Abstract
Gene expression is regulated at many levels including co- or post-transcriptionally, where chemical modifications are added to RNA on riboses and bases. Expression control via RNA modifications has been termed ‘epitranscriptomics’ to keep with the related ‘epigenomics’ for DNA modification. One such RNA modification is the N6-methylation found on adenosine (m6A) and 2′-O-methyladenosine (m6Am) in most types of RNA. The N6-methylation can affect the fold, stability, degradation and cellular interaction(s) of the modified RNA, implicating it in processes such as splicing, translation, export and decay. The multiple roles played by this modification explains why m6A misregulation is connected to multiple human cancers. The m6A/m6Am writer enzymes are RNA methyltransferases (MTases). Structures are available for functionally characterized m6A RNA MTases from human (m6A mRNA, m6A snRNA, m6A rRNA and m6Am mRNA MTases), zebrafish (m6Am mRNA MTase) and bacteria (m6A rRNA MTase). For each of these MTases, we describe their overall domain organization, the active site architecture and the substrate binding. We identify areas that remain to be investigated, propose yet unexplored routes for structural characterization of MTase:substrate complexes, and highlight common structural elements that should be described for future m6A/m6Am RNA MTase structures.
INTRODUCTION
RNA is heavily post-transcriptionally modified (Figure 1A) with the largest diversity of modifications found in transfer RNA (tRNA). The N6-methyladenosine (m6A) modification was discovered in 1974 in messenger RNA (mRNA) from rat (1), and the identification of the N6-2′-O-methyladenosine (m6Am) modification followed shortly after in 1975 (2).
Figure 1.
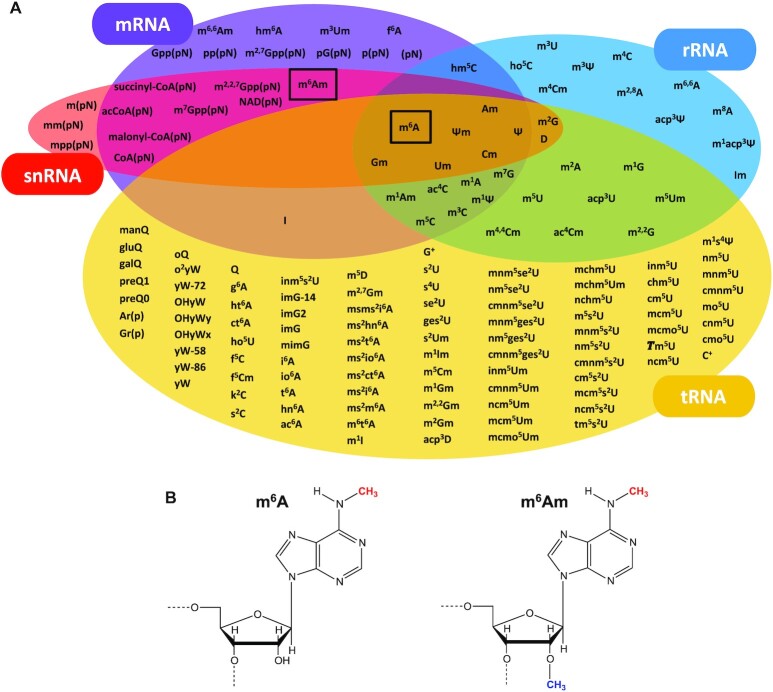
Post-transcriptional RNA modifications in different types of RNA and m6A/m6Am modifications. (A) Overview of the post-transcriptional modifications on mRNA, rRNA, tRNA and snRNA. m6A is found in all of these RNAs whereas m6Am is so far only identified in mRNA and snRNA. In addition to the outlined RNAs, m6A is found in lncRNA and miRNA. (B) m6A and m6Am nucleosides. The N6-methylation is shown in red and the 2′-O-methylation is shown in blue.
m6A (Figure 1B) is the most commonly occurring internal post-transcriptional modification on mRNA from yeast, plants, flies, humans and other mammals (reviewed in (3–5)) and was recently discovered in bacterial mRNA (6) (Figure 1A). In humans, its wide occurrence implicates it in multiple processes including precursor mRNA (pre-mRNA) splicing (7,8), mRNA translation (9–12), stability (8,9,13,14), structure, export and decay (reviewed in (3)). A large variety of roles is also observed in plants (reviewed in (4)), flies (reviewed in (5)) and yeast (15–17). Many functions regulated by m6A on mRNA highlight the involvement of this modification in a range of cellular processes such as cell differentiation and reprogramming (reviewed in (3)), implying an association with numerous human diseases. Accordingly, several studies have demonstrated a link between N6-methylation of human mRNA and progression of different types of cancer by m6A-mediated regulation of expression of tumour-related genes (reviewed in (18)). In addition to mRNA, m6A is also found in ribosomal RNA (rRNA) from both mammals and bacteria, but not in yeast or archaea (19–26) (Figure 1A). In humans, m6A on rRNA affects selective ribosome translation during cellular stress (23,24), and in bacteria it plays a role in rRNA folding and stability (25), translational control (25,27) and cell fitness (26). There are inconclusive reports on the effect of this modification on ribosome and polysome assembly (23,24,27,28) which appears cell line dependent. m6A is also found in bacterial and archaeal tRNA (29,30) where its role is largely unknown, in small nuclear RNA (snRNA) (31–34) (Figure 1A), in microRNA (miRNA) where it affects maturation (35–37) and regulation (38), and in long non-coding RNA (lncRNA) (39–41) where it plays a role in lncRNA-mediated transcriptional repression (41,42).
m6Am (Figure 1B) has, since its discovery, been identified as an internal modification in mRNA (43) and snRNA (44,45) and is found at the 5′-end of most eukaryotic mRNA (2,46), where the modified Am (46,47) is positioned adjacent to the mRNA cap structure (reviewed in (48)) (Figure 1A). The cellular abundance of the N6-methylation at the mRNA cap is context dependent (reviewed in (49)). This has led to strongly divergent results on the role of the modification in translation-promotion or -attenuation and in transcript stabilization (47,50–54), warranting further studies to clarify the biological function of m6Am in the cap.
m6A is a reversible modification in mRNA (reviewed in (49)), meaning that distinct sets of proteins introduce (writers), recognize (readers) and remove (erasers) this mark, allowing it to exert reversible regulation of mRNA metabolism. It has yet to be conclusively determined if such writer/reader/eraser systems exist for m6A on other types of RNA or for m6Am. m6A/m6Am writers are S-adenosyl methionine (SAM)-dependent MTases that perform N6-methylation by transfer of a methyl group from the SAM cofactor to the substrate adenosine. A structure of a bacterial m6A rRNA MTase, RlmJ, was published in 2013, but the recently increased attention on the m6A structural field followed from the 2016 publication of the human m6A ‘writer’ METTL3:METTL14 MTase domain (MTD) complex. All structurally characterized m6A/m6Am RNA MTases are listed in Table 1 and described in detail in this review. A number of m6A/m6Am MTases are functionally characterized but lack structure determination such as human METTL4 that forms m6Am at an internal site in the U2 snRNA (44,45), Escherichia coli RlmF responsible for m6A1618 in the 23S rRNA (26), E. coli tRNA (adenine(37)-N6-)-methyltransferase B6 (TrmB6) that forms m6A37 in tRNA1Val (55) and E. coli tRNA (threonylcarbamoyl adenine(37)-N6-)-methyltransferase O (TrmO) that N6-methylates the adenosine variant; threonylcarbamoyl adenosine (t6A), in tRNA1Thr and tRNA3Thr (56). Currently, the bacterial m6A mRNA MTase(s) lack identification altogether. Crystal structures exist for E. coli TrmO homologues, but m6t6A MTases are not further covered in this review. In addition to m6A, rRNA also carries m26A. This variant of m6A is installed by rRNA dimethyltransferases of the KsgA/Dim1 and Erm families. Many m26A RNA MTases are structurally characterized (57–60) and have similar overall folds to m6A MTases, but vary in catalytic amino acid motifs (57), and are therefore excluded from this review.
Table 1.
Outline of all structurally characterized m6A RNA MTases. mRNA: messenger RNA, rRNA: ribosomal RNA, snRNA: small nuclear RNA, A: adenosine, Am: 2′-O-methylated adenosine, METTL3: methyltransferase-like 3: METTL14: methyltransferase-like 14, METTL16: methyltransferase-like 16, CAPAM: mRNA cap adenosine N6-methyltransferase, METTL5: methyltransferase-like 5, TRMT112: tRNA MTase subunit 11–2, ZCCHC4: zinc finger CCHC-type containing 4, RlmJ: ribosomal RNA large subunit methyltransferase J. FL: full-length, MTD: methyltransferase domain, N: N-terminal extension, ZFD: zinc finger domain, HD: helical domain, VCD: vertebra-conserved domain, WW: Trp-Trp domain. L. pneumophila: Legionella pneumophila, E. coli: Escherichia coli. NP: No publication.
| m6A/m6Am RNA MTase | Substrate(s) | Domain(s) | Ligand | PDB code(s) | References |
|---|---|---|---|---|---|
| Human METTL3:METTL14* | A in mRNA | MTD:N+MTD | None | 5TEY, 5K7M, 5IL0 | (61–63) |
| SAM | 5L6E, 5K7U, 5IL1 | (61–63) | |||
| SAH | 5L6D, 5K7W, 5IL2 | (61–63) | |||
| Sinefungin | 6Y4G | NP | |||
| Compound | 6TTP, 6TTT, 6TU1, 6TTX, 6TTV, 6TTW | NP | |||
| ZFD (METTL3) | none | 5YZ9 | (64) | ||
| Human METTL16 | A in mRNA, snRNA | N+MTD VCD |
None SAH substrate None |
2H00, 6B91, 6GT5 6B92, 6GFK, 6GFN 6DU4, 6DU5 6M1U |
NP, (65, 66) (65, 66) (67) (68) |
| Human CAPAM | Am in mRNA | MTD+HD WW |
None SAH None |
6IRV 6IRW 2JX8 |
(47) (47) NP |
| Zebrafish CAPAM | Am in mRNA | MTD+HD | None SAH Oligo |
6IRX 6IRY 6IS0, 6IRZ |
(47) (47) (47) |
| Human METTL5:TRMT112 | A in rRNA | FL:FL | SAM | 6H2U, 6H2V | (28) |
| Human ZCCHC4 | A in rRNA | MTD+ZFD | SAH | 6UCA | (69) |
| E. coli RlmJ | A in rRNA | FL | None SAM SAH SAH/AMP Bisubstrate |
4BLU 4BLV 6QE5 4BLW 6QE0, 6QDX |
(70) (70) (71) (70) (71) |
| L. pneumophila RlmJ** | Not shown | FL | None | 2OO3 | NP |
*This complex further requires Wilms tumour 1 associating protein (WTAP) for in vivo activity.
**Activity assumed from structure and sequence homology.
In this review, we analyse the structurally and functionally characterized m6A/m6Am RNA MTases to understand their individual and common features in domain/subunit organization and binding of RNA substrate and SAM/SAH cofactor substrate/product. We use this knowledge to identify areas that remain to be investigated, to highlight yet unexplored routes for structural characterization of protein:substrate complexes and to propose common structural elements that should be described when reporting new structures of m6A/m6Am RNA MTases.
The human m6A mRNA MTase METTL3:METTL14
The complex of METTL3:METTL14 makes up the core of a larger complex that installs m6A internally on mRNA when the methylated adenine is located in a specific DRACH consensus motif where A is methylated, D = A, G or U, R = A or G, and H = A, C or U (31). METTL3 was first identified in humans (72) and later shown to associate with METTL14 (73–75). The complex is formed in the cytoplasm (76) and locates to the nucleus via a nuclear localization signal in METTL3 (76). In the nucleus, METTL3 binds to the m6A regulator Wilms tumour 1 associating protein (WTAP) (77). This protein relocates the entire ternary METTL3:METTL14:WTAP complex to nuclear speckles (73,74), which is rich mRNA substrates, ultimately providing WTAP with a role in vivo rather than in vitro (73,74,78). Homologues of all three proteins have been found in mammals, flies, yeast and plants (reviewed in (79)), but not in bacteria (6). Other, less-studied, subunits of the complex include vir-like m6A methyltransferase associated protein (VIRMA), RNA-binding motif protein 15/15B (RBM15/15B) and zinc finger CCCH-type containing 13 (ZC3H13) protein. Like WTAP, these accessory proteins regulate cellular m6A levels in vivo (reviewed in (80)).
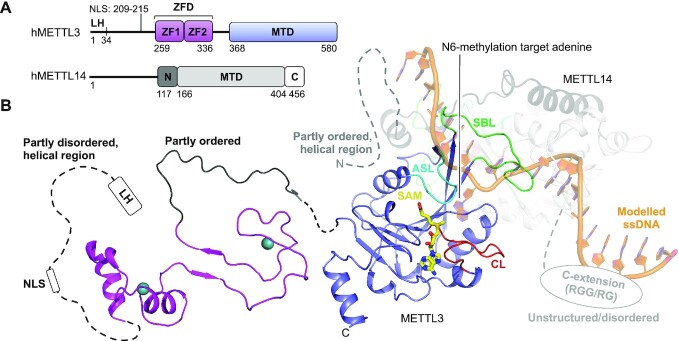
Human METTL3 and METTL14 form a complex with a 1:1 stoichiometry (74). The 64 kDa METTL3 protein covers an N-terminal extension (N-extension) with a leader helix (LH) and a nuclear localization signal (NLS) region. The N-extension is overall predicted to be mainly helical but partly disordered (81,82). This region is followed by a zinc finger domain (ZFD) of two consecutive CCCH zinc finger motifs (ZF1 and ZF2), a partly ordered linker, and a C-terminal MTase domain (MTD) (Figure 2A) (81,82). Human METTL14 of 52 kDa contains an MTD flanked by an N-extension predicted to be partly ordered and mostly helical, and an unstructured, disordered C-terminal extension (C-extension), the latter of which covers arginine/glycine (RGG/RG) motifs (Figure 2A) (81,82). The predicted disorder in the terminal extensions of both proteins explains the reported difficulty in crystallizing full-length METTL3:METTL14 (63). The smallest, stable, complex covers the METTL3 MTD and the METTL14 MTD and N-extension (76), but the METTL3 ZFD is additionally required for full-length activity (61,63,64). The crystal structure of this minimal complex has been solved in the apo form (without ligand), bound with the cofactor substrate SAM (Figure 2B) or product SAH (61–63), with the known MTase inhibitor sinefungin (PDB 6Y4G, no associated publication), or with a high-throughput screening compound (PDB 6TTP, 6TTT, 6TU1, 6TTX, 6TTV, 6TTW, no associated publication) (see Table 1). All crystal structures reveal a pseudo-symmetric heterodimer (Figure 2B) where the subunits interact extensively with residues from all structural elements (83). The tight coupling of the subunits explains why these protein are more stable together than alone in vitro and in vivo (61,75). The MTDs both display the Rossmann fold (reviewed in (84)) that superimpose (PDB 5IL1) with a root-mean square deviation (RMSD) of 2.4 Å over 285 αC-atoms, 34% sequence identity, and a high DALI Z-score of 22.9 (85) (online DALI server, pairwise comparison), highlighting their similarity in structure and sequence. The differences are attributed to three flexible loops present in each subunit, which are generally found across structurally characterized m6A RNA MTases: the catalytic loop (CL: METTL3 D395-L409, METTL14 E192-W211), the substrate binding loop (SBL: METTL3 N461-G479, METTL14 N265-T284) and the active site loop (ASL: METTL3 V507-P514, METTL14 E318-P327) (Figure 2B).
Figure 2.
Structure of the human METTL3:METTL14 complex. (A) Schematic outline of the human METTL3:METTL14 (hMETTL3:hMETTL14) complex protein domains. Crystallized parts are indicated in boxes. (B) Structure of human METTL3 ZFD (light pink) (PDB 5YZ9), SAM-bound (yellow) MTD (blue) (PDB 5IL1) complexed with METTL14 covering the N-extension (black) and MTD (grey) of human METTL14 (PDB 5IL1). The sections not included in the structures are shows as dotted lines, and their structure/order state is indicated. The catalytic loop (CL, red), the active site loop (ASL, cyan) and the substrate binding loop (SBL, green) are shown for METTL3. Single-stranded DNA (ssDNA) (orange) has been modelled from the DNA-bound m6A DNA MTase EcoP15I (PDB 4ZCF).
METTL3 is the catalytic component of the complex and accommodates the SAM cofactor. The cofactor binding pocket is located to one side of the central β-sheet and is enclosed from the sides by the catalytic and active site loop (Figure 2B). Structural analysis identifies many cofactor binding residues in METTL3, five of which are not conserved to METTL14, explaining why METTL14 does not accommodate SAM. It was suggested that the METTL14 loops, corresponding to the METTL3 catalytic and active site loop, could occlude the cofactor pocket (61,63), but the flexibility of these loops suggests that this may not be the case. The METTL3 catalytic loop includes the catalytic DPPW motif (D395-W398), conserved to m6A RNA MTases as D/N-P-P-F/W/Y (reviewed in (86)), where D395 is essential for SAM binding (62) and MTase activity (61), and W398 is thought to π–π stack with the methylated adenine base during substrate binding (61). The DPPW motif is replaced by an EPPL motif (E192-L195) in METTL14 which therefore lacks the catalytically critical D/N residue and the substrate-binding aromatic residue of the catalytic motif, explaining why this subunit has no MTase activity alone in vitro (62). Though METTL14 lacks cofactor binding or activity alone, it increases the affinity of the entire complex for the SAM cofactor by stabilizing the METTL3 structure (87). Importantly, METTL14 cooperates with the METTL3 substrate binding loop to bind RNA in a positively charged groove formed on their protein:protein interface (61–63,87) (Figure 2B), resulting in a dramatic increase of METTL3 MTase activity in vitro when METTL14 is present (62,63). The structure of the METTL14 C-extension is unknown but is rich in RGG/RG motifs that, when deleted, strongly reduce RNA binding of the entire METTL3:METTL14 complex (76). The role of METTL14 is thus three-fold in that it stabilizes the METTL3 structure, increases the overall affinity for the SAM cofactor and facilitates RNA substrate binding.
The METTL3 ZFD has been structurally characterized in solution by NMR (64). The structure shows two zinc finger motifs (ZnF1 and ZnF2) connected by a small β-sheet (Figure 2B). The structure also covers part of the partially ordered linker that connects the ZFD with the METTL3 MTD, which displays a folded-back structure (Figure 2B). The ZFD is essential for METTL3:METTL14 MTase activity (61,63) as it helps recognize the substrate DRACH consensus sequence (64,74). The recognition occurs through residues from ZnF1, ZnF2 and the connecting β-sheet, though with low overall affinity (64). The ZFD thereby functions as a target recognition domain that assists the overall complex in recognizing the DRACH-sequence for methylation. It is interesting to speculate that the poor affinity of the ZFD for the DRACH sequence could enable the entire complex to rapidly scan through many substrates for potential methylation sites, and thereby account for the high abundance of the m6A modification in mRNA.
Methylation by METLL3:METTL14 is sequence specific (31), but the complex shows little preference for particular structural features on mRNAs substrates (74). Biochemical data has mapped RNA interactions with the METTL3 ZFD and MTD and the METTL14 MTD and C-extension, but only shed little light on how the complex specifically recognizes the substrate mRNA DRACH sequence. A model of a substrate-bound complex was generated by superimposing the crystallised heterodimer with the substrate-bound m6A DNA MTase EcoP15I (61) (Figure 2B). This model fits, in part, with the mutation studies on the RNA-binding surface but does not account for interactions with the METTL3 ZFD or METTL14 C-extension. Attempts to achieve METTL3:METTL14:substrate co-crystals have so far only yielded structures without RNA (61).
The human m6A mRNA/snRNA MTase METTL16
METTL16 is a nuclear (88–90) and cytoplasmic (91) protein found in organisms from prokaryotes to eukaryotes (90) and has been functionally characterized in human (METTL16/METT10D), Caenorhabditis elegans (METT-10) (89) and Arabidopsis thaliana (FIONA1) (90). Vertebra homologues of METTL16 form m6A on hairpin 1 (hp1) in the 3′ UTR of methionine adenosyl transferase 2A (MAT2A) mRNA (66,67,92,93). The tethering of METTL16 to hp1 promotes intron splicing, leading to increased production of mature MAT2A mRNA and thus MAT2A protein (67,93). Since MAT2A synthesizes SAM, this regulation acts as a feed-back loop, ensuring optimal production of the SAM synthetase in response to low SAM levels. The underlying mechanism of METTL16-control is independent of its MTase activity (93), suggesting that METTL16 functions as—not only a writer—but also a reader in this system. In addition to hp1, METTL16 further N6-methylates hairpin 2–6 in the same mRNA in vitro (67,94) which may also be the case in vivo (92–94), though the biological relevance of this is yet unknown. METTL16 also N6-methylates A43 in the U6 spliceosomal snRNA (U6 snRNA) in vivo (92,93) and in vitro (66,93), which takes place at an early stage of U6 small nuclear ribonucleoprotein biogenesis (92). METTL16 further binds to a number of pre-mRNAs, mature mRNAs and other ncRNAs and lncRNAs (91,92), among others the cancer-associated MALAT1 lncRNA in vivo and in vitro (88,91), all of which have yet to be confirmed as METTL16 N6-methylation substrates. The E. coli homologue of METTL16 targets rRNA for N6-methylation (26), suggesting a similar target for human METTL16. Interaction between METTL16 and 18S and 28S rRNA has been shown (88,91), but lacks methylation activity data. Though only a few substrates are confirmed for METTL16, the high abundance of this protein in cells (88) indicates that the MTase may have many substrates or play alternative unexplored roles, independent of its MTase activity. Some of these roles are likely attributed to the METTL16 protein located in cytoplasm, where no biological function has yet been mapped (91).
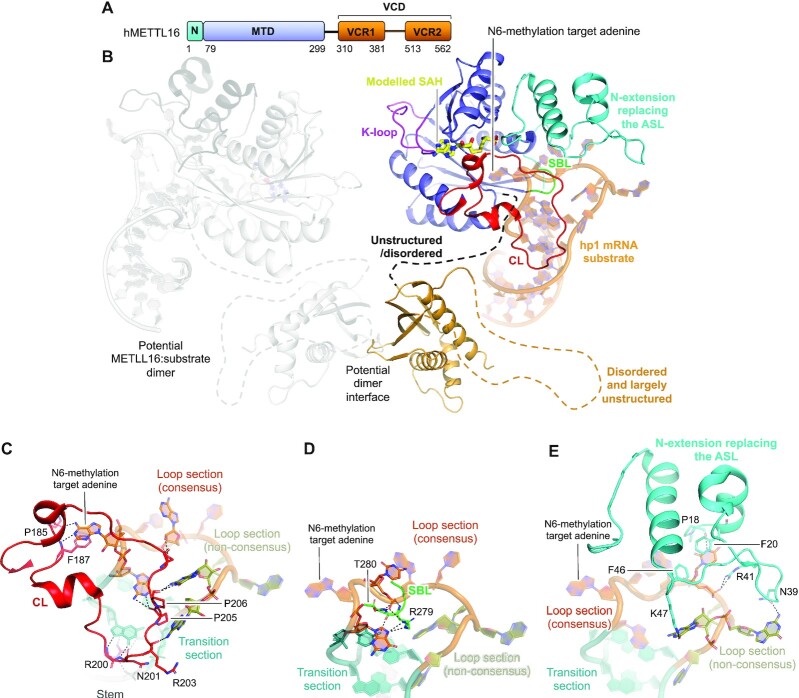
METTL16 proteins cover a small N-extension followed by an MTD. In human METTL16 and its vertebra homologues, the MTD is followed by a vertebra-conserved domain (VCD) (81,82) made up of two vertebra-conserved regions (VCR1 and VCR2), connected by a predicted disordered, largely unstructured region (Figure 3A) (68). The MTD and VCD are connected by a predicted disordered, unstructured linker that, together with the MTD and N-extension, amount to 562 residues in human METTL16 (Figure 3A). Crystal structures are available for the human METTL16 VCD (68), and the MTD in the apo form (65,66) or bound with SAH ((65,66) and PDB 2H00 with no associated publication), with a mutation-stabilized version of the in vivo relevant substrate MAT2A hp1 (Figure 3B), or with the potential substrate MAT2A hp6 (67). Furthermore, a deposited apo structure of a Pyrococcus furiosus MTase with unknown activity (PDB 3EVZ, no associated publication) could be a structural homologue of human METTL16 as it superimposes (PDB 6B91, F46-D291) with an RMSD of 1.9 Å over 221 αC-atoms, 24% sequence identity, and a DALI Z-score of 23.9 (85). However, this enzyme lacks the asparagine/aspartate residue in the m6A RNA MTase catalytic motif, which is instead replaced with an alanine residue, similar to that in METTL14.
Figure 3.
Structure of human METTL16. (A) Schematic outline of the human METTL16 (hMETTL16) protein domains. Crystallized parts are indicated in boxes. (B) Structure of the human METTL16 VCD lacking the linker region connecting VCR1 and VCR2 shown as a dotted line (light orange) (PDB 6M1U), the MTD (blue) and N-terminal extension (cyan) that replaces the active site loop (ASL), co-crystallized with the substrate hairpin, hp1, from the MAT2A mRNA (dark orange) (PDB 6DU4). SAH (yellow) has been modelled from SAH-bound METTL16 crystallized without substrate (PDB 2H00). The catalytic loop (CL, red) and the substrate binding loop (SBL, green) are shown. A second METTL16 molecule is shown in transparent grey with dimerisation through the VCD. The sections not included in the structures are shows as dotted lines, and their structure/order state is indicated. (C) The interactions between the hMETTL16 catalytic loop (CL, red) with the MAT2A mRNA substrate consensus sequence in the substrate loop (orange), non-consensus sequence in the substrate loop (green), transition section (teal) and stem (dark grey). The enzyme residues that interact with the mRNA are shown in sticks and labelled. The N6-methylated target adenine base is highlighted. Hydrogen bonds are shown in black, dashed lines. (D) As in (C), but for the substrate binding loop (SBL, green). (E) As in (C), but for the N-extension/active site loop (ASL, cyan).
The METTL16 MTD accommodates the cofactor SAM (Figure 3B). Like for other m6A RNA MTases, the cofactor binding pocket is surrounded by three protruding loops, all of which are essential for activity (66); the catalytic loop (CL: N184-G223), the substrate binding loop (SBL: Q277-T280) and the active site loop that is replaced by the N-extension (M1-P77), unique to METTL16 (Figure 3B). The long catalytic loop contains the catalytic NPPF motif (N184-F187) (95) where mutation of N184 inactivates METTL16 (66,93). Another loop (K-loop: V160-I177, Figure 3B) has been suggested to act as an intramolecular sensor of cellular SAM levels (67). This loop is found in all structurally characterized m6A RNA MTases, but a regulatory role has thus far only been proposed for METTL16. METTL16 recognizes a UACAGAGAA consensus sequence (66,92–94). The MAT2A hp1 substrate folds into a stem-loop structure in which this consensus sequence lies partly in the loop (UACAG) and partly in a transition section (AGAA) going into the stem (67). The three m6A RNA MTase loops clamp the hp1 substrate to the MTD (Figure 3B). The catalytic loop binds the consensus and non-consensus sections of the substrate loop, and the unusual base pairs formed in the transition section of the RNA (67) (Figure 3C). The numerous substrate interactions stabilize the catalytic loop fold (65,66) but, interestingly, activity rather than RNA binding is affected when the catalytic loop is deleted (66). The aromatic residue from the catalytic motif (F187), located in the catalytic loop, π- π stacks with the target adenine base (Figure 3C), explaining why F187G mutation abrogates MAT2A mRNA binding in vivo (93). The substrate binding loop interacts slightly with the transition section but mainly with the consensus part of the substrate loop (67) (Figure 3D). The substrate binding loop is short compared to that found in other m6A RNA MTases but of equal importance for activity (66). The short substrate binding loop is compensated for by the long N-extension that is positioned where the active site loop is found in other m6A RNA MTases. This N-extension contains many loops and small flexible α-helices and β-strands. It recognizes both the consensus and non-consensus part of the substrate loop (66,67) (Figure 3E), explaining why METTL16 is inactive when lacking this extension (66). Most of the interactions between the m6A RNA MTase loops and the substrate nucleotides from the consensus sequence are base-specific, explaining the METTL16 sequence specificity. The hp1 substrate stem structure is not recognized by the MTD, but is essential for proper folding of the loop/transition regions (67). As such, mutations in the stem hamper complex assembly and in vitro MTase activity (67). A stem-loop structure, similar to the one in hp1, is predicted to form for the U6 snRNA substrate (67,68) and could be similar for other, yet uncharacterized, METTL16 substrates. The importance of both fold and sequence for substrate recognition may be the reason why UACAGAGAA sequences are not widely methylated by METTL16 (66,93).
The other METTL16 domain, the VCD—deleted for an activity-dispensable disordered linker region from P411 to P508—displays a fold of small β-strands flanked by α-helices (68) (Figure 3B). The β-strands from each region make up one large β-sheet, such that the entire VCD is topologically homologous to the RNA-binding kinase-associated 1 (KA1) (68). The METTL16 MTD is monomeric (65,67), but full-length METLL16 has been described as either a monomer (67) or a homodimer formed via the VCD (65). Homodimerization was suggested to be a prerequisite for binding to the potential substrate, MALAT1 triple-helix RNA (65). Interestingly, the possibility of VCD homodimerization was not addressed when the structure of this region was described. Structural analysis of the VCD by PDBePISA (96) reveals a complex formation significance (CSS) score of 1.00 for two symmetry related molecules, implying that their interface could be biologically relevant. The molecules form a symmetrical dimer centred on stacked β-sheets (Figure 3B). The interaction is stabilized by salt bridges between K517 and E366 from each molecule and several intra-subunit hydrogen bonds. It would be interesting to further study this dimer interface and its biological relevance in the full-length METTL16 for MALAT1 mRNA binding. The VCD acts to increase the METTL16 affinity for, and activity towards, the U6 snRNA in vitro (68). When deleted, the overall MTase activity is decreased towards the hp1 substrate (67,93), which is attributed to a MAT2A mRNA splice regulation effect rather than substrate binding (68,93). Thus, the VCD plays a role not only in substrate binding but also in splice regulation.
The human and zebrafish m6Am mRNA MTase CAPAM
The enzyme responsible for the N6-methylation on the mRNA cap-adjacent Am-modified nucleotide was recently identified as the cap-specific MTase, CAPAM (/PCIF1) (47,52,54,97), making this enzyme an m6Am RNA MTase. Homologues of human CAPAM are found in higher organisms, but not in worm and yeast (53,98). Accordingly, m6Am is not detected in yeast, worm, or fly samples, but in mRNA isolated from vertebrate cells (52). CAPAM localizes to the nucleus (52,98), is ubiquitously expressed in most human tissue (98), and knock out of this enzyme induces cell sensitivity to oxidative stress (47).
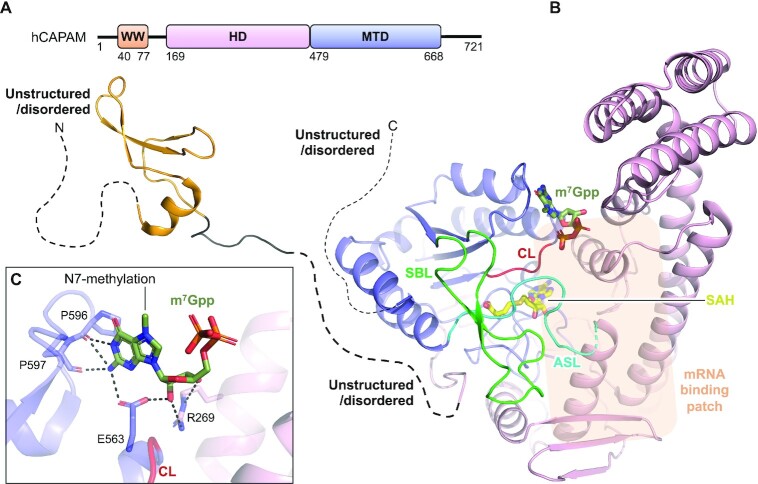
The human CAPAM (hCAPAM) protein of 81 kDa covers a small tryptophane-rich domain (WW), followed by an unstructured stretch of 98 residues, a large helical domain (HD) and a MTD. The ‘WW-linker-HD-MTD’ are together flanked by a short N- and C-extension predicted to be unstructured and disordered (81,82) (Figure 4A). A construct spanning the HD and MTD have been structurally characterized for hCAPAM or for its structural homologue zebrafish CAPAM (zfCAPAM). These structures superimpose with an RMSD <1.5 Å for all aligned Cα atoms and adopt the same fold for both domains and their relative orientation towards each other. hCAPAM and zfCAPAM are both crystallized in the apo form (no ligand) or bound with the cofactor product SAH (Figure 4B) (47). zfCAPAM is further solved in complex with SAH and either of the cap-mimicking oligos m7Gppp-A or m7Gppp-Am-G (Figure 4B), for which density is only visible for the m7Gpp entity (47).
Figure 4.
Structure of human CAPAM. (A) Schematic outline of the human CAPAM (hCAPAM) protein domains. Crystallized parts are indicated in boxes. (B) Structure of human CAPAM with the WW (orange) (PDB 2JX8), the SAH-bound (yellow) MTD (blue), and the HD (pink) (PDB 6IRW). The active site loop (ASL, cyan) that connects the MTD and HD is partly disordered in human CAPAM and is therefore modelled based on zebrafish CAPAM (PDB 6IRX). m7Gpp (dark green) is modelled from zebrafish CAPAM (PDB 6IS0). The catalytic loop (CL, red) and substrate binding loop (SBL, green) are shown. The mRNA substrate binding patch is indicated with a transparent orange box. The sections not included in the structures are shows as dotted lines, and their structure/order state is indicated. (C) Zoom of the active site showing the build part (m7Gppp) of the m7Gpp-derived ligand, co-crystallized with zebrafish CAPAM (PDB 6IS0). The enzyme residues that interact with the ligand are shown in sticks and labelled. Hydrogen bonds are shown in black, dashed lines, none of which are with the N7-methylation.
The MTD of the m6Am RNA MTase, CAPAM, presents with the classical Rossmann fold also seen for all the structurally characterized m6A RNA MTases. It contains the regular substrate binding loop (SBL: P612-N638), the catalytic loop (CL: N553-C557) and the active site loop (ASL: F479-P494), the latter of which connects the MTD with the HD (Figure 4B). The substrate binding loop shows variating structures (PDB 6IRV and 6IRX) that are unrelated to cofactor binding, but could depend on substrate interaction. The catalytic loop covers the catalytic NPPF motif (Figure 4B), the catalytic nature of which is confirmed by inactivation of CAPAM in vivo (47,52,54) and in vitro (54) by mutation of residues therein. The CAPAM HD locates near the MTD catalytic loop and covers two three-helix bundles, a four-helix bundle and two β-sheets (Figure 4B), and bears no structural resemblance to known protein folds (47). The HD contains 15% positively charged residues, most of which are surface exposed and highly conserved in animals (47). Mutation of these residues reduces MTase activity (47), suggesting a role of the HD in substrate binding. CAPAM prefers long substrates (47,52) and the large HD could accommodate such extended substrates. The enzyme shows no sequence specificity (46,47) and the recognition of these long substrates is therefore likely mainly via the RNA backbone rather than with nucleobases. The preference for longer substrates agrees with that capping of nascent mRNA occurs when these are at least 20 nts of length (99). The HD locates close to the active site and, in addition to its potential role in binding long substrates, also assists in cap-structure recognition (Figure 4B,C). The co-crystallized cap-mimicking m7Gpp entity is sandwiched in between the HD and MTD and bound by residues from both the HD and the MTD (Figure 4B,C). Activity studies suggest that CAPAM identifies the capped substrate by specifically recognizing the N7-methylation on m7G (46,47,54,97), but oddly, the structure reveals no residues that specifically recognize this modification (Figure 4B,C). CAPAM does not methylate internal Am (46,97) which is likely a combination of cap-specific recognition where removal of this element abrogates MTase activity (52) and steric hindrance disallowing a mRNA with a sequence 5′ of the methylation site to be accommodated on CAPAM. An internal m6Am modification is found at position A30 in the snRNA splice factor U2 (43) formed by sequence-specific METTL4 (44,45). A structure of METTL4 could help deduce how CAPAM gains structural specificity for methylation of the cap-adjacent Am over the internal counterpart while also lacking the substrate-sequence specificity achieved by METTL4.
A solution structure is available for the hCAPAM WW with terminal extensions (Figure 4B) (PDB 2JX8, no associated publication). This domain plays no role in the N6-methylation reaction (47), but instead functions in CAPAM’s other biological role, as the interaction point between CAPAM and the RNAPII large subunit C-terminal domain (47,98).
The human m6A rRNA MTase complex METTL5:TRMT112
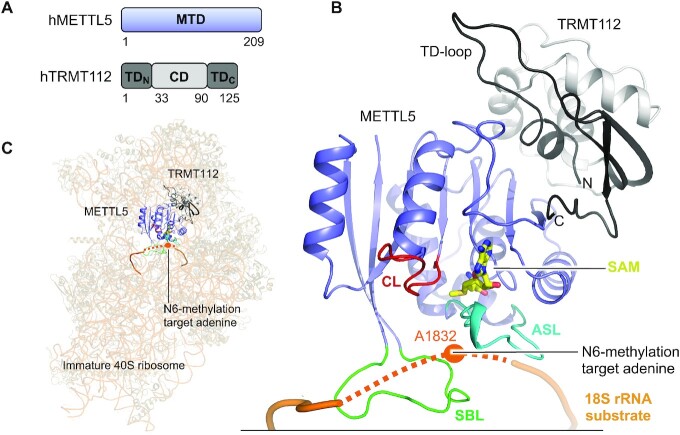
N6-methylation of A1832 (m6A1832) in human 18S rRNA was reported over 30 years ago (100,101). The modification is installed by human METTL5 (24,28) bound to the known activator human TRMT112 (28,102,103), which conveys metabolic stability to METTL5 in vivo and solubility in vitro (28). Homologues of METTL5 are present in metazoans, plants and archaea, but not in yeast and prokaryotes (24,104,105), agreeing with the lack of m6A at A1832-corresponding positions in these species (24). Consistent with a role in rRNA modification, METTL5 localizes to nucleoli (24) where ribosome biogenesis takes place. TRMT112 was first reported as a part of a complex with tRNA MTase Trm11 (106) and has since been shown to interact with many other MTases that target different components of the translation machinery i.e. rRNA, tRNA and release factors (reviewed in (107)). The many interaction partners of TRMT112 reflects its conservation in all three domains of life.
Human METLL5 is a 24 kDa protein covering only a MTD (Figure 5A). Its binding partner, human TRMT112, is a 14 kDa protein divided into two parts; a central helical domain (CD), and a terminal domain (TD) formed by residues from the N-terminus (TDN) and C-terminus (TDC) (Figure 5A). The TRMT112 TD was previously named the zinc-binding domain (ZBD) with reference to other eukaryotic Trm112 proteins that contain a Cys-Cys-Cys-Cys (CCCC) motif coordinating Zn2+ (28). The CCCC-motif is not conserved to human TRMT112 which, accordingly, does not bind Zn2+. We therefore refer to this domain as the TD rather than the ZBD.
Figure 5.
Structure of the human METTL5:TRMT112 complex. (A) Schematic outline of the human METTL5:TRMT112 (hMETTL5:hTRMT112) complex protein domains. Crystallized parts are indicated in boxes. (B) Structure of the SAM-bound (yellow) full-length, human METTL5 (blue) (PDB 6H2V) bound to human TRMT112 (grey/black) (PDB 6H2U). The active site loop (ASL, cyan), catalytic loop (CL, red) and substrate binding loop (SBL, green) are shown. Substrate 18S rRNA (dark orange) is modelled from positioning of METTL5:TRMT112 on the immature 40S ribosome (PDB 6G53). The immature ribosome structure lacks U1830-A1835 from the rRNA (not build), which is here instead indicated as a dotted line and the target base (A1832) is indicated as an orange dot. (C) Positioning of METTL5:TRMT112 into unoccupied density on the surface of the immature 40S ribosome (PDB 6G53). The unbuild part of the rRNA (U1830-A1835) is shown as a dotted line and the target base (A1832) is indicated as an orange dot.
The METTL5:TRMT112 complex has recently been co-crystallized with the SAH cofactor product, and shows a heterodimeric MTase complex associated with a 1:1 stoichiometry (28). METTL5 adopts the Rossmann fold and TRMT112 shows a β-sheet TRMT112 TD formed by folding of the protein extremities, TDN and TDC, around the helical TRMT112 CD (Figure 5A, B). The subunits (TRMT112 from PDB 6H2U, METTL5 from PDB 6H2V) interact tightly (Figure 5B), and similarly to how TRMT112 interacts with other structurally characterized MTase binding partners (103,108–112).
METTL5 is the catalytic components of the complex and binds the SAM cofactor at the end of the central β-sheet through residues from METTL5 only (Figure 5B). METTL5 contains the three m6A RNA MTase loops that surround the active site pocket: the active site loop (ASL: V16-T31), the substrate binding loop (SBL: D185-D199) and the catalytic loop (CL: N126-G136) that contains the catalytic NPPF motif (N126-F129) (Figure 5B). Alanine mutation of the first and last residues in this motif abolishes METTL5:TRMT112 MTase activity in vivo (24) emphasizing the catalytic importance of the motif. METTL5 has detectable in vitro activity alone towards full-length 18S rRNA, but the activity is increased ∼100-fold in the presence of TRMT112 (24). METTL5 and TRMT112 interact tightly via residues from all domains, together concealing a large hydrophobic patch on METTL5 from solvent. This explains why TRMT112 solubilizes METTL5 in vivo and in vitro (28), which is a role often played by TRMT112 in protein complexes (110,113–115). In other complexes, TRMT112 further activates the MTase by increasing its affinity for the SAM cofactor (110,113). TRMT112 could play a similar role with METTL5, as the proteins primarily associate through a segment in METTL5 that contains SAM-binding residues (Figure 5B), some of which are crucial for activity in a METTL5 homologue from yeast (110).
The METTL5:TRMT112 target base, A1832, is buried in the decoding pocket inside the mature 80S ribosome (116) and is therefore likely methylated prior to full maturation of the ribosomal particle. Structures of immature 40S ribosome at different maturation stages (Figure 5C) all show an only partially folded decoding pocket that exposes A1832 to the surface, making it accessible for methylation (Figure 5B,C). Interestingly, one of the later-stage immature ribosome structures (PDB 6G53) show unassigned density of ∼8 Å resolution near A1832 (28), which fits in size with METTL5:TRMT112 (Figure 5C). Modelling of the complex into the density positions the METTL5 active site facing the segment covering A1832 (28) (Figure 5B,C). The substrate binding loop in METTL5, rich in hydrophobic and positively charged RNA-interacting residues (Y190-KFHKK-K196), contacts the rRNA. METTL5:TRMT112 activity towards short 18S rRNA-derived oligos is context dependent (24,28), indicating that the MTase recognizes additional elements of the ribosome. The modelled MTase:ribosome complex proposes an interaction between the TRMT112 CD and rRNA from adjacent regions near ribosomal protein S5, which could serve as such an additional recognition point. The methylation of A1832 in a late-stage immature 40S ribosome places m6A1832 formation as one of the last steps in 40S maturation. It would be interesting to further investigate the immature 40S ribosome using cryoEM, with a focus on METTL5:TRMT112, with the aim to elucidate the binding mode of this MTase to its substrate.
The human m6A rRNA MTase ZCCHC4
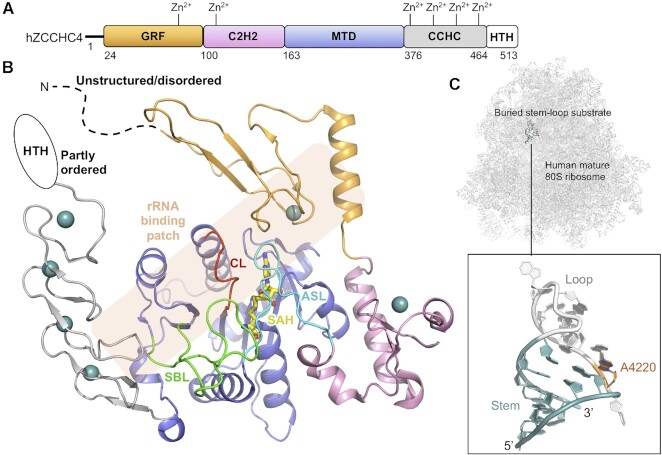
The m6A4220 (alternatively referred to as m6A4190) modification is present in all human 28S rRNA (23,117), and its formation is catalysed by ZCCHC4 (23,27,69). ZCCHC4 is conserved to multicellular organisms, but absent in single-cell eukaryotes, consistent with that yeast lacks m6A-modifications on rRNA (23,27). Human ZCCHC4 localizes to nucleoli where ribosome assembly and maturation take place (23). Consistent with having an rRNA substrate, ZCCHC4 forms direct or RNA-mediated interactions with a number of RNA-binding proteins involved in ribosomal biogenesis or RNA metabolism in nucleoli (23). ZCCHC4 is also implicated in epigenetic gene silencing (118) which appears unrelated to its MTase activity. There are, however, currently no indications of alternative ZCCHC4-substrates (28) that could explain this link.
The 59 kDa monomeric human ZCCHC4 protein contains an MTD and three types of ZFDs (69): a (Gly-Arg-Phe)-type ZFD (GRF), a Cys-Cys-His-His ZFD (C2H2) and a Cys-Cys-His-Cys ZFD (CCHC). These domains are organized as GRF-C2H2-MTD-CCHC, together flanked by an N-extension predicted to be unstructured and a C-extension predicted to cover a partly ordered helix-turn-helix (HTH) motif (Figure 6A) (81,82). ZCCHC4 has been crystallized without its terminal extensions and bound with the SAH cofactor product (69) (Figure 6B). This construct retains in vitro activity comparable to full-length protein, agreeing with that the terminal extensions show no evolutionary conservation (69). The MTD displays a Rossmann fold and the ZFDs display significantly different folds from one another, coordinating either one (GRF and C2H2) or four (CCHC) Zn2+ ions (Figure 6B). The GRF and C2H2 flank one side of the MTD cofactor binding pocket and interact extensively with the MTD (Figure 6B). The CCHC binds to the C-terminal region of the MTD (Figure 6B).
Figure 6.
Structure of human ZCCHC4. (A) Schematic outline of the human ZCCHC4 (hZCCHC4) protein domains. Crystallized parts are indicated in boxes. (B) Structure of the SAM-bound (yellow) human ZCCHC4 showing the GRF (grey), C2H2 (pink), MTD (blue) and CCHC (light orange) domains (PDB 6UCA). Zn2+ ions are shown in teal-coloured spheres. The catalytic loop (CL, red), substrate binding loop (SBL, green) and the active site loop (ASL, cyan) connecting C2H2 with the MTD are shown. The rRNA substrate binding patch is indicated with a transparent orange box. The sections not included in the structures are shows as dotted lines, and their structure/order state is indicated. (C) The mature human 80S ribosome with an insert highlighting the ZCCHC4 substrate stem-loop structure from 28S rRNA with target adenine (A4220, dark orange) placed between the stem (teal) and loop (grey) (PDB 6EK0).
Like other m6A RNA MTases, ZCCHC4 contains three loops around the active site; the active site loop (ASL: L161-F175) here connecting the C2H2 with the MTD, the substrate binding loop (SBL: L334-P357) and the catalytic loop (CL: T275-L282) encompassing the m6A RNA MTase catalytic DPPF motif (D276-F279) (Figure 6B). The catalytic nature of this motif is confirmed by ZCCHC4 inactivation upon D276A mutation (23). For all three loops, both the specific loop residues and the overall loop fold is important for catalysis (69). The fold of each loop is stabilized by an network of internal hydrogen bonds, but also external interactions that link both the catalytic loop and the active site loop to the substrate binding loop. Breakage of the external interactions by mutation either nearly abolish MTase activity or enhances it 1.3-fold, without affecting substrate-binding (69), pointing at a loop-driven, intricate control of catalysis. In agreement with a role for the loops in catalysis, the SAM-bound ZCCHC4 structure shows a cofactor binding pocket that is enclosed by the substrate binding loop and the active site loop (Figure 6B), where structural rearrangements are required to access the cofactor for methylation. This suggests a mechanism where cofactor binding is coupled with RNA substrate recognition, controlled by the loops.
The RNA-binding mode for ZCCHC4 has not been structurally determined, but mutation studies suggest a collaborative effort of, at least, the MTD, GRF and CCHC (69). In the mature 80S ribosome, the target adenine base, A4220, is located in a transition section between a stem and a loop in a buried stem-loop structure (A4205-G4228) (Figure 5C). ZCCHC4 requires an intact stem structure for in vitro activity but not any specific bases herein (69). The adenine base located 5′ of the target adenine base (5′-UAACG-3′, A = target base) is specifically recognized by ZCCHC4 (27). In the stem-loop structure extracted from the 80S ribosome (116) (Figure 5C), this, and the target adenine, are both buried inside the stem-loop and it would therefore require a flip of both for ZCCHC4 to recognize the bases rather than the backbone. This, and the fact that the stem-loop is buried in the ribosome, is consistent with a hypothesis that m6A4220 is formed on immature pre-60S particles. In vivo, ZCCHC4 also associates with another 28S rRNA region (G1854-C1913) (27) located adjacent in space to the A4220-encompassing stem-loop structure in the mature 80S ribosome (116), and potentially therefore also in immature particles. It is possible that interactions with this region is mediated by the ZFDs of ZCCHC4. A structure of ZCCHC4 bound to the immature ribosomal particle would shed light on the potential simultaneous recognition of this region and the substrate stem-loop structure.
The bacterial m6A rRNA MTase RlmJ
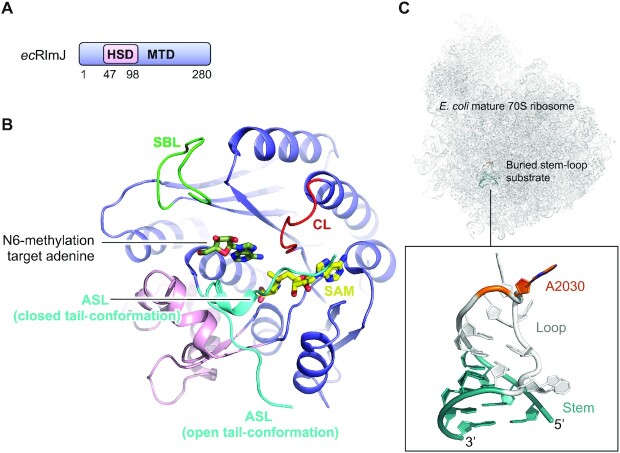
The N6-methylation of A2030 in E. coli 23S rRNA (119,120) is formed by RlmJ both in vivo and in vitro (25). Orthologs of E. coli RlmJ (ecRlmJ) can be found in α-, β- and γ-proteobacteria (25,121,122) E. coli. Inactivation of RlmJ has no particular impact on the proteome or on ribosomal assembly in general (25), but lowers the competitive fitness at long-term growth in the stationary phase (123).
Similar to other m6A RNA MTases, monomeric ecRlmJ of 32 kDa comprises an MTD with a Rossmann fold (124) but has an additional 52 residues long helical subdomain (HSD: R47-P98) inserted between strand β1 and helix α2 of the core Rossman fold (70,71) (Figure 7A,B). The ecRlmJ protein has been crystallized in the apo form (70), with the cofactor SAM (70) (Figure 7B), and with the cofactor product SAH either alone (71) or in combination with the target adenine base analogue AMP (70). ecRlmJ has also been co-crystallized with two bisubstrate analogues (BA) (71) (Figure 7B), each resembling a methylation intermediate between the SAM cofactor and the target adenine, in which these moieties are covalently linked at the point of methylation. An apo structure is also available for an ecRlmJ homologue from Legionella pneumophila (PDB 2OO3, no associated publication) that shares 40% sequence identity with ecRlmJ (PDB 4BLU) and superimposes with an RMSD of 1.6 Å over 279 αC-atoms and a DALI Z-score of 38.2 (85). The biological function of this homologue, however, has yet to be shown.
Figure 7.
Structure of E. coli RlmJ. (A) Schematic outline of the E. coli RlmJ (ecRlmJ) protein domains. Crystallized parts are indicated in boxes. (B) Structure of the ecRlmJ SAM-bound (yellow) MTD (blue) with the inserted HSD (pink) (PDB 4BLV), crystallized with a closed tail-conformation of the active site loop (ASL, cyan). The ASL is further shown from apo RlmJ (PDB 4BLU) in the open tail-conformation. The target adenine (dark green) is modelled from RlmJ co-crystallised with a bisubstrate molecule (BA4) (PDB 6QDX, chain B). The catalytic loop (CL, red) and substrate binding loop (SBL, green) are shown. (C) The mature E. coli 70S ribosome with an insert highlighting the RlmJ substrate stem-loop structure from 23S rRNA with target adenine (A2030, dark orange) placed in the loop (grey) followed by a stem (teal) (PDB 7K00).
RlmJ contains the m6A RNA MTase loops; the substrate binding loop (SBL: V226-T236), the catalytic loop (CL: D164-K170) with the catalytic DPPY motif (D164-Y167) the nature of which is confirmed by loss of MTase activity in vitro upon D164A mutation (70), and the active site loop made up of the entire N-terminal tail (ASL: M1-G11) (Figure 7B). This tail has many homology-conserved residues pointing to a functional role (70). The tail is hugely flexible (70,71) (Figure 7B), and in one position (the closed tail-conformation) it helps form the active site and bind the cofactor (70). Mutation of tail residues results in reduction or loss of MTase activity in vitro (70), suggesting a role in rRNA substrate binding. The RlmJ substrate binding loop contains positively charged residues and is partly disordered in some crystal structures but may be stabilized in the presence of substrate. The substrate binding loop locates near the RlmJ-specific HSD insert (Figure 7B). The HSD is positioned on the RlmJ MTD in a similar location as the N-extension of METTL16 on the MTD. In METTL16, the N-extension binds the RNA substrate, and the HSD could play a similar role in RlmJ. The HSD and substrate binding loop could therefore, together, form an RNA binding platform on RlmJ.
RlmJ targets the A2030 base of 23S rRNA, which is located in a buried stem-loop structure in helix 72 of the 50S ribosomal subunit, and is further buried in the interface between subunits in the mature ribosome (125) (Figure 7C). A2030 is therefore m6A modified in either nascent 23S rRNA (25) or in 23S rRNA at an early stage of 50S ribosomal subunit assembly (25,126). RlmJ resists co-crystallization with the stem-loop RNA, but has been co-crystallized with two bisubstrate analogues, BA2 or BA4 (71). These analogues resemble SAM and the target adenine, covalently linked via the methyl-receiver and -donor atoms, each with a different linker. The linker in BA4 allows positioning of the target adenine in a correct orientation for catalysis, providing clues into the methylation mechanism (Figure 7B). A model including SAH, the target adenine and the active site loop in the closed conformation reveals that the nucleotide is sandwiched in between hydrophobic and positively charged residues from the active site loop and core MTD, some of which are shown to be crucial for in vitro activity (70). In m6A RNA MTases, the hydrophobic residue from the catalytic motif stacks with the target adenine (61,67). In apo RlmJ, this residue (Y167) is rotated away from the active site pocket to a similar position as this residue in apo METTL16. In METTL16, the residue rotates a full 180° upon RNA binding (65) to stack with the target adenine. A similar rotation could be expected in RlmJ when it binds substrate RNA, but is not observed in the RlmJ:BA4 structure (71), suggesting that this rearrangement is induced by binding to longer substrates. In METTL3:METTL14, the aromatic residue from the catalytic motif is a tryptophane, and, interestingly, RlmJ contains such a residue (W195) already in position for stacking with the target adenine, which could alleviate the need for Y167 to rotate. RlmJ was also co-crystallized with AMP, which is, however, not bound in a relevant position for catalysis (70), highlighting that an approach of covalent linkage to the cofactor is required if only the target adenine is investigated.
DISCUSSION
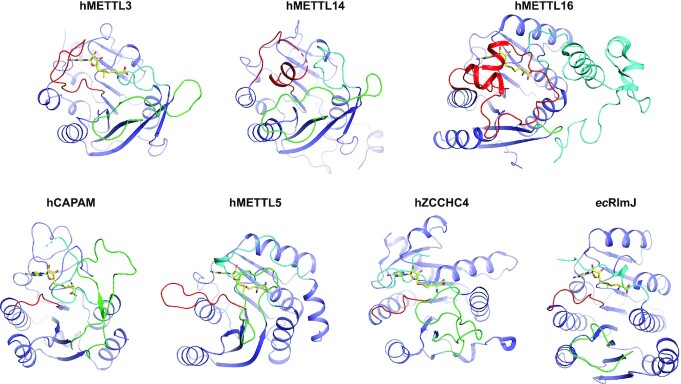
MTases constitute one of the largest enzyme classes in nature, but the cellular substrates and functions of over 200 human candidates remain unknown (28). Here, we review the structurally and functionally characterized m6A RNA MTases. These enzymes target either mRNA or rRNA, or for METTL16 both mRNA and snRNA. Despite their differences in RNA substrates, these MTase enzymes all display the Rossmann fold for their catalytic MTD, and all functionally, but not yet structurally, characterized m6A RNA MTases, are likewise suggested to belong to the Rossmann fold superfamily of proteins. The m6A RNA MTases display low sequence identity, but their MTDs superimpose with good (low) RMSDs between 2.4–4.0 Å (Table 2), demonstrating substantial structural similarities. The cofactor binding pocket and the conserved catalytic motif (DPPW in METTL3, NPPF in METTL16, CAPAM, and METTL5, DPPF in ZCCH4 and DPPY in RlmJ) superimpose particularly well. Superimposition of either of the MTDs with the substrate bound METTL16 reveals steric clashes with the RNA for all enzymes, indicating that their specific substrate recognition differs from that of METTL16. This agrees with that the target adenine in RlmJ is oriented differently to the same base in METTL16 (rotated 180°), but similarly to this base in m6A DNA MTases (71).
Table 2.
Comparison of the methyltransferase domain (MTD) of structurally and functionally characterized methyltransferases (MTases)
| MTase | PDB | MTD residues | Z-score | RMSD (Å) | αC-atoms | Seq. iden. (%) |
|---|---|---|---|---|---|---|
| hMETTL3 | 5IL1 | 369–570 | ||||
| Comparison with the METTL3 MTD | ||||||
| hMETTL14 | 5IL1 | 166–402 | 22.9 | 2.4 | 236 | 34 |
| hMETTL16 | 6DU4 | 78–299 | 3.4 | 3.5 | 213 | 9 |
| hCAPAM | 6IRW | 479–668* | 2.9 | 4.6 | 188 | 7 |
| hMETTL5 | 6H2V | 2–209 | 3.7 | 4.0 | 209 | 9 |
| hZCCHC4 | 6UCA | 163–375 | 2.5 | 3.7 | 214 | 5 |
| ecRlmJ | 4BLV | 1–46+99–280 | 3.2 | 3.5 | 229 | 12 |
*L483-L489 modelled from zfCAPAM (PDB 6IRX). Seq. iden.: sequence identity. Values are deduced from DALI comparison (online DALI server, pairwise comparison) (85). ‘αC-atoms’ denotes the number of αC atoms that have been superimposed to obtain the stated root mean square deviation (RMSD).
Each structurally characterized m6A RNA MTase contains three m6A RNA MTase loops surrounding the active site pocket; the active site loop, the substrate binding loop and the catalytic loop, the latter of which encompasses the conserved catalytic D/N-P-P-F/W/Y motif (Figure 8). These m6A RNA MTase loops play different, but distinct, roles in fine tuning catalysis and RNA-binding. Additional structures of substrate-bound m6A RNA MTases will help further understand the mechanistic interplay between these loops. To allow coherence in the field going forward, we suggest that any future publication of m6A RNA MTase structure(s) address the role of these central loops in enzyme function.
Figure 8.
The m6A RNA MTase loops. The active site loop (cyan), catalytic loop (red) and substrate binding loop (green) is shown on each structurally and functionally characterized m6A RNA MTase described herein. For hMETTL16, the active site loop covers the entire N-terminal extension. For comparison, the MTDs (blue) are positioned with similar cofactor (yellow) orientation. hMETTL14, without cofactor, is oriented as hMETTL3. ec: E. coli, h: human.
The MTDs of the structurally characterized m6A RNA MTases require either a binding partner (METTL5), an auxiliary domain (CAPAM, RlmJ and ZCCHC4), or both (METTL3) to gain their full MTase activity. It would be of strong interest to perform an in-depth bioinformatic analysis of all structurally uncharacterized MTases to pinpoint if auxiliary domains, such as the structurally characterized helical domains and/or zinc-finger domains, are typical additions to the MTD for m6A RNA MTases, and/or if such domains are found generally across MTases. It would also be of interest to study if the currently functionally uncharacterized MTases are, in general, predicted to utilize binding partners to gain MTase activity.
The here-described MTases display significantly different RNA sequence and/or fold recognition specificity. METTL3:METTL14 recognizes a five-nucleotide sequence (DRACH, D = A, G or U, R = A or G, and H = A, C or U) in single stranded RNA, whereas METTL16 recognizes a nine-nucleotide sequence (UACAGAGAA) plus the fold of a stem-loop substrate, explaining why METTL16 does not operate as broadly as METTL3:METTL14 in vivo. Substrate fold, rather than sequence, appears as the main recognition-driver for the remaining MTases. Interestingly, both METTL16, ZCCHC4 and RlmJ recognise a stem-loop RNA, in which the integrity of the stem is important. METTL16 is co-crystallized with a stem-loop substrate that shows no association between enzyme and the substrate stem. The stem, however, bears importance as it shapes the part of the RNA in which the target adenine resides, for optimal positioning in the METTL16 catalytic pocket. It is possible that the stem plays a similar role in shaping and positioning in substrates of ZCCHC4 and RlmJ, rather than directly associating with the MTase.
It can be a strenuous process to obtain crystal structures of proteins with RNA substrates due to the limited number of—often purely electrostatic—interactions between the two components. This is of particular difficulty when using short oligonucleotides encompassing just the sequence binding near, and in, the active site (61) such as a DRACH-sequence RNA for METTL3:METTL14. Recently, we co-crystallized RlmJ with SAM-analogues, covalently linked to the target adenine in substrate RNA, which predicted the substrate binding mode and facilitated mechanistic studies (71). This approach could also serve to deduce the complex between e.g. METTL3:METTL14 and short RNAs encompassing the DRACH sequence. CryoEM is a yet unexplored territory in the field of MTase:substrate complexes, but a cryoEM density map of an immature 40S subunit display unassigned density that could correspond to METTL5:TRMT112 (28). To increase the density of the MTase complex, further studies on this could advantageously be carried out using a recently published method that allows association of surface-binding proteins on ribosomes when these are not initially isolated together from cells (127). This method results in increased the occupancy of the enzyme at the surface of ribosomes and permits deduction of even low-affinity complexes.
Missense mutation in METTL3, METTL14 or METTL16 is directly linked to human cancer (63,64,67,128–132). Missense mutation or homozygous deletion in the METTL5 gene is associated with intellectual developmental disorder autosomal recessive 72 (133,134), expression levels of METTL5 and TRMT112 are upregulated in tumours, particularly solid tumours (24), and a potential role of ZCCHC4 in cancer has been discussed (27,28). Large efforts are currently being made to develop inhibitors against these m6A RNA MTases (reviewed in (135)). Our recently synthesized bisubstrate analogues that combine the structural features of the substrate as well as cofactor, showed potential as inhibitors on a preliminary basis (71). Similar bisubstrate-type molecules have been designed against METTL3 and were suggested to serve as inhibitor design templates (136). It is our hope that further structural studies of these m6A RNA MTases, bound with their RNA substrates and/or protein binding partners, will assist the ongoing efforts in drug development targeting m6A MTases.
Contributor Information
Stephanie Oerum, Expression Génétique Microbienne, UMR 8261, CNRS, Université de Paris, Institut de Biologie Physico-Chimique (IBPC), 75005 Paris, France.
Vincent Meynier, Expression Génétique Microbienne, UMR 8261, CNRS, Université de Paris, Institut de Biologie Physico-Chimique (IBPC), 75005 Paris, France.
Marjorie Catala, Expression Génétique Microbienne, UMR 8261, CNRS, Université de Paris, Institut de Biologie Physico-Chimique (IBPC), 75005 Paris, France.
Carine Tisné, Expression Génétique Microbienne, UMR 8261, CNRS, Université de Paris, Institut de Biologie Physico-Chimique (IBPC), 75005 Paris, France.
FUNDING
Agence Nationale de la Recherche [ANR-19-CE07-0028 to C.T.]; ANR LabEX DYNAMO [ANR-11-LABX-0011]. Funding for open access charge: ANR-19-CE07-0028.
Conflict of interest statement. None declared.
REFERENCES
- 1. Desrosiers R., Friderici K., Rottman F.. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 1974; 71:3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wei C., Gershowitz A., Moss B.. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature. 1975; 257:251–253. [DOI] [PubMed] [Google Scholar]
- 3. Zhao B.S., Roundtree I.A., He C.. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2016; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yue H., Nie X., Yan Z., Weining S.. N6-methyladenosine regulatory machinery in plants: composition, function and evolution. Plant Biotechnol. J. 2019; 17:1194–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lence T., Soller M., Roignant J.Y.. A fly view on the roles and mechanisms of the m6A mRNA modification and its players. RNA Biol. 2017; 14:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deng X., Chen K., Luo G.Z., Weng X., Ji Q., Zhou T., He C.. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015; 43:6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartosovic M., Molares H.C., Gregorova P., Hrossova D., Kudla G., Vanacova S.. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017; 45:11356–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., Zheng H., Klungland A., Yan W.. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 2017; 115:E325–E333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng Q., Hou J., Zhou Y., Li Z., Cao X.. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017; 18:1094–1103. [DOI] [PubMed] [Google Scholar]
- 10. Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millán-Zambrano G., Robson S.C., Aspris D., Migliori V., Bannister A.J., Han N.et al.. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017; 552:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou J., Wan J., Shu X.E., Mao Y., Liu X.M., Yuan X., Zhang X., Hess M.E., Brüning J.C., Qian S.B.. N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cell. 2018; 69:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coots R.A., Liu X.M., Mao Y., Dong L., Zhou J., Wan J., Zhang X., Qian S.B.. m6A Facilitates eIF4F-Independent mRNA Translation. Mol. Cell. 2017; 68:504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vu L.P., Pickering B.F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., Chou T., Chow A., Saletore Y., Mackay M.et al.. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017; 23:1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., Kadiwala J., Hubner N.C., De Los Mozos I.R., Sadée C.et al.. The SMAD2/3 interactome reveals that TGFβ controls m 6 A mRNA methylation in pluripotency. Nature. 2018; 555:256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clancy M.J. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002; 30:4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agarwala S.D., Blitzblau H.G., Hochwagen A., Fink G.R.. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLos Genet. 2012; 8:e1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bodi Z., Bottley A., Archer N., May S.T., Fray R.G.. Yeast m6A methylated mRNAs are enriched on translating ribosomes during meiosis, and under rapamycin treatment. PLoS One. 2015; 10:e0132090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang H., Weng H., Chen J.. m6A Modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020; 37:270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choi Y.C., Busch H.. Modified nucleotides in T1 RNase oligonucleotides of 18S ribosomal RNA of the Novikoff hepatoma. Biochemistry. 1978; 17:2551–2560. [DOI] [PubMed] [Google Scholar]
- 20. Sharma S., Lafontaine D.L.J.. View From A Bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015; 40:560–575. [DOI] [PubMed] [Google Scholar]
- 21. Piekna-Przybylska D.D., Decatur W.A., Fournier M.J.. The 3D rRNA modification maps database: With interactive tools for ribosome analysis. Nucleic Acids Res. 2008; 36:178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sergiev P.V., Aleksashin N.A., Chugunova A.A., Polikanov Y.S., Dontsova O.A.. Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 2018; 14:226–235. [DOI] [PubMed] [Google Scholar]
- 23. Pinto R., Vågbø C.B., Jakobsson M.E., Kim Y., Baltissen M.P., O’Donohue M.F., Guzmán U.H., Małecki J.M., Wu J., Kirpekar F.et al.. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020; 48:830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen H., Liu Q., Yu D., Natchiar K., Zhou C., Hsu C., Hsu P.-H., Zhang X., Klaholz B., Gregory R.I.et al.. METTL5, an 18S rRNA-specific m6Amethyltransferase, modulates expression of stress response genes. 2020; bioRxiv doi:10 June 2020, preprint: not peer reviewed 10.1101/2020.04.27.064162. [DOI]
- 25. Golovina A.Y., Dzama M.M., Osterman I.A., Sergiev P.V., Serebryakova M.V., Bogdanov A.A., Dontsova O.A.. The last rRNA methyltransferase of E. coli revealed: The yhiR gene encodes adenine-N6 methyltransferase specific for modification of A2030 of 23S ribosomal RNA. RNA. 2012; 18:1725–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sergiev P.V., Serebryakova M.V., Bogdanov A.A., Dontsova O.A.. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol. 2008; 375:291–300. [DOI] [PubMed] [Google Scholar]
- 27. Ma H., Wang X., Cai J., Dai Q., Natchiar S.K., Lv R., Chen K., Lu Z., Chen H., Shi Y.G.et al.. N6-methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019; 15:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Tran N., Ernst F.G.M., Hawley B.R., Zorbas C., Ulryck N., Hackert P., Bohnsack K.E., Bohnsack M.T., Jaffrey S.R., Graille M.et al.. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019; 47:7719–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saneyoshi M., Harada F., Nishimura S.. Isolation and characterization of N6-methyladenosine from Escherichia coli valine transfer RNA. BBA Sect. Nucleic Acids Protein Synth. 1969; 190:264–273. [DOI] [PubMed] [Google Scholar]
- 30. Kowalak J.A., Dalluge J.J., McCloskey J.A., Stetter K.O.. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994; 33:7869–7876. [DOI] [PubMed] [Google Scholar]
- 31. Linder B., Grozhik A.V, Olarerin-george A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2016; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shimba S., Bokar J.A., Rottman F., Reddy R.. Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nucelar RNA by HeLa cell extract in vitro. Nucleic Acids Res. 1995; 23:2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gu J., Patton J.R., Shimba S., Reddy R.. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA. 1996; 2:909–918. [PMC free article] [PubMed] [Google Scholar]
- 34. Bringmann P., Ltihrmann R.. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 1987; 213:309–315. [DOI] [PubMed] [Google Scholar]
- 35. Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F.. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015; 519:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berulava T., Rahmann S., Rademacher K., Klein-Hitpass L., Horsthemke B.. N6-adenosine methylation in MiRNAs. PLoS One. 2015; 10:e0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F.. HNRNPA2B1 is a mediator of m6a-dependent nuclear RNA processing events. Cell. 2015; 162:1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen T., Hao Y.J., Zhang Y., Li M.M., Wang M., Han W., Wu Y., Lv Y., Hao J., Wang L.et al.. M6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015; 16:289–301. [DOI] [PubMed] [Google Scholar]
- 39. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M.et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 40. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Recasens M., Gadea M., García B., Panyella D., Ginestar J., Liu N., Parisien M., Dai Q., Zheng G., He C.et al.. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013; 19:1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R.. M6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016; 537:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shibata H., Ro-Choi T.S., Reddy R., Choi Y.C., Henning D., Busch H.. The primary acid sequence of nuclear U-2 ribonucleic. J. Biol. Chem. 1975; 250:3909–3920. [PubMed] [Google Scholar]
- 44. Goh Y.T., Koh C.W.Q., Sim D.Y., ROca X., Goh W.S.S. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 2020; 48:9250–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen H., Gu L., Orellana E.A., Wang Y., Guo J., Liu Q., Wang L., Shen Z., Wu H., Gregory R.I.et al.. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res. 2020; 30:544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keith J.M., Ensinger M.J., Moss B.. HeLa cell RNA(2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 1978; 253:5033–5039. [PubMed] [Google Scholar]
- 47. Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O.et al.. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II–associated methyltransferase. Science (80-.). 2019; 363:eaav0080. [DOI] [PubMed] [Google Scholar]
- 48. Banerjee A.K. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 1980; 44:175–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi H., Wei J., He C.. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019; 74:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q.et al.. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017; 541:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., Shi H., Cui X., Su R., Klungland A.et al.. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018; 71:973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sendinc E., Valle-Garcia D., Dhall A., Chen H., Henriques T., Navarrete-Perea J., Sheng W., Gygi S.P., Adelman K., Shi Y.. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell. 2019; 75:620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hirose Y., Iwamoto Y., Sakuraba K., Yunokuchi I., Harada F., Ohkuma Y.. Human phosphorylated CTD-interacting protein, PCIF1, negatively modulates gene expression by RNA polymerase II. Biochem. Biophys. Res. Commun. 2008; 369:449–455. [DOI] [PubMed] [Google Scholar]
- 54. Boulias K., Toczydłowska-Socha D., Hawley B.R., Liberman N., Takashima K., Zaccara S., Guez T., Vasseur J.J., Debart F., Aravind L.et al.. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell. 2019; 75:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Golovina A.Y., Sergiev P.V., Golovin A.V., Serebryakova M.V., Demina I., Govorun V.M., Dontsova O.A.. The yfiC gene of E. coli encodes an adenine-N6 methyltransferase that specifically modifies A37 of tRNA1 Val(cmo5UAC). RNA. 2009; 15:1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kimura S., Miyauchi K., Ikeuchi Y., Thiaville P.C., De Crécy-Lagard V., Suzuki T.. Discovery of the β-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 2014; 42:9350–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O’Farrell H.C., Scarsdale J.N., Rife J.P.. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli. J. Mol. Biol. 2004; 339:337–353. [DOI] [PubMed] [Google Scholar]
- 58. Schluckebier G., Zhong P., Stewart K.D., Kavanaugh T.J., Abad-Zapatero C.. The 2.2 Å structure of the rRNA methyltransferase ErmC’ and its complexes with cofactor and cofactor analogs: Implications for the reaction mechanism. J. Mol. Biol. 1999; 289:277–291. [DOI] [PubMed] [Google Scholar]
- 59. Guja K.E., Venkataraman K., Yakubovskaya E., Shi H., Mejia E., Hambardjieva E., Karzai A.W., Garcia-Diaz M.. Structural basis for S-adenosylmethionine binding and methyltransferase activity by mitochondrial transcription factor B1. Nucleic Acids Res. 2013; 41:7947–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu L., Petros A.M., Schnuchel A., Zhong P., Severin J.M., Walter K., Holzman T.F., Fesik S.W.. Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide- streptogramin antibiotic resistance. Nat. Struct. Mol. 1997; 4:483–489. [DOI] [PubMed] [Google Scholar]
- 61. Śledź P., Jinek M.. Structural insights into the molecular mechanism of the m6A writer complex. Elife. 2016; 5:e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C.et al.. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016; 534:575–578. [DOI] [PubMed] [Google Scholar]
- 63. Wang P., Doxtader K.A., Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016; 63:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang J., Dong X., Gong Z., Qin L.Y., Yang S., Zhu Y.L., Wang X., Zhang D., Zou T., Yin P.et al.. Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase. Protein Cell. 2019; 10:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ruszkowska A., Ruszkowski M., Dauter Z., Brown J.A.. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 2018; 8:5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mendel M., Chen K.M., Homolka D., Gos P., Pandey R.R., McCarthy A.A., Pillai R.S.. Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 2018; 71:986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Doxtader K.A., Wang P., Scarborough A.M., Seo D., Conrad N.K., Nam Y.. Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol. Cell. 2018; 71:1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aoyama T., Yamashita S., Tomita K.. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 2020; 48:5157–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ren W., Lu J., Huang M., Gao L., Li D., Greg Wang G., Song J.. Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA. Nat. Commun. 2019; 10:5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Punekar A.S., Liljeruhm J., Shepherd T.R., Forster A.C., Selmer M.. Structural and functional insights into the molecular mechanism of rRNA m6A methyltransferase RlmJ. Nucleic Acids Res. 2013; 41:9537–9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Oerum S., Catala M., Atdjian C., Brachet F., Ponchon L., Barraud P., Iannazzo L., Droogmans L., Braud E., Ethève-Quelquejeu M.et al.. Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites. RNA Biol. 2019; 16:798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M.. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997; 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 73. Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S.et al.. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X.et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C.. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014; 16:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schöller E.V.A., Weichmann F., Treiber T., Ringle S., Treiber N., Flatley A., Feederle R., Bruckmann A., Meister G.. Interactions, localization, and phosphorylation of the m6A generating METTL3–METTL14–WTAP complex. RNA. 2018; 24:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Little N.A., Hastie N.D., Davies R.C.. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum. Mol. Genet. 2000; 9:2231–2239. [DOI] [PubMed] [Google Scholar]
- 78. Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D.et al.. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014; 8:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Huang J., Yin P.. Structural insights into N6-methyladenosine (m6A) modification in the transcriptome. Genomics Proteomics Bioinforma. 2018; 16:85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang Y., Hsu P.J., Chen Y.S., Yang Y.G.. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018; 28:616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999; 292:195–202. [DOI] [PubMed] [Google Scholar]
- 82. Ward J.J., McGuffin L.J., Bryson K., Buxton B.F., Jones D.T.. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004; 20:2138–2139. [DOI] [PubMed] [Google Scholar]
- 83. Wallace A.C., Laskowski R.A., Thornton J.M.. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein. Eng. 1995; 8:127–134. [DOI] [PubMed] [Google Scholar]
- 84. Schubert H.L., Blumenthal R.M., Cheng X.. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003; 28:329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Holm L., Laakso L.M.. Dali server update. Nucleic Acids Res. 2016; 44:W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bheemanaik S., Reddy Y.V.R., Rao D.N.. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem. J. 2006; 399:177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang X., Huang J., Zou T., Yin P.. Human m6A writers: Two subunits, 2 roles. RNA Biol. 2017; 14:300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Brown J.A., Kinzig C.G., Degregorio S.J., Steitz J.A.. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:14013–14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dorsett M., Westlund B., Schedl T.. METT-10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans. Genetics. 2009; 183:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kim J., Kim Y., Yeom M., Kim J.H., Nam H.G.. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell. 2008; 20:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nance D.J., Satterwhite E.R., Bhaskar B., Misra S., Carraway K.R., Mansfield K.D.. Characterization of METTL16 as a cytoplasmic RNA binding protein. PLoS One. 2020; 15:e0227647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., Sloan K.E., Bohnsack M.T.. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017; 18:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K.. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017; 169:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., Kumagai S., Ochiai K., Suzuki T., Igarashi K.. S-adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mrna degradation involving METTL16 and YTHDC1. Cell Rep. 2017; 21:3354–3363. [DOI] [PubMed] [Google Scholar]
- 95. Malone T., Blumenthal R.M., Cheng X.. Structure-guided analysis reveals nine sequence motifs conserved among dna amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995; 253:618–632. [DOI] [PubMed] [Google Scholar]
- 96. Krissinel E., Henrick K.. Interference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 97. Sun H., Zhang M., Li K., Bai D., Yi C.. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019; 29:80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fan H., Sakuraba K., Komuro A., Kato S., Harada F., Hirose Y.. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophys. Res. Commun. 2003; 301:378–385. [DOI] [PubMed] [Google Scholar]
- 99. Coppola J.A., Field A.S., Luse D.S.. Promoter-proximal pausing by RNA polymerase II in vitro: transcripts shorter than 20 nucleotides are not capped. Proc. Natl. Acad. Sci. U. S. A. 1983; 80:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Maden B.E.H. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 1986; 189:681–699. [DOI] [PubMed] [Google Scholar]
- 101. Maden B.E.H. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J. Mol. Biol. 1988; 201:289–314. [DOI] [PubMed] [Google Scholar]
- 102. Leismann J., Spagnuolo M., Pradhan M., Wacheul L., Vu M.A., Musheev M., Mier P., Andrade-Navarro M.A., Graille M., Niehrs C.et al.. The 18S ribosomal RNA m6A methyltransferase Mettl5 is required for normal walking behavior in Drosophila. EMBO Rep. 2020; 21:e49443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Van Tran N., Muller L., Ross R.L., Lestini R., Létoquart J., Ulryck N., Limbach P.A., De Crécy-Lagard V., Cianférani S., Graille M.. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 2018; 46:8483–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kowalak J.A., Bruenger E., Crain P.F., McCloskey J.A.. Identities and phylogenetic comparisons of posttranscriptional modifications in 16 S ribosomal RNA from Haloferax volcanii. J. Biol. Chem. 2000; 275:24484–24489. [DOI] [PubMed] [Google Scholar]
- 105. Grosjean H., Gaspin C., Marck C., Decatur W.A., de Crécy-Lagard V.. RNomics and Modomics in the halophilic archaea Haloferax volcanii: Identification of RNA modification genes. BMC Genomics. 2008; 9:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Purushothaman S.K., Bujnicki J.M., Grosjean H., Lapeyre B.. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in Yeast tRNA. Mol. Cell. Biol. 2005; 25:4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bourgeois G., Létoquart J., van Tran N., Graille M.. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules. 2017; 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li W., Shi Y., Zhang T., Ye J., Ding J.. Structural insight into human N6amt1–Trm112 complex functioning as a protein methyltransferase. Cell Discov. 2019; 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Metzger E., Wang S., Urban S., Willmann D., Schmidt A., Offermann A., Allen A., Sum M., Obier N., Cottard F.et al.. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat. Struct. Mol. Biol. 2019; 26:361–371. [DOI] [PubMed] [Google Scholar]
- 110. Liger D., Mora L., Lazar N., Figaro S., Henri J., Scrima N., Buckingham R.H., Van Tilbeurgh H., Heurgué-Hamard V., Graille M.. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011; 39:6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Létoquart J., Huvelle E., Wacheul L., Bourgeois G., Zorbas C., Graille M., Heurgué-Hamard V., Lafontaine D.L.J.. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5518–E5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Létoquart J., Van Tran N., Caroline V., Aleksandrov A., Lazar N., Van Tilbeurgh H., Liger D., Graille M.. Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res. 2015; 43:10989–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bourgeois G., Marcoux J., Saliou J.M., Cianférani S., Graille M.. Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 2017; 45:1971–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Õunap K., Leetsi L., Matsoo M., Kurg R.. The stability of ribosome biogenesis factor WBSCR22 is regulated by interaction with TRMT112 via ubiquitin-proteasome pathway. PLoS One. 2015; 10:e0133841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Heurgué-Hamard V., Graille M., Scrima N., Ulryck N., Champ S., Van Tilbeurgh H., Buckingham R.H.. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 2006; 281:36140–36148. [DOI] [PubMed] [Google Scholar]
- 116. Natchiar S.K., Myasnikov A.G., Kratzat H., Hazemann I.. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017; 551:472–477. [DOI] [PubMed] [Google Scholar]
- 117. Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N.et al.. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018; 46:9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gazin C., Wajapeyee N., Gobeil S., Virbasius C.M., Green M.R.. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007; 449:1073–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Branlant C., Krol A., Machatt A., Ebel J.. The secondary structure of the protein LI binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the LII mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981; 9:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Branlant C., Krol A., Machatt M.A., Pouyet J., Ebel J.P., Edwards K., Kössel H.. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981; 9:4303–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Dehal P.S., Joachimiak M.P., Price M.N., Bates J.T., Baumohl J.K., Chivian D., Friedland G.D., Huang K.H., Keller K., Novichkov P.S.et al.. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2009; 38:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P.et al.. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011; 39:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Palchevskiy V., Finkel S.E.. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J. Bacteriol. 2006; 188:3902–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Punekar A.S., Selmer M.. Purification, crystallization and preliminary X-ray diffraction analysis of the 23S rRNA methyltransferase RlmJ from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013; 69:1001–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Watson Z.L., Ward F.R., Méheust R., Ad O., Schepartz A., Banfield J.F., Cate J.H.D.. Structure of the bacterial ribosome at 2 Å resolution. Elife. 2020; 9:e60482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Siibak T., Remme J.. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA. 2010; 16:2023–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Oerum S., Dendooven T., Catala M., Gilet L., Degut C., Trinquier A., Bourguet M., Barraud P., Cianferani S., Luisi B.F.et al.. Structures of B. subtilis maturation RNases captured on 50S ribosome with Pre-rRNAs. Mol. Cell. 2020; 80:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Giannakis M., Mu X.J., Shukla S.A., Qian Z.R., Cohen O., Nishihara R., Bahl S., Cao Y., Amin-Mansour A., Yamauchi M.et al.. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016; 15:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Muzny D.M., Bainbridge M.N., Chang K., Dinh H.H., Drummond J.A., Fowler G., Kovar C.L., Lewis L.R., Morgan M.B., Newsham I.F.et al.. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S.et al.. COSMIC: Exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015; 43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bell D.W. Novel genetic targets in endometrial cancer. Expert Opin. Ther. Targets. 2014; 18:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tomczak K., Czerwińska P., Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Wspolczesna Onkol. 2015; 19:A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Richard E.M., Polla D.L., Assir M.Z., Contreras M., Shahzad M., Khan A.A., Razzaq A., Akram J., Tarar M.N., Blanpied T.A.et al.. Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am. J. Hum. Genet. 2019; 105:869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hu H., Kahrizi K., Musante L., Fattahi Z., Herwig R., Hosseini M., Oppitz C., Abedini S.S., Suckow V., Larti F.et al.. Genetics of intellectual disability in consanguineous families. Mol. Psychiatry. 2019; 24:1027–1039. [DOI] [PubMed] [Google Scholar]
- 135. Niu Y., Wan A., Lin Z., Lu X., Wan G.. N6-Methyladenosine modification: a novel pharmacological target for anti-cancer drug development. Acta Pharm. Sin. B. 2018; 8:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bedi R.K., Huang D., Eberle S.A., Wiedmer L., Śledź P., Caflisch A.. Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. Chem. Med. Chem. 2020; 15:744–748. [DOI] [PubMed] [Google Scholar]