Abstract
A major stress response influenced by microRNAs (miRNAs) is senescence, a state of indefinite growth arrest triggered by sublethal cell damage. Here, through bioinformatic analysis and experimental validation, we identified miR-340-5p as a novel miRNA that foments cellular senescence. miR-340-5p was highly abundant in diverse senescence models, and miR-340-5p overexpression in proliferating cells rendered them senescent. Among the target mRNAs, miR-340-5p prominently reduced the levels of LBR mRNA, encoding lamin B receptor (LBR). Loss of LBR by ectopic overexpression of miR-340-5p derepressed heterochromatin in lamina-associated domains, promoting the expression of DNA repetitive elements characteristic of senescence. Importantly, overexpressing miR-340-5p enhanced cellular sensitivity to senolytic compounds, while antagonization of miR-340-5p reduced senescent cell markers and engendered resistance to senolytic-induced cell death. We propose that miR-340-5p can be exploited for removing senescent cells to restore tissue homeostasis and mitigate damage by senescent cells in pathologies of human aging.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous, small (∼22-nucleotide) noncoding RNAs that typically suppress gene expression by binding to specific segments of mRNAs, generally in the 3′ untranslated region (UTR), and reducing mRNA stability and/or translation (1,2). There are over 1000 known human miRNAs and they modulate the production of up to 60% of proteins; accordingly, miRNAs regulate many cellular processes including proliferation, apoptosis, development, senescence and differentiation. Given that miRNAs are potent post-transcriptional regulatory factors, they have attracted a great deal of interest as therapeutic agents (3). Most studies on miRNA therapeutics have been directed at diseases such as cancer and cardiovascular disease, two broad pathologies that increase during aging (3–6).
Cellular senescence was described as the finite replicative potential of primary human fibroblasts, first discovered by Leonard Hayflick in 1961 (7). Many decades later, we know that cell senescence is a state of indefinite growth arrest triggered by various internal and external sublethal stresses and characterized by distinct RNA and protein expression patterns. A subset of proteins expressed by senescent cells, including cytokines, growth factors and extracellular matrix-remodeling proteins, comprise the SASP (senescence-associated secretory phenotype), a trait that is believed to profoundly influence tissue and organ function. While senescent cells can be both beneficial and harmful depending on the biological context, the accumulation of senescent cells in older human tissues appears to be detrimental and contributes to pathologies associated with aging, such as cardiovascular disease, neurodegeneration and cancer (8–10). Accordingly, there is great interest in developing therapies directed at senescent cells (senotherapies) and in discovering senescence-associated regulatory proteins, RNAs and signaling pathways. The identification of senescence-associated miRNAs (‘senomiRs’) is an important component of this effort.
Until recently, generating therapeutics to intervene in cellular senescence has been challenging due to a lack of accurate and consistent cellular markers. Senescent cells have been identified using a combination of traits, including evidence of DNA damage (e.g. the presence of γ-H2AX), expression of senescence-associated proteins (e.g. p16/CDKN2A, p21/CDKN1A, etc.) and secretion of some of these proteins (e.g. SASP factors), as well as detection of senescence-associated β-galactosidase (SA-βgal) activity (11). To further characterize senescent cells, recent studies have identified proteomic and transcriptomic signatures of senescence using diverse triggers and cell types (12–15). One such study from our lab revealed 50 elevated RNAs and 18 reduced RNAs, coding and noncoding, shared across senescence paradigms (12). Based on the differentially expressed RNAs, we investigated systematically how the senescence transcriptome is altered by miRNAs.
Bioinformatic analysis followed by experimental validation identified miR-340-5p as a critical miRNA regulator of the senescent transcriptome. Previously implicated in tumor suppression and the antiviral response (16–19), miR-340-5p is found here to foment cellular senescence. miR-340-5p was highly abundant in a range of senescence paradigms, and overexpression of miR-340-5p in proliferating cells rendered them senescent. A key mechanism by which miR-340-5p promoted senescence was through its direct impact on LBR mRNA, a transcript encoding the lamin B receptor (LBR), which shows reduced abundance in senescent cells. Importantly, overexpressing miR-340-5p reduced LBR production and enhanced cell sensitivity to senolytic compounds, while antagonization of miR-340-5p reduced senescent cell markers and conferred resistance to cell death by senolytic drugs. Taken together, these results suggest that miR-340-5p is a novel senomiR that can be exploited to remove senescent cells and thereby mitigate damage from senescent cells in aging human pathologies.
MATERIALS AND METHODS
Cell culture, senescence induction and SA-βgal activity
WI-38 human diploid fibroblasts from fetal lung (Coriell Cell Repositories) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (FBS, Gibco), 1% antibiotics, 1% antimycotics and 1% non-essential amino acids (Invitrogen). Proliferating WI-38 fibroblasts were used at population doubling level (PDL) 20. For ionizing radiation (IR)-induced senescence, proliferating (PDL25) WI-38 cells were exposed to 10 Gy and assayed 10 days after IR or as described. WI-38 were treated with 50 μM etoposide for the times shown. SA-βgal activity was assessed using a senescence-associated β-galactosidase staining kit (Cell Signaling). Primary human coronary artery vascular smooth muscle cells (VSMCs) were obtained as cryopreserved secondary cultures from Lifeline Cell Technology and maintained in the VascuLife® SMC Medium Complete Kit from Lifeline Cell Technology according to the manufacturer’s protocol. hVSMCs were rendered senescent by passaging until replicative exhaustion (passage 12) or exposure to IR (10 Gy). H9C2 rat cardiomyocytes were obtained from ATCC and cultured in DMEM with 10% FBS. H9C2 cells were rendered senescent by treatment with 125 or 250 nM doxorubicin for 7 days.
Cell counting and BrdU incorporation
To assess population growth, cells were transfected to overexpress or antagonize miR-340-5p, as well as to silence or overexpress LBR, and then either counted or subjected to bromodeoxyuridine (BrdU) incorporation analysis. Cell proliferation was assessed using the BrdU Cell Proliferation Assay (Cell Signaling). Briefly, BrdU was added 1 day after transfection of siCtrl or siLBR and the incorporation was detected at days 0, 3 and 6, using the GloMax plate reader (Promega). For miRNA-transfected cells, BrdU was added 3 days after transfection and detected 24 h later. Total cells and dead cells were counted on an automatic cell counter (Bio-Rad) after staining with 0.4% Trypan Blue (Gibco).
Senolytics and caspase 3/7 activity
Cells were transfected as described and treated with senolytics [ABT-737 10 μM (Tocris); dasatinib 20 μM + quercetin 8 μM (Selleckchem)] for 24 h. Viability was monitored by cell counting and represented as the percentage of remaining cells compared to the number of cells at the start of the experiment. Apoptosis was monitored by measuring caspase 3/7 activity using the Caspase-Glo 3/7 Assay System (Promega). Briefly, Caspase-Glo® 3/7 solution was added directly to each well. The plate was then shaken vigorously for 30 s and incubated at 25°C for 30–180 min, whereupon luminescence was measured by the GloMax plate reader (Promega).
miRNAs, siRNAs, site blockers, plasmids and lentiviral expression vectors
To overexpress miRNAs, cells were transfected with precursor miRNAs consisting of a duplex that contained a guide strand and a passenger strand, which improves AGO2 loading and processing. WI-38 cells were transfected with precursor miRNAs from Thermo Fisher Scientific at 25–50 nM: miR-340-5p (PM12670), miR-129-5p (PM10076), miR-19a-3p (PM10649), miR-128-3p (PM11746), miR-124-3p (PM11154), miR-27b-3p (PM10750) and a precursor control miR (miR Ctrl) (AM17110). To antagonize miR-340-5p, we transfected WI-38 cells with anti-miR-340-5p (Thermo Fisher Scientific, AM12670) or anti-miR Ctrl (Thermo Fisher Scientific, AM17010) (25–50 nM). For silencing experiments, WI-38 cells were transfected with siCtrl (Dharmacon D-001810-10-05) or siLBR (25 nM) (Dharmacon L-021505-00-0005), or siLBR (1) targeting the miR site (Qiagen, 1027423). All small RNA transfections were performed using Lipofectamine RNAiMAX (Thermo Fisher Scientific). Target site blockers (SBs) are LNA-enhanced antisense oligonucleotides that bind to the miRNA target site of an mRNA, preventing the miRNA from binding the mRNA, as reported (20,21) and leading to a rise in expression of the protein encoded by the mRNA. The SBs used in this study are miRCURY LNA miRNA Power Target Site Blocker LBR-miR-340-5p (5′-TGCAACAGTTTATAAAGGCA-3′) and miRCURY LNA miRNA Power Target Site Blocker (5′-TAACACGTCTATACGCCCA-3′) from Qiagen.
For plasmids, pLBR, pcDNA and an empty pcDNA plasmid (5 ng) were transfected using the Amaxa Nucleofector following the manufacturer’s protocol. Lentiviral vectors expressing GFP or LBR-GFP (Origene Technologies) were transduced at 3 MOI (multiplicity of infection) with polybrene. Sequences and catalog information for miRNAs and siRNAs are listed in Supplementary Tables S1 and S2, respectively.
Luciferase reporter analysis
To test the impact of miR-340-5p on the 3′UTR of LBR mRNA, dual reporter plasmids psiCHECK2, psiCHECK2-LBR(3′UTR) and psiCHECK2-LBR(mut3′UTR) were generated and studied. The psiCHECK2-LBR(3′UTR) plasmid contains the full-length LBR 3′UTR; the psiCHECK2-LBR(mut3′UTR) contains the full-length LBR 3′UTR, but it has a mutant sequence (ACGATGCC) instead of the wild-type sequence (CTTTATAA) at the miR-340-5p-binding site (positions 2833–2840). Cell cultures (4 × 105 cells/well) were co-transfected with 250 ng of the reporter constructs using Amaxa nucleofection (Lonza Bioscience) following the manufacturer’s instructions and either 50 nM of miR Ctrl or 50 nM of miR-340-5p, along with 50 nM of negative control target site blocker (NC SB) or LBR target site blocker (LBR SB) (Qiagen) using the Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific). Cells were lysed and luciferase activities were assessed using a dual luciferase assay kit (Promega) on a GloMax plate reader (Promega).
RNA isolation and RT-qPCR analysis
RNA was isolated from cells using the TriPure isolation reagent (Roche) and the Direct-zol Mini Kit (Zymo) following the manufacturers’ protocols and including a step of digestion with DNase I. Total RNA was reverse transcribed (RT) into cDNA using Maxima reverse transcriptase (Thermo Fisher Scientific) and random hexamers and measured by quantitative polymerase chain reaction (qPCR) analysis using SYBR Green mix (Kapa Biosystems). The relative mRNA expression levels were calculated by the 2–ΔΔCt method and normalized to ACTB mRNA (22). miRNAs were reverse transcribed using the MiR-X miRNA first-strand synthesis kit (Takara Bio) and quantified by qPCR analysis using the small RNA U6 as normalization control (23). The PCR primers used are listed in Supplementary Table S1.
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer supplemented with Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Lysates were separated by SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were probed with primary antibodies recognizing LBR, HP1α (Abcam), phosphorylated CHK2 (P-CHK2) (Cell Signaling), p21 (CDKN1A), β-Actin (ACTB), HSP90 or p53 (TP53) (Santa Cruz Biotechnology). After incubation with secondary antibodies conjugated with horseradish peroxidase (GE Healthcare), signals were detected using enhanced chemiluminescence and a KwikQuant Imager (Kindle Biosciences, LLC).
Immunofluorescence
LBR immunofluorescence was carried out as previously described (24). Briefly, 500 cells/well were plated in chamber slides (Thermo Scientific Nunc), fixed 24 h later with 4% formaldehyde, permeabilized with 0.2% Triton X-100 and blocked in 10% normal goat serum (Life Technologies) for 1 h at 37°C. After blocking, cells were incubated with antibodies recognizing LBR, p21, HP1α or H3K9me3 in 10% normal goat serum for 1 h at 37°C and then with Alexa Fluor 568 (Life Technologies) in 10% normal goat serum for 30 min at 37°C. After washes with PBS, the nuclei were stained with DAPI (Dojindo Laboratories) for 15 min at 25°C and mounting medium (Life Technologies) was employed to cover the cells. Signals were visualized with a Keyence microscope.
Biotinylated miRNA pulldown
WI-38 fibroblasts were transfected with 50 nM biotinylated miR-340-5p or control biotinylated miR using Lipofectamine RNAiMAX (Thermo Fisher Scientific); 16 h later, cells were washed with PBS and lysed in 1 ml lysis buffer [20 mM Tris (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% NP-40, 100 U of RNaseOUT (Invitrogen) and protease inhibitor (Roche)], and incubated on ice for 10 min. After clearing the cytoplasmic lysates by centrifugation at 12 000 × g (10 min), streptavidin-coated Dynabeads (100 μl) were added and further incubated (8 h, 4°C with rotation). Beads were washed four times with 1 ml ice-cold lysis buffer, RNA was isolated using TriPure isolation solution and the enrichment of LBR mRNA was measured by RT-qPCR analysis and normalized to the levels of GAPDH mRNA.
ChIP-qPCR analysis
The protocol used was adapted from Sen and colleagues (25). Briefly, cells were cross-linked with 1% formaldehyde diluted in PBS for 10 min at 37°C. After glycine quenching, nuclei were prepared from the cell pellets and lysed in buffer containing 10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% sodium lauroylsarcosine and a protease inhibitor cocktail (Thermo Fisher Scientific), and sonicated with a Covaris S220 ultrasonicator, yielding chromatin fragments ∼250 bp long. The supernatant was cleared by addition of 1% Triton X-100 followed by centrifugation and subjected to immunoprecipitation with 5 μg of control IgG, or antibodies recognizing H3 or H3K9me3 at 4°C for 18 h followed by incubation with Dynabeads Protein G (Invitrogen) at 4°C for 18 h. The beads were then washed twice with FA lysis buffer (50 mM HEPES–KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.1% Triton X-100), 1× with FA lysis buffer + 500 mM NaCl, twice with LiCl solution (10 mM Tris–HCl, pH 7.5, 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40) and twice with TE + 0.1% NP-40, and eluted by incubation with elution buffer (50 mM Tris, pH 7.5, 10 mM EDTA, 1% SDS) at 65°C for 30 min with agitation in a thermomixer. The chromatin immunoprecipitation (ChIP) and input samples were then reverse cross-linked by incubation at 65°C overnight and purified using Qiagen columns for qPCR analysis. The primers used for qPCR amplification of gene promoter regions and lamina-bound regions are listed in Supplementary Table S1 (26).
RNA-seq analysis
Next-generation RNA sequencing (RNA-seq) analysis was conducted commercially by LC Sciences and ArrayStar. For sequencing by LC Sciences, the RNA quality was checked using the 6000 Nano LabChip Kit (Agilent Technologies) on a Bioanalyzer 2100 instrument to achieve RIN numbers >7.0; ∼1 μg of RNA was used to construct a small RNA library following the protocol of Illumina’s TruSeq small RNA sample preparation kits. Sequencing was then performed on an Illumina HiSeq2500 instrument with single-end mode (36 bp SE). For sequencing by ArrayStar, total RNA was initially quantified using a Nanodrop ND-1000 instrument, agarose electrophoresis was used to check RNA integrity and ∼1–2 μg total RNA per sample was used for library preparation. Briefly, rRNA was removed from the total RNA with a RiboZero Magnetic Gold Kit and the rRNA-depleted RNA was used for RNA-seq library preparation using the KAPA Stranded RNA-Seq Library Prep Kit (Illumina). The library preparation included (i) fragmentation of the RNA, (ii) reverse transcription to synthesize first-strand cDNA, (iii) second-strand cDNA synthesis incorporating dUTP, (iv) end repair and A-tailing of the double-stranded cDNA, (v) ligation of Illumina-compatible adapters and (vi) PCR amplification and purification of the final RNA-seq library. The completed libraries were evaluated on an Agilent 2100 Bioanalyzer for concentration, fragment size distribution between 400 and 600 bp, and adapter dimer contamination. The amount was determined by absolute quantification qPCR analysis. The barcoded libraries were mixed in equal amounts and used for sequencing using an Illumina HiSeq2500 instrument. For both experiments, the RNA-seq reads were aligned to human genome hg19 using Spliced Transcripts Alignment to a Reference (STAR) software version 2.4.0j and featureCounts (v1.4.6-p5) (27) were used to create gene counts. Differential expression analysis of gene counts was performed in R (version 3.6.3) using the DESeq2 package version 1.26.0 (28). Transcripts were defined as being differentially regulated with Benjamini–Hochberg adjusted P-value <0.05 and absolute log2 fold change >1. Functional analysis of the differentially expressed genes was conducted by Ingenuity Pathways Analysis (IPA Summer Release, June 2020, Qiagen Inc.) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed with clusterProfiler v3.17.3 package in R (29). RNA-seq data are deposited in GSE165469.
ATAC-seq analysis
The library for ATAC-seq (assay for transposase-accessible chromatin followed by deep sequencing) analysis was prepared according to a published protocol (30) using 50 000 intact cells as input. Tn5 transposase tagmentation was used to simultaneously fragment the genome and tag the resulting DNA with Illumina sequencing adapters. DNA fragments were amplified by PCR and subsequently purified using the Qiagen MinElute PCR Purification Kit (Qiagen). The quality and quantity of the final library were analyzed and measured by Agilent Bioanalyzer 2100 (Agilent Technologies) and Life Technologies Qubit 3.0 Fluorometer (Life Technologies), respectively. The final libraries were sequenced using 150-bp paired-end reads on an Illumina HiSeq 2500 Sequencer (Illumina) by Quick Biology. The reads were first mapped to the UCSC hg38 human genome using Bowtie2 version 2.1.0 (31). The mitochondria reads, duplicate reads and non-unique reads were removed before peak calling using MACS2 in the BAMPE mode (32). Differential peaks were identified using the EdgR GLM algorithm embedded in DiffBind program (33) and annotated by using the CHIPpeakAnno package from Bioconductor (34); peaks showing altered abundance with q < 0.05 and >1.5-fold change were considered differentially expressed. Gene annotations of the position of differential peaks were visualized using ChipSeeker (35). The human genome repeat elements of the ATAC-seq peaks were obtained by mapping with the human genome repeat element library at the UCSC database. ATAC-seq data are deposited in GSE165469.
Nuclear analysis
An analysis was performed on immunofluorescent images of DAPI-stained nuclei. The outer boundary of DAPI-stained nuclei was annotated and used as a training set for U-NET (36). After training to detect nuclear boundaries, it was applied to all images. The predicted detection region was extracted and used for morphological analysis of the nuclei. Detected nuclei were reviewed, and overlapping nuclei were excluded from analysis. The morphological properties, area and solidity (the ratio of area to convex hull area), were quantified (37). In addition, the extracted nuclei images were processed by a senescent nuclei prediction model, based on an ensemble of 10 convolutional neural networks, which generated mean probability scores for each nuclei as described by Heckenbach and colleagues (38).
Resources
Details of materials used in this paper, including antibodies, chemical, commercial assays, cell lines, software and algorithms, are provided in Supplementary Table S2.
Statistical analysis
Data are presented as the means ± standard error of the mean (SEM). Significance was tested using two-tailed Student’s t-test; P < 0.05 was considered significant.
RESULTS
Senescent cells express increased levels of miR-340-5p
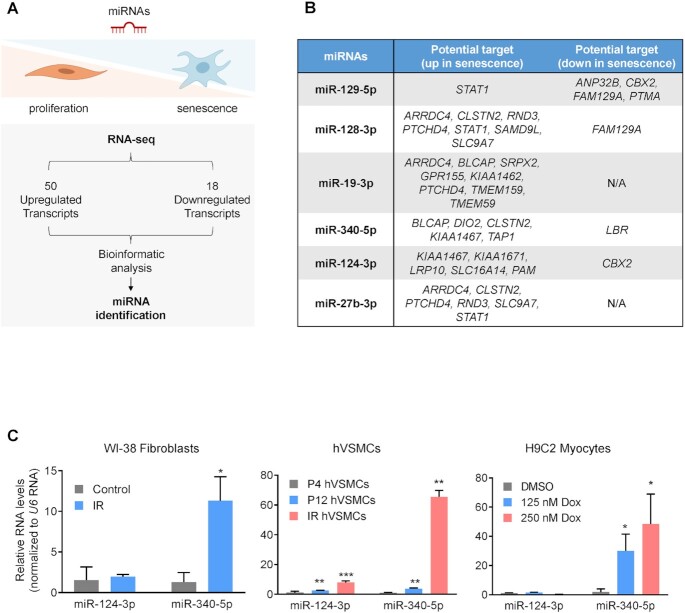
To begin the identification of miRNA regulators of transcriptomic programs in senescence, we analyzed the RNA-seq datasets from Casella and coworkers (12). This study reported RNAs jointly elevated (50 total) and jointly reduced (18 total) across different models of senescence, including fibroblasts, endothelial cells and epithelial cells rendered senescent by replicative passaging or exposure to IR or doxorubicin. Using the bioinformatic prediction tool TargetScan (39), we assembled a list of six candidate miRNAs predicted to bind and regulate those RNAs that were elevated or reduced across senescent models (12), with the goal of identifying ‘master’ senescence-regulatory miRNAs (Figure 1A and B). The miRNAs identified had the potential to bind several senescence-associated RNAs (at least one conserved miRNA-binding site) and hence might control senescence traits. To narrow down the list of interesting miRNAs, we overexpressed these six miRNAs (Supplementary Table S1) in proliferating human diploid WI-38 fibroblasts and measured SA-βgal activity, a well-established characteristic of senescent cells (40) (Supplementary Figure S1A). The results indicated that two miRNAs, miR-124-3p and miR-340-5p, promoted senescence in proliferating cells. We then considered whether the two miRNAs that induced senescence in proliferating cells were changed in senescent cells. We triggered senescence in a range of paradigms, including through exposure of WI-38 fibroblasts to IR [10 Gy (41), collected 10 days later], replicative passaging of hVSMCs for 12 population doublings and doxorubicin treatment (250 nM for 7 days) of H9C2 rat cardiomyocytes. We observed a strong increase in miR-340-5p levels in IR-treated and doxorubicin-treated cells and a modest increase after replicative passaging (Figure 1C; Supplementary Figure S1B). The increase in miR-340-5p expression levels was more prominent in cells with acute and direct DNA damage (IR and doxorubicin) and likely reflects the greater homogeneity of the cells responding to stress. In comparison, miR-124-3p levels were inconsistently altered or showed no change in these diverse senescence models (Figure 1C and data not shown). Given these results, we focused on miR-340-5p as a putative broad regulator of cell senescence. Human miR-340-5p is located in the intronic region of host gene RNF130 (which encodes ring finger protein 130). The levels of RNF130 mRNA in various models of senescence did not change significantly (Supplementary Figure S1C), suggesting that the rise of miR-340-5p in senescence may not be due to overall increased transcription of the host gene.
Figure 1.
miR-340-5p levels increase in senescent cells. (A) Top: schematic of the strategy employed to identify miRNAs either promoting or repressing the senescence-associated transcriptomic program. Bottom: workflow describing the analysis of RNAs highly expressed (50 RNAs) and reduced (18 RNAs) across several models of senescence (12); the miRNAs predicted to bind to and repress these RNAs were selected for analysis. (B) Table of the top miRNA candidates based on the presence of predicted seed sequences complementary to the differentially expressed RNAs (18 + 50) mentioned in panel (A). (C) RT-qPCR analysis of the levels of miR-340-5p and miR-124-3p in proliferating WI-38 human diploid fibroblasts (PDL20) 10 days after treatment with 10 Gy of IR (left), in hVSMCs reaching senescence by passaging to PDL12 or by culture for 10 days after exposure to IR (10 Gy) (middle), and in rat H9C2 cardiomyocytes 10 days after treatment with 125 or 250 nM doxorubicin (right). Values were normalized to the levels of the small RNA U6. Data represent the mean values ± SD from three biological replicates. Significance was established using Student’s t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Elevating miR-340-5p in proliferating cells triggers senescence
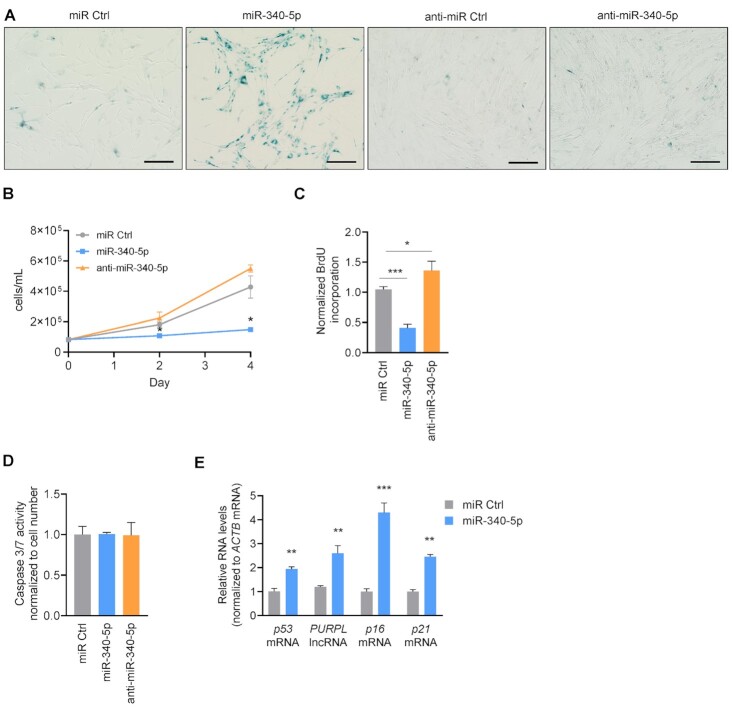
We first investigated the role of miR-340-5p in cell senescence by increasing its abundance in proliferating WI-38 fibroblasts (PDL20). Senescent cells display many distinct phenotypes, including a flattened and enlarged morphology, increased lysosomal activity reflected by enhanced SA-βgal activity (see the ‘Materials and Methods’ section) and irreversible growth arrest that can be measured by assessing proliferation (40,42,43). By 3 days after increasing miR-340-5p levels through transfection, WI-38 fibroblasts displayed an enlarged morphology, elevated SA-βgal activity and reduced cell proliferation relative to control populations (miR Ctrl), as determined by measuring cell numbers and BrdU incorporation (Figure 2A–C; Supplementary Figure S1D), suggesting that miR-340-5p overexpression alone was sufficient to drive proliferating cells into senescence. Conversely, antagonization of miR-340-5p (by transfecting anti-miR-340-5p) reduced SA-βgal activity and enhanced cell proliferation relative to control cells (Figure 2A–C). In additional control experiments, transfecting WI-38 fibroblasts with miR Ctrl did not induce stress pathways, as determined by a lack of significant increase in the stress- and senescence-associated protein p53 (Supplementary Figure S1E). We also compared the effects of miR-340-5p and anti-miR-340-5p to both miR Ctrl and an anti-miR Ctrl and observed no differences in SA-βgal activity, proliferation or morphology between the two control miRNAs; therefore, for the remainder of the experiments in proliferating fibroblasts, we compared miRNAs to miR Ctrl (Figure 2A; Supplementary Figure S1F). Furthermore, neither elevating nor antagonizing miR-340-5p significantly modified caspase 3/7 activity (Figure 2D), suggesting that the changes in population growth were not due to altered cell death. Importantly, overexpressing miR-340-5p in proliferating cells increased the levels of the senescence markers p16 mRNA, p21 mRNA and p53 mRNA, as well as the long noncoding RNA PURPL, which is also elevated in senescent cells (12), relative to control cells (miR Ctrl) (Figure 2E). In sum, heightened levels of miR-340-5p enhanced senescence traits including SA-βgal activity, senescence markers like p16 and p21 mRNAs, and growth arrest.
Figure 2.
miR-340-5p overexpression promotes senescence in proliferating fibroblasts. (A) Micrographs depicting SA-βgal staining of WI-38 fibroblasts at PDL20 4 days after transfection with miR Ctrl, miR-340-5p, anti-miR Ctrl or anti-miR-340-5p. Scale bar: 100 μm. (B) Cell proliferation counts (cells/ml) at days 0, 2 and 4 after transfection with control miRNA (miR Ctrl), miR-340-5p or anti-miR-340-5p, as determined using a Bio-Rad TC20 cell counter. (C) BrdU incorporation analysis was performed 3 days after transfection with miR Ctrl, miR-340-5p or anti-miR-340-5p (see the ‘Materials and Methods’ section). Values were normalized to miR Ctrl. (D) Caspase 3/7 activity was measured 4 days after transfection with miR Ctrl, miR-340-5p or anti-miR-340-5p (see the ‘Materials and Methods’ section). (E) RT-qPCR analysis of the indicated RNAs in early-passage fibroblasts (PDL20) 4 days after transfection with control miRNA (miR Ctrl) or pre-miR-340-5p (miR-340-5p). RNA levels were normalized to the levels of ACTB mRNA. Data in panels (B)–(E) represent the mean values ± SD from three biological replicates. Significance was established using Student’s t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Effects of miR-340-5p on pre-senescent cells
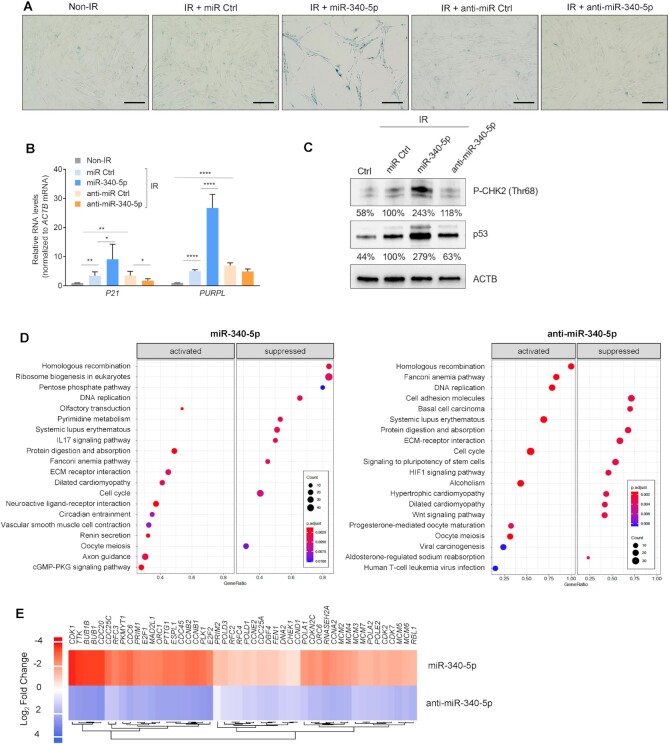
We further analyzed the effects of miR-340-5p overexpression and antagonization in pre-senescent WI-38 fibroblasts; pre-senescent cells typically still proliferate, but have initiated changes in response to damage that have not yet reached the threshold that triggers indefinite cell cycle arrest and other senescence markers (44). Cells were transfected with miR Ctrl, miR-340-5p, anti-miR Ctrl or anti-miR-340-5p, and then either left untreated or exposed to 10 Gy IR and cultured for an additional 4 days to induce a pre-senescent phenotype. At day 4, IR cultures started to show positive SA-βgal activity (Figure 3A) and higher levels of senescent markers p21 mRNA and PURPL lncRNA (Figure 3B) and p53 (Figure 3C). These traits were more accentuated in pre-senescent cultures overexpressing miR-340-5p, as they displayed increased SA-βgal activity and higher levels of senescent markers p21 mRNA, PURPL and p53, while antagonizing miR-340-5p lowered SA-βgal activity and the IR-induced p21 mRNA and p53 protein (Figure 3A–C). The rise in levels of a marker of DNA damage, P-CHK2 (the kinase CHK2 phosphorylated at Thr 68), as well as p53 revealed that overexpression of miR-340-5p exacerbated DNA damage in cells responding to IR-elicited senescence (Figure 3C). In sum, besides promoting senescence when overexpressed, miR-340-5p also augments the cellular response to a senescence-triggering stimulus (Figure 3A–C; Supplementary Figure S2A and B).
Figure 3.
Increasing and antagonizing miR-340-5p have opposing effects on pre-senescent WI-38 fibroblasts. (A) Micrographs depicting SA-βgal staining of WI-38 fibroblasts 4 days after exposure to IR (10 Gy) in the presence of transfected miR Ctrl, miR-340-5p, anti-miR Ctrl or anti-miR-340-5p. Scale bar: 100 μm. (B) RT-qPCR analysis of the levels of p21 mRNA and PURPL (normalized to ACTB mRNA levels) in WI-38 cells treated as explained in panel (A). Data represent the mean values ± SD from three biological replicates. Significance was established using Student’s t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. (C) Western blot analysis of the indicated proteins [CHEK2/CHK2 phosphorylated at threonine 68 and p53/TP53 in cells treated as explained in panel (A)]; ACTB was included as a loading control. The % values under each band represent the densitometric calculations of the signals on western blots, shown as a percentage relative to miR Ctrl and normalized to ACTB signals. (D) Top 20 significantly activated and suppressed KEGG pathways of the mRNAs differentially expressed after miR-340-5p overexpression compared to miR Ctrl (left) and after miR-340-5p antagonization (anti-miR-340-5p) compared to miR Ctrl (right) samples. Gene ratio represents the proportion of the significant mRNAs from the total number of mRNAs implicated in a given pathway. (E) RNA was collected 4 days after transfecting WI-38 (PDL20) fibroblasts with miR-340-5p or anti-miR-340-5p for RNA-seq analysis. Heatmap depicts the differentially expressed RNAs in cell cycle and DNA replication KEGG pathways after transfection with miR-340-5p and with anti-miR-340-5p. Values reflect log2 fold change.
To investigate more deeply the mechanisms and pathways by which miR-340-5p promotes senescence, we performed RNA-seq analysis of WI-38 fibroblasts (PDL20) transfected with miR Ctrl, miR-340-5p or anti-miR-340-5p. We performed functional IPA and KEGG enriched pathway analysis of the differentially abundant transcripts when miR-340-5p was overexpressed and when it was antagonized (Figure 3D; Supplementary Figure S2A and B). IPA and KEGG pathway enrichment showed that the processes highly affected by modulating miR-340-5p included many nuclear events such as DNA replication, cell cycle progression, chromosome maintenance and DNA damage response. In Figure 3E, and Supplementary Figure S2B, we identified a subset of functionally related mRNAs derived from the top enriched pathways by IPA and KEGG analyses that are inversely regulated by overexpressing and antagonizing miR-340-5p and relevant to DNA damage and repair. These results supported the notion that raising miR-340-5p levels triggered a DNA damage response that was opposed by antagonizing miR-340-5p.
miR-340-5p directly binds and suppresses LBR mRNA
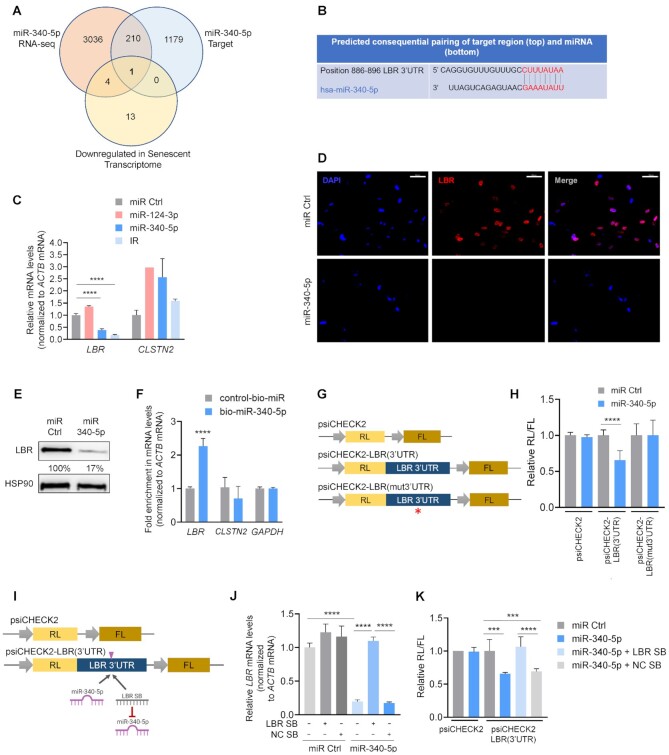
To better understand the mechanism(s) by which miR-340-5p induces cellular senescence, we surveyed the mRNAs predicted computationally (using TargetScan) to be targets of miR-340-5p and the transcripts reduced as a result of miR-340-5p overexpression in WI-38 fibroblasts, identified using RNA-seq analysis (and adjusted P < 0.05). The 211 overlapping transcripts identified were then compared with those decreased in the senescent transcriptome (12), revealing the LBR mRNA as the lone, shared candidate (Figure 4A). LBR mRNA features an 8-mer seed sequence in its 3′UTR that is complementary to miR-340-5p, as demonstrated by TargetScan alignment (Figure 4B). To test whether miR-340-5p regulates LBR production, we overexpressed miR-340-5p in WI-38 fibroblasts, alongside miR Ctrl and miR-124-3p (a control miRNA that is not predicted to target LBR mRNA); cells exposed to IR and collected 7 days later were included as a positive control. As shown (Figure 4C), overexpression of miR-340-5p reduced LBR mRNA abundance in cells, as did IR-induced senescence. The levels of CLSTN2 mRNA, a predicted target of miR-340-5p with similar complementarity, did not change significantly, suggesting that miR-340-5p diminished LBR mRNA levels with some degree of selectivity. The ensuing reduction of LBR protein by miR-340-5p was corroborated by immunofluorescence and western blot analysis (Figure 4D and E; Supplementary Figure S2C). These results indicate that miR-340-5p suppresses the production of LBR mRNA and LBR protein, both abundant in proliferating cells (Figure 4D and E).
Figure 4.
LBR mRNA is a direct target of repression by miR-340-5p. (A) Venn diagram comparing RNAs reduced by miR-340-5p overexpression as determined by RNA-seq analysis, predicted miR-340-5p-target transcripts and transcripts decreased in the senescent transcriptome as identified by RNA-seq analysis. (B) Alignment of miR-340-5p with the LBR 3′UTR seed sequence as predicted by TargetScan. (C) RT-qPCR analysis of the levels of LBR and CLSTN2 mRNAs in WI-38 fibroblasts (PDL20) 4 days after transfection with miR Ctrl, miR-124-3p, miR-340-5p or 10 days after IR. mRNA levels were normalized to ACTB mRNA levels. (D) Immunofluorescence micrographs (40×) to detect nuclei (DAPI, blue) and LBR (red) in WI-38 fibroblasts (PDL20) 4 days after transfecting miRNA Ctrl or pre-miR-340-5p. Scale bar: 50 μm. (E) Western blot analysis of the levels of LBR or loading control HSP90 in WI-38 cells processed as explained in panel (D). The % values represent the densitometric calculations of the signals on the western blots, shown as a percentage relative to miR Ctrl and normalized to HSP90 signals. (F) RT-qPCR analysis of the enrichment of LBR mRNA (as well as controls CLSTN2 mRNA and GAPDH mRNA) in the material pulled down with streptavidin-conjugated magnetic beads 16 h after transfecting cells with either biotinylated miR-340-5p (bio-miR-340-5p) or a control biotinylated RNA (control-bio-miR). (G) Luciferase reporter constructs. The dual reporter vector psiCHECK2 expresses Renilla luciferase (RL) and firefly luciferase (FL) under separate promoters. Plasmid psiCHECK2 is the empty vector control; plasmid psiCHECK2-LBR(3′UTR) has the LBR 3′UTR inserted directly following the RL coding region; plasmid psiCHECK2-LBR(mut3′UTR) has the LBR 3′UTR with a mutation in the miR-340-5p binding site (red asterisk) inserted directly following the RL coding region. (H) Proliferating WI-38 fibroblasts were transfected with psiCHECK2, psiCHECK2-LBR(3′UTR) or psiCHECK2-LBR(mut3′UTR) plasmids; 24 h later, cells were transfected with miR Ctrl or miR-340-5p, and 24 h after that, lysates were prepared and RL activity was normalized to FL activity and further normalized to miR Ctrl transfection for each plasmid. (I) Luciferase reporter constructs depicting the position where miR-340-5p and the LBR SB bind is indicated (pink arrowhead). (J) RT-qPCR analysis of LBR mRNA levels 4 days after co-transfection of either miR Ctrl alone or miR-340-5p in the presence or absence of a negative control site blocker (NC SB) or a site blocker specifically designed to abrogate binding of miR-340-5p to the LBR mRNA (LBR SB). (K) Assessment of LBR 3′UTR activity. Proliferating WI-38 fibroblasts were transfected with psiCHECK2 or psiCHECK2-LBR(3′UTR) plasmids; 24 h later, cells were transfected with miR Ctrl, miR-340-5p, miR-340-5p + LBR site blocker (LBR SB) or miR-340-5p with a negative control site blocker (NC SB). RL activity was normalized to FL activity and further normalized to miR Ctrl transfection for each plasmid. Data in panels (C), (F), (H), (J) and (K) represent the mean values ± SD from three biological replicates. Significance was established using Student’s t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
We sought confirmation that miR-340-5p directly associated with the LBR 3′UTR by transfecting WI-38 fibroblasts with biotinylated miR-340-5p (or biotinylated miR control) and pulling down bound mRNAs to assess their physical interaction. As shown in Figure 4F, biotinylated miR-340-5p was transfected (Supplementary Figure S2D), and LBR mRNA (but not CLSTN2 mRNA or GAPDH mRNA) was successfully pulled down (Figure 4F), supporting their direct association. Next, we prepared reporter constructs derived from psiCHECK2, containing either wild-type full-length LBR 3′UTR or a modified LBR 3′UTR in which the mutant sequence (ACGATGCC) was inserted instead of the wild-type sequence (CTTTATAA) in the miR-340-5p-binding site in order to determine the impact of miR-340-5p on the suppression of LBR mRNA (Figure 4G). We confirmed the ability of miR-340-5p to reduce the expression of psiCHECK2-LBR(3′UTR) construct, but not psiCHECK2-LBR(mut3′UTR), relative to the empty psiCHECK2 control vector (Figure 4H).
To further assess whether miR-340-5p specifically binds the LBR 3′UTR site (Figure 4B), we tested an LBR 3′UTR SB; the LNA modification on the SB (see the ‘Materials and Methods’ section) allows it to compete effectively with the miRNA/RISC complex for the miRNA target site. As shown, use of the LBR SB effectively blocked the reduction of LBR mRNA by miR-340-5p, supporting the specificity of the miR-340-5p site on the LBR 3′UTR (Figure 4I and J). Similarly with the reporter construct, while overexpressing miR-340-5p did not influence expression of the control vector, it did suppress RL expression from psiCHECK2-LBR(3′UTR); this repression was attenuated by the LBR SB, but not the control SB (Figure 4K). Together, these data confirm that miR-340-5p represses LBR production through the miR-340-5p site on the LBR 3′UTR.
Loss of LBR induces cellular senescence and derepresses LAD-containing heterochromatin
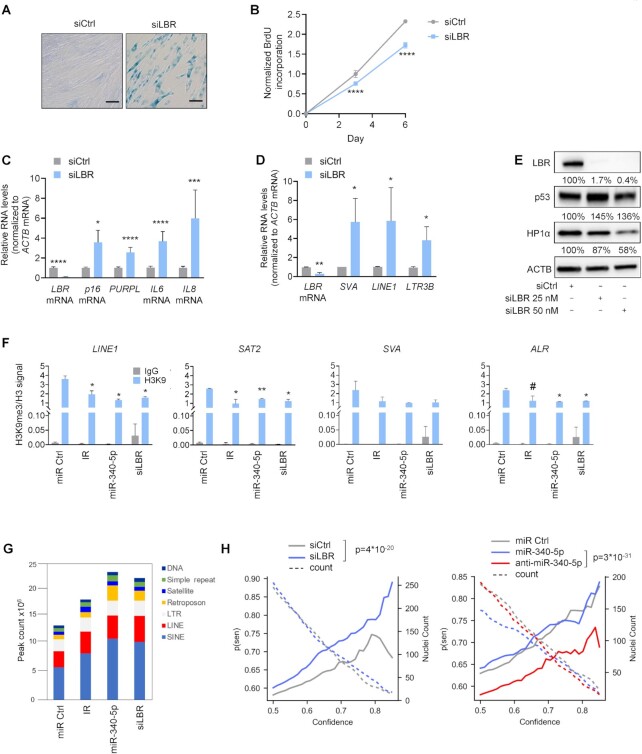
We then sought to determine whether the loss of LBR, which is directly repressed by miR-340-5p, was sufficient to induce cellular senescence in WI-38 cells, given earlier evidence that LBR loss contributed to senescence (45). Compared with transfection of control (siCtrl) siRNA, silencing of LBR using a pool of siRNAs (siLBR; see the ‘Materials and Methods’ section) caused morphological changes consistent with senescence, enhanced SA-βgal activity and reduced cell proliferation (Figure 5A and B). The reduction in the levels of LBR mRNA and LBR was confirmed by RT-qPCR and western blot analyses, respectively, and included positive controls such as the senescence markers p16 mRNA, PURPL, IL6 mRNA and IL8 mRNA, as well as the p53 protein (Figure 5C and E). We tested two other single siRNAs designed to target LBR mRNA, one with extended complementarity to the miR-340-5p seed sequence site in the LBR 3′UTR and one designed to target the coding region of LBR mRNA. The results (Supplementary Figure S3A–C) indicate that both siRNAs strongly elicited changes in cell morphology and proliferation consistent with senescence; however, siLBR directed at the miR site caused long-term toxicity possibly due to off-target effects. Transfection with siLBR targeting the coding region induced the same senescence markers as previously observed (Supplementary Figure S3A–C).
Figure 5.
Loss of LBR promotes senescence and expression of DNA repetitive elements. (A) Micrographs depicting SA-βgal activity in WI-38 fibroblasts (PDL20) 7 days after transfection with control siRNA (siCtrl) or LBR siRNA (siLBR). (B) BrdU incorporation analysis in cells transfected as in panel (A), and collected at 0, 3 and 6 days after transfection. Graph represents normalized BrdU incorporation at each time point. (C) RT-qPCR analysis of LBR mRNA and the senescence-associated RNAs indicated, 7 days after transfection with siCtrl or siLBR; RNA levels were normalized to ACTB mRNA levels. (D) RT-qPCR analysis of the expression of DNA repetitive elements LINE1, SVA and LTR3B, as well as LBR mRNA. Data were normalized to ACTB mRNA levels. (E) Western blot analysis of the proteins indicated (including loading control ACTB) 7 days after transfection with either control siRNA or 25 or 50 nM siLBR. Western blotting signals were analyzed by densitometry and represented as a percentage relative to siCtrl, normalized to ACTB signals. (F) In WI-38 cells (PDL20) that were transfected with miR Ctrl, subjected to IR (10 Gy, 10 days) to induce senescence or transfected with miR-340-5p or siLBR, the relative occupancy of the heterochromatin mark H3K9me3 in the regulatory regions of DNA repetitive elements contained in LADs (LINE1, SVA, SAT2, ALR) and ACTB promoter (Supplementary Figure S3C) was determined by ChIP-PCR analysis using antibodies directed at H3K9me3 (or control IgG), and normalized to input and H3, respectively (n = 2). (G) ATAC-seq sequences were mapped to the hg38 human genome and represented as peak counts associated with the different repeat element classes for each sample. (H) Prediction of senescence in the indicated samples by using an ensemble of deep neural networks trained on the analysis of nuclear morphology of senescent cells (n = 93–175 individual nuclei from two independent experiments). Data in panels (B)–(D) represent the mean values ± SD from three biological replicates. Data in panel (F) represent the mean values ± SD from two biological replicates. Significance was established using Student’s t-test. #P ≤ 0.09; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Given that LBR functions in the maintenance of heterochromatin tethers to the inner nuclear membrane (46), we measured the levels of RNAs encoded by typically repressed DNA repetitive elements preserved in heterochromatin, lamina-associated domains (LADs). By RT-qPCR analysis, we measured the expression levels of several repetitive elements: long interspersed nuclear element 1 (LINE1), SVA elements (SINE/VNTR/Alu) and long terminal repeat 3B (LTR3B) in cells in which LBR was silenced. As shown in Figure 5D, we observed significant increases in the levels of transcripts expressed from these repetitive sequences. We also found decreased levels of HP1α, a heterochromatin-binding protein and scaffold of LBR (Supplementary Figure S4A and B), supporting the notion that there was a loss of heterochromatin when LBR was silenced (26). These findings indicate that the reduction of LBR driven by either a rise in miR-340-5p or ectopic silencing of LBR reduced chromatin stability and promoted the decompaction of LAD regions of DNA.
To further understand the changes in chromatin status resulting from altered miR-340-5p and LBR abundance, we performed ChIP analysis to assess the levels of the heterochromatin mark H3K9me3 in DNA regions bearing LADs. Previous studies suggested that DNA repetitive elements in LADs, such as LINE1, are derepressed in cellular senescence (47); therefore, we tested them by ChIP analysis in fibroblasts rendered senescent by exposure to IR, by overexpressing miR-340-5p or by silencing LBR. As shown in Figure 5F, H3K9me3 occupancy in LINE1, SAT2 (satellite 2 DNA) and ALR (a centromeric satellite DNA element) loci was reduced in the three senescence groups (IR, miR-340-5p, siLBR) relative to proliferating cells; the trend was also seen for SVA elements, although it did not reach significance. These changes were specific to DNA repetitive elements we assessed, as H3K9me3 occupancy on the Actin (ACTB) promoter revealed low heterochromatin levels in all conditions (Supplementary Figure S4C). The ChIP-qPCR results point to the reduction of LBR by miR-340-5p as a mechanism whereby cells promote senescence by reducing chromatin stability, consistent with the observed increased expression of DNA repetitive elements (Figure 5D). In support of this notion, ATAC-seq analysis revealed increased accessible peaks in DNA repetitive elements in cells rendered senescent by IR, miR-340-5p or siLBR compared to proliferating cells (Figure 5G).
Lastly, we examined the nuclei of cells overexpressing miR-340-5p or with silenced LBR to determine whether any changes in nuclear morphology indicative of senescence were detected. We applied a deep neural network model that can predict senescence from nuclear shape with high accuracy (38). We found a consistent increase in predicted senescence based on nuclear morphology after LBR silencing compared to control, while cells treated with anti-miR-340-5p showed a decrease in predicted senescence compared to control (Figure 5H). Additionally, using a neural network to detect nuclei boundaries, differences in miR-340-5p or LBR had no gross effect on nuclear morphology except for convexity, which showed significant difference between miR-340-5p and anti-miR-340-5p (Supplementary Figure S5). In sum, reducing LBR production caused a number of chromatin changes, including loss of heterochromatin associated with LADs, reduction in the heterochromatin mark H3K9me3, heightened transcription of LAD-associated repetitive elements and changes in nuclear morphology predictive of cellular senescence by neural network analysis.
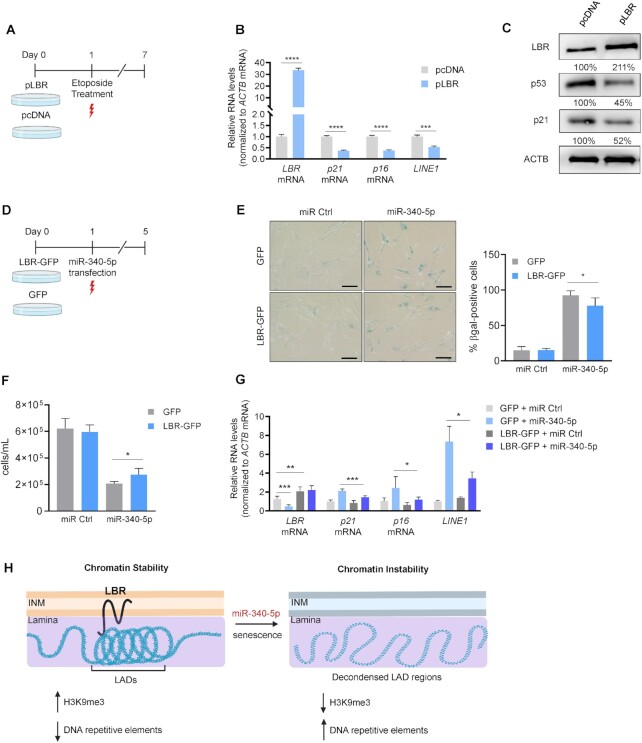
Overexpression of LBR partially reverses the senescent phenotype
Given that repressing LBR production reduced chromatin stability and triggered senescence, we asked whether overexpressing LBR might improve chromatin homeostasis and delay senescence. To this end, we transfected proliferating (PDL20) WI-38 fibroblasts with a plasmid that overexpressed LBR (or a control empty vector, pcDNA) before inducing senescence by exposing cells to etoposide (50 μM) and culturing them for an additional 7 days to establish senescence (Figure 6A). Compared with cells transfected with the empty vector, overexpression of LBR reduced the levels of senescence markers (p21 and p16 mRNAs) as well as LINE1 transcripts (Figure 6B). Overexpression of LBR also reduced p53 and p21 levels (Figure 6C).
Figure 6.
Overexpression of LBR partially rescues the senescent phenotype. (A) Schematic of the experimental design describing transfection of WI-38 PDL20 fibroblasts with pLBR or pcDNA before inducing senescence with etoposide (50 μM) and analyzing cells 6 days later. (B) RT-qPCR analysis of the levels of senescence markers (p16 mRNA, p21 mRNA), and the DNA repetitive element LINE1 (normalized to ACTB mRNA levels) in cells processed as described in panel (A). (C) Western blot analysis of protein levels in cells processed as explained in panel (A). Protein signals were measured by densitometry and quantified as a % value relative to the signals in the pcDNA sample group and normalized to ACTB levels. (D) Schematic of the experimental design describing the transduction of LBR-GFP lentivirus (3 MOI) or GFP lentivirus (3 MOI) in WI-38 fibroblasts (PDL20) before inducing senescence by transfecting miR-340-5p; miR Ctrl was transfected in control populations. (E) Micrographs of SA-βgal activity in WI-38 cells transduced as described in panel (D); right: quantification of the percentage of SA-βgal-positive cells. (F) Cell proliferation was measured 5 days after transduction as described in panel (D); cells were counted using a Bio-Rad TC20 cell counter. (G) RT-qPCR analysis of the levels of LBR mRNA, p21 mRNA, p16 mRNA and LINE1 in cells that were processed as described in panel (D). (H) Schematic of the proposed effects of miR-340-5p and senescence on LBR and the resulting changes in chromatin structure and stability. Data in panels (B) and (E)–(G) represent the mean values ± SD from three biological replicates. Significance was established using Student’s t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Next, we hypothesized that overexpressing LBR might rescue the miR-340-5p-induced senescence in a similar paradigm; for this experiment, we employed a lentivirus to overexpress LBR, given that co-transfection of the pLBR plasmid with the miRNAs for long durations was toxic for WI-38 fibroblasts (Figure 6D). As shown, a lentivirus expressing LBR-GFP partially reversed the senescence program, with a reduction in the percentage of SA-βgal-positive cells and a significant restoration of proliferation (Figure 6E and F) relative to cells transduced with a control, GFP-expressing lentivirus. Moreover, RT-qPCR analysis indicated that the rise in levels of p21 mRNA, p16 mRNA and LINE1 after overexpressing miR-340-5p was significantly suppressed by overexpressing LBR (Figure 6G). These results reflect a modest, but clear rescue of miR-340-5p-induced senescence markers when LBR was overexpressed by 2- to 3-fold. We employed the LBR SB in an additional rescue experiment aimed at testing whether interfering with endogenous miR-340-5p influenced senescence (Supplementary Figure S6A and B). While the LBR SB appeared to delay senescence moderately, as measured 48 h after transfecting SB concentrations capable of reducing LBR levels (10 nM SB), at later times both LBR SB and control SB were highly toxic, and therefore their impact on senescence could not be assessed. Taken together, these data support the hypothesis that senescence triggered by inducers including miR-340-5p is partially mediated by a reduction in LBR levels and the ensuing loss of chromatin stability (Figure 6H).
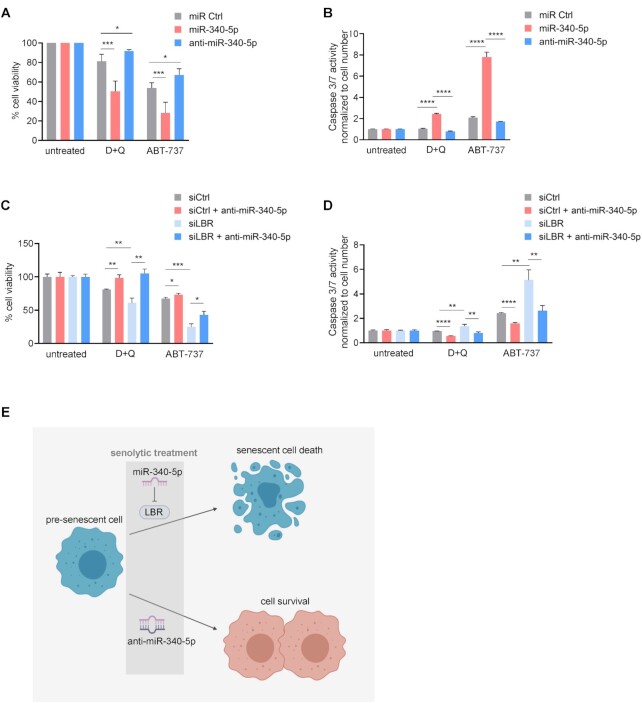
miR-340-5p sensitizes pre-senescent cells to senolytic compounds
Having found that overexpressing miR-340-5p induced senescence while antagonizing it suppressed senescence, we asked whether this miRNA might have therapeutic value by modulating the sensitivity of senescent cells against senolytic drugs (48). First, we established the ability of miR-340-5p to enhance sensitivity of pre-senescent cells to the known senolytic compounds dasatinib and quercetin (D + Q) as well as to ABT-737, all of which preferentially kill senescent cells (49,50). Pre-senescent WI-38 fibroblasts were transfected with miR Ctrl, miR-340-5p or anti-miR-340-5p 24 h before treatment with D + Q, ABT-737 or no treatment for an additional 24 h. Interestingly, pre-senescent cells overexpressing miR-340-5p exhibited reduced viability (Figure 7A), as determined by the percentage of live cells; conversely, cells expressing anti-miR-340-5p were more resistant to senolytic-induced cell death (Figure 7A). The observed effects were more pronounced for ABT-737 than for D + Q, but for both senolytic interventions miR-340-5p overexpression in pre-senescent cells enhanced apoptosis (Supplementary Figure S7A). To further assess apoptosis triggered by senolytics, we measured caspase 3/7 activity in both untreated and treated cells. In agreement with the cytotoxicity data, analysis of pre-senescent cells treated with senolytic compounds revealed that overexpression of miR-340-5p enhanced caspase activity, while expression of anti-miR-340-5p lowered caspase activity (Figure 7B). These observations suggest that by promoting a senescent program, miR-340-5p sensitizes cells to senolytic-driven death by apoptosis, while antagonizing miR-340-5p protects from senolytic-induced cell death. As discussed below, by sensitizing cells to senolysis, miR-340-5p could be harnessed for therapeutic use. On the other hand, antagonizing miR-340-5p appears to prevent or delay the development of senescence and may also be a useful therapeutic tool for maintaining cell homeostasis in aging or age-related pathologies.
Figure 7.
miR-340-5p sensitizes pre-senescent fibroblasts to senolytic death. (A, B) WI-38 fibroblasts (PDL20) were exposed to IR and transfected with miR Ctrl, miR-340-5p or anti-miR-340-5p and cultured for additional 3 days. Cells were then treated with ABT-737 (10 μM) or with the combination of dasatinib (20 μM) + quercetin (8 μM) (D + Q) for 24 h. Cell viability (A) and caspase 3/7 activity (B) were quantified and plotted. (C, D) WI-38 fibroblasts (PDL20) were exposed to IR and transfected with siCtrl alone or siLBR alone or with anti-miR-340-5p and cultured for additional 3 days. Cells were then treated with ABT-737 or the combination D + Q [as in panel (A)] for 24 h. Cell viability (C) and caspase 3/7 activity (D) were quantified and plotted. (E) Schematic illustrating the proposed sensitization of pre-senescent cells to senolytic drugs by miR-340-5p and the proposed protection against senolytics by anti-miR-340-5p. Data in panels (A)–(D) represent the mean values ± SD from three biological replicates. Significance was established using Student’s t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Finally, we determined whether miR-340-5p sensitizes pre-senescent cells to senolytic death specifically by reducing LBR levels. We silenced LBR by transfecting siLBR and performed the same experiment as described above (Figure 7C and D; Supplementary Figure S7B). The results indicate that the loss of LBR is sufficient to sensitize pre-senescent cells to senolytics, and that anti-miR-340-5p attenuated the sensitivity toward senolytics, despite the strong reduction in LBR levels (Figure 4). This result was surprising but suggests other targets of miR-340-5p may also contribute to senescence. When we compared miR-340-5p predicted target mRNAs with all transcripts significantly reduced in WI-38 fibroblasts rendered senescent by exposure to IR, we observed 105 potential transcripts; among them, 16 transcripts have superior miR-340-5p context scores than LBR mRNA (Supplementary Figure S7C). The newly identified 16 transcripts may contribute to the effect of anti-miR-340-5p in this paradigm and will be investigated in future studies. To conclude, our results suggest that the miR-340-5p-mediated reduction of LBR levels, likely by destabilizing lamina-associated chromatin, elicits senescence and thus sensitizes cells to death by senolytics, while anti-miR-340-5p partially protects cells from entering senescence and therefore protects against senolysis (Figure 7E).
DISCUSSION
Toward the elusive goal of finding consistent and specific markers of cellular senescence, we searched for potential noncoding RNAs involved in controlling the senescent cell transcriptome. Bioinformatic mining identified miR-340-5p as a novel regulator of cellular senescence. Previously implicated in tumor suppression and the cellular antiviral response (16–19), here we provide new evidence that miR-340-5p is induced during senescence and promotes the senescent phenotype. In this regard, while senescence is a powerful cancer-preventive trait, whether the tumor-suppressing influence of miR-340-5p in cancer cells is elicited by triggering senescence remains to be studied (51,52).
To date, dozens of miRNAs have been identified that regulate various pathways associated with senescence, many of them directly or indirectly affecting the levels of traditional senescence-regulatory proteins such as p53 or p21 (53–57). Here, an unbiased search for miRNAs influencing gene expression programs in senescence identified miR-340-5p as being elevated during senescence and being able to drive proliferating cells into a state of senescence marked by a significant reduction in proliferation, enhanced SA-βgal activity and increased expression of classical and novel senescence markers (p16, p21, p53 and PURPL). These results suggest that increased expression of miR-340-5p in response to cell damage critically contributes to the implementation of the senescent program. Interestingly, we observed no change in the levels of RNF130 mRNA, the mature transcript from the host pre-mRNA from which miR-340-5p arises, in the various senescence paradigms we tested, as measured by RT-qPCR analysis (Supplementary Figure S1C). Thus, the mechanisms that lead to the specific rise in miR-340-5p levels during senescence are unknown at present, but it will be interesting to identify the regulators of miR-340-5p production, given that suppressing miR-340-5p function (as achieved by using the miR-340-5p antagomir) reversed senescence and restored proliferation.
Whether treatment with the miR-340-5p SB on LBR mRNA (Figure 4H) alone has a similar suppressive influence on senescence remains to be tested. Unfortunately, the SB used in this study caused toxicity when transfected long term. Using lower doses of LBR SB for shorter times (<48 h) suggested a trend of reduced SA-βgal activity (Supplementary Figure S6A and B), but the times were too early to affect the expression of DNA repetitive elements. The introduction of a mutation in the miR-340-5p seed sequence of the LBR gene using CRISPR/Cas9 also failed due to the fact that primary WI-38 fibroblasts become senescent during the selection time needed to generate a stable mutant line. Whether through LBR alone or by including other possible effectors of miR-340-5p actions, our results point to miR-340-5p as a key senomodulatory miRNA, particularly at the decision point when cells may commit to senescence or instead adopt a quiescent or proliferative state.
In agreement with the homeostatic function of LBR to tether heterochromatin to the nuclear envelope, recent studies have discovered that LBR and its ligand, lamin B1 (LMNB1), are strongly reduced in senescence (12,45,58–60). These studies and others point to potential alterations in chromatin architecture and genome function due to lower LBR and LMNB1 expression levels, even though the exact role of LBR in senescence was not elucidated (61,62). Reducing LBR in these earlier studies and in the paradigms studied here enables senescence, supporting the notion that loss of LBR reduces LMNB1 function and leads to the commitment of cells toward senescence [Figure 5 and data not shown (58,63)]. As a consequence of the reduction in LBR and LMNB1, which maintain heterochromatin tethers to prevent aberrant expression of transcriptionally inaccessible DNA, we identified several DNA repetitive elements characteristically repressed in a proliferating cell to be derepressed by the addition of miR-340-5p or the loss of LBR. Through ChIP-PCR and ATAC-seq analyses, we confirmed that the open chromatin states of LAD regions after elevating miR-340-5p or reducing LBR were similar to those in IR-induced senescence. Our results strongly support the emerging concept that decreasing the integrity of the nuclear lamina leads to chromatin instability and ensuing senescence, and align with previous studies in Hutchinson–Gilford progeria syndrome cells where mutations or destabilization of the nuclear lamina-associated chromatin cause premature senescence (64,65). Interestingly, in a recent study, a rise in LINE1 expression levels was associated with late senescence and the late interferon inflammatory response program (47). In our study, LBR abundance is quickly lowered by miR-340-5p or by direct LBR silencing, which may explain why we observe a rapid elevation in the levels of LINE1 and other DNA repetitive elements. Further, the untethering of centromere-specific satellite heterochromatin from lamina and its decompaction in the nucleoplasm have also been identified as a feature of senescent cells (66). These two studies, in conjunction with the finding that cytoplasmic chromatin foments a senescence-associated pro-inflammatory program (67), underscore the important and novel role for LBR in maintaining heterochromatin homeostasis and avoiding senescence.
Among other distinct morphological changes, senescent cells display enlarged nuclei and nucleoli (68,69). Overexpression of miR-340-5p and reduction of LBR cause a disruption of chromatin tethering and changes in chromatin stability but are also linked to distinct changes in the morphology of senescent nuclei. In this study, we used neural networks to identify nuclear boundaries and assessed changes in the morphology of WI-38 cell nuclei with elevated miR-340-5p or reduced LBR. Interestingly, we did not detect any gross changes in nuclear morphology, yet a deep neural network trained to detect senescent cells based on nuclear morphology predicted senescence after LBR silencing and predicted a decrease in senescence by antagonizing miR-340-5p. Taken together, these data suggest that reduction of LBR levels not only contributed to altering chromatin architecture, but also influenced the unique nuclear morphology of senescent cells.
In closing, the finding that overexpressing miR-340-5p and reducing LBR sensitized pre-senescent cells to senolytics, a class of drugs that selectively induces apoptosis in senescent cells, points to a therapeutic value for this miRNA. Moreover, antagonizing miR-340-5p in pre-senescent cells enhanced cell viability and reduced caspase activity. While employing miRNAs alone as therapeutic agents has not been very successful thus far, a more promising new area of research utilizes miRNAs to alter drug sensitivity, as demonstrated for drug resistance in cancer chemotherapy, as well as for diseases such as epilepsy and insulin resistance (70–72). Based on the fact that loss of chromatin stability is a key trigger of senescence, we propose to exploit the therapeutic potential of miR-340-5p to enhance senescence in combination treatments with senolytics to promote senescent cell death and preserve tissue homeostasis.
DATA AVAILABILITY
All data, supplemental data, and data in repositories are available.
Supplementary Material
Contributor Information
Allison B Herman, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Carlos Anerillas, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Sophia C Harris, Confocal Imaging Facility, Laboratory of Cardiovascular Sciences, National Institute on Aging Intramural Research Program, National Institutes of Health, Baltimore, MD 21224, USA.
Rachel Munk, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Jennifer L Martindale, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Xiaoling Yang, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Krystyna Mazan-Mamczarz, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Yongqing Zhang, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Indra J Heckenbach, Center for Healthy Aging, Department of Cellular and Molecular Medicine, University of Copenhagen, Copenhagen, DK-2200, Denmark.
Morten Scheibye-Knudsen, Center for Healthy Aging, Department of Cellular and Molecular Medicine, University of Copenhagen, Copenhagen, DK-2200, Denmark.
Supriyo De, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Payel Sen, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Kotb Abdelmohsen, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
Myriam Gorospe, Laboratory of Genetics and Genomics, National Institute on Aging Intramural Research Program, National Institutes of Health, 251 Bayview Blvd, Baltimore, MD 21224, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [Z01-AG000511]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Krol J., Loedige I., Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chakraborty C., Sharma A.R., Sharma G., Lee S.-S.. Therapeutic advances of miRNAs: a preclinical and clinical update. J. Adv. Res. 2020; 28:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peng Y., Croce C.M.. The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 2016; 1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colpaert R.M.W., Calore M.. MicroRNAs in cardiac diseases. Cells. 2019; 8:737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dimmeler S., Nicotera P.. MicroRNAs in age-related diseases. EMBO Mol. Med. 2013; 5:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayflick L., Moorhead P.S.. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961; 25:585–621. [DOI] [PubMed] [Google Scholar]
- 8. Kovacic J.C., Moreno P., Hachinski V., Nabel E.G., Fuster V.. Cellular senescence, vascular disease, and aging: part 1 of a 2-part review. Circulation. 2011; 123:1650–1660. [DOI] [PubMed] [Google Scholar]
- 9. Martínez-Cué C., Rueda N.. Cellular senescence in neurodegenerative diseases. Front. Cell. Neurosci. 2020; 14:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baar M.P., Brandt R.M.C., Putavet D.A., Klein J.D.D., Derks K.W.J., Bourgeois B.R.M., Stryeck S., Rijksen Y., van Willigenburg H., Feijtel D.A.et al.. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017; 169:132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hernandez-Segura A., Nehme J., Demaria M.. Hallmarks of cellular senescence. Trends Cell Biol. 2018; 28:436–453. [DOI] [PubMed] [Google Scholar]
- 12. Casella G., Munk R., Kim K.M., Piao Y., De S., Abdelmohsen K., Gorospe M.. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019; 47:7294–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Basisty N., Kale A., Jeon O.H., Kuehnemann C., Payne T., Rao C., Holtz A., Shah S., Sharma V., Ferrucci L.et al.. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020; 18:e3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen W., Wang X., Wei G., Huang Y., Shi Y., Li D., Qiu S., Zhou B., Cao J., Chen M.et al.. Single-cell transcriptome analysis reveals six subpopulations reflecting distinct cellular fates in senescent mouse embryonic fibroblasts. Front. Genet. 2020; 11:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hernandez-Segura A., de Jong T.V., Melov S., Guryev V., Campisi J., Demaria M.. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 2017; 27:2652–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S., Choi J.Y., Seok H.J., Park M.-J., Chung H.Y., Bae I.H.. miR-340-5p suppresses aggressiveness in glioblastoma multiforme by targeting Bcl-w and Sox2. Mol. Ther. Nucleic Acids. 2019; 17:245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang L., Men W.-L., Yan K.-M., Tie J., Nie Y.-Z., Xiao H.-J.. MiR-340-5p is a potential prognostic indicator of colorectal cancer and modulates ANXA3. Eur. Rev. Med. Pharmacol. Sci. 2018; 22:4837–4845. [DOI] [PubMed] [Google Scholar]
- 18. Lu G., Zhang Y.. MicroRNA-340-5p suppresses non-small cell lung cancer cell growth and metastasis by targeting ZNF503. Cell. Mol. Biol. Lett. 2019; 24:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao L., Zhang X., Wu Z., Huang K., Sun X., Chen H., Jin M.. The downregulation of microRNA hsa-miR-340-5p in IAV-infected A549 cells suppresses viral replication by targeting RIG-I and OAS2. Mol. Ther. Nucleic Acids. 2019; 14:509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Natarelli L., Geißler C., Csaba G., Wei Y., Zhu M., di Francesco A., Hartmann P., Zimmer R., Schober A.. miR-103 promotes endothelial maladaptation by targeting lncWDR59. Nat. Commun. 2018; 9:2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Louloupi A., Ørom U.A.V.. Inhibiting pri-miRNA processing with target site blockers. Methods Mol. Biol. 2018; 1823:63–68. [DOI] [PubMed] [Google Scholar]
- 22. Hernandez-Segura A., Rubingh R., Demaria M.. Identification of stable senescence-associated reference genes. Aging Cell. 2019; 18:e12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duan Z.-Y., Cai G.-Y., Li J.-J., Bu R., Wang N., Yin P., Chen X.-M.. U6 can be used as a housekeeping gene for urinary sediment miRNA studies of IgA nephropathy. Sci. Rep. 2018; 8:10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Indig F.E., Diaz-Gonzalez F., Ginsberg M.H.. Analysis of the tetraspanin CD9–integrin αIIbβ3 (GPIIb–IIIa) complex in platelet membranes and transfected cells. Biochem. J. 1997; 327:291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sen P., Lan Y., Li C.Y., Sidoli S., Donahue G., Dou Z., Frederick B., Chen Q., Luense L.J., Garcia B.A.et al.. Histone acetyltransferase p300 induces de novo super-enhancers to drive cellular senescence. Mol. Cell. 2019; 73:684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu H., Ji Q., Song M., Ren J., Liu Z., Wang Z., Liu X., Yan K., Hu J., Jing Y.et al.. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 2020; 48:6001–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 28. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu G., Wang L.-G., Han Y., He Q.-Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B., Wilks C., Antonescu V., Charles R.. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2019; 35:421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu D.-Y., Bittencourt D., Stallcup M.R., Siegmund K.D.. Identifying differential transcription factor binding in ChIP-seq. Front. Genet. 2015; 6:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R.. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010; 11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu G., Wang L.-G., He Q.-Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 36. Ronneberger O., Fischer P., Brox T.. Navab N., Hornegger J., Wells W., Frangi A.. U-Net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015. 2015; 9351:Cham: Springer; MICCAI 2015. Lecture Notes in Computer Science. [Google Scholar]
- 37. Filippi-Chiela E.C., Oliveira M.M., Jurkovski B., Callegari-Jacques S.M., Silva V.D., Lenz G.. Nuclear morphometric analysis (NMA): screening of senescence, apoptosis and nuclear irregularities. PLoS One. 2012; 7:e42522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heckenbach I., Ezra M.B., Mkrtchyan G.V., Madsen J.S., Nielsen M.H., Oró D., Mortensen L., Verdin E., Westendorp R., Scheibye-Knudsen M.. Deep learning shows cellular senescence is a barrier to cancer development. 2021; bioRxiv doi:19 March 2021, preprint: not peer reviewed 10.1101/2021.03.18.435987. [DOI]
- 39. Agarwal V., Bell G.W., Nam J.-W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O.. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. U.S.A. 1995; 92:9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y., Boerma M., Zhou D.. Ionizing radiation-induced endothelial cell senescence and cardiovascular diseases. Radiat. Res. 2016; 186:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao H., Halicka H.D., Traganos F., Jorgensen E., Darzynkiewicz Z.. New biomarkers probing depth of cell senescence assessed by laser scanning cytometry. Cytom. Part A: J. Int. Soc. Anal. Cytol. 2010; 77:999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biran A., Zada L., Abou Karam P., Vadai E., Roitman L., Ovadya Y., Porat Z., Krizhanovsky V.. Quantitative identification of senescent cells in aging and disease. Aging Cell. 2017; 16:661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Deursen J.M. The role of senescent cells in ageing. Nature. 2014; 509:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lukášová E., Kovarˇík A., Bacˇíková A., Falk M., Kozubek S.. Loss of lamin B receptor is necessary to induce cellular senescence. Biochem. J. 2017; 474:281–300. [DOI] [PubMed] [Google Scholar]
- 46. Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L.et al.. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013; 152:584–598. [DOI] [PubMed] [Google Scholar]
- 47. De Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., Caligiana A., Brocculi G., Adney E.M., Boeke J.D.et al.. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019; 566:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hanna J., Hossain G.S., Kocerha J.. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019; 10:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu Y., Tchkonia T., Pirtskhalava T., Gower A.C., Ding H., Giorgadze N., Palmer A.K., Ikeno Y., Hubbard G.B., Lenburg M.et al.. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015; 14:644–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yosef R., Pilpel N., Tokarsky-Amiel R., Biran A., Ovadya Y., Cohen S., Vadai E., Dassa L., Shahar E., Condiotti R.et al.. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 2016; 7:11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W.. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997; 88:593–602. [DOI] [PubMed] [Google Scholar]
- 52. Chen Z., Trotman L.C., Shaffer D., Lin H.-K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W.et al.. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005; 436:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suh N. MicroRNA controls of cellular senescence. BMB Rep. 2018; 51:493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Borgdorff V., Lleonart M.E., Bishop C.L., Fessart D., Bergin A.H., Overhoff M.G., Beach D.H.. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1). Oncogene. 2010; 29:2262–2271. [DOI] [PubMed] [Google Scholar]
- 55. Pichiorri F., Suh S.-S., Rocci A., De Luca L., Taccioli C., Santhanam R., Zhou W., Benson D.M., Hofmainster C., Alder H.et al.. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010; 18:367–381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Xiao J., Lin H., Luo X., Luo X., Wang Z.. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011; 30:5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Munk R., Panda A.C., Grammatikakis I., Gorospe M., Abdelmohsen K.. Senescence-associated microRNAs. Int. Rev. Cell Mol. Biol. 2017; 334:177–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arai R., En A., Takauji Y., Maki K., Miki K., Fujii M., Ayusawa D.. Lamin B receptor (LBR) is involved in the induction of cellular senescence in human cells. Mech. Ageing Dev. 2019; 178:25–32. [DOI] [PubMed] [Google Scholar]
- 59. Freund A., Laberge R.-M., Demaria M., Campisi J.. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012; 23:2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shah P.P., Donahue G., Otte G.L., Capell B.C., Nelson D.M., Cao K., Aggarwala V., Cruickshanks H.A., Rai T.S., McBryan T.et al.. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013; 27:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chiang M., Michieletto D., Brackley C.A., Rattanavirotkul N., Mohammed H., Marenduzzo D., Chandra T.. Polymer modeling predicts chromosome reorganization in senescence. Cell Rep. 2019; 28:3212–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kristiani L., Kim M., Kim Y.. Role of the nuclear lamina in age-associated nuclear reorganization and inflammation. Cells. 2020; 9:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lukášová E., Kovařík A., Kozubek S.. Consequences of lamin B1 and lamin B receptor downregulation in senescence. Cells. 2018; 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R.et al.. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. U.S.A. 2004; 101:8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Scaffidi P. Lamin A-dependent nuclear defects in human aging. Science. 2006; 312:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. De Cecco M., Criscione S.W., Peckham E.J., Hillenmeyer S., Hamm E.A., Manivannan J., Peterson A.L., Kreiling J.A., Neretti N., Sedivy J.M.. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013; 12:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dou Z., Ghosh K., Vizioli M.G., Zhu J., Sen P., Wangensteen K.J., Simithy J., Lan Y., Lin Y., Zhou Z.et al.. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017; 550:402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mitsui Y., Schneider E.L.. Increased nuclear sizes in senescent human diploid fibroblast cultures. Exp. Cell Res. 1976; 100:147–152. [DOI] [PubMed] [Google Scholar]
- 69. Bemiller P.M., Lee L.-H.. Nucleolar changes in senescing WI-38 cells. Mech. Ageing Dev. 1978; 8:417–427. [DOI] [PubMed] [Google Scholar]
- 70. Hu W., Tan C., He Y., Zhang G., Xu Y., Tang J.. Functional miRNAs in breast cancer drug resistance. OncoTargets Ther. 2018; 11:1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang J., Tan L., Tan L., Tian Y., Ma J., Tan C.-C., Wang H.-F., Liu Y., Tan M.-S., Jiang T.et al.. Circulating microRNAs are promising novel biomarkers for drug-resistant epilepsy. Sci. Rep. 2015; 5:10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gupta S., Singhal N.K., Ganesh S., Sandhir R.. Extending arms of insulin resistance from diabetes to Alzheimer’s disease: identification of potential therapeutic targets. CNS Neurol. Disord. Drug Targets. 2019; 18:172–184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, supplemental data, and data in repositories are available.