Abstract
The multidomain non-structural protein 3 (Nsp3) is the largest protein encoded by coronavirus (CoV) genomes and several regions of this protein are essential for viral replication. Of note, SARS-CoV Nsp3 contains a SARS-Unique Domain (SUD), which can bind Guanine-rich non-canonical nucleic acid structures called G-quadruplexes (G4) and is essential for SARS-CoV replication. We show herein that the SARS-CoV-2 Nsp3 protein also contains a SUD domain that interacts with G4s. Indeed, interactions between SUD proteins and both DNA and RNA G4s were evidenced by G4 pull-down, Surface Plasmon Resonance and Homogenous Time Resolved Fluorescence. These interactions can be disrupted by mutations that prevent oligonucleotides from folding into G4 structures and, interestingly, by molecules known as specific ligands of these G4s. Structural models for these interactions are proposed and reveal significant differences with the crystallographic and modeled 3D structures of the SARS-CoV SUD-NM/G4 interaction. Altogether, our results pave the way for further studies on the role of SUD/G4 interactions during SARS-CoV-2 replication and the use of inhibitors of these interactions as potential antiviral compounds.
INTRODUCTION
Coronaviruses (CoVs) constitute a large family of viruses belonging to the Nidovirales order. Some can infect humans, most often causing mild cold-like symptoms. Nevertheless, three deadly epidemics have already occurred in the 21st century, including the outbreak of severe acute respiratory syndrome (SARS) and the current COVID-19 (1).
The SARS-CoV genome contains 14 open reading frames, with ORF1a and ORF1b occupying two thirds of the genome and encoding 16 non-structural proteins (Nsp1-16) after auto-proteolytic processing of the polyproteins. Among them, the Nsp3 protein plays a crucial role in the formation and activity of the viral replication/transcription complex (2). Originally, this protein was shown to contain a non-conserved domain called SARS-Unique Domain or SUD, which is absent in less pathogenic coronaviruses infecting humans (3,4) or incomplete in the MERS-CoV Nps3 protein (5). The 3D-structure of this domain, solved by X-ray crystallography and NMR spectroscopy, has revealed the presence of two macrodomains (SUD-N and SUD-M) and a frataxin-like domain (SUD-C), corresponding to the N-terminal, Middle and C-terminal parts of SUD, respectively (6–8). These three domains are also called Mac2, Mac3 and DPUP (9,10), but we chose the SUD-N, SUD-M and SUD-C nomenclature since the combination of these three domains is unique to SARS viruses. SARS-CoV SUD N and M macrodomains have homologous structures and both interact with DNA and RNA oligonucleotides, especially those folded into guanine-quadruplex (G4) structures (6–8,11,12). Although no structure of a SUD-NM/G4 complex has been solved yet, structural models of this complex have been proposed (5,13). These models have been used to identify the amino-acids of SUD putatively involved in G4 binding (5) and to study the conformational dynamics of the SUD-NM/G4 complex (13). At a functional level, using a reverse-genetic system, it has been shown that SUD-M deletion and targeted mutations altering G4 binding largely affect the activity of the SARS-CoV replication/transcription complex (5). SARS-CoV SUD also interacts with cellular proteins. A fusion of SUD and PLpro Nsp3 domains binds to the E3 ubiquitin ligase RCHY1 and this interaction contributes to the virulence of SARS-CoV in human cells (14). More recently, it has been shown that both SARS-CoV and SARS-CoV-2 SUD domains interact with the human poly(A)-binding protein (PABP)-interacting protein A (Paip1) and that this interaction enhances SARS-CoV RNA translation (15). The SARS-CoV SUD domain also modulates the inflammatory response in lung epithelial cells and in a mouse model allowing to study pulmonary inflammation but the molecular mechanisms of this effect have not yet been characterized (16). Altogether, these data strongly support the role of SARS-CoV SUD in viral replication and cell response to infection and suggest that these effects may be mediated in part by interactions of SUD with G4 structures.
G4s are non-canonical nucleic acid structures formed by G-rich RNA or DNA sequences, which result from the stacking of two or more guanine quartets stabilized by a central spine of cations (17,18). These structures are highly polymorphic and have been extensively studied in vitro, using biophysical and structural approaches (19). G4s regulate major eukaryotic cell processes and are enriched in key regulatory regions of their genomes and transcriptomes including telomeres, promoters and 5′UTR of highly transcribed genes (20,21). While DNA G4s regulate the transcription of a number of human genes, especially oncogenes, RNA G4s participate in several mechanisms linked with mRNA metabolism, such as their translation, splicing, stability, polyadenylation and localization (22,23). G4s are also present in the genomes of DNA and RNA viruses and control critical steps of their replication (24–27). For instance, G4s present in the HIV-1 genome regulate reverse transcription and transcription steps (28,29) and these regulations require their interaction with cellular proteins (30,31). Conversely, some viral proteins act on viral replication through their interaction with RNA G4s (32,33).
Intensive research has also been carried out to identify molecules binding to or mimicking these structures. These molecules, called G4-ligands and G4-mimics respectively, can interfere with G4 activity and interaction with other partners (34–36). Two of them have been tested in the clinic against cancer and others exhibit promising antiviral activities in vitro (25,26).
Several algorithms have been developed to predict G4 propensity at the genome-wide level (37,38). One of them, G4Hunter, allows to predict putative quadruplex forming sequences (PQS) with unprecedented accuracy (37,39,40). This algorithm has recently been used to predict PQSs in several Nidovirales genomes (41). The highest density was found in Arteriviridae and the lowest in Coronaviridae. The SARS-CoV-2 genome contains a significantly lower density of G4 motifs than expected, with only one PQS in its negative-strand, using a default threshold (1.2) for G4Hunter search. This default threshold is optimal to detect PQS accurately and exhaustively, while higher thresholds (1.4 or more) decrease the number of false positives; more than 98% of experimentally tested motifs with a score of 1.5 or more form a bona fide G4 in vitro. Interestingly, SARS-CoV-2 does not contain a single G-rich motif with a score above 1.4 (according to G4Hunter) nor a single sequence matching default Quadparser parameters (4 runs of 3 or more G separated by 1–7 nucleotides) (42,43). Relaxing the parameters by using a lower threshold or looking for GG instead of GGG runs gives more hits, but generates a lot of false positives or ‘weak G4s’, as previously shown (37). Using a different algorithm with less stringent parameters (44), 25 PQS were predicted in the SARS-CoV-2 genome but all of them correspond to unstable G4s with only two G-tetrads (45,46). G4 formation was confirmed for two of the predicted RNA sequences but both motifs were thermally unstable with melting temperatures of 30.5 and 31.5°C, suggesting that these quadruplexes were mostly unfolded at physiological temperature (45). More generally, the G4 folding capacity of predicted SARS-CoV-2 PQSs is still under investigation as well as their relevance as antiviral targets (45–48).
These observations prompted us (i) to investigate whether a SUD domain is equally present in SARS-CoV-2 Nsp3; (ii) if this domain is present, to analyze its ability to bind G4s and (iii) to investigate whether these interactions can be modulated by G4-ligands. Our studies revealed that the SARS-CoV-2 Nsp3 protein indeed contains a SUD domain that interacts with specific DNA and RNA G4s but not with the putative G4s predicted in the SARS-CoV-2 genome. These interactions can be disrupted by mutations preventing G4 folding and by G4-ligands. Interestingly, structural models of the SARS-CoV-2 SUD-NM protein, alone or interacting with a DNA or RNA G4 reveal significant differences with the SARS-CoV homologous protein and its G4 complexes. These models are important to help identify antiviral molecules targeting specifically the SARS-CoV-2 SUD/G4 interaction.
MATERIALS AND METHODS
Oligonucleotides
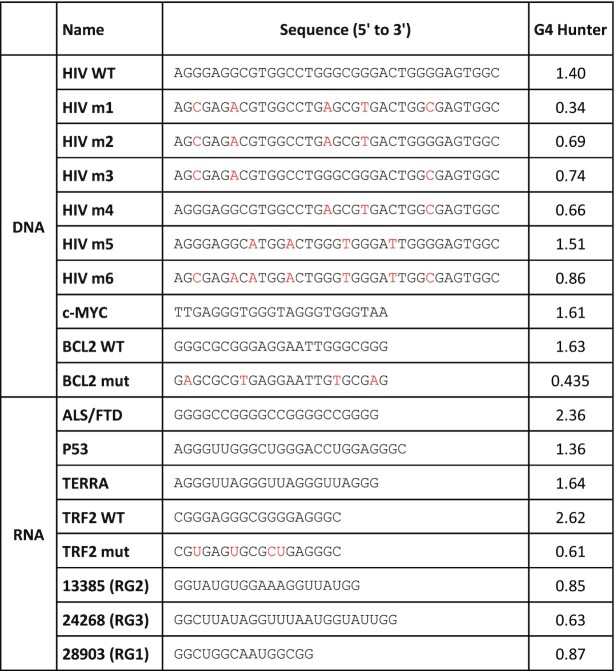
HPLC-purified oligonucleotides forming G4s were purchased from Eurofins Genomics and Eurogentec, in the 10–50 nmol scale. When indicated, they contain a Biotin-triethylene glycol (Biotin-TEG) tag at their 3′ end. Sequences and G4Hunter scores of all oligonucleotides are presented in Table 1. Sequence of oligonucleotides used for cloning and mutagenesis are presented in Supplementary Table SII.
Table 1.
Sequence and G4Hunter score of the oligonucleotides used in this study to generate G4 structures. Mutations introduced in HIV m1-m6, BCL2 mut and TRF2 mut sequences (as compared to HIV WT, BCL2 WT and TRF2 WT oligonucleotides) are shown in red
|
G4 ligands
All tested compounds have been previously described: bisquinolinium derivatives (PhenDC3 (49), PhenDH2 (50) and PDC (51)), phenanthroline derivatives (JG986 and JG1045) (52) and metallated porphyrins (Ni-MA and Au-TMPyP4) (53–55).
Expression and purification of SARS-CoV-2 SUD proteins
Sequences coding for selected regions of SARS-CoV-2 Nsp3 were cloned into the pET28a (NdeI/XhoI) plasmid, allowing the expression of the three recombinant SUD proteins called SUD-M, SUD-NM and SUD-NMC (corresponding to Nsp3 amino acids 549–676, 413–676 and 413–745 respectively) with an N-terminal polyhistidine-Tag. The quadruple mutation MutA (G489A, G490A, T491A, K497A) was introduced on the pET28a WT SUD-NM plasmid using designed oligonucleotides and the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies). pET28a SUD-NM Mut4 (K589A, K592A, E595A) was synthesized by TwistBioscience. The WT and mutated SUD coding sequences were verified by sequencing (Eurofinsgenomics) before being used for protein expression. Escherichia coli BL21(DE23)/pDIA17 (56) were transformed by the constructed pET28a plasmids and grown at 30°C in 2YT medium under kanamycin and chloramphenicol selection. At OD600nm = 1.5, protein expression was induced by 1 mM IPTG for 4h culture at 30°C. Bacterial pellets were resuspended in buffer A (250 mM NaCl, 25 mM Tris-HCl pH 7 (for SUD-M) or pH 8.5 (for SUD-NM and SUD-NMC)), supplemented with protease inhibitors (Complete ULTRA tablets EDTA-free, Roche). The bacterial suspension was lysed by sonication, and then further incubated 60 min at 4°C in the presence of RNAseA (ThermoFisher, EN0531) and Benzonase (Sigma, E1014). After centrifugation at 12 000 rpm and 4°C for 45 min, the supernatant was applied to a TALON® Metal Affinity Resin (TaKaRa) using the batch/gravity-flow column purification procedure at room temperature. Washing steps were performed using the corresponding buffer A with 5 mM imidazole and different concentrations of NaCl (250, 500, 1000, 150 mM). The resin-bound protein was eluted with buffer A complemented with 150 mM NaCl and 150 mM imidazole. SUD-NM and SUD-NMC proteins were further purified by size-exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated in 150 mM NaCl, 25 mM Tris-HCl pH 8.5 or 7.5 respectively. Eluted fractions were dialyzed against 150 mM NaCl, 25 mM Tris–HCl pH 9, 20% glycerol and stored frozen in small aliquots. Quality control of the recombinant WT proteins was performed according to a previously published approach (57) following the ARBRE-MOBIEU/P4EU guidelines (https://arbre-mobieu.eu/guidelines-on-protein-quality-control/). Protocols are detailed in the supplementary section.
Molecular mass measurements by size exclusion chromatography coupled to static light scattering detection (SEC-SLS) and viscometry
The oligomerization state of the SUD proteins was determined by size exclusion chromatography (SEC) coupled to a triple detection (concentration detector: UV detector, refractometer; static light scattering 7°, 90°; viscometer) on a Omnisec resolve and reveal instrument (Malvern Panalytical). Before equilibrating the column and detectors, the 20 mM Tris–HCl pH 9, 500 mM NaCl running buffer was filtered through 0.2 μm filters. All SUD proteins were injected on a Superdex 75 10/300 GL column (GE) at 20°C. Samples were eluted at 0.4 ml/min after injection of a 100 μl sample at 3.4 mg/ml. External calibration was done with bovine serum albumin (BSA) (Sigma) using an injection of 100 μl at 1.4 mg/ml. The refractive index, static light scattering and the viscosity measurements were processed to determine the mass average molecular mass and the intrinsic viscosity using the OMNISEC V11.10 software (Malvern Panalytical, UK). The theoretical hydrodynamic radius and intrinsic viscosity were calculated from the PDB model using the Hydropro software (58).
Isothermal differential spectra (IDS) and circular dichroism (CD)
Oligonucleotides were prepared at a final concentration of 2.5 μM in a 20 mM HEPES pH 7.2 buffer prepared in diethylpyrocarbonate-treated water. Solutions were briefly heated to 70°C, then cooled back to room temperature, and a first absorbance spectrum (unfolded sample) was recorded in black Hellma 10 mm path length quartz cuvettes. KCl was then added to a final concentration of 0.1 M and the sample was incubated at room temperature for 10 min, before recording a second absorbance spectrum (both in the 220–335 nm wavelength range, 400 nm/min, with an autozero at 335 nm). The second spectrum (folded with K+) was first corrected for dilution, and the difference between the two spectra corresponds to the IDS (59) which share similarities with the thermal difference spectra (60). CD experiments were performed using an Aviv CD spectrometer model 215 equipped with a water-cooled Peltier unit. Oligonucleotides at 2.5 μM, prepared as for IDS, were placed at 25°C in a 1 cm path length cells and spectra were recorded from 220 to 320 nm with 1 nm step. Three consecutive scans from each sample were merged to produce averaged spectra, which were corrected using buffer baselines measured under the same conditions.
G4 pull-down
Selected 3′-Biotin-TEG labeled oligonucleotides (Table 1) were folded in 10 mM Tris–HCl pH 8, 100 mM KCl, 0.1 mM EDTA, with a 5 min 95°C denaturation followed by a slow cool down (2°C / min). High-affinity streptavidin magnetic beads (Pierce, 88817) were equilibrated in binding buffer (10 mM Tris–HCl pH 8, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.05% Tween) prior to incubation with the G4-folded oligonucleotides (0.1–0.5 nmoles oligonucleotides/10 μl streptavidin magnetic beads) for 2 h at 4°C. After two washes in the binding buffer (to remove unbound oligonucleotides), the beads were incubated with 2–10 μg of recombinant proteins for 1 h at 4°C. Beads were then washed with binding buffer containing increasing KCl concentrations (200–500 mM) and retained proteins were finally eluted from the beads by a 5 min incubation at 95°C in 2× SDS-PAGE loading buffer. Eluted proteins were analyzed by SDS-PAGE and western blot analysis using a His-HRP antibody (Sigma-Aldrich, A7058).
Surface plasmon resonance (SPR)
Real-time biosensing experiments were performed on a Biacore T200 instrument (GE Healthcare) equilibrated at 25°C in binding buffer (composition described above) containing additional BSA (0.2 mg/ml) to avoid non-specific binding. Biotinylated G4s (20 nM) were first captured on streptavidin-coated CM5 sensorchips (GE Healthcare), reaching a density of 80–100 resonance units (RU ≈ pg mm–2). Serially diluted SUD-NM samples (7.8–1000 nM) were then injected one at a time in triplicate for 2 min at a flow rate of 30 μl/min over the oligonucleotide-derivatized surfaces (association phase), after which buffer was flowed at 30 μl/min for 3 min (dissociation phase). Finally, the chip surfaces were regenerated by stripping the remainder SUD-NM with a 90 s injection of KCl 3M. The association and dissociation profiles were analyzed using two complementary methods to verify self-consistency: (i) the concentration-dependence of the steady-state SPR signal (Req) was analyzed and fitted using the equation Req = (Rmax * C)/(Kd + C), where C is the concentration of SUD-NM and Rmax the maximal binding capacity of the G4 surface and (ii) the kinetic association/dissociation profiles were analyzed using the global fitting software BIAevaluation 4.0 (GE Healthcare) assuming either a single Langmuir binding mechanism or a model involving a conformational change of the SUD/G4 complex after the initial contact between the two partners.
Homogenous time resolved fluorescence (HTRF)
These assays were performed using purified His-Tagged SUD domains, biotinylated G4s and specific antibodies coupled to fluorophores (CISBIO, Saclay, France), namely Streptavidin-d2 (acceptor conjugate) and MAb Anti-6His-Eu cryptate (donor conjugate). Each assay was performed in triplicate in a 96-well low-binding surface 1/2 area AlphaPlate (PerkinElmer). 5 μl of 4× SUD domain solution and 5 μl of 4× biotinylated G4 solution were added to each well, except for the control wells where either one partner or both were replaced by the assay buffer T (25 mM Tris–HCl pH 8, 100 mM KCl, 1 mM DTT, 0.05% Tween, 0.5% BSA). After 1 h incubation at room temperature, 10 μl of donor–acceptor conjugate mixture (1:1) diluted in PBS pH 7.4 and BSA 0.1% was added to each well. After a further 1 h incubation at room temperature, the fluorescence emission was measured at 620 and 665 nm using an excitation wavelength of 340 nm on a microplate reader (Infinite M1000 Pro, TECAN). The HTRF ratio was calculated as follows: (fluorescence intensity at 660 nm/ fluorescence intensity at 620 nm) × 104. For the chase experiments (in the presence of non-biotinylated G4s) or the evaluation of G4 ligands, the assay was modified as follows: 5× solutions of SUD-domain and biotinylated G4 were used, and 4 μl of each as well as 2 μl of the non-biotinylated G4 or the tested compound (in DMSO or water) were added to each well. IC50 values were calculated from the experimental data using the KaleidaGraph software (Synergy Software).
Computational modeling of the SARS-CoV-2 SUD-NM
The three-dimensional structure of SARS-CoV-2 SUD-NM was computed with the Modeller software (61). The most complete and highest resolution (2.2 Å) crystal structure of SARS-CoV SUD-NM (PDB code: 2W2G, (8)) was chosen as template structure to generate this model. Taking into account the slight differences between the structures of monomers A and B in the crystallographic structure (RMSD of 0.7 Å when superposed), the monomer of SARS-CoV-2 SUD-NM was modeled using as templates the two monomers of the 2W2G PDB structure whereas the dimer of SARS-CoV-2 SUD-NM was modeled from the entire 2W2G crystallographic structure. Adding the NMR structure with PDB code: 2JZE (6) or the 2.8 Å resolution crystal structure with PDB code: 2WCT (8) as template structures in the modeling process did not improve the quality of the SARS-CoV-2 SUD-NM models. An energy minimization step for all the modeled structures was then performed using the YASARA force field (62).
Docking computations
The best quality model of SARS-CoV-2 SUD-NM was then used for docking simulations. The docking computations of the major G4 structure formed in the human BCL2 promoter region (PDB code: 2F8U; (63)) on the modeled SARS-CoV-2 SUD-NM structure were performed with autodock-vina (64) which predicts interactions between small molecules and proteins and the poses with the lowest binding energy score were reported. We also performed docking computations with the human telomeric RNA (TERRA) quadruplex (PDB code: 3IBK, (65)). All figures were made with PyMOL (66).
RESULTS
SARS-CoV-2 Nsp3 contains a SARS Unique domain (SUD)
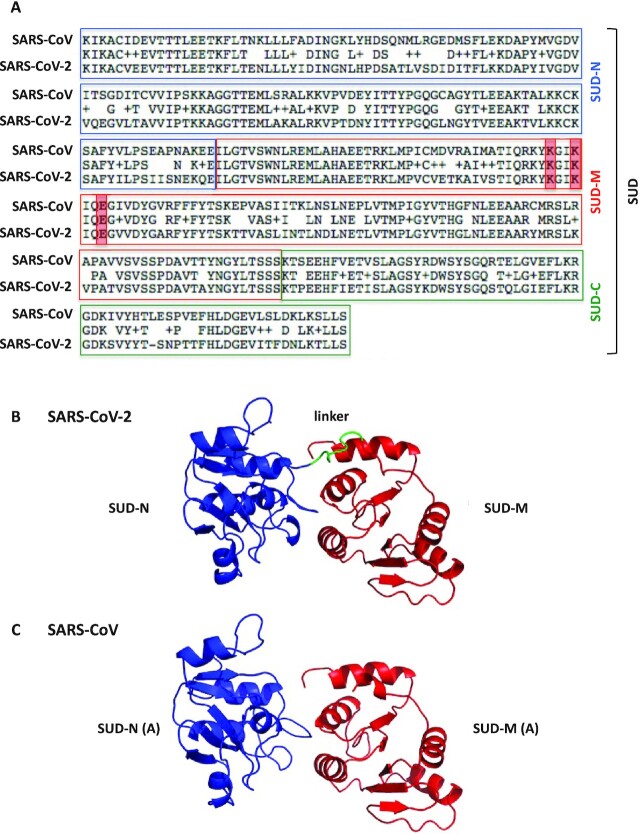
We first determined that the SARS-CoV-2 Nsp3 comprised a SUD domain. SUD-N, SUD-M and SUD-C sequences are indeed present in SARS-CoV-2 strains and are highly similar to the corresponding macrodomain and frataxin-like fold sequences of SARS-CoV (Figure 1A). In addition, amino acids identified as crucial for G4 binding (8) and SARS-CoV replication (5) are conserved in the SARS-CoV-2 SUD-M domain. Therefore, as previously shown for SARS-CoV, the SUD domain is probably a key region of the SARS-CoV-2 virus Nsp3 protein.
Figure 1.
Primary structure alignment of the SARS-CoV and SARS-CoV-2 SUD proteins and homology modeling of the SARS-CoV-2 SUD-NM protein. (A) The alignment was performed using the BLAST algorithm with the Blosum62 scoring matrix between amino acids 389–720 and 413–744 of SARS-CoV (NC004718) and SARS-CoV-2 (BetaCoV_Wuhan_IVDC-HB-01_2019) Nsp3 proteins, respectively. More precisely, the SUD-N and SUD-M macrodomains and the SUD-C frataxin-like domain of these two viruses share 88.2, 96.1 and 91.4% amino-acid sequence similarities (ExPASy LALIGN algorithm). The K565-K568-E571 residues (highlighted in red), present in SARS-CoV SUD-M and important for viral replication (5) are conserved in SARS-CoV-2 SUD-M. (B) Ribbon representation of the modeled structure of the SARS-CoV-2 SUD-NM computed with the Modeller software using the SARS-CoV SUD-NM protein as template structure (PDB code: 2W2G (8)). SUD-N is in blue, SUD-M in red and the 518–524 linker connecting the two macrodomains is in green. (C) Structure of the SARS-CoV SUD-NM monomer A template structure (PDB code: 2W2G) provided for comparison and in the same orientation than in (B).
The three-dimensional structure of SUD-NM from SARS-CoV-2 was predicted thanks to the Modeller software (Figure 1B). The high sequence identity (76%) and similarity (95%) between the SUD-NM proteins from SARS-CoV and SARS-CoV-2 allowed to generate a good quality model that could be used in docking simulations (see below). The structure of the modeled SARS-CoV-2 SUD-NM is very close to the SARS-CoV SUD-NM template structure (RMSD = 0.99 Å) (Figure 1B-C). The linker connecting the two macrodomains, not visible in the template crystallographic structure, and the region [423:432] are less conserved in SUD-NM and the disulfide bridge which connects a SUD-N loop to a SUD-M helix in the SARS-CoV SUD-NM crystal structure does not appear to be preserved in the SARS-CoV-2 SUD model. The absence of this disulfide bridge may allow a greater adaptability of the SARS-CoV-2 SUD-NM protein when binding to G4s, as motions between the two macrodomains should be less constrained.
Recombinant SARS-CoV-2 SUD proteins production and characterization
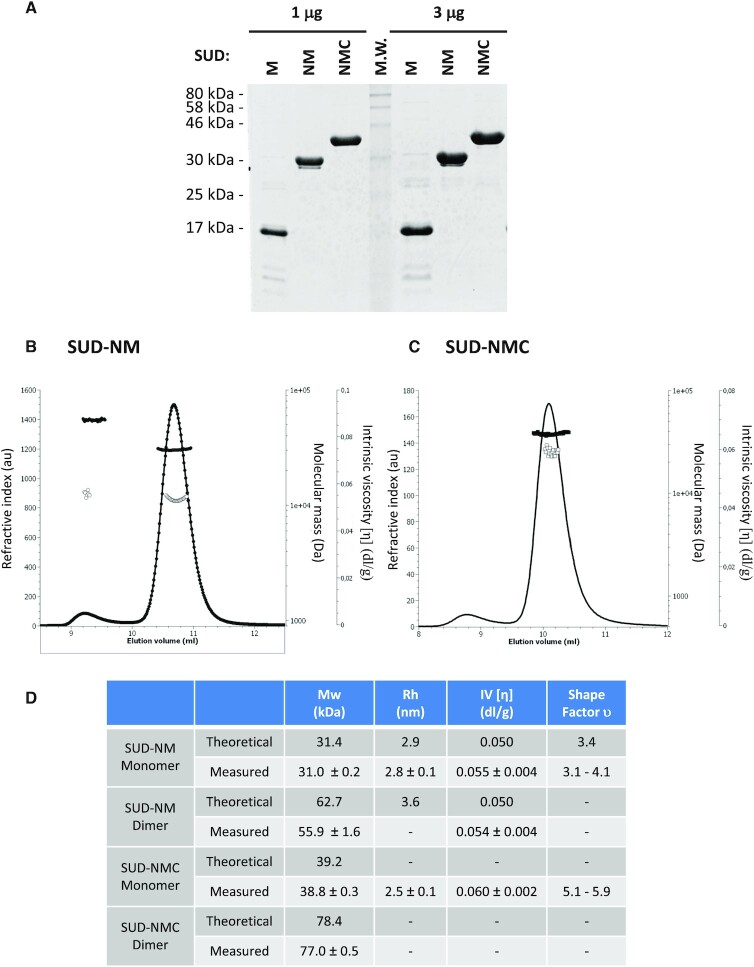
SUD-M, SUD-NM (named SUDcore in (8)) and SUD-NMC of SARS-CoV-2 (corresponding to amino-acids 549–676, 413–676 and 413–745 of SARS-CoV-2 Nsp3 respectively) were overexpressed in E. coli as soluble proteins with an N-terminal polyhistidine-tag and purified using a two-step chromatographic procedure, resulting in more than 95% purity as indicated by SDS-PAGE (Figure 2A, peptidic sequence presented in Supplementary Figure S1). The solubility, integrity and identity of the proteins were checked by dynamic light scattering (DLS), UV–visible spectroscopy and mass spectrophotometry analysis of the samples (Supplementary Figure S2–S4). The protein storage buffer was optimized by DLS and nano differential scanning fluorimetry (nanoDSF) by varying the pH and the salinity of the solutions. All the proteins were optimally stable in 20 mM Tris–HCl pH 9, 500 mM NaCl at 20°C and could be stored for a few days at 4°C or frozen at –80°C after dialysis in the presence of 20% glycerol.
Figure 2.
Characterization of purified SARS-CoV-2 proteins. (A) 1 μg and 3 μg of the three studied proteins were separated on a 15% polyacrylamide-SDS gel and visualized by Coomassie blue staining of the gel. (B, C) Hydrodynamic characterization of SUD-NM (b) and SUD-NMC (C) after size exclusion chromatography separation: refractive index (plain line), molecular mass (plain circle), intrinsic viscosity (open circle). (D) Summary of hydrodynamic characterization and modeling.
The oligomeric state of SUD-NM and SUD-NMC proteins was investigated by static light scattering associated with intrinsic viscosity measurements. SUD-NM and SUD-NMC were mostly monomeric in solution at the concentration measured, but some dimer formation could be observed (Figure 2B–D). Interestingly, the monomers of SUD-NM and SUD-NMC showed an intrinsic viscosity [η] typical of globular assemblies (Figure 2B–D). These values combined with hydrodynamic radius (Rh) measurements performed by DLS, allow to calculate the shape factor υ (67,68). For SUD-NM, υ is comprised between 3.1 and 4.1, which is compatible with a slightly oblate shape of the protein as observed in the computed model (see below). In contrast, the shape factor υ of SUD-NMC has a value between 5.1 and 5.9 showing a more elongated molecule probably due to the C-terminal part. Overall, SUD-NM and SUD-NMC proteins in solution mainly behave as monomers that differ by their elongation. Using the modeled structure of SUD-NM (Figure 1B), we computed the theoretical values of Rh and η (Figure 2D). Comparing the experimental and theoretical values, obtained for the SUD-NM construct, strongly argues in favor of a monomeric state of this protein.
Selection of DNA and RNA G4s
Although SUD probably interacts with cellular RNA G4s, given its expected cytoplasmic location, we tested the capacity of both DNA and RNA G4s to interact with SUD. The sequences and G4 folding scores (calculated with G4Hunter) of the corresponding oligonucleotides are presented in Table 1. HIV-1 LTRIII (hereafter abbreviated as HIV), c-MYC and BCL2 were selected as DNA G4s because of their role in transcriptional regulation and their well-known structural properties (63,69,70). BCL2 G4 was also chosen because its NMR solution structure (PDB: code 2F8U, (63)) has been docked in the SARS-CoV SUD-NM structure (5). Similarly, RNA G4s were selected to cover a diversity of structures and activities in human cells. More precisely, we chose the GGGGCC hexanucleotide repeat associated to neurodegenerative diseases ALS and FTD (71), the RNA telomeric repeat called TERRA (72), a G4 motif present in P53 mRNA contributing to alternative splicing (73), and a G4 motif located in the 5′UTRs of TRF2 mRNA and repressing its translation (74). Mutated sequences of HIV, BCL2 and TRF2 motifs, unable to form a G4 structure (74) were also added as controls in our interaction assays (see Table 1 for the corresponding G4Hunter scores). CD and IDS both confirmed that the BCL2 and TRF2 mutants failed to form a quadruplex (Supplementary Figure S5).
Interaction of SARS-CoV-2 SUD recombinant proteins with DNA and RNA G4s, shown by G4 pull-down assay
The interaction of all three SUD constructs with the selected DNA and RNA G4s was first evaluated using a G4 pull-down assay (Figure 3). For this assay, biotinylated oligonucleotides were folded into G4 following a heat denaturation and a slow renaturation in a potassium buffer and attached to streptavidin-coated magnetic beads. Biotin addition at one extremity of the oligonucleotides did not prevent G4 formation, as shown by CD and IDS (Supplementary Figure S5). SUD proteins were incubated with the G4-covered beads and after several washes with increasing salinity buffers (100–500 mM KCl), the retained proteins were revealed by SDS-PAGE and immuno-detection. This assay was initially performed on HIV, c-MYC and BCL2 DNA G4s (Figure 3A-B). Mutated HIV (m1 to m6) and BCL2 (mut) oligonucleotides unable to form G4s were used in this assay to evaluate the specificity of SUD for G4 structures. Among these mutants, HIV m1 to m4 and BCL2 mut have the lowest G4 folding propensity, as shown by their G4Hunter scores (Table 1). The three SUD recombinant proteins were evaluated for their ability to interact with HIV and c-MYC G4s (Figure 3A). SUD-M interacted with all the different oligonucleotides which suggests that this domain is too short or too positively charged (pI >9), to discriminate between G4 and non-G4 folded structures. Conversely, SUD-NM and SUD-NMC interacted preferentially with HIV and c-MYC G4s. Among the different HIV G4s, both proteins interacted preferentially with the WT, m5 and m6 oligonucleotides which suggests a preferential interaction with stable G4 structures. We also tested the interaction of SUD-NM for the WT and mut sequences of BCL2 DNA G4 and observed a preferential interaction for the WT sequence, as seen for the HIV G4 (Figure 3B). We then tested the capacity of SUD-NM to interact with the five selected RNAs (four of them being able to form G4s, Table 1). As shown in Figure 3C, this protein interacted preferentially with TRF2 WT G4, and mutations destabilizing this structure (TRF2 mut) largely disfavored its interaction with SUD-NM. Even if G4 pull-down is qualitative rather than quantitative, the results obtained with this assay suggests that the SARS-CoV-2 SUD protein binds to G4 structures and that the quadruplex motif found in the TRF2 mRNA can be chosen as a prototypal high-affinity binding partner.
Figure 3.
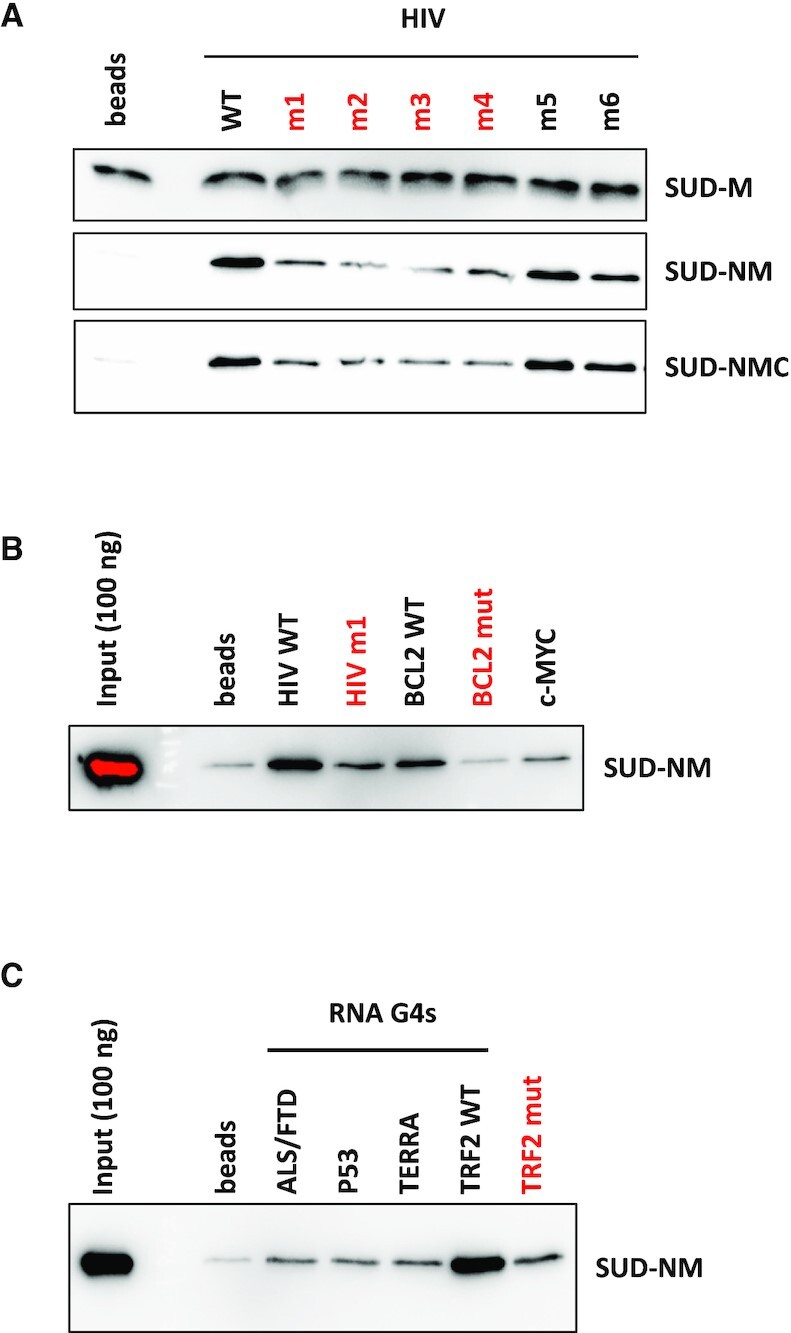
G4 pull-down of SARS-CoV-2 SUD proteins on DNA and RNA G4s. Interaction of SUD-M, NM and NMC proteins with DNA or RNA oligonucleotides folded as G4s and attached to streptavidin magnetic beads. Oligonucleotides labeled in red have the lowest G4Hunter scores. Proteins retained by the G4-coated beads were separated by SDS-PAGE and analyzed by Western blotting. (A) Comparative interaction of SUD proteins with DNA oligonucleotides displaying different G4 folding propensities. (B) Comparative interaction of SUD-NM with WT and mutant DNA G4s. (C) Comparative interaction of SUD-NM with four WT and one mutant RNA G4s. Among the sequences tested here, the TRF2 WT oligo-ribonucleotide appears to be the best RNA G4 substrate for SUD-NM.
SPR analysis of the interaction between SARS-CoV-2 SUD-NM and RNA G4s
Surface plasmon resonance can be used to quantify the affinity of proteins for DNA or RNA G4s (75–77). In this study, we used this technique to quantify the interaction between SARS-CoV-2 SUD-NM and the RNAs previously tested by G4 pull-down assay. The selected biotinylated G4 folded oligonucleotides (Table 1) were attached to a streptavidin coated CM5 sensor chip and serial dilutions of SUD-NM were injected on this surface (association phase) followed by binding buffer (dissociation phase). As a representative example, the association and dissociation profiles obtained by flowing 500 nM of SUD-NM over five RNAs, are presented in Figure 4A, while the profiles corresponding to the injections of increasing concentrations of SUD-NM (7.8–1000 nM) are shown in Figure 4B. In all cases, the real-time profiles are markedly biphasic, both at the level of association and dissociation, whatever the concentration of SUD-NM that is used (Figure 4B). This could indicate that the SUD-NM/G4 interaction involves a slow conformational change between an initial transient complex (fast association/fast dissociation) and a final more stable one.
Figure 4.
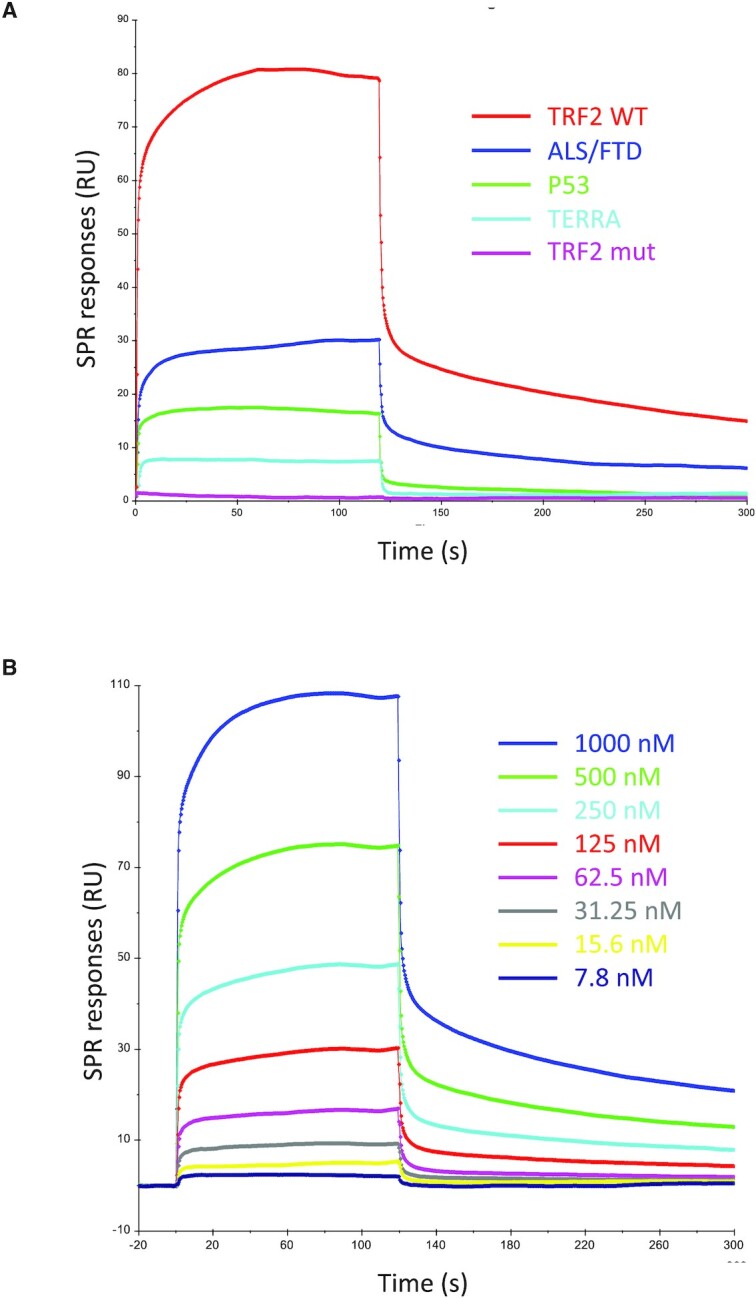
SPR sensorgrams corresponding to the interaction of SARS-CoV-2 SUD-NM with the five selected RNA G4s. (A) Real-time association and dissociation profiles corresponding to the interaction of 500 nM SUD-NM with five different RNA-G4s (TRF2 WT, TRF2 mut, ALS/FTD, P53 and TERRA) (B) Sensorgrams corresponding to the interaction of SUD-NM at eight different concentrations (7.8–1000 nM) with the TRF2 G4. A Kd of ∼500 nM was determined from the analysis of the concentration-dependence of the steady-state SPR response.
The SPR study confirms that a variety of RNA G4s can be good substrates of SUD-NM. Of course, this result is restricted to the selected G4s and does not yet demonstrate that the ribose backbone or a parallel G4 conformation favor binding to SUD-NM. However, these results support our hypothesis that Nsp3 interacts via its SUD domain with RNA G4s present in the cytoplasm during SARS-CoV-2 replication.
Specificity of the SARS-CoV-2 SUD/G4 interaction studied by HTRF
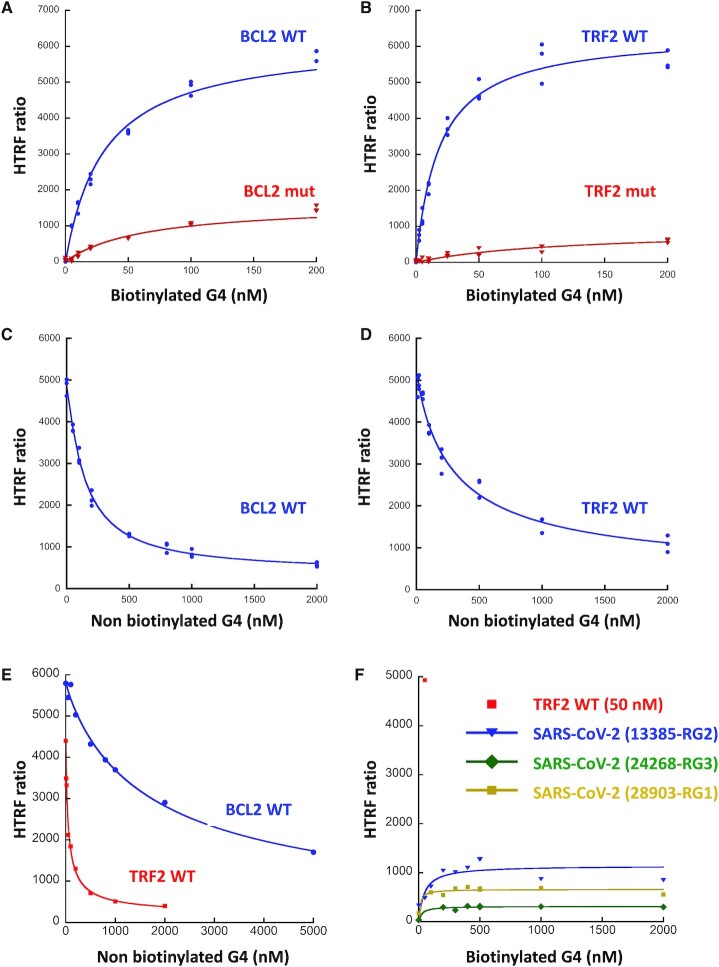
We set up a Homogenous Time Resolved Fluorescence (HTRF) assay to confirm the SUD-G4 interactions observed by G4 pull-down and SPR and to further evaluate their specificity for G4 structures. We used His-tagged SUD-NM and biotinylated G4s, which are recognized by two conjugates, anti-6His-Eu-cryptate (donor) and Streptavidin-d2 (acceptor), respectively. The interaction between the two partners results in a fluorescence energy transfer, which can be expressed as an HTRF ratio (see Material & Methods section). Based on the pull-down and SPR assays, we first selected two DNA G4s (BCL2 and c-MYC) and one RNA G4 (TRF2). For BCL2 and TRF2, we also used oligonucleotides containing mutations abolishing G4 folding (see Table 1). A significant signal is observed on WT BCL2, c-MYC and TRF2, confirming that SUD-NM binds to DNA and RNA G4s (Figure 5 and Supplementary Figure S6A). To find the optimal signal window, the concentrations of each partner were varied. For SUD-NM, the best concentration range was 10–30 nM. For the BCL2, c-MYC and TRF2 WT G4s, Kd values of 31, 64 and 18 nM could be calculated from the curve fitting. Plateau of the HTRF ratio were observed at 100 and 50 nM for BCL2 and TRF2 WT G4s and a hook effect was observed with c-MYC G4 at concentrations higher than 200 nM. These data indicate a slightly better affinity of SUD-NM for the TRF2 RNA G4 than for the BCL2 and c-MYC DNA G4s. Interestingly, these interactions are disrupted by mutations abolishing G4 folding (Figure 5A-B). The specificity of the HTRF assay was also demonstrated using non-biotinylated WT BCL2, c-MYC or TRF2 G4s as competitors. A dose-response experiment, performed for each G4, clearly shows a drastic decrease of the HTRF ratio at the highest concentrations of the non-biotinylated G4s (Figure 5C-D and Supplementary Figure S6B). This displacement does not depend on the concentration of SUD-NM (not shown). Interestingly, non-biotinylated TRF2 G4 displaces the SUD-NM/BCL2 DNA G4 interaction more efficiently, confirming the better affinity of SUD-NM for the TRF2 G4 partner (Figure 5E).
Figure 5.
HTRF assay of the interaction between SARS-CoV-2 SUD-NM and the BCL2, TRF2 and SARS-CoV-2 G4s. (A) His-SUD-NM (30 nM) and biotinylated BCL2 WT or mutant (5 to 200 nM) were incubated for 1 h at room temperature before addition of the donor and acceptor conjugates. (B) Same as in (A) with TRF2 WT or mutant (2.5 to 200 nM). KD of 31 ± 3 and 18 ± 3 nM were calculated for BCL2 WT and TRF2 WT respectively, using the Kaleidagraph software to fit the experimental data. (C) His-SUD-NM (30 nM) and biotinylated BCL2 WT (100 nM) were incubated for 1h at room temperature in the presence of increasing concentrations of non-biotinylated BCL2 WT (0–2 μM). (D) Same as in (C) with His-SUD-NM (30 nM) and biotinylated TRF2 WT (50 nM) in the presence of increasing concentrations of non-biotinylated TRF2 WT (0–2 μM). (E) His-SUD-NM (30 nM) was incubated for 1 h at room temperature with biotinylated BCL2 WT (100 nM) and increasing concentrations of non-biotinylated TRF2 WT (0 to 2 μM) (red curve) or with biotinylated TRF2 WT (50 nM) and increasing concentrations of non-biotinylated BCL2 WT (0–5 μM) (blue curve). (F) His-SUD-NM (30 nM) and biotinylated RNA RG1, RG2 and RG3 from SARS-CoV-2 sequence (5–2000 nM) were incubated for 1h at room temperature before addition of the donor and acceptor conjugates. TRF2 WT (50 nM) was used as a positive control.
Several PQS have been predicted in the SARS-CoV-2 genome but all of them correspond to presumably unstable G4s with only two G-tetrads and relatively low G4Hunter scores ((45,46) and Supplementary Table S1). This lack of stable PQS in a 30 kb genome suggests a strong counter-selection against G4s in SARS-CoV-2 which may prevent deleterious cis-interaction between the SUD domains and the viral genome. To test this hypothesis, we studied by HTRF the interaction of SUD-NM with the three most stable SARS-CoV-2 PQS proposed by these predictions (45,46). They are located at positions 13385, 24268 and 28903 of the SARS-CoV-2 genome and have been respectively called RG1, RG2 and RG3 (48) or N, nsP10 and S-b in (47). Results presented in Figure 5F clearly show the absence of interaction of SUD-NM with these three viral PQS and support the existence of only trans-interactions between the SUD domain and host cell RNA G4s.
Altogether, results obtained by G4-pulldown, SPR and HTRF assays reveal that the SARS-CoV-2 SUD-NM domain can interact with both DNA and RNA G4s and show that this interaction is specific for G4 structures.
G4-ligands inhibit SUD-NM/TRF2 G4 interaction
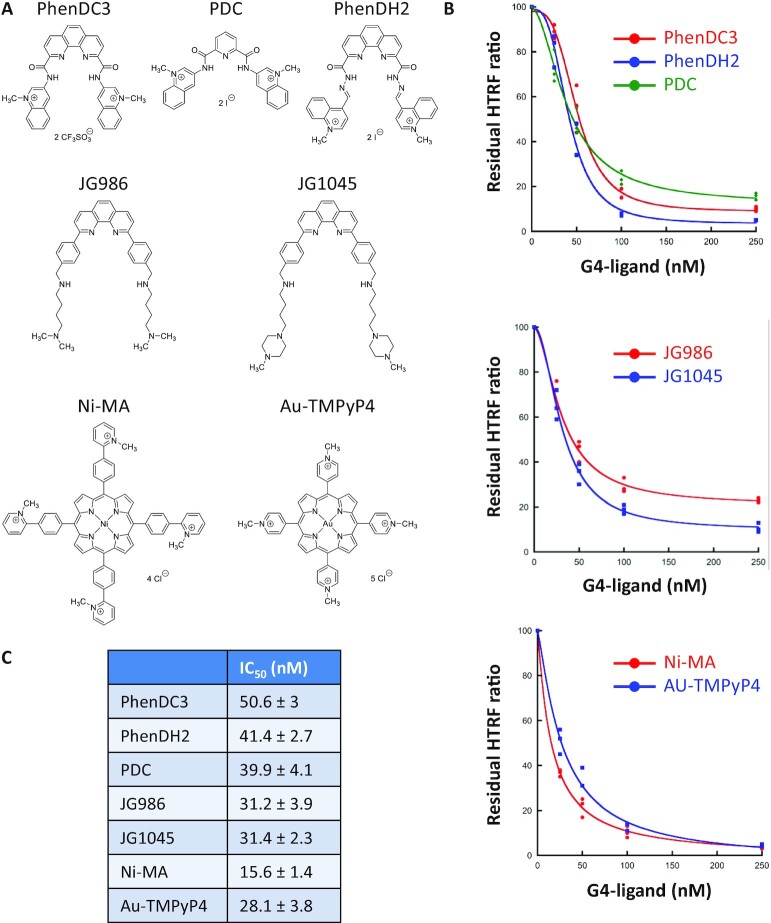
If the SUD/G4 interaction is essential for SARS-CoV-2 replication, as previously shown for SARS-CoV (5), molecules inhibiting or destabilizing this interaction should affect viral replication. To identify such inhibitors, we investigated whether small compounds tightly binding to G-quadruplexes, called G4-ligands, would perturb the SUD/G4 interaction. We selected known, high-affinity G4-ligands (Figure 6A) belonging to different chemical series such as bisquinolinium derivatives (49–51), phenanthroline derivatives (52) or metallated porphyrins (53,54), and performed a competition HTRF assay by targeting the SUD-NM/TRF2 G4 interaction with different concentrations of these molecules (Figure 6B). A decrease of the fluorescence signal close to the background level was observed for most of the G4 ligands at a concentration of 250 nM. This study revealed a clear inhibition of the SUD/G4 interaction by these G4-ligands, with IC50 values between 15 and 50 nM (Figure 6C). No inhibition of this interaction was observed with trimethyl psoralen (TMP), a DNA binder that only weakly interacts with G4 structures (Supplementary Figure S6C) (78,79).
Figure 6.
Inhibition of the SUD-NM / TRF2 G4 interaction by known G4-ligands. (A) Structure of the G4-ligands used in this study and belonging to three different chemical series (B) Competition HTRF assay: His-SUD-NM (10 nM) and biotinylated TRF2 G4 (50 nM) were incubated for 1 h at room temperature in the presence of increasing concentrations (from 25 to 250 nM) of seven G4-ligands. The curves for each chemical series are presented separately. (C) IC50 values calculated using the Kaleidagraph software to fit the experimental data are presented in (B).
Molecular modeling confirms the presence of a G4-binding site in the SARS-CoV-2 SUD-NM protein and suggests a different G4 binding mode from that of the SARS-CoV SUD-NM protein.
The modeled structure of SARS-CoV-2 SUD-NM (Figure 1B) suggests that a partially positively charged depression between the SUD-N and SUD-M macrodomains could be a putative binding site for G4s on a SUD-NM monomer (Figure 7A). Docking at this site of the major G4-DNA structure formed in the human BCL2 promoter region (PDB code: 2F8U, (63)) is presented in Figure 7B-C. Since SARS-CoV SUD-NM was crystallized as a dimer (8) and molecular dynamics studies suggested a G4-induced dimerization of the SARS-CoV SUD-NM domain (13), we also investigated docking of the BCL2 G4 to a putative SARS-CoV-2 SUD-NM dimer in Figure 7D–F. On a dimeric SUD-NM, the large and rather positively charged cleft at the dimer surface might constitute the main G4 binding site (Figure 7D–F). The BCL2 quadruplex (2F8U) could be docked at the center of this site which involves both SUD-M(A) and SUD-M(B) monomers (Figure 7E-F), and comprises Lys592 and Glu595 residues, homologous to the SARS-CoV Lys568 and Glu571 residues (highlighted in red in Figure 1A), that have previously been shown to be involved in SUD-G4 interactions and viral replication (5,8) (Supplementary Figure S7).
Figure 7.
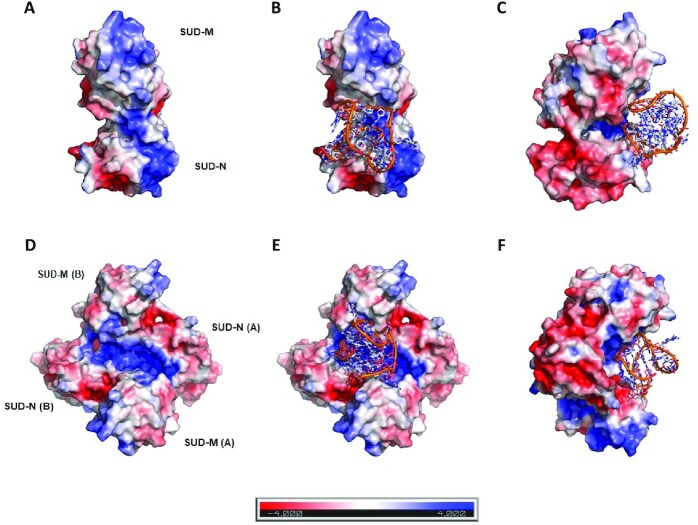
Docking of the human BCL2 WT promoter G4 (PDB code: 2F8U; (63)) on the modeled structure of SARS-CoV-2 SUD-NM. The G4 structure is figured in orange and in sticks representation, whereas the modeled structure of SARS-CoV-2 SUD-NM is presented as an electrostatic surface. (A) Electrostatic surface of the modeled SUD monomer showing positively charged regions (in blue) on the N and M macrodomains. (B) Docking of the 2F8U G4-DNA structure on the monomer of the modeled structure of SARS-CoV-2 SUD-NM. The putative binding site is a depression between the SUD-N and SUD-M macrodomains, which is partially positively charged. (C) Same as B) but turned 90° along the vertical axis. (D) Electrostatic surface of the modeled SUD dimer showing an extended positively charged region running on the dimer surface. (E) The best scoring pose of the 2F8U G4-DNA structure docked on the dimer of the modeled structure of SARS-CoV-2 SUD-NM. (F) Same as E) but turned 90° along the vertical axis. This docking studies reveal that the sugar-phosphate sides of the G-quadruplex are the only involved in the binding to the monomer of SUD-NM whereas it is the tetrad on the top of the G4 structure and the sugar-phosphate sides that are involved in the association of the G4-DNA to the dimeric SUD-NM.
We also performed computations with the human telomeric RNA (TERRA) quadruplex structure (PDB code: 3IBK, (65)) (Supplementary Figure S7). The G4-RNA and G4-DNA bind to the same site on the monomeric SUD-NM. Both G4s also bind to the same site on the dimeric form of SUD-NM. The dockings suggested that the sugar-phosphate sides of the G4-RNA structure could be mainly involved in the binding to the monomer of SUD-NM whereas the association of the G4-RNA to the dimeric SUD-NM could involve the nitrogenous bases and the sugar-phosphate sides of the top half of the G4 structure.
To test if the G4 binding surfaces of SARS-CoV and SARS-CoV-2 SUD domains are similar or not, we purified the SARS-CoV-2 SUD-NM Mut4 protein (K589A, K592A, E595A), homologous to the SARS-CoV SUD Mut4 protein which has been shown to lose its G4 binding capacity (8) (mutated residues highlighted in Figure 1A and Supplementary Figure S8A-B). We also purified the SARS-CoV-2 SUD-NM MutA protein (G489A, G490A, T491A, K497A), with designed mutations that should disrupt SUD-NM dimerization, according to our structural model (Supplementary Figure S8A-B). Using our established HTRF assay, we tested the effect of these mutations on the interaction between BCL2 DNA and TRF2 RNA G4s (Supplementary Figure S8C-D). First, mutations that should perturb the predicted SUD-NM dimerization interface did not affect the G4-binding capacity of this protein. In addition, although the Mut4 mutation largely affects G4-binding by SARS-CoV SUD protein (8), it only had a minor effect on the SARS-CoV-2 SUD/G4 interaction. Combined with the proposed structural models, these results suggest that the SARS-CoV-2 SUD dimerization is not essential for G4-binding and that SARS-CoV and SARS-CoV-2 SUD do not involve the same residues or conformations when they bind to G4 structures.
DISCUSSION
Host–virus interactions represent promising targets for antiviral strategies. This study focuses on the interaction between the SUD domain of the SARS-CoV-2 Nsp3 protein and DNA or RNA G-quadruplexes. This interaction has a high therapeutic potential, and its targeting could contribute to the identification of molecules inhibiting SARS-CoV-2 replication.
In this study, we experimentally demonstrate that the Nsp3 SUD domain of the SARS-CoV-2 virus is functional and is able to interact with G4 structures. We first show a good homology between the SUD macro and frataxin-like subdomains of SARS-CoV and SARS-CoV-2 (Figure 1A). This homology and the published structures of the SARS-CoV SUD N and M subdomains allow us to propose a 3D structural model of SARS-CoV-2 SUD NM (Figure 1B). This model, which has been refined by a molecular dynamics step, is one of the first where all the residues are positioned (see also (13)). Among them, the non-conserved amino acid residues can be displayed and compared in the SUD proteins of the SARS-CoV and the SARS-CoV-2. Using this model, we could perform docking simulations of two G4 structures (BCL2 DNA G4, PDB code: 2F8U and TERRA RNA G4, PDB code: 3BIK respectively) with both monomeric or dimeric forms of the SUD-NM protein (Figure 7 and Supplementary Figure S7). Interestingly, these two G4 structures bind to the same sites of the proteins, but these sites slightly differ on monomeric and dimeric conformations. On the monomer, our simulations indicate that residues from both N (D440, N442, G443, N444, K462, K463, P466) and M (Y605, K610, T611, S615 N618, T619 and D622) macrodomains are involved in G4 recognition (Figure 7B-C and Supplementary Figure S7). Fifty percent of these residues are identical to those in the same positions in the SARS-CoV sequence with a similarity score reaching 78%. The secondary structure elements (SSE) they belong to are the loop [440–444], the alpha-helix [457–464], the beta-strand [603–607], the loop [608–612] and the alpha-helix [613–623]. Differently, the SUD-NM dimer/G4 interaction essentially involves the M macrodomain (K592, Q594, E595, R603, Y605, K610, T611, S615, N618, T619 and D622) and requires fewer residues from the N macrodomain (D440, N442, G443, N444) (Figure 7E-F, Supplementary Figure S7 and S8A-B). Sixty percent of these residues are identical to those in the same positions in the SARS-CoV sequence and the similarity score reaches 80%. The SSEs they belong to are the loop [440–446], the loop [592–597], the beta-strand [603–607], the loop [608–612] and the alpha-helix [613–623]. Interestingly, the proposed putative binding sites, are different from those proposed for SARS-CoV in the literature (5,8), and reveal large cavities and a good complementarity in charge for the G4 both in the monomer and in the dimer, with the linker between the SUD-N and SUD-M subdomains contributing to the interactions. This last feature could not be observed using the crystallographic structure of SARS-CoV SUD-NM as the 3D-coordinates for the linker region are lacking.
We used three in vitro methods to study the SARS-CoV-2 SUD/G4 interaction: G4 pull-down, SPR and HTRF. The initial G4 pull-down assays have revealed three major observations. First, although the three amino-acids essential for G4 interaction are probably located in SUD-M (5,8), this subdomain alone is not sufficient to confer a selective interaction with G4s (Figure 3A), probably as a result of its high isoelectric point which makes SUD-M a polycation at neutral pH. Second, SUD-NM and NMC proteins interact with both DNA and RNA G4s and these interactions are specific for G4 structures, since mutations preventing G4 folding of the oligonucleotide substrates abolish these interactions. Finally, although qualitative, the G4 pull-down assays suggest a better affinity of SUD-NM for TRF2 RNA G4 in comparison to other RNA G4s (Figure 3C) and for DNA G4s (compare Figure 3B-C). SPR assays, performed with the different RNA G4 substrates have confirmed the preferential interaction of SARS-CoV-2 SUD-NM for the TRF2 RNA G4 structure (Figure 4). Finally, HTRF assays performed on WT and mutant BCL2 and TRF2 G4s confirm the specific interaction of SARS-CoV-2 SUD-NM protein for G4 structures and allow to quantify these interactions with Kd of 30 nM and 20 nM for BCL2 DNA and TRF2 RNA G4s, respectively. Competition HTRF assays performed with both substrates also confirm the higher affinity of SUD-NM for the TRF2 G4 partner (Figure 5E). We also used the HTRF assays to investigate the effect of specific mutations of SARS-CoV-2 SUD-NM on its binding to BCL2 DNA and TRF2 RNA G4s. Residues involved in SARS-CoV-2 SUD-NM dimerization (MutA) or homologous to SARS-CoV SUD residues interacting with G4 (Mut4) (5,8) were chosen to initiate this strategy (Supplementary Figure S8A-B). Interestingly, these mutations have very minor effect on SUD/G4 interaction (Supplementary Figure S8C-D), which suggests that the G4 binding sites and protein multimeric conformations associated with G4 binding of SARS-CoV and SARS-CoV-2 SUD are probably different. Further experimental structural investigations are needed to determine the molecular features required for SARS-CoV-2 SUD/G4 interaction. In particular, these studies should allow to precisely define the G4 binding site, the roles of each subdomain (N, M and C) and of their potential multimerization (13) in the G4 binding property (Figure 7, Supplementary Figure S7 and S8)
The conservation of the SUD domain between SARS-CoV and SARS-CoV-2 may play a role in the high pathogenicity of these two viruses. Given that the G4-binding property of this domain is involved in SARS-CoV replication (5), and that the SARS-CoV-2 SUD domain binds to G4s (Figures 3–5), we hypothesize that compounds that could impair this interaction may have an antiviral effect. Using the established HTRF assay reporting the interaction between SARS-CoV-2 SUD-NM and TRF2 RNA G4, we have screened different G4-ligands belonging to four chemical series and interestingly all of them were found to inhibit the SUD/G4 interaction, with IC50 between 15 and 50 nM (Figure 6B). This inhibition was not a given since some G4-ligands, such as PhenDC3, have been shown to actually favor the interaction between a quadruplex and a different G4-binding protein, Nucleolin (80). On the other hand, trimethyl psoralen (TMP), a DNA binder weakly binding G4 structures does not inhibit the SUD/G4 interaction (Supplementary Figure S6C). These results suggest that the G4-binding cleft in the SUD-NM domain does not tolerate the presence of this additional molecule bound to the G4 structure, while other proteins such as Nucleolin can easily accommodate such compound. When the actual RNA target(s) of SARS-CoV2 SUD protein would have been identified (ongoing work), our HTRF assay would be easily modified accordingly and used to screen larger chemical libraries for the identification of efficient inhibitors of the targeted interaction. Drug-like compounds selected by this screen could then be rapidly validated in a cellular assay to monitor their antiviral potency against SARS-CoV-2.
Recently, both SARS-CoV and SARS-CoV-2 SUD-N domains have been shown to interact with human Paip1, a component of the cellular translation machinery (15). Paip1 promotes translation, through its interaction with several translation factors such as PABP, eiF3, eiF-4A and eiF-4G proteins (81,82). In the case of SARS-CoV, the SUD-Paip1 interaction stimulates viral translation but no effect has been observed on translation of cellular mRNAs (15). However, this lack of global effect could hinder more selective ones. Because G4s are enriched in the 5′UTR of mRNAs (83) and regulate different steps of translation (84), we propose that the SUD/Paip1 interaction participates in a selective translational regulation of G4-containing mRNAs of the infected cells. SUD binding to these G4s would recruit Paip1 and interfere, positively or negatively, with the recruitment of the other translation factors. We investigated whether SUD could simultaneously interact with G4 and Paip1 using the solved structure of the SARS-CoV SUD-N/Paip1 M complex ((15), PDB code: 6YXJ) and our structural model of SARS-CoV-2 SUD-NM/G4-RNA complex (Supplementary Figure S7). Using these structures, we computed a model of the SUD-NM/Paip1M/G4-RNA complex (Supplementary Figure S9), which suggest that Paip1 can interact with a region of SUD-N distant from and opposed to that involved in the binding of G4 structures (in the monomer as well as in the dimer of SUD-NM). In this model, the structures of SUD-N in SUD-N-Paip1 and in SUD-NM superpose quite well. We thus hypothesize that the complex established between SARS-Cov2 SUD and human Paip1 should not prevent SUD binding to G4s present in 5′UTR of cellular mRNAs and could participate to their translational regulation. Further biochemical and functional studies are required to validate this hypothesis and should reveal new antiviral targets.
Knowing that SUD-NM can interact with DNA and RNA G4s, it is now of importance to determine the actual DNA or RNA target(s) under physiological conditions. Several PQS have been predicted in the SARS-CoV-2 genome but all of them correspond to presumably unstable G4s ((45,46) and Supplementary Table S1). The three most stable PQSs (RG1, RG2 and RG3 predicted at positions 13 385, 24 268 and 28 903 of the SARS-CoV-2 genome (45,46,48)) were selected to check their potential interaction with the SARS-CoV-2 SUD-NM protein using HTRF assays. These experiments clearly show that none of them interact with the protein (Figure 5F) and confirm our hypothesis that a counter-selection of stable PQS in the SARS-CoV-2 genome enables the preferential interaction of the SUD domain with host cell DNA or RNA targets from the infected cells, with minimal interference of its own viral RNA. Given the predominant cytoplasmic location of Nsp3, we propose that the host partners of SUD should rather be RNAs than genomic DNA. A number of RNA-binding proteins able to interact with G4s have previously been described, and some of them are involved in the replication of other viruses (28,32,33,85,86). Experiments are ongoing to identify the preferential partners of the SARS-CoV-2 SUD domain, whether these are ncRNAs or, more likely, mRNAs possibly coding for proteins involved in immune response, inflammation, or stress response. These SUD/RNA-G4 complexes could also be required for the process of viral replication. Their characterization is therefore crucial for a better understanding of these processes and for the identification of molecules preventing or destabilizing them. Our results, showing the capacity of some G4-ligands to inhibit the SARS-CoV-2 SUD/G4 interaction pave the way for a global screening of molecules able to inhibit this interaction and to test their antiviral properties.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Y. Jacob for sharing reagents (87). We also thank Y. Jacob, C. Demeret, S. van der Werf and P. Arimondo for fruitful discussions and Y. Luo for technical assistance. M.L. thanks S. Pochet for hosting his team in her laboratory during the Covid19 pandemic.
Notes
Present address: Olivier Helynck, Institut Pasteur, Plateforme de criblage Chémogénomique et Biologique, C2RT, Paris, France.
Present address: Hélène Munier-Lehmann, Institut Pasteur, Plateforme de criblage Chémogénomique et Biologique, C2RT, Paris, France.
Contributor Information
Marc Lavigne, Institut Pasteur, Département de Virologie. CNRS UMR 3569, Paris, France.
Olivier Helynck, Institut Pasteur, Unité de Chimie et Biocatalyse. CNRS UMR 3523, Paris, France.
Pascal Rigolet, Institut Curie, Université Paris-Saclay, CNRS UMR 9187, Inserm U1196, Orsay, France.
Rofia Boudria-Souilah, Institut Pasteur, Département de Virologie. CNRS UMR 3569, Paris, France.
Mireille Nowakowski, Institut Pasteur, Plateforme de Production et Purification de Protéines Recombinantes, C2RT, CNRS UMR 3528, Paris, France.
Bruno Baron, Institut Pasteur, Plateforme de Biophysique Moléculaire, C2RT, CNRS UMR 3528, Paris, France.
Sébastien Brülé, Institut Pasteur, Plateforme de Biophysique Moléculaire, C2RT, CNRS UMR 3528, Paris, France.
Sylviane Hoos, Institut Pasteur, Plateforme de Biophysique Moléculaire, C2RT, CNRS UMR 3528, Paris, France.
Bertrand Raynal, Institut Pasteur, Plateforme de Biophysique Moléculaire, C2RT, CNRS UMR 3528, Paris, France.
Lionel Guittat, Université Sorbonne Paris Nord, INSERM U978, Labex Inflamex, F-93017 Bobigny, France; Laboratoire d’optique et Biosciences, Ecole Polytechnique, Inserm U1182, CNRS UMR7645, Institut Polytechnique de Paris, Palaiseau, France.
Claire Beauvineau, Institut Curie, Université Paris-Saclay, CNRS UMR 9187, Inserm U1196, Orsay, France.
Stéphane Petres, Institut Pasteur, Plateforme de Production et Purification de Protéines Recombinantes, C2RT, CNRS UMR 3528, Paris, France.
Anton Granzhan, Institut Curie, Université Paris-Saclay, CNRS UMR 9187, Inserm U1196, Orsay, France.
Jean Guillon, Inserm U1212, CNRS UMR 5320, Laboratoire ARNA, UFR des Sciences Pharmaceutiques, Université de Bordeaux, Bordeaux, France.
Geneviève Pratviel, CNRS UPR 8241, Université Paul Sabatier, Laboratoire de Chimie de Coordination, Toulouse, France.
Marie-Paule Teulade-Fichou, Institut Curie, Université Paris-Saclay, CNRS UMR 9187, Inserm U1196, Orsay, France.
Patrick England, Institut Pasteur, Plateforme de Biophysique Moléculaire, C2RT, CNRS UMR 3528, Paris, France.
Jean-Louis Mergny, Laboratoire d’optique et Biosciences, Ecole Polytechnique, Inserm U1182, CNRS UMR7645, Institut Polytechnique de Paris, Palaiseau, France.
Hélène Munier-Lehmann, Institut Pasteur, Unité de Chimie et Biocatalyse. CNRS UMR 3523, Paris, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Covid Task force of the Institut Pasteur (Paris, France); ANR RA-COVID-19; internal resources of the involved laboratories originating from the Institut Pasteur, Institut Curie, and the French national research institutions (CNRS, Inserm); Université de Bordeaux; Université de Toulouse; Université Paris-Saclay; Université Sorbonne Paris-Nord; Institut Polytechnique de Paris. Funding for open access charge: ANR, RA-COVID-19.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fung T.S., Liu D.X.. Similarities and dissimilarities of COVID-19 and other coronavirus diseases. Annu. Rev. Microbiol. 2021; 10.1146/annurev-micro-110520-023212. [DOI] [PubMed] [Google Scholar]
- 2. Lei J., Kusov Y., Hilgenfeld R.. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018; 149:58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E.. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003; 331:991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W.et al.. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003; 84:2305–2315. [DOI] [PubMed] [Google Scholar]
- 5. Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R.. A G-quadruplex-binding macrodomain within the ‘SARS-unique domain’ is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015; 484:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterjee A., Johnson M.A., Serrano P., Pedrini B., Joseph J.S., Neuman B.W., Saikatendu K., Buchmeier M.J., Kuhn P., Wuthrich K.. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J. Virol. 2009; 83:1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson M.A., Chatterjee A., Neuman B.W., Wuthrich K.. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J. Mol. Biol. 2010; 400:724–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R.. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009; 5:e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y., Savinov S.N., Mielech A.M., Cao T., Baker S.C., Mesecar A.D.. X-ray structural and functional studies of the three tandemly linked domains of non-structural protein 3 (nsp3) from murine hepatitis virus reveal conserved functions. J. Biol. Chem. 2015; 290:25293–25306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 2016; 135:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan J., Kusov Y., Mutschall D., Tech S., Nagarajan K., Hilgenfeld R., Schmidt C.L.. The ‘SARS-unique domain’ (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem. Biophys. Res. Commun. 2007; 364:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R. 3rd, Wuthrich K.et al.. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008; 82:5279–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hognon C., Miclot T., Iriepa C.G., Francés-Monerris A., Grandemange S., A. T., Marazzi M., Barone G., Monari A.. Role of RNA guanine quadruplexes in favoring the dimerization of SARS unique domain in coronaviruses. J. Phys. Chem. Lett. 2020; 11:5661–5667. [DOI] [PubMed] [Google Scholar]
- 14. Ma-Lauer Y., Carbajo-Lozoya J., Hein M.Y., Muller M.A., Deng W., Lei J., Meyer B., Kusov Y., von Brunn B., Bairad D.R.et al.. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E5192–E5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lei J., Ma-Lauer Y., Han Y., Thoms M., Buschauer R., Jores J., Thiel V., Beckmann R., Deng W., Leonhardt H.et al.. The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. EMBO J. 2021; 40:e102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang Y.S., Ko B.H., Ju J.C., Chang H.H., Huang S.H., Lin C.W.. SARS unique domain (SUD) of severe acute respiratory syndrome coronavirus induces NLRP3 inflammasome-dependent CXCL10-mediated pulmonary inflammation. Int. J. Mol. Sci. 2020; 21:3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwok C.K., Merrick C.J.. G-Quadruplexes: prediction, characterization, and biological application. Trends Biotechnol. 2017; 35:997–1013. [DOI] [PubMed] [Google Scholar]
- 18. Malgowska M., Czajczynska K., Gudanis D., Tworak A., Gdaniec Z.. Overview of the RNA G-quadruplex structures. Acta Biochim. Pol. 2016; 63:609–621. [DOI] [PubMed] [Google Scholar]
- 19. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M.et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 21. Hansel-Hertsch R., Spiegel J., Marsico G., Tannahill D., Balasubramanian S.. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018; 13:551–564. [DOI] [PubMed] [Google Scholar]
- 22. Al-Zeer M.A., Kurreck J.. Deciphering the enigmatic biological functions of RNA guanine-quadruplex motifs in human cells. Biochemistry. 2019; 58:305–311. [DOI] [PubMed] [Google Scholar]
- 23. Dumas L., Herviou P., Dassi E., Cammas A., Millevoi S.. G-Quadruplexes in RNA biology: recent advances and future directions. Trends Biochem. Sci. 2021; 46:270–283. [DOI] [PubMed] [Google Scholar]
- 24. Metifiot M., Amrane S., Litvak S., Andreola M.L.. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014; 42:12352–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruggiero E., Richter S.N.. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018; 46:3270–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seifert H.S. Above and beyond watson and crick: guanine quadruplex structures and microbes. Annu. Rev. Microbiol. 2018; 72:49–69. [DOI] [PubMed] [Google Scholar]
- 27. Abiri A., Lavigne M., Rezaei M., Nikzad S., Zare P., Mergny J.L., Rahimi H.R.. Unlocking G-quadruplexes as antiviral targets. Pharmacol. Rev. 2021; 73:897–923. [DOI] [PubMed] [Google Scholar]
- 28. Butovskaya E., Solda P., Scalabrin M., Nadai M., Richter S.N.. HIV-1 nucleocapsid protein unfolds stable RNA G-quadruplexes in the viral genome and is inhibited by G-quadruplex ligands. ACS Infect Dis. 2019; 5:2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perrone R., Doria F., Butovskaya E., Frasson I., Botti S., Scalabrin M., Lago S., Grande V., Nadai M., Freccero M.et al.. Synthesis, binding and antiviral properties of potent core-extended naphthalene diimides targeting the HIV-1 long terminal repeat promoter G-Quadruplexes. J. Med. Chem. 2015; 58:9639–9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scalabrin M., Frasson I., Ruggiero E., Perrone R., Tosoni E., Lago S., Tassinari M., Palu G., Richter S.N.. The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 2017; 7:45244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tosoni E., Frasson I., Scalabrin M., Perrone R., Butovskaya E., Nadai M., Palu G., Fabris D., Richter S.N.. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015; 43:8884–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dabral P., Babu J., Zareie A., Verma S.C.. LANA and hnRNP A1 regulate the translation of LANA mRNA through G-quadruplexes. J. Virol. 2020; 94:e01508-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Norseen J., Johnson F.B., Lieberman P.M.. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009; 83:10336–10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016; 59:5987–6011. [DOI] [PubMed] [Google Scholar]
- 35. Sun Z.Y., Wang X.N., Cheng S.Q., Su X.X., Ou T.M.. Developing novel G-quadruplex ligands: from interaction with nucleic acids to interfering with nucleic acid(-)protein interaction. Molecules. 2019; 24: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Asamitsu S., Obata S., Yu Z., Bando T., Sugiyama H.. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules. 2019; 24:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bedrat A., Lacroix L., Mergny J.L.. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic. Acids. Res. 2016; 44:1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dhapola P., Chowdhury S.. QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 2016; 44:W277–W283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brazda V., Kolomaznik J., Lysek J., Bartas M., Fojta M., Stastny J., Mergny J.L.. G4Hunter web application: a web server for G-quadruplex prediction. Bioinformatics. 2019; 35:3493–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Puig Lombardi E., Londono-Vallejo A.. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020; 48:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bartas M., Brazda V., Bohalova N., Cantara A., Volna A., Stachurova T., Malachova K., Jagelska E.B., Porubiakova O., Cerven J.et al.. In-depth bioinformatic analyses of nidovirales including human SARS-CoV-2, SARS-CoV, MERS-CoV viruses suggest important roles of non-canonical nucleic acid structures in their lifecycles. Front Microbiol. 2020; 11:1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huppert J.L. Hunting G-quadruplexes. Biochimie. 2008; 90:1140–1148. [DOI] [PubMed] [Google Scholar]
- 43. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kikin O., D’Antonio L., Bagga P.S.. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ji D., Juhas M., Tsang C.M., Kwok C.K., Li Y., Zhang Y.. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief. Bioinform. 2021; 22:1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Panera N., Tozzi A.E., Alisi A.. The G-quadruplex/helicase world as a potential antiviral approach against COVID-19. Drugs. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cui H., Zhang L.. G-Quadruplexes are present in human coronaviruses including SARS-CoV-2. Front. Microbiol. 2020; 11:567317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao C., Qin G., Niu J., Wang Z., Wang C., Ren J., Qu X.. Targeting RNA G-quadruplex in SARS-CoV-2: a promising therapeutic target for COVID-19?. Angew. Chem. Int. Ed. Engl. 2020; 59:2–9. [DOI] [PubMed] [Google Scholar]
- 49. De Cian A., Delemos E., Mergny J.L., Teulade-Fichou M.P., Monchaud D.. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007; 129:1856–1857. [DOI] [PubMed] [Google Scholar]
- 50. Reznichenko O., Quillevere A., Prado, Martins R., Loaec N., Kang H., Lista M.J., Beauvineau C., Gonzalez-Garcia J., Guillot R., Voisset C.et al.. Novel cationic bis(acylhydrazones) as modulators of Epstein-Barr virus immune evasion acting through disruption of interaction between nucleolin and G-quadruplexes of EBNA1 mRNA. Eur. J. Med. Chem. 2019; 178:13–29. [DOI] [PubMed] [Google Scholar]
- 51. Pennarum G., Granotier C., Gautier L.R., Gomez D., Hoffschir F., Mandine E., Riou J.F., Mergny J.L., Mailliet P., Boussin F.D.. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005; 24:2917–2928. [DOI] [PubMed] [Google Scholar]
- 52. Guillon J., Cohen A., Das R.N., Boudot C., Gueddouda N.M., Moreau S., Ronga L., Savrimoutou S., Basmaciyan L., Tisnerat C.et al.. Design, synthesis, and antiprotozoal evaluation of new 2,9-bis[(substituted-aminomethyl)phenyl]-1,10-phenanthroline derivatives. Chem. Biol. Drug Des. 2018; 91:974–995. [DOI] [PubMed] [Google Scholar]
- 53. Sabater L., Fang P.J., Chang C.F., De Rache A., Prado E., Dejeu J., Garofalo A., Lin J.H., Mergny J.L., Defrancq E.et al.. Cobalt(III)porphyrin to target G-quadruplex DNA. 2015; 44:3701–3707. [DOI] [PubMed] [Google Scholar]
- 54. Romera C., Bombarde O., Bonnet R., Gomez D., Dumy P., Calsou P., Gwan J.F., Lin J.H., Defrancq E., Pratviel G.. Improvement of porphyrins for G-quadruplex DNA targeting. Biochimie. 2011; 93:1310–1317. [DOI] [PubMed] [Google Scholar]
- 55. Pipier A., De Rache A., Modeste C., Amrane S., Mothes-Martin E., Stigliani J.L., Calsou P., Mergny J.L., Pratviel G., Gomez D.. G-Quadruplex binding optimization by gold(iii) insertion into the center of a porphyrin. Dalton Trans. 2019; 48:6091–6099. [DOI] [PubMed] [Google Scholar]
- 56. Munier H., Gilles A.M., Glaser P., Krin E., Danchin A., Sarfati R., Barzu O.. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 1991; 196:469–474. [DOI] [PubMed] [Google Scholar]
- 57. Raynal B., Lenormand P., Baron B., Hoos S., England P.. Quality assessment and optimization of purified protein samples: why and how?. Microb. Cell Fact. 2014; 13:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ortega A., Amoros D., Garcia de la Torre J.. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011; 101:892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brazda V., Luo Y., Bartas M., Kaura P., Porubiakova O., Stastny J., Pecinka P., Verga D., Da Cunha V., Takahashi T.S.et al.. G-Quadruplexes in the archaea domain. Biomolecules. 2020; 10:1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mergny J.L., Li J., Lacroix L., Amrane S., Chaires J.B.. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005; 33:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Webb B., Sali A.. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics. 2016; 54:5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krieger E., Joo K., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K.. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009; 77(Suppl.9):114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dai J., Chen D., Jones R.A., Hurley L.H., Yang D.. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006; 34:5133–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Trott O., Olson A.J.. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010; 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Collie G.W., Haider S.M., Neidle S., Parkinson G.N.. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010; 38:5569–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. DeLano W.L. The PyMOL Molecular Graphics System. 2002; Version 2.4.0, Schrödinger, LLC. [Google Scholar]
- 67. Harding S.E. The combination of the viscosity increment with the harmonic mean rotational relaxation time for determining the conformation of biological macromolecules in solution. Biochem. J. 1980; 189:359–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chenal A., Guijarro J.I., Raynal B., Delepierre M., Ladant D.. RTX calcium binding motifs are intrinsically disordered in the absence of calcium: implication for protein secretion. J. Biol. Chem. 2009; 284:1781–1789. [DOI] [PubMed] [Google Scholar]
- 69. Perrone R., Nadai M., Frasson I., Poe J.A., Butovskaya E., Smithgall T.E., Palumbo M., Palu G., Richter S.N.. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013; 56:6521–6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R.et al.. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lacroix L., Seosse A., Mergny J.L.. Fluorescence-based duplex-quadruplex competition test to screen for telomerase RNA quadruplex ligands. Nucleic Acids Res. 2011; 39:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Marcel V., Tran P.L., Sagne C., Martel-Planche G., Vaslin L., Teulade-Fichou M.P., Hall J., Mergny J.L., Hainaut P., Van Dyck E.. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011; 32:271–278. [DOI] [PubMed] [Google Scholar]
- 74. Gomez D., Guédin A., Mergny J.L., Salles B., Riou J.F., Teulade-Fichou M.P., Calsou P.. A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010; 38:7187–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen H., Sun H., Chai Y., Zhang S., Guan A., Li Q., Yao L., Tang Y.. Insulin-like growth factor type I selectively binds to G-quadruplex structures. Biochim. Biophys. Acta Gen. Subj. 2019; 1863:31–38. [DOI] [PubMed] [Google Scholar]
- 76. Calabrese D.R., Chen X., Leon E.C., Gaikwad S.M., Phyo Z., Hewitt W.M., Alden S., Hilimire T.A., He F., Michalowski A.M.et al.. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun. 2018; 9:4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. von Hacht A., Seifert O., Menger M., Schutze T., Arora A., Konthur Z., Neubauer P., Wagner A., Weise C., Kurreck J.. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res. 2014; 42:6630–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alcaro S., Musetti C., Distinto S., Casatti M., Zagotto G., Artese A., Parrotta L., Moraca F., Costa G., Ortuso F.et al.. Identification and characterization of new DNA G-quadruplex binders selected by a combination of ligand and structure-based virtual screening approaches. J. Med. Chem. 2013; 56:843–855. [DOI] [PubMed] [Google Scholar]
- 79. Paul S., Samanta A.. Ground- and excited-state interactions of a psoralen derivative with human telomeric G-quadruplex DNA. J. Phys. Chem. B. 2018; 122:2277–2286. [DOI] [PubMed] [Google Scholar]
- 80. Figueiredo J., Miranda A., Lopes-Nunes J., Carvalho J., Alexandre D., Valente S., Mergny J.L., Cruz C.. Targeting nucleolin by RNA G-quadruplex-forming motif. Biochem. Pharmacol. 2021; 189:114418. [DOI] [PubMed] [Google Scholar]
- 81. Martineau Y., Derry M.C., Wang X., Yanagiya A., Berlanga J.J., Shyu A.B., Imataka H., Gehring K., Sonenberg N.. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol. Cell. Biol. 2008; 28:6658–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Derry M.C., Yanagiya A., Martineau Y., Sonenberg N.. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb. Symp. Quant. Biol. 2006; 71:537–543. [DOI] [PubMed] [Google Scholar]
- 83. Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S.. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016; 13:841–844. [DOI] [PubMed] [Google Scholar]
- 84. Kamura T., Katsuda Y., Kitamura Y., Ihara T.. G-quadruplexes in mRNA: a key structure for biological function. Biochem. Biophys. Res. Commun. 2020; 526:261–266. [DOI] [PubMed] [Google Scholar]
- 85. Bian W.X., Xie Y., Wang X.N., Xu G.H., Fu B.S., Li S., Long G., Zhou X., Zhang X.L.. Binding of cellular nucleolin with the viral core RNA G-quadruplex structure suppresses HCV replication. Nucleic Acids Res. 2019; 47:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lista M.J., Martins R.P., Billant O., Contesse M.A., Findakly S., Pochard P., Daskalogianni C., Beauvineau C., Guetta C., Jamin C.et al.. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017; 8:16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim D.K., Knapp J.J., Kuang D., Chawla A., Cassonnet P., Lee H., Sheykhkarimii D., Samavarchi-Tehrani P, Abdouni H., Rayhan A.et al.. A Comprehensive, Flexible Collection of SARS-CoV-2 Coding Regions. G3 (Bethesda). 2020; 10:3399–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.