Dear Editor,
MicroRNAs (miRs) have emerged as cardiovascular biomarkers and myocardial regulators with diagnostic and therapeutic potential. 1 , 2 However, the miR profile of patients with hypertrophic cardiomyopathy (HCM) and miRs role in this genetic disease with heterogeneous phenotype 3 are incompletely determined.
Here a multi‐step strategy provided evidence of differentially expressed miRs (DEmiRs) in plasma and myocardial tissue from HCM (Supplementary Materials: Materials and Methods).
In the first step, a profile of 1128 expressed mature miRs was identified (Figure 1) in 25 plasma samples from a cohort of 36 HCM (Figure S1, Tables S1 and S2) and healthy individuals (CTRL, n = 11) by next generation sequencing (NGS). Principal component analysis of NGS data did not clearly define a separation between HCM and CTRL, while a significant differential expression was found in a subset of 139 plasma miRs. Twenty‐eight of them were DEmiRs significant both for p value and false discovery rate (Figures 1B and 1C, Tables S3–S4), and demonstrated 1626 putative targets by in silico search with miRTargetLink Human engine. Among them, 50 target genes were interconnected with 13 DEmiRs by “strong” experimental methods like reporter gene assay (Figure S2A), corresponded to a complex protein network (Figure S2B) with associated functions/pathways (Supplemental_Enrichment).
FIGURE 1.
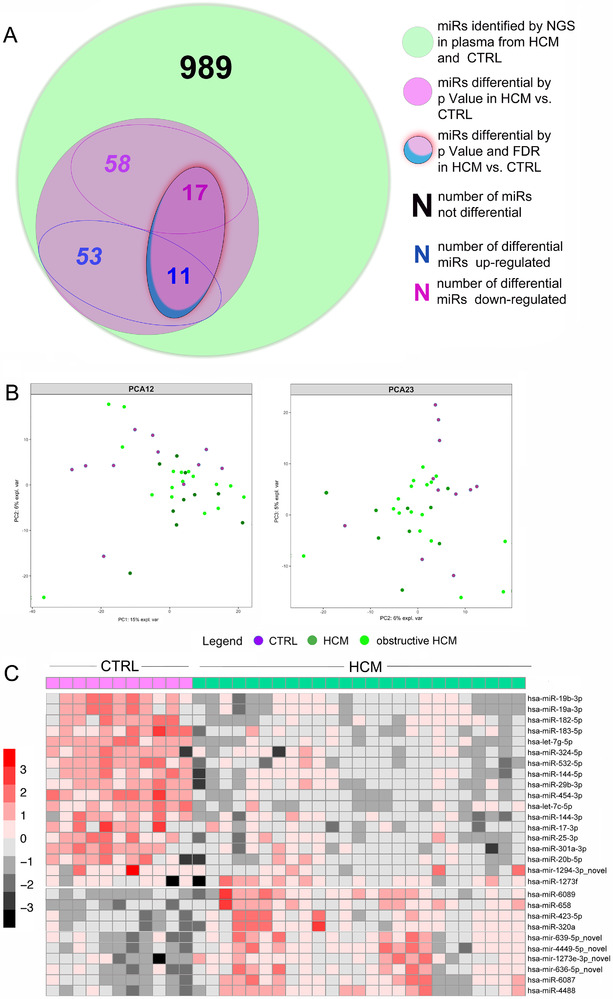
NGS analysis of plasma miRs. Venn diagram representation of the plasma miRs up‐/down ‐regulated in HCM patients vs. CTRL by NGS (A) and corresponding scatterplots of principal component analysis (PCA) of patients and controls screened by NGS (B) showing no remarkable discrete clustering of the non‐obstructive HCM (HCM in the panel) vs. obstructive HCM vs. CTRL. Heatmap of the relative expression of miRs differential by NGS displays the existence of finest difference in miR expressions between HCM and CTRL (C)
To validate NGS performance and exclude false DEmiRs due to high specificity but moderate sensitivity of the assay, 4 37 miRs (16 DEmiRs, 21 identified plasma miRs involved in cardiovascular diseases/proposed as biomarkers for cardiomyopathy or heart failure [HF] by other studies) were determined in 22 HCM, 10 CTRL by quantitative real‐time polymerase chain reaction (RT‐qPCR). Eight plasma DEmiRs were confirmed by RT‐qPCR: hsa‐miR‐19a‐3p, hsa‐miR‐20b ‐5p, hsa‐miR‐29b‐3p, hsa‐miR‐126‐5p, hsa‐miR‐144‐3p, hsa‐miR‐454‐3p and hsa‐miR‐4732‐5p were up‐regulated, and hsa‐miR‐182‐5p was down‐regulated (Figure 2A, Table S5). All, except the hsa‐miR‐454‐3p demonstrated acceptable accuracy by receiver operating characteristic (ROC) analysis (Figure 2B). Conversely, six miRs were undetected in all plasma samples (i.e., hsa‐miR‐1273a, hsa‐miR‐1273c, hsa‐miR‐1285‐3p, hsa‐miR‐363‐5p, hsa‐miR‐658, and hsa‐miR‐6089) and 23 miRs showed a comparable expression in HCM and CTRL (Table S5, Figure S3). Our approach showed an overall agreement but incomplete overlapping of NGS and RT‐qPCR results, highlighting the challenge of an accurate quantification. 4
FIGURE 2.
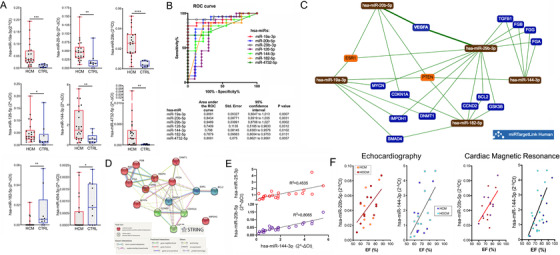
Analysis of plasma miRs by RT‐qPCR and in silico engines and association between plasma miRs by RT‐qPCR and left ventricle ejection fraction. Differential plasma miR expression levels in HCM vs. CTRL by RT‐qPCR are plotted (A). Values are presented as boxes (min to max) and dots indicate single sample values. Mann‐Whitney test is applied and significant differences are shown as *p < 0.05 and **p < 0.01, ***p < 0.001, ****p < 0.0001. Receiver‐operator characteristic (ROC) curve analysis of plasma DEmiRs is shown (B). AUC > 0.7 with significant p values was considered as threshold for good discriminant performance. The network obtained by miR TargetLink Human for strong interactions among DEmiRs and their putative target genes is shown (C). Orange nodes show target genes associated with three or more miRs, blue nodes those shared by less than three miRs, brown nodes indicate miRs. The interactions among the proteins encoded by target genes drawn by STRING v11 are presented (D). Nodes corresponding to clustered proteins are presented in the same color. Linear relations between hsa‐miR‐144‐3p and hsa‐miR‐20b‐5p or hsa‐miR‐25‐3p in the plasma samples of HCM population are shown (E). Linear relation between the hsa‐miR‐144‐3p and hsa‐miR‐20b‐5p expression levels determined by RT‐qPCR and the left ventricle ejection fraction (EF) assessed by routine echocardiography or cardiac magnetic resonance in patients either with non obstructive (HCM) or obstructive (HOCM) hypertrophic cardiomyopathy is shown (F)
The in silico analysis associated with validated plasma DEmiRs 15 predicted target genes (Figure 2C), three interacting protein clusters (Figure 2D), and numerous biological processes (Supplemental_Enrichment 1).
The Spearman's rank coefficient calculation differently correlated pairs of miRs highly‐expressed by RT‐qPCR in HCM and CTRL (Figure S4A), and a significant linear relationship relating hsa‐miR‐144‐3p to both hsa‐miR‐20b‐5p and hsa‐miR‐25‐3p in HCM was found (Figure 2E). These three miRs were interconnected by 93 predicted targets, including PTEN and BCL2L11 genes backed‐up with strong evidence (Figure S4B), and leading to network of 17 proteins involved in pathways of cardiac relevance such as the regulation of cardiac muscle cell proliferation and of response to endoplasmic reticulum stress (Figure S4C). Furthermore, the linear association between plasma levels of hsa‐miR‐144‐3p and hsa‐miR‐20b‐5p and % ejection fraction (EF) found in the HCM population (Figure 2F, Table S6) suggested a role for these two miRs in the regulation of myocardial function.
Thereafter, 20 miRs already validated in the plasma (including all up‐regulated DEmiRs) were determined in septal myectomy samples from 21 obstructive HCM of our cohort and nine donor hearts (ctrl). RT‐qPCR showed three downregulated (hsa‐miR‐144‐3p, hsa‐miR‐451a and hsa‐miR‐223‐3p, Figure 3A) and two up‐regulated (hsa‐miR‐374b‐5p and hsa‐miR‐4485‐3p, Figure 3B) tissue DEmiRs.
FIGURE 3.
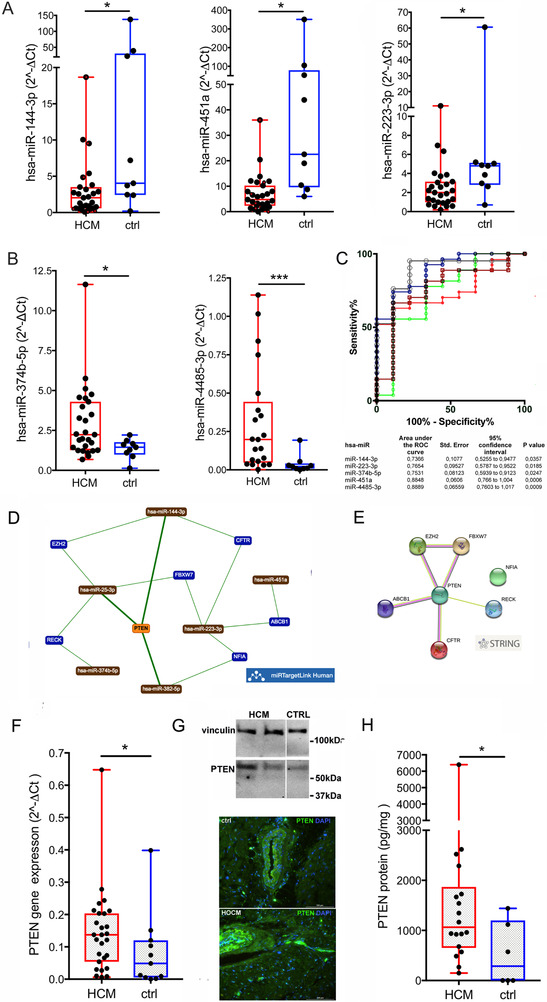
Analysis of myocardial tissue miRs by RT‐qPCR and expression of PTEN. The expression levels of DEmiRs determined by RT‐qPCR in the myocardial tissue samples of HCM vs. ctrl are plotted. The miRs down‐regulated in HCM are in A, those upregulated in B. Receiver‐operator characteristic (ROC) curve analysis of tissue DEmiRs is shown (C). AUC > 0.7 with significant p values was considered as threshold for good discriminant performance. The network obtained by miR TargetLink Human for strong interactions among hsa‐miR‐4451, hsa‐miR‐382‐5p, hsa‐miR‐25‐3p, tissue DEmiRs and their putative target genes is presented (D). Orange nodes show target genes associated with three or more miRs, blue nodes those shared by less than three miRs, brown nodes indicate miRs. The corresponding protein network drawn by STRING v11 is shown (E). The upregulation of PTEN gene determined by RT‐qPCR in HCM vs. ctrl tissues is plotted, (F). Representative qualitative evaluation of the protein presence into myocardial tissues from two HCMs and one ctrl by western blot (G, up) is shown. Immunofluorescence images of HCM and ctrl cryosections are displayed (G, down) and PTEN quantification by specific ELISA is plotted (H). Values In A, B, F, and H plots are presented as boxes (min to max), and dots indicate single sample values. Mann‐Whitney test is applied and significant differences are shown as *p < 0.05
The miR144/451 family was associated with extracellular matrix remodeling, 5 negative regulation of hypertrophy and autophagy, 6 and cardiac diseases by other reports (e.g., the HUNT study 7 ). Our data in HCM showed that hsa‐miR‐144‐3p and hsa‐miR‐451a were linearly related, while hsa‐miR‐4732‐5p undetermined in the majority of samples (Figure S5), confirming a published report in HCM patients, 6 but also suggesting an opposite trend in plasma and tissue for hsa‐miR‐144‐3p expression, due to increased cardiac release/decreased synthesis. This hints to a role in cardiac remodeling in HCM, deserving further investigations.
Moreover, ROC analysis demonstrated the good performance of 4 out of 5 tissue DEmiRs (Figure 3C).
The other miRs tested in tissues showed comparable expression levels in HCM and ctrl (Figure S6), but four of them were related to HCM clinical phenotypes (Figure S7). Specifically, hsa‐miR‐4451 was linearly associated with interventricular septum thickness, both hsa‐miR‐382‐5p and hsa‐miR‐25‐3p to glomerular filtration rate, and hsa‐miR‐382‐5p negatively and non‐linearly to Troponin T.
The calculation of Spearman correlation between miR pairs showed some positive relationships in HCM tissue (>0.60) with significant linear fits (Figure S8A‐SAC).
In silico analysis of myocardial DEmiRs and of miRs associated with clinical parameters predicted networks of targets genes and interacting proteins (Figures 3D 3E, and S8D) partially superimposable to those drawn for validated plasma DEmiRs, and showed PTEN as a shared target in both HCM plasma and myocardial tissue. Notably, a significant up‐regulation of PTEN gene expression was determined in myectomies from HCM vs. ctrl (p = 0.035, Figure 3F) and increased expressed protein amount (Figures 3G and 3H, p = 0.047) was also found in HCM samples. PTEN deletion in mice drove variable in vivo and in vitro effects on cardiomyocyte hypertrophy, 8 and a pro‐hypertrophic signaling pathway involving miR‐20b and PTEN was proposed in conditions of pressure‐overload cardiac hypertrophy. 8 , 9 , 10 To the best of our knowledge PTEN‐related mechanisms in human HCM have not been elucidated. Based on our results we hypothesize a mechanistic role for DEmiRs in the modulation of PTEN in HCM, possibly in relation to metabolic alterations, and suggesting the need for dedicated mechanistic studies.
In summary (Figure 4), starting from the whole plasma miR transcriptome of HCM and CTRL, we determined the relative and absolute abundance of specific miRs and delineated a preliminary panel of plasma/tissue DEmiRs which included hsa‐miR‐182‐5p, hsa‐miR‐126‐5p, hsa‐miR‐19a‐3p, hsa‐miR‐20b‐5p, hsa‐miR‐29b‐3p, hsa‐miR‐144‐3p, hsa‐miR‐223‐3p, and hsa‐miR‐4485‐3p as potential biomarkers setting HCM apart from other diseases (Table 1 for comparison with other human studies). The DEmiR target genes network drawn by in silico analyses hinted to a role for PTEN in HCM pathogenesis.
FIGURE 4.
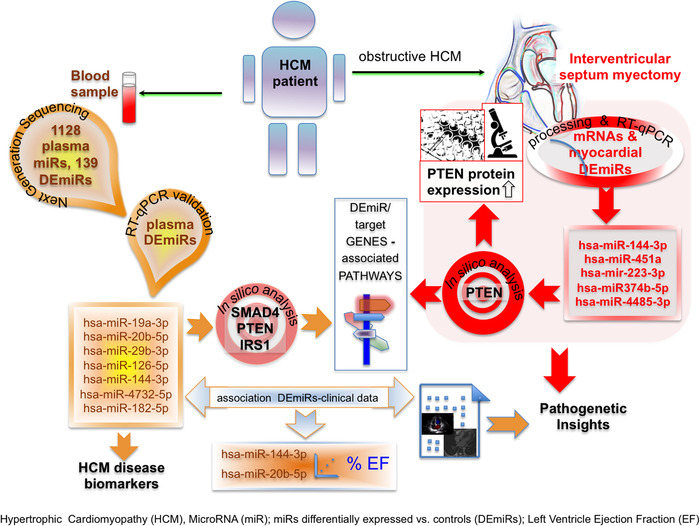
Schematic representation of the study results
TABLE 1.
DEmiRs: Comparison with published studies
| Plasma | Myocardial tissue | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HCM (our study) | CAD | HF | AF | AMI | other pathologies | HCM (our study) | other pathologies | References_DOI | References_full citation | |
| miR‐19a | UP | UP | UP (chronic Chagas disease) | NDE | UP (chronic Chagas disease) |
Nonaka, C.K.V. et al, Circulating miRNAs as Potential Biomarkers Associated with Cardiac Remodeling and Fibrosis in Chagas Disease Cardiomyopathy. International journal of molecular sciences, 20. |
||||
|
Mansouri, F. and Seyed Mohammadzad, M.H. Molecular miR‐19a in Acute Myocardial Infarction: Novel Potential Indicators of Prognosis and Early Diagnosis. Asian Pacific journal of cancer prevention: APJCP, 21, 975–982. |
||||||||||
|
Zhong, J. et al, Circulating microRNA‐19a as a potential novel biomarker for diagnosis of acute myocardial infarction. International journal of molecular sciences, 15, 20355–20364. |
||||||||||
|
Gao, F. et al, Therapeutic role of miR‐19a/19b in cardiac regeneration and protection from myocardial infarction. Nature communications, 10, 1802. |
||||||||||
| miR‐20b | UP | DOWN (T2DM) | NDE |
Wander, P.L. et al, Short Report: Circulating microRNAs are associated with incident diabetes over 10 years in Japanese Americans. Scientific reports, 10, 6509. |
||||||
|
Zampetaki, A. et al, Plasma microRNA profiling reveals loss of endothelial miR‐126 and other microRNAs in type 2 diabetes. Circulation research, 107, 810–817. |
||||||||||
|
Zhou, J.et al, microRNA expression profiling of heart tissue during fetal development. International journal of molecular medicine, 33, 1250–1260. |
||||||||||
| miR‐29b | UP | UP | UP (chronic Chagas disease) | NDE | UP (chronic Chagas disease, arrythmogenic right ventricle cardiomyopathy) |
Nonaka, C.K.V. et al, Circulating miRNAs as Potential Biomarkers Associated with Cardiac Remodeling and Fibrosis in Chagas Disease Cardiomyopathy. International journal of molecular sciences, 20. |
||||
|
Yang, Q. et al, Aberrant expression of miR‐29b‐3p influences heart development and cardiomyocyte proliferation by targeting NOTCH2. Cell proliferation, 53, e12764. |
||||||||||
| miR‐126 | UP | DOWN | UP | UP | UP | DOWN (T2DM, chronic renal disease) | NDE |
Wang, X. et al, Expression of miR‐126 and its potential function in coronary artery disease. African health sciences, 17, 474–480. |
||
|
Li, H.Y. et al, Plasma MicroRNA‐126‐5p is Associated with the Complexity and Severity of Coronary Artery Disease in Patients with Stable Angina Pectoris. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology, 39, 837–846. |
||||||||||
|
Fourdinier, O. et al, Serum levels of miR‐126 and miR‐223 and outcomes in chronic kidney disease patients. Scientific reports, 9, 4477. |
||||||||||
| miR‐144 | UP | UP | UP | UP (arrythmogenic right ventricle cardiomyopathy) | DOWN | UP (arrythmogenic right ventricle cardiomyopathy) |
de Gonzalo‐Calvo, D. et al, Epigenetic Biomarkers and Cardiovascular Disease: Circulating MicroRNAs. Revista espanola de cardiologia (English ed.), 70, 763–769. |
|||
|
Bye, A. et al, Circulating microRNAs predict future fatal myocardial infarction in healthy individuals ‐ The HUNT study. Journal of molecular and cellular cardiology, 97, 162–168. |
||||||||||
|
Abu‐Halima, M. et al, Micro‐RNA signatures in monozygotic twins discordant for congenital heart defects. PloS one, 14, e0226164. |
||||||||||
|
Song, L. et al, MiR‐451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. Journal of cellular and molecular medicine, 18, 2266–2274. |
||||||||||
| miR‐182 | DOWN | UP | UP | UP (arrythmogenic right ventricle cardiomyopathy) | NDE | DOWN (arrythmogenic right ventricle cardiomyopathy) |
Taurino, C. et al, Gene expression profiling in whole blood of patients with coronary artery disease. Clinical science (London, England : 1979), 119, 335–343. |
|||
|
Cakmak, H.A. et al, The prognostic value of circulating microRNAs in heart failure: preliminary results from a genome‐wide expression study. Journal of cardiovascular medicine (Hagerstown, Md.), 16, 431–437. |
||||||||||
| https://doi.org/10.1038/srep21228 |
Li, N. et al, miR‐182 Modulates Myocardial Hypertrophic Response Induced by Angiogenesis in Heart. Scientific reports, 6, 21228. |
|||||||||
| miR‐223 | NDE | DOWN |
Barsanti, C. et al, Differential regulation of microRNAs in end‐stage failing hearts is associated with left ventricular assist device unloading. BioMed research international, 2015, 592512. |
|||||||
| miR‐223 (Cont.) |
Chuang, T.Y. et al, MicroRNA‐223 Expression is Upregulated in Insulin Resistant Human Adipose Tissue. Journal of diabetes research, 2015, 943659. |
|||||||||
|
Lu, H., Buchan, R.J. and Cook, S.A., MicroRNA‐223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovascular research, 86, 410–420. |
||||||||||
| miR‐374b | NDE | DOWN | UP (STEMI vs. NSTEMI) | UP | DOWN (calcific aortic stenosis: valves) |
Ward, J.A. et al, Circulating Cell and Plasma microRNA Profiles Differ between Non‐ST‐Segment and ST‐Segment‐Elevation Myocardial Infarction. Family medicine & medical science research, 2, 108. |
||||
|
Xu, H.X. et al, Differential Expression of MicroRNAs in Calcific Aortic Stenosis. Clinical laboratory, 63, 1163–1170. |
||||||||||
| miR‐451a | NDE | NDE | DOWN | https://doi.org/10.1111/jcmm.12380 | Song, L. et al, MiR‐451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. Journal of cellular and molecular medicine, 18, 2266–2274. | |||||
| miR‐454 | DOWN | UP (DCM in children) *** | NDE | https://doi.org/10.3109/1354750x.2015.1118533 | Enes Coşkun, M. et al, Plasma microRNA profiling of children with idiopathic dilated cardiomyopathy. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals, 21, 56–61. | |||||
| miR‐4485 | NDE | UP | / | / | ||||||
| miR‐4732 | UP | UP (congenital heart defetcs) | NA | https://doi.org/10.1371/journal.pone.0226164 | Abu‐Halima, M .et al, Micro‐RNA signatures in monozygotic twins discordant for congenital heart defects. PloS one, 14, e0226164. | |||||
LEGEND: NDE, not differentially expressed vs. ctrl; UP, up‐regulated; DOWN, down‐regulated; NA, undetermined by RT‐qPCR.
AUTHOR CONTRIBUTIONS
Conceptualization: C.F. and P.G.C.; Data curation: D.L., G.B, I.O. and O.R.; Formal analysis: M.R. and C.F.; Investigation: M.L., D.L., G.B., G.d'A., G.B, F.deC. and C.F.; Methodology: M.L., D.L.; Project administration: C.F.; Supervision: P.G.C.; Visualization,: M.L., M.R. and C.F.; Writing original draft: M.L., D.L., C.F. and P.G.C.; Writing review & editing: I.O., C.F. and P.G.C. All authors have read and agreed to the published version of the manuscript.
Supporting information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
We would like to thank Marco Magnoni, Silvia Del Rio, and Carmem Sartorio for their help in patient's enrolment and samples collection.
Present address
Davide Lazzeroni, Prevenzione e Riabilitazione Cardiovascolare, IRCCS Fondazione Don Carlo Gnocchi, Milano.
Giulia Benedetti, Radiology Department, Cardiothoracic Radiology, Guy's and St Thomas' NHS Foundation Trust, London, UK.
Funding information
This work was supported by the Italian Ministry of Health (grant number: NET‐2011‐02347173).
REFERENCES
- 1. Li Y, Huo C, Pan T, et al. Systematic review regulatory principles of non‐coding RNAs in cardiovascular diseases. Brief Bioinform. 2019;20(1):66–76. [DOI] [PubMed] [Google Scholar]
- 2. de Gonzalo‐Calvo D, Vea A, Bär C, et al. Circulating non‐coding RNAs in biomarker‐guided cardiovascular therapy: a novel tool for personalized medicine?. Eur Heart J. 2019;40(20):1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maron BJ. Clinical course and management of hypertrophic cardiomyopathy. New Engl J Med. 2018;379(7):655–668. [DOI] [PubMed] [Google Scholar]
- 4. Ouyang T, Liu Z, Han Z, Ge Q. MicroRNA detection specificity: recent advances and future perspective. Anal Chem. 2019;91(5):3179–3186. [DOI] [PubMed] [Google Scholar]
- 5. Scrimgeour NR, Wrobel A, Pinho MJ, Høydal MA. microRNA‐451a prevents activation of matrix metalloproteinases 2 and 9 in human cardiomyocytes during pathological stress stimulation. Am J Physiolo CELL Physiol. 2020;318(1):C94–C102. [DOI] [PubMed] [Google Scholar]
- 6. Song L, Su M, Wang S, et al. MiR‐451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014;18(11):2266–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bye A, Røsjø H, Nauman J, et al. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals ‐ The HUNT study. J Mol Cell Cardiol. 2016;97:162–168. [DOI] [PubMed] [Google Scholar]
- 8. Roe ND, Xu X, Kandadi MR, et al. Targeted deletion of PTEN in cardiomyocytes renders cardiac contractile dysfunction through interruption of Pink1‐AMPK signaling and autophagy. Biochim Biophys Acta. 2015;1852(2):290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oudit GY, Kassiri Z, Zhou J, et al. Loss of PTEN attenuates the development of pathological hypertrophy and heart failure in response to biomechanical stress. Cardiovasc Res. 2008;78(3):505–514. [DOI] [PubMed] [Google Scholar]
- 10. Zhang M, Jiang Y, Guo X, et al. Long non‐coding RNA cardiac hypertrophy‐associated regulator governs cardiac hypertrophy via regulating miR‐20b and the downstream PTEN/AKT pathway. J Cell Mol Med. 2019;23(11):7685–7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information


