Abstract
N6-methyladenosine (m6A) is a common modification on endogenous RNA transcripts in mammalian cells. Technologies to precisely modify the RNA m6A levels at specific transcriptomic loci empower interrogation of biological functions of epitranscriptomic modifications. Here, we developed a bidirectional dCasRx epitranscriptome editing platform composed of a nuclear-localized dCasRx conjugated with either a methyltransferase, METTL3, or a demethylase, ALKBH5, to manipulate methylation events at targeted m6A sites. Leveraging this platform, we specifically and efficiently edited m6A modifications at targeted sites, reflected in gene expression and cell proliferation. We employed the dCasRx epitranscriptomic editor system to elucidate the molecular function of m6A-binding proteins YTHDF paralogs (YTHDF1, YTHDF2 and YTHDF3), revealing that YTHDFs promote m6A-mediated mRNA degradation. Collectively, our dCasRx epitranscriptome perturbation platform permits site-specific m6A editing for delineating of functional roles of individual m6A modifications in the mammalian epitranscriptome.
INTRODUCTION
RNA modifications, collectively called epitranscriptomics, offers a complex and dynamic level of regulation that is reflected in alterations of RNA levels and translation that ultimately determines cell fate and function. Highly dynamic covalent modifications, such as N6-methyladenosine (m6A), pseudouridine (Ψ), 5-methylcytosine (m5C) and N1-methyladenosine (m1A) (1), play critical roles in eukaryotic RNA epitranscriptomic pathways and have been implicated in diseases, especially cancer (2). m6A is a frequent base modification in the mammalian transcriptome and is enriched near the stop codon and the untranslated regions (UTRs) of mRNAs (3). m6A signaling modulates RNA secondary structures (4), splicing (5), nuclear localization (6), stability (7,8) and translation efficiency (9). m6A regulates DNA damage responses (10), X-chromosome gene silencing (11), cellular heat shock responses (12), hepatic lipid metabolism (13), long-term memory creation (14), spermatogonia differentiation (15), maternal-to-zygotic transition (16), embryonic stem-cell self-renewal and differentiation (17) tumorigenesis (18) and anti-tumor immune responses (19).
Like chromatin modifications in epigenetics, the summation of the regulation of epitranscriptomics is reflected in the activities of writers, erasers and reader. The core subunits of the m6A installation (‘writer’) complex in eukaryotic cells, contain methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14) (20) and Wilms tumor 1‐associated protein (WTAP) (21). Within this complex, METTL3 catalyzes a methyl group transfer from S-adenosyl methionine (SAM) to adenine within a single-stranded RNA (ssRNA) sequence motif DRACH (D = A, G, or U; R = A or G; H = A, C, or U), whereas METTL14 provides RNA binding sites as scaffolds (20). AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated protein are m6A ‘erasers’ that demethylate m6A from the adenine at the same DRACH motifs (22,23). m6A modifications serve as signals that are decoded by ‘reader’ proteins, such as YT521-B homology domain-containing proteins to impact splicing, stability, translation and localization of mRNAs (24).
The pleiotropic effects of m6A are mediated by m6A readers in both the nucleus and cytoplasm. Investigations into the impact of m6A on various biological processes and phenotypes have relied on the global manipulation of m6A writers, erasers and readers, which results in bulk, non-specific changes in the methylation state of many sites. As a result, causal relationships between specific m6A modifications and downstream phenotypic changes remain obscure. Writers and erasers that are supposed to have opposite roles in regulating cellular behaviors were both found to maintain survival of glioblastoma (GBM) cells (25–27). To elucidate the functional roles of individual m6A modifications in living cells, novel m6A editors that modify specific sites of individual transcripts without altering the global RNA methylation pattern are required.
The development of CRISPR-associated nuclease Cas13 technologies has enabled precise mRNA editing and interrogation of RNA modifications in biological processes (28). The Cas13 family bind and cleave ssRNA targeted by a complementary guide RNA (29). To date, Cas13 effector proteins, including Cas13a (1250 aa), Cas13b (1150 aa), Cas13c (1120 aa) and Cas13d (930 aa) have been reported to show high RNA knockdown efficacy with minimal off-target activity. RfxCas13d (CasRx) is the smallest known enzyme with most substantial targeted knockdown efficiency in the Cas13 family when fused to a nuclear localization sequence (NLS) (30–32). While a CRISPR-Cas9-based editor has been used to manipulate m6A modifications on transcriptomes (33), a CRISPR-Cas13-based platform is better positioned for m6A editing because it does not require an additional synthetic PAMmer oligonucleotide. Based on previous strategies that fused m6A writers (34) or erasers (35,36) with Cas13b, we hypothesized that tethering catalytically inactive CasRx (dCasRx) to m6A writers or erasers could manipulate specific m6A sites targeted by relevant Cas13 guide RNAs. The small sizes of dCasRx epitranscriptome editors allow them to be packaged into lentivirus to study cells that are difficult to transfect using other strategies.
Here, we developed and optimized precise m6A editors through conjugation of dCasRx with METTL3 or ALKBH5, enabling efficient manipulation of individual m6A sites within endo-transcripts and minimal off-target alterations in both normal mammalian cells and cancer cells. While early studies reported that m6A readers YTHDF1, YTHDF2 and YTHDF3 have been reported to have distinct roles in mediating RNA degradation or translation, a recent study demonstrated that YTHDF1, YTHDF2 and YTHDF3 function together to mediate degradation of m6A-containing mRNAs, thus contradicting the prevailing understanding (37). Our dCasRx mRNA editing technology enabled us to further interrogate the functional roles of YTHDF paralogs in cells through controlled manipulation of m6A levels at the YTHDF paralogs-associated m6A sites and measurement of the alteration of mRNA abundance. Our results indicates that high level of methylation levels at YTHDF paralogs-associated m6A sites enhanced degradation of select endo-transcripts, and that all 3 YTHDFs promote m6A-dependent mRNA degradation.
MATERIALS AND METHODS
Design of dCasRx epitranscriptomic editors
dCasRx epitranscriptomic editors were constructed by fusing candidate methyltransferase METTL3 or demethylase ALKBH5 to the C terminus of SV40 NLS-dCasRx (Addgene plasmid, #118634) via a nucleoplasmin NLS (K-R-P-A-A-T-K-K-A-G-Q-A-K-K-K-K)-G-S-S linker. The dCasRx-dMETTL3 conjugate contained a single mutation at D395A to form the dCasRx-dMETTL3. The dCasRx-dALKBH5 conjugate contained a single mutation at H204A to form the dCasRx-dALKBH5. A Flag (D-Y-K-D-D-D-D-K)-G-G-G-G-G-HA (Y-P-Y-D-V-P-D-Y-A) signal was added at the C terminus of all epitranscriptomic editors as a tag for detection.
Design of sgRNAs
Information of distributions of YTHDF paralogs was based on a database GSE78030 (11). We selected adenine sites that have been reported accessible to methylation modifications by m6A in a database GSE63753 (38) as target sites for designing sgRNAs. Designed sgRNAs were checked using the NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to avoid unwanted mRNA off-target bindings in the human genome. Sequence of sgRNAs for dCasRx was provided in Supplementary Table S1, and for dCas13b was provided in Supplementary Table S2.
Cell culture
HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (Gibco, #C11995500CP) supplemented with 10% fetal bovine serum (Gibco, #10099-141C), 1% penicillin/streptomycin (Hyclone, #SV30010) and 1% GlutaMax™ Supplement (Gibco, #35050-061) at 37°C, 5% CO2. GSC 3565 and GSC 468 cells were cultured in Neurobasal™-A Medium (Gibco, #12349-015) supplemented with 2% B-27™ supplement (Gibco, #12587-010), 20 ng/ml recombinant human EGF protein (R&D, #236-EG), 20 ng/ml recombinant human FGF basic protein (R&D, #4114-TC),1% penicillin/streptomycin (P/S, Invitrogen, SV30010), 1% sodium pyruvate (Gibco, #11360-070) and 1% GlutaMax™ supplement (Gibco, #35050-061) at 37°C, 5% CO2.
Plasmid transfection
Plasmid transfection was carried out using LipoD293™ In Vitro DNA Transfection Reagent (SignaGen Laboratories, SL100668) following manufacture's protocol. For six-well assays, HEK293T cells were co-transfected with 1.5 μg dCasRx conjugate plasmid and 1.5 μg sgRNA plasmid per well. Transfected cells were cultured under normal conditions (37°C, 5% CO2) for 36 h before analysis. dCas13b-M3 and dCas13b-M3M14 were gifts from David Liu (Addgene plasmid, #155366 and #155367).
Lentivirus packaging
In 100 mm dishes, HEK293T cells were transfected using 6 μg of the proposed plasmid, 4 μg of psPAX2 (Addgene plasmid, # 12260) and 2 μg of pMD2.G plasmid (Addgene plasmid, #12259) per well with LipoD293™ In Vitro DNA Transfection Reagent. The transfected cells were incubated under normal conditions for 48 h and then harvested. The supernatant of HEK293T cell culture was collected after centrifugation at room temperature (RT). Concentration of virus was quantified using Lentivirus Concentration Solution (Genomeditech, #GM-040801-100) following manufacture's protocol. The lentivirus was stored at −80°C before use.
Constructing GSCs expressing dCasRx epitranscriptomic editors and sgRNAs
To generate dCasRx epitranscriptomic editors edited GSCs, GSCs were first digested as single cells followed by incubation with dCasRx epitranscriptomic editors lentiviruses for 12 h in culture medium. GSCs expressing dCasRx epitranscriptomic editors were selected in medium containing 1 μg/ml Puromycin (Beyotime, #ST551) for 5 days, owing to the Puromycin-resistance of dCasRx conjugate plasmids. Then, GSCs expressing dCasRx epitranscriptomic editors were infected with sgRNAs lentiviruses with Blasticidin (Beyotime, #ST018) resistance. GSCs were selected with Blasticidin S HCl supplemented medium for 3 days to select GSC cells expressing both dCasRx and sgRNAs. The engineered GSCs were used for following experiments.
Protein extraction and western blotting
Cells were washed twice with phosphate-buffered saline (PBS) and lysed with RIPA Lysis Buffer (Beyotime, #P0013C) supplemented with phenylmethanesulfonyl fluoride (Sigma-Aldrich, #p7626) and cOmplete™ (Roche, #4693132001) on ice. Supernatant of cell lysates was collected after centrifugation and denatured at 100°C with loading buffer. Samples were loaded onto an 8% 15-well sodium dodecyl sulphate-polyacrylamide gelelectrophoresis gel, and the gel was transferred to a 0.45-μm polyvinylidene difluoride membrane (Merckmillipore, #IPVH00010) after electrophoresis. Membranes were blocked with 5% non-fat powdered milk (Sangon Biotech, #A600669-0250) and incubated overnight at 4° cold room with anti-HA (C29F4) rabbit mAb (Cell Signaling Technology, #3724), GAPDH (Proteintech, #60004-1-Ig), YTHDF1 (Proteintech, #17479-1-AP), YTHDF2 (Proteintech, #24744-1-AP) and YTHDF3 (Proteintech, #25537-1-AP) antibodies in TBST (TBS + 0.5% Tween-20) with 1% bovine serum albumin (BSA, Sangon Biotech, # A600332-0100). After washing three times with TBST, membranes were incubated with secondary antibodies (Cell Signaling Technology, #7074 and #7076) for 1 h at RT. The membrane was washed and incubated with an enhanced chemiluminescence ECL (Thermo scientific, #34580) for 2 min then imaged by ChemiDoc XRS+ System (BIO RAD, #1708265).
Immunofluorescence microscopy
An HA epitope tag (Y-P-Y-D-V-P-D-Y-A) was cloned onto the C terminus of dCasRx epitranscriptomic editors for detecting the location of dCasRx epitranscriptomic editors. HEK293T and GSC cells expressing dCasRx epitranscriptomic editors were seeded and cultured on cover slips (Solarbio, # YA0350) in 24-well plates. After 36 h of incubation, the culture medium was discarded, and the cover slips were washed once with PBS gently. Cells were fixed with 4% paraformaldehyde (Servicebio, #G1101) for 10 min. Cells were then washed three times with PBS and permeabilized with PBS + 0.2% Triton-X100 (PBST) for 15 min at RT. Cells were blocked in blocking buffer (10% BSA in PBST) for 30 min and stained with HA antibody (CST, #3724) in blocking buffer overnight at 4°C. Cells were then washed three times with PBST and stained with donkey anti-rabbit IgG (H+L) Alexa Fluor Plus 488 (Thermo Fisher Scientific, #A32790) in blocking buffer for 1 h at RT followed by 1 h of DAPI (Roche, # 33495822) staining. Images were acquired using a confocal laser scanning microscope (Olympus, FV3000-IX83).
RNA isolation and RT-qPCR
For RNA extraction, fresh cells were first washed twice with PBS, and then total RNA was extracted using TRIzol™ Reagent (Thermo Fisher Scientific, #15596018) following manufacturer's protocol. For each sample, 1 μg of total RNA was used for reverse transcription to cDNA using Novoscript Plus All in one First Strand cDNA Synthesis SuperMix (Novoprotein, #E047-01S). The cDNA templates were used in reverse transcriptase-quantitative polymerasechain reaction (RT-qPCR) quantification. Each 20 μl qPCR reaction contained 1 μl of cDNA, 1 μM forward and reverse primers and 10 μl of 2× SYBR Green Master Mix (Novoprotein, #E096-01S). Reaction mixture was heated at 95°C for 1 min followed by 40 repeated cycles with the following conditions: 95°C for 20 s and 60°C for 60 s. All assays were repeated with three independent experiments. The primers used in RT-qPCR assays were listed in the Supplementary Table S3.
SELECT technology for detection of m6A
Detection of m6A at targeted sites was based on the SELECT technology modified from a previous protocol (39). For each sample, 1 μg of total RNA was incubated with 40 nM Up primer, 40 nM Down primer and 5 μM dNTP (New England Biolabs, #N0446S) in 17.5 μl 1× CutSmart buffer (New England Biolabs, #B7204S). A progressive annealing cycle was carried out: 1 min each at 90°C, 80°C, 70°C, 60°C, 50°C and 40°C for 6 min. Subsequently, 2.5 μl of enzyme mixture containing 0.01 U Bst 2.0 DNA polymerase (New England Biolabs, #M0537S), 0.5 U SplintR ligase (New England Biolabs, #M0375S) and 10 nmol ATP (New England Biolabs, # P0756S) were added to the 17.5 μl annealing products. The final 20 μl reaction mixtures were incubated at 40°C for 20 min, denatured at 80°C for 20 min and then kept at 4°C. Afterward, 2 μl of final products were transferred to a reaction mixture containing 200 nM SELECT common primers and 2× SYBR Green Master Mix for qPCR analysis. The run cycle was set up as: 95°C for 1 min followed by 40 cycles of (95°C, 20 s; 60°C, 60 s). The SELECT products of targeted sites were normalized to the RNA abundance of corresponding transcripts containing m6A sites. All assays were performed with three independent experiments. Primers used in the SELECT assays were listed in Supplementary Table S4.
Low input m6A-RIP coupled with qPCR
The strategy of low input m6A-RIP was modified from Zeng's protocol (40). Total RNA from cells was extracted using Trizol reagent, and DNA was removed by adding Dnase I (New England Biolabs, #M0303S). A total volume of 5 μg total RNA was resuspended to 18 μl with RNase-free water. A total of 2 μl of 10× RNA Fragmentation Buffer (100 mM Tris–HCl, 100 mM ZnCl2 in nuclease-free H2O) was added to the RNA and incubated in a preheated thermal cycler for 5 min at 95°C for fragmentation. The total RNA was chemically fragmented into ∼200-nt-long fragments. To prepare antibody-bead for me-RNA immunoprecipitation, 15 μl of protein-A magnetic beads (Thermo Fisher Scientific, #10002D) and 15 μl of protein-G magnetic beads (Thermo Fisher Scientific, #10004D) were incubated with 5 μg anti-m6A antibody (Cell Signaling Technology, #56593) at 4°C overnight. The prepared antibody-bead mixture was resuspended in 500 μl of IP reaction mixture containing fragmented total RNA and incubated for 2 h at 4°C. The RNA reaction mixture was then washed twice in 1000 μl of IP buffer, followed by elution buffer with continuous shaking for 1 h at 4°C. Additional phenol–chloroform isolation and ethanol precipitation treatments were performed to purify the RNA. We also used NovoProtein Novoscript Plus All in one First Strand cDNA Synthesis SuperMix followed by qPCR for quantification. All assays were performed with three independent experiments. Primers used in the low-input m6A-RIP assay were listed in Supplementary Table S5.
YTHDF-paralogs-combined RNA immunoprecipitation coupled with qPCR
The strategy of YTHDF-paralogs-combined RNA immunoprecipitation was modified from Keene's protocol (41). Collect more than 5 million HEK293T cells per RIP sample, and it was lysed with polysome lysis buffer (described in Keene's protocol) supplemented with RNase inhibitors and protease inhibitors. Lysate was incubated with protein A/G beads coating with antibodies (anti-YTHDF1, Proteintech, #17479-1-AP; anti-YTHDF2, Proteintech, #24744-1-AP; anti-YTHDF3, Proteintech, #25537-1-AP) 4 h. After washed with 1 ml of ice-cold NT2 buffer (described in Keene's article) five times, the RNP components can be released from bead by incubating with Proteinase K (BBI, #B600169-0002) for 30 min at 55°C. Release the RNP components and isolate the RNA from the immunoprecipitated pellet by adding Trizol reagent. All RNA from RIP or input samples was performed RT-qPCR assay as the previous description. All assays were performed with three independent experiments. Primers used in the YTHDF-paralogs-RIP assay were listed in Supplementary Table S6.
Subcellular fractionation assay
The strategy for subcellular fractionation assay was followed by Zhang's protocol (26). Briefly, HEK293T cells were collected by centrifugation and washed with PBS and resuspended in cold NP-40 lysis buffer (10 mM Tris–HCl pH 7.5, 0.15% NP40, 150 mM NaCl) for 3 min. The lysate was then transferred onto 2.5 volumes of an ice-cold sucrose buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 24% sucrose) and centrifuged at 13 000 rpm for 10 min at 4°C to separate the subcellular fractions. The supernatant (the cytoplasmic fraction) and pellet (nuclear fraction) were collected for RNA extraction by TRIzol, respectively. All RNA from different fractions was performed RT-qPCR assay as the previous description. The primers used in RT-qPCR assay were listed in Supplementary Table S3. All assays were performed with three independent experiments.
MeRIP-seq
m6A-modified RNA was extracted from total RNA using the same strategy as previously described low input m6A-RIP method. me-RNA library was constructed by using SMARTer Stranded Total RNA-Seq Kit version 2 (Takara, # 634413) following the manufacturer's protocol and then sequenced on a NovaSeq 6000 (Illumina) using paired-end reads 150-cycle kit.
MeRIP-seq analysis
MeRIP-seq raw data were trimmed for adapters using the TrimGalore v.0.6.6. Trimmed reads were aligned to human gene with STAR aligner with reference annotation UCSC human genome version 38 (GRCh38). Differentially methylated sites were detected using R package MeTDiff. Each replication was grouped together. Log fold change and corresponding P values were calculated using Python package matplotlib.
Cell proliferation assay
2000 GSC cells were seeded into 96-well plate well with 200 μl medium at the beginning. Before detection, cells were cooled down to RT, followed by adding 40 μl of Cell Titer (Promega, #G7572) per well. The plate was shaken at 120 r.p.m. for 15 min at RT, and 150 μl of the final product was used for proliferation detection on a microplate reader (Thermo, Varioskan LUX). All assays were performed with three independent experiments.
mRNA stability assay
HEK293T cells co-transfected with dCasRx conjugate and relevant sgRNA vectors were treated with 5 μg/ml transcription inhibitor Actinomycin D (Selleck, #S8964) and collected at various time points (0, 2, 4 and 6 h). The total RNA was extracted and reverse transcribed as described before. The cDNA templates were used in RT-qPCR quantification. The primers used in RT-qPCR assay were listed in Supplementary Table S3. All assays were performed with three independent experiments.
shRNA silencing in HEK293T cells
HEK293T were infected with lentiviruses expressing shRNAs against YTHDF1, YTHDF2 and YTHDF3 respectively or together. The cells infected with shRNA were selected with Puromycin (1 µg/ml) for 3 days. shRNA sequences were listed in Supplementary Table S7.
Quantification and statistical analyses
All statistical analyses are described in the figure legends. Two-way repeated-measures ANOVA was used for statistical analysis with Dunnett multiple hypothesis test correction.
RESULTS
Design of programmable m6A editors
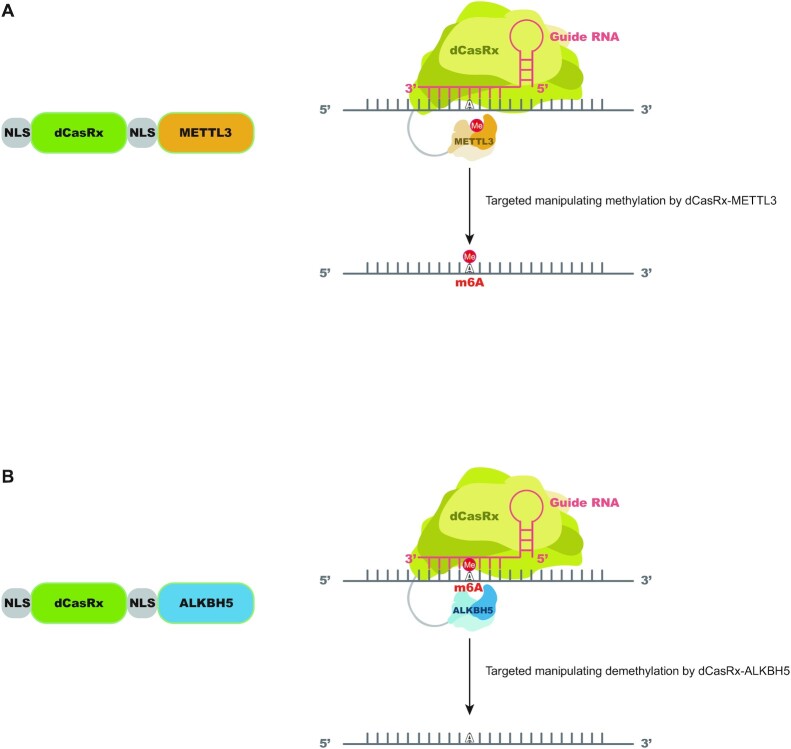
An optimal site-specific m6A regulatory platform should display specificity in location to be modified, enable either deposition of m6A or its removal and be simple to alter. Based on these goals, we fused the dCasRx to an m6A writer, METTL3, or eraser, ALKBH5, to install or remove m6A at targeted sites with single guide RNAs (sgRNAs). Individual sgRNAs were cloned into a vector under a U6 promoter together with CasRx-specific scaffold sequence (Supplementary Figure S1a). Because methylation and demethylation processing of m6A occur primarily in the nucleus (42), we added two segments of NLS to the dCasRx epitranscriptome editors to promote nuclear localization of the editing complex. The resulting m6A editing complexes are designated as NLS-dCasRx-NLS-METTL3 (dCasRx-METTL3) and NLS-dCasRx-NLS-ALKBH5 (dCasRx-ALKBH5) (Figure 1A and B).
Figure 1.
Design of a targeted RNA methylation system. (A) Proposed strategy for NLS-dCasRx-NLS-METTL3 (dCasRx-METTL3). dCasRx was fused to METTL3 and NLS to mediate site-specific methylation of adenosine to m6A with the presence of sgRNA, as well as to ensure dCasRx-METTL3 localization in the nucleus. (B) Proposed strategy for NLS-dCasRx-NLS-ALKBH5 (dCasRx-ALKBH5). dCasRx was fused to ALKBH5 and NLS to mediate site-specific demethylation of m6A to adenosine with the presence of sgRNA, as well as to ensure dCasRx-ALKBH5 localization in the nucleus.
Validation of programmable site-specific m6A writer and eraser activity
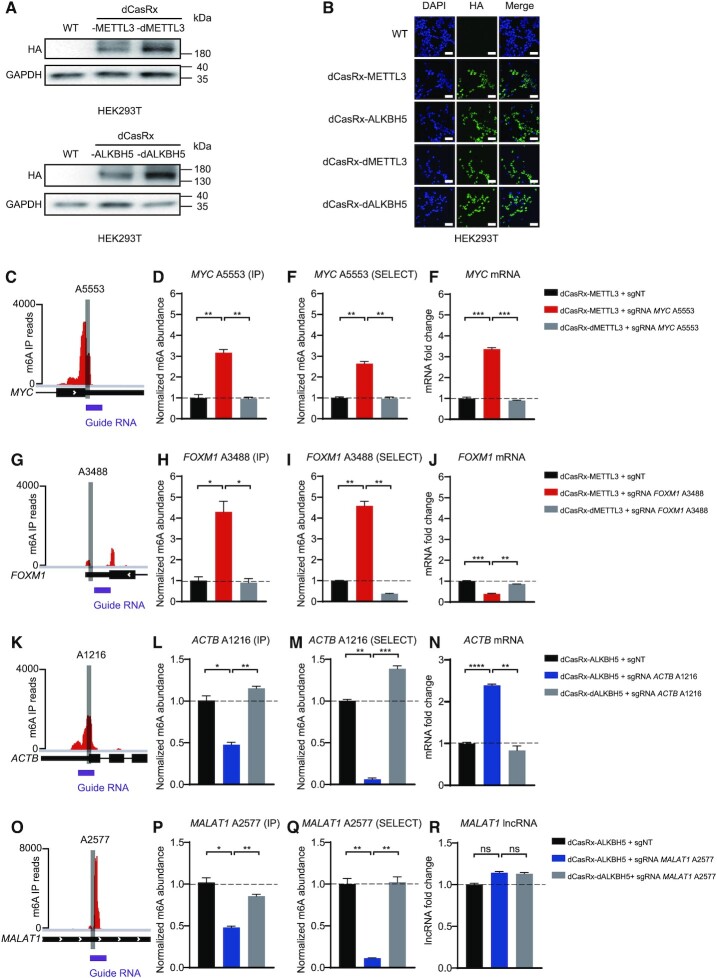
To confirm that dCasRx-METTL3 and dCasRx-ALKBH5 were properly expressed and localized in the nucleus, HEK293T cells were transduced and analyzed by immunoblot and confocal imaging (Figure 2A and B). As controls, we generated dCasRx with methylase-dead METTL3 D395A (dMETTL3) or demethylase-dead ALKBH5 H204A (dALKBH5) expressed in the nuclei to preclude the possibility that effects were caused by presence of non-specific dCasRx epitranscriptome editors.
Figure 2.
Cellular localization of dCasRx epitranscriptomic editors and targeted manipulation of endogenous transcript methylation in HEK293T cell. (A) Western blot results of HA-tag demonstrated expression of dCasRx epitranscriptomic editors in living HEK293T cells when treated with dCasRx-METTL3, dCasRx-ALKBH5, dCasRx-dMETTL3 and dCasRx-dALKBH5. (B) Representative immunofluorescence images of HEK293T cells transfected with HA-tagged dCasRx epitranscriptomic editors. Scale bars, 40 μm. (C) Schematic diagram of m6A distribution in MYC mRNA. (D) Normalized abundance of m6A at MYC A5553 detected by me-RIP. (E) Normalized abundance of m6A at MYC A5553 detected by SELECT. (F) Abundance of MYC mRNA increased after dCasRx-METTL3 editing in HEK293T cell. (G) Schematic diagram of m6A distribution in FOXM1 mRNA. (H) Normalized abundance of altered m6A at FOXM1 A3488 detected by me-RIP. (I) Normalized abundance of altered m6A at FOXM1 A3488 detected by SELECT. (J) Abundance of FOXM1 mRNA decreased after dCasRx-METTL3 editing in HEK293T cell. (K) Schematic diagram of m6A distribution in ACTB mRNA. (L) Normalized abundance of m6A at ACTB A1216 detected by me-RIP. (M) Normalized abundance of m6A at ACTB A1216 detected by SELECT. (N) Abundance of ACTB mRNA increased after dCasRx-ALKBH5 editing in HEK293T cell. (O) Schematic diagram of m6A distribution in MALAT1 RNA. (P) Normalized abundance of altered m6A at MALAT1 A2577 detected by me-RIP. (Q) Normalized abundance of altered m6A at MALAT1 A2577 detected by SELECT. (R) Abundance of MALAT1 lncRNA has not changed after dCasRx-ALKBH5 editing in HEK293T cell. Distributions of m6A was based on a database GSE63753 (49). Data were displayed as mean ± SEM (ANOVA; ns: not significant, *:P < 0.05, **:P < 0.01, ***:P < 0.001, ****:P < 0.0001; n = 3).
As proof-of-principle, we characterized the editing window of dCasRx-METTL3 or dCasRx-ALKBH5 by designing nine different sgRNAs surrounding the selected sites of two transcripts for which m6A modification has been previously reported: the A1216 site of the β-actin (ACTB) transcript (Supplementary Figure S1b) and the A3488 site of the forkhead box protein M1 (FOXM1) (Supplementary Figure S1e). We co-transfected these sgRNAs along with dCasRx-METTL3 or dCasRx-ALKBH5 into HEK293T cells. Eight sgRNAs were tiled across a 20 bp region spanning the targeted site with a 3 nt gap between each sgRNA with one additional sgRNA covered the targeted site. The editing efficiency of our dCasRx epitranscriptomic editors varied for different targeted sites on different transcripts, and demonstrated good editing efficiency within the editing window from −7 to +7 nt window relative to the targeted site (Supplementary Figure S1c, d, f and g). Based on these results, we selected the sgRNAs covering the target site as the sgRNA to test the efficiency of the editing system.
Traditional methods to asses site-specific m6A modification, such as methylated RNA immunoprecipitation (me-RIP) coupled with RT–qPCR or a sequencing-based approach, methylation-individual-nucleotide resolution cross-linking and immunoprecipitation (miCLIP) lack either single-nucleotide resolution or quantitative capability. To circumvent these limitations, we used an established single-base elongation- and ligation-based qPCR amplification method, termed SELECT, to measure m6A level alterations (39). The SELECT technique employs the ability of m6A to hinder the elongation activity of DNA polymerase and the ability of DNA–ligase to selectively catalyze nick ligation between the elongated Up Probe and Down Probe. The final products were quantified by qPCR to reflect the m6A abundance. Using an orthogonal method, we verified that site-specific m6A alterations detected using SELECT reliably reflected methylation level changes at targeted sites, as described by previous studies (40).
We next investigated the ability of nuclear-localized dCasRx epitranscriptomic editors to install or remove m6A modifications on endogenous transcripts in HEK293T cells. We first evaluated the ‘writing’ efficiency of dCasRx-METTL3 on adenine sites that display low degree of native methylation in HEK293T cells. Adenine A5553 within the 3′-UTR of MYC mRNA (Figure 2C) and adenine A3488 within the 3′-UTR of FOXM1 mRNA (Figure 2G) were targeted. With co-transduction of the targeting sgRNA, dCasRx-METTL3 expression increased MYC A5553 methylation (Figure 2D and E) followed by an upregulation of MYC mRNA (Figure 2F). Transduction with a different sgRNA increased FOXM1 A3488 methylation (Figure 2H and I) followed by a decrease of FOXM1 mRNA (Figure 2J). This was consistent with the conclusions reported in previous studies (26,43). These results indicated that specific m6A sites were editable by dCasRx epitranscriptomic editors. In control studies, transduction with methyltransferase-inactive variants (dCasRx-dMETTL3) of these constructs at comparable expression to their active counterparts did not increase methylation level of target sites in the corresponding mRNAs. Another control using dCasRx-METTL3 with non-targeted sgRNA (sgNT) also showed no significant alteration of m6A levels at targeted sites, confirming that expression of dCasRx epitranscriptomic editors alone did not induce m6A changes at targeted sites. In reciprocal studies, we evaluated the efficiency of m6A ‘erasing’ by dCasRx-ALKBH5 using adenine sites that have a high degree of native methylation in HEK293T cells. Adenine A1216 within the 3′-UTR of ACTB mRNA (Figure 2K) and adenine A2577 of Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) RNA (Figure 2O) were targeted. Both SELECT and me-RIP showed a significant decrease of methylation at targeted sites in the dCasRx-ALKBH5 edited cells compared with the NT or dCasRx-dALKBH5 group (Figure 2L, M, P and Q), indicating that dCasRx-ALKBH5 could remove m6A specifically, which induced an increase of total ACTB mRNA (Figure 2N) but had no effect on the MALAT1 long non-coding RNA (Figure 2R).
Site-specific m6A modification informs effects on mRNA processing
As m6A modifications have been linked to mRNA processing, our system empowers the ability to directly interrogate m6A effects on mRNA regulation at a single transcript level. To investigate the reported effects of m6A on mRNA splicing (5,42), we assessed mRNA splicing of specific transcripts by measuring the exclusion rate of the intron closest to the edited m6A site. Increased exclusion rates of a MYC intron following editing, indicating that m6A upregulation promoted MYC mRNA splicing (Supplementary Figure S2a), while altered m6A levels did not alter FOXM1, ACTB and MALAT1 RNA splicing (Supplementary Figure S2b-d).
We next tested m6A regulation RNA nuclear export. Nuclear fractions were extracted and the proportion of selected mRNAs in the nucleus after m6A editing was quantified by qPCR. The nuclear RNA U1 was used as a quality control of nuclear fraction purity (Supplementary Figure S2e). dCasRx-METTL3-mediated m6A upregulation inhibited FOXM1, but not MYC, nuclear mRNA export (Supplementary Figure S2f and g), while dCasRx-ALKBH5-mediated m6A downregulation promoted ACTB but not MALAT1 nuclear mRNA export (Supplementary Figure S2h and i).
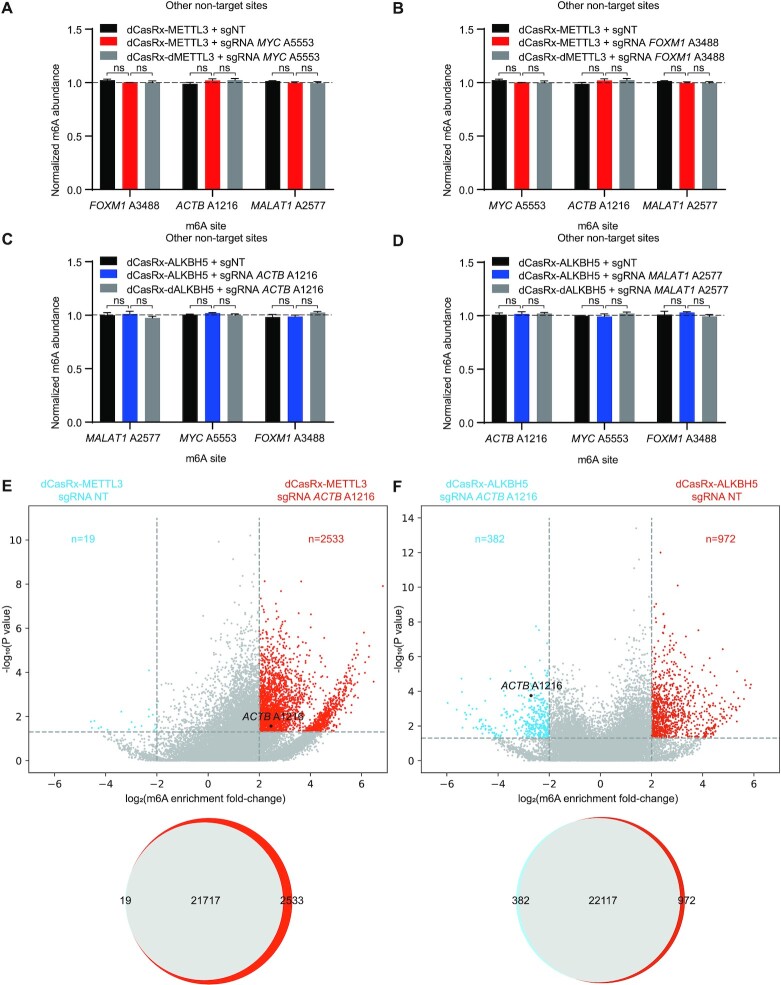
Assessment of off-target effects and editing efficiency of the site-specific writer-eraser platform
To evaluate the off-target activity of dCasRx epitranscriptomic editors, we measured changes in m6A levels in three other non-targeted adenine sites in HEK293T cells. dCasRx-METTL3-transduced or dCasRx-ALKBH5-transduced cells did not alter m6A levels at non-targeted adenine sites (Figure 3A–D), suggesting a low risk of off-target editing of the dCasRx epitranscriptomic platform. To ensure that dCasRx m6A editors do not result in global methylation changes, we performed transcriptome-wide RNA methylation changes by m6A-RNA immunoprecipitation-sequencing (meRIP-seq). We selected ACTB A1216 as the target site because its methylation level could be successfully manipulated by both dCasRx-METTL3 and dCasRx-ALKBH5 in HEK293T cells (Figure 2L and M; Supplementary Figure S4c). Compared to the control sgNT group, dCasRx-METTL3 with sgRNA targeting ACTB A1216 site mildly increased m6A level at 2532 additional sites out of total 24 269 detected (10.4%) (Figure 3E), whereas dCasRx-ALKBH5 only decreases m6A levels at 381 additional sites out of total 23471 detected (1.6%) (Figure 3F).
Figure 3.
Off-target methylation by dCasRx epitranscriptomic editors in HEK293T cells. (A–D) Normalized m6A abundance at non-targeted adenine sites in each specific-site targeted sgRNA group, MYC A5553 targeted sgRNA group (A), FOXM1 A3488 targeted sgRNA group (B), ACTB A1216 targeted sgRNA group (C) and MALAT1 A2577 targeted sgRNA group (D), was detected by SELECT. Data were represented as mean ± SEM. (ANOVA, ns: not significant, n = 3). (E andF) Differential m6A sites in HEK293T cells transfected with dCasRx-METTL3 (E) or dCasRx-ALKBH5 (F) and sgRNA ACTB A1216 or sgNT. Top: volcano graphs depict differential m6A sites between sgRNA ACTB A1216 and sgNT groups. Bottom: Venn diagrams show overlap of all methylated m6A sites for the above comparisons. meRIP-seq analysis was performed with two independent biological replicates.
We also compared the editing efficiency of our dCasRx-METTL3 system to a Cas13b-based m6A editing system developed recently (34). Compared with dCas13b-M3 and dCas13b-M3M14, our dCasRx-METTL3 achieve a similar editing efficiency on the site A1216 and even a higher editing efficiency on site FOXM1 A3488 (Supplementary Figure S3a and b).
Manipulating m6A levels at FOXM1 and MYC mRNAs impact glioblastoma stem cell proliferation
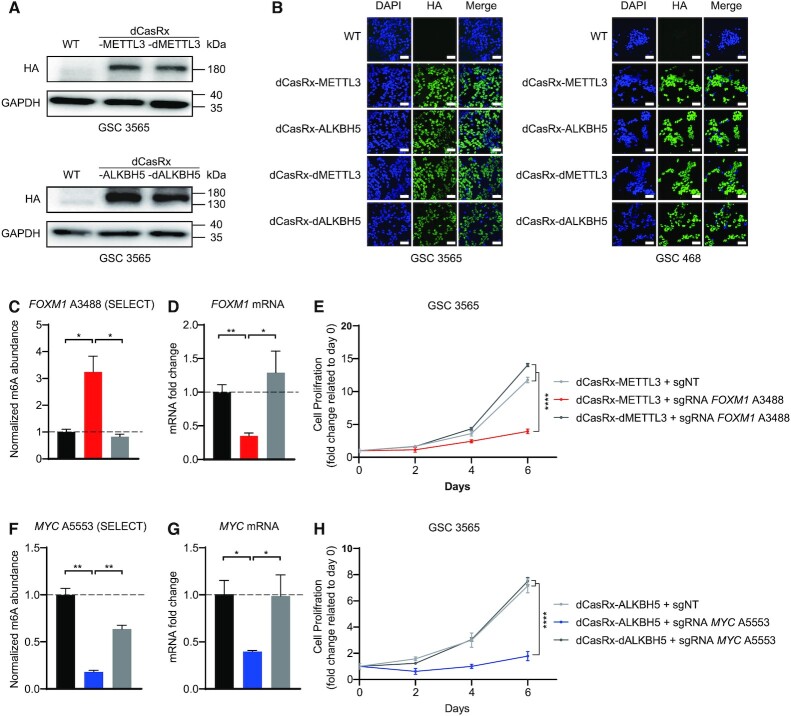
As m6A serves context-specific roles in disease states, we sought to leverage our editing platform to investigate the effects of m6A modifications that have been specifically linked to maintenance of disease states. GBM is the most prevalent primary malignant brain tumor and contains stem-like cells, called glioblastoma stem cells (GSCs). METTL3 and ALKBH5 have both been described to maintain GSCs (25,26). Therefore, we applied the dCasRx epitranscriptomic editors to GSCs. Unlike HEK293T cells, GSCs could not be easily transfected with plasmids encoding dCasRx-METTL3 or dCasRx-ALKBH5. Thus, we developed lentiviral transduction of dCasRx editors in GSCs with enrichment by Puromycin selection. The sgRNAs were packaged into lentiviruses using the same procedure. GSCs expressing dCasRx-METTL3 or dCasRx-ALKBH5 were then transduced with these sgRNA-lentiviruses. Both the dCasRx-METTL3 or dCasRx-ALKBH5 constructs were expressed and localized in nuclei of both GSC 3565 and GSC 468 cells (Figure 4A and B).
Figure 4.
Targeted manipulation of m6A level via dCasRx epitranscriptomic editors on endogenous transcripts affects GSC proliferation. (A) Expression of dCasRx epitranscriptomic editors dCasRx-METTL3 or dCasRx-ALKBH5 in GSC 3565. (B) Representative immunofluorescence images of GSCs transfected with HA-tagged dCasRx epitranscriptomic editors. Scale bars, 40 μm. (C) Methylation at A3488 of endogenous mRNA FOXM1 in GSC 3565 increased by three folds by dCasRx-METTL3 editing. (D) FOXM1 mRNA decreased after dCasRx-METTL3 editing in GSC 3565. (E) Proliferation of GSC 3565 decreased with an increased m6A levels at FOXM1 A3488 mediated by dCasRx-METTL3, compared to NT group and dCasRx-dMETTL3 group. (F) A decrease of methylation at A5553 of endogenous mRNA MYC was mediated by dCasRx-ALKBH5 editing in GSC 3565. (G) mRNA expression level of MYC decreased after dCasRx-ALKBH5 editing in GSC 3565. (H) Proliferation of GSC 3565 decreased with a decreased m6A level at MYC A5553 mediated by dCasRx-ALKBH5, compared to NT group and dCasRx-dALKBH5 group. Data are represented as mean ± SEM. (ANOVA; *, P-value < 0.05; **, P-value < 0.01; ***, P-value < 0.001; ****, P-value < 0.0001, n = 3).
The transcription factors FOXM1 and MYC regulate GSC proliferation, self-renewal and tumorigenicity. FOXM1 and MYC mRNAs contain adenine sites that can be methylated by m6A. Our programmable epitranscriptome writer dCasRx-METTL3 increased the m6A levels at the A3488 site of the FOXM1 transcript (Figure 4C); and the programmable eraser dCasRx-ALKBH5 reduced the m6A level at the A5553 site of the MYC transcript in GSC 3565 cells compared to non-targeting controls and catalytically inactive controls (Figure 4F). Low m6A levels at the 3′UTR on FOXM1 mRNA enhances the expression of FOXM1 nascent transcripts (26), whereas m6A at the 3′UTR in MYC stabilizes its mRNA levels (43,44). Consistent with these reported findings, targeted m6A methylation on the A3488 FOXM1 transcript site using dCasRx-METTL3 decreased FOXM1 mRNA levels (Figure 4D). Targeted m6A demethylation on the A5553 MYC transcript site downregulated MYC mRNA levels (Figure 4G). Non-targeting sgRNAs and catalytically inactive constructs were used as controls. Downregulation of FOXM1 or MYC mRNA induced by dCasRx editing inhibited the proliferation of GSC 3565 (Figure 4E and H). These results demonstrated that our dCasRx epitranscriptomic editors could manipulate m6A levels on endo-transcripts in disease states.
YT521-B homology Domain-containing Family (YTHDF) proteins mediate degradation of m6A-mRNAs
The effects of m6A in cytosolic transcripts are mediated by a complex network of interactions between specific m6A sites and specific members of the YTHDF of m6A-binding proteins. The YTHDF family includes three paralogs, YTHDF1, YTHDF2 and YTHDF3, each with distinct reported functions. Different studies identified distinct roles for each family member in enhancing translation or promoting degradation of different mRNA species (24,37).
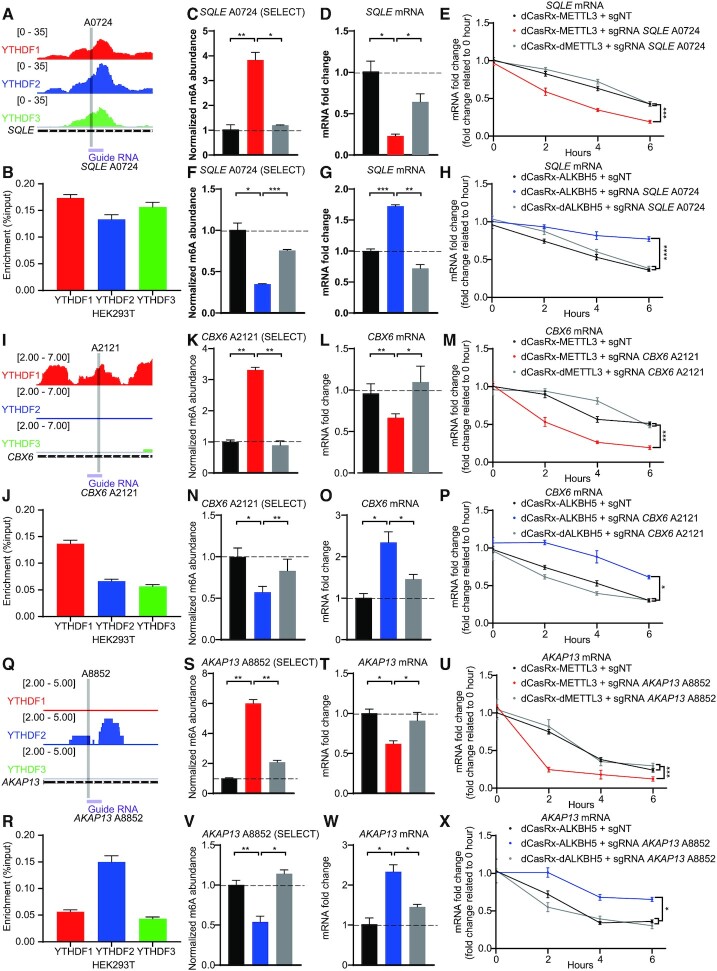
To interrogate the function of the YTHDF proteins at specific genomic sites, we analyzed a database (GSE78030) which presented the distribution of YTHDF paralogs across the endo-transcriptome in HEK293T cells (11). We then selected screened mRNA candidates that contained m6A sites for YTHDF paralog binding. We first performed RIP-qPCR for each YTHDF paralog to confirm the interactions between YTHDF paralogs and selected mRNA candidates. Among the screened mRNAs, m6A sites at SQLE A0724 (Figure 5A and B) and ACTB A1216 (Supplementary Figure S4a and b) presented high-affinity binding sites to all three YTHDF paralogs. The m6A site at SQLE A0724 was located at the 5′UTR, which has potential to bind eIF3 to initiate cap-independent translation (9), while the m6A site of ACTB A1216 was located at the 3′UTR and near the stop codon. To examine whether methylation at a single site contributed to the degradation of these mRNAs, we designed sgRNAs targeting the corresponding m6A sites. m6A installation via dCasRx-METTL3 editing significantly decreased mRNA expression of SQLE (Figure 5C and D) and ACTB (Supplementary Figure S4c and d). Conversely, m6A removal via dCasRx-ALKBH5 editing significantly increased relevant mRNAs (Figures 5F-G and 2M-N). To confirm that the effects of m6A editing were mediated by changes in degradation, we assayed mRNA stability. Consistent with our findings, m6A removal led to accelerated mRNA degradation and thus decreased mRNA levels (Figure 5E and H; Supplementary Figure S4e and f).
Figure 5.
m6A sites binding with DF paralogs control the degradation of endogenous transcripts in HEK293T cells. (A, I andQ) Schematic diagrams of distribution of DF paralogs in endogenous SQLE mRNA (A), CBX6 mRNA (I), AKAP13 mRNA (Q). Distributions of DF paralogs was based on a database GSE78030 (11). (B, J andR) The combination of YTHDF paralogs at SQLE A0724 (B), CBX6 A2121 (J), AKAP13 A8852 (R) in HEK293T cells, quantified by YTHDF paralog RIP coupled with RT-qPCR. Data are displayed as mean ± SEM (n = 3). (C, K andS) Normalized abundance of altered m6A at SQLE A0724 (C), CBX6 A2121 (K), AKAP13 A8852 (S) edited by dCasRx-METTL3. (D, L andT) Abundance of SQLE mRNA (D), CBX6 mRNA (L), AKAP13 mRNA (T) decreased after dCasRx-METTL3 editing. (E, M andU) mRNA degradation measurement of SQLE (E), CBX6 (M), AKAP13 (U) in HEK293T cells edited with dCasRx-METTL3. (F, N andV) Normalized abundance of altered m6A at SQLE A0724 (F), CBX6 A2121 (N), AKAP13 A8852 (V) edited by dCasRx-ALKBH5. (G, O andW) Abundance of SQLE mRNA (G), CBX6 mRNA (O), AKAP13 mRNA (W) increased after dCasRx-ALKBH5 editing. (H, P andX) mRNA degradation measurement of SQLE (H), CBX6 (P), AKAP13 (X) in HEK293T cells transfected with dCasRx-ALKBH5. Data are represented as mean ± SEM. (ANOVA; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n = 3).
We then identified two gene candidates containing m6A sites which have higher binding affinity for specific individual YTHDF paralogs and designed sgRNAs accordingly. CBX6 A2121 (Figure 5I and J) and SERBP1 A3240 (Supplementary Figure S4g) preferentially bound to YTHDF1. Writing at these YTHDF1-preferent m6A sites led to downregulation of each corresponding mRNA (Figure 5K, L, N and O; Supplementary Figure S4h–k). AKAP13 A8852 (Figure 5Q and R) and MLLT3 A1068 (Supplementary Figure S4g) had higher binding affinity to YTHDF2. Writing to these YTHDF2-preferent m6A sites which similarly led to downregulation of the corresponding mRNAs (Figure 5S, T, V and W; Supplementary Figure S4m–p). These results suggest that that installation of m6A at YTHDF1- or YTHDF2-bound sites mediate mRNA degradation (Figure 5M, P, U and X). We next sought to investigate whether inclusion of YTHDF3 would induce different alterations. We therefore screened two m6A sites which have higher binding affinity to both YTHDF1 and YTHDF3 versus YTHDF2: HDGF A1908 (Supplementary Figure S5a and b) and GSTP1 A0659 (Supplementary Figure S5i). Manipulation of the methylation level at these sites by dCasRx-METTL3 or dCasRx-ALKBH5 still promoted mRNA degradation (Supplementary Figure S5c–h and j–m).
To further determine whether the downregulation of selected mRNAs after dCasRx-METTL3 mediated m6A editing was YTHDF-dependent, we performed shRNA-mediated knockdown of YTHDF1, YTHDF2 or YTHDF3 separately or together in HEK293T cells expressing the dCasRx system. Knockdown efficiency was confirmed by western blot (Supplementary Figure S6a). We found that single knockdown of YTHDF1, YTHDF2 or YTHDF3 individually can partially rescue the downregulation of target mRNA caused by dCasRX-METTL3 editing. Triple knockdown of YTHDF1, YTHDF2 and YTHDF3 almost completely restored target mRNA level (Supplementary Figure S6b–e).
By modulating the methylation level of targeted sites recognized by YTHDF paralogs with dCasRx-METTL3 and dCasRx-ALKBH5, we found all three YTHDFs play a similar role in promoting m6A-mediated mRNA degradation at the selected transcripts. These results indicate that our epitranscriptomic editor system can be utilized to investigate the transcriptomic site-specific interactions between m6A modified transcripts and mRNA readers or other modifying factors.
DISCUSSION
The advancement of various gene editors enables functional analysis of epigenomic marks and facilitates investigation of epigenetic control over biological processes (45,46). RNA modifications are essential parts of the epitranscriptome, and the highly dynamic and reversible m6A modifications in mammalian transcriptomes regulate nearly all aspects of RNA metabolism and functionality (47,48). However, much of our current knowledge about m6A was mainly based on genetic perturbations induced by global overexpression or knockout of relevant genes, which modify the whole transcriptome instead of target sites of interest. To interrogate the site-specific effects of m6A interacting with multiple readers, a strategy to manipulate targeted m6A sites within endo-transcripts is essential.
In this work, we developed the first dCas13Rx-mediated epitranscriptomic editors that coupled with RNA methyltransferase METTL3 or demethylase ALKBH5 to achieve bidirectional modulation of targeted m6A sites in living cells. We demonstrated that nucleus-located dCasRx epitranscriptomic editors enabled site-specific m6A installation or removal with low off-target alterations. In addition to being transfected into normal human cells, the dCasRx epitranscriptomic editors could be packaged into lentiviruses, owing to their small sizes and be used to investigate the function of single m6A sites in GBM cells that were difficult to be transfected by other means. We demonstrated that m6A played opposite roles in the regulation of FOXM1 and MYC mRNA—decreased expression of FOXM1 transcripts but increased MYC expression. Alterations in the expression level of FOXM1 and MYC subsequently affected GSCs proliferation. Lastly, using the dCasRx epitranscriptomic editors, we demonstrated that increased methylation of YTHDF paralogs-bound sites induced mRNA degradation. Collectively, these results demonstrate that our dCasRx system can be used to dissect previously unclear interactions and to elucidate the causal relationships between m6A modifications and phenotypes.
This work demonstrates a proof-of-concept dCasRx-based strategy with the potential for high-throughput screening of m6A modifications in whole epitranscriptome using a suitable sgRNA library. Judging from the success of high-throughput functional genomic screening based on CRISPR-Cas9 technology (49) and verified efficiency of dCasRx epitranscriptomic editors, our technology has broad applications in a variety of studies including investigation of epitranscriptome of difficult-to-transfect cells.
DATA AVAILABILITY
All raw sequencing data and selected processed data sets have been deposited to NCBI Sequence Read Archive at the accession number PRJNA727525. All data accessed from external sources and prior publications have been referenced in the text.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Tengfei Xiao for helpful discussion.We thank all technicians from Westlake University facilities for the technical assistance.
Contributor Information
Zhen Xia, Fudan University, Shanghai, 200433, China; Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
Min Tang, Department of NanoEngineering, University of California San Diego, 9500 Gilman Drive, La Jolla, CA,92307, USA.
Jiayan Ma, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
Hongyan Zhang, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
Ryan C Gimple, Department of Pathology, Case Western Reserve University, Cleveland, OH, 44106, USA.
Briana C Prager, Department of Pathology, Case Western Reserve University, Cleveland, OH, 44106, USA; Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, Cleveland, OH,44195, USA.
Hongzhen Tang, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
Chongran Sun, Department of Neurosurgery, 2nd affiliated hospital, school of Medicine, Zhejiang University, Hangzhou, Zhejiang, 310009, China.
Fuyi Liu, Department of Neurosurgery, 2nd affiliated hospital, school of Medicine, Zhejiang University, Hangzhou, Zhejiang, 310009, China.
Peng Lin, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
Yutang Mei, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
Ruoxin Du, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
Jeremy N Rich, University of Pittsburgh Medical Center Hillman Cancer Center, Department of Neurology, University of Pittsburgh, Pittsburgh, PA,15261, USA.
Qi Xie, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang, 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, 310024, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [82073268]; Westlake Education Foundation. Funding for open access charge: National Natural Science Foundation of China [82073268].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hoernes T.P., Erlacher M.D.. Translating the epitranscriptome. Wiley Interdiscip. Rev. RNA. 2017; 8:e1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbieri I., Kouzarides T.. Role of RNA modifications in cancer. Nat. Rev. Cancer. 2020; 20:303–322. [DOI] [PubMed] [Google Scholar]
- 3. Meyer K.D., Jaffrey S.R.. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014; 15:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T.. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiao W., Adhikari S., Dahal U., Chen Y.S., Hao Y.J., Sun B.F., Sun H.Y., Li A., Ping X.L., Lai W.Y.et al.. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 6. Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P.et al.. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife. 2017; 6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G.et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R.. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiang Y., Laurent B., Hsu C.H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S.et al.. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017; 543:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R.. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016; 537:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B.. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong X., Yu J., Frazier K., Weng X., Li Y., Cham C.M., Dolan K., Zhu X., Hubert N., Tao Y.et al.. Circadian clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep. 2018; 25:1816–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z., Wang M., Xie D., Huang Z., Zhang L., Yang Y., Ma D., Li W., Zhou Q., Yang Y.G.et al.. METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018; 28:1050–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu K., Yang Y., Feng G.H., Sun B.F., Chen J.Q., Li Y.F., Chen Y.S., Zhang X.X., Wang C.X., Jiang L.Y.et al.. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017; 27:1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao B.S., Wang X., Beadell A.V., Lu Z., Shi H., Kuuspalu A., Ho R.K., He C.. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017; 542:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K.et al.. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jaffrey S.R., Kharas M.G.. Emerging links between m(6)A and misregulated mRNA methylation in cancer. Genome Med. 2017; 9:2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U.et al.. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019; 566:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X.et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S.et al.. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H.et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G.et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zaccara S., Ries R.J., Jaffrey S.R.. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019; 20:608–624. [DOI] [PubMed] [Google Scholar]
- 25. Visvanathan A., Patil V., Arora A., Hegde A.S., Arivazhagan A., Santosh V., Somasundaram K.. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018; 37:522–533. [DOI] [PubMed] [Google Scholar]
- 26. Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bogler O.et al.. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017; 31:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G.et al.. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017; 18:2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L.et al.. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016; 353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A.et al.. RNA targeting with CRISPR-Cas13. Nature. 2017; 550:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D.. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018; 173:665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wessels H.H., Mendez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E.. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020; 38:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Granados-Riveron J.T., Aquino-Jarquin G.. CRISPR-Cas13 precision transcriptome engineering in cancer. Cancer Res. 2018; 78:4107–4113. [DOI] [PubMed] [Google Scholar]
- 33. Liu X.M., Zhou J., Mao Y., Ji Q., Qian S.B.. Programmable RNA N(6)-methyladenosine editing by CRISPR-Cas9 conjugates. Nat. Chem. Biol. 2019; 15:865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilson C., Chen P.J., Miao Z., Liu D.R.. Programmable m(6)A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat. Biotechnol. 2020; 38:1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li J., Chen Z., Chen F., Xie G., Ling Y., Peng Y., Lin Y., Luo N., Chiang C.M., Wang H.. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 2020; 48:5684–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu J., Dou X., Chen C., Chen C., Liu C., Xu M.M., Zhao S., Shen B., Gao Y., Han D.et al.. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020; 367:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zaccara S., Jaffrey S.R.. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020; 181:1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiao Y., Wang Y., Tang Q., Wei L., Zhang X., Jia G.. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N(6) -methyladenosine modification. Angew. Chem. Int. Ed. Engl. 2018; 57:15995–16000. [DOI] [PubMed] [Google Scholar]
- 40. Zeng Y., Wang S., Gao S., Soares F., Ahmed M., Guo H., Wang M., Hua J.T., Guan J., Moran M.F.et al.. Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 2018; 16:e2006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keene J.D., Komisarow J.M., Friedersdorf M.B.. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006; 1:302–307. [DOI] [PubMed] [Google Scholar]
- 42. Yao W., Han X., Ge M., Chen C., Xiao X., Li H., Hei Z.. N(6)-methyladenosine (m(6)A) methylation in ischemia-reperfusion injury. Cell Death. Dis. 2020; 11:478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L.et al.. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018; 20:285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dixit D., Prager B.C., Gimple R.C., Poh H.X., Wang Y., Wu Q., Qiu Z., Kidwell R.L., Kim L.J.Y., Xie Q.et al.. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 2020; 11:480–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Allis C.D., Jenuwein T.. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016; 17:487–500. [DOI] [PubMed] [Google Scholar]
- 47. Stricker S.H., Koferle A., Beck S.. From profiles to function in epigenomics. Nat. Rev. Genet. 2017; 18:51–66. [DOI] [PubMed] [Google Scholar]
- 48. Lewis C.J., Pan T., Kalsotra A.. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2017; 18:202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W.. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014; 509:487–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data and selected processed data sets have been deposited to NCBI Sequence Read Archive at the accession number PRJNA727525. All data accessed from external sources and prior publications have been referenced in the text.