Abstract
The industrial yeast Pichia pastoris has been harnessed extensively for production of proteins, and it is attracting attention as a chassis cell factory for production of chemicals. However, the lack of synthetic biology tools makes it challenging in rewiring P. pastoris metabolism. We here extensively engineered the recombination machinery by establishing a CRISPR-Cas9 based genome editing platform, which improved the homologous recombination (HR) efficiency by more than 54 times, in particular, enhanced the simultaneously assembly of multiple fragments by 13.5 times. We also found that the key HR-relating gene RAD52 of P. pastoris was largely repressed in compared to that of Saccharomyces cerevisiae. This gene editing system enabled efficient seamless gene disruption, genome integration and multiple gene assembly with positive rates of 68–90%. With this efficient genome editing platform, we characterized 46 potential genome integration sites and 18 promoters at different growth conditions. This library of neutral sites and promoters enabled two-factorial regulation of gene expression and metabolic pathways and resulted in a 30-fold range of fatty alcohol production (12.6–380 mg/l). The expanding genetic toolbox will facilitate extensive rewiring of P. pastoris for chemical production, and also shed light on engineering of other non-conventional yeasts.
INTRODUCTION
Yeasts are used widely as cell factories for production of proteins, chemicals and advanced biofuels. The budding yeast Saccharomyces cerevisiae has been used for brewing and baking for >4000 years, and currently, it is regularly used for bio-ethanol production (1). With the aid of modern molecular biology, S. cerevisiae has been harnessed for the production of many industrially relevant biochemicals and biofuels (2). Recently, a number of non-conventional yeasts have also attracted great attention as potential chassis cells for production of fine chemicals and recombinant proteins. These non-conventional yeasts have unique and excellent properties, such as tolerance to inhibitors and low pH that may have advantages for specific bioprocesses (3). However, it is still challenging to extensively engineer these non-conventional yeasts because of a dearth of genetic editing tools (4), compared with the model yeast S. cerevisiae for which there are numerous advanced genetic tools and biological devices (5).
Among non-conventional yeasts, the methylotrophic yeast Pichia pastoris (syn. Komagataella phaffii) is an established protein production platform, especially in the industrial enzymes and the biopharmaceutical industry (6). In addition, there is much interest in harnessing P. pastoris for production of small molecules and, particularly, in establishing a methanol biotransformation process (7). Compared with protein production that involves overexpression of a single gene, engineering biosynthesis of small molecules always involves optimization of multiple-step pathways and extensive rewiring of cellular metabolism. For example, production of opioids in yeast required the heterologous expression of >30 genes from multiple species (8). Global reconstruction of lipogenesis in S. cerevisiae involved rewiring four categories of pathways with more than 20 genes (9), which were genome-integrated for stable expression.
From the successful engineering of S. cerevisiae cell factories, we can identify three prerequisites for construction of stable cell factories: (i) clear genome information; (ii) efficient and precise genome engineering tools; (iii) sufficient genetic devices including promoters and integration sites for stable gene expression. The genome information of P. pastoris is thoroughly annotated (10) (www.pichiagenome.org). In addition, the CRISPR-Cas9 system was established and optimized in P. pastoris by evaluating diverse codon-optimized Cas9 genes, sgRNA and promoters for expression of Cas9 and the sgRNA, which enabled efficient cutting of the genomic DNA (11,12). However, in P. pastoris, non-homologous end joining (NHEJ) is the dominant pathway for repairing double-strand breaks (DSB). Hence, NHEJ impedes precise genome engineering, in particularly, seamless gene deletion and genome integration that depend on homologous recombination (Figure 1). Furthermore, only a paucity of sites has been identified into which exogenous genes can be integrated without disrupting cell viability. Lastly, the lack of well-characterized promoters makes it challenging in fine tuning of gene expression, since appropriate promoters can mediate dynamic regulation of biosynthesis pathways and maintaining cell robustness (13).
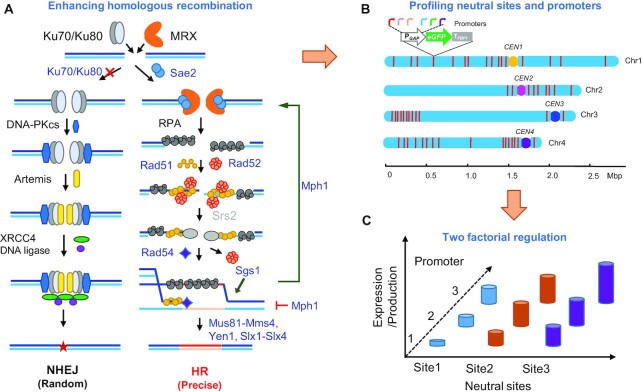
Figure 1.
Engineering recombination machinery and expanding genetic tool box for metabolic engineering of P. pastoris. (A) Enhancing recombination for precise genetic engineering by manipulation of the homologous recombination (HR) process and repressing the nonhomologous end joining (NHEJ). The genes marked as blue were deleted or overexpressed for enhancing HR efficiencies. (B) The neutral sites and promoters were profiled by using eGFP expression cassettes with the established genetic engineering platform. The red bar represents the location of neutral sites in the genome. (C) The variable transcription of promoters and neutral sites enables two-factorial regulation of gene expression and biosynthetic pathways.
To address these challenges in genetic engineering of P. pastoris, we have extensively engineered the recombination machinery of this yeast to enhance homologous recombination, which significantly improved the efficiency of seamless gene disruption and simultaneous integration of multiple genes. With this precise genome engineering platform, we systematically identified 46 genomic integration sites for stable gene expression and temporally profiled 18 promoters for regulation of gene expression. Ultimately, we developed a two-factorial gene regulation strategy by using the identified integration sites and promoters. This strategy enabled a 30-fold range in regulation of fatty alcohol biosynthesis (12–380 mg/l). This expanded genetic tool box (Figure 1) should facilitate the metabolic engineering of this important industrial yeast P. pastoris and other non-conventional yeasts for production of small molecules.
MATERIALS AND METHODS
Strains and cultivation
All strains used in this study are listed in Supplementary Table S1. P. pastoris GS115 was kindly offered by Prof. Menghao Cai from East China University of Science and Technology, and other strains were stored in our lab or constructed in this study. Unless otherwise specified, yeast strains were cultivated in YPD medium containing 20 g/l glucose, 20 g/l peptone and 10 g/l yeast extract. For screening transformants, YPD medium was supplemented with zeocin or G418. Delft basic salt medium (2.5 g/l (NH4)2SO4, 14.4 g/l KH2PO4, 0.5 g/l MgSO4•7H2O, 40 mg/l histidine, 1 ml/l Vitamin solution, 2 ml/l Trace metal solution) (14,15), was used for cell cultivation and fermentation with 20 g/l glucose and/or 10 g/l methanol as carbon sources. Escherichia coli DH5α was grown in LB medium (10 g/l tryptone, 10 g/l NaCl, and 5 g/l yeast extract) with specific antibiotics, such as ampicillin (100 mg/l) to maintain plasmids. All yeast strains were cultivated at 30°C, 220 rpm in a shake incubator (Zhichu Shaker ZQZY-CS8). Pre-cultures were cultivated in 15 ml tube with a working volume of 3 ml, and cells for transformation and fermentation were both cultured in 100 ml flasks with a working volume of 20 ml. For removing gRNA plasmids, the verified strains were cultured in 3 ml YPD medium for about 20 h, and then streaked on YPD plates. Majority of newly grown colonies were plasmid-free and verified on selection plates.
Establishing CRISPR–Cas9 system
The plasmid pPICZ was used as the backbone, which was modiefied from the commercial plasmid pPICZαA by deleting the α-factor secretion signal sequence. Cas9 coding sequence was amplified from plasmid pECAS9-kanMX-gRNA (16) with primers CAS9-F and CAS9-R. Replication origin panARS was amplified from Kluyveromyces lactis genome with primers panARS-F and panARS-R. Bidirectional promoter PHTX1 was amplified from P. pastoris GS115 genome DNA with primers HTX1-F and HTX1-R. The hammerhead ribozyme (HH), GUT1-gRNA, and the hepatitis delta virus ribozyme (HDV) (HH-GUT1-gRNA-HDV) were synthesized by Sangon Biotech (Shanghai). DAS1 terminator (TDAS1) was amplified from P. pastoris GS115 genome DNA by using the primer pair TDAS1-F/TDAS1-R. Supplementary Table S2 lists all the primers used in this study. The genome editing vector pPICZ-Cas9-gGUT1 was constructed by Gibson assembly, which contained Cas9, TDAS1, HH-GUT1-gRNA-HDV, panARS and vector backbone. Restriction enzyme cleavage sequence SpeI was added between Cas9 and the promoter to facilitate subsequent vector construction.
DNA transformation was performed with a condensed electroporation protocol (17), and the transformed cells were grown for three days on YPD plates containing 100 μg/ml Zeocin. Twenty colonies were selected from each plate for identification of positive clones. The genomic manipulations were verified with colony PCR, and the genomic DNA was extracted according to a described protocol (18).
Single gene deletion and integration
Expression cassettes of gRNA were constructed by fusing gRNA promoter sequence PHTX1 and terminator sequence TAOX1, which were cloned by using primers listed at Supplementary Table S3. Plasmid pPICZ-Cas9-gGUT1 was constructed by using the gRNA expression cassette and vector backbone with Gibson assembly. Donor DNA cassettes were constructed by fusing the upstream and downstream homologous arms (with 50–1000 bp length) of targeting gene by overlap extension PCR.
Gene integration expression cassettes were constructed by fusing the upstream HA, promoter sequence, ORF, terminator and downstream HA by overlap extension PCR. The transformations were performed using a condensed electroporation protocol as described above. 8–20 colonies were selected for colony PCR. The positive rates were calculated as the ratio of the positive clone to the total number of clones picked.
Engineering recombination machinery
KU70, MPH1, SGS1, TOP3 and RMI1 genes were seamlessly deleted by CRISPR-Cas9 based genome editing tools. The gRNA plasmids used for targeting KU70, MPH1, SGS1, TOP3 and RMI1 gene were constructed with primers (19P-74/19-160 and 19P-75/19-161), (19P-74/19-134 and 19P-75/19-135), (19P-74/19-76 and 19P-75/19-77), (19P-74/19-78 and 19P-75/19-79) and (19P-74/19-80 and 19P-75/19-81), respectively. The fragments used for deleting KU70, MPH1, SGS1, TOP3 and RMI1 gene were fused by primers 19P-74/19P-75, and finally constructed into gRNA vector backbone by Gibson Assembly® Master Mix.
The upstream and downstream homologous arms (HAs) of targeting gene with a length of 1000 bp, were amplified using primers respectively. These two HAs were fused by overlap extension PCR and used as donors for seamless gene deletion. The S. cerevisiae genes ScRAD51, ScRAD54, and ScSAE2 were amplified from its genome, and the endogenous genes PpRAD51, PpRAD52 and PpRAD54 were amplified from the genome of P. pastoris GS115. ScRAD52, ScRAD54, ScSae2, PpRAD51, PpRAD52 and PpRAD54 were then cloned into the pPICZ plasmid under the control of PGAP promoter. The plasmids were linearized by PmeI and integrated into the AOX1 sites of the GS115 genome by electro-transformation. The MUS81, MMS4, SLX1, SLX4 and YEN1 genes were cloned from P. pastoris and integrated into PNSI-1 site of P. pastoris by using pPICZ-Cas9-gPNSI-1 as the gRNA plasmid. The overexpression cassettes of MUS81-MMS4, SLX1-SLX4 and YEN1 were fused with the PNSI-1 upstream and downstream homologous arms.
Assembly of multiple fragments at single genome loci
The MmCAR, npgA and ADH5 gene expression cassettes of a fatty alcohol biosynthetic pathway were simultaneously integrated into FAA1 site by using the gRNA plasmid pPICZ-Cas9-gFAA1. The promoters PADH2, PTEF1 and PTPI were used to control gene expression of MmCAR, npgA and ADH5, respectively. The gene expression cassettes were constructed by overlap extension PCR, and the homologous regions of the three fragments were set in the promoter or the terminator, with a length of about 500 bp (Figure 3B). The homologous arm length of the segments at both ends and the genome was 1000 bp. About 1 μg fragments were used for transformation with a molar ratio of 1:1:1 for the three fragments. The KanMX expression cassette for motoring the DNA repair process, was constructed using PADH2 as the promoter, the TGAP as the terminator, GUT1 as the integration site with the HA length of 1000 bp. The KanMX expression cassette was divided to three fragments: the first fragment containing upstream HA, PADH2 and upstream of KanMX ORF; the second fragment containing KanMX ORF lacking starting codon ATG; the third fragment contained KanMX downstream ORF, TGAP and downstream HA. The start codon was removed to avoid the improper integration of KanMX with antibiotic resistance.
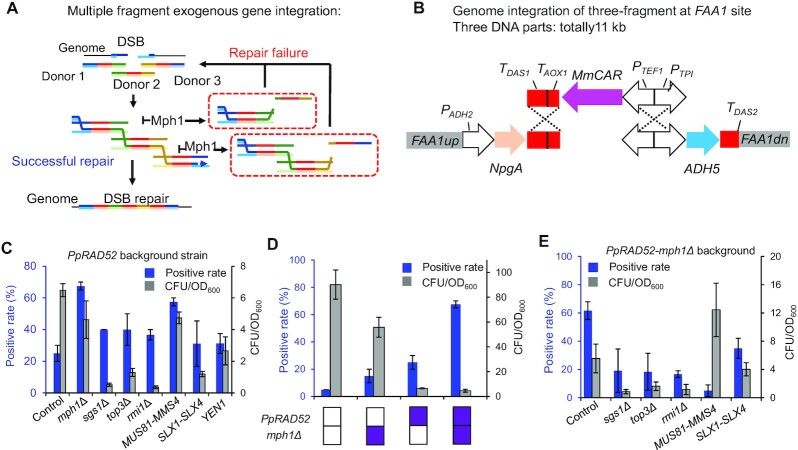
Figure 3.
Engineering multi-invasion-induced rearrangement process for enhancing genome integration of multiple fragments. (A) Schematic diagram of multi-invasion-induced rearrangement process. (B) Three fragments were integrated into FAA1 site for constructing the fatty alcohol biosynthetic pathway. (C) Effect of mph1Δ, sgs1Δ, top3Δ, rmi1Δ and MUS81-MMS4, SLX1-SLX4, YEN1 overexpression on simultaneous genome integration of three fragments on background strain PC110 with PpRAD52 overexpression. Five to twenty clones from each plate were picked for colony PCR analysis, and two plates were used. (D) Effect of MPH1 deletion and/or RAD52 overexpression on simultaneous genome integration of three fragments. Forty clones from two plates were picked for colony PCR analysis. (E) Further engineering HR relating genes in strain XDP11 with mph1Δ and PpRAD52 had a negative effect on the genome integration of multiple fragments.
eGFP expression cassette construction and neutral site integration
To test gene expression at neutral sites, the PGAP promoter was use to drive expression of eGFP, which was amplified from the plasmid pJQ01 (19). The 1 kb upstream and downstream homologous arms of each neutral locus were fused with the eGFP cassette by overlap extension PCR. Supplementary Table S3 lists all the gRNA sequences of neutral sites. For the eGFP integration at neutral sites on Chromosome 1, the CRISPR-Cas9 system was in accordance with previous version (Supplementary Figure S1A). For the neutral sites on Chromosome 2, 3 and 4, the Cas9 gene was integrated into the PNSI-2 site and a simplified version of the sgRNA plasmid was used for integration of the eGFP cassette (Supplementary Figure S1B). 300 ng Cas9–gRNA or gRNA plasmid and 500 ng donor DNA were co-transformed by electroporation for genome integration in the PpRAD52 overexpression strain PC110 (17,20). The transformants were cultivated on YPD plates with antibiotic for three days, then 8–10 random clones were inoculated into 300 μl YPD liquid medium with antibiotic for overnight cultivation. Clones were verified by PCR or fluorescence detection. The seed was cultured in 3 ml YPD medium overnight, and then adjusted to an optical density at 600 nm (OD600) of 0.1 (equivalent to 2 × 106 cells/ml) for cultivation. Samples were harvested and analyzed every 24 h. 200 μl dilution was used for fluorescence detection by microplate reader (Tecan Spark 3). The excitation and emission wavelengths were 485 and 525 nm (gain setting, 80). All fluorescence measurements were normalized to the OD600 values.
Fatty alcohol biosynthesis
For the FFA derived pathway construction, MmCAR, npgA and ScADH5 expression cassettes were integrated into the PNSI-2, PNSI-3 and PNSI-4 sites, respectively. For the fatty acyl-CoA derived pathways construction, ScADH5 and FaCoAR cassettes were integrated into the PNSI-4 and PSNI-5 sites, respectively (Supplementary Figure S2). These genes were simultaneously integrated into corresponding neutral site by using the multiplexed CRISPR technologies. Cultures were inoculated, from overnight precultures, at an initial OD600 of 0.1 in 20 ml Deft-His medium and cultivated at 220 rpm, 30°C for 96 h. The metabolite extraction and analysis procedure were executed following as described (14,15).
Real-time PCR experiment
The yeast strains S. cerevisiae CEN.PK 113-11C and P. pastoris GS115 were cultivated in YPD medium at 30 °C, 220 rpm overnight. The total RNA was extracted by RNA simple Total RNA Kit (DP419, TIANGEN, Beijing, China). 1 μg total RNA of each sample was reversely transcribed to cDNA using the PrimeScript® RT reagent Kit (Takara Bio Inc.) according to the manufacturer's protocol. A two-step PCR reaction was employed by using SYBR® Premix Ex TaqTM II (Takara Bio Inc.). Actin gene was chosen as the endogenous reference gene, and the data analysis was conducted by the method of 2–ΔΔCT as described previously (21). Primers were listed in Supplementary Table S2, and all strains with three biologically independent parallel samples were adopted to guarantee the reproducibility of all the results.
RESULTS
Enhancing the homologous recombination efficiency for single manipulation
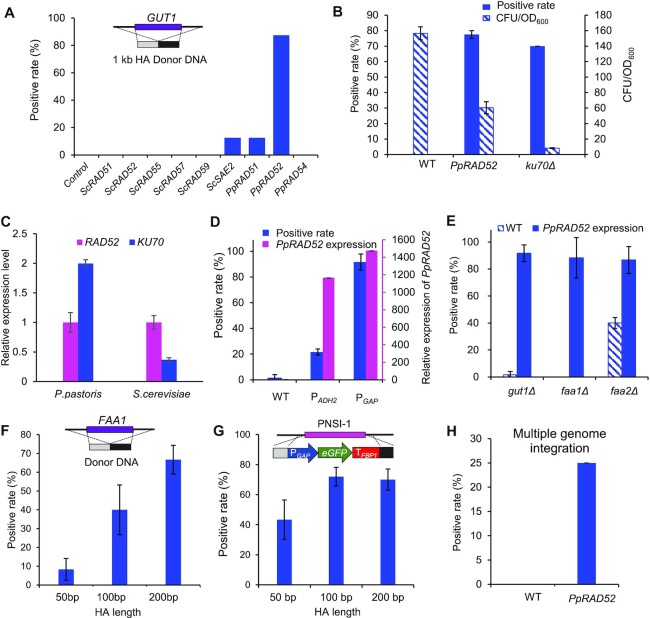
As previously reported (20), a bidirectional promoter PHTX1 (22) was used to express the CAS9 gene and the gRNA. pPICZ was used as the plasmid backbone, and the autonomously replicating sequence panARS, cloned from Kluyveromyces lactis, was used as the replication origin of episomal plasmid (Supplementary Figure S1A). This system enabled a 93% positive rate of genome cutting when targeting the GUT1 gene, which was evaluated by a plate seeing test (Supplementary Figure S3), since GUT1 disruption resulted a growth defect on glycerol plates (11). However, co-transformation of a donor DNA with 1000 bp homologous arms, only resulted in less than 1.67% of seamless gene deletion (0 positive colony from eight picked colonies in Figure 2A, or 1 positive colony from 60 picked colonies in Figure 2E). This low frequency suggested that NHEJ was the dominant mechanism in repairing DSB and HR is extremely repressed in this yeast. Thus, we sought to enhance HR efficiency by engineering the recombination machinery (Figure 1A). Key components of NHEJ-based DNA repair are Ku heterodimer proteins Ku70 and Ku80, which bind to the ends of DNA DSB. Disruption of KU70 repressed NHEJ and resulted in a higher HR efficiency (20,23). We tried to delete KU70 of P. pastoris, however, we obtained much fewer colonies when deleting the fatty acyl-CoA synthase 1 gene FAA1 (Figure 2B). We also found that ku70Δ caused the loss of large genome fragments when integrating genes at PNSI-1 site that localized at the chromosome terminal (Supplementary Figure S4). These data suggested that the HR process of P. pastoris is not strong enough to support the HR-mediated DSB repair, and blocking NHEJ had a negative effect on stability of chromosome terminal. We next tried to enhance HR process in P. pastoris by overexpressing the ScSAE2, ScRAD52 and ScRAD51, which support the highly efficient HR based genome editing in S. cerevisiae even with short homologous arms of 50 bp (24,25). Overexpression of ScSAE2 enabled higher positive rates (12.5%) in seamless deletion of GUT1 compared with the wild-type strain (0%, Figure 2A). Overexpression of endogenous PpRAD51 and PpRAD52 resulted in much higher positive rates (Figure 2A). In particular, PpRAD52 overexpression the highest positive rate of 77.5% which was 6.6-fold higher compared with a KU70 deletion strain (Figure 2A). These results agreed with a previous report that Rad52 can promote capture of the second end of DSB, which offers an alternative way of maturing the intermediates of DSB repair. Rad52 can also accelerate the assembly of a Rad51 filament on ssDNA-RPA complex (26). Overexpression of endogenous PpRAD54 did not improve positive rates of seamless gene deletion, although it was reported that Rad54 protein could overcome the inhibition of DNA strand exchange caused by Rad51 binding to dsDNA, which is beneficial for Rad51 dissociation and DNA strand exchange (27). Relative quantitative analysis of KU70 and RAD family genes expression suggested that RAD52 gene expression was lower than that of KU70 gene in the wild-type P. pastoris, whereas, in S. cerevisiae, RAD52 is expressed at a higher level compared with KU70 (Figure 2C). The expression of RAD52 was also the lowest compared with other three genes in P. pastoris, which further suggested that it might be the limiting factor of homologous recombination (Supplementary Figure S5). We also found that high expression of PpRAD52 was beneficial for HR-mediated GUT1 disruption (Figure 2D). These data suggest that high expression of RAD52 is essential for efficient homologous recombination relative to NHEJ process. PpRAD52 overexpression also significantly improved the seamless deletion efficiency for other genes such as FAA1 and FAA2 (Figure 2E). These data verified our assumption that the HR process of P. pastoris was relatively weak and could be enhanced by overexpression of the HR-related genes. Homologous arms with the lengths of 200 bp were effectively for seamless deletion of FAA1 gene (Figure 2F) and genome integration of a GFP expression cassette (Figure 2G). Furthermore, PpRAD52 overexpression enabled co-integration of three fragments into three genomic loci (Supplementary Figure S2) with a positive rate of 25%, whereas the wild-type strain failed to yield positive clones (Figure 2H). This platform was used successfully for co-integration of expression cassettes of MmCAR, npgA and ScADH5 from a fatty alcohol biosynthetic pathway (14,15).
Figure 2.
Enhancing homologous recombination (HR) for precise genome editing. (A) Screening HR related genes for improving seamless deletion. The heterologous HR related genes from S. cerevisiae or endogenous P. pastoris genes were overexpressed and be followed by deleting GUT1 in P. pastoris. The strains carried the plasmid expressing Cas9 and gRNA for targeting GUT1. The deletion cassette was constructed by fusing the upstream and downstream homologous arms of 1 kb. Wild-type strain GS115 without overexpressing RAD genes was used as a control. Eight colonies were picked for colony PCR analysis with primer pair Check-GUT1-F/Check-GUT1-R. (B) PpRAD52 overexpression improved the seamless deletion of FAA1 compared with KU70 deletion. (C) Relative expression level of KU70 and RAD52 in S. cerevisiae or P. pastoris. The housekeeping actin gene was chosen as baseline for quantification of expression level. Relative expression level of these genes was calculated by normalized the expression level of RAD52 in the respective strains. (D) PpRAD52 overexpression improved the seamless deletion of GUT1 in P. pastoris. The PGAP and PADH2 were used to overexpress RAD52 and the relative expression levels of RAD52 were analyzed by qPCR. (E) PpRAD52 overexpression improved the seamless deletion of GUT1, FAA1, and FAA2. (F) HR rates for FAA1 deletion with various HA lengths in strains of PpRAD52 overexpression. (G) HR rates for eGFP overexpression cassette integration into PNSI-1 site with various HA lengths in strains of PpRAD52 overexpression. (H) The positive rates of integration of three genes at three genome loci. Twenty colonies were picked for colony PCR analysis. 1 kb homologous arms was used in both integration and knockout experiments unless otherwise noted. Mean values and standard deviations of biological triplicates were shown in 2B to 2H.
Enhancing assembly of multiple gene expression cassettes
Rapid assembly of multiple DNA fragments in plasmids or single genome loci is helpful for construction and optimization of biosynthetic pathways composed of multiple genes (28). Thus, we assessed the possibility of in vivo assembly of three gene expression cassettes in fatty alcohol biosynthesis pathway with homologous arms of 500–1000 bp (Figure 3B). PpRAD52 expression had 54-fold higher positive rate (91.7%) when deleting GUT1 in compared to the wild-type strain of 1.67% (Figure 2E). However, integration of multiple fragments had relatively low positive rate (25%), making it difficult and time consuming to obtain the positive clones (Figure 3C). Thus, we revisited the HR recombination machinery.
Unlike the single fragment recombination for replacement or deletion, multiple fragments integration occurs as a multi-invasion-induced rearrangement (MIR) process (Figure 3A). We here thus tried to enhance the over-expression of endogenous structure-selective endonucleases (SSEs) that recognize and cleave the structures formed at the boundaries of DNA strand exchange intermediates and have key functions in the MIR process in S. cerevisiae (29). The synthesis pathway of fatty alcohol was divided into three fragments for multi-fragment recombination at FAA1 site (Figure 3B). Overexpression of endogenous SSE genes MUS81-MMS4, SLX1-SLX4 or YEN1 significantly improved the positive rates of three-fragment integration, but resulted in lower colony numbers of 27.0%, 81.6% and 59.1% compared with the control strain, respectively (Figure 3C). As a pivotal intermediate in the homologous recombination process, nascent displacement loops (D-loops) can be disrupted by the Mph1 and Sgs1-Top3-Rmi1 helicase-topoisomerase complex in S. cerevisiae (30). Consequently, deletion of MPH1 resulted in a 2.7-fold higher positive rate in integration of three fragments, albeit with a bit fewer colony. Although deletion of SGS1, TOP3 or RMI1 resulted in about 1.5 times higher positive rates compared with the control strain, the CFU/OD600 numbers were dramatically reduced to 5.5–19.8% of the control strain (Figure 3C). Combination of the PpRAD52 overexpression and MPH1 deletion (strain XPD11) improved the genome integration efficiency of three fragments at a single genomic locus by 13.5 times in compared to the wild-type strain (Figure 3D). Further, overexpression of SSE genes in strain XPD11 led to lower positive rates of 8.1–56.7%, and further deletion of SGS1, TOP3 or RMI1 resulted in reduced positive rates and CFU/OD600 number (Figure 3E). In the strain XPD11 (PpRAD52, mph1Δ), there was high and no different positive rates in integrating various cargo sizes of expression cassettes when genome integration of three fragments, and a slight decrease of positive rate was observed in integrating larger single DNA fragments (Supplementary Figure S6). These data showed the robustness of this system and should be helpful for integration of multiple genes when constructing long biosynthetic pathways.
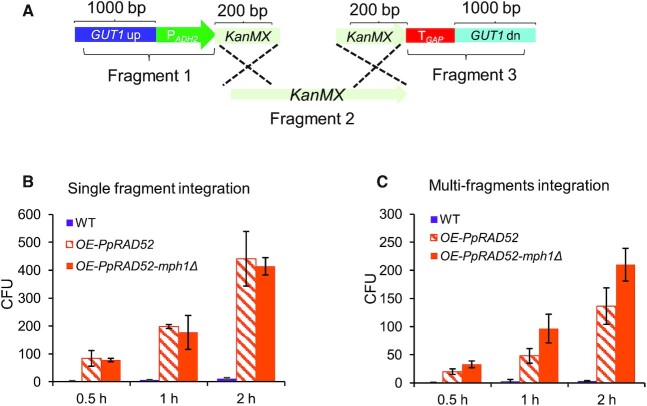
We finally monitored the multi-invasion-induced integration of the three fragments by using a KanMX cassette, which was divided into three DNA fragments: (1) an upstream segment that contained homologous arms to integration sites, promoters and the upstream region of the KanMX open reading frame (ORF); (2) the KanMX ORF lacking the start codon; and (3) the downstream segment containing downstream region of the KanMX ORF, terminator and downstream homologous arms to the integration site (Figure 4A). The recombination process can be tracked on the G418-containing plates because incorrect integration would not confer G418 resistance. The colony number increased with the incubation time, suggesting HR-based repair was time dependent. PpRAD52 overexpression had a higher repair efficiency in integration of a single DNA fragment (Figure 4B), whereas further deleting MPH1 improved the repair process only during multi-invasion-induced integration (Figure 4C). This observation supported our assumption that Rad52 can promote the capture of genome DSB whereas mph1Δ could relieve the repression of nascent D-loops that is essential for repair of multiple fragments with single or non-homologous arms to chromosomes (Figure 3A).
Figure 4.
Monitoring the repair process of multi-invasion-induced integration of multiple DNA fragments. (A) Scheme of genome assembly a KanMX cassette from three fragments for monitoring the repair process of multi-invasion-induced rearrangement process. (B) The CFU number for integration of single fragments in strains with MPH1 deletion and/or PpRAD52 overexpression. (C) The CFU number for integration of three fragments in strains with MPH1 deletion and/or PpRAD52 overexpression. The transformed cells were incubated in recovery medium for 1, 1.5 and 2 h to monitor the repair process.
Neutral site profiling
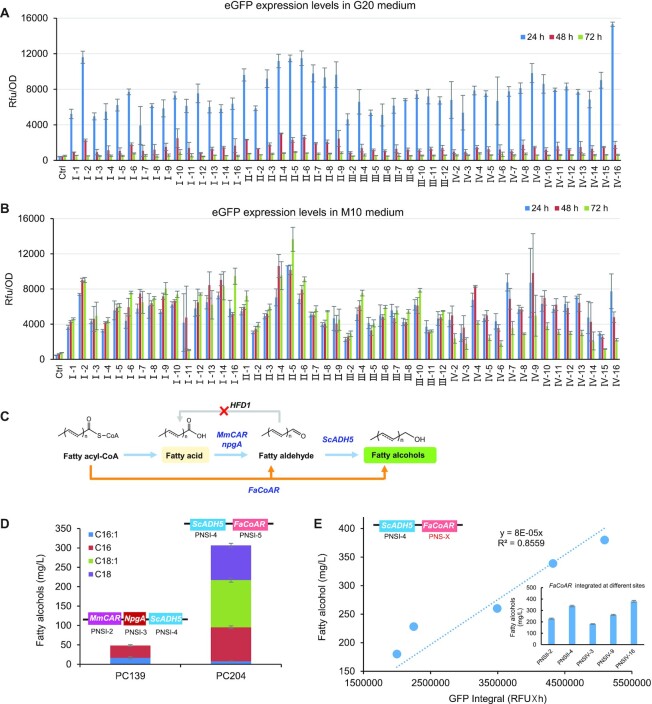
Extensive metabolic rewiring or engineering of biosynthetic pathways with many plasmid-based expression systems would require multiple different genetic markers and likely be unstable due to recombination between plasmids (31). Thus, neutral sites, chromosomal loci that can be disrupted without significant effects on cellular physiology and metabolism, should be identified and adopted as integration targets for extensive metabolic engineering. We screened the annotated P. pastoris genome sequence (10), for regions with >1500 bp between adjacent ORFs. We excluded 750 bp as potential promoter sequence and 500 bp as potential terminator sequence. With these two criteria, 53 potential neutral sites were characterized by integrating an eGFP expression cassette in PpRAD52 overexpression strain PC110 or PC111 with the established CRISPR-Cas9 system (Figure 1B, Supplementary Figure S1B), which enabled high integration efficiencies at almost all neutral sites (Supplementary Table S3). We also inspected the possible influence of 5′ upstream sequences of PNSI-1 site on eGFP expression by inserting the DNA sequence of promoter TAOX1 or 250 bp random sequence of PpHIS4 gene into the upstream of eGFP expression cassette. There was no significant difference in fluorescence intensity compared to that of pure eGFP expression cassette (Supplementary Figure S7), which suggested that the variant upstream sequence did not affect expression of heterologous genes.
To simplify the construction of the gRNA expression plasmid, we integrated the CAS9 gene into the PNSI-2 site and fixed the N6 variable sequences that are the reverse complement of the last six base pairs of the HH part (Supplementary Figure S1A, B). This operation significantly shortened the primer length for plasmid construction without sacrificing the integration efficiencies (Supplementary Figure S1D, E and Supplementary Table S3). The neutral sites PNSI-15, PNSIII-1, PNSIII-1, PNSIII-3, PNSIII-9 and PNSIV-1 were excluded due to the failure in obtaining positive transformants (Supplementary Table S3), which might be attributed to the non-functional of the gRNA and/or off-target effect of the CRISPR system. Although correct recombinants at PNSII-2 and PNSIII-2 site were obtained, they were abandoned due to the low positive rates. In particular, sgRNA target sequence of PNSII-2 was located in the CEN2 gene (32), which might cause genome instability. Integration of eGFP at all potential neutral site did not retard cell growth in either glucose or methanol medium (Supplementary Figure S8). Therefore, we screened 46 neutral sites for further study. We observed varying eGFP signals at different integration sites (Figure 5A and B), which suggested that genome structure or position could affect gene expression. Specifically, integration of eGFP at the PNSIV-16 site showed the strongest fluorescence intensity because there are two bidirectional copies of genomic PNSIV-16 sequence.
Figure 5.
Neutral site profiling by expression of eGFP and production of fatty alcohol. (A) The eGFP expression level from different neutral sites in minimal medium containing 20 g/l glucose. (B) The eGFP expression level from different neutral sites in minimal medium containing 10 g/l methanol. (C) The fatty alcohol biosynthetic pathways. The fatty acid derived pathway consists of the genes encoding carboxylic acid reductase from Mycobacterium marinum (MmCAR), the activation proteins (NpgA) and S. cerevisiae ADH5 (ScADH5). The fatty acyl-CoA derived pathway consists of fatty acyl-CoA reductase gene FacoAR and ScADH5. The endogenous fatty aldehyde dehydrogenase gene HFD1 was deleted to block the fatty aldehyde consumption. (D) The fatty alcohol titers by fatty acid or fatty acyl-CoA derived pathways. (E) The correlation of fatty alcohol titers and eGFP signals with the integration of FacoAR or eGFP expression cassette at various neutral sites.
With these neutral sites, we genomically integrated two types of fatty alcohol biosynthetic pathways (Figure 5C) that derived from free fatty acid (FFA) or fatty acyl-CoA (14). Integration of three codon-optimized genes of an FFA-derived pathway, enabled production of 48 mg/l fatty alcohols (Figure 5D). Integration of codon-optimized FaCoAR encoding fatty acyl-CoA reductase, enabled the production of 306 mg/l fatty alcohols (Figure 5D). Because eGFP signals varied at the different integration sites, we assessed the expression of the fatty alcohol biosynthesis by integration of FaCoAR at different neutral sites. Consistently, fatty alcohol production positively correlated with the integral of eGFP intensities (R2 = 0.86, Figure 5E, Supplementary Figure S9), which suggested that the neutral site can be used for varying gene expression and pathway optimization.
Promoter profiling
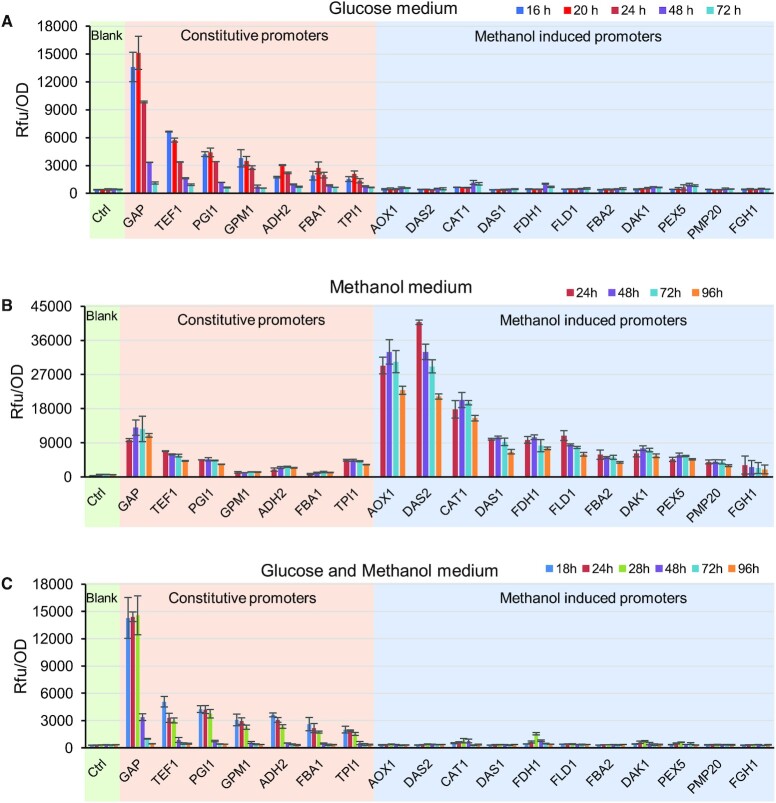
Promoters are essential elements of gene regulation and can be used to balance metabolic pathways (33). A plethora of promoters were recently characterized by microarray analysis (34). However, the complex expression patterns of promoters of P. pastoris have not been fully profiled on different carbon sources (glucose, and methanol) over a time scale, and such profiling could be beneficial for dynamically regulating biosynthetic pathways. Thus, we integrated a collection of promoters at the neutral PNSII-5 site and profiled their activities in a time-dependent manner at different carbon sources. In minimal glucose medium, the promoters related to glucose metabolism produced strong eGFP signals, among which, PGAP was the strongest one (Figure 6A). The eGFP signals were highly induced during exponential growth phase and reached the highest level at 20 h (Figure 6A and Supplementary Figure S10). Then, eGFP signals decreased considerably with glucose consumption, which showed that the promoters PGAP and PTEF1were not strictly constitutive. The promoters related to methanol utilization were completely repressed in minimal glucose medium (Figure 6B), which displayed strong glucose catabolite repression on methanol metabolism. In minimal methanol medium, the methanol utilization promoters were all induced, among which PDAS2 and PAOX1 were classified as strong methanol induced promoters, PCAT1 was classified as moderate methanol-induced promoter and other promoters were assigned as weak methanol-induced promoters (Figure 6B). These results were in accord with the expression profile of these promoters in previous reports (22,34). The glucose metabolism promoters had comparable but more stable expression in methanol medium compared with expression in glucose medium (Figure 6), which suggested that these promoters could be used for construction of biosynthetic pathways for methanol biotransformation. Lastly, we investigated the promoter profile in minimal medium containing 20 g/l glucose and 10 g/l methanol (G20M10). In this case, the glucose metabolism promoters had similar expression profiles in compared to the glucose-only medium. Conversely, the methanol utilization promoters were completely repressed even after depletion of the glucose (Figure 6C), which revealed that glucose catabolites had strong repression on methanol catabolism. There was no strict de-repression phase during the cultivation process, unless a certain proportion of the new medium was added (34).
Figure 6.
Time-dependent profiling of various promoters with glucose or methanol as carbon source. (A) The fluorescence of eGFP controlled by various promoters in minimal glucose medium. (B) The fluorescence of eGFP controlled by various promoters in minimal methanol medium. (C) The fluorescence of eGFP controlled by promoters in minimal medium containing 20 g/l glucose and 10 g/l methanol. Supplementary Figure S8 contains cell growth and carbon source profiles can be found in. All data represent the mean ± s.d. of three independent yeast clones.
Two-factorial regulation of chemical production via neutral sites and promoters
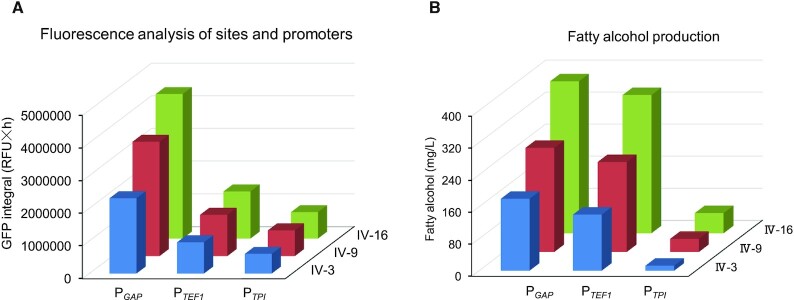
Because the integration sites (Figure 5) and promoters (Figure 6) drove gene expression to varying levels, we investigated a two-factorial regulation strategy for tuning gene expression by combining different neutral sites and promoters. We selected the promoters (PTPI, PTEF1 and PGAP) and the neutral sites (PIV-3, PIV-9 and PIV-16) for tuning gene expression because they showed a large variation in controlling eGFP expression (Figures 5A and 6A). Different combinations of these two elements led to a 7.3-fold range in eGFP expression (Figure 7A). We then used this strategy to regulate FaCoAR expression and observed a 30-fold range in fatty alcohol production (12.6–380 mg/l) on glucose medium, which was similar to the eGFP signal profile (Figure 7B). These data demonstrated that this two-factorial regulation can be used to balance metabolic pathways by tuning the expression of corresponding genes. We also tried to use methanol as a carbon source for fatty alcohol production, which was much lower than that of glucose as a carbon source (Supplementary Figure S11). Further rewiring the cellular metabolism should be required for enhancing the conversion of methanol toward target products.
Figure 7.
Engineering fatty alcohol biosynthesis in P. pastoris with characterized neutral sites and promoters. (A) The corresponding eGFP signal integral with different neutral sites and promoters. (B) Two-factorial regulation of fatty alcohol biosynthesis by tuning the expression of FaCoAR with different neutral sites and promoters. The fatty alcohols were extracted from 72 h cultivated cells at 30°C, 220 rpm. The cells were cultivated in Delft minimal medium with 20 g/l glucose. All data represent the mean±s.d. of three yeast clones.
DISCUSSION
Non-conventional yeasts have been recognized as potential chassis cell factories because of their desirable phenotypes, including Crabtree negativity, thermotolerance, and broad substrate spectrum (35). As a ‘generally regarded as safe’ (GRAS) microorganism, P. pastoris is an industrially important host for heterologous protein production. Recently, P. pastoris has gained considerable interest as a host for metabolic engineering to produce value-added products (6). In particular, P. pastoris’s methanol utilization feature makes it a potential host for methanol biotransformation toward value-added chemicals (7,36). P. pastoris has been also engineered into an autotroph for CO2 utilization with methanol as an electron donor (37).
Convenient genetic tools are prerequisites for extensive metabolic rewiring toward robust microbial cell factories. Here we established a genetic toolbox for P. pastoris that included enhanced HR process for precise genetic manipulation, characterization of genome neutral sites for gene integration and promoter profiling for tuning gene expression. We also established a two-factorial regulation strategy to tune the gene expression by combining adoption of promoters and neutral sites. This genetic toolbox enabled the genomic reconstruction and regulation of fatty alcohol biosynthetic pathways with a high variance.
The CRISPR-Cas9 system revolutionized genome editing with high efficiency, accuracy and convenience, and the system has been adapted to several non-conventional yeasts (4,38). Optimization of the CRISPR-Cas9 system by diverse codon-optimized Cas9 genes and promoters for expression of Cas9 and sgRNA has enabled efficient targeted cleavage of genome DNA in P. pastoris (11). However, the dominance of NHEJ in DSB repair interferes with seamless gene deletion and targeted integration of homologous DNA cassettes (with correct targeting rates of <30%) in P. pastoris (20,23). Repression of NHEJ by deleting KU70/KU80 significantly improved the positive rate of seamless gene deletion and targeted DNA integration in various non-conventional yeasts (20,23,39–43). However, permanent inactivation of NHEJ might not be desirable due to its essential role in general DNA repair process. It was observed that Ku deficient strain had much less colony number(41,43) and silence of heterologous genes (44) in compared to the wild-type strain. We here also observed DNA sequences loss when targeting to the chromosomal terminal position. Alternatively, we showed that overexpression of HR-related genes significantly improved the positive rates of DNA integration and seamless gene deletion. In particular, RAD52 overexpression produced the highest positive rates even with constructs that had relatively short homologous arms; this effect may have been due to improved competition with Ku proteins for binding to DSB (45). Similarly, heterologous expression of S. cerevisiae ScRAD52 significantly improved HR in mammalian cells (46) and Yarrowia lipolytica (47). The greater expression of RAD genes in S. cerevisiae likely explains the high homologous recombination efficiency relative to NHEJ process in S. cerevisiae. These data suggested that the HR process is naturally severely suppressed by NHEJ, and it needed to be enhanced for precise genome engineering in P. pastoris. It should be mentioned that high expression of RAD52 with an episomal plasmid resulted no transformant and chromosomal integrated expression of RAD52 had similar growth profile in compared to the wild-type strain (Supplementary Figure S12), which suggested moderate expression of RAD52 is very essential for cellular fitness. Interesting, we recently found that the expression of RAD51, not RAD52 was limited in HR process of Ogataea polymorpha (48), suggesting the difference expression profile of RAD genes among different organisms.
Simultaneous integration of multiple DNA fragment at one genomic locus is of interest in construction of multi-step biosynthetic pathways, which can be easily created in S. cerevisiae because of its efficient HR system (28). Compared with a single DNA fragment with two flanking homologous arms, multiple DNA fragments have only one (the side fragments) or no homologous arm (the middle fragments) to genome sequence, which makes it more challenging to form the HR pivotal D-loops intermediates (Figure 3A) (30). Disruption of the endogenous helicase gene MPH1 significantly improved the simultaneous assembly of three DNA fragments into the genome, possibly owing to the higher stability of D-loops (30). Mph1 disrupts the nascent D-Loops and represses the D-Loop extension in S. cerevisiae, which funnels HR toward non-crossover-based recombination and improves genome stability (29,30,49). However, multiple DNA fragment assembly relies on crossover-based repair with multi-invasions, and MPH1 disruption might loosen the rigorousness of DNA repair, thereby improving the efficiency of multi-invasion of three DNA fragments. RAD52 overexpression improved cell viability in both single and three DNA fragment assembly, whereas MPH1 disruption improved cell viability only in three DNA fragment assembly. This result suggested that attenuated resection of double-strand break ends was beneficial for stability of D-Loop and assembly of multiple DNA fragments in P. pastoris. Although the Sgs1-Top3-Rmi1 (STR) helicase-topoisomerase complex is also shown responsible for disruption of nascent D-Loops, we found that disruption of SGS1 severely hampered cell viability when assembling three DNA fragments. This negative effect of SGS1 disruption can be explained by the fact that Sgs1 promotes heteroduplex rejection and channels toward break-induced repair, which always occurs with short homologous arms or single-sided homologous arms (49). Further, disruption of STR genes (SGS1, TOP3 and RMI1) or overexpression of SSE genes (SLX1-SLX4 and MUS81-MMS4) with PpRAD52 overexpression and MPH1 deletion reduced the positive rates and CFU numbers, which suggested that balancing SSE and helicase was essential for multiple fragment integration. In general, homologous recombination is a dynamic process that enables high-fidelity DNA repair through multiple metastable and reversible intermediates. This dynamic balance is even more necessary for a multi-fragment repair process (50). Actually, it has been reported that high expression level of Mus81-Mms4 may cause premature termination of D-loop in homologous region and the failure of multi-fragment homologous recombination up on MPH1 deletion in S. cerevisiae (51). The process of DNA homologous recombination repair is complex, which is worthy to be studied further in P. pastoris.
Stable expression is considered a prior factor in constructing robust cell factories for the extended period of industrial fed-batch process. Therefore, genomic integration is more favored during strain development compared with plasmid-based expression (52). The abundance of neutral loci strongly supported the extensive metabolic engineering in S. cerevisiae (31,53). However, there is a limited number of gene integration sites were identified in non-conventional yeasts such as Y. lipolytica (54,55) and O. polymorpha (56). In general, a heterologous gene was integrated into the sites of alcohol oxidase gene AOX1 or gene HIS4 by a single crossover event in P. pastoris (57). Recently, three integration sites were identified and used to construct the 6-methylsalicylic acid biosynthetic pathway in P. pastoris (39). However, the lack of sufficient integration sites is still a major obstacle in constructing P. pastoris cell factories. Availability of abundant neutral sites would facilitate genomic integration of multiple genes for more comprehensive and predictable metabolic engineering (58,59). Furthermore, availability of promoters with various strengths should enable pathway balancing by tuning the expression of corresponding genes. With our high HR-based genetic engineering platform, we screened 46 possible neutral sites and characterized 18 promoters by efficient genomic integration of an eGFP expression cassette. Interestingly, the eGFP signals driven by PGAP of different neutral sites varied 3.9-fold and 4.7-fold after 24 h in glucose and methanol medium at shaker fermentation condition, respectively. Similarly, 8.7-fold and 2.4-fold differential expressions due to chromosomal location were found in S. cerevisiae and E. coli, respectively (52,53). This finding indicated that chromosome location affected gene transcription. Furthermore, the eGFP signals of all neutral sites were more stable in methanol medium than in glucose medium, which suggested that PGAP promoted more stable transcription in methanol medium. Similarly, constitutive promoters, mainly related to glucose metabolism, had decreased strength over time in glucose medium even in the presence of glucose, but they showed stable strength in methanol medium. The methanol-responsive promoters were completely repressed in glucose medium and could not be de-repressed or reactivated by methanol even after depletion of the glucose in mixed glucose and methanol medium. These data suggested methanol metabolism is severely repressed by glucose catabolites. Previous studies also identified or constructed a variety of (artificial) promoters, which provided sufficient regulation elements for balancing the metabolic pathways (22,60,61).
The varying expression from different neutral sites enables both construction and regulation of fatty alcohol biosynthetic pathways. In addition to the diversified promoter profiles, we can optimize metabolic pathways by using combinations of different promoters and neutral sites, leading to dynamic regulation of the fatty alcohol biosynthesis with a 30-fold range. The highest fatty alcohol titer (380 mg/l) was comparable with comprehensively engineered non-oleaginous yeasts (Supplementary Table S4) (14,62–67). This two-factorial regulation strategy should be beneficial for balancing biosynthetic pathways by regulating the expression of corresponding genes.
In summary, we established a genetic engineering platform with high HR efficiency by systematically engineering the recombination machinery in P. pastoris (Table 1). With this platform, we rapidly characterized the neutral sites and profiles several promoters in different cultivation conditions, which provides sufficient genetic elements and also a two-factorial regulation strategy for extensive metabolic engineering of P. pastoris. The knowledge and strategies described here should be conveniently applied to other non-conventional yeasts for construction of robust microbial cell factories.
Table 1.
Genetic engineering tools for P. pastoris
| HR mediate genetic engineering | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain engineering | Cas9 expression | sgRNA expression | Genome cutting efficiency | Seamless deletion | Genome integration | Marker needed | Minimal HA length | Ref. |
| / | PHTA1, episomal | PHTB1, episomal | 43–95% | 2.4% | Single gene (24%) | Yes | 1000 bp | (11) |
| ku70Δ for blocking NHEJ | PHTA1, episomal | PHTB1, episomal | 94% | gut1Δ (100%) | / | No | 1000 bp | (20) |
| ku70Δ for blocking NHEJ | PENO1, episomal | PtRNA3, episomal | 93% | / | Three genes (20%) | No | 500 bp | (12) |
| ku70Δ for blocking NHEJ | PHTA1, episomal | PFLD1, PAOX1, or PGAP, episomal | 75–98% | / | Two genes (58–70%) | No | 1000 bp | (39) |
| Three genes (13–32%) | ||||||||
| Enhance HR by overexpressing RAD52 and/or deleting MPH1 | PHTA1, episomal | PHTB1, episomal | 93% | gut1Δ (90%) | Single gene (43–70%) | No | 50 bp | This study |
| faa1Δ (88%) | Three genes at one site (67.5%) | |||||||
| faa2Δ (88%) | There genes at three sites (25%) | |||||||
HR, homologous recombination; HA, homologous arms.
DATA AVAILABILITY
All plasmids and engineered strains used in this study are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Energy Biotechnology Platform of DICP for providing facility assistance. The authors thank AiMi Academic Services (www.aimieditor.com) for English language editing and revise services.
Author contributions: Y.J.Z. conceived the study; P.C. and X.D. designed and performed all the experiments and analyzed the data; X.W. assisted with the neutral site profiling; L.G. assisted with integration of the multiple fragments; M.Y. assisted with the construction of gRNAs for natural site integration. P.C., X.D. and Y.J.Z. wrote the manuscript; all authors approved the manuscript.
Contributor Information
Peng Cai, Division of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; School of Bioengineering, Dalian University of Technology, Dalian 116024, China.
Xingpeng Duan, Division of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; Dalian Key Laboratory of Energy Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China.
Xiaoyan Wu, Division of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; Dalian Key Laboratory of Energy Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Linhui Gao, Division of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; Dalian Key Laboratory of Energy Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Min Ye, Division of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; Dalian Key Laboratory of Energy Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yongjin J Zhou, Division of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; CAS Key Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; Dalian Key Laboratory of Energy Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China; Laboratory of Synthetic Biology for Biocataysis, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31700082, 21807100, 21922812]; Dalian Science and Technology Innovation Funding [2019J12GX030]; DMTO [DICP DMTO201701] from Dalian Institute of Chemicals Physics, CAS. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. X.D., P.C. and Y.J.Z. have filed two patents for protecting part of the work described herein. All other authors declare no competing financial interests.
REFERENCES
- 1. Van Vleet J.H., Jeffries T.W.. Yeast metabolic engineering for hemicellulosic ethanol production. Curr. Opin. Biotechnol. 2009; 20:300–306. [DOI] [PubMed] [Google Scholar]
- 2. Hong K.K., Nielsen J.. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell. Mol. Life. Sci. 2012; 69:2671–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rebello S., Abraham A., Madhavan A., Sindhu R., Binod P., Karthika Bahuleyan A., Aneesh E.M., Pandey A.. Non-conventional yeast cell factories for sustainable bioprocesses. FEMS Microbiol. Lett. 2018; 365:fny222. [DOI] [PubMed] [Google Scholar]
- 4. Cai P., Gao J., Zhou Y.. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb. Cell Fact. 2019; 18:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lian J., Mishra S., Zhao H.. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications. Metab. Eng. 2018; 50:85–108. [DOI] [PubMed] [Google Scholar]
- 6. Yang Z., Zhang Z.. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: a review. Biotechnol. Adv. 2018; 36:182–195. [DOI] [PubMed] [Google Scholar]
- 7. Zhou Y.J., Kerkhoven E.J., Nielsen J.. Barriers and opportunities in bio-based production of hydrocarbons. Nat. Energy. 2018; 3:925–935. [Google Scholar]
- 8. Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M., Smolke C.D.. Complete biosynthesis of opioids in yeast. Science. 2015; 349:1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu T., Zhou Y.J., Huang M., Liu Q., Pereira R., David F., Nielsen J.. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018; 174:1549–1558. [DOI] [PubMed] [Google Scholar]
- 10. De Schutter K., Lin Y.C., Tiels P., Van Hecke A., Glinka S., Weber-Lehmann J., Rouze P., Van de Peer Y., Callewaert N.. Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 2009; 27:561–566. [DOI] [PubMed] [Google Scholar]
- 11. Weninger A., Hatzl A.M., Schmid C., Vogl T., Glieder A.. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 2016; 235:139–149. [DOI] [PubMed] [Google Scholar]
- 12. Dalvie N.C., Leal J., Whittaker C.A., Yang Y., Brady J.R., Love K.R., Love J.C.. Host-informed expression of CRISPR guide RNA for genomic engineering in komagataella phaffii. ACS Synth. Biol. 2020; 9:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou Y.J., Hu Y., Zhu Z., Siewers V., Nielsen J.. Engineering 1-alkene biosynthesis and secretion by dynamic regulation in yeast. ACS Synth. Biol. 2018; 7:584–590. [DOI] [PubMed] [Google Scholar]
- 14. Zhou Y.J., Buijs N.A., Zhu Z., Qin J., Siewers V., Nielsen J.. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 2016; 7:11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou Y.J., Buijs N.A., Zhu Z., Gomez D.O., Boonsombuti A., Siewers V., Nielsen J.. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J. Am. Chem. Soc. 2016; 138:15368–15377. [DOI] [PubMed] [Google Scholar]
- 16. Zhu Z., Zhou Y.J., Kang M.K., Krivoruchko A., Buijs N.A., Nielsen J.. Enabling the synthesis of medium chain alkanes and 1-alkenes in yeast. Metab. Eng. 2017; 44:81–88. [DOI] [PubMed] [Google Scholar]
- 17. Lin-Cereghino J., Wong W.W., Xiong S., Giang W., Luong L.T., Vu J., Johnson S.D., Lin-Cereghino G.P.. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. BioTechniques. 2005; 38:44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Looke M., Kristjuhan K., Kristjuhan A.. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011; 50:325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao J., Yuan W., Li Y., Bai F., Jiang Y.. Characterization of inulinase promoter from Kluyveromyces marxianus for intensive protein expression in industrial biotechnology. FEMS Yeast. Res. 2017; 17:fox062. [DOI] [PubMed] [Google Scholar]
- 20. Weninger A., Fischer J.E., Raschmanova H., Kniely C., Vogl T., Glieder A.. Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J. Cell Biochem. 2018; 119:3183–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 22. Vogl T., Kickenweiz T., Pitzer J., Sturmberger L., Weninger A., Biggs B.W., Kohler E.M., Baumschlager A., Fischer J.E., Hyden P.et al.. Engineered bidirectional promoters enable rapid multi-gene co-expression optimization. Nat. Commun. 2018; 9:3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naatsaari L., Mistlberger B., Ruth C., Hajek T., Hartner F.S., Glieder A.. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. Plos One. 2012; 7:e39720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krejci L., Altmannova V., Spirek M., Zhao X.. Homologous recombination and its regulation. Nucleic Acids Res. 2012; 40:5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mimitou E.P., Symington L.S.. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008; 455:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kowalczykowski S.C. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2015; 7:a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Solinger J.A., Kiianitsa K., Heyer W.-D.. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002; 10:1175–1188. [DOI] [PubMed] [Google Scholar]
- 28. Shao Z., Zhao H., Zhao H.. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009; 37:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Piazza A., Wright W.D., Heyer W.D.. Multi-invasions are recombination byproducts that induce chromosomal rearrangements. Cell. 2017; 170:760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piazza A., Shah S.S., Wright W.D., Gore S.K., Koszul R., Heyer W.D.. Dynamic processing of displacement loops during recombinational DNA repair. Mol. Cell. 2019; 73:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mikkelsen M.D., Buron L.D., Salomonsen B., Olsen C.E., Hansen B.G., Mortensen U.H., Halkier B.A.. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 2012; 14:104–111. [DOI] [PubMed] [Google Scholar]
- 32. Coughlan A.Y., Hanson S.J., Byrne K.P., Wolfe K.H.. Centromeres of the yeast Komagataella phaffii (Pichia pastoris) have a simple inverted-repeat structure. Genome Biol. Evol. 2016; 8:2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hartner F.S., Ruth C., Langenegger D., Johnson S.N., Hyka P., Lin-Cereghino G.P., Lin-Cereghino J., Kovar K., Cregg J.M., Glieder A.. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 2008; 36:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vogl T., Sturmberger L., Kickenweiz T., Wasmayer R., Schmid C., Hatzl A.M., Gerstmann M.A., Pitzer J., Wagner M., Thallinger G.G.et al.. A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth. Biol. 2016; 5:172–186. [DOI] [PubMed] [Google Scholar]
- 35. Lobs A.K., Schwartz C., Wheeldon I.. Genome and metabolic engineering in non-conventional yeasts: Current advances and applications. Synth. Syst. Biotechnol. 2017; 2:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duan X., Gao J., Zhou Y.J.. Advances in engineering methylotrophic yeast for biosynthesis of valuable chemicals from methanol. Chinese Chem. Lett. 2018; 29:681–686. [Google Scholar]
- 37. Gassler T., Sauer M., Gasser B., Egermeier M., Troyer C., Causon T., Hann S., Mattanovich D., Steiger M.G.. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 2020; 38:210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raschmanova H., Weninger A., Glieder A., Kovar K., Vogl T.. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: Current state and future prospects. Biotechnol. Adv. 2018; 36:641–665. [DOI] [PubMed] [Google Scholar]
- 39. Liu Q., Shi X., Song L., Liu H., Zhou X., Wang Q., Zhang Y., Cai M.. CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Fact. 2019; 18:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rajkumar A.S., Varela J.A., Juergens H., Daran J.G., Morrissey J.P.. Biological parts for Kluyveromyces marxianus synthetic biology. Front. Bioeng. Biotechnol. 2019; 7:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao M., Gao M., Ploessl D., Song C., Shao Z.. CRISPR-Mediated genome editing and gene repression in Scheffersomyces stipitis. Biotechnol. J. 2018; 13:e1700598. [DOI] [PubMed] [Google Scholar]
- 42. Schwartz C., Frogue K., Ramesh A., Misa J., Wheeldon I.. CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnol. Bioeng. 2017; 114:2896–2906. [DOI] [PubMed] [Google Scholar]
- 43. Gao S., Tong Y., Wen Z., Zhu L., Ge M., Chen D., Jiang Y., Yang S.. Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J. Ind. Microbiol. Biotechnol. 2016; 43:1085–1093. [DOI] [PubMed] [Google Scholar]
- 44. Guo F., Dai Z., Peng W., Zhang S., Zhou J., Ma J., Dong W., Xin F., Zhang W., Jiang M.. Metabolic engineering of Pichia pastoris for malic acid production from methanol. Biotechnol. Bioeng. 2020; 118:357–371. [DOI] [PubMed] [Google Scholar]
- 45. Zan H., Tat C., Qiu Z., Taylor J.R., Guerrero J.A., Shen T., Casali P.. Rad52 competes with Ku70/Ku86 for binding to S-region DSB ends to modulate antibody class-switch DNA recombination. Nat. Commun. 2017; 8:14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Di Primio C., Galli A., Cervelli T., Zoppe M., Rainaldi G.. Potentiation of gene targeting in human cells by expression of Saccharomyces cerevisiae Rad52. Nucleic Acids Res. 2005; 33:4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ji Q., Mai J., Ding Y., Wei Y., Ledesma-Amaro R., Ji X.J.. Improving the homologous recombination efficiency of Yarrowia lipolytica by grafting heterologous component from Saccharomyces cerevisiae. Metab. Eng. Commun. 2020; 11:e00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao J., Gao N., Zhai X., Zhou Y.J.. Recombination machinery engineering for precise genome editing in methylotrophic yeast Ogataea polymorpha. iScience. 2021; 24:102168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mehta A., Beach A., Haber J.E.. Homology requirements and competition between gene conversion and break-induced replication during double-strand break repair. Mol. Cell. 2017; 65:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wright W.D., Shah S.S., Heyer W.D.. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018; 293:10524–10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mazon G., Symington L.S.. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol. Cell. 2013; 52:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goormans A.R., Snoeck N., Decadt H., Vermeulen K., Peters G., Coussement P., Van Herpe D., Beauprez J.J., De Maeseneire S.L., Soetaert W.K.. Comprehensive study on Escherichia coli genomic expression: does position really matter. Metab. Eng. 2020; 62:10–19. [DOI] [PubMed] [Google Scholar]
- 53. Flagfeldt D.B., Siewers V., Huang L., Nielsen J.. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009; 26:545–551. [DOI] [PubMed] [Google Scholar]
- 54. Schwartz C., Shabbir-Hussain M., Frogue K., Blenner M., Wheeldon I.. Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica. ACS Synth. Biol. 2017; 6:402–409. [DOI] [PubMed] [Google Scholar]
- 55. Holkenbrink C., Dam M.I., Kildegaard K.R., Beder J., Dahlin J., Domenech Belda D., Borodina I.. EasyCloneYALI: CRISPR/Cas9-based synthetic toolbox for engineering of the yeast Yarrowia lipolytica. Biotechnol. J. 2018; 13:e1700543. [DOI] [PubMed] [Google Scholar]
- 56. Yu W., Gao J., Zhai X., Zhou Y.J.. Screening neutral sites for metabolic engineering of methylotrophic yeast Ogataea polymorpha. Synth. Syst. Biotechnol. 2021; 6:63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vogl T., Gebbie L., Palfreyman R.W., Speight R., Master E.R.. Effect of plasmid design and type of integration event on recombinant protein expression in Pichia pastoris. Appl. Environ. Microbiol. 2018; 84:e02712-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang M., Luan G., Lu X.. Systematic identification of a neutral site on chromosome of Synechococcus sp. PCC7002, a promising photosynthetic chassis strain. J. Biotechnol. 2019; 295:37–40. [DOI] [PubMed] [Google Scholar]
- 59. Mikkelsen M.D., Buron L.D., Salomonsen B., Olsen C.E., Hansen B.G., Mortensen U.H., Halkier B.A.. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 2012; 14:104–111. [DOI] [PubMed] [Google Scholar]
- 60. Portela R.M., Vogl T., Kniely C., Fischer J.E., Oliveira R., Glieder A.. Synthetic core promoters as universal parts for fine-tuning expression in different yeast species. ACS Synth. Biol. 2017; 6:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vogl T., Ruth C., Pitzer J., Kickenweiz T., Glieder A.. Synthetic core promoters for Pichia pastoris. ACS Synth. Biol. 2014; 3:188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tang X., Chen W.N.. Enhanced production of fatty alcohols by engineering the TAGs synthesis pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015; 112:386–392. [DOI] [PubMed] [Google Scholar]
- 63. Runguphan W., Keasling J.D.. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014; 21:103–113. [DOI] [PubMed] [Google Scholar]
- 64. Dahlin J., Holkenbrink C., Marella E.R., Wang G., Liebal U., Lieven C., Weber D., McCloskey D., Ebert B.E., Herrgard M.J.et al.. Multi-Omics analysis of fatty alcohol production in engineered yeasts saccharomyces cerevisiae and Yarrowia lipolytica. Front. Genet. 2019; 10:747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hu Y., Zhu Z., Gradischnig D., Winkler M., Nielsen J., Siewers V.. Engineering carboxylic acid reductase for selective synthesis of medium-chain fatty alcohols in yeast. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:22974–22983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Feng X., Lian J., Zhao H.. Metabolic engineering of Saccharomyces cerevisiae to improve 1-hexadecanol production. Metab. Eng. 2015; 27:10–19. [DOI] [PubMed] [Google Scholar]
- 67. d’Espaux L., Ghosh A., Runguphan W., Wehrs M., Xu F., Konzock O., Dev I., Nhan M., Gin J., Reider Apel A.et al.. Engineering high-level production of fatty alcohols by Saccharomyces cerevisiae from lignocellulosic feedstocks. Metab. Eng. 2017; 42:115–125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids and engineered strains used in this study are available upon request.