Abstract
Background:
Brain metastases are manually identified during stereotactic radiosurgery (SRS) treatment planning, which is time consuming and potentially challenging.
Purpose:
To develop and investigate deep learning (DL) methods for detecting brain metastasis with MRI to aid in treatment planning for SRS.
Materials and Methods:
In this retrospective study, contrast material–enhanced three-dimensional T1-weighted gradient-echo MRI scans from patients who underwent gamma knife SRS from January 2011 to August 2018 were analyzed. Brain metastases were manually identified and contoured by neuroradiologists and treating radiation oncologists. DL single-shot detector (SSD) algorithms were constructed and trained to map axial MRI slices to a set of bounding box predictions encompassing metastases and associated detection confidences. Performances of different DL SSDs were compared for per-lesion metastasis-based detection sensitivity and positive predictive value (PPV) at a 50% confidence threshold. For the highest-performing model, detection performance was analyzed by using free-response receiver operating characteristic analysis.
Results:
Two hundred sixty-six patients (mean age, 60 years ± 14 [standard deviation]; 148 women) were randomly split into 80% training and 20% testing groups (212 and 54 patients, respectively). For the testing group, sensitivity of the highest-performing (baseline) SSD was 81% (95% confidence interval [CI]: 80%, 82%; 190 of 234) and PPV was 36% (95% CI: 35%, 37%; 190 of 530). For metastases measuring at least 6 mm, sensitivity was 98% (95% CI: 97%, 99%; 130 of 132) and PPV was 36% (95% CI: 35%, 37%; 130 of 366). Other models (SSD with a ResNet50 backbone, SSD with focal loss, and RetinaNet) yielded lower sensitivities of 73% (95% CI: 72%, 74%; 171 of 234), 77% (95% CI: 76%, 78%; 180 of 234), and 79% (95% CI: 77%, 81%; 184 of 234), respectively, and lower PPVs of 29% (95% CI: 28%, 30%; 171 of 581), 26% (95% CI: 26%, 26%; 180 of 681), and 13% (95% CI: 12%, 14%; 184 of 1412).
Conclusion:
Deep-learning single-shot detector models detected nearly all brain metastases that were 6 mm or larger with limited false-positive findings using postcontrast T1-weighted MRI.
Summary
Deep learning single-shot detector algorithms identified brain metastases at postcontrast MRI with high sensitivity and moderate positive predictive value, potentially assisting treatment planning of stereotactic radiosurgery for brain metastases.
Brain metastases occur in approximately 20%–40% of patients with cancer, and the incidence rate could rise because of longer survival rates (1). Because whole-brain radiation therapy can cause cognitive decline and may not be necessary for patients with fewer than four brain metastases (2,3), stereotactic radiosurgery (SRS) is the preferred treatment method. With highly focused radiation delivered to small metastasis volumes, SRS treats metastases effectively and minimizes damage to normal brain tissues. Accurate detection of brain metastases and subsequent precise segmentation are essential for SRS treatment planning. Currently, brain metastases are manually identified by neuroradiologists, which can be laborious and time consuming because there is no a priori knowledge about the number or location of the metastases. Furthermore, if the patient has multiple metastases, identification of all the metastases could be challenging.
Deep learning (DL) has seen an explosion of interest and applications across the medical imaging domain. For brain metastasis identification with MRI, a few DL approaches based on semantic segmentation using fully convolutional networks have been proposed (4–7). By mapping images to pixelwise probability distributions of tissue compositions, these attempts have often led to substantial numbers of false-positive findings (up to 200 per patient) (5). In addition, although these implementations can be used to directly segment brain metastases, the segmentation performance is generally low (Dice coefficients of approximately 0.77 and 0.67) (5,6). Furthermore, some of these approaches rely on patch-wise predictions, in which the patch size must be specifically tuned for different patient populations because brain metastases vary considerably in size. Finally, some of these approaches were developed by using multiple MRI acquisitions as inputs, which may reduce their feasibility for broad clinical application.
Recently, DL single-shot detector (SSD) algorithms (8,9) were developed for object detection. Unlike two-stage detectors (10–12), DL SSDs perform detection by matching densely aligned anchor boxes to ground truth boxes. Coordinates and prediction confidences of the matched anchor boxes are then simultaneously regressed on pyramidal feature maps. Attempts have been made to implement DL SSDs for lung or breast lesion detection and malignancy classification in CT or diffusion MRI (13); however, to the best of our knowledge, detection of brain metastases using DL SSDs has not been reported.
In this study, we developed and investigated DL SSDs for brain metastasis detection by using contrast material–enhanced T1-weighted MRI. Variations in encoder architectures and loss functions were explored to compare the detection performance across different model constructs.
Materials and Methods
Institutional review board approval was received for this Health Insurance Portability and Accountability Act–compliant retrospective study. The requirement to obtain written informed consent was waived.
Study Participants
Two hundred sixty-six patients who underwent gamma knife SRS for brain metastases at the University of Texas MD Anderson Cancer Center between January 2011 and August 2018 were included. The inclusion criterion was any patient who had undergone treatment planning for SRS by a board-certified radiation oncologist. We avoided selecting patients with a history of primary brain cancers.
MRI Examination
Each patient underwent imaging with a three-dimensional T1-weighted spoiled gradient-echo MRI sequence after the administration of contrast material (MultiHance; Bracco Diagnostics, Princeton, NJ). Typical imaging parameters were as follows: repetition time msec/echo time msec, 6.9/2.5; number of signal averages, 1.7; flip angle, 12°; matrix size, 256 × 256; field of view, 24 × 24 cm; and voxel size, 0.94 × 0.94 × 1.00 mm. Two hundred forty-six patients underwent imaging with a 1.5-T scanner (Signa HDxt; GE Healthcare, Waukesha, Wis), and 20 patients underwent imaging with a 3.0-T scanner (Discovery MR750w, GE Healthcare).
MRI Analysis
A total of 1147 brain metastases were independently identified by board-certified neuroradiologists (J.M.J., M.K.G., and M.M.C., with 7, 4, and 3 years of experience, respectively) and contoured by the treating radiation oncologist (J.L., with 12 years of experience). On average, each patient had four metastases. The size of a brain metastasis was measured as the largest cross-sectional dimension when projected onto a two-dimensional plane in the craniocaudal direction. The mean metastasis size (±standard deviation) was 10 mm ± 8. The number of metastases smaller than 6 mm and 6 mm or larger was 507 and 640, respectively. The distributions of metastasis sizes and metastases across patients are shown in Figure 1.
Figure 1:
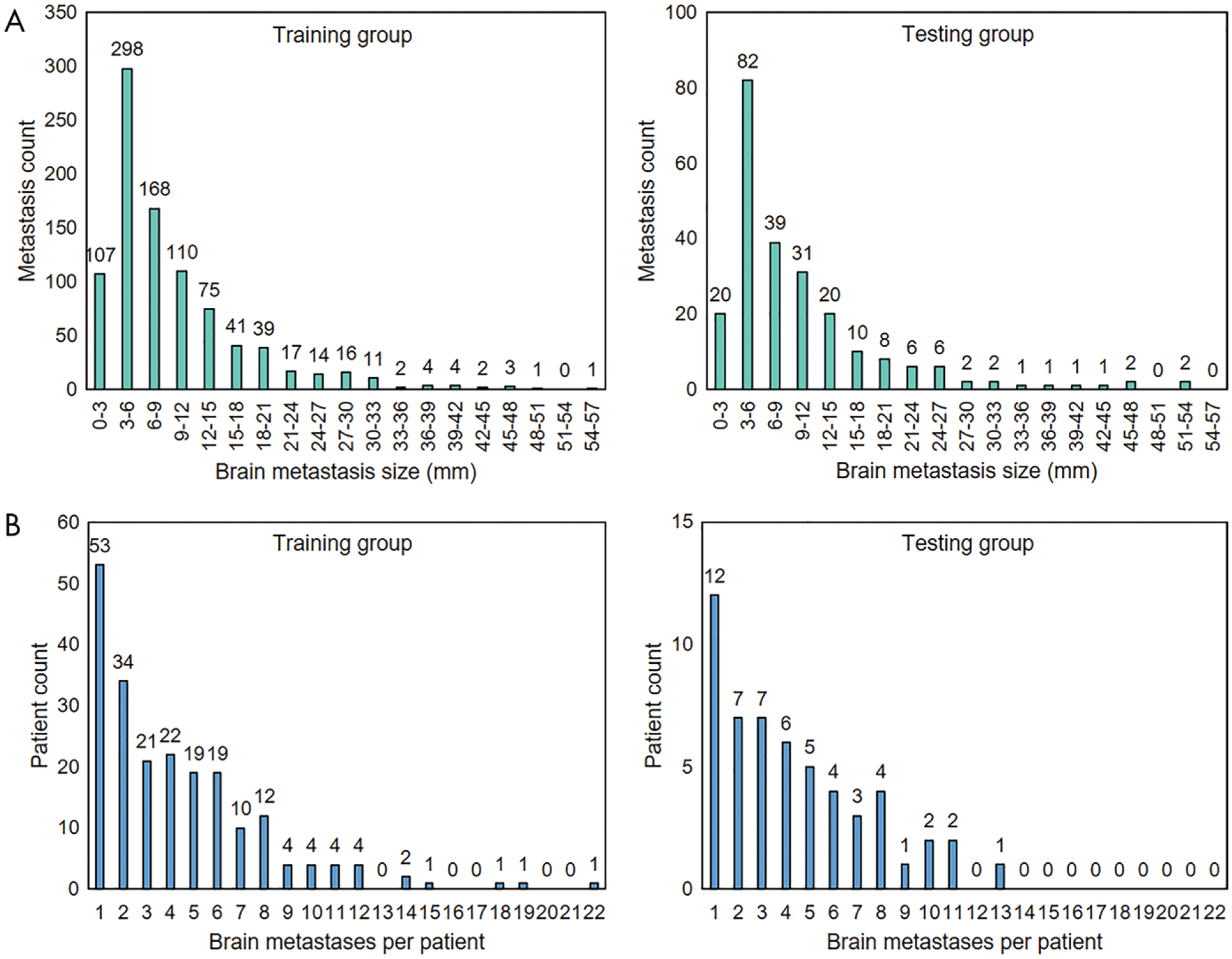
A, Bar charts show distribution of brain metastasis sizes in training group (left) and testing group (right). The mean brain metastasis size was 10 mm ± 8 (standard deviation) in the training group and 10 mm ± 9 in the testing group, and the ratios between the number of metastases smaller than 6 mm and those 6 mm or larger were 405:508 and 102:132, respectively. B, Bar charts show distribution of brain metastases across patients in the training group (left) and testing group (right). Both groups had a mean of four metastases per patient.
Data Curation
The 266 MRI scans were randomly split into two groups with a ratio of approximately 80%:20% (212:54) for model training and testing; none of the training group cases were present in the testing group. The training group scans contained 6711 axial slices showing metastases. The slices were randomly split into training and validation sets with a ratio of 75%:25% (5033:1678), which were used for model training and internal tuning, respectively. All axial slices for each patient (both with and without a lesion present) in the testing group comprised the testing set, which was used for model performance evaluation. Finally, all slices in the training, validation, and testing sets were normalized to the signal intensity range of 0–1.
The ground truth labels for each brain metastasis cross-section were the bounding box coordinates (xmin, ymin, xmax, ymax), which were prepared from the corresponding segmentation mask and represented the start and end positions of the box height and width. The bounding box was computed by using connected components analysis. Because most of the metastasis cross-sections were round, we set all ground truth bounding boxes to have aspect ratios (box height divided by width) equal to 1, in which the maximum dimension was repeated for the shorter dimension. The bounding box therefore tightly encompassed the entire metastasis cross-section and was reliable for model training and evaluation.
Construction of SSDs
SSDs were constructed by using Keras (https://keras.io/) (14). We defined the baseline SSD as an SSD whose context encoder was constructed only with convolutional layers. The baseline SSD consisted of 16 convolutional layers for feature extraction, among which six layers were used for metastasis detection. The six detection layers had feature maps with matrix sizes of 128 × 128, 64 × 64, 32 × 32, 16 × 16, 8 × 8, and 4 × 4. L2 normalization (15) with initial scales of 10 and five was applied to the first and second layers, respectively, because they had different feature scales. A detailed representation of the model’s construction is shown in Figure 2.
Figure 2:
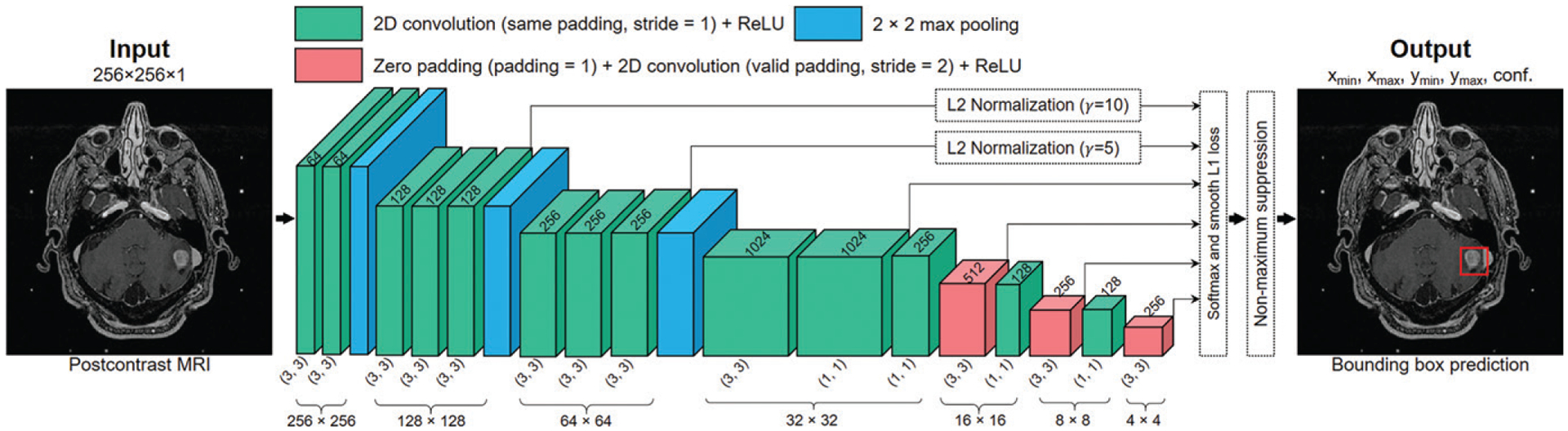
Illustration of the baseline single-shot detector (SSD) algorithm that was constructed and developed. The three-dimensional boxes represent the feature maps extracted from the original image after operations of convolution, maximum pooling, and zero padding, which all change the dimensionality (eg, 256 × 256, represented by the three-dimensional box height and depth) and channels (eg, 64, represented by the three-dimensional box width) of the feature maps. Predictions were made at six resolution scales. L2 normalization was used for the first two detection layers to account for the different scales of the feature maps. The classification and bounding box position regression losses were concatenated to form the SSD loss. The outputs of the model were the bounding box coordinates (xmin, xmax, ymin, ymax) and detection confidences of the brain metastases. conf. = classification value, ReLU = rectified linear activation unit, 2D = two-dimensional.
Because SSDs use pre-aligned anchor boxes for object detection, scales (defined as box height divided by image height) and aspect ratios (defined as box height divided by box width) of the anchor boxes are important hyperparameters. Because the metastasis size was typically small and varied continuously, the scales were determined based on two considerations. First, the anchor boxes should contain a minimum of 0.8 pixels on the corresponding feature maps. Second, the anchor boxes should cover as many sizes as possible. Therefore, in our models the six prediction layers had base scales of 0.008, 0.016, 0.032, 0.064, 0.115, and 0.2. To cover a broader range of metastasis sizes besides the base scale, each layer had another larger detection scale, which was determined by the geometric mean of the base and the adjacent scales. This equated to 0.24 for the last layer. Because all ground truth boxes had aspect ratios of 1, the anchor boxes were set to have the same aspect ratio.
The threshold for SSDs to label the anchor boxes as positive (matched with the ground truth metastases) or negative (matched with the background) was an important consideration. To match more anchor boxes with the metastases, anchor boxes that had an intersection over union of at least 0.2 with the metastasis ground truth box were labeled as positive, whereas anchor boxes that had an intersection over union of 0.1 or less were labeled as negative. Anchor boxes that had an intersection over union between the two thresholds were labeled as neutral and would not affect the loss computation.
The proposed and labeled anchor boxes were regressed by using the SSD loss function, which was a weighted sum of the anchor box classification and localization losses. Because the number of negative anchor boxes largely exceeded the number of positive ones, we set the negative-to-positive ratio to 3 to balance the negative and positive instances. The weight of the localization loss was set to 1. These two hyperparameters of the SSD loss function yielded the highest performance in this study.
Training Configuration and Procedure
Random affine transformations were applied to amplify the number of training set slices by a factor of five. The ground truth bounding boxes were transformed accordingly. No data augmentation was used for the validation set. The minibatch size was set to 16.
Weights of the convolutional kernels were initialized by using a He normal initialization (16), and all convolutional layers were activated by using rectified linear activation units (17). No regularization or dropout layers were used. An Adam optimizer (18) was used to train the network. The initial learning rate was set to 2 × 10−4 and was reduced by 20% if there was no improvement in validation loss after three epochs. Training was terminated if the validation loss did not improve after seven epochs. The models were trained on a DGX-1 workstation (NVIDIA, Santa Clara, Calif).
Ablation Studies
In addition to the baseline SSD, three constructs using different encoder structures or loss functions but the same single-shot detection principles were trained and evaluated to compare their performance. Because performance of residual networks improves with depth (19), we used a ResNet50 convolutional encoder with the original SSD loss for brain metastasis detection. Focal loss has been proposed to improve the detection performance by emphasizing the misclassified objects (9). To evaluate such performance in the context of our study, we incorporated focal loss into the baseline SSD to train a third model. Finally, we tested metastasis detection combining the ResNet50 feature pyramid network (20) and focal loss, an algorithm termed RetinaNet (9). It was reported to reach state-of-the-art detection performance and to effectively detect small objects (9).
Model Evaluation
Although the SSDs were developed for slice-by-slice predictions, the final performances of the models were evaluated in three dimensions. The adjacent output bounding boxes were first stacked to form a detection volume, which was then compared with the ground truth volumes surrounding the metastases. The ground truth volumes were considered true-positive findings if they had at least one voxel detected; otherwise they were false-negative findings. The detection volumes were considered false-positive findings if they had no voxel overlap with any of the ground truth volumes.
Statistical Analysis
Performances of the four models, including the baseline SSD, SSD with a ResNet50 backbone, SSD with focal loss, and RetinaNet, were assessed and compared by using metastasis-based sensitivity and positive predictive value (PPV) at a detection confidence threshold of 50%. Because all MRI scans from test patients were integrated as one data set, the within-patient correlation for patients with multiple lesions was not considered. Detection confidence is the numeric output of the SSD’s classifier and ranges from 0% to 100%. For example, for binary classification (metastasis vs background), 50% confidence is the threshold that determines the object class within the bounding box.
Sensitivity was defined as the ratio between the number of true-positive findings and all metastases of the entire testing set (true-positive findings/[true-positive findings + false-negative findings]). The PPV was defined as the ratio between the number of true-positive findings and all detections (true-positive findings/[true-positive findings + false-positive findings]). For the best-performing model, free-response receiver operating characteristic and PPV versus sensitivity curves with respect to the metastasis size (<3 mm, ≥3 mm to <6 mm, and ≥6 mm) were plotted to inspect the model’s performance for brain metastases of different size ranges separately. The model’s sensitivity was also inspected with respect to the metastasis shape (round vs oval) and type (hemorrhagic vs necrotic).
To evaluate the variance of the models’ predictions, each model was trained five times. The corresponding testing results were averaged and compared by using the independent t test. All code developed for our study can be accessed at https://github.com/Joe15327/brain-metastases-detection.
Results
Patient Characteristics
The mean age of the 266 patients included in our study was 60 years ± 14. There were 118 men and 148 women. The demographic characteristics and cancer types of the patient population are shown in Table 1.
Table 1:
Patient Demographics and Primary Cancer Types in the Training and Testing Groups
| Parameter | Training Group | Testing Group | Total |
|---|---|---|---|
| Patient demographics | |||
| No. of patients | 212 | 54 | 266 |
| Mean age (y)* | 57 ± 14 | 62 ± 12 | 60 ± 14 |
| M/F ratio | 93:119 | 25:29 | 118:148 |
| Magnetic field strength | |||
| 1.5 T | 197 | 49 | 246 |
| 3.0 T | 15 | 5 | 20 |
| Primary cancer types | |||
| Lung | 79 (37) | 23 (43) | 102 (38) |
| Melanoma | 43 (20) | 12 (22) | 55 (21) |
| Breast | 38 (18) | 6 (11) | 44 (16) |
| Renal | 25 (12) | 3 (5.6) | 28 (11) |
| Gastrointestinal | 9 (4.2) | 2 (3.7) | 11 (4.1) |
| Genitourinary | 6 (2.8) | 1 (1.8) | 7 (2.6) |
| Sarcoma | 5 (2.4) | 0 (0.0) | 5 (1.9) |
| Thyroid | 2 (1.0) | 2 (3.7) | 4 (1.5) |
| Testicular | 2 (1.0) | 2 (3.7) | 4 (1.5) |
| Head and neck | 2 (1.0) | 2 (3.7) | 4 (1.5) |
| Neuroendocrine carcinoma | 1 (0.5) | 1 (1.8) | 2 (0.8) |
Note.—Except where indicated, data are numbers of patients, with percentages in parentheses.
Data are means ± standard deviation.
Detection Performance of the Baseline SSD
Inference took approximately 1 second per patient on the DGX-1 workstation. At a confidence threshold of 50%, the baseline DL SSD yielded the best performance among the four detectors and achieved an overall detection sensitivity of 81% (95% confidence interval [CI]: 80%, 82%; 190 of 234) and PPV of 36% (95% CI: 35%, 37%; 190 of 530) for the entire testing set. Specifically, for metastases smaller than 3 mm, the sensitivity was 15% (95% CI: 12%, 18%; three of 20) and the PPV was 100% (95% CI: 100%, 100%; three of three). For metastases measuring 3 mm or larger to smaller than 6 mm, the sensitivity was 70% (95% CI: 68%, 72%; 57 of 82) and the PPV was 35% (95% CI: 32%, 38%; 57 of 161). For metastases 6 mm or larger, the sensitivity was 98% (95% CI: 97%, 99%; 130 of 132) and the PPV was 36% (95% CI: 35%, 37%; 130 of 366). Examples of true-positive, false-positive, and false-negative inferences are shown in Figures 3 and 4.
Figure 3:
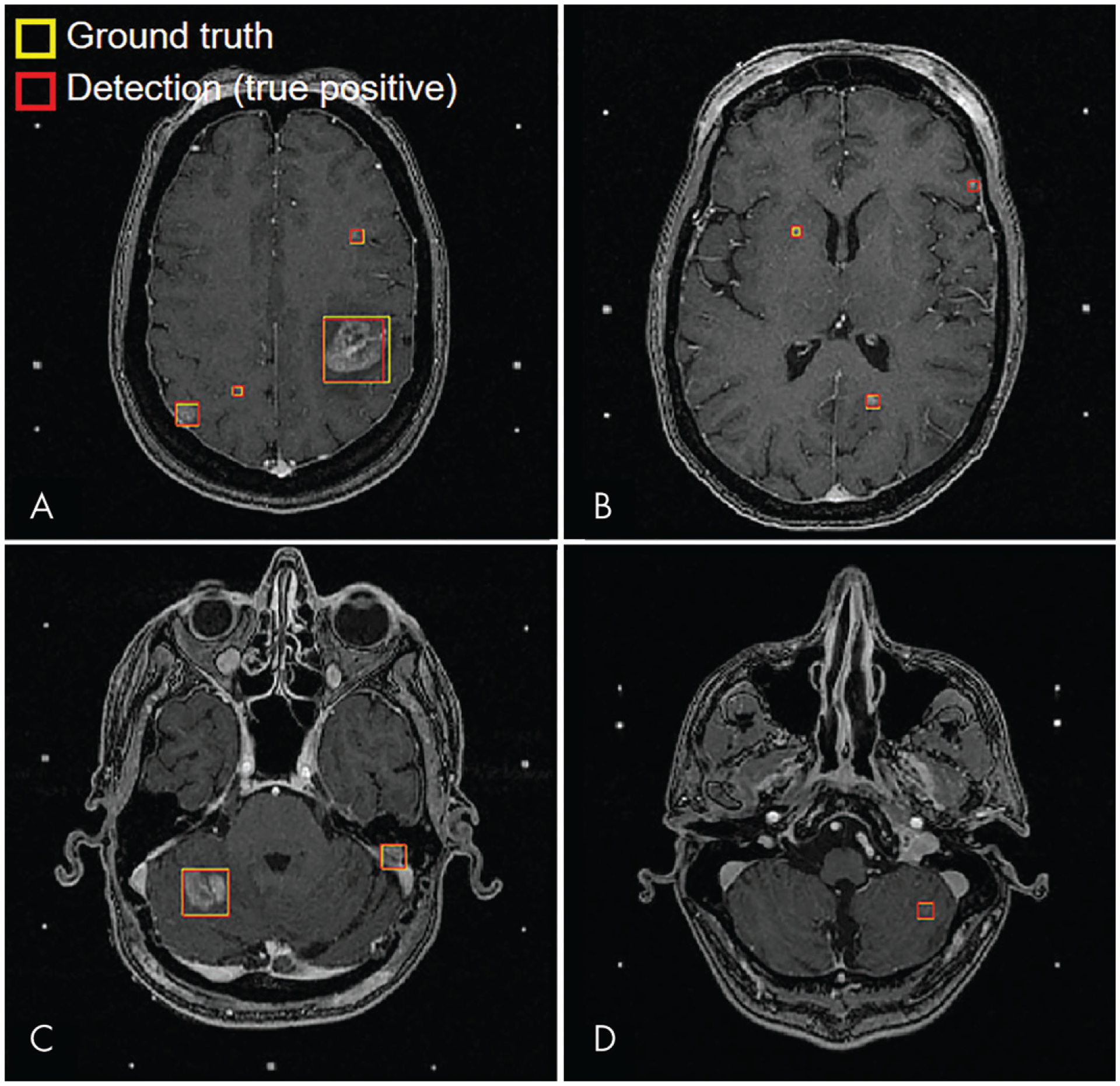
MRI scans show examples of true-positive inferences from baseline single-shot detector at 50% confidence threshold for four representative test patients. The metastasis sizes were, A, 4, 6, 9, and 28 mm (patient 1); B, 3, 5, and 6 mm (patient 2); C, 10 and 20 mm (patient 3); and, D, 7 mm (patient 4).
Figure 4:
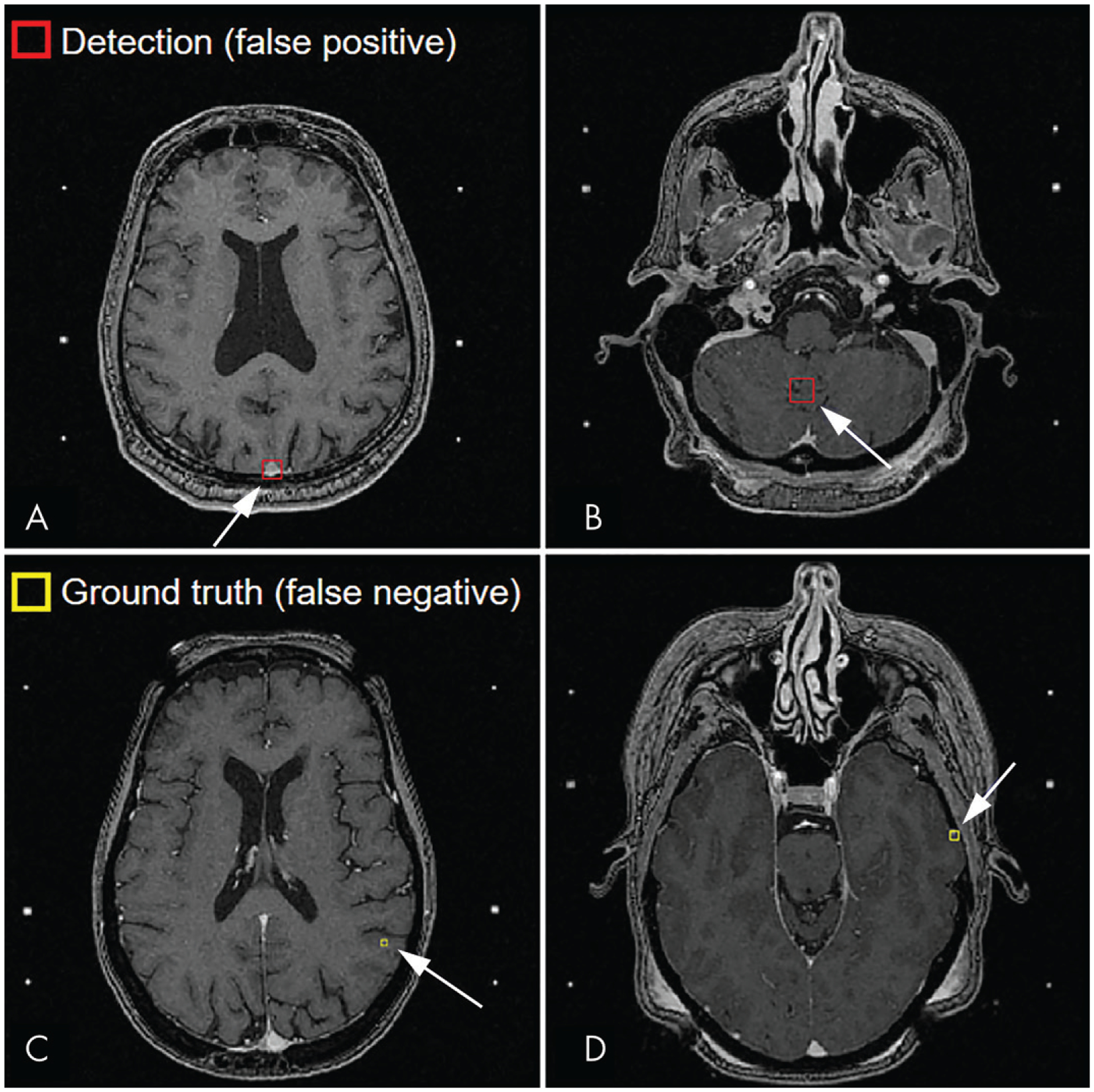
A, B, Examples of false-positive inferences (arrrow) at MRI. False-positive findings were typically, A, blood vessels (patient 5) or, B, variations in brain tissue contrast (patient 6). C, D, Examples of false-negative inferences (arrow) at MRI. The missed metastases were, C, 3 mm (patient 7) and, D, 4 mm (patient 8). Missed metastases were typically located at the gray matter–white matter or brain-skull interfaces of the cerebrum..
Detailed detection performance of the baseline SSD is shown in Figure 5. The free-response receiver operating characteristic analysis showed that although detection of smaller metastases was more challenging, for higher numbers of false-positive findings per patient, sensitivity improved by more than 50% for metastases 3 mm or larger to smaller than 6 mm and more than 100% for metastases smaller than 3 mm. Conversely, the PPV-sensitivity plots showed that lower PPV corresponded to higher sensitivity, and the PPV was higher for smaller metastases than for larger metastases.
Figure 5:
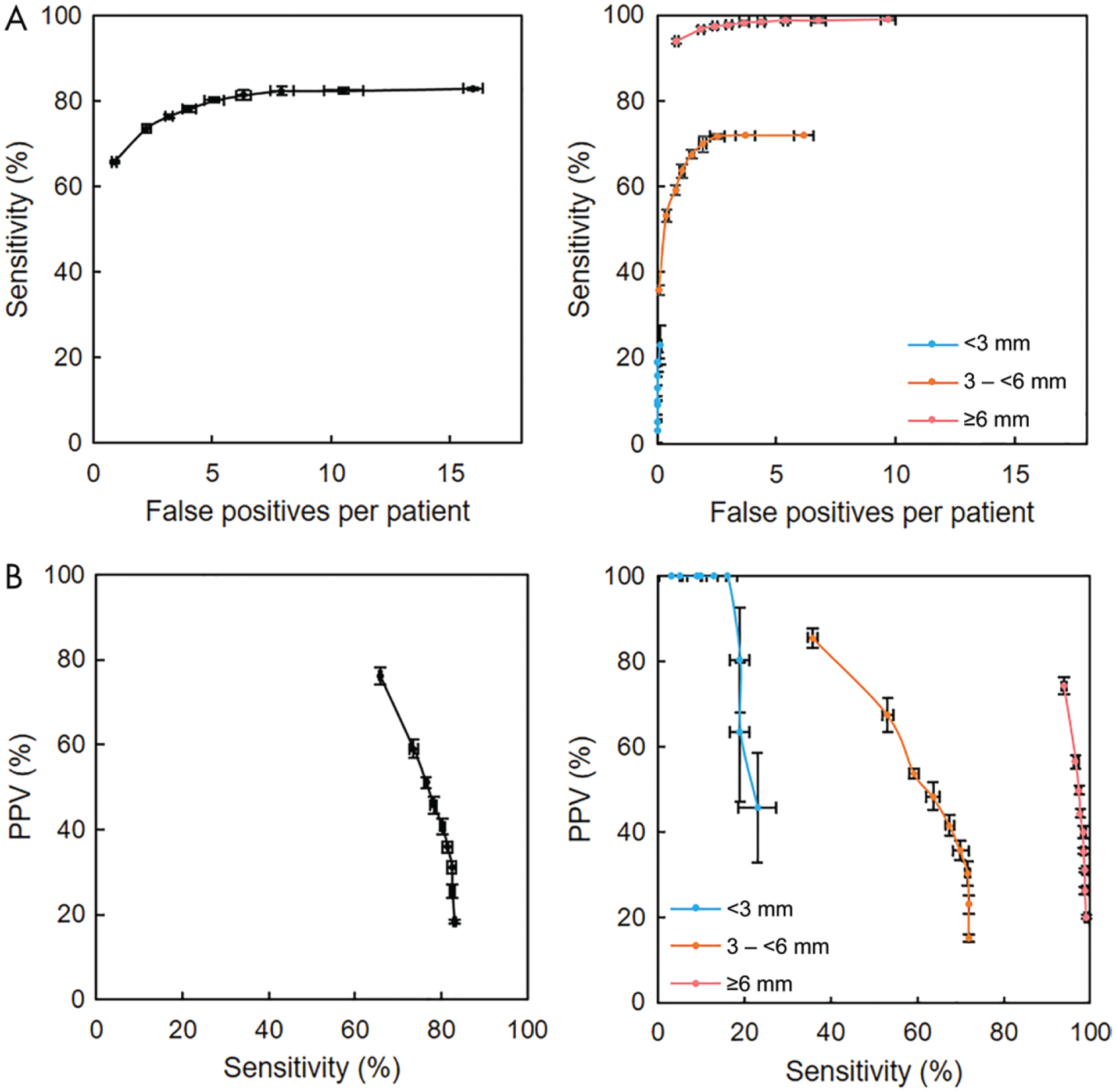
Detection performance of highest-performing detector (baseline single-shot detector). A, Free-response receiver operating characteristic curves for all metastases (left) and for metastases smaller than 3 mm, 3 mm or larger to smaller than 6 mm, and 6 mm or larger (right). Higher sensitivity occurred with a higher number of false-positive findings per patient; however, for metastases measuring 6 mm or larger, high sensitivity could be achieved with fewer false-positive findings per patient. B, Plots of positive predictive value (PPV) versus sensitivity for all metastases (left) and for metastases smaller than 3 mm, 3 mm or larger to smaller than 6 mm, and 6 mm or larger (right). Higher sensitivity occurred with reduced PPV; however, for metastases measuring 6 mm or larger, high PPV could be achieved with high sensitivity. For all plots, data points are averages of predictions from five random initializations of model training, and the horizontal and vertical error bars are 95% confidence intervals.
Detection sensitivity of the baseline SSD showed nearly no difference for metastasis shape (round vs oval: 81% [95% CI: 80%, 82%; 91 of 113] vs 82% [95% CI: 80%, 84%; 99 of 121]; P = .04). However, a significant difference was found between metastasis types (hemorrhagic vs necrotic: 78% [95% CI: 77%, 79%; 135 of 174] vs 92% [95% CI: 90%, 94%; 55 of 60]; P < .01). In addition, the sensitivity for necrotic metastases smaller than 6 mm was 73% (95% CI: 70%, 77%; 11 of 15), whereas that for metastases 6 mm or larger was 98% (95% CI: 96%, 99%; 44 of 45) (P < .01).
Detection Performance across Different Model Constructs
Detection sensitivity and PPV at the 50% confidence threshold for the baseline SSD and the other three models of the ablation study are summarized in Table 2. The overall sensitivities and PPVs of the three models were lower than those of the baseline SSD (P < .05 for all). Nonetheless, RetinaNet achieved more than double the sensitivity of the baseline SSD for metastases smaller than 3 mm. However, the PPV of RetinaNet was low for these metastases.
Table 2:
Sensitivities and PPVs of Different Deep Learning SSDs at a 50% Confidence Threshold for Different Metastasis Sizes
| All Metastases | Metastases <3 mm | Metastases ≥3 mm to < 6 mm | Metastases ≥6 mm | |||||
|---|---|---|---|---|---|---|---|---|
| Technique | Sensitivity (%) | PPV (%) | Sensitivity (%) | PPV (%) | Sensitivity (%) | PPV (%) | Sensitivity (%) | PPV (%) |
| Baseline SSD | 81 (80, 82) [190/234] | 36 (35, 37) [190/530] | 15 (12, 18) [3/20] | 100 (100, 100) [3/3] | 70 (68, 72) [57/82] | 35 (32, 38) [57/161] | 98 (97, 99) [130/132] | 36 (35, 37) [130/366] |
| SSD plus ResNet50 | 73 (72, 74) [171/234] | 29 (28, 30) [171/581] | 10 (7,13) [2/20] | 100 (100, 100) [2/2] | 55 (53, 57) [45/82] | 34 (31, 37) [45/134] | 94 (93, 95) [124/132] | 28 (26, 30) [124/445] |
| SSD plus focal loss | 77 (76, 78) [180/234] | 26 (26, 26) [180/681] | 20 (14, 26) [4/20] | 80 (64, 96) [4/5] | 62 (60, 64) [51/82] | 21 (19, 23) [51/248] | 95 (94, 96) [125/132] | 29 (28, 30) [125/428] |
| RetinaNet | 79 (77, 81) [184/234] | 13 (12, 14) [184/1412] | 40 (33, 47) [8/20] | 2 (1, 3) [8/410] | 66 (61, 71) [54/82] | 9 (8,10) [54/573] | 92 (91, 93) [122/132] | 28 (27, 29) [122/429] |
Note.—Numbers in parentheses are 95% confidence intervals, and numbers in brackets are raw data. The baseline SSD achieved the highest metastasis-wise sensitivity and PPV. PPV = positive predictive value, SSD = single-shot detector.
Discussion
The developed single-shot detector (SSD) algorithms showed promising results to assist stereotactic radiosurgery (SRS) treatment planning for brain metastases. At a 50% confidence threshold, the baseline SSD was able to detect 130 of 132 (98%; 95% confidence interval [CI]: 97%, 99%) brain metastases measuring 6 mm or larger and 60 of 102 (59%; 95% CI: 57%, 61%) metastases smaller than 6 mm, with a positive predictive value (PPV) of 36% (95% CI: 35%, 37%; 190 of 530). The detection sensitivity primarily depended on the metastasis size (P < .01): Small metastases typically had poorly defined boundaries and low contrast. Furthermore, 88% of the false-negative metastases smaller than 6 mm (37 of 42) were located near the brain-skull or gray matter–white matter interfaces of the cerebrum and resembled small blood vessels; 12% (five of 42) were located in the cerebellum. In addition, sensitivity was also higher for necrotic metastases than for hemorrhagic metastases (92% vs 78%, P < .01). Necrotic metastases typically had more prominent features with hypointense regions and larger sizes (75% [45 of 60] were ≥6 mm), making them easier to be differentiated from blood vessels.
Like most detection algorithms, there is a trade-off between sensitivity and PPV in our models. By lowering the confidence threshold, more of the smaller metastases could be detected at a cost of reduced PPV. However, the sensitivity remained similar as the false-positive findings increased to 15 per patient. Therefore, a confidence threshold around 40% may be appropriate for clinical use, wherein 20% of the very small metastases can be detected by the model and the limited false-positive findings can still be easily rejected by neuroradiologists.
Compared with previously reported approaches using DL for detecting brain metastases, our study has two major differences. First, our approach used networks dedicated to object detection. It was able to use the entire image context, therefore avoiding patch-wise inferences, which may lack robustness because of the broad range of brain metastasis sizes. Second, many previous approaches require multiple inputs from different MRI acquisitions (eg, T1-weighted, T2-weighted, and fluid-attenuated inversion recovery MRI) (5,7). Our approach uses only postcontrast T1-weighted MRI to detect brain metastases. The use of a single MRI acquisition reduces the overhead costs and potential complications from interseries MRI co-registration, improving the feasibility for clinical applications.
An unexpected result of our study was that the other three detection models with deeper and more sophisticated constructs than the baseline SSD yielded lower overall performance on the testing data set (P < .05 for all). There are two potential explanations for these findings. First, although the focal loss function handles the issue of positive-negative sample imbalance during training, it may not be as efficient as direct mining of the negative samples. Second, although ResNet (19) and RetinaNet (9) were reported to achieve superior performance over models that used VGG16 (21) as the convolutional encoder (3% improvement of image classification accuracy and 4.5% improvement of object detection PPV), such achievements were realized on natural images, which contain different content than brain MRI scans having a single channel of information.
Our study has several limitations. First, all data used in this study emanated from the same institution and therefore do not account for variabilities in scanning techniques and hardware implementations across hospitals. Evaluation in patients at external locations would be necessary to test the models’ generalizability. Second, although we achieved high sensitivity for larger metastases, detection performance for smaller metastases is limited. Considering that metastases smaller than 6 mm comprise approximately 43% of SRS targets in our sample patients, approximately 17% of the targets could be missed and therefore require inspection by neuroradiologists. In developing SSDs, we achieved improved performance in detecting smaller brain metastases by including more patient MRI scans in the training set. Therefore, collection of more data for training the model may benefit the detection of smaller metastases. In addition, the use of three-dimensional fully convolutional networks may help boost performance, whereby the interslice information can be used to differentiate small metastases and small blood vessels.
Future work will involve developing automatic brain metastasis segmentation methods based on the detection(s). A potential segmentation approach could be training a separate fully convolutional network (eg, U-Net) (22) to segment the brain metastases after detection. With use of the coordinates of the detection bounding boxes, MRI regions containing the metastases can be cropped and input to the segmentation fully convolutional network to generate the segmentation masks. With the brain metastases specifically cropped out, we expect the ensemble of detection and segmentation fully convolutional networks could achieve high segmentation performance with high sensitivity and specificity, assisting treatment planning for stereotactic radiosurgery.
Key Results.
Overall, the deep learning (DL) algorithm using postcontrast T1-weighted MRI only achieved 81% sensitivity and 36% positive predictive value (PPV), with six false-positive discoveries per patient for brain metastasis detection.
For metastases measuring 6 mm or larger, sensitivity of the DL model was 98% and the PPV was 36%, with three to four false-positive findings per patient.
Acknowledgments:
The authors thank the University of Texas MD Anderson Cancer Center’s Department of Scientific Publications for their help editing this article. J.W.S. would like to acknowledge the generous donors of the Pauline Altman-Goldstein Foundation.
J.W.S. is supported by a fellowship from the donors of the Pauline Altman-Goldstein Foundation. Z.Z. and J.W.S. are supported in part by a sponsored research grant from Siemens Healthineers.
Disclosures of Conflicts of Interest:
Z.Z. disclosed no relevant relationships. J.W.S. disclosed no relevant relationships. J.M.J. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a paid consultant for Kura Oncology; has grants/grants pending from Blue Earth Diagnostics. Other relationships: disclosed no relevant relationships. M.K.G. disclosed no relevant relationships. M.M.C. disclosed no relevant relationships. T.M.B. disclosed no relevant relationships. Y.W. disclosed no relevant relationships. J.B.S. disclosed no relevant relationships. M.D.P. disclosed no relevant relationships. J.L. disclosed no relevant relationships. J.M. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a paid consultant for C4 Imaging; has grants/grants pending from GE Healthcare and Siemens Healthineers; receives royalties from GE Healthcare and Siemens Healthineers. Other relationships: disclosed no relevant relationships.
Abbreviations
- CI
confidence interval
- DL
deep learning
- PPV
positive predictive value
- SRS
stereotactic radiosurgery
- SSD
single-shot detector
References
- 1.Patchell RA. The management of brain metastases. Cancer Treat Rev 2003;29(6):533–540. [DOI] [PubMed] [Google Scholar]
- 2.Chang EL, Wefel JS, Hess KR, et al. Neurocognition in patients with brain metastases treated with radiosurgery or radiosurgery plus whole-brain irradiation: a randomised controlled trial. Lancet Oncol 2009;10(11):1037–1044. [DOI] [PubMed] [Google Scholar]
- 3.Brown PD, Jaeckle K, Ballman KV, et al. Effect of radiosurgery alone vs radiosurgery with whole brain radiation therapy on cognitive function in patients with 1 to 3 brain metastases: a randomized clinical trial. JAMA 2016;316(4):401–409. [Published correction in JAMA 2018;320(5):510] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- 5.Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 2018;95:43–54. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Stojadinovic S, Hrycushko B, et al. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One 2017;12(10):e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grøvik E, Yi D, Iv M, Tong E, Rubin DL, Zaharchuk G. Deep learning enables automatic detection and segmentation of brain metastases on multi-sequence MRI. ArXiv:1903.07988 [preprint] https://arxiv.org/abs/1903.07988. Published online March 18, 2019. Accessed June 1, 2019. [Google Scholar]
- 8.Liu W, Anguelov D, Erhan D, et al. SSD: Single shot multibox detector. In: Leibe B, Matas J, Sebe N, Welling M, eds. Computer Vision – ECCV 2016. ECCV 2016. Lecture Notes in Computer Science, vol 9905. Cham, Switzerland: Springer, 2016;21–37. [Google Scholar]
- 9.Lin TY, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. In: Proceedings of the IEEE International Conference on Computer Vision, 2017;2980–2988. [Google Scholar]
- 10.Girshick R, Donahue J, Darrell T, Malik J. Region-based convolutional networks for accurate object detection and segmentation. IEEE Trans Pattern Anal Mach Intell 2016;38(1):142–158. [DOI] [PubMed] [Google Scholar]
- 11.Girshick R Fast R-CNN. In: Proceedings of the IEEE International Conference on Computer Vision, 2015; 1440–1448. [Google Scholar]
- 12.Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. Poster presented at: Advances in Neural Information Processing Systems Conference; December 7–12, 2015; Montreal, Canada. https://papers.nips.cc/paper/5638-faster-r-cnn-towards-real-time-object-detection-with-region-proposal-networks. Accessed June 1, 2019. [Google Scholar]
- 13.Jaeger PF, Kohl SA, Bickelhaupt S, et al. Retina U-Net: embarrassingly simple exploitation of segmentation supervision for medical object detection. ArXiv:1811.08661 [preprint] https://arxiv.org/abs/1811.08661. Published online November 21, 2018. Accessed June 1, 2019. [Google Scholar]
- 14.Keras: the Python deep learning library. https://keras.io/. Accessed June 1, 2019.
- 15.Liu W, Rabinovich A, Berg AC. Parsenet: looking wider to see better. ArXiv:1506.04579 https://arxiv.org/abs/1506.04579. Published online June 15, 2015. Accessed June 1, 2019. [Google Scholar]
- 16.He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In: Proceedings of the IEEE International Conference on Computer Vision, 2015; 1026–1034. [Google Scholar]
- 17.Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: Proceedings of the 27th International Conference on Machine Learning (ICML-10) (paper 432), 2010. https://icml.cc/Conferences/2010/papers/432.pdf. Accessed June 1, 2019. [Google Scholar]
- 18.Kingma DP, Ba J. Adam: a method for stochastic optimization. ArXiv:1412.6980 [preprint] https://arxiv.org/abs/1412.6980. Published online December 22, 2014. Accessed June 1, 2019. [Google Scholar]
- 19.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016; 770–778. [Google Scholar]
- 20.Lin TY, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017; 2117–2125. [Google Scholar]
- 21.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. ArXiv:1409.1556 [preprint] https://arxiv.org/abs/1409.1556. Published online September 4, 2014. Accessed June 1, 2019. [Google Scholar]
- 22.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical image computing and computer-assisted intervention – MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Cham, Switzerland: Springer, 2015. [Google Scholar]


