Abstract
Omics technologies are widely used in biomedical research. Precision medicine focuses on individual-level disease treatment and prevention. Here, we propose the usage of the term ‘precision omics’ to represent the combinatorial strategy that applies omics to translate large-scale molecular omics data for precision disease understanding and accurate disease diagnosis, treatment and prevention. Given the complexity of both omics and precision medicine, precision omics requires standardized representation and integration of heterogeneous data types. Ontology has emerged as an important artificial intelligence component to become critical for standard data and metadata representation, standardization and integration. To support precision omics, we propose a precision omics ontology hypothesis, which hypothesizes that the effectiveness of precision omics is positively correlated with the interoperability of ontologies used for data and knowledge integration. Therefore, to make effective precision omics studies, interoperable ontologies are required to standardize and incorporate heterogeneous data and knowledge in a human- and computer-interpretable manner. Methods for efficient development and application of interoperable ontologies are proposed and illustrated. With the interoperable omics data and knowledge, omics tools such as OmicsViz can also be evolved to process, integrate, visualize and analyze various omics data, leading to the identification of new knowledge and hypotheses of molecular mechanisms underlying the outcomes of diseases such as COVID-19. Given extensive COVID-19 omics research, we propose the strategy of precision omics supported by interoperable ontologies, accompanied with ontology-based semantic reasoning and machine learning, leading to systematic disease mechanism understanding and rational design of precision treatment and prevention.
Short Abstract
Precision medicine focuses on individual-level disease treatment and prevention. Precision omics is a new strategy that applies omics for precision medicine research, which requires standardized representation and integration of individual genetics and phenotypes, experimental conditions, and data analysis settings. Ontology has emerged as an important artificial intelligence component to become critical for standard data and metadata representation, standardization and integration. To support precision omics, interoperable ontologies are required in order to standardize and incorporate heterogeneous data and knowledge in a human- and computer-interpretable manner. With the interoperable omics data and knowledge, omics tools such as OmicsViz can also be evolved to process, integrate, visualize and analyze various omics data, leading to the identification of new knowledge and hypotheses of molecular mechanisms underlying disease outcomes. The precision COVID-19 omics study is provided as the primary use case to illustrate the rationale and implementation of the precision omics strategy.
Keywords: omics, precision omics, ontology, interoperability, COVID-19, precision medicine
Introduction
Omics is a collection of biological disciplines whose names end in the suffix ‘-omics’, such as genomics, transcriptomics, proteomics, epigenomics, metabolomics and interactomics. Omics technologies allow us to integrate multiple associated and interacting components in biological systems to study the mechanisms of complex biological dynamic processes under specific conditions such as diseases and treatments. Omics technologies have been widely used in various applications including biomedical research of diseases such as cancer [1], infectious diseases [2] and autoimmune diseases [3]. The integration of multiple omics technologies (multi-omics) is the trend of omics research [4], which makes the assessment of diseases and treatments more reliable. Multi-omics has been applied to support different areas of research such as molecular typing, treatment and prognosis research [5–8].
Omics technologies have been widely used to study the COVID-19 pandemic [9] as evidenced by over 160 related papers available in PubMed as of 7 May 2021. COVID-19 is caused by the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in lung, kidney, and many other organs. As of 7 May 2021, COVID-19 has resulted in approximately 156 million confirmed cases and 3.2 million deaths worldwide. Multiple omics technologies have also been applied to study host immune status and drug screening against SARS-CoV-2 [10–13]. Even though extensive COVID-19 omics research has been conducted, we do not have a comprehensive understanding of the disease, nor do we have effective drug treatment against SARS-CoV-2. A more effective strategy is required to better study COVID-19.
Precision medicine, also called personalized medicine, is an emergent approach for disease treatment and prevention that considers individual variability in genes, environment and lifestyle [14, 15]. Precision medical research often requires the omics examination of biomedical specimens and cells at different levels, and a multimodal and integrated approach is then applied to integrate the data from biopsy tissue samples with clinical background of human subjects. For example, the NIH-supported Kidney Precision Medicine Project (KPMP) study aims to find the fundamental molecular mechanisms and new treatments for human kidney disease [16, 17]. KPMP involves patient recruitment, clinical data collection, pathological examination, omics assays, integrated analysis with artificial intelligence (AI) and data visualization. In addition, precision medicine has also come with emerging technologies such as machine learning, deep learning and system biology. Omics technologies have greatly supported precision medicine and broader precision biology research. In this article, we propose to use the term ‘precision omics’ to represent the application of omics technologies for precision medicine research.
A major bottleneck in precision omics studies is the disintegrated and non-interoperable big data and knowledge. Big data are characterized by 5 V’s (volume, veracity, velocity, variety and value) [18]. Precision omics research is often accompanied by large, complex, heterogeneous and multidimensional big data generation. Disintegrated and non-interoperable data and knowledge cannot be interpreted by computers, inhibiting computer-assisted reasoning. Unfortunately, omics data are often disintegrated. To support the integration of data from various studies and the rigor and reproducibility of biomedical studies, the FAIR Guiding Principles propose that all research data should be findable, accessible, interoperable and reusable (FAIR) for both machine and human users [19]. To enable data to be FAIR, it is required to add data about the data (metadata), link each data element to a controlled and shared vocabulary, ideally an ontology. Ontology has been proven to be critical to support omics data integration. However, given hundreds of biomedical ontologies developed, it becomes a new silo issue how to integrate different ontologies and make ontologies interoperable.
In this manuscript, we will introduce our proposed ‘precision omics’ strategy, the basics of ontologies, interoperable ontologies and how to use interoperable ontologies to support precision omics research. We argue that precision omics supported by interoperable ontologies would help us to better study COVID-19 disease and allow us to develop a systematic and comprehensive from-molecule-to-phenotype understanding of the disease and rationally develop effective precision treatment and prevention measures at the individual or sub-group level.
The rise of precision omics research
The precision medicine initiative, initiated by the Obama Administration in 2015, marks the beginning of the new era of precision medicine [15]. According to the precision medicine initiative, precision medicine is defined as an emerging approach for treating and preventing diseases by taking into account individual variability for each person [14]. The individual variability covers different areas, including individuals’ genes, phenotypes, environments, behaviors and lifestyles. With a deeper understanding of diseases, it is not necessary to diagnose diseases only based on appearances, but to require more accurate and effective disease classification by considering various variables and conditions.
Figure 1 is provided to illustrate the reasons why we need precision medicine. In typical experiments with inbred mice, we do not usually have to care about many variables (Figure 1). For example, we can just control vaccine or no vaccine in BALB/c mice. Since all the BALB/c mice are genetically the same and the living conditions for all mice are the same as well except for the treatment to be tested, we can effectively identify the results in such a well-controlled experiment. However, such results cannot be directly translated to human. For example, while mouse models have advanced our understanding of sepsis and sepsis-associated kidney diseases, many clinical trials targeting important pathways identified with convincing murine model results have failed to improve human patient survival [20, 21]. There are two main reasons that make it challenging to translate mouse model results to human usage. First, although mouse and human are similar in many different ways, genetically they still differ a lot. Second, unlike the inbred mouse model, humans are so diverse among individuals in terms of genetics, phenotypes, living conditions, medical history, etc. Without considering these differences, it is often difficult to identify the omics patterns linking to the outcome (Figure 1).
Figure 1 .
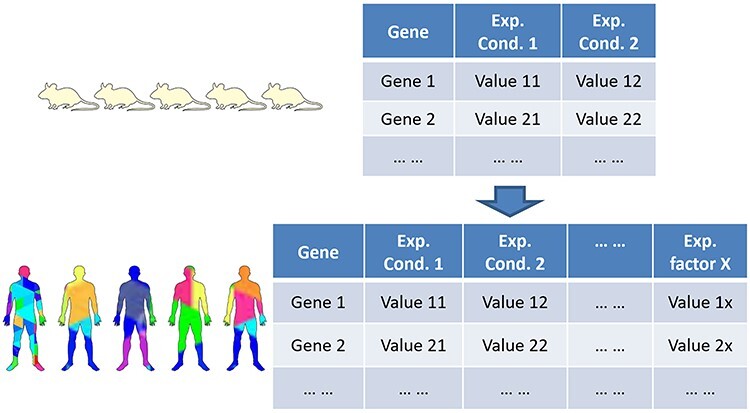
Increasing complexity from single well-controlled omics study to complex multi-factor heterogeneous omics study. Current omics studies have evolved from simplified inbred mouse (or cell line) to complex precision medicine studies. See text for detailed discussion.
Precision medicine is also important in clinical practice. A patient diagnosed with cancer was usually provided with the same treatment as the others having the same type and stage of cancer. However, different patients respond differently to treatment, because different patients may have different tumor diver genes and driver mutations. Meanwhile, the driving mechanisms of different tumors may overlap. It means that we can divide the patients into subgroup levels rather than pure individual levels. Another example is vaccine safety. Currently, licensed vaccines are generally safe. Even so, some severe adverse events may still occur in a small percentage of the population. Understanding the causal relations between personal characteristics and vaccine adverse event occurrence would allow rational design of precision measurement to avoid serious adverse events in susceptible populations [22].
In this article, the term precision omics is formally proposed to represent the application of omics technologies to study precision medicine research questions, with an emphasis on identifying how omics technologies can be efficiently used to support precision medicine research. We argue that the key to precision omics research is the precision classification and integration of various conditions or variables so that the heterogeneous and complex omics data can be interlinked, reproduced, seamlessly integrated and robustly analyzed.
Figure 2 illustrated our envisioned precision omics processes commonly applied in biomedical studies, which is also applied to study COVID-19. The whole pipeline covers many steps, including the human subject recruitment and collection, experimental process, specimen extraction, omics assay, data standardization and integration. Each step requires precision, i.e. all the variables in each step need to be recorded, standardized and integrated into the whole analysis pipeline. Before the sample collection, the human (or other organisms) can be treated in different ways such as drug administration, vaccination, etc. Samples for the omics study include molecular samples, such as DNA, RNA, protein and metabolites, which have originated from cells. We may use purified cells or cell line cells, or directly use tissue samples that include mixed cell types. Sample processing protocols can affect cell composition and further affect data. Omics assays include microarray, RNA-seq, BS-seq, ChIP-seq, whole-genome sequencing, whole-exome sequencing and mass spectrum, etc. The data analysis methods and tools also need to be defined in a precision manner. For example, different omics data analysis programs may be pre-trained with different genome annotations. The usage of different reference genome versions and genome annotations may then affect the data analysis results.
Figure 2 .
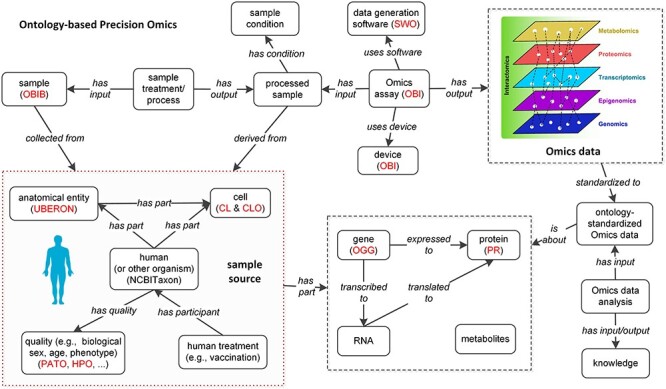
Precision omics data analysis supported by interoperable ontologies. Ontology details are provided in Table 1. To make the data flow interoperable, the ontologies are required to be interoperable.
The major challenges in making the precision omics research reproducible and robust are: (i) the lack of standardized representation of various metadata, clinical and omics data, and prior knowledge needed for effective data analysis. (ii) The lack of computer interpretability of the data, metadata and knowledge, and the relations among individual concepts. The lack of standard representation and computer interpretability makes it impossible to efficiently process, integrate, share and analyze heterogeneous and high volume big data in clinical and basic biomedical research in a standard and reproducible way. Reproducibility will then suffer.
An effective solution to the above challenges is the usage of ontology. In the precision omics pipeline (Figure 2), we have also proposed to use various ontologies to represent and integrate various classes of entities. In the next section, we will introduce the basic concepts of ontology and various specific biological and biomedical ontologies.
Ontologies and their applications in precision omics
Corresponding to various omics technologies, many minimum information standards, such as minimum information about a microarray experiment [23] and minimum information about a single amplified genome [24], have been developed as guidelines for reporting omics data derived by relevant methods in biosciences. These minimum information standards support unambiguous omics experiment data reporting and interpretation. It has also been realized that the minimal information standards need the support of ontology. For example, the Ontology for Biomedical Investigations (OBI) [25, 26] has been developed and used to standardize data and metadata types in different biomedical domains, further supporting reproducibility.
In the informatics field, a formal ontology is a human- and computer-interpretable set of terms and relations that represent entities in a specific domain and how they relate to each other. As stated in the definition, ontology provides a standardization and computer interpretability of the data, metadata and knowledge. Ontologies are widely used in biomedical data and metadata standardization, and robustly support data integration, sharing, reproducibility, and computer-assisted data analysis [27–30]. Currently, BioPortal [31] has stored over 400 ontologies and provided visualization and support for ontology query and analysis. Ontobee [32] stored approximately 200 biomedical ontologies, the majority of which belong to the high-quality Open Biomedical Ontology (OBO) library, which is detailed in the next section.
To illustrate how ontology works, Figure 3 shows the ontological structure for the drug Sirolimus (a.k.a., rapamycin) under the context of the Chemical Entities of Biological Interest (ChEBI), a commonly used ontology of chemical compounds [33]. ChEBI has two main branches: the chemical entity branch in which chemical entities are classified based on shared structural features, and the role branch in which entities are classified based on their roles in the biological or chemical activities or various applications. For example, from the chemical entity branch, Sirolimus (ChEBI:9168) is a ‘macrolide,’ which is a ‘polyketide.’ From the role branch, Sirolimus has role of ‘immunosuppressive agent,’ which is a ‘immunomodulator’ (Figure 3). Sirolimus is an mTOR inhibitor that can inhibit the activity and expression of active cytokines, such as IL-2, IL-6 and IL-10, thereby controlling the cytokine storm. Sirolimus has been found to have the potential of treating COVID-19 [34].
Figure 3 .
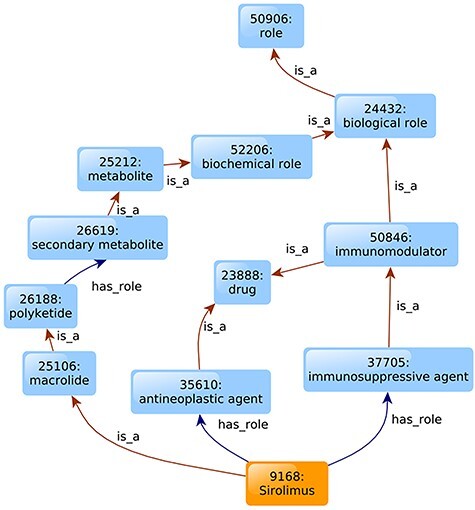
Illustration of Sirolimus ChEBI ontology. Only part of the ChEBI annotations of Sirolimus is shown. The ‘is_a’ relation describes the structural features of the drug. And the ‘has_role’ relation describes drug’s activities in biological or chemical systems or their use in applications.
Ontologies have played an important role in anti-SARS-CoV-2 drug research. Our previous studies identified over 100 experimentally or clinically verified drugs against coronavirus infections [35, 36]. Among these drugs, 99 have their corresponding chemical entities recorded in the ChEBI ontology. Using the semantical information recorded in ChEBI, we were able to cluster these anti-coronavirus chemicals based on a ChEBI semantic similarity analysis (Figure 4). The semantic similarity can be used to evaluate the associations of drugs and help in drug reclassification and repurposing. By clustered 99 coronavirus-related drugs based on the semantic similarity matrix, seven clusters were identified (Figure 4). As ChEBI annotates drugs from two aspects, namely chemical entities and functional roles at a higher level rather than just chemical structure, these similarity clusters may infer potential drug design.
Figure 4 .
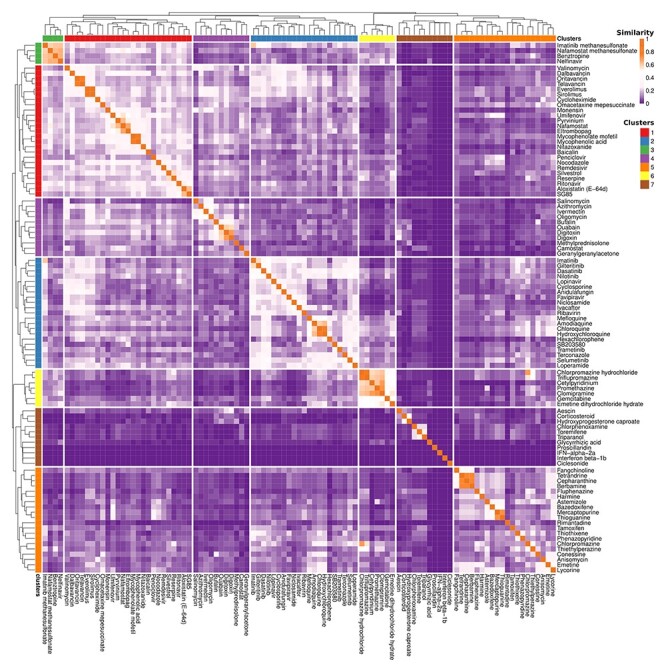
Anti-coronavirus drugs clustering based on ChEBI semantic similarity. The 99 anti-coronavirus drugs with ChEBI annotations were pairwise compared with semantic similarity based on Lin’s method [85]. The heatmap color represents the similarity of drugs. The color on the upper side represents the cluster group. The color scales are on the right upper side.
In addition to hierarchical classification of entities such as chemicals and data analysis such as Gene Ontology (GO) enrichment [37], ontologies have been widely used in many other areas [27, 29]. Ontologies can be applied to support data exchange. For example, BioPAX has been used to standardize how biological pathway data are represented and exchanged [38]. The OBI [25, 26] has been used for integrative representations of data in various areas of life science and clinical investigations. Ontologies can also be used to support natural language processing and metadata standard generation [30, 39, 40].
As shown in Figure 2, various ontologies are required to represent and integrate various classes of entities in precision omics. Table 1 provides more details about these commonly used ontologies in precision omics. Given hundreds of biological and biomedical ontologies being developed, can we just randomly select a list of ontologies for our precision omics studies? The answer is of course no. In the next section, we introduce ontology interoperability and how it is important for precision omics research.
Table 1.
Commonly used interoperable ontologies and their usages in omics-related projects
| Domain | Ontology | Omics-related applications |
|---|---|---|
| Taxonomy | NCBITaxon [86] | LINCS [55] |
| Anatomy | UBERON [87] | ENCODE [39], KPMP [16, 17], LINCS |
| In vivo cells, cell line cells | CL [88] | ENCODE, KPMP |
| CLO [89] | ENCODE, LINCS | |
| Molecular entities | ChEBI [33] | ENCODE |
| OGG [90] | KPMP | |
| PR [91] | KPMP | |
| Biological processes, cellular components and molecular functions of gene products | GO [37] | GEO [92], KPMP, STRING [82] |
| Diseases and phenotypes | DO [93] | CGI [94] |
| MONDO [95] | KPMP | |
| HPO [42] | KPMP | |
| Adverse event | OAE [96] | KMMP |
| Vaccine | VO [44] | ImmPort [97]; KPMP |
| Experimental assays | OBI [25, 26] | ENCODE, KPMP |
Abbreviations: CGI: Cancer Genome Interpreter; CL: Cell Ontology; CLO: Cell Line Ontology; ChEBI: Chemical Entities of Biological Interest; DO: Disease Ontology; ENCODE: Encyclopedia of DNA Elements; GEO: Gene Expression Omnibus; GO: Gene Ontology; HPO: Human Phenotype Ontology; ImmPort: The Immunology Database and Analysis Portal; KPMP: Kidney Precision Medicine Project; LINCS: The Library of Integrated Network-Based Cellular Signatures; MONDO: Mondo Disease Ontology; NCBITaxon: NCBI Taxonomy Ontology; OAE: Ontology of Adverse Events; OBI: Ontology for Biomedical Investigations; OGG: Ontology of Genes and Genomes; PR: Protein Ontology; UBERON: Uber Anatomy Ontology; VO: Vaccine Ontology.
Effective precision omics requires ‘interoperable’ ontologies
To ensure standardized data representation and integration, all ontologies that support data annotation and semantic reasoning are required to be integrated. The key to achieve ontology integration is the interoperability among relevant ontologies.
Ontology interoperability (i.e. interconnection) is essential for data sharing and integration. Interoperability is not just about the packaging of terms (i.e. syntax), but the simultaneous transmission of the meaning with the terms (i.e. semantics). Interoperability is a requirement for data FAIRness. Ontology is a special type of data and it guides data standardization. Therefore, ontology also requires interoperability. Given the foundational role of ontology in data exchange, ontology interoperability is a requirement to enable machine-computable logic, reasoning, and knowledge discovery. Hundreds of ontologies are developed as indicated by >800 biomedical ontologies deposited in BioPortal [31]. While ontologies significantly increase data standardization and reproducibility, a critical issue appears, i.e. a large number of ontologies often overlap each other but are not interoperable.
To foster ontology interoperability and reproducibility, the OBO Foundry (http://obofoundry.org/) was initiated in 2007 by many ontology developers who agree to adopt a set of principles (e.g. collaboration, openness) specifying best practices in ontology development [41]. The OBO ontology library has included approximately 200 ontologies (including GO). The Ontobee program developed by He laboratory is the default ontology repository and linked data server of OBO ontologies [32]. The ontologies listed in Table 1 are all OBO ontologies.
The disease is commonly manifested at the phenotype level; however, omics data (e.g. transcriptomics) are expressed at the molecular level. Many gaps exist from the molecular level to the phenotype level. To fill up the gaps, a precision omics study involves various types of data (e.g. omics data, experimental condition data and sample data) and prior knowledge. Prior knowledge (e.g. protein–protein interactions) helps interpret new omics data and interlink different types of omics data better. To accurately describe the initial state and prognosis, human subject genetics, physiological conditions, and phenotypes deserve careful recording and classification. Additionally, sample types, sample treatment and experimental conditions are required to be standardized. All these can be standardized using interoperable ontologies such as Human Phenotype Ontology (HPO) [42], ChEBI [33], Drug Ontology (DrON) [43] and Vaccine Ontology (VO) [44, 45] (Table 1). Interoperable ontologies would provide solid support for integrating and interlinking the omics data and related knowledge to disease outcomes. The standardized data with the support of interoperable ontologies would transform all the datasets to a novel scale, and the resulting integrated data and knowledge can then be further analyzed using ontology-supported AI technologies, leading to new ways of evaluating the status of organisms.
To further support precision omics research, we propose a Precision Omics Ontology Hypothesis (POOH), which states that the ‘effectiveness’ of precision omics usage in precision phenotype explanation is positively correlated with the interoperability of ontologies to be used for relevant data and knowledge integration:
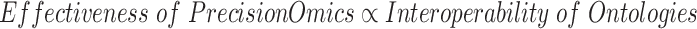 |
The effectiveness of precision omics research can be defined by how effectively an omics study can discover the mechanism of disease outcomes and further predict new disease treatment/prevention at an individual or group level. The general concept of the interoperability of ontologies has been explained above. The POOH hypothesis can be explained by the nature of precision omics. The goal of precision omics requires the integration of complex heterogeneous data types, which then requires the interoperability of different ontologies that systematically represent, standardize and share the omics big data in an interoperable and reproducible way.
As a test case for POOH hypothesis evaluation, acute kidney injury (AKI) occurs in COVID-19 patients with a varied occurrence range from 0–39% in infected patients (http://www.nephjc.com/news/covidaki). Is that possible for us to apply precision omics to identify which variable(s) contribute to the AKI variation in human patients? This study can be performed collaboratively from different laboratories in multiple groups. To be effective in the precision omics study, we will need to classify all possible human subject variables, various experimental conditions from different laboratories, and data processing and analysis methods. To ensure the heterogeneous data able to be analyzed together with the support of prior knowledge, we will require to use interoperable ontologies for data integration and prior knowledge incorporation.
Given the importance of ontology interoperability, how can we efficiently develop interoperable ontologies? For this purpose, the strategy of ‘eXtensible Ontology Development’ (XOD) [46] has been proposed. These XOD principles propose ontology term reuse (XOD1), ontology semantic alignment (XOD2), design pattern-based new term generation (XOD3), and the involvement of community effort for new ontology development (XOD4). These principles promote standardized and interoperable data and knowledge representation and integration. Many tools, such as ROBOT [47] and the list of ‘Ontoanimal’ tools (e.g. Ontofox [48], Ontorat [49], and Ontodog [50]), have also been developed to support extensible ontology development.
Interoperable ontologies for precision kidney and COVID-19 studies
In addition to the general reference ontologies that are applicable across different domains (Table 1), we also need domain-specific ontologies that focus on specific domains. Our specific precision omics research typically focuses on specific domains under the overall umbrella of precision medicine. Here the domain represents a specific research area. Ideally, each domain would need an integrative ontology specifically for the domain. Next, we provide some examples to demonstrate the usage of domain-specific ontologies for precision omics analysis.
The Kidney Tissue Atlas Ontology (KTAO) is a domain ontology specifically for the domain of kidney precision medicine studies [17, 51]. Supported by the Kidney Precision Medicine Project (KPMP, http://kpmp.org), KTAO aims to systematically represent, standardize, and integrate kidney-related cell types, cell states, locations, gene markers, kidney phenotypes and diseases, and the semantic relations among these entities. KTAO supports comprehensive KPMP studies, including precision omics studies, with the aim to understand and treat AKI and chronic kidney disease [51].
Another ontology supported by KPMP is the community-based Ontology of Precision Medicine and Investigation (OPMI) [17, 52]. OPMI provides a general framework and commonly used terms in precision medicine. As an OBO reference ontology for various specific domains across precision medicine, OPMI can be applied to support data interoperability and knowledge presentation in kidney or other precision medicine projects.
The community-based Coronavirus Infectious Disease Ontology (CIDO) is a newly developed ontology for the domain of coronavirus infectious diseases, including COVID-19 and other coronavirus diseases such as SARS and MERS [53]. As an OBO library ontology, CIDO provides standardized annotation and representation of coronavirus infectious diseases, including their etiology, pathogenesis, transmission, epidemiology, diagnosis, prevention and treatment. CIDO incorporates many terms from existing ontologies. For example, CIDO imports COVID-19 vaccine terms defined in the Vaccine Ontology (VO) [44, 45]. A recent study reports the usage of CIDO to systematically model, represent and analyze over 100 experimentally or clinically verified drugs against coronavirus infections [35]. More than simple tables or Excel sheets, the systematical ontology representation allowed advanced classification and linkage of these drugs, chemical compounds, drug targets, biological processes, viruses and the relations among these entities. Derived from this study, a ‘Host-coronavirus interaction (HCI) checkpoint cocktail’ strategy was proposed to interrupt the important checkpoints in the dynamic HCI network, and interoperable ontologies will greatly support related data analysis and rational drug-design process [35].
Not every domain has its own domain ontology. By incorporating terms from reference ontologies with the addition of more specific terms and relationships, we can create application ontologies appropriate for a single project or end-use. The XOD principles and methods as described above can also be used to develop new domain-specific application ontologies. The KTAO and CIDO have been developed this way [17, 51, 53]. In general, the process of their development should also be community-driven and developed by following the OBO Foundry ontology development principles. The process of its construction involves the integration and interoperability of different ontologies.
Interoperable ontology-supported precision COVID-19 omics research
Many omics data resources are using interoperable reference ontologies for their standardized data integration and analysis. Ontologies have been frequently used in popular omics resources such as ArrayExpress [54], ENCODE [39], NIH Library of Integrated Network-based Cellular Signatures (LINCS) [55] and Kidney Precision Medicine Project (KPMP) [16]. For example, ENCODE has used many reference ontologies including the Cell Ontology (CL), Cell Line Ontology (CLO), OBI and UBERON [39] (Table 1). KPMP uses KTAO, a domain ontology that has already incorporated many reference ontologies (Table 1). The introduction of interoperable ontologies supports data/metadata standardization and integration, allowing advanced data sharing and analyses.
Many omics tools have also been developed to use reference ontologies for enhanced performance [56]. For example, the Investigation/Study/Assay (ISA) metadata tracking framework (https://isa-tools.org/) is an open-source system developed to help manage the increasingly diverse data set in life science. The ISA-Tab file format describes experimental metadata types based on the investigation, study and assay categories. LinkedISA then provides ontology-based semantic representation of ISA-Tab experimental metadata [40]. Ometa is a data-driven metadata tracking system that supports configuration, capturing, curation and sharing of ontology-supported metadata types [57]. Onassis is an R package that uses ontology to drive integrative omics data analysis [58]. Onassis uses ontology terms to simplify the annotations of samples from large-scale repositories. Onassis also structures the dataset by the hierarchical organization of samples according to the semantic similarity between their associated ontology terms, supporting the efficient semantic analysis of omics data.
Omics data analysis tools have been developed to support COVID-19 research. For example, Overmyer et al. [59] designed a web-based tool that can interactively explore multi-omics data sets, cross-omics correlation analysis and predict the severity of COVID-19 through machine learning algorithms. Debmalya Barh et al. [12] conducted an integrated analysis of multi-omics data to study several drug candidates for this viral disease. However, although multi-omics data and related web platforms were developed to study COVID-19, ontology technology has rarely been used.
Here, we illustrate the usage of a prototype web software OmicsViz (http://medcode.link/omicsviz/ or http://hegroup.org:8080/omicsviz/) for COVID-19 research. By introducing the Cell Line Ontology, OmicsViz can analyze and visualize the interference of drugs on gene expression of cell line cells that are derived at specific tissues. Using the GO and KEGG pathway enrichment analysis, OmicsViz can dynamically calculate and retrieve enriched pathways of perturbed genes. OmicsViz also stores related interactomics knowledge, specifically, drug–target interactions and protein–protein interactions. The interlinkage of the interactomics knowledge can expand the drug–target network to imply additional related drugs. Next, we demonstrate how OmicsViz could be applied to support possible COVID-19 drug studies.
By simple mouse clicks, OmicsViz allows users to perform dynamic analysis of the LINCS 1000 data and visualization of the effects of thousands of drugs on cell lines [60] (Figure 5). The records of the drugs come from DrugBank [61] (Figure 5). LINCS L1000 profiled gene expression changes following pharmacologic or genetic (knockdown or over-expression) perturbation of cell lines. OmicsViz developed a pipeline to process the data, perform statistical analysis and developed a user-friendly web interface to demonstrate the results. If a user searched a tissue such as ‘lung’ and a drug such as ‘Sirolimus,’ and selected ‘up-regulated’ and the number of 50, OmicsViz would export a heatmap of the first 50 genes with significantly up-regulated gene expression profiles at the 6 and 24 h after Sirolimus treatment (Figure 5A). Using the identified 50 genes as input, a user could also query functional enriched GO terms (Figure 5B). OmicsViz also provides a way for users to identify the target proteins of the drug Sirolimus and the other drugs that share the same target proteins with Sirolimus (Figure 5C).
Figure 5 .
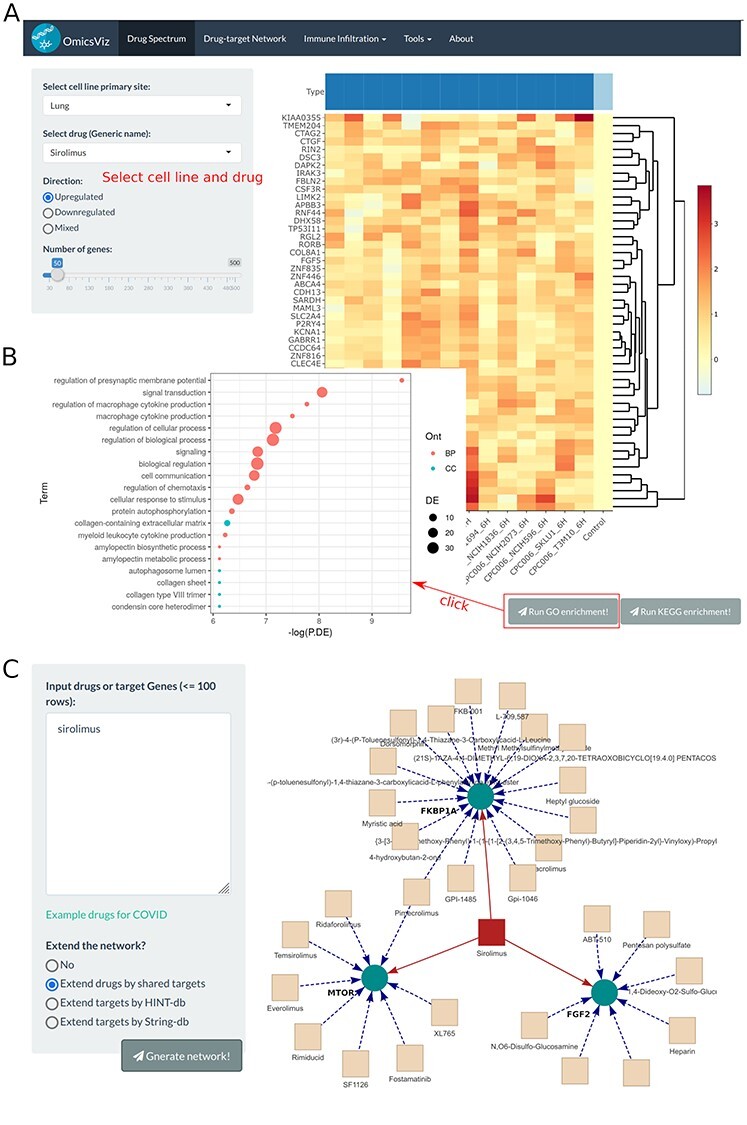
OmicsViz analysis of Sirolimus. (A) Query of perturbed gene expression profiles in lung cell line cells treated by Sirolimus. Red represents up-regulation and blue represents down-regulation. (B) GO enrichment analysis of significantly changed genes identified in (A). The topGO function in limma package was used for the analysis. The x-axis represents the negative natural logarithms of P-values. Different colors indicate different aspects of GO. The size of nodes represents the relative scale of differentially expressed genes in the specified GO term. (C) Interaction of Sirolimus, its associated target proteins, and other drugs that share the target proteins.
The OmicsViz analysis is able to generate new scientific insights. For example, the analysis showed in Figure 5 was performed to evaluate the possibility of using Sirolimus for COVID-19 treatment. Since lung is the primary tissue targeted by the SARS-CoV-2, we examined how Sirolimus affected the gene profiles in lung-derived cell line cells. The top 50 genes affected by Sirolimus were identified (Figure 5A) and their associated genes’ functional enrichment was detected (Figure 5B). The resulting enriched processes, such as cellular response to stimulus, regulation of chemotaxis and autophagosome lumen appear to be required for fighting against SARS-CoV-2. Our OmicsViz analysis also found that Sirolimus has three target proteins, including MTOR, FGF2 and FKBP1A (Figure 5C). Many expanded drugs, such as Everolimus and heparin, were found to share the target proteins with Sirolimus. As a derivative of Sirolimus, Everolimus works similarly to Sirolimus as an mTOR (mammalian target of rapamycin) inhibitor. The finding of heparin in our interactome network is aligned with the recent finding of the hypercoagulable state of blood in COVID-19 patients [62]. Interestingly, Hippensteel et al. [63] reviewed preclinical evidence and established biological plausibility for heparin and synthetic heparin-like drugs as therapies for COVID-19 through antiviral and anti-inflammatory effects Sirolimus and heparin share the same target of FGF2 (Fibroblast growth factor receptor 2), which plays an important role in the regulation of cell survival, cell division, angiogenesis, cell differentiation and cell migration [64].
A new tool called ‘SARS-CoV-2 network’ has been added to OmicsViz (Figure 6). This tool provides the network of SARS-CoV-2 and its host protein–protein interactions (PPIs) based on the results of 481 interactions (Figure 6A) as obtained from a systematic study reported in Nature [65]. Furthermore, the virus–host PPIs can be significantly extended by adding 229 drug–target interactions obtained from DrugBank (Figure 6B). The tool can be applied to identify potential anti-coronaviral drugs. The SARS-CoV-2 Nsp5 protease (a.k.a. 3CLpro) is essential for virus replication [66], and it is also predicted to be an adhesin [67]. Our network shows that Nsp5 interacts with three host proteins (i.e. TRMT1, HDRAC2 and GPX1) (Figure 6B), each of which may be targeted by many drugs. For example, the drug cannabidiol targets glutathione peroxidase 1 (GPX1), a cytosolic selenoenzyme that has known antiviral properties [68]. Cannabidiol is found to induce interferon expression and up-regulate its antiviral signaling pathway [69]. Another drug romidepsin, a histone deacetylase (HDAC) inhibitor, can inhibit the activity of HDAC2. Meanwhile, romidepsin can also interrupt ABCC1 (ATP Binding Cassette Subfamily C Member 1), which interacts with viral protein Orf9c [65] (Figure 6C). Liu et al.[70] demonstrated that romidepsin can effectively block the entry of SARS-CoV-2. Our network also shows many drugs can interact with multiple human proteins targeted by viral proteins. For example, fostamatinib, a tyrosine kinase inhibitor, targets 10 human proteins that interact with 6 viral proteins (i.e. M, Orf9b, N, Nsp9, Nsp12 and Nsp13). Fostamatinib inhibits MUC1 in the respiratory tract and has the potential to treat serious outcomes of COVID-19, including acute respiratory distress syndrome and acute lung injury [71]. Fostamatinib is currently under clinical trial for COVID-19 treatment (Clinicaltrials.gov ID: NCT04579393). In the future, we plan to add more annotations provided by ontologies, such as drug functional components and mechanisms of actions in ontologies including the ChEBI [33] and National Drug File-Reference Terminology (NDF-RT) [72], which would further enhance the data integration and mining capability of OmicsViz, facilitating more efficient drug repurposing.
Figure 6 .
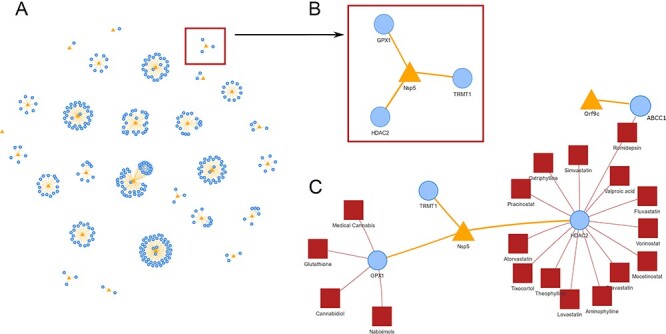
SARS-CoV-2 network analysis in OmicsViz. (A) SARS-CoV-2 and host PPI based on results reported in Nature [65]. (B) Enlarged viral protein Nsp5 and its interacting three host proteins. (C) An extended network including Nsp5, its interacting host proteins, and the known drugs targeting these host proteins. Romidepsin targets both HDAC2 (which interacts with Nsp5) and ABCC1 (which interacts with viral Orf9c). Blue, yellow and red colors represent host protein, viral protein and drug, respectively.
The usage of interoperable ontologies can potentially provide more support in the future development of OmicsViz. Through cell classification by the Cell Line Ontology (CLO) and drug classification by DrON or ChEBI ontologies, we may discover cell responses to drugs from various perspectives. With ontology-standardized interactomics to interlinking omics knowledge, we can further extend our discoveries. The cell line gene expression profiles may correlate with clinical patient gene expression profiles. By studying the responses of different cell lines to different drugs and mapped patient and cell line gene expression profiles, Duan et al. [73] were able to identify specific drugs that effectively treated both cell lines and patients with similar profiles. OmicsViz can now group cells based on their primary sites and analyze their response to drugs. However, the classification of cells can also be performed by cell morphological types such as epithelial cells and fibroblasts. Similarly, drugs can also be grouped based on their biological functions, chemical structures and functional groups. The introduction of known PPI can also expand the drug–target network provided in our Sirolimus network (Figure 5) and the specific SARS-CoV-2 network (Figure 6). All these additional cell, drug and PPI classifications can be defined using interoperable ontologies and incorporated into the future OmicsViz development, which will lead to the identification of more findings and hypotheses of biological relevance.
Challenges and opportunities
The contributions of this paper are multi-fold. First, after reviewing the omics technologies and precision medicine, we propose and define the strategy of ‘precision omics.’ Second, after introducing ontology and ontology interoperability, we propose the POOH hypothesis, stating that there is a positive relationship between the effectiveness of precision omics and the interoperability of ontologies. Third, two interoperable ontologies, KTAO and CIDO, were further introduced to illustrate how interoperable ontologies can be developed to support precision omics studies for kidney and COVID-19. Fourth, we demonstrate and discuss the usage of the OmicsViz tool for precision COVID-19 omics research, in which interoperable ontologies are being used to efficiently integrate knowledge from external resources for enhanced analysis and visualization.
Precision medicine research is easy to say but difficult to do. As defined by precision medicine, we need to examine the relations especially causal relations, between different variables of individuals and the medical diagnosis/treatment/prevention at the individual level. To achieve this goal, omics studies need to identify how different variables, such as human sex, age, genetic conditions, behaviors, external stresses or infections, could possibly change the patterns of DNA mutation, RNA/protein expression, and metabolite production and function, and then affect the clinical outcomes. The sample preparation, experimental assay and data analysis settings may also change the results of omics studies. Due to these difficulties, a huge gap exists between the omics studies and the omics result interpretation for explaining clinical phenotypes and disease outcomes. Unfortunately, current omics research practices cannot meet the demand.
To fill up the gap defined above, we propose the strategy of ‘Precision Omics.’ The essence of this strategy to explore and include all possible variables that would affect the precision medicine outcomes, standardize them using interoperable ontologies, and apply ontology-based data processing and analysis methods to study the relations between different variables and disease outcomes, leading to effective and efficient individual level disease diagnosis, treatment and prevention.
Interoperability between ontologies remains a critical bottleneck. Even though the OBO Foundry is initiated to promote ontology interoperability, and the ontologies in the OBO library are developed to support interoperability, true interoperability among ontologies has not been achieved for most ontologies [74]. We have previously developed an OLOBO program with the goal of interlinking different OBO ontologies, and found that more work is required to make OBO ontologies fully interoperable [75]. Since non-OBO ontologies commonly do not follow the OBO principles that support interoperability, more interoperability issues are expected.
The POOH hypothesis is raised to emphasize the importance of ontology interoperability. It states that there is a positive correlation between the ‘effectiveness’ of precision omics usage in precision phenotype explanation and interoperability of ontologies to be used for relevant data and knowledge integration. Both the effectiveness of precision omics usage and interoperability of ontologies are quantifiable using mathematical formulae, which deserve further exploration in the future.
Although hundreds of ontologies are reported in BioPortal and approximately 200 reported in the OBO library and Ontobee, the number of interoperable ontologies needed for precision medicine studies is still insufficient. As argued in this article, each domain of research deserves its own community-based domain ontology. Such domain ontologies can be classified as application ontologies that are largely developed by collaborating and reusing terms from reference ontologies. An existing challenge is the lack of domain-specific ontologies for many research domains. However, the shortage of domain ontologies also provides an opportunity for researchers in various domains of research.
To support precision omics analysis, it is important to develop and apply interoperable interactome knowledge. Many interactome knowledge resources are available. For example, the Online Mendelian Inheritance in Man (OMIM) provides a continuously updated catalog of human genes and genetic disorders and traits, with a special focus on the gene–phenotype relationships [76]. Examples of well-known interaction and pathway resources include REACTOME [77, 78], KEGG [79], BioGRID [80], Human Reference Interactome (HuRI) atlas [81] and STRING [82]. Unfortunately, these interaction resources are not interoperable. This interactome knowledge from various resources has not been integrated into a well-defined interoperable ontology framework. A solution to this challenge is the development of an ontology-based platform for the ontologization of various interaction knowledge. For example, the Interaction Network Ontology (INO) [83, 84] is an OBO library ontology aimed to ontologically classify biological interactions and the networks among the interactions.
The development and usage of interoperable ontologies provide new perspectives and angles to advance existing omics tools (e.g. OmicsViz) for more advanced omics data analysis. In practice, current omics tools still have a lot of potentials to be improved by incorporating interoperable ontologies for data and knowledge integration and semantic reasoning and analysis. Ontology is a critical basis for knowledge representation and reasoning (KR&R), a key component of AI. We expect that new algorithms and tools based on advanced interoperable ontologies are being developed, lifting the power of ontology-supported AI machine learning technologies.
To fight against COVID-19 pandemic, extensive research including omics research has been conducted, leading to the accumulation of large amounts of omics data and knowledge. Many COVID-19 related databases, such as the BioGRID COVID-19 specific interaction knowledge (https://thebiogrid.org/project/3) and COVID-19 vaccine knowledgebase (http://www.violinet.org/cov19vaxkb), have also been generated. However, we argue that what we have achieved and done is not sufficient. All the omics data and knowledge are not interoperable, and their semantic relations are not established and understood by computers. The community-based Coronavirus Infectious Disease Ontology (CIDO) is now developed to support COVID-19 related data and knowledge representation and integration [53]. By incorporating and integrating with other ontologies, we have demonstrated how CIDO can be used to effectively model, standardize and integrate experimentally or clinically verified drugs against coronavirus infections [35]. Moreover, we need new ontology-based theories and frameworks for a paradigm change. Derived from the CIDO-based drug study, we proposed a ‘Host-coronavirus interaction (HCI) checkpoint cocktail’ strategy that targets to interrupt the important checkpoints in the dynamic HCI network to develop new drug cocktails against COVID-19 [35]. To achieve this goal, precision omics technologies may be employed, and all the resulting data and knowledge can be integrated with interoperable ontologies. Ontology-based novel AI and machine learning methods can also be developed and applied. We believe that interoperable ontology-supported precision omics will not only enhance our understanding of COVID-19 and offer clues to rational treatment/prevention, but will also help us fight other diseases.
Conclusion
Different from inbred mouse studies with well-controlled variables, precision medicine focuses on examining various variables in individuals for precision treatment and prevention of diseases. To more efficiently apply omics for precision medicine study, we propose the strategy of ‘precision omics’ with a goal to measure and standardize various human conditions, sample variables, experimental assay settings, and data analysis methods using community-based computer-interpretable ontologies. Ontology interoperability is key to seamlessly interlink different entities and analyze their semantic relations. We further propose the POOH, proposed to define the positive correlation between the effectiveness of precision omics usage in precision phenotype explanation and interoperability of ontologies to be used for data and knowledge integration. Different ontology-based omics data resources and tools (e.g. OmicsViz) are being developed to support precision omics studies. COVID-19 research is used as a model example to illustrate how precision omics can be performed with the support of interoperable ontologies.
Key Points
Precision omics, a new concept proposed to use in the paper, aims to use omics for precision medicine research.
Precision omics requires interoperable ontologies to standardize and integrate heterogeneous data and knowledge.
POOH hypothesis proposes the positive correlation between the effectivity of precision omics research and interoperability of ontologies.
Omics tools, such as OmicsViz, can be used to support COVID-19 research and enhanced with interoperable ontologies.
Zhigang Wang is an assistant research professor in Peking Union Medical College, Beijing, China. He is focused on biostatistics and big data visualization using omics and ontology data.
Yongqun He is an associate professor in the University of Michigan Medical School, Ann Arbor, MI, USA. He is focused on ontology, microbiology, vaccinology and nephrology.
Contributor Information
Zhigang Wang, Peking Union Medical College, Beijing, China.
Yongqun He, University of Michigan Medical School, Ann Arbor, MI, USA.
Availability of data and material
Not applicable.
Authors’ contributions
Z.W. is biostatistics expert, developed OmicsViz, and focused on omics data analysis. Y.H. is ontology and microbiology expert, and focused on ontology interoperability and applications. Z.W. and Y.H. discussed the precision omics strategies and co-prepared the manuscript.
Acknowledgements
We acknowledge the support by the OBO Foundry ontology community.
Funding
The research was partly supported by National Institutes of Health grants 1UH2AI132931 and 1R01AI081062; and the University of Michigan Medical School Global Reach Fund; Michigan Medicine–Peking University Health Sciences Center Joint Institute for Clinical and Translational Research; National Key R&D Program of China (No. 2017YFC0907505); CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2020-I2M-2-001 and No. 2018-I2M-AI-009).
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
References
- 1. Tang B, Hsu PY, Huang TH, et al. Cancer omics: from regulatory networks to clinical outcomes. Cancer Lett 2013;340:277–83. [DOI] [PubMed] [Google Scholar]
- 2. Mangioni D, Peri AM, Rossolini GM, et al. Toward rapid sepsis diagnosis and patient stratification: what's new from microbiology and omics science. J Infect Dis 2020;221:1039–47. [DOI] [PubMed] [Google Scholar]
- 3. Schena FP, Serino G, Sallustio F, et al. Omics studies for comprehensive understanding of immunoglobulin A nephropathy: state-of-the-art and future directions. Nephrol Dial Transplant 2018;33:2101–12. [DOI] [PubMed] [Google Scholar]
- 4. Graw S, Chappell K, Washam CL, et al. Multi-omics data integration considerations and study design for biological systems and disease. Mol Omics 2021;17:170–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan MM, Ernst O, Manes NP, et al. Multi-omics strategies uncover host-pathogen interactions. ACS Infect Dis 2019;5:493–505. [DOI] [PubMed] [Google Scholar]
- 6. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y, Yang M, Ng DM, et al. Multi-omics data analyses construct TME and identify the immune-related prognosis signatures in human LUAD. Mol Ther Nucleic Acids 2020;21:860–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Way GP, Sanchez-Vega F, La K, et al. Machine learning detects pan-cancer ras pathway activation in the cancer genome atlas. Cell Rep 2018;23:172–180 e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin B, Liu J, Liu Y, et al. Progress in understanding COVID-19: insights from the omics approach. Crit Rev Clin Lab Sci 2020;1–18. https://pubmed.ncbi.nlm.nih.gov/33375876/. [DOI] [PubMed] [Google Scholar]
- 10. Bernardes JP, Mishra N, Tran F, et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity 2020;53:1296–1314 e1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Su Y, Chen D, Yuan D, et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 2020;183:1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barh D, Tiwari S, Weener ME, et al. Multi-omics-based identification of SARS-CoV-2 infection biology and candidate drugs against COVID-19. Comput Biol Med 2020;126:104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Overmyer KA, Shishkova E, Miller IJ, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021;12:23–40.e7. https://pubmed.ncbi.nlm.nih.gov/33096026/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ashley EA. The precision medicine initiative: a new national effort. JAMA 2015;313:2119–20. [DOI] [PubMed] [Google Scholar]
- 15. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El-Achkar TM, Eadon MT, Menon R, et al. A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: guidelines from the kidney precision medicine project. Physiol Genomics 2020;53:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ong E, Wang LL, Schaub J, et al. Modelling kidney disease using ontology: insights from the Kidney Precision Medicine Project. Nat Rev Nephrol 2020;16:686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higdon R, Haynes W, Stanberry L, et al. Unraveling the complexities of life sciences data. Big Data 2013;1:42–50. [DOI] [PubMed] [Google Scholar]
- 19. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Becker GJ, Hewitson TD. Animal models of chronic kidney disease: useful but not perfect. Nephrol Dial Transplant 2013;28:2432–8. [DOI] [PubMed] [Google Scholar]
- 21. Bao YW, Yuan Y, Chen JH, et al. Kidney disease models: tools to identify mechanisms and potential therapeutic targets. Zool Res 2018;39:72–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie J, Zi W, Li Z, et al. Ontology-based precision vaccinology for deep mechanism understanding and precision vaccine development. Curr Pharm Des 2020;27:900–10. [DOI] [PubMed] [Google Scholar]
- 23. Brazma A. Minimum information about a microarray experiment (MIAME)--successes, failures, challenges. ScientificWorldJournal 2009;9:420–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bowers RM, Kyrpides NC, Stepanauskas R, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 2017;35:725–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bandrowski A, Brinkman R, Brochhausen M, et al. The ontology for biomedical investigations. PLoS One 2016;11:e0154556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brinkman RR, Courtot M, Derom D, et al. Modeling biomedical experimental processes with OBI. J Biomed Semantics 2010;1(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoehndorf R, Schofield PN, Gkoutos GV. The role of ontologies in biological and biomedical research: a functional perspective. Brief Bioinform 2015;16:1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform 2008;67–79. https://pubmed.ncbi.nlm.nih.gov/18660879/. [PMC free article] [PubMed] [Google Scholar]
- 29. Schulz S, Balkanyi L, Cornet R, et al. From concept representations to ontologies: a paradigm shift in health informatics? Health Inform Res 2013;19:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dugan VG, Emrich SJ, Giraldo-Calderon GI, et al. Standardized metadata for human pathogen/vector genomic sequences. PLoS One 2014;9:e99979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whetzel PL, Noy NF, Shah NH, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res 2011;39:W541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ong E, Xiang Z, Zhao B, et al. Ontobee: a linked ontology data server to support ontology term dereferencing, linkage, query and integration. Nucleic Acids Res 2017;45:D347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hastings J, Owen G, Dekker A, et al. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res 2016;44:D1214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Husain A, Byrareddy SN. Rapamycin as a potential repurpose drug candidate for the treatment of COVID-19. Chem Biol Interact 2020;331:109282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y, Hur J, Chan WKB, et al. Ontological modeling and analysis of experimentally or clinically verified drugs against coronavirus infection. Sci Data 2021;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Y, Chan W, Wang Z, et al. Ontological and bioinformatic analysis of anti-coronavirus drugs and their implication for drug repurposing against COVID-19. Preprints 2020. doi: 10.20944/preprints202003.0413.v1. [DOI] [Google Scholar]
- 37. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Demir E, Cary MP, Paley S, et al. The BioPAX community standard for pathway data sharing. Nat Biotechnol 2010;28:935–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malladi VS, Erickson DT, Podduturi NR, et al. Ontology application and use at the ENCODE DCC. Database (Oxford) 2015;2015:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gonzalez-Beltran A, Maguire E, Sansone SA, et al. linkedISA: semantic representation of ISA-Tab experimental metadata. BMC Bioinformatics 2014;15(Suppl 14):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007;25:1251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kohler S, Gargano M, Matentzoglu N, et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res 2021;49:D1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanna J, Bian J, Hogan WR. An accurate and precise representation of drug ingredients. J Biomed Semantics 2016;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin Y, He Y. Ontology representation and analysis of vaccine formulation and administration and their effects on vaccine immune responses. J Biomed Semantics 2012;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ozgur A, Xiang Z, Radev DR, et al. Mining of vaccine-associated IFN-gamma gene interaction networks using the Vaccine Ontology. J Biomed Semantics 2011;2(Suppl 2):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He Y, Xiang Z, Zheng J, et al. The eXtensible ontology development (XOD) principles and tool implementation to support ontology interoperability. J Biomed Semantics 2018;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jackson RC, Balhoff JP, Douglass E, et al. ROBOT: a tool for automating ontology workflows. BMC Bioinformatics 2019;20:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiang Z, Courtot M, Brinkman RR, et al. OntoFox: web-based support for ontology reuse. BMC Res Notes 2010;3(175):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiang Z, Lin Y, He Y. Ontorat web server for automatic generation and annotations of new ontology terms. In: The 3rd International Conference on Biomedical Ontologies (ICBO). Graz, Graz, Austria, 2012. http://ceur-ws.org/Vol-897/poster_812.pdf CEUR Workshop Proceedings. [Google Scholar]
- 50. Zheng J, Xiang Z, Stoeckert CJ, Jr, et al. Ontodog: a web-based ontology community view generation tool. Bioinformatics 2014;30:1340–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He Y, Steck B, Ong E, et al. KTAO: A kidney tissue atlas ontology to support community-based kidney knowledge base development and data integration. In: International Conference on Biomedical Ontology 2018 (ICBO-2018). Corvallis, USA: Oregon, 2018, p. 1–6. [Google Scholar]
- 52. He Y, Ong E, Schaub J et al. OPMI: the ontology of precision medicine and investigation and its support for clinical data and metadata representation and analysis. In: The 10th International Conference on Biomedical Ontology (ICBO-2019), July 30–August 2. Buffalo, NY, USA, 2019, p. 1–10. [Google Scholar]
- 53. He Y, Yu H, Ong E, et al. CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci Data 2020;7:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Athar A, Fullgrabe A, George N, et al. ArrayExpress update - from bulk to single-cell expression data. Nucleic Acids Res 2019;47:D711–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stathias V, Turner J, Koleti A, et al. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res 2020;48:D431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wanichthanarak K, Fahrmann JF, Grapov D. Genomic, proteomic, and metabolomic data integration strategies. Biomark Insights 2015;10:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Singh I, Kuscuoglu M, Harkins DM, et al. OMeta: an ontology-based, data-driven metadata tracking system. BMC Bioinformatics 2019;20:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Galeota E, Kishore K, Pelizzola M. Ontology-driven integrative analysis of omics data through Onassis. Sci Rep 2020;10:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Overmyer KA, Shishkova E, Miller IJ, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst 2021;12:23–40 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu C, Su J, Yang F, et al. Compound signature detection on LINCS L1000 big data. Mol Biosyst 2015;11:714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bachler M, Bosch J, Sturzel DP, et al. Impaired fibrinolysis in critically ill COVID-19 patients. Br J Anaesth 2020;126:590–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hippensteel JA, LaRiviere WB, Colbert JF, et al. Heparin as a therapy for COVID-19: current evidence and future possibilities. Am J Physiol Lung Cell Mol Physiol 2020;319:L211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sehgal SN. Sirolimus: its discovery, biological properties, and mechanism of action. Transplant Proc 2003;35:7S–14. [DOI] [PubMed] [Google Scholar]
- 65. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020;583:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stobart CC, Sexton NR, Munjal H, et al. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J Virol 2013;87:12611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ong E, Wong MU, Huffman A, et al. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol 2020;11:1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Seale LA, Torres DJ, Berry MJ, et al. A role for selenium-dependent GPX1 in SARS-CoV-2 virulence. Am J Clin Nutr 2020;112:447–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nguyen LC, Yang D, Nicolaescu V, et al. Cannabidiol inhibits SARS-CoV-2 replication and promotes the host innate immune response. bioRxiv 2021.March 10, 2021. doi: 10.1101/2021.03.10.432967 preprint: not peer reviewed. [DOI]
- 70. Liu K, Zou R, Cui W, et al. Clinical HDAC inhibitors are effective drugs to prevent the entry of SARS-CoV2. ACS Pharmacol Trans Sci 2020;3:1361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kost-Alimova M, Sidhom EH, Satyam A, et al. A high-content screen for mucin-1-reducing compounds identifies fostamatinib as a candidate for rapid repurposing for acute lung injury. Cell Rep Med 2020;1:100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peters LB, Bahr N, Bodenreider O. Evaluating drug-drug interaction information in NDF-RT and DrugBank. J Biomed Semantics 2015;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Duan Q, Kou Y, Clark NR, et al. Metasignatures identify two major subtypes of breast cancer. CPT Pharmacometrics Syst Pharmacol 2013;2:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ghazvinian A, Noy NF, Musen MA. How orthogonal are the OBO Foundry ontologies? J Biomed Semantics 2011;2(Suppl 2):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ong E, He Y. OLOBO: a new ontology for linking and integrating open biological and biomedical ontologies. In: International Conference on Biomedical Ontology 2017, September 13–15, 2017. Newcastle upon Tyne, UK. 2017, p. 1–2. [Google Scholar]
- 76. Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:D789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fabregat A, Sidiropoulos K, Garapati P, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2016;44:D481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Haw R, Loney F, Ong E, et al. Perform pathway enrichment analysis using ReactomeFIViz. In: Canzar S (ed). Protein-Protein Interaction Networks: Methods and Protocols. Springer, 2019, 2018, Submitted by invitation. [Google Scholar]
- 79. Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Oughtred R, Stark C, Breitkreutz BJ, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res 2019;47:D529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Luck K, Kim DK, Lambourne L, et al. A reference map of the human binary protein interactome. Nature 2020;580:402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hur J, Ozgur A, Xiang Z, et al. Development and application of an interaction network ontology for literature mining of vaccine-associated gene-gene interactions. J Biomed Semantics 2015;6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ozgur A, Hur J, He Y. The Interaction Network Ontology-supported modeling and mining of complex interactions represented with multiple keywords in biomedical literature. BioData Min 2016;9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lin D. An information-theoretic definition of similarity. In Proceedings of the 15th International Conference on Machine Learning. Morgan Kaufmann, 1998, 296–304. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.55.1832.
- 86. NCBITaxon : An Ontology Representation of the NCBI Organismal Taxonomy. OBO Technical WG 2021, http://obofoundry.org/ontology/ncbitaxon.html.
- 87. Mungall CJ, Torniai C, Gkoutos GV, et al. Uberon, an integrative multi-species anatomy ontology. Genome Biol 2012;13:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Diehl AD, Meehan TF, Bradford YM, et al. The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J Biomed Semantics 2016;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sarntivijai S, Lin Y, Xiang Z, et al. CLO: The Cell Line Ontology. J Biomed Semantics 2014;5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. He Y, Liu Y, Zhao B. OGG: a biological ontology for representing genes and genomes in specific organisms. In: The 2014 International Conference on Biomedical Ontologies (ICBO 2014). Houston, TX, USA, 2014, p. 13–20. CEUR Workshop Proceedings. [Google Scholar]
- 91. Natale DA, Arighi CN, Blake JA, et al. Protein ontology: a controlled structured network of protein entities. Nucleic Acids Res 2014;42:D415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kibbe WA, Arze C, Felix V, et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res 2015;43:D1071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tamborero D, Rubio-Perez C, Deu-Pons J, et al. Cancer genome interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 2018;10:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shefchek KA, Harris NL, Gargano M, et al. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 2020;48:D704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. He Y, Sarntivijai S, Lin Y, et al. OAE: The Ontology of Adverse Events. J Biomed Semantics 2014;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bhattacharya S, Dunn P, Thomas CG, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data 2018;5:180015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


