Summary
Agouti related protein (AgRP) is a hypothalamic regulator of food consumption in mammals. However, AgRP has also been detected in circulation, but a possible endocrine role has not been examined. Zebrafish possess two agrp genes; hypothalamically expressed agrp1, considered functionally equivalent to the single mammalian agrp, and agrp2, which is expressed in pre-optic neurons and uncharacterized pineal gland cells, and whose function is not well understood. By ablation of AgRP1-expressing neurons and knockout of the agrp1 gene, we show that AgRP1 stimulates food consumption in the zebrafish larvae. Single-cell sequencing of pineal agrp2-expressing cells revealed molecular resemblance to retinal-pigmented epithelium cells and anatomic analysis shows that these cells secrete peptides, possibly into the cerebrospinal fluid. Additionally, based on AgRP2 peptide localization and gene knockout analysis, we demonstrate that pre-optic AgRP2 is a neuroendocrine regulator of the stress axis that reduces cortisol secretion. We therefore suggest that the ancestral role of AgRP was functionally partitioned in zebrafish by the two AgRPs; with AgRP1 centrally regulating food consumption, while AgRP2 acts as a neuroendocrine factor regulating the stress axis.
Introduction
Energy homeostasis is maintained via tight regulation of food consumption versus energy expenditure. Regulation of food consumption in vertebrates is complex, involving numerous neural and endocrine signals that act as orexigenic or anorexigenic inputs [1]. The arcuate nucleus of the mammalian hypothalamus is a site of convergence for these central and peripheral energy store signals, and contains at least two distinct populations of neurons critically involved in the regulation of body weight: anorexigenic neurons that express proopiomelanocortin (POMC), and orexigenic neurons that express Agouti related protein (AgRP) [2].
POMC neurons reduce food intake and increase energy expenditure by releasing α-melanocyte-stimulating hormone (αMSH), a product of POMC processing, which activates melanocortin receptor 4 (MC4R). AgRP neurons have the opposite effect; inhibiting POMC neurons and antagonizing the action of αMSH by the competitive binding of AgRP to MC4R, thereby causing an increase of food consumption and body weight [2].
There is considerable evidence in support of AgRP’s direct role in the regulation of food consumption in rodents. AgRP stimulates feeding when injected into the brain ventricles [3] and constitutive over-expression of AgRP leads to increased food consumption, obesity and metabolic derangements [4]. Activation of AgRP neurons is sufficient to orchestrate feeding behavior [5,6], while ablation of AgRP neurons leads to starvation [7,8].
AgRP neurons also co-release neuropeptide Y (NPY) and γ-aminobutyric acid (GABA) and project to multiple brain regions, including areas in the hypothalamus, thalamus and the hindbrain [9], where each of the molecules act via different pathways to promote feeding and inhibit metabolism. Injection of NPY or GABA receptor antagonists into the paraventricular hypothalamus significantly inhibits food intake during AgRP neuronal activation [10]. Additionally, GABAergic signaling onto the parabrachial nucleus restores appetite after acute AgRP neuron ablation and prevents anorexia [11].
Two paralogs of the agouti family have been identified in the zebrafish brain, agrp1 and agrp2 (annotated as zebrafish agouti signaling protein 2b, asip2b); however, the respective functions of the two proteins remain incompletely understood. AgRP1 is expressed in the hypothalamus, and its ectopic overexpression results in increased linear growth in adult fish [12], while conversely, transient knockdown results in decreased somatic growth during larval development [13]. Accordingly, zebrafish AgRP1 is thought to be a regulator of food consumption, but this has never been directly tested by targeting AgRP1 neurons. Here we test this hypothesis by eliminating the endogenous AgRP1 system. Our results confirm that AgRP1-expressing neurons and the AgRP1 peptide are indeed regulators of food consumption.
The zebrafish AgRP2 is expressed in an uncharacterized group of cells in the pineal gland, as well as in a small group of neurons in the pre-optic area that project towards the pituitary, where they interface with the pituitary vasculature [14], suggesting an endocrine role for this peptide. We show that the pineal AgRP2-expressing cells are retinal-pigmented epithelium (RPE)-like cells that can secrete peptides, possibly into the cerebrospinal fluid (CSF). Moreover, our data reveals that AgRP2 from pre-optic neurons is a neuroendocrine regulator of the stress response system, down-regulating cortisol secretion.
Results
AgRP1 is a regulator of feeding behavior in the larval zebrafish
Based on the stimulation of feeding by exogenous administration of MC4R antagonist in the goldfish [15], the increased linear growth in transgenic zebrafish with ectopic expression of AgRP1 [12], and the conserved neuroanatomy of the hypothalamic AgRP1 system [14,16,17], it has been assumed that AgRP1 neurons stimulate food consumption in fish, as is the case in mammals. However, this assumption has never been directly tested. To this end, we utilized TgBAC(agrp1:Gal4-VP16)tlv04; Tg(UAS:nsfB-mCherry)c264 larvae to ablate AgRP1-expressing neurons. The nfsB gene encodes an E-coli nitroreductase (NTR) enzyme that can render prodrugs such as metronidazole into a cell-impermeable cytotoxic product, enabling cell-specific ablation [18,19]. Transgenic larvae were allowed to develop normally until 6 day post fertilization (dpf) and were then exposed to metronidazole, which led to AgRP1 neuron ablation as indicated by the loss of the mCherry signal (Figure 1A). Food consumption was analyzed by quantifying the amount of fluorescent paramecia consumed by 8-dpf AgRP1 neuron-ablated larvae as compared to the amount consumed by agrp1:Gal4-VP16 larvae (NTR-negative sibling controls) that were exposed to metronidazole (control group). Ablation of AgRP1 neurons resulted in significantly decreased food consumption as compared to the control group (t-test, p<0.01, Figure 1B), suggesting that AgRP1-expressing neurons stimulate food consumption in zebrafish. Reduced feeding after AgRP1 neuron ablation is unlikely to be due to locomotor impairments as kinematic analysis revealed normal motor function (Figure S1). Thus, AgRP1-expressing neurons are involved in the regulation of food consumption in zebrafish.
Figure 1. AgRP1, but not AgRP2, is a regulator of feeding behavior.
(A) Left- dorsal view of 5-dpf larva expressing NTR-mCherry fusion protein in agrp1-expressing neurons. The cell bodies are located in the hypothalamus and project towards the post-optic commissure and the anterior commissure. Right- the same larva at 8-dpf, after treatment with metronidazole. AgRP1 neurons were ablated, and the mCherry signal cannot be identified. Abbreviation: AC; anterior commissure, H; hypothalamus, POC; post-optic commissure. Scale bar represents 50μm.
(B) Quantification of the food consumed by 8-dpf AgRP1 neuron-ablated larvae and NTR-negative sibling controls (t-test, n=6, p<0.01). Values represent the mean fluorescence intensity ± SE. See also Figure S1.
(C) Schematic illustration of the AgRP1 precursor in WT and agrp1 KO fish. The predicted WT AgRP1 contains 127 amino-acids, including the c-terminal Agouti motif. The predicted mutated AgRP1 contains 99 amino-acids and lacks the Agouti motif.
(D) Immunostaining analysis of AgRP1 in WT (left) and agrp1 KO (right) larvae using zebrafish specific AgRP1 polyclonal antibody. In the mutant, as appose to WT, the truncated AgRP1 is not transported along the axons to reach the AC and POC (arrowheads). Ventral view, Abbreviation: H; hypothalamus, AC; anterior commissure, POC; post-optic commissure. Scale bar represents 50μm.
(E) Quantification of the food consumed by 8-dpf agrp1 KO and controls (progeny of WT siblings, t-test, n=9, p<0.001). Values represent the mean fluorescence intensity ± SE.
(F) Dorsal view of 8-dpf agrp1 KO and WT sibling. Scale bar represents 1mm.
(G) Body length measurements of 8-dpf agrp1 KO, their heterozygotes and WT siblings (one-way ANOVA, n=15-26, p<0.01). Circles represent single measurements; horizontal lines represent the mean body length ± SE.
(H) Schematic illustration of the AgRP2 precursor in WT and agrp2 KO fish. The predicted WT AgRP2 contains 136 amino-acids, including the c-terminal Agouti motif. The predicted mutated AgRP2 contains only 17 amino-acids and lacks the Agouti motif.
(I) Immunostaining analysis of AgRP2 in WT (left) and agrp2 KO (right) larvae using the zebrafish specific AgRP2 polyclonal antibody. Ventral view of the pituitary. No signal is detected in pituitaries of mutated larvae. Abbreviation: Pit; pituitary. Scale bar represents 10μm.
(J) Quantification of the food consumed by 8-dpf agrp2 KO and controls (progeny of WT siblings, t-test, n=6, p>0.05). Values represent the mean fluorescence intensity ± SE.
(K) Left- pineal gland of 3-dpf larva expressing NTR-mCherry fusion protein in agrp2-expressing cells and EGFP under the promoter of aanat2 (which marks the pineal photoreceptors [80]). Right- same larva at 8-dpf, after treatment with metronidazole. AgRP2 cells were ablated while the photoreceptors were not damaged. Scale bar represents 10μm.
(L) Quantification of the food consumed by 8-dpf AgRP2 neuron-ablated larvae and NTR-negative sibling controls (t-test, n=6, p>0.05). Values represent the mean fluorescence intensity ± SE.
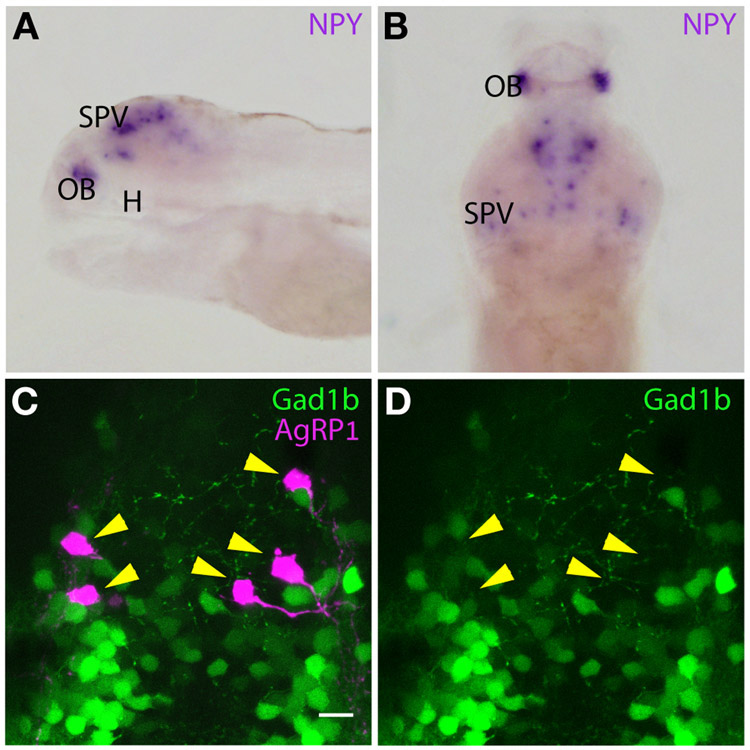
In mice, ablation of AgRP hypothalamic neurons resulted in an anorexia-like phenotype: reduced feeding and loss of body weight [7,8]. This result was attributed to acute and cumulative loss of GABA, NPY and AgRP signaling from AgRP neurons, since AgRP knockout (KO) alone does not result in reduced feeding in mice [20]. Nevertheless, by analyzing the NPY mRNA expression pattern in larval zebrafish we show that NPY is not expressed in AgRP1 neurons (Figure 2A and 2B), as recently reported [21]. Additionally, by crossing agrp1:Gal4-VP16; UAS:nfsB-mCherry with TgBAC(gad1b:EGFP)nns25 fish (an established GABAergic neuron reporter line [22]), we show that AgRP1 neurons are not gad1b-positive (Figure 2C and 2D). This was further verified by co-immunodetection of EGFP and AgRP1, using a custom-made polyclonal antibody targeting the entire zebrafish AgRP1 precursor in TgBAC(gad1b:EGFP) larvae (Video S1). We therefore tested whether KO of agrp1 would lead to the reduced feeding observed in AgRP1 neuron-ablated fish. For this purpose, we generated an agrp1 KO line, which possesses a 34 nucleotide insertion that disrupts the inhibitor cysteine-knot motif (the bioactive agouti motif [23], Figure 1C) and leads to a premature stop codon. The mutated, truncated AgRP1 does not undergo axonal transport and was detected by the polyclonal antibody in cell bodies only (Figure 1D).
Figure 2. Zebrafish AgRP1 neurons do not co-express NPY or Gad1b.
(A) In-situ hybridization (ISH) analysis of NPY in 5-dpf larva, lateral view, eyes were dissected. NPY is expressed in the stratum periventriculare layer of the optic tectum and in the olfactory bulb, but is not detected in the hypothalamus. Abbreviation: H; hypothalamus, OB; olfactory bulb, SPV; stratum periventriculare.
(B) ISH analysis of NPY in 5-dpf larva, dorsal view, eyes were dissected. NPY is expressed in the stratum periventriculare layer of the optic tectum and in the olfactory bulb. Abbreviation: OB; olfactory bulb, SPV; stratum periventriculare.
(C) Hypothalamus of 5-dpf agrp1:Gal4-VP16; UAS:nfsB-mCherry, gad1B:EGFP. The EGFP is not localized to AgRP1 neurons (arrowheads). Scale bar represents 10μm. See also Video S1.
(D) Same as (C), presentation of the EGFP layer only. The localization of the AgRP1 neurons is marked by arrowheads to demonstrate the absence of EGFP.
When tested for food consumption at 8-dpf, agrp1 KO larvae consumed fewer paramecia in comparison to genetically-related wild-type (WT) controls (progeny of WT siblings, t-test, p<0.001, Figure 1E). Thus, in contrast to mammals [20], KO of agrp1 is sufficient to reduce feeding in the zebrafish larvae. In addition, at this stage agrp1 KO larvae show a 2.5% decrease in body length (one-way ANOVA, p<0.01, Figure 1F and 1G). Although we cannot rule out the possibility that the decreased feeding in agrp1 KO larvae is a result of their slightly smaller body size, these results confirm that, in addition to its role in regulation of food consumption, AgRP1 affects metabolism and growth even during pre-feeding larval stages [13].
Thus, by studying the effect of AgRP1 neuron-ablation and agrp1 KO, two complementary models were generated that confirms the central role of the endogenous AgRP1 in regulation of food consumption in zebrafish.
AgRP2 is not required for food consumption
We have previously shown in zebrafish that AgRP2 is expressed in pre-optic neurons that project towards the pituitary and suggested a neuroendocrine role for this peptide [14]. We also observed innervation of AgRP1 neurons by AgRP2 axons and suggested a cross-talk between these two neuronal systems [14]. To explore whether AgRP2 or agrp2-expressing neurons regulate food consumption, possibly by central endocrine regulation or by modifying the activity of AgRP1 neurons, we tested food consumption in agrp2 KO and AgRP2 neuron-ablated larvae. We generated agrp2 KO fish, with a 4 bp deletion near the start of the AgRP2 coding sequence, leading to a frame shift, premature stop codon (Figure 1H), and loss of the peptide (Figure 1I). When tested for food consumption, agrp2 KO larvae consumed a similar quantity of paramecia as their WT controls (t-test, p>0.05, Figure 1J), suggesting that AgRP2 is not required for the regulation of feeding behavior. To test whether AgRP2-expressing cells might be involved in regulation of food consumption independently of AgRP2, for example by modifying AgRP1 neuronal activity, we utilized double transgenic agrp2:Gal4-VP16; UAS:nfsB-mCherry to ablate AgRP2 neurons (Figure 1K). AgRP2 neuron-ablated larvae consumed the same amount of food as their controls (t-test, p>0.05, Figure 1L), suggesting that AgRP2-expressing cells are not required for feeding. It should be noted that upon agrp2 KO or AgRP2 neuron ablation, the pineal AgRP2 system is also affected and therefore it is clear from these results that pineal AgRP2 is not involved in regulation of food consumption as well.
In summary, loss of AgRP1, but not AgRP2, significantly reduces food consumption, suggesting an evolutionary conserved role for AgRP1 in the regulation of feeding.
Single-cell sequencing of the pineal gland revealed that AgRP2 is expressed in retinal epithelium-like cells
A major site of AgRP2 expression in zebrafish is the pineal gland [14,24]. We have previously shown that pineal AgRP2 cells are not photoreceptors or neurons [14], and these cells remained uncharacterized, hindering the investigation of the function of pineal AgRP2. Thus, to molecularly and genetically characterize these cells, pineal gland single-cell sequencing analysis was performed.
Dissected pineal glands of adult transgenic TgBAC(agrp2:Gal4-VP16)tlv05; Tg(UAS:nfsB-mCherry)c264; Tg(foxd3:EGFP)zf104 or Tg(tph2:mCherry)y227; Tg(gfap:EGFP)mi2001, were used in two different single-cell sequencing experiments, termed scSeq1 and scSeq2, respectively. The fluorescent markers were used to aid with both the dissection of the pineal glands and cluster analysis of the sequenced cells. Single-cell barcoded cDNAs were prepared using a droplet-based system (10x genomics), and were sequenced and analyzed as described in the materials and methods section.
Cluster analysis of the sequenced cells revealed seven transcriptionally distinct clusters for scSeq1, and eight for scSeq2 (Figures 3A and 3B). We identified the type of cells represented by each cluster by examining selectively expressed genes present in both scSeq1 and scSeq2 (Data S1 and Figure S2). The eighth cluster in scSeq2 expressed known habenular markers [25], such as kiss1, gng8 and tac3a (Data S1 and Figure S2) likely resulting from dissection of adjacent habenular cells along with the pineal gland. Thus, single-cell RNA sequencing indicates that the pineal gland is composed of at least seven different cell-types. Examination of the unique gene repertoire of each of the seven clusters (Data S1 and Figure S2) revealed that these are (Figure 3): rod-like photoreceptors, cone-like photoreceptors, neurons, retinal-pigmented epithelium-like (RPE-like) cells, Müller-like glia, macrophages/microglia, and an unidentified cell-type (cluster 4).
Figure 3. Single-cell sequencing analysis of zebrafish pineal gland.
(A) 2D visualization (tSNE) of scSeq1 pineal single-cell clusters. Each point represents a single cell, colored according to cell type. The pineal contains seven transcriptionally distinct cell types (see also Figure S2 and Data S1).
(B) 2D visualization (tSNE) of scSeq2 pineal single-cell clusters. Each point represents a single cell, colored according to cell type. Eight transcriptionally distinct cell types are observed. One cluster was identified as habenular cells in addition to the seven types of pineal cells identified in scSeq1 (see also Figure S2 and Data S1).
Agrp2 was expressed in the RPE-like cells in both scSeq experiments, as was mCherry in scSeq1 (in which agrp2:Gal4-VP16; UAS:nfsB-mCherry transgenic fish were used). Thus, the RPE-like cells are the agrp2-expressing cells.
The RPE-like cluster is the second largest cluster of cells (18% of scSeq1 cells and 13% of scSeq2). This cluster expressed several genes that are involved in the retinoid (visual) cycle (Data S1 and Figure S2). For example, rgra (retinal G-protein coupled receptor) and rdh5 (Retinol Dehydrogenase 5) catalyze the conversion of all-trans-retinol to 11-cis-retinal [26,27]. These same genes are expressed in the RPE layer of the retina [26,27]. stra6 (stimulated by retinoic acid 6) and rbp5 (retinol binding protein 5) are associated with transport of retinol, and rpe65 (retinal pigment epithelium-specific 65 KDa protein) is involved in isomerization of retinol [28], and is specifically expressed in the RPE layer of the retina [28]. These results suggest that this cluster participates in the retinoid cycle in the pineal gland and acts similarly to the RPE, therefore we termed this cluster RPE-like. Notably, however, genes related to pigment formation, a distinctive feature of the retina RPE layer [29], were not detected in the RPE-like cluster.
The expression of gfap (glial fibrillary acidic protein), previously assigned to be a marker of the pineal interstitial cells [30], was also found to be specific to this cluster (Data S1), as well as egfp in scSeq2 (as a result of gfap:EGFP usage in scSeq2 experiment). Our previous immunohistochemical analysis on developing 5-dpf larvae failed to co-localize GFAP-immuno-reactive cells and AgRP2-expressing cells [14]. The reasons for these contradictory results may be the early developmental stage of the pineal gland in the immunostaining analysis and/or the use of an antibody directed to mammalian GFAP [31].
Overall, these results show that, consistent with the evolutionary and developmental relationship of the pineal gland and the retina, the pineal gland is composed of cell types that are analogous to the types of cells of the retina. Notably, our data reveals that the pineal includes a previously uncharacterized cell-type with strong molecular similarities to RPE cells that selectively express agrp2.
Pineal AgRP2/RPE-like cells are secretory cells
Developmental analysis of the fluorescently-labeled agrp2-expressing cells in agrp2:Gal4-VP16; UAS:nfsB-mCherry and TgBAC(agrp2:EGFP)tlv06 transgenic lines revealed that, starting at late larval stages, the fluorescent protein is no longer confined to the pineal gland, but is also found above the telencephalic area (Figure 4A). The earliest age in which this phenomenon was observed is 7-dpf. Removal of the top adult skull revealed that the fluorescent protein remained attached to the skull, and was not present in the brain (Figure 4B). To test whether the fluorescent protein is secreted from the pineal gland or locally expressed in that area, we performed ISH for agrp2 mRNA and egfp mRNA on dissected skulls, which include the attached pineal gland (Figure 4C and 4D). This revealed that neither agrp2 nor egfp are expressed outside of the pineal gland, suggesting that the fluorescent proteins are either secreted from the pineal to this area or that pineal AgRP2/RPE-like cells project there. To test whether AgRP2 pineal cells send projections above the telencephalon, we crossed agrp2:Gal4-VP16; UAS:nfsB-mCherry fish with Tg(UAS:mYFP)mde6. In UAS:mYFP, the yellow fluorescent protein (YFP) is fused to a membrane tag, which results in its translocation to the membrane revealing the cellular structure [32]. Interestingly, the mYFP was only localized to the pineal gland (Figure 4E), suggesting that AgRP2/RPE-like cells do not project outside the pineal area, but rather can secrete cytosolic proteins, such as EGFP and mCherry. In addition, branched, microvilli-like structures were revealed by sparse mYFP labeling, protruding from AgRP2 cells (Figure 4F).
Figure 4. Pineal AgRP2/RPE-like cells are secretory cells.
(A) Dorsal view of a 21-dpf agrp2:EGFP fish head. The EGFP is not confined to the pineal and can be observed above the telencephalon (arrowheads).
(B) Dissected skull of adult agrp2:EGPF fish, immunostained with an antibody against EGFP. Ventral view, anterior to top. The EGFP remains bound to the skull. Scale bar, 100μm.
(C) ISH of agrp2 (purple) performed on an adult skull. Ventral view [same as (B)], anterior to top. agrp2 mRNA is localized to the pineal gland.
(D) ISH of egfp (purple) performed on an adult skull. Ventral view [same as (B)], anterior to top. egfp mRNA is localized to the pineal gland.
(E) Dorsal view of a 21-dpf agrp2:Gal4-VP16; UAS:nfsB-mCherry; UAS:mYFP fish, focusing on the pineal gland with sparse mYFP labeling. The membrane-bound YFP (green) is confined to the pineal (arrowheads), while the mCherry can be found outside of the pineal, possibly secreted from the pineal region. Dashed line represents the circumference of the pineal gland. Scale bar, 10μm.
(F) Enlargement of (E) reveals the cellular architecture of AgRP2 cells, bearing branched microvilli-like structure (left arrowhead). Scale bar, 10μm.
(G) Dorsal view of a 7-dpf agrp2:Gal4-VP16; UAS:nfsB-mCherry fish larvae before laser ablation, focusing on the pineal gland and the telencephalic area. The mCherry is mainly detected in the pineal gland. Scale bar, 10μm.
(H) Same larva as (H), at 10-dpf, 3-day post laser-ablation. Dorsal view focusing on the pineal gland and the telencephalic area. The mCherry signal from the pineal is lost and is merely detected above the telencephalic region. Scale bar, 10μm.
(I) Dorsal view focusing on the pineal gland and the telencephalic area of 10-dpf agrp2:Gal4-VP16; UAS:nfsB-mCherry control larva with intact pineal gland. At this age the mCherry signal can already be detected above the telencephalic area (Arrowheads). Scale bar, 10μm.
(J) Dissected skull of adult agrp2:Gal4-Vp16; UAS:Kaede fish. Ventral view, anterior to top. The Kaede signal can only be detected in the pineal area. Scale bar, 100μm.
Abbreviation: e; eye, OE; olfactory epithelium, P; pineal. See also Figures S3 and S4.
To further explore the secretory process from AgRP2/RPE-like cells, we laser ablated the pineal gland of 7-dpf agrp2:Gal4-VP16; UAS:nfsB-mCherry larvae. The laser ablation resulted in the elimination of the mCherry signal from this area above the telencephalon (Figure 4G-5I), strengthening the assertion that AgRP2/RPE-like pineal cell are involved in secretion to this area. Interestingly, a cross between agrp2:Gal4-VP16 and Tg(UAS:Kaede)rk8 fish revealed that unlike cytosolic EGFP or mCherry, the cytosolic Kaede was not secreted from AgRP2/RPE-like cells (Figure 4J). Kaede forms a homotetrameric complex with a molecular weight of 116 kDa [33], which is much larger than EGFP (27 kDa) for example, and it is therefore possible that the secretion process from AgRP2/RPE-like cells is size dependent and that larger protein complexes cannot be secreted. In teleost, the area above the telencephalon is part of the ventricular system, thus, these findings suggest that the pineal AgRP2/RPE-like cells can secrete peptides, possibly into the CSF.
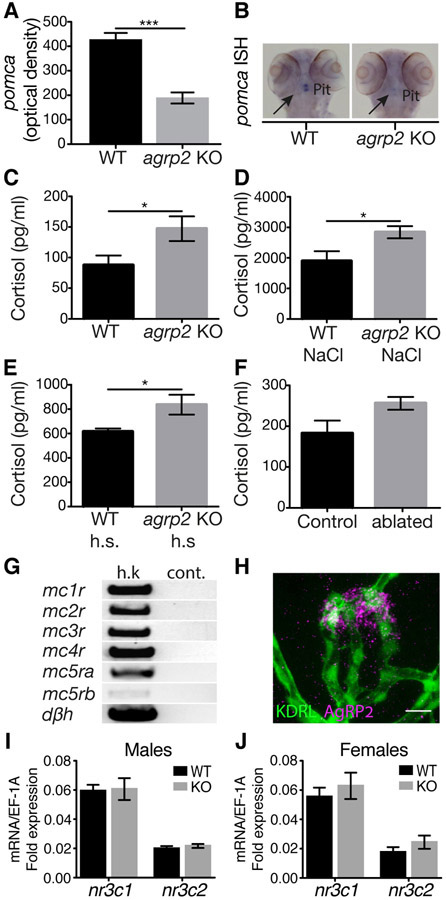
Figure 5. AgRP2 is a regulator of the stress axis.
(A) ISH analysis of pituitary pomca mRNA expression. pomca is significantly decreased in agrp2 KOs in comparison to their WT controls (t-test, n=22, p<0.001). Values represent the mean optical density ± SE.
(B) Representative pomca ISH signals of agrp2 KO and WT control.
(C) Basal state cortisol levels were measured in agrp2 KOs and their WT controls (t-test, n=6, p<0.05). Values represent the mean cortisol levels ± SE.
(D) Cortisol levels of agrp2 KOs and their WT controls after 10 minutes of osmotic shock (250mM NaCl, t-test, n=6, p<0.05). Values represent the mean cortisol levels ± SE.
(E) Cortisol levels of agrp2 KOs and their WT controls after 10 minutes of 36°C heat-shock (h.s, t-test, n=6, p<0.05). Values represent the mean cortisol levels ± SE.
(F) Cortisol levels of AgRP2 ablated larvae and their controls (t-test, n=6, p=0.056). Values represent the mean cortisol levels ± SE.
(G) PCR analysis of melanocortin receptor expression in the head-kidney, where the interrenal tissue is located. Dopamine beta hydroxylase (dβh) was used as a positive control for interrenal tissue cells. Abbreviation: h.k; head-kidney, cont.; negative control.
(H) Immunostaining analysis of the pituitary vasculature using the zebrafish specific AgRP2 antibody (magenta) and EGFP antibody in kdrl:EGFP larvae (green), which expresses EGFP in vascular endothelial cells. Scale bar represents 10μm.
(I) Real-time PCR analysis of glucocorticoid receptor or mineralocorticoid receptor expression in male pituitary glands, encoded by the nr3c1 and nr3c2, respectively (t-test, n=8, p >0.05).
(J) Real-time PCR analysis of glucocorticoid receptor or mineralocorticoid receptor expression in female pituitary glands, encoded by the nr3c1 and nr3c2, respectively (t-test, n=8, p >0.05).
AgRP2/RPE-like cells are not required for circadian rhythms of locomotor activity
The pineal gland is considered a key component of the circadian clock system in fish. Blocking the molecular circadian oscillator, specifically in the melatonin-producing pineal photoreceptor cells, disrupts normal rhythms of circadian locomotor activity [34], and eliminating pineal gland melatonin synthesis disrupts circadian clock regulation of the sleep/wake cycle [35]. In the retina, the RPE layer participates in the visual cycle and maintains photoreceptors integrity. Therefore, we hypothesized that AgRP2/RPE-like cells of the pineal gland may be required for proper circadian control, either directly or by supporting the pineal photoreceptor activity. To this end, we tested circadian rhythms of locomotor activity in agrp2 KO and AgRP2/RPE-like ablated larvae. Both agrp2 KO and RPE-like ablated larvae showed robust circadian rhythms of locomotor activity under constant dim light, similar to control larvae (Kolmogorov-Smirnov test, p>0.05, Figure S3). These results indicate that neither the AgRP2/RPE-like cells of the pineal gland nor the AgRP2 peptide are required for maintaining circadian rhythms of locomotor activity.
KO or neuronal ablation of AgRP2 does not affect background adaptation
The pineal gland has also been associated with pigmentation and transient knockdown of agrp2 has been shown to affect background adaptation in larval zebrafish [36]. Background adaptation is performed by dispersing or aggregating melanosomes. Under a dark background melanosomes disperse, whereas under a white background melanosomes are aggregated and larvae appear lighter. Transient knockdown of agrp2, performed by Morpholino-modified oligonucleotides, resulted in melanosome aggregation defect when larvae were placed on a white background [36]. However, methods of gene knockdown using Morpholinos have been widely criticized for off target effects, toxicity and frequent discrepancies between morphant and mutant phenotypes [37]. Therefore, to test whether KO of the agrp2 gene affects background adaptation, we assessed the melanosome size of 6-dpf agrp2 KO and WT controls (progeny of WT siblings) after prolonged or short adaptation to a black or white background. When placed on black background for prolonged period, agrp2 KOs pigment coverage area was larger in comparison to their WT controls (two-way ANOVA, p<0.01, Figure S4). Moreover, KOs showed normal pigment aggregation when placed on a white background, regardless the adaptation time (Figure S4). Thus, agrp2 KO fish are not impaired in their ability to aggregate or disperse melanosomes in response to background color. These results are in contrast to previously reported Morpholino-based experiments [36], and may be attributed to potential compensatory effects of agrp2 KO or the methodology used [38].
To eliminate this possibility as well as to test whether other factors arising from AgRP2 pineal cells or pre-optic neurons are involved in background adaptation, we also measured pigment coverage area after ablation of AgRP2-expressing cells. Both AgRP2-ablated larvae and their controls were able to aggregate and disperse their melanosomes according to the background color (Figure S4), suggesting that AgRP2-expressing cells are not necessary for background adaptation.
Pre-optic AgRP2 is a regulator of the stress axis
The anatomical properties of pre-optic AgRP2 neurons, which terminate at the adenohypophysis [14], suggest a potential hypophysiotropic or neuroendocrine role. To test these possibilities, we examined whether agrp2 KO affects the expression of pituitary hormones, and in particular hormones that are involved in growth and metabolism. Expression of pomca (proopiomelanocortin a), gh (growth hormone) and tshβ (thyroid stimulation hormone, β subunit) was examined by semi-quantitative ISH in 6-dpf agrp2 KO larvae and WT controls. While gh and tshβ expression did not significantly differ between agrp2 KO and their WT controls (t-test, p>0.05), pomca expression was significantly reduced (t-test, p<0.0001, Figure 5A and 5B). POMC is a polypeptide precursor that undergoes tissue-specific, post-translational processing yielding several biologically active peptides. In zebrafish, the pituitary population of pomca-expressing cells is mainly corticotropic, producing the adrenocorticotropic hormone (ACTH) [39].
ACTH is secreted from the pituitary gland and acts on the adrenal gland (or the interrenal tissue in teleost) to stimulate glucocorticoid secretion and synthesis [40]. Glucocorticoids orchestrate the stress response, as well as attenuating it by negative feedback on the secretion and synthesis of ACTH [40]. To test whether the reduced levels of pomca in the pituitary of agrp2 KO result in changes in glucocorticoid levels, we tested the concentration of cortisol, the principal corticosteroid in teleost, in basal state and under stress [10 minute treatment of osmotic shock (250mM NaCl) or heat-shock (36°C)]. Cortisol was extracted from groups of 15 larvae at 6-dpf, and was measured using enzyme-linked immunosorbent assay (ELISA). Our hypothesis was that since pomca mRNA levels in the pituitary are low, cortisol levels would also be low. However, contrary to our hypothesis, agrp2 KO larvae had significantly higher cortisol levels, both in basal state (t-test, p<0.05) and under stress (t-test, p<0.05), in comparison to their WT controls (Figure 5C-5E). A similar trend was also observed following AgRP2 neuronal ablation although not significant (Figure 5F). However, the antibiotic treatment itself led to increased basal cortisol levels and therefore might partially mask the effects of the ablation.
One plausible explanation for the effects of AgRP2 is that this peptide acts directly on the interrenal tissue to suppress cortisol production and that the higher basal cortisol levels in the agrp2 KO larvae down-regulate pomca mRNA expression by a negative feedback mechanism. To test this possibility, we examined whether the melanocortin receptors are expressed in the interrenal tissue. Indeed, all the zebrafish melanocortin receptors were found to be expressed in the head-kidney, where the interrenal tissue is located (Figure 5G), including mc1r and mc4r, that were previously shown to be antagonized by AgRP2 [36], suggesting AgRP2 can act directly on the interrenal tissue.
AgRP2 pre-optic neuronal terminals interface with the pituitary vasculature, where synaptophysin aggregates can be found [14], suggesting neuroendocrine secretion from AgRP2 pre-optic neurons. To test whether AgRP2 can be secreted from these terminals into the blood, we performed co-immunostaining for AgRP2 and EGFP in Tg(kdrl:EGFP)S843 larvae, which express EGFP in vascular endothelial cells [41]. Indeed, dense staining of AgRP2 was localized adjacent to the pituitary vasculature (Figure 5H), suggesting that AgRP2 is secreted into the general circulation, from which it may reach the interrenal tissue.
To verify that effects of AgRP2 KO on the stress axis are not the result of changes in pituitary glucocorticoid receptor or mineralocorticoid receptor expression (encoded by the nr3c1 and nr3c2, respectively), we quantified their expression using real-time PCR. No differences were found in nr3c1 and nr3c2 pituitary expression in both male and female agrp2 KOs and their WT controls (Figure 5I and 5J).
Therefore, although we cannot rule out the involvement of pineal AgRP2 in this process, it is most likely that pre-optic AgRP2 is a neuroendocrine factor that is secreted into the blood circulation, directly acting on the interrenal tissue to down regulate cortisol secretion.
Discussion
The anatomical and functional properties of the two AgRP systems in zebrafish show different degrees of similarity with those of the single mammalian AgRP system. Mammalian AgRP peptide is expressed in the hypothalamus and acts to stimulate food consumption. This neuronal regulatory system has been considered to be evolutionary conserved across vertebrates, and, accordingly, the zebrafish AgRP1 neurons have been assumed to perform this function. Here we confirm this conserved role of AgRP1 by showing the direct effect of the endogenous AgRP1 neuronal system, as well as of the AgRP1 peptide, on feeding in the larval zebrafish. Similar to the effect of AgRP neuron ablation in mice, ablation of AgRP1 neurons in zebrafish resulted in reduced food consumption (Figure 1B). However, while KO of agrp in mice does not result in reduced feeding [20], likely by developmental compensation mechanisms involving NPY and GABA [10,20,42], KO of agrp1 in zebrafish reduced food consumption (Figure 1E) and growth. These differences are potentially due to the fact that in contrast to mammals, AgRP1 neurons in zebrafish do not appear to co-express either NPY or GABA (Figure 2). Overall, these findings argue for the evolutionary conserved role of AgRP in energy homeostasis.
In mammals, AgRP and POMC arcuate nucleus neurons share similar axonal termination sites [9], as they regulate feeding by opposite effects on these common targets. This co-projection pattern is similar in zebrafish, where AgRP1 neurons and the hypothalamic POMC neurons both innervate the post-optic commissure, the pre-optic area and the anterior commissure [14,43] (Figure 1D), suggesting a high degree of anatomical conservation of the hypothalamic POMC-AgRP pathways.
Despite overall similar anatomical organization and functional properties, zebrafish AgRP1 neurons and mammalian AgRP neurons differ in the extent of their innervation sites. Mammalian AgRP hypothalamic neurons project to several hypothalamic structures and multiple brain areas [9]. AgRP terminals were also found in the median eminence and the posterior pituitary in mammals [9,44,45], and in the infundibulum and median eminence in ducks [46], suggesting that the mammalian and avian AgRP may also act as a neuroendocrine factor. In contrast, the zebrafish AgRP1 neurons are not hypophysiotropic, as revealed here by the immunostaining analysis using zebrafish-specific AgRP1 antibody and by the previous analysis of agrp1:Gal4-VP16; UAS:nfsB-mCherry transgenic fish [14].
On the other hand, pre-optic AgRP2 neurons project towards the pituitary vasculature [14], where AgRP2 congregates (Figure 5), likely in secretory vesicles [14], thus suggesting a neuroendocrine role for AgRP2. This neuroendocrine role is supported by the effect of AgRP2 KO, which resulted in disrupted stress axis: higher cortisol levels under both basal and stressed states, which down regulates pituitary pomca mRNA expression by negative feedback mechanisms (Figure 5F). The expression of melanocortin receptors in the head-kidney, where the interrenal tissue is located (Figure 5G), and previous results demonstrating the lack of those receptors in the pituitary [14] further strengthen the hypothesis that AgRP2 directly acts on the interrenal tissue. Although KO of agrp2 as well as ablation of AgRP2-expressing cells affect both pre-optic and the pineal AgRP2, the anatomical features of the two AgRP2 cellular populations suggest that the effect imposed on the stress axis is the result of the pre-optic neuroendocrine AgRP2 cells. Hence, we propose that pre-optic AgRP2 has a role in neuroendocrine control of cortisol synthesis and secretion.
The presence of AgRP2 in the pineal gland is particularly intriguing as this gland shares homologies with the retina. The pineal gland of non-mammalian vertebrates is a photosensitive organ, which contains an intrinsic circadian oscillator and influences daily rhythms and seasonal changes [30,47,48]. Melatonin, the principal hormone produced by the pineal gland, is synthesized and secreted during the night in order to coordinate physiological and behavioral circadian rhythms. In mammals, melatonin concentrations are 20-fold higher in the third ventricle CSF then in the plasma [49]. In teleost, the lumen of the pineal gland is an extension of the third ventricle, and it was suggested that melatonin may reach the brain by diffusion through the pineal lumen CSF [30,50]. Several types of cells have been previously identified in the teleost pineal gland, including rod and cone photoreceptors, neurons, macrophages and less characterized interstitial cells [30,50]. Accordingly, it has been suggested that the pineal interstitial cells are a type of support cells that form a diffusion barrier with the pineal lumen CSF [30,50]. Our single-cell sequencing results reveal two types of supportive cells, Müller glia-like and RPE-like, which exclusively express agrp2.
In the retina, the RPE layer has several roles: re-isomerization of all-trans-retinol back into 11-cis-retinal, phagocytosis of discarded outer segment portions of rod and cone photoreceptors, pigment synthesis and formation of a retinal-blood barrier [29,51]. Although we did not find genetic evidence for pigment formation or phagocytosis by the pineal AgRP2 cells, the transcriptome of these cells revealed expression of key genes involved in retinal isomerization properties, including RPE-specific genes such as rdh5 and rpe65a, which perform dehydrogenization and isomerization of retinol, respectively. Additionally, there are indications that these cells may form a barrier similar to that of the RPE. The RPE is a tight epithelium, regulating nutrients, ion and water transport between photoreceptors and capillaries. Water transport is facilitated by aquaporin-1, a water channel [29,51]. Consistent with this, the AgRP2 pineal cells express claudin encoding genes, cldn7a and cldn19, which are tight junction adhesion molecules, and aqp1a.1, encoding for the aquaporin water channel (Data S1). The expression of aqp1a.1 in the pineal gland was also previously detected by ISH [52]. These findings suggest that AgRP2 pineal cells are RPE-like cells, participating in the pineal retinoid cycle and possibly forming a barrier that regulates transport of molecules between the pineal photoreceptors and the vasculature and/or the CSF.
Indeed, cytosolically expressed fluorescent proteins in the AgRP2/RPE-like cells are secreted from the pineal gland. These fluorescent proteins were found in an area above the telencephalon (Figure 4), which in teleosts is part of the ventricular system [53]. Thus, it is possible that pineal AgRP2/RPE-like cells form a barrier between the pineal gland and the CSF to regulate the passage of molecules, such as melatonin, between them. As a result, an unbound fluorescent protein that is expressed in the AgRP2/RPE-like cells can be found in the surrounding area of the telencephalon, while membrane-bound fluorescent proteins, such as mYFP, remain within the pineal gland (Figure 4). Accordingly, we propose that AgRP2 is secreted from the RPE-like cells into the CSF, through which it may act on downstream targets. Interestingly, the mammalian arcuate nucleus of the hypothalamus, where AgRP neurons reside, is in contact with the CSF, and a reciprocal movement of compounds between the CSF and the arcuate nucleus has been documented [54]. It is therefore possible that, in addition to its axonal targets, mammalian AgRP might also be secreted into the CSF to affect feeding behavior via distant downstream brain areas. Thus, communication of AgRP-expressing cells through the CSF may represent an unappreciated, evolutionary conserved mode of AgRP action.
The zebrafish agrp1 and agrp2 are considered paralogous genes, although their phylogenetic relationship is a matter of debate [55,56]. Based on the anatomical and functional characteristics and similarities with the single mammalian and avian agrp, we propose that an evolutionary subfunctionalization event has occurred for the zebrafish agrp genes; partitioning the pre-existing functions of the ancestor agrp gene into agrp1 as a neuromodulator involved in feeding behavior and agrp2 is a neuroendocrine factor involved in the stress response. Indeed, despite the fact that the mammalian single AgRP is associated mainly with food consumption, AgRP is also detected in the median eminence [9] and the circulation [57], but its possible neuroendocrine functions have been largely overlooked. Additionally, we propose that the pineal agrp2 represents a case of neofunctionalization; where agrp2 has acquired another yet unknown function, via secretion into the CSF.
In summary, genome duplication has resulted in two agrp genes that are expressed in the zebrafish brain. The hypothalamic agrp1 is conserved, and directly regulates feeding behavior whereas the pre-optic agrp2 is a neuroendocrine modulator of the stress response, down regulating cortisol secretion. AgRP2 is also expressed in RPE-like cells of the pineal gland and is likely to be released into the CSF with an as yet unknown function.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yoav Gothilf (yoavgothilf@gmail.com).
Experimental model and subject details
Adult zebrafish were raised in a recirculation water system under 12-hours light:12-hours dark cycles at 28°C and fed twice a day. Embryos were generated by natural mating, placed in 10 cm Petri dishes with egg water containing methylene blue (0.3 ppm), and raised in a light-controlled incubator at 28°C (light intensity, 1.8 W/m2). To prevent pigmentation for ISH and immunostaining analysis, egg water was supplemented with 0.2 mM N-Phenylthiourea from 1-dpf onward.
Experiments were performed on larvae before sex differentiation, or on adult fish of either sex. The previously described and registered transgenic lines used in this study are: TgBAC(agrp1:Gal4-VP16)tlv04, TgBAC(agrp2:Gal4-VP16)tlv05, TgBAC(agrp2:EGFP)tlv06, Tg(UAS:nsfB-mCherry)c264, TgBAC(gad1b:EGFP)nns25, Tg(foxd3:EGFP)zf104, Tg(tph2:mCherry)y227; Tg(gfap:EGFP)mi2001, Tg(UAS:mYFP)mde6, Tg(kdrl:EGFP)S843, Tg(UAS:kaede)rk8, Tg(wt1b:EGFP)li1, Tg(cdh17:EGFP)zf237. The generation of the KO lines used in this study is described below.
All procedures were approved by the Tel Aviv University Animal Care Committee and conducted in accordance with the requirements of the Council for Experimentation on Animal Subjects, Ministry of Health, Israel.
Methods details
Whole-mount in-situ hybridization
Zebrafish agrp2 mRNA (reference sequence NM_001271291.1, bases 41-544), pomca mRNA (reference sequence NM_181438.3, bases 239-900) gh mRNA (reference sequence NM_001020492.2, bases 51-683), tshβ mRNA (reference sequence NM_181494.2, bases 69-483), npy mRNA (reference sequence NM_131074.2, bases 111-725) and egfp (sequence ID MH048881.1 bases 2785-3504) were cloned into pGEM-T Easy (Promega), linearized, and used as template for in vitro synthesis of digoxigenin (DIG)-labelled anti-sense riboprobes (DIG RNA labelling kit, Roche). WT and agrp2 KO 6-dpf larvae were fixed overnight in 4% paraformaldehyde (PFA) and stored in 100% methanol.
Whole-mount in-situ hybridization (ISH) analysis was carried out according to an established protocol [58]. The larvae were treated with cold acetone (−20°C) for 8 minutes, transferred back to 100% methanol, gradually rehydrated, and transferred to PBS supplemented with 0.1% Tween 20 (PBTw). The samples were then digested with 10μg/ml proteinase K for 40 minutes and fixed in 4% PFA for 20 minutes. The samples were then washed 3 times in PBTw, and pre-hybridized in hybridization buffer (50% formamide, 5×SSC, 5mM EDTA, 0.1% Tween 20, 0.1% CHAPS, 50μg/ml heparin and 1mg/ml yeast RNA) for 3 hours. The samples were then hybridized overnight with 1-2ng probe/μl hybridization buffer at 65°C. The next day the samples were washed twice (30 minutes each wash) in 50% formamide, 0.2×SSC, 0.3% CHAPS at 65°C, followed by wash in 2×SSC, 0.3% CHAPS for 15 minutes at 65°C, and twice in 0.2×SSC, 0.3% CHAPS (30 minutes each wash) at 65°C. Samples were then transferred to MAB buffer [0.1M maleic acid (pH = 7.5) and 150mM NaCl] for 15 minutes at room temperature followed by 1 hour incubation in blocking solution [50% MAB buffer 5% calf serum and 2% blocking reagent (Roche)]. 10μl of anti-DIG conjugated to alkaline-phosphatase antibody (Roche) was then added to the blocking solution and incubated with the samples for 3 hours at room temperature. The antibody was then washed 6 times in PBTw (15 minutes each wash) and the samples were stored at 4°C overnight. In the following day the samples were incubated in staining buffer (0.1M Tris-HCl, 0.1M NaCl, 50mM MgCl2 and 0.1% Tween 20) for 10 minutes and the staining reaction occurred by replacing the staining buffer with the alkaline-phosphatase substrate BM-purple (Roche). The staining reaction was carefully monitored and stopped before signal saturation by washing the BM-purple with PBTw, followed by fixation in 4% PFA for 30 minutes. The samples were then washed in PBS. For image analysis, the ISH-stained larvae were placed in 70% glycerol and photographed under a dissecting microscope (SZX12, Olympus) equipped with a digital camera (DP70, Olympus). The ISH signal was quantified using ImageJ.
Antibody preparation
To prepare customized antibodies against the zebrafish AgRP1 and AgRP2, the cDNAs of agrp1 and agrp2 encompassing the entire precursors (excluding the signal peptides) were amplified from clones containing the corresponding cDNAs using the primers Agrp1F: 5’ AGCATATGTCTCATCCACACCTGAG 3’, Agrp1R: 5’ AGGGATCCAGAACTTGTCTCTATGCATATTCG 3’, Agrp2F: 5’ AGCATATGATCACATGCGAAGCCAGC 3’, Agrp2R: 5’ AGGGATCCGTGATTAGGTCTTCTTGG 3’, and subcloned into pET-15b vector. Each protein was expressed in Rosetta-gami B(DE3)pLysS E. coli cells (Novagen) as N-terminal His-tagged recombinant protein. The proteins from the insoluble fraction were purified with nickel-nitrolotriacetic acid columns (Promega), and run on a 16% SDS-PAGE. Dominant band of ∼15 kDa (AgRP1) and ∼14 kDa (AgRP2) were excised from the gel and used as an antigen for production of antiserum in rabbits (Pacific Immunology Corp.). The final bleed serums were used as the primary antibody for AgRP1 and AgRP2 immunostaining analyses (rabbit anti-zfAgRP1 and rabbit anti-zfAgRP2).
Validation of antibodies specificity
Antibodies specificity was analyzed by western blot analysis (as described below). A 15 kDa band of AgRP1 and 14 kDa band of AgRP2 were identified when each of the recombinants proteins were incubated with their corresponding antibodies (Figure S5), indicating that the antibodies can recognize the denatured recombinant proteins. Pre-adsorption of each of the antibodies with their corresponding antigen (as described below) resulted in disappearance of the signal (Figure S5). Pre-adsorption of anti-zfAgRP1 with the denatured recombinant AgRP2 had no effect on the signal indicating that anti-zfAgRP1 does not recognize the denatured recombinant AgRP2 (Figure S5). Pre-adsorption of anti-zfAgRP2 with the denatured recombinant AgRP1 had no effect on the signal indicating that anti-zfAgRP2 does not recognize the denatured recombinant AgRP1 (Figure S5).
The ability of the antibodies to detect the native endogenous zebrafish protein was examined using whole-mount immunostaining (as described below). Anti-zfAgRP1 signal was detected in cell bodies in the ventral hypothalamus, with immunoreactive fibers projecting towards the post-optic commissure and the anterior commissure (Figure 1D), precisely identical to the previously described anatomy of the AgRP1 system [14]. Importantly, no signal was detected in the pineal gland or pre-optic area, demonstrating the specificity of anti-zfAgRP1 to the endogenous AgRP1.
Anti-zfAgRP2 signal was detected in the pineal gland and axonal terminals in the pituitary (Figure 1D), precisely identical to the previously described anatomy of the AgRP2 system [14]. Importantly, no signal was detected in the hypothalamus, demonstrating the specificity of anti-zfAgRP2 to AgRP2. Pre-adsorption of each of the antibodies with their corresponding recombinant peptide (as described below) resulted in disappearance of the fluorescent signal. Pre-adsorption of anti-zfAgRP1 with the denatured recombinant AgRP2 had no effect on the signal indicating that anti-zfAgRP1 does not recognize AgRP2 (Figure S5). Pre-adsorption of anti-zfAgRP2 with AgRP1 led to a reduction of the fluorescent signal indicating that anti-zfAgRP2 can bind the denatured recombinant AgRP1 (Figure S5). Nevertheless, no signal was observed in the agrp2 KO larvae, indicating that anti-zfAgRP2 does not recognize the endogenous native AgRP1 or any other antigen in the larvae.
Antibody pre-adsorption
Pre-adsorption of rabbit anti-zfAgRP1 and rabbit anti-zfAgRP2 with the recombinant AgRP1 and AgRP2 was performed as follows: the inclusion bodies from 5 ml E.coli cultures expressing recombinant AgRP1 or AgRP2 were cleaned on IMAC columns (HisPur Cobalt Spin columns, Thermo Scientific) under denaturing conditions, eluted with 500 mM Imidazole and ~20 μg total proteins/well were resolved on a 16% SDS-PAGE gel. The proteins were transferred to a Nitrocellulose membrane (0.2μm pore size), followed by blocking with PBS +0.5% Triton x100 (PBTr)+1% milk and incubated overnight at 4°C with rabbit anti-zfAgRP1 and rabbit anti-zfAgRP2 at a 1:100 dilution in PBTr+1% milk for Western blot and PBTr+0.5% Triton x-100, 1% DMSO for immunostaining.
Immunostaining
The immunostaining protocol was performed as previously described [60]. Zebrafish larvae (6-dpf) were anesthetized and fixed overnight in 4% PFA and stored in 100% methanol at −20°C for at least overnight. The larvae were treated with cold acetone (−20°C) for 8 minutes, transferred back to 100% methanol, and then gradually rehydrated to PBS + 0.5% Triton x100 (PBTr). The samples were then digested with 10μg/ml proteinase K in PBTr for 40 minutes and fixed in 4% PFA for 20 minutes. Samples were then washed 3 times with PBTr and incubated for 1 hour in blocking solution (PBTr, 10% calf serum and 1% DMSO). The samples were incubated overnight at 4°C with primary antibody in blocking solution. The primary antibodies used were rabbit anti-zfAgRP1 (1:500), rabbit anti-zfAgRP2 (1:1000), and mouse anti-GFP (1:100, MBL International Cat# M048-3). The next day, the samples were washed 4 times, for 30 minutes each wash, in PBTr and then incubated for 2 hours with the secondary antibodies; Alexa Fluor 488 donkey anti-mouse IgG (1:100, Jackson ImmunoResearch) and Alexa Fluor 594 donkey anti-rabbit IgG (1:100, Jackson ImmunoResearch). The samples were then washed 6 times in PBTr and transferred to PBS for the following imaging steps.
Western Blot
100 ng of IMAC purified recombinant AgRP1 and AgRP2 were run on 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, blocked and incubated with the antibodies and the pre-adsorbed antibodies at a dilution of 1:1000 overnight at 4°C. After washes with PBTr, the membrane pieces were incubated for 1 hour with goat anti-rabbit- HRP (1:5000, Genscript). Signal was obtained with Western Lightning Plus-ECL kit (Perkin Elmer) and visualized using ChemiDoc Touch Imaging System (Bio Rad).
Confocal microscopy imaging
Transgenic 3–8-dpf larvae were anesthetized and placed in low melting agarose for dorsal imaging. For ventral imaging, the larvae were anesthetized and fixed in 4% PFA, washed in PBS, and their eyes and jaws were dissected. All images were obtained using a Leica TCS SP8 confocal laser-scanning microscope equipped with Leica LAS AF image acquisition software.
Preparation of isolated pineal cells for single-cell sequencing
For preparation of isolated pineal cells, 20 adult (0.5–1 years old) double transgenic zebrafish, TgBAC(agrp2:Gal4-VP16)tlv05; Tg(UAS:nfsB-mCherry)c264; Tg(foxd3:EGFP)zf104 which express mCherry in AgRP2 pineal cells and EGFP in pineal neurons, or Tg(tph2:mCherry)y227; Tg(gfap:EGFP)mi2001, which express mCherry in the pineal photoreceptor cells and EGFP in astrocytes and pineal interstitial cells, were used. The 20 transgenic fish of each line were used in two different single-cell sequencing experiments, termed scSeq1 and scSeq2. Fish were anesthetized in 1.5 mM Tricaine (Sigma), sacrificed by decapitation, and pineal glands were removed under a fluorescent dissecting microscope and placed in PBS. Pineal glands were then dissociated to produce isolated cells using papain (Papain Dissociation System, Worthington Biochemical Corporation) according to the manufacturer’s instructions. Several times during the dissociation procedure the cell suspension was analyzed under a microscope to verify dispersion into single cells. When the dissociation procedure was complete, the cell-suspension was centrifuged, supernatants were carefully removed and the cell pellet suspended in PBS containing 0.04% BSA. The cells were then filtered using a 20μm cell strainer to remove possible undissociated cells and debris.
To quantify the cell-suspension concentration, and to verify the cells viability, 20μl of the suspension were mixed with 0.4% Trypan blue, followed by cell-counting (LUNA-II automated cell counter, Logos Biosystems). The final concentrations ranged between 200-300 cells/μl.
Single-cell sequencing
The single-cell suspension was loaded onto the commercially available droplet-based single-cell barcoding system (10x Chromium Controller, 10x Genomics) and a Single Cell 3’ Reagent Kit v2 (10x Genomics) was used to prepare pineal single-cell 3’ barcoded cDNA and Illumina-ready sequencing libraries according to the manufacturer’s instructions. The libraries were then sequenced using Illumina HiSeq 2500.
Computational analysis of single-cell sequencing data
Alignment and quantification.
The Sequenced data was processed by CellRanger (filtering, barcode and UMI counting) using default command line options, as previously described [62], and gene-barcode matrices were generated. The sequenced reads were aligned to the zebrafish GRCz10 genome assembly (Ensembl release 90), including the added custom genes mCherry, Gal4-VP16, egfp and exorh, using STAR [64] implement in CellRanger.
For scSeq1, 224,888,310 reads were obtained with 141,617 mean reads per cell and a total estimated number of 1,588 cells. For scSeq2, 468,279,908 reads were obtained with 151,940 mean reads per cell and a total estimated number of 3,082 cells.
Filtering, scaling and identification of variable genes.
The generated gene-barcode matrices were then processed using the Seurat R package, version 2.2 (including filtration, scaling and clustering) [66,68]. In filtering RNA-seq data, genes were kept if they were expressed in at least 3 cells. In addition, we removed outliers that had unusual low or high number of genes and/or unique molecular identifiers (UMIs), which potentially represent cellular fragments or multiple cells (multiplets), respectively. A lower threshold was set to 200 for genes, and the high threshold was set at 15,000 for UMIs. The filtration resulted in a total number of 1,108 and 2,266 cells for scSeq1 and scSeq2, respectively.
An analysis of different thresholds showed that these values resulted in low number of multiplets, without compromising for the amount of data in the following PCA and clustering analysis.
The data was then normalized, using a global-scaling method (i.e., Seurat’s LogNormalize function) and variable genes were detected; 2,015 and 1,898 for scSeq1 and scSeq2, respectively (i.e., Seurat’s FindfVarGenes function) using default parameters. Finally, before cluster analysis, the data was scaled (i.e., using Seurat’s ScaleData function).
PCA and clustering.
The scaled data matrix was used for principal component analysis (PCA) and clustering. PCA was used for dimension-reduction, with the variable genes detected used as input. To determine how many PCs to include in the downstream analysis we used Seurat’s jackstraw and elbow functions to calculate the change in variance as a function of PC number. Most of the variance could be explained with PCs 1-15 for scSeq1 and PCs 1-14 for scSeq2. Further exploration of the data revealed that PCs 1-10 and PCs 1-12 best clustered the data for scSeq1 and scSeq2, receptively, while higher PCs had a negligible impact on clustering.
Clustering analysis was performed using a graph-based clustering approach implemented in Seurat’s FindClusters command. To visualize the datasets in 2D, t-distributed stochastic neighbor embedding (tSNE) was used with the same input PCs as the clustering analysis, using Seurat’s RunTSNE command.
Generation of KO fish
Genome editing with clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system was used to produce agrp1 KO fish as previously described [70]. To generate guide RNA (gRNA), the sequence 5’ GGGCAGATGATGACCCAGACAGG 3’ was cloned into the pT7-gRNA vector and transcribed using a T7 expression kit (NEB). Cas9 was transcribed using mMessage mMachine T3 kit (Ambion) from the pT3TS-nCas9n vector followed by the Poly-A tailing kit (Ambion) according to the manufacturer’s instructions. Approximately 2nl of a solution containing 300ng/μl Cas9 mRNA and 20-50ng/μl gRNA were injected into one-cell stage embryos (F0). Embryos were raised to adult stage at which time they were crossed with WT fish, and their progeny (F1) were screened using a heteroduplex mobility assay [72], using the primers agrp1KO-F 5’ CCATAACACAGGATCTGGGC 3’ and agrp1KO-R 5’ CTGTGGATTCTCTGTGCGAGT 3’. A F1 fish carrying a 34bp insertion was found and confirmed by sequencing. Mutated F2 heterozygotes were crossed to produce homozygous KO fish and their WT siblings. The progeny of the F3 mutated homozygotes and of their WT sibling were used in following behavioral assays.
Transcription activator-like effector nucleases (TALEN) system was used to create agrp2 KO fish. Specific TALENs were designed using ZiFiT trageter [73] to target the first exon of agrp2 (TALE F target sequence 5’ TGACGACGGCGGTGCTGA 3’, TALE R target sequence 5’ TGAAGAACAAGCAGATG 3’) and to disrupt an existing SapI restriction enzyme site. The TALENs were constructed using REAL assembly kit as previously described [74] and transcribed using mMESSAGE mMACHINE T7 kit (Ambion) followed by the Poly-A tailing kit (Ambion) according to the manufacturer’s instructions. Approximately 2nl of the TALENs mRNA were injected into one-cell embryos (F0) at concentration of 300ng/μl. The embryos were raised and their progeny (F1) were fin-clipped and screened by PCR (using the primers: agrp2KO-F 5’ CCTCGAGCTGATTGAGGACT 3’, agrp2KO-R 5’ TGTTGTAATAGCAGATCCTTGGTG 3’) and SapI restriction analysis for mutation in AgRP2 coding sequence. A F1 fish carrying a 4bp deletion was found and confirmed by sequencing. Mutated F2 heterozygotes were crossed to produce homozygous KO fish and their WT siblings. The progeny of the F3 mutated homozygotes and of their WT sibling were used in following behavioral, genetics and hormonal analysis.
Validation of the KO
To validate the effect of agrp1 KO on the production of the AgRP1 peptide, we performed immunostaining of AgRP1 in WT and agrp1 KO fish. In contrast to WT, in agrp1 KO fish, immunostaining was only detected in the hypothalamic cell bodies (Figure 1D), but not in the axonal projections. The truncated mutant peptide, which does not undergo axonal transport, was detected by the polyclonal antibody in cell bodies. Thus, the immunostaining analysis revealed that a mature AgRP1 is not produced.
To validate the effect of agrp2 KO on the production of the AgRP2 peptide, we generated a polyclonal antibody against the entire zebrafish AgRP2 precursor. Immunostaining analysis showed that in contrast to WT, in the agrp2 KO fish, no signal was detected in any of the expression sites, verifying that the mutation indeed led to lack of AgRP2.
Feeding Assays
Food consumption was measured according to a previously described protocol [75], with several modifications. 5-dpf Larvae were placed in small tanks, each genotype or treatment in separate tanks, and received paramecia twice a day until 7-dpf. At 7-dpf the larvae were transferred into a 6-well plate, 6 larvae of the same genotype per well, and were fed again with paramecia for two hours, after which the wells were washed twice with 6ml of E3 medium (5mM NaCl, 0.17mM KCl, 0.33mM CaCl2, 0.33mM MgSO4). Subsequently, the larvae were not fed for 18 hours, and the feeding assays were performed at 8-dpf. To quantify the amount of food consumed, the paramecia was fluorescently labeled with 25μl/ml of 4-Di-10-ASP (Thermo Fisher scientific) for 20 minutes, after which it was harvested using a 20μm mesh in order to remove remaining dye. The concentration of the paramecia/E3 was manually evaluated under a microscope, and each well received 500 fluorescently labeled paramecia. The larvae were fed with the labeled paramecia for 30 minutes, and then the wells were washed twice with E3 to remove residual paramecia. The larvae were then anesthetized and homogenized. The homogenates were transferred into a 96-well plate for fluorescent measurement, using Synergy HTX plate reader (BioTek) or Synergy HT plate reader (for AgRP1 ablation experiments, BioTek), setup with excitation wavelength of 488 nm and emission wavelength of 528 nm.
Feeding time was calibrated as follows: groups of 6 larvae received 500 fluorescently labeled paramecia and were allowed to feed for different periods of time, after which the larvae were homogenized and the fluorescence intensity was measured. Saturation of the fluorescent signal was observed after 40 minutes, suggesting that this assay reliably reflects food consumption when the larvae are allowed to feed for less than 40 minutes.
The ideal number of labeled paramecia supplied was calibrated, as follows: groups of 6 larva received 600 paramecium with varying portion of fluorescently labeled paramecia and were allowed to feed for 40 minutes, after which the larvae were homogenized and the fluorescence intensity was measured. A linear correlation between the percentage of the labeled paramecia and the fluorescence intensity of the larvae was observed (R2>0.99), validating the reliability and sensitivity of the assay to detect differences in the amount of consumed paramecia.
For testing the effect of neuronal ablation (see below) on food consumption, agrp1:Gal4-VP16; UAS:nfsB-mCherry and agrp2:Gal4-VP16; UAS:nfsB-mCherry larvae were treated with 10mM metronidazole during the nights of 3- and 4-dpf (for AgRP2-neuron ablation experiments only) and during the night of 6-dpf (for AgRP1- or AgRP2-neuron ablation experiments), for which they were transferred back to petri dishes for the period of metronidazole treatment. Ablation was confirmed by the disappearance of the mCherry signal.
Neuronal ablation
Conditional ablation was performed using transgenic lines expressing the nfsB gene that encodes for Nitroreductase fused to the fluorescent protein mCherry. Nitroreductase converts the antibiotic metronidazole into a cytotoxic product, resulting in cellular ablation [18]. Metronidazole (Sigma) was dissolved in E3 medium to a final concentration of 10 mM. The nfsB expressing larvae were place in petri dishes containing metronidazole.
Two-photon laser ablation
Pineal laser ablation was performed with a commercial two-photon microscope (Femtonics), combined with a Ti:Sapphire laser (Chameleon Ultra II) and an Olympus 20x/0.95 NA water-dipping objective on 7-dpf agrp2:Gal4-VP16; UAS:nfsB-mCherry fish. Pineal cells were eliminated by scanning a 760 nm beam for 3 seconds over a 1.5 x 0.2 μm2 area. The laser intensity at the objective focal plane was ~25 mW. The ablated area was imaged before and after the ablation, as well as at 10-dpf with a Zeiss LSM 780 confocal microscope equipped with Zen software.
Background adaptation analysis
For background adaptation experiments, agrp2 KO and their WT controls (WT sibling progeny) were raised on clear petri dishes until 3-dpf or 5-dpf, after which they were transferred to black or white bottom petri dishes for 3 days or overnight. For the medium and short adaptation time, at 6-dpf half of the larvae which were placed overnight on black background were transferred to white bottom petri dish for 2 hours or 30 minutes, after which the larvae were fixed in cold 4% PFA on ice and transferred to 70% glycerol for imaging. For AgRP2 ablation experiments, agrp:Gal4-Vp16; UAS:nfsB-mCherry larvae and their controls (siblings not expressing mCherry) were treated with 10mM metronidazole during the nights of 3- and 4-dpf, and processed by similar protocol.
For pigment analysis, larvae were imaged from dorsal side using Olympus dissecting microscope (SZX12, Olympus) equipped with digital camera (DP70, Olympus). Each image was cropped to the same size and the area of the pigmented upper plane was selected and cropped using Photoshop. The pigmented area of the processed images was measured using ImageJ.
Cortisol extraction and ELISA
agrp2 KO and their WT controls (WT sibling progeny) were raised separately in a 6-well plate with 5 ml E3 medium for 6 days (15 larvae per well, 6 wells per genotype/treatment). At 6-dpf, the larvae were immobilized by filling the wells with ice-cold E3. The larvae were collected into 1.5ml tubes, excess E3 was removed, and the samples were frozen in ethanol/dry-ice bath, after which they were stored in −80°C until processed. To extract cortisol, the samples were thawed and homogenized in 150μl of ddw, 500μl of ethyl acetate were added and vortexed, and the solvent and aqueous phase were separated by centrifugation. The upper organic phase was transferred into a new 1.5ml tube, and the procedure was repeated again using the aqueous phase to extract maximum cortisol. A speed-vacuum concentrator was used to evaporate the solvent, and the precipitant containing the cortisol was dissolved in 200μl (or 1ml for the stressor treated larvae, see below) ELISA-buffer, and preceded immediately for ELISA.
Cortisol concentrations were quantified using cortisol ELISA kit (Cayman Chemical), according to the manufacturer’s instructions.
For stressor treatments AgRP2 KO larvae and their WT controls were subjected to an osmotic shock by incubating in 250mM NaCl for 10 minutes, or subjected to a heat shock at 36°C for 10 minutes, after which they were immediately immobilized in ice-cold E3 and processed as described above.
For AgRP2 ablation experiments, agrp:Gal4-Vp16; UAS:nfsB-mCherry larvae and their controls (siblings not expressing mCherry) were treated with 10mM metronidazole during the nights of 3- and 4-dpf, and processed by similar protocol of cortisol extraction and ELISA.
Head-Kideny cDNA preparation
EGFP expressing head-kidney of an adult Tg(wt1b:EGFP)li1; Tg(cdh17:EGFP)zf237 fish was dissected under a fluorescent microscope and homogenized in TRIzol (Ambion). RNA was extracted using phenol/chlorophorm purification followed by ethanol precipitation. cDNA was prepared using Verso cDNA synthesis kit (Thermo scientific) according to the manufacturer’s instructions. Primers used for melanocortin receptors amplification are listed in Table S1.
Real-Time PCR analysis of pituitary nr3c1 and nr3c2 expression
Adult agrp2 KO and their WT controls (WT sibling progeny) were separated into tanks containing 4 fish of the same sex, 2 fish of each genotype, and were acclimated for two-weeks prior to the dissections. Pituitaries were dissected and placed in QIAzol Lysis Reagent (QIAGEN) and a biopsy of the fins was taken for genotyping. RNA was prepared using Tissue lipid mini kit (QIAGEN) according to the manufacturer’s instructions, and used as a template for cDNA preparation using qScript cDNA sythsis kit (Quantabio), according to the manufacturer’s instructions.
The real-time quantitative PCR procedure was performed as previously described [77]. Serial dilutions (0.2, 0.1, 0.05, 0.025, 0.0125, and 0.00625) were prepared from each pituitary cDNA sample, and the efficiencies of the specific gene amplifications were compared by plotting Ct versus log(template). The PCR mixture consisted of 3μl of diluted cDNA sample, 200nM of each primer (Table S1), and 10μl of Fast SYBR Green Master Mix (Applied Biosystems) in a final volume of 20μl. Amplification was carried out in a LightCycler 96 System (Roche) according to the manufacturer's protocol. The cDNAs of each gene were amplified simultaneously in separate tubes in duplicates. A dissociation-curve was analyzed to confirm the presence of only one product. To eliminate false positives, a reverse-transcriptase negative control was run for each primer pair. To assess the relative expression of nr3c1 and nr3c2 mRNAs, the genes were normalized against an endogenous reference (ef1a) by the comparative threshold cycle method.
Body length measurements
Body length measurements were performed on 8-dpf progeny of incrossed agrp1 KO heterozygote fish. The larvae were anesthetized in 1.5 mM Tricaine (Sigma), photographed under a dissecting microscope (SZX12, Olympus) equipped with a digital camera (DP70, Olympus), followed by DNA extraction and genotyping. Body length was calculated using ImageJ.
Startle response
6-dpf larvae were placed in a custom 9-well plate and tested individually for startle response to acoustic stimulus. 40 Acoustic stimuli were given in two different amplitudes, 29 and 42 m/sec2, in a pseudorandom order. The tracking was performed using a high-speed camera (1000 frames/second), and previously described tracking software [78]. Each stimulus was accompanied with a 120ms recording, 30ms before and 90ms after the stimulus, with a 20 seconds window between stimuli (to prevent desensitization).
For ablation experiments, the larvae were treated with 10mM metronidazole during the nights of 3- and 4-dpf (for AgRP1 and AgRP2 experiments).
Circadian rhythms of locomotor activity
Circadian rhythms of locomotor activity were performed as previously described [79]. 4-dpf larvae were placed in 48-well plates in the observation chamber of the DanioVision Tracking System (Noldus Information Technology) and exposed, for acclimation, to 12-hours light (1.8 W/m2):12-hours dim light (0.013 W/m2 lux) regime followed by 3 days of constant dim light during 6–8-dpf. Live video tracking and analysis was conducted using the Ethovision 11.0 software (Noldus Information Technology). Activity was measured at 6–8-dpf, as the distance moved by a larva in 10 minute time bins. The activity record of each individual was subjected to Fourier analysis, and fitness to a circadian rhythm was scored with a g-factor as previously described [79].
Quantification and statistical analysis
Statistical analysis was performed using Statistica, GraphPad Prism and MATLAB. The statistical parameters are described in the relevant figure legends. All t-tests were 2-tailed, and the n numbers reported represent the number of independent replicates. ISH signals, melanosomes areas, and larvae body length were measured using ImageJ. Single-cell RNA-seq analysis was performed using the R package Seurat [66] as described above.
Data and software availability
The accession number for the scSeq data reported in this paper is GEO: GSE123778.
Supplementary Material
Pineal cells marker genes (Excel file). Related to Figure 3.
Adjusted p-value ranked list of putative markers for the pineal gland cell clusters. The results for both scSeq experiments are presented. Grey background represents a gene that was detected in both scSeq experiments. LogFC represents the logarithmic fold change of the gene expression.
Zebrafish AgRP1 neurons do not co-express Gad1b. Related to Figure 2.
Co-immunstaining of EGFP (green) and AgRP1 (magenta) in 6-dpf TgBAC(gad1b:EGFP)nns25 larva. Ventral view of the hypothalamus. Scale bar represents 10μm.
Acknowledgments
This research was supported by Research Grant Award No. 4892–16 from The United States– Israel Binational Agricultural Research and Development Fund (to Y.G. and R.D.C.), by Grant No. 2013433 from The United States– Israel Binational Science Foundation, Jerusalem, Israel (to Y.G. and H.A.B.), by grant no. 433/16 from the Israel Science Foundation (to Y.G.), by grant No. 2015-67015-23488 from USDA-NIFA (to R.D.C. and M.M.) and by the Recanati Foundation (to Y.G.). We gratefully acknowledge Steven L. Coon, James R. Iben and Tianwei Li from the NICHD Molecular Genomics Core for the technical support in the single-cell sequencing experiments. We thank Krasimir Slanchev for sharing fish lines and anatomical expertise. We thank Suresh J. Jesuthasan for fruitful discussions.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Morton GJ, Cummings DE, Baskin DG, Barsh GS, and Schwartz MW (2006). Central nervous system control of food intake and body weight. Nature 443, 289–295. Available at: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 2.Cone RD (2005). Anatomy and regulation of the central melanocortin system. Nat Neurosci 8, 571–578. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15856065. [DOI] [PubMed] [Google Scholar]
- 3.Rossi M, Kim MS, Morgan DG, Small CJ, Edwards CM, Sunter D, Abusnana S, Goldstone AP, Russell SH, Stanley SA, et al. (1998). A C-terminal fragment of Agouti-related protein increases feeding and antagonizes the effect of alpha-melanocyte stimulating hormone in vivo. Endocrinology 139, 4428–4431. [DOI] [PubMed] [Google Scholar]
- 4.Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, and Barsh GS (1997). Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science (80-. ). 278, 135–138. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9311920. [DOI] [PubMed] [Google Scholar]
- 5.Aponte Y, Atasoy D, and Sternson SM (2011). AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci 14, 351–355. Available at: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, Maratos-Flier E, Roth BL, and Lowell BB (2011). Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest 121, 1424–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luquet S, Perez FA, Hnasko TS, and Palmiter RD (2005). NPY/AgRP Neurons Are Essential for Feeding in Adult Mice but Can Be Ablated in Neonates. Science (80-. ). 310, 683–685. Available at: http://www.sciencemag.org/content/310/5748/683.abstract. [DOI] [PubMed] [Google Scholar]
- 8.Gropp E, Shanabrough M, Borok E, Xu AW, Janoschek R, Buch T, Plum L, Balthasar N, Hampel B, Waisman A, et al. (2005). Agouti-related peptide-expressing neurons are mandatory for feeding. Nat Neurosci 8, 1289–1291. Available at: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- 9.Bagnol D, Kaelin CB, Day HE, Ollmann M, Gantz I, Akil H, Barsh GS, Watson SJ, L. XY (1999). Anatomy of an endogenous antagonist: relationship between Agouti-related protein and proopiomelanocortin in brain. J Neurosci 19, RC26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atasoy D, Betley JN, Su HH, and Sternson SM (2012). Deconstruction of a neural circuit for hunger. Nature 488, 172–177. Available at: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Howell MP, and Palmiter RD (2009). Loss of GABAergic signaling by AgRP neurons to the parabrachial nucleus leads to starvation. Cell 137, 1225–1234. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2729323/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Y, and Cone RD (2007). Creation of a genetic model of obesity in a teleost. FASEB J 21, 2042–2049. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17341684. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C, Forlano PM, and Cone RD (2012). AgRP and POMC Neurons Are Hypophysiotropic and Coordinately Regulate Multiple Endocrine Axes in a Larval Teleost. Cell Metab. 15, 256–264. Available at: http://www.sciencedirect.com/science/article/pii/S1550413111004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shainer I, Buchshtab A, Hawkins TA, Wilson SW, Cone RD, and Gothilf Y (2017). Novel hypophysiotropic AgRP2 neurons and pineal cells revealed by BAC transgenesis in zebrafish. Sci. Rep 7, 44777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerda-Reverter JM, Ringholm A, Schioth HB, and Peter RE (2003). Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: involvement in the control of food intake. Endocrinology 144, 2336–2349. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12746294. [DOI] [PubMed] [Google Scholar]
- 16.Cerda-Reverter JM, and Peter RE (2003). Endogenous melanocortin antagonist in fish: structure, brain mapping, and regulation by fasting of the goldfish agouti-related protein gene. Endocrinology 144, 4552–4561. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12960082. [DOI] [PubMed] [Google Scholar]
- 17.Forlano PM, and Cone RD (2007). Conserved neurochemical pathways involved in hypothalamic control of energy homeostasis. J Comp Neurol 505, 235–248. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17879270. [DOI] [PubMed] [Google Scholar]
- 18.Pisharath H, Rhee JM, Swanson MA, Leach SD, and Parsons MJ (2007). Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev 124, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pisharath H, and Parsons MJ (2009). Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol Biol 546, 133–143. [DOI] [PubMed] [Google Scholar]
- 20.Qian S, Chen H, Weingarth D, Trumbauer ME, Novi DE, Guan X, Yu H, Shen Z, Feng Y, Frazier E, et al. (2002). Neither Agouti-Related Protein nor Neuropeptide Y Is Critically Required for the Regulation of Energy Homeostasis in Mice. Mol. Cell. Biol 22, 5027–5035. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC139785/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong I, Kim E, Kim S, Kim H-K, Lee D-W, Seong JY, and Park H-C (2018). mRNA expression and metabolic regulation of npy and agrp1/2 in the zebrafish brain. Neurosci. Lett 668, 73–79. [DOI] [PubMed] [Google Scholar]
- 22.Satou C, Kimura Y, Hirata H, Suster ML, Kawakami K, and Higashijima S (2013). Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development, dev-099531. [DOI] [PubMed] [Google Scholar]
- 23.Millhauser GL, McNULTY JOEC, Jackson PJ, Thompson DA, Barsh GS, and Gantz I (2003). Loops and Links: Structural Insights into the Remarkable Function of the Agouti-Related Protein. Ann. N. Y. Acad. Sci 994, 27–35. [DOI] [PubMed] [Google Scholar]
- 24.Toyama R, Chen X, Jhawar N, Aamar E, Epstein J, Reany N, Alon S, Gothilf Y, Klein DC, and Dawid IB (2009). Transcriptome analysis of the zebrafish pineal gland. Dev Dyn 238, 1813–1826. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19504458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey S, Shekhar K, Regev A, and Schier AF (2018). Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-seq. Curr. Biol 28, 1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lidén M, and Eriksson U (2006). Understanding retinol metabolism-structure and function of retinol dehydrogenases. J. Biol. Chem [DOI] [PubMed] [Google Scholar]
- 27.Hao W, and Fong HKW (1999). The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium. J. Biol. Chem 274, 6085–6090. [DOI] [PubMed] [Google Scholar]
- 28.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma J-X, Crouch RK, and Pfeifer K (1998). Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet. 20, 344. Available at: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 29.Strauss O (2005). The retinal pigment epithelium in visual function. Physiol. Rev 85, 845–881. [DOI] [PubMed] [Google Scholar]
- 30.Ekstrzm P, and Meissl Hi. (1997). The pineal organ of teleost fishes. Rev. Fish Biol. Fish. 7, 199–284. Available at: 10.1023/A:1018483627058. [DOI] [Google Scholar]
- 31.Blechman J, Anbalagan S, Matthews GG, and Levkowitz G (2018). Genome Editing Reveals Idiosyncrasy of CNGA2 Ion Channel-Directed Antibody Immunoreactivity Toward Oxytocin. Front. cell Dev. Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plucińska G, Paquet D, Hruscha A, Godinho L, Haass C, Schmid B, and Misgeld T (2012). In vivo imaging of disease-related mitochondrial dynamics in a vertebrate model system. J. Neurosci 32, 16203–16212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, and Miyawaki A (2002). An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci 99, 12651–12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Moshe Livne Z, Alon S, Vallone D, Bayleyen Y, Tovin A, Shainer I, Nisembaum LG, Aviram I, Smadja-Storz S, and Fuentes M (2016). Genetically blocking the zebrafish pineal clock affects circadian behavior. PLoS Genet. 12, e1006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gandhi AV, Mosser EA, Oikonomou G, and Prober DA (2015). Melatonin Is Required for the Circadian Regulation of Sleep. Neuron 85, 1193–1199. Available at: http://www.sciencedirect.com/science/article/pii/S0896627315001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Song Y, Thompson DA, Madonna MA, Millhauser GL, Toro S, Varga Z, Westerfield M, Gamse J, Chen W, et al. (2010). Pineal-specific agouti protein regulates teleost background adaptation. Proc Natl Acad Sci U S A 107, 20164–20171. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20980662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kok FO, Shin M, Ni C-W, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, et al. (2015). Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish. Dev. Cell 32, 97–108. Available at: http://www.sciencedirect.com/science/article/pii/S1534580714007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, and Stainier DYR (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230. [DOI] [PubMed] [Google Scholar]
- 39.Liu NA, Huang H, Yang Z, Herzog W, Hammerschmidt M, Lin S, and Melmed S (2003). Pituitary corticotroph ontogeny and regulation in transgenic zebrafish. Mol Endocrinol 17, 959–966. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12576489. [DOI] [PubMed] [Google Scholar]
- 40.Mommsen TP, Vijayan MM, and Moon TW (1999). Cortisol in teleosts: dynamics, mechanisms of action, and metabolic regulation. Rev. Fish Biol. Fish. 9, 211–268. Available at: 10.1023/A:1008924418720. [DOI] [Google Scholar]
- 41.Jin S-W, Beis D, Mitchell T, Chen J-N, and Stainier DYR (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209. Available at: http://dev.biologists.org/content/develop/132/23/5199.full.pdf. [DOI] [PubMed] [Google Scholar]
- 42.Krashes MJ, Shah BP, Koda S, and Lowell BB (2013). Rapid versus delayed stimulation of feeding by the endogenously released AgRP neuron mediators GABA, NPY, and AgRP. Cell Metab 18, 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Löhr H, Hess S, Pereira MMA, Reinoß P, Leibold S, Schenkel C, Wunderlich CM, Kloppenburg P, Brüning JC, and Hammerschmidt M (2018). Diet-induced growth is regulated via acquired leptin resistance and engages a Pomc-Somatostatin-growth hormone circuit. Cell Rep. 23, 1728–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrie H-L, Peilin C, Chien L, Kang C, Smith MS, Judy LC, and Roger DC (1999). Characterization of the Neuroanatomical Distribution of Agouti-Related Protein Immunoreactivity in the Rhesus Monkey and the Rat. Endocrinology 140, 1408–1415. Available at: http://press.endocrine.org/doi/abs/10.1210/endo.140.3.6544. [DOI] [PubMed] [Google Scholar]
- 45.Brent DW, Didier B, Christopher BK, Michael MO, Ira G, Stanley JW, and Gregory SB (1999). Physiological and Anatomical Circuitry between Agouti-Related Protein and Leptin Signaling. Endocrinology 140, 2387–2397. Available at: http://press.endocrine.org/doi/abs/10.1210/endo.140.5.6728. [DOI] [PubMed] [Google Scholar]
- 46.Mirabella N, Esposito V, Squillacioti C, De Luca A, and Paino G (2004). Expression of agouti-related protein (AgRP) in the hypothalamus and adrenal gland of the duck (Anas platyrhynchos). Anat Embryol 209, 137–141. Available at: 10.1007/s00429-004-0431-0. [DOI] [PubMed] [Google Scholar]
- 47.Falcón J, Besseau L, Fuentès M, Sauzet S, Magnanou E, and Boeuf G (2009). Structural and Functional Evolution of the Pineal Melatonin System in Vertebrates. Ann N Y Acad Sci 1163, 101–111. Available at: 10.1111/j.1749-6632.2009.04435.x. [DOI] [PubMed] [Google Scholar]
- 48.Falcón J, Migaud H, Muñoz-Cueto JA, and Carrillo M (2010). Current knowledge on the melatonin system in teleost fish. Gen. Comp. Endocrinol 165, 469–482. Available at: http://www.sciencedirect.com/science/article/pii/S0016648009001907. [DOI] [PubMed] [Google Scholar]
- 49.Skinner DC, and Malpaux B (1999). High melatonin concentrations in third ventricular cerebrospinal fluid are not due to Galen vein blood recirculating through the choroid plexus. Endocrinology 140, 4399–4405. [DOI] [PubMed] [Google Scholar]
- 50.FalcÓn J, Besseau L, and Boeuf GBT-FP (2006). Molecular and Cellular Regulation of Pineal Organ Responses. In Sensory Systems Neuroscience (Academic Press; ), pp. 243–306. Available at: http://www.sciencedirect.com/science/article/pii/S1546509806250064. [Google Scholar]
- 51.Schachat AP, Wilkinson CP, Hinton DR, Sadda SR, and Wiedemann P (2017). Ryan’s Retina E-Book (Elsevier Health Sciences; ). [Google Scholar]
- 52.Thisse B, and Thisse C (2004). Fast release clones: a high throughput expression analysis. ZFIN direct data Submiss. 2004. [Google Scholar]
- 53.Folgueira M, Bayley P, Navratilova P, Becker TS, Wilson SW, and Clarke JDW (2012). Morphogenesis underlying the development of the everted teleost telencephalon. Neural Dev. 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodríguez EM, Blázquez JL, and Guerra M (2010). The design of barriers in the hypothalamus allows the median eminence and the arcuate nucleus to enjoy private milieus: The former opens to the portal blood and the latter to the cerebrospinal fluid. Peptides 31, 757–776. Available at: http://www.sciencedirect.com/science/article/pii/S0196978110000239. [DOI] [PubMed] [Google Scholar]
- 55.Braasch I, and Postlethwait JH (2011). The teleost agouti-related protein 2 gene is an ohnolog gone missing from the tetrapod genome. Proc Natl Acad Sci U S A 108, E47–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schiöth HB, Västermark Å, and Cone RD (2011). Reply to Braasch and Postlethwait: Evolutionary origin of the teleost A2 agouti genes (agouti signaling protein 2 and agouti-related protein 2) remains unclear. Proc Natl Acad Sci U S A 108, E49–E50. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3069206/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J-Y, Finniss S, Yang Y-K, Zeng Q, Qu S-Y, Barsh G, Dickinson C, and Gantz I (2000). Agouti-related protein-like immunoreactivity: characterization of release from hypothalamic tissue and presence in serum. Endocrinology 141, 1942–1950. [DOI] [PubMed] [Google Scholar]
- 58.Westerfield M (2000). The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed. (Eugene: Univ. of Oregon Press; ). [Google Scholar]
- 59.Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, and Parsons MJ (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol 304, 811–824. Available at: http://www.sciencedirect.com/science/article/pii/S0012160607000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner KJ, Bracewell TG, and Hawkins TA (2014). Anatomical Dissection of Zebrafish Brain Development. In Brain Development: Methods and Protocols, Sprecher GS, ed. (Totowa, NJ: Humana Press; ), pp. 197–214. Available at: 10.1007/978-1-62703-655-9_14. [DOI] [PubMed] [Google Scholar]
- 61.Gilmour DT, Maischein H-M, and Nüsslein-Volhard C (2002). Migration and Function of a Glial Subtype in the Vertebrate Peripheral Nervous System. Neuron 34, 577–588. Available at: http://www.sciencedirect.com/science/article/pii/S0896627302006839. [DOI] [PubMed] [Google Scholar]
- 62.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, and Zhu J (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernandes AM, Fero K, Arrenberg AB, Bergeron SA, Driever W, and Burgess HA (2012). Deep brain photoreceptors control light-seeking behavior in zebrafish larvae. Curr. Biol 22, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernardos RL, and Raymond PA (2006). GFAP transgenic zebrafish. Gene Expr. Patterns 6, 1007–1013. [DOI] [PubMed] [Google Scholar]
- 66.Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495. Available at: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hatta K, Tsujii H, and Omura T (2006). Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc 1, 960. [DOI] [PubMed] [Google Scholar]
- 68.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411. Available at: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perner B, Englert C, and Bollig F (2007). The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol 309, 87–96. [DOI] [PubMed] [Google Scholar]
- 70.Jao LE, Wente SR, and Chen W (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013 August 20;110(34)13904–9. doi 10.1073/pnas.1308335110. Epub 2013 Aug 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou W, Boucher RC, Bollig F, Englert C, and Hildebrandt F (2010). Characterization of mesonephric development and regeneration using transgenic zebrafish. Am. J. Physiol. Circ. Physiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ota S, Hisano Y, Muraki M, Hoshijima K, Dahlem TJ, Grunwald DJ, Okada Y, and Kawahara A (2013). Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes to Cells 18, 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sander JD, Zaback P, Joung JK, Voytas DF, and Dobbs D (2007). Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 35, W599–W605. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1933188/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, and Yeh J-RJ (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol 29, 697–698. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3154023/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimada Y, Hirano M, Nishimura Y, and Tanaka T (2012). A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PLoS One 7, e52549. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=23300705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biran J, Ben-Dor S, and Levavi-Sivan B (2008). Molecular identification and functional characterization of the kisspeptin/kisspeptin receptor system in lower vertebrates. Biol. Reprod 79, 776–786. [DOI] [PubMed] [Google Scholar]
- 78.Burgess HA, and Granato M (2007). Sensorimotor gating in larval zebrafish. J. Neurosci 27, 4984–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tovin A, Alon S, Ben-Moshe Z, Mracek P, Vatine G, Foulkes NS, Jacob-Hirsch J, Rechavi G, Toyama R, Coon SL, et al. (2012). Systematic Identification of Rhythmic Genes Reveals camk1gb as a New Element in the Circadian Clockwork. PLoS Genet. 8, e1003116. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3527293/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gothilf Y, Coon SL, Toyama R, Chitnis A, Namboodiri MAA, and Klein DC (1999). Zebrafish serotonin N-acetyltransferase-2: marker for development of pineal photoreceptors and circadian clock function. Endocrinology 140, 4895–4903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pineal cells marker genes (Excel file). Related to Figure 3.
Adjusted p-value ranked list of putative markers for the pineal gland cell clusters. The results for both scSeq experiments are presented. Grey background represents a gene that was detected in both scSeq experiments. LogFC represents the logarithmic fold change of the gene expression.
Zebrafish AgRP1 neurons do not co-express Gad1b. Related to Figure 2.
Co-immunstaining of EGFP (green) and AgRP1 (magenta) in 6-dpf TgBAC(gad1b:EGFP)nns25 larva. Ventral view of the hypothalamus. Scale bar represents 10μm.
Data Availability Statement
The accession number for the scSeq data reported in this paper is GEO: GSE123778.