- MLPA
Multiplex ligation-dependent probe amplification
- MRI
Magnetic resonance imaging
- NGS
Next-generation sequencing
- VUS
Variant of uncertain significance
Authors' contributions
DT and LG conducted the patient follow-up and wrote the draft of the manuscript. YF, CW, JD, AL, SW, KG and HY collected the clinical and laboratory data and followed up on patients. XC analyzed muscle biopsies. SW conducted and analyzed the EMG. XL helped the statistical analysis of the data.YZ analyzed the thigh muscle MRIs of the patients. BM, BJ, YH, XZ, BZ, WZ, CZ, YW and YY provided patients data as multi-center participants. CQ, ZH and CL analyzed the disease-related complications. YJ, AR, CB and XW took part in study design and critical discussions. HX conceived the study, and participated in the study design and coordination, and the manuscript revision. All authors participated in drafting and critically revising the article and approved the final manuscript.
Funding
This study was supported by grants from the Natural Science Foundation of Beijing Municipality (No. 7212116); National Natural Science Foundation of China (No. 81571220); National Key Research and Development Program of China (No. 2016YFC0901505); Beijing Key Laboratory of Molecular Diagnosis and Study on Pediatric Genetic Diseases (No. BZ0317).
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request. All data relevant to the study are included in the article and its supplementary information files.
Declaration
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Peking University First Hospital (No. 2015[916], Beijing, China). All participants and/or their parents signed an informed consent to participate in the study, and they could withdraw from the study anytime without given reasons.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dandan Tan and Lin Ge contributed equally to this work.
References
- 1.Yurchenco PD, McKee KK, Reinhard JR, Rüegg MA. Laminin-deficient muscular dystrophy: molecular pathogenesis and structural repair strategies. Matrix Biol. 2018;71–72:174–187. doi: 10.1016/j.matbio.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliveira J, Gruber A, Cardoso M, Taipa R, Fineza I, Gonçalves A, et al. LAMA2 gene mutation update: toward a more comprehensive picture of the laminin-α2 variome and its related phenotypes. Hum Mutat. 2018;39:1314–1337. doi: 10.1002/humu.23599. [DOI] [PubMed] [Google Scholar]
- 3.Straub V, Murphy A, Udd B. LGMD workshop study group. 229th ENMC international workshop: limb girdle muscular dystrophies—Nomenclature and reformed classification Naarden, the Netherlands, 17–19 March 2017. Neuromuscul Disord. 2018;28:702–10. [DOI] [PubMed]
- 4.Geranmayeh F, Clement E, Feng LH, Sewry C, Pagan J, Mein R, et al. Genotype-phenotype correlation in a large population of muscular dystrophy patients with LAMA2 mutations. Neuromuscul Disord. 2010;20:241–250. doi: 10.1016/j.nmd.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Zambon AA, Ridout D, Main M, Mein R, Phadke R, Muntoni F, et al. LAMA2-related muscular dystrophy: natural history of a large pediatric cohort. Ann Clin Transl Neurol. 2020;7:1870–1882. doi: 10.1002/acn3.51172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sframeli M, Sarkozy A, Bertoli M, Astrea G, Hudson J, Scoto M, et al. Congenital muscular dystrophies in the UK population: clinical and molecular spectrum of a large cohort diagnosed over a 12-year period. Neuromuscul Disord. 2017;27:793–803. doi: 10.1016/j.nmd.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Abdel Aleem A, Elsaid MF, Chalhoub N, Chakroun A, Mohamed KAS, AlShami R, et al. Clinical and genomic characteristics of LAMA2 related congenital muscular dystrophy in a patients’ cohort from Qatar. A population specific founder variant. Neuromuscul Disord. 2020;30:457–471. doi: 10.1016/j.nmd.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Ge L, Zhang C, Wang Z, Chan SHS, Zhu W, Han C, et al. Congenital muscular dystrophies in China. Clin Genet. 2019;96:207–215. doi: 10.1111/cge.13560. [DOI] [PubMed] [Google Scholar]
- 9.Kemaladewi DU, Bassi PS, Erwood S, Al-Basha D, Gawlik KI, Lindsay K, et al. A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene. Nature. 2019;572:125–130. doi: 10.1038/s41586-019-1430-x. [DOI] [PubMed] [Google Scholar]
- 10.Reinhard JR, Lin S, McKee KK, Meinen S, Crosson SC, Sury M, et al. Linker proteins restore basement membrane and correct LAMA2-related muscular dystrophy in mice. Sci Transl Med. 2017;9:eaal4649. doi: 10.1126/scitranslmed.aal4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gawlik KI, Harandi VM, Cheong RY, Petersén A, Durbeej M. Laminin α1 reduces muscular dystrophy in dy2J mice. Matrix Biol. 2018;70:36–49. doi: 10.1016/j.matbio.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 12.Hall TE, Wood AJ, Ehrlich O, Li M, Sonntag CS, Cole NJ, et al. Cellular rescue in a zebrafish model of congenital muscular dystrophy type 1A. NPJ Regen Med. 2019;4:21. doi: 10.1038/s41536-019-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkozy A, Foley AR, Zambon AA, Bönnemann CG, Muntoni F. LAMA2-related dystrophies: clinical phenotypes, disease biomarkers, and clinical trial readiness. Front Mol Neurosci. 2020;13:123. doi: 10.3389/fnmol.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong H, Tan D, Wang S, Song S, Yang H, Gao K, et al. Genotype/phenotype analysis in Chinese laminin-α2 deficient congenital muscular dystrophy patients. Clin Genet. 2015;87:233–243. doi: 10.1111/cge.12366. [DOI] [PubMed] [Google Scholar]
- 15.Ge L, Liu A, Gao K, Du R, Ding J, Mao B, et al. Deletion of exon 4 in LAMA2 is the most frequent mutation in Chinese patients with laminin α2-related muscular dystrophy. Sci Rep. 2018;8:14989. doi: 10.1038/s41598-018-33098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding J, Zhao D, Du R, Zhang Y, Yang H, Liu J, et al. Clinical and molecular genetic analysis of a family with late-onset LAMA2-related muscular dystrophy. Brain Dev. 2016;38:242–249. doi: 10.1016/j.braindev.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Jain MS, Meilleur K, Kim E, Norato G, Waite M, Nelson L, et al. Longitudinal changes in clinical outcome measures in COL6-related dystrophies and LAMA2-related dystrophies. Neurology. 2019;93:e1932–e1943. doi: 10.1212/WNL.0000000000008517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Løkken N, Born AP, Duno M, Vissing J. LAMA2-related myopathy: frequency among congenital and limbgirdle muscular dystrophies. Muscle Nerve. 2015;52:547–553. doi: 10.1002/mus.24588. [DOI] [PubMed] [Google Scholar]
- 19.Magri F, Nigro V, Angelini C, Mongini T, Mora M, Moroni I, et al. The Italian limb girdle muscular dystrophy registry: relative frequency, clinical features, and differential diagnosis. Muscle Nerve. 2017;55:55–68. doi: 10.1002/mus.25192. [DOI] [PubMed] [Google Scholar]
- 20.Chan SH, Foley AR, Phadke R, Mathew AA, Pitt M, Sewry C, et al. Limb girdle muscular dystrophy due to LAMA2 mutations: diagnostic difficulties due to associated perpheral neuropathy. Neuromuscul Disord. 2014;24:677–683. doi: 10.1016/j.nmd.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Fan Y, Tan D, Song D, Zhang X, Chang X, Wang Z, et al. Clinical spectrum and genetic variations of LMNA-related muscular dystrophies in a large cohort of Chinese patients. J Med Genet. 2020;58:326–333. doi: 10.1136/jmedgenet-2019-106671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geis T, Rödl T, Topaloğlu H, Balci-Hayta B, Hinreiner S, Müller-Felber W, et al. Clinical long-time course, novel mutations and genotype-phenotype correlation in a cohort of 27 families with POMT1-related disorders. Orphanet J Rare Dis. 2019;14(1):179. doi: 10.1186/s13023-019-1119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durbeei M. Laminin-α2 chain-deficient congential muscular dystrophy: pathophysiology and development of treatment. Curr Top Membr. 2015;76:31–60. doi: 10.1016/bs.ctm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Vanasse M, Paré H, Zeller R. Medical and psychosocial considerations in rehabilitation care of childhood neuromuscular diseases. Handb Clin Neurol. 2013;113:1491–1495. doi: 10.1016/B978-0-444-59565-2.00019-8. [DOI] [PubMed] [Google Scholar]
- 25.Case LE, Apkon SD, Eagle M, Gulyas A, Juel L, Matthews D, et al. Rehabilitation management of the patient with Duchenne muscular dystrophy. Pediatrics. 2018;142:S17–S33. doi: 10.1542/peds.2018-0333D. [DOI] [PubMed] [Google Scholar]
- 26.Takaso M, Nakazawa T, Imura T, Okada T, Ueno M, Saito W, et al. Surgical correction of spinal deformity in patients with congenital muscular dystrophy. J Orhtop Sci. 2010;15:493–501. doi: 10.1007/s00776-010-1486-9. [DOI] [PubMed] [Google Scholar]
- 27.Apkon SD, Alman B, Birnkrant DJ, Fitch R, Lark R, Mackenzie W, et al. Orthopedic and surgical management of the patient with Duchenne muscular dystrophy. Pediatrics. 2018;142:S82–S89. doi: 10.1542/peds.2018-0333J. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Jones KJ, Vignier N, Morgan G, Chevallay M, Barois A, et al. Congenital muscular dystrophy with primary partial laminin alpha2 chain deficiency: molecular study. Neurology. 2001;57:1319–1322. doi: 10.1212/wnl.57.7.1319. [DOI] [PubMed] [Google Scholar]
- 29.Liang Y, Li G, Chen S, He R, Zhou X, Chen Y, et al. Muscle MRI findings in a one-year-old girl with merosin-deficient congenital muscular dystrophy type 1A due to LAMA2 mutation: a case report. Biomed Rep. 2017;7:193–196. doi: 10.3892/br.2017.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris E, McEntagart M, Topf A, Lochmüller H, Bushby K, Sewry C, et al. Clinical and neuroimaging findings in two brothers with limb girdle muscular dystrophy due to LAMA2 mutations. Neuromuscul Disord. 2017;27:170–174. doi: 10.1016/j.nmd.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Graziano A, Bianco F, D'Amico A, Moroni I, Messina S, Bruno C, et al. Prevalence of congenital muscular dystrophy in Italy: a population study. Neurology. 2015;84:904–911. doi: 10.1212/WNL.0000000000001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen Q, Lim KRQ, Yokota T. Current understanding and treatment of cardiac and skeletal muscle pathology in laminin-α2 chain-deficient cogenital muscular dystrophy. Appl Clin Genet. 2019;12:113–130. doi: 10.2147/TACG.S187481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallgren-Pettersson C, Bushby K, Mellies U, Simonds A. ENMC. 117th ENMC workshop: ventilatory supportin congenital neuromuscular disorders – congenital myopathies, congenital muscular dystrophies, congenital myotonic dystrophy and SMA (II) 4–6 April 2003, Naarden The Netherlands. Neuromuscul Disord. 2004;14:56–69. doi: 10.1016/j.nmd.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Donkervoort S, Dowling JJ, Laporte J, MacArthur D, Bönnemann CG. 214th ENMC workshop participants. 214th ENMC International Workshop: Establishing an international consortium for gene discovery and clinical research for Congenital Muscle Disease, Heemskerk, The Netherlands, 6–18 October 2015. Neuromuscul Disord. 2019;29:644–650. doi: 10.1016/j.nmd.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Jones KJ, Morgan G, Johnston H, Tobias V, Ouvrier RA, Wilkinson I, et al. The expanding phenotype of laminin alpha2 chain (merosin) abnormalities: case series and review. J Med Genet. 2001;38:649–657. doi: 10.1136/jmg.38.10.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang WC, Tian X, Yuo CY, Chen WZ, Kan TM, Su YN, et al. Comprehensive target capture/next-generation sequencing as a second-tier diagnostic approach for congential muscular dystrophy in Taiwan. PLoS ONE. 2017;12:e0170517. doi: 10.1371/journal.pone.0170517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natera-de Benito D, Muchart J, Itzep D, Ortez C, González-Quereda L, Gallano P, et al. Epilepsy in LAMA2-related muscular dystrophy: an electro-clinico-radiological characterization. Epilepsia. 2020;61:971–983. doi: 10.1111/epi.16493. [DOI] [PubMed] [Google Scholar]
- 38.Pini A, Merlini L, Tomé FM, Chevallay M, Gobbi G. Merosin-negative congenital muscular dystrophy, occipital epilepsy with periodic spasms and focal cortical dysplasia. Report of three Italian cases in two families. Brain Dev. 1996;18:316–322. doi: 10.1016/0387-7604(96)00028-9. [DOI] [PubMed] [Google Scholar]
- 39.Vigliano P, Dassi P, Di Blasi C, Blasi CD, Mora M, Jarre L. LAMA2 stop-codon mutation: merosin-deficient congential muscular dystrophy with occipital polymicrogyria, epilepsy and psychomotor regression. Eur J Paediatr Neurol. 2009;13:72–76. doi: 10.1016/j.ejpn.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Philpot J, Bagnall A, King C, Dubowitz V, Muntoni F. Feeding problems in merosin deficient congenital muscular dystrophy. Arch Dis Child. 1999;80:542–547. doi: 10.1136/adc.80.6.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vezain M, Gérard B, Drunat S, Funalot B, Fehrenbach S, N'Guyen-Viet V, et al. A leaky splicing mutation affecting SMN1 exon 7 inclusion explains an unexpected mild case of spinal muscular atrophy. Hum Mutat. 2011;32:989–994. doi: 10.1002/humu.21528. [DOI] [PubMed] [Google Scholar]
- 42.Morbidoni V, Baschiera E, Forzan M, Fumini V, Ali DS, Giorgi G, et al. Hybrid minigene assay: an efficient tool to characterize mrna splicing profiles of NF1 variants. Cancers (Basel) 2021;13:999. doi: 10.3390/cancers13050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kemaladewi DU, Maino E, Hyatt E, Hou H, Ding M, Place KM, et al. Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism. Nat Med. 2017;23:984–989. doi: 10.1038/nm.4367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Clinical findings of patients with LAMA2-related muscular dystrophy.
Additional file 2. The ages of patients with LAMA2-related muscular dystrophy.
Additional file 3. Effect of motor and epilepsy on survival of LAMA2-CMD.
Additional file 4. Genetical analysis of patients with LAMA2-related muscular dystrophy.
Additional file 5. Pathogenicity analysis of 17 missense variants.
Additional file 6. Genotype-phenotype correlations in patients with LAMA2-related muscular dystrophy.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request. All data relevant to the study are included in the article and its supplementary information files.