Abstract
Lipid research is attracting more and more attention as various key roles and novel biological functions of lipids have been demonstrated and discovered in the organism. Mass spectrometry (MS)-based lipidomics approaches are the most powerful and effective tools for analysis of cellular lipidomes with very high sensitivity and specificity. However, the artifacts generated from in-source fragmentation are always present in all kinds of ion sources, even soft ionization techniques (i.e., electrospray ionization and matrix-assisted laser desorption/ionization (MALDI)). These artifacts can cause many problems for lipidomics, especially when the fragment ions correspond to/are isomeric species of other endogenous lipid species in complex biological samples. These commonly observed artifacts could lead to misannotation, false identification, and consequently, incorrect attribution of phenotypes, and will have negative impact on any MS-based lipidomics research including but not limited to biomarker discovery, drug development etc. Liquid chromatography-MS, shotgun lipidomics, and MALDI-MS imaging are three representative lipidomics approaches in which ion source-generated artifacts are all manifested and are comprehensively summarized in this article. The strategies on how to avoid/reduce the artifacts of in-source fragmentation on lipidomics analysis are also discussed in details. We believe that with the recognition and avoidance of ion source-generated artifacts, MS-based lipidomics approaches will provide better accuracy on comprehensive analysis of biological samples and will make greater contribution to the research on metabolism and translational/precision medicine (collectively termed functional lipidomics).
Keywords: artifacts, functional lipidomics, in-source fragmentation, lipidomics, MALDI-MSI, mass spectrometry
1. Introduction
1.1. Complexity of lipids
Lipid species play numerous crucial roles in organism, already attracting considerable attention. With the development of genomics, proteomics, and molecular biology, it has been revealed that lipids and their related metabolites play key roles in numerous very important pathways and networks with more and more novel biological functions (e.g., anti-diabetes and anti-inflammatory effects, regulation of mitochondrial ATP release, and immunological regulation) being discovered (Kong et al., 2018; Wu, Shi, & Xu, 2016; Yore et al., 2014). Up to now, it has been unambiguously demonstrated that aberrant lipid metabolism is associated with the pathogenesis of various major human diseases, such as obesity and diabetes, varieties of cancer, neurodegenerative disorders (e.g., Alzheimer’s disease), and autoimmune diseases (e.g., systemic lupus erythematosus and gout) (Borgquist et al., 2016; Han, 2005; Hu et al., 2016; Lu et al., 2018; Perry et al., 2014). Therefore, determination of lipid alteration (including the levels and/or the composition of lipid species and/or classes) under different patho(physio)logical conditions and/or after perturbation could greatly facilitate understanding of disease pathogenesis, discovery of potential biomarkers, and development of more effective treatment.
However, lipids are extremely complicated, leading to the huge challenge of their qualitative and quantitative analyses. First, the number of cellular lipid species is exceedingly huge. It is anticipated that tens to hundreds of thousands of possible lipids, which are generally categorized into different classes and subclasses depending on the headgroup and the type of linkages between the headgroup and aliphatic chains, are present in a biological system at the levels of amol/mg to nmol/mg protein (Fahy et al., 2005; Shevchenko & Simons, 2010). Secondly, these lipids are dynamically keeping homeostasis of the system via many interwoven pathways and networks. In addition to the distribution and the diversities of lipid species varied from species, cell types, cellular organelles, subcellular membranes, leaflets of membrane bilayers, and membrane microdomains, their content and composition are also dynamically changing with life cycles, environmental conditions, or physiological and/or pathological perturbation (Han, 2016a; Wang et al., 2016). It is difficult to reveal/reflect the underlying mechanism(s) solely based on the determination of one lipid specie or a few lipid species. Thus, the metabolism of the entire lipidome should be investigated in a system biology approach (Dennis, 2009). Thirdly, the lipid species within one class/subclass are very similar. The lipid species within one lipid class or subclass have an identical polar headgroup and backbone, but various aliphatic chains differing in length (such as different number of carbon atoms), degrees of unsaturation, locations of double bonds, potential branches, and linkages connecting to backbones (Hu, Duan, & Han, 2020; Wang, Han, & Han, 2013). This structural characterization sometimes results in isobaric/isomeric mass overlaps, thereby limiting unambiguous lipid identification even between lipid classes/subclasses. To overcome the challenge, lipidomics has been proposed in early 2000 as an independent discipline to investigate all lipids present in a biological system in a large scale and at the levels of intact molecular species (Han & Gross, 2003; Lagarde et al., 2003).
1.2. MS-based lipidomics approaches
With the cooperative efforts of relevant scientists and the rapid development of modern analytical technologies, such as nuclear magnetic resonance techniques, fluorescence spectroscopy, high performance liquid chromatography (HPLC), microfluidic technologies, and particularly mass spectrometry (MS), the progress of lipidomics has been greatly accelerated since its emerging (Armstrong, 2009). Nowadays, MS-based approaches have already become the most powerful and remarkable ones in current lipidomics studies, since they inherently possess very high sensitivity and specificity (Hu, Wang, & Han, 2017; Yang & Han, 2016). In general, based on whether the lipid solution delivered to the ion source chamber is under a condition of constant concentration during the period of lipid analysis, the MS-based approaches can be divided into two major categories: one is LC-MS-based lipidomics, the other is direct infusion-based lipidomics that is also termed “shotgun lipidomics” (Han & Gross, 2005). The former fully takes advantage of the combination of high-separation performance of HPLC and high sensitivity of MS to analyze lipid species including the very low abundance lipid molecular species, while the latter maximally exploits the unique chemical and physical properties of lipid species, such as hydrophobicity, stability, and reactivity in different lipid classes/subclass for analysis of individual lipid species, including those of less ionizable or in very low abundance lipid species (Han, Yang, & Gross, 2012). Moreover, depending on the features and the mass spectrometers employed, both LC-MS-based and shotgun lipidomics could be further classified into different subtypes or variations, respectively (Hu et al., 2019).
Over the last decade, another important methodology termed matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) has been rapidly developed (Harvey, 2012). In comparison with the aforementioned lipidomics platforms, the MALDI-MSI approach enables one to investigate the spatial distribution and relative abundance of biomolecules directly in situ, particularly in tissue sections without any prior labeling or extraction processes. This technology could reveal the presence of changes of the analytes of interest even though the alterations are present in a very small area of tissue/organ. Moreover, MALDI is capable of ionizing molecules across a wide range of molecular weights, ranging from hundreds of Da to beyond 100 kDa. The approach has been widely employed to provide information about the in situ localization and identification of varieties of endogenous/exogenous molecules (such as lipids, proteins, peptides, xenobiotic compounds, and metabolites) in extensive fields including oncology, immunology, pharmacology, plant metabolism, etc. (Abbas et al., 2019; Galli et al., 2016; Yalcin & de la Monte, 2015). The MALDI-MSI on lipids is one of the most important research topics used to reveal the crucial roles of lipid species in organism. Additionally, since other desorption and ionization techniques have been developed, many other MSI platforms also have been formed for MS imaging (for instance, desorption electrospray ionization, laser ablation electrospray ionization, and secondary ion mass spectrometry) (Sturtevant, Lee, & Chapman, 2016). They have their own features and advantages (Han, 2016b).
1.3. Important to recognize and minimize ion source-generated artifacts in lipidomics
As we know, mass spectrometry can be used to identify and quantify molecules by detecting the mass to charge (m/z) ratios and fragment ions of their ionized forms in all of the MS-based lipidomics strategies. An essential step for this purpose is to convert molecules into their ion forms in an ion source. Electrospray ionization (ESI) and MALDI are two of the most prominent ion sources used in lipidomics, although many other technologies for ion generation have been developed and applied for lipid analysis (Han, 2016b). In addition, both of the technologies belong to soft ionization techniques, in which (quasi)molecular ions of analytes are generated with minimal in-source fragmentation (as the name suggests, a fragmentation process that occurs in the ion source during MS analysis). Unfortunately, in-source fragmentation is always present to some extent and even could not be neglected in some conditions. Generally, the degree of in-source fragmentation largely depends on the chemical structure of the analyte (e.g., sugar is more prone to in-source fragmentation to lose a water molecule and/or sugar molecule than others in the positive-ion mode) and harsh ionization conditions (e.g., high ionization temperature, high ionization voltage, very short distance between the spray tip and the inlet that leads to an extremely high electrical capacity, inappropriate setting of the gate voltages, etc.) (Han, 2016b).
Many studies have demonstrated that in-source fragmentation also exists in lipidomics analysis, for example, phosphatidic acid (PA) fragments could be produced from in-source fragmentation of their phosphatidylserine (PS) counterparts; the fragment ions of phosphatidylcholine (PC) and lysophosphatidylcholine (lysoPC) species yielded in source have the same masses as endogenous phosphatidylethanolamine (PE) and lysophosphatidylethanolamine (lysoPE) counterparts, respectively (Gathungu et al., 2018); the in-source fragmentation of lysoPC species also could lead to the observation of lysophosphatidic acid (lysoPA)-like species (Zhao & Xu, 2009); etc. It should be recognized that lipid species may appear in a variety of special ion forms in the presence of different modifiers in the solution or ionic liquids, leading to the production of unique fragmentation pathways after CID. Some characteristic fragment ions could be generated in these ion forms and used to elucidate these fragmentation pathways in the gas phase. However, these ion forms may be rarely present in routine experiments of lipidomics analysis and not practically useful for biological applications. In this review, only the commonly observed ion source-generated artifacts from the ion forms of lipid species widely used for analysis of biological samples are discussed. If the in source-generated artifacts correspond to other endogenous lipids, the ion-source fragmentation could be problematic, especially in shotgun lipidomics and MALDI-MSI analysis, as it largely interferes with identification and subsequent quantitation of lipid species present in complex biological samples. Specifically, the influence of in-source fragmentation on lipidomics is at least in the following three ways: 1) the presence of both precursors and ion source-generated artifacts could further increase spectral complexity and complicate data analysis including feature selection and spectral interpretation; 2) in-source fragmentation could potentially reduce ionization sensitivity and alter their quantification because of the loss of a certain amount of lipids in this process, resulting in falsely low concentrations; and (3) accordingly, the concentration of the endogenous lipids can be reported as falsely higher concentration if ion source-generated artifacts have the same mass with these lipid counterparts (Gathungu et al., 2018; Hu, Duan, & Han, 2020). Furthermore, the rate of in-source fragmentation usually depends on the kinetics and thermodynamics of the fragment ion yielded from the precursor ion (Han, Yang, & Gross, 2012). Therefore, in-source fragmentation could significantly result in missing the biologically relevant lipids and/or incorrect attribution of a phenotype to an artifact species or class.
Consequently, recognition and minimization, if avoidance is inevitable, of the artifacts resulted from in-source fragmentation are of importance during the method development stage for a selected category of lipids. Recently, several reports have been published to address the in-source fragmentation of lipids and proposed some strategies to minimize its influences (Gathungu et al., 2018; Xu et al., 2018; Xu, Lu, & Rabinowitz, 2015). In particular, Criscuolo et al. systemically evaluated the effects of ESI parameters (e.g., source temperature) on three different orbitrap-based platforms in an unprecedented detail to reduce the extent of lipid in-source fragmentation without an obvious loss in sensitivity (Criscuolo, Zeller, & Fedorova, 2020). All of these studies are very valuable and desirable. In this review, these commonly observed artifacts that can be readily mis-identified as endogenous lipids are comprehensively summarized. The chemical mechanisms responsible for the production of the fragmentation are also discussed. Finally, some strategies that can be employed to minimize/eliminate these artifacts in lipidomics analysis are proposed.
2. Commonly observed ion source-generated artifacts in lipidomics analysis
2.1. Artifacts resulted from in-source fragmentation in LC-MS and shotgun lipidomics
A. Fragment ions corresponding to PA species generated from in-source fragmentation of PS counterparts
It is one of the most representative examples for in-source fragmentation in lipidomics analysis that PA ions could be readily generated from ion-source dissociation of PS counterparts (Han, 2016b; Hu et al., 2019). PS is widely present in organism, serving as a structural component of the plasma and secretory membranes and accounting for approximately 5–10 % of the total phospholipids in most cases (Vance, 2015). It has been demonstrated that PS also engages in many important cellular processes, such as being an activator of several enzymes including protein kinase C, Na+/K+ ATPase, and neutral sphingomyelinase, and playing key roles as a critical signal for platelet activation, skeletal muscle development, and macrophage recognition (Leventis & Grinstein, 2010). As one class of phospholipids, PS comprises two acyl chains at the sn-1 and −2 positions of the glycerol moiety with the polar headgroup phosphoserine attached to the sn-3 position of the glycerol (Figure 1A). Thus, PS belongs to the category of negatively charged glycerophospholipids under physiological conditions. Although the acyl chains in PS vary greatly among cell types and organelles, saturated fatty acids of 16 or more carbon atoms are usually located in the sn-1 position, whereas unsaturated fatty acids are generally attached to the sn-2 position. Like PE and PC species, PS also displays asymmetrical distribution between the inner and outer leaflets, that is, PS normally localizes to the inner, cytoplasm-facing leaflet of the plasma bilayer membrane and the cytoplasm-facing leaflets of endosomal and Golgi membranes (Leventis & Grinstein, 2010). This asymmetrical membrane localization directly involves cell-cell or cell-vesicle contacts (Frey & Gaipl, 2011). Therefore, analysis of PS species, including their content, profile, and spatial distribution, could advance our understanding of related biological processes and reveal potential pathogenesis of diseases.
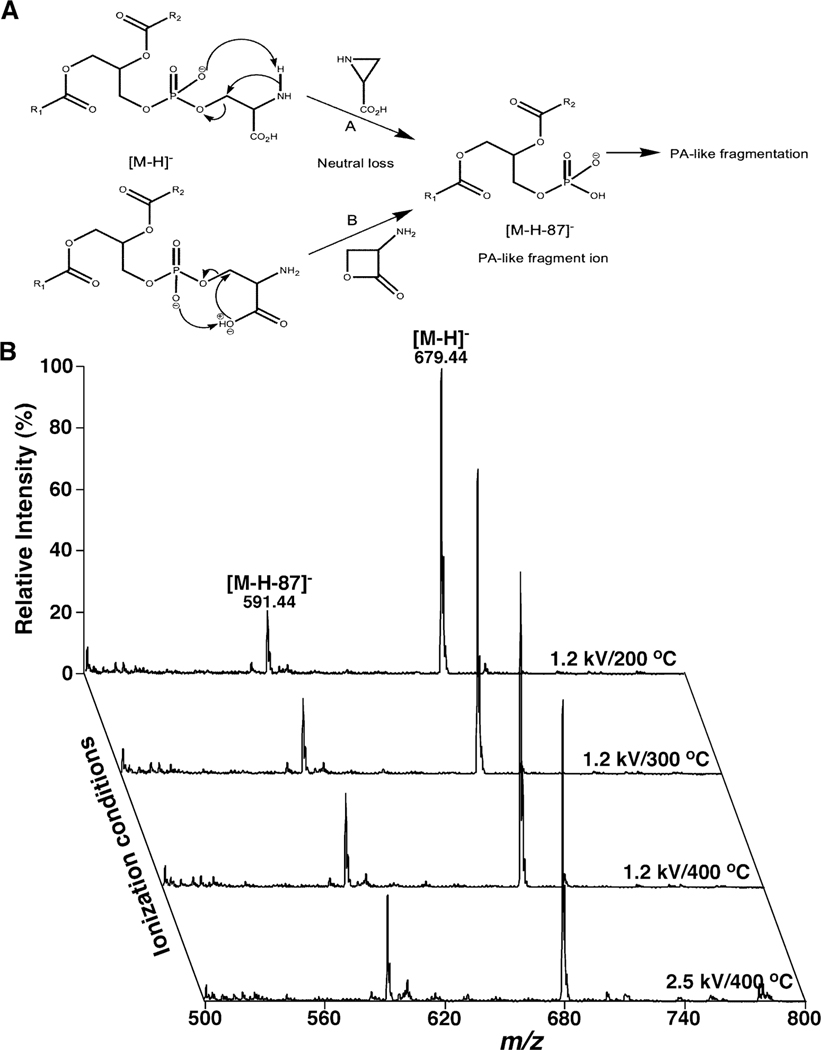
Figure 1.
Representative demonstration of PA artifacts resulted from in-source fragmentation of PS counterparts. (A) The fragmentation pathways proposed for formation of the deprotonated PA ion from PS after CID (Hsu & Turk, 2005). (B) Comparison of the ESI mass spectra of deprotonated di14:0 PS (m/z 679.4) acquired in the negative-ion mode at different conditions of temperature or spray voltage. The spectra were acquired on a QqQ mass spectrometer (Thermo Fisher TSQ Quantiva) equipped with an automated nanospray ion source (TriVersa NanoMate, Advion Bioscience Ltd.) and displayed after normalization to the base peaks. The PS species undergo in-source fragmentation to produce the PA fragment ion at m/z 591.4. The relative abundance of the PA artifact normalized to that of the PS molecular ion reflects the extent of in-source fragmentation. PA and PS denote phosphatidic acid and phosphatidylserine, respectively.
Since PS species are anionic phospholipids, analysis of these species is usually conducted in the negative-ion mode of ESI-MS. The deprotonated PS species are formed as a predominant molecular ion (i.e., [M-H]-). The fragmentation pattern of the deprotonated ions includes an intense fragment ion (i.e., [M-H-87]-) corresponding to a deprotonated PA counterpart yielded from the loss of serine (i.e., 87 amu), and a series of fragment ions that are essentially identical to those ions present in the fragmentation spectra of deprotonated PA counterparts after collision-induced dissociation (CID) (Han, 2016b). Two fragmentation mechanisms have been proposed for the loss of serine of the deprotonated PS species after CID (Figure 1A) (Hsu & Turk, 2005). The characteristic fragmentation pattern has been widely used for identification and quantitation of PS species in lipidomics analysis, particularly in shotgun lipidomics (Han & Gross, 2005).
Unfortunately, the facile loss of serine from PS species could occur in the ion source to generate the PA fragments. This ion-source fragmentation not only directly leads to the decreased ionization sensitivity of PS, but also results in the PA artifacts compounding the analysis of endogenous PA species at varied ionization conditions (e.g., different source temperature and spray voltage) (Figure 1B). Both source temperature and spray voltage are two of the most critical factors that affect ion generation and ionization efficiency in all the ionization technologies. As shown, the higher the source temperature, the more severely occurs the in-source fragmentation (Figure 1B). Similarly, the degree of source fragmentation increases with the higher spray voltage used (Figure 1B). Thus, if ionization condition is not well controlled, PA fragments generated from in-source fragmentation of PS could greatly complicate the quantification of endogenous PA species. In other words, the in-source fragmentation affects the analysis of not only the PS species, but also the true PA species present in lipid extracts from biological samples.
Therefore, it is very important to determine whether they are molecular or fragment ions when measurement of PA species becomes interested in lipidomics analysis. Some strategies could be considered in lipidomics analysis to solve this issue. From the MS standpoint, a PA artifact can be confirmed by examining the presence of the PA fragment ion resulted from the exogenously added internal standard of PS species. The degree of this fragment ion essentially represents the levels of PA counterparts resulted from those endogenously presented PS species. Thus, deduction of these PA fragment ions from the PA profile obtained from a lipid extract is necessary. From the biological point, PA is a critical precursor for biosynthesis of all phospholipids, such as PC, PE, and PS, as well as glycerolipids, such as diacylglycerol (DAG) and triacylglycerol (TAG) species. PA species also implicate in many regulatory processes including signaling pathways in cell growth, proliferation, and reproduction, and respond to hormones and biotic and abiotic stresses (Vance & Vance, 2004; Wang et al., 2006). The concentration of PA species in organism is strictly regulated and it is usually present in very low abundance in biological samples compared with other phospholipids (e.g., PS), accounting for about 1–2% of total phospholipids (Vance, 2015). Finally, PA species are commonly consisted of saturated or monounsaturated fatty acyls, while polyunsaturated fatty acyls are enriched in PS species as previously reported (Han et al., 2007; Hu et al., 2015).
Although these strategies can assist in identifying PA fragment ions, it is still difficult to use these methods in practical analysis of complex biological samples. The best way to solve the problem is to eliminate/minimize this fragmentation process in the ion source. For example, a standard solution of PS species can be further used to fine-tune the instrument after the instrument is tuned with a standard solution recommended by the instrumental manufacturers, and a less harsh ionization condition (e.g., lower source temperature, lower spray voltage, etc.) can be purposely set at the beginning of the further tune/calibration experiment. In any cases, an individual PS which is absent endogenously should always be added to check in the presence of ion source-generated PA fragments. A suitable ionization condition can be examined with the minimal existence of some PA artifacts.
B. Dimethyl PE artifacts yielded from ion-source dissociation of anion adducts of PC counterparts
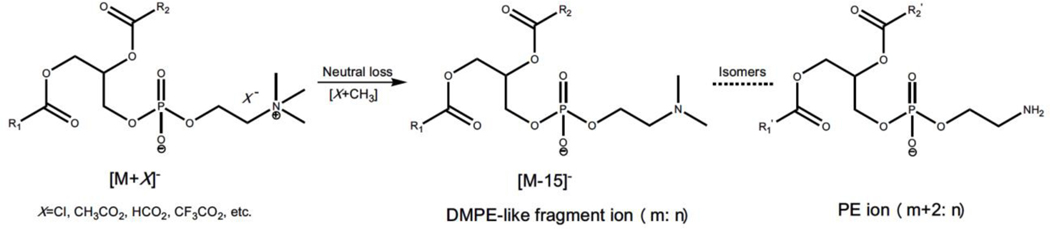
Dimethyl PE (DMPE) artifacts generated from in source-fragmentation of various anion adducts of PC counterparts are also commonly observed in the negative-ion mode of ESI-MS (Xu et al., 2018). It is well-known that PC species are the most abundant phospholipids in the majority of biological samples, such as plasma, the liver, kidneys, and the heart. These species contain a polar headgroup quaternary amine in the form of zwitterions attached to the phosphate (Figure 2). Thus, in the negative-ion mode, PC species can readily form various adduct ions with those small anion(s) present in the matrix (i.e., [M+X]-, where X = Cl, CH3CO2, HCO2, CF3CO2, etc.) (Hsu & Turk, 2009). These adducts tend to generate a [M-15]- ion (i.e., [M+X-CH3X]-) equivalent to DMPE counterparts. These DMPE artifacts even are present as the predominant quasimolecular ions of PC species in many cases, indicating that the neutral loss of CH3X from the anion adducts is very facile (Han & Gross, 1995; Kerwin, Tuininga, & Ericsson, 1994). DMPE artifacts generated from in-source fragmentation of those anion adducts of PC counterparts are easily misidentified as the endogenous DMPE species in lipidomics analysis (Houjou et al., 2004). Thus, the presence of these fragment ions can greatly challenge the identification of true DMPE lipids in lipid extracts as well as their accurate quantification.
Figure 2.
Structural illustration of DMPE fragment ions generated from in-source fragmentation of small anion adducts of PC counterparts and of their PE isomers. In the scheme, m and n denote the total number of carbon atoms and the total number of double bonds in two fatty acyls, respectively. The total number of carbon atoms in R1’ and R2’ in PE species is 2 carbon atoms more than that in R1 and R2 present in DMPE counterparts.
Some methods can be applied to distinguish the molecular ions of endogenous DMPE species from the artifacts resulted from in-source fragmentation of various anion adducts of PC counterparts in the negative-ion mode. Similar to the PA fragment ions discussed above, the presence of DMPE fragments can be readily confirmed by the according anion adducts of a PC standard. In fact, the total concentration of DMPE species present in biological samples is very low in most cases, since they only exist as biosynthetic intermediates of PC species through the methylation pathway from corresponding PE species (Cole, Vance, & Vance, 2012). There are two primary metabolic routes for de novo synthesis of PC species in mammal. One is the pathway through condensation of diglyceride and CDP-choline (also termed the Kennedy pathway). The other is the PE methylation pathway, converting PE to PC via three sequential methylation reactions by PE N-methyltransferase (Cole, Vance, & Vance, 2012). The PE methylation pathway is only significant in the liver, contributing up to 30 % of total hepatic PC synthesis. Therefore, compared with PE and PC species, the total content of DMPE species present in biological samples is much lower. In addition, dynamic lipidomics studies have revealed that PE methylation pathway produces primarily polyunsaturated fatty acid-containing PC species in mammals (Bilgin et al., 2011; DeLong et al., 1999; Pynn et al., 2011). However, the major PC species have two mono-unsaturated fatty acid moieties in yeast (Boumann et al., 2003; Boumann et al., 2004). Hence, the different profiles generated from the PE methylation pathway vs. ion-source fragmentation of PC species could also be a useful information for identification of DMPE artifacts in MS-based lipidomics analysis.
Furthermore, in source-fragmentation of anion adducts of PC counterparts could be obviously reduced/eliminated with a less harsh ionization condition. After the instrument is tuned with a standard solution as recommended, a particular standard of PC species could be used for further tuning to minimize the in-source generated DMPE artifact. However, it should be recognized that the minimal existence of DMPE artifacts is inevitable in a suitable ionization condition.
It should be noticed that DMPE artifacts (m: n) resulted from in-source fragmentation of anion adducts of PC species are also structural isomers of PE (m+2: n) counterparts (Figure 2). Since endogenous DMPE species are present in very low abundance in common lipid extracts, some software programs conducting lipid library research solely based on molecular masses (i.e., m/z) tend to mis-annotate the in source-generated DMPE (m: n) artifacts as PE (m+2: n) counterparts.
Some useful approaches could be applied to avoid the mis-annotation of DMPE artifacts as PE counterparts. First, in addition to using exogenous standards of PC species for further tuning, it is strongly advised that the ions annotated as PE species should be further confirmed through product-ion ESI-MS analysis in the negative-ion mode. Depending on whether the fatty acyl substituents are identical or not, one or two intense fragment ions around m/z 300 corresponding to fatty acyl carboxylate ions are present in the product-ion ESI-MS spectrum of deprotonated PE species. These fragment ions could clearly indicate the total number of carbon atoms in both of two fatty acyl substituents. Moreover, it is obvious that the fragmentation pattern of any DMPE (m: n) artifacts is different from that of mis-annotated deprotonated PE (m+2: n) species. Second, many derivative methods (e.g., Fmoc chloride and 4-(dimethylamino) benzoic acid) have been developed to achieve the analysis of PE species by exploring the specific reactivity of the primary amine group (Han et al., 2005; Zemski Berry et al., 2009). The advantages of chemical derivatization for analysis of PE species (such as introduction of a special group for generation of new fragmentation ions of a selected lipid class, shifting the molecular masses of lipid species of interest from the overlapped region to a new region without overlaps, etc.) have been extensively discussed (Hu, Duan, & Han, 2020; Hu et al., 2019; Wang, Wang, & Han, 2016). Therefore, chemical derivatization is one of the most attractive and effective strategies to remove the interference of DMPE artifacts for the analysis with endogenous PE species present in lipid extracts.
C. LysoPA-like artifacts resulted from in-source fragmentation of other lysophospholipid counterparts
Although it is common that some lysophospholipid-like artifacts (e.g., lysoPC and lysoPE) could be generated from the ion-source dissociation of counterparts of endogenous phospholipid classes (i.e., PC and PE species) in the negative ion mode through neutral loss of fatty acids and/or fatty ketenes in harsh ionization conditions (Hsu & Turk, 2009), they can be identified by using exogenous phospholipid standards and be avoided under a less harsh ionization condition as aforementioned. Incidentally, from the biological standpoint, lipid metabolites are present in a relatively small amount, usually accounting for <5 % of their parent lipid classes under normal physiological conditions (Han, 2016b). Therefore, in-source fragmentation of phospholipids must be taken into consideration when the level of a class of lysophospholipids is over 10 % of that of their parent phospholipid class.
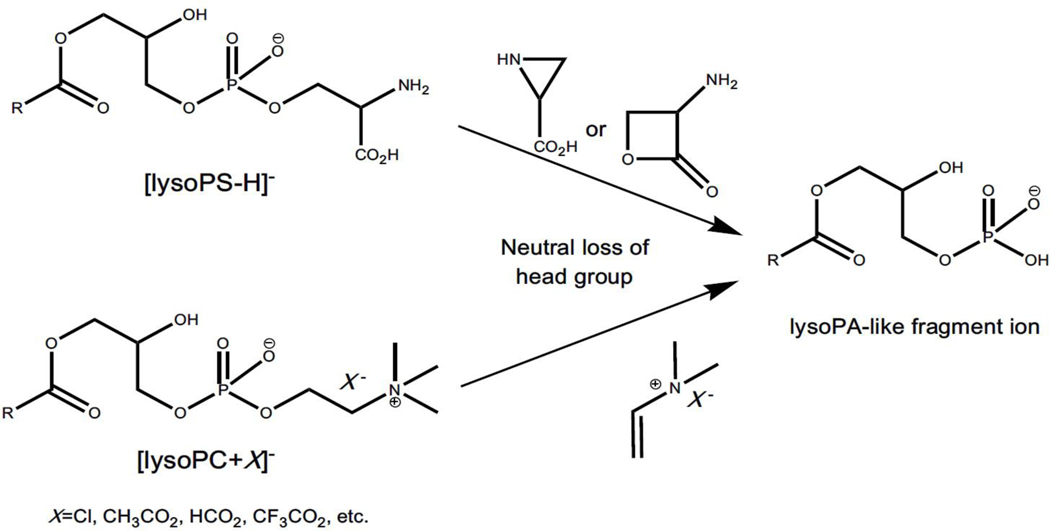
In addition to this type of ion-source generated lysophospholipid-like artifacts described above, it is particularly noteworthy that lysoPA-like artifacts can also be yielded from in-source fragmentation of other lysophospholipid counterparts (e.g., lysoPC and lysoPS species) through the neutral loss of their choline/serine headgroup (Figure 3) (Zhao & Xu, 2009). LysoPA species, which are released from a variety of mammalian cells including platelets, adipocytes, fibroblasts, and some cancer cells, are present in blood circulation. These species are an important class of lipid signaling mediators that exhibit various physiological activities. For instance, they involve regulation of cell cycle progression, the actin cytoskeleton, cellular differentiation, Ca2+ homeostasis, and cell survival via specific receptors, such as LPA1–6, and G-protein-coupled family of receptors (i.e., GPR35 and GPR87) (Contos, Ishii, & Chun, 2000; Noguchi et al., 2009; Pagès et al., 1999; Pagès et al., 2000). The metabolic abnormality of lysoPA species is thought to induce some human diseases including cancer, arteriosclerosis, sepsis, and other pathophysiological conditions. Furthermore, the levels of lysoPA species in plasma/serum have been identified as potential biomarkers for these diseases (Drobnik et al., 2003; Fuchs et al., 2005). Therefore, accurate analysis of lysoPA species in biological samples becomes critically important. However, it has been demonstrated that lysoPA-like artifacts can be generated from the conversion of lysoPC and lysoPS counterparts happened in the ion source through neutral loss of headgroups in the negative-ion mode of ESI-MS analysis (Zhao & Xu, 2009). It should be recognized that the level of endogenous lysoPA individual species is much lower than that of their lysoPC and/or lysoPS counterparts (Wang, Wang, & Han, 2015). The in-source fragmentation of lysoPC and/or lysoPS, even though present in very low content, could easily lead to mis-annotation and falsely high concentration of lysoPA species (Zhao & Xu, 2009). Thus, special attention should be paid to the analysis of lysoPA species.
Figure 3.
Representative scheme of lysoPA-like artifacts resulted from in-source fragmentation of other lysophospholipids classes. The ion-source generated lysoPA-like artifacts are from lysoPS and lysoPC via the neutral loss of their serine and choline headgroups in the negative-ion mode of ESI-MS analysis, respectively.
Similar strategies could be applied to reduce/eliminate these artifacts, such as exogenous standard method or optimizing ionization condition method. Any ion peaks that are identified as lysoPA species and coeluted with other lysophospholipid species in LC-MS analysis should be paid particular attention as this type of artifacts. Moreover, pre-separation of lysoPC and/or lysoPS from lysoPA is seen as the best way for accurate analysis of lysoPA species, particularly in shotgun lipidomics. However, it is extremely difficult to achieve the complete separation between lysoPA species and other lysophospholipids, especially in lipid extracts from complex biological samples.
A novel prefractionation method for comprehensive analysis of all kinds of lysophospholipids by shotgun lipidomics perfectly solving the problem of in-source fragmentation has been developed (Wang, Wang, & Han, 2015). Briefly, on the basis of the unique physical/chemical properties that lysophospholipids tend to be present in the aqueous phase after organic solvent extraction and the Lewis acid-base interaction between HybridSPE zirconia ions and lysophospholipids, the novel strategy of sample preparation separates lysoPA from other lysophospholipid classes and enriches all these lysophospholipid classes from aqueous solution using a HybridSPE column eluted with a basic solution through varied pH conditions (Hu, Duan, & Han, 2020; Wang, Wang, & Han, 2015). The method has been successfully applied for comprehensive analyses of various lysophospholipid classes including lysoPA, lysoPE, lysoPC, lysoPS, lysophosphatidyinositol, and lysophosphatidyglycerol present in individual lipid extracts from biological samples (e.g., mouse liver) by multi-dimensional MS-based shotgun lipidomics with very high accuracy, efficiency, sensitivity, and reproducibility, greatly facilitating the progress of lysophospholipid-related research (Wang, Wang, & Han, 2015).
D. Cholestadiene ion generated from in-source fragmentation of both free cholesterol and cholesteryl esters
Cholesterol is one of the essential lipid components of mammalian cell membranes, especially as a major component of lipid ordered membrane domains, and a precursor for the synthesis of various steroid hormones (Maxfield & Tabas, 2005). It appears mainly as free cholesterol in membranes and as cholesteryl ester (CE) species which are esterified with a variety of fatty acids, and stored in lipid droplet and transported in lipoprotein particles, respectively (van Meer, Voelker, & Feigenson, 2008). Cholesterol has unique biophysical properties. Changes of cholesterol content in cellular membrane can greatly alter the composition and organization of lipid membranes and have profound effects on cellular functions such as signal transduction and membrane trafficking (Lange, Ye, & Steck, 2004). It is not surprising that aberrant metabolism of cholesterol involves in pathogenesis of many human diseases. For example, the role of cholesterol and its metabolites in atherosclerosis has been studied for decades, and many molecular mechanisms have been proposed in considerable detail (Hoeke et al., 2016; Sandesara et al., 2019). Moreover, both free and total cholesterol levels are widely used to predict an individual’s risk of coronary heart disease in clinical and preventive healthcare (Ridker, 2014). Therefore, this feature requires accurate quantitation of free cholesterol and CEs in both research and clinical diagnose.
Cholestadiene ion represents an example that can be readily yielded from in-source fragmentation of ammonium adducts of both free cholesterol and CE species in the positive-ion mode of ESI-MS. The existence of this artifact could complicate the analysis of both free cholesterol and CE species. Many chromatographic methodologies have been developed for quantitative analysis of free cholesterol and CEs including HPLC, thin layer chromatography, and gas chromatography (Hoving, 1995). However, these conventional methods are time-consuming and often lack the sensitivity required for the analysis of lipoproteins or cellular subfractions. Therefore, several MS-based approaches have been established for quantification of cholesterol and its ester species (Briche, Carter, & Webb, 2002; Lembcke et al., 2005). However, direct analyses of free cholesterol and CE species with intact molecular ions by ESI-MS are hampered due to their poor ionization efficiencies, since a prominent charge site is absent in this category of lipids.
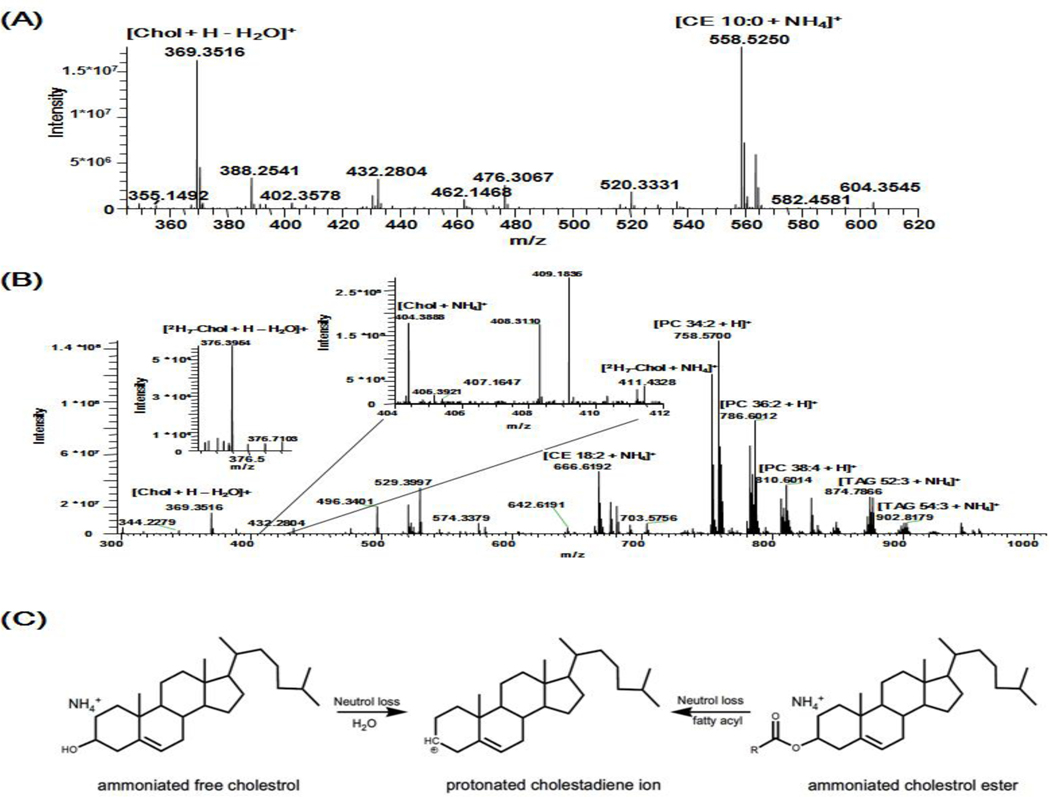
Cholesterol and CE species can be readily ionized as their adducts with a variety of small cations, such as ammonium, lithium, and sodium in the positive-ion mode in the presence of an according modifier in the lipid solution. Thus, an ammonium salt is commonly added as a modifier for sensitive analysis of cholesterol and CEs (Duffin et al., 2000; Gallego, Hojlund, & Ejsing, 2018; Horing et al., 2019). In addition, the ammoniated free cholesterol (e.g., [M+NH4]+) tend to generate protonated cholestadiene ion (m/z 369.3, [M+H-H2O]+) via the neutral loss of H2O in the ion source. Moreover, this fragment ion usually appears as the dominant product ion in the mass range of m/z 50–1000 and might be monitored for quantitation of endogenous free cholesterol instead of its quasimolecular ion (e.g., [M+NH4]+), which is detected in a very low intensity or even buried in the baseline noise under experimental conditions in lipidomics analysis (Figure 4B) (Graessler et al., 2009).
Figure 4.
Schematic illustration of a cholestadiene fragment ion with m/z 369.3516 generated from in-source fragmentation of ammonium adduct ions of free cholesterol and CEs. (A) The spectrum of synthetic CE 10:0 detected as an ammonium adduct at m/z 558.5245 in the positive-ion mode of FTMS. (B) The spectrum of human plasma in the positive-ion mode of FTMS. Cholesterol and 2H7-cholesterol are detected as low abundant ammonium adducts having m/z 404.3887 and 411.4326, respectively. Free cholesterol and CEs undergo in-source fragmentation to produce the fragment ion of m/z 369.3516. Some abundant endogenous lipid species are annotated (Reprinted from the supplemental materials of ref. (Gallego, Hojlund, & Ejsing, 2018) with permission from the American Chemical Society, Copyright 2018). (C) Representative scheme of a cholestadiene fragment ion resulted from in-source fragmentation of ammoniated free cholesterol and CEs.
Unfortunately, protonated cholestadiene ion is not specific to free cholesterol, since it can also be generated from in-source fragmentation of ammonium adduct ions of CEs. In product-ion ESI-MS spectra of ammoniated CE species, the protonated cholestadiene ion corresponding to the neutral loss of a fatty acid is the most abundant product ion after CID. Therefore, it enables one to profile individual molecular species of CEs by precursor-ion scanning of m/z 369.3 sensitively in the presence of an ammonium salt in the lipid solution. Nonetheless, this fragment process could also occur in the ion source due to the facile loss of the fatty acid in CE species (Gallego, Hojlund, & Ejsing, 2018) (Figure 4A). Therefore, detection of a protonated cholestadiene ion for analysis of free cholesterol can result in a falsely high concentration in lipidomics analysis.
In addition to a less harsh ionization condition employed to reduce ion source-generated artifacts, several other measures are effective for resolving this issue. These include 1) improvement of the ionization efficiency of free cholesterol by using chemical derivatization with methoxyacetic acid, sulfur trioxide, or acetyl chloride (Liebisch et al., 2006; Sandhoff et al., 1999); and 2) using more advanced commercial mass instruments. For instance, high resolution full scan FTMS can be employed to monitor cholesterol as intact ammonium adduct (Horing et al., 2019). With the development of more sensitive hybrid quadrupole-Orbitrap-based instruments (e.g., Orbitrap Fusion and Q-Exactive), it is possible to quantify the level of free cholesterol using novel monitoring modes, such as parallel reaction monitoring (PRM) or multiplexed PRM. In these approaches, to achieve the quantitation, selected ammoniated cholesterol and appropriate internal standard are sequentially isolated by a quadrupole mass analyzer, fragmented in a collision cell, and then fragment ions from each of the selected precursor ions are trapped inside the collision cell prior to being routed together to an orbitrap mass analyzer for simultaneous detection (Gallego, Hojlund, & Ejsing, 2018; Horing et al., 2019).
E. Ceramide-like artifacts generated from in-source fragmentation of glycosphingolipids
Incidentally, in-source fragmentation also extensively occurs in analysis of glycosphingolipids containing a ceramide backbone and a glycan moiety (e.g., hexosylceramide and lactosylceramide species) in the positive-ion mode of ESI-MS analysis. This is due to that sugar residue is prone to the neutral loss in an ion source (Levery, 2005). Thus, the fragment artifacts corresponding to ceramide-like ions can lead to a falsely high concentration of endogenous ceramide species in lipid extracts if the in-source fragmentation is not considered, especially of the biological samples that are enriched with glycosphingolipids. The methods aforementioned can also be used to identify and eliminate the ceramide-like artifacts.
2.2. Ion source-generated artifacts in MALDI-MSI
MALDI-MSI technique is becoming a powerful and valuable tool for analysis of lipid spatial distribution, which could provide unique insights into the cellular organization and/or metabolism in biological systems. Since MALDI ion source is relatively harsher than the one in ESI, the ion source-generated artifacts present in LC-MS and shotgun lipidomics not only still exist in MALDI-MSI analysis, but also is much more severe. Therefore, all those artifacts discussed above and their extents resulted should be recognized. The investigators using MALDI-MS should understand the complication caused by these artifacts during the analysis.
In addition to these artifacts, there exist some unique fragment ions which can be readily mis-annotated as endogenously existing lipid species or isomeric to endogenous lipid species. This mis-identification is largely due to two facts. First, simple sample pretreatment for MALDI-MSI of tissue slides compared to ESI-MS analysis of lipid extracts pre-excludes prior labeling or extraction processes. Thus, the complexity of microenvironments (e.g., pH, the differential concentrations of sodium and potassium, etc.), instead of an added modifier, could yield differential effects on ionization efficiency and/or fragmentation. Secondly, in the majority of MALDI-MSI studies, tandem MS analysis following MS imaging is not easy and seldomly performed, and assigned lipid species in almost all reports in the literature are solely annotated from the detected m/z. Unfortunately, incorrect annotation is frequently made.
It is well known that sodium ion always exists in varieties of tissue samples and it is the major small cation in most of the microenvironments. Those facile fragment ions yielded from sodium adducts of a variety of lipids including glycerolipids, glycerophospholipids, and sphingolipid species can be generated from in-source decay. These artifacts are frequently annotated as their endogenous counterparts or other isomeric lipid species, leading to erroneous interpretation of many observations. In this section, these ion source-generated artifacts in MALDI-MSI are summarized.
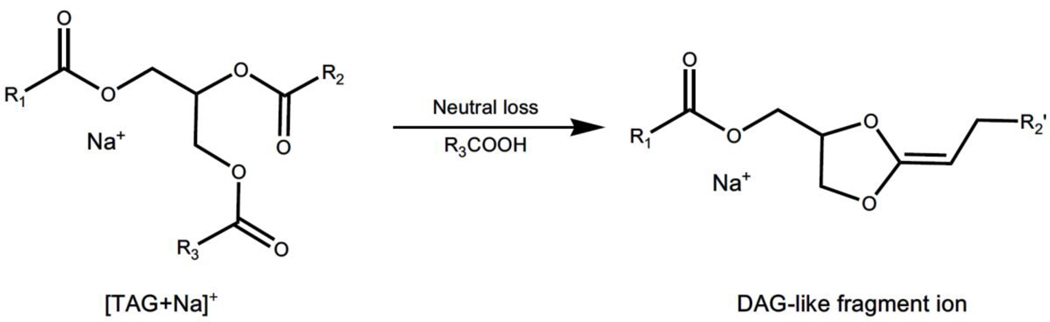
A. Sodiated diacylglycerol-like artifacts generated from in-source decay of sodiated TAG counterparts
TAG and DAG are two of major glycerolipid classes in biological samples. The incidence of metabolic syndrome (e.g., abdominal obesity, hypertension, dyslipidemia, and hyperinsulinemia) increases with the global prevalence of obesity. It is believed that the first step towards developing insulin resistance and metabolic syndrome is associated with the accumulation of lipids in non-adipose tissue, mainly in the form of TAG species (Nascimento-Ferreira et al., 2017; Samuel & Shulman, 2016). Therefore, studies focusing on the deposition of TAG species in different tissues are of importance, especially in the liver, because more than one-fourth of the global population has been affected with non-alcoholic fatty liver disease (NAFLD) caused by abnormal accumulation of hepatic TAGs and DAGs (Younossi et al., 2016). Moreover, it has been demonstrated that the spatial distribution of individual TAG species in different hepatic zone (such as periportal zone, midzone, and pericentral zone) varies with the severity of progress of NAFLD (Alamri et al., 2019). In other words, the distribution of TAG species in different organs, the liver in particular, could reflect the development of related diseases. Thus, the information about the regional distribution of individual TAG and/or DAG species in different organs revealed by MALDI-MSI could greatly contribute to the understanding of the underlying molecular mechanisms and the progress of the diseases.
As well known, it is difficult to ionize TAGs by protonation, even in the presence of organic acids since a prominent charge site is absent in this class of lipids (Duffin, Henion, & Shieh, 1991). However, TAG species can be readily ionized as adducts with many different small cations, for example, ammonium, lithium, or sodium in the positive-ion mode, displaying very strong quasimolecular ion peaks in the spectra. Thus, if there is no additional modifier, MALDI mass spectra of TAG species exclusively display the sodiated species due to common existence of sodium ion in tissues. After CID, tandem MS analysis of sodiated TAGs in the product-ion analysis mode shows some prominent fragment ions corresponding to the neutral loss of paired free fatty acid(s) and their sodium salt (Duffin, Henion, & Shieh, 1991; Herrera, Potvin, & Melanson, 2010). This unique fragmentation pattern has been maximally exploited by our group for identification and quantitation of TAG species present in biological samples using an MDMS-SL approach (Han & Gross, 2001). Unfortunately, these fragment ions (i.e., sodiated DAG-like species) can also be generated in a MALDI ion source because of the facile loss of fatty acid(s), leading to mis-annotation, and thereby totally false regional distribution of endogenous DAG species (Figure 5) (Al-Saad et al., 2003).
Figure 5.
Representative scheme of DAG-like artifacts arising from in-source fragmentation of sodiated TAG counterparts in the positive-ion mode of MALDI-MSI. There is no selective dissociation of a certain fatty acyl chain at the sn-1, −2 or −3 position of the glycerol moiety when the fatty acyl chains are similar in their length and degrees of unsaturation. DAG and TAG represent diacylglycerol and triacylglycerol, respectively.
In addition to the identification of these artifacts using exogenous standards of TAG, there are some methods to eliminate/overcome the issue. First of all, it is well known that the higher the laser power, the more fragment ions are yielded in a MALDI ion source (Yang et al., 2011). The laser power for application in MALDI strongly depends on the sample preparation step (e.g., matrix crystal size), stepper motor accuracy, and laser spot size (Han, 2016b). Thus, reduction of the laser power could effectively decrease the in-source fragmentation of sodiated TAGs through the selection of more suitable matrix and laser spot size (Dreisewerd et al., 1995); Secondly, endogenous DAGs in biological samples are usually present in much lower levels than that of TAGs, since most of DAG species are used for de novo synthesis of TAGs and phospholipids (e.g., PE and PC species). Furthermore, DAG is a key regulator of cell physiology, controlling the membrane recruitment and activation of signaling molecules. Thus its generation and metabolism are strictly controlled, which suggest that the concentration of DAG is kept relatively constant (Perry et al., 2014); Thirdly, the ion source generated DAG profile is usually much complicated than that of TAG in an identical tissue spot since each TAG species could yield up to three or more DAG-like fragment ions. Finally, in theory, using new matrices for selective ionization of DAG species and/or attenuating in-source decay of TAG species is also one of the best methods to prevent this problem. To this end, our group has developed an optimized mixture of matrices for enhanced coverage of lipid analysis and imaging by MALDI-MSI with minimal in-source fragments (Wang, Wang, & Han, 2018). Moreover, it is also desirable that novel approaches of pretreatment of samples for enhancing the sensitivity of DAGs including on-tissue digestion and on-tissue derivatization might be developed in the future. These strategies have been successfully used to facilitate detection and localization of peptides, proteins, and some drugs through modification of analytes in the tissue (Norris & Caprioli, 2013).
B. Sodiated PA artifacts resulted from in-source dissociation of many sodiated glycerophospholipid classes
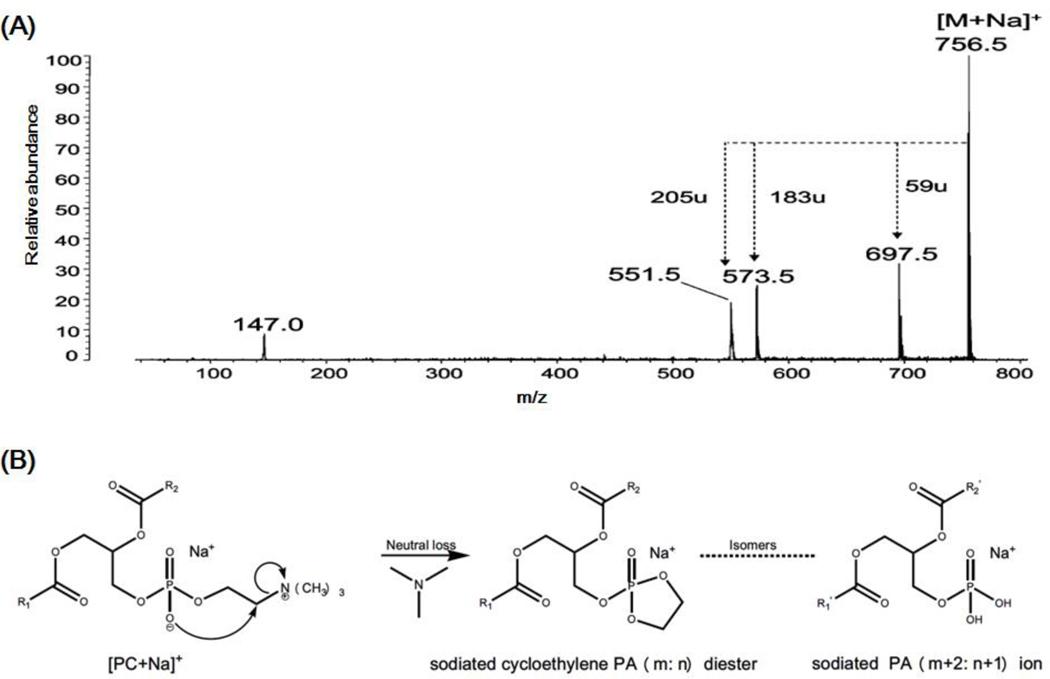
The spatial distribution of glycerophospholipids is also an important subject in MALDI-MSI. Like TAGs, many glycerophospholipids including PC, PE, and PS species are detected mainly in the form of sodium adducts in positive-ion MALDI-MSI. The fragment ions corresponding to the neutral loss of a portion of their each headgroup, such as trimethylamine (m/z 59), possible vinylamine (m/z 43), and serine (m/z 87) are always present in the post-source decay fragmentation spectra of sodiated PC, PE and PS lipids in MALDI-MS, respectively (Stübiger, Pittenauer, & Allmaier, 2008). Moreover, these sodiated lipids could also fragment in the ion source to produce PA-like artifact ions corresponding to the facile loss of a portion of their headgroup. Incidentally, the type of these artifacts generated from in-source fragmentation of PCs and other phospholipids (e.g., PE and PS species) is different due to the fragmentation pathways for their formation are different (Figures 6 and 7). Herein, PA-like artifacts yielded from the in-source fragmentation of PCs are taken as an example.
Figure 6.
Schematic illustration of PA-like artifact generated from in-source fragmentation of sodium adduct of PC in MALDI-MSI. (A) The post-source decay fragmentation spectrum of sodiated di16:0 PC (m/z 756.5) displays three predominant headgroup-related fragment ions resulted from the loss of trimethylamine (m/z 59) cholinephosphate (m/z 183), and sodiated cholinephosphate (m/z 205) (Reprinted from ref. (Stübiger, Pittenauer, & Allmaier, 2008) with permission from the American Chemical Society, Copyright 2008). (B) The fragmentation pathway proposed for formation of the sodiated cycloethylene PA (m: n) diester ion, which is the isomer of sodiated PA (m+2: n+1) ion (Hsu & Turk, 2003).
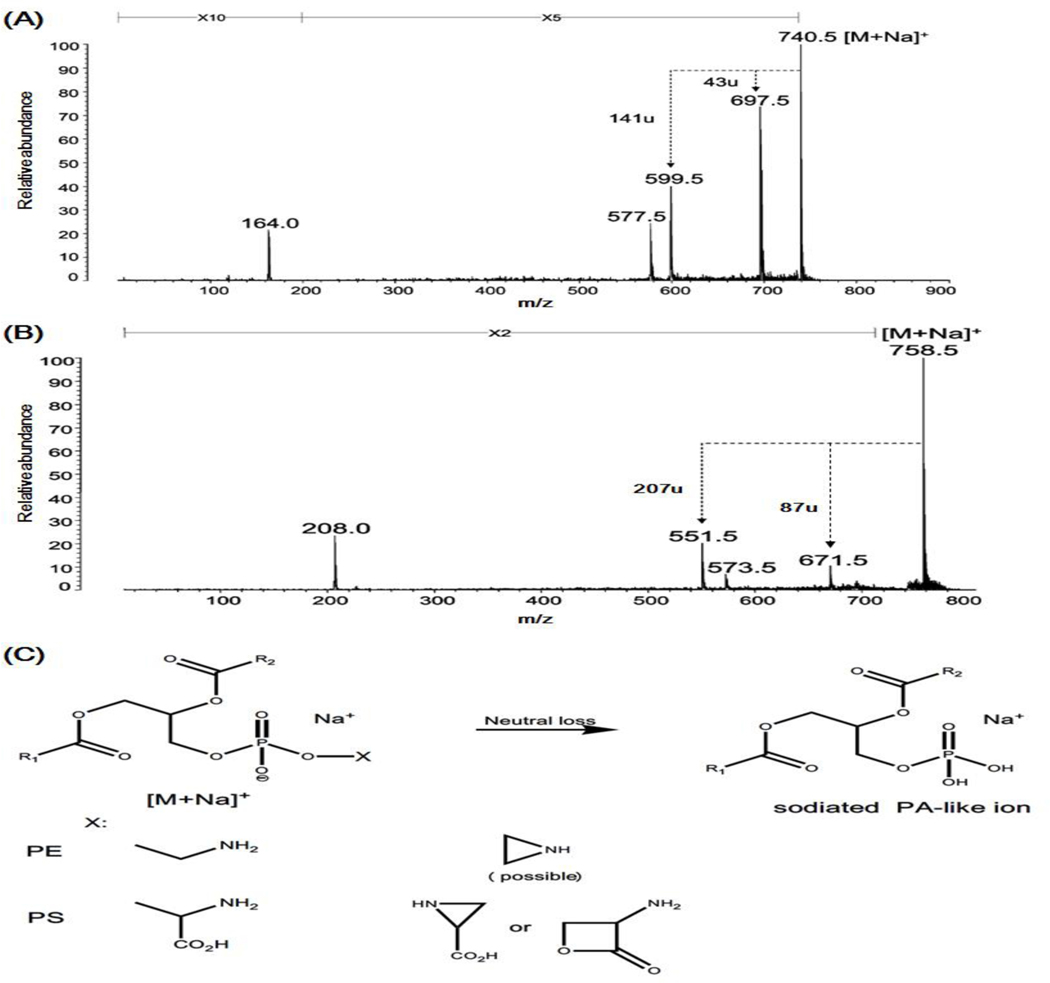
Figure 7.
Schematic illustration of PA-like artifacts yielded from in-source fragmentation of sodium adduct of PE and PS species in MALDI-MSI. (A) and (B) are the post-source decay fragmentation spectra of sodiated 16:0/18:1 PE (m/z 740.5) and di16:0 PS (m/z 758.5) showing the predominant headgroup-related fragment ions resulted from the neutral loss of a portion of their each headgroup (i.e., possible vinylamine (m/z 43) and serine (m/z 87)), respectively. (Reprinted from ref. (Stübiger, Pittenauer, & Allmaier, 2008) with permission from the American Chemical Society, Copyright 2008). (C) Representative scheme of PA-like artifacts resulted from in-source fragmentation of sodiated PE and PS species in MALDI-MSI. PA, PE, and PS denote phosphatidic acid, phosphatidylethanolamine, and phosphatidylserine, respectively.
As aforementioned, in LC-MS and shotgun lipidomics analysis, DMPE/PE-like artifacts can be yielded from the in-source fragmentation of anion adducts of PC counterparts in the negative-ion mode. However, sodiated PC species usually yield the strongest signals in the positive-ion MALDI-MSI due to the stable quaternary ammonium moiety. The post-source decay fragmentation spectra of sodiated PCs display three predominant headgroup-related fragment ions resulted from the losses of trimethylamine (i.e., [M+Na-59]+), phosphocholine (e.g., [M+Na-183]+), and sodiated cholinephosphate (i.e., [M+Na-(Na+182)]+) (Figure 6A), respectively (Stübiger, Pittenauer, & Allmaier, 2008). These fragment ions also exist in fragmentation spectra of sodium adducts of other subclasses of PCs (i.e., plasmenylcholine and plasmanylcholine species) (Hsu & Turk, 2003).
It is worth noted that the fragment ion (i.e., sodiated cycloethylene PA (m: n) diester) corresponding to the neutral loss of trimethylamine can also be yielded from in-source fragmentation of sodiated PC counterpart. The possible mechanism leading to the artifacts is illustrated in Figure 6B. More importantly, the artifacts and true sodiated PA (m+2: n+1) present in biological samples are isomeric, possessing identical molecular weights. Since the concentration of PC species is the most abundant one in the majority of biological samples, in-source fragmentation of sodiated PC species yields intense sodiated cycloethylene PA (m: n) diesters regardless of the efforts to minimize the artifacts. Researchers using MALDI-MSI tend to mis-annotate this type of fragments to their endogenous isomers (i.e., sodiated PA (m+2: n+1) species) due to a lack of tandem MS identification. This type of mis-annotation clearly leads to falsely regional distribution of endogenous PA species in MALDI-MSI analysis.
Similarity, PA-like artifacts could also arise from in-source fragmentation of other sodiated phospholipids (i.e., PE and PS species) in the positive-ion mode of MALDI-MSI (Figure 7A–B). However, it should be pointed out that the chemical structures of PA-like artifacts generated from sodium adduct of these two lipid classes is different from that of PC species. The former has an identical structure with endogenous sodiated PA species (Figure 7C), while the latter is the isomer of true sodiated PAs present in biological samples as aforementioned (Figure 6B). Therefore, in theory, more attention should be paid to this type of PA-like artifact from PE and PS species.
In fact, compared with those mis-annotated artifacts resulted from PC species, the levels of the PA-like artifacts generated from sodiated PE and PS species are much lower, since the signals of these other classes of phospholipids are largely suppressed by the coexisting PC species (Petkovic et al., 2001). Therefore, identification and avoidance of the PA-like artifacts resulted from sodiated PC species become of importance in analysis of spatial distribution of PA species with an MALDI-MSI approach. In fact, it is not difficult to confirm the artifacts using exogenous standards of PC species. In addition, from the standpoint of biological functions, endogenous PA species have unique chemical structures mainly containing saturated or monounsaturated fatty acyls, and their concentrations are much lower than that of PCs as mentioned in Section 2A. In addition to minimizing the artifacts by reducing the laser power, there exist other methods for avoidance of the artifacts as described in the last section. For instance, the only fragment ion cholinephosphate (m/z 184) is shown in post-source decay fragmentation spectra of protonated PC species. Therefore, tissue washing procedures can be used to remove sodium ion prior to application of matrix in MALDI-MSI workflows. For example, a simple and effective method to desalt the tissue sections with ammonium acetate solution that is suitable for MALDI-MSI has been developed (Wang, Liu, & Wu, 2011). The method not only removes alkali metal ions, but also enhances the overall spectral quality, benefitting additionally in profiling of lipids in tissue. Moreover, anionic lipids such as PAs are usually determined in tissue sections by MALDI-MSI in the negative-ion mode (Angel et al., 2012). These ways can essentially eliminate the appearance of PA artifacts described above.
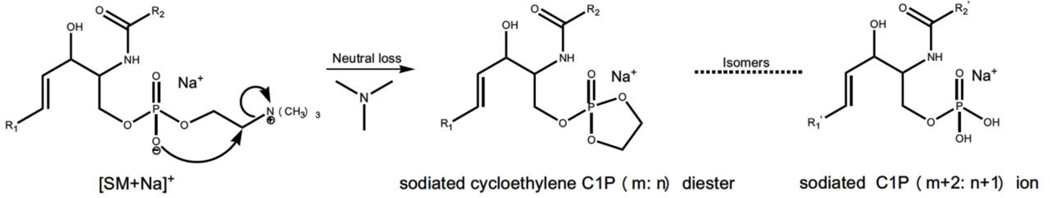
C. Sodiated ceramide 1-phosphate artifacts yielded from in-source decay of sodiated sphingomyelin (SM) counterparts
Since PCs and SMs have an identical polar headgroup, similar in-source fragmentation is also present in ionization of SM species using a MALDI-MSI approach (Stübiger, Pittenauer, & Allmaier, 2008). Specifically, like PC species, the post-source decay spectrum of sodiated SM species mainly includes two headgroup-related fragment ions corresponding to the neutral loss of trimethylamine (i.e., [M+Na-59]+) and the loss of phosphocholine (e.g., [M+Na-183]+), respectively (Han & Gross, 1995). These fragment ions can also be produced from in-source fragmentation of sodiated SM species owing to the facile loss of the headgroup. The yielded intense fragment ion (i.e., sodiated cycloethylene ceramide-1-phosphate (C1P) (m: n) diester) arising from the loss of trimethylamine of sodiated SM counterpart is isomeric to sodiated C1P (m+2: n+1) species (Figure 8). In the MALDI-MSI study, these source-yielded artifacts are easily mis-annotated as the isomeric sodiated C1P (m+2: n+1) species and lead to serious mis-interpretation of the existence and distribution of CIP species. Therefore, these C1P artifacts have to be considered if the spatial distribution of C1P species is of interest in MALDI-MSI analysis.
Figure 8.
The fragmentation pathway proposed for the formation of the sodiated cycloethylene C1P diester. In-source fragmentation of sodiated SM species produces the sodiated cycloethylene C1P (m: n) diester counterparts, which is an isomer of sodiated C1P (m+2: n+1) ion, leading to mis-interpretation of these ion-source yielded artifacts. C1P and SM abbreviate ceramide 1-phosphate and sphingomyelin, respectively. m and n represent the total number of carbon atoms and the total number of double bonds in both the ceramide backbone and the fatty acylamide in C1P.
Accordingly, the strategies (i.e., reducing laser power and tissue washing) that can be employed to avoid the PA artifacts are also effective for prevention of the C1P artifacts. Using exogenous standards of SM species to confirm the existence of C1P artifacts is always useful in MALDI-MSI analysis if possible. Furthermore, as one of the important sphingolipid metabolites, C1P is formed by phosphorylation of ceramide catalyzed by ceramide kinase and acts as a signaling molecule involving a diverse range of cellular processes, such as stimulating DNA synthesis and cell division for fibroblasts, blocking apoptosis in macrophages, controlling phagocytosis in neutrophils, and mediating inflammatory responses (Maceyka & Spiegel, 2014). Thus, its metabolism is precisely regulated and kept in relatively constant in the cellular level, and its concentration is usually much lower than that of SM counterpart in biological samples. Moreover, like PA species, it is suggested that analysis of C1P prefers to be conducted in the negative-ion mode of MALDI-MSI.
3. Summary
Advances of MS-based lipidomics approaches have greatly accelerated the progress on lipid research, including qualitative and quantitative analyses, and spatial distribution. However, the artifacts generated from in-source fragmentation have become a huge problem in lipidomics, especially when the artifacts identical or isomeric to the endogenous lipids. These artifacts can lead to false results, and thereby, incorrect attribution of a phenotype to an artifact species or class if they are not avoided. Therefore, it is greatly important to recognize and eliminate these possible artifacts in MS-based lipidomics approaches. The commonly observed artifacts and the effective approaches employed to identify and prevent/reduce these artifacts are summarized in the review (Table 1).
Table 1.
Summary of commonly observed in-source fragmentation and strategies for recognition and avoidance of these generated artifacts in lipidomics analysis.
| MS-based lipidomics approach | Lipid class | In-source fragmentation | Strategies for recognition and avoidancea |
|---|---|---|---|
|
| |||
| LC-MS and shotgun lipidomicsb | PS - PA | NLS of serine - generation of PA | 1) A less harsh ionization condition; 2) an exogenous standard of PS included; 3) the lower concentration of PA species compared with PS species; and 4) the structural characteristics of endogenous PA and PS species. |
| PC - DMPE/PE | NLS of CH3X from [PC+X]- - generation of DMPE species (mis-annotation as isomeric PE) | 1) A less harsh ionization condition; 2) inclusion of an exogenous standard of PC; 3) very low concentration of DMPE species; 4) the structural characteristics of endogenous DMPE species; 5) further confirmation through product-ion ESI-MS analysis; and 6) chemical derivatization. | |
| Other lysophospholipid - lysoPA | NLS of choline/serine headgroup from lysoPC and lysoPS -generation of lysoPA species | 1) Inclusion of exogenous standards of lysoPC and lysoPS; 2) a less harsh ionization condition; and 3) the novel prefractionation method. | |
| Free cholesterol/CE - cholestadiene ion | NLS of water/fatty acid from free cholesterol/CE, respectively - generation of cholestadiene ion | 1) A less harsh ionization condition employed; 2) chemical derivatization; and 3) using more advanced commercial mass instruments. | |
| Glycosphingolipid - ceramide | NLS of sugar residue - generation of ceramide | The aforementioned methods for lysoPA and cholesterol. | |
| MALDI-MSI | TAG-DAG | NLS of fatty acid/fatty acid salt from TAG - generation of DAG or DAG analogs | 1) Reduction of the laser power; 2) the much lower levels of endogenous DAGs compared with that of TAG species; 3) the more complicated profile of ion source generated DAG in an identical tissue spot; 4) using new matrices; and 5) novel approaches of pretreatment of samples, such as on-tissue digestion and on-tissue derivatization |
| Glycerophospholipids - PA | NLS of trimethylamine/ vinylamine/serine from sodiated glycerophospholipids (e.g., PC, PE and PS) - generation of PA/ isomeric species of PA | 1) Reduction of the laser power; 2) the unique chemical structures of endogenous PA species; 3) much lower concentrations of PA species compared with PC species; 4) tissue washing procedures; and 5) determination of PA species in the negative-ion mode. | |
| SM - C1P | NLS of trimethylamine headgroup from sodiated SM species - generation of isomeric species of C1P | 1) Reduction of the laser power; 2) much lower concentrations of C1P species compared with SM counterparts; 3) tissue washing procedures; and 4) determination of C1P species in the negative-ion mode. | |
The original references from which these strategies were employed are cited in the text. See text for the abbreviations.
All these artifacts discussed in LC-MS and shotgun lipidomics also exist in MALDI-MSI analysis with more severity.
Generally, it is a very useful and simple method, in which an exogenous lipid standard added can be used to confirm the ion source-generated artifacts and further tune the instrument to eliminate/reduce the in-source fragmentation. Thus, we strongly suggest that at least one exogenous lipid standard for each lipid class of interest should be included in samples in MS-based lipidomics analysis. Moreover, to ensure the accuracy of data collected by MS-based lipidomics, the strategies such as a less harsh ionization condition (e.g., low ionization temperature and low ionization voltage), chemical derivatization, novel strategies of sample preparation, suitable matrix, biosystem knowledge on concentrations of selected lipids, etc. should be considered to maximally reduce/eliminate these artifacts, and thus the false results. Finally, an actual result of a class of lipids identified and quantified by MS-based lipidomics must comply with the existence of these lipid species in a biological system and the reasonable relative concentrations/profile of these species in comparison with those of other related lipid species and/or classes. To this end, accurate quantitative analysis of lipid species is crucial for comparison between lipid classes (Burla et al., 2018; Wang, Wang, & Han, 2017).
Highlights.
Overviewed the ion source-generated artifacts relevant to other endogenous lipid species in lipidomics analysis
Summarized the strategies that can be used to avoid/reduce the artifacts of in-source fragmentation to achieve accurate and comprehensive lipidomics analysis
Acknowledgments
The work was partially supported by National Natural Science Foundation of China (No.81803861), Natural Science Foundation of Zhejiang Province of China (No. LY20B050006), National Key R&D Program of China (2018YFC1705500), National Institute on Aging Grant RF1 AG061872, and National Institute of Neurological Disorders and Stroke Grant U54 NS110435, as well as from the UT Health San Antonio intramural institutional research funds, the Mass Spectrometry Core Facility, and the Methodist Hospital Foundation.
Abbreviations
- C1P
ceramide-1-phosphate
- CEs
cholesteryl esters
- CID
collision-induced dissociation
- DAG
diacylglycerol
- ESI
electrospray ionization
- HPLC
high performance liquid chromatography
- MALDI-MSI
matrix-assisted laser desorption/ionization-mass spectrometry imaging
- MS
mass spectrometry
- NAFLD
non-alcoholic fatty liver disease
- PA
phosphatidic acid
- lysoPA
lysophosphatidic acid
- PC
phosphatidylcholine
- lysoPC
lysophophatidylcholine
- PE
phosphatidylethanolamine
- DMPE
dimethyl PE
- lysoPE
lysophosphatidylethanolamine
- PRM
parallel reaction monitoring
- PS
phosphatidylserine
- lysoPS
lysophosphatidylserine
- SM
sphingomyelin
- TAG
triacylglycerol
References
- Abbas I, Noun M, Touboul D, Sahali D, Brunelle A, Ollero M. 2019. Kidney lipidomics by mass spectrometry imaging: a focus on the glomerulus. Int J Mol Sci 20: 1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saad KA, Zabrouskov V, Siems WF, Knowles NR, Hannan RM, Hill HJ. 2003. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of lipids: ionization and prompt fragmentation patterns. Rapid Commun Mass Spectrom 17: 87–96. [DOI] [PubMed] [Google Scholar]
- Alamri H, et al. 2019. Mapping the triglyceride distribution in NAFLD human liver by MALDI imaging mass spectrometry reveals molecular differences in micro and macro steatosis. Anal Bioanal Chem 411: 885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel PM, Spraggins JM, Baldwin HS, Caprioli R. 2012. Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal Chem 84: 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D. 2009. Lipidomics. In: Walker JM, editor. Methods and protocols. New York, USA: Humana Press.pp 1–579. [Google Scholar]
- Bilgin M, et al. 2011. Quantitative profiling of PE, MMPE, DMPE, and PC lipid species by multiple precursor ion scanning: a tool for monitoring PE metabolism. BBA-Mol Cell Biol Lipids 1811: 1081–1089. [DOI] [PubMed] [Google Scholar]
- Borgquist S, et al. 2016. Apolipoproteins, lipids and risk of cancer. Int J Cancer 138: 2648–2656. [DOI] [PubMed] [Google Scholar]
- Boumann HA, Damen MJ, Versluis C, Heck AJ, de Kruijff B, de Kroon AI. 2003. The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species. Evidence for lipid remodeling. Biochemistry 42: 3054–3059. [DOI] [PubMed] [Google Scholar]
- Boumann HA, de Kruijff B, Heck AJ, de Kroon AI. 2004. The selective utilization of substrates in vivo by the phosphatidylethanolamine and phosphatidylcholine biosynthetic enzymes Ept1p and Cpt1p in yeast. FEBS Lett 569: 173–177. [DOI] [PubMed] [Google Scholar]
- Briche C, Carter D, Webb K. 2002. Comparison of gas chromatography and liquid chromatography mass spectrometric measurements for high accuracy analysis of cholesterol in human serum by isotope dilution mass spectrometry. Rapid Commun Mass Spectrom 16: 848–853. [DOI] [PubMed] [Google Scholar]
- Burla B, et al. 2018. MS-based lipidomics of human blood plasma: a community-initiated position paper to develop accepted guidelines. J Lipid Res 59: 2001–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LK, Vance JE, Vance DE. 2012. Phosphatidylcholine biosynthesis and lipoprotein metabolism. BBA-Mol Cell Biol Lipids 1821: 754–761. [DOI] [PubMed] [Google Scholar]
- Contos J, Ishii I, Chun J. 2000. Lysophosphatidic acid receptors. Mol Pharmacol 58: 1188–1196. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Zeller M, Fedorova M. 2020. Evaluation of Lipid In-Source Fragmentation on Different Orbitrap-based Mass Spectrometers. J Am Soc Mass Spectrom 31: 463–466. [DOI] [PubMed] [Google Scholar]
- DeLong CJ, Shen YJ, Thomas MJ, Cui Z. 1999. Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J Biol Chem 274: 29683–29688. [DOI] [PubMed] [Google Scholar]
- Dennis EA. 2009. Lipidomics joins the omics evolution. P Natl Acad Sci USA 106: 2089–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisewerd K, Schürenberg M, Karas M, Hillenkamp F. 1995. Influence of the laser intensity and spot size on the desorption of molecules and ions in matrix-assisted laser desorption/ionization with a uniform beam profile. Int J Mass Spectrom Ion Processes 141: 127–148. [Google Scholar]
- Drobnik W, et al. 2003. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J Lipid Res 44: 754–761. [DOI] [PubMed] [Google Scholar]
- Duffin K, Obukowicz M, Raz A, Shieh J. 2000. Electrospray/tandem mass spectrometry for quantitative analysis of lipid remodeling in essential fatty acid deficient mice. Anal Biochem 279: 179–188. [DOI] [PubMed] [Google Scholar]
- Duffin KL, Henion JD, Shieh JJ. 1991. Electrospray and tandem mass spectrometric characterization of acylglycerol mixtures that are dissolved in nonpolar solvents. Anal Chem 63: 1781–1788. [DOI] [PubMed] [Google Scholar]
- Fahy E, et al. 2005. A comprehensive classification system for lipids. J Lipid Res 46: 839–861. [DOI] [PubMed] [Google Scholar]
- Frey B, Gaipl US. 2011. The immune functions of phosphatidylserine in membranes of dying cells and microvesicles. Semin Immunopathol 33: 497–516. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Schiller J, Wagner U, Häntzschel H, Arnold K. 2005. The phosphatidylcholine/lysophosphatidylcholine ratio in human plasma is an indicator of the severity of rheumatoid arthritis: Investigations by 31P NMR and MALDI-TOF MS. Clin Biochem 38: 925–933. [DOI] [PubMed] [Google Scholar]
- Gallego SF, Hojlund K, Ejsing CS. 2018. Easy, fast, and reproducible quantification of cholesterol and other lipids in human plasma by combined high resolution MSX and FTMS analysis. J Am Soc Mass Spectrom 29: 34–41. [DOI] [PubMed] [Google Scholar]
- Galli M, Zoppis I, Smith A, Magni F, Mauri G. 2016. Machine learning approaches in MALDI-MSI: clinical applications. Expert Rev Proteomics 13: 685–696. [DOI] [PubMed] [Google Scholar]
- Gathungu RM, et al. 2018. Optimization of electrospray ionization source parameters for lipidomics to reduce misannotation of in-source fragments as precursor ions. Anal Chem 90: 13523–13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. 2009. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One 4: e6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. 2005. Lipid alterations in the earliest clinically recognizable stage of Alzheimer’s disease: implication of the role of lipids in the pathogenesis of Alzheimer’s disease. Curr Alzheimer Res 2: 65–77. [DOI] [PubMed] [Google Scholar]
- Han X. 2016a. Lipidomics for studying metabolism. Nat Rev Endocrinol 12: 668–679. [DOI] [PubMed] [Google Scholar]
- Han X 2016b. Lipidomics: comprehensive mass spectrometry of lipids. NJ, USA: Wiley. [Google Scholar]
- Han X, Gross R. 1995. Structural determination of picomole amounts of phospholipids via electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom 6: 1202–1210. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. 2001. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem 295: 88–100. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. 2003. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res 44: 1071–1079. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 24: 367–412. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Yang K, Zhao Z, Abendschein D, Gross R. 2007. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: a shotgun lipidomics study. Biochemistry 46: 6417–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Cheng H, Fikes KN, W GR. 2005. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J Lipid Res 46: 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Gross RW. 2012. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev 31: 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DJ. 2012. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: an update for 2007–2008. Mass Spectrom Rev 31: 183–311. [DOI] [PubMed] [Google Scholar]
- Herrera LC, Potvin MA, Melanson JE. 2010. Quantitative analysis of positional isomers of triacylglycerols via electrospray ionization tandem mass spectrometry of sodiated adducts. Rapid Commun Mass Spectrom 24: 2745–2752. [DOI] [PubMed] [Google Scholar]
- Hoeke G, Kooijman S, Boon MR, Rensen PC, Berbee JF. 2016. Role of brown fat in lipoprotein metabolism and atherosclerosis. Circ Res 118: 173–182. [DOI] [PubMed] [Google Scholar]
- Horing M, Ejsing CS, Hermansson M, Liebisch G. 2019. Quantification of cholesterol and cholesteryl ester by direct flow injection high-resolution fourier transform mass spectrometry utilizing species-specific response factors. Anal Chem 91: 3459–3466. [DOI] [PubMed] [Google Scholar]
- Houjou T, Yamatani K, Nakanishi H, Imagawa M, Shimizu T, Taguchi R. 2004. Rapid and selective identification of molecular species in phosphatidylcholine and sphingomyelin by conditional neutral loss scanning and MS3. Rapid Commun Mass Spectrom 18: 3123–3130. [DOI] [PubMed] [Google Scholar]
- Hoving E. 1995. Chromatographic methods in the analysis of cholesterol and related lipids. J Chromatogr B Biomed Appl 671: 341–362. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J. 2003. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: The fragmentation processes. J Am Soc Mass Spectrom 14: 352–363. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J. 2005. Studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes. J Am Soc Mass Spectrom 16: 1510–1522. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J. 2009. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: mechanisms of fragmentation and structural characterization. J Chromatogr B Analyt Technol Biomed Life Sci 877: 2673–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Duan Q, Han X. 2020. Strategies to improve/eliminate the limitations in shotgun lipidomics. Proteomics 20: 201900070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Wang C, He L, Han X. 2019. Novel strategies for enhancing shotgun lipidomics for comprehensive analysis of cellular lipidomes. TRAC-Trend Anal Chem 120: 115330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Wang M, Han X. 2017. Shotgun lipidomics in substantiating lipid peroxidation in redox biology: Methods and applications. Redox Biol 12: 946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, et al. 2015. Lipidomics revealed idiopathic pulmonary fibrosis-induced hepatic lipid disorders corrected with treatment of baicalin in a murine model. AAPS J 17: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, et al. 2016. Oxidative stress leads to reduction of plasmalogen serving as a novel biomarker for systemic lupus erythematosus. Free Radical Biol Med 101: 475–481. [DOI] [PubMed] [Google Scholar]
- Kerwin JL, Tuininga AR, Ericsson LH. 1994. Identification of molecular species of glycerophospholipids and sphingomyelin using electrospray mass spectrometry. J Lipid Res 35: 1002–1014. [PubMed] [Google Scholar]
- Kong JN, et al. 2018. Novel function of ceramide for regulation of mitochondrial ATP release in astrocytes. J Lipid Res 59: 488–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde M, Géloën A, Record M, Vance D, Spener F. 2003. Lipidomics is emerging. BBA-Mol Cell Biol L 1634: 61. [DOI] [PubMed] [Google Scholar]
- Lange Y, Ye J, Steck TL. 2004. How cholesterol homeostasis is regulated by plasma membrane cholesterol in excess of phospholipids. P Natl Acad Sci USA 101: 11664–11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembcke J, Ceglarek U, Fiedler GM, Baumann S, Leichtle A, Thiery J. 2005. Rapid quantification of free and esterified phytosterols in human serum using APPI-LC-MS/MS. J Lipid Res 46: 21–26. [DOI] [PubMed] [Google Scholar]
- Leventis PA, Grinstein S. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 39: 407–427. [DOI] [PubMed] [Google Scholar]
- Levery SB. 2005. Glycosphingolipid structural analysis and glycosphingolipidomics. Methods Enzymol 405: 300–369. [DOI] [PubMed] [Google Scholar]
- Liebisch G, Binder M, Schifferer R, Langmann T, Schulz B, Schmitz G. 2006. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim Biophys Acta 1761: 121–128. [DOI] [PubMed] [Google Scholar]
- Lu L, et al. 2018. Shotgun lipidomics revealed altered profiles of serum lipids in systemic lupus erythematosus closely associated with disease activity. Biomolecules 8: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M, Spiegel S. 2014. Sphingolipid metabolites in inflammatory disease. Nature 510: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature 438: 612–621. [DOI] [PubMed] [Google Scholar]
- Nascimento-Ferreira MV, et al. 2017. The lipid accumulation product is a powerful tool to predict metabolic syndrome in undiagnosed Brazilian adults. Clinical nutrition 36: 1693–1700. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Herr D, Mutoh T, Chun J. 2009. Lysophosphatidic acid (LPA) and its receptors. Curr Opin Pharmacol 9: 15–23. [DOI] [PubMed] [Google Scholar]
- Norris JL, Caprioli RM. 2013. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev 113: 2309–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès C, Rey A, Lafontan M, Valet P, Saulnier-Blache J. 1999. Ca2+-independent phospholipase A2 is required for alpha2-adrenergic-induced preadipocyte spreading. Biochem Biophys Res Commun 265: 572–576. [DOI] [PubMed] [Google Scholar]
- Pagès G, et al. 2000. LPA as a paracrine mediator of adipocyte growth and function. Ann N Y Acad Sci 905: 159–164. [DOI] [PubMed] [Google Scholar]
- Perry RJ, Samuel VT, Petersen KF, Shulman GI. 2014. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 510: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic M, et al. 2001. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: phosphatidylcholine prevents the detection of further species. Anal Biochem 289: 202–216. [DOI] [PubMed] [Google Scholar]
- Pynn CJ, Henderson NG, Clark H, Koster G, Bernhard W, Postle AD. 2011. Specificity and rate of human and mouse liver and plasma phosphatidylcholine synthesis analyzed in vivo. J Lipid Res 52: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker P. 2014. LDL cholesterol: controversies and future therapeutic directions. Lancet 384: 607–617. [DOI] [PubMed] [Google Scholar]
- Samuel VT, Shulman GI. 2016. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. The Journal of clinical investigation 126: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandesara PB, Virani SS, Fazio S, Shapiro MD. 2019. The forgotten lipids: triglycerides, remnant cholesterol, and atherosclerotic cardiovascular disease risk. Endocr Rev 40: 537–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhoff R, Brügger B, Jeckel D, Lehmann WD, Wieland FT. 1999. Determination of cholesterol at the low picomole level by nano-electrospray ionization tandem mass spectrometry. J Lipid Res 40: 126–132. [PubMed] [Google Scholar]
- Shevchenko A, Simons K. 2010. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol 11: 593–598. [DOI] [PubMed] [Google Scholar]
- Stübiger G, Pittenauer E, Allmaier G. 2008. MALDI seamless postsource decay fragment ion analysis of sodiated and lithiated phospholipids. Anal Chem 80: 1664–1678. [DOI] [PubMed] [Google Scholar]
- Sturtevant D, Lee YJ, Chapman KD. 2016. Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr Opin Biotechnol 37: 53–60. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. 2015. Phospholipid synthesis and transport in mammalian cells. Traffic 16: 1–18. [DOI] [PubMed] [Google Scholar]
- Vance JE, Vance DE. 2004. Phospholipid biosynthesis in mammalian cells. Biochem Cell Biol 82: 113–128. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang M, Han X. 2015. Comprehensive and quantitative analysis of lysophospholipid molecular species present in obese mouse liver by shotgun lipidomics. Anal Chem 87: 4879–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Liu CB, Wu HW. 2011. A simple desalting method for direct MALDI mass spectrometry profiling of tissue lipids. J Lipid Res 52: 840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang C, Han X. 2018. Enhanced coverage of lipid analysis and imaging by matrix-assisted laser desorption/ionization mass spectrometry via a strategy with an optimized mixture of matrices. Anal Chim Acta 1000: 155–162. [DOI] [PubMed] [Google Scholar]
- Wang M, Han RH, Han X. 2013. Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Anal Chem 85: 9312–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang C, Han RH, Han X. 2016. Novel advances in shotgun lipidomics for biology and medicine. Prog Lipid Res 61: 83–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang C, Han X. 2016. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why? Mass Spectrom Rev 36: 693–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang C, Han X. 2017. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why? Mass Spectrom Rev 36: 693–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Devaiah SP, Zhang W, Welti R. 2006. Signaling functions of phosphatidic acid. Prog Lipid Res 45: 250–278. [DOI] [PubMed] [Google Scholar]
- Wu W, Shi X, Xu C. 2016. Regulation of T cell signalling by membrane lipids. Nat Rev Immunol 16: 690–701. [DOI] [PubMed] [Google Scholar]
- Xu L, Wang X, Jiao Y, Liu X. 2018. Assessment of potential false positives via orbitrap-based untargeted lipidomics from rat tissues. Talanta 178: 287–293. [DOI] [PubMed] [Google Scholar]
- Xu YF, Lu W, Rabinowitz JD. 2015. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal Chem 87: 2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin EB, de la Monte SM. 2015. Review of matrix-assisted laser desorption ionization-imaging mass spectrometry for lipid biochemical histopathology. J Histochem Cytochem 63: 762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yu Y, Song F, Liu S. 2011. Structural characterization of neutral oligosaccharides by laser-enhanced in-source decay of MALDI-FTICR MS. J Am Soc Mass Spectrom 22: 845–855. [DOI] [PubMed] [Google Scholar]
- Yang K, Han X. 2016. Lipidomics: techniques, applications, and outcomes related to biomedical sciences. Trends Biochem Sci 41: 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yore Mark M, et al. 2014. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159: 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi Z, Koenig A, Abdelatif D, Fazel Y, Henry L, Wymer M. 2016. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64: 73–84. [DOI] [PubMed] [Google Scholar]
- Zemski Berry KA, Turner W,W, VanNieuwenhze MS, Murphy RC. 2009. Stable isotope labeled 4-(dimethylamino)benzoic acid derivatives of glycerophosphoethanolamine lipids. Anal Chem 81: 6633–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Xu Y. 2009. Measurement of endogenous lysophosphatidic acid by ESI-MS/MS in plasma samples requires pre-separation of lysophosphatidylcholine. J Chromatogr B Analyt Technol Biomed Life Sci 877: 3739–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]